Abstract
Hematopoietic stem cells differentiate into a broad range of specialized blood cells. This process is tightly regulated and depends on transcription factors, micro-RNAs, and long non-coding RNAs. Recently, also circular RNA (circRNA) were found to regulate cellular processes. Their expression pattern and their identity is however less well defined. Here, we provide the first comprehensive analysis of circRNA expression in human hematopoietic progenitors, and in differentiated lymphoid and myeloid cells. We here show that the expression of circRNA is cell-type specific, and increases upon maturation. CircRNA splicing variants can also be cell-type specific. Furthermore, nucleated hematopoietic cells contain circRNA that have higher expression levels than the corresponding linear RNA. Enucleated blood cells, i.e. platelets and erythrocytes, were suggested to use RNA to maintain their function, respond to environmental factors or to transmit signals to other cells via microvesicles. Here we show that platelets and erythrocytes contain the highest number of circRNA of all hematopoietic cells, and that the type and numbers of circRNA changes during maturation. This cell-type specific expression pattern of circRNA in hematopoietic cells suggests a hithero unappreciated role in differentiation and cellular function.
INTRODUCTION
Each day more than 1012 cells are produced in the bone marrow from hematopoietic stem cells (HSCs). HSCs differentiate into various progenitor cells, which in turn generate many types of myeloid and lymphoid cells (1). This process requires a tight regulation of gene expression. Transcription factors, long non-coding RNAs (lncRNAs), and microRNAs (miRNAs) contribute to differentiation (2–4). In addition to miRNA and lncRNA, also other non-coding RNAs emerge as important regulatory factors. Recently, it was shown that an alternative splicing mechanism can give rise to stable circular RNA (circRNA) with distinct regulatory capacity (5–7).
CircRNAs derive from transcripts that are back-spliced and joined head-to-tail at the splice sites (6,8). This covalent circularization of single stranded RNA molecules results in a novel backward fusion of two gene segments that can be of intronic and/or exonic origin (8). The formation of circRNA relies on complementary sequences in flanking introns that bring two splicing sites in close vicinity, and thus facilitate the back-splicing event, a process that can be regulated by DHX9 and ADAR to control the circRNA formation (9,10). This circularization renders circRNA much more stable than linear RNAs (11). CircRNAs do not contain poly-A tails. As a consequence, they are not detected by the most widely used RNAseq methods that are based on poly-A selection. Our current knowledge of circRNA expression is therefore still at its infancy.
Several functions have been attributed to circRNA (12). They can serve as miRNA sponges (13,14), or as transcriptional activators (15,16). Furthermore, circRNA have been shown to segregate RNA binding proteins ((17); BioRxiv: 10.1101/115980), and can even become translated into proteins through cap-independent translation initiation (5,18). CircRNA may also regulate the differentiation of HSCs (19). Indeed, circRNA expression has been described in several blood cells (7,20–22). Together with recent reports on circRNA in neuronal and myocyte differentiation cells and in extracellular vesicles (5,23,24) and other cell types (25,26), these findings prompted us to interrogate which circRNAs are expressed in hematopoietic cells and whether the expression of circRNA alters during hematopoietic differentiation.
CircRNAs can be identified by their unique back-spliced junction, which results in chimeric reads alignment in the RNA-seq data. This feature distinguishes them from linear RNA (6); Figure 1A). Here, we used previously published transcriptome deep-sequencing data on primary human hematopoietic cells to define the expression pattern of circRNA during differentiation. This comprehensive analysis identified >59 000 circRNAs in hematopoietic cells and in enucleated mature myeloid cells combined, of which >14 000 circRNAs were newly annotated. We found that circRNA expression is cell-type specific and alters during differentiation. Furthermore, differentiated cells contain substantially higher levels of circRNA. We conclude that circRNA expression is widespread in hematopoietic cells, which warrants their further functional characterization.
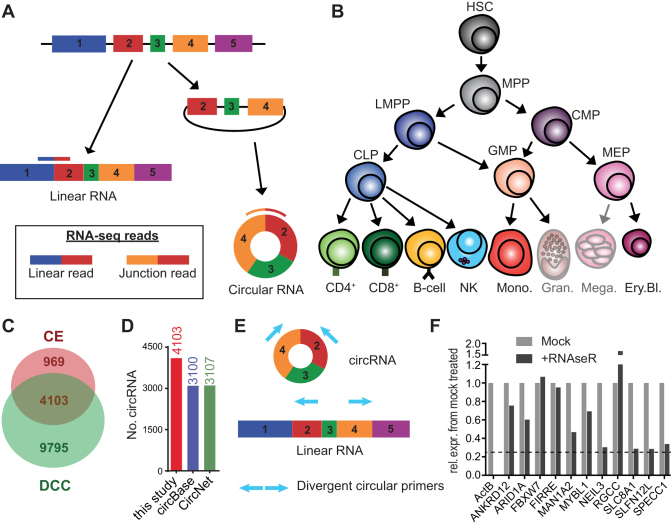
Figure 1.
Circular RNA expression in hematopoietic cells. (A) Diagram presenting the forward-splicing of a linear RNA and the back-splicing of a circular RNA. (B) Diagram of hematopoietic differentiation (inspired by ref (27)). HSC: Hematopoietic stem cell, MPP: Multipotent progenitor, LMPP: lymphoid-primed multipotent progenitor, CLP: common lymphoid progenitor, CMP: common myeloid progenitor, GMP: Granulocyte-macrophage progenitor, MEP: Megakaryocyte-erythrocyte progenitor, CD4: CD4+ T cells, CD8: CD8+ T cells, NK: Natural killer cells, Mono.: Monocytes, Gran.: Granulocytes, Mega.: Megakaryocytes, Ery.Bl.: Erythroblasts. (C) The number of circRNA (≥2 junction reads in at least one sample of a specific cell population) detected in hematopoietic cells using DCC and CircExplorer2 (CE). (D) Numbers of circRNA detected by DCC and CE combined using low confidence cut-off, compared to circRNA annotated in circBase and in CircNet. CircRNA that were not previously annotated were 1003 for circBase and 996 for CircNet. (E) Scheme of primer design for detection by PCR of circular RNA. (F) Graph showing the remaining expression after RNAseR treatment relative to mock treated for different circRNA. β-actin mRNA was used as a linear transcript control. Dashed line depicts the 50-fold higher resistance to RNAseR treatment, compared to the β-actin linear control (as in (6)). Data are representative of 2 donors.
MATERIALS AND METHODS
Data set selection
The SRA repository from NCBI was searched for datasets of human hematopoietic cells that contained paired-end data with long reads (>100nt), at least 20 million reads/sample, and that used random hexamers for cDNA preparation, and no 5′ or 3′ capture methods for library preparation. The selected data set (accession GSE74246/SRP065216) from (27) used total RNA and the TruSeq/NEB Next Ultra library preparation kit. For granulocytes, RBC and platelets, we used datasets that used ribosomal RNA-depleted total RNA for sample preparation. SRA accession numbers for platelets (project: PRJEB4522): ERR335311, ERR335312 and ERR335313 (28); for RBCs: (GEO: GSE63703) SRR2124299, SRR2124300, SRR2124301 and (GEO: GSE69192) SRR2038798 (29,30), and for granulocytes: (project: PRJEB8740) ERR789064, ERR789082, ERR789195 and ERR789201 (31).
CircRNA identification and analysis
Data sets were aligned to the human genome (GRCh37/hg19-release75) using STAR version 2.5.2b allowing for chimeric detection (see Supplementary Methods). The output file of STAR was analysed with DCC 0.4.4 (32) and CircExplorer2 (CE) 2.01 (33) to detect, filter and annotate circRNA (see supplementary method). DCC was used for detection of linear reads at the circRNA coordinates (using the option –G). For both CE and DCC, we used GRCh37/hg19 genome annotations.
CircRNA expression was considered low confidence detection when at least 2 junction reads were found in at least one sample by both DCC and CE, and high confidence detection when at least two junction reads were found in all biological replicates of one specific cell type, by both tools. Junction read counts were normalized to reads per million mapped reads (RPM). The maximal circRNA length was calculated with the exon length information provided by the CE annotations. These annotations were then used to calculate the first and last circularized exon.
For differential expression analysis of high confidence circRNA in hematopoietic cells, we used DESeq2 (34) with P-adjusted <0.05. Kmeans clustering was performed with pheatmap 1.0.8 (35) with option kmeans_k. The number of 14 clusters was chosen after manual inspection of the circRNA clusters.
Circular over linear ratios (CLR) were calculated for the coordinates of high confidence circRNA (n = 489) with the linear counts obtained with DCC (see Supplemental method). Linear counts were corrected for sequencing depth to obtain linear RPM. CLR was calculated by using circular RPM/linear RPM. Data analysis was performed in R (3.4.1) and R-Studio (1.0.143).
Plots and graphs
Heatmaps were generated in R using pheatmap 1.0.8. Plots and graphs were generated with ggplot2 (36) in R, or Graphpad PRISM version 7.0. Venn diagrams were produced with http://bioinformatics.psb.ugent.be/beg/tools/venn-diagrams from the University of Gent.
Cell population isolation
Peripheral blood mononuclear cells (PBMCs) were obtained according to the Declaration of Helsinki (seventh revision, 2013) from healthy volunteers with written informed consent (Sanquin, Amsterdam, the Netherlands). PBMCs of 3–4 donors were isolated by Lymphoprep density gradient separation (Stemcell Technologies). Specific cell types were isolated with CD4+ and CD8+ MACS beads for T cells, CD14+ beads for monocytes, CD56+ beads for NK cells, and CD34+ beads for CD34+ progenitors (purity > 98%, as determined by flow cytometry) according to the manufacturer's protocols (Miltenyi). B cells were isolated with CD19+ Dynabeads and detach-a-beads (Invitrogen). Red blood cell fractionation was performed with a Percoll-Urografin gradient as previously described (37). Pellets were frozen and kept at −80°C.
Flow cytometry
Red blood cell fractions were washed in PBS+1% bovine serum albumin (BSA, Sigma-Aldrich), and incubated for 30 min at 4°C with anti-CD71-APC (Miltenyi; Clone AC102) and Thiazole orange (TO) (Sigma-Aldrich, 100 ng/ml). Cells were washed once with PBS + 1% BSA and acquired on LSR Fortessa (BD). Data analysis was performed with FlowJo version X (Tree Star).
RNA extraction
Pellets were thawed on ice for 5 min and RNA was extracted with Trizol (Life Technologies) according to the manufacturer's protocol. RNA was resuspended in RNAse-free water and measured on a Nanodrop (ThermoFisher). Half of the isolated RNA was treated for 15 min at 37°C with 8 units/μg RNAseR (Epicentre) in water supplemented with the digestion buffer provided by the manufacturer. Both RNAseR-treated and untreated control samples were purified with mini Quick-spin columns (Roche). cDNA preparation was performed with random hexamers using Super Script III reverse transcription (Invitrogen) according to the manufacturer's protocol.
Primers, PCR and qPCR
Primers were designed with CircInteractome (17) and with the NCBI primer blast tool (38), and manufactured by Sigma. Alternatively, the first and last 100nt of the circRNA junction were joined and used to blast for primer pairs. Primer sequences used to detect linear and circular RNA are found in Supplementary Table S5. PCRs were performed on two to three biological replicates with ReddyMix PCR Master-Mix (Thermo-Fischer), and products were run on 2% agarose gels. PCR reactions were loaded with the same volume of RNAseR and of mock treated sample, which both underwent Quick spin column purification. Primer pairs that yielded one specific band and that showed standard curves with r2 >0.98, and one peak temperature in the melting curves were used for RT-qPCR. Lower limit of detection was assessed with serial dilutions of cDNA for the standard curve. RT-qPCRs were performed with technical duplicates and of two to three biological replicates using Power SYBR-green (ThermoFisher) with the standard protocol (Tm = 60°C for 1 min) on a 7500 Real-time qPCR system (Applied Biosystems). DNAse/RNAse-free water was used as non-template control. RT-qPCR data were analysed using 7500 Software v2.3 (Applied Biosystems). None of the samples showed possible contamination in melting curve. Ct thresholds were determined by the software. The expression was normalized using the 2e(delta-Ct) method using 18S as a reference gene and plotted using GraphPad PRISM.
RESULTS
CircRNAs are broadly expressed in hematopoietic cells
To determine which circRNAs are expressed in hematopoietic cells, we used the RNA-seq data set of Corces et al. (27) that contained a broad range of hematopoietic progenitors and differentiated immune cells (Figure 1B). Because this dataset was generated with random hexamers in combination with oligo-dT primers, a slight 3’bias was observed for linear RNAs. Yet, this did not impede the identification and quantification of circular RNAs (see below). Samples were included in the analysis when they reached a sequencing depth of at least 20M reads and when paired-end reads had a length of at least 100 bases. This cut-off resulted in 2–3 replicates per cell type, with an average of 29 million total reads and an average of 26.5 million mapped reads per sample. Each population contained at least one replicate with over 30 million mapped reads.
We used two different tools for circRNA detection, i.e. DCC (32) and CircExplorer2 (CE) (33). Both tools allow for fast and accurate detection of circRNAs by detecting back-splice junction reads of circRNA from RNA-seq data with a low false positive rate (reviewed in (39,40)). We used a cut-off for circRNA detection when at least two junction reads were detected in at least one sample of the 2–3 biological replicates from one specific population, according to the study of Maass et al. (7) (Figure 1C, Supplementary Table S1). 13 898 different circRNAs were identified with DCC, and 5072 circRNAs with CE. To reduce false positive detection rate (40), we only considered the 4103 circRNAs for further analysis that were detected with both tools (hereafter called ‘low confidence’ cut-off). Of those 4103 circRNAs, 1003 and 996 were not yet annotated in circBase (41) and CircNet (42) respectively (Figure 1D). 428 circRNAs (10.4%) were newly identified circRNAs that were not annotated in either data base (figure 1D). Only 16 circRNAs (0.4%) of the 4103 circRNAs derived from intronic junctions (Supplementary Figure S1A). The vast majority of the 4103 circRNAs (4087 circRNA; 99.6%) contained exon–exon junctions.
We validated the expression of the identified circRNAs by RT-PCR (Figure 1E) in human peripheral blood mononuclear cells (PBMC), including highly expressed and lowly expressed circRNAs. 39/40 tested circRNAs were detected, of which 13 circRNAs were novel (Supplementary Table S5). The identity of circRNAs was further confirmed by the absence of sensitivity to exonuclease ribonuclease R (RNAse R) (Figure 1F; Supplementary Table S5), and by Sanger sequencing (Supplementary Figure S2A).
CircRNA generation, size and distribution in hematopoietic cells
We next determined which genes and which exons were used for the generation of circRNA. The distribution of the identified circRNA per chromosome was similar to the distribution of all coding and non-coding genes (Figure 2A). When analyzing the biological processes of circRNA-generating genes, they were enriched in housekeeping functions (Supplementary Figure S1B). The predicted numbers of exons used by circRNA is on average 5.96 exons as determined by CE annotation based on the known linear exon usage, with a minimum of one exon and a maximum of 56 exons (Figure 2B). The calculated length of the circRNA was on average 1057 nucleotides (Figure 2C).
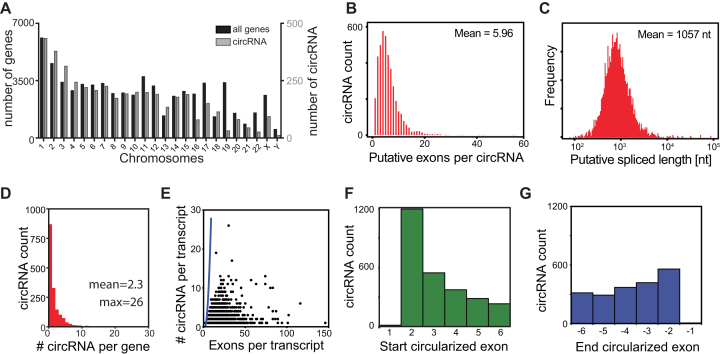
Figure 2.
Characterization of circRNA detected in hematopoietic cells. (A) Distribution of circRNA (in grey) and known genes from hg19/GRCh37 (in black) per chromosome. (B, C) Putative exon usage (B) and maximal putative length (C) of the 4103 circRNA detected in hematopoiesis, as determined by the start and end position of circRNA and splicing annotations of the RNA linear transcript. (D) Number of detected circRNA per gene. (E) Relation between the number of circRNA per transcript and the number of exons of that transcript. Blue line represents the calculated maximum circRNA number per transcript, if all exon combinations were used for circularization. (F, G) Distribution of the first (F) and the last (G) 6 exons used for circularization from the linear RNA in circRNA. 1197/4103 (29.2%) circRNA use the second exon as start exon, and 559/4103 (13.6%) use the one but last exon as end exon.
We next interrogated how many circRNA variants derive from one gene, by defining the different exon usage of circRNAs. To this end, we calculated all different back-splice junction reads found per gene in hematopoietic cells. This analysis revealed that 930 genes (53%) generated only one circRNA variant. However, the overall expression of circRNA variants is between 1 and 26 variants per gene with a mean of 2.3 circRNA/gene (Figure 2D). If circRNA would be a random event, the maximum possible number of circRNA should increase near-exponentially as the number of exons per transcripts increases (Figure 2E, blue line). There was, however, no overt correlation between the detected number of circRNA variants per gene and the number of exons per transcripts, suggesting that the generation of circRNA is not a random event (Figure 2E; compare the data points with blue line).
Finally, we questioned which exon of the linear transcript is used by circRNA as the starting exon for circularization. In line with the canonical splicing rules (43), circRNAs in hematopoietic cells barely use the first exon (Figure 2F). Rather, circRNAs favor the second exon of a transcript as starting exon, which is found in 1197/4103 circRNAs (29.2%) (Figure 2F). We also determined which exon is used for back-splicing, referred to as the end circularized exon. Again, the last exon of a linear transcript was not used (Figure 2G). However, we could not detect a clear preference for a specific end exon (Figure 2G).
CircRNA expression in hematopoiesis alters upon differentiation
We next interrogated whether the expression of circRNA alters during hematopoietic differentiation. We focused on circRNAs that followed the following definition for high confidence: circRNA detected at least two junction reads in each biological replicate of one specific cell population (as opposed to low confidence that required two reads only in one biological replicate). High confidence circRNAs were again considered only when identified with by both DCC and CE. This analysis yielded 489 high confidence circRNAs, of which 13 circRNAs were newly identified (Figure 3A, Supplementary Table S1). The number of exon usage per circRNA and length of high confidence circRNA were comparable to those identified with the low-confidence cut-off (Supplementary Figure S1C, D).
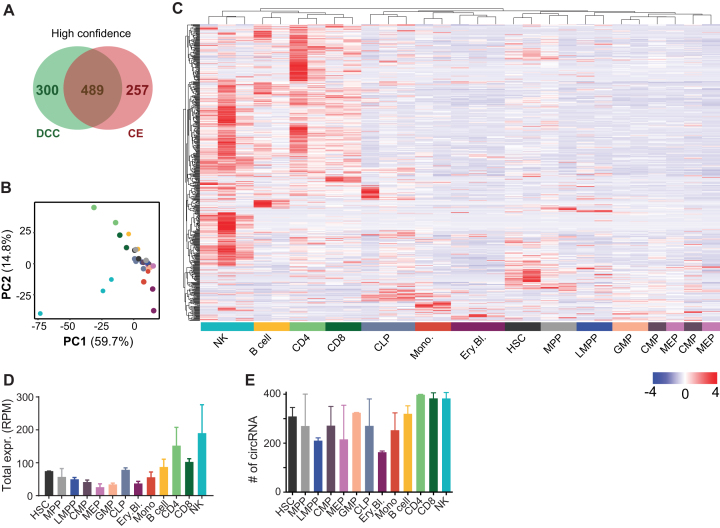
Figure 3.
circRNA expression in hematopoietic cells. (A) Number of high confidence circRNA detection, as defined by ≥ 2 junction reads in each replicate of one specific cell population). 33 out of the 489 circRNA were newly identified. (B, C) PCA plot (B) and (C) unsupervised Pearson correlation clustering (row-scaled RPM) of the circRNA expression in the hematopoietic populations depicted in a heat map (n = 2–3 per cell type). (D, E) Sum of different circRNA expressed per cell type (D), and of the overall number of circRNA expressed per population (E). n = 2–3, error bar depicts standard deviation. RPM: reads per million mapped reads.
Comparable to the population-specific distribution of linear mRNA (27), principal component analysis (PCA) of circRNA expression recapitulated the cellular differentiation status of HSC populations (Figure 3B). This finding indicates that circRNA may follow specific gene expression during differentiation. Indeed, HSC and progenitor cells are located at the intersection of the differentiated myeloid and lymphoid cells (Figure 3B). Differentiated myeloid cells clustered with each other, away from the progenitors, and in opposite directions as the mature lymphoid cells (Figure 3B). Unsupervised clustering based on circRNA expression also showed a clear clustering of HSCs with progenitors in a heat map, away from differentiated cell populations (Figure 3C). We observed that lymphocytes had the highest levels of circRNA expressed, based on the sum of all circRNA RPM (Figure 3D). The number of distinct circRNA, however, was similar in various cell populations, indicating that the abundance of circRNA, but not the variety of circRNA, was increased in B cells, CD4+ and CD8+ T cells, and in NK cells (Figure 3D, E).
CircRNA expression during hematopoietic differentiation is cell-type specific
To determine the cell-type specific expression of circRNA, we performed differential expression analysis on the high confident circRNA that were detected in Figure 3 (n = 489). This analysis performed with DESeq2, identified 102 differentially expressed circRNAs with a P-adjusted <0.05 (Figure 4A, Supplementary Table S2). We then clustered these 102 circRNAs by pattern of expression using K-means clustering (k = 14). This analysis revealed that circRNAs are expressed in a cell-type specific manner (Figure 4A, B).
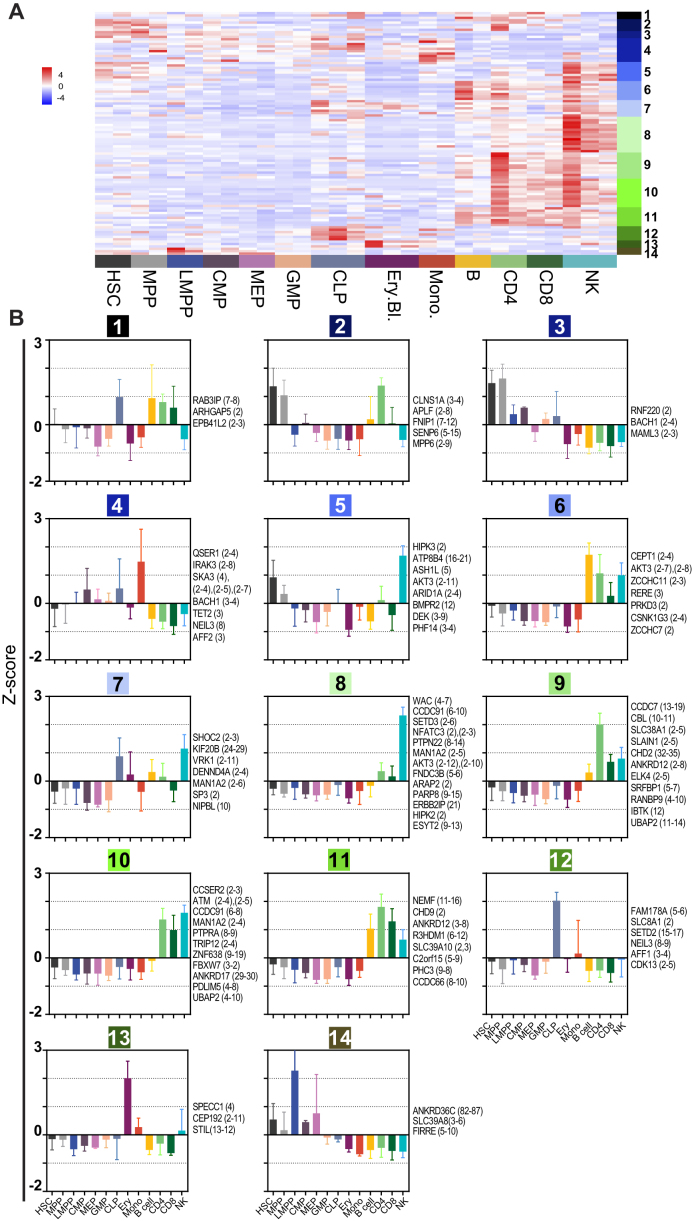
Figure 4.
CircRNA expression is cell-type specific. (A) Heatmap of differentially expressed circRNAs, as determined with DESeq2 (P-adj.<0.05). On the right, the 14 K-means clusters are depicted (n = 102, row-scaled RPM). (B) K-means clusters (k = 14) depicting the relative expression (Z-score) across the hematopoietic cell populations. CircRNA gene names with the exon usage in brackets are depicted for each cluster on the right of the graph.
Clusters 2, 3, 5, 12 and 14 include circRNA expressed by the progenitors (Figure 4B). CircRNAs in clusters 3 and 14 were mainly expressed in early progenitors, i.e. HSCs, MMPs, and LMPPs. Cluster 2 circRNAs were more restricted to HSCs and MPPs, but also showed circRNA expression in lymphoid cells. Clusters 12 showed the signature of CLPs, concomitant with low expression in differentiated lymphoid cells. Cluster 5 circRNAs were shared between HSC and MPPs and with NK cells.
Lymphoid cells specific circRNAs were represented by clusters 1, 6, 8, 9, 10 and 11 (Figure 4B). Cluster 6 showed higher circRNA expression in B cells, cluster 9 for CD4+ T cells, and cluster 8 for NK cells. Cluster 10 and 11 showed a mixed lymphoid signature, with the exception of B cells in cluster 10. Cluster 1 showed a mixed signature with high expression in CLP and lymphoid cells except NK cells. Myeloid cell-specific circRNAs were present in clusters 4 and 13 (Figure 4B). Cluster 13 was erythroid specific and 4 mostly monocyte-specific with some background expression in CMP and CLP. We validated the distribution of several circRNAs in blood cell subsets by RT-PCR and RT-qPCR (Supplementary Figure S1E, G). Indeed, circ-FNDC3B (exon 5-6) was broadly expressed with the highest expression levels in NK cells. The newly identified circ-ELK4 (exon 4-3), circ-MYBL1 (exon 12-7), and circ-SLFN12L (exon 2-3) show the highest expression in T cells and NK cells (Supplementary Figure S1E, G).
Interestingly, different circular junctions were generated from the same gene, and thereby different exon usage was detected in different clusters (Figure 4B). For example, circ-BACH1 (exon 3-4) was preferentially expressed in monocytes (cluster 4), whereas circ-BACH1 (exon 2-4) was increased in HSCs and MPPs (cluster 3). This finding was confirmed by RT-PCR (Supplementary Figure S1F). Likewise, different variants of circ-AKT3 (clusters 5, 6 and 8) and circ-CCDC91 circRNA (clusters 8 and 10) were expressed across lymphoid cells. In conclusion, our analysis shows that the expression of circRNA is cell-type specific.
The ratio of circRNA over linear RNA alters with differentiation
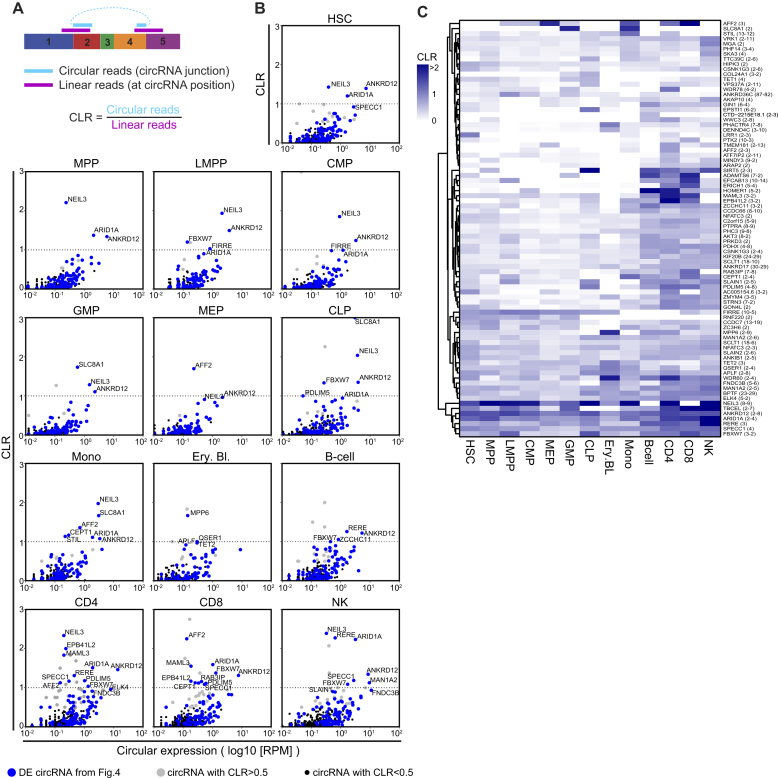
We next investigated how the expression levels of circRNA compares with that of the corresponding linear RNA. Because circRNA detection is restricted to the junction reads, the levels of linear mRNA expression are also defined by comparing the number of reads at the start and at the end position of the circRNA (Figure 5A). DCC analysis corrects for the double counting of linear reads (start + end; (32)), and allows us to calculate the circular-over-linear ratio (CLR) on the 489 circRNAs expressed in hematopoietic cells (Figures 3 and 5A).
Figure 5.
CircRNA usage alters during hematopoietic differentiation. (A) Scheme showing how the circular-to-linear ratio (CLR) is computed using DCC (32). (B) CLR of circRNA from Figure 3 for each population against the number of reads of the circRNA (log10 (RPM)). CLR >1 indicates that the sample contains more transcripts for the circRNA than the corresponding linear transcript. Blue dots are the 102 differentially expressed circRNA from Figure 4, gray dots are circRNA with a CLR >0.5, and black dots are circRNA with CLR <0.5. (C) Heatmap showing the average CLR within a population, of circRNA having CLR >0.5 in at least one of the hematopoietic populations.
We plotted the CLR against the circRNA expression for each hematopoietic population. 82 circRNAs showed a CLR >0.5, indicating that these circRNAs reached at least 50% of the levels of the linear RNA (Figure 5B, gray dots). The 102 circRNAs used to determine cell-type specific circRNA expression in Figure 4 followed the same expression pattern (Figure 5B, blue dots). The expression level for several circRNAs reached even higher expression levels than the corresponding linear RNAs (CLR >1; Figure 5B, Supplementary Table S3). The number of circRNA with a CLR >1 was highest in differentiated lymphoid cells, i.e. B cells, T cells and NK cells.
We next determined if the ratio between circular and linear RNAs alters during hematopoietic differentiation. To this end, we plotted all circRNA with a CLR >0.5 (n = 82) in a heat map (Figure 5C). Some circRNA like circ-NEIL3 (exon 8–9) or circ-ANKRD12 (exon 2–8) retained a high CLR in all cell populations. In contrast, the CLR of circ-SLC8A1 (exon 2) was only high in GMPs and in monocytes. The CLR of circ-AFF2 (exon 3) was high in MEPs and monocytes as well as in T cells (Figure 5C). Circ-RERE (exon 3) showed the highest CLR in NK cells, but was also found in B cells and T cells, albeit with lower CLRs (Figure 5C). Combined, these findings show that the use of circRNA and their correlation to the linear RNA is cell-type specific and changes during hematopoiesis.
Red blood cells and platelets have high expression levels of circRNA
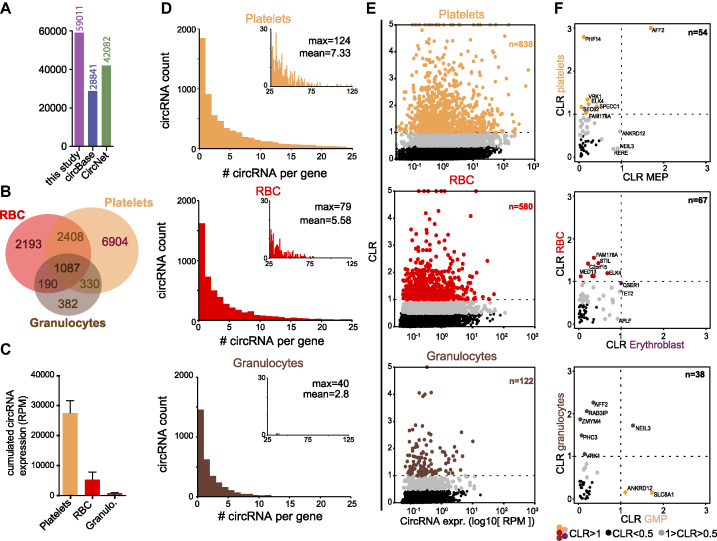
Maturation of red blood cells is accompanied with an increase in heterochromatin and condensation of the nucleus, followed by extrusion of the nucleus. Platelets are shed from megakaryocytes. Both red blood cells and platelets undergo their final steps of maturation in absence of active gene transcription (44). To investigate the expression levels of circRNA in RBC and platelets, we first used publicly available RNA expression data of platelets, red blood cells (RBC), and granulocytes isolated from peripheral blood (28–31). By combining DCC and CE and using the low-confidence cut-off of two junction reads in at least 1 replicate of one specific cell population, we identified 59 011 circRNA, of which 28 841 and 42 082 were annotated in circBase and CircNet, respectively (Figure 6A). Over 14 100 circRNA (23.9%) were thus newly identified in this data set. Of the 59 011 identified circRNA, platelets express the highest numbers of circRNA (47 654), followed by RBCs (27 409) and granulocytes (8 925) (Supplementary Figure S3A, Supplementary Table S4). With the high confidence cut-off of two junction reads in each biological replicate of one cell type, we detected 10 729 circRNA in platelets, 5 878 circRNA in RBCs, and 1 989 circRNA in granulocytes (Figure 6B, Supplementary Table S4). Of these, both shared and cell-type specific circRNAs were detected (Figure 6B). Platelets not only contained the highest numbers of different circRNA, but also clearly outnumbered RBCs and granulocytes with the overall RPM of circRNA RPM (Figure 6C).
Figure 6.
Red blood cells and platelets express high levels of circRNA. (A) CircRNA detection (low confidence; i.e. ≥2 junction reads in at least one sample) in granulocytes, red blood cells (RBC) and platelets, compared to annotations in circBase and CircNet databases. (B) circRNA expression with high confidence (≥2 junction reads in all replicate of one population). (C) The sum of circRNA detected with high confidence per cell type (RPM). (D) Number of low-confidence circRNA detected per gene in platelets (top panel), RBC (middle panel) and granulocytes (lower panel). The insert histograms show circRNA found with more than 25 circRNA/gene. Numbers depict Maximum circRNA per gene (max) and the average circRNA per gene (mean). (E) Circular-to-linear ratio (CLR) plotted against the circRNA expression (log1(RPM)) of high-confidence circRNA. Black dots show circRNA with CLR<0.5 and colored dots show circRNA with CLR >1, for platelets (top panel), RBC (middle panel) and granulocytes (lower panel). (F) CLR of high confidence circRNA detected in both progenitor cells and differentiated cells for platelets with MEP (n = 54 circRNA, top panel), RBC with erythroblasts (n = 67 circRNA, middle panel) and granulocytes with GMP (n = 38 circRNA, lower panel).
We next determined the characteristics of circRNA in differentiated myeloid cells. With the low-confidence cut-off, we calculated a median 4 and an average of 4.43 exons/circRNA for all three cell types combine. Using the high confidence cut-off, we calculated an average of 4.04 in platelets, 3.58 in RBC and 3.07 exons/circRNA in granulocytes (exon information extracted from Supplementary Table S4). We also estimated the number of circRNA variants that are expressed per gene in the three different cell types (Figure 6D). Granulocytes expressed on average 2.8 circRNA per gene with a maximum of 40 circRNA/gene, which resembled the numbers of circRNA measured per gene in hematopoietic cells (Figure 2D). Conversely, RBCs and platelets express on average 5.58 and 7.33 circRNA per gene, respectively, with a maximum of 79 for RBCs and 124 for platelets (Figure 6D). Of note, none of the three cell types were included in the data set of Figures 1–5. Thus, the relative expression levels of circRNA and the number of variants expressed per gene in enucleated RBCs and platelets exceed those of all other hematopoietic cell types.
The high prevalence of circRNA in platelets and RBCs is also reflected by a high CLR (Figure 6E). Concomitant with the loss of de novo transcription in enucleated cells, RBCs and platelets contain high levels of circRNA, and a significant number of circRNA have a high CLR. 7.8% of the circRNA expressed in platelets had a CLR > 1 (n = 838). Similarly, RBCs had 9.87% circRNA with a CLR >1 (n = 580), and 6.13% (n = 122) in granulocytes (Figure 6E).
We then determined how the circRNA expression pattern of these three cell types corresponded to the circRNA usage in the respective progenitor. We found that Erythroblasts shared only 177 circRNAs with RBC, MEP shared 225 circRNAs with platelets, and GMP shared 151 circRNAs with granulocytes (Supplementary Figure S3B–D). We then compared the CLRs of shared circRNA between progenitors and differentiated cells. The hematopoiesis dataset (27) and the differentiated myeloid cells (28–31) were different data sets generated with different methods, i.e. total RNA and with ribosomal RNA depletion, respectively. We included the circRNA and the corresponding linear mRNAs in this analysis that had at least 0.1 RPM each (Figure 6F; Supplementary Table S4). The circRNA that have a CLR >0.5 (gray) or CLR >1 (colored) are more prevalent in mature platelets, RBCs and granulocytes than in the precursor populations (Figure 6F). In addition, the circRNA with a CLR >0.5 in mature cells barely overlap with those of the precursor cells (Figure 6F). This finding suggests that these circRNAs are generated during differentiation and do not merely reflect an accumulation of stable circRNAs from the progenitor cell. Combined, we conclude that circRNAs are highly expressed in differentiated myeloid cells, and that the highest relative levels are detected in the enucleated RBCs and platelets.
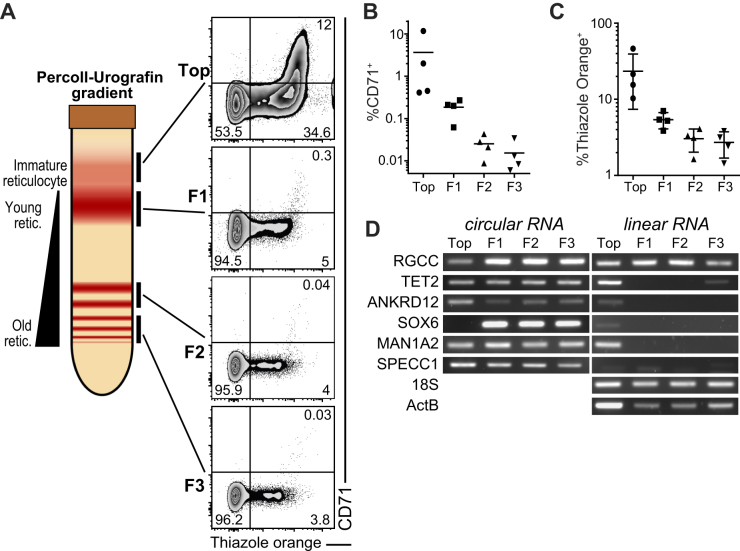
CircRNA expression is retained in aging RBCs
RBCs have a life span of 120 days, and during this period RBCs age (45). We therefore analysed if and how the linear and circular RNA expression altered during RBC maturation and ageing. To separate reticulocytes and young erythrocytes from old erythrocytes we used a Percoll-Urografin gradient ((37), Figure 7A). The maturation status of reticulocytes and the erythrocytes was confirmed with CD71 expression and the nucleic acid content with Thiazole Orange (TO) by flow cytometry (Figure 7A). Upon ageing, the expression of CD71 is lost (Figure 7B) and the TO staining (Figure 7C) is decreased.
Figure 7.
CircRNA are conserved throughout red blood cell aging.(A) Left panel shows a representation of a Percoll-Urografin gradient separation after centrifugation. The immature reticulocyte fraction is found in the top fraction (bellow a neutrophil-rich ring), the lower fractions (F1, F2, F3) separate the aging red-blood cells from young to old (top to bottom). Right panel shows a flow-cytometry measurement of the fractions for CD71 expression and nucleic acid (thiazole orange) staining (representative of 4 donors). (B, C) Graph showing the percentage of CD71+ (B) and thiazole orange + (C) in the fractions from (A) (n = 4). (D) RNA extracted from the fractions in (A) were reverse transcribed and circRNA and linear RNA detection was performed using PCR. 18S and Beta-actin (ActB) were used as a linear control. (D) is representative of 3 donors.
We used these four fractions of red blood cells to detect RBC-specific circRNA with relatively high expression and CLR over 0.5 and their linear mRNAs by RT-PCR (Figure 7D; Supplementary Figure S2B). During ageing, only the highly stable β−ACTIN mRNA, 18S RNA, and RGCC mRNA is detected in mature erythrocytes (Figure 7D, right panel). All other measured linear mRNAs lose their expression upon differentiation (Figure 7D, right panel). In contrast, circ-TET2 (exon 3), circ-ANKRD12 (exon 2–8), circ-MAN1A2 (exon 2–5) and SPECC1 (exon 4) remained stably detectable in young, and in old erythrocytes (Figure 7D, left panel). Interestingly, whereas SOX6 mRNA was only detectable in young reticulocytes, circ-SOX6 (exon 8–9) was specifically detected in mature reticulocytes (Figure 7D). Thus, circRNA expression is stably maintained during RBC ageing.
DISCUSSION
CircRNA can drive a subset of cellular functions (5,13,46), yet their role in gene regulation and expression in human hematopoietic cells is not well understood. To our knowledge, we here provide the first comprehensive analysis of circRNA expression during hematopoiesis. We identified over 14 000 novel circRNAs, which significantly extends the list of currently annotated circRNAs in the human transcriptome. How specific these newly identified circRNA are for hematopoietic cells is yet to be determined. Because the identified circRNAs are formed both from genes with general functions and from hematopoietic specific genes, and because the CLR indicates a cell-type specific enrichment of circRNA, we postulate that at least some of the identified circRNA should be cell-type specific.
Of note, the library for the analysis of the data set we used to study circRNAs in hematopoietic cells (27) was prepared with oligo-dT and with random hexamers. This method should not hamper circRNA detection that is only detected with random hexamers. However, both oligo-dT and random hexamer anneal to linear RNA. This can result in an over-estimation of the linear RNA expression, and consequentially, in an underestimation of the actual CLR. Even with this limitation, our data show that circRNA can be abundantly expressed in hematopoietic cells, and some circRNA at even higher levels than the corresponding linear RNA.
Similar to differentiating neurons and myocytes (5,23) the expression levels of circRNA increase during hematopoietic differentiation. This increase could be a mere result of accumulation of stable circRNA during differentiation. However, we show here that some circRNAs are preferentially expressed in progenitors. For example, the expression levels of circ-FIRRE (exon 5–10) was high in all progenitors except from CLPs, and was significantly reduced upon differentiation. In particular in LMPPs, circ-FIRRE (exon 5–10) was expressed at higher levels than the linear FIRRE RNA (Figures 4B and 5B). Another product of the FIRRE gene, the lncRNA FIRRE was shown to regulate part of the pluripotency of embryonic stem cells (ESC) via Repeat RNA Domains (RRD). Intriguingly, we observed that circ-FIRRE (exon 5–10) also contains all RRDs described for the LncRNA FIRRE. Together with recent work on ESC that indicated the dominant expression of circ-FIRRE over the linear RNA (47), it is tempting to speculate that some of the effects that are attributed to the lncRNA FIRRE could also be exerted by the circular FIRRE RNA.
Interestingly, the exon usage for circRNA generation—as we describe for circ-BACH1—can alter during differentiation. Several studies showed that alternative splicing during hematopoietic differentiation is primarily driven by exon skipping (2,44). The original hypothesis was that skipped exons were degraded via non-sense mediated decay. However, these exons can also be used for the formation of circRNA (48). As exon skipping is increased during late erythropoiesis (44), and circRNA expression is then increased, it is tempting to speculate that exon skipping contributes to the production of circRNA in erythrocytes. However, exon skipping cannot be the sole and perhaps even not the major source of circRNA. Many circRNA contain several exons, and the generation thereof appears cell type specific and differentiation dependent. In addition, a subset of circRNA display an increased expression over the linear variant. Combined, these data suggest a highly orchestrated production of circRNA. Future investigations should reveal how the formation of circRNA relates to the exon skipping, or to other regulatory events.
Similar to published work on cardiac circRNA (49) we find an average of 5.96 exons per circRNA. Other studies in myocytes, total blood and neurons report that circRNA contain 2–3 exons (5,21,23). This discordance may stem from different algorithms used, or from differently prepared data sets. However, and possibly more likely, this difference in exon usage may be a biological difference of different cell types. In line with that, also we detect differences, in high-confidence data, in exon usage in RBC (mean 3.58) and platelets (mean 4.04) when compared to granulocytes (mean 3.07).
Platelets and erythrocytes contain high levels of circRNA. This can at least in part be explained by the degradation of linear transcripts (29). Interestingly, the circRNA expression of erythroblast and MEP does not fully overlap with the differentiated RBCs and platelets. This again suggests that in addition to accumulation of stable circRNA in enucleated cells, also the generation of circRNA may alter during differentiation. Furthermore, platelets release specific circRNA in extracellular vesicles (24), which may alter the composition of the remaining circRNA. This specific release also suggests that circRNA may be involved in platelet-associated cellular processes. Whether the high prevalence of circRNA in anucleated cells is merely a result of their high stability, or whether the circRNA can be generated outside of the nucleus is yet to be determined. The high prevalence of circRNA could possibly be used as template for translation in these cells, in particular in RBCs that have a life span of 120 days.
The function of circRNA is to date not well understood (50). Whereas some circRNA can serve as a miRNA sponge (51), this does not apply to most of the currently known circRNA (14,52). CircRNA may also serve as sponges for RNA-Binding Proteins (12) (BioRxiv: DOI:10.1101/115980), or cross-talk with the transcriptional machinery (15,16). Recent studies show that circRNA can be also translated into functional protein (5,18,53). Thus, the function of circRNA is most probably diverse. Our analysis indicates that genes from which circRNA arise, are enriched in housekeeping functions (Supplementary Figure S1B). Whether this enrichment points to a functional role of the circRNA in regulating HSC differentiation, or whether this solely reflects the abundance of housekeeping genes and thus the increased probability to detect circRNAs from housekeeping genes is yet to be determined.
Here, we provide a comprehensive analysis of circRNA in hematopoietic cells. Future studies will reveal how circRNA can contribute to the regulatory processes in hematopoietic cells, and possibly in the make up of the proteome. Furthermore, circRNA may be a useful tool to distinguish diseased from healthy hematopoietic cells and be used as potential biomarkers, as previously suggested for several other types of cancers (54).
DATA AVAILABILITY
All bioinformatics tools used in this study are described in the supplementary method. The following datasets were used in this study: SRP065216, ERR335311, ERR335312, ERR335313, SRR2124299, SRR2124300, SRR2124301, SRR2038798, ERR789064, ERR789082, ERR789195, ERR789201, and are available on SRA (https://www.ncbi.nlm.nih.gov/sra).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank S. Heshusius and K. Moore for input on bioinformatics, E. Heideveld, M. Hansen, P-P. Unger, and R. Temming for help in isolating monocytes, platelets, B-cells, and NK cells, respectively, and the department of hematopoiesis at Sanquin (Amsterdam) for providing isolated CD34+ cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Landsteiner Foundation of Blood Transfusion Research; Dutch Science Foundation (LSBR-Fellowship 1373 and VIDI grant) [ 917.14.214 to M.C.W.]. Funding for open access charge: Landsteiner Foundation of Blood Transfusion Research; Dutch Science Foundation (LSBR-Fellowship 1373 and VIDI grant) [917.14.214 to M.C.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Orkin S.H., Zon L.I.. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008; 132:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goode D.K., Obier N., Vijayabaskar M.S., Lie-A-Ling M., Lilly A.J., Hannah R., Lichtinger M., Batta K., Florkowska M., Patel R. et al. . Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell. 2016; 36:572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo M., Jeong M., Sun D., Park H.J., Rodriguez B.A.T., Xia Z., Yang L., Zhang X., Sheng K., Darlington G.J. et al. . Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell. 2015; 16:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bissels U., Bosio A., Wagner W.. MicroRNAs are shaping the hematopoietic landscape. Haematologica. 2012; 97:160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. et al. . Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017; 66:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al. . Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 7. Maass P.G., Gla P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O.. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017; 95:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016; 17:205–211. [DOI] [PubMed] [Google Scholar]
- 9. Aktaş T., Ilik I.A., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A.. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017; 544:115–119. [DOI] [PubMed] [Google Scholar]
- 10. Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C. et al. . Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015; 10:170–177. [DOI] [PubMed] [Google Scholar]
- 11. Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E.. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013; 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hentze M.W., Preiss T.. Circular RNAs: Splicing's enigma variations. EMBO J. 2013; 32:923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. et al. . Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017; 8526:eaam8526. [DOI] [PubMed] [Google Scholar]
- 14. Guo J.U., Agarwal V., Guo H., Bartel D.P.. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014; 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L.. Circular intronic long noncoding RNAs. Mol. Cell. 2013; 51:792–806. [DOI] [PubMed] [Google Scholar]
- 16. Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. et al. . Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015; 22:256–264. [DOI] [PubMed] [Google Scholar]
- 17. Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M.. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016; 13:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. et al. . Translation of CircRNAs. Mol. Cell. 2017; 66:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia P., Wang S., Ye B., Du Y., Li C., Xiong Z., Qu Y., Fan Z.. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-Mediated exhaustion. Immunity. 2018; 48:688–701. [DOI] [PubMed] [Google Scholar]
- 20. Durek P., Nordström K., Gasparoni G., Salhab A., Kressler C., de Almeida M., Bassler K., Ulas T., Schmidt F., Xiong J. et al. . Epigenomic profiling of human CD4+T cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity. 2016; 45:1148–1161. [DOI] [PubMed] [Google Scholar]
- 21. Memczak S., Papavasileiou P., Peters O., Rajewsky N.. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015; 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y., Yu X., Luo S., Han H.. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun. Ageing. 2015; 12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. et al. . Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015; 58:870–885. [DOI] [PubMed] [Google Scholar]
- 24. Preußer C., Hung L.-H., Schneider T., Schreiner S., Hardt M., Moebus A., Santoso S., Bindereif A.. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles. 2018; 7:1424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C.. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017; 8:1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kristensen L.S., Okholm T.L.H., Venø M.T., Kjems J.. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018; 15:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corces M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L., Snyder M.P., Pritchard J.K., Kundaje A., Greenleaf W.J. et al. . Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 2016; 48:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kissopoulou A., Jonasson J., Lindahl T.L., Osman A.. Next generation sequencing analysis of human platelet polyA+ mRNAs and rRNA-depleted total RNA. PLoS One. 2013; 8:e81809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alhasan A.A., Izuogu O.G., Al-Balool H.H., Steyn J.S., Evans A., Colzani M., Ghevaert C., Mountford J.C., Marenah L., Elliott D.J. et al. . Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016; 127:e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doss J.F., Corcoran D.L., Jima D.D., Telen M.J., Dave S.S., Chi J.T.. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genomics. 2015; 16:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kornienko A.E., Dotter C.P., Guenzl P.M., Gisslinger H., Gisslinger B., Cleary C., Kralovics R., Pauler F.M., Barlow D.P.. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016; 17:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng J., Metge F., Dieterich C.. DCC – specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2015; 32:1–13. [DOI] [PubMed] [Google Scholar]
- 33. Zhang X.O., Wang H. Bin, Zhang Y., Lu X., Chen L.L., Yang L.. Complementary sequence-mediated exon circularization. Cell. 2014; 159:134–147. [DOI] [PubMed] [Google Scholar]
- 34. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kolde R. Pheatmap: pretty heatmaps. 2012.
- 36. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2009; NY: Springer. [Google Scholar]
- 37. Ovchynnikova E., Aglialoro F., Bentlage A.E.H., Vidarsson G., Salinas N.D., Von Lindern M., Tolia N.H., Van Den Akker E.. DARC extracellular domain remodeling in maturating reticulocytes explains Plasmodium vivax tropism. Blood. 2017; 130:1441–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L.. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012; 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeng X., Lin W., Guo M., Zou Q.. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017; 13:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hansen T.B. Improved circRNA identification by combining prediction algorithms. Front. Cell Dev. Biol. 2018; 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Glažar P., Papavasileiou P., Rajewsky N.. circBase: a database for circular RNAs. RNA. 2014; 20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y.C., Li J.R., Sun C.H., Andrews E., Chao R.F., Lin F.M., Weng S.L., Hsu S. Da, Huang C.C., Cheng C. et al. . CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016; 44:D209–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pimentel H., Parra M., Gee S., Ghanem D., An X., Li J., Mohandas N., Pachter L., Conboy J.G.. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2014; 42:4031–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rifkind J.M., Nagababu E.. Hemoglobin redox reactions and red blood cell aging. Antioxid. Redox Signal. 2013; 18:2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. et al. . Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Izuogu O.G., Alhasan A.A., Mellough C., Collin J., Gallon R., Hyslop J., Mastrorosa F.K., Ehrmann I., Lako M., Elliott D.J. et al. . Analysis of human ES cell differentiation establishes that the dominant isoforms of the lncRNAs RMST and FIRRE are circular. BMC Genomics. 2018; 19:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kelly S., Greenman C., Cook P.R., Papantonis A.. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015; 427:2414–2417. [DOI] [PubMed] [Google Scholar]
- 49. Tan W.L.W., Lim B.T.S., Anene-Nzelu C.G.O., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D.A. et al. . A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017; 113:298–309. [DOI] [PubMed] [Google Scholar]
- 50. Greene J., Baird A.-M., Brady L., Lim M., Gray S.G., McDermott R., Finn S.P.. Circular RNAs: Biogenesis, function and role in human diseases. Front. Mol. Biosci. 2017; 4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 52. Jeck W.R., Sharpless N.E.. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014; 32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.-L., Wang Y. et al. . Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017; 27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kristensen L.S., Hansen T.B., Venø M.T., Kjems J.. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018; 37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All bioinformatics tools used in this study are described in the supplementary method. The following datasets were used in this study: SRP065216, ERR335311, ERR335312, ERR335313, SRR2124299, SRR2124300, SRR2124301, SRR2038798, ERR789064, ERR789082, ERR789195, ERR789201, and are available on SRA (https://www.ncbi.nlm.nih.gov/sra).