Abstract
Faithful chromosome segregation depends on correct spindle microtubule-kinetochore attachment and requires certain spindle-associated proteins (SAPs) involved in regulating spindle dynamics and chromosome segregation. Little is known about the spindle-associated proteome in the early divergent Trypanosoma brucei and its roles in chromosome segregation. Here we report the identification of a cohort of divergent SAPs through localization-based screening and proximity-dependent biotin identification. We identified seven new SAPs and seventeen new nucleolar proteins that associate with the spindle, and demonstrated that the kinetochore protein KKIP4 also associates with the spindle. These SAPs localize to distinct subdomains of the spindle during mitosis, and all but one localize to nucleus during interphase and post-mitotic phases. Functional analyses of three nucleus- and spindle-associated proteins (NuSAPs) revealed distinct functions in chromosome segregation. NuSAP1 is a kinetoplastid-specific protein required for equal chromosome segregation and for maintaining the stability of the kinetochore proteins KKIP1 and KKT1. NuSAP2 is a highly divergent ASE1/PRC1/MAP65 homolog playing an essential role in promoting the G2/M transition. NuSAP3 is a kinetoplastid-specific Kif13-1-binding protein maintaining Kif13-1 protein stability and regulating the G2/M transition. Together, our work suggests that chromosome segregation in T. brucei requires a cohort of kinetoplastid-specific and divergent SAPs with distinct functions.
INTRODUCTION
Faithful chromosome segregation is crucial for maintaining genome integrity and for cell viability in all organisms. Chromosome segregation is triggered by the activation of the anaphase-promoting complex/cyclosome (APC/C), which is inhibited by the spindle assembly checkpoint (SAC) that functions as a surveillance system by monitoring kinetochore-spindle microtubule attachment errors and delaying anaphase onset (1). Once activated, the APC/C ubiquitinates securin, an inhibitor of the cysteine protease separase, leading to the degradation of securin by the 26S proteasome. Consequently, separase cleaves cohesin, thereby releasing the bound sister chromatids for segregation through the elongation of the mitotic spindle and the separation of spindle poles towards the cell cortex (2,3).
Assembly of a bipolar mitotic spindle is necessary for accurate segregation of sister chromatids during mitosis, and involves the microtubule nucleation pathways mediated by the centrosome, the chromatin, and the microtubule (4). The mitotic spindle is a macromolecular machine composed of microtubule bundles, non-motor microtubule-associated proteins (MAPs), and motor proteins that orchestrate forces to organize and separate the chromosomes (5). Numerous (>200) spindle-associated proteins (SAPs) have been identified in animals (5), and they play diverse functions in regulating spindle dynamics and chromosome segregation (6).
Trypanosoma brucei belongs to the kinetoplastida group of flagellated protists, which also include Trypanosoma cruzi and Leishmania spp., and is the causative agent of sleeping sickness in humans and nagana in cattle in sub-Saharan Africa. T. brucei has a complex life cycle by alternating between the tsetse fly vector and the mammalian hosts. Trypanosoma brucei proliferates as the procyclic (insect) form in the midgut of the tsetse fly and as the bloodstream form in the mammalian bloodstream, and there are considerable differences in cell cycle control between the two life cycle forms. Trypanosoma brucei undergoes a closed mitosis by assembling an intranuclear mitotic spindle, and constructs the kinetochores with 20 kinetoplastid-specific proteins and a divergent Ndc80 homolog (7–9). It is noteworthy that the number of kinetochore-like plaques detected in T. brucei is less than the number of the mega-base chromosomes (10). T. brucei also contains ∼100 mini-chromosomes, which do not have centromere activity (11) but are segregated through a spindle microtubule-dependent mechanism during mitosis (12). Distinct structures of the centriole have not been detected at the spindle poles, despite the presence of a ring-like structure near the nuclear envelop at the spindle poles (13). This ring-like structure at the two spindle poles may function as intranuclear microtubule-organizing centers to nucleate spindle microtubules (13). Notably, the well-recognized microtubule nucleation machinery, known as the γ-tubulin complex, is not detected at the spindle poles and is not required for spindle assembly in T. brucei (14), raising the question of how spindle microtubule nucleation is controlled.
A number of SAPs have been identified and functionally characterized in T. brucei. These SAPs include the Aurora B kinase TbAUK1 (15) and its associated chromosomal passenger proteins TbCPC1 and TbCPC2 (16), the TbAUK1-interacting kinesins KIN-A and KIN-B (16), the nucleolar protein TbNOP86 (17), the MCAK family kinesin Kif13-1 (18,19), XMAP215 (20), TbMlp2/TbNup92 (21,22), Kharon1 (23), MAP103 (24) and a spindle pole-localizing Tousled-like kinase homolog TbTLK1 (25). Some of these SAPs appear to be required for spindle assembly and chromosome segregation (15,16,25). However, the mechanism and the regulation of spindle assembly and chromosome segregation in T. brucei remain poorly understood.
In this report, we set out to identify new SAPs in T. brucei by localization-based screening and proximity-dependent biotin identification (BioID) (26) using some known SAPs as baits, and to characterize the function of selected SAPs in chromosome segregation. Our work suggests that faithful chromosome segregation in T. brucei requires a cohort of kinetoplastid-specific SAPs and divergent SAPs that localize to distinct subdomains of the mitotic spindle and play distinct functions in regulating chromosome segregation.
MATERIALS AND METHODS
Trypanosome cell culture
Two procyclic (insect) forms of T. brucei cell lines were used in this work. The 29-13 cell line (27), which was used for RNA interference, was cultured in SDM-79 medium containing 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Inc), 15 μg/ml G418 and 50 μg/ml hygromycin. The 427 cell line, which was used for protein tagging, was cultured at 27°C in SDM-79 medium supplemented with 10% fetal bovine serum. Cells were diluted once the cell density reaches 5 × 106 cells/ml.
Identification of SAPs by localization-based screening
The 30 hypothetical proteins whose transcripts were highly enriched in S phase (Supplementary Table S1), the 21 orphan kinesins and kinetoplastid-specific kinesins (Supplementary Table S2), and the T. brucei Fidgetin homolog (Tb927.5.1870) were each endogenously tagged with a triple HA epitope by the PCR-based gene tagging method (28) in the 427 cell line. Transfectants were selected with 1 μg/ml puromycin and cloned by limiting dilution in a 96-well plate containing SDM-79 medium supplemented with 20% heat-inactivated fetal bovine serum. Western blotting was carried out with anti-HA antibody (clone# HA-7, Catalog # H3663, Sigma-Aldrich) to confirm the tagging of proteins. Cells were then immunostained with FITC-conjugated anti-HA antibody (1:400 dilution, Clone# HA-7, Catalog# H7411, Sigma-Aldrich) and imaged under an inverted fluorescence microscope.
Bioinformatics and sequence analyses of spindle-associated proteins
The search for homologs of T. brucei SAPs was performed with Hidden Markov Models (HMM) by using the jackhammer algorithm for iterative search against reference proteomes, UniProtKB, and SwissProt databases (www.hmmer.org) (29). The search for conserved motifs in the new SAPs was performed using the NCBI conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (30). The search for coiled-coil motifs in the new SAPs was performed using the COILS algorithm (https://embnet.vital-it.ch/software/COILS_form.html) (31).
Expression of BirA*-fused SAPs in T. brucei and BioID
To express BirA*-fused spindle-associated proteins in T. brucei, the full-length coding sequence of NuSAP1, NuSAP2, Kif13-1, TbMlp2 and TbAUK1 was cloned into pLew100-BirA*-HA-BLE vector (32). The resulting plasmids were each linearized with NotI and then transfected into T. brucei 29-13 cell line. Transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution. Expression of BirA*-fusion proteins was induced with 0.5 μg/ml tetracycline, and verified by western blotting with anti-HA antibody and immunofluorescence microscopy with FITC-conjugated anti-HA antibody.
BioID was carried out essentially as described in our previous publications (32–34). BirA*-HA-fused proteins were each overexpressed by induction with 0.5 μg/ml tetracycline for 24 h, and cells (∼3 × 109) were incubated with 50 μM biotin for an additional 24 h. Cells were washed three times with PBS and treated with lysis buffer (0.4% SDS, 500 mM NaCl, 5 mM EDTA, 1 mM DTT, 50 mM Tris–HCl, pH 7.4). Cleared lysate was incubated with 500 μl of streptavidin-coated Dynabeads (Invitrogen) at 4°C for 4 h. As the negative control, non-induced control cells (∼3 × 109) were similarly treated and subject to purification with the same amount of Dynabeads. Dynabeads were washed five times with 50 mM ammonium bicarbonate and re-suspended in 100 mM ammonium bicarbonate. To reduce the disulfide bond, 10% DTT was added, followed by adding 50% iodoacetamide for alkylation. Subsequently, 5% DTT was added to the solution, and bound proteins were digested with trypsin at 37°C overnight. Trypsin digestion was stopped by adding trifluoroacetic acid to approximately pH2.0. The protein digests were desalted and analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) interfaced with an Eksgent nano-LC 2D plus chipLC system (Eksigent Technologies) at the Proteomics Core Facility of the University of Texas Health Science Center at Houston.
Data analysis was performed according to our published methods (22,28). Raw data files were searched against the T. brucei genome database using the Mascot search engine. The search conditions used peptide tolerance of 10 ppm and MS/MS tolerance of 0.8 Da with the enzyme trypsin and two missed cleavages.
RNA interference
To generate RNAi cell lines, a 578-bp DNA fragment (nucleotides 1453-2030) from the coding region of the NuSAP1 gene, a 500-bp DNA fragment (nucleotides 1-500) from the coding region of the NuSAP2 gene, a 580-bp DNA fragment (nucleotides 509-1088) from the coding region of the NuSAP3 gene, a 567-bp DNA fragment (nucleotides 1–567) from the coding region of the Fidgetin gene, and a 435-bp DNA fragment (nucleotides 350-784) from the coding region of the SPB1 gene were each cloned into the pZJM vector (35). A 448-bp fragment (nucleotides 1149–1696) from the coding region of the Kif13-1 gene was cloned into the stem-loop RNAi vector pSL (36). The resulting plasmids were each linearized with NotI and transfected into the 29-13 cell line by electroporation according to our published procedures (37). Transfectants were selected with 2.5 μg/ml phleomycin and cloned by limiting dilution. RNAi was induced by 1.0 μg/ml tetracycline. Cell growth was monitored daily by counting the cells with a hemacytometer under an inverted light microscope.
In situ epitope tagging of proteins
Epitope tagging of SAPs from their respective endogenous locus was carried out using the PCR-based method (28), and transfectants were selected with appropriate antibiotics (40 μg/ml G418 or 1 μg/ml puromycin) and cloned by limiting dilution as described above. For co-localization of SAPs with the spindle, NuSAP1, NuSAP2, NuSAP4 and SPB1 were tagged with a C-terminal PTP epitope (neomycin resistance) in the cell line that expresses the N-terminal 3HA-tagged β-tubulin (puromycin resistance), whereas NuSAP3, KKIP4, KIN-F, and Fidgetin were tagged with a C-terminal triple HA epitope (neomycin resistance) in the cell line that expresses the C-terminal PTP-tagged Kif13-1 (puromycin resistance). For co-localization of SAPs with kinetochores, NuSAP1-NuSAP4, Fidgetin and KIN-F were each tagged with a C-terminal PTP epitope (neomycin resistance) in the cell line that expresses the C-terminal triple HA-tagged KKT2 (puromycin resistance). KKT2 was tagged with a C-terminal PTP epitope (puromycin resistance) and all other kinetochore proteins described in Figure 7 were tagged with a C-terminal triple HA epitope (puromycin resistance) in pZJM-NuSAP1 RNAi cell line. β-Tubulin was endogenously tagged with an N-terminal triple HA epitope (puromycin resistance) in pZJM-NuSAP1 RNAi cell line. Kif13-1 and NUP89 (38) were each endogenously tagged with a C-terminal triple HA epitope (puromycin resistance) in pZJM-NuSAP3 RNAi cell line. NuSAP3 was endogenously tagged with a C-terminal triple HA epitope (puromycin resistance) in pZJM-Kif13-1 RNAi cell line.
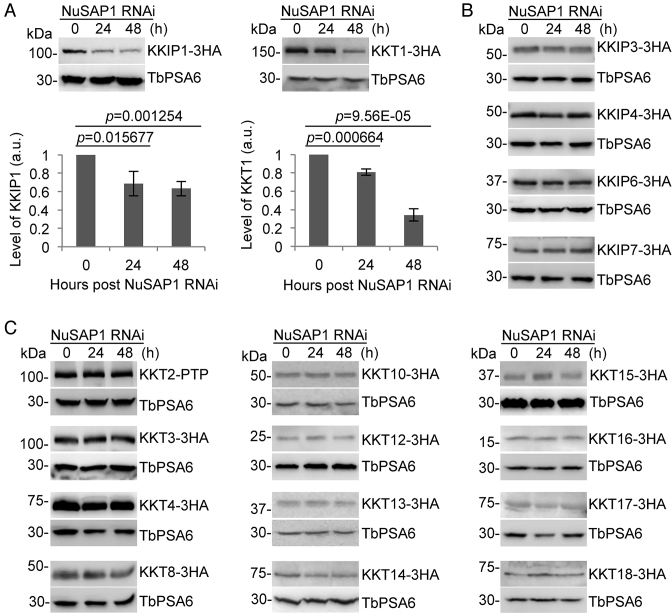
Figure 7.
Depletion of NuSAP1 destabilizes the kinetochore proteins KKIP1 and KKT1. (A) Effect of NuSAP1 depletion on the stability of the outer kinetochore protein KKIP1 and the inner kinetochore protein KKT1. KKIP1 and KKT1 were each endogenously tagged with a triple HA epitope at their respective C-terminus in NuSAP1 RNAi cell line. Western blotting was carried out with anti-HA antibody and anti-TbPSA6 antibody, which served as the loading control. The graphs below the western blots are the quantitation of KKIP1 and KKT1 protein intensity from three independent experiments. Error bars indicate S.D. (B) Effect of NuSAP1 depletion on the stability of other outer kinetochore proteins (KKIP3, KKIP4, KKIP6 and KKIP7). KKIP proteins were endogenously tagged with a C-terminal triple HA epitope and detected by western blotting with anti-HA antibody. TbPSA6 served as the loading control. (C) Effect of NuSAP1 RNAi on the stability of other inner kinetochore proteins (KKT2-KKT4, KKT8-KKT10 and KKT12-KKT18). KKT2 was endogenously tagged with a PTP epitope, and the other KKT proteins were endogenously tagged with a triple HA epitope and detected by anti-Protein A antibody or anti-HA antibody, respectively. TbPSA6 served as the loading control.
Co-immunoprecipitation and western blotting
Co-immunoprecipitation was carried out according to our previous procedures (33). Briefly, cells (5 × 107) expressing endogenously PTP-tagged Kif13-1 and 3HA-tagged NuSAP3 were lysed by incubating with 1 ml immunoprecipitation buffer (25 mM Tris–HCl, pH7.6, 100 mM NaCl, 1 mM DTT, 1% NP-40, and protease inhibitor cocktail) on ice for 30 min. Cleared lysate was incubated with 50 μl IgG Sepharose 6 fast flow affinity resin (Catalog # 17096901, GE Healthcare) at 4°C for 1 h, and immunoprecipitates were washed six times with the immunoprecipitation buffer. Proteins bound to the IgG resin were eluted with 10% SDS, separated by SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with anti-HA antibody to detect 3HA-tagged NuSAP3 and with anti-Protein A antibody (catalog # P3775, Sigma-Aldrich) to detect PTP-tagged Kif13-1. Cells expressing Kif13-1-PTP alone and NuSAP3-3HA alone were included as negative controls. The experiment was repeated three times.
Immunofluorescence microscopy
Cells were washed once with PBS, adhered to the coverslips for 30 min at room temperature, fixed with cold methanol (–20°C) for 30 min, and then rehydrated with PBS for 10 min at room temperature. Cells adhered on the coverslips were blocked with 3% BSA in PBS for 1 h at room temperature, and incubated with the primary antibody for 1 h at room temperature. The following primary antibodies were used: FITC-conjugated anti-HA monoclonal antibody for 3HA-tagged proteins, and anti-Protein A polyclonal antibody for PTP-tagged proteins (1:400 dilution, catalog # P3775, Sigma-Aldrich). Subsequently, cells were washed three times with PBS, and then incubated with Cy3-conjugated anti-rabbit IgG (1:400 dilution, Catalog# C2306, Sigma-Aldrich) for 1 h at room temperature. Cells on the coverslips were washed three times with PBS, mounted with DAPI-containing VectaShield mounting medium (Vector Labs), and imaged under an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60× 1.42-NA lens. Images were acquired using the Slidebook 5 software.
Fluorescence in situ hybridization (FISH)
To detect chromosome segregation defects in NuSAP1 RNAi cells, the coding sequence of M5 ribosomal RNA (chromosome 8) and the minichromosomal 177-bp DNA repeat (12) were used as markers for mega-base chromosome and mini-chromosomes, respectively. The coding sequence of M5 ribosomal RNA gene was amplified by PCR with the primers (Forward: 5′-GGGTACGACCATACTTGGC-3′ and Reverse: 5′-AGAGTACAACACCCCGGGTT-3′). The mini-chromosomal 177-bp DNA repeat was amplified with the primers (Forward: 5′-TAAATGGTTCTTATACGAATG-3′ and Reverse: 5′AACACTAAAGAACAGCGTTG-3′). The two DNA fragments were directly labeled with digoxigenin-dUTP (Sigma) during PCR and purified. Prior to hybridization, digoxigenin-labeled DNA probes were precipitated with salmon sperm DNA (10 mg/ml) and yeast tRNA (10 mg/ml), and re-suspended in hybridization buffer (50% formamide, 2× saline-sodium citrate (SSC), 10% dextran sulfate). FISH was performed according to the protocol published previously (12) with minor modifications. Cells were collected by centrifugation, washed twice with PBS, and then attached to glass coverslips for 30 min. Cells were then fixed with cold methanol (–20°C) for 30 min, rehydrated in PBS for 10 min, and then pre-hybridized in hybridization buffer for 60 min. Prior to hybridization, digoxigenin-labeled probes and fixed cells were denatured at 95°C for 5 min. Cells were incubated with digoxigenin-labeled probes at 37°C for 16 h in a humidified chamber. After hybridization, cells were washed three times in 50% formamide, 2× SSC at 37°C, three times in 0.2% SSC at 50°C, three times in 4× SSC at room temperature. For detection of digoxigenin-labeled DNA probes, cells were blocked with 4× SSC, 3% BSA for 30 min, and then incubated with FITC-conjugated anti-digoxigenin antibody (1:400 dilution, Clone# DI-22, Catalog# F3523, Sigma-Aldrich) for 1 hour. After washing three times with 2× SSC, cells were mounted in DAPI-containing VectaShield (Vector Labs) mounting medium, and imaged under an inverted fluorescence microscope as described above.
Statistical analysis
Statistical analysis was performed using the t-test in the Microsoft Excel software. Detailed n values for each panel in the figures were stated in the corresponding legends. For immunofluorescence microscopy, images were randomly taken and all cells in each image were counted.
RESULTS
Identification of spindle-associated proteins in T. brucei
Previous studies showed that certain proteins involved in chromosome segregation and spindle assembly were up-regulated from the S phase of the cell cycle (39). Thus, we anticipated that some SAPs might also be up-regulated from S phase. To identify these SAPs, we selected 30 hypothetical proteins (Supplementary Table S1) that are highly up-regulated during S phase, tagged them with a triple HA epitope in the procyclic form of T. brucei, and examined their subcellular localization by immunofluorescence microscopy. Two hypothetical proteins were found to be localized to the nucleus during interphase and to the spindle during mitosis (Supplementary Figure S1A), and we named them NuSAP1 (Tb927.11.8370) and NuSAP2 (Tb927.9.6110) for Nucleus- and Spindle-Associated Protein 1 and 2, respectively. Using the COILS program (31), four coiled-coil motifs were detected in NuSAP1 (Supplementary Figure S1B). Further analysis using Hidden Markov Models (HMM) (29) identified NuSAP1 homologs in the kinetoplastid parasites only (Supplementary Figure S2A), suggesting that NuSAP1 is a kinetoplastid-specific protein. By searching the conserved domain database (CDD) (30), we found that NuSAP2 contains a C-terminal ∼277-aa (a.a. 695–971) sequence homologous to the MAP65/ASE1 superfamily (Supplementary Figure S1B). This superfamily of proteins include the Arabidopsis thaliana microtubule-associated protein MAP65 (40), the Sacchromyces cerevisiae ASE1 (Anaphase Spindle Elongation 1) protein (41), and the human PRC1 (Protein Regulator of Cytokinesis 1) protein (42), all of which are involved in anaphase spindle elongation and cytokinesis. Analysis of NuSAP2 by HMM identified close homologs in kinetoplastid parasites and yeast ASE1 as a distant homolog (Supplementary Figure S2B). It is noteworthy that the MAP65/ASE1-like domain in NuSAP2 is only about half of the conserved MAP65/ASE1 domain found in ASE1, MAP65 and PRC1 (Supplementary Figure S3A, B). Alignment of this 277-aa sequence in NuSAP2 with the corresponding sequences in ASE1, PRC1 and MAP65 showed an overall sequence identity of only ∼10% (Supplementary Figure S3C), and phylogenetic analysis placed the NuSAP2 homologs from the kinetoplastid parasites in a distinct clade (Supplementary Figure S3D). These results suggest that NuSAP2 is a highly divergent homolog of ASE1/PRC1/MAP65.
We recently also attempted to characterize the over-represented kinetoplastid-specific kinesins and orphan kinesins in T. brucei (43). We epitope-tagged 22 such kinesins (Supplementary Table S2), and immunofluorescence microscopy showed that one orphan kinesin (Tb927.3.2020) localizes to the nucleus during interphase and to the spindle during mitosis (Supplementary Figure S1A). We named this kinesin KIN-F, following our nomenclature of the T. brucei orphan kinesins (44–46). KIN-F is a new spindle-associated kinesin in T. brucei, in addition to KIN-A, KIN-B, and Kif13-1. Additionally, since the Leishmania major Fidgetin homolog localizes to the spindle (47), we asked whether the T. brucei Fidgetin homolog (Tb927.5.1870) also localizes to the spindle, and we found that it localizes to the nucleus during interphase and to the spindle during mitosis (Supplementary Figure S1A). Together, the localization-based approach allowed the identification of four nucleus- and spindle-associated proteins, NuSAP1, NuSAP2, KIN-F and Fidgetin.
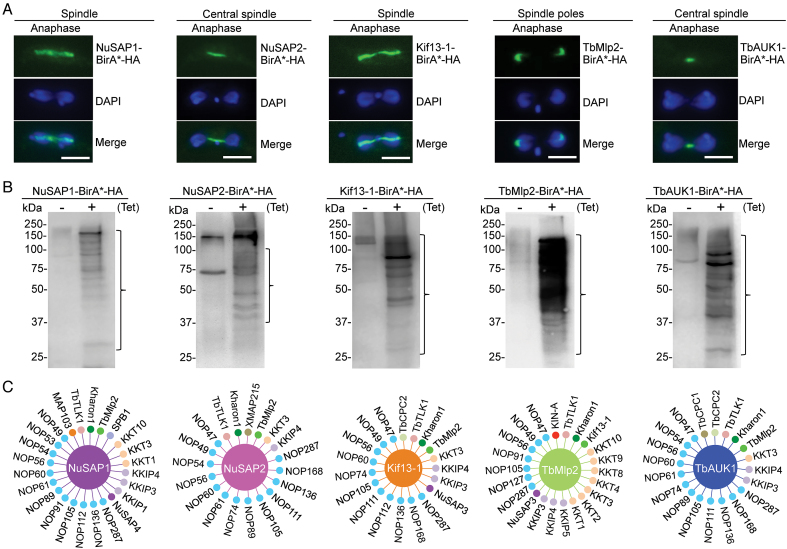
We next employed the BioID approach to identify new SAPs, using NuSAP1, NuSAP2 and some known SAPs (Kif13-1, TbMlp2 and TbAUK1), which localize to distinct sub-domains of the spindle, as the baits (Figure 1A). NuSAP1 and Kif13-1 localize to the entire spindle during anaphase, NuSAP2 and TbAUK1 localize to the central spindle during anaphase, and TbMlp2 localizes to the spindle poles during anaphase (Figure 1A). Expression of BirA*-fused NuSAP1, NuSAP2, Kif13-1, TbMlp2 and TbAUK1 all resulted in biotinylation of numerous proteins, as detected by western blotting with anti-streptavidin antibody (Figure 1B). Biotinylated proteins were purified and analyzed by LC–MS/MS, and the proteins identified by mass spectrometry were first searched for the SAPs published previously. Collectively, BioID experiments with the five baits were able to identify seven previously known SAPs, TbCPC1 (16), TbCPC2 (16), KIN-A (16), TbTLK1 (25), Kharon1 (23), XMAP215 (20) and MAP103 (24) (Figure 1C), thus validating the feasibility of BioID for identifying SAPs.
Figure 1.
Identification of spindle-associated proteins by BioID. (A) Confirmation of the subcellular localization of BirA*-fused spindle-associated proteins. Cells expressing ectopically BirA*-HA-tagged NuSAP1, NuSAP2, Kif13-1, TbMlp2 and TbAUK1 were immunostained with FITC-conjugated anti-HA antibody and counterstained with DAPI. Scale bar: 5 μm. (B) Detection of biotinylated proteins (outlined with brackets) by western blotting with anti-Streptavidin antibody. (C) Summary of spindle-associated proteins identified by BioID with the five spindle-associated proteins as baits. The bait proteins are each depicted as a large circle in the middle, and their respective proximity partners are depicted as small circles surrounding the bait proteins. MAP103, TbTLK1, Kharon1, TbCPC1, TbCPC2, KIN-A and XMAP215 have been demonstrated as SAPs previously.
To identify new SAPs from the BioID experiments, we epitope-tagged the hypothetical proteins that have high Mascot scores (Supplementary Tables S3–S7), and examined their subcellular localizations by immunofluorescence microscopy. Two proteins, identified by Kif13-1 BioID and NuSAP1 BioID, respectively (Figure 1C), were found to localize to the nucleus during interphase and to the spindle during mitosis (Supplementary Figure S1A) and were thus named NuSAP3 (Tb927.7.4450) and NuSAP4 (Tb927.4.1730). NuSAP3 contains a VHS (domain present in VPS-27, Hrs and STAM) motif at the N-terminus (Supplementary Figure S1B), and NuSAP4 contains a coiled-coil motif at the C-terminus (Supplementary Figure S1B). Sequence analysis by HMM identified their homologs only in the kinetoplastid parasites (Supplementary Figure S2C, D), suggesting that NuSAP3 and NuSAP4 are kinetoplastid-specific proteins. One protein, identified by NuSAP1 BioID (Figure 1C), was found to localize to kinetochores during interphase and to the spindle during mitosis (Supplementary Figure S1A), and was recently reported as an outer kinetochore protein named KKIP4 (9). Another protein, also identified by NuSAP1 BioID (Figure 1C), was found to localize to the spindle poles during mitosis (Supplementary Figure S1A), and was named SPB1 (Tb927.11.9690) for Spindle Pole Body protein 1. SPB1 contains eight MORN (Membrane Occupation and Recognition Nexus) repeats (Supplementary Figure S1B), and sequence analysis by HMM identified numerous MORN domain-containing proteins throughout all three domains of life (data not shown). Seventeen proteins, identified by a collective BioID with the five baits (Figure 1C), were found to localize to the nucleolus during interphase and to the spindle during mitosis (Supplementary Figure S4). These proteins are nucleolar proteins that associate with spindle microtubules during mitosis for segregation into the nuclei. Thus, they were named NOP47 to NOP287 for Nucleolar Protein of XXX kDa (XXX denotes numbers). Three proteins were found to localize to kinetochores, which were recently reported as outer kinetochore proteins named KKIP1, KKIP3, and KKIP5 (9). A further analysis of the mass spectrometry data detected seven other kinetochore proteins, KKT1-KKT4 and KKT8-KKT10, among which KKT4 was recently reported to additionally localize to the spindle during metaphase (Llauro, A., Hayashi, H., Bailey, M.E., Wilson, A., Ludzia, P., Asbury, C.L. and Akiyoshi, B., 2017 BioRxiv, https://doi.org/10.1101/216812). These kinetochore proteins were identified by a collective BioID experiments with the five baits (Figure 1C).
Altogether, through localization-based screening and proximity interactions with known SAPs as baits, we have identified a total of eight new SAPs, including six nucleus- and spindle-associated proteins (NuSAP1-NuSAP4, KIN-F, and Fidgetin), a spindle pole-associated protein (SPB1), and a kinetochore- and spindle-associated protein (KKIP4) (Supplementary Table S8). The BioID experiments also identified seventeen new nucleolar proteins (NOPs) that associate with the spindle, many of which may be involved in ribosome biogenesis or nucleolus-related biological processes based on the functions of their close homologs in yeast and humans (Supplementary Table S8).
Subcellular localizations of SAPs during the cell cycle
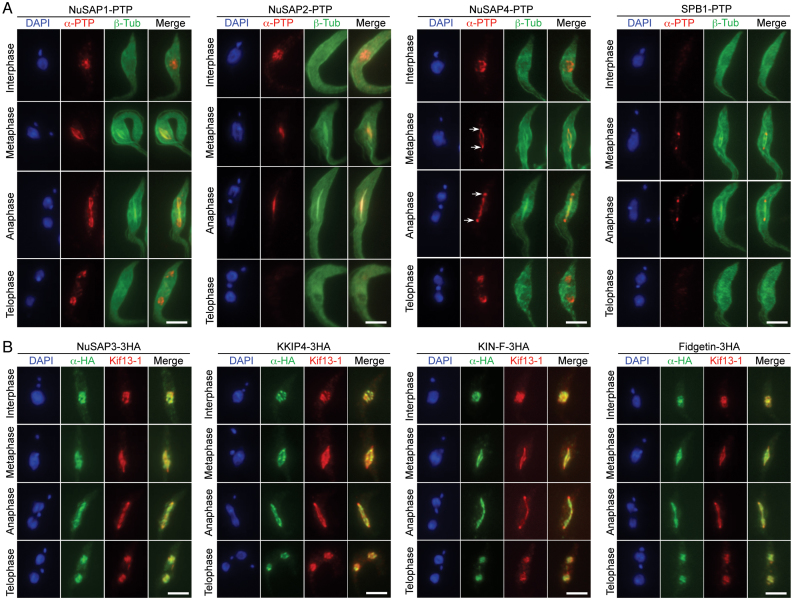
The localization of the newly identified SAPs, excluding the nucleolar proteins NOP47 to NOP287, during the cell cycle was investigated, and their localization to spindle microtubules was confirmed by co-immunostaining with the spindle marker. We endogenously tagged β-tubulin with an N-terminal triple HA epitope in cells expressing endogenously PTP-tagged NuSAP1, NuSAP2, NuSAP4 and SPB1 at their C-terminus (Figure 2A). Another spindle marker, Kif13-1 (18,19), was endogenously tagged with a C-terminal PTP epitope and used for co-localization with 3HA-tagged NuSAP3, KKIP4, KIN-F and Fidgetin (Figure 2B).
Figure 2.
Subcellular localizations of spindle-associated proteins during the cell cycle. (A) Determination of the subcellular localization of NuSAP1, NuSAP2, NuSAP4 and SPB1 by co-immunofluorescence microscopy with β-tubulin to label spindle microtubules. Each of these proteins was tagged with a C-terminal PTP epitope, and β-tubulin was tagged with an N-terminal triple HA epitope. Scale bars: 5 μm. (B) Determination of the subcellular localization of NuSAP3, KKIP4, KIN-F, and Fidgetin by co-immunostaining with Kif13-1 as a spindle marker. NuSAP3, KKIP4, KIN-F and Fidgetin were each endogenously tagged with a C-terminal triple HA epitope, and Kif13-1 was endogenously tagged with a C-terminal PTP epitope. Scale bars: 5 μm.
By immunofluorescence microscopy, five SAPs, NuSAP1, NuSAP3, KKIP4, KIN-F and Fidgetin, were found to display similar subcellular localization patterns during the cell cycle (Figure 2A, B). They were each detected as punctate dots within the nucleus in interphase and telophase cells, and were detected on the spindle microtubules during metaphase and anaphase (Figure 2A, B). KKIP4 was recently reported as an outer kinetochore protein (9), but our result showed that KKIP4 was distributed on the entire spindle during metaphase and anaphase (Figure 2B). Given that KKIP4 was tagged with a smaller triple HA epitope at the C-terminus in our work and was tagged with YFP at the N-terminus in the previous work (9), the distinction in KKIP4 localization might be attributed to the different size and location of the epitope tag used for determining KKIP4 localization.
The rest three SAPs, NuSAP2, NuSAP4 and SPB1, appeared to have distinct subcellular localization patterns. NuSAP2 was detected as punctate dots within the nucleus in interphase cells (Figure 2A), but it was detected at the central portion of the spindle during metaphase and anaphase and was not detectable in telophase cells (Figure 2A). NuSAP4 was detected as punctate dots in the nucleus during interphase and telophase, and was detected on the spindle microtubules and was somewhat enriched at the spindle poles during metaphase and anaphase (Figure 2A, arrows). SPB1 was not detectable in interphase and telophase cells, but it was detected at the spindle poles during metaphase and anaphase (Figure 2A).
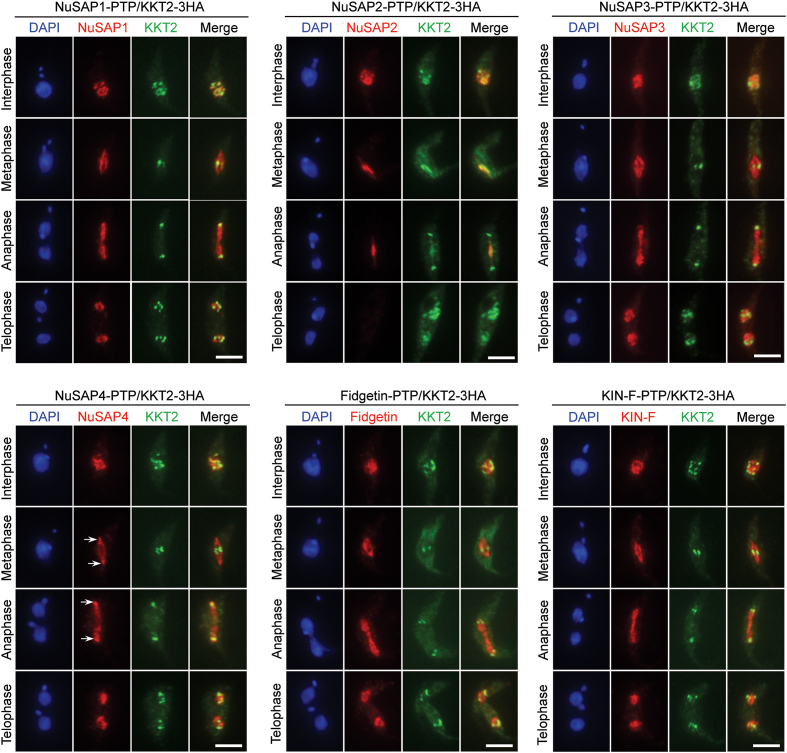
Partial co-localization of SAPs with kinetochores during the cell cycle
The observation that NuSAP1, NuSAP3, NuSAP4, Fidgetin and KIN-F were all detected as punctate dots in the nucleus during interphase and telophase and that NuSAP2 as punctate dots in the nucleus in interphase (Figure 2) prompted us to examine whether they localize to kinetochores, as in the case of KKIP4 (Figure 2B). To this end, these SAPs were each endogenously tagged with a PTP epitope in cells co-expressing 3HA-tagged KKT2 for co-immunofluorescence microscopy. NuSAP1 and NuSAP4 appeared to co-localize with KKT2 at almost all of the punctate dots in interphase, but the two proteins only partly overlapped with KKT2 in telophase (Figure 3). During mitotic phases, NuSAP1 was on the entire spindle, but it overlapped with KKT2 at the equatorial plate of the metaphase spindle and the poles of the anaphase spindle (Figure 3), whereas the spindle pole-enriched NuSAP4 overlapped with KKT2 in anaphase (Figure 3). NuSAP2 also co-localized with KKT2 at almost all of the punctate dots in interphase and at the equatorial plate of the anaphase spindle (Figure 3). NuSAP3, Fidgetin and KIN-F, however, only partly overlapped with KKT2 in the nucleus during interphase and telophase, at the equatorial plate of the metaphase spindle, and at the poles of the anaphase spindle (Figure 3). These results indicate that NuSAP1, NuSAP2 and NuSAP4 likely associate with kinetochores during interphase.
Figure 3.
Partial co-localization of SAPs with kinetochores during the cell cycle. Cells co-expressing PTP-tagged SAPs (NuSAP1-NuSAP4, Fidgetin and KIN-F) and 3HA-tagged kinetochore protein KKT2 were co-immunostained with anti-Protein A polyclonal antibody and FITC-conjugated anti-HA monoclonal antibody. Cells were counterstained with DAPI for DNA. Scale bars: 5 μm.
NuSAP1 is a kinetoplastid-specific protein required for faithful chromosome segregation
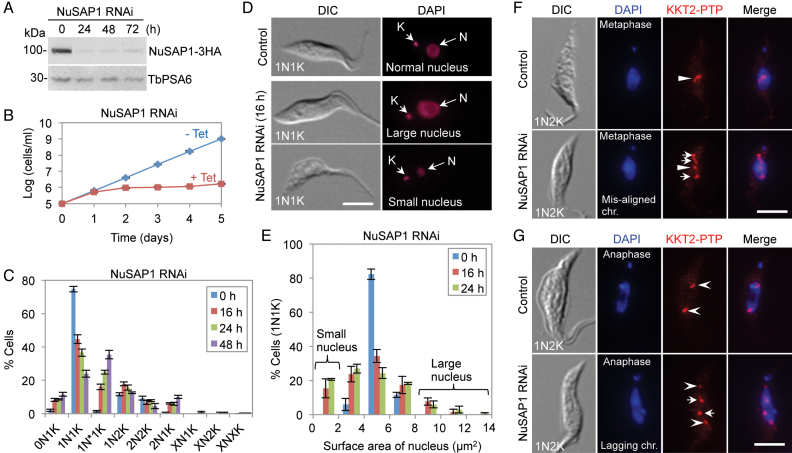
To investigate the function of these newly identified SAPs, we carried out RNAi in the procyclic form of T. brucei to knock down the seven SAPs except KKIP4, as KKIP4 has been demonstrated to be nonessential (9). We successfully generated the RNAi cell line for NuSAP1-NuSAP3, Fidgetin, and SPB1, but failed to obtain the RNAi cell line for NuSAP4 and KIN-F due to low knockdown efficiency of these two proteins. Depletion of Fidgetin and SPB1 did not cause any growth defects in the procyclic form (Supplementary Figure S5). Therefore, our functional analysis of SAPs was focused on NuSAP1, NuSAP2 and NuSAP3, as all of them are essential in the procyclic form (see below). Western blotting confirmed the knockdown of NuSAP1, which was tagged with a C-terminal triple HA epitope at one of its endogenous loci, after RNAi induction for 24 h (Figure 4A). Depletion of NuSAP1 caused a severe growth defect (Figure 4B), suggesting that NuSAP1 is essential for cell proliferation. To analyze potential cell cycle defects caused by NuSAP1 RNAi, we counted the cells of different numbers of nucleus (N) and kinetoplast (K), the cell's unique mitochondrial genome. The different cell cycle stages in T. brucei can be distinguished by the numbers of nucleus and kinetoplast in each cell. Cells at G1 and S phases contain one nucleus and one kinetoplast (1N1K), cells at G2 and early mitosis (prometaphase to early anaphase) contain one nucleus and two kinetoplasts (1N2K), and cells from late anaphase to cytokinesis contain two nuclei and two kinetoplasts (2N2K). We found that after NuSAP1 RNAi, there was a significant decrease of 1N1K cells and a significant increase of abnormal 1N1K cells (1N*1K), in which the nucleus is either larger or smaller than the control nucleus (Figure 4C–E). These 1N*1K cells constituted ∼35% of the total cell population after RNAi induction for 48 h (Figure 4C). They were very likely derived from the division of the cells that had undergone unequal nuclear division (see below). Moreover, abnormal 2N1K and 0N1K (zoid) cells also emerged (Fig 4C), suggesting that aberrant cytokinesis occurred in some 2N2K cells to produce 2N1K and 0N1K cells. Defective chromosome segregation was also detected in many of the 1N2K cells, as shown by chromosome mis-alignment in metaphase-like 1N2K cells (Figure 4F) and lagging chromosomes (anaphase lag) in early anaphase-like 1N2K cells (Figure 4G). The mis-aligned chromosomes and lagging chromosomes were labeled by immunostaining of the kinetochores with PTP-tagged KKT2 (Figure 4F, G).
Figure 4.
RNAi-mediated ablation of NuSAP1 causes mitotic defects. (A) Western blotting to detect NuSAP1 protein level before and after NuSAP1 RNAi. NuSAP1 was endogenously tagged with a triple HA epitope. TbPSA6 served as the loading control. (B) Effect of NuSAP1 RNAi on cell proliferation. (C) Effect of NuSAP1 RNAi on cell cycle progression examined by quantification of cells with different numbers of nucleus (N) and kinetoplast (K). Error bars indicated S.D. from three independent experiments. (D) Depletion of NuSAP1 produced cells with abnormal-sized nucleus. N, nucleus; K, kinetoplast DNA. Scale bar: 5 μm. (E) Nucleus size of 1N1K cells in control and NuSAP1 RNAi cells. Surface area (μm2) of nucleus was measured by ImageJ and plotted as the percentage of cells with different sizes of nucleus. 100 1N1K cells were measured for each time point. Error bars indicated S.D. from three independent experiments. (F) NuSAP1 RNAi caused chromosome mis-alignment at metaphase. Chromosomes were represented by KKT2 immunostaining. Arrowheads indicate the chromosomes aligned at the equatorial plate, whereas arrows indicate mis-aligned chromosomes. Scale bar: 5 μm. (G) Depletion of NuSAP1 caused anaphase chromosome lag. Chromosomes were represented by KKT2-labeled kinetochores. Open arrowheads indicate the normally segregated chromosomes at the spindle poles, whereas arrows indicate the lagging chromosomes in the middle of the segregating nucleus. Scale bar: 5 μm.
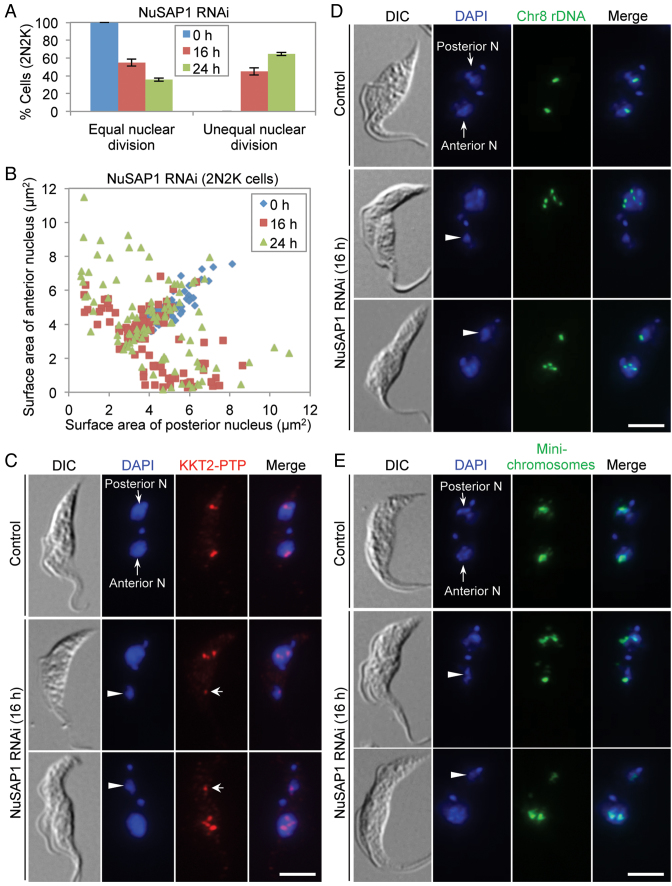
The 2N2K cells were not increased after NuSAP1 RNAi, but after RNAi induction for 24 h, ∼60% of the NuSAP1-deficient 2N2K cells underwent unequal nuclear division (Figure 5A), leading to the production of a larger nucleus and a smaller nucleus (Figure 5B–E). The smaller nucleus (Figure 5C–E, arrowheads) was segregated to either the posterior of the cell or the anterior of the cell. Unequal nuclear division was confirmed by immunostaining of kinetochores with PTP-tagged KKT2 (Figure 5C) and by fluorescence in situ hybridization (FISH) to label the mega-base chromosomes (Figure 5D) and the mini-chromosomes (Figure 5E). Altogether, the unequal nuclear division detected in 2N2K cells and the chromosome mis-alignment and anaphase lag detected in 1N2K cells after NuSAP1 RNAi suggest that NuSAP1 is required for faithful chromosome segregation.
Figure 5.
NuSAP1 is required for equal chromosome segregation. (A) Quantification of the 2N2K cells with equal and unequal nuclear division in control and NuSAP1 RNAi cells induced for 16 and 24 h. A total of 100 2N2K cells were counted for each time point, and three repeats were performed. Error bars indicated S.D. (B) Measurement of the surface area (μm2) of the anterior nucleus and the posterior nucleus of the 2N2K cells in control and NuSAP1 RNAi cells. The same set of the 100 2N2K cells used for counting in panel A was used for measurement. (C) NuSAP1 RNAi caused unequal chromosome segregation. Chromosomes were represented by detecting the PTP-tagged KKT2. Arrowheads indicate the small nucleus at either the anterior of the cell or the posterior of the cell, whereas arrows show the KKT2-labeled kinetochores in the small nucleus. Scale bar: 5 μm. (D, E) Unequal segregation of mega-base chromosomes (D) and mini-chromosomes (E) in NuSAP1 RNAi cells. Shown are the FISH results using the M5 rRNA gene on chromosome #8 and the mini-chromosomal 177-bp DNA repeat as probes. Cells were counterstained with DAPI for DNA. Arrowheads indicate the small nucleus at either the anterior of the cell or the posterior of the cell in NuSAP1 RNAi cells. Scale bars: 5 μm.
Depletion of NuSAP1 does not affect spindle formation
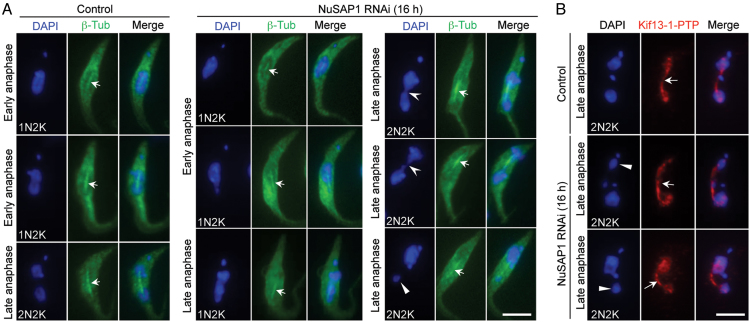
The defects in chromosome segregation by NuSAP1 RNAi prompted us to examine whether the mitotic spindle was still formed in NuSAP1 RNAi cells. Two spindle markers, the triple HA-tagged β-tubulin and the PTP-tagged Kif13-1, were used. In control anaphase cells, a bar-shaped spindle labeled by 3HA-β-tubulin was consistently observed (Figure 6A, arrows). In NuSAP1 RNAi cells induced for 16 h, a bar-shaped spindle labeled by 3HA-β-tubulin was also consistently detected in cells that either did not have visible chromosome segregation defects or had a chromosome bridge connecting the two segregating nuclei or undergoing unequal nuclear segregation (Figure 6A). Similar results were obtained with Kif13-1 as the spindle marker. The NuSAP1-deficient 2N2K cells undergoing unequal nuclear division still assembled a bar-shaped spindle (Figure 6B, arrows). These results suggest that NuSAP1 is not required for spindle assembly. Instead, NuSAP1 may regulate kinetochore-spindle microtubule attachment.
Figure 6.
Spindle formation is not affected in NuSAP1 RNAi cells. (A) Spindle structure in control and NuSAP1 RNAi cells visualized by immunostaining of 3HA-tagged β-tubulin. The solid arrowhead indicates the small nucleus, the open arrowheads indicate the chromosome bridge connecting the two segregating nuclei, and the arrows show the anaphase spindle. Scale bar: 5 μm. (B) Spindle structure in control and NuSAP1 RNAi cells visualized by immunostaining of PTP-tagged Kif13-1. The solid arrowheads indicate the small nucleus, whereas the arrows indicate the anaphase spindle. Scale bar: 5 μm.
Depletion of NuSAP1 destabilizes the kinetochore proteins KKIP1 and KKT1
The localization of NuSAP1 to kinetochores during interphase prompted us to investigate whether NuSAP1 depletion has any effect on the stability and/or localization of kinetochore proteins. To this end, we tagged five of the seven outer kinetochore proteins (KKIP1, KKIP3, KKIP4, KKIP6, and KKIP7) and 15 out of the 20 inner kinetochore proteins (KKT1-KKT4, KKT8, KKT10 and KKT12-KKT18) with a triple HA epitope from their respective endogenous locus in the NuSAP1 RNAi cell line. Western blotting showed that the levels of KKIP1 and KKT1 decreased significantly after NuSAP1 RNAi from 24 h and onward (Figure 7A). However, the other kinetochore proteins were not affected by NuSAP1 depletion (Figure 7B, C). These results suggest that NuSAP1 depletion destabilized the outer kinetochore protein KKIP1 and the inner kinetochore protein KKT1. The underlying mechanism for this specific effect on these two kinetochore proteins remains unclear.
NuSAP2 is an MAP65/ASE1/PRC1 domain-containing protein required for the G2/M transition
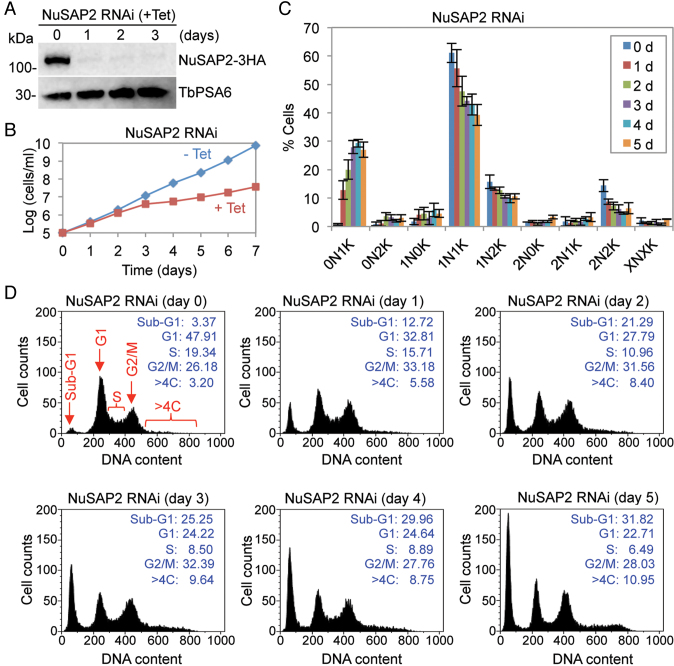
The physiological function of NuSAP2 was also investigated in the procyclic form. RNAi-mediated depletion of NuSAP2, which was endogenously tagged with a C-terminal triple HA epitope in NuSAP2 RNAi cell line, was confirmed by western blotting (Figure 8A). RNAi of NuSAP2 caused a severe growth defect (Figure 8B), suggesting that NuSAP2 is essential for cell proliferation. Analysis of the cells at different cell cycle stages before and after NuSAP2 RNAi induction showed an accumulation of 0N1K (zoid) cells up to ∼29% of the total population after 4 days of RNAi and a significant decrease of 1N1K cells from ∼60% to ∼40% (Figure 8C), indicating aberrant cytokinesis occurred in some 1N2K cells to produce a 1N1K cell and a 0N1K cell. Flow cytometry analysis detected a significant decrease of G1 cells from ∼48% to ∼24% and of S-phase cells from ∼19% to ∼8% after RNAi induction for 3 days, followed by an increase of G2/M cells from ∼26% to ∼32% and of sub-G1 (zoid) cells from ∼3% to ∼25% (Figure 8D). Moreover, cells with greater than 4C DNA content was also increased from ∼3% to ∼10% after NuSAP2 RNAi for 3 days (Figure 8D). These results suggest that NuSAP2 RNAi caused a moderate G2/M defect. Together with the findings that NuSAP2 contains a MAP65/ASE1-like motif at the C-terminus (Supplementary Figure S3), these results suggest that NuSAP2 is a highly divergent MAP65/ASE1/PRC1 homolog in T. brucei and plays a role in promoting G2/M progression.
Figure 8.
NuSAP2 is a MAP65/ASE1/PRC1 domain-containing protein required for the G2/M cell cycle transition. (A) Knockdown of NuSAP2 by RNAi. Shown are the western blots of endogenous 3HA-tagged NuSAP2 and TbPSA6, which served the loading control, before and after RNAi induction. (B) RNAi of NuSAP2 caused a severe growth defect. (C) Effect of NuSAP2 depletion on cell cycle progression. Shown is the quantification of cells at different cell cycle stages of non-induced control and NuSAP2 RNAi cells induced for up to 5 days. Cells were stained with DAPI, and more than 180 cells at each time point were counted for the numbers of nucleus (N) and kinetoplast (K). Errors bars indicate S.D. calculated from three repeats. (D) Flow cytometry analysis of NuSAP2 RNAi cells. The insets show the percentage of cells at different cell cycle stages.
NuSAP3 is a Kif13-1-binding protein and maintains Kif13-1 stability for the G2/M transition
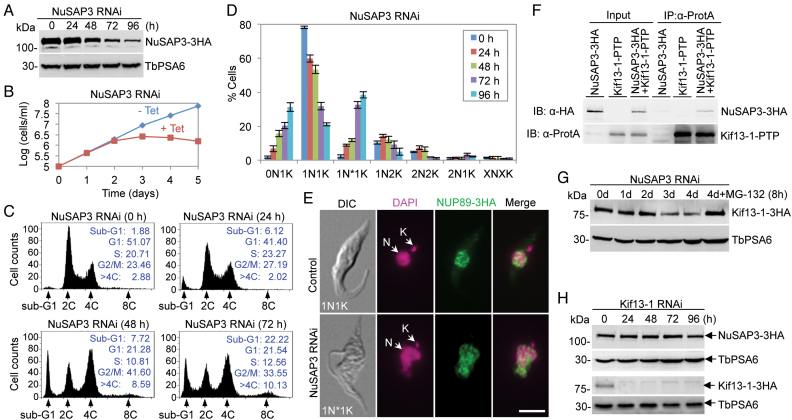
To study the function of NuSAP3, RNAi was carried out in the procyclic form. Western blotting showed that NuSAP3 protein, which was endogenously tagged with a triple HA epitope, was gradually decreased from 48 h of RNAi induction but was not depleted from the cells (Figure 9A). The knockdown of NuSAP3, however, caused a severe growth defect from day 3 of RNAi (Figure 9B), indicating that NuSAP3 is essential for trypanosome proliferation. To examine whether cell cycle progression was affected by NuSAP3 knockdown, flow cytometry was carried out. The results showed that after NuSAP3 RNAi induction, there was a gradual decrease of G1 cells from ∼51% to ∼22% and of S-phase cells from ∼21% to ∼13%, which was accompanied by a gradual increase of G2/M cells from ∼23% to ∼42% after 48 h (Figure 9C), suggesting a G2/M defect. The G2/M cells dropped to ∼34% after 72 h of RNAi (Figure 9C), likely due to the increase of the cells with greater than 4C DNA content (>4C) from ∼3% to ∼10% (Figure 9C). The sub-G1 (0N1K) cells increased from ∼2% to ∼22% of the total population (Figure 9C), indicating that aberrant cytokinesis occurred in some 1N2K cells to produce a 1N1K cell and a 0N1K cell. There was also an accumulation of abnormal 1N1K cells (1N*1K) cells that contained a nucleus of larger size and somewhat irregular shape (Figure 9D, E). Together, these results suggest that NuSAP3 is required for G2/M transition.
Figure 9.
NuSAP3 is a Kif13-1-interacting protein required for promoting chromosome segregation and for maintaining Kif13-1 stability. (A) RNAi knockdown of NuSAP3 in T. brucei, as demonstrated by western blotting of endogenously 3HA-tagged NuSAP3 in NuSAP3 RNAi cells induced for up to 4 days. TbPSA6 served as the loading control. (B) Depletion of NuSAP3 caused a severe growth defect. (C) Flow cytometry analysis of NuSAP3 RNAi cells. Insets show the percentage of cells at different cell cycle stages. (D) Effect of NuSAP3 depletion on cell cycle progression. Shown is the quantification of cells at different cell cycle stages as determined by counting the numbers of cells with different numbers of nucleus (N) and kinetoplast (K), which was stained with DAPI. A total of 200 cells were counted for each time point, and three repeats were carried out. Error bars indicate S.D. 1N*1K cells represent the cells with an abnormal-shaped nucleus and one kinetoplast. (E) Morphology of the nucleus shape of 1N1K cells in control and NuSAP3 RNAi cell line. Nuclear envelope was labeled by immunostaining of the endogenously 3HA-tagged nuclear pore protein NUP89, and DNA was stained with DAPI. N, nucleus; K, kinetoplast. Scale bar: 5 μm. (F) NuSAP3 interacts with Kif13-1 in vivo in T. brucei. Shown is the co-immunoprecipitation of PTP-tagged Kif13-1 and 3HA-tagged NuSAP3 from T. brucei cell lysate. Kif13-1-PTP alone and NuSAP3-3HA alone served as the negative controls. The co-IP was repeated three times, and similar results were obtained. (G) Effect of NuSAP3 depletion on Kif13-1 protein stability. Kif13-1 was endogenously tagged with a C-terminal triple HA epitope in NuSAP3 RNAi cell line. TbPSA6 served as the loading control. The experiment was repeated three times, and similar results were obtained. (H) Effect of Kif13-1 depletion on the level of NuSAP3 protein. NuSAP3 was endogenously tagged with a C-terminal triple HA epitope in Kif13-1 RNAi cell line. TbPSA6 served as the loading control. Similar results were obtained from three repeats.
NuSAP3 was identified by Kif13-1 BioID (Figure 1), indicating that it may interact with Kif13-1. To test this possibility, we carried out co-immunoprecipitation experiments. NuSAP3 was endogenously tagged with a triple HA epitope and co-expressed with PTP-tagged Kif13-1 in T. brucei. Immunoprecipitation of Kif13-1-PTP was able to pull down NuSAP3-3HA from trypanosome cell lysate (Figure 9F), confirming that the two proteins interact in vivo in trypanosomes. To understand the functional relationship between NuSAP3 and Kif13-1, we investigated the effect of NuSAP3 knockdown on Kif13-1 protein stability. Western blotting showed that the level of Kif13-1 protein, which was endogenously tagged with a triple HA epitope in NuSAP3 RNAi cell line, started to decrease from day 3 of RNAi induction, but the protein was not depleted even after RNAi induction for longer times (Figure 9G). Treatment of the NuSAP3 RNAi cells with the proteasome inhibitor MG-132 stabilized Kif13-1 (Figure 9G), indicating that NuSAP3 depletion caused degradation of Kif13-1 by the proteasome. Thus, one of the molecular functions of NuSAP3 is to maintain Kif13-1 stability. Conversely, the effect of Kif13-1 depletion on NuSAP3 protein stability was also investigated. Western blotting showed that Kif13-1 depletion did not affect NuSAP3 protein stability (Figure 9H), demonstrating that Kif13-1 is not required for maintaining NuSAP3 stability.
DISCUSSION
Spindle-associated proteins in animals include non-motor MAPs and motor proteins, which regulate spindle microtubule dynamics, spindle orientation and positioning, and spindle microtubule-kinetochore attachment (6). Only a limited number of SAPs have been discovered in the protozoan parasite T. brucei; therefore, it is necessary to identify and characterize all SAPs in T. brucei for a comprehensive understanding of the mechanisms for spindle assembly and chromosome segregation. As the first step towards this goal, our current work identified eight new SAPs in T. brucei, bringing the total number of T. brucei SAPs to nineteen. Although our BioID baits covered almost every subdomains of the spindle, the number of SAPs identified in the current work is very small, compared to the over 200 SAPs in animals (5). This may be attributed to the inherent limitation of the BioID technique, which detects only the neighboring proteins within a radius of ∼10 nm (48). It could also be attributed to the lack of many binding partners and near neighbors of the five bait proteins, despite that they localize to almost every subdomain of the spindle. Additional BioID experiments using the new SAPs as baits (Supplementary Table S8) and a proteome-wide localization screening (49) may identify more SAPs in T. brucei.
Many SAPs identified in yeast and animals are microtubule-associated proteins, which are either motor proteins (dyneins and kinesins) or non-motor proteins (6). Among the new SAPs identified in T. brucei, KIN-F and NuSAP2 are potentially able to bind to microtubules. KIN-F possesses a well-conserved microtubule-binding motor domain at its N-terminus (Supplementary Figure S1B), and NuSAP2 contains a ∼277-aa sequence (Supplementary Figure S3) that is conserved in the MAP65/ASE1/PRC1 family proteins, which are all microtubule-binding proteins (40–42). Fidgetin belongs to a family of microtubule-severing and depolymerizing enzymes that also include Katanin and Spastin (50). Spastin and Katanin appear to be capable of binding to microtubules (51), but whether Fidgetin can also bind to microtubules is unclear. NuSAP1, NuSAP3, and NuSAP4 have no homology to any known MAPs in other eukaryotes; whether they contain a microtubule-binding motif, such as a stretch of charged residues (52,53), and are capable of binding to microtubules remains to be determined. Alternatively, they may associate with the spindle indirectly through interacting with other MAPs, as in the case of NuSAP3 (Figure 9F), which interacts with Kif13-1, a microtubule depolymerizing kinesin functioning on the spindle (18,19).
Two kinetochore proteins, KKT4 and KKIP4 (Figure 2B), have been shown to localize to the spindle during mitosis in T. brucei. While KKT4 localizes to the spindle poles in addition to the kinetochores during metaphase (Llauro, A., Hayashi, H., Bailey, M.E., Wilson, A., Ludzia, P., Asbury, C.L. and Akiyoshi, B., 2017 BioRxiv, https://doi.org/10.1101/216812), KKIP4 localizes to the entire spindle in metaphase and anaphase (Figure 2B) and to the kinetochores during interphase and telophase (9). Nevertheless, KKT4 binds to microtubules directly, and this interaction requires several charged residues (R123, K132, and R154) at the N-terminus of KKT4 (Llauro, A., Hayashi, H., Bailey, M.E., Wilson, A., Ludzia, P., Asbury, C.L. and Akiyoshi, B., 2017 BioRxiv, https://doi.org/10.1101/216812). Surprisingly, a BLAST search with KKT4 as the bait identified a 122-aa sequence (a.a. 362–483) that is homologous (E-value: 1.09e-03) to the N-terminal domain of the Schizosaccharomyces pombe kinetochore protein Spc7 (Supplementary Figure S6). Spc7 is a component of the NMS (Ndc80-MIND-Spc7) complex and belongs to the Spc105/KNL1 family, which binds to microtubules through its N-terminal domain (54). The NMS complex in S. pombe is equivalent to the highly conserved tripartite KMN (KNL1-Mis12-Ndc80) complex in other organisms, which constitutes the microtubule-binding interface of the kinetochore (54). However, it appears that binding of KKT4 to microtubules does not require the 122-aa sequence that is homologous to the Spc7 N-terminal domain, suggesting distinct mechanisms of microtubule binding between KKT4 and Spc7. A thorough bioinformatics analysis of KKIP4 using HMM does not identify sequence similarity to any known kinetochore proteins and MAPs; whether KKIP4 is capable of binding to microtubules remains to be investigated.
The finding that seventeen nucleolar proteins associate with the mitotic spindle (Supplementary Figure S4) is in line with the previous observation that the nucleolus segregates along the mitotic spindle during mitosis in T. brucei (13). During metaphase, the nucleolus assumes a bar-shaped form, extending between the spindle poles, and during anaphase, the nucleolus is divided into two equal-sized clusters (13). Among the seventeen nucleolar proteins, NOP54, NOP105, NOP112, and NOP127 appeared to divide into two equal-sized clusters during anaphase, similar to the pattern of the nucleolar protein FYRP (Supplementary Figure S4). Notably, the rest thirteen nucleolar proteins remain a bar-shaped form, extending between the two segregating nuclei during anaphase (Supplementary Figure S4). This localization pattern of those other nucleolar proteins resembles that of TbNOP86 (17), but differs from that of Nopp140, which displays a typical nucleolus segregation pattern (17,55). Thus, it appears that some nucleolar proteins remain associated with the anaphase spindle, while some other nucleolar proteins have already divided into the two segregating nuclei during anaphase in T. brucei. The nucleolus is a sub-nuclear compartment for ribosome biogenesis, and is also a storage site for a number of proteins that do not have roles in ribosome biogenesis (56). Profiling of the HeLa cell nucleolar proteome revealed that more than half of the 700 nucleolar proteins are unrelated to ribosome biogenesis (57,58). Some nucleolar proteins apparently have an essential role in mitotic progression (56). The best-characterized nucleolar protein with a role in mitosis is the Cdc14 phosphatase, which is tethered in a nucleolar complex prior to anaphase, and upon entry into anaphase, it is released from the nucleolus and dephosphorylates Cdh1 for activation of APC/C (59,60). Other nucleolar proteins that have a mitotic role include the human nucleolar protein NUSAP, which binds to spindle microtubules and is required for spindle organization and chromosome segregation (61), and the T. brucei nucleolar protein TbNOP86, which is required for mitotic progression in both the procyclic and the bloodstream forms (17). We speculate that some of the spindle-associated nucleolar proteins identified in this work may also play roles in mitosis. Future efforts will be directed to investigate their potential roles in regulating spindle dynamics and/or chromosome segregation.
Depletion of the three NuSAPs caused distinct mitotic defects (Figures 4, 5, 8 and 9). Knockdown of NuSAP1 caused chromosome mis-alignment, anaphase lag, and unequal nuclear division (Figures 4–6), but did not affect spindle assembly (Figure 6), suggesting that NuSAP1 deficiency may cause kinetochore-spindle microtubule attachment defects. The underlying mechanisms remain unknown, but the specific effect of NuSAP1 depletion on the stability of the kinetochore proteins KKIP1 and KKT1 (Figure 7) suggests that NuSAP1 may be involved in maintaining the stability of KKIP1 and KKT1. Given that NuSAP1 localizes to the kinetochores during interphase (Figure 3), it suggests that NuSAP1 may exert the function of stabilizing KKIP1 and KKT1 at the kinetochores prior to the onset of mitosis. Depletion of NuSAP2, however, appeared to cause a moderate defect on mitosis, despite a severe growth defect (Figure 8). The most prominent phenotype of NuSAP2 RNAi is the accumulation of anucleate (zoid) cells (Figure 8C, D), which apparently was produced by an aberrant cytokinesis in cells that have a mitotic defect, as observed in the RNAi of a mitotic cyclin in the procyclic form of T. brucei (62,63). Thus, NuSAP2 appears to promote the G2/M transition or the mitotic onset without playing a role in cytokinesis. This function of NuSAP2 distinguishes it from ASE1, PRC1, and MAP65, which are involved in anaphase spindle microtubule bundling and are required for cytokinesis (40–42). Depletion of NuSAP3 also produced zoid cells (Figure 9C, D), but the mitotic defects, as shown by the increase of G2/M cells and the accumulation of the 1N*1K cells with a large nucleus (Figure 9D, E), appeared to be more severe than that caused by NuSAP2 RNAi. Given that NuSAP3 interacts with Kif13-1 (Figure 9F) and NuSAP3 depletion destabilizes Kif13-1 (Figure 9G), NuSAP3 may execute its function in chromosome segregation by maintaining Kif13-1 stability.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Kyle Roux of Sanford Research for providing the BirA*-HA plasmid (Addgene) and Ms Li Li of the Proteomics Core Facility of University of Texas Health Science Center at Houston for assistance with LC–MS/MS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01AI101437, R01AI118736 to Z.L.]. Funding for open access charge: NIH [R01AI101437, R01AI118736].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lara-Gonzalez P., Westhorpe F.G., Taylor S.S.. The spindle assembly checkpoint. Curr. Biol. 2012; 22:R966–R980. [DOI] [PubMed] [Google Scholar]
- 2. Saunders W.S. Mitotic spindle pole separation. Trends Cell Biol. 1993; 3:432–437. [DOI] [PubMed] [Google Scholar]
- 3. Roostalu J., Schiebel E., Khmelinskii A.. Cell cycle control of spindle elongation. Cell Cycle. 2010; 9:1084–1090. [DOI] [PubMed] [Google Scholar]
- 4. Prosser S.L., Pelletier L.. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell. Biol. 2017; 18:187–201. [DOI] [PubMed] [Google Scholar]
- 5. Petry S. Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 2016; 85:659–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maiato H., Sampaio P., Sunkel C.E.. Microtubule-associated proteins and their essential roles during mitosis. Int. Rev. Cytol. 2004; 241:53–153. [DOI] [PubMed] [Google Scholar]
- 7. Akiyoshi B., Gull K.. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014; 156:1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nerusheva O.O., Akiyoshi B.. Divergent polo box domains underpin the unique kinetoplastid kinetochore. Open Biol. 2016; 6:150206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Archivio S., Wickstead B.. Trypanosome outer kinetochore proteins suggest conservation of chromosome segregation machinery across eukaryotes. J. Cell Biol. 2017; 216:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Solari A.J. Mitosis and genome partition in trypanosomes. Biocell. 1995; 19:65–84. [PubMed] [Google Scholar]
- 11. Obado S.O., Bot C., Nilsson D., Andersson B., Kelly J.M.. Repetitive DNA is associated with centromeric domains in Trypanosoma brucei but not Trypanosoma cruzi. Genome Biol. 2007; 8:R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ersfeld K., Gull K.. Partitioning of large and minichromosomes in Trypanosoma brucei. Science. 1997; 276:611–614. [DOI] [PubMed] [Google Scholar]
- 13. Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., Gull K.. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 2000; 108:501–513. [DOI] [PubMed] [Google Scholar]
- 14. Zhou Q., Li Z.. gamma-Tubulin complex in Trypanosoma brucei: molecular composition, subunit interdependence and requirement for axonemal central pair protein assembly. Mol. Microbiol. 2015; 98:667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tu X., Kumar P., Li Z., Wang C.C.. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 2006; 281:9677–9687. [DOI] [PubMed] [Google Scholar]
- 16. Li Z., Lee J.H., Chu F., Burlingame A.L., Gunzl A., Wang C.C.. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS ONE. 2008; 3:e2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boucher N., Dacheux D., Giroud C., Baltz T.. An essential cell cycle-regulated nucleolar protein relocates to the mitotic spindle where it is involved in mitotic progression in Trypanosoma brucei. J. Biol. Chem. 2007; 282:13780–13790. [DOI] [PubMed] [Google Scholar]
- 18. Chan K.Y., Matthews K.R., Ersfeld K.. Functional characterisation and drug target validation of a mitotic kinesin-13 in Trypanosoma brucei. PLoS Pathog. 2010; 6:e1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wickstead B., Carrington J.T., Gluenz E., Gull K.. The expanded Kinesin-13 repertoire of trypanosomes contains only one mitotic Kinesin indicating multiple extra-nuclear roles. PLoS ONE. 2010; 5:e15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wheeler R.J., Scheumann N., Wickstead B., Gull K., Vaughan S.. Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: implications for microtubule-based morphogenesis and mutant analysis. Mol. Microbiol. 2013; 90:1339–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holden J.M., Koreny L., Obado S., Ratushny A.V., Chen W.M., Chiang J.H., Kelly S., Chait B.T., Aitchison J.D., Rout M.P. et al. Nuclear pore complex evolution: a trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol. Biol. Cell. 2014; 25:1421–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morelle C., Sterkers Y., Crobu L., DE M.B.-B., Kuk N., Portales P., Bastien P., Pages M., Lachaud L.. The nucleoporin Mlp2 is involved in chromosomal distribution during mitosis in trypanosomatids. Nucleic Acids Res. 2015; 43:4013–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchez M.A., Tran K.D., Valli J., Hobbs S., Johnson E., Gluenz E., Landfear S.M.. KHARON is an essential cytoskeletal protein involved in the trafficking of flagellar membrane proteins and cell division in african trypanosomes. J. Biol. Chem. 2016; 291:19760–19773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayashi H., Akiyoshi B.. Degradation of cyclin B is critical for nuclear division in Trypanosoma brucei. Biol. Open. 2018; 7:bio.031609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z., Gourguechon S., Wang C.C.. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J. Cell Sci. 2007; 120:3883–3894. [DOI] [PubMed] [Google Scholar]
- 26. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wirtz E., Leal S., Ochatt C., Cross G.A.. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 28. Shen S., Arhin G.K., Ullu E., Tschudi C.. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 2001; 113:171–173. [DOI] [PubMed] [Google Scholar]
- 29. Krogh A., Brown M., Mian I.S., Sjolander K., Haussler D.. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994; 235:1501–1531. [DOI] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z. et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005; 33:D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lupas A., Van Dyke M., Stock J.. Predicting coiled coils from protein sequences. Science. 1991; 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- 32. Hu H., Zhou Q., Li Z.. SAS-4 Protein in Trypanosoma brucei controls life cycle transitions by modulating the length of the flagellum attachment zone filament. J. Biol. Chem. 2015; 290:30453–30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Q., Hu H., Li Z.. An EF-hand-containing protein in Trypanosoma brucei regulates cytokinesis initiation by maintaining the stability of the cytokinesis initiation factor CIF1. J. Biol. Chem. 2016; 291:14395–14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dang H.Q., Zhou Q., Rowlett V.W., Hu H., Lee K.J., Margolin W., Li Z.. Proximity interactions among basal body components in Trypanosoma brucei identify novel regulators of basal body biogenesis and inheritance. MBio. 2017; 8:e02120–e02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Z., Morris J.C., Drew M.E., Englund P.T.. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000; 275:40174–40179. [DOI] [PubMed] [Google Scholar]
- 36. Wei Y., Hu H., Lun Z.R., Li Z.. Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat. Commun. 2014; 5:4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Z., Zou C.B., Yao Y., Hoyt M.A., McDonough S., Mackey Z.B., Coffino P., Wang C.C.. An easily dissociated 26 S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J. Biol. Chem. 2002; 277:15486–15498. [DOI] [PubMed] [Google Scholar]
- 38. DeGrasse J.A., DuBois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., Chait B.T.. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics. 2009; 8:2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Archer S.K., Inchaustegui D., Queiroz R., Clayton C.. The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS ONE. 2011; 6:e18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muller S., Smertenko A., Wagner V., Heinrich M., Hussey P.J., Hauser M.T.. The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 2004; 14:412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pellman D., Bagget M., Tu Y.H., Fink G.R., Tu H.. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 1995; 130:1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang W., Jimenez G., Wells N.J., Hope T.J., Wahl G.M., Hunter T., Fukunaga R.. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell. 1998; 2:877–885. [DOI] [PubMed] [Google Scholar]
- 43. Wickstead B., Gull K.. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol. Biol. Cell. 2006; 17:1734–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Z., Umeyama T., Wang C.C.. The chromosomal passenger complex and a mitotic kinesin interact with the Tousled-like kinase in trypanosomes to regulate mitosis and cytokinesis. PLoS ONE. 2008; 3:e3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu H., Hu L., Yu Z., Chasse A.E., Chu F., Li Z.. An orphan kinesin in trypanosomes cooperates with a kinetoplastid-specific kinesin to maintain cell morphology by regulating subpellicular microtubules. J. Cell Sci. 2012; 125:4126–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu L., Hu H., Li Z.. A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol. Microbiol. 2012; 83:565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Casanova M., Crobu L., Blaineau C., Bourgeois N., Bastien P., Pages M.. Microtubule-severing proteins are involved in flagellar length control and mitosis in Trypanosomatids. Mol. Microbiol. 2009; 71:1353–1370. [DOI] [PubMed] [Google Scholar]
- 48. Kim D.I., Birendra K.C., Zhu W., Motamedchaboki K., Doye V., Roux K.J.. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2453–E2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dean S., Sunter J.D., Wheeler R.J.. TrypTag.org: a trypanosome genome-wide protein localisation resource. Trends Parasitol. 2017; 33:80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sharp D.J., Ross J.L.. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 2012; 125:2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eckert T., Le D.T., Link S., Friedmann L., Woehlke G.. Spastin's microtubule-binding properties and comparison to katanin. PLoS ONE. 2012; 7:e50161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P. et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008; 133:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M. et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 2012; 23:968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A.. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006; 127:983–997. [DOI] [PubMed] [Google Scholar]
- 55. Kelly S., Singleton W., Wickstead B., Ersfeld K., Gull K.. Characterization and differential nuclear localization of Nopp140 and a novel Nopp140-like protein in trypanosomes. Eukaryot. Cell. 2006; 5:876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pederson T., Tsai R.Y.. In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 2009; 184:771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I.. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002; 12:1–11. [DOI] [PubMed] [Google Scholar]
- 58. Scherl A., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J.. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002; 13:4100–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W., Jang J., Charbonneau H., Deshaies R.J.. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999; 97:233–244. [DOI] [PubMed] [Google Scholar]
- 60. Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., Deshaies R.J.. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004; 305:516–519. [DOI] [PubMed] [Google Scholar]
- 61. Raemaekers T., Ribbeck K., Beaudouin J., Annaert W., Van Camp M., Stockmans I., Smets N., Bouillon R., Ellenberg J., Carmeliet G.. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 2003; 162:1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li Z., Wang C.C.. A PHO80-like cyclin and a B-type cyclin control the cell cycle of the procyclic form of Trypanosoma brucei. J. Biol. Chem. 2003; 278:20652–20658. [DOI] [PubMed] [Google Scholar]
- 63. Hammarton T.C., Clark J., Douglas F., Boshart M., Mottram J.C.. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J. Biol. Chem. 2003; 278:22877–22886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.