Abstract
Despite advances in XNA evolution, the binding capabilities of artificial genetic polymers are currently limited to protein targets. Here, we describe the expansion of in vitro evolution techniques to enable selection of threose nucleic acid (TNA) aptamers to ochratoxin A (OTA). This research establishes the first example of an XNA aptamer of any kind to be evolved having affinity to a small-molecule target. Selection experiments against OTA yielded aptamers having affinities in the mid nanomolar range; with the best binders possessing KD values comparable to or better than those of the best previously reported DNA aptamer to OTA. Importantly, the TNA can be incubated in 50% human blood serum for seven days and retain binding to OTA with only a minor change in affinity, while the DNA aptamer is completely degraded and loses all capacity to bind the target. This not only establishes the remarkable biostability of the TNA aptamer, but also its high level of selectivity, as it is capable of binding OTA in a large background of competing biomolecules. Together, this research demonstrates that refining methods for in vitro evolution of XNA can enable the selection of aptamers to a broad range of increasingly challenging target molecules.
INTRODUCTION
Life is controlled on a molecular level by the unparalleled ability of nucleic acids to store and process genetic information. This has inspired significant research effort focused on understanding how nucleic acids evolve and function, and how these capabilities can be harnessed for applications in biotechnology. In vitro evolution has proven to be a powerful tool for exploring these questions, and simultaneously provides a method for generating nucleic acid polymers having novel molecular recognition functionality. These polymers, known as aptamers, are single stranded oligonucleotides that can be evolved through a process called SELEX (Systematic Evolution of Ligands by EXponential enrichment) to bind to a wide variety of target molecules with high affinity and specificity (1–3). Since their discovery, aptamers have shown promise in a variety of applications including therapeutics, affinity purification, and biosensing, where antibodies have long been considered the gold standard. Aptamers provide numerous advantages over antibodies including increased thermal stability and tolerance to surfactants. Additionally, aptamers can be chemically synthesized, enabling more cost-effective production and allowing for the incorporation of a wide range of labels and non-natural functional groups (4). However, aptamers comprised of native DNA and RNA retain some limitations due to their instability in biological fluids and cellular environments (5).
Recently, there has been a growing interest in backbone-modified nucleic acids, or xeno nucleic acids (XNA), as these polymers are not readily recognized and degraded by nucleases, and thus are well-suited for in vivo applications (6,7). Arguably, the most famous XNA aptamer to date is Macugen, a backbone-modified aptamer that binds vascular endothelial growth factor isoform VEGF165 to prevent the overgrowth of blood vessels associated with macular degeneration (8). In the development of Macugen, aptamer sequences were evolved using native RNA, then the native nucleotides were systematically replaced with 2′-OMe or 2′-F nucleotides to generate a biostable XNA aptamer that retained the necessary binding affinity to VEGF165. While this example supports the utility of XNA aptamers, the post-selection modification strategy is expensive and time-consuming. In contrast, the direct selection of aptamers from XNA libraries offers a more robust approach. Fortuitously, many XNAs are capable of Watson–Crick base pairing with DNA and RNA, which has provided encouragement for their use as alternative genetic systems (9). Recent efforts in this field have generated replication technologies that permit information transfer between DNA and different XNAs, establishing the capacity for in vitro evolution using these artificial genetic systems (10). While the direct selection of XNA aptamers holds significant promise for generating biopolymers having novel functions, these artificial genetic systems can present challenges for in vitro selection experiments. An initial challenge is the limited availability of the building blocks and polymerases needed to create XNA polymers. For many non-natural nucleic acids, the phosphoramidites and triphosphates needed for solid phase and enzymatic synthesis, respectively, are neither commercially available nor easily synthesized. Additionally, while recent advances in enzyme engineering have provided polymerases capable of XNA transcription and reverse transcription, they are often characterized by lower fidelity and processivity (11). Consequently, transcription efficiency is often reduced, which can lead to undersampling of library diversity and introduce sequence bias that negatively impacts the efficacy of the selection (12). Additionally, the difficulty of generating and utilizing XNA primers has limited transcription to techniques where the polymer is extended from a DNA primer to yield a DNA–XNA chimera. While aptamer selections using these DNA-primed XNA libraries have been successful, the DNA portion is sometimes involved in target binding and can be difficult to completely remove or convert to XNA post-selection. Under biological conditions, this vital region of the aptamer may then be degraded, compromising the binding affinity of the aptamer. To date, direct selection using DNA-primed XNA libraries has provided hexitol nucleic acid (HNA) aptamers to hen-egg lysozyme and HIV trans-activating response RNA element (11), a (3′,2′)-α-ι-threose nucleic acid (TNA) aptamers to human thrombin and HIV-reverse transcriptase (13,14), a 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamer to HIV-1 reverse transcriptase (15), and a 2′-fluoro-modified aptamer for human neutrophil elastase (HNE) (16).
These examples demonstrate that despite the challenges associated with in vitro selection using XNA, aptamers comprising a variety of backbone structures can be generated for protein targets. However, successful selection of an XNA aptamer to a small-molecule target has yet to be reported. It is widely accepted that small molecules are more difficult targets for aptamer selection compared to proteins. In general, small molecules present significantly less surface area and fewer functional groups for the aptamer to interact with to gain favorable binding interactions (17–21). These factors also complicate the partitioning of bound from unbound sequences, as selection methods typically rely on immobilization of the target molecule. This inherently compromises one functional group on the target, which carries a significantly greater risk in the case of small-molecule targets (18). Furthermore, the relatively small change in size between free XNAs and those bound to the small-molecule target presents complications in the screening and characterization of putative aptamer sequences. Given these challenges, it is not surprising that only ∼25% of all reported selection experiments are for small-molecule targets (18). However, many small molecules including toxins and biomarkers play key roles in biology and the environment, therefore developing receptors that can sensitively and selectively detect these targets is of high utility.
Given the exciting capabilities provided by XNA in the context of protein recognition, we sought to extend these by selecting the first XNA aptamer to a small-molecule target. As our target molecule, we chose ochratoxin A (OTA), a food-contaminating mycotoxin which causes nephrotoxic effects when consumed (Figure 1A) (22). OTA has a negative immunogenic response, which makes it an interesting target for aptamer development, as antibodies are difficult to generate. In addition, several DNA aptamers have been evolved for OTA, enabling comparison and functional validation of our newly generated XNA aptamers. We anticipated that TNA would be well-suited for in vitro evolution because of the availability of enzymes that support efficient in vitro transcription and reverse transcription (23,24), as well as its superb stability in the presence of nucleases (Figure 1B) (25). Given the increased difficulty of generating aptamers to small-molecule targets, we introduced new capabilities in XNA SELEX aimed at increasing enrichment efficiency and ensuring binding of the free target molecule. Central among these was the use of a TNA primer to give a fully non-native library. Using our improved SELEX protocol, we generated seven sequences having affinity for OTA. Analysis using microscale thermophoresis (MST) revealed that five of these sequences have KD values in the nM range, with the best being 92 ± 2 nM. Importantly, the use of MST to quantify the binding affinity of our aptamer sequences confirms that despite the use of immobilized target during the selection experiments, our aptamers are capable of binding to the free target in solution. Using secondary structure predictions, we were able to minimize two of our aptamer sequences to a functional core <35 nt, a length that enables chemical synthesis using phosphoramidite monomers available in our lab. Aptamer A04T.2, our shortest sequence at 31 nt, was found to have a KD of 71 ± 8 nM. This represents tighter binding than even the best DNA aptamer to OTA, and is the first example of a TNA aptamer to be validated by chemical synthesis. Using this truncated aptamer sequence, we demonstrate the impressive resistance of TNA to degradation by DNase I, human liver microsomes, and human blood serum. Furthermore, we highlight the selectivity of our TNA aptamer in that it retains binding affinity for OTA in the presence of 50% human serum. Together, these experiments demonstrate that XNA aptamers having high affinity and selectivity can be generated for small-molecule targets, and provide a toxin-binding aptamer for potential use in diagnostics and therapeutics.
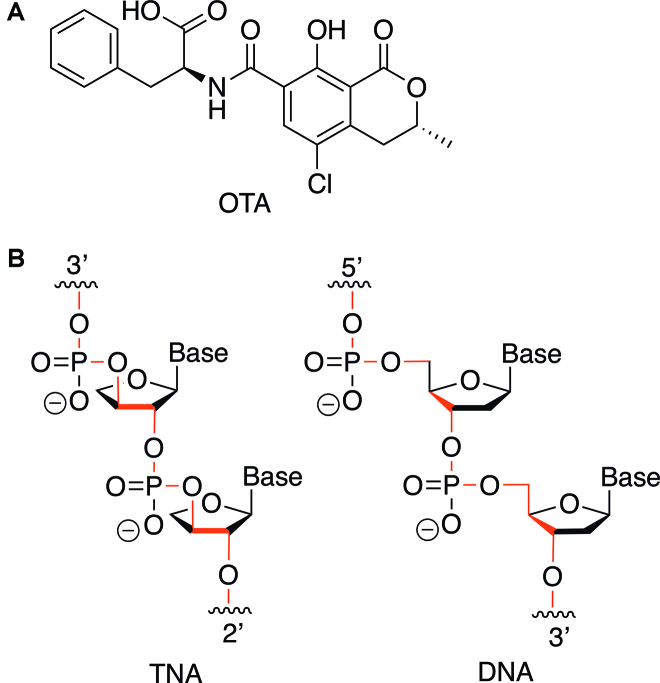
Figure 1.
Chemical structures of (A) ochratoxin A (OTA) and (B) TNA and DNA polymers. TNA has a five-atom backbone perodicity and phosphodiester linkages at the 3′ and 2′ positions of the furanose ring.
MATERIALS AND METHODS
Oligonucleotides synthesis
TNA triphosphates used for primer extension reactions were synthesized as described previously (26,27). TNA phosphoramdites were synthesized by published methods for solid phase synthesis of TNA polymers (28). KOD RI polymerase used for TNA transcription was expressed and purified by previously developed techniques (29). All DNA and chemically synthesizable TNA sequences were purchased from the University of Utah DNA/Peptide Synthesis Core Facility and further information can be found in Supplementary Figure S1. All oligonucleotides were purified by denaturing PAGE and the desired band was excised and incubated in crush and soak buffer (300 mM ammonium acetate, 1 mM EDTA, pH 8.0) at 40°C overnight. Samples were separated from gel pieces and buffer exchanged into water using Amicon Ultra-0.5 Centrifugal Unit with Ultracel 10 membrane (EMD Millipore).
Attachment of ochratoxin A to magnetic beads
300 μg of ochratoxin A (Enzo Life Sciences) was suspended in 500 μl of 0.1 M MES buffer, pH 5. After equilibration to room temperature, 0.4 mg EDC and 1.1 mg sulfo-NHS were added to the ochratoxin A solution and allowed to react for 15 min. 1.2 μl of 2-mercaptoethanol was added to the reaction to quench the remaining EDC. 500 μl of M-270 amine beads (Life Technologies) were washed three times with 1 ml 1× PBS. After each wash, the beads were separated from the supernatant using a magnetic separation stand (DynaMag-Spin, Life Technologies). The beads were re-suspended in 500 μl 1× PBS and added to the solution of ochratoxin A. After allowing the reaction to proceed for 1 h, hydroxylamine was added to a final concentration of 10 mM to quench the remaining NHS ester. The functionalized beads were washed three times with 1 ml binding buffer and re-suspended in 500 μl binding/selection buffer (100 mM NaCl, 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2, 0.02% Tween 20, 20 mM Tris–HCl, pH 7.6) for storage at 4°C. Successful attachment of ochratoxin A was validated by monitoring fluorescence intensity (λex = 380 nm, λem = 444 nm).
PCR amplification of ssDNA library
A single stranded DNA library having the sequence 5′-GCGCATACCAGCTTATTCAATT-N50-AGATAGTAAGTGCAATCTCGGC-3′ (20 pmol) was amplified in 50 μl PCR reactions containing 0.2 μM template, 0.2 μM each primers, 200 μM dNTPs, and 2.5 U Taq DNA polymerase in 1× Thermopol buffer (20 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM KCl. 2 mM MgSO4, 0.1% Triton X-100, pH 8.8, New England Biolabs). The forward primer had the sequence 5′-TTTTTTTTTTTTTTTTTTTT/Sp9/GCGCATACCAGCTTATTCAATT-3′ and the reverse primer had the sequence 5′-/FAM/GCCGAGATTGCACTTACTATCT-3′. PCR was carried using a program with an initial denaturation at 95°C for 3 min, 20 cycles of (95°C for 30 s, 54°C for 30 s, and 72°C for 20 s), and a final extension 72°C for 2 min. The amplified double stranded DNA was purified using a PCR cleanup column (Qiagen) and the PEG-functionalized strand was separated from the FAM labeled strand on a denaturing 10% polyacrylamide gel. The gel was stained with ethidium bromide and the desired band was excised and incubated in crush and soak at 40°C overnight. Samples were recovered using size-exclusion centrifugal units as described previously.
Transcription of TNA library
The TNA library was generated by primer extension using 75 pmol of purified PEGylated ssDNA library which was aliquoted into multiple reactions. Primer extension reactions were performed in 10 μl volumes containing 100 uM tNTPs with 500 nM primer-template complex. The FAM labeled TNA primer having sequence 3′-/FAM/GCCGAGATTGCACTTACTATCT-2′ was annealed to the PEGylated ssDNA in 1× Thermopol buffer by heating to 95°C for 5 min, cooling to 4°C for 10 min, and incubating at 50°C until addition of the enzyme. KOD RI TNA polymerase was pretreated with MnCl2 (1 mM) in a 1:1 (v/v) ratio and added to the mixture. tNTPs were added last at a final concentration of 100 μM, and the reactions were incubated at 50°C for 10 h. Primer extension reactions were quenched using TNA stop buffer (50% (w/v) urea, 10 mM EDTA) and denatured at 95°C for 5 min. Reactions were pooled and purified on a denaturing 10% polyacrylamide gel as described above.
TNA SELEX
The purified TNA library (∼1013 sequences) was folded in 1× selection buffer in a total volume of 500 μl by heating to 95°C for 5 min, cooling at 4°C for 10 min, and a final incubation at 25°C for 10 min. 70 μl (1.8 × 108) of OTA functionalized beads were washed three times with selection buffer and then re-suspended in the 500 μl of folded TNA library. After incubation for 1 h at 25°C, the unbound sequences were removed by washing three times with 100 μl binding buffer, using the magnetic stand to separate the supernatant from the beads. The beads were re-suspended in 100 μl binding buffer and the bound sequences were recovered by elution with heat at 95°C for 5 min. The heat elution step was repeated twice. Starting at round 5, bound sequences were recovered by target elution using excess OTA: after the three washes, the beads were re-suspended in 500 μl of OTA (100 μM) for 1 h. After incubation, the supernatant containing the bound sequences was separated from the beads and concentrated using a 10 kDa spin filter. The amount of recovered TNA was quantified by comparison to a calibration curve created using a FAM-labeled TNA standard of the same size. The standard and SELEX output were analyzed for fluorescence intensity using a BioTek Synergy 2 multi-mode plate reader.
DNase digestion to remove DNA contaminants
The recovered TNA was divided into 10 μl portions and 1.5 U DNase I (New England Biolabs) was added to each aliquot. The solutions were incubated for 45 min at 37°C. The DNase was denatured for 10 min at 75°C and the TNA solutions left on ice until further use.
TNA reverse transcription and regeneration of ssDNA library
Reverse transcription reactions were performed in a 20 μl reaction volume. 7.4 μl of the TNA/DNase mixture was combined with dNTPs (500 μM) and MgCl2 (3 mM) in 1× Thermopol buffer. The recovered TNA was annealed with 5 pmol of unmodified forward primer in 1× Thermopol buffer as previously described. 1.6 U of Bst DNA polymerase I large fragment (New England Biolabs) was added to the primer-template complex and the reaction incubated at 55°C for 3.5 h. The RT reactions were divided into 5 μl aliquots and PCR amplified as previously described for the DNA library using unmodified versions of the forward and reverse PCR primers and an annealing temperature of 50°C. This material was purified using MinElute PCR clean up columns and re-amplified using the PEG-forward and FAM-reverse PCR primers to enable strand separation. The recovered PEG-functionalized DNA template was then used to initiate transcription of the TNA library for the subsequent round of selection.
Initial screening of putative aptamer sequences
After nine rounds of selection, cDNA from rounds 7, 8 and 9 were amplified in separate PCR reactions using unmodified primers and cloned into a pCR™4-TOPO® TA vector using a TOPO TA cloning kit for sequencing (Life Technologies). The vector was amplified in Escherichia coli and plasmid DNA was recovered from 25 colonies per selection round using a Miniprep Kit (Quiagen). These DNA plasmids were sequenced at the University of Utah DNA sequencing core facility. Sequences were aligned using Multalin and secondary structure analysis was performed using the DNA folding form in Mfold (30). DNA templates corresponding to the 10 potential TNA aptamer sequences were ordered having a PEG-T20 modification, enabling them to be directly used for TNA synthesis. The PEGylated templates were annealed to the FAM-labeled TNA reverse primer as previously described to generate TNA for each potential aptamer.
MST analysis of binding interactions
Initial MST experiments to assess initial aptamer candidates were performed by 2bind GmbH. Briefly, a serial dilution of OTA in the appropriate binding buffer was carried out to provide solutions having a range of concentrations between 1.832 nM and 60 μM. 5 μl of each solution was mixed with 5 μl of the FAM-labeled aptamer, which was held at a constant concentration of 10 nM. The final concentrations of OTA in each capillary ranged from 916 pM to 30 μM. For the selectivity studies, OTA was replaced by either ochratoxin B, ochratoxin C, warfairin, citrinin, or aflatoxin B1 (Cayman Chemical). In the cases of ochratoxin B and aflatoxin B1, final concentrations of each target were increased to a range of 15 nM to 500 μM in order to obtain significant signal to evaluate binding. Each sample was analyzed on a Monolith NT.115 Pico at 25°C, with 40% LED power and 80% laser power. Data were fitted using Origin Lab analysis software to determine the aptamer KD values.
Sequence minimization and evaluation
To qualitatively assess binding of enzymatically-synthesized truncated sequences A04T and B29T, OTA functionalized magnetic beads were washed three times with binding buffer and separated into aliquots of 20 μl (2 × 107) beads. 4 pmol of each aptamer sequence was folded in binding buffer and then added to an aliquot of beads and incubated for 1 h at 25°C. Unbound sequences were removed using three washes with binding buffer and then the bound fraction was eluted into 100 μl buffer by heating to 95°C for 10 min. Fluorescence was analyzed using a plate reader as described above to determine the percentage of aptamer bound to the beads. Binding affinities for truncated aptamer sequences were determined by MST as described above.
Fluorescence polarization
The fluorescence polarization (FP) binding assay was performed using the inherent fluorescence of OTA. Binding could be determined by monitoring polarization changes of OTA free in solution and OTA bound to aptamer. Prior to FP analysis, a serial dilution was performed to provide 12 different aptamer solutions in 1× binding buffer within the range of 10 μM to 4.8 nM. 50 μl aliquots of each aptamer solution were mixed with 50 μl of OTA held at a constant concentration of 50 nM. Polarization measurements were acquired after a short 5 min incubation at room temperature. Polarization was measured in a Biotek Cytation 5 multi-mode plate reader configured with the Blue/UV FP filter cube. For all measurements, a 360/40 excitation and 460/40 emission filters were used in conjunction with a 400 nm cut off dichroic mirror.
TNA stability assay
FAM-labeled DNA aptamer A08 and TNA aptamer A04T.2 were prepared in three nuclease challenge conditions: 1.5 U DNase I (RNase-free, New England Biolabs) in 1× DNase I reaction buffer (10 mM Tris–HCl, 2.5 mM MgCl2, 0.5 mM CaCl2, pH 7.6), 50% (v/v) human blood serum (normal pool, Fisher BioReagents) in 1× Dulbecco's PBS, 0.5 mg/ml pooled human liver microsomes (XenoTech) in 1× selection buffer. TNA and DNA aptamer (4 pmol) were each folded in the respective buffer for each condition and DNase I, human blood serum, and human liver microsomes were added to the desired concentration. These samples were then incubated at 37°C for 3 days, separated on a denaturing 10% polyacrylamide gel, and analyzed using a GE Typhoon laser gel scanner. TNA and DNA aptamers folded in selection buffer were used for comparison in these experiments. The samples were also assessed for their ability to retain OTA binding using a bead-binding assay. Samples were prepared as described above and incubated with an aliquot of 20 μl (2 × 107) OTA-functionalized beads for 30 min at 25°C. Bound sequences were recovered after three washes using heat elution and quantified by fluorescence. For the MST experiments, Cy5-labeled DNA aptamer A08 and TNA aptamer A04T.2 were folded in 1× Dulbecco's PBS at a final concentration of 100 nM. An equal volume of human blood serum was added and the mixture was incubated at 37°C for either 3 or 7 days. Binding affinities were measured for the aptamers in serum using the MST method described above.
RESULTS AND DISCUSSION
Transcription of a fully XNA library using a TNA primer
Typical XNA SELEX experiments utilize a DNA primer, upon which an XNA library is synthesized using enzyme-mediated primer extension. This can create challenges post-selection, as removing the primer sequence or converting it to XNA may lead to a decrease in binding affinity of the aptamer. Additionally, because the engineered polymerases used to transcribe and reverse transcribe XNA still have a strong bias toward DNA, small amounts of DNA contamination can undermine the enrichment of aptamer sequences. We anticipated that this would be especially problematic in the selection of aptamers for small-molecule targets, but recognized that if we could use a TNA primer to provide fully TNA library, then DNase digestion could be employed to eliminate unwanted DNA contamination during the selection rounds (Supplementary Figure S2).
Thus, before beginning our aptamer selection, we performed initial experiments to determine whether TNA transcription would be possible using a TNA primer. Given the significant structural differences between TNA and DNA/RNA, a key challenge in TNA SELEX is the availability of polymerases capable of transcribing DNA into TNA and reverse transcribing TNA into DNA. The Chaput lab has recently made a number of exciting advances in this field, and while completely unbiased libraries still remain somewhat problematic due to G:G mispairing during TNA transcription, libraries having all four letters can now be transcribed efficiently (29,31,32). In exploring transcription from a TNA primer, we anticipated that a library having an N50 random region composed of 38.5% each of A and T and 11.5% each of C and G would allow for sufficient diversity and secondary structural elements, while also permitting efficient and faithful transcription. To evaluate the fidelity of transcribing this four-letter library, we first needed to optimize conditions for the primer extension reaction. An initial transcription reaction using our SELEX library and TNA primer revealed that the original incubation temperature for the primer extension reaction (55°C) yielded no full-length product. We performed a temperature screen, and found that an incubation temperature of 50°C and time of 10 h was optimal for reactions using the TNA primer (Supplementary Figure S3). The yield of this reaction is slightly less than what we observe using a DNA primer, but we hypothesized that this would not be problematic if the sequences that were being transcribed had the expected nucleotide content, indicating that transcription from the TNA primer does not introduce unwanted sequence bias (Supplementary Figure S4). To determine nucleotide distribution and evaluate the fidelity of the transcription of our library, TNA from a scaled up primer extension reaction was recovered and reverse transcribed into the corresponding DNA library. The DNA library was amplified using the appropriate primers and inserted into a TOPO vector, cloned into E. coli, and DNA from 20 clones was submitted for Sanger sequencing. Analysis of the library sequences revealed that the average amount of each nucleotide in the random region was 32.3% A, 10.3% G, 39.8% T and 17.2% C (Supplementary Figure S5). While these percentages differ slightly from the starting composition of our library, the retention of all four nucleobases suggests that any sequence bias is limited, and given the transcription efficiency achieved, we concluded that primer extension using the TNA primer under our optimized conditions would be suitable for use in our selection experiments.
TNA SELEX for small-molecule targets
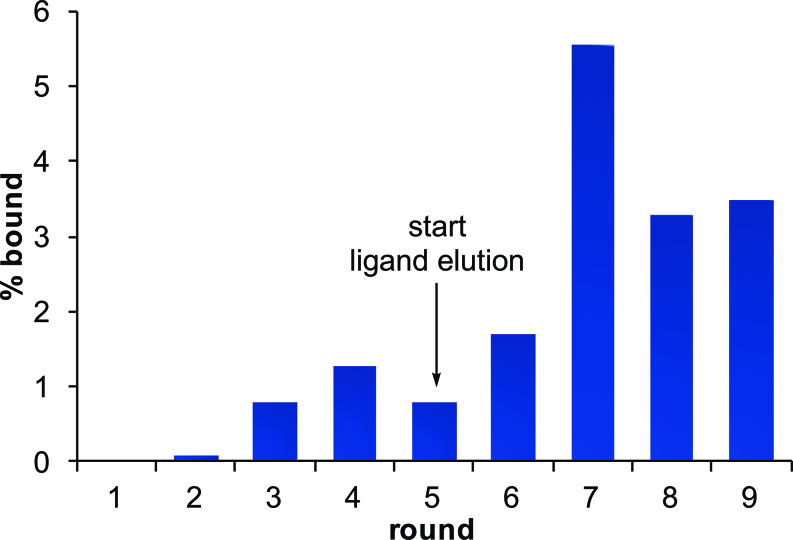
To initiate selection experiments, our ssDNA library was amplified using modified PCR primers (Supplementary Figure S1), which allow strand separation by gel electrophoresis. The PEGylated ssDNA library was recovered and annealed to a FAM-labeled TNA primer, then extended using KOD RI polymerase and tNTPs to generate the TNA library (Figure 2). We began our selections with approximately 15 pmol (∼1013 sequences) of TNA, which we incubated with OTA-functionalized magnetic beads. In rounds 1–4, bound sequences were recovered by incubation at 95°C. While this strategy maximizes recovery in early rounds, a key risk is that the aptamers generated will only bind to the immobilized version of the target molecule. Thus, for rounds 5–9, we partitioned bound sequences from the beads by elution with a solution containing an excess of OTA, as this ensures that all sequences recovered are capable of binding to the unmodified target molecule. Additionally, since the attachment of the OTA to amine beads renders the phenylalanine region of the molecule less accessible, we expected that elution with free target molecule would allow for more interaction with this region of the molecule, and therefore enrich tighter binders. The recovered sequences from each round were incubated with DNase I prior to reverse transcription to eliminate any DNA template that was not effectively removed during the gel purification step. If this step is not performed, even a small amount of residual DNA will efficiently amplify during RT-PCR, compromising the ability to enrich the desired target-binding sequences. We chose to perform the digestion step after the selection step in order to eliminate unnecessary purification steps and negate the possibility of generating aptamers to the DNase, both of which would negatively impact the efficiency of enrichment. Importantly, this step was only possible because of our choice to utilize a TNA primer to generate a fully TNA library. Additionally, the use of a FAM-labeled TNA primer to generate our library enabled us to analyze the fraction of library recovered in each round using fluorescence spectroscopy. From these data, we monitored enrichment throughout the selection experiment. We observed a gradual increase in enrichment until round 5, at which point the slight decrease in percentage of library recovered was expected due to initiation of the ligand elution (Figure 3). This presumably results because there is a fraction of the sequences that have been enriched to bind tightly to the immobilized target, but they do not bind to free OTA in solution.
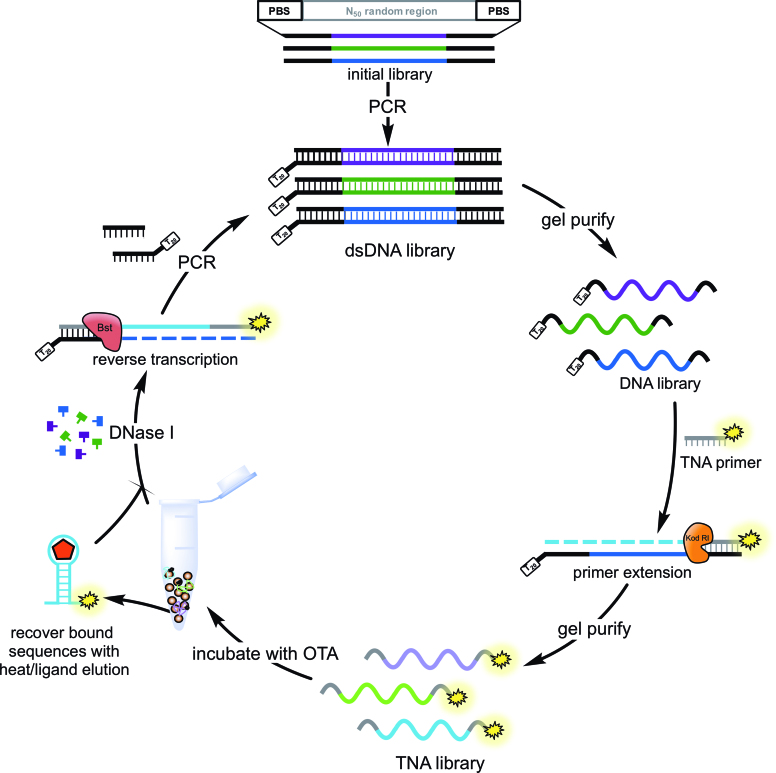
Figure 2.
TNA SELEX to generate OTA-binding aptamers. The initial ssDNA library is amplified using a forward primer modified with a PEG spacer and polyT tail to enable separation and recovery by denaturing PAGE. The PEGylated DNA template is then annealed to the FAM-labelled TNA primer and extended using KOD RI polymerase to generate the TNA library for each selection round. The TNA library is incubated with OTA-functionalized magnetic beads, and bound sequences recovered by either heat (rounds 1–4) or ligand elution (rounds 5–9). These sequences are then treated with DNase I to digest any remaining DNA template. The TNA is then reverse transcribed back into DNA using Bst DNA polymerase and PCR amplified for the next round of selection.
Figure 3.
Enrichment of OTA aptamers during TNA SELEX. Percent bound for each round was quantified by monitoring the fluorescence of the FAM-labeled TNA library.
Sequence recovery and screening of initial aptamer candidates
We observed a considerable increase in library recovery at round 7, and carried out two additional selection rounds to determine whether these would result in further enrichment. However, recovery appeared to show a slight decrease and then plateau with rounds 8 and 9. Despite the lower recovery in the last two rounds, we hypothesized that it would be valuable to compare the sequencing results across rounds 7, 8 and 9, as sequences that persisted through multiple rounds would have a higher probability of being genuine hits. Using Sanger sequencing, 25 clones from each round were analyzed, with the goal of identifying consensus within and between the final rounds of selection (Supplementary Figure S6). We identified 10 sequences which were represented by multiple copies in a single round, were conserved over multiple rounds of selection, or were closely related to well-represented sequences (Table 1). We purchased PEGylated DNA templates corresponding to each of these sequences, enabling their synthesis using enzymatic primer extension. With these sequences in hand, we next turned to characterizing their binding affinity with the OTA target. In choosing analysis method, a primary consideration was to avoid immobilization of the target molecule. As described earlier, immobilization of small-molecule targets can result in dramatic changes to chemical structure, and we were especially wary of carrying out of analysis using the same immobilization chemistry that had been employed in the selection rounds, as this could lead to false positive results. MST appeared to be a promising approach, as it relies on the change in thermophoretic movement of a molecule in response to the addition of a binding partner, and does not require immobilization of either the receptor or target (33). Additionally, MST has sufficient sensitivity to detect even minor perturbations, such as those caused by binding of a small-molecule target to a receptor, enabling us to place the fluorescent tag on the TNA strand and avoid modification of the OTA target (34). The MST data revealed that seven of the 10 TNA sequences showed binding to OTA, with five of these sequences having binding affinities in the nM range (Table 1 and Supplementary Figure S7). Impressively, the two best TNA aptamers, A04 and A07, were found to have KD values of 92 ± 2 and 96 ± 4 nM, respectively, representing higher affinity for OTA than the best full length DNA aptamer for this target (KD = 148 ± 3) (35).
Table 1.
TNA sequences of potential aptamers and their corresponding KD values determined by MST. M, C and R denoted after the sequence corresponds to sequences for which there were multiple copies, conserved over multiple rounds, or were closely related.
| TNA Sequence (3′→2′) | KD (nM) | |
|---|---|---|
| A04 | GCCGAGATTGCACTTACTATCTCAATAGGGTAAAAAAAAAAAGTTGGTCCTATGGGTGGGTTGTTCAGTAATTGAATAAGCTGGTATGCGC (R) | 92 ± 2 |
| A07 | GCCGAGATTGCACTTACTATCTGATGTTATTGTGTTTATTGGAATTTAAAGTATATGGGATGTTTGGTTAGTAATTGAATAAGCTGGTATGCGC (C) | 96 ± 4 |
| A10 | GCCGAGATTGCACTTACTATCTATTAGGAATAAGGTATGAATAGAGATGTATAAGTTGTGTGGACGGTGTAATTGAATAAGCTGGTATGCGC (M) | 175 ± 29 |
| B02 | GCCGAGATTGCACTTACTATCTCAAGAAGGGTAGTAAGAGGAGTGGGTGATTATGGGTAAGTTGTATGAGGTAATTGAATAAGCTGGTATGCGC (M) | 1968 ± 139 |
| B17 | GCCGAGATTGCACTTACTATCTGAAAGGGTGGAGATTGATAGGTATAGAGAAACGATAAGGGGGTGTAGAGTAATTGAATAAGCTGGTATGCGC (M) | – |
| B29 | GCCGAGATTGCACTTACTATCTAAAGAAGAGTAGTAAGAGGCGTGGGTGATTATGGGTAAGTTGTATGAGGTAATTGAATAAGCTGGTATGCGC (R) | 184 ± 7 |
| C06 | GCCGAGATTGCACTTACTATCTGTTGAGGGAAGAAGTGGTGTAATGTGGAATAATTGTTGTAATAGTAATTGATAAGCTGGTATGCGC (M) | 1414 ± 30 |
| C07 | GCCGAGATTGCACTTACTATCTCATCAATAAAGATTATAGGGTGAAGGTGATAGTAGGTGGTATATAGGGTAATTGAATAAGCTGGTATGCGC (R) | – |
| C14 | GCCGAGATTGCACTTACTATCTAAAGAAAGGTAGTAAGAGTGGGTGATTATGGGTAAGTTGTATGAGGTAATTGAATAAGCTGGTATGCGC (R) | – |
| C22 | GCCGAGATTGCACTTACTATCTCAAGAAGAGTAGTAAGAGGAGTGGGTGGTTATGAGTAAGTTGTATGTGGTAATTGAATAAGCTGGTATGCGC (R) | 406 ± 79 |
| DNA Sequence (5′→3′) | ||
| DNA aptamer | GCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTG | 148 ± 3 |
Determination of the minimal binding region for TNA aptamers
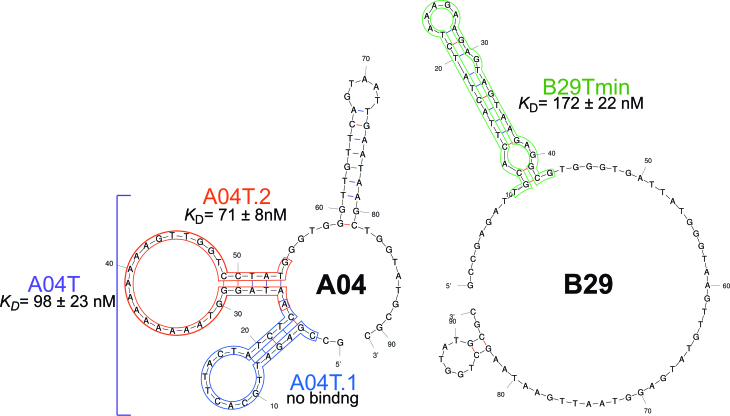
We next sought to determine the minimal binding region of the aptamers having the highest affinities. A key goal in these experiments was to identify sequences that could be generated using chemical oligonucleotide synthesis, as this method is significantly more scalable than enzymatic synthesis, and thus is far preferable for future practical applications of our TNA aptamers. In contrast to DNA synthesis, TNA synthesis presents a unique challenge in that both the phosphoramidite and alcohol coupling partners are attached to secondary carbon atoms. As a result, steric hindrance significantly decreases the coupling yield for each step, limiting the length of oligonucleotides that can be practically synthesized. In the case of DNA oligonucleotides, sequences of up to 200 nt can be routinely synthesized, but chemical synthesis of TNA is currently limited to ∼40 nt. Thus, of the five aptamers having nM binding affinities, aptamers A04 and B29 were chosen for our minimization studies, as secondary structure prediction suggested putative binding regions of <40 nt (Supplementary Figure S8). In both of these sequences, the main structural elements are near the 3′ terminus (which is analogous to the 5′ terminus of DNA), allowing us to design initial truncations that could be enzymatically synthesized using the same TNA primer that was employed in the SELEX experiments. For sequence A04, 37 nt were removed from the 2′ terminus to give A04T (54 nt), and for sequence B29, 51 nucleotides were removed from the 2′ terminus give B29T (43 nt). Initial screening using OTA-functionalized beads showed that both truncated sequences were capable of binding to the target. This provided the necessary encouragement to pursue chemical synthesis of sequences having further truncations, and we synthesized the stem loop of B29 (B29Tmin, 35 nt) and loops 1 (A04T.1, 22 nt) and 2 (A04T.2, 31 nt) of A04, each having one additional flanking nucleotide on either side of the predicted structural element (Figure 4). Using MST analysis, we measured a KD value of 172 ± 22 nM for the 35 nt truncation B29Tmin, which is similar to that of the full length B29 aptamer, confirming that only the main stem loop is required for binding to OTA. In the case of the A04-derived sequences, we found that A04T.1 showed no binding to OTA, but the 31 nt sequence A04T.2 bound OTA with a KD value 71 ± 8 nM. This establishes that loop 2 of the sequence is the binding site for OTA, and it is interesting and fortuitous that the truncated sequence shows higher affinity than either the parent A04 aptamer or the A04T truncation containing both stem loops. Additionally, we were able to validate the binding affinity of the best minimized sequence A04T.2 using FP analysis. We hypothesized that using the inherent fluorescence of OTA would provide the best sensitivity, as OTA polarization should be directly impacted by the binding of the much larger TNA aptamer sequence. We were pleased to observe a similar KD value of 120 ± 4 nM using this method (Supplementary Figure S9). The ability to generate the highest affinity aptamer using solid-phase synthesis provides a scalable route to acquiring material for use in diagnostic or therapeutic applications. Additionally, the ability to replicate affinity data between enzymatically and chemically synthesized oligonucleotides validates the faithfulness of information transfer between DNA and TNA in the selection and sequencing experiments. To our knowledge, this is the first example demonstrating this transition from enzymatic to chemical synthesis for a fully XNA aptamer. For further validation, we demonstrate that the corresponding DNA sequences for TNA aptamer A04T.2 and B29Tmin show no binding to OTA (Supplementary Figure S10). This result is in accordance with the common observation that aptamers do not exhibit cross-reactivity when the backbone structure is changed (36).
Figure 4.
Rational truncation of TNA aptamers A04 and B29 based on secondary structure predictions generated using Mfold (30). Outlined stem loop regions show minimized structures that were chemically synthesized using phosphoramidites.
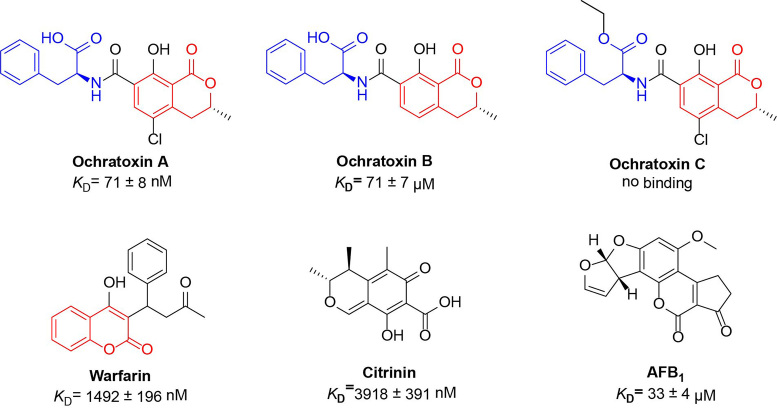
After establishing the affinity of TNA aptamer A04T.2, we next assessed the selectivity of this aptamer against relevant analogues using MST analysis (Supplementary Figure S11). Our selectivity screen included ochratoxin B (OTB), ochratoxin C (OTC), warfarin, and related mycotoxins citrinin and aflatoxin B1 (AFB1) (Figure 5). We observed a 1000-fold weaker affinity of A04T.2 with OTB and no binding with OTC. This high level of selectivity was unexpected, as OTA and its structural analogues differ by only single chloro or ethyl functional groups. A04T.2 showed more modest selectivity (20-fold weaker affinity) for warfarin, which shares a common isocoumarin structure. We observed moderate to high selectivity of A04T.2 against other mycotoxins, with a 50- and 500-fold reduction in affinitiy for citrinin and AFB1, respectively. These levels of selectivity are extremely advantageous, as some fungi simultaneously produce different mycotoxins which can co-contaminate foods (37). The selectivity of A04T.2 in binding to OTA over structurally related analogues is surprising, given that negative selections were not performed in our SELEX method. We hypothesize that the unique rigidity of the TNA backbone may be responsible for this fortuitous result.
Figure 5.
Analogues utilized to evaluate selectivity of TNA aptamer A04T.2. Key phenylalanine (blue) and isocoumarin (red) regions are highlighted for reference.
Biostability comparison of TNA and DNA aptamers with affinity for OTA
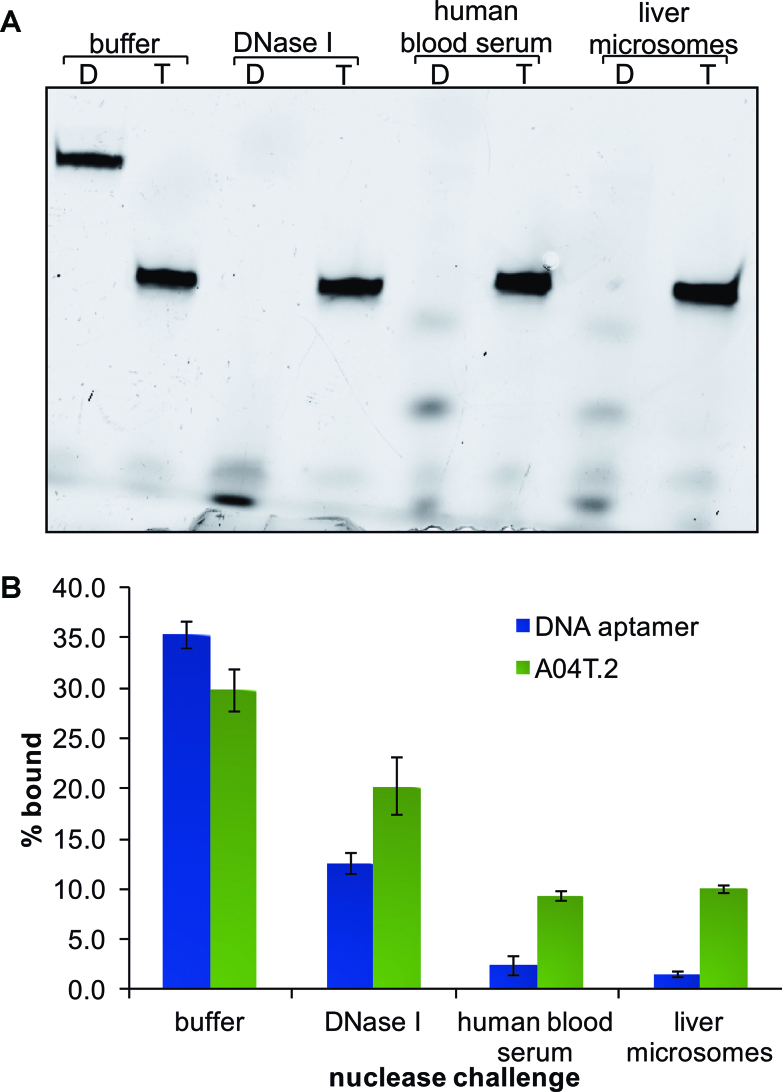
In addition to quantifying binding affinity, we also sought to assess the stability of our aptamer in a variety of nuclease rich environments. The Chaput lab has shown that TNA has superior stability compared to RNA and other XNAs such as FANA when challenged with nucleases in biologically relevant matrices (25). Additionally, they have recently shown that TNA aptamers can function in the presence of nucleolytic enzymes, however, there have been no reports that evaluate the ability of TNA aptamers to bind to their target molecule in nuclease-rich biological mixtures such as human blood serum (14). For these experiments, we challenged the highest affinity DNA aptamer and our best TNA aptamer (A04T.2) to increasingly harsh conditions including DNase I, 50% human serum in phosphate buffered saline (PBS), and 0.5 mg/mL human liver microsomes. Of these conditions, microsomes provide the most stringent test of nucleic acid stability due to the abundance of nucleases having differing activities and sequence preferences (25). As shown in Figure 6A, after incubation at 37°C for 3 d in these harsh conditions, the DNA aptamer is fully degraded, while the TNA remains intact with no degradation products detectable by denaturing PAGE. After the nuclease treatment, each sample was incubated with OTA-functionalized beads to qualitatively assess whether the target-binding capability of the TNA and DNA aptamers was retained. The binding data in Figure 6B support the PAGE results showing degradation of the DNA aptamer, as we observe significantly less binding after incubation in all three nuclease-rich conditions in comparison to the percent bound by a control DNA sample incubated only in buffer. In comparison, A04T.2 shows a smaller change in binding capacity after incubation in DNase I, human serum, or liver microsomes. Given the high biological stability of TNA, we hypothesize that the decreased binding that we do observe may be attributable to interference from the proteins and other molecules present in the serum and microsome matrices.
Figure 6.
Comparison of the biostability of FAM-labeled TNA aptamer A04T.2 and DNA aptamer A08. (A) Denaturing PAGE analysis of the TNA (T) and DNA (D) aptamers after incubation in conditions of increasing nuclease stringency: selection buffer (control), 1.5 U DNase I, 50% human blood serum in PBS, and 0.5 mg/mL human liver microsomes. Samples were incubated under these conditions for 3 days at 37°C. (B) Bead-binding assay to determine retention of aptamer binding in the presence of nucleases. Each column and error bar represents the average and standard deviation of two trials.
Retention of OTA binding in the presence of nucleases
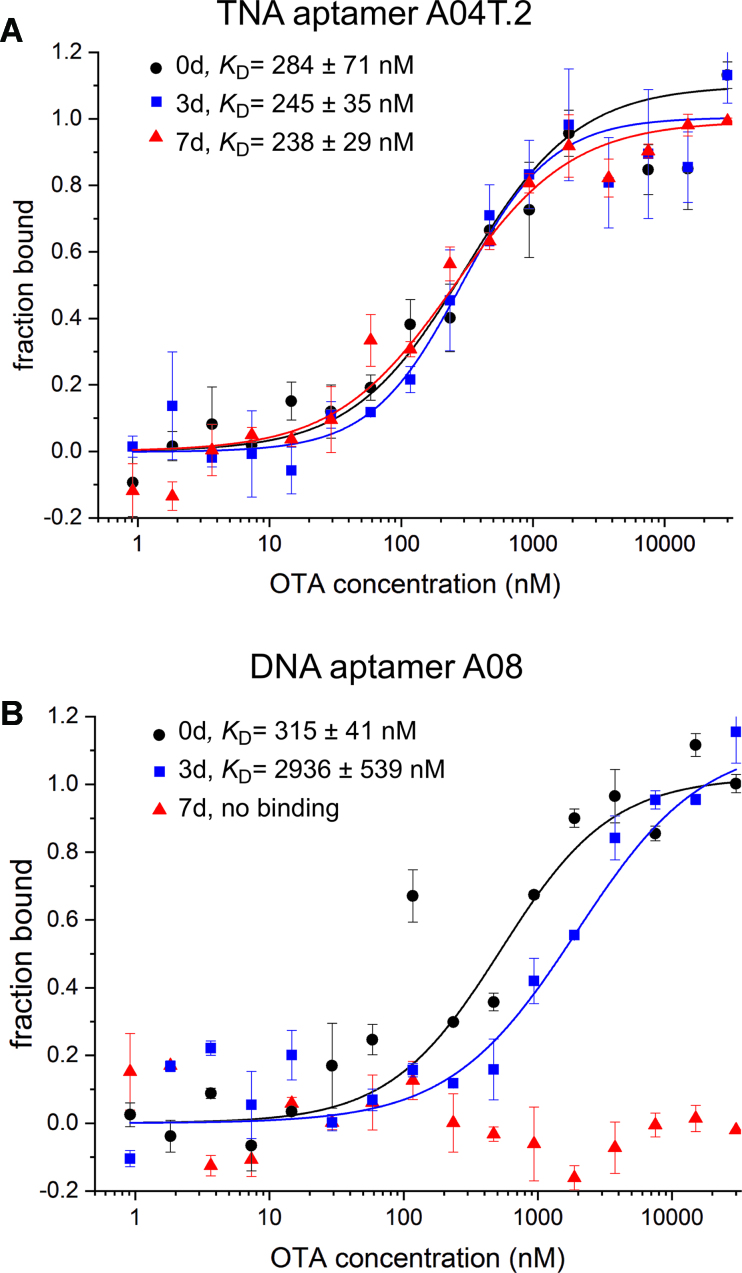
To test this hypothesis, we returned to MST analysis to quantify binding affinity of the aptamers under biological conditions. Human serum is currently regarded as the standard for evaluating oligonucleotide stability and aptamer selectivity, and therefore we performed all experiments using 50% human serum in PBS. To avoid spectral overlap with the background fluorescence of the serum, Cy5 was employed in place of FAM for labeling the DNA and TNA aptamers in these experiments. Both aptamers were incubated in 50% human serum at 37°C for 0, 3 or 7 days, then MST measurements were acquired for each sample and compared to results obtained earlier for each aptamer in buffer alone (Figure 7). After incubation in serum for 3 days, our TNA aptamer A04T.2 shows a no reduction in binding affinity (KD = 245 ± 35 nM), while the DNA aptamer shows a 10-fold reduction in affinity (KD = 2396 ± 539 nM) in comparison to the 0d control. We were surprised to not observe a greater reduction in binding affinity of the DNA aptamer after 3 days in serum, as our PAGE and bead-based experiments suggested near complete degradation of the DNA. However, there is evidence that terminal functionalization of nucleic acids can protect against degradation in serum, and given the sensitivity of MST, a binding isotherm can still be acquired even if only a small amount of intact aptamer remains (38). This notion is supported by the data acquired after seven days of incubation in serum, as the binding affinity of aptamer A04T.2 remains unchanged (KD = 238 ± 29 nM), but the DNA aptamer shows no detectable binding to OTA, suggesting complete degradation. The ability of our TNA aptamer to maintain target binding capability under such harsh conditions highlights the exceptional nuclease resistance of TNA polymers.
Figure 7.
MST analysis of (A) TNA and (B) DNA aptamers binding to OTA after incubation in 50% human blood serum in PBS at 37°C for 0, 3 or 7 days. For both aptamers, the binding affinity measured in selection buffer was used as the control. Error bars represent the standard deviation of two independent trials.
CONCLUSIONS
In summary, we have demonstrated in vitro evolution of the first XNA aptamers capable of binding to a small molecule target. Our best TNA aptamers have mid nM affinity for the OTA target, surpassing the affinity of the best reported DNA aptamer for the same target. Achieving success in these current experiments required the expansion of XNA selection technology to enhance enrichment by suppressing the background arising from DNA contamination. We were able to successfully minimize two of the aptamer sequences to lengths of ≤35 nt, enabling them to be generated using solid-phase oligonucleotide synthesis. The 31 nt truncated sequence A04T.2 exhibited the highest affinity of any sequence tested, and validates the integrity of information transfer between enzymatically and chemically synthesized XNA. We also show that the TNA aptamer is capable of binding to OTA after a seven-day incubation in human serum, whereas the best DNA aptamer no longer functions after exposure to these conditions. This suggests that these aptamers will be particularly advantageous for in vivo applications such as treatment of poisoning though sequestration of toxins. While OTA served as a convenient proof of concept to validate the efficacy of the selection process, we anticipate that this selection approach will be amenable to other small-molecule toxins that pose a greater threat to human health.
In addition to the high performance of our TNA aptamer, the demonstration that the sequence can be minimized to a length that is compatible with chemical synthesis provides access to significantly greater quantities of material, enabling new applications that were previously not feasible for XNA aptamers due to the limited practical scale of enzymatic synthesis. For example, efforts are currently underway in our laboratory to engineer modified versions of our TNA aptamers into biosensors for use in toxin detection. We envision that these sensors will find high utility in field-based testing, where accurate quantitation of a desired target must proceed with minimal sample preparation and using assays capable of enduring rigorous shipment and storage conditions. Together, the ability of XNA aptamers to simultaneously provide high affinity, selectivity, and stability suggests a bright future for these biopolymers in diverse applications in biotechnology.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. John Chaput and his laboratory for the generous gift of KOD RI plasmids and technical advice. We would also like to thank Dr. Mike Hanson from the University of Utah DNA/peptide core facility for chemical synthesis of the TNA polymers and Dr. Thomas Schubert from 2bind GmbH for performing the MST experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DARPA Folded Non-Natural Polymers with Biological Function (Fold F(x)) Program [N66001-14-2-4054]. Any opinions, findings and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of DARPA. Funding for open access charge: DARPA Folded Non-Natural Polymers with Biological Function (Fold F(x)) Program [N66001-14-2-4054]; National Science Foundation [CBET 1818476 to J.M.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ellington A.D., Szostak J.W.. In vitro selelction of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 2. Roberston D.L., Joyce G.F.. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990; 344:467–468. [DOI] [PubMed] [Google Scholar]
- 3. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 4. Peterson A.M., Jahnke F.M., Heemstra J.M.. Modulating the substrate selectivity of DNA aptamers using surfactants. Langmuir. 2015; 31:11769–11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor A.I., Arrangundy-Franklin S., Holliger P.. Towards applications of synthetic genetic polymers in diagnosis and therapy. Curr. Opin. Chem. Biol. 2014; 22:79–84. [DOI] [PubMed] [Google Scholar]
- 6. Herdewijn P., Marliere P.. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009; 6:791–808. [DOI] [PubMed] [Google Scholar]
- 7. Steele F.R., Gold L.. The sweet allure of XNA. Nat. Biotech. 2012; 30:624–625. [DOI] [PubMed] [Google Scholar]
- 8. Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjić J.. 2′-Fluoropyrimidine RNA-based aptamers to the 165-Amino acid form of vascular endothelial growth factor (VEGF165): inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998; 273:20556–20567. [DOI] [PubMed] [Google Scholar]
- 9. Pinherio V.B., Holliger P.. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 2012; 16:245–252. [DOI] [PubMed] [Google Scholar]
- 10. Meek K.N., Rangel A.E., Heemstra J.M.. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods. 2016; 106:29–36. [DOI] [PubMed] [Google Scholar]
- 11. Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S-Y., McLaughlin S.H. et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012; 336:341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taylor A.I., Holliger P.. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat. Protoc. 2015; 10:1625–1642. [DOI] [PubMed] [Google Scholar]
- 13. Yu H., Zhang S., Chaput J.C.. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012; 4:183–187. [DOI] [PubMed] [Google Scholar]
- 14. Mei H., Liao J.Y., Jimenez R.M., Wang Y., Bala S., McCloskey C., Switzer C., Chaput J.C.. Synthesis and evolution of a TNA aptamer bearing 7-Deaza-7-Substituted guanosine residues. J. Am. Chem. Soc. 2018; 140:5706–5713. [DOI] [PubMed] [Google Scholar]
- 15. Alves Ferreira-Bravo I., Cozens C., Holliger P., DeStefano J.J.. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015; 43:9587–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thirunavukarasu D., Chen T., Hongdilokkul N., Romesburg F.E.. Selection of 2′-Fluoro-Modified aptamers with optimized properties. J. Am. Chem. Soc. 2017; 139:2892–2895. [DOI] [PubMed] [Google Scholar]
- 17. Mckeague M., DeRosa M.C.. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids. 2012; 2012:748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruscito A., DeRosa M.C.. Small-Molecule binding Aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016; 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spill F., Weinstein Z.B., Shemirani A.I., Ho N., Desai D., Zaman M.H.. Controlling uncertainty in aptamer selection. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12076–12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blind M., Blank M.. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids. 2015; 4:e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruscito A., McConnell E.M., Koudrina A., Velu R., Mattice C., Hunt V., McKeague M., DeRosa M.C.. In vitro selection and characterization of DNA aptamers to a small molecule target. Curr. Protoc. Chem. Biol. 2018; 4:233–268. [DOI] [PubMed] [Google Scholar]
- 22. Cruz-Aguado J.A., Penner G.. Determination of ochratoxin a with a DNA aptamer. J. Agric. Food Chem. 2008; 56:10456–10461. [DOI] [PubMed] [Google Scholar]
- 23. Ichida J.K., Zou K., Horhota A., Yu B., McLaughlin L.W., Szostak J.W.. An in vitro selection system for TNA. J. Am. Chem. Soc. 2005; 127:2802–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu H., Zhang S., Dunn M.R., Chaput J.C.. An efficient and faithful in vitro replication system for threose nucleic acid. J. Am. Chem. Soc. 2013; 135:3583–3591. [DOI] [PubMed] [Google Scholar]
- 25. Culbertson M.C., Temburnikar K.W., Sau S.P., Liao J.Y., Bala S., Chaput J.C.. Evaluating TNA stability under stimulated physiological conditions. Bioorg. Med. Chem. Lett. 2016; 26:2418–2421. [DOI] [PubMed] [Google Scholar]
- 26. Sau S.P., Fahmi N.E., Liao J-Y., Bala S., Chaput J.C.. A scalable synthesis of a-L-Threose nucleic acid monomers. J. Org. Chem. 2016; 8:2302–2307. [DOI] [PubMed] [Google Scholar]
- 27. Sau S.P., Chaput J.C.. A One-Pot synthesis of a-L threofuranosyl nucleoside triphosphates (tNTPs). Bioorg. Med. Chem. Lett. 2016; 26:3271–3273. [DOI] [PubMed] [Google Scholar]
- 28. Zhang S., Chaput J.C.. Synthesis of Threose Nucleic Acid (TNA) phosphoramidite monomers and oligonucleotide polymers. Curr. Protoc. Nucleic Acids Chem. 2012; 4:doi:10.1002/0471142700.nc0451s50. [DOI] [PubMed] [Google Scholar]
- 29. Larsen A.C., Dunn M.R., Hatch A., Sujay S.P., Youngbull C., Chaput J.C.. A general strategy for expanding polymerase function by droplet microfluidics. Nat. Commun. 2016; 7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dunn M.R., Larsen A.C., Zahurancik W.J., Fahmi N.E., Meyers M., Suo Z., Chaput J.C.. DNA polymerase-mediated synthesis of unbiased threose nucleic acid (TNA) polymers requires 7-Deazaguanine to suppress G:G mispairing during TNA transcription. J. Am. Chem. Soc. 2015; 137:4014–4017. [DOI] [PubMed] [Google Scholar]
- 32. Dunn M.R., Chaput J.C.. Reverse transcription of threose nucleic acid by a naturally occurring DNA polymerase. ChemBioChem. 2016; 17:1804–1808. [DOI] [PubMed] [Google Scholar]
- 33. Duhr S., Braun D.. Thermophoretic depletion follows Boltzmann distribution. Phys. Rev. Lett. 2006; 96:168301. [DOI] [PubMed] [Google Scholar]
- 34. Entzian C., Schubert T.. Studying small molecule-aptamer interactions using MicroScale Thermophoresis (MST). Methods. 2016; 97:27–34. [DOI] [PubMed] [Google Scholar]
- 35. McKeague M., Velu R., Hill K., Bardóczy V., Mészáros T., De Rosa M.C.. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins. 2014; 6:2435–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., Lu Y.. Functional Nucleic Acids for Analytical Applications. 2009; NY: Springer Science+Business Media; 67–68. [Google Scholar]
- 37. Bazin I., Faucet-Marqis V., Monje M-C., El Khoury M., Marty J.-L., Pfohl-Leszkowicz A.. Impact of pH on the stability and the cross-reactivity of ochratoxin A and citrinin. Toxins (Basel). 2013; 5:2324–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shaw J.P., Kent K., Bird J., Froehler B.. Modified deoxynucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991; 19:747–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.