Abstract
SWI/SNF complexes associate with genes and regulate transcription by altering the chromatin at the promoter. It has recently been shown that these complexes play a role in pre-mRNA processing by associating at alternative splice sites. Here, we show that SWI/SNF complexes are involved also in pre-mRNA 3′ end maturation by facilitating 3′ end cleavage of specific pre-mRNAs. Comparative proteomics show that SWI/SNF ATPases interact physically with subunits of the cleavage and polyadenylation complexes in fly and human cells. In Drosophila melanogaster, the SWI/SNF ATPase Brahma (dBRM) interacts with the CPSF6 subunit of cleavage factor I. We have investigated the function of dBRM in 3′ end formation in S2 cells by RNA interference, single-gene analysis and RNA sequencing. Our data show that dBRM facilitates pre-mRNA cleavage in two different ways: by promoting the association of CPSF6 to the cleavage region and by stabilizing positioned nucleosomes downstream of the cleavage site. These findings show that SWI/SNF complexes play a role also in the cleavage of specific pre-mRNAs in animal cells.
INTRODUCTION
SWI/SNF is an ATP-dependent chromatin remodeling complex that consists of ∼10 highly conserved subunits (1,2). The SWI/SNF complex of Drosophila melanogaster contains only one ATPase subunit, Brahma (dBRM), which interacts with the core subunits Snf5-related 1 (SNR1) and Moira (MOR). It interacts also with several accessory subunits. The human SWI/SNF contains one of two ATPase subunits: either human BRM (hBRM) or Brahma-related gene 1 (hBRG1). The SWI/SNF complexes regulate transcription by remodeling nucleosomes at promoter regions and gene bodies (3,4). Genome-wide studies have revealed that SWI/SNF core subunits co-localize extensively with RNA polymerases and show high occupancy levels at promoters, enhancers and CTCF motifs (5). These studies have also revealed high levels of SWI/SNF occupancy at transcription termination sites (5–7).
SWI/SNF regulates not only transcription but also pre-mRNA splicing and 3′ end formation. In human cells, hBRM contributes to the inclusion of alternative exons in the CD44 pre-mRNA by decreasing the rate of elongation of RNA polymerase II (RNAPII) through a mechanism that involves the RNA-binding protein Sam68 (8,9). Moreover, the hBRM regulates the alternative splicing of 3′ terminal exons by controlling the stability of CSTF1, a factor involved in pre-mRNA 3′ end formation (10). In D. melanogaster, dBRM influences alternative pre-mRNA processing through unknown mechanisms (11–13). Interestingly, a fraction of dBRM is associated with the ribonucleoprotein (RNP) fractions in the nucleus of D. melanogaster S2 cells, and the BRM protein of the dipteran Chironomus tentans is associated with nascent pre-mRNP particles (11), which suggests that SWI/SNF plays a direct role in the regulation of pre-mRNA processing.
The 3′ end of mRNAs is formed co-transcriptionally through a complex process that involves the cleavage of the nascent pre-mRNA and the polyadenylation of the resulting 3′ end. This process requires cis-acting elements that recruit the cleavage and polyadenylation machineries: an upstream sequence element (USE, usually a UGUA sequence), the poly(A) signal (usually AAUAAA or AUUAAA) located 10–30 bases upstream of the mRNA cleavage site (CS), and a downstream sequence element (DSE, U/GU rich) (14,15). The processing of the pre-mRNA 3′ end is catalyzed by multiprotein complexes that are evolutionarily conserved: the cleavage and polyadenylation specificity factor (CPSF), the cleavage stimulation factor (CstF), and the cleavage factors I and II (CFIm and CFIIm). CPSF consists of six polypeptides: CPSF1-4, FIP1L1 and WDR33, and recognizes the poly(A) signal through its largest subunit CPSF1 (reviewed in 16). CstF is composed of CSTF1-3 and binds to the DSE. The CFIm complex is a tetramer that contains two small CPSF5 subunits and two large subunits of either CPSF7 or CPSF6. The binding of CFIm to the USE in the pre-mRNA is one of the earliest steps in the assembly of the cleavage and polyadenylation machinery. CFIm plays an important role in the recruitment of other 3′ end processing factors, even in the absence of a consensus poly(A) signal (17), and regulates alternative polyadenylation and the length of the 3′ UTR in human cells (18).
More than 70% of mammalian genes and approximately half of the genes in D. melanogaster have multiple CS, and the use of alternative CSs is regulated through several mechanisms (19). The abundance of 3′ end-processing factors (20–22), the rates of transcription elongation by RNAPII (23) and the nucleosome landscape at the 3′ end of the gene (24) are some of the factors that influence CS selection. Apart from generating mRNA diversity through alternative polyadenylation, changes in pre-mRNA cleavage and polyadenylation are also associated with cellular responses to stress (10,25).
We have investigated the role of SWI/SNF in pre-mRNA 3′ end processing in D. melanogaster at a genome-wide scale. We have shown that depletion of dBRM has specific effects on the processing of different subsets of pre-mRNAs, and we have identified a group of genes for which correct 3′ end-processing depends on dBRM. These genes are characterized by high dBRM levels and an open chromatin structure downstream of the CS. SWI/SNF associates with nascent pre-mRNPs (11), and this led us to pose the hypothesis that the SWI/SNF-mediated regulation of 3′ end processing relies on interactions between SWI/SNF and factors associated with nascent transcripts. We have carried out a comparative proteomics study aimed at elucidating the molecular basis for the role of SWI/SNF in pre-mRNA 3′ end formation to test this hypothesis. We have identified cleavage and polyadenylation factors that are bound to the SWI/SNF ATPases in both human and fly cells. Moreover, we have shown that dBRM facilitates the association of the CFIm factor CPSF6 to the 3′ end of genes of D. melanogaster. Our results provide experimental evidence for two different mechanisms by which dBRM promotes pre-mRNA 3′ end processing in metazoans.
MATERIALS AND METHODS
Cell culture
Drosophila melanogaster S2 cells were cultured at 28°C in Schneider's Drosophila medium (Invitrogen) containing 10% heat-inactivated FBS, 50 U/ml penicillin and 50 μg/ml streptomycin. Human HeLa and C33A cells (26) were cultured at 37°C and 5% CO2 in high-glucose DMEM (HyClone) medium supplemented with 10% FBS, 50 U/ml penicillin and 50 μg/ml streptomycin.
Antibodies
The antibody used to immunoprecipitate hBRG1 was the anti-ratBRG1 rabbit polyclonal antibody raised and characterized by Östlund Farrants et al. (27). The anti-ratBRG1 was also used for IP of endogenous dBRM in S2 cells. The cross-reactivity of this antibody against dBRM was shown by Tyagi et al. (11). Western blot analysis of endogenous dBRM was performed using a rabbit antibody raised against the C-terminal part of ct-BRM by Tyagi et al. (11). The rabbit anti-SNR1 and anti-MOR antibodies were raised and characterized by Dingwall et al. (28) and Mohrmann et al. (29), respectively. We also used the following commercial antibodies from Abcam: anti-hBrm (ab15597), anti-RNAPII CTD (ab5408), anti-tubulin (ab7291), anti-hCPSF1 (ab81552), anti-hCPSF2 (ab126760), anti-HA tag (ab9110), anti-V5 tag (ab9116) and anti-IgG (ab46540). Secondary antibodies for Western blotting were horseradish peroxidase conjugates purchased from DakoCytomation.
RNA interference in S2 cells
RNAi experiments in S2 cells were carried out as described by Tyagi et al. (11). Double-stranded RNAs (dsRNAs) complementary to dBRM or GFP were synthesized by in vitro transcription using the MegaScript RNAi kit (Ambion) from gene-specific PCR fragments with incorporated T7 promoters at both ends. The sequences of the PCR primers used for dsRNA synthesis are provided as additional text in the supplementary information. 3 × 106 S2 cells were cultured in six-well plates overnight, washed with serum-free and antibiotic-free Schneider's medium, and treated with 30 μg of dsRNA per well. The cells were harvested 48 h after the addition of dsRNA. Cells treated with the same amount of GFP-dsRNA were used as a control. The efficiency of the RNAi treatment was analyzed by Western blotting and quantified by densitometry as described below.
Overexpression of recombinant dBRM in S2 cells
A stably transfected cell line for the expression of V5-tagged recombinant dBRM has been described in Yu et al. (30). The expressions of the recombinant dBRM was under the control of the metallothionein promoter of the pMT vector (Invitrogen). Expression of the recombinant protein was induced by treating the cells with 200 μM copper sulfate for 24 h before harvesting and analysis.
Expression of hBRG1 in C33A cells
C33A cells were transiently transfected with the pBJ5-BRG1 plasmid for the expression of hBRG1 (31) and harvested 48 h after transfection. The pOPRSVI vector was used in parallel for control purposes.
RNA isolation, reverse transcription and qPCR
Total RNA was isolated from S2 cells using the RNAqueous kit (Ambion). DNA contamination was removed by subsequent treatment with 1 U DNase I (Thermo Scientific) for 30 min at 37°C. Human C33A cells were harvested 2 days after transfection with pOPRSVI (control) or pBJ5-BRG1 with a final confluence of 80%. RNA was isolated from C33A cells using Qiazol (Qiagen). In all cases, RNA purity, yield and integrity were analyzed by spectrophotometry using a NanoDrop instrument (Thermo Scientific). Total RNA was reverse-transcribed with Superscript III (Thermo Fisher Scientific) using random primers (Thermo Fisher Scientific) and following the recommendations of the manufacturer. Synthesized cDNAs were stored at –20°C and used for qPCR in duplicate reactions using a KAPA SYBR Fast qPCR Kit (KAPA Biosystem) in a QIAGEN Rotor-Gene Q system. The qPCR assays followed the MIQE guidelines for primer design. All primer pairs fulfilled quality criteria according to amplification efficiency and melting curves. The results presented are compiled data from multiple independent biological replicates, each analyzed in duplicate. For each experiment, the number of independent replicates is provided in the figure legend. In the cleavage assays, the level of uncleaved pre-mRNA for a given transcript was normalized to the 3′ UTR level of the transcript in the same sample. In other RT-qPCR experiments, the relative abundance of each transcript was normalized to the abundance of Act5C mRNA in the same sample. All primer sequences are provided as additional text in the supplementary information. The oligonucleotides were manufactured by Life Technologies Europe BV.
RNA-seq
Total RNA was isolated from S2 cells using the RNAqueous kit (Ambion) and used to construct poly(A) libraries for massive parallel sequencing on a Illumina HiSeq2000 sequencer, high-output mode, to a depth of at least 6 M paired reads per sample (2 × 125 bp). Library construction and sequencing were carried out at the Science for Life Laboratory (Stockholm, Sweden). High-quality reads were mapped with TopHat2 (32) to the Drosophila melanogaster genome assembly, build BDGP6 (Dm3). The number of reads that mapped to genes was extracted with the featureCounts tool from the package Rsubread (33), using the Dm3 Ensembl genome annotation as a reference. Differential gene expression analysis using the read counts obtained from the two independent replicates was carried out using the Limma package (34). Correlation analysis of biological replicates showed high reproducibility (Supplementary Figure S1). Reads from both experiments were analyzed together for metagene and heatmap representations, but used as separate replicates for differential expression analysis. Heatmap plots and metagene plots were generated with the ngs.plot package (35), and k-means clustering was analysed using the same package. The RNA-seq data are available at GEO with accession number GSE95236 (datasets ‘empty dsGFP’ and ‘empty dsBRM’ biological replicates 1 and 2).
Chromatin immunoprecipitation (ChIP-qPCR)
Drosophila S2 cells were fixed at room temperature with 1% formaldehyde, and the chromatin was extracted from formaldehyde-treated cells as described by Hessle et al. (36). The chromatin was fragmented by sonication using a Bioruptor (Diagenode) with 60 pulses of 30 s. The lysate was cleared by centrifugation at 17 000 g for 30 min. A DIG–DNA complex was added to the chromatin extracts and used as external standard for quantification, as described by Eberle et al. (37). Sheared chromatin was incubated with one of the following antibodies at 4°C overnight: anti-V5 (Abcam, ab9116), anti-HA (Abcam, ab9110), anti-CTD (Abcam, ab5408). Anti-IgG (Abcam, ab46540) was used as a negative control. Dynabeads Protein G was incubated with samples for 1 h at 4°C, washed five times with RIPA buffer (10 min each time), and then washed once with 0.05 M Tris–HCl (pH 8), 2 mM EDTA. The crosslinking was reversed in TE buffer containing 0.5% SDS, 0.1 mg/ml RNase A and 0.2 mg/ml proteinase K (Thermo Scientific) at 55°C for 3 h, and subsequently at 65°C overnight. The immunoprecipitated DNA was finally extracted with a mixture of phenol, chloroform and isoamylalcohol (Sigma). For qPCR analysis, the KAPA SYBR Fast qPCR Kit (KAPA Biosystem) was used in a Rotor-Gene Q system (Qiagen). Standard curves for each primer pair were used to quantify the immunoprecipitated DNA. The ChIP-qPCR signals were normalized using a DIG–DNA antibody complex (37). The qPCR assays followed the MIQE guidelines as described above.
ChIP-seq data analysis
ChIP-seq experiments were carried out by Jordán-Pla et al. (7) using an antibody against the endogenous dBRM protein (27). The ChIP-seq data are available at GEO with accession number GSE95236 (datasets ‘endogenous BRM input replicates 1 and 2’ and ‘endogenous BRM IP replicates 1 and 2). For metagene and heatmap plotting purposes, the bam files for two biological replicates were merged into one file and compared to a merge of the two biological replicates of their corresponding inputs. The replicates were treated independently for enrichment calculations. The ngs.plot package (35) was used to calculate dBRM enrichment levels throughout the genome, and to plot metagene and heatmap representations. Statistical significance between different metagene distributions was calculated using the Kolmogorov–Smirnov (K–S) test in GraphPad. A fragment size of 200 bp was selected as input parameter to calculate the read coverage, based on the average sonicated DNA fragment size in our Bioanalyzer runs. Coverage tracks for visual inspection of the Bowtie2 alignments with genome browsers were generated with the bamCompare tool from the DeepTools package (33) with default parameters. Average values of dBRM enrichment over the genebody region, and up to 200 bp upstream of the TSS and downstream of the CS, were calculated.
Chromatin environment downstream of cluster 1 genes
The overlaps between a region extending 1kb downstream of the CS of genes in clusters 1, 4 and entire genome, and the chromatin-domain classification from modENCODE (38) were found using the join intervals tool, as implemented in Galaxy (https://usegalaxy.org/). We used the Peak Annotation and Visualization tool (PAVIS; 39) to identify genomic features in a window of 1 kb downstream of genes in clusters 1 and 4, choosing between strand-specific and unstranded annotation options to detect features downstream of the genes in the same strand (in tandem, or sense) or in the opposite strand (convergent or antisense).
SDS-PAGE and western blot
Cells were spun down and resuspended in Laemmli sample buffer containing 8 M Urea. Samples were denatured by heating to 100°C. Proteins were separated by SDS-PAGE in gels containing 7.5% polyacrylamide using the Mini-Protean II system (BioRad). The proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were probed with antibodies diluted in PBS containing 0.05% Tween-20 and 1% milk. Protein signals were detected using the ECL system (GE Healthcare) in a Gel Doc EZ system (BioRad). Quantitative analyses of RNAi and overexpression treatments were carried out by blot densitometry using the band identification and quantification tools of the Image Lab 6.0.0 Software (BioRad). Tubulin was used as a loading reference. Three independent blots were analyzed for each type of treatment, and the results are expressed as average fold change ±1 standard deviation.
Coimmunoprecipitation
Chromosomal RNP fractions were prepared from S2 cells and HeLa cells as described by Tyagi et al. (11) and incubated at 4°C overnight in the presence of primary antibody. An antibody produced by Östlund Farrants et al. (27) was used to precipitate hBRG1 from HeLa cells and dBRM from S2 cells. In the case of hBRM, a commercial anti-hBrm antibody (ab15597, Abcam) was used. Negative control immunoprecipitations were carried out in parallel using an anti-IgG antibody (ab46540). A slurry of Sepharose protein A/G (GE Healthcare) was used to pull down the bound complexes. The immunoprecipitated material was washed three times with PBS containing 0.1% Nonidet P40 and 1 mM PMSF, and was eluted from the Sepharose beads with 1% SDS for 10 min at room temperature. The proteins were finally precipitated with acetone and resolved by SDS-PAGE.
High-throughput mass spectrometry (LC–MSMS)
Liquid chromatography and mass spectrometry were carried out at Proteomics Karolinska, Karolinska Institute (PK/KI). The immunoprecipitated proteins from three independent experiments were resolved in SDS-PAGE. Entire gel lanes were digested with trypsin and the peptides were extracted, resolved and analyzed by mass spectrometry using an ESI-FTICR instrument. Proteins immunoprecipitated in the negative controls were analyzed in parallel. Mascot 2.3.02 was used for the identification of the peaks, and the significance threshold was set at 0.05. Only proteins with at least two different peptides detected, at least one unique peptide, score above 20, and score at least twice as high as in the negative control were considered positive. The lists of proteins identified in each IP experiment were compared using Ensembl Genes 91, GRCh38.p12 for human samples and BDGP6 for D. melanogaster, using the human nomenclature as a reference. For this purpose, the human orthologs for the Drosophila proteins identified were obtained from Biomart. The Euler diagram presented in Figure 6 was drawn using the EulerAPE software (40). The Gene Ontology (GO) enrichment tests were performed using the PANTHER Overrepresentation test (41) released 2017-12-05 and GO Ontology database released on 2018-02-02 using the Fisher's exact test with FDR multiple test correction. Only GO terms with a P-value < 0.05 were considered.
Figure 6.
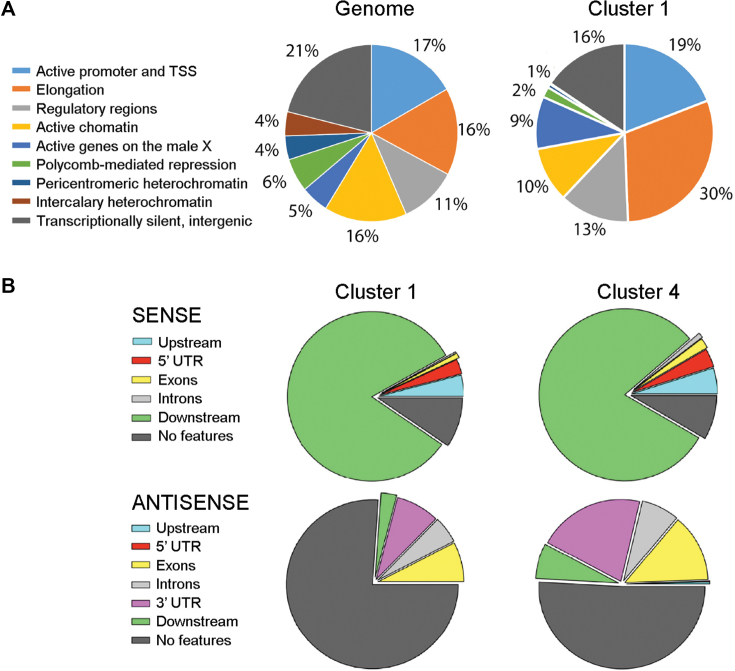
The sequences downstream of the genes in cluster 1 are gene-poor and enriched in open chromatin features. (A) The sequences downstream of the CS were classified according to the 9-state chromatin model of Karchenko et al. (38). The pie diagrams show the occurrences of each type of chromatin state in the average of the S2 genome and in the genes of cluster 1. (B) The pie diagrams show the occurrences of annotated gene features in a window of 350 bp downstream of the CS of the genes in clusters 1 and 4. Features in the ‘sense’ direction refer to genes in tandem, whereas features in the ‘antisense’ direction reveal the presence of convergent transcription units. ‘No features’ denotes the absence of annotated gene features in the region of interest.
Cloning and expression of recombinant dBRM and CPSF6 in S2 cells
A stably transfected cell line that expresses V5-tagged recombinant dBRM was constructed and described in Yu et al. (30). Stably transfected cells that express recombinant CPSF6 with a V5 tag were constructed as follows. The open reading frame of CPSF6 was amplified by PCR with Pfu polymerase (Agilent) using the plasmid LD25239 (Drosophila Genomics Resource Center) as a template. Forward and reverse primers contained SpeI and ApaI cloning sites, respectively. The recombinant CPSF6 PCR product was cloned into a pMT-puromycin vector containing a V5 tag (Addgene). The pMT-CPSF6-V5 plasmid was purified using QIAprep Spin Miniprep Kit (Qiagen) and transfected into S2 cells using a calcium phosphate transfection kit (Invitrogen). Stably transfected cells were selected with 5 μg/ml puromycin and further maintained in medium containing 2 μg/ml puromycin. The pMT-puromycin vector without any inserted sequence was also transfected into S2 cells, and the resulting cells were used as a control (‘empty’ control). The stably transfected cells co-expressing recombinant dBRM with V5 tag and recombinant CPSF6 with HA tag were constructed from the recombinant CPSF6 plasmid LD25239 (Drosophila Genomics Resource Center). The open reading frame of CPSF6 was amplified by PCR using Pfu polymerase (Agilent) with a forward primer containing the SpeI cloning site, and a reverse primer containing the HA sequence and the ApaI cloning site. The recombinant CPSF6-HA PCR product was cloned into a pMT/V5-His vector (Invitrogen). The pMT-CPSF6-HA plasmid was purified using a QIAGEN Plasmid Midi Kit (QIAGEN) and transfected into cells that expressed recombinant dBRM using a calcium phosphate transfection kit (Invitrogen). The pCoHygro plasmid (Invitrogen) was also co-transfected with the pMT-CPSF6-HA plasmid for hygromycin selection. Stably transfected cells were selected and cultured in a medium that contained 300 μM hygromycin B (Invitrogen). The pMT-CPSF6-HA and pCoHygro plasmids were co-transfected into the cells expressing pMT-puromycin vector as a control.
ATAC-qPCR
RNAi was carried out as described above with dsRNAs complementary to either dBRM or GFP (control). The cells were analysed 48 h after the start of the dsRNA treatment. The ATAC-seq protocol of Buenrostro et al. (42) was adapted for S2 cells. For each experiment, 5 × 104 cells were immediately placed in 50 μl ice cold ATAC lysis buffer (10 mM Tris–HCl, pH 7.4,10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and tagmented using the Nextera Library Preparation Kit (Illumina) (FC-121-1030). The tagmented DNA was purified using Qiagen MinElute Kit (Qiagen) and amplified through five PCR cycles following the manufacturer's instructions. Chromatin accessibility at specific loci was analyzed by qPCR using the Nextera forward adapter primer and a gene specific reverse primer. The qPCR signals in control and treated samples were normalized to Hsp70 promoter region and the results were expressed as fold change control vs. treated. The qPCR assays followed the MIQE guidelines as described above. Primer sequences are provided as Supplementary Data.
Statistical analysis
Histograms show average values and error bars represent standard deviations. The numbers of biological replicates are indicated in the figure legends. One-sample or two-sample t-tests were used for statistical testing of results from single gene analyses. The one-sample test was used to establish whether fold changes were significantly different from 1. The Kolmogorov–Smirnov (K–S) test was used for statistical testing of differences in metagene distributions. Statistical testing of GO overrepresentation was carried out by Panther using the Fisher's exact test with correction for multiple testing. The tests used in each case are indicated in the figure legends.
RESULTS
dBRM and hBRG1 facilitate 3′ end cleavage in fly and human cells
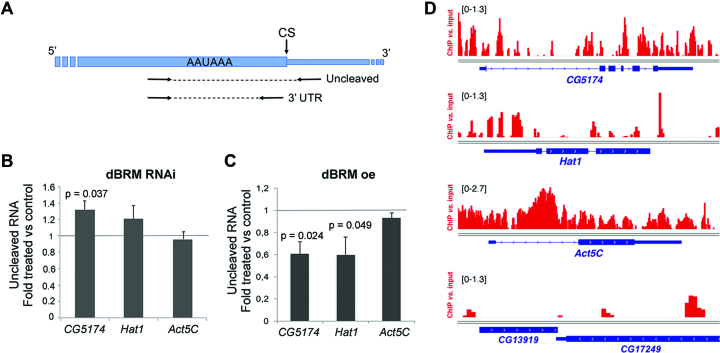
We carried out RNA interference (RNAi) experiments in S2 cells to deplete dBRM. Previous studies showed that depletion of dBRM for four days blocks cell cycle progression and has profound effects on the gene expression patterns of S2 cells (44). We chose to harvest the cells after only two days of dsRNA treatment to minimize indirect, long-term effects caused by proliferation arrest. In the conditions of our experiments, the levels of dBRM were reduced to 28% as assessed by Western blotting (Supplementary Figure S2A). We used a PCR-based 3′ end cleavage assay to analyze the effects of the dBRM depletion on pre-mRNA 3′ end processing. The assay involved two pairs of PCR primers to amplify either the 3′ UTR sequence or the uncleaved pre-mRNA (Figure 1A). These primers were used in quantitative RT-PCR experiments (qRT-PCR) to quantify the relative abundances of uncleaved pre-mRNA products and, in this way, measure the cleavage efficiency at specific CSs. We used this assay to analyze the effect of dBRM depletion on the processing of two pre-mRNAs that had been identified as dBRM targets in previous studies: CG5471 and Hat1 (12). The CG5174 gene codes for the highly expressed tumor protein D52-like. According to the gene model available at FlyBase (http://flybase.org/reports/FBgn0034345), the CG5174 pre-mRNA has two alternative CSs and we focused on the distal one, which is used in nine out of ten annotated isoforms. We also analyzed Hat1 (also known as CG2051), a moderately expressed gene that codes for histone acetyltransferase 1. This gene has two alternative promoters and a single CS (http://flybase.org/reports/FBgn0037376). The levels of uncleaved CG5174 and Hat1 pre-mRNA were approximately 20% higher in dBRM-depleted cells than in GFP control cells, whereas the cleavage of an unrelated pre-mRNA, Act5C, was not affected (Figure 1B). In a reciprocal experiment, overexpression of recombinant dBRM caused a significant reduction of uncleaved CG5174 and Hat1 pre-mRNAs, but did not affect Act5C (Figure 1C and Supplementary Figure S2B). These results suggest that dBRM favors the 3′ end cleavage of specific pre-mRNAs in S2 cells.
Figure 1.
The cleavage of CG5174 and Hat1 pre-mRNAs is regulated by dBRM. (A) Drawing showing the position of the primers used for RT-qPCR in the cleavage assay. The thick box represents the 3′ UTR. The thin stretch represents genomic sequences downstream of the cleavage site (CS). The relative amount of uncleaved pre-mRNA was quantified using the uncleaved primers and normalized to the amount of 3′ UTR (using the 3′ UTR primers). (B) S2 cells were treated with dsRNA complementary to either dBRM or GFP (control) for 48 h. The relative levels of uncleaved pre-mRNAs were quantified by RT-qPCR, and normalized to the 3′ UTR levels of each transcript. The bars show the average ratio between dsBRM and dsGFP samples. The error bars are standard deviations from three biological replicates. (C) Stably transfected S2 cells were treated with 200 μM CuSO4 for 24 h to induce the expression of recombinant dBRM. The relative levels of uncleaved pre-mRNAs were quantified by RT-qPCR as in C. The error bars are standard deviations from three biological replicates. (D) dBRM occupancy in four selected genes visualized in the IGV browser. The Y-axis shows the enrichment of the ChIP signal relative to the input. The bracketed numbers on each panel show the y-axis values. CG13919 is shown as an example of a gene that is not occupied by dBRM. The dBRM ChIP-seq data was from Jordán-Pla et al. (7). In B and C, one sample t-tests were used for statistical testing. Significant probability (p) values are shown in the figure.
A fraction of dBRM associates co-transcriptionally with nascent pre-mRNAs (11) and we were interested in understanding if SWI/SNF acts locally to promote pre-mRNA processing. We analyzed the ChIP-seq data of Jordán-Pla et al. (7) using an antibody against the endogenous dBRM in S2 cells (Supplementary Figure S3) and we detected the presence of dBRM at CG5174 and Hat1 (Figure 1D), which suggests that the observed effects of dBRM on pre-mRNA 3′ processing are direct. Interestingly, Act5C was also bound by dBRM but the cleavage of the Act5C pre-mRNA was not affected by dBRM depletion or overexpression. This observation indicates that SWI/SNF is not necessarily involved in the 3′ processing of all the dBRM-bound genes (see Discussion).
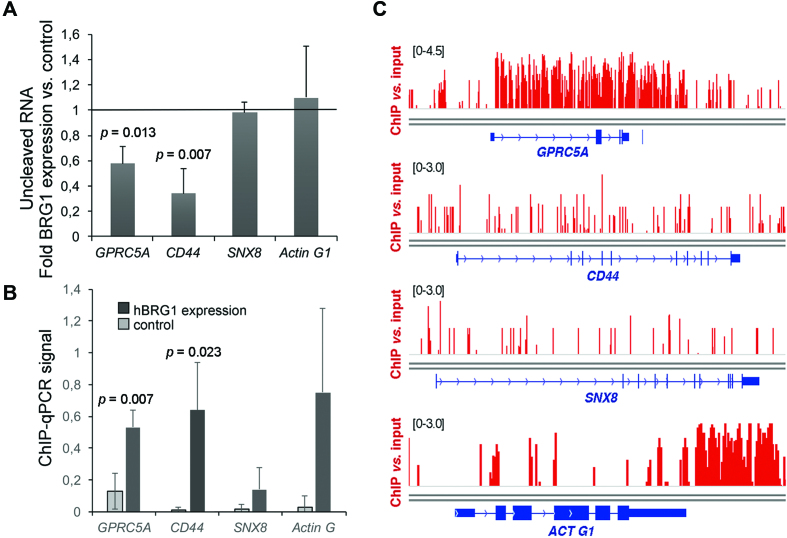
We also investigated the possible impact of SWI/SNF levels on 3′ end cleavage in human cells, and we chose to analyze hBRG1 since we showed in a previous study that hBRG1 is closely associated with RNA-binding proteins (11). We expressed recombinant wild-type hBRG1 in C33A cells, which lack SWI/SNF ATPases (26) (Supplementary Figure S2C), and we quantified the cleavage of transcripts from the CD44 and GPRC5A genes, two genes that are known SWI/SNF targets (45,46), in the presence or absence of hBRG1. For both genes, we observed significantly higher cleavage efficiencies (i.e. less uncleaved pre-mRNAs) in cells that expressed hBRG1 than in control C33A cells that did not express any SWI/SNF ATPase (Figure 2A). The effect of hBRG1 expression on 3′ end cleavage was gene-specific, as shown by the fact that expression of hBRG1 did not affect the cleavage of transcripts derived from two unrelated genes: SNX8 and ACTG1 (Figure 2A). ChIP-qPCR experiments showed that hBRG1 was recruited to CD44 and GPRC5A (Figure 2B). Analysis of ChIP-seq data from previous studies (5) showed that endogenous hBRG1 is recruited to these loci also in HeLa cells, which normally express the SWI/SNF ATPases (Figure 2C).
Figure 2.
The cleavage of GPRC5A and CD44 pre-mRNAs is regulated by hBRG1. (A) C33A cells were transfected with a plasmid for the expression of hBRG1, and harvested 48 h after transfection. Cells transfected with the empty vector pOPRSVI were used as control. Relative levels of uncleaved pre-mRNAs were quantified by RT-qPCR and normalized to the 3′ UTR of each transcript. The figure shows the average ratio between overexpression and control samples. The error bars are standard deviations from four biological replicates. One sample t-test was used for statistical testing. Significant probability (p) values are shown in the figure. (B) ChIP-qPCR using an antibody against hBRG1 to analyze the presence of the protein at the CS of the analyzed genes. ChIP was performed in C33A cells expressing hBRG1 48 h after transfection. An IgG antibody was used in parallel as a negative control. ChIP results were quantified by qPCR using the ‘uncleaved primers’ shown in Figure 1A. ChIP signals are represented as percentage of input after subtracting IgG values from positive signals. The error bars are standard deviations from three biological replicates. Two sample, two-tailed t-test was used for statistical testing. Significant probability (p) values are shown in the figure. (C) hBRG1 occupancy in the genes analyzed in A and B, based on ChIP-seq data from Euskirchen et al. (5).
The results reported above suggest that the SWI/SNF ATPases regulate the 3′ end cleavage of specific pre-mRNAs in both fly and human cells. ChIP experiments revealed that SWI/SNF could be detected in the analyzed genes, which suggests that SWI/SNF acts locally to promote pre-mRNA 3′ end processing. Moreover, our results also show that SWI/SNF does not affect the pre-mRNA processing of all the SWI/SNF-bound genes.
dBRM is required for the efficient 3′ end cleavage of hundreds of pre-mRNAs in S2 cells
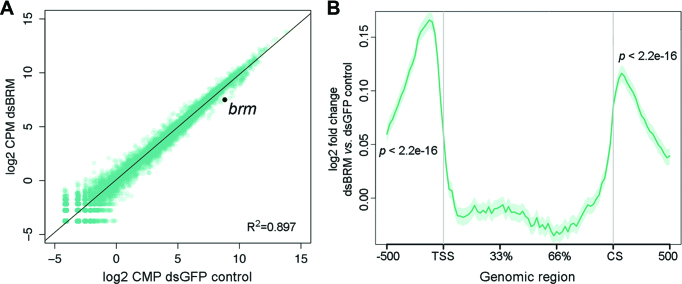
We carried out RNA-seq in S2 cells depleted of dBRM to study the role of SWI/SNF in 3′ end processing at a genomic scale. Differential expression analysis of cells depleted of dBRM for two days revealed mild effects on gene expression compared to control cells (Figure 3A). Sixty six genes, including brm itself, showed expression levels decreased by at least 50% and 98 genes showed expression increased by at least 50% (Supplementary Tables S1 and S2), but a differential gene expression analysis carried out using the Limma package (see Materials and Methods) did not identify any genes with significantly changed expression levels. However, a metagene analysis of RNA levels in gene flanking sequences revealed that dBRM depletion caused a very significant increase of average RNA levels upstream of the TSS and downstream of the CS (Figure 3B). As expected, the average RNA levels were lower in the gene flanking regions than in the gene body (Supplementary Figure S4).
Figure 3.
RNA-seq reveals genome-wide effects of dBRM depletion in gene flanking regions. S2 cells were treated with dsRNA complementary to dBRM for 48 h to deplete dBRM. Control cells were treated with dsGFP and analyzed in parallel. The transcriptomes of dBRM-depleted and control cells were analyzed by RNA-seq. (A) The scatter plot shows normalized RNA levels (expressed as log2 counts per million, CPM) in control cells (X-axis) and in dBRM-depleted cells (Y-axis). The Pearson's correlation coefficient is provided in the figure. (B) Metagene representation of the average effect of dBRM depletion on RNA levels in the gene bodies and flanking regions of all genes in S2 cells (n = 13924). The Y-axis shows log2 value of fold change in cells depleted of dBRM compared to control cells. The X-axis indicates genomic regions including the gene body, 0.5 kb upstream of the TSS and 0.5 kb downstream of the CS. A one-sample t-test was used for testing the statistical significance of the change in RNA levels observed in the 5′ and 3′ flanking regions.
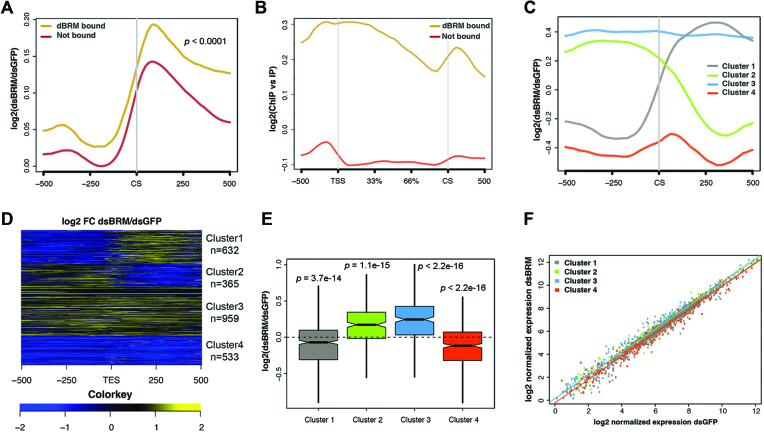
The role of SWI/SNF in chromatin regulation at gene promoters is well established (47), but its function in downstream gene regions is less understood. Based on the effects of dBRM depletion on 3′ end cleavage reported above, we hypothesized that the accumulation of RNA downstream of the CS in dBRM-depleted cells could be due to defects in 3′ end processing. We were interested in understanding direct regulatory events, and we restricted our study to dBRM-bound genes recently identified by ChIP-seq in S2 cells (7) (Supplementary Table S3). Interestingly, the increase of RNA levels that we observed downstream of the CS in dBRM-depleted cells was more pronounced in the set of dBRM-bound genes than in the rest of the transcriptome (Figure 4A). Moreover, as reported by Jordán-Pla et al. (7), the average gene profile of ChIP-seq reads along the dBRM-bound genes revealed that dBRM is preferentially located at the TSSs and downstream of the CSs (Figure 4B). These observations link SWI/SNF to transcription termination and/or 3′ end processing. It is possible that the increased RNA levels downstream of the CS result from a switch in alternative CS usage (alternative polyadenylation) or from defects in pre-mRNA 3′ end processing. Figure 4A shows strand-specific RNA levels relative to the most distal CS annotated for each gene, and therefore the observed changes in RNA levels downstream of the CS cannot be explained by switches in alternative polyadenylation. Moreover, our analysis also excludes the influence of antisense cryptic transcripts.
Figure 4.
RNA-seq reveals global effects of dBRM depletion on different subsets of genes in S2 cells. (A) Metagene representation of the effect of dBRM depletion on RNA levels 0.5 kb around the annotated CSs. The Y-axis shows log2 value of fold change in cells depleted of dBRM compared to control cells. Genomic regions upstream and downstream of CS are indicated on the X-axis. The plot shows an average increase of RNA reads downstream of the CS. The increase is more pronounced in the dBRM-bound genes (brown) than in the non-bound genes (red). Statistical testing of the difference between the two metagene distribution was done using the Kolmogorov-Smirnov test. The figure shows RNA reads from the sense strand only. (B) Metagene analysis of dBRM occupancy along the gene body and flanking sequences (X-axis). The Y-axis shows the log2 of the ChIP signal normalized to the input. The dBRM-bound genes are shown in brown (n = 2521 genes) and the non-bound genes in red (n = 11 403 genes). (C) Clustering of dBRM-bound genes according to the changes induced by dBRM-depletion on RNA levels in the CS region. (D) Heatmap showing the effect of dBRM depletion on the RNA level along each of the genes in the four clusters shown in B. (E) Boxplot showing the effects of dBRM depletion on the average expression of the genes in each of the four clusters. Statistical testing was done using a Wilcoxon test. (F) Scatter plot comparing the levels of expression of each gene in control cells (X-axis) and in cells depleted of dBRM (Y-axis).
K-means clustering of transcripts from the dBRM-bound genes into four groups according to the RNA-seq read distribution at the CS revealed that dBRM affects different subsets of transcripts differently (Figure 4C and Supplementary Table S4), and heatmap analyses revealed that the response to dBRM depletion was relatively homogeneous within each cluster (Figure 4D). Clusters 1 (gray) and 4 (red) comprised genes that were positively regulated by dBRM, as shown by the fact that the average expressions of these genes are lower in dBRM-depleted cells than in control cells, whereas clusters 2 (green) and 3 (blue) comprised genes that were negatively regulated by dBRM (Figure 4E). The range of expression levels in all clusters covered a wide range (Figure 4F).
In clusters 3 and 4, the changes in RNA levels downstream of the CS paralleled the changes in the gene bodies, and were presumably a consequence of the changed transcription level. For example, in the case of cluster 4, lower transcription rates would result in fewer RNAPII molecules reaching the end of the gene and, consequently, fewer downstream transcript sequences being produced. Instead, for genes in clusters 1 and 2, the changes in RNA levels downstream of the CS were opposite to the changes in the gene body. Cluster 1 was particularly interesting because the RNA levels downstream of the CS were increased upon dBRM depletion. Many of the genes in cluster 1 lacked alternatively polyadenylated isoforms, which strongly suggests that the increased RNA levels downstream of the CS reflect 3′ end processing defects.
We searched for enriched GO terms in the different gene clusters and we found differential enrichment of biological processes and molecular functions. For example, cluster 1 was significantly enriched in genes related to growth factor receptor binding (FDR q-value = 2–88E–03), behavior (FDR q-value = 8.83E–04) and positive regulation of cell proliferation (FDR q-value = 4.58E–03), among others, whereas cluster 4 was more enriched in genes involved in the immune response (FDR q-value = 1.83E–3). The fact that gene clusters that are differentially affected by dBRM depletion are linked to different GO terms suggests that the role of SWI/SNF in 3′ end processing is physiologically relevant and specialized.
In summary, we have identified a group of dBRM-bound genes (cluster 1) with impaired 3′ end processing in dBRM-depleted cells, and our GO analysis shows that the differential effects of dBRM depletion on pre-mRNA processing correlate with specific biological functions.
Sequence determinants and chromatin features of genes with dBRM-dependent 3′ end processing
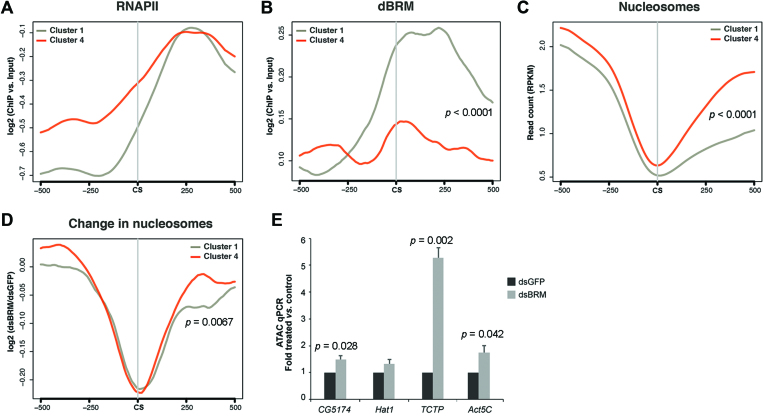
We were interested in understanding why the genes in cluster 1, which show 3′ end processing defects in dBRM-depleted cells, were particularly dependent on SWI/SNF for pre-mRNA 3′ end processing, and we looked for relevant sequence and chromatin features in this group of genes. The genes in cluster 4, which were also positively regulated by dBRM but did not show defects in 3′ end processing in dBRM-depleted cells, were analyzed in parallel as a reference. We analyzed publically available RNAPII ChIP-seq data (43) and observed that the average levels of RNAPII downstream of the CS were similar in the two groups of genes (Figure 5A). In contrast, the levels of dBRM at the CS and in the region immediately downstream of the CS were significantly higher in the genes of cluster 1 than in the genes of cluster 4 (Figure 5B). The average nucleosome density in the gene body was similar in both clusters, as revealed by analysis of MNase-seq data produced by Shi et al. (4), but the genes of cluster 1 were characterized by a lower nucleosome density downstream of the CS (Figure 5C), which suggests that the genes of cluster 1 generally have a more open chromatin conformation in the downstream region. We also used the data from Shi et al. (4) to analyze the effect of dBRM depletion on nucleosome density, and found that the density of nucleosomes around the CS was reduced in the two groups of genes, in agreement with previous observations (4) (Figure 5D). However, the region that showed reduced nucleosome occupancy in dBRM-depleted cells was significantly broader for the genes of cluster 1. In summary, our genome-wide analyses support the conclusion that the genes in cluster 1 are characterized by high dBRM levels downstream of the CS and require dBRM to maintain the structure of the chromatin downstream of the CS, a region that in these genes is characterized by a relatively low nucleosome density. In agreement with this conclusion, ATAC-qPCR analysis of the downstream region of selected genes, including CG5174 and Hat1, confirmed significantly increased chromatin accessibility in dBRM-depleted cells for three out of four analyzed loci (Figure 5E).
Figure 5.
The genes in cluster 1 are characterized by high dBRM occupancy and low nucleosome density downstream of the CS. (A–C) Metagene analyses of RNAPII, dBRM and nucleosome occupancy 0.5 kb around the CSs in the genes of cluster 1 (gray, n = 632 genes) and cluster 4 (red, n = 533 genes). The RNAPII and dBRM ChIP-seq data were from Lam et al. (43) and Jordán-Pla et al. (7), respectively. The nucleosome density data was from Shi et al. (4). (B) The distribution of dBRM in the CS/+500 region differs significantly between the two groups. (C) The average nucleosome density downstream of the CS is significantly lower in cluster 1. (D) Metagene analysis of changed nucleosome occupancy upon dBRM depletion based on data from Shi et al. (4). The reduced nucleosome occupancy downstream of the CS in dBRM-depleted cells is significantly more pronounced for the genes of cluster 1 than for the genes of cluster 4. (E) Chromatin accessibility in the downstream regions of four selected genes was analyzed by ATAC-qPCR. TCTP is a Cluster 1 gene. Statistical testing of differences between metagene distributions was done using the Kolmogorov–Smirnov test and one-sample t-test was used for statistical testing of differences in the ATAC-qPCR experiments. Significant p values are shown in the figure.
The differences in chromatin structure described above prompted us to further characterize the downstream region of the cluster 1 genes, and we made use of the 9-state chromatin classification elaborated by Kharchenko et al. (38). This classification, which is the result of systematic profiling of chromatin signatures based on ChIP-seq data for eighteen histone modifications and non-histone chromatin proteins, divides the chromatin landscape in nine functional domains that are specified in the left side of Figure 6A. Our analysis revealed that the sequences downstream of the genes in cluster 1 were enriched in features of transcribed chromatin (state 2, Elongation) (Figure 6A), which suggested the existence of neighboring genes located downstream of the CS. However, the analysis of annotated genomic features in a region of length 350 bp located immediately downstream of the CS showed that UTRs, exons and introns constituted a low fraction of the downstream sequences (7.7% and 20.9% in the sense and antisense strands, respectively) (Figure 6B). Analysis of larger regions (1.1 and 3.1 kb downstream of the CS) gave similar results and we concluded that neighboring transcription units downstream of the cluster 1 genes are not abundant. In summary, the 3′ flanking regions of the genes in cluster 1 are gene-poor but have an open chromatin conformation that resembles that of transcribed chromatin.
We also looked for sequence elements that are important for 3′ end processing in the genes of clusters 1 and 4. The poly(A) signal AAUAAA was present in the genes of both clusters, but was more frequent in the genes of cluster 1 (66.5% versus 54%, see Table 1). The canonical USE motif UGUA, which is the binding site of the CFIm complex, was also more frequent in cluster 1 than in cluster 4 (40% versus 34%). The higher frequency of potential CFIm binding sites in the genes of cluster 1 suggests that these genes depend particularly strongly on CFIm function.
Table 1.
Frequency of cis-regulatory elements in the 3′ region of the genes of clusters 1 and 4
| Frequency (% genes) | ||||
|---|---|---|---|---|
| Motif | Sequence | Region | Cluster 1 | Cluster 4 |
| Poly(A) signal | AAUAAA | -50 to CS | 66.5 | 54 |
| AUUAAA | -50 to CS | 13 | 15 | |
| Poly(A) signal | AAUAAA | CS to +500 | 43 | 42 |
| AUUAAA | CS to +500 | 38 | 37 | |
| USE | UGUA | -60 to -10 | 40 | 34 |
| DSE | UUUGUU | CS to +50 | 11 | 10 |
| UUUUUU | CS to +50 | 12.5 | 11 | |
The table shows the percentage of genes in each cluster that contains the motif at least once within the region analyzed.
dBRM and hBRG1 interact with components of the 3′ end processing machinery
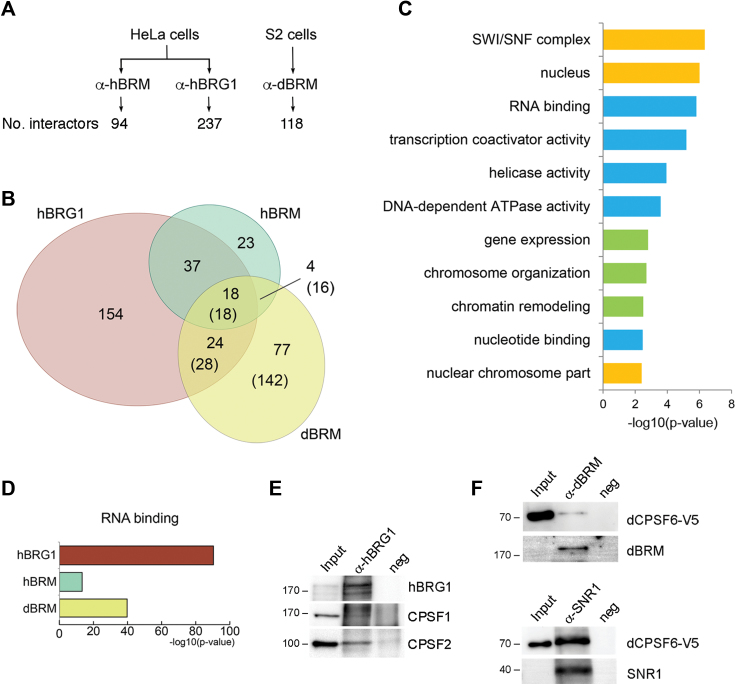
Based on the results presented above and on the reported association of SWI/SNF with nascent pre-mRNPs (11), we hypothesized that the role of dBRM in 3′ pre-mRNA processing relies on physical interactions between SWI/SNF and proteins associated with nascent transcripts. We used immunoprecipitation (IP) and high-throughput mass spectrometry methods to characterize the SWI/SNF RNP interactome in S2 cells, to identify such proteins. The human SWI/SNF RNP interactome was analyzed in parallel for comparative purposes. Chromosomal RNP fractions were prepared from S2 and HeLa cells, and used for IP experiments using antibodies against hBRG1, hBRM or dBRM (Figure 7A). The immunoprecipitated proteins were identified by label-free mass spectrometry and are listed in the Supplementary Tables S5–S7. Negative control immunoprecipitations were processed in parallel (see Materials and Methods) and the proteins detected in the negative control samples are listed in the Supplementary Table S8. As expected, SWI/SNF subunits were among the identified interactors: 12 SWI/SNF subunits were associated with hBRG1 (GO:0016514, P-value = 1.98e–15), 12 with hBRM (GO:0016514, P-value = 1.01e–19) and four with dBRM (GO:0035060, P-value = 2.04e–04). These results strongly suggest that hBRG1, hBRM and dBRM are part of SWI/SNF complexes when associated with RNPs, as initially proposed by Tyagi et al. (11).
Figure 7.
SWI/SNF interacts with pre-mRNA processing factors in fly and human cells. Endogenous dBRM, hBRM and hBRG1 were immunoprecipitated from S2 and HeLa cells, respectively, and the immunoprecipitated proteins were identified by mass spectrometry. (A) The figure shows the number of interactors identified for each of the SWI/SNF ATPases by IP and label-free mass spectrometry. (B) Euler diagram showing the overlaps among the proteins coimmunoprecipitated with each ATPase. The diagram includes only hits with annotated orthologs in D. melanogaster. For D. melanogaster proteins, the number of human orthologues is given in parentheses. (C, D) PANTHER overrepresentation tests using the complete GO database. The p values shown in the figures were calculated using a Fisher's exact test with FDR correction and are expressed –log10(P-value). (C) Selected GO terms enriched in the group of 18 proteins from the intersection between hBRG1, hBRM and dBRM human orthologs. Biological processes, cellular compartments and molecular functions are shown in green, yellow and blue, respectively. (D) Overrepresentation of the GO term ‘RNA binding’ in the interactomes of hBRG1, hBRM and dBRM human orthologs. (E) Western blot analysis of coimmunoprecipitated proteins showing the interaction of human CPSF1 and CPSF2 with hBRG1. (F) Endogenous dBRM or SNR1 were immunoprecipitated from S2 cells that expressed V5-tagged CPSF6. CPSF6 is interacts with dBRM and SNR1. The approximate mobility of molecular mass standards is shown to the left of the blots, in kDa.
To compare the interactors detected in human and fly samples, we searched for the human orthologs of the dBRM interactors, which resulted in a total of 204 proteins. When the three interactomes were compared with each other, the interactome of dBRM was more similar to that of hBRG1 than to that of hBRM (Figure 7B). The GO terms in the three interactomes and in their intersections were significantly enriched, which suggests that some of the interactions and their function are conserved in man and fly. Figure 7C shows 11 selected GO terms that were significantly enriched in the group of 18 proteins that were common to hBRM, hBRG1 and dBRM. Apart from terms related to the function of SWI/SNF in ATP-dependent chromatin remodeling, the term ‘RNA binding’ (GO:0003723) was significantly enriched in each of the individual interactomes: 145 RNA-binding proteins were associated with hBRG1 (P-value = 3.20e–91), 37 associated with hBRM (P-value = 4.23e–14), and 32 with dBRM (P-value = 5.56e–14) (Figure 7D).
Many proteins detected in our proteomics study belong to complexes that are involved in regulation of RNAPII transcription (GO:0006357; 48 hBRG1 interactors, 25 hBRM interactors and 7 dBRM interactors) and mRNA splicing (GO:0000398; 65 hBRG1 interactors, 17 hBRM interactors and 11 dBRM interactors) (Supplementary Tables S5–S7), which suggests that SWI/SNF plays multiple roles in mRNA biogenesis. Interestingly, the SWI/SNF interactomes of S2 and HeLa cells shared cleavage and polyadenylation specificity factors. In particular, hBRG1 interacted with CPSF1 and CPSF2, and dBRM interacted with CPSF1 and CPSF6, one of the components of the CFIm complex and a reported regulator of 3′ end processing (18). Some of these interactions were validated by co-IP and Western blotting (Figure 7E–F).
In summary, our results suggest that SWI/SNF plays a direct role in 3′ end processing through protein-protein interactions with components of the cleavage and polyadenylation machinery.
dBRM promotes the interaction of CPSF6 with the cleavage site
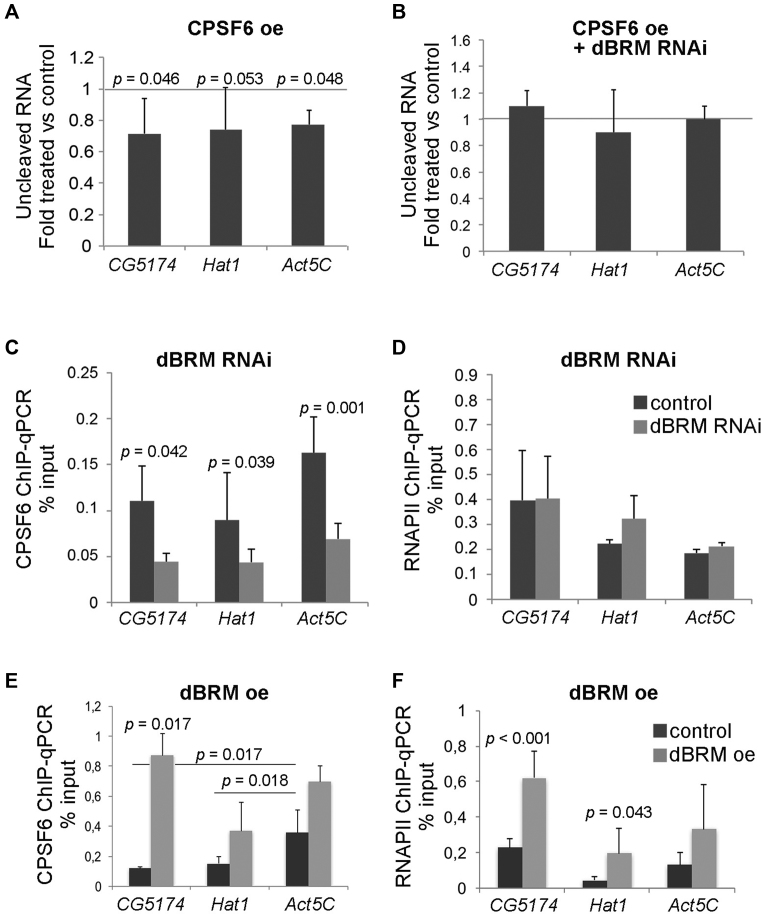
We focused on the CPSF6-dBRM interaction because this interaction was also reported in the protein interaction map of D. melanogaster (48) and because CFIm is a key regulator of 3′ end processing (18). We constructed a V5-tagged version of CPSF6, expressed it in S2 cells (Supplementary Figure S2D), and used the PCR-based cleavage assay to analyze the effect of CPSF6 levels on the cleavage of CG5174 and Hat1 pre-mRNAs. CPSF6 overexpression increased the cleavage efficiency of the three pre-mRNAs analyzed (Figure 8A), which suggests that CPSF6 is a rate-limiting factor in pre-mRNA cleavage. Interestingly, dBRM depletion abolished the effect of CPSF6 overexpression on pre-mRNA cleavage, which shows that CPSF6 facilitates 3′ end cleavage in a dBRM-dependent manner (Figure 8B).
Figure 8.
Cleavage of the CG5174 and Hat1 pre-mRNAs is regulated by CPSF6 in a dBRM-dependent manner. (A) The effect of CPSF6 on 3′ end cleavage was analyzed using the PCR-based cleavage assay described in Figure 1 in cells that overexpressed V5-tagged recombinant CPSF6. The bars in the histogram represent the average ratio between overexpression and control samples. The error bars represent standard deviations from three biological replicates. (B) The effect of CPSF6 on 3′ end cleavage was analyzed in S2 cells depleted of dBRM and in control cells (dsGFP-treated). (C) ChIP-qPCR analysis was carried out in cells that expressed V5-tagged CPSF6 with an antibody against the V5 tag to analyze the occupancy of CPSF6 at the CS of CG5174, Hat1 and Act5C in control cells and in dBRM-depleted cells (dsBRM). (D) ChIP-qPCR analysis was carried out in parallel with an antibody against the CTD of RNAPII. (E and F) Reciprocal ChIP-qPCR experiments to the ones shown in C-D showing the effect of dBRM overexpression in the occupancy of CPSF6 (E) and RNAPII (F) with the CS of target genes. Antibodies against the HA tag were used to immunoprecipitate recombinant CPSF6 (HA-tagged) in S2 cells that also expressed recombinant dBRM (V5-tagged). The ChIP-qPCR signals in C–F are expressed as percentage of input. In all cases, the histograms show average values and the error bars represent standard deviations from three biological replicates. One sample t-test were used for testing statistical significance.
We hypothesized that dBRM is important for the association of CPSF6 with the 3′ UTRs, and we carried out ChIP-qPCR to test this possibility. Due to the lack of antibodies against the endogenous CPSF6 protein, we knocked down dBRM in S2 cells that expressed V5-tagged CPSF6 and we used an anti-V5 antibody for ChIP-qPCR experiments. The ChIP-qPCR signals for CPSF6 near the CS were lower in cells depleted of dBRM than in control cells (Figure 8C). The levels of CPSF6 protein were not affected by the RNAi treatment (Supplementary Figure S2E), nor were the levels of expression of the mRNAs that code for the components of the cleavage and polyadenylation machinery (Supplementary Table S9). The results suggest that indirect effects due to a downregulation of CPSF, CstF or CFIm subunits are not occurring. We also carried out ChIP experiments with an antibody against the largest subunit of RNAPII, and we showed that the levels of RNAPII in the analyzed regions were not affected by the dBRM depletion (Figure 8D). This observation ruled out indirect effects due to decreased RNAPII occupancy and decreased nascent pre-mRNA density. The fact that dBRM depletion resulted in decreased levels of CPSF6 at the 3′ UTRs of the analyzed genes suggests that dBRM is required for the efficient binding of CPSF6 to the CS region.
Next we carried out reciprocal experiments in which we overexpressed dBRM and analyzed the binding of CPSF6 to the CG5174, Hat1 and Act5C 3′ UTRs. We constructed S2 cell lines to express simultaneously recombinant CPSF6 (HA-tagged) and dBRM (V5-tagged) (Supplementary Figure S2F). Elevated dBRM levels increased significantly the recruitment of CPSF6 to the CSs of the CG5174 gene. A reproducible increase was also observed for Hat1 and Act5C, but the changes were below the significance threshold (Figure 8E). Interestingly, in control conditions, the association of CPSF6 to the CS region of the Act5C gene was significantly higher than in CG5174 and Hat1.
RNAPII at the 3′ UTRs of the analyzed genes was also relatively higher in the cells that overexpressed dBRM (Figure 8F), but the magnitude of the RNAPII change was much lower than that observed for CPSF6 (compare Figure 8E and F). We further confirmed the contribution of dBRM to the interaction of CPSF6 with the 3′ region of four selected genes from cluster 1 (Supplementary Figure S5). These results support the conclusion that dBRM promotes 3′ end formation by facilitating the interaction of CPSF6 with the 3′ UTR. We also conclude that not all pre-mRNAs require dBRM for efficient 3′ end processing, as shown by results on the Act5C gene (see Discussion).
CPSF6 interacts with the transcription machinery at an early stage of transcription, and travels together with RNAPII along the gene body (49). We asked whether dBRM affects the binding of CPSF to the gene promoters, and we measured CPSF6 abundance at the TSSs of the CG5174 and Hat1 genes. Overexpression of dBRM had a very small effect on the binding of CPSF6 to the TSSs (Supplementary Figure S6). These results indicate that SWI/SNF promotes 3′ end formation by facilitating the function of CPSF6 in the pre-mRNA 3′ region, without affecting the initial loading of CPSF6 to the transcription machinery.
DISCUSSION
SWI/SNF is a sophisticated and highly complex chromatin remodeler. It has a broad interaction network (5 and our present data), it regulates gene expression through ATP-dependent and ATP-independent mechanisms (7,50), and it controls both gene transcription and pre-mRNA processing (8,11–13). We have shown using RNA-seq that depletion of dBRM in S2 cells causes significant changes in RNA levels upstream of the TSSs and downstream of the CSs. The increase of average RNA levels observed upstream of the TSSs suggests that dBRM might play a global role in regulating alternative promoter usage or transcription start fidelity. The changes observed at the CS have been the focus of the present study and are linked to the role of dBRM in pre-mRNA maturation.
We have shown that the experimental modulation of dBRM levels leads to changes in the 3′ end processing of a subset of pre-mRNAs in S2 cells. Comprehensive RNA-seq studies available at modENCODE show that the levels of dBRM expression are remarkably different in different organs and developmental stages of D. melanogaster (51), which suggests that dBRM also regulates 3′ end processing in vivo. SWI/SNF is a master regulator of animal development (reviewed in 52) and our findings suggest that the regulation of gene expression by dBRM not only affects transcription and alternative splicing, as previously reported, but also pre-mRNA 3′ end cleavage.
The results presented here suggest that the role of SWI/SNF in 3′ end processing is direct. Changes in the levels of dBRM did not affect the levels of transcripts that code for the components of the 3′ end processing machinery, which argues against indirect effects due to changes in the expression of processing factors. The proteomics analysis we carried out showed that SWI/SNF interacts physically with cleavage and polyadenylation factors. We cannot establish whether the interaction between CPSF6 and SWI/SNF is direct or indirect, but the IP experiments show that both factors interact with each other in the chromosomal RNP fraction. In addition, dBRM is associated with its target genes. These observations suggest that SWI/SNF acts locally to regulate the 3′ end cleavage of a subset of pre-mRNAs. Furthermore, the human hBRG1 protein also interacts with CPSF factors and regulates the 3′ end processing of genes to which it is recruited, which suggests that the mammalian SWI/SNF complex plays a direct role in pre-mRNA 3′ end maturation.
In mammalian cells, cellular stress induces defects in pre-mRNA 3′ end processing and results in the synthesis of readthrough ‘downstream-of-gene’ transcripts (DoGs) through a calcium-regulated pathway (25). It remains to be investigated whether DoGs are also produced in Drosophila cells. The downstream transcripts that we have detected in dBRM-depleted S2 cells are only a few hundred nucleotides long and differ in this respect from DoGs, many of which are longer than 100 kb (25).
SWI/SNF is physically linked to the 3′ end processing machinery
The hBRM protein participates in the regulation of the alternative splicing of the human CD44 pre-mRNA through a mechanism that involves the RNA-binding protein Sam68 (8). A fraction of SWI/SNF is associated with RNP fractions of the cell nucleus in both human and insect cells, and immuno-electron microscopy has shown that a fraction of dBRM is associated with nascent pre-mRNP particles in the salivary glands of the dipteran Chironomus tentans (11). However, the only protein known to mediate the interactions between SWI/SNF and the nascent pre-mRNAs was Sam68. Therefore, a systematic analysis of the RNP interactome of the SWI/SNF catalytic subunits was important, and our study has revealed the existence of physical and functional links between SWI/SNF and cleavage and polyadenylation factors in both human and insect cells.
Our co-IP experiments revealed that not only dBRM but also SNR1, another subunit of the SWI/SNF core of D. melanogaster, interacts with CPSF6. This suggests that the SWI/SNF complex is involved in 3′ end processing, not dBRM alone. Interestingly, the interaction between SNR1 and CPSF6 was stronger than that between dBRM and CPSF6, which suggests that CPSF6 interacts with SNR1 directly and with dBRM indirectly via SNR1.
The mechanism of cleavage regulation by SWI/SNF
The impact of alternative polyadenylation on the outcome of gene expression and on the coding capacity of the genome has been extensively documented (reviewed recently in 53), and SWI/SNF probably influences alternative polyadenylation choices. However, many of the 3′ end processing events that are facilitated by dBRM concern genes that lack alternatively polyadenylated isoforms, and our study suggests that dBRM favors the constitutive 3′ end maturation of a subset of pre-mRNAs.
We have identified differential features of genes that rely on dBRM for efficient 3′ end processing. One of these features is the relatively high occurrence of the USE consensus motif, which is the binding site for CFIm. We have also found that the levels of dBRM around the CS are higher than average in this group of genes, and that dBRM and SNR1 interact physically with the CFIm subunit CPSF6. Moreover, our single-gene studies show that CPSF6 favors 3′ end cleavage in a dBRM-dependent manner. Altogether, these observations suggest a mechanism in which dBRM, or more probably a SWI/SNF complex that contains dBRM, interacts with CPSF6 and promotes its function at the CS (Figure 9). The CFIm complex is recruited to transcribed genes during the initial stages of transcription together with CPSF and CstF (49,54), but our ChIP experiments showed that dBRM does not significantly affect the association of CPSF6 with the TSSs. Therefore, we suggest that the CPSF6-SWI/SNF interaction either provides an alternative recruitment pathway for CFIm to the proximity of the downstream region of the gene, or acts by enhancing the binding to the USE of CFIm complexes that were already associated with the elongating RNAPII complex.
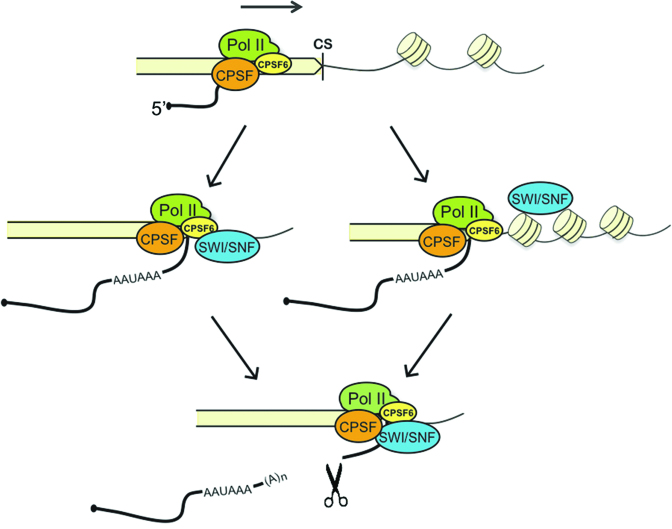
Figure 9.
A model for the role of SWI/SNF in pre-mRNA 3′ end cleavage in Drosophila melanogaster. SWI/SNF is recruited to the 3′ end region of the gene, where it facilitates pre-mRNA processing by acting at two levels. On the one hand, dBRM enhances the association of CPSF6 with the CS. On the other hand, SWI/SNF contributes to the packaging of the chromatin downstream of the CS.
Another differential feature of the genes that rely on dBRM for efficient 3′ end processing is that they are flanked by gene-poor regions that show features of open chromatin. Moreover, the chromatin of this group of genes becomes more open than average (more reduced nucleosome occupancy) in dBRM-depleted cells. This feature is relevant because the chromatin context influences the rate of RNAPII elongation and the efficiency of pre-mRNA 3′ end cleavage (55). Expressed genes are typically characterized by a strong nucleosome depletion around the CSs and by the existence of positioned nucleosomes immediately downstream of the CS (4,53,56). Interestingly, strong polyadenylation sites have better positioned downstream nucleosomes (57), which probably slows down RNAPII elongation downstream of the CS. This, in turn, promotes CS recognition (23). In this context, our results provide evidence for a mechanism in which SWI/SNF facilitates pre-mRNA 3′ end processing by stabilizing nucleosome positioning in the downstream region of genes that are intrinsically flanked by relatively open chromatin domains. These genes, as discussed above, depend more strongly on the function of CFIm for the efficient assembly of the cleavage and polyadenylation complex.
Our study also shows that not all transcripts depend on SWI/SNF for efficient 3′ end formation, as illustrated by the analysis of Act5C. Depletion of dBRM increases chromatin accessibility and reduces the association of CPSF6 with the downstream region of the Act5C gene, but does not affect the cleavage of the Act5C pre-mRNA. Interestingly, in control conditions, the levels of CPSF6 in the downstream region of Act5C are higher than in CG5174 and Hat1, and this difference may explain why the Act5C pre-mRNA cleavage is not dependent on dBRM levels. Based on these observations, it is tempting to speculate that housekeeping genes such as Act5C might be able recruit 3′ end processing factors efficiently through mechanisms that do not require SWI/SNF.
DATA AVAILABILITY
The results of the mass spectrometry experiments are included as supplementary data (Tables S5–S8). The RNA-seq and ChIP-seq data are available at the NCBI Gene Expression Omnibus (GEO) under accession GSE95236. GFP_control and dsBRM MNase-seq datasets from Shi et al. (4) were downloaded from ArrayExpress (E-MTAB-1967). RNAPII ChIP and input files from Lam et al. (43) were downloaded from ArrayExpress (E-MTAB-1084).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank George Farrants for English editing. The authors would like to acknowledge support from the Science for Life Laboratory, the National Genomics Infrastructure, NGI and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure. The authors would also like to acknowledge support from BILS (Bioinformatics Infrastructure for Life Sciences) and from Proteomics Karolinska (PK/KI) for help in the analysis of RNA-seq data and LC–MSMS, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council [2015-04553 to N.V.]; Swedish Cancer Society [CAN 2016/460]; Department of Molecular Biosciences; Wenner-Gren Institute at Stockholm University [doctoral support to S.Y. and A.G., and postdoctoral support to A.J.P and A.R.]. Funding for open-access charge: the Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dingwall A.K., Beek S.J., McCallum C.M., Tamkun J.W., Kalpana G.V., Goff S.P., Scott M.P.. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell. 1995; 6:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mohrmann L., Verrijzer C.P.. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. BBA - Gene Struct. Expr. 2005; 1681:59–73. [DOI] [PubMed] [Google Scholar]
- 3. Bouazoune K., Brehm A.. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006; 14:433–449. [DOI] [PubMed] [Google Scholar]
- 4. Shi J., Zheng M, Ye Y., Li M., Chen X., Hu X., Sun J., Zhang X., Jiang C.. Drosophila Brahma complex remodels nucleosome organizations in multiple aspects. Nucleic Acids Res. 2014; 42:9730–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., Snyder M.. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011; 7:e1002008-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Attanasio C., Nord A.S., Zhu Y., Blow M.J., Biddie S.C., Mendenhall E.M., Dixon J., Wright C., Hosseini R., Akiyama J.A. et al. Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 2014; 24:920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jordán-Pla A., Yu S., Waldholm J., Källman T., Östlund Farrants A.K., Visa N.. SWI/SNF regulates half of its targets without the need of ATP-driven nucleosome remodeling by Brahma. BMC Genomics. 2018; 19:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Batsché E., Yaniv M., Muchardt C.. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:22–29. [DOI] [PubMed] [Google Scholar]
- 9. Kornblihtt A.R. Chromatin, transcript elongation and alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:5–7. [DOI] [PubMed] [Google Scholar]
- 10. Fontana G.A., Rigamonti A., Lenzken S.C., Filosa G., Alvarez R., Calogero R., Bianchi M.E., Barabino S.M.. Oxidative stress controls the choice of alternative last exons via a Brahma-BRCA1-CstF pathway. Nucleic Acids Res. 2017; 45:902–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tyagi A., Ryme J., Brodin D., Östlund Farrants A.K., Visa N.. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009; 5:e1000470-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waldholm J., Wang Z., Brodin D., Tyagi A., Yu S., Theopold U., Farrants A.K., Visa N.. SWI/SNF regulates the alternative processing of a specific subset of pre-mRNAs in Drosophila melanogaster. BMC Mol. Biol. 2011; 12:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zraly C.B., Dingwall A.K.. The chromatin remodeling and mRNA splicing functions of the Brahma (SWI/SNF) complex are mediated by the SNR1/SNF5 regulatory subunit. Nucleic Acids Res. 2012; 40:5975–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Retelska D., Iseli C., Bucher P., Jongeneel C.V., Naef F.. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics. 2006; 7:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Millevoi S., Vagner S.. Molecular mechanisms of eukaryotic pre-mRNA 3′-end processing regulation. Nucleic Acids Res. 2010; 38:2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elkon R., Ugalde A. P., Agami R.. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013; 14:496–506. [DOI] [PubMed] [Google Scholar]
- 17. Dettwiler S., Aringhieri C., Cardinale S., Keller W., Barabino S.M.L.. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 2004; 279:35788–35797. [DOI] [PubMed] [Google Scholar]
- 18. Gruber A.R., Martin G., Keller W., Zavolan M.. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol. 2012; 9:1405–1412. [DOI] [PubMed] [Google Scholar]
- 19. Tian B., Manley J.L.. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013; 38:312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S., Yamamoto J., Chen Y., Aida M., Wada T., Handa H., Yamaguchi Y.. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells. 2010; 15:1003–1013. [DOI] [PubMed] [Google Scholar]
- 21. Martin G., Gruber A.R., Keller W., Zavolan M.. Genome-wide analysis of pre-mRNA 3′-end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports. 2012; 1:753–763. [DOI] [PubMed] [Google Scholar]
- 22. Yao C., Biesinger J., Wan J., Weng L., Xing Y., Xie X., Shi Y.. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:18773–18778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinto P.A., Henriques T., Freitas M.O., Martins T., Domingues R.G., Wyrzykowska P.S., Coelho P.A., Carmo A.M., Sunkel C.E., Proudfoot N.J. et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011; 30:2431–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ji Z., Luo W., Li W., Hoque M., Pan Z., Zhao Y., Tian B.. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol. Syst. Biol. 2011; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vilborg A., Passarelli M.C., Yario T.A., Tycowski K.T., Steitz J.A.. Widespread Inducible Transcription Downstream of Human Genes. Mol. Cell. 2015; 59:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang W., Côté J., Xue Y., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M. et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996; 15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 27. Östlund Farrants A.K., Blomquist P., Kwon H., Wrange O.. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol. 1997; 17:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dingwall A.K., Beek S.J., McCallum C.M., Tamkun J.W., Kalpana G.V., Goff S.P., Scott M.P.. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell. 1995; 6:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lisette Mohrmann L., Langenberg K., Krijgsveld J., Kal A.J., Heck A.J.R., Verrijzer C.P.. Differential targeting of two distinct SWI/SNF-Related Drosophila Chromatin-Remodeling complexes. Mol. Cell. Biol. 2004; 24:3077–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu S., Waldholm J., Böhm S., Visa N.. Brahma regulates a specific trans-splicing event at the mod(mdg4) locus of Drosophila melanogaster. RNA Biol. 2014; 11:134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khavari P.A., Peterson C.L., Tamkun J.W., Mendel D.B., Crabtree G.R.. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993; 366:170–174. [DOI] [PubMed] [Google Scholar]
- 32. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hessle V., Björk P., Sokolowski M., de Valdivia E.G., Silverstein R., Artemenko K., Tyagi A., Maddalo G., Ilag L., Helbig R. et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol. Biol. Cell. 2009; 20:3459–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eberle A.B., Böhm S., Östlund Farrants A.-K., Visa N.. The use of a synthetic DNA-antibody complex as external reference for chromatin immunoprecipitation. Anal. Biochem. 2012; 426:147–152. [DOI] [PubMed] [Google Scholar]
- 38. Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Minoda A., Riddle N.C., Ernst J., Sabo P.J., Larschan E., Gorchakov A.A., Gu T. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011; 471:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang W., Loganantharaj R., Schroeder B., Fargo D., Li L.. PAVIS: a tool for Peak Annotation and Visualization. Bioinformatics. 2013; 29:3097–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Micallef L., Rodgers P.. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One. 2014; 9:e101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D.. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways,and data analysis tool enhancements. Nucleic Acids Res. 2017; 45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J.. ATAC-seq: A method for assaying chromatin accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015; 109:doi:10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lam K.C., Mühlpfordt F., Vaquerizas J.M., Raja S.J., Holz H., Luscombe N.M., Manke T., Akhtar A.. NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012; 8:e1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moshkin Y.M., Mohrmann L., van Ijcken W.F.J., Verrijzer C.P.. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 2007; 27:651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K.. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001; 106:309–318. [DOI] [PubMed] [Google Scholar]
- 46. Reisman D.N., Strobeck M.W., Betz B.L., Sciariotta J., Funkhouser W. Jr, Murchardt C., Yaniv M., Sherman L.S., Knudsen E.S., Weissman B.E.. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002; 21:1196–1207. [DOI] [PubMed] [Google Scholar]
- 47. Peterson C.L., Workman J.L.. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000; 10:187–192. [DOI] [PubMed] [Google Scholar]
- 48. Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O. et al. A protein complex network of Drosophila melanogaster. Cell. 2011; 147:690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Venkataraman K., Brown K.M., Gilmartin GM.. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005; 19:1315–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kwok R.S., Li Y.H., Lei A.J., Edery I., Chiu J.C.. The catalytic and non-catalytic functions of the Brahma chromatin-remodeling protein collaborate to fine-tune circadian transcription in Drosophila. PLoS Genet. 2015; 11:e1005307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O., Booth B.W., Wen J., Park S., Suzuki A.M. et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014; 512:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yaniv M. Chromatin remodeling: from transcription to cancer. Cancer Genet. 2014; 207:352–357. [DOI] [PubMed] [Google Scholar]
- 53. Tian B., Manley J.L.. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017; 18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L.. The carboxy-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997; 385:357–361. [DOI] [PubMed] [Google Scholar]
- 55. Spies N., Nielsen C.B., Padgett R.A., Burge C.B.. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009; 36:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang C., Pugh B.F.. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009; 10:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang H., Chen J., Liu H., Sun X.. The nucleosome regulates the usage of polyadenylation sites in the human genome. BMC Genomics. 2013; 14:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results of the mass spectrometry experiments are included as supplementary data (Tables S5–S8). The RNA-seq and ChIP-seq data are available at the NCBI Gene Expression Omnibus (GEO) under accession GSE95236. GFP_control and dsBRM MNase-seq datasets from Shi et al. (4) were downloaded from ArrayExpress (E-MTAB-1967). RNAPII ChIP and input files from Lam et al. (43) were downloaded from ArrayExpress (E-MTAB-1084).