Abstract
CRISPR/Cas9 is an attractive platform to potentially correct dominant genetic diseases by gene editing with unprecedented precision. In the current proof-of-principle study, we explored the use of CRISPR/Cas9 for gene-editing in myotonic dystrophy type-1 (DM1), an autosomal-dominant muscle disorder, by excising the CTG-repeat expansion in the 3′-untranslated-region (UTR) of the human myotonic dystrophy protein kinase (DMPK) gene in DM1 patient-specific induced pluripotent stem cells (DM1-iPSC), DM1-iPSC-derived myogenic cells and DM1 patient-specific myoblasts. To eliminate the pathogenic gain-of-function mutant DMPK transcript, we designed a dual guide RNA based strategy that excises the CTG-repeat expansion with high efficiency, as confirmed by Southern blot and single molecule real-time (SMRT) sequencing. Correction efficiencies up to 90% could be attained in DM1-iPSC as confirmed at the clonal level, following ribonucleoprotein (RNP) transfection of CRISPR/Cas9 components without the need for selective enrichment. Expanded CTG repeat excision resulted in the disappearance of ribonuclear foci, a quintessential cellular phenotype of DM1, in the corrected DM1-iPSC, DM1-iPSC-derived myogenic cells and DM1 myoblasts. Consequently, the normal intracellular localization of the muscleblind-like splicing regulator 1 (MBNL1) was restored, resulting in the normalization of splicing pattern of SERCA1. This study validates the use of CRISPR/Cas9 for gene editing of repeat expansions.
INTRODUCTION
Myotonic dystrophy type 1 (DM1, also known as Steinert's disease) is an autosomal dominant disease and one of the most common dominant neuromuscular disorders in adults with a prevalence ranging from 0.5 to 18.1 per 100,000 individuals (1,2). Typically, DM1 patients suffer from progressive myopathy and myotonia, cardiac conduction defects and cognitive impairments (1). DM1 is caused by the presence of expanded trinucleotide CTG repeats in the 3′ untranslated region (UTR) of DMPK (myotonic dystrophy protein kinase) gene on chromosome 19q13.3 (3,4). Disease severity correlates with the number of CTG repeats, varying between 50 to >2000 repeats in severely afflicted DM1 patients. In contrast, healthy individuals have a limited number of repeats in the DMPK 3′ UTR (i.e. 5–37 CTG repeats). The repeat length increases during the patient's lifetime with an intra- or inter-tissue variability (5). In addition, the repeat size also increases with successive generations ultimately giving rise to increasingly more severe disease phenotypes, a phenomenon defined as anticipation (6). The corresponding DMPK RNA transcripts contain expanded CUG repeats (designated as CUGexp-RNA) that contribute directly to the DM1 pathogenesis (7). This occurs primarily through nuclear sequestration of the so-called muscle blind like (MBNL) family of splicing factors by the pathogenic CUGexp RNA. Consequently, this results in the accumulation of ribonuclear foci (NF) in the nuclei of patient's cells, one of the quintessential characteristics of DM1 (8,9).
Since DM1 patients suffer from skeletal muscle dysfunction, it is widely considered as the prime target tissue to assess novel targeted therapies. In the current study, we therefore investigated the potential of the clustered, regularly interspaced, short palindromic repeats (CRISPR)/Cas9 system to eliminate the pathogenic CTG repeats expansion from the DMPK gene in DM1 patient-specific myogenic cells (Figure 1A). The CRISPR/Cas9 system was initially discovered as a naturally occurring microbial defense system that recognizes and cleaves foreign DNA in a sequence-specific manner (10–12). Since then, it has been adapted successfully as a versatile RNA-guided gene-editing tool for mammalian cells (13–15). CRISPR/Cas9-based gene editing has been shown to enable correction of both recessive and autosomal dominant disorders (16–25). Typically, gene editing using CRISPR/Cas9 can be achieved by co-expression of the CRISPR-associated (Cas9) gene with a chimeric single guide RNA (gRNA) composed of the CRISPR RNA (crRNA) and the trans-activating crRNA (tracrRNA).
Figure 1.
Generation of DM1-iPS cells (DM1-iPSCs) and DM1-iPSC derived inducible myogenic cells (DM1-iPSC-Myo). (A) Schematic overview showing CRISPR/Cas9 based correction of DM1 patient iPSCs derived myogenic cells (DM1-iPSC-Myo). (B) Representative image of DM1-iPSC clones and healthy control iPSCs stained for RNA foci by fluorescent in situ hybridization (FISH). An antisense Cy3-labeled probe was used against trinucleotide CUG expanded repeat. Arrowheads indicated ribonuclear foci. Upper panel represents stained nuclei at lower magnification (scale bar = 20μm) and lower panel represents higher magnification of selected region (scale bar = 2μm). Nuclei were counter-stained with DAPI. (C) Southern blot analysis to detect the length of trinucleotide CTG repeats in five DM1-iPSC clones from two DM1 patients (L22, L81 and L23; FL8 and FL5) and healthy control iPSCs. EcoRI digested genomic DNA was subjected to agarose gel electrophoresis and probed to detect the DMPK locus. (mut = mutant; wt = wild type). (D) Representative image of FISH staining on DM1-iPSC-Myo for detection of ribonuclear foci. Arrowheads indicate multiple RNA foci in nuclei of DM1-iPSC-Myo. Healthy iPSC-Myo were used as a negative control. Upper panel represents stained nuclei at lower magnification (scale bar = 20 μm) and lower panel represents higher magnification of selected region (scale bar = 2 μm). Nuclei were counter-stained with DAPI. (E) Myogenic conversion of DM1-iPSC-Myo (L81 and L23) and healthy iPSC-Myo post MyoD induction were stained for a mature muscle marker, myosin heavy chain (MyHC) (scale bar = 100 μm). Nuclei were counter-stained with DAPI. (F) Southern blot analysis of trinucleotide CTG repeats length in DM1-iPSC-Myo (L81 and L23; FL8 and FL5) and healthy-iPSC-Myo to check the length of triplet repeats post-differentiation (mut = mutant; wt = wild type).
In the current study, we have proven the ability of CRISPR/Cas9 to selectively excise trinucleotide repeat expansions and phenotypically correct DM1 patient-specific induced pluripotent stem cells (DM1-iPSC) and non-transformed DM1 patient-specific cells capable of undergoing myogenic differentiation, consistent with the robust correction of the cellular phenotype. In particular, the CRISPR/Cas9-based correction was validated in DM1 patient-specific myoblasts and myogenic cells derived from DM1-iPSCs (26,27). These DM1-iPSCs were subsequently induced to undergo lineage-specific differentiation to obtain DM1 patient-specific myogenic cells, myocytes and myotubes. Primary myoblasts and iPSC-derived myogenic cells have a distinctive capacity towards myogenic differentiation and are attractive target cells to validate the potential of CRISPR/Cas9 for gene editing of DM1. Myoblasts are derived from satellite cells that are muscle stem/progenitor cells capable of generating multinucleated muscle fibres through cell fusion. Whereas, the iPSC-derived myogenic cells (iPSC-Myo) used in the present study resemble primary mesoangioblasts (Mabs) with respect to their ability to form mature myocytes and myotubes, and based on their surface markers and transcriptome expression patterns (26,27). Mesoangioblasts are pericyte-derived cells and regarded as non-canonical precursors of myogenesis. Such non-canonical myogenic precursors typically do not express PAX7, MYF5 or MYOD during their proliferation stage until they are terminally differentiated (28–31). Mesoangioblasts and iPSC-Myo can extravasate from the circulation allowing repair of the afflicted degenerating muscle tissue. Since myoblasts and iPSC-Myo cells are non-transformed and non-tumorigenic as opposed to immortalized cell lines, they eventually undergo cellular senescence, consistent with the Hayflick limit that is a characteristic of bona fide primary cells. Though it is typically more challenging to achieve efficient gene editing in non-transformed cells, we observed robust reduction of ribonuclear foci in the DM1 myoblasts and DM1-iPSC-derived myogenic cells with up to 40–50% efficiency after CRISPR/Cas9 based gene correction. Consequently, the normal intracellular localization of the muscleblind-like splicing regulator 1 (MBNL1) was restored resulting, in turn, in the normalization of the splicing pattern of the sarco/endoplasmic reticulum Ca2+-ATPase 1 (SERCA1), that is associated with DM1 pathology. Our study also showed that by using ribonuclear protein (RNP) transfections in DM1-iPSC it is possible to achieve 90% correction efficiencies without the need for any selective enrichment step.
This proof-of-concept study validates the use of CRISPR/Cas9 to genetically correct nucleotide repeat expansions associated with dominant genetic disorders like DM1, that cause severe human pathologies. Moreover, in the current study, we also validated the use of single molecule real-time (SMRT) sequencing as a comprehensive and sensitive assay to characterize the efficiency of trinucleotide repeat editing by CRISPR/Cas9. The increased sensitivity of SMRT sequencing overcomes the limitations of some conventional strategies. This has implications to characterize the outcome of gene editing for other nucleotide repeat disorders.
MATERIALS AND METHODS
Generation of DM1 patient- specific induced pluripotent stem cells (DM1-iPSCs)
Myoblasts and fibroblasts of DM1 patients were obtained from a 46-year and 11-year-old female DM1 donors respectively, with their informed consent. The patients displayed clinical traits of ptosis, atrophy, muscle weakness, neck flexors, myotonia, cataract, electrocardiogram conduction abnormalities and daytime somnolence. Fibroblasts from a healthy donor were used as controls. The fibroblasts were cultured in Dulbecco's modified Eagle's medium with high glucose (DMEM-HG, Thermo Scientific) containing 10% inactivated fetal bovine serum (FBS, Thermo Scientific), 100 IU/ml penicillin and 0.1 mg/ml streptomycin (Pen-Strep, Thermo Scientific), 1 mM sodium pyruvate (Thermo Scientific), 1% (v/v) MEM non-essential amino acids (Thermo Scientific) and 1 mM l-glutamine (Thermo Scientific). The myoblast cells were cultured in ready to use skeletal muscle cell growth medium with supplements (Cat # C-23060, Promo-cell). Non-self inactivating retroviral vectors expressing OCT4, SOX2, KLF4 and cMYC with MoMuLV promoter were used to transduce 1 × 105 cells per well of a six-well dish (32). At 16 h post transduction, media containing the retroviral particles were removed and replaced with fresh media, followed by another media change at 48 h. At day 4, the transduced cells were passaged and plated on a 0.1% gelatin-coated plate. On day 5, the culture media was changed to hES medium containing ‘knockout’ Dulbecco's modified Eagle's medium (KO DMEM, Thermo Scientific), 20% ‘knockout’ serum replacement (KOSR, Thermo Scientific), 1% (v/v) MEM-Non Essential Amino Acids (MEM NEAA, Thermo Scientific), 2 mM l-glutamine (Thermo Scientific), 50 μM β-mercaptoethanol, 100 IU/ml penicillin and 0.1 mg/ml streptomycin (Pen-Strep, Thermo Scientific) and 0.5 mM valproic acid (VPA, Sigma Aldrich), with subsequent media change every alternate day up to 15 days. Well-grown distinct colonies were then individually picked by mechanical passaging and transferred onto murine feeder cells (Globalstem; GSC-6001) inactivated with mitomycin C (10 μg/ml, Santa Cruz Biotechnology) for further expansion and were referred to as iPSC clones. At this stage, they were at passage 0 and maintained in culture till passage 13–14, before these iPSC clones were characterized further for pluripotency markers and teratoma formation (as described below). These iPSC were subsequently cultured on feeder-free Geltrex™ matrix (Thermo Scientific) and in Essential 8™medium (Thermo Scientific). Dulbecco's phosphate bufferred saline (DPBS) containing 50 mM EDTA (ethylene-diamine-tetra-acetic acid, Thermo Scientific) was used to detach the iPSC, which were then passaged at a split ratio between 1:4 to 1:6.
Immunocytochemistry
The cells were washed with phosphate bufferred saline (PBS) (Sigma Aldrich) and fixed with 4% paraformaldehyde (PFA; Sigma Aldrich) at room temperature (RT) for 10 min. Post-fixation cells were permeabilized with 0.2% Triton X-100 (Sigma Aldrich) and 1% bovine serum albumin (BSA, Sigma Aldrich) in PBS for 15 min at room temperature (RT). Donkey serum (10%) or goat serum (10%) (Sigma Aldrich) was used to reduce potential background to the secondary antibody. Cells were incubated overnight at 4°C with the following primary antibodies: mouse anti-myosin heavy chain (MyHC; MF20, Developmental Studies Hybridoma Bank, USA; 1:2 dilution), mouse anti-lamin A/C (Novocastra; NCL-LAM-A/C; 1:250 dilution), rabbit anti-Sox2 (Abcam, USA; ab97959; 1:200 dilution), mouse monoclonal anti-Oct3/4 (Santa Cruz Biotechnology, USA; SC5279; 1:100 dilution), mouse monoclonal anti-spCas9 (Abcam, USA; ab210571; 1:200 dilution). After primary antibody incubation, samples were washed with 0.2% Triton X100 (Sigma Aldrich) containing 1% BSA (Sigma Aldrich) in PBS (Sigma Aldrich) and then incubated with the corresponding 488, 594 and/or 647 nm fluorochrome conjugated IgGs (Molecular Probes; 1:500 dilution) together with DAPI (Molecular Probes; 1:1000 dilution) for 1 h at RT in 0.2% Triton X100 in PBS. After three final washes, dishes or slides were mounted using fluorescent mounting medium (Dako) and observed under fluorescent microscope (Leica, Olympus). Images were analyzed using ImageJ (NIH) (https://imagej.nih.gov/ij/). To assess expression of alkaline phosphatase, the cells were washed with PBS and fixed in 4% PFA (Sigma Aldrich) at RT for 10 min. The cells were then stained with Sigma-Fast BCIP/NBT staining kit (Sigma Aldrich; B5655–5TAB), as per manufacturer's protocol.
Teratoma formation assay
All mouse experiments were carried out according to protocols approved by the university animal research ethical committee of the Vrije Universiteit Brussel (VUB). For teratoma formation, sub-confluent human iPSC were harvested and re-suspended in a 2:1 mixture of Essential 8™ medium (Life Technologies) and Matrigel (BD Biosciences) with 10 μM Rho-associated, coiled-coil containing protein kinase (ROCK) inhibitor (Y-27632, Stem cell Technologies). iPSC (106) were injected subcutaneously between the shoulder blades of CB17 SCID mice (Taconic, Denmark). The resulting tumors were harvested from the injected mice 8 to 9 weeks after transplantation. Samples were fixed in 10% neutral buffered formalin, embedded in paraffin and sections were prepared for staining with hematoxylin and eosin.
Array comparative genomic hybridization (aCGH)
Full human genome coverage aCGH was performed using a 180K Cytosure ISCA v2 array (Oxford Gene Technology, UK). It is based on 180K DNA oligonucleotides spread over the genome with a minimal resolution of 100 kb and with a higher resolution at the loci for recognized micro-deletions, micro-duplication syndromes, specific genes and the subtelomeric regions. Genomic DNA from healthy donor iPSC and DM1-iPSC was extracted using Qiagen Blood &Tissue Kit (Qiagen, Germany) as per manufacturer's protocol. iPSC samples and commercially available reference sample (EA-100F; Kreatech; the Netherlands) were differentially labeled by random prime labeling system using Cy3- and Cy5-labeled deoxycytidine triphosphates (dCTPs) (Amersham Biosciences). Post-labeling and hybridization, slides were scanned using a high resolution microarray scanner (Agilent). aCGH analysis was performed using CytoSure Interpret Software (Oxford Gene Technology, UK).
Myogenic differentiation of iPSC
The differentiation protocol of DM1-derived or healthy iPSC to muscle cells was based on our previously described procedure (26,27). According to the four-stage differentiation protocol (Supplementary Figure S4B), the iPSCs were induced to differentiate into mesoangioblast-like cells, which were designated as iPSC-Myo or DM1-iPSC-Myo for the healthy and DM1-derived cells, respectively (iPSC-Myo are equivalent to so-called HIDEMs or Human Induced pluripotent stem cell-DErived Mesoangioblast-like cells) (26). Each stage of the 4-stage protocol lasted 1 week and required hypoxic culture conditions (3% O2). Firstly, the healthy donor iPSCs or DM1-iPSCs were dissociated into single cell suspension with an dissociation medium containing 0.5 mM EDTA (Thermo Scientific), 0.1 mM β-mercaptoethanol (Thermo Scientific) and 1% FBS in DPBS without CaCl2 and MgCl2 (Thermo Scientific) and replated on Matrigel matrix (BD Biosciences) at a density of 3 × 105 cells/cm2 in α-MEM nucleosides basal media (Thermo Scientific; Cat. No. 22571–020) containing 10% FBS (Thermo Scientific), 100 IU/ml penicillin and 0.1 mg/ml streptomycin (Pen-Strep, Thermo Scientific), 2 mM l-glutamine and 0.1 mM β-mercaptoethanol for 1 week at 37°C, 5% CO2 and 2 to 3% O2. After one week of culture, the cells were again dissociated as in step 1 with gentle scraping, if required. The cells were then replated on Matrigel-coated surface at a density of 2.5 × 104 cells /cm2 in the same media as in stage 1. In the third stage, the cells were trypsinized and replated on Matrigel at higher cell density (∼2 × 104 cells/cm2 to obtain 80% confluency) with iPSC-Myo medium composed of MegaCell medium (Sigma Aldrich), 100 IU/ml penicillin and 0.1 mg/ml streptomycin (Pen-Strep, Thermo Scientific), 5% FBS (Thermo Scientific), 2 mM l-glutamine (Thermo Scientific), 1% (v/v) MEM-Non Essential Amino Acid, 5 ng/ml basic fibroblast growth factor (bFGF, Thermo Scientific) and 0.05 mM β-mercaptoethanol (Thermo Scientific). In the fourth stage, cells were trypsinized and plated on uncoated surface (without Matrigel) in iPSC-Myo medium and passaged when confluent. From this stage, the iPSC-Myo cells obtained were maintained like mesangioblast (Mabs) cells. After the fourth stage, the cells were characterized for membrane marker expression by flow cytometric analysis (as described below). The final myogenic differentiation step involved transducing the iPSC-Myo or DM1-iPSC-Myo with a tamoxifen-inducible lentiviral vector expressing MyoD (with multiplicity of infection, MOI = 20), as described in Kimura et al. (33). Incubating the transduced cells with 1 mM tamoxifen (Sigma Aldrich) allowed for the inducible, synchronous and efficient differentiation of the iPSC-Myo and DM1-iPSC-Myo into mature, multinucleated skeletal myotubes.
Flow cytometry
The cells were harvested as mentioned above and resuspended in PBS containing 1% FBS along with 0.5 mM EDTA. The cell suspension was incubated with specific antibodies for 1 h at 4°C. The following monoclonal antibodies were used: FITC (fluorescein isothiocyanate)-conjugated anti-CD 13 (ID Labs, # IDAC1071), FITC-conjugated anti-CD31 (Immunostep, # 31F-100T), FITC-conjugated anti-CD44 (BD Biosciences, # 560977), FITC-conjugated anti-CD 49b (BD Biosciences, # 555498), PE (phycoerythrin)-conjugated anti-CD 45 (BD Biosciences, # 555483) and PE-conjugated anti-CD146 (Biocytex, # 5050-PE100T). Thereafter, the cells were washed with PBS and fixed in 2% PFA. Analysis was performed on at least 10,000 events per sample using a FACS caliber flow cytometer (BD Biosciences). The acquisition was performed using CELLQUEST software (BD Biosciences) and analyzed using FCS-express software (DeNovo).
Southern blot analysis
Genomic DNA of healthy iPSCs, healthy iPSC-Myo, DM1-iPSCs, DM1-iPSC clones and DM1-iPSC-Myo was extracted using Qiagen DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's protocol. After digestion of 10 μg DNA with EcoRI (Roche Diagnostics) and agarose gel electrophoresis, the DNA was subjected to Southern blot analysis using a DIG-labelled probe that was generated with the PCR DIG Probe Synthesis Kit (Roche Diagnostics). This probe was generated with forward primer 5′-TTATTGTCTGTCCCCACCTA-3′ and reverse primer 5′-GGGATCACAGACCATTTCTTTCT-3′ specific to 3′UTR region of human DMPK gene (Supplementary Figure S4A).
Fluorescent in situ hybridization (FISH)
Healthy iPSCs or DM1-iPSCs were pre-treated with 10 μM Y-27632 rock inhibitor (RI, Stem cell Technologies) for 1 h prior to their dissociation with TrypLE Express (Life Technologies). These cells were then plated on Geltrex (Life Technologies)-coated chambered slides (LabTek) in Essential 8™ Medium (Life Technologies) containing Y-27632. After overnight incubation, fresh Essential 8™ media was provided without Y-27632. In the case of iPSC-Myo, DM1-iPSC-Myo and DM1 primary myoblasts 0.05% trypsin-EDTA (Thermo Scientific) was used to dissociate the cells and plate them on 0.1% gelatin (Millipore)-coated chambered slides (Thermo Scientific). On the day of staining, the media was removed and cells were washed thrice with PBS and fixed with 4% PFA (Sigma Aldrich) at RT for 15 min. Thereafter, the cells were washed twice for 5 min each using 70% ethanol. After ethanol wash, the cells were rehydrated with two 10 min washes with 0.5 mM MgCl2 in PBS. After the rehydration step, the cells were hybridized in a hybridization chamber with custom Peptide Nucleic Acid (PNA)-probe (CY3–5′-CAGCAGCAGCAGCAG-3′) (Eurogentec) at a dilution of 1:500 in hybridization buffer [0.1% BSA (Sigma Aldrich) in water, 50% volume of formamide (Thermo Scientific) and 25% volume of SSC Buffer (Life Technologies)] for 90 min at 37°C. In the next step, the cells were washed with 0.1% tween 20 (Sigma Aldrich) in PBS for 5 min at RT. Thereafter the cells were incubated with 0.1% Tween 20 in PBS at 45°C for 30 min. The incubation was followed by three washes with PBS and DAPI staining (Thermo Scientific) (1:500 dilution) for 7 min. After DAPI staining, the slides were mounted using Prolong Anti-fade mounting media (Thermo Scientific) and observed under an inverted fluorescent microscope (Olympus). It is well established that genomic DNA denaturation is critically important to enable efficient hybridization of the PNA FISH probe to the DNA target (34–36). Hence, in the current RNA FISH specific protocol we avoided any DNA denaturation step following previously published reports, rendering any potential DNA target inaccessible to the PNA FISH probe (34–37).
Combined FISH and immunofluorescence-specific for the muscleblind protein 1 (MBNL1) was subsequently performed according to Holt et al. (38). A monoclonal antibody specific for MBNL1 (# MB1a; CIND) (38) was used as primary antibody at a dilution of 1:4 followed by Alexa Fluor 647-conjugated donkey anti-mouse (Life Technologies) secondary antibody. After immunofluorescence, FISH was performed as described above. Microscopy images were captured using Nikon Eclipse 80i, Olympus-IX81 microscope and Z stack acquisition was performed by Olympus-IX81 microscope. Analysis of microscopy images was performed by ImageJ software (NIH, https://imagej.nih.gov/ij/).
CRISPR/Cas9 engineering of DM1-iPSC-Myo cells
The CMV-hspCas9-EF1-GFP lentiviral vector plasmid was purchased from System Biosciences (Catalog # CASLV105PA-1) and expressed the human codon-usage optimized S. pyogenes Cas9 from the human cytomegalovirus (CMV) promoter (Supplementary Figure S4C). It also expressed the green fluorescent protein (GFP) as reporter from the elongation factor 1α promoter (EF1α). The DNA encoding the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA (Supplementary Table S1) were synthesized as oligonucleotides, annealed and cloned into MLM3636 (Addgene plasmid ID # 43860). The choice of gRNAs was based on the presence of PAM consensus sequences that were in close proximity to the expanded CTG repeat. Moreover, gRNA were selected based on computational strategies to also minimize the risk of off-target effects, as described in Fu et al. (39,40). The gRNA expression cassette composed of the pol III U6 promoter-gRNA-scaffold, which was then PCR-amplified with BsiWI and SpeI flanking restriction enzyme sites and cloned into a lentiviral vector backbone (41) along with Blue fluorescent protein (BFP; Evrogen; FP172) a reporter expressed under cytomegalovirus (CMV) promoter (Supplementary Figures S4C and S5A). The scrambled gRNA cassette was amplified from commercially available plasmid (Origine, GE100021) and cloned into the lentiviral vector backbone as described above.
Lentiviral transduction
Lentiviral transduction of the CMV-hSpCas9-EF1-GFP, 5′& 3′-CTGrepeat-gRNA-CMV-BFP and scrambled gRNA-CMV-BFP was carried out with MOI = 25 per construct. The lentiviral particles were produced as previously described (42). A detailed protocol has been mentioned in supplementary methods. Transduction cocktail was prepared using iPSC-Myo medium (see above) with polybrene (8 μg/ml; Santa Cruz; sc134220) and the respective concentrated viral vector amount. The cells were incubated for 16 h with 7 ml of the vector-containing medium. Subsequently, the medium was replaced with fresh iPSC-Myo medium.
Nucleofection and clonal isolation of DM1-iPSCs
The nucleofection of DM1-iPSCs were carried out using Lonza P3 primary cell 4D nucleofection kit according to manufacturer's protocol (Lonza). Prior to nucleofection, the DM1 –iPSCs were maintained in feeder free culture condition as described previously. For the nucleofection reaction mix, a ribonucleoprotein (RNP) complex was prepared by mixing 60 pmol Cas9 protein (Integrated DNA Technologies) and 150 pmol of each sgRNA (5′-CTGrepeat-sgRNA, 3′-CTGrepeat-sgRNA) (Synthego) followed by a 10 min incubation at room temperature (control conditions comprised the following: 60 pmol Cas9 and 300 pmol scrambled gRNA; 150 pmol 5′-CTGrepeat-sgRNA, 150 pmol 3′-CTGrepeat-sgRNA and no Cas9) mixes. Thereafter, DM1–iPSCs were dissociated into single cells with TrypLE express (Thermo Scientific). Post dissociation, 5 × 105 DM1– iPSCs were resuspended in 20 μl P3 nucleofection buffer and mixed with previously prepared RNP complexes and nucleofected using the ‘CA137’ program (using the Nucleofector 4D). Post-nucleofection, the cells were plated on geltrex-coated surface, in Essential 8™ medium (Thermo Scientific) with 10μM rock inhibitor (Y-27632, Stem cell Technologies). A day post-nucleofection, cells were supplemented with fresh Essential 8™ medium (Thermo Scientific) and cultured for further downstream experiments. DM1–iPSCs treated with Cas9 protein and sgRNA (5′-CTGrepeat-sgRNA, 3′-CTGrepeat-sgRNA) were cloned by limiting dilution. Dissociated DM1–iPSCs were plated as single cells on vitronectin (Stemcell Technologies) coated 96-well plate with Clone R supplement and mTeSR culturing medium (Stemcell Technologies). After 10–15 days of culturing with regular media change, single clones were established. These DM1-iPSC clones obtained were transferred to larger surface area and maintained as iPSCs clonal lines until further analysis.
qRT-PCR and RT-PCR analysis
RNA was extracted using Qiagen RNeasy Mini kit (Qiagen) according to manufacturer's instruction. The extracted RNA was used for cDNA synthesis using Superscript III-RT cDNA synthesis Kit (Life technologies) according to manufacturer's protocol. 1μg RNA was taken as starting amount for the cDNA synthesis. Upon cDNA synthesis, it was stored at −20°C until used for quantitative reverse transcription (qRT)-PCR. qRT-PCR analysis for DMPK expression was performed using SYBR green (Thermo Scientific) with specific primers for DMPK (5′- GGATGAAACAGCTGAAGTGGC -3′ and 5′- TGCCTCTAGGTCCCGGTTC -3′) and hGAPDH housekeeping gene (5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′) as control for normalization. For the analysis of the alternative-splicing patterns of SERCA1 mRNA, 1 μg of extracted mRNA was reverse transcribed to cDNA using Superscript III-RT cDNA synthesis Kit (Thermo Scientific). The synthesized cDNA was diluted 1:5 with nuclease-free H2O and used for RT-PCR. PCR mix consisted of 2 μl of diluted cDNA, 0.2 μl Phusion Hot Start II DNA Polymerase (Thermo Scientific), 0.4 μl 10 mM dNTP Mix (Thermo Scientific), 4 μl 5x Phusion GC Buffer (Thermo Scientific), 2 μl each of 5 μM SERCA1-specific forward primer 5′-GCTCATGGTCCTCAAGATCTCAC-3′ and reverse primer 5′-AGCTCTGCCTGAAGATGTGTCAC-3′. PCR amplification was performed by an initial denaturation for 2 min at 98°C; followed by 25–35 cycles consisting of 10 s at 98°C, 30 s at an annealing temperature of 59°C and 30 s at 72°C which was then concluded by a final extension of 2 min at 72°C. PCR reactions were performed using an s1000 Thermal Cycler (Bio-Rad). PCR amplicons were electrophoretically resolved on 3% agarose gels containing EtBr and visualized by an Alpha Imager HP (Cell Biosciences). The band-density was quantified using ImageJ software analysis tool (NIH, https://imagej.nih.gov/ij/) as described in previously published reports (43–45). A detailed analysis procedure is mentioned in Supplementary Methods section.
Triplet repeat Primed PCR (TP-PCR)
Triplet-repeat primed PCR was performed as described in Philippa et al. (46). Each 25 μl PCR comprised FastStart Reaction Buffer without MgCl2 (Roche), 2 mM MgCl2 (Roche), GC-rich solution (Roche), 10 mM dNTP mix (Roche), 20 μM forward primer (FAM) 5′-AACGGGGCTCGAAGGGTCCTTGTAGC-3′ and reverse primer 5′-GGCGGTGGCGGCTGTTGCCAGCAGCAGCAGCAG-3′, 1U FastStart Taq DNA Polymerase (Roche) and 50 ng of genomic DNA. The PCR amplification conditions comprised of an initial denaturation of 94∘C for 5 min followed by 35 cycles of denaturation at 94∘C for 45 s, annealing at 70∘C for 30 s with extension at 72∘C for 30 s and a final extension at 72∘C for 10 min. PCR amplicons were subjected to fragment length analysis by capillary electrophoresis on an Applied Biosystems DNA-Analyzer (ABI 3730) with ROX as internal size marker. The data was analyzed using Peak-Scanner software (Thermo Scientific).
Single-molecule real-time (SMRT) sequencing of the target locus
Genomic DNA of CRISPR/Cas9-corrected DM1-iPSC-Myo, untreated DM1-iPSC-Myo, non-corrected DM1-iPSC-Myo control conditions (i.e. Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9) and wild-type iPSC-Myo was extracted using Qiagen DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's protocol. Genomic DNA (20–40 ng) was used for PCR. The target locus was amplified using the LongAmp Taq PCR kit (New England Biosciences) based on the 5′-ATCTTCGGGCAGCCAATCAAC-3′ (DM-PAC, forward primer) and 5′-CGTGGAGGATGGAACACGGAC -3′ (DM-BR, reverse primer) that match a target sequence 571 basepair (bp) upstream and 62 bp downstream of the respective CRISPR/Cas9 PAM target sites flanking the DMPK CTG repeat (Figure S6A) PCR amplification was performed by an initial denaturation for 2 min at 94°C, followed by 30 cycles consisting of 30 s at 94°C, 30 s at the 60°C, prolonged 8 min extension at 65°C and concluded by a final extension of 10 min at 65°C. PCR reactions were performed using an s1000 Thermal Cycler (Bio-Rad). Thereafter, library preparation of the amplicons was carried out using PacBio Barcoded Adapters for Multiplex SMRT Sequencing using SMRT bell Barcoded Adapter Complete Prep Kit (Pacific Biosciences, Menlo Park, CA, USA). During the library preparation each amplicon was labeled with a different barcode, which was introduced via the PacBio adapter. Each library was subsequently sequenced on a PacBio RS II sequencer using the DNA/polymerase binding Kit P6 v2 (Pacific Biosciences) for a 360-min movie. All sequencing runs used PacBio's DNA Sequencing reagent Kit 4.0 v2. Post sequencing the long reads generated by SMRT sequencing cross each molecule multiple times generating highly accurate reads. Demultiplexed Circular Consensus (CCS) reads were therefore generated with the RS_ReadsOfInsert.1 protocol from the PacBio's SMRT portal (v2.3.0) with a minimum of one full pass, a minimum predicted accuracy of 90% and demultiplexing with symmetric barcodes. Afterwards a custom python script was used to extract the input read size of on-target reads and to determine the size and structure of the repeat when present in the read.
On-target and off-target analysis by DNA sequencing
The CRISPR/Cas9-targeted locus was subjected to Sanger sequencing. The target locus was amplified using 5′-CTGGTCTCCCCGTCCAGATA-3′ (forward primer) and 5′-GAACCAACGATAGGTGGGGG-3′ (reverse primer). PCR amplification was performed by an initial denaturation for 2 min at 98°C; followed by 35 cycles consisting of 10 s at 98°C, 30 s at the 59°C, 30 s at 72°C and concluded by a final extension of 8 min at 72°C. PCR reactions were performed using an s1000 Thermal Cycler (Bio-Rad). PCR products were cloned into Zero Blunt TOPO vector (K2875-J10, Life Technologies) and transformed into bacteria. Plasmids containing PCR amplicon inserts were extracted using the plasmid mini-prep system (Promega) and sequenced using a T3 sequencing primer (5′-ATTAACCCTCACTAAAGGGA–3′) by the Sanger sequencing (GATC, Germany). The sequences were analyzed using Ape software (http://biologylabs.utah.edu).
Off-target sites for the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA (Supplementary Table S2), were predicted using the validated algorithm from the Zhang lab (http://crispr.mit.edu/). From the list of 20 predicted off-target sites identified by this algorithm, we selected the four most probable off-target sites for the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA, that shared significant homology with the actual on-target sites (Supplementary Table S2). These selected off-target sites were PCR-amplified and were subjected to deep sequencing. Briefly, PCR amplification was performed by an initial denaturation for 2 min at 98°C; followed by 35 cycles consisting of 10 s at 98°C, 30 s at the 58°C, 30 s at 72°C and concluded by a final extension of 8 min at 72°C using Phusion Hot Start II high-fidelity DNA polymerase (F549S; Life Technologies) and the respective primers (Supplementary Table S3) to generate PCR products of sizes 226–387 bp. After PCR and agarose gel electrophoresis, the PCR products were purified using QIAquick Gel Extraction Kit (Qiagen). The PCR products were sequenced and analyzed using the MiSeq PE150 platform (BGI, Europe). Raw image files were processed by Illumina pipeline for base calling with default parameters and the sequences of each individual sample were generated as 150-bp paired-end reads. The 150-bp paired end reads were subjected to bioinformatics analysis.
The raw data were quality controlled by removing the Illumina adapter sequences and low quality reads were discarded using SOAP (Short Oligonucleotide Alignment Program; BGI, Hongkong). Subsequently, the read data were aligned to the human reference genome sequence (Homo sapiens GRCh38/hg38) using Burrows-Wheeler Aligner (BWA) in the Biostar Galaxy (http://usegalaxy.org) resulting in BAM format files. The BAM files were imported into Geneious R9.1.2 (Biomatters Ltd, New Zealand) to visualize the alignment of reads on human chromosomes. The analysis of small insertions and deletions (indels) was performed using FreeBayes variation/SNP analysis [(Garrison E, M.G. 2012) Haplotype-based variant detection from short-read sequencing. arXiv.org. arXiv: 1207.3907 [q-bio.GN]], which is the Bayesian genetic variant detector designed to find small polymorphisms. The percentage of insertions/deletions (indels) were obtained by polymorphisms with the following parameters: mapping quality, alternate allele frequency, ploidy of 2, minimum alternate count of 2, minimum alternate fraction of 0.2, and combines nearby variants of 3. Afterward, the annotation of polymorphism was also generated as vcf files and the data were exported as CSV format. The % InDels of the nucleotide position around PAM sequence were plotted using GraphPad Prism 6 (GraphPad Software Inc., USA).
Study design
The data sets were based on three independent CRISPR/Cas9-based correction experiments in four different DM1-iPSC-derived myogenic cell cultures. The data are based on a total of four DM1-iPSC cell lines from two DM1 patients and their respective DM1-iPSC-Myo derivatives. Each correction experiment was independently performed at a separate time point (n = 3). Quantification of the nuclear foci was performed in a blinded fashion. Consequently, all of the downstream experiments were also repeated accordingly, based on these independently generated DM1-iPSC-Myo cells. In particular, this implies that the nuclear foci quantification, SERCA1 splicing analysis, MBNL co-localization, DMPK mRNA expression analysis were based on four DM1-IPSC-Myo cultures (designated as L23, L81 from DM1 patient #1; FL5, FL8 from DM1 patient #2) and three independent CRISPR/Cas9 correction experiments (total n = 12). The cells used in this study are of human origin which was authenticated with aCGH for genomic integrity. The cultures were negative for mycoplasma. Statistical methods such as unpaired Student's t-test and one-way ANOVA were performed as indicated with statistical significance level (α) set at 0.05: *P < 0.05, **P < 0.01, ***P < 0.001. Ribonuclear foci were quantified either by manual multifocal acquisition or by a z-stack acquisition method, as specified accordingly. Bland–Altman statistical plot confirmed that the two methodologies were in agreement and could be considered interchangeable.
RESULTS
Characterization and myogenic differentiation of DM1 patient-specific induced pluripotent stem cells (DM1-iPSCs)
Both primary myoblasts and fibroblasts from DM1 patients were reprogrammed into iPSCs (designated as DM1-iPSCs) by retroviral transduction of four reprogramming factors OCT4, SOX2, KLF and c-MYC (32). Mouse embryonic fibroblasts (MEFs) were briefly used as feeder cells during the generation of iPSCs. The individual DM1-iPSC clones (designated as L22, L23, L81; patient #1; FL8, FL5; patient #2) were subsequently expanded in feeder-free conditions (Supplementary Figures S1A and S3A). The pluripotency of the DM1-iPSC clones was ascertained by assessing the pluripotency maker expression, i.e. OCT4, SSEA4, SSEA3, TRA 1–60 and alkaline phosphatase (AP) (Supplementary Figure S1B) and by demonstrating their ability to generate teratomas consisting of ectoderm, endoderm and mesoderm tissue structures after subcutaneous transplantation into CB17 SCID mice (Supplementary Figure S1D). Similarly, the pluripotency was also confirmed for DM1-iPSC clones FL5 and FL8 (patient #2). Molecular karyotyping of the newly generated DM1-iPSC clones (L22, L23, L81; FL8, FL5) by array comparative genome hybridization (aCGH) with a 100 kb resolution revealed a normal karyotype (Supplementary Figures S1C and S3C).
The CTG trinucleotide repeat expansion was confirmed by Southern blot analysis of the DMPK locus with a probe specific for the DMPK 3′ UTR region (Figure 1C; Supplementary Figure S4A). Southern blot analysis revealed that the DM1-iPSC clones were heterozygous and contained a mutated DMPK allele with a characteristic CTG repeat expansion and a wild-type DMPK allele. CTG trinucleotide repeat expansion in the DM1-iPSC clones derived from patient #1 (L22 and L23) corresponded to ∼1000 and ∼1300, respectively, whereas clone L81 exhibited two expanded populations having ∼1400 and ∼1600 repeats (Figure 1C). Triplet-repeat length in the DM1-iPSCs FL8 clone from patient #2 was ∼1700, whereas the FL5 clone exhibited two repeat sizes corresponding to ∼1400 and ∼1600 repeats (Figure 1C). The difference in repeat length among the different DM1-iPSC clones derived from the same patient may reflect somatic mosaicism in the original DM1 patient-derived primary cells from which the iPSCs were derived consistent with somatic CTG instability (47). This is in line with the occurrence of somatic CTG repeat instability in DM1 mouse models in vivo. However, it cannot be excluded that this CTG repeat instability may arise upon cell expansion in vitro (48–51).
Analysis of our Southern blots for the healthy control iPSCs show two bands ∼1 kb apart (Figure 1C, Supplementary Figure S4A). The experimental observation (in Figure 1C) is in agreement with known Alu polymorphisms seen in the DMPK locus (4,52,53). Therefore, considering our observation and the previous studies (4,52,53), we could infer that the two distinct bands for the healthy control DMPK alleles represent two polymorphic DMPK alleles. The upper band of the healthy control iPSC contain the ∼1 kb Alu repeat (designated as Alu+) whereas the lower band did not (designated as Alu-). Southern blot analysis revealed that the wild-type DMPK allele of the DM1-iPSC clones L81 and L23 is Alu+ (Figure 1C and Supplementary Figure S4A), whereas the wild-type DMPK allele of the DM1-iPSC clones FL8 and FL5 is Alu- (Figure 1C and Supplementary Figure S4A). Subsequent sequencing confirmed that the wild-type DMPK allele contained a normal number of CTG repeats (patient #1: 5 CTG repeats; patient #2: 13 CTG repeats) (Supplementary Figures S7A and B).
One of the hallmarks of DM1 is the emergence of ribonuclear foci in the nuclei of the affected patient cells, due to the expanded CUG repeats in the 3′UTR region of DMPK transcript (54). We therefore confirmed the presence of the ribonuclear foci in the DM1-iPSC clones by fluorescence in situ hybridization (FISH) (Figure 1B, Supplementary Figure S3D). The number of ribonuclear foci in DM1-iPSCs typically ranged from three to five per nucleus with a maximum of eight to nine foci per nucleus. As expected, no ribonuclear foci were detected in the iPSCs derived from a healthy donor (Figure 1B).
Since DM1 patients suffer from muscle dysfunction, we subsequently induced the DM1-iPSCs to differentiate into myogenic cells using an established myogenic differentiation protocol (Supplementary Figure S4B), we have previously reported (26,27). As expected the four-stage differentiation process yielded a population of iPSC-derived induced myogenic cells (designated as DM1-iPSC-Myo) having robust in vitro expansion potential (Supplementary Figures S1E and S3A). Cytofluorimetric analysis of the DM1-iPSC-Myo revealed expression of mesodermal/vascular cell surface markers such as CD13, CD44, CD49b and CD146 but absence of CD31 and CD45 (Supplementary Figure S2A). The DM1-iPSC-Myo also displayed alkaline phosphatase (AP) expression, detectable using enzymatic activity assays (Supplementary Figure S2C). Importantly, apart from being a marker of pluripotent stem cells, AP is also a characteristic marker of DM1-iPSC-Myo cells (26,27), since they are similar to AP-positive mesoangioblasts (29). The expression of these markers on DM1-iPSC-Myo is consistent with the expression pattern on both normal healthy iPSC-Myo and primary human skeletal muscle pericyte-derived mesoangioblasts (Supplementary Figures S2C and A) (26,27) (29). The differentiation of DM1-iPSCs into DM1-iPSC-Myo (or control healthy iPSCs into iPSC-Myo) was relatively robust, as we could not detect presence of any residual undifferentiated OCT4+ or SOX2+ DM1-iPSCs (Supplementary Figure S2B). The DM1-iPSC-Myo and control iPSC-Myo were stained for human lamin A/C and the analysis of staining showed uniform positive expression of human lamin A/C in these differentiated cells. We could not detect any human Lamin A/C negative residual murine feeder cells that were employed during the initial stages of iPSCs generation (Supplementary Figure S1F).
Subsequently, we analyzed the expanded CTG repeat length in the DM1-iPSC-Myo cells upon myogenic differentiation of the DM1-iPSC clones. Triplet-repeat length in DM1-iPSC-Myo L23 was ∼1300, whereas L81 had two expanded populations having ∼1200 and ∼1400 repeats. This is consistent with the repeat length in the respective original DM1-iPSC clones derived from patient #1. In the second DM1 patient, the DM1-iPSC-Myo FL5 population contained ∼1400 and ∼1600 repeats (Figure 1F). This is also in accordance with the number of repeats in the original DM1-iPSC clone from patient #2. However, a slight reduction in repeat length was apparent in the FL8 population, compared to the original DM1-iPSC clone (i.e. ∼1500 versus ∼1700) (Figure 1C and F). This could possibly be attributed to CTG repeat instability upon cell expansion in vitro as reported in previous studies (48–51). In contrast, the control healthy iPSC-Myo showed the expected pattern of 5 CTG repeats as in their iPSCs counterparts, from which they were derived (Figure 1F).
Most importantly, the presence of CTG repeats in the DMPK 3′ UTR was consistent with the emergence of the characteristic ribonuclear foci in the DM1-iPSC-Myo, whereas no foci could be detected in control healthy iPSC-Myo (Figure 1D). It is noteworthy that these ribonuclear foci were larger and brighter in the DM1-iPSC-Myo than in the undifferentiated DM1-iPSCs, in accordance with previous studies (55). Finally, to complete the myogenic differentiation process, we subjected the DM1-iPSC-Myo to MyoD-induced myogenic differentiation into skeletal myotubes and myocytes, as described previously (26,27). The widespread expression of myosin heavy chain (MyHC), a mature muscle marker, in the differentiated multinucleated myotubes was consistent with robust differentiation of DM1-iPSC-Myo (or healthy control iPSC-Myo) with >80% efficiency (Figure 1E, Supplementary Figure S3B). Hence, a patient-specific DM1-iPSC-derived cellular model was hereby established capable of robust myogenic differentiation into DM1-patient specific mesoangioblast-like cells, myocytes and myotubes. Hence, this served as a suitable DM1 in vitro disease model to further examine the consequences of CRISPR/Cas9-mediated excision of the CTG repeats in the mutant DMPK allele.
CRISPR/Cas9-mediated targeting of CTG trinucleotide repeats in DM1-iPSC-derived myogenic cells
The validation of the myogenic DM1-iPSC-Myo cellular platform allowed us to subsequently assess the potential of CRISPR/Cas9 to specifically excise the CTG trinucleotide repeats. We therefore generated lentiviral vectors that expressed the S. pyogenes Cas9 protein and a green fluorescent protein (GFP) reporter gene (Supplementary Figure S3C). In addition, lentiviral vectors that expressed gRNAs specifically designed to target sequences just 5′ upstream and 3′ downstream of the CTG repeat expansion in the DMPK 3′UTR (designated as 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA) along with a blue fluorescent reporter (BFP) gene were produced (Figure 2A, Supplementary Figures S5A and B). As controls, we used vectors that were identical in design but encoded scrambled gRNAs instead. Fluorescence microscopy analysis and quantification of GFP+ and BFP+ DM1-iPSC-Myo at 72 h post transduction revealed relatively robust transduction efficiency corresponding to 84 ± 5% for Cas9 (GFP) and 93 ± 6% for both gRNAs combined (i.e. 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA) (BFP) (Supplementary Figures S5A and B). To directly assess the Cas9 protein expression in DM1-iPSC-Myo cells transduced with the Cas9 vector an immunofluorescence staining was performed using Cas9-specific monoclonal antibodies. A total of 82.7 ± 3.2% to 91.0 ± 0.3% of the transduced DM1-iPSC-Myo cells (derived from three independent DM1-iPSC clones L23, FL8 and FL5) expressed the Cas9 protein (average: 85.1 ± 4.5%) (Figure 2B; Supplementary Figure S5C). This is consistent with the assessment of transduction efficiency based on GFP expression (Supplementary Figures S5A and B). These results indicated that most cells expressed Cas9 and that the transduction efficiency and expression of the Cas9 was relatively robust. Nevertheless, some variation in fluorescence intensity was apparent, indicating that Cas9 expression did vary among individual cells.
Figure 2.
A dual gRNA approach for CRISPR/Cas9-mediated correction of DM1-iPSC Myo and evidence for trinucleotide CTG repeat excision. (A) Diagrammatic representation for targeting of the 3 ‘UTR region of the DMPK gene using a dual gRNA approach for CRISPR/Cas9-mediated gene correction. The dual gRNAs (5′& 3′-CTGrepeat-gRNA) target Cas9 on either side of the CTG repeat region for excision of the expanded trinucleotide repeat. (B) Cas9 immunofluorescence staining of CRISPR/Cas9 treated DM1-iPSC-Myo cells, at 1-week post transduction. The upper panel shows representative images of DM1-iPSC-Myo cells stained for Cas9 (in red) and co-stained with DAPI for nuclei (in blue) (scale bar = 50 μm). The lower panel shows the graph for the quantitation of microscopy data for Cas9 positive cells. (C) Representative electropherograms of Triplet Repeat Primed PCR (TP) products from DM1-iPSC-Myo after CRISPR/Cas9-mediated gene editing from three independent experiments for each of the three treatments (Cas9 and 5′ & 3′-CTGrepeat-gRNA; Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9) and untreated control conditions (WT-iPSC-Myo and DM1-iPSC-Myo). (D) Sanger sequencing results of on-target activity. The DMPK target locus was amplified by primers flanking the 2 SNPs [C>T; G>A: mutant > wild-type allele] and the CTG repeat region [(CTG)∼1371 /(CTG)5]. The SNPs allowed discrimination of mutant (C&G) and wild-type alleles (T&A). Analysis of CRISPR/Cas9 activity on the targeted mutant allele showed a large deletion [(–) ∼4188 bp] between the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA target sites. CRISPR/Cas9 activity on wild type allele was also detected by deletions between the corresponding gRNA target sites. Representative sequences of the wild-type allele with commonly found deletions and insertions are depicted in the figure. SNPs marked in red are seen in the mutant allele and those in blue are present in the wild type allele. Insertions are indicated by (+) and deletions are indicated by (–). Small letters represent the inserted nucleotides.
To assess whether CRISPR/Cas9 can be used to excise the CTG repeats in DM1 patient's cells, a rapid triplet repeat primed PCR (TP-PCR) assay was conducted on the genomic DNA isolated from the CRISPR/Cas9 transduced DM1-iPSC-Myo (Figure 2C) (13). Capillary electrophoresis of the PCR amplicons and analysis of the pattern of PCR amplicons revealed a pronounced difference in the signal intensity in the CRISPR/Cas9-treated cells compared to controls. In particular, DM1-iPSC-Myo engineered with Cas9 and the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA exhibited a marked decrease in the frequency (signal intensity) of PCR amplicons compared to control DM1-iPSC-Myo cells transduced with Cas9 and the scrambled gRNA or only with the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA without Cas9. The disappearance of the characteristic CTG repeat pattern after gene editing with Cas9 and the dual 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA resemble that of healthy control iPSC-Myo and was consistent with the removal of CTG repeats in the DMPK 3′UTR. Conversely, the CTG repeat pattern of the control conditions either without Cas9 or with scrambled gRNAs (and Cas9) resembled that of non-transduced DM1-iPSC-Myo control cells, indicating that the CTG repeats remained intact within the DMPK 3′ UTR locus for those control conditions. Taken together, these findings strongly suggested that treating DM1-iPSC-Myo with CRISPR/Cas9 resulted in a relatively robust excision of the CTG repeats from the DMPK 3′ UTR. This obviated the need for selective enrichment of gene-edited cells subsequent to CRISPR/Cas9 correction. The TP-PCR analysis supported the hypothesis that the expanded CTG repeat was excised by the CRISPR/Cas9 system in the DM1-iPSC-Myo cells.
Subsequently, we demonstrated by Sanger sequencing of the junction area post-excision, that the CTG repeat of both wild-type and mutant DMPK alleles could be excised by CRISPR/Cas9. We therefore exploited the presence of single nucleotide polymorphisms (SNPs) in the wild-type DMPK allele which is distinct from the mutant DMPK allele. The DM1 patient's cells were heterozygous with respect to the CTG repeats in the DMPK 3′UTR yielding >1000 repeats in the mutant allele and only 5 CTG repeats in the wild-type allele (Figure 1C–F and Supplementary Figure S4A). In addition, we identified two small nucleotide polymorphisms (SNPs) further distinguishing this mutant from the normal allele. The corresponding T→C and A→G SNPs were located respectively at position 835 and 640 nucleotides upstream from the 5′ end of the CTG repeat (Figure 2D). The presence of these two SNPs was exploited to independently confirm excision of the expanded CTG repeats by Sanger sequencing on amplicons, amplified by primers flanking the SNPs and the expanded CTG repeats (Figure 2D). Deletion of ∼ 4188 bp fragment resulted in the excision of the entire full-length CTG repeat expansion in the CRISPR/Cas9-corrected DM1-iPSC-Myo. This deletion was flanked by the respective 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA target sites and was consistent with the requirement of a protospacer adjacent motif (PAM) near the Cas9 cleavage site. Taken together, these results confirm the TP-PCR data (Figure 2C) and demonstrate CRISPR/Cas9-mediated editing of the CTG trinucleotide repeat expansion in DM1-derived myogenic cells. The presence of the SNPs further distinguished wild-type from mutant DMPK alleles also permitted the identification of deletions of the (CTG)5 repeat region and small insertions/deletions (indels) at the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA target sites in the wild-type DMPK locus. Since the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA target sites were located in the 3′ UTR of the DMPK gene, this design avoids altering the DMPK protein-coding sequence itself. Although due to the design of the gRNAs that targeted a sequence in proximity of the PAM consensus sites upstream and downstream of the CTG repeat, some flanking non-repeat sequences were also excised, as expected.
To better characterize the molecular consequences of the CRISPR/Cas9-mediated excision of the CTG repeat, an emerging new sequencing technology was employed based on single-molecule real time (SMRT) sequencing, given its high accuracy and ability to sequence even through extended trinucleotide repeat regions (56,57). Prior to SMRT library preparation and sequencing, the 3′ UTR DMPK target region of the CRISPR/Cas9-corrected DM1-iPSC-Myo, untreated DM1-iPSC-Myo and non-corrected DM1-iPSC-Myo control conditions (i.e. Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9) was PCR-amplified (Figure 3A–C, Supplementary Figure S6A and B). The amplicons were subsequently analyzed by gel electrophoresis. In the case of the non-edited control cells (i.e. untreated DM1-iPSC-Myo, non-corrected DM1-iPSC-Myo cells), we expected two different sizes of the amplified products; a ∼ 723 bp amplicon from the uncut wild-type DMPK allele and a >4000 bp amplicon from the uncut mutant DMPK allele containing the CTG repeat expansion (Figure 3A, Supplementary Figure S6A and B). Similarly, the wild-type iPSC-Myo cells yielded only the ∼723 bp amplicon.
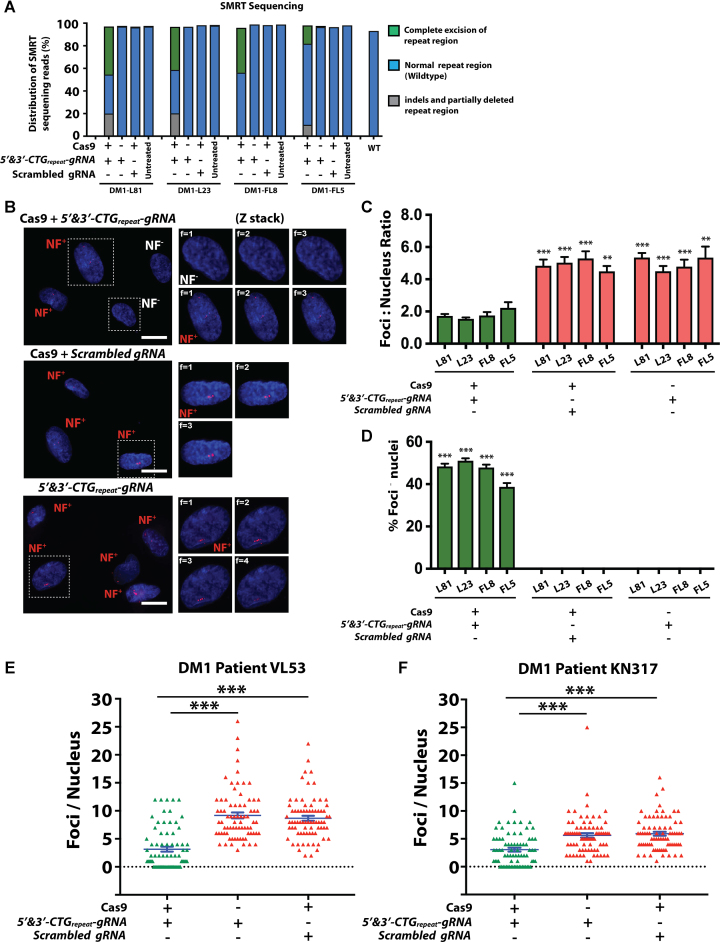
Figure 3.
Analysis of target region in the CRISPR/Cas9-corrected DM1-iPSC-Myo and ribonuclear foci staining of corrected DM1-iPSC-Myo and DM1 primary myoblasts. (A) Graph representing distribution of SMRT sequencing reads based on the various amplicon sizes ∼633 bp (excised fragments) and ∼723bp (WT fragments). The sequences ranging between ∼723 bp and ∼4000 bp were fragments with indels and partially deleted repeat regions. Each bar represents distribution of reads from each of the three conditions (Cas9 + 5′ & 3′-CTGrepeat-gRNA, Cas9 control and gRNA control) and untreated DM1-iPSC-Myo control. (B) Representative image of CRISPR/Cas9-corrected DM1-iPSC-Myo (L81) stained for ribonuclear foci. Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9 were used as negative controls. An antisense Cy3-labeled probe was used to detect the presence of ribonuclear foci (NF). The ribonuclear foci negative and positive nuclei were denoted as NF− (white) and NF+ (red), respectively. Each representative image is a maximum intensity z projection of the z slice images. For all the conditions (Cas9 + 3′ & 5′-CTGrepeat-gRNA, scrambled gRNA and no Cas9) enlarged z slices of selected ribonuclear foci negative (NF-) and positive (NF+) nucleus are represented. Nuclei were counter-stained with DAPI (scale bar = 20 μm). (C) Quantification of ribonuclear foci (NF) in CRISPR/Cas9-corrected DM1-iPSC Myo. The total number of ribonuclear foci per total number of nuclei was calculated. Total of nuclei counted is 6500. The data is represented as mean ± SEM. The statistics were performed using two-tailed unpaired Student's t-test (***P <0.001; **P < 0.01; *P < 0.05). (D) Graph shows the % of ribonuclear foci-negative nuclei in the CRISPR/Cas9-corrected DM1-iPSC-Myo to determine the overall efficiency of CRISPR/Cas9 correction. A total number of 4915 nuclei were examined by FISH staining. The data is represented as mean ± SEM. The statistics were performed using one-way ANOVA (***P <0.001; **P < 0.01; *P < 0.05). The quantitation was done in a blinded fashion. (E and F) Graphs showing quantification of ribonuclear foci (NF) in CRISPR/Cas9 corrected DM1 primary myoblasts from two DM1 patients (VL53 and KN317). The foci per nucleus was calculated and represented by each data point in the graph. The data is represented as mean ± SEM. The statistics were performed using two-tailed unpaired Student's t-test (***P <0.001; **P < 0.01; *P < 0.05).
The CRISPR/Cas9-treated DM1-iPSC-Myo cells contained a mixed population of both edited and non-edited cells, consistent with the nuclear foci results (Figure 3B–D; Supplementary Figures S8A–C) Complete excision of the CTG repeat expansion, along with the flanking sequences containing the PAM consensus sites, resulted in an amplified product of ∼633 bp. This ∼633 bp amplicon corresponded to both wild type and mutated DMPK alleles from the DM1-iPSC-Myo cells subjected to CRISPR/Cas9-mediated editing with Cas9 and 5′ & 3′-CTGrepeat-gRNAs (Figure 3A, Supplementary Figure S6A and B). In contrast, this 633 bp amplicon was absent in all of the control conditions (i.e. untreated DM1-iPSC-Myo cells, DM1-iPSC-Myo cells treated with Cas9 and scrambled gRNA; DM1-iPSC-Myo cells treated with 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9). Since the CRISPR/Cas9-treated DM1-iPSC-Myo cell population also contained unedited cells, we could detect the ∼723 bp amplicon corresponding to the unedited wild-type allele. Interestingly, the DM1-iPSC-Myo cells subjected to CRISPR/Cas9-mediated editing with Cas9 and 5′&3′-CTGrepeat-gRNAs did not only show the expected ∼633 bp amplicon but also displayed amplicons with varying sizes that fell in the range of 723–4000 bp. The occurrence of such a heterogeneous amplicon pattern would be consistent with a partial deletion of the CTG repeats at either the 5′ or 3′ end of the repeat expansion. This could be explained by a single gRNA-mediated Cas9 cleavage either upstream or downstream of the CTG repeat expansion in the 3′ DMPK UTR. This is consistent with the SMRT sequence data (Supplementary Figures S7A and B) and previous reports (24).
The amplicons were then subjected to SMRT library preparation with the appropriate barcoded adaptors followed by SMRT sequencing. SMRT sequencing enabled us to distinguish between three different categories of DMPK alleles corresponding to: (i) complete CRISPR/Cas9-dependent excision of the CTG repeat, (ii) partial CRISPR/Cas9-dependent excision of the CTG repeat and (iii) non-edited alleles (either wild-type or mutant). The distribution of these distinct categories of SMRT sequence reads has been graphically represented in (Figure 3A, Supplementary Figure S6A and B). Representative SMRT sequence-reads corresponding to these three different scenarios were shown in Supplementary Figure S6A (DM1-iPSC-Myo L81/L23) and S6b (DM1-iPSC-Myo FL5/FL8). The comprehensive SMRT sequencing analysis corroborated our observations based on the DMPK 3′ UTR–specific PCR and the disappearance of the ribonuclear foci after CRISPR/Cas9-mediated correction (Figures 3A–D and 4A; Supplementary Figures S8A–C and S9). Excision of the CTG repeat expansion could only be detected in CRISPR/Cas9-corrected DM1-iPSC-Myo in the presence of 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and Cas9. Most of the CRISPR/Cas9-corrected DM1-iPSC-Myo lines (DM1-L81, DM1-L23, DM1-FL8) yielded ∼40% sequence reads consistent with excision of the CTG repeat expansion, though this was somewhat lower (∼10%) in just one of the line (DM1-FL5). The differences correlated well with the differences in nuclear foci between the various DM1-iPSC-Myo populations (Figure 3B–D; Supplementary Figure S8A–C). Excision of the CTG repeat expansion was consistent with the presence of indels at the respective target sites located in proximity of the PAM sites. In contrast, excision of the entire CTG repeat expansion or indels could not be detected in the controls (i.e. untreated DM1-iPSC-Myo cells, DM1-iPSC-Myo cells treated with Cas9 and scrambled gRNA or DM1-iPSC-Myo cells treated with 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9). This excludes possible contamination or false positives. SMRT sequencing confirmed the presence of unedited wild-type alleles with normal CTG repeats (i.e. 5 CTG repeat in DM1-L81, DM1-L23, patient #1; 13 CTG repeat in DM1-FL8, DM1-FL5, patient #2). Interestingly, partial deletions of the CTG repeats were also apparent, but again only in the presence of Cas9 and 5′ & 3′-CTGrepeat-gRNA and not in the controls without Cas9 or with scrambled gRNA and Cas9. These partial CTG repeat deletions were consistent with the presence of indels in proximity of the respective PAM sites and reflect the heterogeneous 3′ DMPK UTR PCR-amplification pattern (Figure 3A, Supplementary Figure S6A and B). It is likely that these partial deletions in the CTG repeat are due to a single CRISPR/Cas9-mediated cut either upstream or downstream of the CTG repeat resulting in repeat instability. Indeed, such partial repeat deletions have also been observed after single target cleavage with other designer nucleases such as TALENs or ZFNs (58–60) which results in 5′-3′ resection of the ends followed by either a replication slippage or homologous recombination pathway.
Figure 4.
Biological effects of CRSIPR/Cas9 mediated correction of DM1-iPSC-Myo. (A) Dual staining for MBNL1 and ribonuclear foci co-localization in the CRSIPR/Cas9-corrected versus control conditions (Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9). Representative image of DM1-iPSC-Myo stained for MBNL1 and Ribonuclear foci by combined immunostaining-FISH staining. Each representative image is a maximum intensity z projection of the z slices. For control conditions, enlarged image of selected nuclei are represented under different filters. For the condition (Cas9 + 3′& 5′-CTGrepeat-gRNA) enlarged z slices of selected ribonuclear foci negative (NF-) and positive (NF+) nucleus are represented under different filters. Nuclei were counterstained with DAPI. (B) Quantification of the microscopy data is represented in term of ratio between the total dual positive (MBNL1+RNA+ foci)/total number of nuclei observed in each condition for the L23, L81, FL8 and FL5 DM1-iPSC-Myo cells. The data is represented as mean ± SEM. The statistics were performed using two-tailed unpaired Student's t-test (***P <0.001; **P < 0.01; *P < 0.05). (C and D) Reversal of defective alternative splicing pattern of SERCA in CRISPR/Cas9-corrected DM1-iPSC- derived muscle cells (myocytes/myotubes). Muscle cells obtained from Cas9 and 5′& 3′-CTGrepeat-gRNA treated DM1-iPSC-Myo was analyzed for alternative splicing with primers specific to exon 21 and exon 23. The fraction of splice variants either including or excluding exon 22 was quantified based on the band intensity and then plotted. Healthy control sample (WT), scrambled gRNA and no Cas9 conditions were used as controls. GAPDH was used for normalization. The data is represented as mean ± SEM. The statistics were performed using two-tailed unpaired Student's t-test (***P <0.001; **P < 0.01; *P < 0.05).
CRISPR/Cas9 has been shown to demonstrate a variable degree of ‘off-target’ cleavage activity (61,62). We therefore investigated the extent of potential off-target activity in the CRISPR/Cas9-corrected DM1-iPSC-Myo. Two putative off-target sites for each gRNA (5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA) (Supplementary Figure S10A and B) were predicted using a validated computational algorithm from the Zhang lab (http://crispr.mit.edu/). MiSeq deep sequencing was performed on PCR amplicons containing the target region surrounding these putative computationally predicted off-target sites. Overall, no detectable off-target activity above background (i.e. in control condition without Cas9 or with scrambled gRNAs) or only very low level potential off-target activity ranging between 0.2% and 0.3% was detected for each of the putative off-target sites of either 5′-CTGrepeat-gRNA or 3′-CTGrepeat-gRNA (Supplementary Figure S10A and B) consistent with the presence of indels at the putative off-target sites (Supplementary Table S4).
Biological effects of CRISPR/Cas9 mediated correction of DM1-iPSC-Myo cells
We subsequently assessed whether the CRISPR/Cas9-mediated excision of the CTG repeat expansion resulted in functional correction of the DM1-associated cellular pathology in DM1-iPSC-Myo. The expression of the pathogenic CUGexp RNA in DM1-iPSCs or DM1-iPSC-Myo is associated with accumulation of the ribonuclear foci (Figure 1B and F). FISH analysis revealed a pronounced difference in ribonuclear foci accumulation in the nuclei of CRISPR/Cas9-treated cells compared to controls, based on nearly 6500 individual nuclei counted (Figure 3B–D; Supplementary Figure S8A–C). In particular, four DM1-iPSC-Myo lines derived from two different DM1 patients (DM1 patient 1: L81 and L23; DM1 patient 2: FL8 and FL5) were engineered with the Cas9 and the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA vectors and exhibited a significant decrease in ribonuclear foci. The foci: nucleus ratio ranged between 1.54 ± 0.09 to 2.2 ± 0.36 in cells treated with Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA which was significantly decreased (***P < 0.001; **P < 0.01; *P < 0.05) compared to control DM1-iPSC-Myo cells engineered with Cas9 and scrambled gRNA (5.29 ± 0.45 to 4.49 ± 0.33) or engineered with only the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA without Cas9 (5.35 ± 0.28 to 4.50 ± 0.33) (Figure 3B and C; Supplementary Figure S8A–C). Similarly, a significant proportion of DM1–iPSC-Myo nuclei that underwent gene-editing with CRISPR/Cas9 contained no ribonuclear foci (% ribonuclear foci-negative nuclei ranged from 38.7 ± 1.7% to 51.9 ± 1.22% between different DM1–iPSC-Myo lines) which was significantly increased (***P < 0.001) compared to control DM1–iPSC-Myo transduced with Cas9 and scrambled gRNA (% ribonuclear foci-negative nuclei = 0%) or transduced with 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA but no Cas9 (% ribonuclear foci-negative nuclei = 0%) (Figure 3B and D; Supplementary Figure S8A–C). The reduction in ribonuclear foci after gene editing with CRISPR/Cas9 was consistent with the excision of CTG repeats in the DMPK 3′ UTR (Figures 2C, D and 3A–C) and with the absence of such foci in healthy control iPSC or its myogenic iPSC-Myo derivative (Figure 1B and D). Conversely, the presence of ribonuclear foci in the control conditions (i.e. without Cas9; with scrambled gRNAs and Cas9) resembled that of uncorrected DM1-iPSC and DM1-iPSC-Myo cells (Figure 1B and D), consistent with the presence of the CTG repeat expansion in the DMPK 3′ UTR locus (Figure 1C and F).
In addition to DM1-iPSC-Myo, we also corrected non-transformed DM1 patient primary myoblasts, which are distinct from DM1-iPSC-Myo based on their origin from satellite cells as discussed previously. These CRISPR/Cas9 treated myoblasts were stained for ribonuclear foci to assess the functional outcome. In accordance with our previous observation in DM1-iPSC-Myo, a significant decrease (***P < 0.001) in the average foci/nucleus was also detected in DM1 patient-derived primary myoblasts that were transduced with Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA vectors as compared to the non-corrected control conditions (i.e. Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9) (Figure 3E and F; Supplementary Figure S6C). Nuclei devoid of ribonuclear foci could only be detected in the DM1 patient-derived primary myoblasts that were treated with the Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA vectors and not in the non-corrected control conditions (i.e. Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9) (Figure 3E and F; Supplementary Figure S6C). It is therefore particularly encouraging that the reduction in ribonuclear foci after CRISPR/Cas9-mediated gene editing was consistently observed in DM1 patient-derived primary myoblasts (from two different DM1 patients) and in all the four DM1-iPSC-Myo lines (also from 2 different DM1 patients) (Figure 3C and D). Overall, the functional correction was relatively efficient in both DM1-iPSC-Myo and DM1 myoblasts, obviating the need for selective enrichment of these gene-corrected non-transformed cells.
After demonstrating effective correction of the characteristic DM1 genotype and cellular phenotype by CRISPR/Cas9-mediated excision of the CTG repeats in the DMPK 3′ UTR, we subsequently examined other downstream pathways characteristic of DM1, in particular the MBNL1 sequestration into ribonuclear foci and SERCA1 alternative splicing. It is known that the pathogenic CUGexpRNA sequesters the muscleblind-like protein 1 (MBNL1) in the CUGexpRNA-protein nuclear aggregates of DM1 patient's cells (38,63). We therefore assessed the localization of MBNL1 by FISH / immunostaining experiments in CRISPR/Cas9-corrected versus non-corrected DM1-iPSC-Myo. MBNL1 co-localized with ribonuclear foci in non-corrected DM1-iPSC-Myo (Figure 4A and B; Supplementary Figure S9). The reduction in ribonuclear foci after gene editing with CRISPR/Cas9 is consistent with the concomitant delocalization of the MBNL1 protein. In particular, DM1-iPSC-Myo cells engineered with Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA exhibited a significant decrease in ratio of total number of co-localized MBNL1+ RNA+ foci to total nuclei across all four distinct DM1-iPSC-Myo cell populations (MBNL1+Ribonuclear+ foci: nucleus ratio ranging from 0.41 ± 0.04 to 0.73 ± 0.09) compared to control DM1-iPSC-Myo cells engineered with Cas9 and scrambled gRNA (MBNL1+ RNA + foci: nucleus ratio ranging from 1.36 ± 0.08 to 1.93 ± 0.26) or engineered with only the 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA but without Cas9 (MBNL1+ RNA + foci: nucleus ratio ranging from 1.46 ± 0.03 to 1.69 ± 0.25) (Figure 4A and B; Supplementary Figure S9).
We subsequently determined whether correction of the CTG repeat expansion by CRISPR/Cas9 and the concomitant disappearance of ribonuclear foci could reverse the aberrant alternative splicing pattern that is characteristic of DM1. The CRISPR/Cas9-corrected DM1-iPSC-Myo were therefore induced to differentiate into myocytes/myotubes (Figure 1E) and the differential splicing pattern of SERCA1 was checked. SERCA1 is one of the known biomarkers of DM1 pathology and exhibits a characteristic abnormal splicing pattern in DM1 patients. The exclusion of SERCA1 exon 22 consistently emerged as the dominant splicing pattern of SERCA1 in DM1 in contrast to the predominant inclusion of the SERCA1 exon 22 in wild-type cells (33,45,64). Our current results are consistent with this differential splicing pattern of SERCA1 in DM1 patient versus healthy cells. The DM1-iPSC-Myo-derived myocytes/myotubes showed predominant exclusion of SERCA1 exon 22 as opposed to its predominant inclusion in healthy iPSC-Myo-derived myocytes/myotubes (Figure 4C and D). Particularly in DM1-iPSC-Myo-derived myocytes/myotubes, only 0.77 ± 0.25 to 8.74 ± 2.64% of the SERCA1 transcripts correspond to the exon 22 + containing splice variant whereas this increased to 75.29 ± 6.34 to 82.34 ± 2.34% in myocytes/myotubes derived from healthy iPSC-Myo cells. CRISPR/Cas9-mediated excision of the CTG repeats expansion from the DMPK 3′ UTR in DM1-iPSC-Myo-derived myocytes/myotubes resulted in the normalization of the SERCA1 splicing pattern, more similar to that of healthy iPSC-Myo-derived myocytes/myotubes. This is consistent with a significant increase in exon 22 included splice variant after gene editing of the DMPK gene with Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA (7.46 ± 0.53% to 40.39± 3.34%) compared to controls without Cas9 (1.55 ± 0.11% to 3.2 ± 0.87%) or with scrambled gRNA and Cas9 (2.2 ± 0.10% to 4.2 ± 2.3%). Conversely, in the absence of Cas9 or when scrambled gRNAs were employed, the exclusion of exon 22 emerged as the predominant splice variant, similar to the splicing pattern of non-treated DM1-iPSC-Myo-derived myocytes/myotubes (Figure 4C and D). Taken together, these results demonstrate that the CRISPR/Cas9-mediated excision of the CTG trinucleotide repeat expansion in DM1-derived myogenic cells was effective in reversing the DM1 phenotype in the DM1-iPSC-Myo cells and their differentiated derivatives (myocytes/myotubes).
After demonstrating effective functional corrections in the CRISPR/Cas9-treated cell populations, we assessed the stability of DMPK transcripts. To demonstrate that the CRISPR/Cas9-medated excision of the CTG repeat in the 3′ UTR region did not affect the DMPK mRNA stability, a quantitative reverse transcriptase PCR (qRT-PCR) was performed to detect DMPK mRNA levels in the CRISPR/Cas9-corrected DM1-iPSC-Myo cells after differentiation into myocytes/myotubes and in its respective non-corrected DM1-iPSC-Myo-derived myocytes/myotubes controls (i.e. Cas9 and scrambled gRNA; 5′-CTGrepeat-gRNA, 3′-CTGrepeat-gRNA and no Cas9). The qRT-PCR was designed based on primers that amplify the junction of exon 10–11 until exon 12 of the DMPK gene. Our results demonstrated that there was no significant difference in the DMPK mRNA expression levels in the CRISPR/Cas9-corrected DM1-iPSC-Myo cells as compared to the non-corrected DM1-iPSC-Myo control cells (Supplementary Figure S8D).
CRISPR/Cas9-correction of DM1-iPSCs
The CRISPR/Cas9-corrected DM1-iPSC-Myo population is heterogeneous with respect to the expanded CTG repeat excision pattern, consistent with the SMRT analysis (Figure 3A, Supplementary Figure S7A and B). Different scenarios can be envisaged that account for this heterogeneity. This may possibly reflect CTG repeat excision of the mutant, wild-type or even both DMPK alleles, since the CRISPR/Cas9 strategy cannot distinguish between both. Alternatively, in some cells none of the alleles would be affected. Ideally, to formally demonstrate this, it would be required to analyze CTG repeat excision on individual clones derived from the CRISPR/Cas9-treated DM1-iPSC-Myo population by Southern blot analysis. However, we could never obtain individual clones by limiting dilution, which likely reflects the non-transformed nature and intrinsic biology of these differentiated mesoangioblast-like cells. To overcome this limitation, we decided to therefore conduct Southern blot analysis on individual iPSC clones derived from CRISPR/Cas9-corrected DM1-iPSCs. To introduce the CRISPR/Cas9 components into the DM1-iPSCs we initially relied on the use of lentiviral vectors as was used successfully for the DM1-iPSC-Myo cells. However, lentiviral transduction of the DM1-iPSCs was not as efficient as in the case of DM1-iPSC-Myo cells. Moreover, to deliver the CRISPR/Cas9 components into the DM1-iPSCs it may be desirable to use a non-integrating non-viral delivery method instead, to eliminate the risk of insertional oncogenesis associated with random genomic integration. We therefore explored the use of a non-viral gene correction method based on nucleofection of ribonucleoprotein (RNP) complexes composed of Cas9 proteins and synthetic single guide gRNA (sgRNA).
As mentioned previously, a total of four DM1-iPSC lines were generated from 2 different DM1 patients (designated as L23, L81 from DM1 patient #1; FL5, FL8 from DM1 patient #2). These DM1-iPSC lines (n = 4) were transfected with RNP-CRISPR/Cas9 complexes consisting of Cas9 protein, 5′-CTGrepeat-sgRNA and 3′-CTGrepeat-sgRNA. This was repeated in 3 independent experiments (i.e. 4 × 3 = 12). Delivery of RNP complex consisting of Cas9 protein and the corresponding synthetic single gRNA (sgRNA) into the DM1-iPSCs consistently resulted in a significant decrease in ribonuclear foci (based on ≈1500 nuclei counted). In particular, the foci: nucleus ratio ranged between 0.92 ± 0.37 to 1.23 ± 0.38 which was significantly decreased (***P < 0.001; **P < 0.01; *P < 0.05) compared to control DM1-iPSCs cells transfected with RNP complexes containing Cas9 and scrambled sgRNA (5.44 ± 0.78 to 6.40 ± 0.41) or transfected with 5′-CTGrepeat-sgRNA and 3′-CTGrepeat-sgRNA without Cas9 protein (4.64 ± 0.26 to 5.88 ± 0.15) (Figure 5A; Supplementary Figure S11A–D). Similarly, a significant proportion of DM1–iPSCs nuclei that underwent gene-editing with the RNP-CRISPR/Cas9 complexes contained no ribonuclear foci (% ribonuclear foci-negative nuclei ranged from 59.84 ± 3.9% to 67.9 ± 2.35% between different DM1–iPSCs lines) which was significantly increased (***P < 0.001) compared to control DM1–iPSCs transfected with RNP complexes containing Cas9 and scrambled sgRNA (% ribonuclear foci-negative nuclei = 0%) or only sgRNA 5′-CTGrepeat-sgRNA and 3′-CTGrepeat-sgRNA but no Cas9 protein (% ribonuclear foci-negative nuclei = 0%) (Figure 5B; Supplementary Figure S11A–D). The reduction in ribonuclear foci after gene editing with CRISPR/Cas9 was consistent with the previous data on excision of CTG repeats in DM1-iPSC-Myo cells and DM1 myoblasts. Conversely, the presence of ribonuclear foci in the control conditions (i.e. without Cas9 protein; or with scrambled sgRNA and Cas9 protein) resembled that of uncorrected DM1-iPSCs and DM1-iPSC-Myo cells (Figure 1B and D), consistent with the presence of the CTG repeat expansion in the DMPK 3′ UTR locus (Figure 1C and F).
Figure 5.
Analysis of CRISPR/Cas9 corrected DM1-iPSCs and isolated DM1-iPSC clones by Southern blot assay, target region sequencing, TP-PCR and ribonuclear foci staining. (A) Quantification of ribonuclear foci (NF) in CRISPR/Cas9-corrected DM1-iPSCs. The total number of ribonuclear foci per total number of nuclei was calculated. Total of nuclei counted is 1500. The data is represented as mean ± SEM. The statistics were performed using two-tailed unpaired Student's t-test (***P <0.001; **P < 0.01; *P < 0.05). (B) Graph shows the % of ribonuclear foci-negative nuclei in the CRISPR/Cas9-corrected DM1-iPSCs to determine the overall efficiency of CRISPR/Cas9 correction. A total number of 1500 nuclei were examined by FISH staining. The data is represented as mean ± SEM. The statistics were performed using one-way ANOVA (***P <0.001; **P < 0.01; *P < 0.05). (C) Southern blot analysis to detect the presence of expanded trinucleotide CTG repeat region in 23 DM1-iPSC clones isolated from four DM1-iPSC lines derived from two DM1 patients (L81 and L23; FL8 and FL5) and healthy control iPSCs. EcoRI digested genomic DNA was subjected to agarose gel electrophoresis and probed to detect the DMPK locus. (mut = mutant; wt = wild type). (D) Representative sequences obtained from sanger sequencing of target locus in each of the four selected DM1-iPSC clones (L23: C03, L81:C10, FL8:C09 and FL5:C12). For ease of alignment we have placed a reference sequence with 5′ and 3′-CTGrepeat-gRNA target sites (green and blue) and protospacer adjacent motif (PAM) region (red) highlighted. In the sequence alignment deleted region has been marked with (–) and insertions highlighted in lowercase. (E) Representative electropherograms of Triplet Repeat Primed PCR (TP) products from DM1-iPSC clones (corrected clone: L23 C03; non-corrected clone: L23 C32) isolated after CRISPR/Cas9-mediated gene editing. (F) Representative images of a corrected (L23 C03) and non-corrected (L23 C32) DM1-iPSC clone stained for ribonuclear foci. An antisense Cy3-labeled probe was used to detect the presence of ribonuclear foci (NF). The ribonuclear foci negative and positive nuclei were denoted as NF− (white) and NF+ (red), respectively. Nuclei were counter-stained with DAPI (scale bar = 20 μm). Each representative image is a maximum intensity z projection of the z slices. For both clones enlarged z slices of selected ribonuclear foci negative (NF–) or positive (NF+) nucleus are represented.
Triplet repeat primed PCR and ribonuclear foci analysis of individual CRISPR/Cas-corrected DM1-iPSC clones
Single-cell clones were obtained by limiting dilution from the DM1-iPSC populations derived from each of the four lines (L23, L81 from DM1 patient #1; FL5, FL8 from DM1 patient #2) nucleofected with the RNP-CRISPR/Cas9 complexes. Ribonuclear foci staining and triplet repeat primed PCR (TP-PCR) analysis of a total of 161 individual clones isolated from the four DM1-iPSCs lines transfected with the RNP-CRISPR/Cas9 complexes revealed that 142 clones were corrected and 19 were non-corrected. In particular, TP-PCR analysis revealed a pronounced difference in the TP-PCR signal intensity plots (i.e. electropherograms) of the corrected DM1-iPSC clones compared to untreated DM1-iPSC controls. In contrast, the electropherograms of the non-corrected DM1-iPSC clones were similar to the electropherograms obtained from untreated DM1-iPSC controls. Representative electropherograms are shown (Figure 5E; Supplementary Figure S12A and B). Analysis of ribonuclear foci staining revealed that the corrected DM1-iPSC clones were devoid of ribonuclear foci as compared to non-corrected clones, where we could detect presence of ribonuclear foci. Representative ribonuclear foci FISH staining (maximum intensity z projection of z slices) of corrected vs. non-corrected DM1-iPSC clones are shown (Figure 5F; Supplementary Figure S13). The ribonuclear foci staining of the individual clones were entirely consistent with the electropherogram results.
Southern blot analysis and target region sequencing of individual CRISPR/Cas-corrected DM1-iPSC clones
Subsequently, based on the TP-PCR and ribonuclear foci analysis using FISH (Figure 5E and F; Supplementary Figures S13 and S12A, B), we selected a total of 23 (17 corrected and 6 non-corrected) DM1-iPSC clones for Southern blot analysis to confirm the excision of the expanded CTG repeat. A probe specific for the DMPK 3′ UTR region that harbors the expanded CTG repeat was employed (Supplementary Figure S4A).
The Southern blot analysis demonstrated that the corrected DM1-iPSC clones (FL8: c14, c10, c09, c07; FL5: c17, c13, c12, c05; L81: c25, c19, c14, c10, c06; L23: c30, c10, c03, c01) lacked the characteristic mutated expanded allele (mut) containing the expanded CTG repeat as compared to their respective non-corrected DM1-iPSC clones (FL8: c35, c01; FL5: c28; L81: c08; L23: c32, c06) (Figure 5C). The disappearance of the large-sized bands in the corrected DM1-iPSC clones is consistent with the lack of large-sized bands in healthy iPSC controls. There is a well-known polymorphism in the DMPK locus that can be attributed to the presence or absence of an Alu repeat (4,52,53). This Alu polymorphism accounts for the presence of two distinct bands in the healthy iPSC controls. Hence, one of the wild-type DMPK alleles in the healthy iPSC control contains an Alu repeat (designated as Alu+) whereas the Alu repeat was absent in the other wild-type allele (designated as Alu-). Similarly, in the corrected DM1-iPSC clones derived from FL8 (c14, c10, c09, c07) and FL5 (c17, c13, c12, c05) DM1-iPSCs, we could detect two distinct bands corresponding to the polymorphic Alu+ and Alu- wild-type DMPK alleles, consistent with the same two bands in healthy iPSCs (Figure 3C). The corrected DM1-iPSC clones derived from L81 DM1-iPSCs (c25, c19, c14, c10, c06) and L23 DM1-iPSCs (c30, c10, c03, c01) had a single band that corresponded to the upper (Alu+) band from healthy iPSC controls (Figure 5C). The non-corrected DM1-iPSC clones (L81: c08; L23: c32, c06; FL8: c35, c01; FL5: c28) were heterozygous and contained a mutated DMPK allele (denoted as mut) with a characteristic CTG repeat expansion and a wild-type DMPK allele (denoted as wt) (Figure 5C). CTG repeat expansion in the non-corrected DM1-iPSC clones derived from FL8 and FL5 (FL8: c35, c01; FL5: c28) ranged from ∼1400 to ∼1700 repeats, whereas in case of L81 and L23 (L81: c08; L23: c32, c06) the range was between ∼1100 to ∼1800 repeats. The L23-c32 exhibited two repeat sizes (Figure 5C), which may be attributed to repeat instability incurred during the clonal expansion, as reported previously (48–51). The Southern blot analysis was consistent with the results obtained by TP-PCR and ribonuclear foci analysis, providing direct proof that the CRISPR/Cas9-mediated excision of the expanded CTG repeats in DM1-iPSC cells resulted in phenotypic correction.
Subsequently, we selected one corrected clone from each of the four DM1-iPSCs (L81: c10; L23: c03; FL8: c09, c07; FL5: c12) for target region sequencing of both DMPK alleles in each clone. The target region was PCR amplified, cloned and sequenced by Sanger sequencing. Typically, up to 16 cloned amplicons were sequenced for each clone. The sequencing results (Figure 5D) confirmed excision of the expanded CTG repeat in the CRISPR/Cas9-corrected DM1-iPSC clones. This is consistent with the CRISPR/Cas9-induced double-strand DNA cut and indel formation in close proximity to the consensus NGG PAM sequences that are located upstream and downstream of the expanded CTG repeat (Figure 5D). Two possible sequences could be retrieved for each of the clones, indicating that CRISPR/Cas9 targeted both the mutant and wild-type DMPK alleles. The consequences of excising the CTG repeat from the 3′UTR of the wild-type DMPK allele are unknown. If future studies would reveal some unintended consequences, then further fine-tuning of the editing strategy may ultimately be needed to achieve allele-specific gene editing. Overall, the consistency of the data obtained by Southern blot analysis, Sanger sequencing, TP-PCR and ribonuclear foci analysis (FISH) validates each of these methods and underscores the robustness of this comprehensive analysis.
DISCUSSION
Advances in genome engineering have led to gene editing tools being increasingly used for addressing monogenic genetic disorders. This has revolutionized the field of gene therapy in making important strides forward in the treatment of recessive genetic diseases, which has recently witnessed several gene therapy products receiving regulatory approval (65–67). Monogenic recessive diseases that result in a loss-of-function associated with a specific mutation can be corrected by the addition of a functional wild-type copy of the cognate mutated gene. However, treating dominant genetic disorders by addition of a functional gene copy will neither suffice to compensate for the genetic defect nor reverse the underlying pathology. Instead, specific inactivation of the dominant gain-of-function allele is required, which is more challenging. The emergence of programmable, nucleotide-specific gene-editing technologies based on CRISPR/Cas9 opens new perspectives to correct such dominant genetic disorders.
In the current study, we established proof-of-concept using a cellular disease model whereby a dominant genetic defect (i.e. DM1) could be specifically corrected using CRISPR/Cas9. Skeletal muscle dysfunction represents one of the main clinical manifestations of DM1. To validate CRISPR/Cas9 for gene editing of DM1, we therefore relied not only on DM1 patient-specific primary myoblasts but also on DM1 patient-specific iPSCs and DM1-iPSC-derived myogenic cells (27). DM1 myoblasts, DM1-iPSCs and DM1-iPSC-Myo cells distinguish themselves from healthy cells by the nuclear accumulation of ribonuclear foci. This was consistent with the abnormal sequestration of MBNL1 and the aberrant splicing pattern (e.g. SERCA1) in case of the terminally differentiated myotubes and myocytes. Consequently, these DM1 patient-specific myogenic cellular models replicates many of the key features of DM1 in vitro and may therefore prove useful to validate new therapeutic strategies (in casu gene editing) or to ultimately gain a deeper understanding of the underlying pathophysiological mechanisms of DM1. One of the main strengths of the current proof-of-concept study is that it is based on DM1 patient-derived myogenic cells, capable of muscle tissue regeneration and replicating muscle function in vitro. They could potentially be used to replace dysfunctional and degrading muscle tissue in an autologous setting (26,27). These DM1 patient-derived myogenic cells are non-transformed and offer an attractive model system to better understand and characterize the genetic and cellular consequences of CRISPR/Cas9-mediated targeting at the DMPK locus, as shown in the current study. Though transformed cell lines could be considered as alternatives, it is well established that they typically tend to skew the overall gene targeting efficacies and often have abnormal DNA repair characteristics, raising questions over their clinical relevance (68,69). Moreover, the cellular consequences of gene correction may differ compared to when non-transformed cells are employed (70,71). The availability of this DM1-iPSC-Myo cellular model therefore complements the use of alternative DM1 disease-specific transformed cell lines (68,69,72–76).
To correct the mutant DMPK allele encoding the pathogenic CUGexpRNA, we designed a strategy that relied on a dual gRNA approach to target the flanking regions just 5′-upstream and 3′-downstream of the CTG repeat expansion. This likely caused a Cas9-mediated double-strand DNA break at the respective sites in the DMPK locus in close proximity to the PAM consensus sequence, followed by the subsequent excision of the entire ∼ 4.1 kb CTG repeat and re-ligation of the juxtaposed DNA fragments, in agreement with the sequencing data obtained by Sanger and SMRT sequencing. In particular, we also demonstrate that the large >1200 trinucleotide (CTG) repeat expansion could be excised with relatively high efficiency and fidelity by CRISPR/Cas9, obviating the need for further selective enrichment of gene-edited cells (69). Most importantly, by excising the CTG expanded repeats in the DMPK 3′ UTR, the DMPK protein coding sequence itself remained unchanged and the DMPK mRNA expression level remained unchanged. Moreover, instead of relying on a single gRNA as this would increase the likelihood of obtaining mainly partial deletions of the CTG repeat, our strategy ensured complete excision of trinucleotide repeat region. This distinguishes the current approach from previous gene editing attempts based on homologous directed repair (HDR) using TALENs that resulted in a truncated DMPK protein and were overall less efficient compared to the current approach (77,78). Consequently, CRISPR/Cas9-mediated deletion of the CTG repeat results in a functional correction of the quintessential DM1 disease manifestations at the cellular and molecular level, consistent with the disappearance of the ribonuclear foci, restoration of MBNL1 sequestration and correction of the spliceopathy based on SERCA1 splicing. SERCA1 in particular has been found to be associated with impaired Ca2+ homeostasis in DM1 muscle that may account, at least in part, to the abnormal muscle function (33).
The molecular consequences of the CRISPR/Cas9-mediated excision of the CTG repeat in the DMPK 3′UTR were analyzed by TP-PCR, Sanger sequencing, single-molecule real-time (SMRT) sequencing and Southern blot analysis, providing direct evidence of CTG repeat excision in the DMPK target locus. The results were consistent across the different assays used. SMRT sequencing bypasses critical limitations of conventional technologies in the context of highly repetitive conditions such as trinucleotide expansions, even enabling detection of minor allelic variants that occur at lower frequency. In particular, measurements from isolated molecules overcome the problems of sample heterogeneity and diminishing resolution inherent in bulk sequencing approaches (56,79). Moreover, SMRT sequencing has a higher accuracy and is able to sequence through large (tandem) repeats compared to existing ‘next generation’ sequencing platforms (56,57). Indeed, we have previously used and validated this method to detect AGG interruptions in the quintessential CGG repeat expansion of the FMR1 gene (79). To our knowledge, our current study is the first to implement SMRT sequencing technology to characterize the CTG repeat expansion in DM1 after CRISPR/Cas9-mediated gene editing. We believe that this represents a technological advancement over existing methodologies with broader implications for the gene-editing field at large and for triplet nucleotide repeat expansion in particular. Moreover, SMRT also obviates concerns associated with the use of internal primers that bind within the CTG repeat sequence, as in the case of TP-PCR.
SMRT sequencing allowed us to readily distinguish wild-type uncut alleles from targeted DMPK alleles (wild-type or mutant). In some cases, it was possible to unambiguously demonstrate targeting of the wild-type allele due to the presence of indels in proximity of the respective PAM sites located upstream or downstream of the normal CTG repeat with the characteristic low number of repeats (wild-type allele in 2 patients: 5 or 13 repeats) (Supplementary Figures S7A and S7B). Similarly, it was also possible to demonstrate targeting of the mutant allele due to the presence of indels in proximity of the respective PAM sites located upstream or downstream of the CTG repeat expansion (Supplementary Figures S7A and S7B). Using SMRT sequencing we could not discriminate between excisions of the CTG repeat in mutant versus wild-type alleles. However, only excision of the pathogenic expanded CTG repeat (and not of the wild-type CTG repeat) would have resulted in ribonuclear foci negative nuclei or an overall decrease in ribonuclear foci: nucleus ratio. Hence, the number of ribonuclear foci-negative cells provide a reliable means to assess the net effect of the CRISPR/Cas9-mediated CTG repeat excision of the pathogenic allele and does not take into account the effect of CRISPR/Cas9-medaited excision of the CTG repeat in wild-type allele.
To better understand the consequences of the CRISPR/Cas9 editing on CTG repeat excision of wild-type vs. mutant DMPK alleles, we conducted Southern blot analysis on individual clones derived from CRISPR/Cas9-corrected DM1-iPSCs. To introduce the CRISPR/Cas9 components into the DM1-iPSCs we explored the use of a non-viral gene correction method based on nucleofection of ribonucleoprotein (RNP) complexes composed of Cas9 proteins and synthetic single guide gRNA (sgRNA). This overcomes the limitations of using lentiviral vectors for delivery of CRISPR/Cas9 components and therefore eliminates the risk of insertional oncogenesis. Remarkably, robust correction efficiencies upto 90% could be attained in DM1-iPSCs as confirmed at the clonal level by ribonuclear foci analysis following ribonucleoprotein (RNP) transfection of CRISPR/Cas9 components without the need for selective enrichment. Most importantly, Southern blot analysis confirmed that the phenotypic correction was due to excision of the expanded CTG repeat.
Southern blot analysis, SMRT sequencing, allele-specific PCR and Sanger sequencing of target region (Figures 3A, 2D, 5C and D; Supplementary Figures S6B–C, S7A–B) experimentally also demonstrated that both the mutant and wild-type DMPK 3′UTR had undergone specific editing by CRISPR/Cas9 in both cellular models (i.e. DM1-iPSCs or DM1-iPSC-Myo). This is consistent with the fact that the gRNAs used to target the sequences upstream and downstream of the CTG repeat were identical between the wild type and mutant DMPK alleles. Hence, based on the current CRISPR/Cas9 strategy, it was not possible to obtain specific excision of the CTG repeat only in the mutant DMPK allele and not in the wild-type allele. However, it is reassuring that excision of the CTG repeat in either the wild type or mutant DMPK locus does not result in any mutation of the DMPK protein itself, since the repeat is located in the 3′UTR. Moreover, qRT-PCR demonstrated that CRISPR/Cas9-mediated excision of the CTG repeat did not affect the steady-state DMPK mRNA levels (Supplementary Figure S8D).
In conclusion, CRISPR/Cas9-mediated excision of the CTG repeat sequences in the wild type or mutant DMPK alleles might not result in any qualitative or quantitative differences in DMPK protein expression minimizing the risk of untoward side effects. Nevertheless, a targeting strategy specific for the mutant DMPK allele would still be preferred. This could potentially be achieved by exploiting the presence of single nucleotide polymorphisms (SNPs) that discriminate the wild type from mutant DMPK alleles. In particular, the occurrence of SNPs in the NGG consensus PAM site provides a particularly attractive strategy to develop an allele-specific targeting approach that discriminates between wild type and mutant alleles (80). One limitation of this approach however, in contrast to our current strategy, is that it would not be generally applicable to all DM1 patients since it would depend on specific SNPs. Targeting the gRNAs to the CTG repeat region itself or to the junction would not have made sense since it would likely also result in the targeting of many other genes with CTG repeats, resulting in potential off-target effects.
Taken together, our results suggest that CRISPR/Cas9 is an attractive platform to potentially correct dominant genetic diseases by robust gene editing, including other disorders caused by large nucleotide repeat expansion, such as Huntington's disease, spinocerebellar ataxia and fragile X syndrome. To our knowledge, there are several novel aspects associated with our study. Our study achieves triplet repeat excision in patient-derived iPSCs and differentiated myogenic cells (as opposed to immortalized cell lines). Substantially higher correction efficiencies (i.e. at least 40-fold higher) were attained in the present study (up to 90% using CRISPR/Cas9-sgRNA RNP complex transfection of DM1-iPSCs) compared to the relatively modest correction efficiencies recently reported in fragile X syndrome (FXS) iPSCs or ES cells (24). In that study only 2 to 3 edited iPSC or ESC-derived clones with CGG repeat deletions were established out of approximately 100 clones (2–3% efficiency). In our study relatively high correction efficiencies obviate the need to screen and selectively pick individual clones (69), which is not always possible with primary cells, without inducing cellular transformation (as in the case of mesangioblasts or iPSC-Myo cells). Moreover, the correction of abnormal CTG repeat expansion in DM1 was achieved without the use of a wild-type allele or the use of an exogenous donor sequence, but rather through NHEJ induced by the Cas9 nuclease, eliminating the need for selective enrichment of homologous recombinants (81).
The aforementioned Park et al. study (24) focuses on fragile X syndrome (FXS), an X-linked neurological disorder, resulting from a CGG repeat expansion in the fragile X mental retardation 1 (FMR1) gene. It is clearly distinct in terms of its underlying pathological mechanisms, disease manifestations and affects distinct target tissues and organs compared to DM1. Indeed, our current study is complementary to the study of Park and colleagues (24) as it shows the impact of gene editing of triplet repeats on the spliceopathy, intrinsic to DM1 in muscle cells, as opposed to the reversion of FMR1 promoter methylation in the selected clones upon neuronal differentiation (24). We demonstrated CRISPR/Cas9-mediated excision of trinucleotide repeat expansions that extend upto >1200 repeats (corresponding to >3600 nucleotides). To achieve this, we had designed a dual gRNA strategy to target the flanking regions just 5′-upstream and 3′-downstream of the CTG repeat expansion. The approach described by Park et al., relied on just a single gRNA upstream of the CGG repeat sequence of the FMR1 gene which is a much more contracted repeat expansion (i.e. 200–450 repeats) (24). It seems unlikely that this single gRNA-based strategy described by Park et al. would have sufficed to obtain efficient excision of the very large extended repeat expansions in the DMPK locus. The deletion triggered by just a single Cas9-induced double-strand DNA break, would have to encompass the entire CTG repeat region (i.e. > 3600 nucleotides).
Based on the relative transduction efficiencies of the Cas9 and gRNA lentiviral vectors, we estimated that the percentage of DM1-iPSC-Myo cells containing the 3 vectors corresponded to ∼70–80%. Hence, the requirement for triple co-transduction to obtain efficient CTG repeats excision explains, at least in part, why the decrease in ribonuclear foci could not attain 100%. Nevertheless, considering overall gene correction efficiencies over 50% (Figure 3B,C and D; Supplementary Figure S8A-C) and taking the actual transduction efficiencies into account, this would imply that nearly 80% of the DM1-iPS-Myo cells that contained the 3 vectors (i.e. Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA vectors) were actually phenotypically corrected. To further improve the overall gene editing efficiency, future studies should ideally incorporate the Cas9, 5′-CTGrepeat-gRNA and 3′-CTGrepeat-gRNA expression cassettes into a single vector backbone, obviating the need for triple lentiviral co-transductions or by using CRISPR/Cas9-sgRNA RNP transfections as shown in the current study.
Although our results with excising repeats show great promise, an important question for future studies is the need to address the induction of off-target mutations. This would need to be addressed prior to use of any CRISPR-Cas9 based strategy for therapeutics. Recent work has described a number of genome-wide methods for identifying off-target effects of CRISPR-Cas9 nucleases (82–87). In this study, we examined relatively small number of potential off-target sites predicted by an in silico program for mutations as judged by targeted next-generation sequencing. Although this approach yielded encouraging result that no indel mutations were found at these sites, we acknowledge and are aware of limitations in the capability of the in silico program we used to identify bona fide off-target mutations (83). Our future work will therefore focus on implementing genome-wide strategies, like GUIDE-seq or CIRCLE-seq (83,88), which will more comprehensively assess for off-target effects induced by pairs of gRNAs used in our studies. Alternatively, whole-genome sequencing (WGS) could be used to assess the presence of indels and single nucleotide variants (SNVs). Indeed, a recent study demonstrated that WGS analysis of both indels and SNVs as the most thorough method for identifying off-target mutations and showed an unexpectedly high number of SNVs compared to the widely accepted assumption that CRISPR causes mostly indels at regions homologous to the gRNA (89). Nevertheless, this study is somewhat controversial since published sequence data were used as reference rather than use of a control mice not injected with the Cas9 protein [(Wilson et al.- The experimental design and data interpretation in ‘Unexpected mutations after CRISPR Cas9 editing in vivo’ by Schaefer et al. are insufficient to support the conclusions drawn by the authors. BioRxiv, 2017; doi: https://doi.org/10.1101/153338)]. Moreover, that study was underpowered as it was based on a limited number of animals. Given this caveat it is challenging to tease out which variants are due to the natural genetic variation caused by genetic drift that pre-existed prior to the CRISPR treatment and which ones are CRISPR-related. Additionally, we note that it may be possible to further enhance on-target editing and minimize the risk of putative off-target effects by implementing our strategy using the latest generation of higher-fidelity CRISPR/Cas9 systems (90,91), which were developed and described as we were carrying out the current study.
It has been shown that CRISPR/Cas9-mediated cleavage could potentially result in chromosomal translocations (92–94). Hence, it cannot be excluded that cleavage of the wild type and mutant DMPK allele could trigger translocations between homologous chromosomes carrying either the wild type or mutant DMPK allele. This would recreate the wild type or mutant DMPK allele, and would dilute off any net benefit. Indels could potentially be incorporated at the translocation site but since this site is expected to be located in the 3′UTR it is unlikely that this would trigger serious adverse consequences. Nevertheless, reciprocal chromosomal translocations could potentially also occur between the actual target site and any off-target sites that may result in unexpected, potentially adverse effects.
Targeting the CTG repeat in the 3′UTR of the DMPK gene by CRISPR/Cas9 complements previous therapeutic strategies that are typically focusing on inhibiting the CUGexp RNA instead (95–100). Prior to moving towards clinical applications in DM1 patients it is important to first demonstrate that DM1 patient-derived muscle cells could be corrected by CRISPR/Cas9. This has now been demonstrated in the current study using both primary myoblasts as well as myogenic cells derived from DM1 patient-specific iPSCs. Future in vivo gene editing approaches in DM1 patients will likely result in the editing of resident myogenic stem cells in the muscle tissues (21). Hence, it is particularly important to establish proof of concept that CRISPR/Cas9 can phenotypically correct myogenic cells from DM1 patients since they could contribute significantly to muscle repair and potentially enhance the overall efficacy of the gene editing strategy. Therefore, it is particularly encouraging that relatively robust gene correction efficiencies could be attained in such non-transformed DM1 cell lines, which are more representative of what could be expected in primary cells or myogenic progenitor cells in human. This would allow us to investigate the consequences of transient expression of the CRISPR/Cas9 components on the overall targeting efficiency of the expanded CTG repeat compared to what can be achieved with bona fide integrating vectors as a benchmark. Moreover, we could substitute the use of retroviral vectors for reprogramming with potentially safer methods based on non-integrating vectors instead (101–103). The nature of the reprogramming protocol is unlikely to have any effect on the outcome of the DMPK targeting itself with CRISPR/Cas9.
In the next stage, it will be equally important to demonstrate that CRISPR/Cas9 could be used to correct the DM1 disease phenotype in vivo in a preclinical animal model that mimics the cognate human disease. Several transgenic DM1 mouse models have been generated based on the inclusion of a CTG repeat expansion in the DMPK locus (104–106). These preclinical models replicate some of the pathological features of DM1 observed in patients and are therefore well suited to assess the efficacy of CRISPR/Cas9-based excision of the CTG repeat in vivo. It is also promising that recent studies have exhibited that the triplet repeat expansion in the huntington (HTT) locus could be excised by CRISPR/Cas9-based gene editing in a mouse model of Huntington's disease (107). This further accentuates the potential of using CRISPR/Cas9 for in vivo gene editing of DM1, which we are now going to assess in a DM1 mouse model. This may ultimately lead to a new clinically relevant approach for DM1 by in vivo muscle-directed CRISPR/Cas9-mediated gene editing (18,19,21,108).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Anneleen Bogaerts, Sabrina D’Haese and Mia Vercammen for technical assistance and Wilfried Cools for assistance with statistical analysis.
Author Contributions: S.D. designed and performed experiments, analyzed data and wrote the manuscript. S.A., K.S., N.N., E.S., D.M., W.D., T.J., J.T., W.T., H.W. and Y.C.C. performed experiments and analyzed data. S.S., P.I., J.R.V. designed experiments and analyzed data. A.F.K. and D.F. provided samples, optimized experimental conditions and provided critical comments on manuscript. M.F.M.G. and F.S.T. performed experiments, analyzed data and provided critical comments on manuscript. Y.F., D.R. and J.K.J. designed and performed experiments and helped in drafting the manuscript. M.K.C. and T.V. designed experiments, conceived and coordinated the project and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds voor Wetenschappelijke Onderzoek (FWO); Koning Boudewijn Stichting – Walter Pyleman Fund; VUB/UZ Willy Gepts grant, VUB Strategic Research Program ‘Groeier’; VUB Industrieel Onderzoeksfonds (Groups of Expertise in Applied Research (GEAR), AFM grant (to M.C. and T.V.); National Institutes of Health (NIH) Director's Pioneer Award DP1 [GM105378 to J.K.J.]. Funding for open access charge: FWO.
Conflict of interest statement. J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Beacon Genomics, Editas Medicine, Poseida Therapeutics and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
REFERENCES
- 1. Udd B., Krahe R.. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012; 11:891–905. [DOI] [PubMed] [Google Scholar]
- 2. Theadom A., Rodrigues M., Roxburgh R., Balalla S., Higgins C., Bhattacharjee R., Jones K., Krishnamurthi R., Feigin V.. Prevalence of muscular dystrophies: a systematic literature review. Neuroepidemiology. 2014; 43:259–268. [DOI] [PubMed] [Google Scholar]
- 3. Fu Y.H., Pizzuti A., Fenwick R.G. Jr., King J., Rajnarayan S., Dunne P.W., Dubel J., Nasser G.A., Ashizawa T., de Jong P. et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992; 255:1256–1258. [DOI] [PubMed] [Google Scholar]
- 4. Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O’Hoy K. et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992; 255:1253–1255. [DOI] [PubMed] [Google Scholar]
- 5. Savic Pavicevic D., Miladinovic J., Brkusanin M., Svikovic S., Djurica S., Brajuskovic G., Romac S.. Molecular genetics and genetic testing in myotonic dystrophy type 1. Biomed. Res. Int. 2013; 2013:391821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Groh W.J., Groh M.R., Shen C., Monckton D.G., Bodkin C.L., Pascuzzi R.M.. Survival and CTG repeat expansion in adults with myotonic dystrophy type 1. Muscle Nerve. 2011; 43:648–651. [DOI] [PubMed] [Google Scholar]
- 7. Mulders S.A., van Engelen B.G., Wieringa B., Wansink D.G.. Molecular therapy in myotonic dystrophy: focus on RNA gain-of-function. Hum. Mol. Genet. 2010; 19:R90–R97. [DOI] [PubMed] [Google Scholar]
- 8. Day J.W., Ranum L.P.. RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord. 2005; 15:5–16. [DOI] [PubMed] [Google Scholar]
- 9. Klein A.F., Dastidar S., Furling D., Chuah M.K.. Therapeutic approaches for dominant muscle Diseases: Highlight on myotonic dystrophy. Curr. Gene Ther. 2015; 15:329–337. [DOI] [PubMed] [Google Scholar]
- 10. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S.. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010; 468:67–71. [DOI] [PubMed] [Google Scholar]
- 13. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. Elife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young C.S., Hicks M.R., Ermolova N.V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J.A. et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-Derived muscle cells. Cell Stem Cell. 2016; 18:533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan Y., Ma Y., Li Q., Sun Z., Ma L., Wu L., Wang L., Zeng L., Shao Y., Chen Y. et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol. Med. 2016; 8:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N.. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016; 351:400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016; 351:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park C.Y., Kim D.H., Son J.S., Sung J.J., Lee J., Bae S., Kim J.H., Kim D.W., Kim J.S.. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a Patient-Derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015; 17:213–220. [DOI] [PubMed] [Google Scholar]
- 21. Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016; 351:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016; 34:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016; 34:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park C.Y., Halevy T., Lee D.R., Sung J.J., Lee J.S., Yanuka O., Benvenisty N., Kim D.W.. Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep. 2015; 13:234–241. [DOI] [PubMed] [Google Scholar]
- 25. DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Munoz D.P., Boffelli D. et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016; 8:360ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tedesco F.S., Gerli M.F., Perani L., Benedetti S., Ungaro F., Cassano M., Antonini S., Tagliafico E., Artusi V., Longa E. et al. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci. Transl. Med. 2012; 4:140ra189. [DOI] [PubMed] [Google Scholar]
- 27. Maffioletti S.M., Gerli M.F., Ragazzi M., Dastidar S., Benedetti S., Loperfido M., VandenDriessche T., Chuah M.K., Tedesco F.S.. Efficient derivation and inducible differentiation of expandable skeletal myogenic cells from human ES and patient-specific iPS cells. Nat. Protoc. 2015; 10:941–958. [DOI] [PubMed] [Google Scholar]
- 28. Cossu G., Bianco P.. Mesoangioblasts–vascular progenitors for extravascular mesodermal tissues. Curr. Opin. Genet. Dev. 2003; 13:537–542. [DOI] [PubMed] [Google Scholar]
- 29. Dellavalle A., Sampaolesi M., Tonlorenzi R., Tagliafico E., Sacchetti B., Perani L., Innocenzi A., Galvez B.G., Messina G., Morosetti R. et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat. Cell Biol. 2007; 9:255–267. [DOI] [PubMed] [Google Scholar]
- 30. Loperfido M., Steele-Stallard H.B., Tedesco F.S., VandenDriessche T.. Pluripotent stem cells for gene therapy of degenerative muscle diseases. Curr. Gene Ther. 2015; 15:364–380. [DOI] [PubMed] [Google Scholar]
- 31. Loperfido M., Jarmin S., Dastidar S., Di Matteo M., Perini I., Moore M., Nair N., Samara-Kuko E., Athanasopoulos T., Tedesco F.S. et al. piggyBac transposons expressing full-length human dystrophin enable genetic correction of dystrophic mesoangioblasts. Nucleic Acids Res. 2016; 44:744–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S.. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872. [DOI] [PubMed] [Google Scholar]
- 33. Kimura T., Nakamori M., Lueck J.D., Pouliquin P., Aoike F., Fujimura H., Dirksen R.T., Takahashi M.P., Dulhunty A.F., Sakoda S.. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005; 14:2189–2200. [DOI] [PubMed] [Google Scholar]
- 34. Gunkel M., Chung I., Worz S., Deeg K.I., Simon R., Sauter G., Jones D.T., Korshunov A., Rohr K., Erfle H. et al. Quantification of telomere features in tumor tissue sections by an automated 3D imaging-based workflow. Methods. 2017; 114:60–73. [DOI] [PubMed] [Google Scholar]
- 35. Parker M., Chen X., Bahrami A., Dalton J., Rusch M., Wu G., Easton J., Cheung N.K., Dyer M., Mardis E.R. et al. Assessing telomeric DNA content in pediatric cancers using whole-genome sequencing data. Genome Biol. 2012; 13:R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taneja K.L. Localization of trinucleotide repeat sequences in myotonic dystrophy cells using a single fluorochrome-labeled PNA probe. Biotechniques. 1998; 24:472–476. [DOI] [PubMed] [Google Scholar]
- 37. Francois V., Klein A.F., Beley C., Jollet A., Lemercier C., Garcia L., Furling D.. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat. Struct. Mol. Biol. 2011; 18:85–87. [DOI] [PubMed] [Google Scholar]
- 38. Holt I., Mittal S., Furling D., Butler-Browne G.S., Brook J.D., Morris G.E.. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells. 2007; 12:1035–1048. [DOI] [PubMed] [Google Scholar]
- 39. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singh K., Evens H., Nair N., Rincón M.Y., Sarcar S., Samara-Kuko E., Chuah M.K., VandenDriessche T.. Efficient In Vivo Liver-Directed Gene Editing Using CRISPR/Cas9. Mol. Ther. 2018; 26:1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. VandenDriessche T., Thorrez L., Naldini L., Follenzi A., Moons L., Berneman Z., Collen D., Chuah M.K.. Lentiviral vectors containing the human immunodeficiency virus type-1 central polypurine tract can efficiently transduce nondividing hepatocytes and antigen-presenting cells in vivo. Blood. 2002; 100:813–822. [DOI] [PubMed] [Google Scholar]
- 42. Matrai J., Cantore A., Bartholomae C.C., Annoni A., Wang W., Acosta-Sanchez A., Samara-Kuko E., De Waele L., Ma L., Genovese P. et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011; 53:1696–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Du H., Cline M.S., Osborne R.J., Tuttle D.L., Clark T.A., Donohue J.P., Hall M.P., Shiue L., Swanson M.S., Thornton C.A. et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010; 17:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fugier C., Klein A.F., Hammer C., Vassilopoulos S., Ivarsson Y., Toussaint A., Tosch V., Vignaud A., Ferry A., Messaddeq N. et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011; 17:720–725. [DOI] [PubMed] [Google Scholar]
- 45. Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A.. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006; 15:2087–2097. [DOI] [PubMed] [Google Scholar]
- 46. Dryland P.A., Doherty E., Love J.M., Love D.R.. Simple Repeat-Primed PCR analysis of the myotonic dystrophy type 1 gene in a clinical diagnostics environment. J. Neurodegener. Dis. 2013; 2013:857564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lavedan C., Hofmann-Radvanyi H., Shelbourne P., Rabes J.P., Duros C., Savoy D., Dehaupas I., Luce S., Johnson K., Junien C.. Myotonic dystrophy: size- and sex-dependent dynamics of CTG meiotic instability, and somatic mosaicism. Am. J. Hum. Genet. 1993; 52:875–883. [PMC free article] [PubMed] [Google Scholar]
- 48. De Temmerman N., Seneca S., Van Steirteghem A., Haentjens P., Van der Elst J., Liebaers I., Sermon K.D.. CTG repeat instability in a human embryonic stem cell line carrying the myotonic dystrophy type 1 mutation. Mol. Hum. Reprod. 2008; 14:405–412. [DOI] [PubMed] [Google Scholar]
- 49. Gomes-Pereira M., Fortune M.T., Monckton D.G.. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 2001; 10:845–854. [DOI] [PubMed] [Google Scholar]
- 50. Gomes-Pereira M., Hilley J.D., Morales F., Adam B., James H.E., Monckton D.G.. Disease-associated CAG.CTG triplet repeats expand rapidly in non-dividing mouse cells, but cell cycle arrest is insufficient to drive expansion. Nucleic Acids Res. 2014; 42:7047–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seriola A., Spits C., Simard J.P., Hilven P., Haentjens P., Pearson C.E., Sermon K.. Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum. Mol. Genet. 2011; 20:176–185. [DOI] [PubMed] [Google Scholar]
- 52. Mahadevan M.S., Amemiya C., Jansen G., Sabourin L., Baird S., Neville C.E., Wormskamp N., Segers B., Batzer M., Lamerdin J. et al. Structure and genomic sequence of the myotonic dystrophy (DM kinase) gene. Hum. Mol. Genet. 1993; 2:299–304. [DOI] [PubMed] [Google Scholar]
- 53. Zerylnick C., Torroni A., Sherman S.L., Warren S.T.. Normal variation at the myotonic dystrophy locus in global human populations. Am. J. Hum. Genet. 1995; 56:123–130. [PMC free article] [PubMed] [Google Scholar]
- 54. Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H.. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995; 128:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wojciechowska M., Krzyzosiak W.J.. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011; 20:3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loomis E.W., Eid J.S., Peluso P., Yin J., Hickey L., Rank D., McCalmon S., Hagerman R.J., Tassone F., Hagerman P.J.. Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 2013; 23:121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McFarland K.N., Liu J., Landrian I., Godiska R., Shanker S., Yu F., Farmerie W.G., Ashizawa T.. SMRT sequencing of long tandem nucleotide repeats in SCA10 reveals unique insight of repeat expansion structure. PLoS One. 2015; 10:e0135906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mittelman D., Moye C., Morton J., Sykoudis K., Lin Y., Carroll D., Wilson J.H.. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Richard G.F., Viterbo D., Khanna V., Mosbach V., Castelain L., Dujon B.. Highly specific contractions of a single CAG/CTG trinucleotide repeat by TALEN in yeast. PLoS One. 2014; 9:e95611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang W., Zheng J., He Y., Luo C.. Tandem repeat modification during double-strand break repair induced by an engineered TAL effector nuclease in zebrafish genome. PLoS One. 2013; 8:e84176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cradick T.J., Fine E.J., Antico C.J., Bao G.. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013; 41:9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S.. A muscleblind knockout model for myotonic dystrophy. Science. 2003; 302:1978–1980. [DOI] [PubMed] [Google Scholar]
- 64. Dixon D.M., Choi J., El-Ghazali A., Park S.Y., Roos K.P., Jordan M.C., Fishbein M.C., Comai L., Reddy S.. Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci. Rep. 2015; 5:9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aiuti A., Cattaneo F., Galimberti S., Benninghoff U., Cassani B., Callegaro L., Scaramuzza S., Andolfi G., Mirolo M., Brigida I. et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 2009; 360:447–458. [DOI] [PubMed] [Google Scholar]
- 66. Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011; 365:2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Boztug K., Schmidt M., Schwarzer A., Banerjee P.P., Diez I.A., Dewey R.A., Bohm M., Nowrouzi A., Ball C.R., Glimm H. et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010; 363:1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. van Agtmaal E.L., Andre L.M., Willemse M., Cumming S.A., van Kessel I.D.G., van den Broek W., Gourdon G., Furling D., Mouly V., Monckton D.G. et al. CRISPR/Cas9-Induced (CTGCAG)n repeat instability in the myotonic dystrophy type 1 Locus: Implications for therapeutic genome editing. Mol. Ther. 2017; 25:24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Provenzano C., Cappella M., Valaperta R., Cardani R., Meola G., Martelli F., Cardinali B., Falcone G.. CRISPR/Cas9-Mediated Deletion of CTG Expansions Recovers Normal Phenotype in Myogenic Cells Derived from Myotonic Dystrophy 1 Patients. Mol Ther Nucleic Acids. 2017; 9:337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014; 510:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C.. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005; 435:646–651. [DOI] [PubMed] [Google Scholar]
- 72. Gonzalez-Barriga A., Mulders S.A., van de Giessen J., Hooijer J.D., Bijl S., van Kessel I.D., van Beers J., van Deutekom J.C., Fransen J.A., Wieringa B. et al. Design and analysis of effects of triplet repeat oligonucleotides in cell models for myotonic dystrophy. Mol. Ther. Nucleic Acids. 2013; 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ketley A., Chen C.Z., Li X., Arya S., Robinson T.E., Granados-Riveron J., Udosen I., Morris G.E., Holt I., Furling D. et al. High-content screening identifies small molecules that remove nuclear foci, affect MBNL distribution and CELF1 protein levels via a PKC-independent pathway in myotonic dystrophy cell lines. Hum. Mol. Genet. 2014; 23:1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dansithong W., Jog S.P., Paul S., Mohammadzadeh R., Tring S., Kwok Y., Fry R.C., Marjoram P., Comai L., Reddy S.. RNA steady-state defects in myotonic dystrophy are linked to nuclear exclusion of SHARP. EMBO Rep. 2011; 12:735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ravel-Chapuis A., Belanger G., Yadava R.S., Mahadevan M.S., DesGroseillers L., Cote J., Jasmin B.J.. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012; 196:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hoskins J.W., Ofori L.O., Chen C.Z., Kumar A., Sobczak K., Nakamori M., Southall N., Patnaik S., Marugan J.J., Zheng W. et al. Lomofungin and dilomofungin: inhibitors of MBNL1-CUG RNA binding with distinct cellular effects. Nucleic Acids Res. 2014; 42:6591–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xia G., Gao Y., Jin S., Subramony S., Terada N., Ranum L.P., Swanson M.S., Ashizawa T.. Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 ips-cell derived neural stem cells. Stem Cells. 2015; 33:1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gao Y., Guo X., Santostefano K., Wang Y., Reid T., Zeng D., Terada N., Ashizawa T., Xia G.. Genome therapy of myotonic dystrophy type 1 iPS cells for development of autologous stem cell therapy. Mol. Ther. 2016; 24:1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ardui S., Race V., Zablotskaya A., Hestand M.S., Van Esch H., Devriendt K., Matthijs G., Vermeesch J.R.. Detecting AGG interruptions in male and female FMR1 premutation carriers by single-molecule sequencing. Hum. Mutat. 2017; 38:324–331. [DOI] [PubMed] [Google Scholar]
- 80. Shin J.W., Kim K.H., Chao M.J., Atwal R.S., Gillis T., MacDonald M.E., Gusella J.F., Lee J.M.. Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016; 25:4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. An M.C., O’Brien R.N., Zhang N., Patra B.N., De La Cruz M., Ray A., Ellerby L.M.. Polyglutamine disease modeling: epitope based screen for homologous recombination using CRISPR/Cas9 system. PLoS Curr. 2014; 6:doi:10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W.. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015; 33:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015; 33:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S.. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015; 12:237–243. [DOI] [PubMed] [Google Scholar]
- 85. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang X., Wang Y., Wu X., Wang J., Qiu Z., Chang T., Huang H., Lin R.J., Yee J.K.. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015; 33:175–178. [DOI] [PubMed] [Google Scholar]
- 87. Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G.. COSMID: A Web-based tool for identifying and validating CRISPR/Cas Off-target sites. Mol. Ther. Nucleic Acids. 2014; 3:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K.. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods. 2017; 14:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schaefer K.A., Wu W.H., Colgan D.F., Tsang S.H., Bassuk A.G., Mahajan V.B.. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods. 2017; 14:547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Blasco R.B., Karaca E., Ambrogio C., Cheong T.C., Karayol E., Minero V.G., Voena C., Chiarle R.. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014; 9:1219–1227. [DOI] [PubMed] [Google Scholar]
- 93. Maddalo D., Manchado E., Concepcion C.P., Bonetti C., Vidigal J.A., Han Y.C., Ogrodowski P., Crippa A., Rekhtman N., de Stanchina E. et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014; 516:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Torres R., Martin M.C., Garcia A., Cigudosa J.C., Ramirez J.C., Rodriguez-Perales S.. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat. Commun. 2014; 5:3964. [DOI] [PubMed] [Google Scholar]
- 95. Kole R., Krainer A.R., Altman S.. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012; 11:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Langlois M.A., Boniface C., Wang G., Alluin J., Salvaterra P.M., Puymirat J., Rossi J.J., Lee N.S.. Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J. Biol. Chem. 2005; 280:16949–16954. [DOI] [PubMed] [Google Scholar]
- 97. Mulders S.A., van den Broek W.J., Wheeler T.M., Croes H.J., van Kuik-Romeijn P., de Kimpe S.J., Furling D., Platenburg G.J., Gourdon G., Thornton C.A. et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Phylactou L.A., Darrah C., Wood M.J.. Ribozyme-mediated trans-splicing of a trinucleotide repeat. Nat. Genet. 1998; 18:378–381. [DOI] [PubMed] [Google Scholar]
- 99. Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A.. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012; 488:111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang W., Wang Y., Dong S., Choudhury R., Jin Y., Wang Z.. Treatment of type 1 myotonic dystrophy by engineering site-specific RNA endonucleases that target (CUG)(n) repeats. Mol. Ther. 2014; 22:312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jia F., Wilson K.D., Sun N., Gupta D.M., Huang M., Li Z., Panetta N.J., Chen Z.Y., Robbins R.C., Kay M.A. et al. A nonviral minicircle vector for deriving human iPS cells. Nat. Methods. 2010; 7:197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K.. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009; 458:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Takahashi K., Yamanaka S.. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016; 17:183–193. [DOI] [PubMed] [Google Scholar]
- 104. Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A.. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000; 289:1769–1773. [DOI] [PubMed] [Google Scholar]
- 105. Seznec H., Agbulut O., Sergeant N., Savouret C., Ghestem A., Tabti N., Willer J.C., Ourth L., Duros C., Brisson E. et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet. 2001; 10:2717–2726. [DOI] [PubMed] [Google Scholar]
- 106. Wang G.S., Kearney D.L., De Biasi M., Taffet G., Cooper T.A.. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007; 117:2802–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yang S., Chang R., Yang H., Zhao T., Hong Y., Kong H.E., Sun X., Qin Z., Jin P., Li S. et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease. J. Clin. Invest. 2017; 127:2719–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. VandenDriessche T., Chuah M.K.. CRISPR/Cas9 flexes its Muscles: In vivo somatic gene editing for muscular dystrophy. Mol. Ther. 2016; 24:414–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.