Abstract
According to the traditional view, GTPases act as molecular switches, which cycle between distinct ‘on’ and ‘off’ conformations bound to GTP and GDP, respectively. Translation elongation factor EF-Tu is a GTPase essential for prokaryotic protein synthesis. In its GTP-bound form, EF-Tu delivers aminoacylated tRNAs to the ribosome as a ternary complex. GTP hydrolysis is thought to cause the release of EF-Tu from aminoacyl-tRNA and the ribosome due to a dramatic conformational change following Pi release. Here, the crystal structure of Escherichia coli EF-Tu in complex with a non-hydrolysable GTP analogue (GDPNP) has been determined. Remarkably, the overall conformation of EF-Tu·GDPNP displays the classical, open GDP-bound conformation. This is in accordance with an emerging view that the identity of the bound guanine nucleotide is not ‘locking’ the GTPase in a fixed conformation. Using a single-molecule approach, the conformational dynamics of various ligand-bound forms of EF-Tu were probed in solution by fluorescence resonance energy transfer. The results suggest that EF-Tu, free in solution, may sample a wider set of conformations than the structurally well-defined GTP- and GDP-forms known from previous X-ray crystallographic studies. Only upon binding, as a ternary complex, to the mRNA-programmed ribosome, is the well-known, closed GTP-bound conformation, observed.
INTRODUCTION
Proteins are highly dynamic in nature, and most often, their motions are tightly coupled to functionality. The P-loop NTPase (nucleoside triphosphatase) superfamily (1) presents numerous examples of proteins that undergo substantial structural transitions as part of their functional cycles, which are related to diverse cellular processes such as transport, signal transduction and translation to mention a few.
Among the NTPases, the GTPase superfamily has been subject to thorough structural studies since the 1980s (2). These studies established that the identity of the bound guanine nucleotide (i.e. GDP versus GTP) gives rise to distinct structural conformations (3). The guanine-nucleotide binding domains (or G-domains) of all family members display a conserved overall structure and five characteristic consensus sequence motifs with individual roles in GDP/GTP binding (4). The switching between an active, GTP-bound form and an inactive, GDP-bound form is common to the GTPases and is often regulated by guanine-nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). Nucleotide binding and hydrolysis are accompanied by structural changes in two regions, switch I and II, which are thought to be a direct consequence of the presence of either GDP or GTP. A more detailed scheme has however been suggested, in which both structural forms are able to bind both nucleotides (5). In the case of translation elongation factor Tu (EF-Tu), this was recently highlighted by the finding that EF-Tu·GDP may dissociate from the ribosome in either an open or a closed conformation (6).
Elongation factor Tu is a GTPase with an essential role in prokaryotic protein synthesis (7). During the translation elongation cycle, aminoacylated tRNAs (aa-tRNA) are brought to the A site of the mRNA-programmed ribosome in a ternary complex consisting of aa-tRNA, EF-Tu, and GTP. Cognate codon–anticodon interaction induces conformational changes in the ribosome, which stimulates the GTPase activity of EF-Tu. GTP hydrolysis and release of Pi result in a conformational rearrangement of EF-Tu leading to release from the ribosome. Reactivation of EF-Tu via guanine-nucleotide exchange is facilitated by elongation factor Ts (EF-Ts).
Numerous structures have been determined of EF-Tu from various organisms covering most states during its catalytic cycle. The initial structure of E. coli EF-Tu in complex with GDP showed that EF-Tu consists of three domains organized with a characteristic open cleft in the middle (8–15). Subsequently, crystal structures of EF-Tu from T. aquaticus and T. thermophilus in complex with the GTP analogue guanosine 5′-[β,γ-imido]triphosphate (GDPNP) revealed that EF-Tu adopts a more compact or closed conformation in the GTP-bound state as opposed to the open GDP-bound state (16,17). The relative orientation of domains II and III is essentially the same in the open and the closed conformation. In contrast, domain I is rotated approximately 90° relative to domains II and III between the GTP- and GDP-bound state and the switch 1 and 2 regions (residues 40–62 and 80–100, respectively, in E. coli EF-Tu) are extensively rearranged. A part of the switch 1 region (residues 52–59) changes from a β-hairpin in the EF-Tu·GDP structure to an α-helix in the EF-Tu·GDPNP structure, while the position of an α-helix of the switch 2 region is shifted in the primary sequence. Crystal structures of various ternary complexes show that EF-Tu has a closed conformation, similar to the free EF-Tu·GDPNP structure, when bound to aa-tRNA. The aa-tRNA recognition is mediated by residues from all three domains of EF-Tu ((18–20) and PDB ID 1OB2). The structure of the ternary complex trapped on the 70S ribosome shows that EF-Tu essentially adopts the same overall closed conformation as in the free ternary complex (21,22).
Here, we present the first crystal structure of E. coli EF-Tu in complex with the non-hydrolysable GTP analogue, GDPNP, at a resolution of 2.50 Å. Remarkably, EF-Tu displays an open conformation similar to the GDP-bound form of the factor. This finding is complemented by a study of the conformational dynamics of EF-Tu in solution. Pairs of fluorescent labels were placed in domains I and III to enable fluorescence resonance energy transfer (FRET) as a measure of interdomain distance to allow distinction between the open and closed forms of EF-Tu and possibly provide evidence for any intermediate or alternate conformations. FRET measurements in the presence of various ligands revealed that EF-Tu in solution exists in a dynamic equilibrium of conformations with similar FRET distributions, irrespective of the nature of the bound guanine nucleotide, even in the ternary complex with aa-tRNA. The GTP-bound form is driven towards the previously described closed conformation only upon binding to the ribosome.
MATERIALS AND METHODS
A more detailed description of the applied methods is included in the online Supplementary Data.
Purification of E. coli EF-Tu for crystallographic studies
E. coli EF-Tu was purified from Rosetta (DE3) cells by anion chromatography. The protein was eluted with a linear gradient of 50–300 mM KCl. Eluted fractions containing EF-Tu were precipitated in 2.8 M ammonium sulphate. The precipitated protein was further purified by hydrophobic interaction chromatography, with elution by a linear gradient of 1.5–0.75 M ammonium sulphate. The eluted fractions containing EF-Tu were dialyzed and subjected to a second round of anion exchange chromatography.
Nucleotide exchange and crystallization
Purified EF-Tu was dialyzed against nucleotide exchange buffer (200 mM ammonium sulphate, 30 mM Tris pH 8.0, 1 mM DTT and 10% glycerol). Subsequent additions brought MgCl2 to a final concentration of 1 mM, GDPNP to a molecular ratio of 5:1 with respect to EF-Tu, and alkaline phosphatase to 2 units per mg EF-Tu. The solution was incubated on ice and subsequently clarified by centrifugation. The EF-Tu·GDPNP complex was purified by gel filtration. E. coli EF-Tu·GDPNP was concentrated to 8 mg/ml prior to crystallization experiments.
Crystallization trials were carried out at 20°C utilizing the sitting drop vapor diffusion technique. The initial crystallization conditions were established through commercial crystal screens. Subsequently, the crystallization buffer was optimized to 2 M ammonium sulphate, 0.1 M HEPES pH 7.5, 2% (v/v) PEG 400 and 1 mM DTT and single needle crystals were obtained by streak-seeding the drops with serial dilutions of crushed crystals. Single crystals were isolated from the mother liquor and rapidly transferred to cryoprotection buffer (2 M ammonium sulphate, 0.1 M HEPES 7.5 (NaOH) and 20% glycerol) and immediately flash frozen. Data were collected at Maxlab beamline I911-2 (Lund, Sweden).
Data processing and structure determination
Data processing and scaling were performed with the XDS package (23). Phases were determined by molecular replacement with the Phaser application of Phenix (24). Search models were prepared from the structure of E. coli EF-Tu bound to GDP (PDB ID 1EFC) (10). The guanine nucleotide and all water molecules were removed and the individual domains were separated and used in combination as search models. Model building was performed in Coot (25) and the fitted model was refined with Phenix refine (24). The final model were validated with Procheck (26) and MolProbity (27).
Preparation of EF-Tu for solution FRET studies
We sought to detect structural transitions between the open and closed conformations of EF-Tu by introducing a FRET donor (Cy3) and acceptor (Cy5) in each of the domains I and III, at positions which are predicted to move relative to each other during the structural transitions. Sites for introducing Cys residues for labeling with maleimide dyes were selected based on the analysis of the following structures: 70S with stalled Thermus thermophilus EF-Tu·GDP·kirromycin·aa-tRNA (PDB codes 1WRN, 1WRO), E. coli EF-Tu·GDPNP·kirromycin·aa-tRNA (PDB code: 1OB2) and EF-Tu·GDP (PDB code: 1DG1). Three pairs of amino acid residues, T33/M351, D47/D314 and D165/D314 (Supplementary Figure S1), were chosen for labeling, and cysteines were introduced at these positions in a variant of EF-Tu, denoted EF-TuAV, which contains only one native cysteine, as described in the accompanying paper (28). The resulting mutants, denoted EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314, were expressed, purified, labeled and characterized as described elsewhere (28).
FRET states of dual-labeled EF-Tu mutants monitored by single-molecule confocal fluorescence burst detection
Freely diffusing EF-Tu molecules labeled with both fluorescent donor and acceptor (Cy3 and Cy5, respectively) gave rise to bursts of fluorescence on traversing the detection volume of a confocal optical system built around an inverted microscope (29,30). Low concentrations (50 pM) of the relevant dual-labeled complexes were used to minimize the observation of more than one EF-Tu molecule at any given time. Individual fluorescence photons from freely diffusing complexes of labeled EF-Tu were detected as they passed through the confocal volume using avalanche photodiodes (29,30), and binned into donor (Cy3) and acceptor (Cy5) channels by a Flex 03-LQ-01 correlator adapter card with a 500 μs integration time. A thresholding scheme applied to the sum of the two channels was used to determine when a molecule was occupying the detection volume. Adjacent time bins which were part of the same event were merged (see Supplement). The resulting counts on the donor and acceptor channels, nCy3 and nCy5, were used to calculate an apparent FRET efficiency 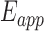 = (nCy5 – χnCy3) / (nCy5 – χnCy3 + nCy3) for each EF-Tu molecule. Here, the bleed-through parameter, χ, was estimated using molecules with only a donor label. The data were not corrected for differential detector sensitivity or quantum yields, so FRET values should be interpreted as reporting on relative distances. Histograms representing the distribution of FRET efficiencies for individual EF-Tu molecules reveal different populations of conformations. The histograms were fitted by the sum of three beta distributions by maximum likelihood estimation with the software application MEMLET (31). Uncertainties of each fit parameter were estimated via boot-strapping (see Supplement). For each component beta distribution, we report the average apparent FRET efficiency, EApp, total counts for each population, A, and beta component widths, w along with their 95% confidence intervals [C.I.] (Supplementary Table S1). The standard errors, δ, reported in the text, are defined as the span of the 95% confidence interval divided by four. Equations for the beta distribution, EApp, and w are given in the Supplement. One of these sub-populations, with a transfer efficiency near zero (Figure 3), is a robust artefact resulting from molecules without the acceptor dye or with a bleached acceptor dye. We identify the remaining two subpopulations as distinct conformational states of EF-Tu.
= (nCy5 – χnCy3) / (nCy5 – χnCy3 + nCy3) for each EF-Tu molecule. Here, the bleed-through parameter, χ, was estimated using molecules with only a donor label. The data were not corrected for differential detector sensitivity or quantum yields, so FRET values should be interpreted as reporting on relative distances. Histograms representing the distribution of FRET efficiencies for individual EF-Tu molecules reveal different populations of conformations. The histograms were fitted by the sum of three beta distributions by maximum likelihood estimation with the software application MEMLET (31). Uncertainties of each fit parameter were estimated via boot-strapping (see Supplement). For each component beta distribution, we report the average apparent FRET efficiency, EApp, total counts for each population, A, and beta component widths, w along with their 95% confidence intervals [C.I.] (Supplementary Table S1). The standard errors, δ, reported in the text, are defined as the span of the 95% confidence interval divided by four. Equations for the beta distribution, EApp, and w are given in the Supplement. One of these sub-populations, with a transfer efficiency near zero (Figure 3), is a robust artefact resulting from molecules without the acceptor dye or with a bleached acceptor dye. We identify the remaining two subpopulations as distinct conformational states of EF-Tu.
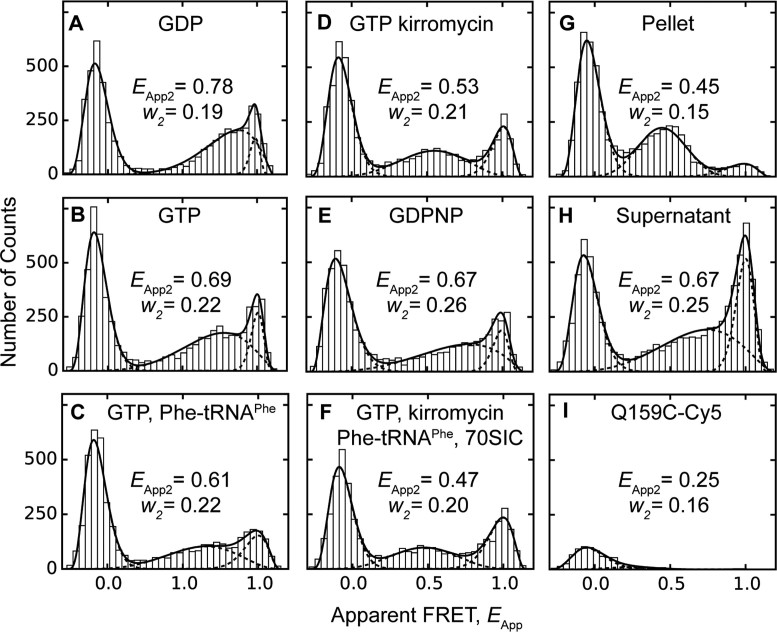
Figure 3.
FRET distributions of dual-labeled EF-TuAV-33/351 on and off the ribosome. Dual-labeled EF-TuAV-33/351 was observed for 10 min under a confocal microscope at 50 pM concentration to detect fluorescence bursts under different conditions: (A) 1 mM GDP; (B) 1 mM GTP, 3 mM PEP, 0.005 mg/ml pyruvate kinase; (C) 2 μM Phe-tRNAPhe, 1 mM GTP, 3 mM PEP, 0.005 mg/ml pyruvate kinase; (D) 1 mM GTP, 3 mM PEP, 0.005 mg/ml pyruvate kinase, 30 μM kirromycin; (E) 1 mM GDPNP; (F) 2 μM Phe-tRNAPhe, 500 pM 70SIC, 1 mM GTP, 3 mM PEP, 0.005 mg/ml pyruvate kinase, 30 μM kirromycin; (G) pellet of EF-TuAV-33/351·GDP·kirromycin·Phe-tRNAPhe·70SIC obtained after ultracentrifugation through a sucrose cushion; (H) supernatant from (G). (I) A negative control containing the Cy5-labeled EF-Tu mutant Q159C showed no fluorescence bursts in the high-FRET region, when illuminated with a 514-nm laser. The common peak below EApp = 0.2 is an artefact resulting from molecules with no or inactive acceptor fluorophores. Fitted apparent FRET values, EApp, total counts for each population, A, and beta distribution widths, w, are provided in Supplementary Table S1. A statistical comparison of EApp2 and w2 values obtained for the mid-FRET peaks under different conditions are provided in Supplementary Table S2. The EApp3 values of the high FRET peaks falling in the range 0.95–0.97 displayed no significant differences. Individual beta function fits are shown as dashed lines, while the solid line corresponds to their sum.
RESULTS
Overall structure of E. coli EF-Tu·GDPNP
Full length E. coli EF-Tu (residues 1–393) was purified and crystallized in complex with GDPNP. X-ray diffraction data were collected to 2.5 Å resolution (Table 1). The crystals belong to the space group P21212 with the unit cell parameters a = 61.24 Å, b = 243.59 Å and c = 67.13 Å. The structure was determined by molecular replacement using coordinates extracted from the E. coli EF-Tu·GDP structure (PDB: 1EFC; (10)) as the search model.
Table 1.
Data collection and refinement statistics. Statistics for the highest-resolution shell are shown in parentheses
| E. coli EF-Tu·GDPNP | |
|---|---|
| Wavelength (Å) | 1.0379 |
| Resolution range (Å) | 29.43–2.47 (2.56–2.47) |
| Space group | P 21 21 2 |
| Unit cell (Å) | 61.24 243.60 67.13 |
| Total reflections | 283 684 (24 406) |
| Unique reflections | 36 491 (3509) |
| Multiplicity | 7.8 (7.0) |
| Completeness (%) | 98.29 (95.64) |
| Mean I/sigma(I) | 18.1 (4.2) |
| Wilson B-factor | 34.03 |
| R-merge | 10.4 (76.3) |
| CC1/2 | 99.6 (79.9) |
| Reflections used in refinement | 36 409 (3468) |
| Reflections used for R-free | 2068 (196) |
| R-work | 0.1708 (0.2892) |
| R-free | 0.2099 (0.3192) |
| Number of non-hydrogen atoms | 6341 |
| Macromolecules | 5942 |
| Ligands | 149 |
| Solvent | 250 |
| Protein residues | 772 |
| RMS(bonds) (Å) | 0.003 |
| RMS(angles) (°) | 0.61 |
| Ramachandran favored (%) | 97.66 |
| Ramachandran outliers (%) | 0.00 |
| Average B-factor | 40.12 |
| Macromolecules | 39.97 |
| Ligands | 44.46 |
| Solvent | 41.28 |
The crystal unit cells contained two copies of EF-Tu·GDPNP, two Mg2+ ions and 362 water molecules. Both protein chains were unambiguously traced in the electron density from Thr8 to Ser393. Structure statistics for the model of E. coli EF-Tu·GDPNP are provided in Table 1.
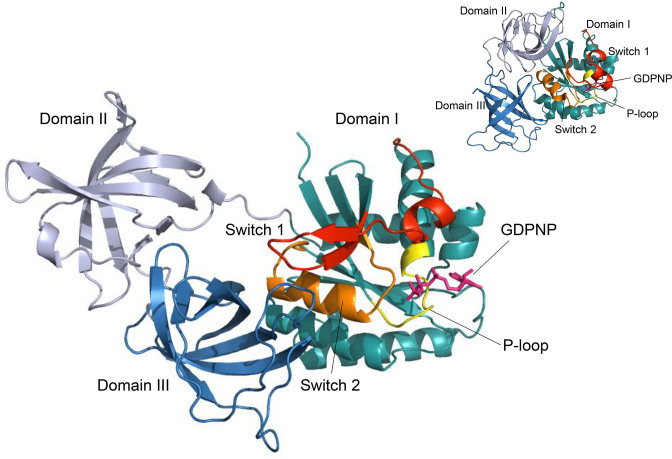
Each chain of EF-Tu·GDPNP in the final model consists of three domains (Figure 1) in agreement with previously determined structures. The three domains have a mutual orientation resembling the open, GDP-bound conformation of other known EF-Tu structures (8–11,15) despite unambiguous electron density for the γ-phosphate of the GDPNP molecule (Figure 2B).
Figure 1.
The open structure of EF-Tu·GDPNP. The cartoon representation is colour coded as follows: Domain I (residues 1–200) is cyan except for the Switch 1 region (red), the switch 2 region (orange), and the P-loop (yellow). Domain II (residues 201–300) is coloured light blue and Domain III (residues 301–393) is dark blue. GDPNP is shown as magenta sticks. In the upper right corner, the closed conformation of EF-Tu (PDB: 1OB2) is shown for comparison. The G domains are shown in the same orientation for the two conformations.
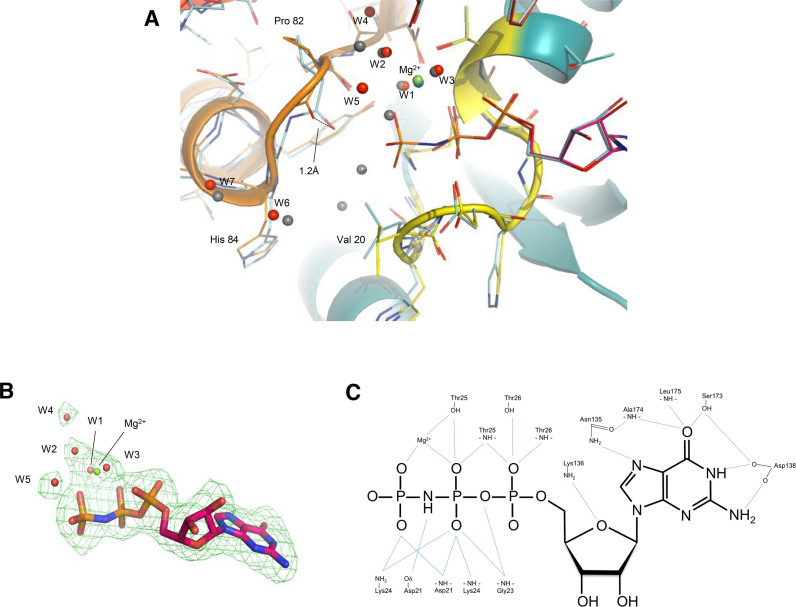
Figure 2.
Details of GDPNP binding. (A) Superpositioning of E. coli EF-Tu·GDPNP and E. coli EF-Tu·GDP (PDB:1EFC). EF-Tu·GDPNP is shown in cartoon format and residues of the P-loop and switch 2 regions are shown as lines. The colour coding is the same as in Figure 1. Water molecules are shown as red spheres, and the Mg2+ ion as a green sphere. For EF-Tu·GDP, residues of the P-loop and switch I and II are shown as light blue lines, while water molecules and Mg2+ are shown as grey spheres. (B) Omit map density for GDPNP. The Fo-Fc omit map density was contoured at 0.319 e-/Å3 corresponding to 3.0σ with a cushion of 4 Å. (C) Details of GDPNP binding. The hydrogen bonds (distance: <3.2 Å) between GDPNP and EF-Tu as well as the coordination of Mg2+ by GDPNP and EF-Tu are indicated by light blue lines. Interactions involving water molecules are not shown.
The space group and the unit cell parameters are essentially identical to those of E. coli EF-Tu·GDP determined by Song et al. (10). The overall conformation is likewise nearly identical. Superimposing the E. coli EF-Tu·GDPNP and EF-Tu·GDP (PDB:1EFC) structures yields a root-mean-square deviation (rmsd) of 0.200 Å (Figure 2A).
Guanine-nucleotide binding and structure of the switch regions of EF-Tu·GDPNP
Overall, the GDP part of GDPNP is bound as in the previously reported structures of EF-Tu·GDP (8–11,15) via a network of hydrogen bonds involving amino acids located in four loop regions connecting β-strands with α-helices (Figure 2C). In short, the binding of the guanine moiety is mediated by hydrogen bonds to the amino acid residues Asn135, Asp138, Ser173, Ala174 and Leu175 and the aliphatic part of the Lys136 side chain stacks with the guanine ring. The ribose interacts with the amino group of Lys136 and through a water molecule with Thr26. The α-phosphate interacts with Thr26 and a water molecule, which in turn is coordinating the Mg2+ ion. The oxygens of the β-phosphate interact with amides of the P-loop (residues 18–25), the side chain of Thr25, and the amino group of Lys24. One of the β-phosphate oxygens chelates the Mg2+ ion. The only difference in coordination of the Mg2+ ion is the replacement of a water molecule in the GDP-structure with a γ-oxygen of GDPNP in the present structure. Positions of water molecules coordinating the Mg2+ ion are maintained as compared to the GDP structure, while other waters observed near the phosphate binding loop are either absent or have shifted position.
The β,γ-imido group of GDPNP interacts with the carboxylate group of Asp21 as previously observed in EF-Tu·GDPNP structures. The presence of the additional phosphate group causes only minor changes in the structure around the binding site; the side chain of Val20 rotates 120° around chi1, the carbonyl oxygen of Pro82 moves 1.2 Å and this movement propagates as an overall small movement of Cα atoms in the entire switch II region of ∼0.5 Å (Figure 2A). The rotation of Val20 and the interaction of the β,γ-imido group with Asp21 is also seen in structures of EF-Tu in complex with GDPNP in the closed form e.g. in EF-Tu·GDPNP·Phe-tRNAPhe·kirromycin (PDB: 1OB2). In contrast, the interactions observed in the structures of T. aquaticus EF-Tu·GDPNP (17) and T. thermophilus EF-Tu·GDPNP (16) between the γ-phosphate oxygens and the main chain at residues Gly83 and Thr62 as well as the side chain of Thr62 are not present in our structure of E. coli EF-Tu·GDPNP.
Dynamics of freely diffusing EF-Tu analysed by fluorescence burst detection microscopy
The surprising finding that E. coli EF-Tu bound to GDPNP displays the open structure known from the GDP-bound form of the factor was complemented by a study of the dynamic distribution of E. coli EF-Tu conformations in solution using a FRET approach. FRET requires the covalent attachment of fluorescent donor and acceptor fluorophores at positions expected to efficiently report on structural changes. Commonly, the covalent attachment of fluorescent probes occurs via native or engineered cysteine residues (32).
Previously, we have prepared a mutant of E. coli EF-Tu, denoted EF-TuAV, in which the native cysteines at positions 137 and 255 were replaced by alanine and valine, respectively, while the cysteine at position 81 was retained to avoid loss of activity (33). In the EF-TuAV background, pairs of cysteines were introduced at either positions 33 and 351, positions 47 and 314 or positions 165 and 314 to report on relative distance changes between domains I and III after labeling with the FRET reporter pair Cy3 and Cy5 (Supplementary Figure S1). The resulting mutants were named EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314, respectively. Single-molecule total internal reflection fluorescence (TIRF) microscopy of dual-labeled EF-TuAV-33/351 allowed the detection of a structural transition of EF-Tu on the ribosome, which was dependent on GTP hydrolysis. In contrast, the dual-labeled mutants EF-TuAV-47/314 and EF-TuAV-165/314 reported little or no distance change (28).
Here, dual-labeled EF-TuAV-33/351 is subject to a detailed, fluorescence burst detection microscopy analysis to monitor the structural distribution free in solution (29,30), while dual-labeled EF-TuAV-47/314 and EF-TuAV-165/314 are described in less detail because their distance changes were so small.
All mutants were simultaneously labeled with Cy3 and Cy5 maleimides to give a mixture of EF-Tu labeled with both Cy3 and Cy5 (dual-labeled EF-Tu) as well as EF-Tu labeled with Cy3 or Cy5 only. Nearly equal incorporation of Cy3 and Cy5 was observed. Under the applied labeling conditions, the background labeling of Cys81 was ∼0.1 dye/protein, while the total labeling of EF-TuAV-33/351, EF-TuAV-47/314 and EF-TuAV-165/314 were 1.0–1.2 dye/protein (data not shown). In the accompanying study, the three labeled EF-Tu mutants were found to have activities comparable to wild-type EF-Tu with respect to poly(Phe) synthesis, protection of the ester bond of Phe-tRNAPhe from spontaneous hydrolysis, and accommodation of Phe-tRNAPhe into the ribosomal A site (28). The expected distances between the labels in the GDP- and GTP-bound forms (based on PDB entries 1DG1 and 1OB2, respectively) as well as the corresponding theoretical FRET values are given in Supplementary Figure S1. The calculation is based on Ro = 5.0 nm and the assumption that κ2 = 2/3, which is common for relative distance measurements (34).
Fluorescence burst detection of freely diffusing complexes was applied to measure FRET values of dual-labeled EF-TuAV-33/351 both free and bound to the ribosome (Figure 3 and Supplementary Table S1). Distributions of apparent FRET values, EApp, in Figures 3A–H were fitted by a mixture of three beta distributions, showing a peak at the zero position due to EF-Tu molecules containing only Cy3 labels or bleached Cy5, and two non-zero peaks. The higher peak has an essentially constant EApp around 0.97 (EApp3, Supplementary Table S1), while the EApp2 value and width (w2) of the middle peak display significant differences, depending on the EF-Tu complex (Supplementary Tables S1 and S2). The higher EApp peak is not due to direct excitation of Cy5, since an EF-Tu mutant labeled site-specifically with Cy5 at position 159 (Figure 3I) displayed only the peak at zero FRET due to cross-talk between the excitation and detection channels. Thus, the non-zero peaks represent dual-labeled EF-Tu molecules belonging to populations with different FRET efficiencies.
In assessing the significance of the results presented in Figure 3, the three parameters to be considered are the mean FRET efficiency, EApp2, and width, w2, of the intermediate FRET peak, as well as f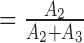 , the ratio of events in the mid-FRET peak to the total number of events in the mid- and high-FRET peaks (Supplementary Table S1). The GDP-bound form of EF-TuAV-33/351 has an intermediate peak corresponding to an apparent FRET efficiency of 0.78 ± 0.01 (EApp2 ± δ2) with width 0.186 ± 0.004 (w2 ± δw2), where δ2 and δw2 are the S.E.M. uncertainties of EApp2 and w2, respectively (Figure 3A, Supplementary Table S1). This is consistent with the expected value of 0.74 based on the structure of the EF-Tu·GDP complex (Supplementary Figure S1; 8,15). In contrast, the EApp2 values related to GTP-like forms of EF-Tu (0.53–0.69, Figures 3B–E) are higher than the expected value of 0.41 based on the structures of the EF-Tu·GDPNP complex (16,17), the ternary complex (18), or the aurodox (a kirromycin analogue) complex of EF-Tu·GDP (35), which is similar to EF-Tu·GTP. The central FRET peaks in the GTP-like forms (panels B-E) are also significantly wider (w2 ≥ 0.21 ± 0.01) than the GDP state (w2 = 0.186 ± 0.004) (Supplementary Table S2). These w2 values are larger than would occur due to the statistics of photon counting in the fluorescence bursts (36) suggesting variability and/or dynamics of the inter-probe distance in the GTP-like states.
, the ratio of events in the mid-FRET peak to the total number of events in the mid- and high-FRET peaks (Supplementary Table S1). The GDP-bound form of EF-TuAV-33/351 has an intermediate peak corresponding to an apparent FRET efficiency of 0.78 ± 0.01 (EApp2 ± δ2) with width 0.186 ± 0.004 (w2 ± δw2), where δ2 and δw2 are the S.E.M. uncertainties of EApp2 and w2, respectively (Figure 3A, Supplementary Table S1). This is consistent with the expected value of 0.74 based on the structure of the EF-Tu·GDP complex (Supplementary Figure S1; 8,15). In contrast, the EApp2 values related to GTP-like forms of EF-Tu (0.53–0.69, Figures 3B–E) are higher than the expected value of 0.41 based on the structures of the EF-Tu·GDPNP complex (16,17), the ternary complex (18), or the aurodox (a kirromycin analogue) complex of EF-Tu·GDP (35), which is similar to EF-Tu·GTP. The central FRET peaks in the GTP-like forms (panels B-E) are also significantly wider (w2 ≥ 0.21 ± 0.01) than the GDP state (w2 = 0.186 ± 0.004) (Supplementary Table S2). These w2 values are larger than would occur due to the statistics of photon counting in the fluorescence bursts (36) suggesting variability and/or dynamics of the inter-probe distance in the GTP-like states.
The free, GTP-bound forms of EF-TuAV-47/314 (Supplementary Figures S2B and S2C) and EF-TuAV-165/314 (Supplementary Figures S2G and S2H) did not show a prominent wide mid-FRET peak similar to that of the free, GTP-bound species of EF-TuAV-33/351 (Figures 3B–E). This may partly be due to the fact that the predicted FRET differences between the structural extremes of EF-TuAV-47/314 and EF-TuAV-165/314 are much smaller than for EF-TuAV-33/351 (Supplementary Figure S1). No mid-FRET peak of the GDP-form of EF-TuAV-47/314 (Supplementary Figure S2A) could be distinguished, possibly due to its merger with the ubiquitous high-FRET peak. For EF-TuAV-165/314, the histogram of the GDP-bound complex (Supplementary Figure S2F) did reveal a small broad peak indicative of a distinct, open conformation, which is notably absent in the histograms of the GTP-bound complexes of this mutant (Supplementary Figures S2G and H).
Ribosome-binding shifts the conformational equilibrium of EF-Tu·GTP towards the active conformation
Our expectation of detecting a distinct closed conformation of EF-Tu bound to small ligands was not realized, since none of the free GTP or GTP analogue forms of EF-TuAV-33/351 (Figures 3B–E) displayed the predicted EApp2 value of 0.41. Likewise, EF-TuAV-47/314 bound to GTP did not show a distinct peak at 0.77 (Supplementary Figures S2B and S2C), while EF-TuAV-165/314 turned out to be a poor reporter of the structural changes due to the omnipresent high-FRET peak that would obscure the expected peak around 0.99.
In order to drive EF-Tu into the closed conformation known from structural studies, the ternary complexes of EF-TuAV-33/351 and EF-TuAV-47/314 were added to a five-fold excess of the 70S initiation complex (70SIC) in the presence of kirromycin. For EF-TuAV-33/351, this caused a reduction in the Eapp2 value to 0.47 ± 0.01 (w2 = 0.20 ± 0.01) (Figure 3F and Supplementary Table S1). Isolation of ribosome-bound complexes in this sample by ultracentrifugation gave a more distinct, narrower (w2 = 0.15 ± 0.004) FRET peak with a similar Eapp2 of 0.45 ± 0.01 (Figure 3G), and a large increase in the area of the intermediate relative to higher FRET peak (f in Supplementary Table S1). Concomitantly, this ratio was decreased in the supernatant resulting after ultracentrifugation (Figure 3H), indicating that the higher FRET (Eapp3 ∼ 0.98) form of dual-labeled EF-TuAV-33/351 is bound to the ribosome more weakly than the intermediate FRET form and may correspond to less active material. Notably, the EApp2 and w2 values observed for ribosome-bound EF-TuAV-33/351 are significantly different from those observed in complexes that are not ribosome-bound (Supplementary Table S2). Importantly, all of these observations are independent of both the fitting procedure employed and the instrument calibration.
For EF-TuAV-47/314, 70SIC binding (Supplementary Figure S2D) cause the appearance of EF-Tu molecules with intermediate FRET efficiencies averaging lower (EApp2 = 0.52 ± 0.03) than the predicted value of 0.77. This discrepancy may reflect a variable position of residue 47, which resides in the flexible switch I region.
DISCUSSION
Our X-ray crystallographic study shows that E. coli EF-Tu bound to GDPNP can take up an open conformation equivalent to the inactive, GDP-bound form of the factor. Single-molecule FRET studies of E. coli EF-Tu bound to various ligands support this finding and show that in solution, EF-Tu samples a set of conformations, which are driven towards the well-known GTP-bound conformation upon binding to the ribosome.
Accommodation of GDPNP in the open conformation of EF-Tu
The accommodation of the additional phosphate of GDPNP in the GDP conformation appears to require small movements of the switch 2 region. These movements do not propagate to the switch 1 region, as observed for Thermus EF-Tu·GDPNP, so that the three domains remain in the open conformation. This indicates that partitioning among the conformations differs for EF-Tu isolated from different sources. A similar observation has been made for members of the Ras-family of small GTPases, which exist in a dynamic equilibrium between an active and an inactive conformation when bound to GDPNP, as observed here for E. coli EF-Tu (Figure 3E). In ras, the conversion between the active and inactive GDPNP-bound form involves mainly the switch regions (37–39). Three substates have been defined for the inactive state of ras, of which substate 1 (H-Ras; PDB: 3KKN) is characterized by the dissociation of Thr35 and Gly60 from the γ-phosphate (40). This is similar to the absence of an interaction between the γ-phosphate and the equivalent Thr61 and Gly83 in the present structure. Thus, the open and closed conformations of EF-Tu·GDPNP may be analogous to the inactive and active forms of GDPNP-bound Ras p21, respectively.
It may appear puzzling why E. coli EF-Tu·GDPNP crystallize in the open conformation as opposed to the closed conformation observed upon crystallization of Thermus EF-Tu·GDPNP. A plausible explanation is that the equilibrium constant for the transition between closed and open GDPNP-bound conformations differs between EF-Tu from different species, and during crystallization different structural states are captured. For Ras isoforms (41) and members of the Ras family of GTPases (42,43), the equilibrium constants governing the structural interconversion between the active and inactive GDPNP-bound states, which correspond to the closed and open forms of GDPNP-bound EF-Tu, respectively, have been shown to differ markedly, likely due to residues flanking the switch 1 region that may play a role in balancing the structural equilibrium (41,44). The sequence alignment of EF-Tu from 897 different species reveals a ‘hotspot’ of variability (residues 37–48 in E. coli EF-Tu) located prior to and in the N-terminal part of the switch 1 region (Supplementary Figure S3). Moreover, the variation is particularly pronounced when comparing E. coli EF-Tu with EF-Tu of thermophilic origin. This diversity may, along with differences in crystallization conditions, account for the distinct states taken up by EF-Tu·GDPNP complexes of different origin upon crystallization.
In the active conformation, EF-Tu is capable of hydrolyzing GTP through activation of a water molecule by His84 (45). In the present structure, this histidine is 2.3 Å further away from the γ-phosphate than observed in the active conformation. Furthermore, we do not observe any water molecules in the structure positioned for a nucleophilic attack. Thus, it is unlikely that GTP hydrolysis occurs in the open conformation. Similarly, the two water molecules essential for intrinsic GTP hydrolysis by H-Ras were found to be absent in the inactive, GDPNP-bound state (46).
Conformational dynamics of EF-Tu in solution
For decades, the accepted view of the superfamily of GTPases has been that the members of this family switch between structurally distinct conformations depending on the nature of the bound nucleotide (2). In particular, X-ray crystallographic studies of EF-Tu (7) off the ribosome have defined two different conformations: the closed, active, GTP-bound form and the open, inactive, GDP-bound form. Recently, the role of conformational dynamics as a regulator of molecular recognition has been highlighted (47). In particular, the model of ‘conformational selection’ (or ‘population shift’) suggests that ligand-binding proteins exist in a dynamic equilibrium of conformations of which only a subset binds ligand. This strategy of ligand binding is opposed to the ‘induced fit’ strategy according to which ligand binding induces a new protein conformation, which leads to tighter binding. For EF-Tu as well as other GTPases, the rigid reaction scheme entailing switching between two structural extremes has become more flexible and dynamic (5), and our data indicate that the dynamics of EF-Tu fit better to a ‘conformational selection’ scheme rather than an ‘induced fit’ model.
Our fluorescence burst detection studies of dual-labeled EF-TuAV-33/351 support the view by Ehrenberg and coworkers (5) that effector binding rather than nucleotide binding defines the conformational state of a GTPase. The EF-TuAV-33/351·GDP complex in solution has an apparent FRET value (0.78 ± 0.01, Figure 3A) consistent with that predicted for the EF-Tu·GDP crystal structures (0.74, Supplementary Figure S1). In contrast, the EF-TuAV-33/351 complexes in solution considered to be ‘EF-Tu·GTP’-like, based on crystal structures (EF-Tu·GDPNP (17), EF-Tu·GDPNP·aa-tRNA (18), EF-Tu·GDP·kirromycin (35)), appear to exist in a dynamic equilibrium of conformations, with apparent FRET values falling on a continuum between those expected for ‘EF-Tu·GTP’ and ‘EF-Tu·GDP’ (0.53–0.69; Figure 3B–E), and with central FRET peaks which are significantly broader (Supplementary Table S2) than that of the GDP state. Indeed, it is only when EF-TuAV-33/351 is bound to its effector, the ribosome, as a kirromycin-inhibited ternary complex, that it displays an apparent FRET value (0.47, with a narrow w2 of 0.15, Figure 3G) similar to the value expected from the EF-Tu·GTP crystal structure (0.41; Supplementary Figure S1). A similar tendency is observed for EF-TuAV-47/314 (Supplementary Figure S2D). Other ribosome-related GTPases exhibit a similar behaviour: EF-G·GTP switches from an elongated to a compact conformation upon binding to the pretranslocation ribosome (48); the GTP-bound form of eIF5B has a higher affinity for the ribosome than the GDP-bound form (49) despite strong similarity in the structures of the free, binary complexes (50); IF2, which appears to be flexible in solution, converts into an extended, well-defined conformation on binding to the ribosome (51); and the conformation of the ribosome-bound stress-response factor BipA differs from that of isolated BipA (52). The conformational equilibrium of GTP-bound Ras-p21 is also shifted towards the active structure upon effector binding (37,41,53–55).
The modest differences within the group of conformations exhibited by the GDP- and GTP-bound forms of EF-TuAV-33/351 reported by FRET (Figure 3A and B) cannot easily explain the difference in aa-tRNA affinities of two orders of magnitude between the two binary complexes in solution (56). However, the apparently more dynamic behaviour of the GTP-bound conformation, as manifested by a higher w2 value, along with a slightly more closed conformation, as measured by EApp2 reporting on the distance between positions 33 and 351, may explain the difference. Upon binding of Phe-tRNAPhe to EF-TuAV-33/351·GTP (Figure 3C), the structural equilibrium is driven further towards the closed conformation as indicated by a decrease in EApp2. Notably, EF-TuAV-33/351 in the ternary complex is quite dynamic (it has the same w2 as the GTP-bound complex) and the fully closed conformation is reached only upon interaction with the 70SIC (Figure 3G) i.e., crystallization and ribosome binding bring EF-Tu·GTP·aa-tRNA into similar low-fluctuation minima (57).
Our observation that EF-Tu adopts more GTP-like conformations on the ribosome may imply a role of EF-Tu during accommodation. Noel and Whitford (58) have shown that on the ribosome, EF-Tu in its GTP-like state reduces the number of conformations available to the transported tRNA molecule and thereby facilitates accommodation of the tRNA elbow. This is in agreement with our accompanying study of conformational dynamics of EF-Tu during aa-tRNA delivery (28), which also highlights a possible involvement of EF-Tu during the accommodation process.
CONCLUSION
Traditionally, the activation of G-proteins has been thought to occur via an induced-fit model, where the presence or absence of the γ-phosphate of GTP determines the functional state of the protein (59). Here, we challenge this view by showing that EF-Tu·GDPNP can take up an open, GDP-like conformation in crystals and in solution. In solution, EF-Tu·GTP adopts a broad spectrum of conformations, whereas EF-Tu·GDP has a more rigid, open conformation. Our data suggest that, in binding EF-Tu·GTP in an active conformation, the ribosome selects from a pool of EF-Tu conformations spanning a continuum from the open to the closed conformation.
DATA AVAILABILITY
The structure has been deposited in the PDB under ID 6EZE.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Karen Margrethe Nielsen and Xiaonan Cui for skilled technical assistance, and Prof. Feng Gai and Dr Lin Guo for providing access to their single-molecule confocal microscope. Recombinant TEV protease was kindly donated by Dr Ditlev E. Brodersen.
Author Contributions: Y.E.G., B.S.C., S.S.T. and C.R.K. conceived the project and advised on experimental design. J.S.J., M.B. and S.S.T. determined the structure of E. coli EF-Tu·GDPNP. D.K. designed, produced, labeled and characterized mutants of EF-Tu. D.K. and S.P. performed single-molecule studies and analysed the data. D.K., S.P., B.S.C., Y.E.G., S.S.T. and C.R.K. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Leo Pharma Research Foundation (to D.K. and C.R.K.); Novo Nordisk Foundation (to C.R.K.); Danish Council for Independent Research (Natural Sciences) (to C.R.K.); National Institutes of Health [R01-GM080376 and R35-GM118139 to Y.E.G. and B.S.C.]. Funding for open access charge: Private Danish Foundations.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sareste M., Sibbald P., Wittinghofer A.. The P-loop - a common motif in ATP-and GTP-binding proteins. Trends Biochem. Sci. 1990; 15:430–440. [DOI] [PubMed] [Google Scholar]
- 2. Bourne H., Sanders D., McCormick F.. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991; 349:117–127. [DOI] [PubMed] [Google Scholar]
- 3. Kjeldgaard M., Nyborg J., Clark B.F.. The GTP binding motif: variations on a theme. FASEB J. 1996; 10:1347–1368. [PubMed] [Google Scholar]
- 4. Wittinghofer A., Vetter I.R.. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011; 80:943–971. [DOI] [PubMed] [Google Scholar]
- 5. Hauryliuk V., Hansson S., Ehrenberg M.. Cofactor dependent conformational switching of GTPases. Biophys. J. 2008; 95:1704–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu W., Chen C., Kavaliauskas D., Knudsen C.R., Goldman Y.E., Cooperman B.S.. EF-Tu dynamics during pre-translocation complex formation: EF-Tu.GDP exits the ribosome via two different pathways. Nucleic Acids Res. 2015; 43:9519–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kavaliauskas D., Nissen P., Knudsen C.R.. The busiest of all ribosomal assistants: elongation factor tu. Biochemistry. 2012; 51:2642–2651. [DOI] [PubMed] [Google Scholar]
- 8. Kjeldgaard M., Nyborg J.. Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol. 1992; 223:721–742. [DOI] [PubMed] [Google Scholar]
- 9. Abel K., Yoder M., Hilgenfeld R., Jurnak F.. An alpha to beta conformational switch in EF-Tu. Structure. 1996; 4:1153–1159. [DOI] [PubMed] [Google Scholar]
- 10. Song H., Parsons M., Rowsell S., Leonard G., Philips S.. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol. 1999; 285:1245–1256. [DOI] [PubMed] [Google Scholar]
- 11. Heffron S., Moeller R., Jurnak F.. Solving the structure of Escherichia coli elongation factor Tu using a twinned data set. Acta Cryst. D Biol. Cryst. 2006; 62:433–438. [DOI] [PubMed] [Google Scholar]
- 12. Andersen G., Thirup S., Spremulli L., Nyborg J.. High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP. J. Mol. Biol. 2000; 297:421–436. [DOI] [PubMed] [Google Scholar]
- 13. Vitagliano L., Masullo M., Sica F., Zagari A., Bocchini V.. The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with GDP reveals novel features in nucleotide binding and exchange. EMBO J. 2001; 20:5305–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vitagliano L., Ruggiero A., Masullo M., Cantiello P., Arcari P., Zagari A.. The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with magnesium and GDP. Biochemistry. 2004; 43:6630–6636. [DOI] [PubMed] [Google Scholar]
- 15. Polekhina G., Thirup S., Kjeldgaard M., Nissen P., Lippmann C., Nyborg J.. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996; 4:1141–1151. [DOI] [PubMed] [Google Scholar]
- 16. Kjeldgaard M., Nissen P., Thirup S., Nyborg J.. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993; 1:35–50. [DOI] [PubMed] [Google Scholar]
- 17. Berchtold H., Reshetnikova L., Reiser C.O., Schirmer N.K., Sprinzl M., Hilgenfeld R.. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993; 365:126–132. [DOI] [PubMed] [Google Scholar]
- 18. Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B.F., Nyborg J.. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995; 270:1464–1472. [DOI] [PubMed] [Google Scholar]
- 19. Parmeggiani A., Krab I.M., Watanabe T., Nielsen R.C., Dahlberg C., Nyborg J., Nissen P.. Enacyloxin IIa pinpoints a binding pocket of elongation factor Tu for development of novel antibiotics. J. Biol. Chem. 2006; 281:2893–2900. [DOI] [PubMed] [Google Scholar]
- 20. Nissen P., Thirup S., Kjeldgaard M., Nyborg J.. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999; 7:143–156. [DOI] [PubMed] [Google Scholar]
- 21. Schmeing T.M., Voorhees R.M., Kelley A.C., Gao Y.G., Murphy F.V.T., Weir J.R., Ramakrishnan V.. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009; 326:688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Voorhees R.M., Schmeing T.M., Kelley A.C., Ramakrishnan V.. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010; 330:835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993; 26:795–800. [Google Scholar]
- 24. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D, Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D, Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 26. Laskowski R., Macarthur M., Moss D., Thornton J.. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993; 26:283–291. [Google Scholar]
- 27. Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. 3rd, Snoeyink J., Richardson J.S. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007; 35:W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kavaliauskas D., Chen C., Liu W., Cooperman B.S., Goldman Y.E., Knudsen C.R.. Structural dynamics of translation elongation factor Tu during aa-tRNA delivery to the ribosome. Nucleic Acids Res. 2018; doi:10.1093/nar/gky651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo L., Chowdhury P., Glasscock J.M., Gai F.. Denaturant-induced expansion and compaction of a multi-domain protein: IgG. J. Mol. Biol. 2008; 384:1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schuler B., LIpman E., Eaton W.. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002; 419:743–747. [DOI] [PubMed] [Google Scholar]
- 31. Woody M.S., Lewis J.H., Greenberg M.J., Goldman Y.E., Ostap E.M.. MEMLET: An Easy-to-Use tool for data fitting and model comparison using maximum-likelihood estimation. Biophys. J. 2016; 111:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kapanidis A., Weiss S.. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J. Chem. Phys. 2002; 117:10953–10964. [Google Scholar]
- 33. Liu W., Kavaliauskas D., Schrader J.M., Poruri K., Birkedal V., Goldman E., Jakubowski H., Mandecki W., Uhlenbeck O.C., Knudsen C.R. et al. Labeled EF-Tus for rapid kinetic studies of pretranslocation complex formation. ACS Chem. Biol. 2014; 19:9519–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dale R., Eisinger J., Blumberg W.. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 1979; 26:161–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vogeley L., Palm G.J., Mesters J.R., Hilgenfeld R.. Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. Crystal structure of the complex between EF-Tu.GDP and aurodox. J. Biol. Chem. 2001; 276:17149–17155. [DOI] [PubMed] [Google Scholar]
- 36. Gopich I.V., Szabo A.. Theory of the statistics of kinetic transitions with application to single-molecule enzyme catalysis. J. Chem. Phys. 2006; 124:154712. [DOI] [PubMed] [Google Scholar]
- 37. Geyer M., Schweins T., Herrmann C., Prisner T., Wittinghofer A., Kalbitzer H.R.. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996; 35:10308–10320. [DOI] [PubMed] [Google Scholar]
- 38. Ito Y., Yamasaki K., Iwahara J., Terada T., Kamiya A., Shirouzu M., Muto Y., Kawai G., Yokoyama S., Laue E.D. et al. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry. 1997; 36:9109–9119. [DOI] [PubMed] [Google Scholar]
- 39. Geyer M., Assheuer R., Klebe C., Kuhlmann J., Becker J., Wittinghofer A., Kalbitzer H.R.. Conformational states of the nuclear GTP-binding protein Ran and its complexes with the exchange factor RCC1 and the effector protein RanBP1. Biochemistry. 1999; 38:11250–11260. [DOI] [PubMed] [Google Scholar]
- 40. Lu S., Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R., Zhang J.. Ras conformational ensembles, allostery, and signaling. Chem. Rev. 2016; 116:6607–6665. [DOI] [PubMed] [Google Scholar]
- 41. Spoerner M., Wittinghofer A., Kalbitzer H.R.. Perturbation of the conformational equilibria in Ras by selective mutations as studied by 31P NMR spectroscopy. FEBS Lett. 2004; 578:305–310. [DOI] [PubMed] [Google Scholar]
- 42. Liao J., Shima F., Araki M., Ye M., Muraoka S., Sugimoto T., Kawamura M., Yamamoto N., Tamura A., Kataoka T.. Two conformational states of Ras GTPase exhibit differential GTP-binding kinetics. Biochem. Biophys. Res. Commun. 2008; 369:327–332. [DOI] [PubMed] [Google Scholar]
- 43. Spoerner M., Hozsa C., Poetzl J.A., Reiss K., Ganser P., Geyer M., Kalbitzer H.R.. Conformational states of human rat sarcoma (Ras) protein complexed with its natural ligand GTP and their role for effector interaction and GTP hydrolysis. J. Biol. Chem. 2010; 285:39768–39778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shima F., Ijiri Y., Muraoka S., Liao J., Ye M., Araki M., Matsumoto K., Yamamoto N., Sugimoto T., Yoshikawa Y. et al. Structural basis for conformational dynamics of GTP-bound Ras protein. J. Biol. Chem. 2010; 285:22696–22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voorhees R.M., Schmeing T.M., Kelley A.C., Ramakrishnan V.. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010; 330:835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsumoto S., Miyano N., Baba S., Liao J., Kawamura T., Tsuda C., Takeda A., Yamamoto M., Kumasaka T., Kataoka T. et al. Molecular mechanism for conformational dynamics of Ras.GTP elucidated from In-Situ structural transition in crystal. Sci. Rep. 2016; 6:25931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Michielssens S., de Groot B.L., Grubmuller H.. Binding affinities controlled by shifting conformational equilibria: opportunities and limitations. Biophys. J. 2015; 108:2585–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin J., Gagnon M.G., Bulkley D., Steitz T.A.. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015; 160:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shin B.S., Maag D., Roll-Mecak A., Arefin M.S., Burley S.K., Lorsch J.R., Dever T.E.. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell. 2002; 111:1015–1025. [DOI] [PubMed] [Google Scholar]
- 50. Roll-Mecak A., Cao C., Dever T.E., Burley S.K.. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000; 103:781–792. [DOI] [PubMed] [Google Scholar]
- 51. Simonetti A., Marzi S., Billas I.M.L., Tsai A., Fabbretti A., Myasnikov A.G., Gualerzi C.O., Klaholz B.P.. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15656–15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumar V., Chen Y., Ero R., Ahmed T., Tan J., Li Z., Wong A., Bhushan S., Gao Y.. Structure of BipA in GTP form bound to the ratcheted ribosome. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:10944–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Linnemann T., Geyer M., Jaitner B.K., Block C., Kalbitzer H.R., Wittinghofer A., Herrmann C.. Thermodynamic and kinetic characterization of the interaction between the Ras binding domain of AF6 and members of the Ras subfamily. J. Biol. Chem. 1999; 274:13556–13562. [DOI] [PubMed] [Google Scholar]
- 54. Gronwald W., Huber F., Grunewald P., Sporner M., Wohlgemuth S., Herrmann C., Kalbitzer H.R.. Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe. Structure. 2001; 9:1029–1041. [DOI] [PubMed] [Google Scholar]
- 55. Geyer M., Herrmann C., Wohlgemuth S., Wittinghofer A., Kalbitzer H.R.. Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling. Nat. Struct. Biol. 1997; 4:694–699. [DOI] [PubMed] [Google Scholar]
- 56. Pingoud A., Block W., Wittinghofer A., Wolf H., Fischer E.. The elongation factor Tu binds aminoacyl-tRNA in the presence of GDP. J. Biol. Chem. 1982; 257:11261–11267. [PubMed] [Google Scholar]
- 57. Loveland A.B., Demo G., Grigorieff N., Korostelev A.A.. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017; 546:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Noel J.K., Whitford P.C.. How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat. Commun. 2016; 7:13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vetter I.R., Wittinghofer A.. The guanine nucleotide-binding switch in three dimensions. Science. 2001; 294:1299–1304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure has been deposited in the PDB under ID 6EZE.