Abstract
Objective:
A comprehensive review of the network regulation of exosomes and microRNAs (miRNAs) in neurodegenerative diseases was done, centering on the mechanism of the formation of exosomes and miRNAs and the sorting mechanism of exosomal miRNAs, with the aim to provide a theoretical basis in the search of biomarkers and the treatment of neurodegenerative diseases.
Data Sources:
The comprehensive search used online literature databases including NCBI PubMed, Web of Science, Google Scholar, and Baidu Scholar.
Study Selection:
The study selection was based on the following keywords: exosomes, miRNAs, central nervous system (CNS), and neurodegenerative diseases. The time limit for literature retrieval was from the year 2000 to 2018, with language restriction in English. Relevant articles were carefully reviewed, with no exclusions applied to study design and publication type.
Results:
Exosomes are the smallest nanoscale membranous microvesicles secreted by cells and contain important miRNAs, among other rich contents. In the CNS, exosomes can transport amyloid β-protein, α-synuclein, Huntington-associated protein 1, and superoxide dismutase I to other cells. These events relieve the abnormal accumulation of proteins and aggravating neurological diseases. In some neurodegenerative diseases including Alzheimer's disease, Parkinson's disease, Huntington's disease, and amyotrophic lateral sclerosis, miRNAs are pathologically altered as an inexorable course, suggesting that miRNAs may contribute neurodegeneration. Exosomes and miRNAs form a network to regulate the homeostasis of the CNS, both synergistically and individually.
Conclusion:
The network of exosomes and miRNAs that regulates CNS homeostasis is a promising biomarker for the diagnosis and treatment of neurodegenerative diseases.
Keywords: Central Nervous System, Exosomes, microRNAs, Neurodegenerative Diseases
摘要
目的:
围绕外泌体和microRNAs的形成机制,以及外泌体microRNAs的分选机制,综述了外泌体和microRNAs在神经退行性疾病中的网络调控作用,旨在为神经退行性疾病的治疗、生物标志的寻找提供理论基础。
数据源:
基于在线文献数据库的全面检索,包含NCBI PubMed,科学引文索引,Google学术搜索和百度学术搜索等。
文献筛选:
文献筛选基于外泌体、microRNAs、中枢神经系统和神经退行性疾病关键词进行检索,检索年限从2000年到2018年,所有检索内容语言均为英文。相关文章已进行仔细筛选,没有因研究设计和出版类型而排除文献。
结果:
外泌体是细胞分泌的纳米级别膜性小泡,包含丰富的内容物,其中较为重要的就是microRNAs。在中枢神经系统中,外泌体能转运ß样淀粉蛋白、a-突触核蛋白、亨廷顿相关蛋白1以及超氧化物岐化酶I到其它细胞,即能缓解蛋白的异常积聚,也可能加重神经系统疾病。在神经退行性疾病,包括AD、PD、HD、ALS,microRNAs会发生病理上的改变,且是一个必然的过程,暗示着microRNAs可能是导致神经退行性疾病的因素。不仅如此,在中枢神经系统中,外泌体和microRNAs既有协同调控也有单独调控,两者构成网络共同调控着中枢神经系统稳态。
结论:
外泌体和microRNAs形成网络共同调控中枢神经系统的稳态,有望成为诊断和治疗神经退行性疾病的有效工具。
INTRODUCTION
An exosome is an extracellular vesicle with a phospholipid bilayer membrane structure and a diameter ranging from 30 to 100 nm.[1] The surface membrane proteins and exosome contents provide a rich source of biomarkers for various pathological conditions and antigens presented to immune cells through participation in cellular signaling.[2,3,4] The contents of exosomes are closely associated with their parent cells and include diversified proteins, lipids, noncoding RNA (which include circular RNA and microRNA [miRNA]), and other molecules, which may transport the exosome contents to neighboring or more distant cells.[5] Thus, exosomes represent a novel form of intercellular communication among cells without cell-to-cell direct contact.[6,7] With the help of exosomes, cells may change their levels of proteins, lipids, and nucleic acids.[8]
Exosomes are secreted from diverse sites and are found in various body fluids such as blood,[9] urine,[10] saliva,[11] breast milk,[12] amniotic fluid,[13] and cerebrospinal fluid.[14] In the central nervous system (CNS), both neurons and neuroglial cells can secrete and release exosomes into the extracellular environment,[15] suggesting a diversified and important functions of exosomes.
Protein aggregation and the formation of inclusion bodies in selected areas of the nervous system due to neuronal cell death is an important cause of neurodegenerative diseases.[5] MiRNAs regulate the level of proteins by regulating levels of mRNAs.[16] Alterations in the expression profile of miRNAs in Alzheimer's disease (AD) patients,[17] Parkinson's disease (PD) patients,[18] amyotrophic lateral sclerosis (ALS) patients,[19] and Huntington's disease (HD) prodromal patients[20] illustrate the important role of miRNAs in neurodegenerative diseases. Dysregulation of miRNAs is closely related with AD, PD, HD, and ALS.[21,22,23]
Understanding the network regulation of exosomes and miRNAs in neurodegenerative diseases will aid in the understanding of the pathogenesis of neurodegenerative diseases and identification of valuable biomarkers.
EXOSOME BIOGENESIS
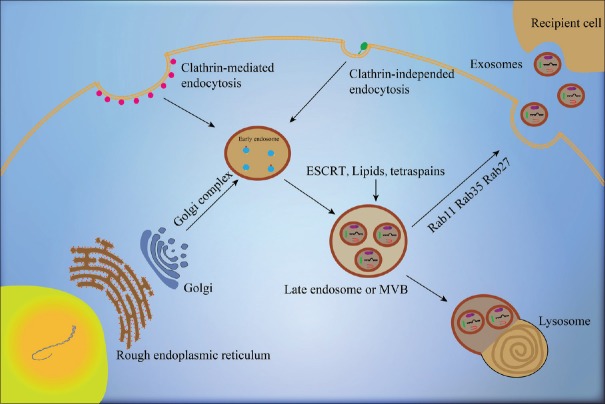
Exosomes' formation involves a series of complex processes. Understanding the generation of exosomes may help better understand their functions [Figure 1]. The generation of exosomes is initiated by early endosomes (EEs) formed from internal budding of the plasma membrane. EEs interact with the Golgi complex to form late endosomes (LEs) or multivesicular bodies (MVBs), which further form intraluminal vesicles (ILVs, i.e., exosomes) through internal budding of the plasma membrane.[1,8] During this process, molecules including the endosomal sorting complex required for transport, lipids (such as ceramide), and tetraspanins participate in ILV formation.[24] The MVBs formed can either be degraded by fusing with lysosomes or released into the extracellular space as exosomes by fusing with the plasma membrane. RAB proteins including RAB11, RAB27, and RAB35 participate in the trafficking of MVBs to the plasma membrane and in the secretion of exosomes.[8,24,25]
Figure 1.
Mechanism of exosome biogenesis and secretion. EEs are generated through internal budding of the plasma membrane, which may be inward invaginations mediated or not mediated by clathrin. EEs interact with the Golgi complex and form LEs, which then form ILVs, i.e., exosomes, through internal budding of the plasma membrane. ESCRT, lipids, and tetraspanins participate in the formation of ILVs. Some of the MVBs formed are transported by associated RAB proteins (RAB11, RAB35, and RAB27) to fuse with the plasma membrane and are then released into the extracellular space as exosomes. Some other MVBs fuse with lysosome for degradation. The red points represent clathrin and the green points represent lipid raft-associated GPI-anchored proteins. EEs: Early endosomes; LEs: Late endosomes; ILVs: Intraluminal vesicles; ESCRT: Endosomal sorting complex required for transport; MVBs: Multivesicular bodies; GPI: Glycosylphosphatidylinositol.
BIOGENESIS OF MIRNAS AND SORTING MECHANISM OF EXOSOMAL MIRNAS
Genesis of miRNAs
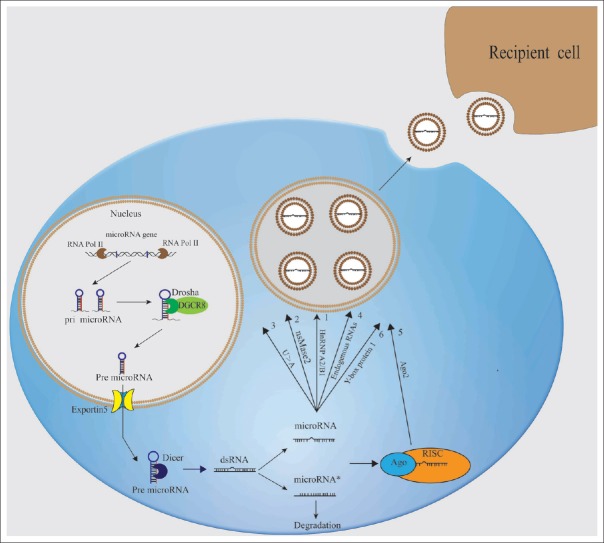
The formation of mature miRNAs involves several steps [Figure 2]. First, by interacting with RNA polymerase II, the miRNA genes are transcribed to primary miRNA transcript (pri-miRNA), which has long sequences and presents some transcription characteristics of RNA polymerase II such as 5' cap structure and 3' poly (A) tail.[26] Pri-miRNA is cleaved by intranuclear RNase III Drosha to produce precursor miRNA (pre-miRNA), which have a length of 70 nt and feature a stem-loop structure. With no or little enzymatic activity, Drosha needs the help of DGCR8 to catalyze its RNA substrate.[27] The pre-miRNA produced is transported into the cytoplasm by the Ran-GTP-dependent nucleocytoplasmic transporter Exportin 5, which is located in the cell membrane. Exportin 5 can recognize and avidly bind pre-miRNA containing a protruding 3' arm and protects the integrity of this 3' region during the transport of pre-miRNA from the nucleus to the cytoplasm.[28] In the cytoplasm, the pre-miRNA is further cleaved by RNase III Dicer to form mature miRNA with 21-23 nt.[29] The miRNA is then conveyed to the RNA-induced silencing complex (RISC) to form miRNA RISC (miRISC), which recognizes and combines with the 3' untranslated region (3'UTR) of mRNA through a miRNA-specific sequence. miRISC also mediates the degradation of mRNA or inhibits its translation. Ago 1 and Ago 2 have vital roles in the miRISC-mediated silencing.[30,31,32]
Figure 2.
Mechanism of miRNA formation and sorting mechanism for exosomal miRNAs. miRNA genes are transcribed by RNA polymerase II as pri-miRNAs, which are processed using two cleavage events to produce mature miRNA. The primary cleavage of animal pri-miRNA is located in the nucleus and produces miRNA precursors with a length of 70 nt and a stem-loop structure, which are referred to as pre-miRNA. The secondary cleavage is located in cytoplasm where pre-miRNA is cleaved to mature miRNAs with 21–23 nt. The cleavages in the maturation process of miRNA are catalyzed by two types of RNase III, i.e., Drosha and Dicer, respectively. Then, the mature miRNAs incorporate into RISC to form a miRISC complex. Mature miRNAs are sorted into exosomes through six mechanisms: (1) the miRNA motif and hnRNPA2B1-dependent pathway, (2) nSMase2-dependent pathway, (3) 3'-end of the miRNA sequence-dependent pathway, (4) endogenous RNA-mediated pathway, (5) Ago2-dependent pathway, and (6) (YBX1)-dependent pathway. pri-miRNA: Primary miRNA transcript; miRISC: miRNA RISC; hnRNPA2B1: Heterogeneous nuclear ribonucleoprotein A2/B1; nSMase2: Neutral sphingomyelinase 2; YBX1: Y-box binding protein 1; miRNAs: microRNAs.
The mechanism of micro RNAs sorting to exosomes
According to current studies, mature miRNAs are sorted into exosomes through six potential mechanisms [Figure 2].
The first is the miRNA motif and heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1)-dependent pathway. The short sequence motifs over-represented in miRNAs (EXOmotifs) guide miRNA sorting into exosomes. Mutation of EXOmotifs will affect miRNA content in the exosomes. HnRNPA2B1 is a RNA-binding protein that can regulate the transport and subcellular localization of mRNA in neurons. HnRNPA2B1 is sumoylated in the cytoplasm and specifically combines with the EXOmotifs (GGAG) to regulate sorting of miRNAs into exosomes.[33]
The second is neutral sphingomyelinase 2 (nSMase2)-dependent pathway. Blocking or overexpressing the activity of nSMase2 reduces and increases the quantity of miRNAs sorted into exosomes, respectively.[34]
The third is the 3' end of the miRNA sequence-dependent pathway. Koppers-Lalic et al.[35] studied the exosomes secreted by maternal B cells and found that the 3' end-rich poly (A) of a miRNA is more abundant in B cells, while the 3' end-rich poly (U) of a miRNA is more abundant in the exosomes originating from B cells. This indicates a potential exosome sorting pathway for miRNA.
The fourth is the endogenous RNA-mediated pathway. Squadrito et al.[36] described that the altered expression of miRNA target mRNA in macrophages can regulate miRNA sorting into exosomes by facilitating the transport of miRNA from the cytoplasm to MVBs.
The fifth is the Ago2-dependent pathway. Ago2 is a component of the RISC. McKenize et al.[37] found that the overactive KRAS genes of KRAS-mutant cells inhibit the localization of Ago2 in the multivesicular endosomes and reduce the secretion of Ago2 into the exosomes, where the level of Ago2 regulates the sorting of miRNA into exosomes.
The sixth is the Y-box binding protein 1 (YBX1)-dependent pathway. YBX1 is a RNA-binding protein. Shurtleff et al.[38] reported that YBX1 can bind mi-233 and help it sort into exosomes. However, the authors did not find evidence of any role of Ago2 in the sorting of mi-233 into exosomes.
CRITICAL ROLES OF EXOSOMES AND MIRNAS IN NEURODEGENERATIVE DISEASES
Exosomes: mediators of neurodegeneration, neuroprotection, and therapeutics
Neuroglial cells include different subsets, which jointly maintain nerve homeostasis, signal transduction, and cellular communication, implying a close cooperation between neurons and neuroglial cells.[39,40] In the CNS, neurons, microglia, astrocytes, and oligodendrocytes can secrete microvesicles into the extracellular environment.[41] These microvesicles or exosomes play a vital role in the occurrence and progression of neurodegenerative diseases, in addition to regulating normal brain functions such as the development and repair of neurons and maintenance of synaptic function [Figure 3].[42]
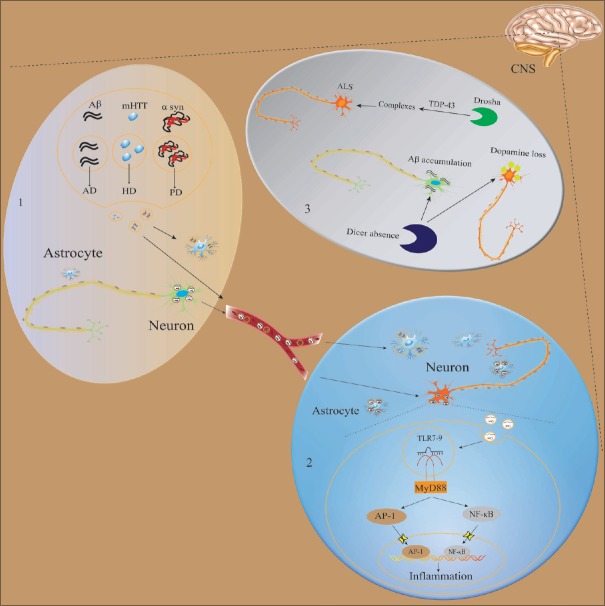
Figure 3.
Exosomes and miRNA regulatory network in neurodegenerative diseases. (1) Aberrant aggregation of Aβ, α-syn, and mHTT in neurons is the primary cause of AD, PD, and HD. Neurons and neuroglial cells can release exosomes into the extracellular space or transport them to the neighboring cells through blood. (2) Exosomes contain miRNA. After the exosomes fuse with the membrane and release miRNAs into the intracellular plasm, TLRs are activated. TLR7-9 activates the myeloid differentiation factors (MyD88) and then activates nuclear factors nuclear factor-kappa B and transcription factors activator protein-1, leading to neuroinflammation and neuronal death. (3) miRNA formation disorder is closely associated with neurodegenerative diseases. Deficiency of Dicer is related to the accumulation of Aβ and reduction of dopamine. In ALS patients, the mutated TDP-43 forms a complex with heterogeneous nuclear ribonucleoproteins family proteins and Drosha, implying that Drosha is an essential enzyme for the formation of miRNA. Aβ: Amyloid-β peptide; α-syn: α-synuclein; mHTT: Mutated Huntingtin; AD: Alzheimer's disease; PD: Parkinson's; HD: Huntington's disease; TLR: Toll-like receptor; ALS: Amyotrophic lateral sclerosis; TDP-43: TAR DNA-binding protein 43; miRNAs: microRNAs.
The main pathological characteristics of AD are excessive accumulation of extracellular senile plaques (composed of amyloid-β peptide [Aβ]) and intracellular neurofibrillary tangles (NFTs) containing hyperphosphorylated tau protein, which can degenerate neurons and activate microglia and astrocytes to produce pro-inflammatory cytokines. The inflammatory environment triggers alteration in the nervous system and breaches the blood–brain barrier (BBB), thus inducing AD.[43] β cleavage of amyloid precursor protein (APP) occurs in EEs to produce Aβ, which enters MVBs and is secreted into the extracellular space in the form of exosomes.[44] Aβ can be transported by the exosomes to lysosome where it is degraded. Disorder of this clearance pathway will cause accumulation of Aβ, leading to the occurrence of AD.[45]
PD is the second most common neurodegenerative disorder after AD. PD is characterized by degeneration and death of dopaminergic (DA) neurons in the substantia nigra pars compacta and formation of intracytoplasmic eosinophilic inclusion bodies (termed Lewy bodies) in the surviving neurons, where α-synuclein (α-syn), as the primary component, abnormally accumulates.[46,47] Although most cases of PD are sporadic, familial cases have also been associated with different genes that include SNCA, leucine-rich receptor kinase 2 (LRRK2), PARKIN, PTEN-induced kinase 1, and DJ-1.[46] As a pathogenic factor, α-syn has a vital association with exosomes. Emmanouilidou et al.[48] found that α-syn monomers and polymers produced by neuroblastoma cells are secreted to the extracellular space through exosomes using a calcium-dependent mechanism and cause the death of neuron recipient cells. Alvarez et al.[49] subsequently reported a similar phenomenon where cultured SY5Y neuroblastoma cells released α-syn through exosomes secreted to the extracellular space. Exosomes play a critical role in PD. They transport α-syn to lysosomes for degradation. Disorder of this clearance pathway results in the escape and degradation of α-syn and the exosome-mediated transport of the components to recipient cells.[49,50] LRRK2 plays a vital role in the secretion of exosomes and fusing of MVBs with the membrane. LRRK2 R1441C mutation induces the formation of aberrant MVBs and disrupts the balance between the formation and clearance of MVBs, consequently leading to PD.[51] In addition, the autophagic system will also help α-syn escape degradation. Accumulated α-syn will form MVBs, followed by exosome secretion to the extracellular space. Adjoining cells take up exosomes by phagocytosis, followed by the intracellular release and accumulation of α-syn, resulting in cytotoxicity.[52] Exosomes can serve as a biomarker for the diagnosis of PD.
HD is an autosomal dominant-inherited neurodegenerative disease caused by mutation of Huntingtin (HTT). Mutated HTT (mHTT) will cause alterations in protein conformation, resulting in aberrant aggregation in cells, neuronal death, and progressive neurodegeneration.[53] Huntington-associated protein 1 (HAP1) and mHTT have high affinity and can interact with hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), which can recognize and recruit ubiquitinated substrate and aid its entry into the MVBs-mediated sorting and degradation pathway.[54] Overexpressed HAP1 can effectively block the transport of epidermal growth factor receptor (EGFR) from EEs to LEs, inhibit the degradation of internalized EGFR, enhance the activity of the signal pathway of EGFR, and reduce the cytotoxicity caused by mHTT.[54,55] Recent studies described that exosomes from adipose-derived stem cells (ASC-exo) can significantly reduce the accumulation of mHTT in R6/2 mouse-derived neurons,[56] indicating a potential effect of ASC-exo in the therapy of HD.
Cytoplasmic TAR DNA-binding protein 43 (TDP-43) aggregation is an important pathological hallmark of ALS and frontotemporal lobar degeneration.[57,58] The TDP-43 protein is present in blood and cerebrospinal fluid.[59] Nonaka et al.[57] introduced insoluble TDP-43 protein from ALS patients into SH-SY5Y cells and reported aggregation of the protein in a self-template manner. The findings demonstrated that the accumulation of pathological TDP-43 protein has prion-like properties. Related studies demonstrated that protein aggregation and autophagy inhibition promote the secretion of exosome TDP-43 from Neuro2a cells and primary neurons.[58] Exposure of Neuro2a cells to exosomes from patients with ALS causes TDP-43 to accumulate in Neuro2a cells, suggesting that exosomes are involved in the propagation of TDP-43. However, inhibition of exosome secretion by inactivation of nSMase2 with GW4869 or by silencing with RAB27A also produced accumulation of TDP-43 in Neuro2a cells,[58] suggesting that exosomes containing pathological TDP-43 play an important role not only in the transmission of TDP-43 but also in the transport and clearance of pathological TDP-43.
Neurodegenerative diseases involve a common molecular and cellular mechanism of misfolding and accumulation of proteins and formation of inclusion bodies in specific brain regions.[60] Under normal conditions, exosomes can transport the accumulated proteins to lysosomes or extracellular plasma for degradation. Under pathological conditions, clearance is disrupted. Knowledge of the pathogenesis of this clearance pathway is necessary to understand the potential therapy of neurodegenerative diseases.
Effect of regulatory dysregulation of micro RNAs on neurodegenerative diseases
MiRNAs are a type of small regulatory RNA molecules with a length of 21–23 nt; those participate in various physiological processes associated with neurodegenerative diseases such as AD, PD, and HD.[61,62,63] MiRNAs contribute to neurodegenerative diseases primarily by three pathways: (1) inhibition of the translation of proteins or degrading proteins through target regulation-related gene mRNA; (2) participation in neuroinflammation by directly combining with toll-like receptor (TLR) or regulating TLR mRNA expression; and (3) miRNA formation disorder [Figure 3].
Micro RNA-targeting genes in neurodegenerative diseases
Aβ and hyperphosphorylated tau protein are the key factors that cause AD. They are individually regulated by APP, presenilin-1 (PS1), PS2, and tau protein genes. The genes can be regulated by miRNA [Table 1].[64]
Table 1.
miRNAs in neurodegeneration diseases
| miRNA | Neurodegeneration diseases | Target genes | Effect on target genes | References |
|---|---|---|---|---|
| miR-101 | AD | APP | Downregulation | [86] |
| miR-193b | AD | APP | Downregulation | [75] |
| miR-16 | AD | APP | Downregulation | [87] |
| miR-20a miR-17-5p miR-106b | AD | APP | Downregulation | [67] |
| miR-200b | AD | APP | Downregulation | [66] |
| miR-16 | AD | APP | Downregulation | [68] |
| miR-219 | AD | MAPT | Downregulation | [88] |
| miR-34 | AD | MAPT | Downregulation | [73] |
| MiR-7 | PD | SNCA | Downregulation | [81] |
| miR-4639-5p | PD | DJ-1 | Downregulation | [82] |
| miR-125b miR-146a miR-150 miR-214 | HD | HTT | Downregulation | [89] |
| miR-141/200a | ALS | FUS | Downregulation | [90] |
| miR-218 | ALS | Motoneurons 218DKO | Downregulation | [91] |
miRNA: MicroRNAs; ALS: Amyotrophic lateral sclerosis; HD: Huntington's disease; PD: Parkinson's disease; AD: Alzheimer's disease; APP: Amyloid precursor protein; HTT: Huntingtin.
Increase in Aβ level caused by increased APP will lead to synaptic function disorder and degeneration of neurons and eventually will cause cognitive decline.[65] Recent studies have found that the miR-200b, miR-429, and miR-20a family (including miR-20a, miR-106b, and miR-17-5p), miR-16, and miR-101 can downregulate the expression of APP.[66,67,68] The expression of proteins translated from the PS1 gene and PS2 gene is the components of γ-secretase. Mutations in these proteins cause increased γ-secretase activity, which is closely related with the generation of Aβ.[64] A study of the expression profile of miRNA in PS1-knockout mouse brain tissue found that downregulation of miR-9 expression was related with neurodegenerative diseases.[69] Other studies further found that miR-9 also participates in regulating the neurogenesis of zebrafish and mice.[70,71] The tau protein, which is abundant in neurons, is a kind of microtubule-associated protein (MAP). Tau protein molecule with normal biological function contains two to three phosphate groups. However, in AD patients, tau protein is abnormally hyperphosphorylated and contains five to nine phosphate groups, and is dissociated from microtubules and aggregates in neuron soma and dendrites to form NFTs.[72] In M17D cells, miR-34a directly combines with the 3'-UTR of MAP-tau to inhibit the expression of endogenous tau protein.[73] In addition, miR-125b activates kinases, such as ERK1/2 and CDK5/P35, by inhibiting the expression of DUSP6 and PPP1CA, thus leading to hyperphosphorylation of tau protein.[74] Related clinical studies have found that the blood of mild cognitive impairment (MCI) and dementia of Alzheimer-type (DAT) patients contains lower levels of exosomal miR-193b compared with that in controls, with lower content in DAT patients compared to that in patients with MCI. In addition, the exosomal miR-193b in the cerebrospinal fluid of DAT patients is lower than that of the controls, indicating that miR-193b can be used as a biomarker for the diagnosis of AD.[75]
The aberrant expression and accumulation of α-syn are crucial in the pathogenesis of PD. Overexpressed α-syn can be toxic to dopamine neurons.[76] Shamsuzzama et al.[77] developed and studied the transgenic Caenorhabditis elegans (C. elegans) and miRNA let-7 functionally deleted transgenic C. elegans models and found that let-7 functional deletion decreased α-syn expression but did not influence dopamine neurons. In sporadic PD cases, several genome-wide association studies had reported that the α-syn gene (SNCA) is one of the genomic loci posing the highest risk in patients.[78,79,80] McMillan et al.[81] found that miR-7 can combine with the 3'-UTR of SNCA mRNA to inhibit its transcription and regulate the expression of α-syn. The authors further found that the expression of miR-7 was decreased in the substantia nigra of the brain of PD patients,[81] suggesting a potential action of miR-7 in the therapy of PD. Moreover, Chen et al.[82] found significantly upregulated expression of hsa-miR-4639-5p in the blood plasma of PD patients and reported the downregulation of the DJ-1 protein level by negatively regulating the posttranscription level of the PD-associated gene DJ-1, eventually causing severe oxidative stress and neuronal death. The authors considered that hsa-miR-4639-5p can serve as a biomarker for early PD diagnosis.[82] In addition, Hoss et al.[18] studied the miRNA expression profiles in the prefrontal cortex of controls and PD patients screened for miR-10b-5p. The significant decrease of miR-10b-5p expression in PD patients suggested that miR-10b-5p could be used as a molecular marker for the clinical diagnosis of PD [Table 1].
HD is an autosomal dominant-inherited neurodegenerative disease that results from the encoded HTT polyglutamine sequence lengthened by the CAG trinucleotide repeat mutations in exon 1 of the IT15 gene.[53] The mHTT gene exon 1 (mHTT-Exon-1) can directly bind genomic DNA and alter its conformation, leading to altered gene expression.[83] When mHTT-Exon-1 is overexpressed in NT2 cells, the miR-34b expression level will significantly increase. Inhibiting the miR-34b expression will reduce the distribution of mHTT in cytoplasm and its in vitro toxicity.[84] However, miR-27a can increase the expression level of multidrug resistance protein-1 (MDR-1) when used to transfect R6/2 HD mouse-derived NSC cells. MDR-1 can transport mHTT and eventually reduce the accumulation of mHTT in cells [Table 1].[85]
Activation of toll-like receptors by micro RNAs in neuroinflammation
Many neurodegenerative diseases are related with inflammation, which can increase cell injury and cause neuronal death.[92] TLR is a type of innate immune receptor that when activated can activate the downstream signal molecules through the MyD88-dependent or independent pathway, eventually causing the release of inflammatory factors.[93] TLRs of microglia and neurons can also cause CNS injury even in the absence of an obvious pathogen, indicating that the ligand of activated TLRs probably originates from the host.[94] In unaffected or undamaged cells, miRNA can activate TLRs as a physiological ligand and also as damage-associated molecular patterns to trigger an immune response.[95,96,97] TLR7 is localized in endosomes, where it can recognize RNA viruses and is expressed in the neuronal cells.[96,98] Lehmann et al.[96] found that miRNA let-7 can activate TLR7 in neuronal cells and consequently causes neurodegenerative diseases, while TLR7-deleted mice can resist the influence of neurodegenerative diseases [Figure 3]. Later, data have more convincingly demonstrated that neurons can activate the TLR7 located in endosomes and further activate MyD88 and the downstream signal molecules through the uptake of the exosomes containing let-7, eventually causing death of neurons.[99] In general, TLRs display a wide range of expressions in the CNS. Examples include the expressions of TLR3, 4, 7, and 9 in human neuronal cells, expressions of TLR2 in human oligodendrocytes, expression of TLR3-4 in human astrocytes, and expression of TLR1-4 in human microglia.[21] We can speculate that miRNAs play a critical role in activating TLRs to cause inflammation and consequently cause neurodegenerative diseases. Supporting data are needed.
MiRNA-generating disorders in neurodegenerative diseases
Drosha and Dicer are the two types of RNase III enzymes. They are critical to the formation of miRNAs. They individually catalyze the sequence cleaving in the process of miRNAs maturation. The deficiency or formation disorder of these two enzymes is closely related to neurodegenerative diseases.[100] TDP-43 protein is vital for the maturation of miRNAs. In ALS patients, mutated TDP-43 will combine with heterogeneous nuclear ribonucleoproteins family proteins and Drosha to form a complex.[101] Similarly, the deficiency of Dicer can impair the expression of minor miRNAs in adult forebrain and eventually cause hyperphosphorylation of tau proteins, consequently leading to neuron death.[102] The deficiency of Dicer will also cause the accumulation of Aβ and loss of dopamine [Figure 3].[100] For example, miR-133b, which is highly expressed in the midbrain under normal conditions, is deficiently expressed in PD patients and in an animal model of PD owing to deficiency of Dicer.[103]
The collective results that have been presented show that miRNAs play a vital regulation role in the CDN under normal and pathological conditions. Moreover, the occurrence of neurodegenerative diseases caused by miRNA-activated TLRs implies that miRNA may have many other regulatory functions, which require further research.
CONCLUSIONS
Exosomes act as a conduit for cell-to-cell communication. Their components can reflect the function status of the cells of origin, contain cytopathic molecular information, and aid in the diagnosis of diseases. Owing to their cargo function, nanometer size, and ability to cross the BBB, advantages in therapeutic effect, targeting ability, immune response, and safety, exosomes provide a novel approach for treatment of neurodegenerative diseases and brain-targeted drug delivery. This review highlights a wide range of regulatory functions of miRNAs in the CNS. Exosomes can transport let-7 to activate TLR7. This implies a novel regulation mechanism for neuroinflammation. Furthermore, abnormal expression of miRNA has been confirmed in a patient with neurodegenerative disease.[82,104] miRNAs are a type of small RNA molecule those extensively regulate physiological and pathological processes. They can be used as biomarkers for early diagnosis of neurodegenerative diseases. In CNS regulatory processes, exosomes and miRNAs form a network to regulate CNS homeostasis, both synergistically and individually.
Great progress has been made in the studies on neurodegenerative diseases such as AD, PD, HD, and ALS. However, there are still many problems to be solved and therapies for these diseases are mostly conservative at present. These diseases remain incurable. Taking AD as an example, there is no drug to delay or prevent the progression of AD. Exosomes and miRNAs offer the potential for the diagnosis and treatment of neurodegenerative diseases.
Financial support and sponsorship
This study was supported by a grant of Institute of Environmental Science of China West Normal University, Environmental Education Center of China West Normal University and Research Projects for Young Teachers of China West Normal University (No. 416535).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yuan-Yuan Ji
REFERENCES
- 1.Kourembanas S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 2.Müller G. Microvesicles/exosomes as potential novel biomarkers of metabolic diseases. Diabetes Metab Syndr Obes. 2012;5:247–82. doi: 10.2147/DMSO.S32923. doi: 10.2147/DMSO.S32923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–8. doi: 10.1038/sj.cdd.4402237. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 4.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 5.Kalani A, Tyagi A, Tyagi N. Exosomes: Mediators of neurodegeneration, neuroprotection and therapeutics. Mol Neurobiol. 2014;49:590–600. doi: 10.1007/s12035-013-8544-1. doi: 10.1007/s12035-013-8544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramachandran S, Palanisamy V. Horizontal transfer of RNAs: Exosomes as mediators of intercellular communication. Wiley Interdiscip Rev RNA. 2012;3:286–93. doi: 10.1002/wrna.115. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Horizontal transfer of microRNAs: Molecular mechanisms and clinical applications. Protein Cell. 2012;3:28–37. doi: 10.1007/s13238-012-2003-z. doi: 10.1007/s13238-012-2003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–87. doi: 10.1093/intimm/dxh267. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 10.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–73. doi: 10.1073/pnas.0403453101. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, Kawakami H, et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull. 2011;34:13–23. doi: 10.1248/bpb.34.13. doi: 10.1248/bpb.34.13. [DOI] [PubMed] [Google Scholar]
- 12.Admyre C, Johansson SM, Qazi KR, Filén JJ, Lahesmaa R, Norman M, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–78. doi: 10.4049/jimmunol.179.3.1969. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 13.Asea A, Jean-Pierre C, Kaur P, Rao P, Linhares IM, Skupski D, et al. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol. 2008;79:12–7. doi: 10.1016/j.jri.2008.06.001. doi: 10.1016/j.jri.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF, et al. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582–90. doi: 10.1002/path.2145. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 15.De Toro J, Herschlik L, Waldner C, Mongini C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. doi: 10.3389/fimmu.2015.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jager PL, Bennett DA. An inflection point in gene discovery efforts for neurodegenerative diseases: From syndromic diagnoses toward endophenotypes and the epigenome. JAMA Neurol. 2013;70:719–26. doi: 10.1001/jamaneurol.2013.275. doi: 10.1001/jamaneurol.2013.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leidinger P, Backes C, Deutscher S, Schmitt K, Mueller SC, Frese K, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14:R78. doi: 10.1186/gb-2013-14-7-r78. doi: 10.1186/gb-2013-14-7- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers RH. MicroRNA profiles in Parkinson's disease prefrontal cortex. Front Aging Neurosci. 2016;8:36. doi: 10.3389/fnagi.2016.00036. doi: 10.3389/fnagi.2016.000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Si Y, Cui X, Crossman DK, Hao J, Kazamel M, Kwon Y, et al. Muscle microRNA signatures as biomarkers of disease progression in amyotrophic lateral sclerosis. Neurobiol Dis. 2018;114:85–94. doi: 10.1016/j.nbd.2018.02.009. doi:.1016/j.nbd.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed ER, Latourelle JC, Bockholt JH, Bregu J, Smock J, Paulsen JS, et al. MicroRNAs in CSF as prodromal biomarkers for Huntington disease in the PREDICT-HD study. Neurology. 2018;90:e264–72. doi: 10.1212/WNL.0000000000004844. doi: 1212/WNL.0000000000004844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JJ, Zhao B, Zhao J, Li S. Potential roles of exosomal micrornas as diagnostic biomarkers and therapeutic application in Alzheimer's disease. Neural Plast. 2017;2017:1–12. doi: 10.1155/2017/7027380. doi: 10.1155/2017/7027380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–8. doi: 10.1126/science.1059581. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 23.Cardo LF, Coto E, de Mena L, Ribacoba R, Moris G, Menéndez M, et al. Profile of microRNAs in the plasma of Parkinson's disease patients and healthy controls. J Neurol. 2013;260:1420–2. doi: 10.1007/s00415-013-6900-8. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- 24.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–25. doi: 10.1016/j.ceb.2014.05.004. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–94. doi: 10.1016/j.bcp.2011.12.037. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–5. doi: 10.1016/j.molcel.2004.12.002. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN, et al. The drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by exportin 5. Nucleic Acids Res. 2004;32:4776–85. doi: 10.1093/nar/gkh824. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 30.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 31.Tang G. SiRNA and miRNA: An insight into RISCs. Trends Biochem Sci. 2005;30:106–14. doi: 10.1016/j.tibs.2004.12.007. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–66. doi: 10.1101/gad.1210204. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T, et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–59. doi: 10.1074/jbc.M112.446831. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–58. doi: 10.1016/j.celrep.2014.08.027. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 36.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–46. doi: 10.1016/j.celrep.2014.07.035. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 37.McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, et al. KRAS-MEK signaling controls ago2 sorting into exosomes. Cell Rep. 2016;15:978–87. doi: 10.1016/j.celrep.2016.03.085. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016:5. doi: 10.7554/eLife.19276. pii: e19276. doi: 10.7554/eLife19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113(Pt 19):3365–74. doi: 10.1242/jcs.113.19.3365. doi: 10.7554/eLife.19276. [DOI] [PubMed] [Google Scholar]
- 40.Frühbeis C, Fröhlich D, Krämer-Albers EM. Emerging roles of exosomes in neuron-glia communication. Front Physiol. 2012;3:119. doi: 10.3389/fphys.2012.00119. doi: 10.3389/fphys.2012.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, et al. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci. 2011;46:409–18. doi: 10.1016/j.mcn.2010.11.004. doi: 10.1016/j.mcn.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med. 2010;362:329–44. doi: 10.1056/NEJMra0909142. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 44.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, et al. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103:11172–7. doi: 10.1073/pnas.0603838103. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuyama K, Sun H, Mitsutake S, Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J Biol Chem. 2012;287:10977–89. doi: 10.1074/jbc.M111.324616. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson's disease. J Chem Neuroanat. 2012;42:279–84. doi: 10.1016/j.jchemneu.2011.01.005. doi: 10.1056/NEJMra0909142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lees AJ, Hardy J, Revesz T. Parkinson's disease. Lancet. 2009;373:2055–66. doi: 10.1016/S0140-6736(09)60492-X. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 48.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, et al. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30:6838–51. doi: 10.1523/JNEUROSCI.5699-09.2010. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, Sargent IL, Wood MJ, et al. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis. 2011;42:360–7. doi: 10.1016/j.nbd.2011.01.029. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. doi: 10.1186/1750-1326-7- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022–34. doi: 10.1093/hmg/ddp346. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grozdanov V, Danzer KM. Release and uptake of pathologic alpha-synuclein. Cell Tissue Res. 2018;373:175–82. doi: 10.1007/s00441-017-2775-9. doi: 10.1007/s00441-017-2775-9. [DOI] [PubMed] [Google Scholar]
- 53.Xu M, Wu ZY. Huntington disease in Asia. Chin Med J. 2015;128:1815–9. doi: 10.4103/0366-6999.159359. doi: 10.4103/0366-6999.159359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Chin LS, Levey AI, Li L. Huntingtin-associated protein 1 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate and functions in endosomal trafficking. J Biol Chem. 2002;277:28212–21. doi: 10.1074/jbc.M111612200. doi: 10.1074/jbc.M111612200. [DOI] [PubMed] [Google Scholar]
- 55.Li SH, Yu ZX, Li CL, Nguyen HP, Zhou YX, Deng C, et al. Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington's disease. J Neurosci. 2003;23:6956–64. doi: 10.1523/JNEUROSCI.23-17-06956.2003. doi: 10.1523/JNEUROSCI.23-17-06956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M, Liu T, Im W, Kim M. Exosomes from adipose-derived stem cells ameliorate phenotype of Huntington's disease in vitro model. Eur J Neurosci. 2016;44:2114–9. doi: 10.1111/ejn.13275. doi: 10.1111/ejn.13275. [DOI] [PubMed] [Google Scholar]
- 57.Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 2013;4:124–34. doi: 10.1016/j.celrep.2013.06.007. doi: 10.1016/j.celrep.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Iguchi Y, Eid L, Parent M, Soucy G, Bareil C, Riku Y, et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain. 2016;139:3187–201. doi: 10.1093/brain/aww237. doi: 10.1093/brain/aww237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feneberg E, Steinacker P, Lehnert S, Schneider A, Walther P, Thal DR, et al. Limited role of free TDP-43 as a diagnostic tool in neurodegenerative diseases. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15:351–6. doi: 10.3109/21678421.2014.905606. doi: 10.3109/21678421.2014.905606. [DOI] [PubMed] [Google Scholar]
- 60.Li XJ, Li S. Influence of species differences on the neuropathology of transgenic Huntington's disease animal models. J Genet Genomics. 2012;39:239–45. doi: 10.1016/j.jgg.2012.05.002. doi: 10.1016/j.jgg.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–8. doi: 10.1111/j.1750-3639.2007.00120.x. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Mol Neurobiol. 2011;44:359–73. doi: 10.1007/s12035-011-8211-3. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Reddy PH, Williams J, Smith F, Bhatti JS, Kumar S, Vijayan M, et al. MicroRNAs, aging, cellular senescence, and Alzheimer's disease. Prog Mol Biol Transl Sci. 2017;146:127–71. doi: 10.1016/bs.pmbts.2016.12.009. doi: 10.1016/bs.pmbts.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Nicolas M, Hassan BA. Amyloid precursor protein and neural development. Development. 2014;141:2543–8. doi: 10.1242/dev.108712. doi: 10.1242/dev.108712. [DOI] [PubMed] [Google Scholar]
- 66.Liu CG, Wang JL, Li L, Xue LX, Zhang YQ, Wang PC, et al. MicroRNA-135a and -200b, potential biomarkers for Alzheimer's disease, regulate β secretase and amyloid precursor protein. Brain Res. 2014;1583:55–64. doi: 10.1016/j.brainres.2014.04.026. doi: 10.1016/j.brainres.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 67.Hébert SS, Horré K, Nicolaï L, Bergmans B, Papadopoulou AS, Delacourte A, et al. MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–8. doi: 10.1016/j.nbd.2008.11.009. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 68.Liu W, Liu C, Zhu J, Shu P, Yin B, Gong Y, et al. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer's-associated pathogenesis in SAMP8 mice. Neurobiol Aging. 2012;33:522–34. doi: 10.1016/j.neurobiolaging.2010.04.034. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 69.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–81. doi: 10.1261/rna.5980303. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jing L, Jia Y, Lu J, Han R, Li J, Wang S, et al. MicroRNA-9 promotes differentiation of mouse bone mesenchymal stem cells into neurons by notch signaling. Neuroreport. 2011;22:206–11. doi: 10.1097/WNR.0b013e328344a666. doi: 10.1097/WNR.0b013e328344a666. [DOI] [PubMed] [Google Scholar]
- 71.Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–72. doi: 10.1038/nrn2194. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 73.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. J Neurochem. 2013;127:739–49. doi: 10.1111/jnc.12437. doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banzhaf-Strathmann J, Benito E, May S, Arzberger T, Tahirovic S, Kretzschmar H, et al. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J. 2014;33:1667–80. doi: 10.15252/embj.201387576. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu CG, Song J, Zhang YQ, Wang PC. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer's disease. Mol Med Rep. 2014;10:2395–400. doi: 10.3892/mmr.2014.2484. doi: 10.3892/mmr.2014.2484. [DOI] [PubMed] [Google Scholar]
- 76.Du JJ, Chen SD. Current nondopaminergic therapeutic options for motor symptoms of Parkinson's disease. Chin Med J. 2017;130:1856–66. doi: 10.4103/0366-6999.211555. doi: 10.4103/0366-6999.211555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shamsuzzama, Kumar L, Nazir A. Modulation of alpha-synuclein expression and associated effects by microRNA let-7 in transgenic C. Elegans. Front Mol Neurosci. 2017;10:328. doi: 10.3389/fnmol.2017.00328. doi: 10.3389/fnmol.2017.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–9. doi: 10.1038/nature17939. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–7. doi: 10.1038/ng.485. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 81.McMillan KJ, Murray TK, Bengoa-Vergniory N, Cordero-Llana O, Cooper J, Buckley A, et al. Loss of microRNA-7 regulation leads to α-synuclein accumulation and dopaminergic neuronal loss in vivo. Mol Ther. 2017;25:2404–14. doi: 10.1016/j.ymthe.2017.08.017. doi: 10.1016/j.ymthe.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y, Gao C, Sun Q, Pan H, Huang P, Ding J, et al. MicroRNA-4639 is a regulator of DJ-1 expression and a potential early diagnostic marker for Parkinson's disease. Front Aging Neurosci. 2017;9:232. doi: 10.3389/fnagi.2017.00232. doi: 10.3389/fnagi.2017.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benn CL, Sun T, Sadri-Vakili G, McFarland KN, DiRocco DP, Yohrling GJ, et al. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28:10720–33. doi: 10.1523/JNEUROSCI.2126-08.2008. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Björkqvist M, et al. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum Mol Genet. 2011;20:2225–37. doi: 10.1093/hmg/ddr111. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 85.Ban JJ, Chung JY, Lee M, Im W, Kim M. MicroRNA-27a reduces mutant hutingtin aggregation in an in vitro model of Huntington's disease. Biochem Biophys Res Commun. 2017;488:316–21. doi: 10.1016/j.bbrc.2017.05.040. doi: 10.1016/j.bbrc.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 86.Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285:18344–51. doi: 10.1074/jbc.M110.112664. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang B, Chen CF, Wang AH, Lin QF. MiR-16 regulates cell death in Alzheimer's disease by targeting amyloid precursor protein. Eur Rev Med Pharmacol Sci. 2015;19:4020–7. [PubMed] [Google Scholar]
- 88.Santa-Maria I, Alaniz ME, Renwick N, Cela C, Fulga TA, Van Vactor D, et al. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J Clin Invest. 2015;125:681–6. doi: 10.1172/JCI78421. doi: 10.1172/JCI78421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sinha M, Ghose J, Bhattarcharyya NP. Micro RNA -214,-150,-146a and-125b target huntingtin gene. RNA Biol. 2011;8:1005–21. doi: 10.4161/rna.8.6.16035. doi: 10.4161/rna.8.6.16035. [DOI] [PubMed] [Google Scholar]
- 90.Dini Modigliani S, Morlando M, Errichelli L, Sabatelli M, Bozzoni I. An ALS-associated mutation in the FUS 3'-UTR disrupts a microRNA-FUS regulatory circuitry. Nat Commun. 2014;5:4335. doi: 10.1038/ncomms5335. doi: 10.1038/ncomms5335. [DOI] [PubMed] [Google Scholar]
- 91.Amin ND, Bai G, Klug JR, Bonanomi D, Pankratz MT, Gifford WD, et al. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science. 2015;350:1525–9. doi: 10.1126/science.aad2509. doi: 10.1126/science.aad2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Properzi F, Ferroni E, Poleggi A, Vinci R. The regulation of exosome function in the CNS: Implications for neurodegeneration. Swiss Med Wkly. 2015;145:w14204. doi: 10.4414/smw.2015.14204. doi: 10.4414/smw.2015.14204. [DOI] [PubMed] [Google Scholar]
- 93.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 94.Okun E, Griffioen KJ, Lathia JD, Tang SC, Mattson MP, Arumugam TV, et al. Toll-like receptors in neurodegeneration. Brain Res Rev. 2009;59:278–92. doi: 10.1016/j.brainresrev.2008.09.001. doi: 10.1016/j.brainresrev.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:12278–9. doi: 10.1073/pnas.1209414109. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: Let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–35. doi: 10.1038/nn.3113. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 97.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 98.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. doi: 10.1126/science.1093620. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 99.Winkler CW, Taylor KG, Peterson KE. Location is everything: Let-7b microRNA and TLR7 signaling results in a painful TRP. Sci Signal. 2014;7:pe14. doi: 10.1126/scisignal.2005407. doi: 10.1126/scisignal.2005407. [DOI] [PubMed] [Google Scholar]
- 100.Tan L, Yu JT, Tan L. Causes and consequences of microRNA dysregulation in neurodegenerative diseases. Mol Neurobiol. 2015;51:1249–62. doi: 10.1007/s12035-014-8803-9. doi: 10.1007/s12035-014-8803-9. [DOI] [PubMed] [Google Scholar]
- 101.Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, et al. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107:13318–23. doi: 10.1073/pnas.1008227107. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, et al. Genetic ablation of dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–69. doi: 10.1093/hmg/ddq311. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 103.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang KH, Wu YR, Chen CM. Down-regulation of miR-9* in the peripheral leukocytes of Huntington's disease patients. Orphanet J Rare Dis. 2017;12:185. doi: 10.1186/s13023-017-0742-x. doi: 10.1186/s13023-017-0742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]