Abstract
The current deluge of newly identified RNA transcripts presents a singular opportunity for improved assessment of coding potential, a cornerstone of genome annotation, and for machine-driven discovery of biological knowledge. While traditional, feature-based methods for RNA classification are limited by current scientific knowledge, deep learning methods can independently discover complex biological rules in the data de novo. We trained a gated recurrent neural network (RNN) on human messenger RNA (mRNA) and long noncoding RNA (lncRNA) sequences. Our model, mRNA RNN (mRNN), surpasses state-of-the-art methods at predicting protein-coding potential despite being trained with less data and with no prior concept of what features define mRNAs. To understand what mRNN learned, we probed the network and uncovered several context-sensitive codons highly predictive of coding potential. Our results suggest that gated RNNs can learn complex and long-range patterns in full-length human transcripts, making them ideal for performing a wide range of difficult classification tasks and, most importantly, for harvesting new biological insights from the rising flood of sequencing data.
INTRODUCTION
Deep sequencing technology has yielded a torrent of new transcript annotations, creating a need for fresh approaches to unlock the full information potential of these vast datasets. Existing state-of-the-art methods for classification of long RNAs as protein-coding RNAs (mRNAs) or long noncoding RNAs (lncRNAs) rely on human-engineered features, such as the coverage and length of a predicted open reading frame (ORF). These features predispose such models to misclassification of mRNAs encoding small proteins and of lncRNAs with long, un-translated ORFs. Nucleotide hexamer frequency is another commonly used feature, but while it can capture the frequency of codon pairs, it does not benefit from the larger sequence context. These limitations and the annotation challenges ahead demand new approaches to biological sequence classification that are capable of detecting complex, variable-length patterns.
In contrast to conventional machine learning methods, ‘deep learning’—the application of multi-layered artificial neural networks to learning tasks—can discover useful features independently, avoiding biases introduced by human-engineered features (1). Deep learning methods have repeatedly outperformed state-of-the-art ‘shallow’ machine learning algorithms, such as support vector machines (SVM) and logistic regression, as approaches to biological problems in recent years. Multiple bioinformatics applications of deep convolutional neural networks (CNNs) have been published (2–4); however, while CNNs adeptly learn spatial information, recurrent neural networks (RNNs) are better suited for learning sequential patterns because of their serialized structure and ability to handle variable-length inputs (1). RNN-based approaches have had success in the fields of natural language and music (5), and information extraction from biomedical texts (6–8). Researchers have recently begun to apply RNNs to biological sequences for the identification of splice sites (9), microRNA target sites (10), DNA binding sites (11) and the prediction of methylation states (12), as well as to microRNA precursor prediction using inputs of RNA sequences merged with secondary structure predictions (13). While basic RNNs are challenged by most biologically relevant input sequence lengths due to the ‘vanishing gradient problem,’ a difficulty encountered during training due to the multiplication of many small terms when computing the gradient of an error function by the chain rule (14), several recent adaptations addressed this issue. Among the most popular of these modified RNNs are long-short-term-memory (LSTM) RNNs (15) and gated recurrent unit (GRU) RNNs (aRxiv: https://arxiv.org/abs/1409.1259v2), which manage memory to improve the learning of long-range dependencies. Recent studies demonstrated superior performance of GRUs compared to LSTMs for bioinformatics tasks (10,16). We report the successful implementation of a GRU network to accurately predict protein-coding potential of complete, variable-length transcripts. Our method, ‘mRNA RNN’ (mRNN), not only performs as well, if not better than, existing state-of-the-art classifiers, but also learns complex biological rules in the process.
MATERIALS AND METHODS
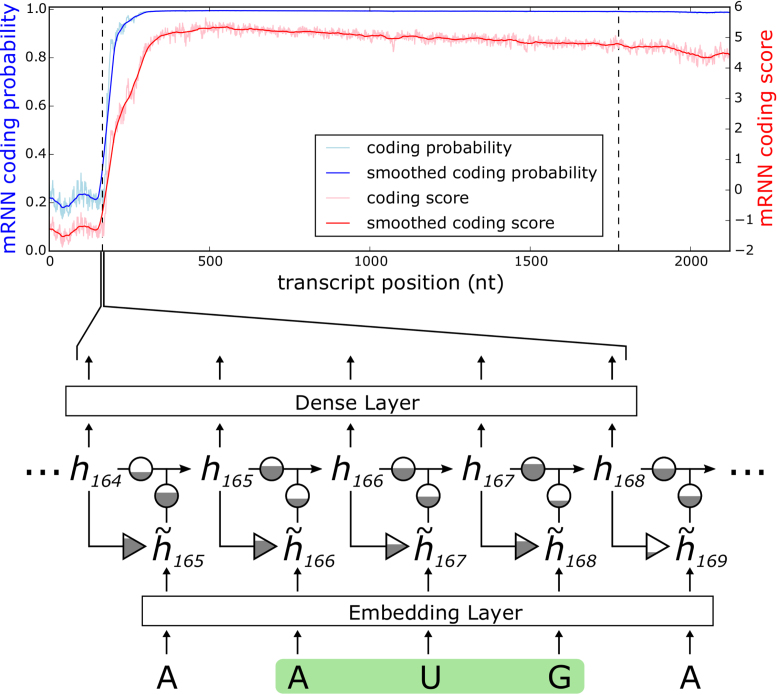
The structure of the GRU used in mRNN is depicted in Figure 1. The GRU is composed of a candidate hidden state layer and a hidden state layer of dimension 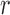 . While RNN applications to biological sequences use one-hot encoding where each input character (A,C,G,U) is encoded as a binary vector with a single non-zero entry
. While RNN applications to biological sequences use one-hot encoding where each input character (A,C,G,U) is encoded as a binary vector with a single non-zero entry 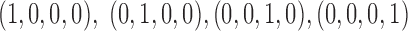 , we achieved better performance with an embedding layer. The embedding layer maps each input character (A, U, G, C or N) to a higher-dimensional representation. This map is linear, so it can be viewed as a multiplication of a
, we achieved better performance with an embedding layer. The embedding layer maps each input character (A, U, G, C or N) to a higher-dimensional representation. This map is linear, so it can be viewed as a multiplication of a 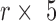 matrix to a one-hot vector. The candidate hidden state
matrix to a one-hot vector. The candidate hidden state 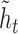 at position
at position 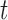 is computed from the input to the network at
is computed from the input to the network at 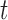 and the previous position's hidden state
and the previous position's hidden state 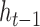 , scaled by the reset gates represented here as triangles. The hidden state
, scaled by the reset gates represented here as triangles. The hidden state 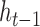 is computed from the previous position’s hidden state
is computed from the previous position’s hidden state 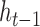 and the current position’s candidate hidden state
and the current position’s candidate hidden state 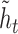 , scaled by the update gates represented here as circles. The gates and hidden states are real-valued vectors of the same dimension, where the dimension is determined via hyper-parameter tuning.
, scaled by the update gates represented here as circles. The gates and hidden states are real-valued vectors of the same dimension, where the dimension is determined via hyper-parameter tuning.
Figure 1.
mRNN Output and Model Schematic. Coding probability and coding potential score is shown at nucleotide-level resolution for the transcript ENST00000371732.9, which encodes caspase recruitment domain family member 9. Values at position 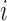 correspond to the mRNN coding probability or
correspond to the mRNN coding probability or 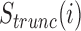 , the mRNN output for the truncated sequence from 1 to
, the mRNN output for the truncated sequence from 1 to 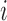 . Vertical dashed lines demarcate the annotated start and end of the CDS. A schematic of the gated RNN is shown below. Equilateral triangles signify reset gates, and the height of the gray fill represents the proportional contribution of the previous hidden state (
. Vertical dashed lines demarcate the annotated start and end of the CDS. A schematic of the gated RNN is shown below. Equilateral triangles signify reset gates, and the height of the gray fill represents the proportional contribution of the previous hidden state (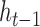 ) to the new candidate hidden state (
) to the new candidate hidden state (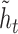 ). The update gate is shown as two circles representing the proportional contributions of the previous hidden state (
). The update gate is shown as two circles representing the proportional contributions of the previous hidden state (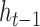 ) and the new candidate hidden state (
) and the new candidate hidden state (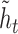 ) to the new hidden state (
) to the new hidden state (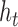 ). Arrows represent matrix products. The embedding layer maps nucleotides to 128-dimnensional vectors.
). Arrows represent matrix products. The embedding layer maps nucleotides to 128-dimnensional vectors.
For training, we provided mRNN with a dataset containing full-length human transcript sequences labeled as mRNAs or lncRNAs. All training and test sets were selected from GENCODE Release 25 (17). This resource for transcript data was recently used to train and test an existing state-of-the-art lncRNA classification tool called FEELnc (18). The data set is challenging for protein-coding potential assessment because 10% of these transcripts lack a start codon and 25% lack a stop codon in the annotated CDS. However, the presence of these incomplete sequences in the data set allows us to train a model that is robust enough to classify sequences in a transcriptome newly built from RNA-sequencing data, which often contains incomplete transcripts. We evaluated mRNN’s performance using a test set—an unbiased random sample of human transcripts composed of 500 mRNAs and 500 lncRNAs selected from the full GENCODE annotation. We also selected a secondary test set—the ‘challenge’ set—of more challenging transcripts, including 500 mRNAs with short CDSs (≤50 codons in GENCODE annotation) and 500 lncRNAs with long (untranslated) ORFs (≥50 codons). Transcripts for training and testing are separated by their associated genes, and transcripts associated with genes used in the test and challenge set are excluded from the training set. At last, we evaluated performance on the entire mouse transcriptome, composed of 77 725 transcripts more than 200 nt long.
From the sequences remaining after the removal of the test and challenge sets, we selected a validation set equal in size to the test set for mRNN’s hyper-parameter tuning and model selection (Supplementary Figure S1). We evaluated several different training strategies, and all decisions for training and hyper-parameters were based on minimizing loss or maximizing accuracy of predictions made on the validation set (Supplementary Figures S2 and 3). We found that mRNN’s performance was improved significantly by pre-training it with augmented data (19) consisting of several mutated copies of each sequence (aRxiv: https://arxiv.org/abs/1601.03651v2). mRNN showed higher validation accuracy when the dataset was augmented by random 1-nt point insertions than by random point mutations. (Supplementary Methods and Figure S4A). Moreover, we found that length-filtered augmented training data (transcripts between 200- and 1000-nt long) yielded lower validation loss than augmented training data unrestricted by length (Supplementary Figure S4B). Therefore, rather than training on the full available training data, we trained on a set of 16 000 mRNAs and 16 000 lncRNAs selected from the sequences between 200- and 1000-nt long. The augmented data set used for pre-training was built from this same training set. In addition to ‘data augmentation,’ we implemented ‘early stopping,’ which exits training if loss on the training set decreases while validation loss does not; both of these strategies help prevent over-fitting during training. We also used ensemble testing using the uniformly weighted predictions of five models (20). We used embedding vectors to represent each nucleotide because this yielded higher validation accuracy than did one-hot encoding when using ensemble testing for the RNN library Passage (Supplementary Figure S3), and we computed the cosine distance between embedding vectors to quantify any learned similarity between nucleotides (Supplementary Figure S5). We also used dropout, which randomly sets network inputs to zero in Passage's GRU implementation, because when combined with an embedding layer, it improved validation accuracy (Supplementary Figure S2). For detailed methods see Supplementary Methods.
For comparison, we used the same test set to assess performance of three non-comparative classifiers considered to be state-of-the-art in speed and performance: CPAT, which is a logistic regression model based on hexamer frequencies and other features computed from the transcript (21); FEELnc, which is a random forest model, also based on human-defined features of the input sequences (18); and longdist, which is an SVM model using principal component analysis (PCA) to reduce the dimensionality of the features that include di-, tri- and tetra-nucleotide patterns, and other pre-defined features computed from ORF lengths (22). No length restrictions were imposed on sequences in the training set for CPAT, FEELnc and longdist, giving these classifiers substantially more training data than mRNN.
We applied a number of approaches to evaluate what mRNN learned. In each case, we computed the change in the mRNN score due to perturbations of the mRNA sequence, and examined sequence features at locations where perturbations significantly changed the predicted coding score. First, we examined the effect of randomly shuffling different regions of mRNAs. Next, we performed a point-mutation analysis where we computed the mRNN score for all possible single-nucleotide mutations to all GENCODE mRNAs fewer than 2000 nt in length (59 133 in total), and examined the distribution of significant score changes. We also performed a pairwise mutation analysis by computing the mRNN score for all possible pairs of mutations, and identified dependent positions with combined score changes significantly higher than the sum of the score changes due to the individual mutations. We defined a measure of ‘synergy’ or dependence, 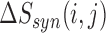 , for a pair of mutations at positions
, for a pair of mutations at positions 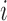 and
and 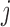 with the equation:
with the equation:
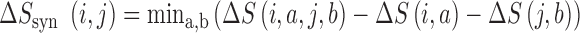 |
where 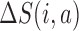 and
and 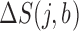 are the change in score due to mutating position
are the change in score due to mutating position 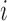 to nucleotide
to nucleotide 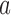 and
and 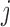 to
to 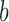 , respectively, and
, respectively, and 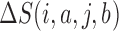 is the change in score from making both mutations. We defined a similar score for compensatory score changes (see Supplementary Methods). At last, we performed a coding score trajectory analysis where we defined a smoothed coding score
is the change in score from making both mutations. We defined a similar score for compensatory score changes (see Supplementary Methods). At last, we performed a coding score trajectory analysis where we defined a smoothed coding score 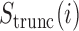 for the truncated transcript from position 1 to
for the truncated transcript from position 1 to 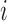 , which is depicted in Figure 1. We then studied the score change
, which is depicted in Figure 1. We then studied the score change 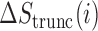 as a function of position for each sequence in our test set, using the same window size
as a function of position for each sequence in our test set, using the same window size 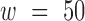 ,
,
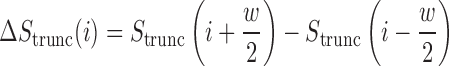 |
To locate regions containing important predictive information, we identified coding score ‘spikes,’—sharp increases in the coding score trajectory. We identified common sequence features present in a region of length 50 nt around spike positions 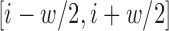 . We found codons that are statistically enriched in the spike regions by comparing the frequencies of each codon in the spike regions to codon frequencies upstream of the spike regions, using the frame defined by the annotated CDS. We computed a P-value using a t-test for each codon comparing the number of occurrences in the spike regions (the spike position ±25 nt) for each significant spike to the 50-nt region immediately before the spike, and applied a Benjamini–Hochberg correction using a false discovery rate (FDR) threshold of 0.01.
. We found codons that are statistically enriched in the spike regions by comparing the frequencies of each codon in the spike regions to codon frequencies upstream of the spike regions, using the frame defined by the annotated CDS. We computed a P-value using a t-test for each codon comparing the number of occurrences in the spike regions (the spike position ±25 nt) for each significant spike to the 50-nt region immediately before the spike, and applied a Benjamini–Hochberg correction using a false discovery rate (FDR) threshold of 0.01.
RESULTS
Training and testing
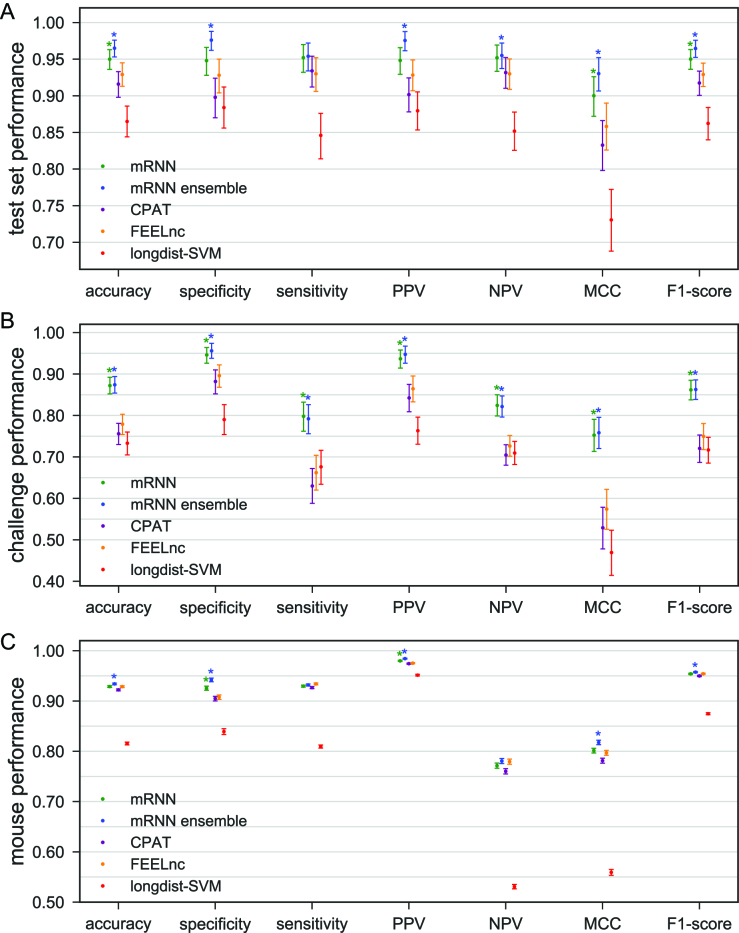
The best resulting mRNN model after training selected by accuracy on the validation set is referred to hereafter as ‘mRNN’. We also implemented an ensemble testing method called ‘mRNN ensemble,’ which uses the weighted average of the five best mRNN models. While the single best mRNN model matched or outperformed CPAT, FEELnc and longdist on the test set, the mRNN ensemble method showed significant improvements in performance over these methods in accuracy, specificity and other metrics at an FDR of 0.05 (Figure 2A). We also compared the classifiers using the challenge set of atypical transcripts (Figure 2B). Both mRNN and mRNN ensemble methods significantly outperformed CPAT, FEELnc and longdist in all metrics on this challenge set. Notably, CPAT, FEELnc and longdist showed low sensitivity for the challenge set (63, 66.2 and 67.6%, respectively), indicating a bias against classification of mRNAs with short ORFs as protein-coding, while mRNN ensemble achieved a sensitivity of 79.2%, demonstrating its superior predictive power for these atypical transcripts.
Figure 2.
Comparison of Classifier Performance. (A–C) Performance of four classifiers trained with human transcript sequences. Error bars are 95% confidence intervals computed from 100 000 bootstrap trials. Asterisks above mRNN or ensemble mRNN indicate the method’s improvement over CPAT, FEELnc and longdist-SVM with an empirical P-value < 0.05 computed from the bootstrap trials. (A) Performance on human test set transcripts, consisting of 500 mRNAs and 500 lncRNAs. (B) Performance on human challenge set transcripts, including 500 mRNAs with ORFs < 50 codons and 500 lncRNAs with ORFs > 50 codons. (C) Performance on GENCODE mouse transcripts greater than or equal to 200nt, including 61 834 mRNAs and 15 891 lncRNAs, using models trained with human data.
As a final test, we compared the generalizability of these methods trained with human data by evaluating their performance on the entire set of mouse GENCODE transcripts >200 nt. This dataset includes 61 834 mRNAs and 15 891 lncRNAs. The mRNN ensemble method performed best in all metrics except sensitivity, with statistically significant (P-value < 0.05) improvements in accuracy, specificity, PPV, MCC and F1-score, showing that it can be used for classification of long RNAs in a new transcriptome when trained on transcripts from a related species (Figure 2C).
Our results demonstrate that mRNN performed at least as well as existing state-of-the-art machine learning applications at the task of classifying mRNAs and lncRNAs, and it achieved this without the aid of user-defined features related to known mRNA characteristics. We next turned our attention to uncover what mRNN learned during training.
Point mutation analysis
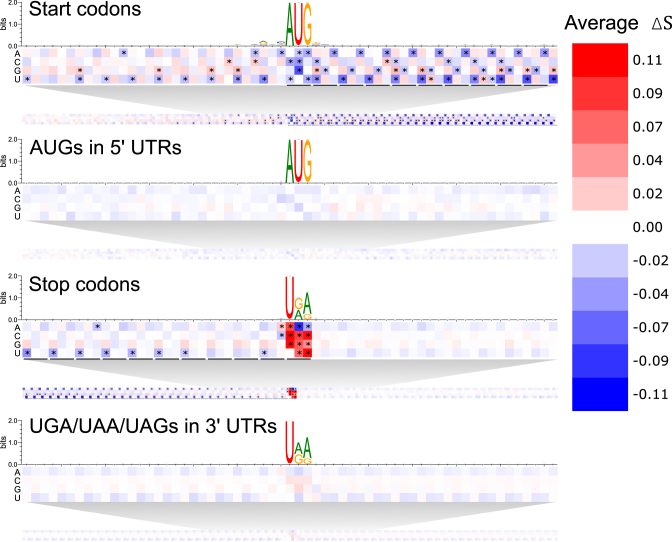
To begin deducing what mRNN learned, we conducted sequence perturbation analyses (Supplementary Methods). Score changes for sequences with shuffled coding sequence (CDS) regions compared to those with shuffled 3′ or 5′ untranslated regions (UTRs) demonstrate that mRNN primarily utilizes organized sequence information in the CDS (Supplementary Figure S6). We next conducted a point-mutation analysis to evaluate changes in score resulting from every possible single-nucleotide substitution for all GENCODE mRNA transcripts under 2000 nt in length (Figure 3). We analyzed score changes resulting from these mutations in thousands of transcripts at positions relative to start codons and stop codons or control AUGs and UGA/UAA/UAG trinucleotides. While the colors of the cells in the heat map in Figure 3 represent the average of a set of score changes resulting from mutations to that position and base over all transcripts examined, the asterisks indicate statistical significance (FDR of 1e-4) of a t-test comparing this set to a set of score changes resulting from mutations to each instance of the same base in the same region (5′ UTR, CDS or 3′ UTR). The annotated start codon marked a clear boundary, with low score changes preceding it and strong changes following it, indicating that sequence perturbations early in the CDS erase more predictive information than perturbations in the 5′ UTR. In contrast, score changes around non-start AUGs in the 5′ UTR were more symmetric before and after the AUG and significantly lower on average. Strikingly, the pattern of average score changes in the true CDS exhibited three-nucleotide periodicity with a persistent aversion to mutations that made codons more similar to in-frame stop codons. This pattern was not observed upstream of the annotated start codon (5′ UTR), nor in the regions flanking either AUGs in the 5′ UTR or control CUGs (Supplementary Figure S7). An aversion to stop codon-like trinucleotides was also observed preceding, but not following, annotated stop codons, suggesting that mRNN recognizes the end of the CDS. This pattern was not observed in regions preceding UGA/UAA/UAG trinucleotides in the 3′ UTR. Notably, mutation of the annotated stop codon significantly increased the coding potential score, showing that mRNN displays a preference for longer ORFs.
Figure 3.
Transcript Point Mutation Maps. Heat maps representing the average change in coding score for 10 s of thousands of transcripts due to point mutations at positions relative to the following elements (from top to bottom): annotated start codons, AUGs in 5′ UTRs, annotated stop codons, and UGA/UAA/UAGs in 3′ UTRs. Sequence logos present the nucleotide composition of the sequences analyzed around the same windows. Asterisks represent cells that are statistically significant at an FDR of 0.0001 using a two-tailed t-test comparing score changes over all transcripts with mutations at a given position to all score changes from mutations of the same base in the corresponding background region. Background regions are 5′ UTRs for the start codons and AUGs, or 3′ UTRs for the stop codons and UGA/UAA/UAGs.
Pairwise analysis
To evaluate whether mRNN learned relationships between distinct features, we performed a pairwise-mutation analysis. To select transcripts for analysis, we first identified those having a 5′ UTR of at least 100 nt, at least 100 codons in the CDS and at least 50 nt in the UTR. We then examined the shortest two transcripts with these properties for convenience of visual data representation. We define 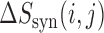 as the minimum of the difference between the coding score change resulting from mutations at two positions
as the minimum of the difference between the coding score change resulting from mutations at two positions 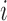 and
and 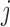 and the sum of score changes associated with the individual mutations. Therefore,
and the sum of score changes associated with the individual mutations. Therefore, 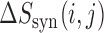 quantifies the ‘score change synergy’ of the pair of mutations, and is strongly negative for highly related positions. We examined both transcripts by altering every possible combination of two nucleotides within the sequence, 945 999 and 987 713 pairs in total.
quantifies the ‘score change synergy’ of the pair of mutations, and is strongly negative for highly related positions. We examined both transcripts by altering every possible combination of two nucleotides within the sequence, 945 999 and 987 713 pairs in total.
Score changes resulting from pairs of mutations made to the shorter transcript, encoding parathyroid hormone 2, PTH2, (Supplementary Figure S8A), illustrate that mRNN learned rules governing stop codons. Pairs of mutations that result in an early stop codon when combined significantly reduce the coding score (Supplementary Figure S8B). When one mutation creates a stop codon, a second mutation creating a second, earlier stop codon renders the latter mutation moot; the total score change for these two mutations is smaller than the sum of individual mutation score changes (Supplementary Figure S8C).
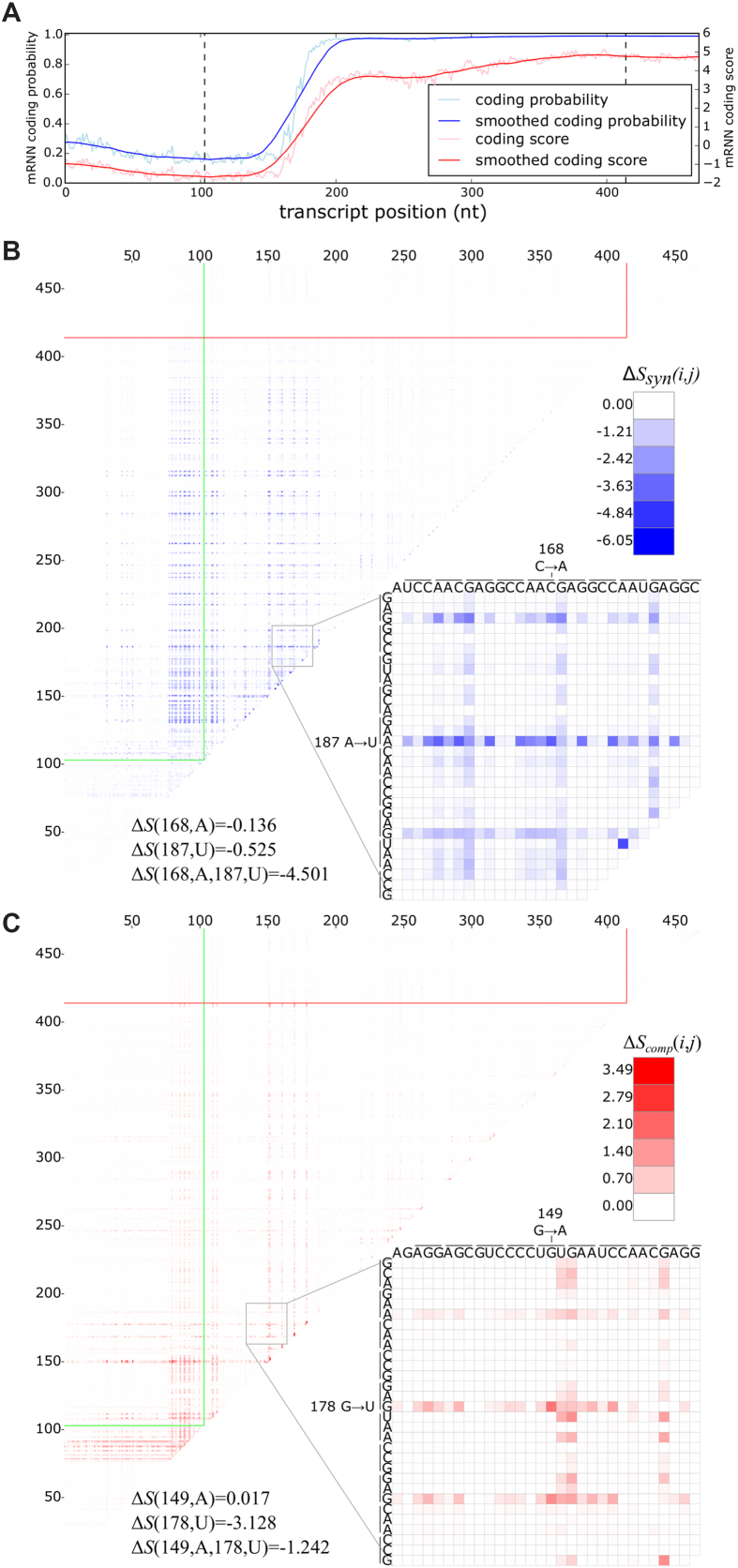
The second shortest transcript, encoding a cancer/testis-specific antigen SPANXB1, has a coding trajectory with a strong spike shortly after the start codon (Figure 4A). We identified several pairs of synergistic mutations, including a point mutation that changed an AAC codon to AAA and another that changed an AAG codon to UAG, which, when combined, resulted in the 12th largest score change synergy, resulting in a reduction in score 6.8-times the sum of the individual score changes (Figure 4B). In other examples, we identified compensatory changes, such as a decrease in score from a nonsense mutation that was significantly diminished when another mutation changed a UGU codon to UAU (Figure 4C). In both cases described, the two mutated positions are located within the coding score spike, despite being separated by 18 and 29 bases, respectively.
Figure 4.
Pair-wise mutation analysis. (A) The mRNN coding trajectory (as in Figure 1), for ENST00000449283.1, a transcript encoding SPANXB1. (B) Pair-wise mutation heat map of synergistic score changes for the same transcript. Values are the score change synergy for a pair of mutated bases at positions 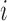 and
and 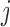 , where
, where 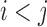 . Score change synergy is the minimum difference between the resulting change in score when the pair of bases is mutated and the sum of the score changes from individual mutations of each base in the pair. (C) Pair-wise mutation heat map of compensatory score changes for the same transcript. Values are the compensatory score change for a pair of mutated bases at positions
. Score change synergy is the minimum difference between the resulting change in score when the pair of bases is mutated and the sum of the score changes from individual mutations of each base in the pair. (C) Pair-wise mutation heat map of compensatory score changes for the same transcript. Values are the compensatory score change for a pair of mutated bases at positions 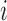 and
and 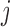 , where
, where 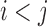 . Compensatory score change is the maximum difference between the resulting change in score when the pair of bases is mutated and the sum of the score changes from individual mutations of each base in the pair. (B and C) Bottom-right of each heat map shows a zoomed-in view of a position pair with a highly compensatory or synergistic score change. Each line spanning three nucleotides represents a codon.
. Compensatory score change is the maximum difference between the resulting change in score when the pair of bases is mutated and the sum of the score changes from individual mutations of each base in the pair. (B and C) Bottom-right of each heat map shows a zoomed-in view of a position pair with a highly compensatory or synergistic score change. Each line spanning three nucleotides represents a codon.
The examples above show that some mutations can exacerbate or compensate for the effects of other, ORF-truncating mutations. Notably, we found prediction-flipping pairs of synergistic mutations as far apart as 200 nt in the top 10 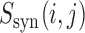 values (out of close to 500 000 negative values). Taken together, these results demonstrate that mRNN learned complex and long-range sequence information dependencies, and can leverage these rules for classification.
values (out of close to 500 000 negative values). Taken together, these results demonstrate that mRNN learned complex and long-range sequence information dependencies, and can leverage these rules for classification.
Coding trajectory analysis
To visualize mRNN’s decision-making process, we computed the coding trajectory 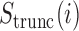 for all transcripts in the human test set (Figure 5A). Remarkably, we found several examples of mRNAs with long 5′ UTRs that mRNN classified as coding only after observing the CDS more than 4000 nt from the transcript start (Supplementary Figure S9). Thus, mRNN remains sensitive to information toward the end of transcripts longer than sequences previously used in any bioinformatics RNN applications that we are aware of, despite being trained only on sequences shorter than 1000 nt long.
for all transcripts in the human test set (Figure 5A). Remarkably, we found several examples of mRNAs with long 5′ UTRs that mRNN classified as coding only after observing the CDS more than 4000 nt from the transcript start (Supplementary Figure S9). Thus, mRNN remains sensitive to information toward the end of transcripts longer than sequences previously used in any bioinformatics RNN applications that we are aware of, despite being trained only on sequences shorter than 1000 nt long.
Figure 5.
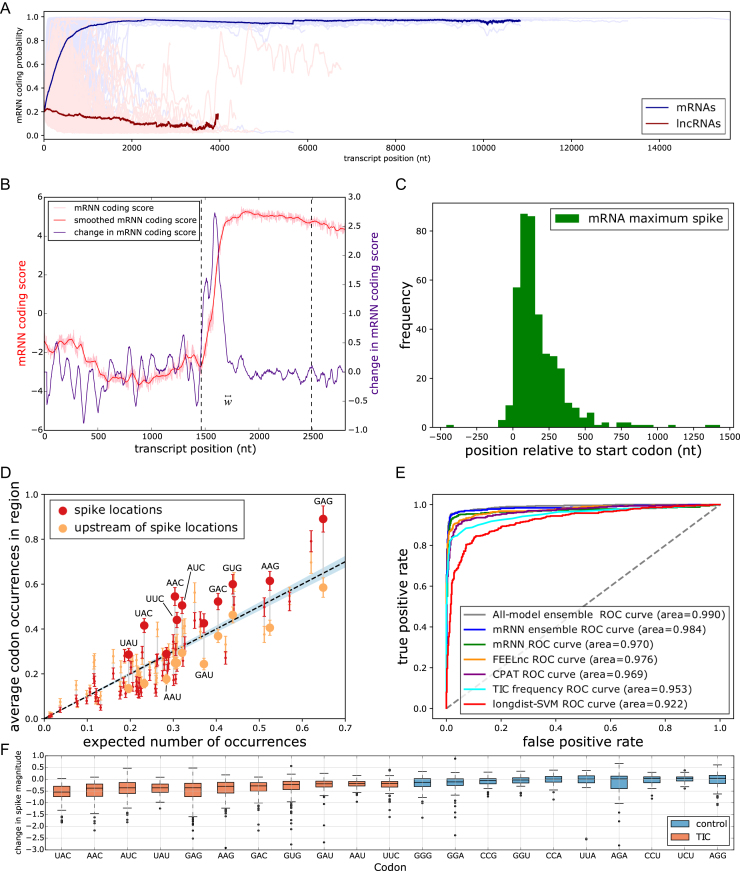
Model Interrogation for Feature Discovery. (A) mRNN coding score trajectories without smoothing for each transcript in the test set. Blue, protein-coding; red, non-coding. Bold lines represent average coding probability when five or more transcripts had lengths at of least 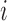 nt. (B) Coding score trajectory for transcript ENST00000458629.1, which encodes C-X-C motif chemokine receptor 6. Vertical dashed lines mark CDS boundaries. (C) Histogram of significant spike locations in test set mRNAs relative to true CDS start positions. (D) Scatterplot showing codons enriched in the spike regions (±25 nt around most significant spike position) compared to 50-nt regions upstream of the spikes. The x-axis is the frequency of each codon in the full set of GENCODE annotated coding regions. The y-axis represents the frequency of the codon in the indicated region. Each pair of points represents a codon. Large, labeled points are TICs—codons statistically enriched (FDR ≤ 0.05) in spike regions compared to the regions upstream of spikes. The dashed line corresponds to global codon frequency, and the blue band is the range of standard error computed from a binomial model. (E) Receiver operator characteristic analysis for five prediction methods including our mRNN ensemble, the best single mRNN model, FEELnc, CPAT, longdist-SVM, TIC frequency and all-model ensemble (a uniformly weighted ensemble of 5 mRNN models, FEELnc and CPAT). TIC frequency is the number of occurrences of TICs within 1000 nt of, and in-frame with, an upstream AUG, but not after an in-frame UGA/UAA/UAG. AUROC values for each method are presented in the legend. (F) mRNN coding score changes resulting from in silico TIC mutations. While the majority of mutations to TICs lead to a decrease in coding score, mutations to control codons (the codons least enriched in the spike regions) result in smaller score changes on average.
nt. (B) Coding score trajectory for transcript ENST00000458629.1, which encodes C-X-C motif chemokine receptor 6. Vertical dashed lines mark CDS boundaries. (C) Histogram of significant spike locations in test set mRNAs relative to true CDS start positions. (D) Scatterplot showing codons enriched in the spike regions (±25 nt around most significant spike position) compared to 50-nt regions upstream of the spikes. The x-axis is the frequency of each codon in the full set of GENCODE annotated coding regions. The y-axis represents the frequency of the codon in the indicated region. Each pair of points represents a codon. Large, labeled points are TICs—codons statistically enriched (FDR ≤ 0.05) in spike regions compared to the regions upstream of spikes. The dashed line corresponds to global codon frequency, and the blue band is the range of standard error computed from a binomial model. (E) Receiver operator characteristic analysis for five prediction methods including our mRNN ensemble, the best single mRNN model, FEELnc, CPAT, longdist-SVM, TIC frequency and all-model ensemble (a uniformly weighted ensemble of 5 mRNN models, FEELnc and CPAT). TIC frequency is the number of occurrences of TICs within 1000 nt of, and in-frame with, an upstream AUG, but not after an in-frame UGA/UAA/UAG. AUROC values for each method are presented in the legend. (F) mRNN coding score changes resulting from in silico TIC mutations. While the majority of mutations to TICs lead to a decrease in coding score, mutations to control codons (the codons least enriched in the spike regions) result in smaller score changes on average.
To identify regions of the sequence that most strongly impact mRNN’s decision, we performed unweighted sliding-average smoothing of the coding potential trajectories, then computed the change in score 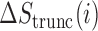 across the sequence for a window
across the sequence for a window 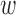 of 50 nt (Figure 5B). Statistically significant spikes (Supplementary Figure S10) were identified in 412 of the 500 test mRNAs, and in only 47 of the 500 lncRNAs by fitting a Gaussian to the distribution of lncRNA spike magnitudes, and computing a P-value for the mRNA spike values with these parameters. The distribution of the significant spike positions for mRNAs peaked within the CDS, shortly after the start codon (Figure 5C; Supplementary Figures S11 and 12A-B).
of 50 nt (Figure 5B). Statistically significant spikes (Supplementary Figure S10) were identified in 412 of the 500 test mRNAs, and in only 47 of the 500 lncRNAs by fitting a Gaussian to the distribution of lncRNA spike magnitudes, and computing a P-value for the mRNA spike values with these parameters. The distribution of the significant spike positions for mRNAs peaked within the CDS, shortly after the start codon (Figure 5C; Supplementary Figures S11 and 12A-B).
To identify the sequence elements associated with significant spikes in coding potential score, we compared the frequencies of in-frame codons in 50-nt windows centered at the spike, to the codon frequencies in the 50-nt windows preceding these spikes. We found 11 significantly enriched codons using a t-test and an FDR of 0.05 (Figure 5D and Supplementary Table S1); we named these translation-indicating codons (TICs). 9 of the 11 TICs were also significantly enriched in spike regions of an independent set of mRNAs with long 5′ UTRs (Supplementary Figure S12). Notably, two codons in the synergistic and compensatory pairwise mutation examples above (AAC and UAU) are TICs.
To assess the predictive power of TICs, we defined a TIC-score as the maximum number of TICs occurring within 1000 nt downstream of an in-frame AUG, and preceding the first in-frame stop codon. This TIC-score was able to accurately predict coding potential in the test set with an AUROC of 0.953, just below that of CPAT at 0.969 and above longdist (Figure 5E). The same rule distinguished mRNAs from lncRNAs in the full mouse GENCODE dataset with an AUROC of 0.939 (Supplementary Figure S13). We next computed the reduction in the spike magnitude—the change in 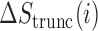 —resulting from the mutation of a given TIC codon in silico. Mutation of TICs resulted in spike height decreases 94.7% of the time, while mutations to the least enriched codons in the spike regions decreased spike height only 59.9% of the time (Figure 5F), demonstrating that TICs are an important part of mRNN’s classification process. We also identified a frame-biased, 12-mer motif enriched in spike regions, which possesses some predictive power (Supplementary Figure S14 and Tables S2-3). Some of the TICs are enriched in GENCODE CDSs relative to UTRs and out-of-frame triplets, demonstrating that mRNN learned the complex sequence context that gives these codons predictive power (Supplementary Figure S15).
—resulting from the mutation of a given TIC codon in silico. Mutation of TICs resulted in spike height decreases 94.7% of the time, while mutations to the least enriched codons in the spike regions decreased spike height only 59.9% of the time (Figure 5F), demonstrating that TICs are an important part of mRNN’s classification process. We also identified a frame-biased, 12-mer motif enriched in spike regions, which possesses some predictive power (Supplementary Figure S14 and Tables S2-3). Some of the TICs are enriched in GENCODE CDSs relative to UTRs and out-of-frame triplets, demonstrating that mRNN learned the complex sequence context that gives these codons predictive power (Supplementary Figure S15).
DISCUSSION
In this study, we have shown that GRU networks can successfully model full-length human transcripts. Previous bioinformatics applications of RNNs restricted input sequence length to 2000 nt or fewer by one of three strategies: filtering the dataset on a length threshold (aRxiv: https://arxiv.org/abs/1701.08318v1), dividing input sequences into segments of a fixed size (23,24), or truncating input sequences (25). However, one important advantage that deep RNNs have over other deep learning methods is the ability to interpret context and long-range information dependencies. In order to exploit the full power of our GRU network, we did not truncate or segment our training sequences, and we did not constrain our test set inputs by sequence length in any way. Our model showed no impairment in classifying long transcripts when evaluated on the entire mouse transcriptome, or even the longest sequences in human, which exceeded 100 000 nt.
Despite mRNN’s featureless architecture, which precluded it from integrating human knowledge of mRNA structure into its learning process, mRNN was able to learn true defining features of mRNAs, including trinucleotide patterns and depletion of in-frame stop codons after the start of an open-reading frame. In addition to surpassing state-of-the-art accuracy in assessment of transcript coding potential, we demonstrate that the GRU network can be harnessed for identifying specific biological attributes, such as the TICs, that distinguish sequence classes. Many TICs are statistically enriched in coding regions and may affect mRNA structure and translation efficiency. Interestingly, some TIC mutations are known to affect protein expression in human disease contexts; for example, in a study of a disease-causing mutant of the cystic fibrosis transmembrane conductance regulator, Bartoszewski et al. found that a single synonymous mutation of Ile507ATC (a TIC) to Ile507ATT (a non-TIC) altered the mRNA structure and reduced expression of the protein (26). Future work is needed to assess whether TICs play a more general role in mRNA structure and protein expression.
At last, we showed that the recurrent nature of mRNN enabled it to leverage long-range information dependencies for classification, as evidenced by the pairwise mutation analysis. This analysis identified many compensatory and synergistic relationships between distant codons, which may be generally important for protein function conservation (27) and adaptation (28), respectively. In agreement with a previous study of intragenic epistasis in prokaryotes, eukaryotes and viruses, we observed that the majority of compensatory mutations occurred between nearby codons (29); however, mRNN was also able to identify long-range compensatory mutations.
We anticipate that GRU-based approaches will be highly useful for future bioinformatics classification tasks, as well as for uncovering new biological insights in the vast amounts of available sequencing data.
DATA AVAILABILITY
Source code implementing data preprocessing, training and downstream analysis is available in the package mRNN from http://github.com/hendrixlab/mRNN.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Stephen Ramsey, Prof. Christopher K. Mathews, Prof. Liang Huang, Prof. Colin Johnson, Prof. P. Andy Karplus and Prof. Michael Freitag for feedback on the manuscript and helpful discussions. The authors thank Mike Tyka for the suggestion to use data augmentation.
Authors’ contribution: S.H., R.K., E.M., A.T. and D.H. wrote the software. S.H., R.K., A.T., P.D. and D.H. did the bioinformatics analysis. R.K., D.H. and S.H. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R56 AG053460, R21 AG052950]; Oregon State University (start-up grant). Funding for open access charge: NIH [R56 AG053460].
Conflict of interest statement. None declared.
REFERENCES
- 1. Goodfellow I., Bengio Y., Courville A.. Deep Learning. 2016; MIT Press; http://www.deeplearningbook.org/. [Google Scholar]
- 2. Zhou J., Troyanskaya O.G.. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015; 12:931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alipanahi B., Delong A., Weirauch M.T., Frey B.J.. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015; 33:831–838. [DOI] [PubMed] [Google Scholar]
- 4. Kelley D.R., Snoek J., Rinn J.L.. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016; 26:990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung J., Gulcehre C., Cho K., Bengio Y.. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. NIPS Deep Learn. Workshop. 2014; http://arxiv.org/abs/1412.3555. [Google Scholar]
- 6. Wang Y., Shen F., Elayavilli R.K., Liu S., Rastegar-Mojarad M., Liu H.. MayoNLP at the BioCreative VI PM Track: Entity-enhanced Hierarchical Attention Neural Networks for Mining Protein Interactions from Biomedical Text. Proceedings of the BioCreative VI Challenge Evaluation Workshop. 2017; 127–130. [Google Scholar]
- 7. Zhang Y., Zheng W., Lin H., Wang J., Yang Z., Dumontier M.. Drug–drug interaction extraction via hierarchical RNNs on sequence and shortest dependency paths. Bioinformatics. 2018; 34:828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rastegar-Mojarad M., Elayavilli R., Wang Y., Liu S., Shen F., Liu H.. Semantic Information Retrieval: Exploring Dependency and Word Embedding Features in Biomedical Information Retrieval. Proceedings of the BioCreative VI Challenge Evaluation Workshop. 2017; 74–77. [Google Scholar]
- 9. Lee B., Lee T., Na B., Yoon S.. DNA-Level splice junction prediction using deep recurrent neural networks. CoRR. 2015; abs/1512.05135:https://dblp.org/rec/bib/journals/corr/LeeLNY15. [Google Scholar]
- 10. Lee B., Baek J., Park S., Yoon S.. deepTarget: End-to-end Learning Framework for microRNA Target Prediction using Deep Recurrent Neural Networks. Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. 2016; Seattle: ACM; 434–442. [Google Scholar]
- 11. Hassanzadeh H.R., Wang M.D.. DeeperBind: Enhancing prediction of sequence specificities of DNA binding proteins. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2016; Shenzhen: IEEE; 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angermueller C., Lee H.J., Reik W., Stegle O.. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017; 18:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park S., Min S., Choi H.-S., Yoon S.. Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, Garnett R. Deep Recurrent Neural Network-Based Identification of Precursor microRNAs. Advances in Neural Information Processing Systems. 2017; Long Beach: Neural Information Processing Systems Foundation, Inc; 2895–2904. [Google Scholar]
- 14. Hochreiter S., Bengio Y., Frasconi P., Schmidhuber J.. Gradient flow in recurrent nets: the difficulty of learning long-term dependencies. A field guide to dynamical recurrent neural networks. 2001; IEEE Press. [Google Scholar]
- 15. Hochreiter S., Schmidhuber J.. Long short-term memory. Neural Comput. 1997; 9:1735–1780. [DOI] [PubMed] [Google Scholar]
- 16. Zhang J.M., Kamath G.M.. Learning the Language of the Genome using RNNs. [Google Scholar]
- 17. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S.. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wucher V., Legeai F., Hedan B., Rizk G., Lagoutte L., Leeb T., Jagannathan V., Cadieu E., David A., Lohi H.. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017; 45:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Dyk D.A., Meng X.-L.. The art of data augmentation. J. Comput. Graph. Stat. 2001; 10:1–50. [Google Scholar]
- 20. Perrone M.P., Cooper L.N.. When Networks Disagree: Ensemble Methods for Hybrid Neural Networks. Neural Networks for Speech and Image processing. 1993; Chapman-Hall; 126–142. [Google Scholar]
- 21. Wang L., Park H.J., Dasari S., Wang S., Kocher J.-P., Li W.. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013; 41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schneider H.W., Raiol T., Brigido M.M., Walter M.E.M., Stadler P.F.. A Support Vector Machine based method to distinguish long non-coding RNAs from protein coding transcripts. BMC Genomics. 2017; 18:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hochreiter S., Heusel M., Obermayer K.. Fast model-based protein homology detection without alignment. Bioinformatics. 2007; 23:1728–1736. [DOI] [PubMed] [Google Scholar]
- 24. Zhang S., Hu H., Jiang T., Zhang L., Zeng J.. TITER: predicting translation initiation sites by deep learning. Bioinformatics. 2017; 33:i234–i242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sønderby S.K., Sønderby C.K., Nielsen H., Winther O.. Dediu A-H, Hernández-Quiroz F, Martín-Vide C, Rosenblueth DA. Convolutional LSTM Networks for Subcellular Localization of Proteins. International Conference on Algorithms for Computational Biology. 2015; Mexico city: Springer; 68–80. [Google Scholar]
- 26. Bartoszewski R.A., Jablonsky M., Bartoszewska S., Stevenson L., Dai Q., Kappes J., Collawn J.F., Bebok Z.. A synonymous single nucleotide polymorphism in ΔF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 2010; 285:28741–28748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y., Meng X., Yang Y., Li H., Wang X., Yang B., Zhang J., Li C., Millar N.S., Liu Z.. Synergistic and compensatory effects of two point mutations conferring target-site resistance to fipronil in the insect GABA receptor RDL. Sci. Rep. 2016; 6:32335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dickinson W.J. Synergistic fitness interactions and a high frequency of beneficial changes among mutations accumulated under relaxed selection in Saccharomyces cerevisiae. Genetics. 2008; 178:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davis B.H., Poon A.F., Whitlock M.C.. Compensatory mutations are repeatable and clustered within proteins. Proc. R. Soc. Lond. B Biol. Sci. 2009; 276:1823–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code implementing data preprocessing, training and downstream analysis is available in the package mRNN from http://github.com/hendrixlab/mRNN.