Abstract
Transcriptional memory is critical for the faster reactivation of necessary genes upon environmental changes and requires that the genes were previously in an active state. However, whether transcriptional repression also displays ‘memory’ of the prior transcriptionally inactive state remains unknown. In this study, we show that transcriptional repression of ∼540 genes in yeast occurs much more rapidly if the genes have been previously repressed during carbon source shifts. This novel transcriptional response has been termed transcriptional repression memory (TREM). Interestingly, Rpd3L histone deacetylase (HDAC), targeted to active promoters induces TREM. Mutants for Rpd3L exhibit increased acetylation at active promoters and delay TREM significantly. Surprisingly, the interaction between H3K4me3 and Rpd3L via the Pho23 PHD finger is critical to promote histone deacetylation and TREM by Rpd3L. Therefore, we propose that an active mark, H3K4me3 enriched at active promoters, instructs Rpd3L HDAC to induce histone deacetylation and TREM.
INTRODUCTION
Post-translational modifications, including acetylation, methylation, phosphorylation and ubiquitination, of histone tails play important roles in eukaryotic transcription (1,2). Histone acetylation directly promotes RNA polymerase II (RNA Pol II) transcription by disrupting histone–DNA interactions and by recruiting additional factors that control local chromatin structure. Histone acetylation is dynamically controlled by both histone acetyltransferases (HATs) and histone deacetylases (HDACs).
A conserved HDAC, Rpd3 is the catalytic subunit of at least two distinct HDACs (3,4). Rpd3 large (Rpd3L) HDAC includes more than 10 subunits and presumably deacetylates histones at promoter regions (5,6). Rpd3L is targeted to inactive promoters by sequence-specific repressors including Ume6 (7). Interestingly, genome-wide chromatin immunoprecipitation-DNA microarray (ChIP-ChIP) analyses revealed that Rpd3L also associates with actively transcribed genes (5,8). Consistent with this data, the loss of Rpd3L increases histone acetylation at active promoters (6,9). In contrast, Rpd3 small (Rpd3S) HDAC primarily deacetylates histones within coding regions to suppress initiation from cryptic promoters, and histone exchange within coding regions, and RNA Pol II elongation (3,4,10–12).
Histone deacetylation by Rpd3L or Rpd3S at distinct regions is likely mediated by co-transcriptional methylations on histone tails (13). During the early stage of transcription, the Set1 methyltransferase interacts with the serine 5 phosphorylated C-terminal domain (CTD) of Rpb1, the largest subunit of RNA Pol II, and/or nascent RNA transcripts to localize H3K4me3 in promoter regions, followed by H3K4me2 in 5′ regions and H3K4me1 in 3′ ends of genes (14–16). Two subunits of Rpd3L, Pho23 and Cti6/Rxt1, can directly bind to trimethylated K4 of histone H3 in vitro (17,18), but whether these interactions contribute to chromatin binding and/or enzyme activity of Rpd3L has not yet been determined. Both Set1 and Rpd3L mediate repression of genes involved in ribosome biogenesis suggesting a link between H3K4 methylation and Rpd3L (19). During transcription elongation, the Set2 methyltransferase interacts with phosphorylated CTD at both serines 5 and 2 to target H3K36me2/3 in the body of genes (20–22). This modification is recognized by the chromodomain of Eaf3 within Rpd3S HDAC. An additional subunit, Rco1, interacts with unmodified histone tails. These two interactions are critical for chromatin binding and histone deacetylation by Rpd3S (3,4,23,24).
During natural growth, cells are exposed to rapidly changing environments and must accordingly reprogram their gene expression patterns for cellular differentiation, development, and adaptation. In addition, they are often re-exposed to the same or different stimuli and can rapidly re-induce the genes required for cellular functions. This response is known as ‘transcriptional memory’ and increases the kinetics of reactivation (25,26). In yeast, transcriptional memory involving inducible GAL genes and INO1 has been extensively studied and the factors involved in have been identified. Multiple mechanisms appear to contribute. For example, transcriptional memory of GAL genes requires the SWI/SNF chromatin remodeling complex and the Htz1 histone variant and is negatively regulated by the Set1 methyltransferase (25,27–29). In addition, long-term transcriptional memory of GAL genes which persists for >6 cell divisions, is positively regulated by Gal1 and Gal3 metabolic proteins (27,30). INO1 memory is also positively regulated by Htz1 (29). Furthermore, the nuclear pore complex that targets active genes to the nuclear periphery also contributes to transcriptional memory of GAL genes and INO1 (31,32).
Although cells must repress unnecessary genes upon environmental changes, whether a gene remembers its previous transcriptionally inactive state remains unclear. In this study, we show that more than 540 yeast genes exhibit ‘memory’ of their preceding inactive states during carbon-source shifts. These genes were slightly repressed during the first galactose incubation, but strong and rapid suppression was seen upon the second galactose pulse. This novel response has been named ‘Transcriptional REpression Memory’ (TREM). Interestingly, Rpd3L bound to active promoters plays important roles in TREM. Whereas loss of Rpd3L had a lesser effect on the kinetics of the first repression, Rpd3L mutants, but not other HDAC mutants tested, displayed a significant delay in the kinetics of the second repression. Surprisingly, the interaction between the Pho23 plant homeodomain (PHD) finger and an active mark, H3K4me3 is critical for histone deacetylation by Rpd3L and is sufficient to promote TREM.
MATERIALS AND METHODS
Yeast strains and culture conditions
Yeast strains used in this study are listed in Supplementary Table S1. The time course experiments were carried out with the indicated strains as previously described (33,34). To generate pho23 (W305A) and cti6 (W98A) strains, the kanMX4 and KlURA3 CORE cassette was integrated 915 or 294 bp downstream from the PHO23 ATG or the CTI6 ATG, respectively. The resulting strains were transformed with the 100 bp oligonucleotides to introduce pho23 (W305A) or cti6 (W98A) mutation.
Reverse transcription and qPCR analysis
RNA was isolated from cells with hot acidic phenol/chloroform (1:1) (Sigma). 0.78 μg of total RNAs treated with RNase-free DNaseI (Thermo Fisher Scientific) were reverse-transcribed to complementary DNA (cDNA) using PrimeScript II first-strand cDNA Synthesis Kit (TaKaRa). The resulting cDNAs were analyzed by real-time quantitative PCR (RT-qPCR) using CFX96 cycler (Bio-rad) and iQ SYBR Green Supermix (Bio-rad). The sequences of oligonucleotides used in this study are listed in Supplementary Table S2.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitations (ChIP) was carried out as previously described (9) with oligonucleotides as listed in Supplelmentary Table S2. 2.0 μl of anti-Rpb3 (BioLegend 626802), 1.0 μl of anti-acetyl H4 (Upstate 06-598), 1.0 μl of anti-H3 (Abcam 1791), or 2.0 μl of anti-myc (BioLegend 65004) was bound to either Protein G or Protein A agarose beads. Binding for anti-Rpb3, anti-H3 and anti-myc or for anti-acetyl H4 was done overnight in FA lysis buffer containing 275 mM NaCl or in FA lysis buffer with 1 M NaCl, respectively. Precipitates were washed with the same buffer, once with FA lysis buffer containing 500 mM NaCl for anti-Rpb3, anti-H3 and anti-myc or with FA lysis buffer containing 1.5 M NaCl for anti-acetyl H4, once with 10 mM Tris–HCl (pH 8.0), 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate, and once with TE (10 mM Tris–HCl [pH 8.0], 1 mM EDTA). Precipitated DNAs were analyzed by real-time quantitative PCR using iQ SYBR Green Supermix (Bio-rad) and CFX96 cycler (Bio-Rad).
Western blot analysis
Cells expressing TAP-tagged or myc-tagged proteins were grown in YPD at 30°C or SC-glucose at 22°C to mid-log phase. Cell were lysed using lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40) with protease inhibitors (Pepstatin A 1 mM, Aprotinine 0.3 mM, Leupeptin 1 mM, PMSF 100 mM) and glass beads. Protein concentration was quantitated by Bradford assay. For SDS-PAGE and western blot analyses, 15–30 μg of protein was used. Proteins were separated in SDS-PAGE and transferred onto a PVDF membrane (Millipore). Membranes were incubated with primary antibody for 3 h and then washed with PBST. After incubation with HRP-conjugated secondary antibody for 1 h, membranes were washed three times with PBST. The blots were visualized on film with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Northern blot analysis
Total RNA was isolated from cells with hot acidic phenol/chloroform (1:1) (Sigma) and 10 μg of total RNA was separated on agarose gel. The RNA was transferred onto membranes and hybridized to radioactive probes as described previously (35). Strand-specific radioactive probes were generated by unidirectional PCR with [α-32P] dATP with only one primer. Hybridization was carried out in a buffer containing 1% BSA, 7% SDS, 1 mM EDTA (pH 8.0) and 300 mM sodium phosphate buffer (pH 7.2).
Histone peptide pulldown assay
Cells expressing myc-tagged or TAP-tagged proteins were grown at 30°C to mid-log phase in YPD. Total proteins were extracted using binding buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 0.05% NP-40) with protease inhibitors (Pepstatin A 1 mM, Aprotinine 0.3 mM, Leupeptin 1 mM, PMSF 100 mM). The extracts were diluted to 150 or 300 mM NaCl and then incubated with 50 μl of dynabeads and 1 μg of biotinylated histone peptides at 4°C overnight. Beads were washed five times with 1 ml of binding buffer and separated by SDS-PAGE followed by western blot analysis.
RNA sequencing and data analysis
Sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina) after ribosomal RNA was depleted using the Ribo-Zero yeast kit (Epicenter), as recommended by manufacturers. Paired-end sequencing of 101-mer read length was carried out on an Illumina NextSeq 500 system. Raw sequencing data were preprocessed using the fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) which included fastq_quality_filter to remove low quality reads and fastx_artifacts_filter to remove homopolymeric sequencing artifacts. Resulting reads were then aligned to the S. cerevisiae S288C reference genome (EF4.74 downloaded from Ensembl) using TopHat version 2.0.13 (https://ccb.jhu.edu/software/tophat/index.shtml) (36). For strand-specific alignment, we used the option of –fr-firststrand and supplied the sample-specific values of insert size (i.e. average and standard deviation) that were obtained from the Bowtie alignment of paired-end reads on the same scaffold with concordant directions. Transcript abundance in FPKM (Fragments Per Kilobase of exon per Million mapped reads) was calculated by Cufflinks v2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/) (37) followed by differential expression analysis using Cuffdiff with the FDR value < 0.05.
Gene ontology (GO) analysis
TREM genes were analyzed for their functional enrichment in the GO terms using ‘GOSlim Process’ annotation in the GeneCodis3 (http://genecodis.cnb.csic.es/) (38). Significant terms were obtained with FDR < 0.05.
ChIP–ChIP data analysis
The raw ChIP-ChiP data on myc-tagged Rpd3 and Sds3 were downloaded from the GEO database (accession number: GSE22636) (5). Ringo (R Investigation of ChIP-chip Oligoarrays) (39) and limma (Linear Models for Microarray Data) R/Bioconductor packages (https://bioconductor.org/) were used for background intensity correction and lowess normalization, respectively. The normalized intensity profiles across the gene from −500 bp from the transcription start site to +500 bp from the transcription end site were obtained by fitting the probe intensities using the lowess fit function in R for each gene. The normalized intensities at 11 points over the gene were estimated using fitted function and the averaged binding profiles for total genes, Rpd3L-senstive genes, or Rpd3L-insensitive genes were shown in Figure 4C and Supplementary Figure S4D.
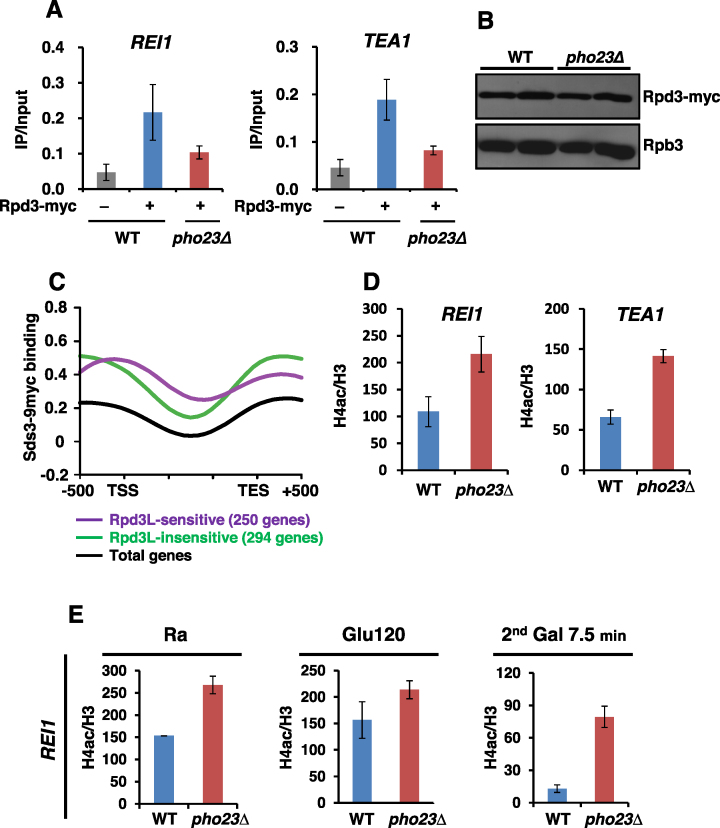
Figure 4.
Rpd3L binds to active promoters to deacetylate histones. (A) Rpd3 binds to promoters of TREM genes. Crosslinked chromatin from the indicated strains grown in SC medium containing glucose was precipitated with an anti-myc antibody. PCR analysis of the precipitated DNA was carried out on the promoter regions of REI1 and TEA1. The signals for Rpd3-myc were quantitated and normalized to the input signal, and the ratios were graphed. Error bars show the S.D. calculated from two biological replicates, each with three technical replicates. (B) Deletion of PHO23 did not affect Rpd3 protein levels. Total proteins extracted from wild type and pho23Δ cells grown in YPD were separated by SDS-PAGE and probed with antibodies indicated on the right. Rpb3 was used as a loading control. (C) Rpd3L occupancy of TREM genes was analyzed using the data sets from Drouin et al. (5). The graph shows the average occupancy of Sds3, a component of Rpd3L, for total genes (black), for 250 Rpd3L-sensitive genes (purple), and for 294 Rpd3L-insensitive genes (green). (D) Rpd3L deacetylates histones at TREM gene promoters. Crosslinked chromatin from the indicated strains grown in SC medium containing glucose was precipitated with anti-H3 or anti-acetyl H4 as indicated. PCR analysis of the precipitated DNA was carried out on the promoters of REI1 and TEA1 as in (A). A non-transcribed region near the telomere of chromosome VI was used for an internal control. The signals for acetyl H4 were quantitated and normalized to the total H3 signal, and the ratios were graphed. Error bars show the standard deviation (S.D.) calculated from two biological replicates, each with three technical replicates. (E) Loss of Pho23 delays deacetylation at the REI1 promoter upon the second galactose incubation. Histone acetylation at the indicated time points was analyzed as in (D).
ChIP-Seq analysis for H3K4me3 pattern
The raw MNase ChIP-Seq data on histone modifications were downloaded from the GEO database (accession number: GSE61888) (40). All sequencing reads were mapped to the S. cerevisiae genome using Bowtie2 ver. 2.2.3 (41) with the default parameters except the ‘–very-sensitive’ option. DANPOS2 ver. 2.2.2 (https://sites.google.com/site/danposdoc/) (42) was used to identify the nucleosome positions enriched with H3K4me3 relative to the input DNA. Specifically, the nucleosome positions were initially identified using the ‘Dpos’ function of DANPOS and then H3K4me3 profiles were retrieved around the transcription start site (from −500bp to +1500bp) using the ‘profile’ function of DANPOS (Figure 5B).
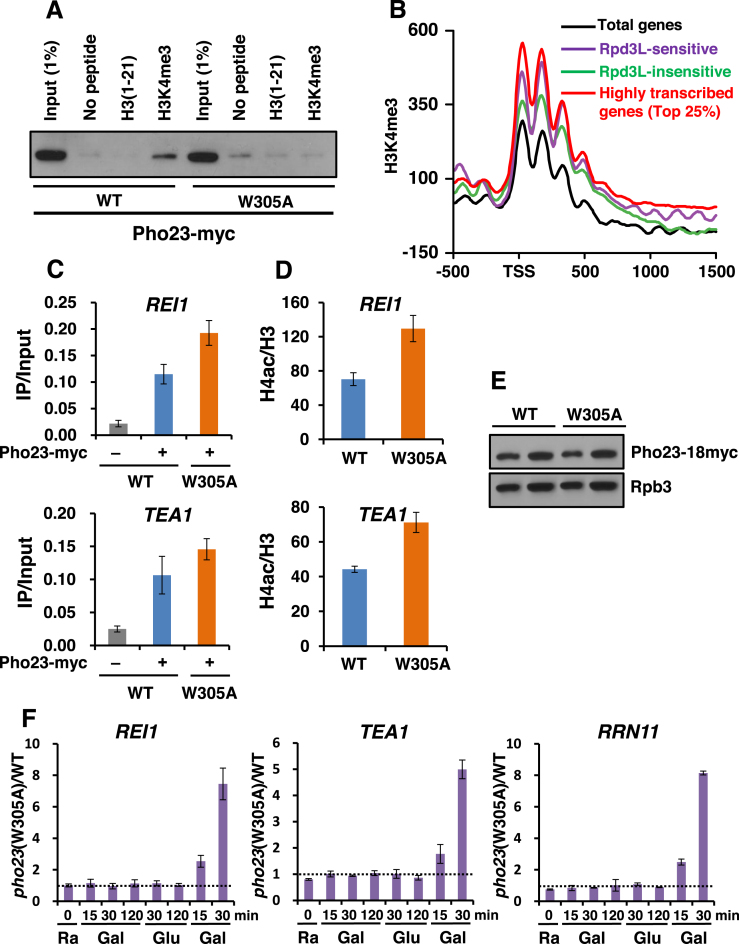
Figure 5.
The interaction between the Pho23 PHD finger and H3K4me3 induces deacetylation and TREM. (A) The Pho23 PHD finger binds to H3K4me3. Histone peptide pulldown assays were performed with whole cell extracts expressing Pho23-myc or a PHD finger mutant, pho23W305A-myc, and 1 μg of the indicated histone peptides immobilized on magnetic beads. Precipitated Pho23 protein was analyzed by immunoblot analyses with an anti-myc antibody. (B) H3K4me3 patterns of TREM genes were analyzed using the data sets from Weiner et al., (40). The graph shows the average enrichment of H3K4me3 for total genes (black), for Rpd3L-sensitive genes (purple), for Rpd3L-insensitive genes (green), and for top 25% highly transcribed genes (red). (C) Pho23 occupancy to two TREM genes was analyzed as in Figure 4A. Crosslinked chromatin from the indicated strains grown in SC medium containing glucose was precipitated with an anti-myc antibody. PCR analyses of the precipitated DNA was carried out on the promoter regions of REI1 and TEA1. (D) The interaction between the Pho23 PHD finger and H3K4me3 promotes deacetylation by Rpd3L. Histone H4 acetylation was analyzed as in Figure 4D. (E) A mutation in the Pho23 PHD finger does not affect its protein levels. Total proteins extracted from wild type (WT) and a PHD finger mutant, pho23 (W305A) cells grown in YPD were separated by SDS-PAGE and probed with the antibodies indicated on the right. Rpb3 was used as a loading control. (F) Pho23 binding to H3K4me3 is crucial for TREM. Transcript levels from PHO23 and pho23 (W305A) cells during carbon-source shifts were determined by RT-PCR as in Figure 2B. The ratio of transcript levels in pho23 (W305A) cells over those in PHO23 cells were plotted. Error bars show the standard deviation (S.D.) calculated from two biological replicates, each with three technical replicates.
RESULTS
Transcriptional REpression Memory: TREM
To investigate global transcriptional memory, the high-density tiled array datasets generated from yeast cells undergoing a series of carbon source shifts were analyzed (33,34). These datasets have been generated from cells that were initially grown in synthetic complete (SC) medium containing raffinose (0 time point), shifted to a medium containing galactose (for 15, 30, 60 and 120 min), then shifted to a medium containing glucose (for 15, 30, 60 and 120 min), and then shifted back to a medium containing galactose (for 15 and 30 min) (Figure 1A). Under these conditions, ∼1000 genes responded differentially to the distinct carbon sources (33). Many genes including TKL2, STL1 and HXT5, were induced by galactose and then repressed gradually when shifted to medium containing glucose. Interestingly, stronger and faster reactivation was seen upon the second exposure to galactose (Figure 1B). Although these genes are not directly involved in galactose metabolism, they showed memory of their prior active states during the first galactose incubation.
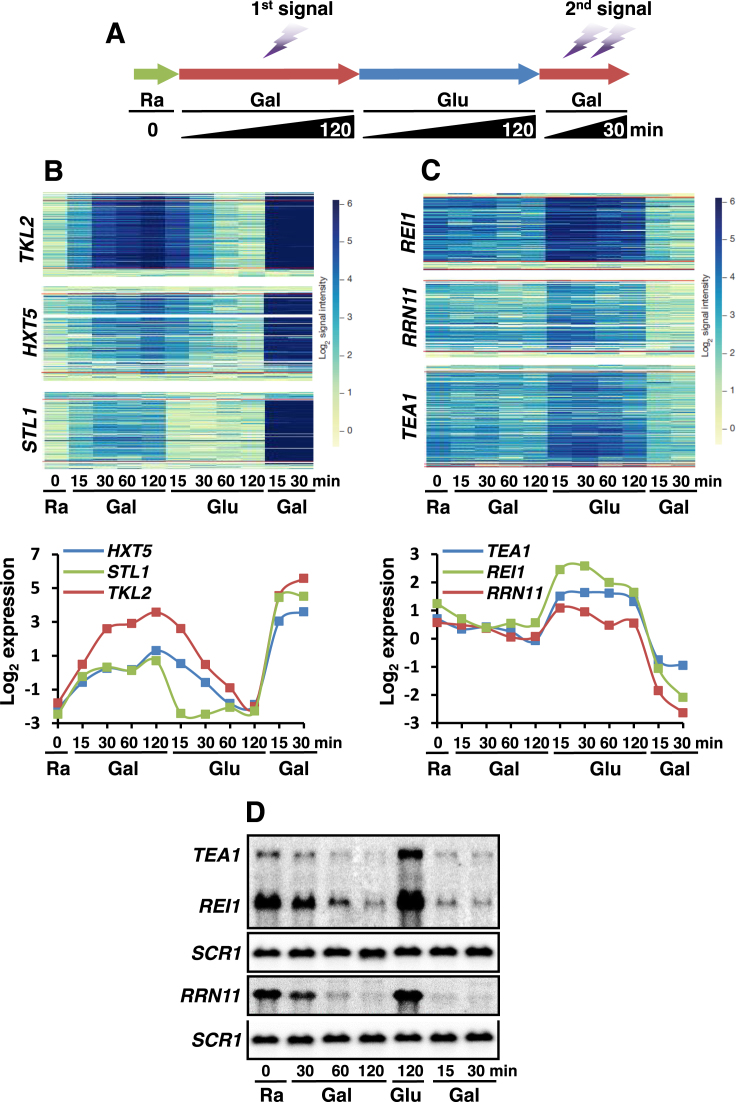
Figure 1.
Transcriptional activation memory versus transcriptional repression memory. (A) Schematic representation of the time course experiments to monitor changes in transcript levels during carbon-source shifts. (B) Transcriptional activation memory. The microarray hybridization signals for each probe arrayed at positions along the galactose-induced genes, TKL2, HXT5 and STL1 from Kim et al. (33). Increased blue color indicates more transcription. Red lines show the annotated start and stop of the mRNAs. The time course as in (A) is arrayed from left to right. Bottom graph shows normalized, log2-transformed mRNA expression levels at each time point. (C) Transcriptional repression memory. The microarray hybridization signals and log2-transformed expression levels are shown for the three TREM genes, REI1, RRN11 and TEA1 as in (B). (D) Northern blot analyses of the TREM genes, TEA1, REI1 and RRN11 upon carbon-source shifts. Cells were grown in SC-raffinose media and then shifted to galactose, glucose, and then back to galactose media for the indicated times. SCR1 was used for a loading control.
More than 500 genes were down-regulated during the first galactose incubation and re-induced in medium containing glucose (33). Surprisingly, the rate of transcriptional repression for some of these genes was much more faster upon the second galactose pulse (Figure 1C). REI1, TEA1 and RRN11 were slightly repressed during the first galactose exposure. In contrast, the second repression was not only faster, but much stronger (Figure 1C). To further confirm the microarray data, we performed northern blot analyses. SCR1 transcript levels were not changed during carbon-source shifts. TEA1, REI1 and RRN11 transcript levels were down-regulated for 120 min during the first galactose incubation and were almost completely repressed within 15–30 min of the second galactose pulse (Figure 1D). Since the rate of transcriptional repression could differ when cells were transferred from raffinose (the first repression) or from glucose (the second repression) to galactose, we monitored the kinetics of repression when cells were shifted from raffinose to galactose for the second repression. REI1 and TEA1 still exhibited memory of their prior inactive states (Supplementary Figure S1A and B). Furthermore, two genes, TKL2 and HXT5 repressed by glucose also showed more rapid repression during the second glucose incubation (Supplementary Figure S1C and D). Although Set3 HDAC and Set2 HMT have been shown to delay mRNA and long non-coding RNA (lncRNA) induction under the same conditions (33,34), the kinetics of repression for REI1 and TEA1 was not affected by the loss of these factors (Supplementary Figure S1E and F).
These findings indicate that the genes remember their previous transcriptionally inactive states so that they are more efficiently repressed upon re-exposure to the same stimuli, which is termed TREM. Furthermore, our results suggest that there are the two distinct types of transcriptional memory: one for activation and the other for repression.
Rpd3L HDAC induces TREM
TREM may be induced by rapid dissociation of activators from promoters. Alternatively, repressors that bind to active promoters could induce TREM. Since Rpd3 is known to associate with active genes, it may induce TREM (5,8). To test this, we therefore analyzed TREM in RPD3 and rpd3Δ cells by quantitative real-time PCR (Figure 2A). The internal control SCR1 was not changed during carbon-source shifts or in the presence or absence of Rpd3. Steady-state transcript levels of three TREM genes, REI1, TEA1 and RRN11, were not affected strongly by the loss of Rpd3 (Supplementary Figure S2A). Furthermore, the kinetics of gene repression during the first galactose incubation changed only slightly in rpd3Δ cells. Surprisingly, the rate of the second repression was significantly delayed in rpd3Δ cells when compared to wild type cells (Figure 2A and Supplementary Figure S2A). The ratio of transcript levels in rpd3Δ over those in wild type showed a strong increase in the mutant during the second repression indicating that the loss of Rpd3 delays TREM (Figure 2A).
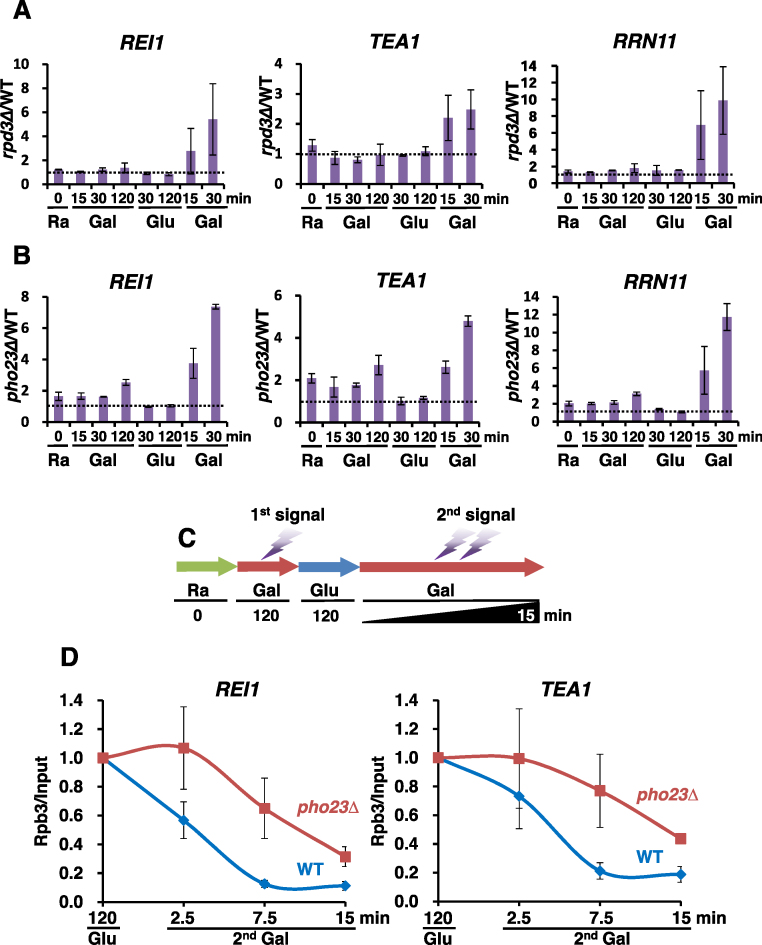
Figure 2.
Rpd3L induces transcriptional repression memory. (A) RPD3 and rpd3Δ strains were grown in SC medium containing raffinose and then sequentially shifted to SC media containing the indicated carbon sources for the times specified in (A). The mRNA levels were determined by RT-PCR with two independent RNA samples. The ratios of transcript levels in rpd3Δ over those in RPD3 were plotted. The original data appear in Supplementary Figure S2A. Error bars show the standard deviation (S.D.) calculated from two biological replicates, each with three technical replicates. (B) pho23Δ shows a significant delay of TREM. Transcript levels of TREM genes were analyzed as in (B). (C) Schematic representation of the time course experiments to monitor changes in RNA Pol II occupancy at TREM genes during carbon-source shifts. (D) Loss of Pho23 delays dissociation of RNA Pol II. PHO23 and pho23Δ strains were grown in SC medium containing raffinose and then sequentially shifted to SC media containing the indicated carbon sources for the times specified under the graphs. Crosslinked chromatin was precipitated with anti-Rpb3 and PCR analysis of the precipitated DNA was carried out on promoters of REI1 and TEA1. A non-transcribed region near the telomere of chromosome VI was used for an internal control. The signals for Rpb3 were quantitated and normalized to the input signal, and the ratios were graphed. Error bars show the standard deviation (S.D.) calculated from two biological replicates, each with three technical replicates.
Rpd3 is the catalytic subunit of both Rpd3S and Rpd3L HDACs (3,4). To determine which HDAC induces TREM, we analyzed gene expression patterns in the Rpd3L mutants, dep1Δ and pho23Δ, and in the Rpd3S mutant, rco1Δ. Loss of the Rpd3L-specific subunits, Pho23 and Dep1 significantly delayed TREM as seen in rpd3Δ cells (Figure 2B and Supplementary Figure S2B), but no effect was seen in rco1Δ cells (Supplementary Figure S2C) suggesting that Rpd3L specifically promotes TREM. We then carried out RNA Pol II ChIP during the second galactose pulse to determine whether Rpd3L promotes dissociation of RNA Pol II. Wild type and pho23Δ cells were grown in raffinose medium and then shifted to a galactose medium for 120 min, then to a glucose medium for 120 min, and then back to a galactose medium for 2.5, 7.5 or 15 min (Figure 2C). Consistent with changes in transcript levels, Rpb3 occupancy decreased rapidly in wild type cells during the second galactose incubation (Supplementary Figure S2A and Figure 2D). In contrast, dissociation of Rpb3 from REI1 and TEA1 genes was delayed in pho23Δ cells (Figure 2D). These data therefore suggest that Rpd3L, but not Rpd3S, induces TREM by promoting dissociation of RNA Pol II or by preventing RNA Pol II recruitment.
Rpd3L HDAC controls global TREM
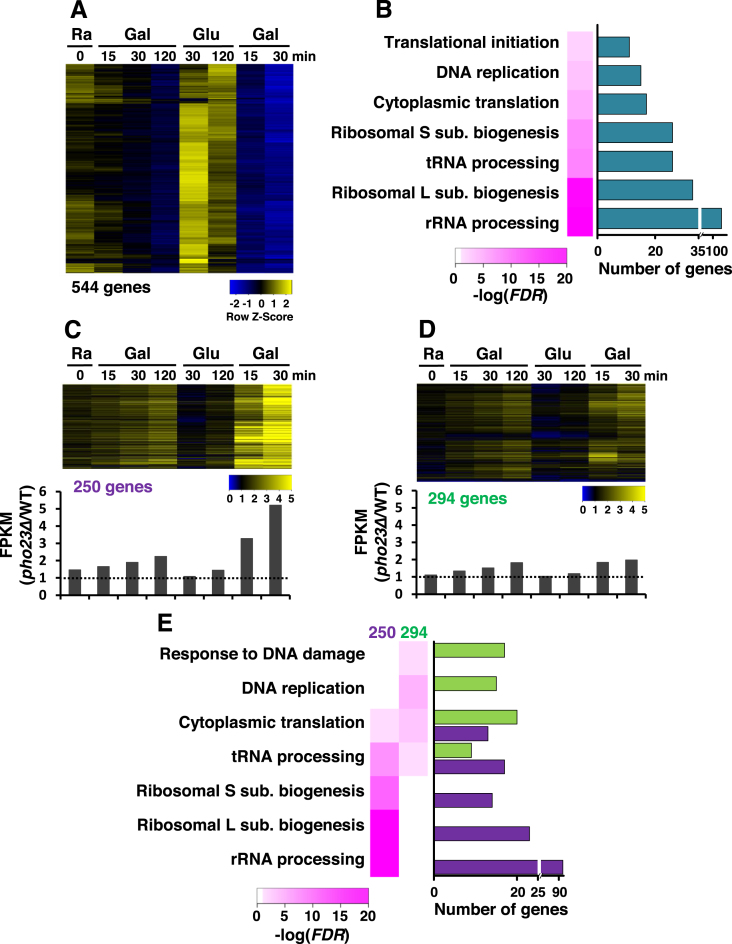
To further investigate the function of Rpd3L in TREM, total RNAs isolated from wild type and pho23Δ cells during carbon-source shifts were analyzed by strand-specific RNA sequencing (Figure 2B). We initially identified TREM genes that satisfied the following three requirements: (i) transcript levels in raffinose > transcript levels after 120 min of the first galactose incubation, (ii) transcript levels after 30 or 120 min of incubation in glucose > transcript level after 15 or 30 min of the second galactose incubation, (iii) transcript level after 15 or 30 min of the first galactose incubation > transcript level after 15 or 30 min of the second galactose pulse. 544 TREM genes were slightly downregulated during the first 120 min galactose incubation but were strongly or completely repressed during the second galactose pulse (Figure 3A). Gene ontologies (GO) of these TREM genes were then analyzed to categorize their cellular functions. Signatures of TREM genes were enriched for rRNA processing, ribosome biogenesis, tRNA processing, or DNA replication (Figure 3B). Furthermore, ∼60% (142 out of 236) of genes involved in ribosome biogenesis (RiBi) and only 19% (26 out of 137) of ribosomal protein (RP) genes appear to show TREM (Supplementary Figure S3A).
Figure 3.
Rpd3L is required for global TREM. (A) Expression profiles of 544 TREM genes. RNA samples from PHO23 and pho23Δ strains in Figure 2C were analyzed by strand-specific RNA sequencing. The TREM genes identified in PHO23 strains are visualized. (B) GO analysis of TREM genes. The number of genes and false discovery rates (FDRs) obtained from GeneCodis3 (http://genecodis.cnb.csic.es/) showed that many TREM genes are involved in ribosomal RNA processing and ribosomal large or small subunit biogenesis. (C) TREM of 250 genes is significantly delayed in pho23Δ cells. The ratio of transcript levels for 250 genes in pho23Δ over those in PHO23 are visualized. Bottom panel shows the averaged expression ratios at each time point. A strong difference in transcript levels is seen during the second galactose pulse. (D) TREM of 294 genes is not regulated by Pho23. The ratio of transcript levels is shown as in (C). (E) GO analysis of Pho23-sensitive and Pho23-insensitive genes. Whereas Pho23-sensitive genes are enriched for rRNA processing and ribosomal subunit biogenesis, Pho23-insensitive genes are mostly involved in DNA replication and DNA damage response.
We next identified Pho23-regulated TREM genes that satisfied the following two conditions: (i) a 2-fold increase of transcript levels in pho23Δ at 30 min of the second galactose incubation, (ii) (transcript levels in pho23Δ/transcript levels in wild type) at 30 min of the second galactose pulse/those at 120 min of the first >1.5. The second requirement was used to identify the genes strongly repressed by Rpd3L during the second galactose incubation. Although all of the TREM genes showed slightly delayed repression during the first repression in pho23Δ cells, the maximum differences in transcript levels in wild type and pho23Δ cells were ∼2-fold at 120 min of the first galactose incubation (Figure 3C and D). In contrast, 46% (250 out of 544 genes) of the TREM genes showed an ∼6-fold increase at 30 min of the second repression indicating that Pho23 is important for TREM of these genes (Figure 3C). The kinetics of the first and second repression of the other 294 genes were very similar in pho23Δ cells suggesting that TREM of these genes is not directly regulated by Rpd3L (Figure 3D). GO analysis of the Pho23-sensitive and Pho23-insensitive gene categories revealed their distinct functions in cellular processes. Whereas the 250 Pho23-sensitive genes were enriched for rRNA processing and ribosome biogenesis, the other 294 genes were enriched for DNA replication and DNA damage response (Figure 3E). Rpd3L is known to repress transcription of RiBi and RP genes, but ∼50% of the target genes (127 out of 250 genes) do not encode RiBi proteins or RP suggesting additional roles for Rpd3L (Supplementary Figure S3A and B) (19,43,44).
Since the Set2 HMT and Set3 HDAC affected the induction kinetics of mRNAs and lncRNAs under the same conditions (33,34), we therefore analyzed the tiling-array datasets to determine whether Set2 or Set3 was involved in the regulation of TREM. Loss of Set2 resulted in rapid and strong repression during the first galactose incubation, but no difference in TREM was seen (Supplementary Figure S3C). In addition, Set3 did not play a role in the induction of TREM (Supplementary Figure S3D). These results indicate that Rpd3L is a key regulator in global TREM.
RNA sequencing also identified 408 genes that are induced more quickly during the second galactose pulse than during the first galactose incubation (Supplementary Figure S3E and F). Although most of these genes are not directly involved in galactose metabolism and were only transiently induced by galactose, they appeared to show memory of their preceding active states under these conditions (45,46). Interestingly, loss of Rpd3L resulted in defects of optimal gene activation (Supplementary Figure S3E and F), but the mechanism by which Rpd3L promotes gene activation remains to be determined.
Rpd3L deacetylates histones at active promoters
Rpd3L is known to bind to both active and inactive promoters (5,8). To test whether Rpd3L directly binds to promoters of TREM genes, a ChIP assay was carried out using a Rpd3-18myc strain. Compared to an untagged strain, an approximately four-fold increase in Rpd3 binding was seen at the promoters of two TREM genes, REI1 and TEA1 (Figure 4A). This binding was also observed at the INO1 promoter, a known target of Rpd3 (Supplementary Figure S4A). Although previous studies and our results indicate that Rpd3 directly binds to promoters of actively transcribed genes, the factors that recruit this complex to active genes have not yet been identified. Interestingly, although the Rpd3 protein level was not affected, Rpd3 occupancy was slightly reduced upon loss of Pho23 (Figure 4A and B). Within Rpd3L HDAC, Pho23 is known to interact directly with Rxt2 and Rxt3. Upon deletion of PHO23, these two proteins dissociated from Rpd3L (47). These findings suggest that Pho23, Rxt2, and Rxt3 proteins may contribute to Rpd3L binding to active genes. Rpd3 occupancy was also slightly reduced when SET1 was deleted (Supplementary Figure S4B), but loss of Set2 had no effect on Rpd3 binding (Supplementary Figure S4C).
We then analyzed a published dataset to monitor global promoter occupancy by Rpd3L (5). Sds3, a component of Rpd3L, strongly bound to the 250 Rpd3L-sensitive genes in comparison to total yeast genes or to the 294 Rpd3L-insensitive genes (Figure 4C and Supplementary Figure S4D). Whereas Sds3 occupancy was the strongest at promoter regions, Rpd3 binding was seen throughout the genes, perhaps because Rpd3S binds downstream transcribed regions. To further explore the function of Rpd3L on TREM genes, acetylation of histone H4 (with an antibody that recognizes tetra-acetyl H4) was assayed by ChIP. Levels of acetylation were normalized to total histone content as measured by H3 levels. Deletion of PHO23 increased acetylation of histone H4 at the promoters of two TREM genes, REI1 and TEA1 (Figure 4D). These data suggest that Rpd3L binds directly to the promoters of TREM genes to deacetylate histones.
Next, histone acetylation patterns were analyzed in cells undergoing carbon-source shifts (Figure 4E). In wild type cells, histone acetylation at REI1 and TEA1 promoters was similar in raffinose and in glucose (after 120 min). At 7.5 min after the second galactose pulse, a strong reduction in histone acetylation was observed suggesting that histone deacetylation at promoters rapidly prevents recruitment or induces dissociation of RNA Pol II (Figures 2D, 4E and Supplementary Figure S4E). In raffinose and in glucose (after 120 min), acetylation at REI1 and TEA1 promoters was slightly higher in pho23Δ cells (Figure 4D, E and Supplementary Figure S4E). Interestingly, an ∼6-fold increase in acetylation was seen at 7.5 min after the second galactose pulse in pho23Δ cells (Figure 4E and Supplementary Figure S4E). These results suggest that histone deacetylation by Rpd3L rapidly prevents RNA Pol II binding to promoters to induce TREM.
The Pho23 PHD finger-H3K4me3 interaction induces deacetylation and TREM by Rpd3L
The Pho23 subunit of Rpd3L is known to preferentially bind H3K4me3 via its PHD finger domain in vitro (17,18). Peptide pulldown assays using yeast total extracts were done to confirm this interaction. Total extracts prepared from Pho23-18myc cells were incubated with beads coupled with H3 tail peptides methylated on K4 or not. Pho23 from wild type cells bound to H3K4me3, but this binding was abrogated when a tryptophan within the methyl-binding pocket of the PHD finger was mutated to alanine (W305A) indicating that the Pho23 PHD finger directly recognizes H3K4me3 (Figure 5A). We then analyzed the H3K4me3 pattern of TREM genes using a published dataset (40). Consistent with previous reports, H3K4me3 was enriched at transcription start sites. Interestingly, the 250 TREM genes regulated by Rpd3L showed high levels of H3K4me3 relative to total genes or the 294 Rpd3L-insensitive genes (Figure 5B). Although transcript levels of the 250 Rpd3L-sensitive genes were lower than those of highly transcribed genes (top 25%, 1545 genes), H3K4me3 levels were comparable at these two gene sets (Supplementary Figure S5A and Figure 5B). This suggests that elevated levels of H3K4me3 at TREM genes may contribute to Rpd3L function.
To test whether the interaction between the Pho23 PHD finger and H3K4me3 affects chromatin binding and/or the deacetylase activity of Rpd3L, ChIP assay was carried out with a Pho23-18myc or a pho23 (W305A)-18myc strain. Interestingly, although deletion of PHO23 reduced Rpd3 binding to TREM genes, no effect or slightly increased Rpd3 binding was seen in the pho23 (W305A) mutant (Figure 5C). Surprisingly, increased acetylation was still observed in this mutant even though the mutation did not affect Pho23 protein levels suggesting that the interaction between the Pho23 PHD finger and H3K4me3 may stimulate the deacetylase activity of Rpd3L (Figure 5D and E). We then analyzed gene expression patterns during carbon-source shifts to investigate the effect of the Pho23 PHD finger–H3K4me3 interaction on TREM. As seen in pho23Δ cells, there was a significant delay in TREM in the pho23 (W305A) mutant indicating that Pho23 binding to H3K4me3 via its PHD finger is critical for TREM (Figures 2B and 5F).
An additional subunit, Cti6 of Rpd3L, also binds H3K4me3 via its PHD finger, but disruption of this interaction by the cti6 (W98A) mutation had no effect on TREM (Supplementary Figure S5B and C). A recent study revealed that Rpd3L was globally re-localized to suppress RNA Pol II transcription during the entry into quiescence by gene specific repressors, including Stb3, Tod6, and Xbp1 (48). However, these factors were not required for Rpd3L-mediated TREM (Supplementary Figure S5D). Taken together, our findings indicate that the interaction between the Pho23 PHD finger and H3K4me3 induces histone deacetylation and TREM by Rpd3L.
DISCUSSION
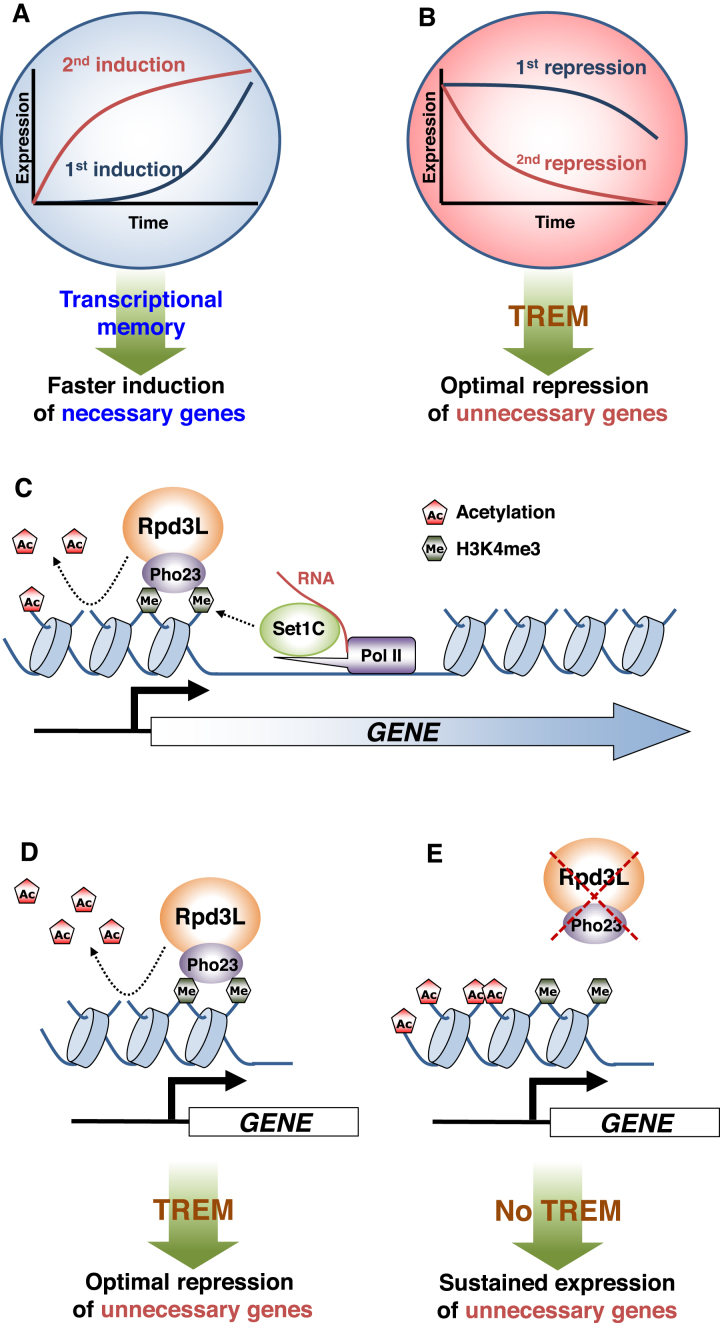
Upon environmental changes, cells must fine-tune their gene expression patterns to adapt. If cells have been previously exposed to the same stimulus, transcriptional memory promotes faster reactivation of required genes (Figure 6A). In addition, it is also critical for cells to effectively suppress the expression of unnecessary genes. In this study, we show, for the first time, that many yeast genes exhibit ‘memory’ of their preceding transcriptional inactive state, and this phenomenon is termed TREM (Figure 6A and B). Enhancement of transcriptional repression via TREM may save cellular energy and resources including transcription machinery by rapidly turning off unnecessary genes for cellular functions. How Rpd3L is recruited to the promoters of TREM genes remains elusive, but we found that the interaction between the Pho23 PHD finger and H3K4me3 enriched at active promoters enhanced deacetylation by Rpd3L to induce TREM (Figure 6C and D). Loss of this interaction may result in hyperacetylation and sustained expression of unnecessary genes (Figure 6E). Recent studies have suggested that transcriptional/epigenetic memory plays an important role in innate immunity. Innate Lymphoid Cells (ILCs) mediate non-specific responses to allergens and were found to be hyperactivated upon re-exposure to the same or unrelated allergens (49). Importantly, a small portion of previously trained cells will become naïve cells once the allergen is eliminated. Repression of unnecessary or hyperactivated genes by TREM is likely important to generate naïve cells from trained cells.
Figure 6.
Transcriptional memory and models for regulation of TREM by Rpd3L. (A) When cells are exposed to a stimulus, the genes necessary for cellular functions are induced (first induction). Transcriptional memory upon re-exposure to the same stimulus significantly increases the rate of gene activation (second induction). (B) In contrast, unnecessary genes are repressed by a stimulus (first repression). TREM upon re-exposure to the same stimulus promotes optimal gene repression (second repression). (C) During early stage of transcription, Set1C, also termed Set1/COMPASS, is recruited by RNA Pol II and/or mRNA transcripts to methylate histone H3 at K4. The Pho23 PHD finger binds to H3K4me3 to enhance histone deacetylation by Rpd3L. (D) Hypoacetylation mediated by the Rpd3L–H3K4me3 interaction results in optimal repression of unnecessary genes by TREM. (E) Hyperacetylation by the loss of Rpd3L or by the loss of the Pho23 PHD finger-H3K4me3 interaction result in sustained expression of unnecessary genes.
Many chromatin regulators, including histone acetyltransferases (HATs), HDACs, or histone methyltransferases (HMTs), do not significantly affect the steady-state levels of transcripts (50). Instead, they likely function to modulate the kinetics of gene activation or repression (13). Although Rpd3L HDAC downregulates some genes in steady-state conditions, delayed repression of many genes was observed in mutants for Rpd3L during carbon-source shifts (Figure 3C and D). Consistent with these findings, Rpd3L HDAC has been shown to affect global gene expression dynamics upon stresses. Furthermore, mutants for this complex exhibited sensitivity to heat shock and salt stress (51–53). In this study, we showed an additional role of Rpd3L inducing TREM that effectively suppresses the transcription of unnecessary genes. The greatest differences in transcript and acetylation levels between wild type and mutants for Rpd3L were seen only when the cells had been exposed previously to the same carbon-source (Figures 2, 3C and 4E).
A recent study revealed that the Rpd3L complex was required for global shut-down of transcription during quiescence entry (48). Sequence-specific transcription factors, including Xbp1 and Stb3 mediate global relocalization of Rpd3 and transcriptional shutoff. Interestingly, these factors were not required for TREM (Supplementary Figure S5D) indicating that Rpd3L uses a distinct mechanism involving the interaction between H3K4me3 and the Pho23 PHD finger to induce TREM. Although the interaction between the Pho23 PHD finger and H3K4me3 did not affect Rpd3L occupancy, the level of acetylation was higher in a Pho23 PHD finger mutant. Furthermore, loss of the interaction delayed TREM suggesting that this interaction enhances TREM by promoting HDAC activity of Rpd3L (Figure 5). Interestingly, disruption of the interaction between the Cti6/Rxt1 PHD finger and H3K4me3 had no effect on TREM (Supplementary Figure S5B and S5C). This finding suggests that the two proteins (Pho23 and Cti6) within the Rpd3L complex that bind to the same histone mark may have distinct functions in transcription.
Although H3K4me3 correlates highly with transcription frequency and is enriched at active promoters, requirement of H3K4me3 for TREM and histone deacetylation by Rpd3L suggests its negative role in transcription (15,54,55). Several previous reports have also supported a repressive role for H3K4me3 in transcription. ING2, a human homologue of Pho23, directly binds to H3K4me3 and contributes to active gene repression by recruiting mSin3-HDAC1 (56). Loss of Set1 also increases the rate of GAL1 reactivation and derepresses RiBi and RP genes during diamide stress (19,28).
H3K4me3 also acts as a binding site for the positive regulators of transcription. TAF3, a general transcription factor, binds to H3K4me3 via its PHD finger to promote transcription (57). Furthermore, several HATs, including NuA3, NuA4, and SAGA in yeast and HBO1 HAT in mammals, have H3K4me3 binding modules (17,58). Yng1 and Yng2, yeast homologues of human inhibitor of growth (ING) proteins, are subunits of NuA3 and NuA4 HATs, respectively, that have PHD fingers that specifically binds to H3K4me3 (13,17). The interaction between H3K4me3 and the Yng1 PHD finger is required for H3K14 acetylation by NuA3 and transcription of a subset of genes (59). ING4 and ING5 are subunits of HBO1 HAT that also binds to H3K4me3 via their PHD fingers to increase histone H3 acetylation (58). The dual functions of H3K4me3 in histone acetylation/deacetylation suggest that TREM may be modulated by the antagonistic functions of the HATs and HDACs that bind to H3K4me3.
An important question is why Rpd3L more actively represses transcription during the second galactose pulse compared to the first. We showed clearly that deletion of PHO23 had a much more stronger effect on histone acetylation during the second repression (Figure 4E and Supplementary Figure S4E), perhaps due to on-going transcription in pho23Δ as a result of the delay in RNA Pol II dissociation (Figure 2D). Alternatively, it is also possible that Rpd3L activity could be modulated by post-translational modifications resulting from environmental changes. A recent study showed that at least two components, Sds3 and Rxt2 of Rpd3L, were extensively sumoylated upon KCl or Sorbitol stress (60). It will be interesting to determine whether this modification modulates Rpd3L function and TREM.
Studies on transcriptional memory have mostly focused on highly inducible genes including GAL genes and INO1. Here we showed that 408 genes exhibited memory of their previous active states upon galactose exposure (Supplementary Figure S3E and F). Most of these genes seem to be transiently induced by galactose as only 10 genes, GAL1, GAL2, GAL3, GAL7, GAL10, GAL80, GCY1, MTH1, PCL10 and FUR4, are constitutively activated by the Gal4 activator in the presence of galactose (46). Therefore, transcriptional memory for most of the genes identified in this study likely require a distinct mechanism. Interestingly, Rpd3L promotes gene repression upon stresses and functions to induce many stress response genes (51,53). In this study, we identified 408 genes that required Rpd3L for optimal induction during the first and the second galactose incubations (Supplementary Figure S3E and F). It will be interesting to determine how Rpd3L differentially affects the two distinct transcriptional responses.
Although transcriptional memory is important for optimal induction of necessary genes or repression of unnecessary ones, some genes should not be hyperactivated or hyperrepressed by transcriptional memory. For example, circadian clock genes and cell cycle regulators are periodically induced and then repressed with the exact same kinetics. Dysregulation of these genes by transcriptional memory may results in abnormal cell cycle patterns, thus it is important to understand the mechanism that removes and/or erases transcriptional memory of these genes.
DATA AVAILABILITY
RNA sequencing data that support the findings of this study have been deposited in ArrayExpress with the accession code E-MTAB-6551.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wonja Choi (Ewha Womans University) and Jung-Shin Lee (Kangwon National University) for strains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation [NRF-2017M3A9B5060887, NRF-2017M3A9G7073033, NRF-2017M3C9A5029980, NRF-2012R1A5A1048236 to T.K.]. Funding for open access charge: By our research grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 2. Li B., Carey M., Workman J.L.. The role of chromatin during transcription. Cell. 2007; 128:707–719. [DOI] [PubMed] [Google Scholar]
- 3. Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005; 123:581–592. [DOI] [PubMed] [Google Scholar]
- 4. Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J. et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005; 123:593–605. [DOI] [PubMed] [Google Scholar]
- 5. Drouin S., Laramee L., Jacques P.E., Forest A., Bergeron M., Robert F.. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLos Genet. 2010; 6:e1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weinberger L., Voichek Y., Tirosh I., Hornung G., Amit I., Barkai N.. Expression noise and acetylation profiles distinguish HDAC functions. Mol. Cell. 2012; 47:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kadosh D., Struhl K.. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997; 89:365–371. [DOI] [PubMed] [Google Scholar]
- 8. Kurdistani S.K., Robyr D., Tavazoie S., Grunstein M.. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002; 31:248–254. [DOI] [PubMed] [Google Scholar]
- 9. Kim T., Buratowski S.. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009; 137:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li B., Gogol M., Carey M., Pattenden S.G., Seidel C., Workman J.L.. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007; 21:1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J.L.. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007; 316:1050–1054. [DOI] [PubMed] [Google Scholar]
- 12. Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., Natarajan K., Workman J.L.. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012; 489:452–455. [DOI] [PubMed] [Google Scholar]
- 13. Woo H., Dam Ha S., Lee S.B., Buratowski S., Kim T.. Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp. Mol. Med. 2017; 49:e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng H.H., Robert F., Young R.A., Struhl K.. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003; 11:709–719. [DOI] [PubMed] [Google Scholar]
- 15. Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E. et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005; 122:517–527. [DOI] [PubMed] [Google Scholar]
- 16. Luciano P., Jeon J., El-Kaoutari A., Challal D., Bonnet A., Barucco M., Candelli T., Jourquin F., Lesage P., Kim J. et al. Binding to RNA regulates Set1 function. Cell Discov. 2017; 3:17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi X., Kachirskaia I., Walter K.L., Kuo J.H., Lake A., Davrazou F., Chan S.M., Martin D.G., Fingerman I.M., Briggs S.D. et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007; 282:2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang S.S., Zhou B.O., Zhou J.Q.. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol. Cell. Biol. 2011; 31:3171–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S. et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012; 10:e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J., Moazed D., Gygi S.P.. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002; 277:49383–49388. [DOI] [PubMed] [Google Scholar]
- 21. Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D.P., Beattie B.K., Emili A., Boone C. et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003; 23:4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li B., Howe L., Anderson S., Yates J.R 3rd, Workman J.L.. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003; 278:8897–8903. [DOI] [PubMed] [Google Scholar]
- 23. Joshi A.A., Struhl K.. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005; 20:971–978. [DOI] [PubMed] [Google Scholar]
- 24. McDaniel S.L., Fligor J.E., Ruan C., Cui H., Bridgers J.B., DiFiore J.V., Guo A.H., Li B., Strahl B.D.. Combinatorial histone readout by the dual plant homeodomain (PHD) fingers of Rco1 mediates Rpd3S chromatin recruitment and the maintenance of transcriptional fidelity. J. Biol. Chem. 2016; 291:14796–14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kundu S., Horn P.J., Peterson C.L.. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007; 21:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gialitakis M., Arampatzi P., Makatounakis T., Papamatheakis J.. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 2010; 30:2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kundu S., Peterson C.L.. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol. Cell. Biol. 2010; 30:2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou B.O., Zhou J.Q.. Recent transcription-induced histone H3 lysine 4 (H3K4) methylation inhibits gene reactivation. J. Biol. Chem. 2011; 286:34770–34776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., Brickner J.H.. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007; 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zacharioudakis I., Gligoris T., Tzamarias D.. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 2007; 17:2041–2046. [DOI] [PubMed] [Google Scholar]
- 31. Light W.H., Freaney J., Sood V., Thompson A., D’Urso A., Horvath C.M., Brickner J.H.. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013; 11:e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., Brickner J.H.. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 2010; 12:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim T., Xu Z., Clauder-Munster S., Steinmetz L.M., Buratowski S.. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012; 150:1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim J.H., Lee B.B., Oh Y.M., Zhu C., Steinmetz L.M., Lee Y., Kim W.K., Lee S.B., Buratowski S., Kim T.. Modulation of mRNA and lncRNA expression dynamics by the Set2-Rpd3S pathway. Nat. Commun. 2016; 7:13534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marquardt S., Hazelbaker D.Z., Buratowski S.. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription. 2011; 2:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nogales-Cadenas R., Carmona-Saez P., Vazquez M., Vicente C., Yang X., Tirado F., Carazo J.M., Pascual-Montano A.. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009; 37:W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toedling J., Skylar O., Krueger T., Fischer J.J., Sperling S., Huber W.. Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007; 8:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weiner A., Hsieh T.H., Appleboim A., Chen H.V., Rahat A., Amit I., Rando O.J., Friedman N.. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell. 2015; 58:371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen K., Xi Y., Pan X., Li Z., Kaestner K., Tyler J., Dent S., He X., Li W.. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013; 23:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P., Tyers M., Rougemont J., Beyer A.L., Loewith R.. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011; 30:3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lippman S.I., Broach J.R.. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19928–19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M.. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009; 457:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E. et al. Genome-wide location and function of DNA binding proteins. Science. 2000; 290:2306–2309. [DOI] [PubMed] [Google Scholar]
- 47. Sardiu M.E., Gilmore J.M., Carrozza M.J., Li B., Workman J.L., Florens L., Washburn M.P.. Determining protein complex connectivity using a probabilistic deletion network derived from quantitative proteomics. PLoS One. 2009; 4:e7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McKnight J.N., Boerma J.W., Breeden L.L., Tsukiyama T.. Global promoter targeting of a conserved lysine deacetylase for transcriptional shutoff during quiescence entry. Mol. Cell. 2015; 59:732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Netea M.G., Latz E., Mills K.H., O’Neill L.A.. Innate immune memory: a paradigm shift in understanding host defense. Nat. Immunol. 2015; 16:675–679. [DOI] [PubMed] [Google Scholar]
- 50. Lenstra T.L., Benschop J.J., Kim T., Schulze J.M., Brabers N.A., Margaritis T., van de Pasch L.A., van Heesch S.A., Brok M.O., Groot Koerkamp M.J. et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011; 42:536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alejandro-Osorio A.L., Huebert D.J., Porcaro D.T., Sonntag M.E., Nillasithanukroh S., Will J.L., Gasch A.P.. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 2009; 10:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruiz-Roig C., Vieitez C., Posas F., de Nadal E.. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 2010; 76:1049–1062. [DOI] [PubMed] [Google Scholar]
- 53. De Nadal E., Zapater M., Alepuz P.M., Sumoy L., Mas G., Posas F.. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004; 427:370–374. [DOI] [PubMed] [Google Scholar]
- 54. Liu C.L., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J.. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005; 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 56. Shi X., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Pena P., Lan F., Kaadige M.R. et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006; 442:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vermeulen M., Mulder K.W., Denissov S., Pijnappel W.W., van Schaik F.M., Varier R.A., Baltissen M.P., Stunnenberg H.G., Mann M., Timmers H.T.. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007; 131:58–69. [DOI] [PubMed] [Google Scholar]
- 58. Hung T., Binda O., Champagne K.S., Kuo A.J., Johnson K., Chang H.Y., Simon M.D., Kutateladze T.G., Gozani O.. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol. Cell. 2009; 33:248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taverna S.D., Ilin S., Rogers R.S., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T. et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006; 24:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lewicki M.C., Srikumar T., Johnson E., Raught B.. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J. Proteomics. 2015; 118:39–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data that support the findings of this study have been deposited in ArrayExpress with the accession code E-MTAB-6551.