Abstract
A significant fraction of mRNAs are degraded by the nuclear exosome in normal cells. Here, we studied where and when these exosome target mRNAs are sorted away from properly exported ones in the cells. We show that upon exosome inactivation, polyA RNAs are apparently accumulated in nuclear foci that are distinct from nuclear speckles (NSs), and provide several lines of evidence supporting that these polyA RNAs mainly correspond to accumulating exosome target mRNAs. These results suggest that exosomal mRNA degradation mostly occurs outside of NSs. In support of this possibility, targeting exosome target mRNAs to NSs stabilizes them by preventing exosomal degradation. Furthermore, inhibiting mRNA release from NSs does not attenuate exosomal degradation in normal cells, and results in polyA RNA accumulation both inside and outside of NSs in exosome inactivated cells, suggesting that passage through NSs is not required for sorting mRNAs for degradation or export. Indeed, exosome target mRNAs that normally do not enter NSs are exported upon exosome inactivation. Together, our data suggest that exosome target mRNAs are mainly degraded in the nucleoplasm before entering NSs and rapid removal of these mRNAs is important for preventing their nuclear export.
INTRODUCTION
The production of export-competent mRNPs is under the surveillance of quality control steps, where aberrant mRNPs resulting from improper or inefficient processing and assembly are subject to exosomal degradation. The RNA exosome (exosome) is a critical component of the mRNA surveillance system (1–4). In vivo activity of the exosome requires multiple cofactors, among which the RNA helicase MTR4 is critical for every aspect of nuclear exosome functions. MTR4 forms into different complexes that link the nuclear exosome to different classes of target RNAs. In mammalian cells, MTR4, together with RBM7 and ZCCHC8, form the NEXT complex that is mainly involved in the degradation of promoter upstream transcripts (PROMPTs) (5). MTR4 also associates with PAPD5 and ZCCHC7 to form the counterpart of the yeast TRAMP complex that functions in the adenylation of rRNA processing intermediates (5,6). In addition, MTR4 associates with ZFC3H1 and together functions in the degradation of long transcripts, such as snoRNA host transcripts, as well as short unstable RNAs including PROMPTs transcribed in the antisense direction (also called uaRNAs) and prematurely terminated RNAs (ptRNAs) (7,8).
For most nuclear mRNAs, the final destiny is either exported to the cytoplasm or degraded in the nucleus. A fundamental question is how these two distinct mRNA pools are sorted. The competition of MTR4 with the mRNA export adaptor ALYREF for associating with the nuclear cap-binding complex (CBC) provides an important mechanism for sorting export-defective mRNAs away from export-competent ones (9). Up-to-date, it remains unknown when mRNA sorting occurs in the cells. If this sorting does not occur in a timely manner, aberrant mRNAs could occupy nuclear factors and also have better chance to be exported to the cytoplasm. Indeed, a recent study reported that normally unstable RNAs subject to exosomal degradation are detected in the polysomes upon exosome inactivation (8).
The nucleus is highly organized and contains multiple sub-nuclear structures, which concentrate-specific proteins that carry out similar processes. In the nucleus, many mRNA export factors, including TREX components (e.g. ALYREF), are mainly concentrated in the sub-nuclear structure, nuclear speckles (NSs) (10–13). Multiple studies suggest that most mRNAs pass through NSs prior to nuclear export (14–19). Thus, if exosomal mRNA degradation occurs before entering NSs, the chances for exosome target mRNAs to recruit nuclear export factors could be limited. However, up-to-date, when and where mRNA fate for export or degradation is determined in the cells remain unknown.
Here, we found that upon exosome inactivation, its target mRNAs are mainly accumulated in nuclear foci outside of NSs, suggesting that exosomal degradation does not occur in these sub-nuclear structures. In support of this view, driving exosome target mRNAs to NSs results in their stabilization due to the prevention of exosomal degradation. Further, by blocking mRNA release from speckles, or by examining export-deficient reporter mRNAs that are known not to enter speckles in normal cells, we provide evidence that mRNA sorting for export or degradation does not require mRNA passage through NSs. Together, our work suggests that mRNA fate for export or degradation is mainly determined in the nucleoplasm before entering NSs.
MATERIALS AND METHODS
Plasmids and antibodies
To construct the Flag-MTR4, Flag-RBM7 and Flag-ZCCHC7, the coding sequence of the corresponding gene was inserted into p3xFlag-CMV-10 (Sigma). Mutagenesis was used to obtain Flag-MTR4 mutant expression plasmids. Plasmids encoding β-globin cDNA (cG), Smad cDNA (cS) were described previously (20,21). Speckle-targeting element (STE) sequence was inserted into the 3′ of β-globin cDNA to construct β-globin cDNA-STE (cG-STE).
Antibody to UAP56, CBP80 and ARS2 were described previously (9,20). The rabbit polyclonal antibodies against MTR4 and MTR3 were purchased from ABclonal Technology. The Tubulin, RRP6, RRP40, Flag and SC35 antibodies were purchased from Sigma, the PAPD5, digoxin, GAPDH, ZCCHC8 and Coilin, PSP1 and PML antibodies were purchased from Protein tech, Roche, Abcam, Dundee cell, SANTA CRUZ, respectively.
Cell culture, transfections and RNAi
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) (Biochrom). Lipofectamine 2000 (Invitrogen) was used for DNA transfection. For RNAi, siRNA transfection was carried out with Lipofectamine 2000 (Invitrogen) or Lipofectamine RNAiMax (Invitrogen) following manufacture’s protocol. The siRNA targeting sequences are shown in Supplementary Table S1. It is of note that both UAP56 and its homolog URH49 must be knocked down to observe a robust export block (22).
DNA micro-injection, FISH and immunofluorescence
Microinjection was carried out as previously described (23). polyA RNA in situ hybridization (FISH) and immunofluorescence (IF) were performed as previously described (20). To detect the HSPA1A, cG, and cS mRNA, an HPLC-purified Alexa 548 conjugated probe that hybridizes to pcDNA3 vector sequence was used. The vector probe targeting sequence is shown in Supplementary Table S2. Images were captured with a DP72-CCD camera (Olympus) on an inverted microscope using DP-BSW software (Olympus). FISH quantitation was carried out using Image J software (National Institutes of Health), and N/C ratios were calculated as described (24). For IF, the SC35, Coilin, PSP1 and PML antibodies were diluted 1: 500, 1:200, 1:200 and 1:50 in blocking buffer, respectively.
To detect the endogenous RHOC and DDX39B mRNA, HeLa cells treated with siRNAs were fixed with 3.6% formaldehyde plus 10% acetic acid in phosphate-buffered saline (PBS) for 20 min, followed with three washes with 1 × PBS and permeabilized with 1 × PBS/0.1% Triton/2 mM VRC for 15 min. The cells were incubated with Dig-labeled-specific probes in hybridization buffer at 55°C for 12–16 h. The targeting sequences of the transcript-specific probes were shown in Supplementary Table S2. After extensively wash, cells were incubated with digoxin antibody for 1 h. After three washes with 1 × PBS for three times, the cells were incubated with the Alexa-488 labeled anti-sheep antibody for 1 h, followed by DAPI staining.
RNA isolation, reverse transcription and PCR analysis
Total RNAs were extracted using TRIzol. RNAs were treated with the RNase-free RQ DNase I (Promega) for 2 h at 37°C, and cDNAs were synthesized from 1 μg of RNAs with random primer using M-MLV reverse transcriptase (Promega). For quantitative PCRs, cDNAs were amplified using GoTaq qPCR Master Mix (Promega) according to the manufacturer’s protocol. Primer sequences are listed in Supplementary Table S3.
PolyA RNA preparation
To examine the levels of polyadenylated PROMPTs in RRP40 and NEXT component knockdown cells, 7.5 μg total RNA was heated to 65°C for 2 min and then placed on ice. The RNA was rotated with Oligo (dT)25 Dynabeads in 10 μl of binding buffer (20 mM Tris at pH7.5, 1M LiCl, 2 mM ethylenediaminetetraacetic acid (EDTA)) for 5 min at room temperature. After two washes with 20 μl washing buffer (10 mM Tris at pH7.5, 0.15M LiCl, 1 mM EDTA), polyA RNA was eluted from the beads using 10 μl of elution buffer (10 mM Tris at pH7.5).
Sub-cellular fractionation
Sub-cellular fractionation was carried out as described (9). 1 × 107 HeLa cells were washed with 1 × PBS and suspended in hypotonic buffer (10 mM HEPES, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.2 mM phenylmethylsulfonyl fluoride (PMSF)/0.5 mM dithiothreitol (DTT)) and incubated for 10 min on ice. The swollen cells were dounced followed by centrifuge. The supernatant was collected as the cytoplasmic extract and the packed nuclear volume (PNV) was estimated. The nuclei were re-suspended slowly in 1/2 PNV of low salt buffer (20 mM HEPES, pH 7.9/1.5 mM MgCl2/20 mM KCl/0.2 mM EDTA/25% Glycerol/0.2 mM PMSF/0.5 mM DTT) followed by adding 1/2 PNV of high salt buffer (20 mM HEPES, pH 7.9/ 1.5 mM MgCl2/ 1.4 M KCl/ 0.2 mM EDTA/25% Glycerol/ 0.2 mM PMSF/ 0.5 mM DTT) and mixed quickly. The mixture was rotated for 30 min at 4°C followed by centrifuge and the supernatant is the nuclear extract. Total RNAs of nuclear or cytoplasmic extract were extracted using TRI Reagent (Sigma).
Bulk polyA RNA analysis
Nuclear RNAs from cells transfected with indicated siRNAs were incubated at 37°C for 30 min in a 30 μl reaction containing 20 mM Tris–HCl (pH 6.8), 8 U RNasin and 150 U RNase T1. After PCA extraction and ethanol precipitation, products were fractionated on 8% urea-polyacrylamide gel electrophoresis and transferred to positively charged nylon membrane (GE). After cross-linking by UV irradiation, the membrane was hybridized with PerfectHyb plus hybridization buffer (Toyobo) containing 32P-end-labeled DNA oligonucleotide probe to polyA RNA at 42°C overnight. After three washes with 2 × SSC/ 0.1% sodium dodecyl sulphate for five times, then visualized by Phosphor Imager.
RESULTS
Exosome inactivation results in nuclear accumulation of polyA RNA foci
MTR4 competes with the mRNA export adaptor ALYREF for associating with CBC, and this competition results in specific exosomal degradation of mRNAs that fail to assemble into export-competent mRNPs (9). Previous studies showed that ALYREF and MTR4 were both localized in the nucleus, concentrating in NSs and nucleoli, respectively (5,12). Different from these proteins, the CBC components CBP80 and ARS2 diffuse in the nucleoplasm (Supplementary Figure S1). These protein localization data raised the possibility that the competition between MTR4 and ALYREF might mainly occur in the nucleoplasm.
To determine where exosomal target mRNAs are accumulated upon exosome inactivation, we examined the distribution of polyA RNAs. We carried out fluorescence FISH using an oligo(dT) probe in cells depleted of MTR4 or exosome components, including the core proteins, RRP40 and MTR3, as well as the associating ribonucleases DIS3 and RRP6. These proteins were efficiently knocked down as revealed by western blotting or semi-quantitative RT-PCR analyses (Figure 1A). Significantly, compared to that in control cells, the nuclear polyA RNA FISH signals were far brighter in exosome- and MTR4-depleted cells (Figure 1B and C). Although knockdown of DIS3 and RRP6 individually did not have significant effect, in co-knockdown cells, polyA RNA signals turned much stronger in the nucleus (Figure 1B and C), suggesting that these two proteins might play redundant roles in nuclear degradation of mRNAs.
Figure 1.
Exosome inactivation results in the accumulation of polyA RNAs in specific nuclear foci. (A) Western and RT-PCR data showing knockdown efficiencies of the exosome components and MTR4. Different amount of cells or RT products of control knockdown cells were used to estimate the knockdown efficiencies. (B) FISH analysis of polyA RNA distribution in exosome and MTR4 knockdown cells. Same exposure was taken for all images. DAPI staining served as nucleus marker. (C) Quantification of nuclear polyA RNA FISH signals. Nuclear polyA RNA FISH signals quantified from 30 cells in each experiment by Image J. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01. (D) Wild-type, but not helicase core mutant MTR4 repressed the nuclear polyA RNA accumulation phenotype in MTR4 knockdown cells. Domain schematic representation of MTR4 is shown on the top. Functional domains are indicated and point mutations of D252A and E253A are marked. The MTR4 siRNA was transfected into HeLa cells using Lipofectamine 2000. Forty-eight hours post-transfection, siRNA sensitive or resistant WT Flag-MTR4, or siRNA resistant helicase core mutant MTR4 expression plasmid, was transfected to MTR4 siRNA treated cells. Twenty-four hours post-transfection, FISH analysis was carried out to observe the distribution of polyA RNAs. IF with the Flag antibody was performed to examine exogenous MTR4 expression. DAPI staining served as nucleus marker. The arrows indicate cells for which nuclear polyA RNA accumulation phenotype was repressed by exogenously expressed MTR4. (E) Nuclear accumulated polyA RNAs do not co-localize with NSs, paraspeckles, Cajal bodies or PML bodies in RRP40 knockdown cells. FISH was carried out using the 70 (nt) oligo-dT probe and IF using indicated antibodies were carried out. DAPI staining served as the nucleus marker. Confocal microscopy was used to visualize the cells.
Considering that knockdown of different exosome components all led to similarly increased nuclear polyA RNA signals, these phenotypes seem unlikely to be caused by siRNA off-target effect. To examine the possible off-target effect of the MTR4 siRNA, we carried out rescue experiment. Expression of the siRNA-resistant wild-type MTR4, but not the siRNA-sensitive wild-type or siRNA-resistant helicase DExH core mutant MTR4 repressed the increased polyA RNA signals (Figure 1D). This result indicates that the increased polyA RNA signals are a direct effect of MTR4 knockdown and the helicase activity might be required for MTR4 functioning in exosome-mediated mRNA degradation.
Nuclear polyA RNAs mainly localize in NSs in normal mammalian cells (14,17–19). However, co-localization analyses showed that in exosome knockdown cells, most nuclear polyA RNA foci do not overlap with SC35, a standard marker for NSs (Figure 1E). We also examined whether these polyA foci were accumulated in Cajal bodies, paraspeckles or PML bodies, but no apparent co-localization was observed (Figure 1E). These data suggest that exosome target mRNAs might not be mainly degraded in these major sub-nuclear domains.
Exosome inactivation does not apparently affect mRNA export
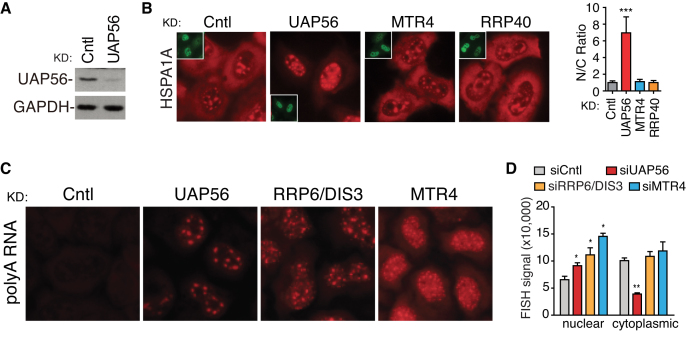
It was possible that exosome inactivation inhibited mRNA export, resulting in nuclear polyA RNA accumulation. However, this possibility was not supported by the following data. First, knockdown of the mRNA export factor UAP56 blocked nuclear export of the HSPA1A mRNA that is not an exosome target, whereas depletion of neither RRP40 nor MTR4 had an apparent effect (Figure 2A and B; Supplementary Figure S2). Second, as expected for mRNA export inhibition, UAP56 knockdown resulted in enhanced nuclear and reduced cytoplasmic polyA RNA signals. In contrast, in nuclear exosome and MTR4 knockdown cells, nuclear FISH signals significantly increased with no apparent change in the cytoplasmic ones (Figure 2C and D). Note that here nucleus-specific exosome components RRP6 and DIS3 were depleted to specifically inhibit nuclear exosome functions. Although it remains possible that nuclear export of some particular mRNAs has been disrupted, these data together indicate that the increased nuclear polyA signals upon exosome inactivation is not a result of mRNA export inhibition.
Figure 2.
mRNA nuclear export is not apparently affected by exosome inactivation. (A) Western blots to examine the knockdown efficiency of UAP56. GAPDH is used as a loading control. (B) Nuclear export of the HSPA1A reporter mRNA is not affected in exosome and MTR4 knockdown cells. The HSPA1A reporter mRNA construct was injected into the nuclei of control, UAP56, RRP40 and MTR4 knockdown cells, followed by FISH to detect the distribution of HSPA1A mRNA at 4 h after injection. Inset images show the injection marker. Same exposure was taken for all images. Quantification of nuclear and cytoplasmic FISH signals of the HSPA1A mRNA. N/C ratios were determined for 30 cells in each experiment. C and N indicate cytoplasmic and nuclear FISH signals, respectively. Error bars, standard deviations (n = 3). Statistical analysis was performed using Student’s t-test. ***P < 0.001. (C) PolyA RNA distribution in UAP56, RRP6/DIS3 and MTR4 knockdown cells. Indicated siRNAs were transfected into HeLa cells. Seventy-two hours post-transfection, FISH was carried out to observe the distribution of polyA RNAs. DAPI staining served as nucleus marker. Note that to accurately quantify the polyA signals in all samples, same exposures were taken for all FISH images. (D) Quantification of nuclear and cytoplasmic polyA RNA FISH signals. Nuclear and cytoplasmic polyA RNA FISH signals quantified from 50 cells in each experiment by Image J. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01.
Elevated nuclear polyA signals upon exosome inactivation are not mainly due to the lengthened polyA tail
Previous work reported that exosome inactivation results in the accumulation of hyperadenylated RNAs in PABPN1-dependent manner (25). Thus, it was possible that the increased polyA RNA signals upon exosome or MTR4 depletion might reflect the lengthened polyA tail, rather than the elevated transcript levels. To test this possibility, we first examined the polyA tail length of nuclear RNAs in control and MTR4 knockdown cells (Figure 3A and B). PABPN1 knockdown cells were used as the control showing altered polyA tail length. Consistent with the previous report (25), the polyA tail of nuclear RNAs was longer in MTR4 knockdown cells and co-knockdown of PABPN1 abolished this lengthening (Figure 3C). If the increased polyA RNA signals upon exosome inactivation had mostly resulted from lengthened polyA tail, one would expect eliminating this lengthening diminish the increased signals. However, compared to those in MTR4 knockdown cells, the polyA FISH signals were not apparently reduced in MTR4/PABPN1 co-knockdown cells (Figure 3D and E). Together, these data suggest that the increased polyA RNA signals upon exosome inactivation do not mainly result from the extended polyA tail, but probably reflect increased transcript levels.
Figure 3.
Increased nuclear polyA signals upon exosome inactivation is not mainly due to the lengthened polyA tail. (A) Western blotting to examine the knockdown efficiencies of MTR4 and PABPN1. HeLa cells were transfected with siRNAs targeting the indicated genes. Seventy-two hours post-transfection, western blotting was carried out with indicated antibodies. GAPDH is used as a loading control. (B) Western blots to examine the purities of nuclear and cytoplasmic fractions prepared from control, MTR4, PABPN1 and MTR4/PABPN1 knockdown cells. UAP56 and tubulin are used as nuclear and cytoplasmic marker, respectively. (C) Nuclear polyA RNA tail analysis from cells transfected with indicated siRNAs. (D) PolyA RNA distribution in MTR4, MTR4/PABPN1 knockdown cells. HeLa cells transfected with indicated siRNAs were used for FISH analysis to examine the distribution of polyA RNAs. DAPI staining was used to indicate the nuclei. Note that to accurately quantify the polyA signals in all samples, same exposures were taken for all FISH images. (E) Quantification of total polyA RNA FISH signals. Total polyA RNA FISH signals quantified from 30 cells in each experiment by Image J. Error bar represent standard deviations from three biological replicates. Statistical analysis was performed using Student’s t-test. ***P < 0.001. (F) Nuclear RNA-seq signal shows that the spliced RHOC mRNA is accumulated in RRP40 knockdown cells. (G) (Left) FISH signals of the endogenous RHOC mRNA in control and RRP40 knockdown cells. (Right) Quantification of nuclear RHOC mRNA FISH signals. Nuclear RHOC mRNA FISH signals were quantified from 30 cells in three experiment by Image J. Error bar represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. ***P < 0.001.
To determine whether increased transcript levels could actually lead to elevated FISH signals, we compared the transcript-specific FISH signal of an exosome target mRNA, RHOC, in control and RRP40 knockdown cells (Figure 3F and G). Consistent with the RNA-seq result, the nuclear RHOC FISH signals indeed became apparently stronger in RRP40-depleted cells. Together, these data suggest that the elevated polyA FISH signals upon exosome inactivation mostly result from increased RNA levels.
Exosome target mRNAs are accumulated in polyA RNA foci
We next sought to examine what kinds of RNAs are accumulated in polyA foci upon exosome inactivation. To this end, we analyzed our previous nuclear polyA RNA-seq data for control and MTR4 knockdown cells (9). Among RNAs that are stabilized more than 1.5-fold upon MTR4 knockdown, 60 and 35% were mRNAs and lncRNAs/PROMPTs, respectively (Figure 4A). To determine what type of RNAs contribute the most to the accumulated polyA RNAs, we computed sequencing reads mapped to these accumulated RNAs. Significantly, 84% of them were from mRNAs (Figure 4B). This analysis reveals that most of accumulated polyA RNAs are mRNAs and suggests that nucleoplasmic polyA foci might be mainly formed by stabilized nuclear exosome target mRNAs. Consistent with the previous finding that exosome inactivation results in stabilization of different parts of individual mRNAs (9), apparent accumulation were detected in the 5′, middle, and 3′ fragments of mRNAs at the genome-wide scale (Figure 4C). Thus, although it is possible that certain part of some transcripts are more sensitive to exosome inactivation, i.e. ptRNAs (8), full-length transcripts of significant fractions of mRNAs are likely stabilized.
Figure 4.
Nuclear polyA foci might be mainly formed by exosome target mRNAs. (A) The pie chart represents quantitative distribution of the RNAs whose RPM were elevated more than 1.5-fold in MTR4 knockdown relative to control knockdown. Each category represents RNAs unique to that category and non-overlapping with previous categories, with the initial category designated as ‘short ncRNA’ and proceeding clockwise. (B) Same as (A), except that the read distributions of the accumulated RNAs were shown. (C) Boxplots represent the distribution of nuclear accumulated reads along different parts of the mRNA in MTR4 knockdown cells at the genome-wide scale. Each mRNA is divided into 5′, middle, 3′ parts with the equal length. (D) Localization of the RHOC mRNA in control and RRP40 knockdown cells. SC35 and DAPI staining served as a marker for NS and nucleus, respectively. Confocal microscopy was used to visualize the cells. Arrowheads indicate RHOC mRNA that co-localized with polyA RNAs. Examples of polyA RNA foci that do not apparently co-localize with the RHOC mRNA are indicated by arrows. (E) Quantification of punctate FISH signal of the RHOC mRNA that localized in nuclear foci formed by polyA RNAs. The bars in the graph indicate the percentage of polyA-positive RHOC punctate FISH signal. (F and G) Same as (D and E), except that the DDX39B mRNA was detected in control and MTR4 knockdown cells.
Are nuclear exosome target mRNAs actually accumulated in the polyA foci? To answer this question, we carried out co-localization analysis for the exosome target mRNAs with polyA RNAs. In control cells, the RHOC mRNA co-localized with both polyA RNAs and SC35. In contrast, in RRP40 knockdown cells, it was not apparently detected in NSs, but at least partially co-localized with polyA RNAs (Figure 4D, arrowheads, and 4E). PolyA RNA foci that do not contain this mRNA might be formed by other exosome targets (Figure 4D, arrows). To examine the localization of exosome target mRNAs in MTR4-depleted cells, the DDX39B mRNA that is sensitive to MTR4 depletion was used for co-localization analysis (Supplementary Figure S3). Indeed, it was localized in some of the polyA foci accumulated in MTR4 knockdown cells (FIgure 4F and G). These results suggest that the specific polyA RNA foci observed upon exosome inactivation might be primarily formed by exosome target mRNAs.
PolyA foci in exosome inactivated cells is not mainly due to accumulated TRAMP, NEXT or ZFC3H1 substrates
In mammalian cells, MTR4 forms into distinct complexes that link the exosome to different kinds of substrate RNAs (5,7,8). Although our RNA-seq data show that mRNA is the most abundant accumulating polyA RNA upon exosome inactivation, the possibility that other kinds of exosome targets also make important contributions still remains. PROMPTs are polyadenylated and are more significantly stabilized than mRNAs upon exosome inactivation (5,9). To examine whether stabilized PROMPTs resulted in the accumulated polyA foci, we depleted NEXT components RBM7 and ZCCHC8 individually or in combination (Figure 5A). In these knockdown cells, although the accumulation of all polyadenylated PROMPTs we tested was comparable to that in RRP40 knockdown, no polyA RNA accumulation was observed (Figure 5B–D). This is probably due to the low abundance of PROMPTs even post-stabilization. In support of this possibility, in exosome inactivated cells, the PROMPTs read population is only one-tenth of that of the mRNA (Figure 5E). These data suggest that the accumulated PROMPTs do not account for a major reason for the increased polyA signals we observed in exosome inactivated cells.
Figure 5.
Formation of polyA foci upon exosome inactivation is not mainly due to accumulation of TRAMP, NEXT or ZFC3H1 substrates. (A) Western blotting show that NEXT components were efficiently knocked down. Tubulin is used as a loading control. (B) PROMPTs accumulate similarly in RRP40 and NEXT knockdown cells. HeLa cells were transfected with control, RRP40, RBM7, ZCCHC8 and RBM7/ZCCHC8 siRNA, 72 h post-transfection, polyA RNAs were prepared followed by RT-qPCRs with primer sets that specifically amplify the indicated PROMPTs. R7 and Z8 represent the RBM7and ZCCHC8, respectively. The bars show RNA levels relative to GAPDH. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. (C) PolyA RNA distribution is not affected in NEXT knockdown cells. HeLa cells transfected with indicated siRNAs were used for FISH analysis to observe the distribution of polyA RNAs. DAPI staining served as nucleus marker. (D) Quantification of nuclear polyA RNA FISH signals from 30 cells in each experiment by Image J. Error bar represent standard deviations from three biological replicates. ***P < 0.001. (E) The relative read abundance of PROMPTs and mRNAs in MTR4 knockdown cell. (F) Western blotting results show that TRAMP components were efficiently knocked down. Tubulin is used as a loading control. (G) Same as (C), except that TRAMP knockdown cells were used for this experiment. (H) Quantification of nuclear polyA RNA FISH signals from 30 cells in each experiment by Image J. Error bar represent standard deviations from three biological replicates. Statistical analysis was performed using Student’s t-test. ***P < 0.001. (I) RT-qPCR to examine the knockdown efficiency of ZFC3H1. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. ***P < 0.001. (J) RT-qPCR to examine SNHG transcript levels in control, MTR4 and ZFC3H1 knockdown cells. The bars show RNA levels relative to 18S rRNA. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. **P < 0.01, ***P < 0.001. (K) FISH analysis of polyA RNA distribution in ZFC3H1 knockdown cells. (Left panel) HeLa cells transfected with ZFC3H1 siRNAs were used for FISH analysis to observe the distribution of polyA RNAs. DAPI staining served as nucleus marker. (Right panel) Quantification of nuclear polyA RNA FISH signals from 30 cells in each experiment by Image J. Error bar represent standard deviations from three biological replicates. ***P < 0.001. (L) Nuclear accumulated polyA RNAs in ZFC3H1 knockdown cells co-localize with NSs, FISH was carried out using the 70 (nt) oligo-dT probe and IF using the SC35 antibody were carried out. Confocal microscopy was used to visualize the cells.
It was also possible that PAPD5, which is thought to be the polyA polymerase in the human TRAMP complex, adds a short polyA tail to exosome targets that are normally not polyadenylated, resulting in increased polyA signals upon exosome inactivation. However, knockdown of PAPD5 did not affect nuclear polyA RNA accumulation in MTR4-depleted cells (Figure 5F–H). Consistently, single or double knockdown of PAPD5 and another TRAMP component, ZCCHC7, did not lead to nuclear polyA RNA accumulation (Figure 5F–H). These results suggest that TRAMP substrates do not make important contribution to the increased polyA signals.
Recent studies reported that MTR4, together with ZFC3H1, functions in the degradation of long transcripts and unstable short RNAs (7,8). To examine whether these RNAs could have caused polyA foci accumulation upon exosome inactivation, we depleted ZFC3H1. As expected, ZFC3H1 depletion resulted in apparent SNHG RNA accumulation (Figure 5I and J) (7). Although nuclear polyA RNA FISH signals were elevated in these knockdown cells, distinct from those detected in exosome depleted cells, these RNAs were apparently co-localized with SC35 (Figure 5K and L). The reason for this elevated nuclear polyA RNA signals remains unclear. One possibility is that stabilized SNHG transcripts and unstable RNAs compete with mRNAs for nuclear export machinery, resulting in partial nuclear mRNA retention. Alternatively, the increased fluorescence might result from ZFC3H1 targets that accumulate in NSs. Nevertheless, this result does not support the possibility that the polyA foci detected upon exosome inactivation are mainly formed by ZFC3H1 targets. Together with the polyA RNA-seq data, exosome target mRNAs likely make the most important contribution to the nuclear polyA foci accumulated upon exosome inactivation, although it remains possible that some NEXT, TRAMP and ZFC3H1 target RNAs are also accumulated in these foci.
Exosome target mRNAs are not mostly degraded in NSs
Accumulating evidence indicates that most mRNAs pass through NSs prior to nuclear export (14–19). The observation that exosome target mRNAs are not accumulated in NSs raised the possibility that these mRNAs might not be mainly degraded in these domains. However, it was also possible that exosomal degradation actually occurs within NSs, and when the exosome is inactivated, the accumulated targets are released from NSs to the nucleoplasm. In this case, one would expect that targeting an exosome target mRNA to NSs decrease its stability. To test this, we used an RNA element (speckle-targeting element, STE) that targets the cG reporter mRNA (β-globin cDNA transcript), an exosome target that otherwise does not associate with NSs (14), into these sub-nuclear domains. Significantly, speckle targeting did not reduce, but apparently increase the level of the cG mRNA (Figure 6A, left panel), suggesting the notion that speckle targeting stabilizes exosome target mRNAs. To examine whether this stabilization was due to the prevention of exosomal degradation, we compared the sensitivity of the cG and cG-STE mRNAs to MTR4 knockdown. Importantly, the cG-STE mRNA (1.4-fold increase) was apparently less sensitive than the cG mRNA (2.7-fold increase), indicating that speckle-targeting enhances mRNA stability by preventing exosomal degradation (Figure 6A, right panel). The remaining sensitivity of the cG-STE mRNA might be due to its incomplete speckle targeting. Together, our data support the notion that exosome target mRNAs are mainly degraded in the nucleoplasm, but not in NSs.
Figure 6.
Exosome target mRNAs are mainly degraded before passing through NSs. (A) Speckle targeting of the cG mRNA prevents exosome degradation. RT-qPCRs to examine the levels of cG mRNA or cG -STE mRNAs in normal HeLa cells (left panel), or in control and MTR4 knockdown cells (right panel). (B) Knockdown of PABPN1 and UAP56 inhibit mRNA release from NSs. Confocal microscopy was used to examine distribution of polyA RNAs in control, PABPN1 and UAP56 knockdown cells. SC35 was used to mark NSs. (C) RT-qPCRs to examine levels of indicated mRNAs from HeLa cells treated with control, MTR4, PABPN1 or UAP56/URH49 siRNAs. The relative levels of indicated RNAs to 18S rRNA were quantified and indicated in the graph. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. (D) Two possibilities in accumulation sites of exosome target mRNAs when mRNA release from NSs is inhibited. (I) They would accumulate in both inside and outside of NSs if the degradation occurs before entering NSs; (II) they would accumulate in exclusively inside of NSs if the degradation occurs after being released from NSs. (E) (Top) Confocal microscopy was used to examine the co-localization of polyA RNAs with NSs in control, MTR4, MTR4/PABPN1 and MTR4/UAP56 knockdown cells. (Bottom) Quantification of polyA foci that do not co-localize with SC35 in each cell. Thirty cells were used for the analysis for each sample. (F) MTR4 depletion results in cytoplasmic accumulation of the cG and cS transcripts. (Top) The cG or cS reporter construct was injected into the nuclei of control and MTR4 knockdown cells, followed by FISH using vector probe to detect the distribution of corresponding mRNA at 2 h later after injection. Same exposure was taken for all images. Inset images show the injection marker. DAPI staining served as nucleus marker. (Bottom) Quantification of nuclear and cytoplasmic FISH signals of the corresponding mRNA. N/C ratios were determined for 30 cells in each experiment. N and C indicate nuclear and cytoplasmic FISH signals, respectively. Error bars, standard deviations (n = 3). ***P < 0.001. (G) (Top) The cS reporter construct was injected into the nuclei of MTR4 and MTR/UAP56 knockdown cells, followed by FISH to detect the distribution of corresponding mRNA at 2 h post-injection. Same exposure was taken for all images. Inset images show the injection marker. DAPI staining served as nucleus marker. (Bottom) Quantification of nuclear and cytoplasmic FISH signals of the cS mRNA. N/C ratios were determined for 30 cells in each experiment. N and C indicate nuclear and cytoplasmic FISH signals, respectively. Error bars, standard deviations (n = 3). ***P < 0.001. (H) Nuclear accumulated cS mRNA in MTR4/UAP56 knockdown cells partially co-localize with NSs. FISH with vector probe and IF using the SC35 antibody were carried out.
Exosomal degradation mainly occurs before mRNAs pass through NSs
We next asked when exosomal mRNA degradation occurs, prior to entering NSs or after being released? In the latter case, blocking mRNA release from NSs would attenuate exosomal mRNA degradation. Previous studies reported that PABPN1 is required for polyA RNA nuclear export (26,27). Consistent with these studies, polyA RNAs were accumulated in the nuclei of PABPN1 depleted cells (Figure 6B). Different from the study showing that accumulated polyA RNAs are only partially detected in NSs in U2OS cells (27), we found that these RNAs were well co-localized with SC35 in HeLa cells (Figure 6B), suggesting that mRNA release from NSs was inhibited. The reason for this discrepancy might be due to cell line difference. In PABPN1 knockdown cells, seven out of eight exosome target RNAs we examined showed unaltered levels (Figure 6C). Further, to examine the effect of PABPN1 knockdown on exosome target mRNA levels at the genome-wide scale, we carried out RNA-seq in RRP40 and MTR4 depleted total cells and compared with published RNA-seq data from PABPN1 knockdown (28). Only ∼7% of exosome target mRNAs were apparently accumulated following PABPN1 knockdown (Supplemental Dataset S1). These data suggest that blocking mRNA release from NSs does not affect exosomal mRNA degradation. However, it was also possible that mRNA release was not tightly blocked by PABPN1 depletion. We thus depleted UAP56 that results in almost complete blockage of mRNAs in NSs and apparently enlarged NSs (14) (Figure 6B). Significantly, UAP56 depletion did not stabilize exosome target mRNAs, but results in reduction in their levels (Figure 6C). Considering the important role of UAP56 in ALYREF recruitment (20), this reduction was probably due to inefficient ALYREF recruitment and enhanced MTR4 binding followed by exosomal degradation. In support of this possibility, co-depletion of RRP40 with UAP56 restored these reduced mRNA levels (Supplementary Figure S4). These results together indicate that blocking mRNA release does not attenuate exosome target mRNA degradation and suggest that exosome target mRNAs might be degraded before entering NSs.
We reasoned when mRNA release from NSs is inhibited, exosome target mRNAs would be detected (i) both inside and outside of NSs if degradation occurs before NSs; (ii) exclusively inside NSs if degradation occurs after NSs (Figure 6D). To further distinguish these possibilities, we examined the distribution of polyA RNAs in cells depleted or co-depleted of control, MTR4, MTR4/PABPN1 and MTR4/UAP56. Consistent with exosome knockdown, upon MTR4 depletion, polyA RNAs were apparently accumulated in nuclear foci that do not co-localize with SC35 (Figure 6E). Interestingly, in MTR4/PABPN1 co-knockdown cells, although most polyA RNAs were accumulated in NSs, in average 12 polyA foci do not co-localize with SC35 in each cell (Figure 6E). Similar observation was obtained with MTR4/UAP56 co-knockdown cells. The relative less apparent polyA foci outside of NSs in these co-knockdown cells compared to MTR4 knockdown could be possibly due to the partial overlap with the enlarged NSs. Nevertheless, these results together indicate that exosomal degradation mainly occurs before mRNAs pass through NSs.
Export-defective mRNAs that do not pass through NSs are accumulated in the cytoplasm upon exosome inactivation
The data described above suggest that prior to entering NSs, mRNA fate for nuclear export and degradation has been determined. If true, entering NSs would not be required for fate switching of exosome target mRNAs from degradation to export upon MTR4 depletion. To test this possibility, we examined the effect of MTR4 depletion on the nucleocytoplasmic distribution of the cG mRNA, which cannot enter NSs or be efficiently exported in normal cells (14,24). As expected, the cG mRNA was mostly distributed in the nuclei of control cells (Figure 6F). In contrast, in MTR4 knockdown cells, apparent cytoplasmic accumulation was detected (Figure 6F), suggesting that nuclear export had occurred. When the cS mRNA that is also export-defective and does not enter NSs was used for this analysis, similar results were obtained (Figure 6F) (14,24). These results indicate that passage through NSs is not required for export-defective mRNAs being sorted away from properly exported ones in normal cells. To determine whether exosome target mRNAs are exported via the regular export pathway upon MTR4 knockdown, we co-knocked down MTR4/UAP56. As shown in Figure 6G, the cS mRNA was completely blocked in the nuclei of these co-knockdown cells. Interestingly, in a significant subset (75%) of these cells, the cS mRNAs were partially accumulated in NSs (Figure 6H). These speckle retained cS mRNAs in MTR4/UAP56 co-knockdown cells might correspond to those exported to the cytoplasm in MTR4 knockdown cells. This result indicates that exosome target mRNAs can be exported through NSs in exosome inactivated cells. Together, our data support the view that mRNA fate for nuclear export or degradation is determined before entering NSs.
DISCUSSION
mRNPs assembled in the nucleus is under tight quality control, resulting in rapid degradation of improperly/inefficiently processed and assembled mRNPs and nuclear export of efficiently processed and assembled ones. The competition between mRNA export adaptor ALYREF and the exosome cofactor MTR4 provides an important mechanism for sorting aberrant mRNPs away from export-competent ones. An important remaining question is where and when the mRNP fate for nuclear export and degradation is determined in the nucleus.
In this work, our data suggest that exosomal mRNA degradation mainly occurs in the nucleoplasm before mRNAs enter NSs (See model in Figure 7). We show that upon exosome inactivation, its target mRNAs are accumulated in nucleoplasmic foci outside of NSs, suggesting that exosomal mRNA degradation does not mainly occur in these sub-nuclear structures. Indeed, driving exosome target mRNAs into NSs inhibits their exosomal degradation. Furthermore, multiple lines of evidence support the notion that exosome target mRNAs are degraded before entering NSs and upon exosome inactivation, these mRNAs could slowly enter NSs and get exported.
Figure 7.
mRNA fate determination for export and degradation mainly occurs before mRNAs enter NSs. Left, in normal cells, on the mRNA, ALYREF and MTR4 competes for binding with CBC. If ALYREF outcompetes MTR4, the mRNA then enters NSs, where it might further recruit ALYREF via other mechanisms as well as other TREX components. Following its release from NSs, the mRNA is exported to the cytoplasm. In the case that MTR4 outcompetes ALYREF, the mRNA is then degraded in the nucleoplasm. Right, in exosome inactivated cells, exosome target mRNAs are mostly detected in the nucleoplasm due to their inefficient recruitment of ALYREF. A part of these stabilized target mRNAs are gradually exported to the cytoplasm through or not through NSs.
During revision of the work, Silla et al. (29) reported similar nuclear polyA foci accumulation in exosome inactivated cells. They showed that this accumulation is dependent on ZFC3H1, and provided evidence that ZFC3H1 functionally competes with ALYREF for sorting RNAs for export or degradation. Consistent with this view, Ogami et al., (8) demonstrated that exosome target uaRNAs and ptRNAs are accumulated in the polysomes in MTR4/ZFC3H1 depleted cells. Thus, it seems likely that exosome takes advantage of different cofactors to compete for binding with the mRNA on both 5′ and 3′ ends to ensure best efficiencies for capture of ‘aberrant’ mRNAs. There are similarities as well as differences between our results and those of Silla et al. (29). For example, we found that in addition to exosome components, depletion of MTR4 also resulted in apparent polyA RNA accumulation outside of NSs. Further, upon MTR4 depletion, although part of accumulated exosome target RNAs were exported to the cytoplasm, significant portion are still detected in the nucleus, suggesting that these accumulated mRNAs cannot efficiently recruit nuclear export factors. These results were consistent with Ogami et al., (8) showing that unstable RNAs were accumulated in both the nucleus and the cytoplasm. Also, we found that depletion of PABPN1, which together with MTR4 and ZFC3H1 forms the PAXT complex, resulted in partial relocalization of exosome target mRNAs into NSs. Finally, our data suggest that exosome target mRNAs are sorted for degradation before entering NSs, and this rapid removal is important for preventing exosome target mRNAs from nuclear export.
At the first glance, mRNA accumulation in the nucleus upon MTR4 depletion may seem to conflict with the notion that MTR4 and ALYREF compete to determine their fate for export or degradation (9). We reason that this seemingly conflictive results might be due to the following reasons. On mature mRNAs, ALYREF recruitment is likely to be the default mode, probably due to its coupling to pre-mRNA processing (10,30,31). In normal cells, mRNAs that have been efficiently processed and/or have strong export elements rapidly recruit ALYREF and are efficiently exported. mRNAs that are improperly or inefficiently processed or do not have strong export element cannot efficiently recruit ALYREF, resulting in MTR4 binding and subsequent exosome recruitment for degradation. In the case of MTR4 knockdown, the low abilities of these ‘defective’ mRNAs in recruiting mRNA export factors remain unchanged. As exosomal degradation is inhibited, these mRNAs gain much more time and chances to be bound by ALYREF and get slowly exported. However, this export cannot be very efficient and a significant fraction of mRNAs are probably still retained in the nucleus. Consistently, exosome target RNAs were accumulated in both the nucleus and the cytoplasm upon exosome inactivation (8,9). Further, although nuclear export of the transiently transcribed exosome targets, the cG and cS mRNAs, appear significantly enhanced upon MTR4 knockdown, they were still apparently detected in the nucleus (Figure 6F). It is also possible that ZFC3H1 functions in nuclear retention of exosome target mRNAs like it does for unstable RNAs (8,29). Further studies are required to study how MTR4-CBC and ZFC3H1/MTR4-PABPN1 cooperate with each other to sort exosome target RNAs from properly exported ones.
Considering that mRNAs have been committed to export before entering NSs, namely ALYREF has been recruited via CBC, why do they pass through these domains? Although early-stage fate determination for export ensures prevention of exosomal mRNA degradation, the assembly for export-competent mRNPs might possibly mainly occur in NSs (See model in Figure 7). In support of this view, ALYREF and many other mRNA export factors are concentrated in NSs (10–13). In the cells, ALYREF is not only recruited to the 5′ region via CBC, but also binds the 3′ region in PABPN1-dependent way and the middle region in a sequence-dependent manner (21,30,32). It is possible that ALYREF binding to these regions might occur in NSs (see model in Figure 7). Another possibility is that entering NSs could avoid continuous competition of MTR4 with ALYREF for the mRNAs whose fate has been determined for nuclear export.
The rapid removal of exosome target RNAs might be important for avoiding wasting nuclear RNA processing factors for functional RNAs and limiting the chances for aberrant RNAs to be exported. In support of this, upon exosome inactivation, export-defective mRNAs as well as normally unstable RNAs could be exported in the cytoplasm (Figure 6F) (8). On one side, these RNAs could also compete with functional mRNAs for translation machinery in the cytoplasm, like Ogami et al., reported for unstable RNAs (8). On the other side, aberrant processed mRNAs with disrupted reading frames could cause cytoplasmic toxicity through their translation product.
The nature of the nuclear foci where the exosome target mRNAs are accumulated remains unclear. One possibility is that they are the degradation sites for exosome target mRNAs. An alternative possibility is that exosome targets mRNAs are not degraded in these foci, but form aggregates in the nucleoplasm when the exosome is inactivated. Future studies are required to decipher the formation mechanism for these polyA foci.
DATA AVAILABILITY
The data associated with the manuscript are available in GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE89123.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Cheng Lab members for useful discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0504400]; National Natural Science Foundation of China [21625302, 31570822, 91540104, 31770880]; China Postdoctoral Science Foundation [BX20180333, 2018M630481]. Funding for open access charge: National Natural Science Foundation of China [21625302].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lebreton A., Tomecki R., Dziembowski A., Seraphin B.. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008; 456:993–996. [DOI] [PubMed] [Google Scholar]
- 2. Schneider C., Anderson J.T., Tollervey D.. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007; 27:324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P.. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 1999; 13:2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D.. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997; 91:457–466. [DOI] [PubMed] [Google Scholar]
- 5. Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H.. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011; 43:624–637. [DOI] [PubMed] [Google Scholar]
- 6. Shcherbik N., Wang M., Lapik Y.R., Srivastava L., Pestov D.G.. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010; 11:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meola N., Domanski M., Karadoulama E., Chen Y., Gentil C., Pultz D., Vitting-Seerup K., Lykke-Andersen S., Andersen J.S., Sandelin A. et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell. 2016; 64:520–533. [DOI] [PubMed] [Google Scholar]
- 8. Ogami K., Richard P., Chen Y., Hoque M., Li W., Moresco J.J., Yates J.R. 3rd, Tian B., Manley J.L.. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017; 31:1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan J., Kuai B., Wu G., Wu X., Chi B., Wang L., Wang K., Shi Z., Zhang H., Chen S. et al. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 2017; 36:2870–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R.. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005; 19:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gatfield D., Le Hir H., Schmitt C., Braun I.C., Kocher T., Wilm M., Izaurralde E.. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001; 11:1716–1721. [DOI] [PubMed] [Google Scholar]
- 12. Zhou Z., Luo M.J., Straesser K., Katahira J., Hurt E., Reed R.. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000; 407:401–405. [DOI] [PubMed] [Google Scholar]
- 13. Kataoka N., Yong J., Kim V.N., Velazquez F., Perkinson R.A., Wang F., Dreyfuss G.. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000; 6:673–682. [DOI] [PubMed] [Google Scholar]
- 14. Dias A.P., Dufu K., Lei H., Reed R.. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010; 1:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akef A., Zhang H., Masuda S., Palazzo A.F.. Trafficking of mRNAs containing ALREX-promoting elements through nuclear speckles. Nucleus-Austin. 2013; 4:326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mor A., White A., Zhang K., Thompson M., Esparza M., Munoz-Moreno R., Koide K., Lynch K.W., Garcia-Sastre A., Fontoura B.M.. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 2016; 1:16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carter K.C., Taneja K.L., Lawrence J.B.. Discrete nuclear domains of Poly(a) Rna and their relationship to the Functional-Organization of the nucleus. J. Cell Biol. 1991; 115:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang S., Deerinck T.J., Ellisman M.H., Spector D.L.. In-vivo analysis of the stability and transport of nuclear Poly(a)+Rna. J. Cell Biol. 1994; 126:877–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Visa N., Puviondutilleul F., Harper F., Bachellerie J.P., Puvion E.. Intranuclear distribution of Poly(a) Rna determined by Electron-Microscope In-Situ hybridization. Exp. Cell Res. 1993; 208:19–34. [DOI] [PubMed] [Google Scholar]
- 20. Chi B., Wang K., Du Y., Gui B., Chang X., Wang L., Fan J., Chen S., Wu X., Li G. et al. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res. 2014; 42:7305–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., Cheng H.. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2013; 41:1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapadia F., Pryor A., Chang T.H., Johnson L.F.. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 2006; 384:37–44. [DOI] [PubMed] [Google Scholar]
- 23. Shi M., Zhang H., Wang L., Zhu C., Sheng K., Du Y., Wang K., Dias A., Chen S., Whitman M. et al. Premature termination codons are recognized in the nucleus in a Reading-Frame dependent manner. Cell Discov. 2015; 1:15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valencia P., Dias A.P., Reed R.. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3386–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bresson S.M., Conrad N.K.. The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 2013; 9:e1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Apponi L.H., Leung S.W., Williams K.R., Valentini S.R., Corbett A.H., Pavlath G.K.. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 2010; 19:1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hurt J.A., Obar R.A., Zhai B., Farny N.G., Gygi S.P., Silver P.A.. A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. J. Cell Biol. 2009; 185:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beaulieu Y.B., Kleinman C.L., Landry-Voyer A.M., Majewski J., Bachand F.. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012; 8:e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Silla T., Karadoulama E., Makosa D., Lubas M., Jensen T.H.. The RNA exosome adaptor ZFC3H1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep. 2018; 23:2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi M., Zhang H., Wu X.D., He Z.S., Wang L.T., Yin S.Y., Tian B., Li G.H., Cheng H.. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 2017; 45:9640–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson S.A., Cubberley G., Bentley D.L.. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell. 2009; 33:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R.. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006; 127:1389–1400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with the manuscript are available in GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE89123.