Abstract
The aim of the present study was to investigate the association between Ki-67 expression and radiomics features of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) in patients with invasive breast cancer. A total of 53 cases with low-Ki-67 expression (Ki-67 proliferation index <14%) and 106 cases with high-Ki-67 expression (Ki-67 proliferation index >14%) were investigated. A systematic approach was applied that focused on the automated segmentation of lesions and extraction of radiomics features. For each lesion 5 morphology, 4 gray-scale histogram and 6 texture features were obtained, and statistical analyzes were performed to assess the differences in these features between the low- and high-Ki-67 expressions. One morphology metric (area), 3 gray-scale histogram indexes (standard deviation, skewness and kurtosis) and 3 texture features (contrast, homogeneity and inverse differential moment) demonstrated a significant difference (P<0.05), with low-Ki-67 expression lesions tending to be smaller, clearer and heterogeneous when compared with the high-Ki-67 expressed cases. These results may provide a noninvasive means to better understand the proliferation of breast cancer.
Keywords: magnetic resonance imaging, proliferation, Ki-67 expression, radiomics, invasive breast cancer
Introduction
As in most other countries, breast cancer is now the most common cancer in Chinese women, cases in China account for 12.2% of all newly diagnosed breast cancers and 9.6% of all deaths from breast cancer worldwide (1). Ki-67 is one of the major markers of tumor proliferation, assessed by immunohistochemistry (IHC) with the anti-Ki-67 antibody called MIB-1 (2). Many investigations have reported that Ki-67 is an independent predictive and prognostic marker in patients with operable breast cancer (3,4). Thus, assessment of Ki-67 is already introduced into daily practice in order to discriminate breast cancer subtypes, predict oncological outcomes, or decide on indications for adjuvant treatment (5). Current preoperative assessment of Ki-67 is mostly based on IHC, which requires tissue specimens typically obtained by needle biopsy. Due to the relatively small tissue sample size and tumor heterogeneity, the Ki-67 expression assessment performed on a needle biopsy sample may not be representative of the tumor entirety. Moreover, Ki-67 assessment can be unavailable in many critical cases where biopsy is in feasible.
Recently, several studies have hypothesized that tumor characteristics at the cellular and genetic levels are reflected in the phenotypic patterns and can be captured with medical imaging (6–8). On this theoretical basis, the radiomics methodology has been proposed that objectively characterizes tumor phenotypes using the advanced quantitative features of medical images. These features, referred to as the radiomics features, are extracted from medical images using advanced mathematical algorithms in a high-throughput way, and can discover tumor characteristics that may fail to be appreciated by the naked eye (9–12).
To date, the previous research on the radiomics features of breast cancer has focused majorly on the separation of the benign and malignant (13,14), molecular receptor status of estrogen receptor (ER)+ and ER−, progesterone receptor (PR)+ and PR−, human epidermal growth factor receptor 2 (HER2)+ and HER2− for dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) (15–20). No report has been released on analyzing the Ki-67 expression with radiomics features. Therefore, the goal of this study is to explore the association of extracted radiomics features with Ki-67 expression on breast DCE-MRI, which could provide a noninvasive means to better understand the proliferation of breast cancer and further providing valuable information for personalized therapy.
Materials and methods
Clinical data
The present retrospective study was approved by the Institutional Review Board of Tianjin Medical University (Tianjin, China). Three hundred seventy-seven (377) Chinese women with invasive breast cancer that were confirmed by pathology and underwent breast DCE-MRI were divided into two groups based on their Ki-67 proliferation index: Low-Ki-67 group-Ki-67 proliferation index less than 14%; high-Ki-67 group-Ki-67 proliferation index more than 14% (5). This breast DCE-MRI data was prospectively collected from January 2015 to September 2015 at our institution. In the entire data set, the low-Ki-67 group was composed of 53 low-Ki-67 expressed lesions that accounts for 14.06% of the total cases. For a preliminary analysis, 106 high-Ki-67 expressed lesions (double of the low-Ki-67 expressed group) were selected at random from the total data set as the high-Ki-67 group. The ages of the patients ranged from 30 to 68 years old (49±10, median 48) in the low-Ki-67 group and from 29 to 72 years old in high-Ki-67 group (47±9, median 50). There was no statistically significant difference in age between the two groups (P=0.483).
Imaging data
The DCE-MRI examinations were performed on a 3.0T scanner using a dedicated 8-channel phased-array breast coil (Discovery MR750; GE Healthcare, Shanghai, China). Sagittal data was obtained by the volume imaging for breast assessment bilateral breast imaging technique, with TR=6.1 ms, TE=2.9 ms, flip angle=15°, matrix size=256×128, field of view=26×26 cm, NEX=1, slice thickness=1.8 mm. Before injection of the contrast agent, serial mask images were obtained. Successively, the contrast agent (Gd-DTPA, 0.2 ml/kg body weight, flow rate 2.0 ml/sec) was manually injected using an automatic MR-compatible power injector, and thereafter flushed with the same total dose of saline solution. The dynamic MRI acquisitions were started immediately after the injection. The acquisition was repeated five times, and each phase took 90–100 sec. The radiomics analysis was conducted at the first post-contrast time-point of the MRI.
Lesion segmentation
The contour of the lesion region of interest (ROI) in the largest DCE-MRI slice of each lesion was automatically constructed using the localizing region-based active contours algorithm (21). Figs. 1 and 2 show the segmented results of two MRI images.
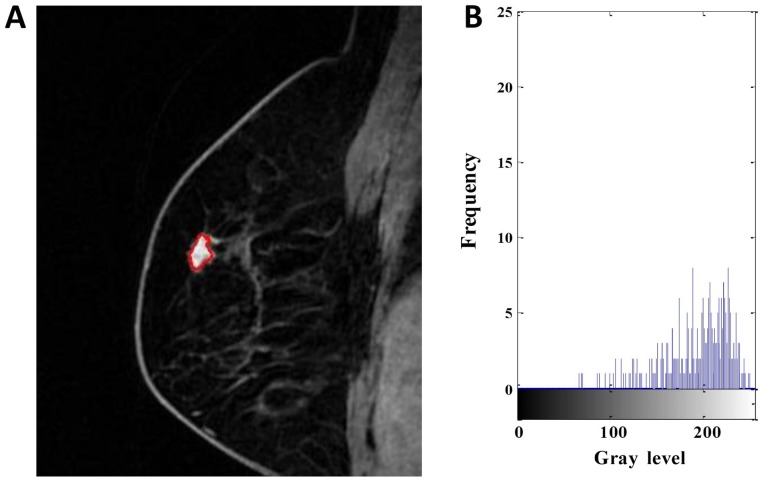
Figure 1.
DCE-MRI of a 63-year-old woman with low-Ki-67 expression. (A) Segmentation outlines obtained from the active contours segmentation method. (B) The gray-scale histogram of the region of interest. The values of the area, SD, skewness, kurtosis, entropy, contrast, homogeneity and inverse differential moment were 436, 48.784, −0.535, 2.242, 6.120, 4.597, 0.529 and 0.079, respectively. DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging. SD, standard deviation.
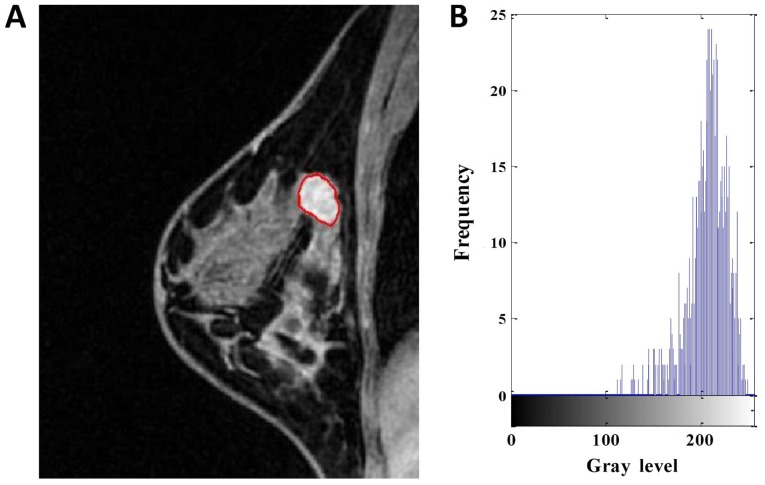
Figure 2.
DCE-MRI of a 43-year-old woman with high-Ki-67 expression. (A) Segmentation outlines obtained from the active contours segmentation method. (B) The gray-scale histogram of the region of interest. The values of the area, SD, skewness, kurtosis, entropy, contrast, homogeneity and inverse differential moment were 810, 31.746, −1.231, 3.985, 4.738, 1.704, 0.675 and 0.076, respectively. DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging. SD, standard deviation.
Radiomics features
Radiomics features provide an objective and quantitative metrics to assess tumor phenotype. In this work, 15 features were extracted from each ROI, including 5 morphological features, 4 gray-scale histograms and 6 texture features, as explained below.
Morphological features
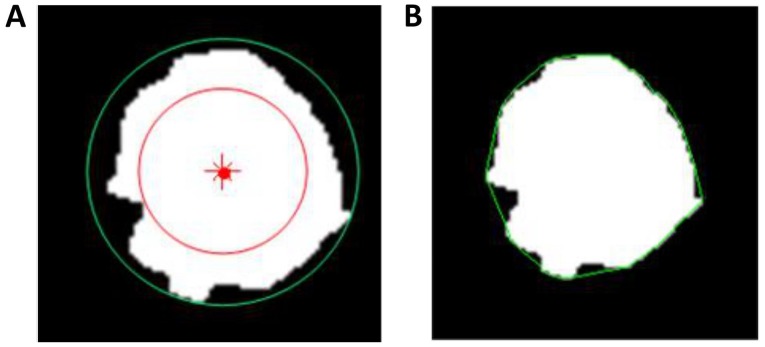
Five metrics, including area, normalized radial length (NRL), roundness, compactness and concavity rate, were calculated for the morphological description of the images. Area is one of the most basic characteristics to describe image ROI, and normally defined as the number of the pixels in the ROI. NRL defined as the Euclidean distance from the center of the lesion ROI to each of its contour pixels and normalized to the maximum radial length of the lesion. Roundness is the measure of how closely the shape of an object approaches that of a mathematically perfect circle, and defined as the ratio of the circumcircle radius to the inscribed circle radius of the lesion ROI. The circum and inscribed circles are defined as circles with their radii being the maximum and minimum distances from a boundary point to the center of the lesion ROI, respectively, as shown in Fig. 3A. Compactness and concavity are associated with shape and margin of the lesion. Compactness is defined as the 4πxA/P2, where A is the cross-sectional area of the tumor and P is the measured length of the perimeter of the lesion. Concavity rate is defined as abs (A-B)/B, where B is the area of the convex hull calculated for the lesion region, as illustrated in Fig. 3B, and ‘abs’ denotes the absolute value.
Figure 3.
Schematics of the (A) roundness and (B) concavity-rate calculations. In the roundness calculation, the red dot is the center of the lesion, and the red and green line circles are the inscribed circle and the circumcircle, respectively. In the concavity-rate calculation, the green circle is the convex hull.
Gray-scale histograms features
Four features were computed for each lesion according to the definitions of the gray-scale histogram: Mean, standard deviation (SD), skewness and kurtosis. Their definition can be found in literatures (22).
Texture features
Texture is a repeating pattern of local variations in image intensity, and is characterized by the spatial distribution of intensity levels in a neighborhood. Six gray-level co-occurrence matrix (GLCM) texture features were obtained for each lesion as defined by Haralick et al (23), including energy, entropy, contrast, correlation, homogeneity and inverse differential moment (IDM). The ROI extraction and radiomics feature calculations were performed using programs written with MATLAB 2014b.
Statistical methods
To compare the differences in morphologic, gray-scale histograms and texture features of low- and high-Ki-67 expression by using radiomics analysis method, the statistical method used was the Mann-Whitney U test, with the significance level set as α=0.05. Statistical analysis was performed using R 3.4.3 software (www.R-project.org). P<0.05 was considered to indicate a statistically significant difference.
Results
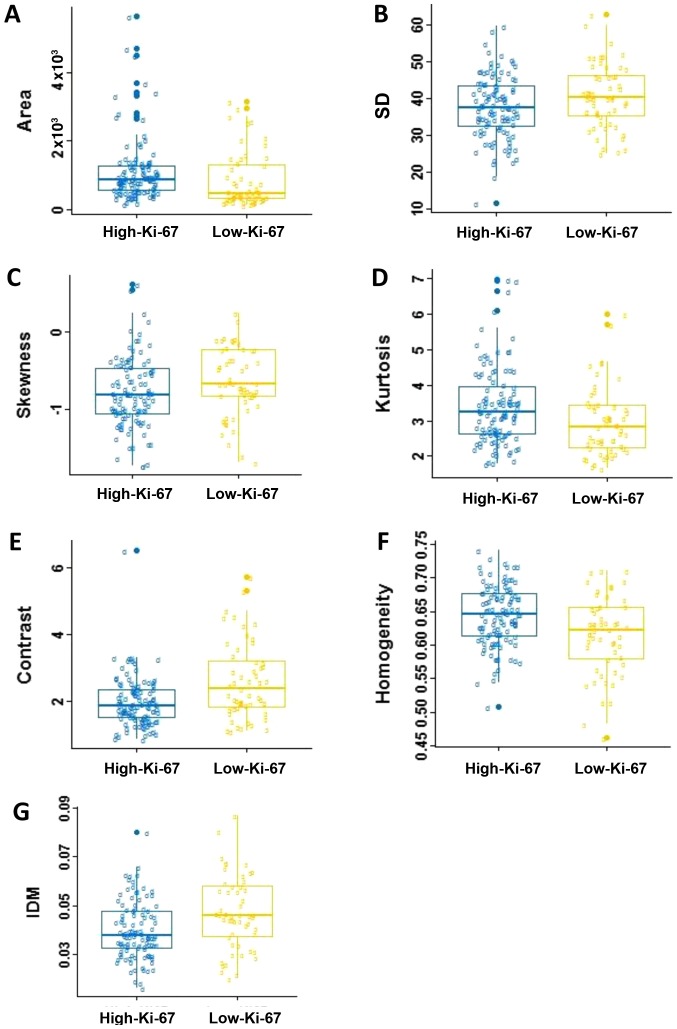
The average values and SD of the radiomics features are compared in Table I. No statistically significant differences were observed in roundness, NRL, compactness, concavity rate, mean, energy, entropy and correlation between two groups (P>0.05). Fig. 4 shows the box and whisker plots of the radiomics parameters values of two groups for P<0.05. It displays the center and spread of a numeric variable in a format.
Table I.
Feature parameters in association with Ki-67 expression.
| Mean ± SD | |||
|---|---|---|---|
| Feature parameters | High-Ki-67 | Low-Ki-67 | P-value |
| Morphologic | |||
| Area | 1,125.7±936.2 | 868.6±814.1 | 0.002 |
| NRL | 0.275±0.040 | 0.271±0.035 | 0.261 |
| Roundness | 0.745±0.148 | 0.732±0.146 | 0.619 |
| Compactness | 0.707±0.135 | 0.705±0.157 | 0.991 |
| Concavity rate | 0.112±0.075 | 0.128±0.106 | 0.449 |
| Gray-level histogram | |||
| Mean | 167.164±22.689 | 170.095±23.550 | 0.342 |
| SD | 37.713±8.495 | 40.874±8.686 | 0.043 |
| Skewness | −0.781±0.449 | −0.636±0.438 | 0.021 |
| Kurtosis | 3.423±1.084 | 2.980±0.970 | 0.005 |
| GLCM | |||
| Energy | 0.044±0.015 | 0.041±0.016 | 0.075 |
| Entropy | 5.238±0.403 | 5.342±0.384 | 0.061 |
| Contrast | 1.976±0.754 | 2.627±1.151 | <0.001 |
| Correlation | 0.847±0.054 | 0.832±0.055 | 0.053 |
| Homogeneity | 0.644±0.045 | 0.616±0.583 | 0.005 |
| IDM | 0.040±0.012 | 0.047±0.015 | 0.002 |
SD, standard deviation; NRL, normalized radial length; GLCM, gray-level co-occurrence matrix; IDM, inverse differential moment.
Figure 4.
Box and whisker plots for the associations between Ki-67 expressions and the DCE-MRI-based phenotypes. (A) Area, (B) SD, (C) skewness, (D) kurtosis, (E) contrast, (F) homogeneity and (G) IDM. SD, standard deviation; IDM, inverse differential moment. DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging.
In the five morphological parameters, only one feature, area, which indicating statistical significance in differentiating the low-Ki-67 expression from the high-Ki-67 expression (P=0.002). As shown in Table I and Fig. 4A, the high-Ki-67 cases tended to have larger lesion size. There was no significant difference in roundness, NRL, compactness and concavity between two groups (P>0.05).
For the gray-scale histograms features, SD, skewness and kurtosis showed statistical significance between the two groups (P=0.043, 0.021 and 0.005). The values of the SD and skewness in the low-Ki-67 group is larger than those in the high-Ki-67 group (Fig. 4B and C), and it is on the contrary for the kurtosis (Table I; Fig. 4D). The histogram is an effective graphical technique for showing both the skewness and kurtosis in the data set (Figs. 1 and 2). Notice that the shape of the two histograms is quite different. It seems that the histograms of the high-Ki-67 cases tend to densely distribute in a narrow area.
For texture features, contrast, homogeneity and IDM showed statistical significance between two groups (P=0.0004, 0.005 and 0.002). The value of contrast, and IDM in the low-Ki-67 group was significantly larger than those in the high-Ki-67 group (Table I; Fig. 4E-G). The homogeneity in the low-Ki-67 group was smaller than that in the high-Ki-67 group (Fig. 4F). As can be seen in Figs. 1 and 2, the kurtosis of the high-Ki-67 case was larger than that of the low-Ki-67. On the contrary, the contrast of the high-Ki-67 case was smaller than that of the low-Ki-67 case (High-Ki-67: Contrast index=1.704, IDM index=0.076, entropy index=4.738, homogeneity index=0.675; Low-Ki-67: Contrast index=4.597, IDM index=0.079, entropy index=6.120, homogeneity index=0.529). The spread of IDM in the low-Ki-67 group is more spread out than that in the high-Ki-67 group (Fig. 4G). This phenomenon can also be found in Fig. 4E.
Discussion
In oncology, biomarkers describe the characteristics of a malignancy on various levels (clinical, histological, molecular and so on) and predict patient's outcome and treatment response, which is the reason why they are increasingly integrated into the clinical routine. Based on the radiomics theory, multiple features can be extracted and linked to clinical, genomic, and histopathological data. Extracted traits describe radiological shape, grey intensity and texture characteristics and can be analyzed on routinely performed images. In this study, these specific radiomic markers and patterns were developed for discriminating between low- and high-Ki-67 expressions of breast in DCE-MRI for the first time. Breast cancer with high-Ki-67 expression responds better to chemotherapy but is associated with poor prognosis (6,24).
In our study a total of 5 morphological features, 4 gray-scale histograms and 6 texture features were extracted to characterize each lesion. The Mann-Whitney test were performed to assess the statistical significance of the difference between the low- and high-Ki-67 expressions. The result shows that the lesion area, SD, skewness, kurtosis, homogeneity and IDM are significantly associated with the Ki-67 expression level.
Morphology is a theory and technique for the analysis and processing of geometrical structures. Clinically, the doctor relies to a large extent on the morphology of the lesion for diagnosis. Li et al (25) indicated that the Ki-67 expression level in breast cancer tissue significantly correlated with the tumor size. This is consistent with the results of this study that the lesion area is significantly associated with the Ki-67 expression. We can find that the values of the area in the low-Ki-67 expression were much smaller than that in the high-Ki-67 expression. This indicated that the increased expression of Ki-67 may predict the increased proliferation of breast cancer cells, enhanced invasiveness, and faster growth of the tumor. Nevertheless, since the definition diagnosis times of the patient cases were difficulty to accurately control in present, the above observation needs to be further validated under an identical condition or by using dynamic analysis in the future investigation.
Skewness describes asymmetrical properties of pixel distribution. A distribution is symmetric if it looks the same to the left and right of the center point. The skewness for a normal distribution is zero, and any symmetric data should have a skewness near zero. Negative values for the skewness indicate data that are skewed left and positive values for the skewness indicate data that are skewed right. We can find in Fig. 4C that two groups almost all data skewed left, and the high-Ki-67 expression was more to the left. Kurtosis is a measure of whether the data are heavy-tailed or light-tailed relative to a normal distribution. We can find in Fig. 4D that the high-Ki-67 group had higher kurtosis. In general, this means that lesions with many voxels of similar uptake are likely to be more biologically proliferating.
Table I indicates that the high-Ki-67 expressed lesions were likely to show more homogeneous. This can be explained that high-Ki-67 lesions have more biologically proliferating, and hence have more voxels of similar uptake that appears to be more homogenous. Most of the GLCM texture features were highly correlated with each other. A homogenously enhanced lesion has lower entropy and higher energy compared to a heterogeneously enhanced lesion. The larger the enhancement texture entropy, the more heterogeneous the tumor. As shown in Figs. 1 and 2, a homogeneously enhanced lesion has lower entropy compared to the heterogeneously enhanced one. Entropy quantifies complexity of the image. The higher values in entropy of the low-Ki-67 expression lesions may suggest that these images are more complex than the high-Ki-67 expression images. The contrast reflects the clarity of the image and the texture of the groove depth. The deeper the groove depth and the greater the contrast, the image is clearer. On the other hand, the contrast value is small, the image is vaguer. IDM reflects the sharpness of the image. A higher value of IDM indicates the image texture is clearer. Fig. 4G indicates that there is a little difference in image clarity in the high-Ki-67 group and the value in the low-Ki-67 group larger than high-Ki-67 group overall. Together, the representation of these features indicates the high-Ki-67 expressed lesions are more likely to be heterogeneous.
Despite our encouraging results, some limitations exist in the present work. First, the automatically extracted features were investigated on the largest axial slice and the value was used to represent the whole lesion. The true texture analysis relies on 3D isotropic image acquisition. However, the 3D whole lesion analysis is computationally more complex and time-consuming. Second, we have only considered the tumor images at the 2nd enhancement phase, and not analyzed the pre-contrasts, other enhancement images and mammary gland tissues. Third, we did not analyze the kinetic features that are important parameters in diagnosis of benign and malignant breast masses. Finally, the patient sample set was relatively small, and hence results of this pilot study are somewhat preliminary. In future study we will work on a larger data set and address these limitations to verify the preliminary results.
In conclusion, our study illustrates the feasibility of the use of radiomics analysis in evaluating the Ki-67 expression level. The low-Ki-67 expression cases tend to be smaller and more heterogeneous than the high-Ki-67 expression cases. Identification of the Ki-67 expression helps provide complementary information for precision medicine to aid clinical decision making. Based on this study, the next aim is to classify the lesion for the low and high-Ki-67 expressions by introducing the machine learning method.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Key Project of Tianjin Science and Technology Committee Foundation grant (grant no. 12ZCDZSY16000) and The Tianjin Municipal Government of China (grant no. 15JCQNJC14500).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MWJ designed the study and edited the manuscript. JY and LJJ performed the statistical analysis. GXP and SPF collected the imaging data. LPF analyzed and interpreted the data.
Ethics approval and consent to participate
The present retrospective study was approved by the Institutional Review Board of Tianjin Medical University (Tianjin, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 2.Gerdes J, Li L, Schlueter C, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, Flad HD. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138:867–873. [PMC free article] [PubMed] [Google Scholar]
- 3.de Azambuja E, Cardoso F, de Castro G, Jr, Colozza M, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M. Ki-67 as prognostic marker in early breast cancer: A meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504–1513. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colozza M, Azambuja E, Cardoso F, Sotiriou C, Larsimont D, Piccart MJ. Proliferative markers as prognostic and predictive tools in early breast cancer: Where are we now? Ann Oncol. 2005;16:1723–1739. doi: 10.1093/annonc/mdi352. [DOI] [PubMed] [Google Scholar]
- 5.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ. Panel members: Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Ealy Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JR, DiGiovanna MP, Killelea B, Lannin DR, Rimm DL. Quantitative assessment Ki-67 score for prediction of response to neoadjuvant chemotherapy in breast cancer. Lab Invest. 2014;94:98–106. doi: 10.1038/labinvest.2013.128. [DOI] [PubMed] [Google Scholar]
- 7.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl AcadSci USA. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu S, Kwee TC, Gatenby R, Saboury B, Torigian DA, Alavi A. Evolving role of molecular imaging with PET in detecting and characterizing heterogeneity of cancer tissue at the primary and metastatic sites, a plausible explanation for failed attempts to cure malignant disorders. Eur J Nucl Med Mol Imaging. 2011;38:987–991. doi: 10.1007/s00259-011-1787-z. [DOI] [PubMed] [Google Scholar]
- 9.Rutman AM, Kuo MD. Radiogenomics: Creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, et al. Radiomics: The process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, Schernberg A, Paragios N, Deutsch E, Ferté C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28:1191–1206. doi: 10.1093/annonc/mdx034. [DOI] [PubMed] [Google Scholar]
- 14.Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol. 2008;15:1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SG, Freed M, Leite APK, Zhang J, Seuss C, Moy L. Separation of benign and malignant breast lesions using dynamic contrast enhanced MRI in a biopsy cohort. J Magn Reson Imaging. 2017;45:1385–1393. doi: 10.1002/jmri.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang RF, Chen HH, Chang YC, Huang CS, Chen JH, Lo CM. Quantification of breast tumor heterogeneity for ER status, HER2 status, and TN molecular subtype evaluation on DCE-MRI. Magn Reson Imaging. 2016;34:809–819. doi: 10.1016/j.mri.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Fan M, Li H, Wang S, Zheng B, Zhang J, Li L. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS One. 2017;12:e0171683. doi: 10.1371/journal.pone.0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou SS, Gombos EC, Chikarmane SA, Giess CS, Jayender J. Computer-aided heterogeneity analysis in breast MR imaging assessment of ductal carcinoma in situ: Correlating histologic grade and receptor status. J Magn Reson Imaging. 2017;46:1748–1759. doi: 10.1002/jmri.25712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agner SC, Rosen MA, Englander S, Tomaszewski JE, Feldman MD, Zhang P, Mies C, Schnall MD, Madabhushi A. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: A feasibility study. Radiology. 2014;272:91–99. doi: 10.1148/radiol.14121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton EJ, Dashevsky BZ, Oh JH, Veeraraghavan H, Apte AP, Thakur SB, Morris EA, Deasy JO. Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging. 2016;44:122–129. doi: 10.1002/jmri.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lankton S, Tannenbaum A. Localizing region-based active contours. IEEE Trans Image Process. 2008;17:2029–2039. doi: 10.1109/TIP.2008.2004611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodenacker K, Bengtsson E. A feature set for cytometry on digitized microscopic images. Anal Cell Pathol. 2003;25:1–36. doi: 10.1155/2003/548678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haralick R, Shanmugam K, Dinstein I. Texture parameters for image classification. IEEE Trans SMC. 1973;3:610–621. [Google Scholar]
- 24.Matsubara N, Mukai H, Masumoto M, Sasaki M, Naito Y, Fujii S, Wada N. Survival outcome and reduction rate of Ki-67 between pre- and post-neoadjuvant chemotherapy in breast cancer patients with non-pCR. Breast Cancer Res Treat. 2014;147:95–102. doi: 10.1007/s10549-014-3084-6. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Han X, Liu Y, Liu G, Dong G. Ki67 as a predictor of poor prognosis in patients with triple-negative breast cancer. Oncol Lett. 2015;9:149–152. doi: 10.3892/ol.2014.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.