Abstract
Long non-coding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) has been reported to be overexpressed in prostate cancer cells and associated with tumorigenesis in various types of cancer. However, the biological function of lncRNA PVT1 remains largely unknown. The aim of the present study was to investigate the effect of lncRNA PVT1 expression on the proliferation and migration of prostate cancer cells. Stably transfected prostate cancer cells with downregulated expression of lncRNA PVT1 were constructed by an efficient siRNA fragment, followed by confirmation by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Proliferation was assessed using CCK-8, colony formation and xenograft assays, and cell migration was evaluated using a wound healing assay. The PathScan® Intracellular Signaling Array kit was utilized to explore the underlying molecular mechanisms of lncRNA PVT1 expression in prostate cancer cells. RT-qPCR results confirmed that the lncRNA PVT1 expression level was successfully knocked down in prostate cancer cells. When lncRNA PVT1 expression was downregulated in prostate cancer cells, proliferation and migration were significantly inhibited, compared with the control lncRNA PVT1 group. Furthermore, PVT1 knockdown decreased the phosphorylation of p38 in DU145 cells. Therefore, the present study demonstrated that lncRNA PVT1 downregulation inhibits the proliferation and migration of prostate cancer cells, and is associated with p38 phosphorylation.
Keywords: prostatic neoplasms, RNA, long noncoding, proliferation, cell movement
Introduction
Prostate cancer has been reported as the second leading cause of cancer-associated male mortality in the United States (1,2). The majority of patients with early stage prostate cancer can be treated by radical prostatectomy and androgen deprivation therapy (3). It has been indicated that after 1–3 years of treatment, the majority of prostate cancer cases ultimately progress to hormone refractory prostate cancer, which is classified as an incurable disease (4). However, the underlying mechanism associated with the carcinogenesis and progression of prostate cancer requires further investigation. Therefore, it the development of a novel, target-specific effective treatment strategy for patients with prostate cancer exhibiting androgen-resistance is urgently required.
Long non-coding RNAs (lncRNAs) are 200 nucleotides in length (5) and serve crucial roles in the progression of malignant tumors (6–9). Plasmacytoma variant translocation 1 (PVT1), an oncogenic lncRNA, is located in the chromosomal region 8q24 adjacent to MYC (10). Overexpression of lncRNA PVT1 has been reported in various types of tumor (11–16), including prostate cancer (17). However, the biological function and underlying mechanism of lncRNA PVT1 in prostate cancer requires further investigation. In the present study, the effect of downregulation of lncRNA PVT1 expression on proliferation and migration was investigated, in addition to its potential underlying mechanism in prostate cancer.
Materials and methods
Cell culture
Human prostate cancer cell lines, PC-3 and DU145, were obtained from American Type Culture Collection (Manassas, VA, USA) and cultivated in Roswell Park Memorial Institute 1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Bio-Sciences, Logan, UT, USA). The cells were maintained at 37°C in 5% CO2.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Total RNA was extracted from PC-3 and DU145 cells, using TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan) from 1 µg of total RNA in a 20 µl reaction volume, according to the manufacturers' protocols. The qPCR reaction mixture was assessed using the ABI 7500 Fast Real Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) utilizing SYBR Premix Ex Taq™ (Takara Bio, Inc.). The GAPDH fragment was used as an internal control gene for standardizing the expression of target genes. The primers were as follows: lncRNA PVT1 forward, 5′-TGAGAACTGTCCTTACGTGACC-3′ and reverse, 5′-AGAGCACCAAGACTGGCTCT-3′; GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′ amd reverse, 5′-GCCCAATACGACCAAATCC-3′. Each experiment was repeated three times. The reaction conditions were 95°C for 10 min, followed by 95°C for 10 sec for 40 cycles and 60°C for 60 sec. The relative mRNA expression level of each sample compared with GAPDH expression was derived using the 2−ΔΔCq method (18,19).
RNA interference
PC-3 and DU-145 cells (1×105 cells) were cultured to 60% confluency in 35-mm culture dishes. Subsequently, the cells were transfected with small interfering (si)-lncRNA PVT1-1# (10 nM) sense, 5′-GCUUGGAGGCUGAGGAGUUTT-3′ and antisense, 5′-AACUCCUCAGCCUCCAAGCTT3′; si-lncRNA PVT1-2# (10 nM) sense, 5′-CCCAACAGGAGGACAGCUUTT-3′ and antisense, 5′-AAGCUGUCCUCCUGUUGGGTT-3′; si-lncRNA PVT1-3# (10 nM) sense, 5′-CCUGUUACACCUGGGAUUUTT-3′ and antisense, 5′-AAAUCCCAGGUGUAACAGGTT-3′; and si-negative control (NC; 10 nM) sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After 24 h, cells were harvested for RNA isolation. The knockdown efficiency was assessed using RT-qPCR. All siRNA senses were supplied from Shanghai Jima Company (Shanghai, China). The most efficient siRNA fragment was subsequently packaged into recombinant lentivirus vectors to construct stably transfected cell lines exhibiting lncRNA PVT1 downregulation, which was performed and produced by the Shanghai Jima Company. ShPVT1 was the same sequence as siPVT1 and packaged into recombinant lentivirus vectors by the Shanghai Jima Company.
Cell proliferation assay
A CCK-8 cell proliferation assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess proliferation, according to the manufacturer's protocol. PC3 and DU145 cells were seeded in 96-well plates at 100 cells per well. Cell were cultured in Dulbecco's modified Eagle's medium, supplemented with 12% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 for 5 days. Absorbance of each well was subsequently measured at 450 nm using a spectrophotometer (xMark; Bio-Rad Laboratories, Inc., Hercules, CA, USA). All assays were performed at least in triplicate.
Wound healing assay
PC-3 and DU145 cells (1×104) were seeded with Dulbecco's modified Eagle's medium supplemented with 12% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc.) into 6-well plates at 80% confluency and incubated overnight at 4°C. Wounds were created with a sterile 10-µl pipette tip. Subsequently, detached cells were washed twice and removed with phosphate-buffered saline (PBS). Migration and movement of cells was assessed by measuring the same section of the wound at 0 and 24 h. This test was repeated three times under the same conditions.
Colony formation assay
The PC-3 and DU145 cells stably transfected with shRNA lncRNA PVT1 and NC were seeded in 6-well plates at a density of 500 cells per well. The medium was replaced with fresh culture medium every 4 days. After 14 days, plates were washed twice with PBS, fixed with 4% paraformaldehyde for 30 min at 4°C, and stained with Wright-Giemsa (Ruigen Co., Beijing, China) for 30 min at room temperature. Subsequent to washing with PBS, colonies with >50 cells were counted under an Olympus confocal laser scanning microscope (Olympus Corporation, Tokyo, Japan).
In vivo xenograft studies
A total of 8 male nude mice (4–6 weeks old; weight, 10–12 g) were purchased from the Model Animal Research Center of Southern Medical University (Guangzhou, China). All animal procedures and experiments were approved by the Southern Medical University Animal Research Ethics Committee (Guangzhou, China). To establish a prostate cancer xenograft model, 1×107 PC3 cells in 200 µl PBS were injected subcutaneously into the left and right flanks of mice. The mice were maintained in a pathogen-free environment (Pa 0.65 cm H2O at 26–28°C, with light for 10 h), with access to sterile food, and tumor length and width were measured every 3 and 4 days after the 13th day. After 33 days, tumor weight and volume were calculated with the formula: Tumor volume=(tumor width)2 × (tumor length) ×0.5.
Intracellular signaling array
The lysates of DU145 cells (1×107 cells) were prepared for detecting modifications of cellular proteins using the PathScan® Intracellular Signaling Array kit (cat. no. 7323; Cell Signaling Technology, Inc., Danvers, MA, USA), according to the manufacturer's instructions. Each sample was repeated in triplicate.
Western blot analysis
Total protein was obtained from DU145 and PC3 cells using protease buffer and inhibitors containing phenylmethanesufonyl fluoride (100 nM; Biyuntian Co. Shanghai, China). Protein concentration was measured by bicinchoninic acid assay. Protein lysates (50 µg) were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Membranes were blocked using 5% skimmed milk in PBST (0.1% triton in PBS) for 2 h at room temperature. Next, membranes were cultured with polyclonal anti-p-P38 (dilution, 1:1,000; cat. no. BS6381; Bioworld Technology, Inc., St. Louis Park, MN, USA) and monoclonal anti-GAPDH (dilution, 1:2,000; cat. no. 5174; Cell Signaling Technology, Inc.) primary antibodies overnight at4°C. Subsequent to being incubated with a horseradish peroxidase-conjugated secondary antibody (cat. no. PV-6000; OriGene Technologies, Inc., Beijing, China) at room temperature for 30 min, signals were developed using enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Statistical analysis
All analyses were performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). All data are expressed as mean ± standard deviation. Differences between two groups were determined using two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of the most effective siRNA
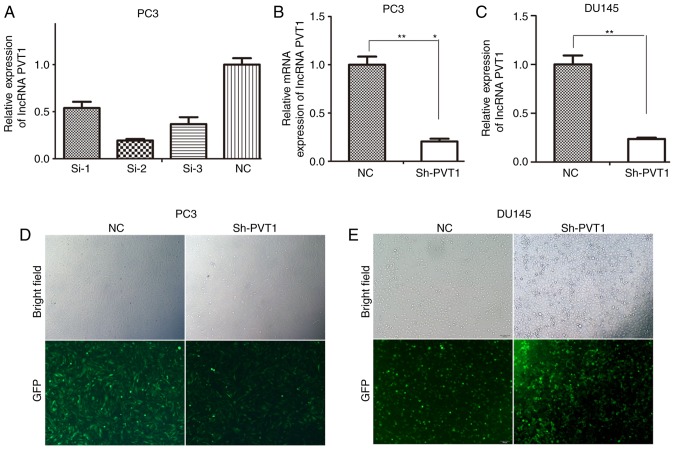
To identify the most effective siRNA for lncRNA PVT1-knockdown, PC3 cells were transfected with three lncRNA PVT1 siRNAs and one NC si-RNA. si-RNA-2# was confirmed to be the most effective siRNA for lncRNA PVT1-knockdown compared with other siRNAs (Fig. 1A). si-RNA-2# was subsequently recombined into a lentivirus vector. To obtain stable cell lines exhibiting lncRNA PVT1-knockdown, the lentivirus vectors were transfected into PC3 cells (Fig. 1D) and DU145 cells (Fig. 1E). Subsequently, RT-qPCR results indicated significant knockdown of lncRNA PVT1 expression levels in PC3 and DU145 cells (Fig. 1B and C).
Figure 1.
Identification of the most effective siRNA for lncRNA PVT1-knockdown. (A) si-RNA-2# was the most effective for knockdown of lncRNA PVT1 expression. (B) lncRNA PVT1 expression was significantly knocked down in PC3 cells. (C) lncRNA PVT1 expression was significantly knocked down in DU145 cells. (D) Efficiency of lentivirus-mediated RNAi was tested in PC3 cells (magnification, ×100). (E) Efficiency of lentivirus-mediated RNAi was tested in DU145 cells (magnification, ×100). *P<0.05 vs. NC group, **P<0.01 vs. NC group. siRNA, small interfering RNA; lncRNA, long non-coding RNA; NC, negative control; RNAi, RNA interference; GFP, green fluorescent protein.
Downregulation of lncRNA PVT1 expression inhibits the proliferation, mobility and colony formation abilities of prostate cancer cells
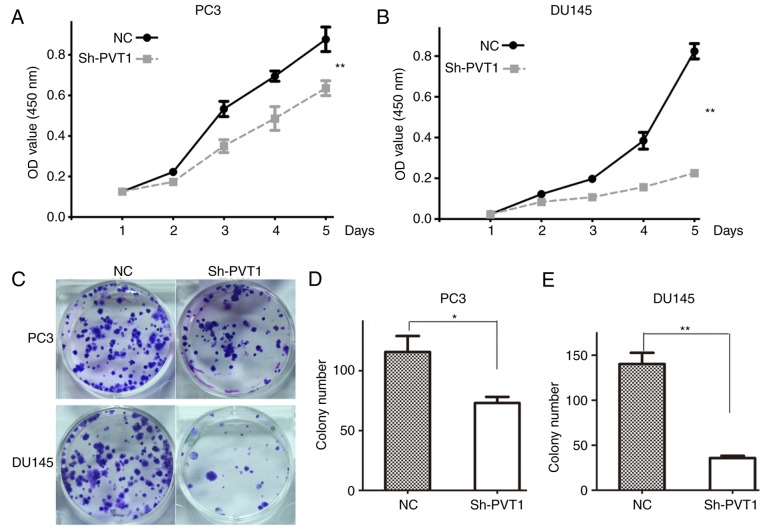
To investigate the effect of lncRNA PVT1 on the proliferation of prostate cancer cells, a CCK-8 assay was performed in PC3 and DU145 cells. The results indicated that lncRNA PVT1 downregulation significantly inhibited the proliferation of PC3 (Fig. 2A) and DU145 (Fig. 2B) cells, compared with the NC group.
Figure 2.
LncRNA PVT1-knockdown inhibits proliferation of prostate cancer cells. (A) Proliferation curves of PC3 cells subsequent to shPVT1 infection. (B) Proliferation curves of DU145 cells subsequent to shPVT1 infection. (C) LncRNA PVT1-knockdown suppressed the colony-formation ability of prostate cancer cells compared with the NC group. (D) Quantification analysis of colony number in PC3 cells. (E) Quantification analysis of colony number in DU145 cells. *P<0.05 vs. NC group, **P<0.01 vs. NC group. lncRNA, long non-coding RNA; NC, negative control; OD, optical density.
The effect of lncRNA PVT1 was also examined on the colony formation ability of prostate cancer cells. The colony number was significantly decreased in shPVT1 PC3 and DU145 cells compared with NC cells (Fig. 2C and D). These results indicated that downregulation of lncRNA PVT1 expression inhibited the colony formation ability of prostate cancer cells.
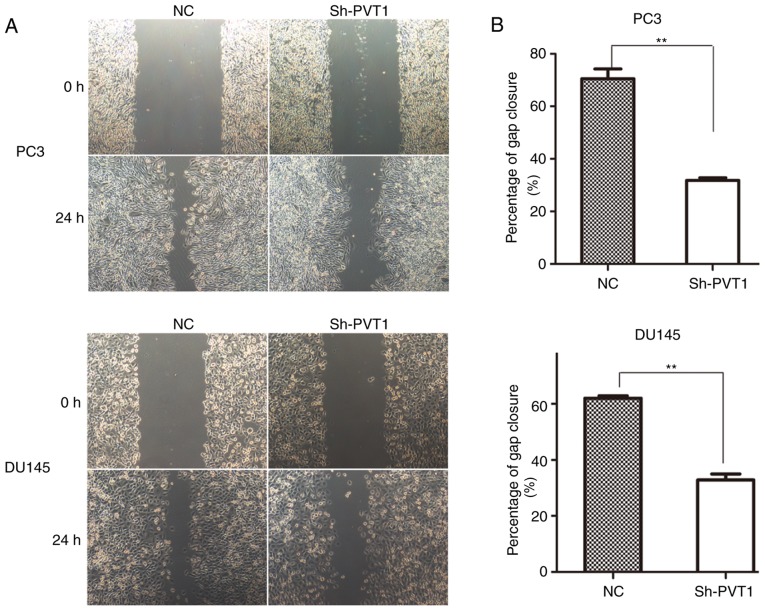
A wound-healing assay was conducted to assess the effect of lncRNA PVT1 expression on the mobility of prostate cancer cells. Downregulation of lncRNA PVT1 expression significantly inhibited the migration of PC3 cells (Fig. 3A) and DU145 (Fig. 3B) cells compared with the NC group.
Figure 3.
lncRNA PVT1-knockdown inhibits migration of prostate cancer cells. (A) Migration capacity of PC3 and DU145 cells was significantly reduced in lncRNA PVT1-knockdown groups at 24 h, compared with the negative control groups. (B) Migrated distance in PC3 and DU145 cells. **P<0.01 vs. control groups. lncRNA, long non-coding; NC, negative control; shRNA, short hairpin RNA.
LncRNA PVT1 downregulation inhibited tumorigenesis in a tumor xenograft model
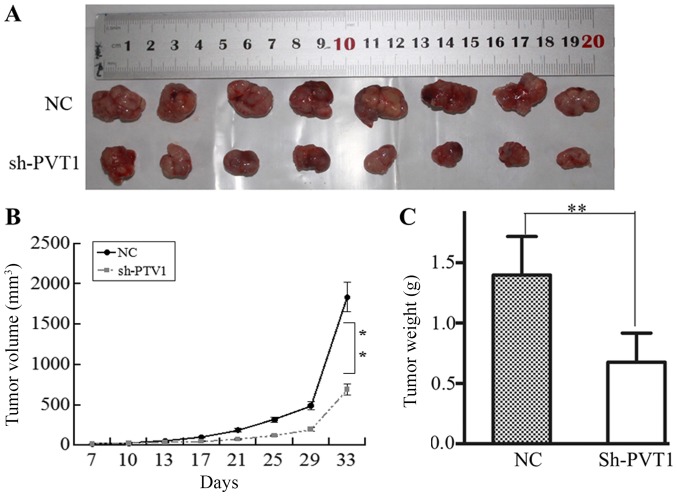
To confirm the effect of lncRNA PVT1 on tumorigenesis in prostate cancer cells, a xenograft tumor formation assay was performed by injecting PC3 cells into mice. After 33 days, all mice were sacrificed, and the size and weight of subcutaneous tumors were calculated (Fig. 4A). Compared with the NC group, the growth rate of xenograft tumors was decreased in the shPVT1 group (Fig. 4B). The mean tumor weight of the shPVT1 group was significantly reduced compared with that of the NC group (Fig. 4C). The results of the present study indicated that lncRNA PVT1 downregulation inhibited tumorigenesis in prostate cancer cells.
Figure 4.
lncRNA PVT1-knockdown inhibits proliferation of xenograft tumors in vivo. (A) Photographs of the solid tumors. (B) Tumor growth. (C) Mean weight of solid tumors. lncRNA, long non-coding; NC, negative control. Tumors in (A) are presented to show their best aspect. **P<0.01.
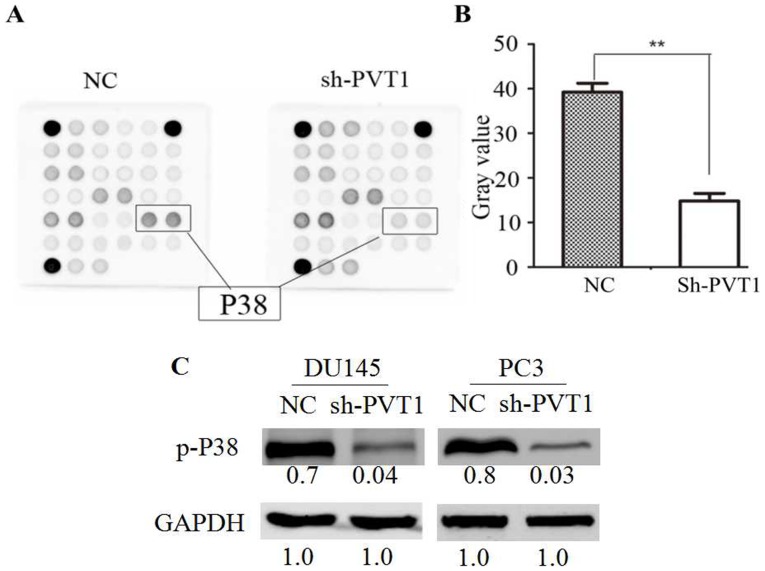
Downregulation of lncRNA PVT1 expression inhibits p38 activation
To explore the underlying molecular mechanisms of lncRNA PVT1 downregulation in prostate cancer cells, the PathScan® Intracellular Signaling Array kit (Chemiluminescent Readout) was utilized in DU145 cells. The phosphorylation level of p38 (Thr180/Tyr182) was demonstrated to be significantly reduced in the shPVT1 group compared with the NC group (Fig. 5A and B). Western blot analysis indicated that the phosphorylation levels of p38 cells were significantly reduced in the DU145 and PC3 cells with downregulated PVT1 expression compared with the NC group (Fig. 5C). Therefore, the data of the present study indicated that lncRNA PVT1 downregulation suppressed p38 phosphorylation.
Figure 5.
lncRNA PVT1-knockdown suppresses p38 phosphorylation in prostate cancer cells. (A) Decreased phosphorylation of p38 was observed in DU145 cells by intracellular signaling array following shPVT1 infection. (B) The gray value of p38 phosphorylation was statistically analyzed. (C) Decreased phosphorylation of p38 was validated in DU145 and PC3 cells by western blotting. **P<0.01 vs. control. lncRNA, long non-coding; NC, negative control; shRNA, short hairpin RNA; p-phosphorylated.
Discussion
In previous studies it has been reported that lncRNA PVT1 expression is upregulated in various types of cancer, including non-small cell lung cancer (NSCLC) (20,21), gastric cancer (22,23), renal cell carcinoma (24), cervical cancer (25), bladder cancer (26), hepatocellular carcinoma (HCC) (27), ovarian cancer (28), acute promyelocytic leukemia (29) and colorectal carcinoma (30). It has been indicated that lncRNA PVT1 is overexpressed in prostate cancer cells and may be a potential biomarker for prostate cancer (31). In previous studies, lncRNA PVT1 expression has been identified as a potential oncogene in multifarious types of cancer. For example, in HCC, high lncRNA PVT1 expression levels have been reported to be associated with a poor clinical prognosis (32). In addition, lncRNA PVT1 has been reported to promote HCC proliferation, the cell cycle, and the acquisition of stem cell-like properties (33). In another study, ectopic lncRNA PVT1 expression was indicated to significantly accelerate proliferation in NSCLC by downregulating p15 and p21 expression (34). In cervical cancer, it has been demonstrated that lncRNA PVT1 expression promoted cervical cell proliferation and migration depending on miR-200b silencing (25). Guan et al (35) reported that lncRNA PVT1-knockdown inhibited proliferation and enhanced apoptosis in breast and ovarian cancer cell lines. Downregulation of lncRNA PVT1 expression has been demonstrated to decrease proliferation and colony formation abilities in vitro and to suppress xenograft tumor proliferation in vivo by regulating p15 and p16 expression (36). The aforementioned findings indicate that lncRNA PVT1 could be a potential therapeutic target for these types of cancer (37).
However, the functional role of lncRNA PVT1 in prostate cancer cells requires further investigation. To examine the detailed underlying mechanism of lncRNA PVT1, lncRNA PVT1 expression was knocked down in PC3 and DU145 cell lines by lentivirus-mediated shRNA. The effect of lncRNA PVT1 on proliferation and migration of prostate cancer cells was evaluated by CCK-8, colony formation, and wound healing assays. The results of the present study indicated that lncRNA PVT1-knockdown inhibited prostate cancer cell proliferation and mobility in vitro. Furthermore, to determine the impact of lncRNA PVT1 on in vivo proliferation, a PC3 cell line stably transfected with lncRNA PVT1 was used to establish tumorigenicity in a tumor xenograft model. The results of the present study indicated that the tumorigenic capacity and weight in the shPVT1 group were decreased compared with the control group. Furthermore, knockdown of lncRNA PVT1 expression decreased the phosphorylation of p38, which has been identified as a mitosis-associated molecule, involved in the modulation of proliferation and migration of cancer cells (38,39). Therefore, lncRNA PVT1 expression may regulate prostate cancer proliferation through phosphorylation of p38. The results of the present study demonstrated that lncRNA PVT1 downregulation inhibited proliferation and migration of prostate cancer cells, which are associated with p38 expression. Therefore, lncRNA PVT1 may represent a novel diagnostic marker and a therapeutic target for prostate cancer. However, further investigation is required.
Acknowledgements
The authors would like to thank the Department of Pathology (Southern Medical University) for technical support.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81773277 and 81270597; Beijing, China), Science and Technology Projects of Guangdong (grant no. c1430611500534; Guandong, China) and Science and Technology Projects of Shenzhen (grant no. 201404153000668; Shenzhen, China).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XMM conceived and designed the experiments. BW, HYW, DJL and XMZ performed the experiments. LRZ, BL and SBZ acquired and analyzed the data. XMM and BW contributed to drafting and revising the manuscript.
Ethics approval and consent to participate
All animal procedures and experiments were approved by the Southern Medical University Animal Research Ethics Committee (Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
References
- 1.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2395. [DOI] [PubMed] [Google Scholar]
- 2.Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62(6 Suppl 1):S3–S12. doi: 10.1016/S0090-4295(03)00776-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, Yang XQ, Han CT, Dai B, Zhang HL, Shi GH, Wang CF, Ye DW. Pathological features of localized prostate cancer in China: A contemporary analysis of radical prostatectomy specimens. PLoS One. 2015;10:e0121076. doi: 10.1371/journal.pone.0121076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellerstedt BA, Pienta KJ. The current state of hormonal therapy for prostate cancer. CA Cancer J Clin. 2002;52:154–179. doi: 10.3322/canjclin.52.3.154. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Tsai M-C, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38–38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, Qian X, Tang Y, Lau YL, de Vries N, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding J, Li D, Gong M, Wang J, Huang X, Wu T, Wang C. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625–1630. doi: 10.2147/OTT.S68854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng C, Cheng S, Xie H, Zhou L, Wu J, Zheng S. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9:955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-dependent Induction of PVT1 and miR-1204. J Biol Chem. 2012;287:2509–2519. doi: 10.1074/jbc.M111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman MH, Tidswell R, Dooley JS, Sandanayake NS, Cerec V, Deheragoda M, Lee AJ, Swanton C, Andreola F, Pereira SP. Whole genome RNA expression profiling of endoscopic biliary brushings provides data suitable for biomarker discovery in cholangiocarcinoma. J Hepatol. 2012;56:877–885. doi: 10.1016/j.jhep.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63:977–983. doi: 10.4149/neo_2016_617. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XW, Bu P, Liu L, Zhang Xz, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232. doi: 10.1016/j.bbrc.2015.04.121. [DOI] [PubMed] [Google Scholar]
- 17.Ilboudo A, Chouhan J, McNeil BK, Osborne JR, Ogunwobi OO. PVT1 Exon 9: A potential biomarker of aggressive prostate cancer? Int J Environ Res Public Health. 2016;13 doi: 10.3390/ijerph13010012. ijerph13010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Lei B, Zhou X, Lv D, Wan B, Wu H, Zhong L, Shu F, Mao X. Apoptotic and nonapoptotic function of caspase 7 in spermatogenesis. Asian J Androl. 2007;719:47–51. doi: 10.4103/1008-682X.169563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 21.Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R, Xu TP, Huang MD, Wang ZX. Long noncoding RNA PVT1 Promotes promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15:1082–1094. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 22.Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63:442–449. doi: 10.4149/314_150825N45. [DOI] [PubMed] [Google Scholar]
- 23.Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Wang YQ, Weng WW, Zhang QY, Yang XQ, Gan HL, Yang YS, Zhang PP, Sun MH, Xu MD, Wang CF. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5:e192. doi: 10.1038/oncsis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124:649–658. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang C, Li J, Liu Y, Chen M, Yuan J, Fu X, Zhan Y, Liu L, Lin J, Zhou Q, et al. Tetracycline-inducible shRNA targeting long non-coding RNA PVT1 inhibits cell growth and induces apoptosis in bladder cancer cells. Oncotarget. 2015;6:41194–41203. doi: 10.18632/oncotarget.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan H, Yang Y, Zhang L, Tang G, Wang Y, Xue G, Zhou W, Sun S. Characterization of the genotype and integration patterns of hepatitis B virus in early- and late-onset hepatocellular carcinoma. Hepatology. 2015;61:1821–1831. doi: 10.1002/hep.27722. [DOI] [PubMed] [Google Scholar]
- 28.Liu E, Liu Z, Zhou Y, Mi R, Wang D. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med. 2015;8:20565–20572. [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng C, Yu X, Lai J, Yang L, Chen S, Li Y. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. doi: 10.1186/s13045-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bawa P, Zackaria S, Verma M, Gupta S, Srivatsan R, Chaudhary B, Srinivasan S. Integrative analysis of normal long intergenic non-coding rnas in prostate cancer. PLoS One. 2015;10:e0122143. doi: 10.1371/journal.pone.0122143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Han J, Zhang J, Li G, Liu H, Cui X, Xu Y, Li T, Liu J, Wang C. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltimore) 2016;95:e4436. doi: 10.1097/MD.0000000000004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Yang N, Zhou WP, Li WL, Li W, Sun SH. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 34.Cui D, Yu CH, Liu M, Xia QQ, Zhang YF, Jiang WL. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol. 2016;37:4127–4134. doi: 10.1007/s13277-015-4261-x. [DOI] [PubMed] [Google Scholar]
- 35.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 36.Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia P, Zhang R, Ge G. C/EBPβ mediates tnf-α-induced cancer cell migration by inducing mmp expression dependent on p38 MAPK. J Cell Biochem. 2015;116:2766–2777. doi: 10.1002/jcb.25219. [DOI] [PubMed] [Google Scholar]
- 39.Cheng G, Gao F, Sun X, Bi H, Zhu Y. Paris saponin VII suppresses osteosarcoma cell migration and invasion by inhibiting MMP-2/9 production via the p38 MAPK signaling pathway. Mol Med Rep. 2016;14:3199–3205. doi: 10.3892/mmr.2016.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.