Abstract
Colorectal cancer (CRC) is the most common malignant tumor type and has become resistant to 5-fluorouracil (5-FU) in recent decades, which is one of the most popular therapies. Recently, microRNA (miRNA or miR) has been investigated as a potential therapeutic strategy for CRC. However, there has been little investigation of the underlying mechanism of the association between expression of miRNA and chemosensitivity. The present study aimed to investigate the effect of miR-1260b inhibitor on CRC cells, and their chemosensitivity to 5-FU, by treating them with the miR-1260b inhibitor. miR-1260b inhibitor was demonstrated to significantly promote the proliferation and invasion of the CRC cell line, HCT116, and to increase the apoptotic rate. Furthermore, it was validated that programmed cell death 4 (PDCD4) was a direct target of miR-1260b inhibitor in CRC with bioinformatics tools and a luciferase assay. Western blot analysis revealed that miR-1260b inhibitor could significantly decrease PDCD4 expression, and downregulate the expression of phosphorylated-Akt (p-Akt) and phosphorylated-extracellular-signal-regulated kinase (p-ERK). In conclusion, it was confirmed that the anti-tumor effect of the miR-1260b inhibitor was conducted by blocking the phosphorylated 3-kinase/Akt pathway as dysregulated protein expression induced by miR-1260b inhibitor was rescued by insulin-like growth factor. Notably, miR-1260b inhibitor could significantly enhanced the chemoresponse of HCT116 cells to 5-FU via reduced proliferation, increased apoptosis, and downregulation of PDCD4, p-Akt and p-ERK protein expression. In summary, the present study may provide a novel direction for future clinical therapy to enhance the chemosensitivity of tumor cells.
Keywords: colorectal cancer, chemosensitivity, microRNA-1260b, phosphorylated-Akt, programmed cell death 4
Introduction
Colorectal cancer (CRC) is the most common gastric and intestinal malignant tumor type. Epidemiological studies of CRC have demonstrated that the incidence and mortality rates of CRC are comparable with those of gastric cancer, esophageal cancer and primary liver cancer, all of which affect the digestive system (1). CRC is also a common cause of cancer-associated mortality and is responsible for ~10% of cancer cases worldwide (2,3). CRC, along with other types of cancer, is defined by different stages depending on the presence of cancer-positive lymph nodes. The 5-year overall survival rate of CRC patients at stage I–II of the disease is ~90%, but for patients with stage III of the disease it is ~60% and for stage IV only ~8% (4,5). Currently, surgical resection and chemotherapy remain the most popular options for CRC treatment (6). Chemotherapy is not suitable for certain patients, due to their age, complications or side effects. Therefore, numerous patients suffer from relapse in stage III of CRC (6). Extensive investigations have been performed to identify diagnostic biomarkers for CRC or novel molecular therapies to replace certain treatments prone to chemoresistance (7–9). It has been reported that a substantial number of molecules and complicated signaling pathways are involved in the occurrence and development of CRC, including the activation of oncogenes, inactivation of tumor suppressor genes and epigenetic modifications (10–12). The elucidation of CRC pathogenesis, examining the molecular dynamics and targeting of molecular abnormalities in CRC are popular areas of research. The role of microRNAs (miRNAs or miRs) has attracted the attention of numerous researchers (13).
miRNAs are a class of endogenous short non-coding RNAs that consist of 19–25 nucleotides. Bioinformatics and cloning studies have demonstrated that miRNAs serve key functions in regulating the expression and function of various genes and proteins by binding to their target genes. It has been estimated that miRNAs may regulate 1/3 of all human genes and control hundreds of target genes (14,15). Previous evidence has indicated that miRNAs regulate proliferation, invasion, migration, angiogenesis and epithelial-mesenchymal transition through degradation of target mRNAs via binding to the 3′ untranslated region (3′UTR) (16,17). Data from microarray profiling of stage III CRC tissues have demonstrated that certain cancer-associated miRNAs are upregulated, while others are downregulated, compared with adjacent noncancerous colorectal tissues. These include miR-18a, miR-1260b and miR-21 (18–22). However, extensive investigation is required to clarify the underlying mechanism and function of these miRNAs in CRC.
Numerous decades ago, PDCD4 was identified as a novel tumor suppressor gene. Overexpressed PDCD4 was sufficient to inhibit the neoplastic transformation induced by tissue plasminogen activator (23,24). Researchers have demonstrated that one of the major functions of PDCD4 is to inhibit the translation process by interacting with the translation initiation factors, eukaryotic initiation factor-4A and the eukaryotic translation initiation factor-4 G, partly by inhibiting their helicase activity (24,25). Furthermore, studies in cultured ovarian cancer cells have suggested that PDCD4 suppresses proliferation and progression of the cell cycle, in addition to inducing apoptosis (25–27). Notably, phosphorylated Akt (p-Akt) is functionally associated with the shuttling of PDCD4 between the cytoplasm and the nucleus through its ability to phosphorylate PDCD4 at positions Ser67 and Ser457 (27,28). PDCD4 is an effector of the phosphorylated 3-kinase (PI3K)/Akt signal pathway, which activates tumorigenesis (28–32). To the best of our knowledge, the present study is the first to demonstrate that PDCD4 is a target of miR-1260b.
In the present study, the effect of miR-1260b inhibitor on the proliferation and migration of the CRC cell line HCT116 was investigated. PDCD4 was demonstrated to be a direct target of miR-1260b through bioinformatics and dual-luciferase reporter assay. In addition, the present study demonstrated that miR-1260b inhibitor could enhance the chemosensitivity of CRC cells to 5-fluorouracil (5-FU). Finally, the current study may provide the basis for a novel therapeutic strategy for CRC by elucidating the underlying mechanism of combined therapy.
Materials and methods
Patient recruitment and tissue sample collection
A total of 30 pairs of fresh CRC and adjacent normal tissues were collected from the Changzhi People's Hospital (Changzhi, China) and the Hospital of Chinese People's Liberation Army (Taiyuan, China) from January 2017 to January 2018. Clinical information, including diagnosis, tumor size, pathological stage and lymphatic metastasis, was reviewed for all enrolled patients (n=30). Patients who had previously received chemotherapy or radiotherapy were excluded from the present study. Among the 30 patients with CRC, the age was between 45–60 years old (median age of 56 years) with a 0.54: 1, male to female ratio. For all patients, samples were collected intraoperatively and flash frozen in liquid nitrogen, then stored at −80°C until subsequent RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. The use of human tissue for mRNA detection was approved by the Changzhi People's Hospital Ethics Committee (Changzhi, China). All recruited patients provided informed consent for participation.
Cell culture
The human colorectal cancer cell lines, HCT116 and SW480, were purchased from Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPM-1640 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and ampicillin (20 µg/ml; Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2 in a humidified cell culture incubator (Sanyo Tokyo Manufacturing Co., Ltd, Tokyo Japan).
Cell transfection
HCT116 cells (3×105) or SW480 cells (2×105) were plated in 6-well plates and, once the cells reached 70% confluency, they were transfected with 100 nM miR-1260b inhibitor (sequence: 5′-AUGGUGGCAGUGGUGGGAU-3′) or negative control (sequence: 5′-UACAAACGUCACGGCGUA-3′) for 4 h (GenePharma, Shanghai, China) using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 in a humidified cell culture incubator, according to the manufacturer's protocol. Following transfection, cells were cultured at 37°C until sampling. The cells were sampled at the indicated time points for the following experiments analysis, including the validation of cell proliferation at 24, 48 and 72 h, and the measurement of protein or RNA expression at 72 h.
MTT assay
The reduction of MTT by metabolically active cells was used as a measure of proliferation. An MTT assay kit (Beyotime Institute of Biotechnology, Shanghai, China) was used to measure proliferation from day 1–3, following the manufacturer's protocol. Briefly, 5×103 HCT116 or SW480 cells in the logarithmic phase were seeded into 96-well plates in triplicate. A volume of 200 µl/well miR-1260b, 5-FU or insulin-like growth factor 1 (IGF1) was added to the plate when cells were completely adherent at 24 h, and the cells were cultured at 37°C (5% CO2) for a further 48 h. MTT (20 µl; 5 mg/ml; (Beyotime) solution was added to each well at 24, 48 and 72 h post miR-1260b inhibitor or negative control transfection. The cells were incubated for an additional 4 h. At the end of incubation, the supernatants were removed with a pipette. Prior to reading under a microplate reader, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added into each well. The proliferation rate was calculated by measuring optical density (OD) at a wavelength of 480 nm.
Transwell cell migration assay
The in vitro cell migration assay was performed according to the method used by Justus et al (33). Briefly, 100 µl HCT116 cells (3×106 cells/well) transfected with miR-1260b inhibitor or negative control were plated in the upper chamber of a transwell (Corning Incorporated, Corning, NY, USA) inserted in a 24-well plate. When the cells had settled, 600 µl in RPMI-1640 media supplemented with 30% fetal bovine serum, was added into the lower chamber of the 24-well plate. At 48 h, the cells that had not migrated from the top of the membrane were removed carefully and the migrated cells were fixed with 4% paraformaldehyde at room temperature for 30 min. Removing the paraformaldehyde, the cells were stained with 1 mg/ml crystal violet solution (Thermo Fisher Scientific) at room temperature for 4 h. Following washing 3 times with pure water, the cells in 4 different fields of view were counted under a light microscope at the magnification of ×10 and the average number of cells was determined. Since HCT116 cells were more sensitive to miR-1260b inhibitor than SW480 cells, HCT116 cells were selected for subsequent experiments.
Flow cytometric analysis of apoptosis
Apoptotic rate was analyzed using the Annexin V-FITC kit and propidium iodide (PI) (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, 1×106 HCT116 cells were plated in 100-mm dishes (Corning Incorporated, Corning, NY, USA) and then transfected with miR-1260b inhibitor or mock vector or 5-FU. At 48 h, the cells were lysed and collected following two washes with cold PBS and re-suspended in 500 µl binding buffer. Then, cells were stained with annexin V-fluorescein isothiocyanate (FITC) and PI for an additional 5 min incubation at room temperature prior to assessment with a flow cytometer (BD, Biosciences, Inc). The data analysis was used ModFit LT™ (version 5.0, Verity Software House, Topsham, ME, USA). For each group, the samples were measured in triplicate.
Predicted target analysis of miR-1260b
MiRecords (http://c1.accurascience.com/miRecords/) is an online database of animal miRNA-target interactions containing validated and predicted targets. The predicted targets are based on the results of multiple miRNA target predication tools, including DIANA-microT, MicroInspector and miTarget. The potential targets of miR-1260b were predicted using two algorithms on MiRecords (version 4).
3′UTR-luciferase reporter gene assay
PDCD4-WT (wildtype) and PDCD4-MT (mutant) were purchased from Genewiz and carried the PDCD4 sequence containing the wild-type or mutant 3′UTR predicted of the miR-1260b binding sites. A total of 1×105 HCT116 cells were seeded to each well in a 24-well plate and cultured at 37°C (5% CO2) overnight. HCT-116 cells were then transfected with 2.5 ng/µl PDCD4 vector or mutant vector using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After 4 h, 50 nM of miR-1260b mimic, control or 5-FU was transfected into the cells which were at ~50% confluence. After 48 h, cells were lysed and the luciferase activities were analyzed using a dual luciferase assay kit (Promega, Madison, WI, USA) and the normalized luciferase activity was calculated with the ratio of firefly luciferase activity to Renilla luciferase activity.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
HCT116 cells were collected and total RNA was isolated from these cells or from tumor tissues using A PicoPure™ RNA Isolation kit (Arcturus, Sunnyvale, CA, USA), according to the manufacturer's protocol. First, 2 µg of mRNA was reverse transcribed into cDNA using SuperScript™ III first-strand synthesis system kit (Thermo Fisher Scientific, Inc.), according to manufacturer's protocols. Then, qPCR was performed with TaqMan (Thermo Fisher Scientific, Inc.) primers for miR1260b (Cat# 4426961, Thermo Fisher Scientific, Inc.) and GAPDH (cat#4331182, Thermo Fisher Scientific, Inc.). The Vii™ 7 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to run the qPCR procedure, according to the manufacturer's protocol. The qPCR procedure includes two stages: Hold stage and PCR stage. The briefly procedure as follows: Step 1: Heating from 25 to 95°C at a rate of 1.6°C/s, holding for 2 min at 50°C. Step 2: Heating from 50 to 95°C at a rate of 1.6°C/s, holding for 10 min at 95°C. For qPCR stage: Step 1: Initial denaturation at 95°C for 15 s. Step 2: Annealing extension at 60°C for 1 min. The temperature was reduced from 95 to 60°C at the rate of 1.6°C/s; the denaturation and extension stages were repeated for 40 cycles. The expression of mRNA was quantified using the 2−ΔΔCq method (34) and normalized to the internal reference gene, GAPDH.
Western blot analysis
Following treatment with miR1260b inhibitor with or without 5-FU for 48 h, HCT116 cells were collected. The cells were lysed in radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck KGaA). Protein concentration was quantified using a BCA Protein Assay kit (Beyotime Institute of Biotechnology). Total protein (30 µg) from each group was loaded and resolved by 10% Tris-SDS-PAGE. Following electrophoresis, the gel was electro-transferred to polyvinylidene fluoride membranes (Thermo Fisher Scientific, Inc.). The following blocking and antibody incubations were performed according to the iBind kit (Thermo Fisher Scientific, Inc.) manufacturer's protocol. Briefly, primary antibodies were incubated at room temperature for overnight. Following 3 times washing with iBind™ Flex/iBind™ Flex FD solution (Cat#SLF2020, Thermo Fisher Scientific, Inc.), the secondary antibodies were used to incubate the membranes at room temperature for 4 h. The primary antibodies were as follows: Anti-p-Akt (cat# 4060, dilution, 1:300; Cell Signalling Technology, Inc., Danvers, MA, USA), anti-PDCD4 (cat# 9535, dilution, 1:300; Cell Signaling Technology, Inc.) anti-phosphorylated-extracellular-signal-regulated kinase (p-ERK). (cat# sc-81492, dilution, 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and anti-β-actin (cat# 58673, dilution, 1:1,000; Santa Cruz Biotechnology, Inc.). Horseradish peroxidase-conjugated IgG secondary antibodies were purchased from Santa Cruz Biotechnology with the dilution of 1:3,000 and the detail information of secondary antibodies as following: Anti-rabbit (cat# sc-2004), anti-mouse (cat# sc-2005) and anti-goat (cat# sc-2020). The signals were detected with SuperSigal® west femto maximum sensitivity substrate (Thermo Fisher Scientific, Inc), and the specific protein bands were captured with Bio-Rad Lab™ 2.0 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
GraphPad Prism7 (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform the two-tailed Student's t-test or one-way ANOVA followed by Dunnett's test to determine the difference between groups. P<0.01 and P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate and data are presented as the mean ± standard deviation.
Results
miR-1260b inhibitor inhibits CRC-cell proliferation and migration
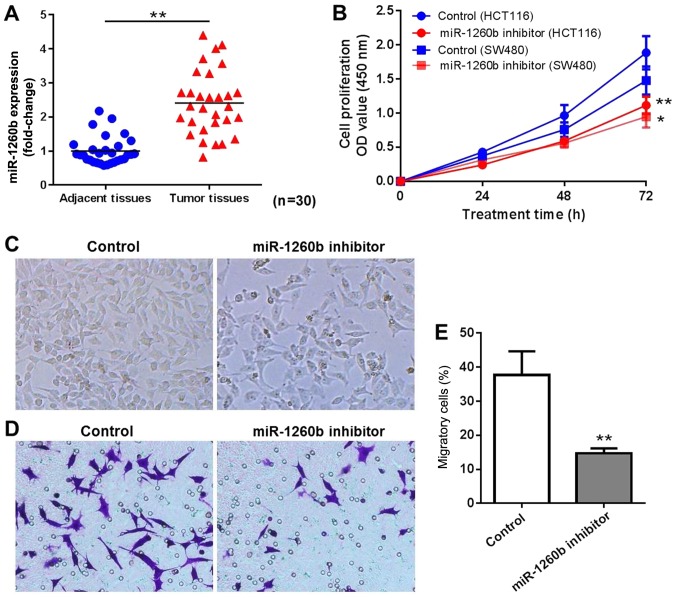
A total of 30 patients diagnosed with CRC were enrolled in the present study. The miR-1260b expression level in CRC tissues and adjacent normal tissues was determined by RT-qPCR. The results indicated that miR-1260b was overexpressed in CRC tissues compared with adjacent normal tissues (P<0.01; Fig. 1A). In order to explore the biological role of miR-1260b in CRC cells, miR-1260b inhibitor or negative control were transiently transfected into HCT116 or SW480 cells. Subsequently, the proliferation and migration of HCT116 cells was detected at 24, 48 and 72 h using an MTT assay and migration assay, respectively. As indicated in Fig. 1B, miR-1260b inhibitor significantly inhibited proliferation of HCT116 and SW480 cells, particularly at 72 h. Since HCT116 cells were more sensitive to miR-1260b inhibitor compared with SW480 cells, HCT116 cells were selected for subsequent experiments. The cell density was decreased in miR-1260b inhibitor transfected HCT-116 cells group compared with that in the control group (Fig. 1C). Similarly, a reduced number of cells was observed in the migration assays following transfection with miR-1260b inhibitor (Fig. 1D and E). These results indicated that miR-1260b inhibitor exerted anti-proliferation and anti-migration effects on HCT116 cancer cells.
Figure 1.
Proliferation and migration of colorectal cancer cells was inhibited by miR-1260b inhibitor. (A) miR-1260b was significantly overexpressed in CRC tissues compared with adjacent normal tissues (**P<0.01). (B) miR-1260b inhibitor exhibited anti-proliferation activity in HCT116 cells, particularly at 72 h. (*P<0.05, **P<0.01 vs. control group). (C) Light microscopy indicating reduced cell density (magnification, ×40). (D) Migration assay indicating a reduced number of migratory cells in the miR-1260 inhibitor group compared with control (magnification, ×40). (E) Quantification rate of cell migration (**P<0.01 vs. control group). miR, microRNA; OD, optical density.
miR-1260b induces CRC-cell proliferation by targeting PDCD4
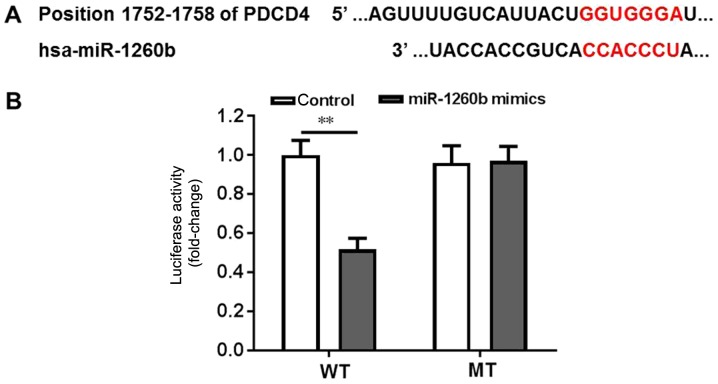
Bioinformatics tools were used to clarify the underlying mechanisms of anti-proliferation and anti-migration effects of miR-1260b on human CRC cells. PDCD4 exhibited the highest prediction score. It is well known that PDCD4 is a suppressor of the apoptosis signalling pathway (35,36). Therefore, it was important to validate whether PDCD4 is a direct target of miR-1260b. Wild-type and mutant PDCD4-3′UTR expression plasmids were constructed, which were fused with a luciferase reporter gene (Fig. 2A). The results indicated that miR-1260b could significantly inhibit the luciferase activity of wild-type PDCD4 compared with the control (P<0.01). However, there was no difference between the control group and the miR-1260 treatment group when PDCD4 was mutated (Fig. 2B). Thus, these findings suggested that PDCD4 was a direct target of miR-1260b.
Figure 2.
PDCD4 is a direct binding target of miR-1260b. (A) Gene structure of PDCD4 at the position of 1752–1758 of PDCD4, indicating the predicted target site of miR-1260b in its 3′UTR, with a sequence of CCACCCU. (B) HCT116 cells were transfected with a luciferase reporter plasmid containing the wild-type or mutant PDCD4 3′UTR sequence. The miR-1260b inhibitor could significantly block the luciferase activity of HCT116 cells transfecte d with the wild-type PDCD4 3′UTR sequence (**P<0.01 vs. control group). WT, wild type; MT, mutated; UTR, untranslated region; PDCD4, programmed cell death receptor 4.
miR-1260b inhibitor enhances 5-FU-induced apoptotic rate in HCT116 cells
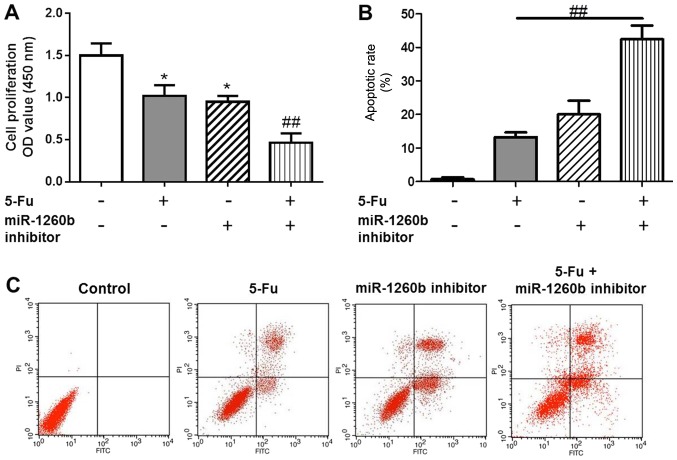
Apoptotic cells were detected with PI/Annexin V staining, in order to investigate the biological function of the miR-1260b inhibitor on CRC-cell proliferation and migration. In order to determine whether there was a synergistic effect with treatment with of miR-1260b inhibitor and 5-FU on the anti-proliferation and induction of apoptosis effects, the MTT and flow cytometry assays were conducted following 5-FU treatment alone or in combination with miR-1260b inhibitor. The results indicated that 5-FU alone or miR-1260b inhibitor alone significantly decreased the proliferation rate of HCT116 cells compared with the vehicle control group. However, the proliferation rate was lowest in the combination group (P<0.05 vs. control group, P<0.01 vs. 5-FU alone treatment group (Fig. 3A). This synergistic effect was also observed in apoptosis. These data indicated that miR-1260b inhibitor with 5-FU treatment significantly induced apoptosis of HCT116 cells, which may be due to miR-1260b enhancing the chemosensitivity of HCT116 cells to 5-FU [P<0.01 vs. 5-FU alone treatment group (Fig. 3B)]. Flow cytometry results demonstrated that miR-1260b inhibitor could induce apoptosis of HCT116 cells (Fig. 3C).
Figure 3.
miR-1260b inhibitor combined with 5-FU exhibits an enhanced anti-proliferation effect on HCT116 cells. (A) Proliferation rate of HCT116 cells with 5-FU, miR-1260b or combination (*P<0.05 vs. control group, ##P<0.01 vs. 5-FU alone treatment group). (B) HCT116-cell apoptotic rate (##P<0.01 vs. 5-FU alone treatment group). (C) Flow cytometry analysis for the distribution of apoptotic cells following different treatments. miR, microRNA; UTR, untranslated region; PI, propidium iodide; OD, optical density.
miR-1260b inhibitor enhances the chemosensitivity of HCT-116 cells to 5-FU due to downregulation of PDCD4 expression
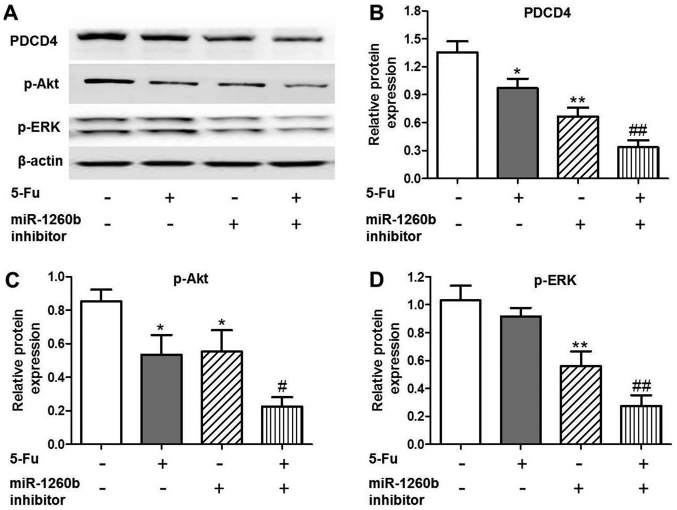
In order to investigate the anti-proliferation mechanisms of miR-1260b inhibitor in HCT116 cells, the expression of apoptosis-associated proteins was evaluated, as well as that of the target protein PDCD4. It was identified that PDCD4 expression was significantly decreased in the 5-FU (P<0.05), miR-1260b (P<0.01) or combination (P<0.01) groups compared with the vehicle control group (Fig. 4A and B). In addition, downregulation trends were identified for the apoptosis-associated proteins, p-Akt and p-ERK, following treatment with miR-1260b inhibitor or combined miR-1260b and 5-FU (Fig. 4A, C and D). These results indicated that miR-1260b increased HCT116-cell apoptotic rate via blocking the p-Akt and p-ERK pathways.
Figure 4.
PDCD4, p-Akt and p-ERK protein expression levels are decreased in HCT116 cells treated with miR1260b inhibitor and/or 5-FU. (A) Protein expression levels of PDCD4, p-AKT and p-ERK following treatment with 5-FU and/or miR-1260b inhibitor, as detected by western blotting. (B) Quantification of the protein expression levels of PDCD4 (*P<0.05, **P<0.01 vs. control group; ##P<0.01 vs. 5-Fu alone treatment group). (C) Quantification of the protein expression levels of p-Akt (*P<0.05, **P<0.01 vs. control group, #P<0.01 vs. 5-FU alone treatment group). (D) Quantification of the protein expression levels of p-ERK (**P<0.01 vs. control group, ##P<0.01 vs. 5-FU alone treatment group). PDCD4, programmed cell death receptor 4; p-phosphorylated; ERK, extracellular-signal-regulated kinase; 5-FU, 5-fluorouracil; miR, microRNA.
miR-1260b inhibitor enhances the chemosensitivity of HCT116 cells to 5-FU via downregulation of the PI3K/Akt pathway
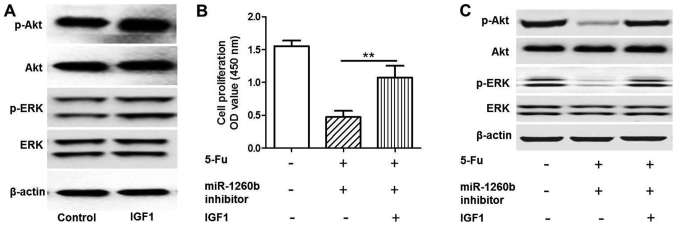
As indicated in Fig. 5, a rescue experiment was conducted by adding insulin-like growth factor 1 (IGF1) to activate the PI3K/Akt pathway. Protein expression levels of p-Akt and p-ERK were detected when HTC116 cells were treated with IGF1 by western blot analysis (Fig. 5A). The MTT assay demonstrated that IGF1 could significantly increase HCT116 proliferation rate compared with the 5-FU and miR-1260b combination treatment group (P<0.01; Fig. 5B). Western blotting indicated that the p-Akt and p-ERK protein expression levels in the IGF-1 group were increased compared with the 5-FU and miR-1260b combination group (Fig. 5C). These results demonstrated that IGF1 rescued the effect induced by miR-1260b inhibitor and 5-FU. Therefore, it was revealed that the underlying mechanism of miR-1260b inhibitor induced suppression of HCT116 proliferation involved in blocking the PI3K/Akt pathway.
Figure 5.
miR-1260b inhibitor enhances the sensitivity of HCT-116 cells to 5-FU via downregulation of the IGF1 pathway. (A) Protein expression levels of p-Akt and p-ERK were detected when HTC116 cells were treated with IGFI by western blot analysis. (B) Quantification of proliferation of cells treated with miR-1260b inhibitor and 5-FU or without IGF1 indicates that IGF1 significantly increased HCT116-cell proliferation rate compared with the 5-FU and miR-1260b combination treatment group (**P<0.01 vs. miR-120b inhibitor with 5-FU group). (C) Protein expression of p-Akt and p-ERK protein expression in HCT116 cells treated with miR-1260b inhibitor and 5-FU with or without IGF1, as detected by western blot analysis. IGF1, insulin-like growth factor 1; OD, optical density; miR, microRNA; 5-FU, 5-fluorouracil; p-phosphorylated; ERK, extracellular-signal-regulated kinase.
Discussion
Previous research has been conducted regarding CRC prevention and diagnosis, as well as the biological, genetic or epigenetic changes of CRC, in order to develop novel biomarkers or therapies for CRC treatment (7,11). Amounting evidence has revealed that aberrant miRNA expression serves a functional role in CRC initiation and progression (16). In a previous review of miRNA expression studies, it was reported that ≥164 miRNAs have been demonstrated to be significantly dysregulated in CRC compared with adjacent normal tissues, including miR-17-92 cluster, miR-31, miR181b, miR-21, miR-135a/b and miR-224 (16). Multiple miRNAs have been reported to be associated with CRC initiation, development and progression, including miR-21, which is expressed during the transition from adenoma to advanced carcinoma (37). Therefore, miR-21 has been identified as an important oncogenic miRNA with roles in tumor initiation, progression and metastasis. However, an association between miR-1260b and CRC has also been reported (21). Navarro-Quiroz et al (38), reported that miR-1260b was overexpressed in human blood dendritic cells. Hirata et al (22) demonstrated that miR-1260b could promote renal cancer cell proliferation and invasion. In addition, the group identified that genistein could achieve its anti-tumor effect via downregulation of miR-1260b expression and targeting of Smad4 in prostate carcinoma (39). It has been reported that miR-1260b is associated with the development of non-small-cell lung cancer (40). In addition, miR-1260b expression has been demonstrated to be increased in CRC tissues, particularly in patients with positive lymph nodes (21). Therefore, miR-1260b may be involved in a variety of cancer cell activities. Furthermore, Nordentoft et al (41) studied the association between miRNA and chemosensitivity in advanced bladder cancer by RNA profiling, and indicated that overexpression of miR-138 could increase the chemosensitivity of RT4 and CLR2169 bladder cancer cells to cisplatin. Valeri et al (42) reported that elevated miR-21 could induce chemoresistance to 5-FU in colon cancer cell lines. In summary, the patients of miR-1260 in chemotherapy response provides a novel direction for CRC investigation.
The present study focused on the underlying mechanism of miR-1260b regulation of CRC-cell proliferation and invasion, as well as its function in the chemosensitivity of CRC to 5-FU. It was identified that miR-1260b was expressed at higher levels in CRC tissues compared with adjacent normal tissues. Furthermore, inhibition of HCT116-cell proliferation and invasion by the miR-1260b inhibitor demonstrated, as well as revealed an increased apoptotic rate. In addition, PDCD4 was identified as a validated target of miR-1260b. PDCD4 protein expression level was markedly decreased by miR-1260b inhibitor, along with downregulated p-Akt and p-ERK protein. PDCD4 is a 64 kDa protein that is preferentially expressed in tumor promoter-resistant cells, but has been indicated to be suppressed in tumor promoter-sensitive cells undergoing neoplastic transformation (23). PDCD4 protein expression has been suggested to be increased during apoptosis in response to different inducers, including retinoic acid, and has been demonstrated to be regulated by topoisomerase inhibitors, cyclooxygenase-2 inhibitors, Myb and Akt (43–46). Lankat-Buttgereit et al (46) demonstrated that the suppression of PDCD4 protein expression could enhance the release of CgA and SgII, which have been identified as diagnostic markers of neuroendocrine tumors. The stimulation of these markers by low levels of PDCD4 was demonstrated to mediated by activation of Akt via the PI3K pathway. Notably, Mudduluru et al (47) reported that p-Akt expression and translocation of PDCD4 from the nucleus to the cytoplasm was inversely correlated with PDCD4 levels in colon carcinoma samples. However, above phenomena was not demonstrated in the normal adjacent tissue. These findings indicate that the interplay between PDCD4 and p-Akt serves a critical function in tumor suppression.
In the present study, it was identified that miR-1260b inhibitor could enhance the chemotherapy response of HCT116 cells to 5-FU via blocking the PI3K/Akt pathway. These trends were disrupted by treatment with IGF1, which serves as an activator of the PI3K/Akt pathway. Depending on the cell type, previous studies have demonstrated that IGF1 promotes neuronal survival, maturation and cell cycle progression, by activating the PI3K/Akt and/or Ras/MAPK pathway (48,49). These findings may provide a novel combination therapy for future CRC treatment.
Acknowledgements
None.
Funding
No funding received.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ, JC, LZ, YD and WY managed the experimental design, tissue collection and experimental execution. XZ, BY, YG, YW and NM analyzed and interpreted the data and executed the experiments. JZ reviewed and approved the final draft of this manuscript before submission. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval for the study was given by The Changzhi People's Hospital ethics committee. All recruited patients provided informed consent for participation.
Patient consent for publication
All patients agreed to publication and a written informed consent was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kraus S, Nabiochtchikov I, Shapira S, Arber N. Recent advances in personalized colorectal cancer research. Cancer Lett. 2014;347:15–21. doi: 10.1016/j.canlet.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 2.O Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer 6th edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mather C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Nagtegaal ID, Quirke P, Schmoll HJ. Has the new TNM classification for colorectal cancer improved care? Nat Rev Clin Oncol. 2011;9:119–123. doi: 10.1038/nrclinonc.2011.157. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T. Biomarker for high-risk patients with stage II colon cancer. Lancet Oncol. 2013;14:1247–1248. doi: 10.1016/S1470-2045(13)70527-8. [DOI] [PubMed] [Google Scholar]
- 6.Chua YJ, Zalcberg JR. Progress and challenges in the adjuvant treatment of stage II and III colon cancers. Expert Rev Anticancer Ther. 2008;8:595–604. doi: 10.1586/14737140.8.4.595. [DOI] [PubMed] [Google Scholar]
- 7.Grady WM, Pritchard CC. Molecular alterations and biomarkers in colorectal cancer. Toxicol Pathol. 42:124–139. doi: 10.1177/0192623313505155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye JJ, Cao J. MicroRNAs in colorectal cancer as markers and targets: Recent advances. World J Gastroenterol. 2014;20:4288–4299. doi: 10.3748/wjg.v20.i15.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 10.Oqin S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: An emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, Tsuchiya KD, ll Park D, Fausel R, Kanngurn S, Welcsh P, Dzieciatkowski S, Wang J, Grady WM. RET is a potential tumor suppressor gene in colorectal cancer. Oncogene. 2013;32:2037–2047. doi: 10.1038/onc.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiemer EA. The role of microRNAs in cancer: No small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Hernando E. microRNAs and cancer: Role in tumorigenesis, patient classification and therapy. Clin Transl Oncol. 2007;9:155–160. doi: 10.1007/s12094-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 15.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Boil Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou JJ, Zheng S, Sun LF, Zheng L. MicroRNA regulation network in colorectal cancer metastasis. World J Biol Chem. 2014;5:301–307. doi: 10.4331/wjbc.v5.i3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilarr E, Tabernero J, Gruber SB. Micromanaging the classification of colon cancer: The role of the microRNAome. Clin Cancer Res. 2011;17:7207–7209. doi: 10.1158/1078-0432.CCR-11-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummins JM, He Y, Leary RJ, Paqliarini R, Diaz LA, Jr, Sjiblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs protential utility in colon cancer: Early detection, prognosis and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Leva G, Cheung DG, Croce CM. miRNA clusters as therapeutic targets for hormone-resistant. Expert Rev Endocrinol Metab. 2015;10:607–617. doi: 10.1586/17446651.2015.1099430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu DR, Guan QL, Limin X, Gao MT, Jiang L, Kang HX. MiR-1260b is a potential prognostic biomarker in colorectal cancer. Med Sci Monit. 2016;22:2417–2423. doi: 10.12659/MSM.898733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata H, Ueno K, Nakajima K, Tabatatai ZL, Hinoda Y, Ishii N, Dahiya R. Genistein downregulates onco-miR-1260b and inhibits Wnt-signalling in renal cancer cells. Br J Cancer. 2013;108:2070–2078. doi: 10.1038/bjc.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonia O, Juste D, Das S, Matsuhashi S, Samuels HH. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antagonist in breast cancer cells. Oncogene. 2004;23:8135–8145. doi: 10.1038/sj.onc.1207983. [DOI] [PubMed] [Google Scholar]
- 24.Jansen AP, Camalier CE, Stark C, Colburn NH. Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol Cancer Ther. 2004;3:103–110. [PubMed] [Google Scholar]
- 25.Wei N, Liu SS, Chan KK, Ngan HY. Tumour suppressive function and modulation of programmed cell death 4 (PDCD4) in ovarian cancer. PLoS One. 2012;7:e30311. doi: 10.1371/journal.pone.0030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin K, Liu MH, Zhang M, Wang F, Fen M, Liu ZJ, Yuan Y, Gao S, Yang L, Zhang W, et al. miR-208a-3p suppresses cell apoptosis by targeting PDCD4 in gastric cancer. Oncotarget. 2016;7:67321–67332. doi: 10.18632/oncotarget.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen F, Mo M, Chen L, An S, Tan X, Fu Y, Rezaei K, Wang Z, Zhang L, Fu SW. MicroRNA-21 down-regulates Rb1 expression by targeting PDCD4 in retinoblastoma. J Cancer. 2014;5:804–812. doi: 10.7150/jca.10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foley NH, Bray IM, Tivnan A, Bryan K, Murphy DM, Buckley PG, Ryan J, O'Meara A, O'Sullivan M, Stallings RL. MicroRNA-184 inhibits neuroblastoma cell survival through targeting the serine/threonine kinase AKT2. Mol Cancer. 2010;9:83. doi: 10.1186/1476-4598-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao N, Budhraja A, Cheng S, Liu EH, Chen J, Yang Z, Chen D, Zhang Z, Shi X. Phenethyl isothiocyanate exhibits antileukemic activity in vitro and in vivo by inactivation of Akt and activation of JNK pathways. Cell Death Dis. 2011;2:e140. doi: 10.1038/cddis.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, Zhen Y, Yang H, Wang H, Zhou Y, Wang E, Marincola FM, Mai C, Chen Y, Wei H. Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e634. doi: 10.1038/cddis.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang WQ, Zhang H, Wang HB, Sun YG, Peng ZH, Zhou G, Yang SM, Wang RQ, Fang DC. Programmed cell death 4 (PDCD4) enhances the sensitivity of gastric cancer cells to TRAIL-induced apoptosis by inhibiting the PI3K/Akt signaling pathway. Mol Diagn Ther. 2010;14:155–161. doi: 10.1007/BF03256368. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov VN, Krasilnikov M, Ronai Z. Regulation of Fas expression by STAT3 and c-Jun is mediated by phosphatidylinositol 3-kinase-AKT signaling. J Biol Chem. 2002;277:4932–4944. doi: 10.1074/jbc.M108233200. [DOI] [PubMed] [Google Scholar]
- 33.Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assay. J Vis Exp. 2014 Jun 1; doi: 10.3791/51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmitten TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2011;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Liwak U, Thakor N, Jordan LE, Roy R, Lewis SM, Pardo OE, Seckl M, Holcik M. Tumor suppressor PDCD4 repressed internal ribosome entry site-mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol Cell Biol. 2012;32:1818–1829. doi: 10.1128/MCB.06317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Li J, Jiang Y, Xu Y, Oin C. Programmed cell death 4 (Pdcd4) suppresses metastatic potential of human hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2009;28:71. doi: 10.1186/1756-9966-28-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 38.Navarro-Quiroz E, Pacheco-Lugo L, Navarro-Quiroz R, Lorenzi H, España-Puccini P, Díaz-Olmos Y, Almendrales L, Olave V, Gonzalez-Torres H, Diaz-Perez A, et al. Profiling analysis of circulating microRNA in peripheral blood of patients with class IV lupus nephritis. PLoS One. 2017;12:e0187973. doi: 10.1371/journal.pone.0187973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirata H, Hinoda Y, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Dahiya R. Genistein down regulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer. 2014;110:1645–1654. doi: 10.1038/bjc.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, Li L, Li J, Li H, Shen Q, Ping J, Ma Z, Zhong J, Dai L. Overexpression of miR-1260b in non-samll cell lung cancer is associated with lymph node metastasis. Aging Dis. 2015;6:478–485. doi: 10.14336/AD.2015.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordentoft I, Birkenkamp-Demtroder K, Agerbak M, Thedorescu D, Ostenfeld MS, Hartmann A, Borre M, Ørntoft TF, Dyrskjøt L. miRNAs associated with chemo-sensitivity in cell lines and in advanced bladder cancer. BMC Med Genomics. 2012;5:40. doi: 10.1186/1755-8794-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107:20198–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, DuBois RN. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene. 2001;20:4450–4456. doi: 10.1038/sj.onc.1204588. [DOI] [PubMed] [Google Scholar]
- 44.Palamarchuk K, Efanov A, Maximov V, Aqeilan RI, Croce CMP. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282–11286. doi: 10.1158/0008-5472.CAN-05-3469. [DOI] [PubMed] [Google Scholar]
- 45.Bitomsky N, Bohm M, Klempnauer KH. Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene. 2004;23:7484–7493. doi: 10.1038/sj.onc.1208064. [DOI] [PubMed] [Google Scholar]
- 46.Lankat-Buttgereit B, Muller S, Schmidt H, Parhofer KG, Gress TM, Göke R. Knockdown of Pdcd4 results in induction of proprotein convertase 1/3 and potent secretion of chromogranin A and secretogranin II in a neuroendocrine cell line. Bio Cell. 2008;100:703–715. doi: 10.1042/BC20080052. [DOI] [PubMed] [Google Scholar]
- 47.Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH, Post S, Jansen A, Colburn NH, Allgayer H. Loss of Pdcd4 expression marks adenoma-carcinoma transition, correlates inversely with pAkt and is a new and independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–1707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 48.Otaegi G, Yusta-Boyo MJ, Vergaño-Vera E, Méndez-Gómez HR, Carrera AC, Abad JL, González M, de la Rosa EJ, Vicario-Abejón C, de Pablo F. Modulation of the PI 3-kinase-Akt signalling pathway by IGF-I and PTEN regulates the differentiation of neural stem/precursor cells. J Cell Sci. 2006;119:2739–2748. doi: 10.1242/jcs.03012. [DOI] [PubMed] [Google Scholar]
- 49.Cui QL, Almazan G. IGF-1 induced oligodendrocyte progenitor proliferation requires PI3K/Akt, MEK/ERK and Src-like tyrosine kinases. J Neurochemistry. 2009;217:361–370. doi: 10.1111/j.1471-4159.2006.04329.x. [DOI] [PubMed] [Google Scholar]