Abstract
Breast Cancer (BC) is one of the most common primary malignant tumors, which is life threatening. Previous studies have demonstrated that microRNAs (miRNA) may regulate or affect the incidence of BC. However, results of these studies are inconsistent, due to factors including the different sequencing platforms and sample selection methods used. To explore the key miRNAs involved in the pathogenesis of BC, and to use these miRNAs to monitor the tumor progression of BC, a systematic review was performed on the previous studies examining BC miRNA; the function of the target genes that were modulated by these key miRNAs were also analyzed. A total of 8 representative miRNA datasets examining the pathogenesis of BC were selected. Key miRNAs were identified by comparing the overlap between these datasets. Then, the target genes of these key miRNAs were predicted through TargetScan. Furthermore, functional enrichment analysis of target genes and transcription factor (TF) binding analysis was also performed using the Database for Annotation, Visualization and Integrated Discovery and Tfacts database, respectively. A total of 6 key miRNAs were identified by comparing the differentially expressed miRNAs datasets in the pathogenesis of BC. Compared with normal tissues, 3 miRNAs were upregulated: Hsa-miR-21b; hsa-miR-29b; and hsa-miR-155; and 3 miRNAs were downregulated: Hsa-miR-10b; hsa-miR-125; and hsa-miR-145. The target genes regulated by the up- and downregulated miRNAs were significantly enriched in the biological processes of ‘transcriptional regulation’, and these target genes depended on RNA polymerase II promoter and DNA template, respective to the up- and downregulated genes. The downregulated key miRNAs were specifically enriched in the biological processes of ‘ephrin receptor signaling pathway’ (GO: 0048013) and ‘axon guidance’ (GO: 0007411). TF analysis of the key miRNA target genes revealed that 104 TFs interacted with the 319 target genes of the upregulated miRNAs, while the 92 TFs interacted with the 254 target genes of the downregulated miRNAs. In total, there were 133 TFs and 63 (47.3%) TFs shared by the 2 types (up- and downregulated) of target genes. In summary, 6 key miRNAs in BC were identified by systematic review; the corresponding target genes and TFs that bind to these target genes were also identified, and the potential functions of target genes were revealed. These data may be beneficial to increasing the accuracy of BC treatment through monitoring miRNA.
Keywords: breast cancer, microRNA, systematic analysis, function and pathways enrichment
Introduction
Breast cancer (BC) is one of the top five malignant cancer types in women worldwide in 2017 (1). According to the World Health Organization, BC resulted in 521,000 mortalities in 2012 (2); additionally, ~1.7 million cases are diagnosed every year (3,4), only 1% of which were diagnosed in males (5). The most common primary symptom of BC is a painless lump, and the diagnosis may be confirmed by biopsy (5). BC is the most frequently occurring cancer in women of reproductive age (15–49 years) (6). In addition, genetic factors account for 5–10% of all primary BC cases (7). Women with either a mother or sister diagnosed with BC prior to 50 years of age have a 1.7% higher risk compared with patients with no family history of BC (8). Prognoses of BC cases are different and depend on a number of factors; therefore, outcomes vary between different cases. Generally, the later the disease stage at point of diagnosis, the poorer the prognosis (5). Therefore, it is important to understand the process of BC pathogenesis.
MicroRNAs (miRNAs/miR) are important in activating and modulating gene expression in a number of critical cellular processes (7). They are small, ubiquitous, endogenous single-chain molecules, 21–25 nucleotides in length, and are non-protein coding. They may inhibit the translation of the target mRNA, or reduce the activity of mRNA through the complete or incomplete complementary binding to the 3′-untranslated region of the target gene and forming a complex then inducing silence of the target gene (9). This may reflect a mechanistic potential, enabling the bound complexes to avoid the mRNA clearing activity of the ribosome (10). Several studies have indicated that differential miRNAs expression in cancer tissues is associated with human BC: For example, the downregulated miRNAs miR-125b, miR-145, miR-21 and miR-155 were closely associated with the biopathological features of different cancer types (11). Overexpressed has-miR-10b is also significantly associated with BC; it promotes cancer cell migration and invasion (12).
Although a series of studies were conducted on the miRNAs of BC, the results of the analysis of miRNAs expression datasets of BC in previous studies remain inconsistent (11–13). These inconsistencies may be due to a number of factors, including the type of sequencing platform, sample selection, study design and exposure assessment (13). By analyzing the key miRNAs in the pathogenesis of BC, the aim of the present study was to explore the association between miRNA and the prevention and treatment of BC.
In the present study, a systematic review of the miRNAs in BC identified in previous studies was conducted; the function of the target genes regulated by the key miRNAs was also analyzed to clarify the potential associations between the miRNAs and the risk of BC. Specifically, results from the prediction of the target genes of a number of BC-associated miRNAs were analyzed. In addition, the target genes and corresponding transcription factors (TF) associated with differentially expressed miRNAs were identified, in order to gain an improved understanding of the interaction of target genes and TF to the differentially expressed miRNAs in BC. Additional studies are required to apply these key miRNAs as biomarkers in BC diagnosis and site-specific treatment.
Materials and methods
Identification of BC miRNA datasets and screening of differentially expressed miRNAs
‘Breast Cancer’ and ‘miRNA’ were set as keywords to screen previously published studies on Google Scholar (https://scholar.google.com/; date of assess, June 25th 2018). Following careful reading of original articles from the records, a total of eight datasets that contained the expression of miRNAs between BC and control tissues were determined. Then, the information concerning all differentially expressed miRNAs in BC was obtained. Subsequently, the details of up- and downregulated miRNAs in BC, as compared with the control normal samples, were also obtained from the reports of associated articles. The frequencies of the up- and downregulated miRNAs in separate datasets were calculated, and the miRNAs that appeared in at least 3 datasets were selected in the present study.
Indication of target genes of differential expressed miRNA in BC
The sequence information of differentially expressed miRNAs was obtained from the miRBase database (http://www.mirbase.org; version 21; June 1st 2017) (14,15); then this sequence information was transferred to a FASTA file (https://molbiol-tools.ca/Convert.htm). Using TargetScan software (version 6.0; http://www.targetscan.org/mamm_31) (16,17), the target genes associated with differentially expressed miRNA were indicated. The parameter for maximum number of target genes for each miRNA was set at 300, while the maximum non-target genes for each miRNA were excluded.
Functional enrichment analysis of target genes of differentially expressed miRNAs in BC
The predicted target genes of the differentially expressed miRNAs were mapped into the Database for Annotation, Visualization and Integrated Discovery (DAVID) (18) to perform the Gene Ontology (GO) (19) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (20) analyses. Then, the top 10 GO terms of the ‘biological processes’ from the GO annotation results were screened out, identified as those with P<0.05.
TF analysis of target genes regulated by up- and downregulated miRNA in BC
The up- and downregulated target genes of BC miRNAs were uploaded to the TFactS database (21,22) at (http://www.tfacts.org). The TFs were predicted using the P-value, q-value, E-value and false discovery rate (http://www.tfacts.org/); only TFs that were <0.05 for all 4 indexes were considered to be reliable. The TFs of target genes, which were mediated by up- and downregulated miRNAs, were counted separately. Subsequently, the number of shared TFs and different TFs of the target genes regulated by the up- and downregulated miRNAs were calculated.
Results
Method of screening
To analyze the key miRNAs and their target genes in BC pathogenesis, and to explore the pathogenesis of BC, a systematic review was performed to first identify the key up- and downregulated miRNAs in BC. The target genes were predicted, and functional and TF analyses of the target genes were subsequently performed. The process is summarized in Fig. 1.
Figure 1.
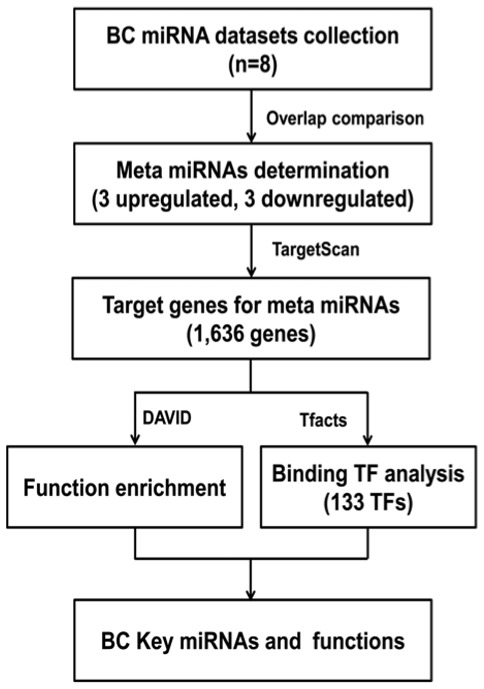
Process of analysis of the present study. By comparing previously published sequencing results of BC miRNAs, a total of 6 miRNAs supported by multiple datasets were identified. Then the target genes regulated by these meta-miRNAs were predicted. The meta-miRNAs functions on pathogenesis of breast cancer were enriched through functional enrichment analysis of the target genes of the meta-miRNAs, and transcription factor binding analysis. The numbers in brackets represent the number of processes, miRNAs or genes obtained by analysis. miRNA, microRNA; BC, breast cancer; DAVID, Database for Annotation, Visualization and Integrated Discovery; TF, transcription factor.
miRNAs identified through screening
A total of 8 independent miRNA expression datasets were identified from previous studies into miRNA expression profiles in BC, which provided details of miRNA profiles in BC tissues compared with in control tissues. In the present study, the 8 BC miRNA datasets were named based on the initials of the authors, as follows: i) SY (23); ii) IM (11); iii) VS (24); iv) HH (25); v) LA (26); vi) KL (27); vii) YL (28); and viii) SH (29). The primary features of the 8 datasets used in the analysis are summarized in Table I; these 8 datasets differed according to the sample type, number of samples, chip types and the number of probes on the chip. By using SVG mapping, the distribution of the differentially expressed miRNAs in the 8 datasets was analyzed, and it was identified that there were notable differences among the up- and downregulated miRNAs in each database (Fig. 2). As depicted in Fig. 2, the count and composition of differentially expressed miRNAs exhibited notable differences between different datasets, and only a limited number of overlapped miRNAs were determined between datasets. There were also notable differences among the differentially expressed miRNAs in the distinct datasets. A total of 99 non-redundant differential miRNAs were identified in the 8 BC miRNA datasets. Table II indicates the number of differentially expressed miRNAs, and the results of the statistical analysis of the differentially expressed miRNAs identified in the 8 BC miRNA datasets.
Table I.
Basic characteristics of the breast cancer datasets.
| Dataset | Sample type | Assay type | miRNA probes, n | Method of validation |
|---|---|---|---|---|
| SY (23) | Three human breast tumors | Multiplex Real-time PCR Assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) | b- | Breast cancer cell colony formation assay |
| IM (11) | 10 normal and 76 neoplastic breast tissues | miRNA microarray chip (KCI version 1.0) and Perkin-Elmer ScanArray XL5K | 368 | Northern blotting |
| HH (25) | 148 patients with BC and 44 age-matched and disease-free controls | TaqMan (Applied Biosystems; Thermo Fisher Scientific, Inc.) | – | RT-qPCR |
| LA (26) | – | – | – | – |
| KL (27) | MCF-7 and MDA-MB-231 cell lines | RT2 miRNA PCR array system (SABiosciences™ Corporation, Frederick, MD, USA) | 376 | RT-qPCR |
| YL (28) | 113 BC and 40 paired non-cancerous NATs | CapitalBio (http://www.capitalbio.com) | 509 | RT-qPCR |
| SH (29) | 48 tissue and 100 serum samples of patients with primary BC and 20 control samples | – | 10 | RT-qPCR |
| VS (40) | 63 primary tumors and 177 normal tissues | Bead-based microRNA profiling platform | 40-mer | Northern blotting |
BC, breast cancer; RT-qPCR, reverse transcription quantitative polymerase chain reaction; miRNA, microRNA; NATs, normal adjacent tumor tissues; -, data not available from the orignal datasets.
Figure 2.
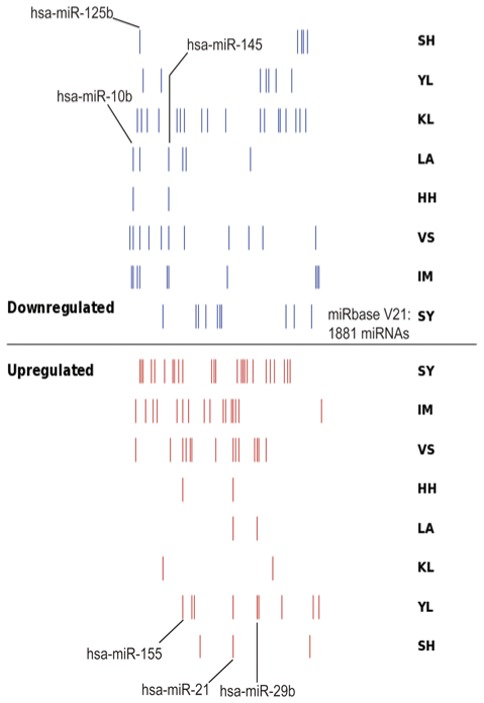
Distribution of differentially expressed miRNAs in 8 datasets. The red lines represent the upregulated miRNAs in the datasets compared with the normal tissues, while the blue lines represent the downregulated miRNAs in the datasets compared with the normal tissue. The distribution pattern of this figure indicates that there are large differences in the numbers and distributions of differentially expressed miRNAs in different data sets. The 6 miRNAs labeled in the figure represent the identified meta-signature miRNAs in the present study, which were differentially expressed in at least three studies among all eight datasets. hsa, homo sapiens; miR/miRNA, microRNA.
Table II.
GO annotation results of the target genes of the up- and downregulated microRNAs in breast cancer samples.
| GO Term | Target gene count | P-value | Fold enrichment |
|---|---|---|---|
| A, Upregulated | |||
| GO:0045944: Positive regulation of transcription from RNA polymerase II promoter | 93 | 1.09×10−12 | 2.19 |
| GO:0006366: Transcription from RNA polymerase II promoter | 59 | 1.79×10−11 | 2.66 |
| GO:0000122: Negative regulation of transcription from RNA polymerase II promoter | 67 | 5.70×10−9 | 2.15 |
| GO:0045893: Positive regulation of transcription, DNA-templated | 50 | 1.85×10−7 | 2.25 |
| GO:0045669: Positive regulation of osteoblast differentiation | 14 | 1.29×10−6 | 5.40 |
| GO:0006351: Transcription, DNA-templated | 127 | 2.03×10−6 | 1.50 |
| GO:0001525: Angiogenesis | 27 | 4.15×10−6 | 2.80 |
| GO:0008284: Positive regulation of cell proliferation | 43 | 5.60×10−6 | 2.13 |
| GO:0021542: Dentate gyrus development | 7 | 5.22×10−5 | 9.52 |
| GO:0006355: Regulation of transcription, DNA-templated | 97 | 5.85×10−5 | 1.49 |
| B, Downregulated | |||
| GO:0045944: Positive regulation of transcription from RNA polymerase II promoter | 81 | 5.22×10−8 | 1.88 |
| GO:0048013: Ephrin receptor signaling pathway | 18 | 1.61×10−7 | 4.76 |
| GO:0006351: Transcription, DNA-templated | 130 | 1.06×10−6 | 1.51 |
| GO:0007411: Axon guidance | 21 | 2.19×10−5 | 3.01 |
| GO:0000122: Negative regulation of transcription from RNA polymerase II promoter | 55 | 8.54×10−5 | 1.74 |
| GO:0006366: Transcription from RNA polymerase II promoter | 42 | 1.60×10−4 | 1.86 |
| GO:0045821: Positive regulation of glycolytic process | 6 | 2.31×10−4 | 9.75 |
| GO:0045893: Positive regulation of transcription, DNA-templated | 41 | 3.40×10−4 | 1.81 |
| GO:0006357: Regulation of transcription from RNA polymerase II promoter | 36 | 5.37×10−4 | 1.86 |
| GO:0043524: Negative regulation of neuron apoptotic process | 16 | 6.83×10−4 | 2.76 |
GO, gene ontology.
There were 4 datasets that had >20 differentially expressed miRNAs: Dataset SY, 37; dataset IM, 29; dataset VS, 27; and dataset KL, 21. Among the upregulated miRNAs, the largest number for one dataset was 26 (SY), followed by dataset IM (17); the HH, LA and KL datasets all had the lowest numbers (all 2). Among the downregulated miRNAs, the largest number for one dataset was 19 (KL), followed by the IM (17) and VS (12) datasets; the HH dataset had the lowest number (2).
Screening and identification of differentially expressed miRNAs
Data from the differentially expressed miRNAs, compared with the control samples, were collected from the previous studies, as presented in Fig. 3. It was identified that 99 non-redundant miRNAs were identified from the 8 independent data sets, 55 of which were upregulated and 54 of which were downregulated. This also indicates the requirement for a systematic review of differential miRNA expression datasets in BC.
Figure 3.
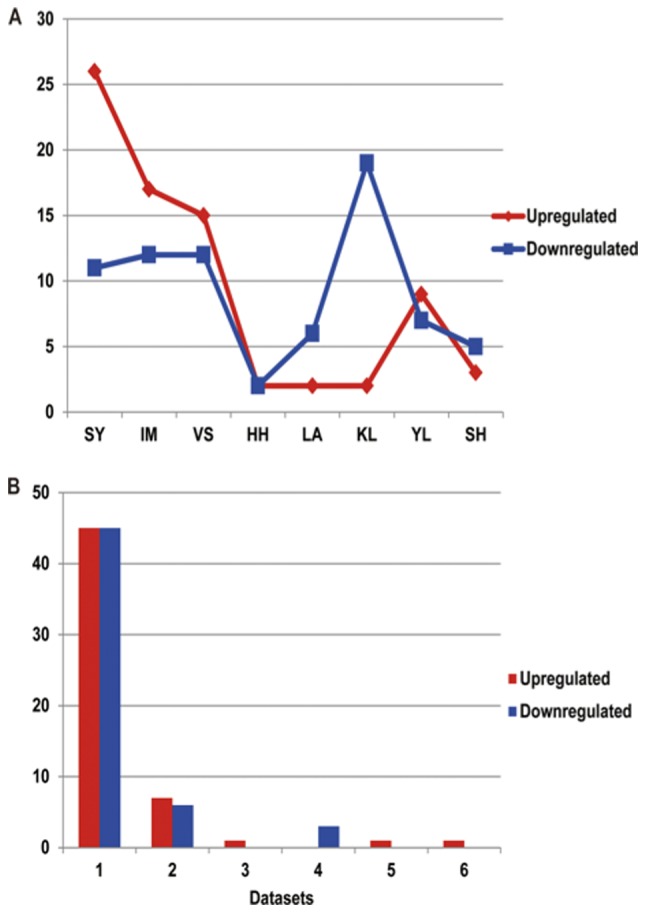
Number of differentially expressed miRNAs, and the statistics of different differentially expressed miRNA supported by the 8 different breast cancer miRNAs datasets. (A) The difference between the number of upregulated miRNAs and downregulated miRNAs compared with the control groups. The x-axis represents the acronyms of the authors and the y-axis represents the number of miRNAs. The red lines represent the upregulated miRNAs in the datasets compared with in the normal tissues, while the blue lines represent the downregulated miRNAs in the datasets compared with in the normal tissues. (B) miRNAs identified by different datasets. The x-axis represents the number of datasets, and the y-axis represents the number of miRNAs. There are 3 upregulated and 3 downregulated cases of differential expressed miRNAs which were supported by the datasets. miRNAs, microRNAs.
The miRNAs expressed in at least 3 databases were selected as the most likely to be correct miRNAs for subsequent analysis. Consequently, a total of 6 meta miRNAs were identified in the differential miRNA datasets of BC. The results indicated that a total of 3 meta miRNAs were upregulated compared with the normal tissues, which were hsa-miR-21b, hsa-miR-29b and hsa-miR-155, and a total of 3 meta miRNAs were downregulated compared with the normal tissues, which were hsa-miR-10b, hsa-miR-125b and hsa-miR-145. These six were considered to be the most likely to be correct differentially expressed miRNAs, and key factors affecting the pathogenesis of BC.
In addition, information regarding miRNA location was collected from the miRBase database, and the specific differential miRNA expression profile data are summarized in Table II. The chromosomal distribution of these six meta miRNAs are relatively dispersed. Specifically, two meta miRNAs (hsa-miR-155, hsa-miR-125b) were located on chromosome 21, and the other four were located separately on 4 different chromosomes: Chromosome 1 (hsa-miR-29b); chromosome 2 (hsa-miR-10b); chromosome 5 (hsa-miR-145); and chromosome 17 (hsa-miR-21). This sporadic distribution indicates that the key miRNAs involved in the development of BC may have large differences in biological function (30).
Prediction of target genes in differentially expressed miRNAs
The target genes of the six meta-miRNAs (summarized in Table II) were predicted using TargetScan software. Although the default parameters were used, the prediction results varied widely. In order to accurately identify the target genes regulated by differentially expressed miRNAs, the maximum number of target genes was set as 300. According to the results, the number of target genes of all six differentially expressed miRNAs reached 300.
Functional analysis results of target genes of differentially expressed miRNAs
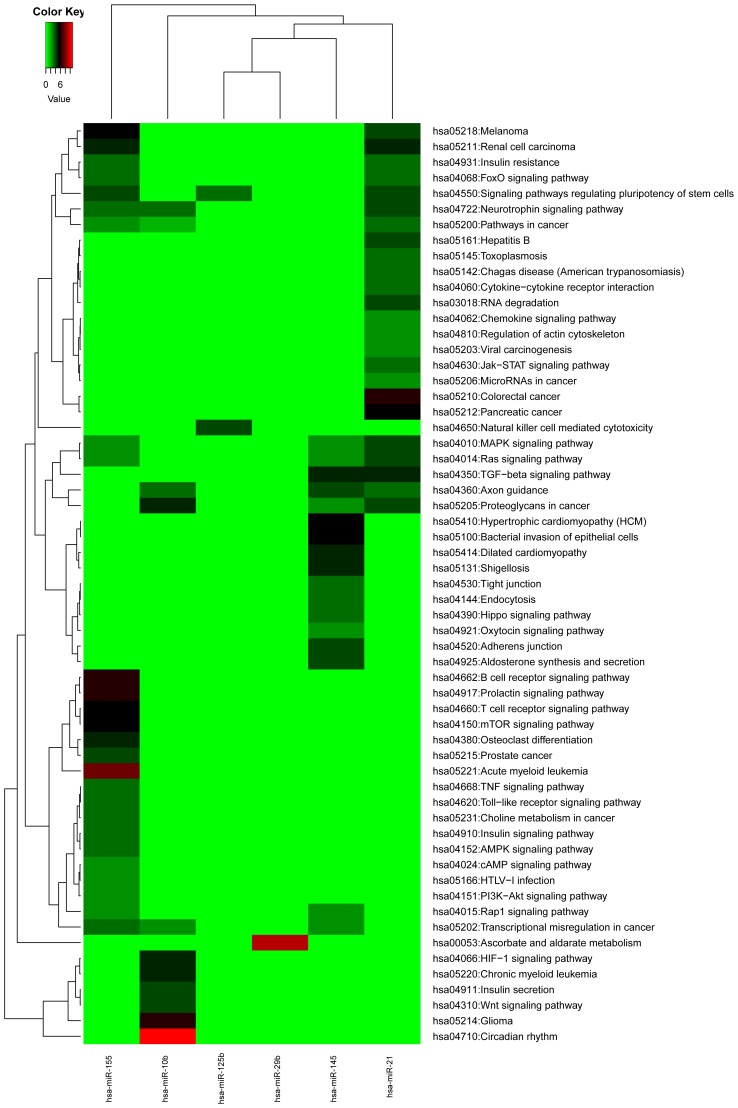
The metabolic pathway enrichment results of each of the target genes regulated by meta miRNAs were obtained, which are presented in Fig. 4. According to this figure, the KEGG Pathway functional enrichment results were identified in all six differentially expressed miRNAs, but the enrichment levels differed. Of these six miRNAs, the target genes of hsa-miR-155 had the widest range of pathway enrichment results, involving 26 KEGG terms; followed by hsa-miR-21, involving 24 KEGG terms; then hsa-miR-145 (17 KEGG terms), hsa-miR-10b (11 KEGG terms), and hsa-miR-125b (2 KEGG terms); and hsa-miR-29b (1 term).
Figure 4.
Metabolic pathway enrichment results of target genes of meta-miRNAs. This figure shows the Kyoto Encyclopedia of Genes and Genomes pathway functional enrichment results were identified in all six differentially expressed miRNAs. The different colors represent the different levels of enrichment levels of the pathway terms in the miRNA target genes; the darker the color, the higher the degree of enrichment. hsa, homo sapiens; miR/miRNA, microRNA.
The target genes identified to be associated with the up- and downregulated miRNAs were uploaded to the DAVID website for functional annotation analysis. Then, the GO enrichment results of the most significant 10 biological processes (Biology Process) corresponding to the target genes regulated by the up- and downregulated miRNAs were collected, and the Pathway results with a P<0.05 were identified. As summarized in Table III, upregulated miRNAs in BC were predominantly enriched in the transcriptional regulation process. Respectively, they depend on the RNA polymerase II promoter (GO: 0045944: Positive regulation of transcription from RNA polymerase II promoter; GO: 000636: Transcription from RNA polymerase II promoter; and GO: 0006366: Transcription from RNA polymerase II promoter) and DNA template (GO: 0045893: Positive regulation of transcription, DNA-templated). The target genes of downregulated miRNAs in BC were enriched in the transcriptional regulation process (GO: 0045944: Positive regulation of transcription from RNA polymerase II promoter; GO: 0006351: Transcription, DNA-templated; GO: 0000122: Negative regulation of transcription from RNA polymerase II promoter; GO: 0045893: Positive regulation of transcription, DNA-templated; and GO: 0006357: Regulation of transcription from RNA polymerase II promoter). Additionally, they were specifically enriched in the ephrin receptor signaling pathway (GO: 0048013: ephrin receptor signaling pathway) and the biological process of axon guidance (GO: 0007411: Axon guidance).
Table III.
List of key meta miRNAs in breast cancer.
| miRNA | Number of supported datasets | Chr no. | Beg | End | Strand sense | Sequence |
|---|---|---|---|---|---|---|
| A, Upregulated | ||||||
| hsa-miR-21 | 6 | 17 | 59841273 | 59841294 | + | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-29b | 3 | 1 | 207802450 | 207802472 | – | UAGCACCAUUUGAAAUCAGUGUU |
| hsa-miR-155 | 5 | 21 | 25573983 | 25574005 | + | UUAAUGCUAAUCGUGAUAGGGGU |
| B, Downregulated | ||||||
| hsa-miR-10b | 4 | 2 | 176150329 | 176150351 | + | UACCCUGUAGAACCGAAUUUGUG |
| hsa-miR-125b | 4 | 21 | 16590290 | 16590311 | + | UCACAAGUCAGGCUCUUGGGAC |
| hsa-miR-145 | 4 | 5 | 149430661 | 149430683 | + | GUCCAGUUUUCCCAGGAAUCCCU |
hsa, homo sapiens; miR/miRNA, microRNA; chr, chromosome.
The results of KEGG metabolic pathway enrichment (Table IV) indicated that the target genes of the upregulated miRNAs were specifically enriched in cancer pathways: (hsa05200: Pathways in cancer) and several signaling pathways [hsa04010: Mitogen-activated protein kinase (MAPK) signaling pathway; hsa04014: Ras signaling pathway; hsa04550: Signaling pathways regulating pluripotency of stem cells; and hsa04151: Phosphoinositide 3-kinase/RAC-alpha serine/threonine protein kinase signaling pathway]. Compared with this, the range enrichment pathways of target genes in the downregulated miRNAs was relatively varied, including: Proteoglycans in cancer (hsa05205); axon guidance (hsa04360); circadian rhythm (hsa04710); transcriptional misregulation in cancer (hsa05202); and MAPK signaling pathway (hsa04010). This result further proved the significance of axon guidance (hsa04360) in the pathogenesis of BC and complexity of the pathogenesis of BC caused by multiple metabolic pathways.
Table IV.
Kyoto Encyclopedia of Genes and Genomes metabolic pathway enrichment results of the upregulated miRNAs target genes in breast cancer samples.
| A, Upregulated miRNAs | |||
|---|---|---|---|
| Term | Target gene count | P-value | Fold enrichment |
| hsa05200: Pathways in cancer | 39 | 1.93×10−6 | 2.29 |
| hsa04010: MAPK signaling pathway | 27 | 3.66×10−5 | 2.44 |
| hsa04014: Ras signaling pathway | 25 | 3.81×10−5 | 2.55 |
| hsa04550: Signaling pathways regulating pluripotency of stem cells | 17 | 3.33×10−4 | 2.80 |
| hsa04151: PI3K-Akt signaling pathway | 30 | 4.16×10−4 | 2.00 |
| B, Downregulated miRNAs | |||
| hsa05205: Proteoglycans in cancer | 24 | 1.17×10−5 | 2.81 |
| hsa04360: Axon guidance | 17 | 8.68×10−5 | 3.14 |
| hsa04710: Circadian rhythm | 8 | 2.56×10−4 | 6.04 |
| hsa05202: Transcriptional misregulation in cancer | 18 | 7.52×10−4 | 2.51 |
| hsa04010: MAPK signaling pathway | 23 | 1.19×10−3 | 2.11 |
miRNA, microRNA; hsa, homo sapiens; MAPK, mitogen activated protein kinase; PI3K, phosphoinositide 3-kinase; Akt, RAC-alpha serine/threonine protein kinase.
Transcription factor analysis of target genes regulated by differentially expressed miRNAs
In the present study, the TFs corresponding to the target genes of up- and downregulated miRNAs (Table II) were also analyzed, and the similarities and differences between the TFs of the two types of target genes identified. As indicated in Fig. 5A, 104 TFs interacted with 319 target genes of the upregulated miRNAs, while 92 TFs interacted with 254 target genes of the downregulated miRNAs. According to the similarities and differences of TFs between the two types of target genes, demonstrated in Fig. 5, it was identified that there were 63 TFs shared by the two types of target genes, accounting for 47.3% of the total numbers of TFs. Fig. 5B indicated the crossover ratio, which is the most significant TF. According to this figure, the E-value was <0.05, and the highest crossover ratio value of transcription factors (SRF) was 42%, which indicated that the results of identified transcription factors were relatively reliable.
Figure 5.
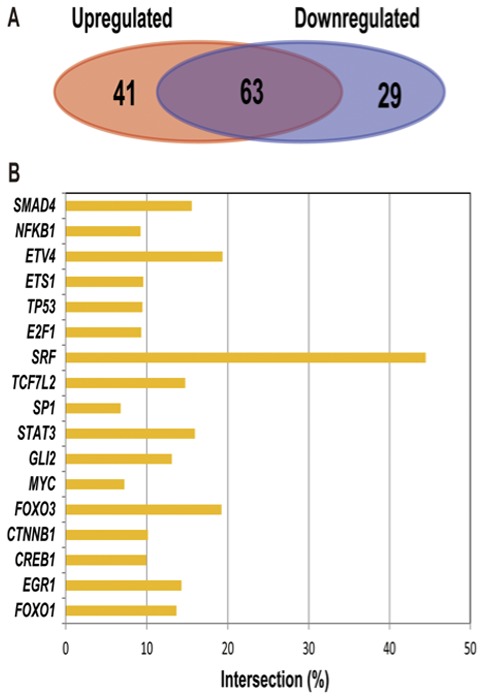
Analysis of the transcription factors in the up- and downregulated miRNAs target genes. (A) Comparison of the shared and unique transcription factors in the up- and downregulated miRNAs. The red background represents the number of transcription factors that were upregulated in the target gene of the miRNA. The blue background represents the number of transcription factors that downregulate the target genes of the miRNA. (B) Cross-rate statistics for important transcription factor regulation associated with target genes of up- and downregulated miRNAs.
Discussion
In the present systematic review, the results of previous studies investigating the association between key miRNAs and the progression of BC were analyzed. A total of six key miRNAs were identified. The purpose of the present study was to analyze the roles of miRNA in BC that may affect the prognosis of BC, then to perform prognostic risk classification on patients, or identify novel therapeutic methods for BC.
Upregulated hsa-miR-21 was identified as closely associated with BC in the present study. In previous studies, upregulated hsa-miR-21 has been demonstrated to be associated with neuroblastoma (31) and lung cancer (30). It was identified that the consistent overexpression of hsa-miR-21 in tumors indicates oncogenic activity, and may be classed as an oncomiR (31). These studies indicate that hsa-miR-21 may act as a universal indicator of cancer, instead of only for BC. Patients with lung cancer and high hsa-miR-155 expression levels exhibited poorer survival rates, compared with those with low hsa-miR-155 expression levels (30). It was also demonstrated that the role of hsa-miR-155 in BC involved the promotion of tumor angiogenesis through targeting the von Hippel-Lindau pathway, and was closely associated with poor prognosis (32). Hsa-miR-29 is a tumor promotor, which promotes metastasis in breast and colon cancer (33). These data suggest that miRNAs are important diagnostic biomarkers for different types of cancer.
Of the three downregulated meta-miRNAs, hsa-miR-145 was identified to consistently exhibit decreased levels at certain cancer stages of colorectal neoplasia (34). It was revealed previously that the decreased expression of has-miR-10b may lead to tumor invasion and metastasis through suppressing homeobox D10 and indirectly activating the pro-metastatic gene Ras homolog family member C (12). Hsa-miR-125b has been demonstrated to function as a tumor suppressor in breast (35), bladder (36) and ovarian cancer (37). It may decrease expression of the Mucin-1 oncoprotein, thereby promoting DNA damage-induced apoptosis (38), suggesting that this miRNA is also an important biomarker for cancer. All the six key miRNAs identified in the present study have been demonstrated to be closely associated with the progress of not only BC, but also a range of cancer types. While five miRNAs indicated consistent roles in different types of cancer, hsa-miR-29 exhibited various functions (33). This may also be a reason for the inconsistent outcomes of biomarker-associated miRNAs in different miRNA studies (35). Hsa-miR-29 and hsa-miR-125b were regarded as novel diagnostic and prognostic biomarkers (33,38).
The KEGG pathway functional enrichment results were calculated for all six differentially expressed miRNAs. The three upregulated BC miRNAs were significantly enriched in the transcriptional regulation process, in particular the progress of RNA polymerase II promoter and DNA template. The target genes of the downregulated BC miRNAs were also enriched in the transcriptional regulation process. This may indicate that these biological transcriptional regulation processes serve a vital role in the pathogenesis of BC. Additionally, the three downregulated miRNAs were specifically enriched in the ephrin receptor signaling pathway (GO: 0048013) and the biological process of axon guidance (GO: 0007411). It has been suggested previously that ephrin receptors (Eph) and ephrins serve as tumor suppressors and negatively regulate tumor growth (39). EphB6 is downregulated in invasive and metastatic BC (39). The roles of Ephs and ephrins are multi-faceted; the up- or downregulated ephrin receptor signaling pathway also indicates a positive and negative effect on the metastasis of BC (24). Additional studies are required to explore the complex function of this entire process. The axon guidance molecules, including ephrin, are important for cell proliferation and the adhesion of normal tissue (40). The downregulated process of axon guidance in the present study indicated that it serves as a tumor suppressor in BC (40). It has potential to serve as a biomarker in BC diagnosis and targeted therapeutic strategies.
Early diagnoses are vital to decreasing the mortality rate of cancer, and specific biomarkers are important for diagnosis, disease prediction and prognosis. Although there is evidence that miRNAs have critical effects on the pathology of BC, these remain incompletely characterized.
In the present study, a systematic review identified six miRNAs and the vital KEGG pathways, which suggests the potential of selecting specific and typical biomarkers for BC, as well as using the pathways and GO functions to further explore effective therapeutics strategies.
In conclusion, miRNAs are important in the development and progression of cancer. In the present study, 6 key miRNAs involved in the pathological process of BC were identified, including the regulated target genes and TFs. The systematic review results indicated the potential for the prediction of BC prognosis and metastasis. Additional exploration of the differential expression of miRNAs and their associated pathways may provide insight into additional useful biomarkers for the diagnosis and prognosis of BC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
FL, XZ and NT conceived and designed the study. MY, JF, YP and JL performed the data analysis and interpretation of data. FL and XZ wrote the paper. MY, JF and JL reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare there are no competing interests.
References
- 1.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 2.Nowbar AN, Howard JP, Finegold JA, Asaria P, Francis DP. 2014 global geographic analysis of mortality from ischaemic heart disease by country, age and income: Statistics from world health organisation and united nations. Int J Cardiol. 2014;174:293–298. doi: 10.1016/j.ijcard.2014.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart BW, Wild CP, editors. World Cancer Report 2014. World Health Organization; Geneva: 2014. The global and regional burden of cancer; p. 16. [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. v1.0. International Agency for Research on Cancer; Lyon, France: 2013. [Decembet;2016 ]. [Google Scholar]
- 5.Kosir MA. https://www.msdmanuals.com/home/women-s-health-issues/breast-disorders/breast-cancer%C2%A0. [Decembet;2016 ];Breast Cancer. MSD manual consumer version. [Google Scholar]
- 6.Reeder JG, Vogel VG. Breast cancer prevention. Cancer Treatment and Res. 2008;141:149–164. doi: 10.1007/978-0-387-73161-2_10. [DOI] [PubMed] [Google Scholar]
- 7.Gage M, Wattendorf D, Henry LR. Translational advances regarding hereditary breast cancer syndromes. J Surg Oncol. 2012;105:444–451. doi: 10.1002/jso.21856. [DOI] [PubMed] [Google Scholar]
- 8.Colditz GA, Kaphingst KA, Hankinson SE, Rosner B. Family history and risk of breast cancer: Nurses' health study. Breast Cancer Res Treat. 2012;133:1097–1104. doi: 10.1007/s10549-012-1985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Dalmay T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013;54:29–38. doi: 10.1042/bse0540029. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Xie SH, Xu B, Lu S, Liu P. Digitalis use and the risk of breast cancer: A systematic review and meta-analysis. Drug Saf. 2017;40:285–292. doi: 10.1007/s40264-016-0484-z. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Shih Ih, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Essaghir A, Toffalini F, Knoops L, Kallin A, van Helden J, Demoulin JB. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res. 2010;38:e120. doi: 10.1093/nar/gkq149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Essaghir A, Demoulin JB. A minimal connected network of transcription factors regulated in human tumors and its application to the quest for universal cancer biomarkers. PLoS One. 2012;7:e39666. doi: 10.1371/journal.pone.0039666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets; Proc Natl Acad Sci USA; 2006; pp. 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 26.Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: Potential effect on breast cancer management. Clin Cancer Res. 2008;14:360–365. doi: 10.1158/1078-0432.CCR-07-0992. [DOI] [PubMed] [Google Scholar]
- 27.Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat. 2012;131:445–454. doi: 10.1007/s10549-011-1424-3. [DOI] [PubMed] [Google Scholar]
- 28.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, Hu C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 31.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 32.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H, Zhang G, Wu JH, Jiang CP. Diverse roles of miR-29 in cancer (review) Oncol Rep. 2014;31:1509–1516. doi: 10.3892/or.2014.3036. [DOI] [PubMed] [Google Scholar]
- 34.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific MicroRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 35.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, Dong W, Huang J, Lin T. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2011;128:1758–1769. doi: 10.1002/ijc.25509. [DOI] [PubMed] [Google Scholar]
- 37.Cowden Dahl KD, Dahl R, Kruichak JN, Hudson LG. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–1215. doi: 10.1593/neo.09942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajabi H, Jin C, Ahmad R, McClary AC, Joshi MD, Kufe D. Mucin 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer. 2010;1:62–68. doi: 10.1177/1947601909357933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaenel P, Mosimann M, Andres AC. The multifaceted roles of Eph/ephrin signaling in breast cancer. Cell Adh Migr. 2012;6:138–147. doi: 10.4161/cam.20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harburg GC, Hinck L. Navigating breast cancer: Axon guidance molecules as breast cancer tumor suppressors and oncogenes. J Mammary Gland Biol Neoplasia. 2011;16:257–270. doi: 10.1007/s10911-011-9225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.