One Sentence Summary:
Inspection of the naive B cell repertoire specific for an HIV vaccine immunogen provides actionable information for human vaccine design and advancement.
Traditional vaccine development to prevent some of the worst current pandemic diseases has been unsuccessful, so far. Germline-targeting immunogens have potential to prime protective antibodies (Abs) via more targeted immune responses. Success of germline-targeting vaccines in humans will depend on the composition of the human naive B cell repertoire, including the frequencies and affinities of epitope-specific B cells. However, the human naive B cell repertoire remains largely undefined. Assessment of antigen-specific human naive B cells among hundreds of millions of B cells from multiple donors may be used as pre-Phase I ex vivo human testing to potentially forecast B cell and Ab responses to new vaccine designs. VRC01 is an HIV broadly neutralizing Ab (bnAb) against the Env CD4 binding site (CD4bs). We characterized naive human B cells recognizing eOD-GT8, a germline-targeting HIV-1 vaccine candidate immunogen designed to prime VRC01-class Abs. Several distinct subclasses of VRC01-class naive B cells were identified, sharing sequence characteristics with inferred precursors of known bnAbs VRC01, VRC23, PCIN63, and N6. Multiple naive B cell clones exactly matched mature VRC01-class bnAb L-CDR3 sequences. Non-VRC01-class B cells were also characterized, revealing recurrent public light chain sequences. Unexpectedly, we also identified naive B cells related to the IOMA-class CD4bs bnAb. These different subclasses within the human repertoire had strong initial affinities (KD) to the immunogen, up to 13 nM, and represent encouraging indications that multiple independent pathways may exist for vaccine-elicited VRC01-class bnAb development in most individuals. The frequencies of these distinct eOD-GT8 B cell specificities give insights into antigen-specific compositional features of the human naive B cell repertoire and provide actionable information for vaccine design and advancement.
INTRODUCTION:
Rational immunogen design holds promise for solving long-standing challenges in developing vaccines to pathogens that inflict high disease burdens. Most human vaccines are dependent on neutralizing antibody (nAb) responses for effectiveness, but for some pathogens, protective nAbs are difficult to generate via a vaccine (1, 2). The “reverse vaccinology 2.0” approach aims to design vaccines that elicit protective Ab responses by working backward from known protective Abs (3-5). Design templating from HIV-1 broadly neutralizing Abs (bnAbs) derived from humans has led to novel “germline-targeting” immunogens that have succeeded in binding inferred germline versions of bnAbs, activating transgenic B cells encoding bnAb inferred germline B cell receptors (BCRs), and generating Ab responses in BCR transgenic mice (6-12). A key gap is that it is unknown how the human B cell repertoire will respond to a given germline-targeting immunogen before human clinical trials. Initiating a clinical trial is an expensive and time-consuming process. Although human immunoglobulin (Ig) transgenic mice (such as Kymab mice) are one alternative, their BCR repertoire can be quite different from humans; for example, the frequency of VRC01-class naive B cells is 150- to 900-fold lower in these mice compared to humans (13). We have developed strategies to query the naive B cell repertoire directly in humans to identify antigen-specific and epitope-specific B cells. This strategy is particularly well suited to assessing germline-targeting immunogens but can be used for any candidate immunogen. This ex vivo method aims to identify, early in a preclinical development pathway, the antigen-specific naive B cells that may respond in a human Phase I vaccine trial. A second goal of this strategy is to identify off-target B cells that may inadvertently cripple a candidate vaccine due to immunodominant off-target responses (14).
VRC01-class bnAbs are among the most broad and potent of HIV-1 bnAbs and are therefore a major target for immunogen design (15). VRC01-class bnAbs bind to the CD4-binding site (CD4bs) of fully glycosylated trimeric HIV-1 Envelope (Env). Unlike most Abs, VRC01-class bnAbs make little use of the heavy chain complementarity determining region 3 (H-CDR3) generated by VDJ recombination and instead predominantly use the VH1-2 H-CDR2 region, encoded entirely by the V segment, for major interactions with the CD4bs. In addition, all VRC01-class bnAbs have an unusually short light chain complementarity determining region 3 (L-CDR3) length of 5 amino acids (aa). A critical problem for generating VRC01-class bnAbs is that VRC01-class naive B cells have essentially no detectable affinity for native Env from most HIV-1 clinical isolates. This is a likely explanation for why VRC01-class bnAbs have only been found in a few HIV+ individuals. A first step in the generation of any HIV-1 bnAb response is the activation of appropriate naive B cells. Overcoming that challenge for VRC01-class B cells accomplishes one key step that may normally preclude VRC01-class bnAb development during HIV-1 infection. The eOD-GT8 immunogen has been designed to specifically bind to human inferred germline versions of VRC01-class bnAbs (6, 16). Immunization with eOD-GT8 60-mer nanoparticles was able to successfully prime VRC01-class B cell responses in transgenic mice engineered to express the inferred germline VRC01 heavy chain (HC) (6, 17). However, it remained unclear if eOD-GT8 had sufficient affinity for binding authentic human VRC01-class naive B cells (in contrast to inferred germline-reverted bnAbs) and if such VRC01-class naive B cells were present in the human repertoire at frequencies plausible for responding to an eOD-GT8 immunization. VRC01-class B cells are defined here [and previously (16)] as BCR VH1-2+ B cells with a 5-aa L-CDR3 and confirmed binding specificity for the HIV-1 Env CD4bs based on flow cytometry probes or biochemical measurements using any of several HIV-1 CD4bs-containing proteins. VRC01-class bnAbs are the subset of VRC01-class Abs with an ability to neutralize virus after affinity maturation. Previously, we identified VRC01-class naive B cells within the human B repertoire capable of binding eOD-GT8, whereas VRC01-class naive B cells were not identified by the previous generation immunogen eOD-GT6 (16), indicating that screening of the human naive B cell repertoire can be useful as one parameter for assessing whether an immunogen is ready to advance to clinical trial. The VRC01-class naive Ab VRC01c-HuGL2 bound to eOD-GT8 in a manner virtually identical to the binding of mature VRC01 bnAb to the CD4bs of gp120, confirming the success of the eOD-GT8 design concept. Frequencies of human VRC01-class naive B cells were determined to be about 1 in 400,000 naive B cells and were concluded to be present in most individuals (16). Using an inferred germline VRC01 mouse model, we recently determined that both cell frequency and BCR affinity affect competitive fitness of naive antigen-specific B cells in an immune response. Naive inferred germline VRC01 B cells present at precursor frequencies of 1 in 1 million were highly fit for immune responses to analogs of eOD-GT8 60-mer under affinity conditions designed to match that of known human naive VRC01-class B cells (12).
The human naive B cell repertoire is vast and has been minimally characterized for antigen-specificities, in large part due to the low affinity interactions involved and the size of the repertoire. Here we have further characterized the human naive B cell repertoire specific for the eOD-GT8 CD4bs, defining both VRC01-class and non-VRC01-class specificities and frequencies, with a diversity of affinities spanning a 10,000-fold KD range. Several distinct subclasses of VRC01-class naive B cells were identified, corresponding to potential precursors of known bnAbs VRC01, VRC23, PCIN63, and N6. In addition, we also identified naive B cells similar to the HIV CD4bs bnAb IOMA, which has characteristics similar to VRC01-class bnAbs (18). Together, analyses of the human antigen-specific naive B cell repertoire to the germline-targeting immunogen eOD-GT8 indicate that distinct subclasses of VRC01-class naive B cells likely represent multiple different advantageous starting points for vaccine-elicited bnAb lineages.
RESULTS
Numerous VRC01-class naive B cells from an individual donor
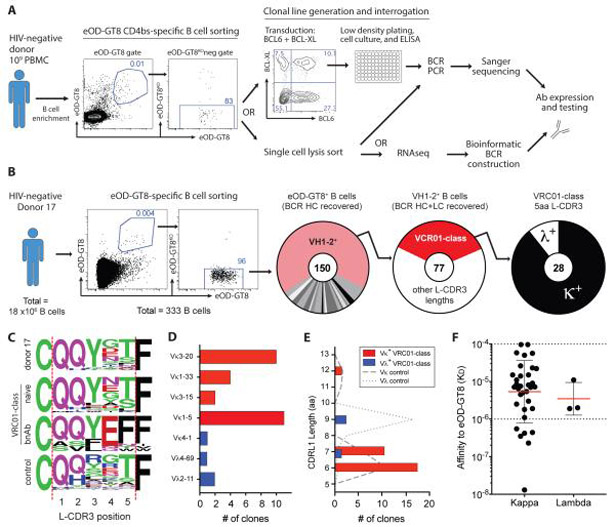
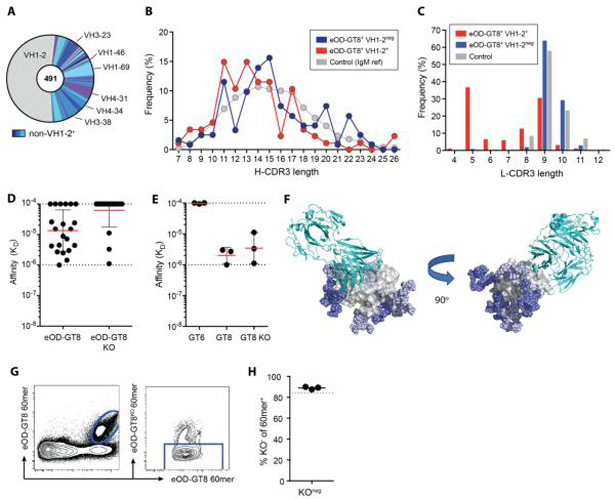
We isolated and characterized a diversity of eOD-GT8-binding (eOD-GT8+) circulating B cells from HIV-1 seronegative individuals by using multiple fluorescently labeled eOD-GT8 tetramer probes in combination with fluorescence-activated cell sorting (FACS) (Figure 1A). CD4bs epitope specific B cells were identified by costaining with eOD-GT8-KO (knock out), which contains mutations within the CD4bs pocket abrogating VRC01-class B cell binding, thereby defining the epitope site. Here, we focused on characterizing large numbers of cells from a small number of donors (three donors), whereas the previous focus was to characterize fewer cells from more donors (15 donors). From one intensively studied individual, 18 × 106 total B cells were purified and 333 individual eOD-GT8 CD4bs-specific B cells (bound eOD-GT8 in two probe colors, and negative for eOD-GT8-KO binding) were sorted for BCR sequencing (Figure 1B). VH1-2+ B cells were highly enriched among eOD-GT8+ B cells. Of the 150 B cells for which HC genes were recovered by PCR, 67% expressed VH1-2*02 (100 of 150, Figure 1B). In contrast, the general human B cell repertoire contains ~4% of VH1-2+ B cells (16, 19). Light chain (LC) sequences were recovered from 77 of 100 VH1-2+ B cells. All HC and light chain (LC) V gene sequences matched germline reference sequences; no somatic mutations were observed, consistent with the cells being naive B cells. Twenty-eight new VRC01-class naive B cell clones were identified from this one individual (Figure 1B and Table S1).
Figure 1. eOD-GT8-binding VRC01-class naive B cells from an individual donor.
A) Schematic of isolation and characterization of eOD-GT8-specific naive human B cells using biotinylated eOD-GT8 monomers tetramerized via fluorescent streptavidin. PBMCs, peripheral blood mononuclear cells; RNA-seq, RNA sequencing.
B) Isolation of eOD-GT8+ VRC01-class naive B cells from an individual HIV-seronegative donor (donor 17). Frequency of VH1-2+ cells (pink) among eOD-GT8+ B cells, VRC01-class naive cells (VH1-2+ + 5 aa L-CDR3, red) among total VH1-2+ B cells, and VRC01-class naive B cells expressing kappa (κ) or lambda (λ) LCs among total VRC01-class naive B cells. Total B cell clones are indicated at the center of each donut plot.
C) L-CDR3 sequence of κ+ VRC01-class naive B cells from donor 17 compared to L-CDR3 sequences from previously isolated VRC01-class naive B cells (16), VRC01-class bnAbs, and unsorted control B cells expressing a L-CD3 of 5 aa.
D) LC V gene usage of VRC01-class naive B cells from donor 17 (n = 28). Red indicates use of V genes observed in VRC01-class bnAbs.
E) L-CDR1 sequence length (International Immunogenetics Information System, IMGT) of VRC01-class naive B cells from donor 17, n =28. Dashed (Vκ) and dotted (Vλ) lines show L-CDR1 lengths of control B cells.
F) SPR determined monovalent affinities (KD) to eOD-GT8 of VRC01-class Abs derived from VRC01-class naive B cells (n=37). Bars represent geometric mean (red) with geometric SD (black). Horizontal dotted line at 10−4 is the limit of detection; the line at 10−6 indicates a reference affinity of 1 μM.
Among this donor’s VRC01-class naive B cells, 89% expressed a kappa variable (Vκ) LC (25 of 28, Figure 1B). κ+ VRC01-class bnAbs use a consensus Gln-Gln-Tyr-Glu-Phe (QQYEF) L-CDR3 sequence (20, 21). The vast majority of the VRC01-class naive B cells isolated here contained a partial VRC01-class bnAb consensus L-CDR3 motif of QQYxx (88%, 22 of 25, Figure 1C). Strikingly, 95% (24/25) of the VRC01-class naive B cells encoded LCs derived from Vκ genes used by VRC01-class bnAbs (Vκ3-20, Vκ1-33, Vκ3-15, and Vκ1-5, Figure 1D). VRC01-class bnAbs contain a shortened or flexible L-CDR1 sequence (20, 21), and selection for short L-CDR1 B cells was a design goal of eOD-GT8. All but one of the Vκ+ VRC01-class naive B cells expressed a short CDR1 region of 6 or 7 aa (95%, 24/25, Figure 1E).
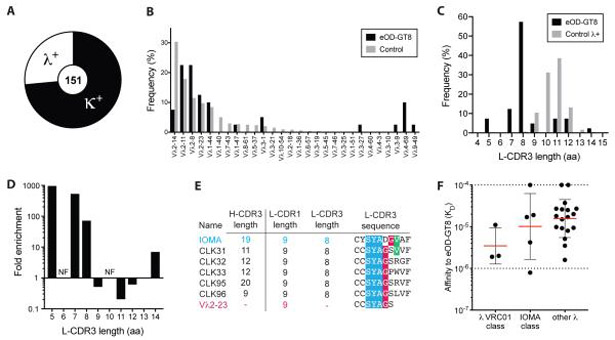
In addition to κ+ VRC01-class naive B cells, λ+ cells were also isolated from this individual. Only three Vλ+ VRC01-class naive B cells were identified among eOD-GT8+ cells (Figure 1). Vλ+ VRC01-class naive B cells were derived from Vλ2-11 and Vλ4-69 (Figure 1D). They contained longer CDRL1 lengths (Figure 1E), as do the known λ+ VRC01-class bnAbs VRC-PG19/20. The L-CDR3 sequences were mostly distinct from known λ+ VRC01-class bnAbs, although two clones contained the important L-CDR3 Glu found in VRC-PG19/20 bnAbs (Figure S1A). In summary, by screening 18 million B cells from a single individual [approximating the number of naive B cells present in a medium-sized human lymph node (LN), see Materials and Methods for calculation], we isolated 25 Vκ+ and 3 Vλ+ eOD-GT8+ VRC01-class naive B cells.
Fourteen additional VRC01-class naive B cells were identified among 6.8 million B cells in two additional donors, for a total of 42 new VRC01-class naive B cell Ab sequences (Table S1). Naive Abs were expressed from these naive B cell sequences. The geometric mean KD of Abs from the newly isolated Vκ+ VRC01-class naive B cells to eOD-GT8 was 5.3 μM (Figure 1F and S1B) and the geometric mean KD of Abs from the Vλ+ VRC01-class naive B cells was 3.5 μM (Figure 1F). Mean affinities were similar to Abs from previously characterized Vκ+ VRC01-class naive B cells (geometric mean KD = 3.4 μM, 15 donors, Figure S1B-C, (16)). To verify the epitope specificity of the newly isolated VRC01-class naive human B cells, we assessed binding to eOD-GT8-KO. In each case, VRC01-class naive B cell Abs did not have measurable affinity to eOD-GT8-KO by surface plasmon resonance (SPR), and ELISA binding assays with eOD-GT8-KO or an irrelevant protein (KLH) confirmed the SPR results of CD4bs specificity (Figure S1D-E). Overall, the sequence characteristics and affinities of VRC01-class naive B cells isolated were indicative of successful germline-targeting by the eOD-GT8 immunogen design.
High affinity N6-subclass human naive B cells targeted by eOD-GT8
We considered that VRC01-class naive B cells encoding different LC characteristics may best be understood as different subclasses, because each subclass may have distinct frequencies and BCR affinity characteristics that could affect their likelihood to respond to eOD-GT8 60-mer immunizations and/or their ability to develop into a bnAb. Thus, the total collection of eOD-GT8+ VRC01-class naive B cells identified across all donors [this study and (16)] was separated into subclasses based on LC V gene, naming each subclass for a representative bnAb member.
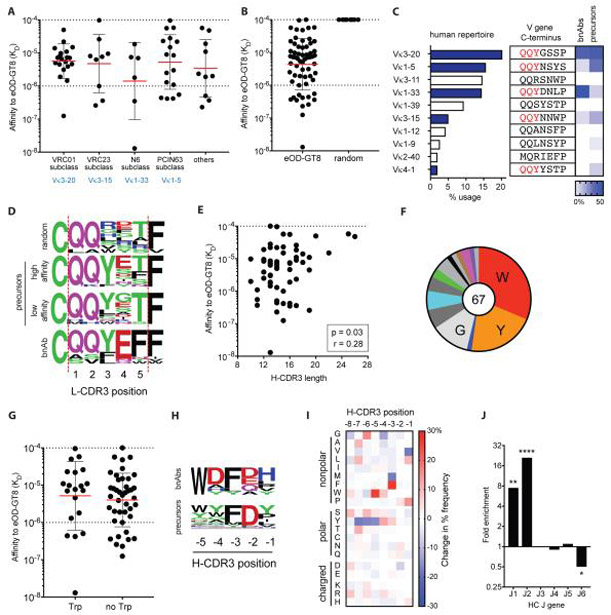
N6 is a potent VRC01-class bnAb that can neutralize 98% of HIV-1 isolates, making it the VRC01-class bnAb with the greatest breadth of any CD4bs-directed Ab (22). N6-subclass (Vκ1-33+ VRC01-class, with other bnAb subclass members including 12A21, 12A12, and VRC-CH31) naive human B cells are a highly desirable starting point for potential bnAb development, both because of the potency and breadth of N6 and the nature of the development pathway of an N6-like bnAb in comparison to VRC01. The bnAb VRC01 is different than N6 in that it required deletions in the L-CDR1 loop to avoid steric clashes with Env loop D and the N276 glycan to achieve potency and breadth (21-23). Deletions are relatively rare in somatic hypermutation (SHM) and may represent a critical bottleneck for bnAb development. It is likely that VRC01-subclass (Vκ3-20+) naive B cells would require deletions in L-CDR1 to acquire bnAb attributes in a human HIV-1 vaccine regimen. In contrast, N6-subclass bnAb maturation can occur via Vκ1-33 CDR1 glycine mutations to enable flexibility to accommodate HIV Env N276 (21, 22, 24). Six N6-subclass naive B cells were identified during B cell repertoire screening (Figure 2A, Table S1, Figure S1F). Two N6-subclass B cells isolated were of particular interest. B cell CLK20 had an exceptional monovalent Ab affinity for eOD-GT8 (KD = 13 nM), the highest affinity for eOD-GT8 of any naive B cell Ab identified. The CLK20 L-CDR3 sequence is QQYDN (Table S1). In addition, N6-subclass B cell CLK9 had a high affinity for eOD-GT8 (KD = 350 nM) and encoded a QQYEF L-CDR3 sequence, which is an exact match to the mature L-CDR3 of bnAb VRC01 (Table S1).
Figure 2. Subclasses and sequence requirements of eOD-GT8+ VRC01-class naive B cells.
Characterization and subclass assignment of VRC01-class B cells [n=18 donors, including VRC01-class naive B cells from (16)].
A) Monovalent affinities of VRC01-class Abs derived from eOD-GT8+ VRC01-class naive B cells separated by bnAb subclass (LC V gene usage), n = 62. Bars represent geometric mean (red) with geometric SD (black).
B) Monovalent affinities of VRC01-class Abs derived from eOD-GT8+ VRC01-class naive B cells (n = 62) and Abs derived from random VH1-2+ + 5-aa L-CDR3 B cells (n = 8).
C) κ V gene usage among the human B cell repertoire (blue bars indicate the presence of a QQYxx motif) and the terminus of the V gene sequence (beginning of the L-CDR3) with the QQYxx motif highlighted in red. The heat map (right) indicates the relative frequency of κ V gene usage among VRC01-class bnAbs or eOD-GT8+VRC01-class naive B cells (n = 66).
D) L-CDR3 sequence of 8 randomly selected VH1-2+ + 5 aa L-CDR3 B cells (eOD-GT8neg), the 12 highest-affinity eOD-GT8+ VRC01-class naive B cells, the 12 lowest-affinity eOD-GT8+ VRC01-class naive B cells, and VRC01-class bnAbs.
E) Pearson correlation analysis of VRC01-class Ab eOD-GT8 affinities and H-CDR3 length (IMGT). n = 62.
F) Amino acid usage at the −5 position from the C-terminal end of the H-CDR3 (also known as Trp100B in VRC01) of VRC01-class naive B cells. Single amino acid letter abbreviations. n = 67.
G) Monovalent affinities of VRC01-class Abs derived from eOD-GT8+ VRC01-class naive B cells that do and do not contain a Trp at the −5 position of H-CDR3 (n = 62). Bars represent geometric mean (red) with geometric SD (black).
H) Amino acid sequence at the C-terminal end of the H-CDR3 of bnAbs used to guide design of eOD-GT8 compared to VRC01-class naive B cells.
I) Heat map of the difference in amino acid usage at the C-terminal end of the H-CDR3 of VRC01-class naive B cells (n = 67) with control (eOD-GT8+ VRC01-class B cells, n = 119). The % aa usage among VRC01-class naive B cells minus % aa usage among non-VRC01-class naive B cells.
J) Fold enrichment of HC J gene usage among eOD-GT8+ VRC01-class naive B cells versus control B cells. Fisher’s exact test (*P < 0.05; **P < 0.005; and ****P < 0.0001).
Two other subclasses of clones were identified: PCIN63-subclass (Vκ1-5+) and VRC23-subclass (Vκ3-15+) (Figure 2A). Identification of these eOD-GT8-binding subclasses was notable, because neither PCIN63-subclass nor VRC23-subclass inferred germline Abs were used in the eOD-GT8 design process (16). Those bnAbs were not known at the time of eOD-GT8 design (25). PCIN63 is the first Vκ1-5+ VRC01-class bnAb identified (26). PCIN63-subclass B cells were the second-most frequent subclass of VRC01-class naive B cells (Figure 2A). It is interesting to note that Vκ1-5+ VRC01-class naive B cells were isolated from healthy donors using eOD-GT8 before an example Vκ1-5+ bnAb was known. These findings demonstrate the success of the deep mutational scanning and multitarget optimization used to focus eOD-GT8 on conserved paratope-epitope features across VRC01-class bnAbs.
A number of Abs from PCIN63-subclass and VRC23-subclass naive B cells had high affinities for eOD-GT8 (KD ≤ 3 μM, Figure 2A). BnAbs VRC23 and PCIN63 also lack L-CDR1 deletions, similar to N6 (25). One potential bnAb precursor, VRC01c-HuGL19, was a VRC23-subclass with an exact match to the VRC23 bnAb L-CDR3 sequence (QQYET) and had a high affinity for eOD-GT8 (KD = 260 nM, Figure S1G, (16)). In summary, we identified N6-subclass, VRC23-subclass, and PCIN63-subclass VRC01-class naive human B cells. Of the donors that were sampled deeply enough to isolate more than one VRC01-class naive B cell, subclasses were identified in seven of nine donors, indicating that subclasses are not restricted to individual donors (Figure S1F) (16). These subclasses appear to be present in most individuals and represent favorable starting points for vaccine elicitation of potential VRC01-class bnAb lineages.
To assess features of eOD-GT8 responsible for binding human VRC01-class naive B cells, we first calculated the expected frequency of VH1-2+ naive B cells coexpressing an LC with a 5 aa L-CDR3 (0.01%) and compared it with the frequency of naive VRC01-class B cells isolated (0.0006%, see Materials and Methods for calculations). Only 6% (0.0006/0.01) of potential VRC01-class naive B cells (VH1-2+ co-expressing a 5 aa L-CDR3 LC) bound to eOD-GT8 in the flow cytometry screens. Thus, eOD-GT8 is selective for VRC01-class features beyond VH1-2+ and a 5 aa L-CDR3. To further examine the selectivity of eOD-GT8, we expressed eight randomly selected VH1-2+ + 5 aa L-CDR3 Abs obtained from paired BCR sequencing of unsorted B cells (19). None of these Abs bound detectably to eOD-GT8 (Figure 2B), confirming that there are sequence requirements for eOD-GT8-binding VRC01-class naive B cells beyond the VH1-2+ + 5 aa L-CDR3 criteria.
Analyzing the full VRC01-class data set, we found that subclasses of VRC01-class B cells were isolated at different frequencies (Figure 2C). To investigate why this may be the case, we compared the Vκ gene usage from VRC01-class bnAbs and eOD-GT8+ VRC01-class naive B cells. We then cross-referenced the Vκ usage against the overall human BCR repertoire, restricted to B cells expressing 5-aa L-CDR3s (24). Finally, noting that nearly all VRC01-class naive B cells that used Vκ genes also contained a 5-aa L-CDR3 CQQYxxF motif, we tabulated the Vκ genes with a germline-encoded QQYxx motif (Figure 2C). VRC01-class bnAbs and eOD-GT8+ VRC01-class naive B cells used the four most common Vκ genes containing an L-CDR3 QQYxx motif (Figure 2C). None of the random VH1-2+ + 5 aa L-CDR3 Abs contained a QQYxx L-CDR3 motif (Figure 2D). To further test the importance of the L-CDR3 sequence for binding to eOD-GT8, we compared L-CDR3 sequences of subsets of VRC01-class naive Abs with the highest and lowest affinities. The QQYxx motif was enriched in the high affinity group (Figure 2D). Furthermore, 5 out of 12 of the highest affinity naive VRC01-class Abs contained a Glu at position 4, matching that position in the mature VRC01 bnAb QQYEF motif (Figure 2D). Three high affinity Abs contained a Phe at position 5. The lowest affinity VRC01-class naive Abs contained less L-CDR3 sequence similarity to mature VRC01 bnAbs. These data show strong LC sequence parallels between eOD-GT8-binding naive human B cells and mature VRC01-class bnAbs.
We then assessed VRC01-class naive B cell HC sequence motifs. Because the major Env CD4bs-binding contacts of VRC01-class bnAbs are made with the H-CDR2 region, VRC01-class bnAbs and VRC01-class naive B cells can accommodate a variety of H-CDR3 lengths and sequences (Table S1). Nevertheless, the H-CDR3 regions of mature VRC01-class bnAbs do provide productive contacts with the Env CD4bs, contributing 10-30% of the HC buried surface area in the Ab-gp120 complex (23, 27). Here we found a modest inverse correlation between H-CDR3 length and affinity of naive VRC01-class B cells to eOD-GT8, suggesting slight preferential binding by BCRs with shorter H-CDR3s (p = 0.03, r = 0.28; Figure 2E). A tryptophan (Trp) at the −5 position from the end of the H-CDR3 is present in most VRC01-class bnAbs (also known as Trp100B in VRC01) (20, 28). Although eOD-GT8 was not specifically designed to favor this VRC01-class feature, 31% of VRC01-class naive B cells did contain a Trp at this position (Figure 2F, 21 of 67 with complete HC sequences). VRC01-class naive B cells with or without Trp at the −5 position of the H-CDR3 had similar affinities to eOD-GT8 (Figure 2G). Thus, although the presence of this H-CDR3 Trp makes one fewer affinity maturation step needed for bnAb maturation, it appears that this residue is not a requirement in the VRC01-class preimmune repertoire for eOD-GT8 binding and could plausibly be converted during SHM post-immunization.
Similarities in the H-CDR3 C-terminal sequences of VRC01-class bnAbs were observed in VRC01-class naive B cells (Figure 2H). To identify amino acids in the H-CDR3 that favor binding to eOD-GT8, we computationally compared amino acid residue usage by VRC01-class naive B cells versus other B cells. Phe at position −3 was enriched, comparably to the well-described Trp100B (Figure 2I). The H-CDR3 enrichment of Trp at position −5 and Phe at position −3 was due to enriched usage of J1 and J2 among eOD-specific VRC01-class naive B cells (p =0.002 and <0.0001, Figure 2J and S1H). In summary, specific L-CDR3 and H-CDR3 motifs were advantageous for eOD-GT8 binding and these eOD-GT8+ sequences are also favorable for VRC01-class bnAb development.
Non-VRC01-class naive B cells
VRC01-class naive B cells are not the only B cells in the naive human repertoire that are able to bind to eOD-GT8. In addition to VRC01-class B cells, three other categories of eOD-GT8-specific B cells were identified based on BCR sequence and the epitope bound: i) non-VRC01-class VH1-2+ B cells that bind to the CD4bs, ii) non-VH1-2 B cells that bind to the CD4bs, and iii) B cells that do not bind to the CD4bs (eOD-GT8-KO binding B cells). It is useful to identify and characterize the diversity of eOD-GT8-specific naive human B cells, because these other cells may compete with the VRC01-class B cells during a response to immunization (14).
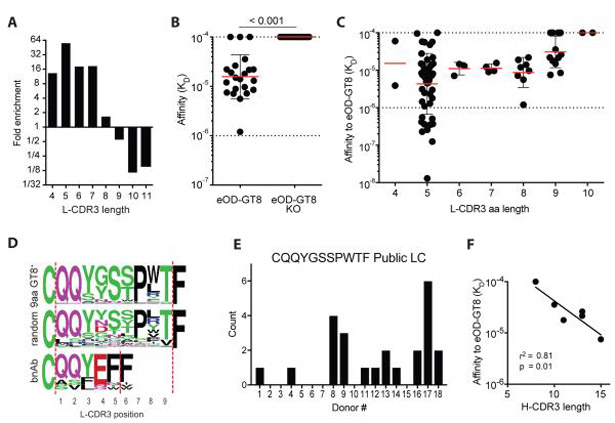
In addition to VRC01-class B cells (VH1-2+ with a 5 aa L-CDR3), we identified eOD-GT8-specific VH1-2+ naive B cells with both shorter and longer L-CDR3 lengths (VH1-2+ non-VRC01-class cells, Figure 3A and S2A). Vκ+ B cells were analyzed first. VH1-2+ B cells with an L-CDR3 length of 5 aa were the most enriched compared to control VH1-2+ B cells (>50-fold, Figure 3A). L-CDR3 lengths of 4, 6, and 7 aa were also enriched (>10-fold, Figure 3A). L-CDR3 lengths of 8 or 9 aa were not enriched (< 2-fold) and L-CDR3 lengths of ≥10 aa were reduced (>6-fold lower frequency, Figure 3A). Most VH1-2+ non-VRC01-class B cells had detectable affinity for eOD-GT8 and none bound to eOD-GT8-KO, confirming their recognition of the CD4bs epitope and thus their potential for direct epitopic competition with VRC01-class naive B cells (Figure 3B). To gain insight into how the competition between B cells targeting the same epitopic site may resolve in vivo after eOD-GT8 60-mer immunization, we measured VH1-2+ non-VRC01-class B cell affinities for eOD-GT8 (Figure 3C). VRC01-class B cells had tighter geometric mean affinity for eOD-GT8 by a factor of two or more compared to VH1-2+ B cells with L-CDR3 lengths other than 5 aa [analysis of variance (ANOVA), p < 0.0001, Figure 3C).
Figure 3. Non-VRC01-class eOD-GT8-specific VH1-2+ naive human B cells.
A) Fold enrichment of eOD-GT8+ VH1-2+ κ+ naive B cells with varying L-CDR3 lengths (IMGT) in comparison to control B cells.
B) Monovalent affinities to eOD-GT8 and eOD-GT8-KO of Abs derived from non-VRC01-class eOD-GT8+ VH1-2+ naive B cells (n = 19). Bars represent geometric mean (red) with geometric SD (black).
C) Monovalent affinities to eOD-GT8 of Abs derived from eOD-GT8+ VH1-2+ naive B cells. Abs were categorized by L-CDR3 length. VRC01-class naive B cells from (16) were included. Bars represent geometric mean (red) with geometric SD (black).
D) L-CDR3 sequences of eOD-GT8+ VH1-2+ naive B cells with a 9 aa L-CDR3, control B cells with a 9 aa L-CDR3, and VRC01-class bnAbs.
E) Number of B cells with a Vκ3-20+ CQQYGSSPWTF L-CDR3 public LC among eOD-GT8+ B cells in multiple donors (n =18).
F) Correlation analysis of H-CDR3 length and eOD-GT8 affinity for VH1-2+ Abs paired with the Vκ3-20+CQQYGSSPWTF L-CDR3 public LC sequence (n = 6).
VH1-2+ eOD-GT8-binding B cells with 9 aa L-CDR3 length were relatively common compared to VRC01-class naive B cells (Figure S2A). How does a B cell with a 9 aa CDRL3 bind eOD-GT8? One possibility was that these B cells bound eOD-GT8 not using the VRC01-class binding mode but rather using alternative binding modes with greater dependence on their H-CDR3 loops, akin to the way most Abs bind cognate antigen. Although 9 aa L-CDR3s are very common in the repertoire, they were expected to be too long to be accommodated in the CD4bs in a VRC01-class binding mode, because of potential clashes between L-CDR3 and Env loop D. However, most of these B cells use Vκ3-20, like VRC01-class naive B cells (Figure S2B). In addition, VH1-2+ 9-aa L-CDR3 Abs use a QQY motif at the beginning of the L-CDR3, similar to in VRC01-class naive B cells (Figure 3D). This L-CDR3 QQY motif was consistently observed regardless of length (Figure S2C). Only 6% of VH1-2+ 9-aa L-CDR3 cells isolated contained a Trp at the H-CDR3 −5 position (Figure S2D, 2 of 34), arguing against improved H-CDR3 interactions as a major reason these cells bind eOD-GT8 using a VRC01-class mode. To assess the potential structural basis of eOD-GT8 binding by VH1-2+ Abs with L-CDR3s of length >5 aa, we modeled eOD-GT8 bound to 40 different VH1-2+ Abs using Rosetta, starting from the crystal structure of eOD-GT8 bound to the VRC01-class human naive B Ab, VRC01c-HuGL2. This analysis of binding energetics in the modeled complexes predicted that L-CDR3s of 7, 8, or 9 aa could be sterically accommodated by a VRC01-class binding mode, but affinity would be reduced compared to the 5 aa L-CDR3 due to unfavorable positioning of the additional LC residues and the need for eOD-GT8 Tyr30 to occupy an unfavorable rotamer conformation to avoid a steric clash with L-CDR3 (Figure S2E). Overall, the pattern of predicted binding energies was similar to the measured variation of KDs with L-CDR3 length (Figure S2F compared to Figure 3C).
Thirty-three percent of VH1-2+ 9-aa L-CDR3 B cells used the exact same Vκ3-20 + Jκ1-01 recombination and an L-CDR3 of QQYGSSPWTF (Figure 3D-E, 20 of 61). Examination of the nucleotide (nt) sequence revealed single nucleotide differences in a number of sequences that all coded for the same amino acid sequence, demonstrating that the LCs were not cloning artifacts (Figure S3A-B). eOD-GT8 was bound by B cells expressing VH1-2 and this public LC in 11 of 18 donors and in multiple instances within 6 donors (Figure 3E). Whereas public LCs have been described in repertoire studies (19, 29, 30) (31), these data demonstrated epitope-specific public LC usage in humans. The length of the VH1-2 H-CDR3s correlated strongly with the eOD-GT8 affinity for Vκ3-20+Jκ1-01+ public LC Abs (Figure 3F). Two other public LCs paired with VH1-2 were found in the human naive B cell repertoire for eOD-GT8 (Figure S3C), demonstrating the potential for multiple, distinct public LCs in an epitopic naive B cell repertoire. Overall, an eOD-GT8 CD4bs-specific repertoire of VH1-2+ non-VRC01-class B cells exists, including those with that use public LCs, but their mean affinities were weaker than VRC01-class naive Abs, suggesting that VRC01-class naive B cells would have a competitive advantage after eOD-GT8 60-mer immunization.
Non-VH1-2 B cell CD4bs competitors and a distinct eOD-GT8 epitopic site
Although VH1-2+ B cells are enriched among isolated eOD-GT8-specific B cells (Figure 1B and Figure 4A), non-VH1-2 B cells represented 47% of the total eOD-GT8-binding B cell repertoire (Figure 4A, 233 of 491). The non-VH1-2 B cells used diverse HCs and similar length distributions of both H-CDR3 and L-CDR3 sequences compared to control B cells (Figure 4B-C). These HC and LC characteristics suggested that eOD-GT8 recognition occurred via conventional H-CDR3 loop-dependent binding. VH1-69 and VH3-23-utilizing bnAbs have been previously described as recognizing the CD4bs via H-CDR3 loop binding (32). Although VH1-69-type and VH3-23-type CD4bs-specific bnAb epitopes were not used to develop eOD-GT8, these were the two most common VH genes used after VH1-2 among eOD-GT8+ cells (Figure 4A). Most of the Abs tested from these non-VH1-2 cells had measurable affinity to eOD-GT8 (Figure 4D), but not eOD-GT8-KO, consistent with recognition of the CD4bs epitope and the potential to compete with VRC01-class naive B cells. CD4bs bnAbs using VH1-46 can use the H-CDR2 region to mimic CD4, but have a distinct binding mode (32) and were not used in the design templating of eOD-GT8. There were few VH1-46+ B cells among eOD-GT8 specific cells (1%, Figure 4A).
Figure 4. Non-VH1-2 eOD-GT8-specific naive human B cells.
A) VH usage of eOD-GT8+ naive human B cells.
B) Frequencies of H-CDR3 lengths (IMGT) of VH1-2+ eOD-GT8+ naive B cells, non-VH1-2 eOD-GT8+ naive B cells, and control B cells.
C) κ L-CDR3 lengths (IMGT) of VH1-2+ eOD-GT8+ naive B cells, non-VH1-2 eOD-GT8+ naive B cells, and control B cells. Panels A to C include VRC01-class naive B cells from (16).
D) Monovalent affinities to eOD-GT8 and eOD-GT8-KO of Abs derived from non-VH1-2 eOD-GT8+ naive B cells. Bars represent geometric mean (red) with geometric SD (black).
E) Monovalent affinities to eOD-GT6, eOD-GT8, and eOD-GT8KO of Abs derived from eOD-GT8KO-binding non-VH1-2 eOD-GT8+ naive B cells. Bars represent geometric mean (red) with geometric standard deviation (black).
F) Site-specific occupancy and composition of glycosylation motifs in free eOD-GT8 monomers compared to eOD-GT8 protomers within the 60-mer nanoparticle. Model of the crystal structure [Protein Data Bank (PDB) ID: 5IES] of VRC01c-HuGL2 Fab (cyan) in complex with eOD-GT8 (gray), in which Man9 glycans (blue and light blue) have been built at every position with >50% occupancy in eOD-GT8 monomer using RosettaCarbohydrate (46). Glycans with >50% occupancy on eOD-GT8 60-mer are light blue.
G) Flow cytometric analysis of naive human B cells with eOD-GT8 60mer and eOD-GT8-KO 60-mer fluorescently labeled B cell probes. Double positive eOD-GT8 60-mer binding cells (eOD-GT8-60-mer-Alexa Fluor 488+ eOD-GT8-60-mer-Alexa Fluor 647+) in the left plot were gated and shown in the right plot.
H) Frequency of eOD-GT8-KO-60-merneg cells among eOD-GT8-60-mer+ cells. Black bar indicates mean (n=3). Red dotted line indicates average frequency of eOD-GT8-KOneg cells among eOD-GT8+ cells when probed with eOD-GT8 streptavidin tetramers instead of eOD-GT8 60-mers.
Off-target B cells are valuable to consider when evaluating the readiness of germline-targeting immunogens. Non-CD4bs-specific (eOD-GT8+eOD-GT8-KO+) B cells were infrequently identified by flow cytometry (Figure 1A-B). Three eOD-GT8-specific non-VH1-2 Abs bound to both eOD-GT8 and eOD-GT8-KO, categorizing these Abs with a specificity to alternative, non-CD4bs epitopes on eOD-GT8 (Figure 4E). The eOD-GT8 and eOD-GT8-KO proteins used as sorting probes each had a His tag, suggesting that the His tag could be the targeted epitope. However, these three Abs failed to bind eOD-GT6, which also has the same His tag sequence, indicating that these Abs are not His tag specific and the epitope is elsewhere on eOD-GT8.
The observed immunofocusing of the B cell repertoire to the eOD-GT8 CD4bs is likely due to several design features of eOD-GT8, including the relatively small size of eOD-GT8 (175 aa), its compact structure, and the shielding by up to ten glycans that cover an appreciable fraction of the eOD-GT8 monomer surface outside the CD4bs. There are, however, unshielded regions of eOD-GT8 outside the CD4bs, particularly the region that is covered by the inner domain within Env gp120 or trimer. Site-specific glycan composition and occupancy analysis of eOD-GT8 monomer and eOD-GT8 60mer nanoparticle revealed that most (8 of 10) glycosylation sites on the monomer are glycosylated, but only four are highly glycosylated on 60-mer nanoparticles (Figure 4F and S3D). We considered that non-CD4bs epitopes on eOD-GT8 may have increased exposure on the eOD-GT8 60-mer nanoparticle due to the incomplete glycan occupancy. In some of our studies we used eOD-GT8 60-mer nanoparticles as sorting probes, and we observed that non-CD4bs specific (eOD-GT8+eOD-GT8+eOD-GT8-KO+) B cell frequencies were low to both eOD-GT8 monomer and 60-mer (10% of each population, Figures 1B, 4G and H,) [additional monomer data in (16)]. This suggested that the human B cell repertoire specific for eOD-GT8 is dominated by BCR specificities recognizing the CD4bs epitope, irrespective of monomer and 60-mer glycan occupancy variation.
IOMA-class naive human B cells
Vλ+ B cells accounted for 26% of the VH1-2+ eOD-GT8-specific human naive B cell repertoire (Figure 5A), consistent with the overall κ:λ ratio in the human repertoire (33). Multiple Vλ genes were used in combination with VH1-2 by eOD-GT8-specific naive B cells (Figure 5B). The distribution of L-CDR3 lengths of Vλ+ VH1-2+ eOD-GT8-specific cells was distinct from unsorted control B cells (Figure 5C). λ+ VRC01-class naive B cells were strongly enriched among Vλ+ eOD-GT8-specific cells (>900 fold compared to control B cells), as well as L-CDR3s of 7 aa and 8 aa (Figure 5D). Unexpectedly, an L-CDR3 length of 8 aa was found in 58% of the Vλ+ VH1-2+ eOD-GT8-specific cells (23 of 40 sequences). All were eOD-GT8-KO negative (Figure S4A-B). Recently, a VH1-2+ CD4bs bnAb named IOMA was discovered that was encoded by a λ2-23 LC with an 8 aa L-CDR3 (18). IOMA is a potentially attractive target for vaccine development because the L-CDR1 affinity maturation is simpler than that of VRC01, consisting only of point mutations. IOMA also contains only ~50% of the total SHM burden of mature VRC01, although IOMA exhibits considerably less breadth and potency of neutralization compared to VRC01 (18). Five eOD-GT8-specific naive B cells were isolated with features similar to IOMA (VH1-2+ + λ2-23+ with an 8 aa L-CDR3, Figure 5E and Table S1). Intriguingly, L-CDR3 amino acid positions 2, 3, 4, and 7 in CLK31 matched that of the mature IOMA bnAb (Figure 5E). In addition, the sequence encoding Asp-Gly at the fifth and sixth positions, which is each only important for IOMA bnAb activity, is each only one nucleotide change away from the naive B cell sequences isolated containing the germline λ2-23 sequence (CCSYAGSxxF) and thus could likely be readily selected during SHM after priming. Whereas the IOMA bnAb and inferred germline IOMA Ab did not show appreciable affinities to eOD-GT8 (Figure S4C), IOMA-class naive Abs (VH1-2+ + λ2-23+ with an 8 aa L-CDR3) bound to eOD-GT8 with strong affinity (Figure 5F).
Figure 5. IOMA-class naive B cells revealed by eOD-GT8.
A) Frequency of Vλ+ and Vκ+ BCRs among eOD-GT8+ naive B cells (n = 151; N = 3).
B) Vλ usage among eOD-GT8+ naive B cells versus control B cells (n = 40; N = 3). Control B cell sequences from (24).
C) L-CDR3 lengths (IMGT) of Vλ+ eOD-GT8+ naive B cells versus λ+ control B cells (n = 40, N = 3).
D) Fold enrichment of eOD-GT8+ VH1-2+ λ+ naive B cells with varying L-CDR3 lengths in comparison to control B cells. NF, not found.
E) CDR lengths and sequences of IOMA bnAb, IOMA-class naive B cells, and the λ2-23 V gene.
F) Monovalent affinities of Abs derived from Vλ+ eOD-GT8+ naive B cells to eOD-GT8. (n = 26). Bars represent geometric mean (red) with geometric SD (black).
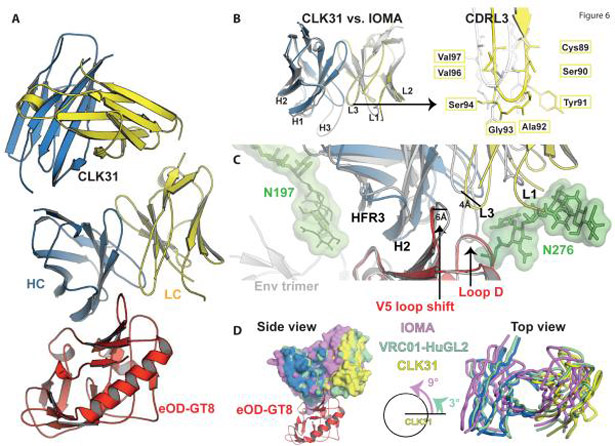
The binding of IOMA-class naive B cells to eOD-GT8 led us to investigate the structure of an IOMA-class naive B cell Ab. We solved the crystal structure of CLK31 in complex with eOD-GT8 at 2.6 Å resolution (Figure 6A and Table S2). Structural alignment of the Fv domains of CLK31 and IOMA reflects high similarity across much of the Fv domain outside of H-CDR3 and L-CDR1 with a Ca root-mean-square deviation (RMSD) of 0.8 Å over 205 atoms (Figure 6B). However, CLK31 lacks the short α-helix in L-CDR1 that is believed to be a key attribute of IOMA (18). Superimposition of eOD-GT8 (in complex with CLK31) on the BG505 HIV Env illustrates a ~6 Å shift in the V5 loop of eOD-GT8 bound to CLK31 versus IOMA bound to Env and a relative displacement of ~4 Å between the L-CDR3s (Figure 6C). Nonetheless, the distance between the L-CDR3 and the V5 loops in the bound conformations of both CLK31 and IOMA is similar (10 Å). CLK31 retains IOMA-like interactions with Env, including a hydrogen bond between Arg71HC and Asp368Env, and H-CDR2 interacts with the CD4-binding loop in a manner that mimics the C″ strand of CD4 (Figure S5A, 6C). Another feature of the IOMA-Env interaction is L-CDR3 penetrating between loop D and V5, allowing Asp93 at the tip of L-CDR3 to make hydrogen bonds to the conserved Arg456Env at the base of V5 and Asn280 (18). CLK31 has Gly rather than Asp at LC position 93 and hydrogen bonding occurs between the backbone oxygen of Gly93CLK31 and Arg26eOD-GT8, the equivalent of Arg456Env (Figure S5A). Thus, at least in part, it appears that Gly93CLK31 can mimic to some extent the function of Asp93IOMA. CLK31 binds to eOD-GT8 with an angle similar to the naive VRC01c-HuGL2 Ab (3° different) (16), whereas IOMA binds the CD4bs at a 9° rotation compared to CLK31 (Figure 6D, Figure S5B). The altered Env-binding orientation of CLK31 compared to IOMA, combined with the lack of a short α-helix in L-CDR1, appears to position CLK31 L-CDR1 in a location that would clash with the 276 glycan were it present (Figure 6C). Even if the L-CDR1 were to form a short helical segment during Ab maturation, the Env-binding orientation is such that it appears that L-CDR1 would still interact unfavorably with the 276 glycan, although the high B-values at the tip of the L-CDR1 in the CLK31+eOD-GT8 complex (Figure S5C) indicate that this region has a higher structural flexibility when compared to its surrounding residues and may be capable of substantial conformation change.
Figure 6. Structure of a naive B cell antibody similar to IOMA.
A) Crystal structure of the IOMA-related Ab CLK31 from a naive B cell in complex with eOD-GT8 at 2.6 Å resolution (HC: blue, LC: yellow, eOD-GT8: red).
B) Similarity between the variable regions of CLK31 and IOMA Abs and their CDRs. Right: Detailed comparison of the structural positions of the L-CDR3 residues in CLK31 and IOMA.
C) Comparison of the binding modes of CLK31 and IOMA combining with eOD-GT8 and BG505 SOSIP (PDB ID: 5T3Z), respectively, at the CD4bs.
D) Rotational angle of approach of CLK31, IOMA bnAb, and VRC01-HuGL2 (naive VRC01-class). Side and top views are shown.
IOMA also has a longer H-CDR3 than CLK31 (19 aa versus 11 aa). In IOMA H-CDR3, Trp100F is positioned to interact with Asn279Env and Asn280Env, analogous to Trp100B of VRC01-class bnAbs, but CLK31 does not contain a Trp to make this interaction (Table S1), and the altered Env-binding orientation of CLK31 does not offer an alternate residue position that could provide this interaction upon mutation. Thus, the structure of CLK31 bound to eOD-GT8 indicates that CLK31 has HC and LC similarities and differences from IOMA, and the degree to which this Ab has potential to develop into an IOMA-class bnAb remains uncertain.
Global analysis of human eOD-GT8-specific B cell repertoire features
Naive B cell precursor frequency is a key factor to consider for reverse vaccinology 2.0 strategies (12, 14). The frequency of eOD-GT8-specific VRC01-class naive B cells can now be calculated as 1 in 0.3 million B cells (Table S3). VRC01-class naive B cells were identified in 14 of 18 total donors, but it is likely that all donors screened had these cells at relatively similar frequencies, based on simple Poisson analysis (16). The few donors in whom VRC01-class naive B cells were not identified only had a limited number of total B cells examined, and thus simply insufficient cells were screened. Analyzing a combination of all naive B cells by sequence and affinity revealed extensive diversity within the eOD-GT8 specific human B cell repertoire.
The eOD-GT8-specific cells can be divided into subclasses, as described above, and other epitope subgroupings relevant for vaccine immunology (Figure 7). VRC01-class naive B cells have a broad range of affinities for eOD-GT8 (Figure 2D). To gauge what the physiologically relevant affinity range may be for B cells in the context of eOD-GT8 60-mer immunization, we recently constructed a mouse model testing a range of affinities and precursor frequencies (12). Upon immunization, VRC01-class naive B cells with high affinity (SPR monovalent KD ≈ 0.5 μM) competed successfully against other (non-VRC01-class) cells in germinal centers and generated VRC01-class memory B cells, even when the VRC01-class naive cells were present at precursor frequencies of 1 in 1 million cells. VRC01-class naive B cells with intermediate affinities (SPR KD ≈ 14 μM) competed effectively only when present at higher frequencies (12). Thus, we conservatively define a subgroup of “high affinity VRC01-class” human naive B cells with KD ≤ 3 μM (the geometric mean between high-affinity and intermediate-affinity groups). This high-affinity VRC01-class subgroup repertoire was present at a frequency of 1 in 0.9 million human naive B cells, representing about one-third of VRC01-class naive B cells identified (Figure 7, and Table S3). The frequency of human high affinity VRC01-class naive B cells is sufficiently high to project substantial VRC01-class responses to eOD-GT8 60-mer immunization in humans, assuming similar B cell biology between mice and humans. “Intermediate affinity VRC01-class” naive B cells are also likely to participate in human immune responses to eOD-GT8 60-mer, based on their affinities and their frequency of 1 in 0.5 million B cells (Figure 7). In the mouse model, VRC01-class naive B cells with weak affinities (KD > 40 μM) were activated by eOD-GT 60-mer in vitro, and recognized and activated by eOD-GT 60-mers in vivo, but were very poorly competitive against other cells when present at precursor frequencies rarer than 1 in 10,000 cells (12). This category of cells (“weak affinity VRC01-class”; Figure 7) is likely underrepresented in our data set because of the difficulty of isolating very low affinity cells by FACS. On the basis of the available data and the frequencies of intermediate affinity VRC01-class B cells, weak affinity VRC01-class naive B cells appear unlikely to be a substantial fraction of the human B cell response to eOD-GT8 60-mer immunization, again assuming similar B cell biology between mice and humans.
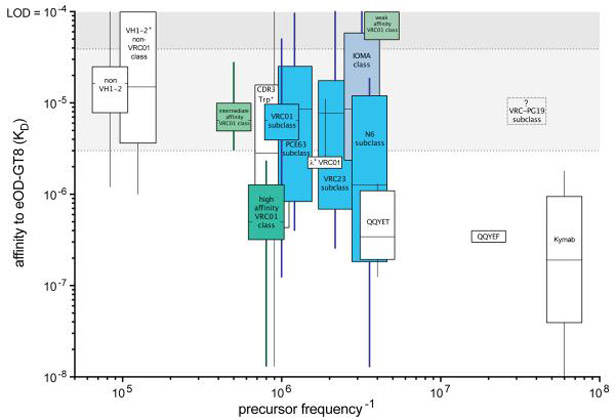
Figure 7. The human naive B cell repertoire specific for eOD-GT8.
The frequencies and affinities of classes and subclasses of eOD-GT8-specific human naive B cells. Box and whisker plots of affinities show mean, quartiles, minimum and maximum range. The inverse of naive B cell frequency is plotted (that is, 1 in x number of B cells). Non-VH1-2 B cells are the most common eOD-GT8-specific B cells; Kymab VRC01-class B cells are the rarest. Gray shading represents the continuum between “unlikely to respond” (darker) and “likely to respond” (lighter) based on tested affinities (KD’s of 0.5, 14, and 40 μM) (12). VRC01-class naive B cells subsets categorized by affinity (green) are shown as high (KD < 3 μM; the geometric mean of in vivo tested B cells with KD’s of 0.5 and 14 μM) (12), intermediate (KD’s between 3 and 40 μM), and weak affinity (KD >40 μM). Subclasses of VRC01-class naive B cells are highlighted in blue. VRC01-class naive B cells with L-CDR3 sequences matching mature bnAbs are indicated with “QQYEF” and “QQYET.” λ+ VRC01-class naive B cells are indicated. VRC01-class naive B cells containing a Trp at the −5 position of the H-CDR3 (also known as Trp100B in VRC01) are indicated as “CDR3 Trp+.” Human VRC-PG19 (λ2-14+) subclass naive human B cells are yet to be identified and denoted with a dashed box containing a “?.” “Kymab” indicates frequencies and affinities of VRC01-class B cells isolated from Kymab mice after one immunization (13). Box width is constant and acknowledges some uncertainty in the frequency calculations. Some boxes were slightly offset for clarity. LOD, limit of detection.
Consideration of frequencies of naive B cell VRC01-subclasses is also of interest, given their different characteristics and bnAb potential. All four subclasses were relatively frequent (Figure 7). The N6-subclass was less frequent (1 in 3.5 × 106 B cells) but included B cells that have high affinities to eOD-GT8. The PCIN63-subclass and VRC01-subclass naive B cells were the most common (each at ~1 in 1 × 106 B cells). VRC23-subclass naive B cells were present at 1 in 2.2 × 106 B cells. VRC-PG20-subclass (λ2-14+) (21) human naive B cells have not yet been isolated, indicating that they exist at a very low frequency. VRC01-class B cells using other λ LCs are less common than κ+ VRC01-class B cells, and have little affinity advantage as a group. We also identified a subgroup of VRC01-class naive B cells with a QQYET L-CDR3 sequence exactly matching mature bnAbs VRC27 and VRC-CH31 that is relatively common (1 in 3.5 × 106 B cells) and had a relatively high mean affinity for eOD-GT8. IOMA-class naive B cells are present at 1 in 3 × 106 B cells. In conclusion, the frequencies and affinities of VRC01-class naive B cells suggested that these cells will be favorably poised to respond to eOD-GT8 immunization in humans.
DISCUSSION
The data reported here characterize an antigen-specific naive B cell repertoire in humans. These data have implications for VRC01-class HIV-1 broadly nAb vaccine efforts, germline-targeting vaccine concepts, and an understanding of B cell diversity. This study focused on characterization of features and properties of VRC01-class bnAbs that are shared by some of the VRC01-class naive B cells identified, with the assumption that the more VRC01-class bnAb sequence features shared by a given VRC01-class naive B cell, the more likely it is that the naive B cell has the potential to develop into a VRC01-class bnAb in an appropriate immunization regimen. In total, 42 new VRC01-class naive B cells were identified, including (i) several subclasses of VRC01-class naive human B cells corresponding to potential precursors of VRC01, VRC23, PCIN63, and N6 bnAbs; (ii) five naive B cells similar to IOMA-class bnAbs; (iii) multiple naive B cells with an exact sequence match to the L-CDR3 of mature bnAbs VRC01 and VRC-CH31; and (iv) naive B cells with high eOD-GT8 binding affinities (CLK20; KD = 13 nM). The frequencies and affinities of each type of eOD-GT8-specific B cell will likely influence their ability to succeed in a vaccine immune response. The relative ranking of high affinity VRC01-class naive B cells within the hierarchy of all eOD-GT8 specific B cells in the human repertoire predicts that these cells are likely to be successfully engaged in humans immunized with the germline-targeting immunogen eOD-GT8 60-mer. With an estimated 1010 to 1011 B cells in the human body, 104 to 105 high affinity VRC01-class naive B cells are predicted to be present. At a frequency of 1 in 0.9 million B cells (Figure 7), an average LN of 50 million total cells would be estimated to contain 23 high affinity VRC01-class naive B cells.
Human naive B cell epitope-specific repertoire data may represent an innovative means of assessing the readiness of candidate immunogens for advancement to clinical trial. A major obstacle to inducing a VRC01-class bnAb by immunization could be a necessity for rare “indels” (insertions and deletions). Many VRC01-class bnAbs have indels necessary for neutralization activity (21, 23); among VRC01-subclass bnAbs, VRC08 is the only exception to the indel requirement (34). Indels are the product of intense SHM in the germinal center response and may be difficult affinity maturation features to induce consistently via immunization (35-37). Some VRC01-class bnAbs do not require indels, including the N6-subclass, PCIN63-subclass, and VRC23-subclass, making these potentially more attainable bnAbs for elicitation by immunization (24). It is encouraging that N6-subclass, PCIN63-subclass, VRC23-subclass, and IOMA-class naive B cells are present in the repertoire and were bound by eOD-GT8. These different subclasses in the human repertoire represent encouraging indications that multiple independent pathways may exist for vaccine-elicited VRC01-class bnAb development in most individuals. The various subclasses in the repertoire can each be conceptualized as independent departure points for starting VRC01-class bnAb lineages via immunization. Athough this study is limited to the naive B cell repertoire found circulating in the blood of North American donors, further studies of the immunogen-specific naive B cell repertoire in donors from around the world and in lymphoid tissue can be conducted. Nonetheless, the eOD-GT8 B cell repertoire frequency and affinity data predict that these subclasses are reasonable candidates to respond in eOD-GT8 60-mer immunized humans.
Additional design modifications of eOD-GT8 could open a potential new immunization path to induction of IOMA-class bnAbs. One advantage for IOMA-class bnAb targeting is that IOMA bnAb does not require deletions or insertions and contains less SHM than VRC01. However, as a vaccine target, IOMA bnAb is considerably less broad and potent than VRC01-class bnAbs and thus less preferred. Only one IOMA-class bnAb has been isolated from HIV-1+ individuals (18); identification of additional IOMA-class bnAbs from HIV-1+ donors would help determine the breadth and potency potential of this bnAb class and would clarify whether the naive B cells harboring BCRs similar to IOMA have characteristics key for potential maturation into IOMA-class bnAbs. In contrast, the N6-subclass naive B cells have clear similarities to multiple N6-subclass bnAbs, indicating their potential for maturation into N6-subclass VRC01-class bnAbs. An additional open question is whether distinct classes of bnAbs specific for the same paratope (such as VRC01-class and IOMA-class bnAbs for the CD4bs) can be elicited in combination using the same or related immunogens. Still, the potential to initiate VRC01-class (including multiple distinct subclasses) and IOMA-class bnAb lineages either individually or together provides multiple potential paths for the induction of a CD4bs bnAb by immunization.
Two groups of potential CD4bs competitor B cells were identified in the eOD-GT8 screening. VH1-2+ non-5-aa L-CDR3 expressing B cells represent not only potential competitors for VRC01-class naive B cells, but also potential bnAb precursors themselves. However, somatic deletion to attain the requisite 5 aa length may be unlikely to be a sufficiently common event to be relevant, because VH1-2+ non-5-aa L-CDR3 cells are not considerably more numerous than VRC01-class naive B cells and have weaker affinities for eOD-GT8 (Figure 7). These cells may best be viewed as unwanted competitor B cells. Many of these VH1-2+ non-5-aa L-CDR3-expressing B cells used public LCs that may potentially have features that could be “designed against” to eliminate those portions of the competitor B cell repertoire from binding eOD-GT8. The other potential CD4bs competitors, non-VH1-2 naive B cells specific for eOD-GT8, represent a group of protein-specific naive human B cells for which the protein was not specifically designed to bind. These lower affinity naive B cells (KD range of 1 to 100 μM) for eOD-GT8 may be outcompeted by VRC01-class naive B cells. They may also represent the typical range of affinities that naive human B cells have for complex, nondesigned protein antigens (Figure 7). This observation implies an initial benchmark affinity (KD of 1 μM) for protein design so that on-target B cells have an affinity advantage versus other competing B cells. A limitation of the techniques used in this study is a likely undersampling of naive B cells with low affinity (KD > 20 μM) to the immunogen, which may make contributions to the B cell response after immunization. Clinical testing of the eOD-GT8 60-mer immunogen will ultimately provide the best answers about the repertoire of responding human B cells and will provide an opportunity to refine interpretations of this B cell repertoire screening approach.
Recruitment and activation of appropriate B cell precursors for HIV bnAbs are thought to be a major impediment to the generation of bnAb responses in humans (38). B cell competition and immunodominance are critical factors that affect the outcome of the Ab response (12, 14, 39, 40). Encouragingly, many parallel results are found between the mouse immunization models and human repertoire analysis; VH1-2+ naive B cells expressing a 5 aa L-CDR3 bind to eOD-GT8 (6, 13, 16). In additional, both in vivo mouse experiments and direct screening of human B cells found eOD-GT8-specific VH1-2+ HCs paired with LCs using V genes of known VRC01-class bnAbs (13) and L-CDR3 sequences showing selection for the QQYxx motif used by VRC01-class bnAbs (6, 11, 13). Although VRC01-class responding cells were rare in Kymab mice, PCIN63-subclass, VRC23-subclass, and N6-subclass B cells were all found in the Kymab model (13), similar to the subclasses identified in the naive human B cell repertoire.
Immunogen-specific naive human B cell analysis can optimize the candidate immunogen pipeline by providing direct human B cell repertoire data in advance of human clinical studies. In the case of eOD-GT8, the human B cell repertoire analysis supports Phase I clinical testing.
MATERIALS AND METHODS
Study design
This study was designed to examine the repertoire of human naive B cells able to bind to the germline-targeting immunogen eOD-GT8. Success of germline-targeting vaccines in humans will depend on the composition of the human naive B cell repertoire, but the immunogen-specific human naive B cell repertoire remains largely undefined. We have used single cell flow cytometry sorting combined with BCR sequencing to isolate and categorize a diversity of eOD-GT8 specific naive human B cells. Affinities and structures were determined to further characterize the binding attributes of the naive B cell Abs with eOD-GT8. Deep eOD-GT8 specific repertoire sequencing was completed for three donors, whereas global eOD-GT8-specific repertoire characterization included naive B cells isolated from donors studied in (16), for a total of N=18 donors.
Human samples, PBMC and B cell isolation
LRS (leukoreduction) tubes were obtained from the San Diego Blood Bank from healthy, HIV-seronegative human donors. These studies do not constitute human subjects research as determined by the La Jolla Institute Institutional Review Board. Peripheral blood mononuclear cells (PBMCs) were isolated by diluting blood about 1:1 with room temperature PBS and layered over Histopaque-1077 (Sigma-Aldrich). Tubes were centrifuged to produce a buffy coat that was then aspirated and moved to a clean tube. PBMC were washed three times with PBS and then frozen in 9:1 fetal bovine serum/dimethyl sulfoxide. More than one billion PBMC were regularly recovered. Frozen cells were thawed before sorting and washed once in R10 medium (RPMI, 10% FBS, 1% L-glutamine, 1% penicillin-streptomycin) and once in MACS buffer (1% BSA (bovine serum albumin) in PBS). CD19+ B cells were isolated using a positive selection magnetic bead separation kit (Miltenyi Biotec). Isolated B cells were then washed and resuspended in R10 medium before sorting. The number of total CD19+ IgG− B cells screened and sequences obtained from donors 16, 17, and 18 are listed in Table S4. Additional materials and methods information is listed in the Supplementary Materials.
Probe generation
Avi-tagged eOD-GT8 and eOD-GT8KO2 monomers were biotinylated and purified (13). For simplicity, eOD-GT8KO2 is referred to as eOD-GT8-KO in the main text and figures. eOD-GT8 biotinylated monomers were individually premixed with fluorescently labeled streptavidin (SA-Alexa Fluor 647 or SA-Brilliant Violet 421) to form eOD-GT8 Alexa Fluor 647 and eOD-GT8 Brilliant Violet 421 tetramers and eOD-GT8KO2 phycoerythrin (PE) tetramers. eOD-GT8KO2 biotinylated monomers were premixed with fluorescently labeled streptavidin-PE. Two “positive” probes and one “negative” probe and three independent fluorochromes were chosen to avoid false positives due to isolation of B cells specific for non-eOD-GT8 epitopes present on the probes such as epitopes present on biotin, streptavidin, and the fluorochromes. The positive probes were labeled with fluorochromes that would give bright signals and would be likely to provide fewer potential epitopes for B cell recognition. eOD-GT8 60-mer nanoparticles nanoparticles were separately labeled with Amine Reactive Alexa Fluor 488 and Alexa Fluor 647 kits (A10235 or A20173, respectively, Thermo Fisher Scientific). eOD-GT8KO 60-mer nanoparticles (6, 24) were labeled with a Pacific Blue protein labeling kit (P30012, Thermo Fisher Scientific).
Sorting
Cells were incubated with eOD-GT8 probes (eOD-GT8-biotin-streptavidin-Alexa Fluor 647, eOD-GT8-biotin-streptavidin-BV421, and eOD-GT8KO2-biotin-streptavidin-PE) for 20 minutes at 4°C. Without washing, anti-CD19 as well as anti-IgG, CD3, CD14, CD16, and Live/Dead for exclusion (Table S5), was added for an additional 20 minutes. A BD FACSAria was used for all cell sorting. Cells were sorted at a flow rate of 1500 events/second using an 85 μm nozzle. Sorting stringency was set to a strict setting to obtain one cell per well. eOD-GT8-specific clonal B cell lines were generated and interrogated in some experiments as in (16).
SPR affinity measurements
All SPR experiments were carried out as described previously (6). All CLK Abs were tested for affinity to eOD-GT8 and eOD-GT8KO2.
Crystallization and data collection
For crystallography, a minimally glycosylated eOD-GT8 [His6x-tagged eOD-GT8minglyc (16)] was produced by transiently transfected 293S (GnTI−/−) cells and then purified using a HisTrap nickel column (GE Healthcare), followed by size exclusion chromatography using a Superdex75 column (GE Healthcare). The Fab constructs of the naive Abs were first affinity purified with human lambda resin (Life Technologies), followed by cation exchange chromatography (GE Healthcare). After incubating the purified Fab with a molar excess of purified eOD-GT8minglyc, the complex was deglycosylated by an Endo H treatment (New England Biolabs), followed by purification over Superdex200 size exclusion chromatography (GE Healthcare). The deglycosylated complex entered crystallization trials using an automated CrystalMation robotic system (Rigaku) at concentrations between 5 and 15 mg/ml. Crystals of the complex of the IOMA-class naive B cell CLK31 Fab with eOD-GT8minglyc (PDB ID: 6D2P; 2.60 Å resolution) were obtained at 20°C in a solution at pH 5.0 of 20% w/v PEG6000, and 0.1 M citric acid (pH 4.0). Crystals were cryoprotected in 25% glycerol followed by fast plunging in liquid nitrogen.
X-ray data collection, data processing and structure determination
Crystals of the Fab combining with eOD-GT8minglyc that had undergone Endo H treatment for partial deglycosylation were subjected to high energy x-ray radiation at Advanced Photon Source (at Argonne National Laboratory) beamlines 23ID-D and 23ID-B. Data processing was performed using HKL2000 (41). Molecular replacement for CLK31 in complex with eOD-GT8minglyc was performed using coordinates from VRC01cHuGL2+eOD-GT8minglyc (PDB ID: 5IES, 2.16Å resolution) as search models in Phaser (42). The structures were refined in PHENIX (43) and model building was done in Coot (44). MolProbity was used for structure validation (45). The refinement statistics are reported in Table S2.
Statistical analysis
Prism 7 (GraphPad) was used for all statistical analyses. A Pearson correlation test was used to assess H-CDR3 vs affinity to eOD-GT8 (Figure 2). Fisher’s exact test was used to determine enrichment of HC J gene usage (Figure 2). One-way ANOVA test was used to compare affinity to eOD-GT8 over different L-CDR3 lengths (Figure 3).
Supplementary Material
ACKNOWLEDGEMENTS:
We thank the La Jolla Institute (LJI) Flow Cytometry Core for expert cell sorting assistance. We thank the LJI Sequencing Core, as well as J. Ashton and his team at the Genomics Research Center, University of Rochester, for single cell RNA sequencing. We thank J. Jardine, B. Dekosky, and G. Georgiou for early assistance with this project.
Funding: This work was supported by NIH National Institute of Allergy and Infectious Disease (NIAID)UM1 AI100663 (Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery) (to S.C., W.R.S., I.A.W., and J.C.P.), NIAID R01 Al113867 (to J.C.P and W.R.S.), and NIAID U24 Al120134 (to S.E.B.); by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium and Center (NAC) (to W.R.S. and I.A.W.) and through The Collaboration for AIDS Vaccine Discovery (CAVD) funding for the IAVI NAC Center (W.R.S., I.A.W.). IAVI’s work is made possible by generous support from many donors including the Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan in partnership with The World Bank, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation (NORAD), the UK Department for International Development (DFID), and the U.S. Agency for International Development (USAID). The full list of IAVI donors is available at http://www.iavi.org. The NIH Office of the Director supported purchase of a BD FACSAria II (NIH S10RR027366) and an Illumina HiSeq 2500 (NIH S10OD016262).
Footnotes
Competing interests: IAVI and the Scripps Research Institute have filed a patent relating to the eOD-GT8 immunogens in this manuscript, which included inventors D.W.K. and W.R.S. W.R.S. is a cofounder and stockholder in CompuVax Inc., which has programs in non-HIV vaccine design that might benefit indirectly from this research.
Data and materials availability: BCR sequences for HCs and LCs have been deposited at the National Center for Biotechnology Information with GenBank accession codes MH349542 to MH349725 and MH500107-MH500190. Coordinates and structure factors have been deposited with the PDB with accession code 6D2P. Materials and information are available by material transfer agreement from the LJI or the Scripps Research Institute.
REFERENCES and Notes
- 1.Plotkin SA, Correlates of protection induced by vaccination, Clin. Vaccine Immunol 17, 1055–1065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koff WC, Burton DR, Johnson PR, Walker BD, King CR, Nabel GJ, Ahmed R, Bhan MK, Plotkin SA, Accelerating next-generation vaccine development for global disease prevention, Science 340, 1232910–1232910 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton DR, What Are the Most Powerful Immunogen Design Vaccine Strategies? Reverse Vaccinology 2.0 Shows Great Promise, Cold Spring Harb Perspect Biol 9, a030262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, What Are the Most Powerful Immunogen Design Vaccine Strategies? A Structural Biologist's Perspective, Cold Spring Harb Perspect Biol 9, a029470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rappuoli R, Bottomley MJ, D'Oro U, De Gregorio O, Finco E, Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design, Journal of Experimental Medicine 213, 469–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, Menis S, Jones M, Kubitz M, Spencer S, Adachi Y, Burton DR, Schief WR, Nemazee D, HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen, Science 349, 156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jardine J, Julien J-P, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR, Rational HIV immunogen design to target specific germline B cell receptors, Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, Briney B, Schiffner T, Garces F, Freund NT, Gitlin AD, Menis S, Georgeson E, Kubitz M, Adachi Y, Jones M, Mutafyan AA, Yun DS, Mayer CT, Ward AB, Burton DR, Wilson IA, Irvine DJ, Nussenzweig MC, Schief WR, HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies, Immunity 45, 483–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamatatos L, Pancera M, McGuire AT, Germline-targeting immunogens, Immunol. Rev 275, 203–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, Lowe S, Chen ST, Heinemann J, Yao K-H, Georgeson E, Gazumyan KL, Saye-Francisco A, Adachi Y, Kubitz M, Burton DR, Schief WR, Nussenzweig MC, Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice, Cell 166, 1445–1458.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, Le KM, Ramos A, Jones M, Saye-Francisco KL, Blane TR, Spencer S, Georgeson E, Hu X, Ozorowski G, Adachi Y, Kubitz M, Sarkar A, Wilson IA, Ward AB, Nemazee D, Burton DR, Schief WR, Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies, Cell 166, 1459–1464.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, Schief WR, Nemazee D, Crotty S, Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens, Immunity 48, 133–146.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sok D, Briney B, Jardine JG, Kulp DW, Menis S, Pauthner M, Wood A, Lee E-C, Le KM, Jones M, Ramos A, Kalyuzhniy O, Adachi Y, Kubitz M, MacPherson S, Bradley A, Friedrich GA, Schief WR, Burton DR, Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice, Science 353, 1557–1560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havenar-Daughton C, Lee JH, Crotty S, Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem, Immunol. Rev 275, 49–61 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Burton DR, Hangartner L, Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design, Annu. Rev. Immunol 34, 635–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereño-Orbea J, Kalyuzhniy O, Deresa I, Hu X, Spencer S, Jones M, Georgeson E, Adachi Y, Kubitz M, deCamp AC, Julien J-P, Wilson IA, Burton DR, Crotty S, Schief WR, HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen, Science 351, 1458–1463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, Pietzsch J, Gray MD, Cupo A, van Gils MJ, Yao K-H, Liu C, Gazumyan A, Seaman MS, Bjorkman PJ, Sanders RW, Moore JP, Stamatatos L, Schief WR, Nussenzweig MC, Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice, Cell 161, 1505–1515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gristick HB, von Boehmer L, West AP, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fätkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ, Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site, Nat. Struct. Mol. Biol 23, 906–915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G, In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire, Nat Med 21, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 20.West AP, Diskin R, Nussenzweig MC, Bjorkman PJ, Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120, Proceedings of the National Academy of Sciences 109, E2083–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang G-Y, Joyce MG, Kwon YD, Longo NS, Louder MK, Luongo T, McKee K, Schramm CA, Skinner J, Yang Y, Yang Z, Zhang Z, Zheng A, Bonsignori M, Haynes BF, Scheid JF, Nussenzweig MC, Simek M, Burton DR, Koff WC, NISC Comparative Sequencing Program, Mullikin JC, Connors M, Shapiro L, Nabel GJ, Mascola JR, Kwong PD, Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies, Immunity 39, 245–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Wu F, Doria-Rose NA, Zhang B, O'Dell S, Chuang G-Y, Georgiev IS, Darko S, Migueles SA, Bailer RT, Louder MK, Alam SM, Kelsoe G, Von Holle T, Haynes BF, Douek DC, Hirsch V, Seaman MS, Shapiro L, Mascola JR, Kwong PD, Connors M, Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth, Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD, Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01, Science 329, 811–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jardine JG, Sok D, Julien J-P, Briney B, Sarkar A, Liang C-H, Scherer EA, Henry Dunand CJ, Adachi Y, Diwanji D, Hsueh J, Jones M, Kalyuzhniy O, Kubitz M, Spencer S, Pauthner M, Saye-Francisco KL, Sesterhenn F, Wilson PC, Galloway DM, Stanfield RL, Wilson IA, Burton DR, Schief WR, Trkola A, Ed. Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design, PLoS Pathog 12, e1005815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang G-Y, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD, Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization, Science 340,751–756 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Landais E, Broadly neutralizing antibodies to HIV-1: Lessons from Protocol C Studies, presented at the HIV Vaccine Keystone Symposia, Steamboat Springs, CO USA, 26–30 March 2017. [Google Scholar]
- 27.Diskin R, Scheid JF, Marcovecchio PM, West AP, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ, Increasing the potency and breadth of an HIV antibody by using structure-based rational design, Science 334, 1289–1293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yacoob C, Pancera M, Vigdorovich V, Oliver BG, Glenn JA, Feng J, Sather DN, McGuire AT, Stamatatos L, Differences in Allelic Frequency and CDRH3 Region Limit the Engagement of HIV Env Immunogens by Putative VRC01 Neutralizing Antibody Precursors, Cell Rep 17, 1560–1570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson KJL, Wang Y, Gaeta BA, Pomat W, Siba P, Rimmer J, Sewell WA, Collins AM, Divergent human populations show extensive shared IGK rearrangements in peripheral blood B cells, Immunogenetics 64, 3–14 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Hoi KH, Ippolito GC, Intrinsic bias and public rearrangements in the human immunoglobulin Vλ light chain repertoire, Genes Immun 14, 271–276 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Arentz G, Thurgood LA, Lindop R, Chataway TK, Gordon TP, Secreted human Ro52 autoantibody proteomes express a restricted set of public clonotypes, J Autoimmun 39, 466–470 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O'Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng H-P, Yang A-S, NISC Comparative Sequencing Program, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD, Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 donors, Cell 161, 1280–1292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bräuninger A, Goossens T, Rajewsky K, Küppers R, Regulation of immunoglobulin light chain gene rearrangements during early B cell development in the human, Eur. J. Immunol 31, 3631–3637 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Zhang Z, Schramm CA, Joyce MG, Do Kwon Y, Zhou T, Sheng Z, Zhang B, O'Dell S, McKee K, Georgiev IS, Chuang G-Y, Longo NS, Lynch RM, Saunders KO, Soto C, Srivatsan S, Yang Y, Bailer RT, Louder MK, Program NCS, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S-L, Legaspi R, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Mullikin JC, Connors M, Kwong PD, Mascola JR, Shapiro L, Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection, Cell 161, 470–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Victora GD, Nussenzweig MC, Germinal centers, Annu. Rev. Immunol 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Crotty S, T follicular helper cell differentiation, function, and roles in disease, Immunity 41, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer CT, Gazumyan A, Kara EE, Gitlin AD, Golijanin J, Viant C, Pai J, Oliveira TY, Wang Q, Escolano A, Medina-Ramírez M, Sanders RW, Nussenzweig MC, The microanatomic segregation of selection by apoptosis in the germinal center, Science 358, eaao2602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang M-Y, Longo NS, Dimitrov DS, Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens, Biochem. Biophys. Res. Commun 390, 404–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L, HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies, Science 346, 1380–1383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, McDermott AB, Yewdell JW, Defining B cell immunodominance to viruses, Nat Immunol 18, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode, Meth. Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, MolProbity: all-atom structure validation for macromolecular crystallography, Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labonte JW, Adolf-Bryfogle J, Schief WR, Gray JJ, Residue-centric modeling and design of saccharide and glycoconjugate structures, J Comput Chem 38, 276–287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H, Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning, Journal of Immunological Methods 329, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brochet X, Lefranc M-P, Giudicelli V, IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis, Nucleic Acids Res 36, W503–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooks GE, Hon G, Chandonia J-M, Brenner SE, WebLogo: a sequence logo generator, Genome Res 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M, Innate-like functions of natural killer T cell subsets result from highly divergent gene programs, Nat Immunol 17, 728–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Upadhyay AA, Kauffman RC, Wolabaugh AN, Cho A, Patel NB, Reiss SM, Havenar-Daughton C, Dawoud RA, Tharp GK, Sanz I, Pulendran B, Crotty S, Lee FE-H, Wrammert J, Bosinger SE, BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data, Genome Med 10, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao L, Diedrich JK, Kulp DW, Pauthner M, He L, Park S-KR, Sok D, Su CY, Delahunty CM, Menis S, Andrabi R, Guenaga J, Georgeson E, Kubitz M, Adachi Y, Burton DR, Schief WR, Yates JR III, Paulson JC, Global site-specific N-glycosylation analysis of HIV envelope glycoprotein, Nat Commun 8, 14954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gatlin CL, Eng JK, Cross ST, Detter JC, Yates JR, Automated identification of amino acid sequence variations in proteins by HPLC/microspray tandem mass spectrometry, Anal. Chem 72, 757–763 (2000). [DOI] [PubMed] [Google Scholar]
- 54.He J, Boegli M, Bruzas I, Lum W, Sagle L, Patterned Plasmonic Nanoparticle Arrays for Microfluidic and Multiplexed Biological Assays, Anal. Chem 87, 11407–11414 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Park SK, Venable JD, Xu T, Yates JR, A quantitative analysis software tool for mass spectrometry-based proteomics, Nat Meth 5, 319–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban Y-EA, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P, ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules, Meth. Enzymol 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mandell DJ, Coutsias EA, Kortemme T, Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling, Nat Meth 6, 551–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.