Abstract
Over the past decade, the field of molecular biology has rapidly incorporated epigenetic studies to evaluate organism-environment interactions that can result in chronic effects. Such responses arise from early life stage stress, the utilization of genetic information over an individual’s life time, and transgenerational inheritance. Knowledge of epigenetic mechanisms provides the potential for a comprehensive evaluation of multigenerational and heritable effects from environmental stressors, such as contaminants. Focused studies have provided a greater understanding of how many responses to environmental stressors are driven by epigenetic modifiers. We discuss the promise of epigenetics and suggest future research directions within the field of aquatic toxicology, with a particular focus on the potential for identifying key heritable marks with consequential impacts at the organism and population levels.
Keywords: Aquatic toxicology, Contaminants, Ecotoxicogenomics, Epigenetics, Stressors
Introduction
The pivotal role played by the epigenome in orchestrating the expression of genes that drive cellular differentiation and the response to environmental change (e.g., temperature, nutrition, pollution) is now undisputed. The epigenome is a suite of mitotically and/or meiotically heritable changes in gene function that occur via mechanisms other than direct changes in deoxyribonuclease (DNA) sequence. Epigenetic regulation of gene function is associated with a wide variety of mechanisms that both positively or negatively impact gene transcription or the production of protein products. Some of the more well‐known mechanisms include DNA methylation and modification of histones through the methylation or acetylation of lysine residues. Functional groups added to specific regulatory regions of DNA (either upstream of genes or bound to the tails of histones that provide DNA’s framework) dictate which regions of coding DNA are accessible to transcriptional machinery. Epigenetics involves the reduction or prevention of transcription via methylation of cytosine nucleobases in regulatory regions, as well as either decreased or increased transcription via methylation or acetylation of lysine in histone tails, respectively. The processes of histone modification and DNA methylation are directed by, and transcripts can be deactivated prior to, translation by short‐stranded noncoding ribonucleic acids (RNAs)1.
We now understand that these 3 major epigenetic mechanisms, DNA methylation, histone modification, and noncoding RNAs (ncRNAs), are shared across most taxa, and that the response to environmental stressors, including aquatic pollutants, is influenced by epigenetic modifiers 2. Thus there is great interest in the study of epigenetics in the many model and nonmodel organisms used for experimentation in the field of aquatic toxicology. In many cases organisms with rapid developmental life stages greatly facilitate investigations into somatic epigenetic change as well as the potential for epigenetic transgenerational inheritance.
Much of the current thinking about epigenetics and its role in adaptation and disease manifestation is derived from research with mammals and plants. Technical advances have greatly expanded our ability to interrogate the epigenome in new species, with greater resolution and on an unprecedented scale (epigenome wide). Moreover, much has been learned through the combination of epigenome‐wide studies and whole transcriptome studies3. Recent research has challenged the most fundamental understanding of the relationship between epigenetic mechanisms and phenotypic plasticity and has forced researchers to consider the degree to which results can be extrapolated across taxa4-6. Despite these unknowns, it is clear that epigenetics is a major engine driving adaptation, which has implications in many areas of basic and applied ecological research. In the present study we build on a previous Focus publication7, discussing new examples of epigenetic research specifically within the field of aquatic toxicology, along with suggested directions for new research. We also give an overview of emerging questions regarding recently recognized links between epigenetic change and classical Darwinian mechanisms of inheritance.
Epigenetic Mechanisms
Epigenetics includes both transgenerational (i.e., germline transmission of information) and nontransgenerational (i.e., somatic, mitotic stability) factors. Epigenetic information can be maintained after cell division, can spread between cells, and can even pass via the germline from one generation to the next. Because of transgenerational factors, DNA methylation is the most broadly assessed mechanism in the study of multigenerational impacts resulting from environment–organism interactions. Nontransgenerational epigenetics can be used to evaluate disease progression during the lifetime of an individual, and to associate events from early life stages that chronically affect growth, development, and reproduction8. However, so‐called nontransgenerational, somatic cell effects can become transgenerational, most noticeably in parthenogenetic individuals (e.g., some cladoceran species), because this form of asexual reproduction results in direct germline transmission of epigenetic information.
By far the best‐studied type of epigenetic mark is DNA methylation. Most commonly this involves the addition of a methyl group to the pyrimidine ring of a cytosine base, giving rise to a 5‐methylcytosine (5‐mC). Methylation of cytosines, often those contained within a (cytosine‐phosphodiester‐guanine [CpG]) island upstream of a gene promoter, usually results in decreased expression of the affected gene because 5‐mCs prevent or inhibit access by transcription factors9. However, as the number of genome‐wide association studies and now epigenome‐wide association studies continue to grow, it is becoming evident that, at least on the genome scale, the relationship between gene expression and methylation levels is not as clear as once believed.
Although DNA methylation can repress gene transcription by blocking the binding of the transcriptional machinery to DNA, the primary mechanism for modulating gene activity is through the modification of chromatin structure (Figure 1). Chromatin, the framework around which DNA is wrapped, exists in 2 distinct formations: euchromatin or heterochromatin. Loosely wound euchromatin is accessible by RNA polymerase II and transcription factors that can range from general coactivators or corepressors to specific nuclear receptor dimers or monomers (i.e., bound estrogen or glucocorticoid receptors). Heterochromatin, around which DNA is tightly assimilated, generally cannot be easily accessed by the multitude of proteins that regulate gene expression. Evidence is mounting that ncRNAs, DNA methylation, and histone modification act in concert to direct chromatin structure and the expression of associated genes1.
Figure 1.
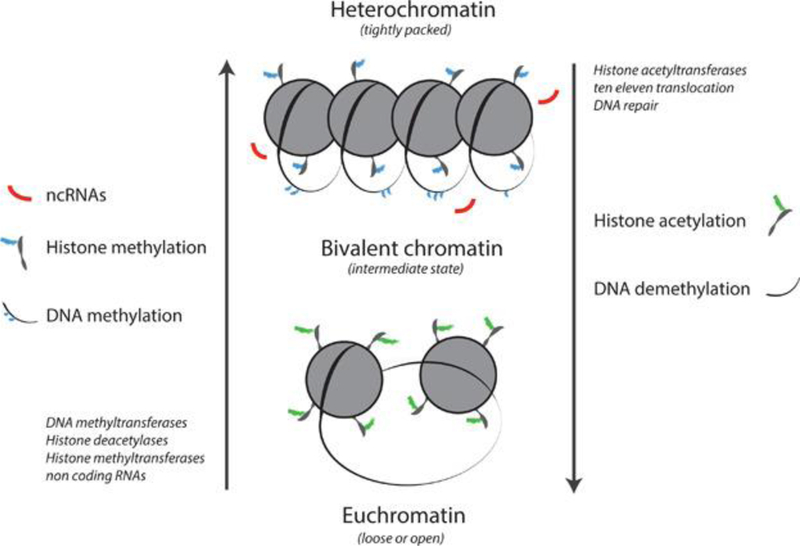
Chromatin exists as euchromatin or heterochromatin, with an intermediate bivalent state. Loosely wound euchromatin is accessible by RNA polymerase II and transcription factors. The genes contained within heterochromatin generally cannot be expressed. DNA methyltransferases, histone deacetylases, and methyltransferases produce a heterochromatic state, whereas histone acetyltransferases and ten‐eleven translocation, along with some types of DNA repair, contribute to a euchromatic state. Evidence suggests that noncoding RNAs, DNA methylation, and histone acetylation and histone methylation act together to influence chromatin structure and the expression of associated genes.
Histone acetylation and methylation, a highly conserved process, involves the addition of one of these functional groups to a lysine (K) on the tail of a specific histone7. It is widely established that lysine 9 on histone H3 (H3K9) is one of the most common sites for methylation (which can be heritable), whereas H3K27 is frequently acetylated. Methylation or acetylation at one of these sites induces a heterochromatic or euchromatic state, respectively. In contrast, methylation of H3K4 results in increased association with active promoter elements. Thus, in terms of histone modifications, the relationship between chromatin structure and acetylation or methylation status may be even less clear‐cut than 5‐mC tags on DNA. In fact, recent research has confirmed the existence of so‐called bivalent chromatin, which is intermediate between euchromatin and heterochromatin1. Histone variants, which differ slightly in amino acid sequence from their commonly present siblings, also appear to play a large and highly conserved role in the remodeling of chromatin. For example, the variant H3.3 is present in actively transcribed chromatin, whereas macroH2A.1 and the isoform H2A.2 can have either positive or negative effects on transcription depending on the gene of interest. These variants appear to be conserved across phylogenetic groups2,10.
While known to also be an important component of the epigenetic machinery, the role of ncRNAs is currently the least well‐understood mechanism. These ncRNAs govern 2 different types of interrelated activities, on the one hand interfering with already transcribed messenger RNA by binding to it and preventing translation (microRNAs [miRNAs]), and on the other directing the remodeling of chromatin to silence specific genes11. These changes appear to be capable of persisting through many rounds of mitosis. Studies that have used injectable ncRNAs have found that the miRNA possessed by sperm can reduce maternal transcripts in early zygotes, reprogramming the phenotype for stress response12.
Modifications of histones and DNA are initiated, removed, and maintained through the action of a number of proteins and protein complexes. Methylation status of genes across the genome is established by the de novo DNA methyltransferases DNMT3a and b, which methylate previously unmethylated DNA, and maintained by DNMT1, which copies pre‐existing methylation during replication13. Mechanisms of demethylation are less well understood and were, until recently, considered a passive process, whereby methylation was lost because of reduced DNMT1 activity. However, mechanisms of active demethylation have been identified. One such mechanisms relies on the ten‐eleven translocation family of proteins, which, through a series of reactions converts 5‐mC to 5‐hydroxymethylcytosine and eventually to 5‐formylcytosine and 5‐carboxylcytosine, which are subsequently converted to unmethylated cytosine through thymine–DNA glycosylase activity14. Alternative demethylation pathways involve DNA repair pathways and associated proteins (base excision repair glycosylases and Gadd45)15. Histone methylation is accomplished through the action of the histone lysine methyltransferases, such as the SET‐domain protein methyltransferase family16.
Acetylation is another histone modification process associated with epigenetic regulation, where the presence of acetyl groups signifies active genes. Acetylation is accomplished through the actions of a diverse group of histone acetytransferases that usually occur in large multi‐subunit complexes, where the constituents of the complex target the histone acetytransferase activity17. Acetylation of histones is reversible, and transcriptional levels of acetylated genes are modulated through the addition and removal of acetyl groups through an interplay of histone acetytransferases and histone deacetylases. The DNA methylation and histone modification are linked through proteins containing methyl‐binding domains. The methyl‐binding domain proteins bind methylated DNA and recruit multiprotein complexes resulting in histone modification and remodeling. The functional significance of this is that methyl‐binding domain proteins are able to either activate or repress gene activity18.
Early Life Exposures
It has been established, at least in vertebrates, that organisms begin life with an epigenetic blueprint. Shortly after fertilization, however, prior epigenetic marks are erased, and the genome undergoes de novo methylation. This occurs again at approximately the time of birth or hatching in primordial germ cells, which will eventually mature and develop into gametes that may pass this information to the next generation. However, some of the similarities between what is established in mammals and what has been observed in the organisms (i.e., fish) used to study the effects of water‐borne pollutants may end there (Figure 2). Much of what is known regarding epigenetics in aquatic organisms results from the study of zebrafish (Danio rerio). For example, in zebrafish it appears that genomic imprinting does not occur as it does in mammals19. Furthermore, CpG island number is lower compared with mammalian genomes, whereas the methylation levels of these regions is comparatively higher in zebrafish19.
Figure 2.
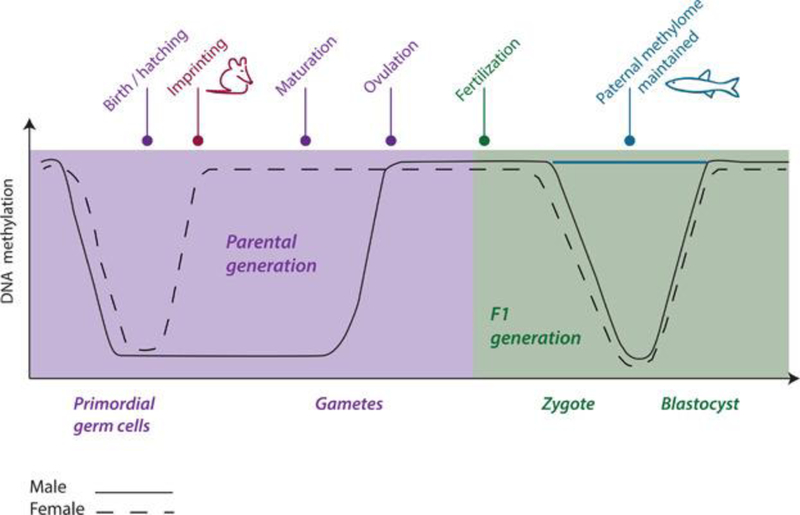
A comparison of time points for erasure and reprogramming of the DNA methylome prior to fertilization and during embryogenesis in mammals and fish. The methylome is erased and reprogrammed in primordial germ cells at approximately the time of birth or hatching. Imprinting follows in mammals, but has not been established in fish. Following fertilization (F1) the methylome is again erased and reprogrammed between the zygote and blastocyststage in male and female mammals, but the paternal methylome appears to be maintained in fish at this stage. Adapted from Head, 2014 23.
The developmental origins of health and disease is a widely accepted paradigm that can be applied across taxa. This expression refers to early life exposures, during the sensitive periods of embryogenesis or the young juvenile period, that result in adverse or diseased adult phenotypes (i.e., metabolic disorders, infertility) as a result of somatic epigenetic transmission. Environmental chemicals are hypothesized to interfere with the process of methylation in primordial germ cells and/or to cause the addition of de novo methyl tags. A recent study in zebrafish measured global methylation and site‐specific addition or removal of 5‐mCs following exposure to several classes of compounds (endocrine disruptors, heavy metals, pharmaceuticals) from 0 to 72 d post fertilization. It was demonstrated that while all exposures resulted in morphological deformities, only one compound (5‐azacytidine), a known inhibitor of DNA methylation, caused significant changes in global methylation. However, several endocrine‐disrupting compounds ([EDCs], bisphenol A [BPA], diethystilbestrol, and 17α‐ethinylestradiol [EE2]) caused locus‐specific alterations in methylation activity, particularly at the vasa gene, which is important for germ cell development20. Other studies19 have shown changes in global methylation levels in zebrafish following exposure to environmental toxicants, specifically for genes involved in early development, metabolism, reproduction, and oncogenesis. Nonmodel species such as the self‐fertilizing mangrove killifish (Kryptolebias marmoratus; Figure 3), and the inland silverside (Menidia beryllina), a euryhaline group spawner, are also sensitive to early life exposure, which results in negative phenotypic outcomes in adults or F1 offspring exposed as gametes21,22. As mentioned above, fish in general have a higher percentage of global DNA methylation in comparison with other vertebrates, and in addition it appears that the paternal methylome plays a stronger role than that of the mother during embryogenesis (Figure 2)23. Thus continued study of the epigenome in zebrafish and other ecotoxicologically relevant fish species is of great importance, because it is evident there are vast differences between higher and lower vertebrates.
Figure 3.

Nonmodel species such as the self‐fertilizing mangrove killifish (Kryptolebias marmoratus) are being used to investigate epigenetic mechanisms. Image sourced from F. Silvestre, Laboratory of Evolutionary and Adaptive Physiology.
Two of the best studied invertebrates with regard to epigenetics are Daphnia species (invertebrate cladoceran) and the Pacific oyster (Crassostrea gigas). Although invertebrate methylation levels are variable compared with the more highly methylated genomes of vertebrates9, there are many similarities between aquatic invertebrates and fish. Similar to fish, the embryonic stage of daphnid species appears to be most sensitive to the alteration of epigenetic tags via the influence of environmental cues, such as temperature changes or pollutant exposure. Typically, progeny are genetically identical, because under normal conditions offspring are produced via parthenogenesis, which makes cladocerans an ideal organism for the study of epigenetically influenced phenotypes24. It has already been established that Daphnia magna possesses the same DNA methytransferases as humans and fish and that DNA methylation occurs. Global methylation levels are altered by exposure to chemicals such as 5‐azacytidine (also in zebrafish), vinclozolin, genistein, and zinc. Furthermore, it has been demonstrated that histones H3 and H4 are both methylated and acetylated, also similar with respect to vertebrates2.
In oysters, specifically C. gigas, the methylome has been evaluated in many tissue types and developmental stages9. Thus it is known that DNA methylation occurs and is correlated with gene expression25, and that (as with other organisms) the sensitivity of early embryonic stages should be emphasized in future studies. Investigations into the nature and timing of epigenetic reprogramming in the early life of invertebrates and comparisons with what is known in fish and mammals in the context of response to environmental stressors are greatly needed.
Transgenerational Effects
Epigenetic transgenerational inheritance is described as transmission of epigenetic tags via the germline, without direct exposure of the affected generation26. In mammals it is established that EDCs such as phthalates and diethylstilbestrol (now banned) cause increased DNA methylation, and that these epigenetic tags lead directly to adverse reproductive health outcomes. However, the large body of work on transgenerational epigenetic effects is based on the placental model of development, which allows for the exposure of both the F1 (fetus) and F2 (fetal germ cells) generations during mammalian gestation. In the majority of nonmammalian models, such as those used in aquatic toxicology research, transgenerational effects manifest in the F2 generation of F0 exposed individuals, because it is only possible for the F1 generation to be directly exposed (as unfertilized eggs or preejaculated sperm). However, in organisms that host the development of their young, such as live bearing fish or cladocerans (i.e., D. magna), true transgenerational effects cannot be observed until the F3 generation (Figure 4), because even the F2 generation can be directly exposed while the germ cells or offspring, respectively, develop inside the mother24. Although carrying studies out to the F3 generation and beyond can pose a challenge, typical test species used in aquatic toxicology are short‐lived, with relatively brief generation times, making such experiments more feasible.
Figure 4.
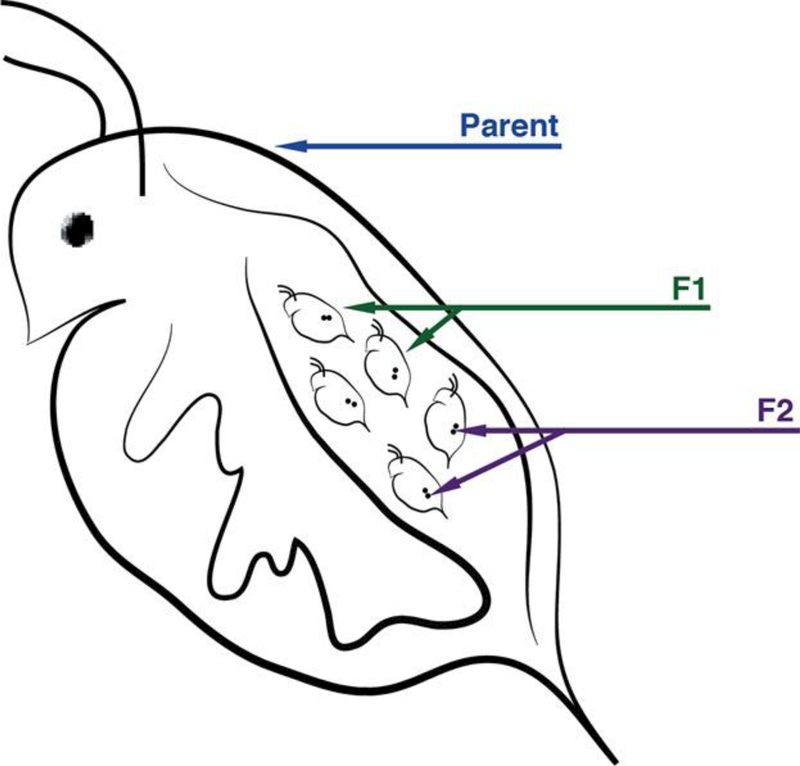
Cladoceran species (e.g., Ceriodaphnia dubia, Daphnia magna) hold great promise as model organisms for epigenetic research. Progeny are typically genetically identical, because offspring are normally produced via parthenogenesis, and thus F2 generation germ cells can be directly exposed during F1 generation development in the brood pouch. Environmentally induced phenotypes have already been documented, as has DNA methylation.
Transgenerational studies have been performed in relatively few species, but existing studies suggest that early life exposures (discussed in the section Early life exposures) can result in altered phenotypes for several subsequent generations. In medaka fish (Oryzias latipes), exposure of embryos to BPA or EE2 for the first 7 d following fertilization resulted in reduced fertilization rates in the F2 and F3 adults and hence reduced survival rates in the F3 and F4 embryos, but no effects were observed in F0 or F1. Notably, analytical chemistry deduced that each F0 embryo absorbed a mere pg/mg (part per trillion) concentration of contaminant on average27. Parental early life exposure to benzo[a]pyrene in zebrafish resulted in deformities that continued through to the F2 and F3 generations28. What is missing from these studies is the mechanism by which these phenotypic abnormalities were transferred to generations not directly exposed to the respective toxicants. Furthermore, in fish, it appears that the paternal exposure history may be more important than that of the mother, because in zebrafish paternal methylation marks are maintained while maternal methylation is erased23. Studies that compare transgenerational effects between maternal and paternal exposures across aquatic test species are greatly needed.
In invertebrates such as Daphnia species, it has been posited that intersex in offspring produced during environmental challenges (altered temperature or other stressors) results from transgenerational inheritance of epigenetic modifications, but this has yet to be proved experimentally24. As mentioned previously, what makes this model unique is that offspring are usually genetically identical, and thus any phenotypic differences between siblings produced via parthenogenesis would be the result of epigenetic modifications. Although data are currently lacking, evidence that daphnids undergo transgenerational epigenetic inheritance exists. For example, daphnids do produce intersex offspring under certain environmental conditions. Mothers that are removed from the conditions that triggered intersex progeny continue to produce them for some time. Other daphnid phenotypes such as the formation of helmets and neckteeth, which are induced by predator cues (e.g., kairomones) and some synthetic chemicals, also appear to be inherited across generations rather than after direct exposure24. Future studies should evaluate the presence of inducible phenotypes out to at least the F3 and F4 generations, which is highly feasible in an organism with a generation time ranging from a few days to a few weeks. It is notable that plants, which are by far the most extensively studied group of living organisms when it comes to epigenetics, can pass phenotypically inherited traits for hundreds of generations26. We have likely only begun to scratch the surface of the potential for comprehending such transmission in relatively understudied aquatic organisms.
Rapid Evolution
Transgenerational inheritance of epigenetic marks appears to facilitate rapid evolution via increasing phenotypic plasticity. However, most studies still consider epigenetically triggered phenotypic change as being separate from classical Darwinian inheritance. Recent research suggests that epigenetic change often precedes and may even trigger or increase the likelihood for mutations resulting in altered DNA sequence. Thus epigenetic variation is the mechanism by which organisms can adapt to rapid environmental perturbations, such as pollution, but these modifications on the surface of DNA or histones can in turn increase the probability that a genomic region might mutate29. This has been demonstrated with transposons, the activity of which may be suppressed by a high degree of DNA methylation but mobilized via other epigenetic states. Furthermore, the frequency of both translocations and inversions can be influenced by all 3 previously described main epigenetic mechanisms. In fact, it has been noted by evolutionary biologists that there is a mismatch between rates of mutation at the genotypic and phenotypic levels, and that epigenetic change appears to be the missing puzzle piece between the two26. Such changes may be beneficial to the species at hand if the parental environment matches that of the offspring or grand‐offspring and beyond, but considering the often pulse‐like or seasonal nature of pollutant introduction in the wild, this may not hold true for the study of long term transgenerational effects induced via toxicant exposure and could result in a so called epigenetic trap29. Furthermore, existing studies on the long‐term adaptation of aquatic species to pollutants or exposure to other stressors show that trade‐offs or evolutionary constraints may be inherent in the process of evolving to tolerate toxicants30, but this may be dependent on the species, the length of exposure, and the type of stressor.
The evolutionary implications of this hypothesis are colossal, considering that the underpinnings of Darwinian inheritance (changes in DNA sequence) relied on a rejection of Lamarck’s hypothesis, which was that traits could be inherited from one’s environment. In essence this implies that the Lamarckian basis for inheritance may facilitate rapid evolution while providing the stimulus for mutations that enable long‐term adaptation26 (Figure 5). Put simply, the two processes appear to be highly integrated with and potentially reliant on each other, and thus should be studied together when possible, particularly in the context of exposure to environmental stressors.
Figure 5.

Recent research 26 suggests that epigenetic change often precedes and may even trigger or increase the likelihood for mutations resulting in altered DNA sequence. Thus epigenetic variation is the mechanism by which organisms can adapt to rapid environmental perturbations, such as pollution, but these modifications on the surface of DNA or histones may in turn increase the probability that a genomic region might mutate, facilitating adaptation over the long term. This suggests that the Lamarckian basis for inheritance facilitates rapid evolution while providing the stimulus for mutations that enable long‐term Darwinian adaptation. Images sourced from Wikimedia.
Analytical Approaches
Standard molecular biology and biochemical techniques, and more recently high‐throughput sequencing approaches, can be used to determine epigenetic modifications of DNA, histones, and ncRNAs. Powerful approaches such as the low‐cost reduced representation bisulfite sequencing and chromatin immunoprecipitation assays that use antibodies for specific histone modifications are commonly used for investigating DNA methylation and histone modification, respectively31. Because promoter regions are relatively free of nucleosomes, there are now a number of genome‐wide approaches that allow the identification of these regions, as well as techniques that detect protein‐mediated DNA interaction sites and DNA binding proteins32. These techniques, as with most high‐throughput molecular approaches, are developing rapidly alongside bioinformatics approaches that integrate different analytical platforms, such as the analysis of DNA methylation data alongside gene expression31. We recommend the recent publications by Yong et al.31, and Kagohara et al.32, which provide thorough reviews of these techniques, as well as their utility for transgenerational and nontransgenerational studies31,32.
Epigenetics in an Ecotoxicological Framework
It has become clear that epigenetic mechanisms allow organisms to adapt to local environments and environmental cues. Epigenetic markers have been shown to be altered following chemical exposure. There is a well‐established linkage among epimutations, dysregulation of epigenetic enzymes (e.g., DNMTs), and the manifestation of disease states33. All these findings suggest that epigenetics is obviously relevant to our understanding of the impacts of pollutant release into environmental media, an area that remains relatively poorly studied. Epigenetics may be useful for understanding and predicting differences in susceptibility within and among populations. For example, hypermethylation of CYP enzymes may result in increased sensitivity to chemical exposure. This has been observed in the human pharmacological arena, where it has clear clinical relevance34. The ubiquity of epigenetic modification and disease manifestation strongly suggests that they have clear relevance to the development of adverse outcome pathways35. For example, epigenetic changes have been found in all cancer types studied and are often found in early stages of tumorigenesis34. As more detailed information on epigenetic mechanisms comes to light across taxa, it is possible that DNA methylation, histone modification, or ncRNAs could eventually be considered causal molecular initiating events. To this end, epigenetics, used in combination with other omics modalities and traditional endpoints focused on different levels of the biological hierarchy, may be useful in unraveling causes of ecological impairment in complex exposure scenarios, particularly those involving multiple stressors. Different endpoints may provide information on different temporal scales, which may be useful in understanding when the critical exposure occurred. Moreover, differentially methylated genes may provide insights into impacted biological pathways, providing clues as to additional potential drivers of adverse outcomes. The role of epigenetics in understanding the dynamics of ecosystem function and response to stressors remains poorly characterized. It is clear, however, that epigenetics provides the foundation for local and short‐term adaptation to environmental change and that epigenetic dysregulation is a key player in the development of adverse outcomes. The breadth of applications for epigenetics within ecotoxicology and associated environmental regulation are just now being considered, and it is up to the aquatic research community to fill in the many existing gaps in knowledge for model and nonmodel species alike.
Questions for future research.
How direct is the connection between epigenetic tags in the promoter region of a gene (DNA methylation) and reduced gene expression?
Is transgenerational inheritance and the potential for rapid evolution driven by pollutant exposure caused by integrated changes between the epigenome and DNA mutations?
Does transgenerational inheritance consistently persist beyond the F2 or F3 generation (depending on reproductive mode) in aquatic toxicology model and nonmodel species?
How do multiple stressors (temperature, acidification, hypoxia) influence epigenetic and hence genetic change, in the context of pollutant exposure under global climate change?
How quickly can aquatic organisms adapt to pollutant or stressor exposure? Are there trade‐offs or evolutionary constraints?
What are the implications of parental exposure of the father, versus the mother? In fish, for example, is paternal exposure history of more importance across species?
Acknowledgment
Funding from the Environmental Protection Agency (EPA STAR # 835799, to S.M. Brander and R.E. Connon) made this work possible.
Disclaimer
The views expressed in this Focus article are those of the authors and do not necessarily represent the views or policies of the US Environmental Protection Agency.
Definitions
- Adverse outcome pathway (AOP)
A structured representation of biological events leading to adverse effects that are relevant to risk assessment.
- Bivalent chromatin
Chromatin that is intermediate between heterochromatin and euchromatin
- Blastocyst
A mammalian blastula in which some differentiation of cells has occurred, the inner cell mass of the blastocyst will become an embryo.
- CpG island
Regulatory regions of DNA where cytosine (C) nucleobases occur adjacent to a guanine (G) in the linear sequence of bases (sharing a phosphodiester bond – p) at a high frequency, often in a promoter region where transcription factors bind.
- DNA methylation
Methyl groups added by DNA methyltransferases to cytosine nucleobases, which typically cause a decrease in the expression of upstream genes.
- Endocrine‐disrupting compound
A chemical capable of imitating, inhibiting, or interfering with the action or production of endogenous hormones.
- Epigenetic transgenerational inheritance
The transmission of epigenetic tags or states via the germline without direct exposure of the affected generation.
- Epigenetic trap
A maladaptive epigenetic response caused when evolutionarily novel factors (e.g., pollutants, other temporary stressors) disrupt epigenetic machinery.
- Epigenome
A suite of mitotically or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence.
- Epimutation
An aberrant chromatin state leading to altered gene expression patterns that occurs in the absence of a change in DNA sequence.
- Euchromatin
Chromatin that is loosely held together resulting in DNA being accessible by transcription factors and RNA polymerase.
- Heterochromatin
Chromatin that is tightly held together resulting in DNA being inaccessible by transcription factors and RNA polymerase.
- Histone acetylation
Acetyl groups added by histone acetyltransferases to lysine residues present in histone tails, generally resulting in open or euchromatin and increased expression of associated genes.
- Histone variant
Histones that differ slightly in amino acid sequence from common varieties and play a role in the remodeling of chromatin (e.g., H3.3).
- Methyl‐binding domain protein
Proteins that bind methylated DNA and recruit multiprotein complexes resulting in histone modification and remodeling, activating or repressing gene activity.
- Methylome
The set of nucleic acid methylation modifications in an organism’s genome or in a particular cell.
- ncRNA
Noncoding RNAs that bind to and thus interfere with the translation of mRNA transcripts into proteins or direct chromatin remodeling.
- Nucleosome
A structural unit of a eukaryotic chromosome that consists of a length of DNA coiled around a core of histones.
- Parthenogenesis
A form of asexual reproduction in an embryo develops from an unfertilized egg, resulting in a genetically identical copy of the parent.
- Primordial germ cells
Cells in an embryo that develop into stem cells that generate reproductive gametes (sperm and eggs) at reproductive maturation.
- RNA polymerase II
A protein–protein complex that catalyzes the transcription of DNA to generate pre‐mRNA and noncoding RNA.
- Somatic epigenetic transmission
The transmission of epigenetic tags or states during mitosis, within an organism’s lifetime.
- Zygote
A diploid cell resulting from the fusion of two haploid gametes; a fertilized egg (ovum).
References
- 1.Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17:487–500. [DOI] [PubMed] [Google Scholar]
- 2.Vandegehuchte M, Janssen C. 2011. Epigenetics and its implications for ecotoxicology.Ecotoxicology 20:607–624. [DOI] [PubMed] [Google Scholar]
- 3.Lou S, Lee HM, Qin H, Li JW, Gao Z, Liu X, Chan LL, Kl Lam V, So WY, Wang Y, Lok S, Wang J, Ma RC, Tsui SK, Chan JC, Chan TF, Yip KY. 2014. Whole‐genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol 15:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome‐wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. [DOI] [PubMed] [Google Scholar]
- 5.Lee TF, Zhai J, Meyers BC. 2010. Conservation and divergence in eukaryotic DNA methylation.Proc Natl Acad Sci U S A 107:9027–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Hoshida Y, Sadler KC. Comparative epigenomic profiling of the DNA methylome in mouse and zebrafish uncovers high interspecies divergence. Front Genet 7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Head JA, Dolinoy DC, Basu N. 2012. Epigenetics for ecotoxicologists. Environ Toxicol Chem 31:221–227. [DOI] [PubMed] [Google Scholar]
- 8.Walter J, Hümpel A. 2017. Introduction to epigenetics. In Epigenetics. Springer Fachmedien, Wiesbaden, Germany, pp 11–29. [Google Scholar]
- 9.Hofmann GE. 2017. Ecological epigenetics in marine metazoans. Front Mar Sci 4:4. [Google Scholar]
- 10.Rivera‐Casas C, Gonzalez‐Romero R, Cheema MS, Ausió J, Eirín‐López JM. 2016. The characterization of macroH2A beyond vertebrates supports an ancestral origin and conserved role for histone variants in chromatin. Epigenetics 11:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou L, Wang D, Baccarelli A. 2011. Environmental chemicals and microRNAs. Mutat Res/Fundamen Mol Mech Mutagen 714:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodgers AB, Morgan CP, Leu NA, Bale TL. 2015. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A 112:13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodenhiser D, Mann M. 2006. Epigenetics and human disease: Translating basic biology into clinical applications . Can Med Assoc J 174:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohli RM, Zhang Y. 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillon SC, Zhang X, Trievel RC, Cheng X. 2005. The SET‐domain protein superfamily: Protein lysine methyltransferases . Genome Biol 6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KK, Workman JL. 2007. Histone acetyltransferase complexes: One size doesn’t fit all. Nat Rev Mol Cell Biol 8:284–295. [DOI] [PubMed] [Google Scholar]
- 18.Bogdanović O, Veenstra GJC. 2009. DNA methylation and methyl‐CpG binding proteins: Developmental requirements and function . Chromosoma 118:549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamstra JH, Aleström P, Kooter JM, Legler J. 2015. Zebrafish as a model to study the role of DNA methylation in environmental toxicology. Environ Sci Pollut Res 22:16262–16276. [DOI] [PubMed] [Google Scholar]
- 20.Bouwmeester MC, Ruiter S, Lommelaars T, Sippel J, Hodemaekers HM, van den Brandhof E‐J, Pennings JLA, Kamstra JH, Jelinek J, Issa J‐PJ, Legler J, van der Ven LTM. 2016. Zebrafish embryos as a screen for DNA methylation modifications after compound exposure. Toxicol Appl Pharmacol 291:84–96. [DOI] [PubMed] [Google Scholar]
- 21.Voisin A‐ S, Fellous A, Earley RL, Silvestre F. 2016. Delayed impacts of developmental exposure to 17‐α‐ethinylestradiol in the self‐fertilizing fish Kryptolebias marmoratus. Aquat Toxicol 180:247–257. [DOI] [PubMed] [Google Scholar]
- 22.DeCourten BM, Brander SM. 2017. Combined effects of increased temperature and endocrine disrupting pollutants on sex determination, survival, and development across generations. Sci Rep, in press. DOI: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Head JA. 2014. Patterns of DNA methylation in animals: An ecotoxicological perspective . Integr Comp Biol 54:77–86. [DOI] [PubMed] [Google Scholar]
- 24.Harris KDM, Bartlett NJ, Lloyd VK. 2012. Daphnia as an emerging epigentic model organism.Genet Res Int 2012:147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavery MR, Roberts SB. 2013. Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc. PeerJ 1:e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skinner MK. 2015. Environmental epigenetics and a unified theory of the molecular aspects of evolution: A neo‐Lamarckian concept that facilitates neo‐Darwinian evolution. Genome Biol Evol 7:1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhandari RK. 2016. Medaka as a model for studying environmentally induced epigenetic transgenerational inheritance of phenotypes. Environ Epigenet 2:dvv010–dvv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrales J, Thornton C, White M, Willett KL. 2014. Multigenerational effects of benzo[a]pyrene exposure on survival and developmental deformities in zebrafish larvae. Aquat Toxicol 148:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Dea RE, Noble DWA, Johnson SL, Hesselson D, Nakagawa S. 2016. The role of non‐genetic inheritance in evolutionary rescue: Epigenetic buffering, heritable bet hedging and epigenetic traps. Environ Epigenet 2:dvv014–dvv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid NM, Proestou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR, Karchner SI, Hahn ME, Nacci D, Oleksiak MF, Crawford DL, Whitehead A. 2016. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science 354:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yong W‐ S, Hsu F‐ M, Chen P‐ Y. 2016. Profiling genome‐wide DNA methylation. Epigenet Chromatin 9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kagohara L, Stein O, Brien G, Kelley D, Flam E, Wick H, Danilova L, Easwaran H, Favorov A, Qian J, Gaykalova D, Fertig E. 2017. Epigenetic regulation of gene expression in cancer: Techniques, resources, and analysis. bioRxiv. 10.1101/114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirbhahai L, Yin G, Bignell JP, Li N, Williams TD, Chipman JK. 2011. DNA methylation in liver tumorigenesis in fish from the environment. Epigenetics 6:1319–1333. [DOI] [PubMed] [Google Scholar]
- 34.Lam Y‐W, Cavalarri LH. eds. 2013. Pharmacogenomics: Challenges and Opportunities in Therapeutic Implementation. Academic Press, Cambridge, MA, USA [Google Scholar]
- 35.Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL. 2010. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem 29:730–741. [DOI] [PubMed] [Google Scholar]


