Abstract
Sensory hair-cell development, function, and regeneration are fundamental processes that are challenging to study in mammalian systems. Zebrafish are an excellent alternative model to study hair cells because they have an external auxiliary organ called the lateral line. The hair cells of the lateral line are easily accessible, which makes them suitable for live, function-based fluorescence imaging. In this chapter, we describe methods to perform functional calcium imaging in zebrafish lateral-line hair cells. We compare genetically encoded calcium indicators that have been used previously to measure calcium in lateral-line hair cells. We also outline equipment required for calcium imaging and compare different imaging systems. Lastly, we discuss how to set up optimal imaging parameters and how to process and visualize calcium signals. Overall, using these methods, in vivo calcium imaging is a powerful tool to examine sensory hair-cell function in an intact organism.
INTRODUCTION
The zebrafish model system has many attributes that make it an ideal system for live, physiology-based fluorescence imaging. Zebrafish larvae are easy to genetically manipulate and are optically transparent (Patton & Zon, 2001). These features have allowed researchers to use the zebrafish system to express genetically encoded calcium indicators (GECIs) to visualize functional activity in vivo (Amsterdam, Lin, & Hopkins, 1995; Higashijima, Masino, Mandel, & Fetcho, 2003). While these advantages are now being rapidly applied to explore whole brain imaging in zebrafish, they are also applicable to studying hair cells of the auditory and vestibular organs in zebrafish (Ahrens, Orger, Robson, Li, & Keller, 2013).
Zebrafish are particularly well suited for studying hair cells because they have an external vestibular organ, the lateral line, which is easily accessible and readily amenable to live imaging. The lateral line is composed of groups of sensory hair cells arranged in rosette structures known as neuromasts. Neuromasts have hair bundlesd apical projections from hair cellsdthat protrude from the surface of the body and detect water movements which is important for schooling, mating, and predator avoidance behaviors (Freeman, 1928). The accessibility of neuromast hair cells in vivo offers a significant advantage over mammalian auditory and vestibular systems, where the sensory organs are encased in a bony labyrinth and must be excised for study.
As a result of its easy accessibility, researchers have used the lateral line to study a diversity of topics, including the genetics of hearing loss, hair-cell development, function, ototoxicity, and regeneration (Kindt, Finch, & Nicolson, 2012; Lush & Piotrowski, 2014; Owens et al., 2008; Sarrazin et al., 2010). However, relatively few studies have comprehensively examined hair-cell function. Many studies have relied on styryl dyes such as DASPEI and FM 1–43 to indicate whether hair cells have mechanotransduction channels and are viable (Lush & Piotrowski, 2014; Owens et al., 2008). FM-dye labeling is a simple and straightforward method, but it lacks detailed and quantifiable information regarding hair-cell function. Alternatively, extracellular microphonic measurements or whole-cell recording of hair-cell currents have been used to examine hair-cell function (Corey et al., 2004; Olt, Johnson, & Marcotti, 2014; Ricci et al., 2013; Trapani & Nicolson, 2010). Although extremely powerful, these techniques are technically challenging and require highly specialized equipment and expertise. In the case of microphonics, measurements represent the summed response of all hair cells in a neuromast, and in the case of whole-cell recordings, measurements are from a single hair cell. On the other hand, techniques that use GECIs provide better spatial resolution. Specifically, calcium imaging methods enable measurements of mechanically evoked activity in all of the individual hair cells in a neuromast simultaneously in a reproducible and quantifiable manner. In addition, an imaging-based approach can be a more straightforward way to examine hair-cell activity for researchers more familiar with microscopy than electrophysiology.
In this chapter, we describe methods used to image calcium activity in hair cells of the zebrafish lateral line. We also discuss how to select a GECI and compare several indicators that have been used previously for calcium imaging in the lateral line. We then outline several suitable optical imaging setups and how to optimize settings for capturing calcium signals. Finally, we address how to process the captured images. We hope our description of these methods will extend the possibilities for hair-cell research in zebrafish.
1. CALCIUM INDICATOR SELECTION AND COMPARISON
1.1. SELECTING A CALCIUM INDICATOR
GECIs are a powerful tool for simultaneous measurement of mechanically evoked calcium responses in multiple hair cells. To appreciate the implications of these signals in the context of a hair cell, it is important to consider what a mechanically evoked calcium response represents. In hair cells, a mechanically evoked calcium response is the sum of many different calcium sources, mainly sites of mechanotransduction and synaptic transmission (Beurg et al., 2010, 2010; Gillespie & Müller, 2009; Jaalouk & Lammerding, 2009; LeMasurier & Gillespie, 2005; Vollrath, Kwan, & Corey, 2007). In response to mechanical hair-bundle deflection, calcium and other cations enter apically through mechanotransduction channels located on the sensory hair bundle. This apical influx of cations changes the receptor potential and triggers activation of voltage-gated calcium channels that then allow the calcium influx required for synaptic transmission at the basal end of the cell. Thus, calcium measurements can offer a good representation of the significant, activity-dependent signals in hair cells.
Currently there are two main types of GECIs. One type includes the fluorescence resonance energy transfer (FRET)-based indicators that rely on two spectrally distinct fluorescent proteins such as yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP). The second type of GECIs is non-FRET-based and relies on a single fluorescent protein. Both types of indicators include a calmodulinecalcium binding domain that alters the fluorescence of the indicator in response to calcium concentration changes (Tian, Hires, & Looger, 2011). For FRET-based calcium indicators, such as cameleons, an increase in the YFP/CFP ratio indicates an increase in calcium (Miyawaki et al., 1997). For non-FRET-based indicators, such as GCaMPs, calcium binding directly alters fluorescence of the single fluorescent protein in a dose-dependent manner. FRET indicators require additional equipment beyond non-FRET-based indicators so that the two fluorescent proteins (eg, YFP and CFP) can be recorded simultaneously. These devices include a “beam splitter” or a dual-chip camera to capture the two emission spectra simultaneously, or alternatively, a high-speed filter turret for sequential image capture. Despite these equipment requirements, FRET-based indicators can be very powerful when measuring fluorescent signals in a live organism. The main advantage is that FRET-based indicators make it easy to identify and discard image acquisitions with movement artifacts. With FRET-based indicators symmetrical changes of both fluorescent signals represent movement, while reciprocal changes in the fluorescent signals indicate true calcium responses.
When choosing a calcium indicator, it is important to consider the calcium concentration that is being measured, including calcium levels at rest and peak calcium levels upon stimulation. Previous work has estimated baseline calcium levels to be approximately 100 nM within the hair-cell cytosol (Ikeda, Sunose, & Takasaka, 1993). Upon stimulation, calcium levels can transiently reach the micromolar range; the calcium concentration varies dramatically according to subcellular location, such as in the hair bundle and the ribbon synapse where peak calcium levels are estimated to be much higher than the cytosol (Bortolozzi, Lelli, & Mammano, 2008; Lumpkin & Hudspeth, 1998). Various reviews have discussed in detail how to appropriately select a GECI (Pérez Koldenkova & Nagai, 2013; Tian et al., 2011). In general, it is important to choose an indicator with a dynamic range appropriate to the calcium signals under investigation. We recommend trying more than one indicator as it is often unclear from in vitro work how a particular indicator will translate in vivo. In this chapter, we focus specifically on GECIs suitable for measuring cytosolic calcium responses in lateral-line hair cells.
For GECI selection in general, in addition to understanding expected changes in calcium concentration upon stimulation, it is also important to understand the calcium dynamics of a biologically relevant stimulus. Recent advances in calcium indicators have improved the speed and sensitivity of the indicators to allow detection of single action potentials (Chen et al., 2013). Although mature hair cells do not fire action potentials, indicator speed and sensitivity could be informative in detecting receptor potential oscillations, which can resonate at high frequency speeds similar to those of action potentials. Lateral-line hair cells are required to sense local water movement along the fish and function as velocity sensors important for rheotactic behaviors (Montgomery, Carton, Voigt, Baker, & Diebel, 2000). Some of these stimuli are likely to be relatively long and slow (seconds), similar to a stimulus in mammalian vestibular organs where hair cells are activated by fluid flow through the semicircular canals. In addition, the lateral line has been shown to encode frequencies up to 100e200 Hz, which is beyond the range current GECIs can reliably encode (Trump & McHenry, 2008; Weeg & Bass, 2002). Therefore, GECIs are suitable for some but not all relevant lateral-line stimuli. Overall, GECIs enable simultaneous activity measurements from populations of hair cells with reasonably high spatial and temporal efficiency.
1.2. COMPARISON OF CALCIUM INDICATORS
To date, four different GECIs have been expressed transgenically in zebrafish to measure calcium activity in hair cells: the FRET-based indicator cameleon D3 and the non-FRET-based indicators GCaMP3, RGECO (a red-shifted variant based on GCaMP3), and GCaMP7a (newer GCaMP variant) (Esterberg, Hailey, Coffin, Raible, & Rubel, 2013; Kindt et al., 2012; Maeda et al., 2014; Pujol-Martí et al., 2014). In these studies, the myosinVIb or pou4f3 (brn3c) promoters were used to express the calcium indicators in hair cells. In the case of the brn3c promoter, a pou4f3:GAL4 driver line was used in combination with a UAS:GCaMP7a transgenic (Pujol-Martí et al., 2014).
Here we provide a side-by-side comparison of three (D3, RGECO, GCaMP3) of these indicators to assist researchers in GECI selection (Fig. 1). Each of these three indicators has a similar dissociation constant (Kd) (approximately 600, 480, and 540 nM respectively), and dynamic range (ratio of calcium-saturated fluorescence to calcium-free fluorescence: 10, 12, and 10, respectively) (Palmer et al., 2006; Tian et al., 2009; Zhao et al., 2011). To examine the activity dependence of each of these different indicator lines, we mount, immobilize, and mechanically stimulate fish identical to the methods described in a later chapter “Physiological recordings from the zebrafish ear and lateral line” in this volume as well as in a previously published methods chapter detailing electrophysiological recordings in lateral-line hair cells (Trapani & Nicolson, 2010). For our preparation, we anesthetize and pin larvae to a Sylgard recording chamber. Larvae are then paralyzed by cardiac injection of alpha-bungarotoxin. Proper pinning and paralysis are essential for high-quality image acquisitions. To ensure reproducible and quantifiable mechanically evoked calcium responses, it is important to prepare and calibrate the fluid jet for each sample. We set our stimulus intensity to produce a near-saturating stimulus. To do this, we position the fluid jet pipette 100 μm away from the edge of the neuromast and limit the pipette tip opening to 30–40 μm. Then, using differential interference contrast optics we locate the tips of the kinocilia and apply a short (200 ms) fluid stimulus and measure the displacement of the kinocilia tips using a camera and micrometer. For these measurements, on our imaging systems we utilize a built in scale bar option within the software to measure the distance deflected. Alternatively, we calibrate our imaging field with a stage micrometer slide to measure deflection distances. The fluid jet should displace the tips of the kinocilia ~2–3 μm. This deflection distance is sufficient to produce a near-saturating stimulus with respect to hair-bundle deflection. Distance and intensity (see Section 2.4) of the fluid jet can be adjusted slightly to achieve the proper tip-deflection distance. We caution against using stronger stimuli which are potentially damaging to hair cells and can cause motion artifacts that lead to false fluorescence intensity changes. For our comparison of these indicators, we chose three different stimulus durations:0.1-, 2-, and 4-s step stimuli (static hair-bundle deflection for the stimulus duration). Hair cells in neuromasts of the primary posterior lateral line are oriented to respond to either an anterior- or a posterior-directed stimulus. For simplicity, in Fig. 1 and Fig. 4 we have only presented data for an anterior stimulus (stimulus towards the anterior of the fish). The range of stimuli was selected to show the low (0.1 s) and high (4 s) end of the dynamic range of the indicators. In general it is good practice to choose stimuli that fall within the dynamic range of the indicator to ensure that the majority of responses are detected and the indicator is not saturated. We also assayed the photostability of each indicator after repeated image acquisitions.
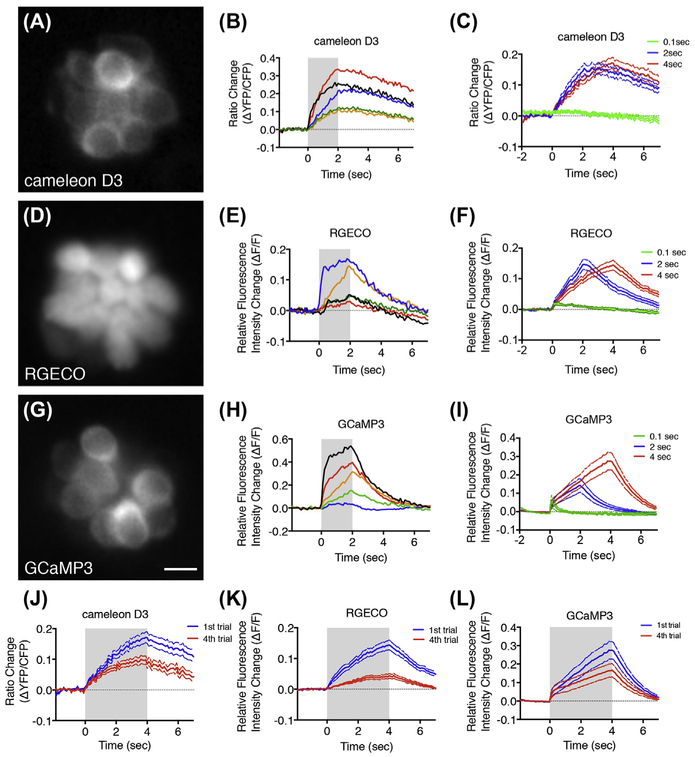
FIGURE 1. Comparison of cameleon D3, RGECO, and GCaMP3 calcium signals in zebrafish hair cells.
(A, D, G) Widefield images of neuromasts expressing cameleon D3, RGECO, and GCaMP3, respectively. (B, E, H) Traces of five representative, individual hair cells for each indicator. (C, F, I) Average calcium response for each indicator in response to a 0.1, 2, and 4 s stimulus, n = 10–15 hair cells. (J, K, L) Calcium signals in response to a 4-s stimulus using each indicator after the first and fourth trial. Larvae were imaged at day 4. Gray boxes denote stimulus period. Scale bar = 5 μm. (See color plate)
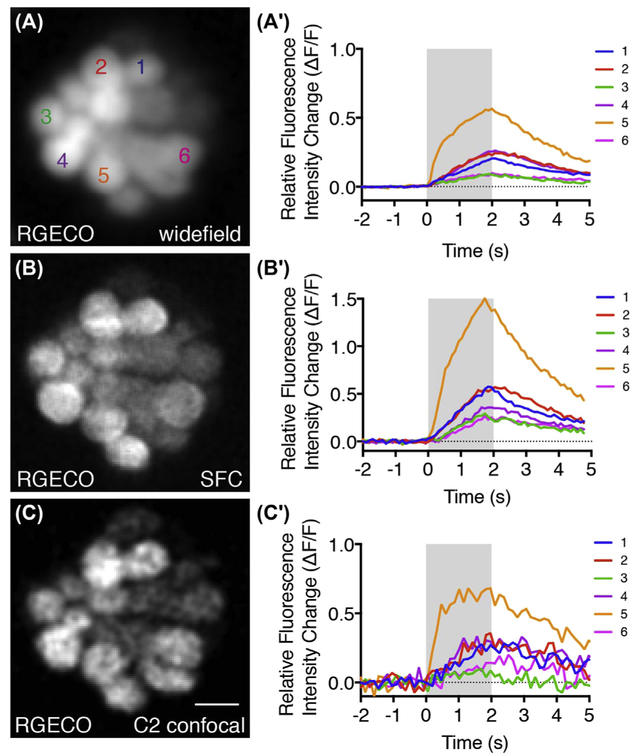
FIGURE 4. Comparison of hair-cell evoked calcium responses on three imaging systems.
(A, B, C) Tg[myo6b:RGECO] images from the same neuromast acquired on a Nikon widefield microscope, Bruker SFC confocal microscope, and Nikon C2 confocal microscope, respectively. (A′, B′, C′) Evoked calcium responses in response to a 2-s step deflection measured on each system using the parameters outlined in Table 1. Larvae were examined at day 5, scale bar = 5 μm. (See color plate)
For our indicator comparison, images were acquired on an upright fixed-stage Nikon widefield microscope with settings and equipment similar to those described in Section 2 and Table 1. A Hamamatsu ORCA-D2 dual-chip camera was used to simultaneously image CFP and YFP for the D3 cameleon indicator. One commonality between all indicators is that the calcium signals detected are extremely heterogenous (Fig. 1B, E, and H) (Kindt et al., 2012; Olt et al., 2014). This heterogeneity is likely due to a combination of the developmental stage and the functional properties of each individual hair cell, as well as the exact imaging plane examined. Using these indicators on average, we found that GCaMP3 was the most sensitive and offered the best dynamic range (Fig. 1C, F, and I). GCaMP3 was able to detect the 0.1 s stimulus reliably and did not saturate even after 4 s of stimulation. Lastly we found that GCaMP3 and D3 are more suitable for repeated imaging than RGECO (Fig. 1JeL). RGECO suffers the same poor photostability as the red fluorescent proteins upon which it is based and is more susceptible to photobleaching than the GCaMP series (Akerboom et al., 2013; Drobizhev, Makarov, Tillo, Hughes, & Rebane, 2011). However, RGECO remains a very useful calcium indicator because it is red-shifted and therefore can be used in combination with other GFP lines to provide additional subcellular information (Maeda et al., 2014). From this comparison we conclude that all of the previously described indicators are suitable for measuring mechanically evoked calcium responses in lateral-line hair cells.Of the three, GCaMP3 is the most sensitive, has the best dynamic range, and is extremely photostable.
Table 1.
Parameters for Three Zebrafish Hair-Cell Calcium Imaging Systems
| Nikon Widefield | Bruker Sweptfield Confocal | Nikon C2 Line- Scanning Confocal | |
|---|---|---|---|
| Detector | Camera - Hamamatsu | Camera - Rolera | PMT - Nikon |
| ORCA-R2 | EM-C2 | Nikon C2 | |
| Gain 50/100 | EM gain 3700/4000 | Gain 75/255 | |
| Bin | 2 × 2 | 2 × 2 | − |
| ROI | 50 × 50 μm | 50 × 50 μm | 50 × 50 μm |
| X-Y resolution | 0.43 μm/pixel | 0.53 μm/pixel | 0.33 μm/pixel |
| Slit size | − | 35 μm slit | − |
| Light source power | Intensilight (mercury) 25% with cy3 filter | Solid state 561 nm laser 5% | Solid state 561 nm laser 5% |
| Speed | 10 fps | 10 fps | 7.5 fps |
| Optical section | Largest, worst | Variable depending on pinhole or slit size | Thinnest, best |
| Max speed | 100 fps | 100 fps | 10 fps |
| Main advantages | Inexpensive | Fast, high optical resolution | Highest optical resolution |
| Main disadvantages | Low optical resolution | Cost | Slow speed, cost, photobleaching |
1.3. VALIDATING A RELEVANT CALCIUM SIGNAL
A primary consideration when measuring calcium signals in a live organism is to ensure that changes in fluorescence intensity are not due to movement artifacts but rather represent genuine activity-dependent changes in calcium (Greenberg & Kerr, 2009). This represents one of the major sources of error in measuring calcium levels in all cells. As mentioned above, the use of FRET-based indicators can help solve this problem, but if using a FRET-based indicator is not an option, there are several ways to determine whether fluorescence changes in an image acquisition sequence represent an activity-dependent calcium signal or movement artifacts.
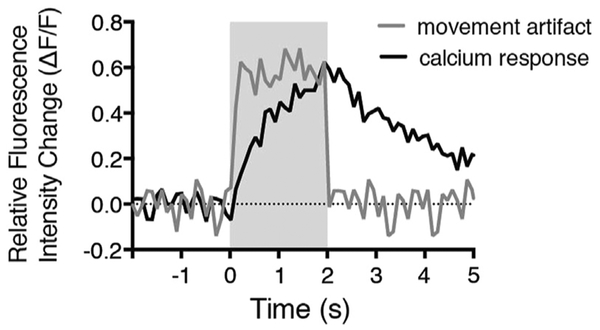
Many variables can cause movement artifacts. In most cases, movement occurs because larvae are not sufficiently immobilized, leading to unwanted drift in X, Y, or Z directions. In other cases, debris from a clogged fluid jet blown over the neuromast during stimulus and image acquisition can cause false changes in fluorescence. Regardless of how well the sample is prepared, some movement will be unavoidable. Therefore, it is important to carefully examine each image acquisition sequence. Acquisitions with excessive Z-movements cannot be corrected by postprocessing and must be eliminated. Acquisitions with minor X-Y movement can be corrected (see Section 3). When evaluating image acquisitions, it is also important to observe the shape and kinetics of the apparent response. If fluorescent signals are the result of movement artifacts, the rise and fall of the fluorescent signal will be abrupt and plateau sharply (this can also occur if the indicator is saturated) (Fig. 2). By contrast, in the case of a true mechanically evoked calcium response for measurements made in the cytosol, the fluorescent signal will rise for the duration of the stimulus and decline slowly after the stimulus has ended (Fig. 2).
FIGURE 2. Distinguishing artifacts from functional calcium signals.
Example of traces resembling a movement artifact is shown in gray, while a true, mechanically evoked hair-cell calcium response is shown in black. Note the slow rise and decay in the black traces versus the abrupt rise and fall in signal in the gray trace. Gray box denotes stimulus period.
In addition to careful sample preparation and acquisition evaluation, it is also possible to verify the legitimacy of an observed signal by repeating the stimulus while the fish is bathed in a mechanotransduction channel blocker such as amiloride or dihydrostreptomycin, or agents that disrupt the tip links that are required for mechanosensitive function (Assad, Shepherd, & Corey, 1991; Ricci, 2002). A 10-min incubation with 5 mM BAPTA tetrasodium salt, and subsequent washout is sufficient to break all tip links and eliminate true mechanically evoked calcium responses (Kindt et al., 2012). It is important to note that 30 mins after BAPTA washout, mechanosensitive responses will start to return as the tip links reform. Other agents, such as amiloride (1 mM) or dihydrostreptomycin (1 mM), may be used to block mechanotransduction but must be added to the solution in the fluid jet and maintained in the bath during image acquisition. Fluorescence changes resulting from movement artifacts will be unaffected by treatment with mechanotransduction channel blockers, and associated image acquisitions can be discarded. Overall, diligent sample preparation, careful examination of image acquisitions, and drug controls will help ensure that any fluorescent signals measured are true, activity-dependent calcium changes in lateral-line hair cells.
2. IMAGING SYSTEMS AND OPTIMAL PARAMETERS
It is ideal but not necessary to have a microscope dedicated exclusively to imaging fluorescence activity of zebrafish hair cells. The options for microscopes and associated accessories are numerous. In this section, we outline three microscope systems suitable for imaging-evoked calcium responses in zebrafish hair cells: widefield, swept-field confocal (SFC), and point-scanning confocal systems. In addition we describe the pros and cons of each imaging system, as well as recommended parameters. Despite the differences in the microscope setups listed above, there are many components in common and the general concepts underlying parameter optimization are the same. Below in Section 2.1 and in Fig. 3, we describe, in general terms, equipment components that are required for all imaging systems. The equipment listed below highlights only a subset of the many options available and the provided parameters are designed to offer a starting point for optimizing imaging settings.
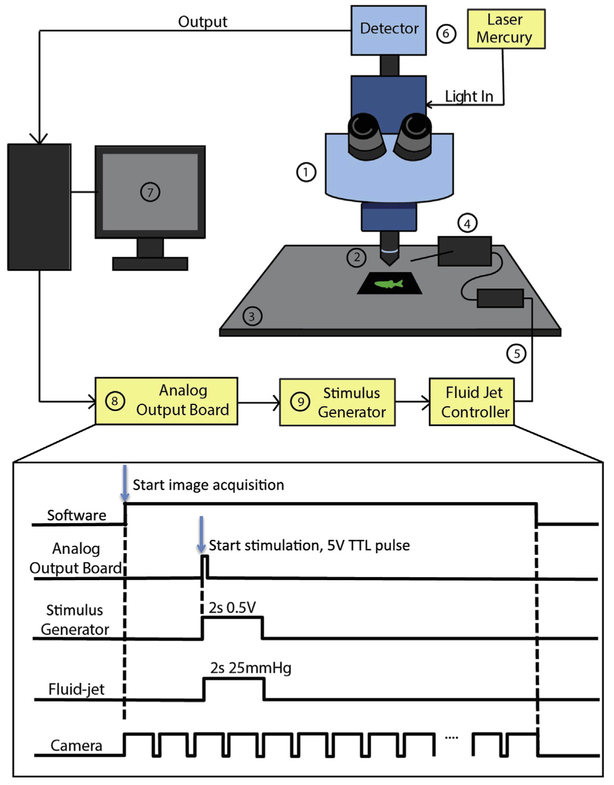
FIGURE 3. Imaging setup for calcium imaging and hair-cell stimulation.
Schematic of the imaging system requirements outlined in Section 2.1. (1) upright microscope; (2) water immersion objective; (3) fixed stage; (4) manipulator to position stimulation pipette; (5) fluid jet (and controller) to stimulate hair cells; (6) light source and detector; (7) imaging software; (8) analog output board; (9) stimulus generator. The bottom panel outlines the signals used to coordinate imaging with hair-cell stimulation. The software starts the image acquisition. This triggers the camera to capture at a given frame rate. During the time series to start hair-cell stimulation, a 5 V TTL pulse is sent from the software at a defined frame rate to the stimulus generator using an analog output board. The stimulus generator converts the TTL pulse into an analog voltage signal with a defined duration and magnitude that is then sent to the fluid jet controller. The fluid jet controller converts the voltage signal into fluid flow by applying positive or negative pressure on the fluid within the stimulating pipette. This, in turn, deflects hair bundles and activates hair cells.
2.1. GENERAL MICROSCOPE AND EQUIPMENT REQUIREMENTS
-
Upright rather than inverted microscope body
Olympus BX51W1, Nikon Eclipse FN1, Zeiss Axioexaminer, Leica DM6FS
60x water immersion objective with a high NA (0.9–1.1) and suitable working distance of ~2 mm
-
Fixed stage
Sutter MT-1000 or MT1078, Burleigh Gibraltar Platforms, Prior Z-Deck
-
Manipulator to position fluid jet or piezo pipette for stimulation
Sutter MP-225, Scientific Patchstar, Sensapex triple axis, Burleigh PCS 6200
-
Fluid jet or piezo to stimulate hair cells
Fluid jet: ALA HSPC-1 and HSPV-P
Piezo: PI instruments PICMA P-882.11–888.11 series plus controller
-
Light source and detector (see Table 1 for detector examples)
Widefield: LED or mercury light (Nikon Intensilight, Excelitas X-Cite
XLED1 and X-cite120Q) and camera for detection
Confocal: Lasers and photomultiplier tubes (PMTs) or camera for detection
-
Imaging software
Molecular Devices Metamorph, Micro-Manager, Zeiss ZEN, Nikon Elements
-
Digital to analog output board to send a transistor–transistor logic (TTL) trigger from the imaging software to a stimulus generator
Consult your microscope software representative
Example: National Instruments PCI 6700 series
-
Stimulus generator to trigger fluid jet or piezo
Tektronix AFG1000–3000 series, BK Precision 4000 series
Vibration isolation table
2.2. CHOOSING A SPECIFIC IMAGING SYSTEM
In addition to these essential components that are required for all imaging systems, a specific imaging system must be chosen. This chapter highlights three different imaging systems: widefield, SFC, and point- or line-scanning confocal systems (Table 1). A widefield imaging system is the simplest and least-expensive system, but the main drawback is the low optical or axial resolution. Point- or line-scanning confocal systems are the most expensive and offer the highest optical resolution, but at the expense of speed. SFC imaging systems, on the other hand, can scan samples using pinholes or slits to balance optical resolution and speed respectively (Castellano-Muñoz, Peng, Salles, & Ricci, 2012). This is similar in some aspects to spinning-disk systems, except the pinhole and therefore optical resolution and light collection are fixed on a spinning-disk system while the pinhole is adjustable on an SFC system.
Widefield and SFC systems both require a camera to detect signals, and the overall speed of image acquisition is limited mainly by camera choice. Current camera-based systems are faster than point-scanning confocal systems that use PMTs to detect signals. Camera choices are ever expanding and many options are available. Commonly used CMOS, EM-CCD, and CCD cameras are all adequate for the imaging described in this chapter. It is best practice to request a demonstration of a specific camera from the manufacturer or vendor to determine if it is suitable for your imaging requirements. When selecting a camera, a relatively high frame rate (10–100 fps) and low noise is desirable, similar to other calcium imaging applications. In the case of zebrafish hair cell imaging, the pixel size is another important consideration. Zebrafish hair cells are quite small: lateral-line hair cells are approximately 5 μm across × 10 μm apex-to-base (magnified 60 × 300 × 600 mm). Many cameras are designed for whole brain, full-field imaging and have a relatively large pixel size ~16 μm. Using a camera with a 16 mm pixel, each hair cell would be represented by (300/16 × 600/16) 18.74 × 37.5 pixels. Cameras with 6–8 μm pixel size offer a more suitable with (300/6 × 600/6) 50 × 100 pixels per hair cell. This is especially true for calcium imaging, as binning is almost always a required imaging parameter and will result in even fewer pixels represented per hair cell. If necessary, large pixel size can be overcome using a magnifying coupling lens to decrease pixel size and increase resolution.
In addition to camera-based imaging systems, an alternative is a point- or line-scanning confocal system. Although these systems offer the best optical sectioning, they use PMTs to detect signals, which have a slower frame rate than cameras. Although faster and more sensitive PMTs are being rapidly developed, camera-based imaging currently offers the fastest acquisition speeds. Additionally, hair-cell calcium imaging is possible with 2-photon systems although due to the superficial location of lateral-line hair cells, the depth advantages gained using a 2-photon system are not utilized and these systems are not discussed further here. With rapid advances in optics and image processing, we expect an ever-greater variety of imaging components, beyond the systems we have discussed, that will satisfy the resolution and speed requirements for hair-cell calcium imaging.
2.3. DETERMINING OPTIMAL IMAGING PARAMETERS
Any imaging setup, including the three systems described here, is useless without determining proper parameters. For a given system, there are optimal settings that balance the amount of light (laser, mercury, or LED) reaching the sample, resolution at the detector (camera, PMT), and speed of acquisition. In all systems, light intensity and detector settings must be identified that are able to detect changes in fluorescent signals at a frame rate relevant for hair-cell activity while minimizing photobleaching. To aid in this process, we have compared the three imaging systems listed in Table 1 using Tg[myo6b:RGECO] larvae. For this comparison, we fixed the exposure time at 100–150 ms. For spontaneous and evoked calcium responses in zebrafish hair cells, a frame rate of at least 10 Hz (100 ms exposure time) is required to detect calcium-dependent activity. Light intensity should be sufficiently high to have adequate signal above noise yet low enough to prevent pixel saturation. For this comparison, an 8-s streaming time series was acquired on each system. A 2-s step stimulus (0.5 V, 25 mmHg fluid jet, 2 μm kinocilia tip deflection) was triggered 2 s into the acquisition. Traces from all three systems are shown in Fig. 4.
Comparing the three imaging systems, it is apparent that patterns of calcium signals are similar: all of the same hair cells respond and the overall pattern of heterogeneity is conserved among systems. For example, on all three systems cell 5 has the largest calcium response, while cell 3 has one of the smallest calcium responses. Despite these similarities, there are differences in the overall magnitude of the calcium signals, the signal-to-noise ratio (SNR), and the kinetics of the calcium signals.All these differences stem mainly from differences in optical resolution between the systems, which increases from widefield to SFC to point-scanning systems (Fig. 4A′-C′). With higher optical resolution, it is more likely that the calcium signals observed are in the imaging plane, rather than due to out-of-focus light from other planes. Increased optical resolution comes at a cost because it decreases the amount of calcium indicator imaged per pixel.
With regards to overall signal magnitude, calcium imaging using the SFC system revealed the largest signals. Both the SFC and widefield systems have a high SNR while the SNR is the lowest on the point-scanning systems when the optical section is smallest (Fig. 4C′). The SNR is high on the widefield system because there is more indicator per optical section compared to the point-scanning confocal system. A large optical section boosts the signal detection but also collects a lot of out-of-focus light which can negatively impact signal magnitude. The increase in signal and high SNR gained by a thicker optical section in widefield (Fig. 4A′) and SFC (Fig. 4B′) systems is at a cost. It is less likely that the signals detected are restricted to the imaging plane. This is the main advantage of point-scanning confocal systems with high optical resolution. Although thinner sectioning and higher optical resolution can impair signal detection, it is advantageous for detecting calcium signals restricted to a particular imaging plane. This trade-off is most apparent in Fig. 4, cell 5. It is very likely that the large calcium signal in cell 5 is because the optical section contains synaptic ribbons, a site of rapid and sizable calcium concentration changes (Bortolozzi et al., 2008). As the optical section tightens from Fig. 4A′ to B′ to C′, the onset time of the calcium signal becomes faster and more representative of a synaptic plane where calcium signals are large and occur rapidly. Overall, of the three systems, SFC offers an optimal compromise between optical resolution and signal detection.
2.4. SYNCHRONIZING A STIMULUS WITH IMAGE ACQUISITION
One major challenge to examining activity while imaging is coordinating image acquisition with stimulus delivery. Major imaging companies are now aware of this gap, especially with the upsurge of neuroscience research and are working to address this issue. The main limitation lies in constraints built into imaging software. We outline a method for image acquisition and stimulus coordination in Fig. 3. In this example the imaging software is programmed to send a 5 V TTL output during the time series image acquisition. This generally requires a digital to analog output board in order for the imaging software to send the 5 V TTL output. The analog output board relays this 5 V TTL output to an external device, such as a stimulus generator. The stimulus generator accepts the 5 V signal as an input or trigger and generates a new output or stimulus waveform. For example, the stimulus generator can be configured to create a variety of stimuli such as steps and ramps, and to control the intensity and duration of the stimulus. The stimulus configured in the stimulus generator is then output as an analog voltage signal which is sent to the fluid jet or piezo controller where the voltage levels and durations are converted into fluid flow or piezo movement and hair-cell stimulation. This pathway and timing of communication are outlined in Fig. 3.
3. IMAGE PROCESSING
There are numerous methods for processing and analyzing raw calcium imaging data. In this section, we discuss the importance of image registration and outline an approach to do this using the free, open-source image processing software ImageJ (Section 3.1, Schneider, Rasband, & Eliceiri, 2012). Then we describe two approaches for visualizing calcium responses. The first is a simple and straightforward method to detect calcium signals using ImageJ (Section 3.2; Schneider et al., 2012). The second method describes algorithms to visualize the spatial dynamics of calcium signals that can be accomplished using MATLAB R2014b software (Section 3.3; The MathWorks, Natick, MA, USA).
3.1. IMAGE REGISTRATION USING IMAGEJ
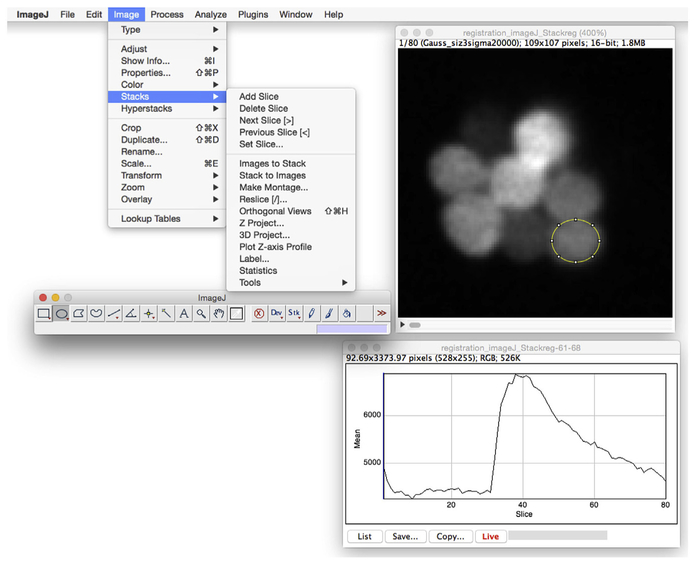
During image acquisition in a live organism, some movement is unavoidable (example in Fig. 5A). While it is possible to correct for movement in the lateral (X-Y) direction, it is difficult to correct for large changes in the Z direction. When evaluating the quality of the acquired images, it is important to discard acquisitions with excessive Z-drift and perform proper controls (see Section 1) to ensure the signals measured are true calcium signals. Lateral movements (X-Y) can also cause artifacts that misrepresent calcium signals, but can be corrected through image registration. Because measurements of fluorescence changes are made throughout the image acquisition, at the same hair cell within a region of interest (ROI), it is essential to perform image registration to remove lateral movements and keep the hair cell within the specified ROI (see example in Fig. 5). One simple method to perform image registration is to utilize the StackReg plugin in ImageJ (Thévenaz, Ruttimann, & Unser, 1998). StackReg functions by performing recursive alignment of the acquired image sequence. The StackReg plugin requires that a second plugin, named TurboReg, is installed, which can be downloaded from the website, http://bigwww.epfl.ch/thevenaz/turboreg/. The underlying principle is to use each slice within a time series as the template to align the next slice, so that the alignment proceeds iteratively. An example of this registration is shown in Fig. 5, where the three images indicate the onset, midpoint, and end of an image acquisition. After image registration, the upward movement (Y-direction) is corrected and the ROI marked by the red circle maintains its original position over time. Fig. 5B′ demonstrates the temporal curve of the fluorescence intensity after registration. As indicated in Fig. 5, without image registration, the signal will be greatly degraded (compare Fig. 5A′ vs. Fig. 5B′) or provide inaccurate information regarding the calcium signals.
FIGURE 5. Live imaging of neuromast RGECO calcium signals with and without image registration.
(A) Representative images captured at the onset, midpoint, and end of image acquisition period before image registration. (A′) Temporal curve of the fluorescence intensity before image registration. (B) Representative images captured at the onset, midpoint, and end of image acquisition period after ImageJ StackReg image registration. (B′) Temporal curve of the fluorescence intensity after StackReg image registration. Red circles (gray in print versions) indicate the same ROI used for calculating the fluorescence intensity. Gray bars in (A′) and (B′) indicate the 2-s fluid jet stimulus. Scale bar = 5 μm.
3.2. SIGNAL DETECTION AND REPRESENTATION IN IMAGEJ
After image registration, to obtain a quick readout of the temporal changes of the calcium signals in ImageJ, select an ROI and go to Image > Stacks > plot z-axis Profile. A new window will appear, showing the mean of the fluorescence intensity of the selected ROI over the time series (Fig. 6). If the Live (marked by the arrow) option is selected, the displayed temporal curve will change as the ROI is moved. The values can be copied (select copy option) and saved for further processing.
FIGURE 6. Plotting the temporal curve of a selected ROI using ImageJ.
To view the intensity changes over time within a ROI, draw a ROI, then under Image-Stacks, select Plot z-axis profile. Select Live to refresh the graph while moving the ROI between cells. Select save to save the intensity values within an ROI.
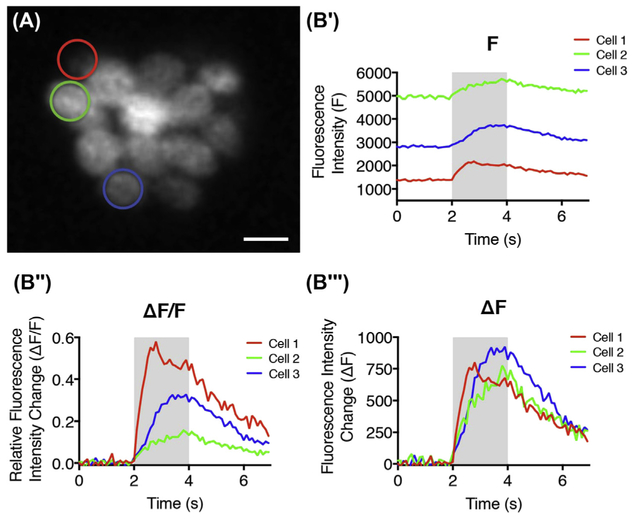
The temporal curve of the fluorescence intensity shows how the signals change during the time series. To have a better comparison among different ROIs or fluorescence intensities obtained with different imaging systems, the relative fluorescence intensity change (ΔF/F0) is more commonly used. However, when ROIs have different baseline fluorescence intensity (F0), that reflects resting calcium availability rather than indicator concentration, ΔF/F0 may not be an appropriate measure. As shown in Fig. 7, three different ROIs with varied baseline intensity have a similar fluorescence intensity change ΔF (Fig. 7B″), but their relative values (ΔF/F0) vary considerably. Therefore, we suggest taking both of these measurements (ΔF/F0 and ΔF) into consideration for signal magnitude comparison.
FIGURE 7. Temporal representation of RGECO calcium signal at three selected ROIs.
(A) A representative RGECO neuromast image with three ROIs (hair cells) indicated by red, green, and blue circles respectively. (B′) Temporal curve of the fluorescence intensity. (B″)Temporal curve of the relative fluorescence intensity change. (B‴) Temporal curve of the fluorescence intensity change. Scale bar = 5 μm. (See color plate)
3.3. SPATIAL DETECTION AND VISUALIZATION USING MATLAB
A major advantage of optical imaging of calcium signals is high spatial resolution. In the case of genetically encoded indicators expressed in lateral-line hair cells, it is possible to examine the responses of all hair cells within a neuromast at once. To better visualize the signal distribution patterns of calcium within the whole neuromast upon fluid jet stimulus, we have outlined the following image processing algorithms that can be used in MATLAB (Yao & Zhao, 2008):
1. Create the baseline image
The images obtained before applying fluid jet stimulus are averaged pixel by pixel over the whole prestimulus period. If there are m frames of prestimulus images, then in the baseline image the fluorescence intensity F0 at the pixel (x,y) is calculated as:
| [1] |
2. Temporally bin the entire image sequence
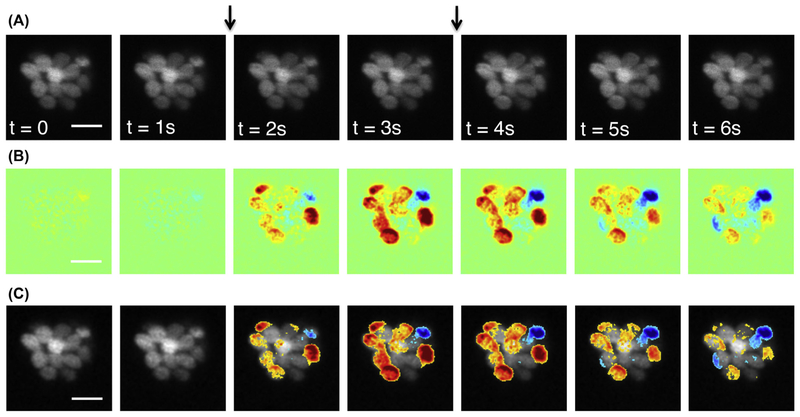
To increase the SNR, the whole image sequence is binned temporally. For example, if there are n frames of images in the whole sequence, including the prestimulus, stimulus, and poststimulus periods. We bin every k images into one to get n/k images, as shown in Fig. 8A and as follows:
| [2] |
FIGURE 8. Spatiotemporal patterns of calcium signal using RGECO.
Temporal binning of the whole image sequence; 7-s time series, acquired at 10 Hz, binned every five frames. A 2-s fluid jet stimulus was applied 2 s into the time series. Half of the binned frames are displayed. (B) Baseline-subtracted, colorized fluorescent-signal images. (C) Combined images created by overlaying the colorized fluorescent-signal images in B onto the baseline grayscale image. The calcium signal distributions can be visualized over the entire neuromast and in individual hair cells. The two arrows indicate the fluid jet stimulation duration. The scale bar = 10 μm. Images were processed in MATLAB. (See color plate)
3. Generate images of fluorescence intensity changes
The baseline image is subtracted from each frame of binned images to obtain images that represent the change in fluorescent signal from baseline. In this way, the frames prior to the stimulus are defined to have no signal, as in:
| [3] |
To better visualize the fluorescence intensity changes, the changes can be scaled and encoded as a lookup table with blue indicating decreased fluorescence intensity and red indicating an increase in intensity, as shown in Fig. 8B. By overlaying the baseline grayscale image with colorized signal images (Fig. 8C) it is possible to determine the fluorescence intensity changes within a neuromast and each individual hair cell.
SUMMARY
In this chapter, we have described methods for measuring mechanically evoked calcium responses in zebrafish lateral-line hair cells. We have compared the three published transgenic zebrafish lines that have been previously used to examine calcium responses in zebrafish hair cells. In addition, we have identified components required to set up a suitable imaging system for these calcium measurements. Then we performed a side-by-side comparison of three suitable imaging systems and provided suggested parameters for each system. Lastly, we have outlined two methods (one simple and one more complex) to visualize and measure calcium signals obtained from lateral-line hair-cell recordings. Overall this information should benefit any researcher looking to examine calcium responses in hair cells of the zebrafish.
DISCUSSION
The live-imaging methods described in this chapter represent an extremely valuable tool in assessing hair-cell function in zebrafish. Advances in imaging and development of faster and more sensitive calcium indicators, along with new transgenic zebrafish lines continue to push these techniques forward (Chen et al., 2013).
In addition to calcium indicators with increased sensitivity, recent work has begun to take advantage of the fact that GECIs can be targeted subcellularly. Here, activity can be examined in distinct subcellular domains, as has been accomplished in zebra-fish to study the role of the mitochondria in hair-cell ototoxicity (Esterberg, Hailey, Rubel, & Raible, 2014). It is possible that localized calcium indicators will provide critical information regarding function of the apical hair bundle and the ribbon synapse, two subcellular compartments where local calcium dynamics are thought to play a critical role in hair-cell function (Ceriani & Mammano, 2012).
Recent work has created calcium indicators that function at different wavelengths, such as the blue-shifted BGECO and the red-shifted RGECO and RCaMP (Akerboom et al., 2013; Zhao et al., 2011). Using these new indicators, it is possible to produce double transgenic larvae that express two different colored calcium indicators in different structures, as has been done to examine concurrent calcium changes in the hair-cell cytosol and in the mitochondria in response to ototoxic insults (Esterberg et al., 2014). For faster stimulus-dependent hair-cell activity, this two-color imaging can be accomplished using the systems described in this chapter using a beam splitter (Photometrics dual-view) or a dual-chip camera (Hamamastu ORCA-D2) which allows imaging of two channels simultaneously during a stimulus (Akerboom et al., 2013). Two-color imaging will also be useful in future work where activity measurements will likely extend past the sensory hair cells to analyses of the hair-cell afferent and efferent neurons. For example, a double transgenic zebrafish that expresses a red indicator in hair cells and green indicator in the afferent fiber could be used to image pre- and postsynaptic calcium simultaneously. Such an approach could provide critical information about how sensory hair cells encode information within the context of a sensory circuit and will further our knowledge of auditory and vestibular systems.
In addition to 2-color imaging and circuit analysis, future work will also benefit from rapid imaging in the Z-axis; in this case, data can be acquired with high spatial information in 3D and also over time, resulting in 4D image acquisitions. This can be accomplished using a piezoelectric motor attached to an objective to allow rapid imaging in multiple Z-planes so that differences in activity across the Z-axis can be recorded (Göbel, Kampa, & Helmchen, 2007). In addition to piezoelectric motors, light-sheet microscopy now allows large volumes to be imaged at relatively fast speeds at single-cell resolution (Ahrens et al., 2013). Light-sheet microscopy is particularly attractive because samples are protected from phototoxicity by low laser intensities; this feature will provide a powerful way to study functional activity repeatedly over large developmental windows.
Finally, in addition to the collection of calcium indicators currently available, there are a wealth of indicators now available to examine other functional hair-cell properties; for example, indicators to detect glutamate, vesicle release, and voltage signals (Marvin et al., 2013; Odermatt, Nikolaev, & Lagnado, 2012; St-Pierre et al., 2014). Using the general methods described in this chapter, these valuable optical tools could be used to provide critical information regarding how hair cells develop, function, succumb to ototoxic insults, and regenerate, all in an intact organism.
ACKNOWLEDGMENTS
We would like to acknowledge Alisha Beirl, Elyssa Monzack, Alain Silk, Josef Trapani, and Michael Waltman for their comments on the manuscript. This work was supported by the Intramural Research Program of the NIH, NIDCD 1ZIADC000085–01.
REFERENCES
- Ahrens MB, Orger MB, Robson DN, Li JM, & Keller PJ (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature Methods, 10, 413–420. [DOI] [PubMed] [Google Scholar]
- Akerboom J, Carreras Calderón N, Tian L, Wabnig S, Prigge M, Tolö J … Looger LL (2013). Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in Molecular Neuroscience, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Lin S, & Hopkins N (1995). The Aequorea victoria Green fluorescent protein can be used as a reporter in live zebrafish embryos. Developmental Biology, 171, 123–129. [DOI] [PubMed] [Google Scholar]
- Assad JA, Shepherd GMG, & Corey DP (1991). Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron, 7, 985–994. [DOI] [PubMed] [Google Scholar]
- Beurg M, Michalski N, Safieddine S, Bouleau Y, Schneggenburger R, Chapman ER … Dulon D (2010). Control of exocytosis by synaptotagmins and otoferlin in auditory hair cells. The Journal of Neuroscience, 30, 13281–13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolozzi M, Lelli A, & Mammano F (2008). Calcium microdomains at presynaptic active zones of vertebrate hair cells unmasked by stochastic deconvolution. Cell Calcium, 44, 158–168. [DOI] [PubMed] [Google Scholar]
- Castellano-Muñoz M, Peng AW, Salles FT, & Ricci AJ (2012). Swept field laser confocal microscopy for enhanced spatial and temporal resolution in live-cell imaging. Microscopy and Microanalysis, 18, 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani F, & Mammano F (2012). Calcium signaling in the cochlea e molecular mechanisms and physiopathological implications. Cell Communication and Signaling, 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A … Kim DS(2013). Ultra-sensitive fluorescent proteins for imaging neuronal activity. Nature, 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DP, García-Añoveros J, Holt JR, Kwan KY, Lin S-Y, Vollrath MA … Zhang DS (2004). TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature, 432, 723–730. [DOI] [PubMed] [Google Scholar]
- Drobizhev M, Makarov NS, Tillo SE, Hughes TE, & Rebane A (2011). Two-photon absorption properties of fluorescent proteins. Nature Methods, 8, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterberg R, Hailey DW, Coffin AB, Raible DW, & Rubel EW (2013). Disruption of intracellular calcium regulation is integral to aminoglycoside-induced hair cell death. The Journal of Neuroscience, 33, 7513–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterberg R, Hailey DW, Rubel EW, & Raible DW (2014). ERemitochondrial calcium flow underlies vulnerability of mechanosensory hair cells to damage. The Journal of Neuroscience, 34, 9703–9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman W (1928). The function of the lateral line organs. Science, 68, 205. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, & Müller U (2009). Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell, 139, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel W, Kampa BM, & Helmchen F (2007). Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nature Methods, 4, 73–79. [DOI] [PubMed] [Google Scholar]
- Greenberg DS, & Kerr JND (2009). Automated correction of fast motion artifacts for two-photon imaging of awake animals. Journal of Neuroscience Methods, 176, 1–15. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Masino MA, Mandel G, & Fetcho JR (2003). Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. Journal of Neuro-physiology, 90, 3986–3997. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Sunose H, & Takasaka T (1993). Effects of free radicals on the intracellular calcium concentration in the isolated outer hair cell of the Guinea pig cochlea. Acta Otolaryngologica (Stockholm), 113, 137–141. [DOI] [PubMed] [Google Scholar]
- Jaalouk DE, & Lammerding J (2009). Mechanotransduction gone awry. Nature Reviews.Molecular Cell Biology, 10, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt KS, Finch G, & Nicolson T (2012). Kinocilia mediate mechanosensitivity in developing zebrafish hair cells. Developmental Cell, 23, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMasurier M, & Gillespie PG (2005). Hair-cell mechanotransduction and cochlear amplification. Neuron, 48, 403–415. [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, & Hudspeth AJ (1998). Regulation of free Ca2þ concentration in hair-cell stereocilia. The Journal of Neuroscience, 18, 6300–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush ME, & Piotrowski T (2014). Sensory hair cell regeneration in the zebrafish lateral line. Developmental Dynamics, 243, 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R, Kindt KS, Mo W, Morgan CP, Erickson T, Zhao H … Nicolson T (2014). Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proceedings of the National Academy of Sciences of the United States of America, 111, 12907–12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J … Looger LL (2013). An optimized fluorescent probe for visualizing glutamate neurotransmission. Nature Methods, 10, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, & Tsien RY (1997). Fluorescent indicators for Ca2þ based on green fluorescent proteins and calmodulin. Nature, 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Carton G, Voigt R, Baker C, & Diebel C (2000). Sensory processing of water currents by fishes. Philosophical Transactions of the Royal Society of London B Biological Sciences, 355, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt B, Nikolaev A, & Lagnado L (2012). Encoding of luminance and contrast by linear and nonlinear synapses in the retina. Neuron, 73, 758–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olt J, Johnson SL, & Marcotti W (2014). In vivo and in vitro biophysical properties of hair cells from the lateral line and inner ear of developing and adult zebrafish. The Journal of Physiology, 592, 2041–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens KN, Santos F, Roberts B, Linbo T, Coffin AB, Knisely AJ … Raible DW (2008). Identification of genetic and chemical modulators of zebrafish mechanosensory hair cell death. PLoS Genetics, 4, e1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, & Tsien RY (2006). Ca2þ indicators based on computationally redesigned calmodulin-peptide pairs. Chemistry & Biology, 13, 521–530. [DOI] [PubMed] [Google Scholar]
- Patton EE, & Zon LI (2001). The art and design of genetic screens: zebrafish. Nature Reviews Genetics, 2, 956–966. [DOI] [PubMed] [Google Scholar]
- Pérez Koldenkova V, & Nagai T (2013). Genetically encoded Ca2þ indicators: properties and evaluation. Biochimica et Biophysica Acta BBA e Molecular Cell Research, 1833, 1787–1797. [DOI] [PubMed] [Google Scholar]
- Pujol-Martí J, Faucherre A, Aziz-Bose R, Asgharsharghi A, Colombelli J, Trapani JG, & López-Schier H (2014). Converging axons collectively initiate and maintain synaptic selectivity in a constantly remodeling sensory organ. Current Biology, 24, 2968–2974. [DOI] [PubMed] [Google Scholar]
- Ricci A (2002). Differences in mechano-transducer channel kinetics underlie tonotopic distribution of fast adaptation in auditory hair cells. Journal of Neurophysiology, 87, 1738–1748. [DOI] [PubMed] [Google Scholar]
- Ricci AJ, Bai J-P, Song L, Lv C, Zenisek D, & Santos-Sacchi J (2013). Patch-clamp recordings from lateral line neuromast hair cells of the living zebrafish. The Journal of Neuroscience, 33, 3131–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin AF, Nuñez VA, Sapède D, Tassin V, Dambly-Chaudière C, & Ghysen A (2010). Origin and early development of the posterior lateral line system of zebrafish. The Journal of Neuroscience, 30, 8234–8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, & Lin MZ (2014). High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nature Neuroscience, 17, 884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, & Unser M (1998). A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing, 7, 27–41. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, & Looger LL (2011). Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harbor Protocols. 10.1101/pdb.top069609. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH … Looger LL (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods, 6, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani JG, & Nicolson T (2010). Physiological recordings from zebrafish lateral-line hair cells and afferent neurons. Methods in Cell Biology, 100, 219–231. [DOI] [PubMed] [Google Scholar]
- Trump WJV, & McHenry MJ (2008). The morphology and mechanical sensitivity of lateral line receptors in zebrafish larvae (Danio rerio). The Journal of Experimental Biology, 211, 2105–2115. [DOI] [PubMed] [Google Scholar]
- Vollrath MA, Kwan KY, & Corey DP (2007). The micromachinery of mechanotransduction in hair cells. Annual Review of Neuroscience, 30, 339–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeg MS, & Bass AH (2002). Frequency response properties of lateral line superficial neuromasts in a vocal fish, with evidence for acoustic sensitivity. Journal of Neurophysiology, 88, 1252–1262. [DOI] [PubMed] [Google Scholar]
- Yao XC, & Zhao YB (2008). Optical dissection of stimulus-evoked retinal activation. Opt Express, 16, 12446–12459. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang Y-F, Nakano M … Campbell RE (2011). An expanded palette of genetically encoded Ca2þ indicators. Science, 333, 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]