Abstract
A dual-wavelength UV-C LED unit, emitting at peaks of 260 nm, 280 nm, and the combination of 260|280 nm together was evaluated for its inactivation efficacy and energy efficiency at disinfecting Escherichia coli, MS2 coliphage, human adenovirus type 2 (HAdV2), and Bacillus pumilus spores, compared to conventional low-pressure and medium-pressure UV mercury vapor lamps. The dual-wavelength unit was also used to measure potential synergistic effects of multiple wavelengths on bacterial and viral inactivation and DNA and RNA damage.
All five UV sources demonstrated similar inactivation of E. coli. For MS2, the 260 nm LED was most effective. For HAdV2 and B. pumilus, the MP UV lamp was most effective. When measuring electrical energy per order of reduction, the LP UV lamp was most efficient for inactivating E. coli and MS2; the LP UV and MP UV mercury lamps were equally efficient for HAdV2 and B. pumilus spores. Among the UV-C LEDs, there was no statistical difference in electrical efficiency for inactivating MS2, HAdV2, and B. pumilus spores. The 260 nm and 260|280 nm LEDs had a statistical energy advantage for E. coli inactivation.
For UV-C LEDs to match the electrical efficiency per order of log reduction of conventional LP UV sources, they must reach efficiencies of 25–39% or be improved on by smart reactor design. No dual wavelength synergies were detected for bacterial and viral inactivation nor for DNA and RNA damage.
Keywords: Combined wavelengths, Electrical energy per order, Human adenovirus type 2, Bacillus pumilus spores, Nucleic acid damage
Graphical abstract
1. Introduction
Ultraviolet (UV) light emitting diodes (LEDs) are an emerging technology for water and wastewater disinfection. Deep UV LEDs emitting UV-C irradiation have proven effective in inactivating bacterial, viral and protozoan pathogen surrogates and have been demonstrated for point-of-use water disinfection (Chatterley and Linden, 2010, Bowker et al., 2011, Lui et al., 2016). UV-C LEDs have enormous potential since they are smaller, lighter, and less fragile than traditional mercury vapor lamps (Vilhunen, 2010). Additionally, they are mercury-free and provide the capability to be turned on and off instantaneously. Given their small size, less than 1 mm2, multiple diodes can emit from different angles as opposed to traditional tubular UV light sources, allowing more options for unique reactor design (Lui et al., 2016, Oguma et al., 2016).
Considerable research has evaluated UV-C LEDs at various wavelengths for pathogen inactivation. Several studies have evaluated the efficacy of germicidal UV LED irradiation, emitted in relatively narrow bandwidths (nominal full width at half maximum, FWHM, of 10–12 nm) at or near 255 nm, 265 nm, 269 nm, 275 nm, 280 nm, and 285 nm for inactivating Escherichia coli (Chatterley and Linden, 2010, Vilhunen, 2010, Bowker et al., 2011, Oguma et al., 2013, Oguma et al., 2016, Lui et al., 2016). At least two studies evaluated UV LEDs emitting at or near 250 nm, 270 nm, and 282 for inactivating Bacillus subtilis spores (Wurtele et al., 2011, Morris, 2012). Other research evaluated UV LEDs emitting at 255 nm and 275 nm for inactivating coliphage MS2 and T7 (Bowker et al., 2011). A recent study evaluated UV LEDs emitting at 285 nm for inactivating adenovirus 5, MS2, and QB (Oguma et al., 2016).
As an emerging technology, LEDs are constantly improving in power output, energy efficiency, lifespan, and economic viability, all of which will make them more practical for widespread use (SETi, 2012, Song et al., 2016). The typical UV-C LED wall plug efficiency, which currently measures at 1–3%, is projected to improve to at least 10% within the decade, following similar improvements seen in visible, UV-A and UV-B LEDs (SETi, 2012, Harris et al., 2013). Additionally, enhancements in thermal management, optical configuration, and hydrodynamic design are consistently improving the system performance and lifespan of commercially available flow-through UV-C LED disinfection systems (Harris et al., 2013). With this rapid research and development, broad potential exists for using UV-C LEDs in sustainable, electrical, photovoltaic, or battery-powered point-of-use water and wastewater disinfection technologies (Harris et al., 2013, Lui et al., 2014).
Among the primary advantages of UV LEDs is that given their compact size, different wavelength outputs can be combined to optimize pathogen inactivation; and, given their low power consumption and improving efficiencies, this inactivation can potentially occur at a low energy cost. Some disinfection studies have evaluated the combination of multiple UV LED wavelengths on pathogens and non-pathogenic surrogate or challenge microorganisms. Chevremont et al. combined LEDs emitting in the UV-A and UV-C range for microbial disinfection and chemical degradation for wastewater treatment (Chevremont et al., 2012). Oguma et al. combined LEDs emitting in the germicidal range, measuring their collective inactivation of E. coli (Oguma et al., 2013). Chevremont concluded that combined UV-A and UV-C wavelengths synergistically enhanced E. coli and Enterococcus faecalis inactivation; however, this work calculated synergy using time-based inactivation kinetics (2012). Oguma reported no synergistic effects from combined wavelengths from fluence-based inactivation data. These conflicting results as well as an interest in the industry to combine UV LEDs for water disinfection and a general knowledge gap regarding the efficacy of combined UV-C wavelengths on bacteria and viruses revealed an opportunity for more synergy research (Song et al., 2016).
Ideally, a tailored UV disinfection system would target bacteria and viruses by combining a wavelength from the dominant germicidal region (250 nm - 280 nm) with a wavelength from the polypeptide absorbance region below 240 nm (i.e. 220–230 nm). This combination could simulate UV emissions from a medium-pressure (MP) mercury lamp, which has been shown to be more effective than low-pressure (LP) UV lamps at inactivating certain pathogens (Malley et al., 2004, Hijnen et al., 2006). In particular, the polychromatic emission from MP UV lamps is more effective than LP UV lamps at inactivating adenovirus due to damage to viral proteins (Linden et al., 2007, Eischeid and Linden, 2011, Beck et al., 2014). To date, 220–230 nm LEDs are not at a practical stage of development for this application. However, LEDs emitting at 280 nm are widely available. UV absorbance at 280 nm is commonly used for protein quantitation (Aitken and Learmonth, 2001). At 280 nm, proteins exhibit a relative peak in UV absorbance due to the absorbance of the aromatic amino acids tyrosine and tryptophan, as well as the cystine disulfide bond (Jagger, 1967, Schmid, 2001). Therefore, it’s reasonable to infer that LEDs emitting specifically at 280 nm could damage proteins, resulting in increased efficacy, relative to LP UV lamps, for inactivating certain viral pathogens.
This research utilized a UV disinfection unit that combined UV-C LEDs emitting at 260 nm and 280 nm, which are near the relative peak UV absorption of nucleic acids and proteins respectively, to target genomic and protein-based regions of bacterial and viral organisms. It was hypothesized that this LED-based polychromatic UV source could attain similar levels of bacterial and viral inactivation as the MP mercury vapor lamp, but at a lower energy cost. The efficacy of a germicidal UV system combining 260 nm and 280 nm LEDs was compared with the efficacy of MP and LP UV systems, for inactivating E. coli, MS2 coliphage, human adenovirus type 2 (HAdV2), and Bacillus pumilus spores. E. coli was chosen as a common microorganism and fecal indicator frequently used to evaluate UV LED disinfection systems. HAdV2 was chosen as one of the most resistant pathogens to UV irradiation, which drives the UV disinfection requirements for all viruses and has demonstrated enhanced inactivation due to protein damage (USEPA, 2006, Eischeid and Linden, 2011, Beck et al., 2014). MS2 coliphage and B. pumilus spores were chosen because of their frequent use for validating polychromatic UV disinfection systems for obtaining adenovirus credit (Linden et al., 2015).
The experimental set-up used in this study provided a unique opportunity to investigate potential synergistic effects of multiple wavelengths on microorganisms. The log inactivation of microorganisms irradiated individually by 260 nm and 280 nm UV LED units was summed together and compared to the log inactivation achieved from the combined 260|280 nm irradiation to examine potential synergies at a given fluence. To provide insight into the mechanisms of possible dual-wavelength synergy, the direct genome damage of HAdV2 and MS2 coliphage was also quantified.
2. Materials and methods
2.1. UV irradiations
Bacterial and viral suspensions were irradiated with a prototype UVinaire™ dual-wavelength UV-C LED unit supplied by AquiSense Technologies (Erlanger, KY, Figure S1). The unit was set up in a collimated beam apparatus (Bolton and Linden, 2003) and operated in three modes, powering the 260 nm LEDs (39 W), 280 nm LEDs (31 W), or the combination of 260 nm and 280 nm together (termed 260|280 nm, 66 W). The LED spectra (Fig. 1), measured with a Maya 2000 Pro spectrometer (Ocean Optics, Dunedin, FL), exhibited peak wavelength emissions at 259.6 nm and 276.6 nm with FWHM bandwidths of 12.6 nm and 9.8 nm, respectively. These spectra are compared to the emission from an MP UV mercury vapor lamp (Rayox, 1 kW, Calgon Carbon Corporation, Pittsburgh, PA) and an LP UV system consisting of 4 × 15 W mercury vapor lamps (Fig. 1).
Fig. 1.
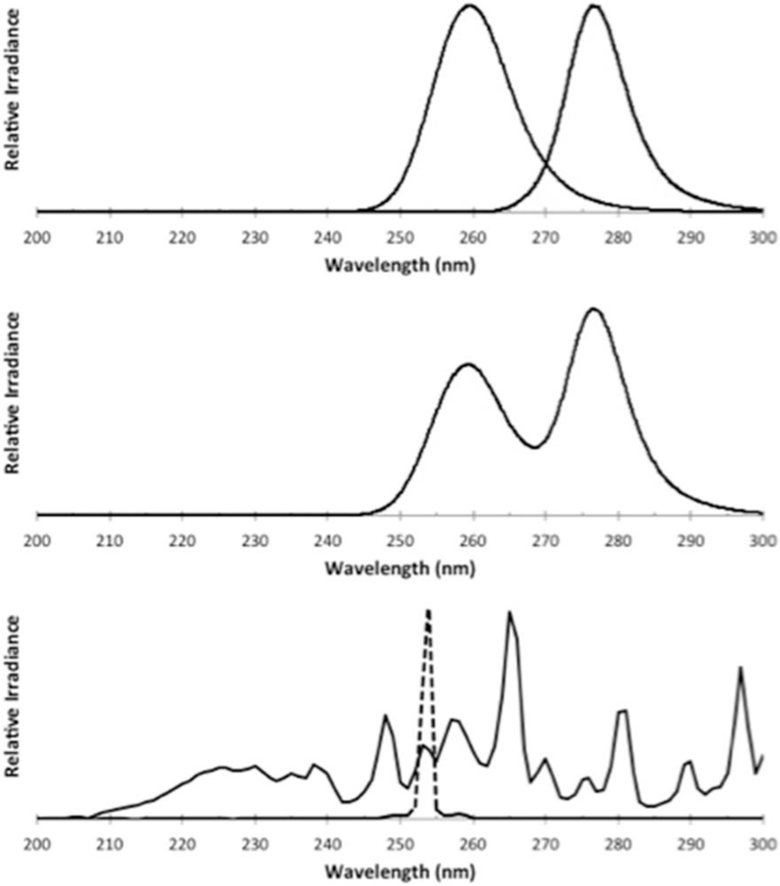
Emission spectra from (top) the 260 nm and 280 nm LEDs when illuminated separately, (middle) the unit with 260 nm and 280 nm LEDs illuminated together (260|280 nm) and (bottom) a medium-pressure (solid) and low-pressure (dashed) mercury vapor lamp
For each UV source, four collimated beam exposures were conducted in triplicate to generate a UV dose response curve up to 3-log inactivation for each microorganism. Stirred suspensions of 5 mL (0.6 cm depth, 3.5 cm diameter) were irradiated at 4 cm from the UV-C LED source. Irradiance was measured at the water surface with an IL-1700 radiometer, SED 240 detector, and W-diffuser (International Light, Peabody, MA). Incident irradiance varied from 0.19 to 0.55 mW/cm2 for the UV-C LEDs, from 0.35 to 1.17 mW/cm2 for the MP UV and from 0.3 to 0.75 mW/cm2 for the LP UV experiments.
Average UV doses for the collimated beam tests were determined as described previously (Bolton and Linden, 2003), adjusting for reflection off the water surface, UV absorption (Cary 100 spectrophotometer, Agilent Technologies, Santa Clara, CA), depth of the water sample, and the uniformity of the distribution of light across the surface of the sample. Petri factors ranged from 0.9 to 0.95 for the UV-C LED work and were approximately 1.0 for MP UV and LP UV exposures.
UV doses for the polychromatic UV-C LEDs and MP UV lamp also accounted for the relative lamp emission (RLE) of each source (Maya 2000 Pro spectrometer) and the wavelength-specific sensitivity of the radiometer detector (given by the radiometer manufacturer) yielding a sensor factor correction (Bolton and Linden, 2003, Linden and Darby, 1997). For the UV-C LED exposures, the RLE and radiometer sensitivity were taken relative to the weighted average wavelength of each LED (i.e. 261 nm for the 260 nm LED, 278 nm for the 280 nm LED, and 271 nm for the 260|280 nm LED combination). For the MP UV, the RLE and radiometer sensitivity were taken relative to 254 nm.
As light emitted from polychromatic sources does not have an equal effect on microorganisms, it is usually weighted germicidally to account for germicidal differences in the wavelength emission (Linden and Darby, 1997). To compare the LED efficacy to MP UV, the average irradiance of each polychromatic light source was weighted germicidally, using the absorption of DNA, which is the industry standard for polychromatic UV sources (Linden and Darby, 1997, USEPA, 2006). For the LED synergy calculations, however, UV doses for the 260 nm, 280 nm, and 260|280 nm combined irradiation were determined using calculations of average irradiance throughout the water sample for simplicity. The average and germicidal irradiances used in this study are given in Table S1 and S2 in the Supporting Information (SI).
Irradiations were conducted at room temperature. The LED array was mounted to a heatsink with a rear-mounted fan for heat dissipation; temperature was not a concern. Immediately after exposure, the irradiated HAdV2 and B. pumilus samples were shipped overnight on icepacks to the US Environmental Protection Agency (EPA) (Cincinnati, OH) and analyzed the following day. E. coli and MS2 coliphage samples were refrigerated at 4 °C prior to same-day analysis at the University of Colorado. Aliquots of each HAdV2 and MS2 coliphage sample were placed in a −80 °C freezer for subsequent DNA and RNA analyses, respectively.
2.2. Energy
Electrical energy per order, EEO is a parameter for characterizing the electrical energy efficiency of disinfection systems. It has been used for interpreting collimated beam data to estimate electrical efficiencies of LP UV and MP UV lamps for large-scale treatment of chemical contaminants (Sharpless and Linden, 2005). In the present work, EEO was used to estimate electrical efficiencies of biological inactivation to compare the performance of the relatively new UV-C LED technology with the more prevalent mercury vapor lamps. Derived previously (Bolton and Stefan, 2002, Sharpless and Linden, 2005), the EEO defines the amount of energy (kWh/m3) required to decrease the concentration of a contaminant or a microorganism by one order of magnitude:
| [1] |
A is the irradiated surface area in cm2. V is the sample volume in liters. kD is the log10fluence-based rate constant in cm2/mJ. C is the wall plug efficiency given by the manufacturer (0.35 for LP UV, 0.15 for MP UV, and 0.004, 0.005, and 0.00444 for the 260 nm, 280 nm and 260|280 nm LEDs respectively). WF is the water factor, accounting for the UV absorbance and depth of the water. The factor 3.6 × 106 is to convert between hrs and sec, mW and kW, and L and m3 (Sharpless and Linden, 2005).
In the case of HAdV2 and B. pumilus spores, where the UV dose response results either followed a linear inactivation not originating at the origin or when the data was best fit with a 2nd-order polynomial, the electrical energy for a specific log reduction, N, of each sample, EEL,N (in kWh/m3) was calculated. The UV dose (mJ/cm2) required for obtaining n-log reduction, DN, was substituted in Eqn. (1), as the inverse of the fluence-based inactivation rate constant:
| [2] |
It is important to note that the results for electrical energy per order and electrical energy for a specific log reduction, EEO and EEL,N, for the exposed samples represent the current state of efficiency for the disinfection technologies compared.
2.3. Bacteria and virus propagation and enumeration
2.3.1. E. coli propagation and enumeration
An overnight culture of E. coli K12 (ATCC 29425) was inoculated into 100 mL of sterile tryptic soy broth (TSB) and incubated at 37 °C until reaching log phase, determined by optical density. The cells were washed with sterile phosphate buffered saline (PBS) three times by centrifugation (5000 rpm, 5 min) and resuspended in PBS for a working concentration of approximately 106 CFU/mL.
For E. coli enumeration, irradiated samples were serially diluted in PBS before using the spread plate technique within 2 h. Volumes of 100 μL of the diluted samples were spread on tryptic soy agar (TSA) and incubated inverted at 37 °C for 18–24 h. Samples were plated in duplicate. Plates yielding 0 to 200 colonies were included in the analysis.
2.3.2. MS2 coliphage propagation and enumeration
For MS2 coliphage, TSB supplemented with ampicillin/streptomycin was inoculated with log-phase host bacteria, E. coli F amp (ATCC 700891), and MS2 coliphage (ATCC 15597-B1). The suspension was incubated with constant shaking for 5 h at 37 °C. After the bacterial debris was removed by centrifugation, the clarified supernatant was decanted to sterile containers and stored at −20 °C.
Coliphage stock was diluted to a working concentration of 5 × 106 PFU/mL. After UV exposure, samples were serially diluted and enumerated in duplicate following EPA Method 1601 (USEPA, 2001). Briefly, 100 μL of each dilution was added to a soft agar containing the log-phase host bacteria. The inoculated soft agar was poured over an agar plate and allowed to harden. Plates were inverted and incubated at 37 ± 0.5 °C for 16–24 hr. Viral plaques were counted to determine the concentration of coliphage.
2.3.3. Adenovirus propagation and enumeration
Human adenovirus type 2 (HAdV2) (ATCC VR-846, Manassas, VA) was propagated in A549 human lung carcinoma cells (ATCC CCL-185) as described in Ryu et al. (2015), resulting in stock titers of 1010 MPN/ml. Viral stocks were stored at −80 °C until UV exposure.
HAdV2 was enumerated using the total culturable virus assay (TCVA). Test cultures of A549 cells were grown in plug-capped 25 cm2 flasks (Greiner) in Dulbecco’s Minimum Essential Medium (DMEM) (Life Technologies, Frederick, MD), with antibiotic-antimycotic solution (Invitrogen, Grand Island, NY), and incubated in ambient air at 37 °C. Test cultures were inoculated 3–4 days after planting. Prior to sample inoculation, A549 cells were washed with 7–10 ml of Earle’s Balanced Salts Solution. Samples were diluted five-fold in PBS, and three to four dilution series per sample were divided among 10 replicate A549 cell culture flasks (10 flasks per dilution). One ml of inoculum was inoculated into each flask; flasks were placed on a rocker at room temperature for a minimum of 90 min to ensure viral attachment before adding 10 ml of media, consisting of DMEM with 2% serum and antibiotic-antimycotic liquid (Life Technologies). Flasks were incubated for 21 days at 37 °C and checked each week for cytopathogenic effects (CPE). A Most Probable Number (MPN) approach was used to estimate the number of infectious units in each sample based on the number of CPE-positive replicates in each of the five-fold dilution series. Each MPN sample was examined for CPE at 2 weeks and 3 weeks post-infection. Log-removal was determined from MPN data.
HAdV2 was also enumerated by Integrated Cell Culture Quantitative PCR (ICC-qPCR), described by Ryu et al. (2015), which quantifies infectious adenoviruses from viral DNA harvested from a cell culture monolayer. Briefly, test cultures were inoculated with HAdV2 samples and incubated for 48 hr at 37 °C in a 5% CO2 incubator. After incubation, cell monolayers were washed with PBS to remove extracellular viruses and harvested by scraping after a freeze-thaw cycle. To reduce the impact of potential false-positive signals, rinse control replicates were processed along with a set of ICC-qPCR standards, and their mean copy numbers were subtracted for respective samples. Viral DNA from cells harvested with infectious viruses was extracted, purified, stored, and then analyzed by qPCR (quantitative polymerase chain reaction) as described previously (Ryu et al., 2015).
2.3.4. B. pumilus propagation and enumeration
B. pumilus (ATCC 27142) was obtained from Mesa Laboratories (Omaha, NE). Spores were produced by inoculating vegetative cells of B. pumilus into half strength (0.5x) Columbia broth (Remel, Lenexa, KS) supplemented with 0.1 mM of MnSO4. Cultures were then incubated for 5 days at 35 °C and 100 rpm. Spores were purified by gradient separation using 58% (v/v) RenoCal-76 (Bracco Diagnostics, Princeton, NJ). Spore preparations were stored in 40% (v/v) ethanol at 4 °C until UV experiments were conducted.
Prior to UV exposure, spores were washed with Butterfield’s buffer (Hardy Diagnostics, Santa Clara, CA) 3 times by centrifugation (15 min at 5184 g) as outlined in Standard Method 9050C section 1.A (APHA et al., 2012). Spores were resuspended and diluted with Butterfield’s buffer for a final, working concentration of 105 CFU/mL. Spore enumeration followed the membrane filtration method outlined in Standard Method 9218B section 3.C (APHA et al., 2012). Filters were placed onto nutrient agar (BD, Sparks, MD) supplemented by trypan blue dye and incubated at 35 °C for 18–24 hr.
2.4. Statistical analysis
Log inactivation was calculated as log (No/Nt). For E. coli and MS2 coliphage, the UV dose response data were fit linearly, and the log10 fluence-based inactivation rate constant, kD(cm2/mJ) was determined as follows:
| [3] |
where No is the number of colony forming units (CFU/mL) or plaque forming units (PFU/mL) of the unirradiated control, Nt is the CFU/mL or PFU/mL for each sample at time t, and F is the fluence at the given wavelength, λ, determined as described above. Data for B. pumilusspores also exhibited linear inactivation kinetics and was fit with the above equation; however, the B. pumilus spore UV dose response exhibited a shoulder and therefore, a constant term was included in the equation above. HAdV2 data, which exhibited curvature and a shoulder, in the case of the total culturable virus assay (TCVA), was fit with a second order polynomial. All data were collected in triplicate and are presented with error bars representing one standard deviation.
2.5. Synergy of inactivation
UV inactivation is fluence-based; therefore, potential synergistic effects of combined wavelengths on microorganism inactivation efficacy were determined for a constant fluence. The 260 nm and 280 nm LEDs supplied irradiance to the combined 260|280 nm LED system in slightly unequal amounts, with the 260 nm UV-C LEDs contributing 0.475 and the 280 nm UV-C LEDs contributing 0.525 of the total irradiance emitted by the combined 260|280 nm LED unit. Therefore, the UV dose response results from the 260 nm and 280 nm LEDs were multiplied by 0.475 and 0.525 respectively and summed together for comparison with the UV dose response results from the 260|280 nm LEDs combined. An independent, two-tailed paired t-test, was used to determine significance (<0.05).
2.6. Synergistic damage to the viral DNA and RNA
This study also measured potential synergistic damage to the viral genomes after exposure to UV irradiation from the combined 260|280 nm emissions. DNA damage of adenovirus, measured as the log-reduction in amplification of the adenoviral genome, was measured by analyzing damage to a 1.1-kilobase pair fragment using a published long-range quantitative polymerase chain reaction (LR-qPCR) method (Beck et al., 2014). RNA damage of MS2 coliphage, measured as the log-reduction for each 1185-base-pair amplicon, was measured following a published Reverse Transcription (RT) PCR method (Beck et al., 2016).
As described above, potential synergy was measured by comparing the log reduction in amplification after exposure to the 260|280 nm LED unit with the sum of log reduction in amplification from the respective contributions of the 260 nm and 280 nm LEDs. A two-tailed independent paired t-test was used to detect significance (p < 0.05).
3. Results and discussion
3.1. UV inactivation
UV dose response results from the collimated beam trials with the UV LEDs and MP UV and LP UV lamps are given in Fig. 2.
Fig. 2.
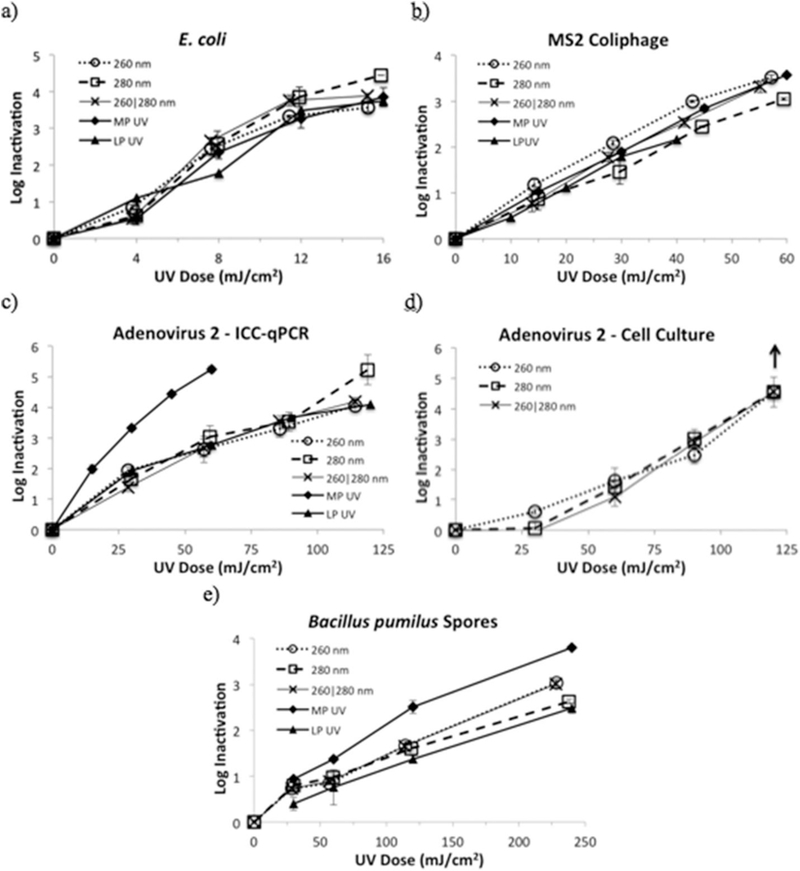
UV dose response of a) E. coli, b) MS2 coliphage, c) HAdV2 as measured by ICC-qPCR, d) HAdV2 as measured by cell culture, and e) B. pumilus spores to LP irradiation from UV LEDs emitting at 260 nm, 280 nm and 260|280 nm combined. Results are shown in comparison to MP UV and LP UV irradiation, when available. Error bars represent ± 1 standard deviation. The up-down in Fig. 2d illustrates that the assay detection limit was reached.
3.1.1. E. coli
UV inactivation of E. coli (Fig. 2a) showed tailing at high doses, arising, in part, from the starting concentration of E. coli and the dilutions used for the experiments. All five UV sources attained over 3-log reduction at UV doses of 12 mJ/cm2 or lower. Fluence-based inactivation rate constants, kD, are given in Table 1.
Table 1.
Inactivation rate constants (log10), kD, of E. coli and MS2 coliphage. For HAdV2 and B. pumilus, UV dose required for 2-log inactivation as well as log inactivation (y) as a function of UV dose (x).
| kD (cm2/mJ) ± 1SD | UV dose for 2-log reduction (mJ/cm2) (Range for ± 1SD) |
||||
|---|---|---|---|---|---|
| UV Source | E. coli | MS2 coliphage |
HAdV2 | B. pumilus spores | |
| ICC-qPCR | Cell culture | ||||
| 260 nm LED | 0.29 ± 8.1E- 3 |
0.066 ± 0.0017 |
38.3 (30.2–47.1) | 69.2 (63.2–74.9) | 143.1 (134.8–151.4) |
| 280 nm LED | 0.31 ± 0.016 | 0.052 ± 0.0013 |
41.0 (30.0–51.2) | 73.5 (64.9–79.0) | 175.9 (159.2–192.6) |
| 260|280 nm LED |
0.32 ± 0.016 | 0.061 ± 0.0010 |
41.1 (37.0–45.5) | 74.2 (65.8–79.8) | 143.6 (135.1–152.1) |
| MP UV Lamp | 0.27 ± 0.013 | 0.061 ± 0.0020 |
16.1 (14.2–17.6) | -- | 101.4 (86.8–116.0) |
| LP UV Lamp | 0.27 ± 0.010 | 0.056 ± 0.0017 |
36.9 (31.9–40.5) | -- | 188.8 (188.8–209.9) |
| Log Inactivation Curves | |||||
| UV Source | HAdV2 ICC-qPCR | HAdV2 cell culture | B. pumilus spores | ||
| 260 nm LED | y = −0.000232×2 + 0.0611x | y = 0.000219×2 + 0.0137x | y = 0.012x + 0.287 | ||
| 280 nm LED | y = −8.375E-5×2 + 0.0523x | y = 0.000272×2 + 0.00719x | y = 0.008x + 0.538 | ||
| 260|280 nm LED | y = −0.000164×2 + 0.0554x | y = 0.000353×2 + 0.00080x | y = 0.0118x + 0.311 | ||
| MP UV Lamp | y = −0.000855×2 + 0.1381x | – | y = 0.0137x + 0.608 | ||
| LP UV Lamp | y = −0.000246×2 + 0.0632x | – | y = 0.0098x + 0.143 | ||
For the UV LEDs, the rate constant at 280 nm (kD = 0.31 cm2/mJ) was similar to that reported by Oguma et al. (2013) of 0.29 cm2/mJ. E. coli inactivation efficacy by the 260 nm and 280 nm UV LEDs were not statistically different, which is interesting given that 260 nm is closer than 280 nm to the relative peak of the UV action spectrum and the relative peak of the UV absorbance of a 0.8 μm layer of E. coli on an agar surface (Gates, 1930).
The MP UV and LP UV germicidal weighted inactivation rates of E. coli were equal and 3-log inactivation was reached at 10–11 mJ/cm2 similar to that reported by Guo et al. (2009).
3.1.2. MS2 coliphage
The UV inactivation of MS2 coliphage (Fig. 2b, Table 1) exhibited linear inactivation kinetics. The 260 nm LEDs were statistically more effective at inactivating MS2 than the 280 nm LEDs, which is expected given that the UV absorbance of MS2 RNA and the MS2 action spectrum both have a relative peak near 260 nm (Rauth, 1965, Mamane-Gravetz et al., 2005). The MS2 virus and its RNA are more sensitive to UV light at 260 nm than at 280 nm (Strauss and Sinsheimer, 1963, Mamane-Gravetz et al., 2005, Beck et al., 2016). The combination of 260|280 nm LEDs together was statistically less effective than the 260 nm LED and statistically more effective than the 280 nm LED; which was also expected.
The UV dose response of MS2 coliphage to the 260 nm and 280 nm LEDs (Table 1) corresponds to 2-log inactivation at UV doses of 30.3 mJ/cm2 and 38.5 mJ/cm2 respectively. These doses are lower than those reported for 255 nm and 275 nm LEDs which caused 2-log inactivation at 50 mJ/cm2 and 55 mJ/cm2 respectively (Bowker et al., 2011). One factor that may account for the differences is that the Bowker study treated the LEDs as monochromatic sources, whereas this study treats them as polychromatic sources and weights the irradiance at each wavelength by the DNA absorbance, which would alter the UV dose. Small differences in UV irradiation between 255 nm and 260 nm and between 275 nm and 280 nm could have also played a role. On the other hand, these results agree very well with those from a recent study of MS2 inactivation by monochromatic irradiation from a tunable laser, in which 2-log reduction was achieved between 24 and 30 mJ/cm2 at 260 nm and between 38 and 53 mJ/cm2 at 280 nm (Beck et al., 2015).
The UV dose response of MS2 coliphage to MP UV irradiation was almost identical to that from the combined 260|280 nm LED unit, corresponding to 2-log reduction at a UV dose of 32.8 mJ/cm2. This MP UV kD was lower than that reported in the literature, requiring a higher UV dose for 2-log inactivation than those previously shown (Malley et al., 2004, Hijnen et al., 2006, Beck et al., 2015).
3.1.3. Adenovirus
Dose response results of HAdV2 inactivation are given in Fig. 2 and Table 1. For the UV-C LEDs, the ICC-qPCR method (Fig. 2c) measured 3-log reduction at UV doses between 64 and 68 mJ/cm2 and 4-log reduction at an estimated 122, 89, and 105 mJ/cm2 for the 260 nm, 280 nm, and 260|280 nm LEDs respectively. The most notable difference between the UV-C LED dose-response curves, was the higher log reduction from the 280 nm LED at the highest dose tested. The dose response was of the same order of magnitude for the first three UV doses, below 30, 60, and 90 mJ/cm2, however, at the highest UV dose tested, between 114 and 119 mJ/cm2, the 280 nm LED caused one full log reduction more inactivation than the 260 and 260|280 nm LEDs. Although this was detected in all three replicate experiments, it warrants further research as it is only one data point. The additional log inactivation detected from the 280 nm LEDs could be due to protein damage from higher energy inputs. In past research, protein damage accelerated at higher UV doses (∼300 mJ/cm2) while being minimally expressed at UV doses below 186 mJ/cm2 (Eischeid and Linden, 2011). The change in kinetics of protein damage with increasing UV dose could be the result of structural changes occurring as the proteins broke down (Rexroad et al., 2003).
When measuring HAdV2 inactivation by the total culturable virus assay (Fig. 2d), results from the three UV-C LED configurations were also similar. A UV dose between 91 and 93 mJ/cm2 was required for 3-log inactivation and an estimated UV dose of 105–109 mJ/cm2for 4-log inactivation. These doses are lower than those reported in the literature. When irradiated with a tunable laser emitting at 260 nm, 103.4 mJ/cm2 and 137.9 mJ/cm2 were required for 3-log and 4-log inactivation respectively; at 280 nm, 93.8 and 125 mJ/cm2 were required for 3- and 4-log inactivation, respectively (Beck et al., 2014). These differences could be attributed to differences in UV sources. The tunable laser was a monochromatic (<1 nm bandwidth) UV light source whereas the LEDs have a full width at half maximum (FWHM) bandwidth of approximately 10–12 nm. Past research has shown that polychromatic irradiation is more effective at inactivating a microorganism or virus than monochromatic light emitting at the weighted average wavelength (Mamane-Gravetz et al., 2005, Wright et al., 2007).
When exposed to MP UV irradiation, HAdV2 inactivation by 2-log, 3-log, and 4-log, as measured by ICC-qPCR, occurred at doses of 16 mJ/cm2, 26 mJ/cm2, and 38 mJ/cm2, respectively. This agrees with past studies showing 2-log and 3-log reduction at MP UV doses of 19 mJ/cm2 and 34 mJ/cm2, respectively, and approximately 4.3-log reduction at a MP UV dose of 40 mJ/cm2 (Linden et al., 2007, Beck et al., 2014). The LP UV dose response of HAdV2 was 2-log, 3-log, and 4-log at 37, 63, and 112 mJ/cm2 respectively, which agrees with previous results using the same method that reported 4-log inactivation at 116 mJ/cm2 (Gerrity et al., 2008). However, these LP UV doses, obtained using ICC-qPCR, were lower than the literature derived from cell culture alone, which reported approximately 1.6-log and 2.25-log at 40 mJ/cm2 and 60 mJ/cm2 respectively or 4-log reduction between 120 and 168 mJ/cm2 (Gerba et al., 2002, Thompson et al., 2003, Shin et al., 2005, Beck et al., 2014). Variations in log-reduction could arise from differences in assays used.
3.1.4. B. pumilus
UV inactivation of B. pumilus spores (Fig. 2e, Table 1) exhibited linear inactivation kinetics with a shoulder and was therefore fit linearly with lines not originating at the origin. B. pumilus spores exhibited almost identical inactivation from the 260 nm and the 260|280 nm UV irradiation, both of which were statistically more effective than the 280 nm UV irradiation for 2-log reduction. This aligns well with the action spectrum of B. pumilusspores, which shows a greater sensitivity to 260 nm irradiation than to 280 nm (Rochelle et al., 2010, Beck et al., 2015). The resistance of Bacillus spores varies greatly with the concentration of MnSO4 used in the spore propagation medium and incubation time. This has been especially evident with B. subtilis, but also has an effect on B. pumilus spore data (Rochelle et al., 2010, Boczek et al., 2015). Given the differences in propagation methods between this study and past studies, it is difficult to compare the B. pumilus spore data with past results for inactivation at 260 nm and 280 nm.
When exposed to MP UV, B. pumilus spores were inactivated by 2-log and 3-log at doses of 102 and 175 mJ/cm2 respectively. These spores were less sensitive than those of a different strain reported in the literature from a similar propagation protocol (Rochelle et al., 2010). The UV dose response of B. pumilus spores to LP UV irradiation showed 2-log reduction at 189 mJ/cm2, which agrees with the literature for different strains of B. pumilus spores propagated with the same method (Rochelle et al., 2010). MP UV was consistently more effective than LP UV at inactivating B. pumilus spores, which was also shown previously for a different strain of B. pumilus (Rochelle et al., 2010). MP UV emits at low wavelengths, including from 220 to 228 nm, where B. pumilus spore inactivation is high, presumably due to damage to small acid-soluble proteins bound to the spore DNA (Setlow, 2006, Rochelle et al., 2010).
3.2. Energy
Comparisons of electrical energy per order, EEO, and the electrical energy required for 2-log reduction, EEL,2, are given in Fig. 3. The LP UV lamp, which is the most efficient of the five UV sources (Autin et al., 2013), had the lowest EEO, corresponding to the least amount of energy required per log reduction of E. coli and MS2, followed by the MP UV lamp. Of the UV-C LED sources, for E. coli inactivation, the 280 nm LEDs and the 260|280 nm LED combination required statistically less energy per log reduction than the 260 nm LEDs. For MS2 coliphage, the 260|280 nm LED combination appeared slightly more energy efficient than the other UV-C LEDs; however, the results were not statistically significant.
Fig. 3.
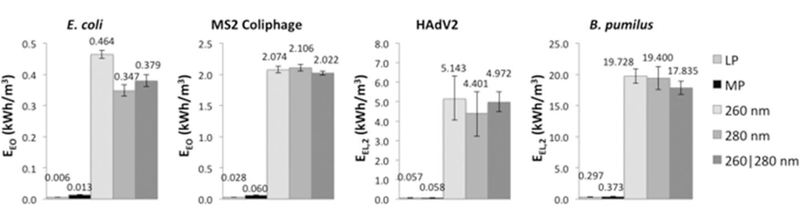
Electrical energy per order (EEO) of reduction of E. coli and MS2 coliphage and electrical energy per 2-log reduction(EEL,2) for HAdV2 and B. pumilus for the five UV sources. Error bars represent ± 1 standard deviation. Note the different y-axis values.
For HAdV2, using the ICC-qPCR data, the electrical energy required for 2-log reduction, EEL,2, was equally low for both the LP UV and MP UV lamps. Enhanced inactivation from the low UV wavelengths (<240 nm) made the MP UV approximately 2.3 times more effective than the LP UV at inactivating HAdV2 (with the standard DNA-weighted dose calculation). However, the LP UV source is approximately 2.3 times more efficient than the MP UV, so the EEL,2 are essentially equal for HAdV2 inactivation. The MP UV had a lower EEL at 3-log reduction whereas the LP UV was lowest at 1- and 2-log reduction (not shown). Of the UV-C LEDs, the 280 nm LED unit appeared to require slightly less energy; however, the results were not statistically significant.
Similarly, for B. pumilus, the electrical energy required for 2-log reduction, EEL,2, was lowest for the LP UV and MP UV lamps; EEL,1 (not shown) was lowest for MP UV and EEL,2 was lowest for LP UV. As with HAdV2, enhanced inactivation by the MP UV at low UV wavelengths balanced out its lower lamp efficiency in the EEL,N calculation. For the UV-C LEDs, the 260|280 nm LED combination appeared to require less electrical energy per log reduction, but the results were not statistically significant.
Table 2 compares the electrical energy per order of log reduction (EEO) between the polychromatic UV sources (MP UV and UV-C LEDs) and the LP UV source. Given the current state of the electrical efficiency for UV-C LED technology, for inactivating these four microorganisms, the UV-C LEDs would have to become approximately 50–90 times more efficient to achieve the same EEO or EEL,N as the LP UV lamp. Given their measured efficiencies of 0.004–0.005, this means that UV-C LEDs with efficiencies of between 0.25 and 0.39 (see Table 2) would be as efficient per order of log reduction as the LP UV mercury vapor lamp for inactivating E. coli, MS2 coliphage, HAdV2 and B. pumilus.
Table 2.
The ratio of EEO and EEL,N for each source to that for the LP UV, as well as the target efficiencies for each source to achieve the same EEO or EEL,N as the LP UV lamp. For HAdV2 and B. pumilus, the number given is the average of the EEL,N to EEL,LP ratios calculated for 1-, 2-, and 3-log reduction.
| UV Source | E. coli | MS2 coliphage | HAdV2 | B. pumilus | ||||
|---|---|---|---|---|---|---|---|---|
| EEO/EEO,LP | Target efficiency | EEO/EEO,LP | Target efficiency | Average EEL,N/EEL,N,LP | Target efficiency | Average EEL,N/EEL,N,LP | Target efficiency | |
| 260 nm LED | 81.5 | 0.33 | 74.2 | 0.30 | 90.8 | 0.36 | 63.1 | 0.25 |
| 280 nm LED | 61.0 | 0.30 | 75.4 | 0.38 | 76.9 | 0.38 | 54.9 | 0.27 |
| 260|280 nm LED | 66.5 | 0.30 | 72.4 | 0.32 | 87.3 | 0.39 | 56.5 | 0.25 |
| MP UV | 2.33 | 0.35 | 2.14 | 0.32 | 1.01 | 0.15 | 1.01 | 0.15 |
| LP UV | 1 | – | 1 | – | 1 | – | 1 | – |
3.3. Synergy
Data evaluating potential UV-C LED synergy is shown in Fig. 4. When the UV dose response results from the 260 nm and 280 nm LEDs are weighted by their respective average irradiance percentages and summed together, visually, they equal the UV dose response results of the 260|280 nm LEDs combined for all four microorganisms.
Fig. 4.
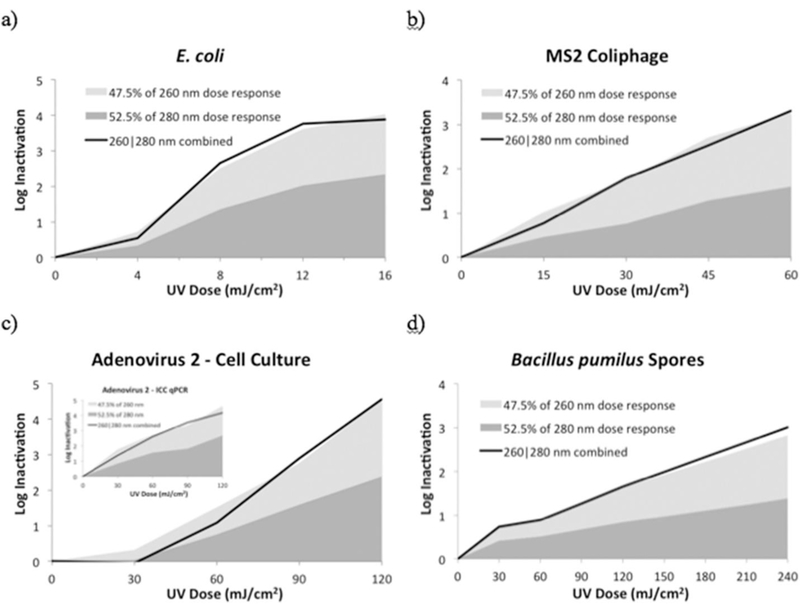
UV dose response of a 260|280 nm combined LED unit (solid line) compared with sum of its UV dose response from separate LED exposures on inactivating a) E. coli, b) MS2 coliphage, c) HAdV2 as measured by cell culture and ICC-qPCR (inset), and d) B. pumilus spores.
Statistically, using a two-tailed paired t-test, the results show that the UV dose response of all four microorganisms to 260|280 nm UV LED irradiation was not statistically (p < 0.05) greater than the sum of the weighted UV dose response from the 260 nm and 280 nm LED irradiation separately, for any microorganism or dose tested. For MS2 coliphage at 45 mJ/cm2 only, and HAdV2 at 30 mJ/cm2 only, the results were statistically significant; however, they differed in the opposite direction, showing that the sum of the log inactivation proportions from 260 nm to 280 nm were greater than the inactivation from those wavelengths combined in the 260|280 nm LED unit.
This work indicates that there is no synergy from a hybrid unit combining 260|280 nm irradiation when compared with the sum of 260 nm and 280 nm wavelengths acting separately for inactivating E. coli, MS2 coliphage, HAdV2, and B. pumilus spores.
3.4. Synergistic damage to the viral DNA and RNA
The log reduction in amplification of the adenoviral genome fragment, which is an indication of DNA damage, after exposure to 260 nm, 280 nm, and 260|280 nm UV LEDs is illustrated in Fig. 5. The inactivation rate constants, k, from the 260 nm and 280 nm LEDs were 0.045 cm2/mJ and 0.032 cm2/mJ respectively, agreeing well with our previous reported values for genome damage following exposure to a monochromatic laser (Beck et al., 2014). When both wavelengths were combined, the inactivation rate constant was slightly higher at 0.049 cm2/mJ. However, this caused no significant synergy as analyzed using the paired t-test at UV doses of 30 mJ/cm2 (p = 0.18), 60 mJ/cm2 (p = 0.07), or 90 mJ/cm2 (p = 0.39).
Fig. 5.
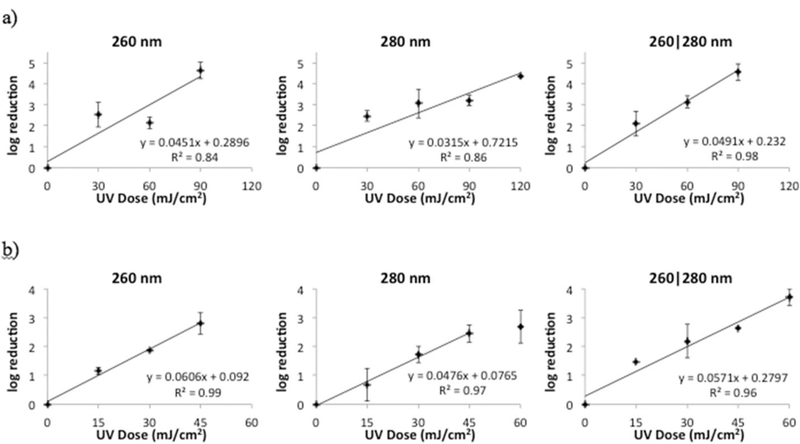
Log reduction in amplification of a) HAdV2 DNA and b) MS2 coliphage RNA to UV light from a UV LED unit emitting at 260 nm, 280 nm and 260|280 nm.
The log reduction in amplification of the MS2 coliphage genome, which is an indication of RNA damage, after exposure to the UV-C LEDs is shown in Fig. 5. The inactivation rate constants, k, from the 260 nm and 280 nm LEDs were 0.061 cm2/mJ and 0.048 cm2/mJ respectively. For the combined 260|280 nm emission, k was 0.057 cm2/mJ. No significant synergy was detected in RNA damage, as analyzed using the paired t-test at UV doses of 15 mJ/cm2 (p = 0.45), 30 mJ/cm2 (p = 0.19), or 45 mJ/cm2 (p = 0.81).
These results expanded on the previous work measuring fluence-based synergy of combined UV-C LED emissions on E. coli (Oguma et al., 2013). The lack of dual-wavelength synergy detected in inactivation of E. coli, MS2 coliphage, HAdV2 and B. pumilus spores, as well as in damage to viral DNA and RNA, agrees with the Second Law of Photochemistry. Photochemical effects of different wavelengths on a molecule should be independent of each other, achieving only as much inactivation or genome damage as the sum of the photonic response from those wavelengths emitting separately. These results confirmed that expectation.
4. Conclusions
This research demonstrates that UV-C LEDs are as effective as common mercury vaporlamps for inactivating bacterial and viral pathogens and pathogen surrogates important to the water treatment industry. Additionally, it shows that multiple wavelengths from 260 nm and 280 nm LEDs acting simultaneously on a microorganism do not cause dual-wavelength synergy for bacterial and viral inactivation nor for DNA or RNA damage. In order to match the electrical efficiencies (energy per log reduction) of conventional LP UV sources, UV-C LEDs must reach efficiencies of 25–39%; however, this finding represents only a snapshot in time of this emerging technology. Advancements in semiconductors and reactor design will continue to improve UV-C LED reactor performance, making the technology more efficient per log reduction for more practical use within the industry. This study will serve as a benchmark for those future comparisons.
Supplementary Material
Acknowledgments
The U.S. Environmental Protection Agency (EPA), through its Office of Research and Development, partially funded and managed the research described herein. S. Beck was partially supported by the U.S. EPA Fellowship Assistance Agreement FP91709801. This work has been subjected to the agency’s administrative review and has been approved for external publication. Any opinions expressed do not reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use. We thank Emma Huff, Jill Hoelle, and Dr. Jennifer Pagan for technical assistance. We also appreciate Jeff Adams and Dr. Nichole Brinkman for their critical review of the manuscript.
References
- Aitken A, Learmonth MP, 2001. Protein Determination by UV Absorption in Walker. J.M. The Protein Protocols Handbook Third Edition. Springer, pp. 3e6. APHA (American Public Health Association), AWWA (American Water Works Association), WEF (Water Environment Federation), 2012. Standard Methods for the Examination of Water and Wastewater, 22nd ed. American Public Health Association, Washington, DC. [Google Scholar]
- Autin O, Romelot C, Rust L, Hart J, Jarvis P, MacAdam J, Parsons SA, Jefferson B, 2013. Evaluation of a UV-light emitting diodes unit for the removal of micropollutants in water for low energy advanced oxidation processes. Chemosphere 92 (6), 745–751. [DOI] [PubMed] [Google Scholar]
- Beck SE, Rodriguez RA, Linden KG, Hargy TM, Larason TC, Wright HB, 2014. Wavelength dependent UV inactivation and DNA damage of adenovirus as measured by cell culture infectivity and long range quantitative PCR. Environ. Sci. Technol 48, 591–598. [DOI] [PubMed] [Google Scholar]
- Beck SE, Wright H, Hargy T, Larason TC, Linden KG, 2015. Action spectra for validation of pathogen disinfection in medium-pressure ultraviolet (UV) systems. Water. Res 70, 27–37. [DOI] [PubMed] [Google Scholar]
- Beck SE, Rodriguez RA, Hawkins M, Linden KG, Hargy TM, Larason TC, 2016. Comparison of loss of infectivity and RNA damage in MS2 coliphage across germicidal UV wavelengths. Appl. Environ. Microbiol 82, 1468–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczek L, Rhodes E, Cashdollar J, Ryu J, Popovici J, Hoelle J, Hayes S, Rodgers M, Ryu H, 2015. Applicability of UV resistant Bacillus pumilus spore as a human adenovirus surrogate for evaluating the effectiveness of virus inactivation in low-pressure UV treatment systems. J. Microbiol. Methods 122, 43–49. [DOI] [PubMed] [Google Scholar]
- Bolton JR, Stefan MI, 2002. Fundamental photochemical approach to the concepts of fluence (UV dose) and electrical energy efficiency in photochemical degradation reactions. Res. Chem. Intermed 2002 28, 857–870. [Google Scholar]
- Bolton JR, Linden KG, 2003. Standardization of methods for fluence (UV dose) determination in bench-scale UV experiments. J. Env. Eng 129 (3), 209–215. [Google Scholar]
- Bowker C, Sain A, Shatalov M, Ducoste J, 2011. Microbial UV fluence-response assessment using a novel UV-LED collimated beam system. Wat. Res 2011 45, 2011–2019. [DOI] [PubMed] [Google Scholar]
- Chatterley C, Linden KG, 2010. Demonstration and evaluation of germicidal UV-LEDs for point-of-use water disinfection. J. Wat. Health 08 (3), 479–486. [DOI] [PubMed] [Google Scholar]
- Chevremont AC, Farnet AM, Sergent M, Coulomb B, Boudenne JL, 2012. Multivariate optimization of fecal bioindicator inactivation by coupling UV-A and UV-C LEDs. Desalination 285, 219–225. [Google Scholar]
- Eischeid AC, Linden KG, 2011. Molecular indications of protein damage in adenoviruses after UV disinfection. Appl. Environ. Microbiol 77 (3), 1145–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates FL, 1930. A study of the bactericidal action of ultraviolet light, III: the absorption of ultraviolet light by bacteria. J. Gen. Physiol 13, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerba CP, Gramos DM, Nwachuku N, 2002. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol 68 (10), 5167–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D, Ryu H, Crittenden J, Abbaszadegan M, 2008. UV Inactivation of Adenovirus Type 4 measured by integrated cell culture qPCR. J. Environ. Sci. Health, Part A Toxic/Hazardous Subst. Environ. Eng 43 (14), 1628–1638. [DOI] [PubMed] [Google Scholar]
- Guo MT, Hu YY, Bolton JR, El-Din MG, 2009. Comparison of low and medium pressure ultraviolet lamps: photoreactivation of Escherichia coli and total coliforms in secondary effluents of municipal wastewater treatment plants. Wat. Res 43 (3), 815–821. [DOI] [PubMed] [Google Scholar]
- Harris TR, Pagan J, Batoni P, 2013. Optical and fluidic co-design of a UV-LED water disinfection chamber. ECS Trans 45 (17), 11–18. [Google Scholar]
- Hijnen WA, Beerendonk EF, Medema GJ, 2006. Inactivation credit of UV radiation for viruses, bacteria, and protozoan (oo)cysts in water: a review. Wat. Res 40 (1), 3–22. [DOI] [PubMed] [Google Scholar]
- Jagger J, 1967. Introduction to Research in Ultraviolet Photobiology Prentice-Hall, Englewood Cliffs, N.J, p. 164. [Google Scholar]
- Linden KG, Darby JL, 1997. Estimating effective germicidal dose from medium pressure UV lamps. J. Environ. Eng 123 (11), 1142–1149. [Google Scholar]
- Linden KG, Thurston J, Schaefer R, Malley JP Jr., 2007. Enhanced UV inactivation of adenovirus under polychromatic UV lamps. Appl. Environ. Microbiol 73 (23), 7571–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden KG, Wright HB, Collins J, Cotton C, Beck SE, 2015. Guidance for Implementing Action Spectra Correction Factors with Medium Pressure UV Disinfection. Final Report, Project #4376 Water Research Foundation, Denver CO. [Google Scholar]
- Lui GY, Roser D, Corkish R, Ashbolt N, Jagals P, Stuetz R, 2014. Photovoltaic powered ultraviolet and visible light emitting diodes for sustainable point-of-use disinfection of drinking waters. Sci. Total Environ 493, 185–196. [DOI] [PubMed] [Google Scholar]
- Lui GY, Roser D, Corkish R, Ashbolt N, Stuetz R, 2016. Point-of-use water disinfection using ultraviolet and visible light-emitting-diodes. Sci. Total. Environ 553, 626–635. [DOI] [PubMed] [Google Scholar]
- Malley JP, Ballester NA, Margolin AB, Linden K, Mofidi A, Bolton JR, Crozes G, Laine JM, Janex ML, 2004. Inactivation of Pathogens with Innovative UV Technologies. American Research Foundation and American Water Works Association
- Mamane-Gravetz H, Linden KG, Cabaj A, Sommer R, 2005. Spectral sensitivity of Bacillus subtilis spores and MS2 coliphage for validation testing of ultraviolet reactors for water disinfection. Environ. Sci. Technol 39 (20), 7845–7852. [DOI] [PubMed] [Google Scholar]
- Morris J, 2012. Disinfection of Bacillus Subtilis Spores Using Ultraviolet Light Emitting Diodes. Electronic Thesis or Dissertation Retrieved from. https://etd.ohiolink.edu/.
- Oguma K, Rattanakul S, Bolton J, 2016. Application of UV light emitting diodes to adenovirus in water. J. Environ. Eng 142 (3). [Google Scholar]
- Oguma K, Kita R, Sakai H, Murakami M, Takizawa S, 2013. Application of UV light emitting diodes to batch and flow-through water disinfection systems. Desalination 328, 24–30. [Google Scholar]
- Rauth A, 1965. The physical state of viral nucleic acid and the sensitivity of viruses to ultraviolet light. Biophys. J 5, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroad J, Wietoff CM, Green AP, Kierstead TD, Scott MO, Middaugh CR, 2003. Structural ability of adenovirus type 5. J. Pharm. Sci 92, 665–678. [DOI] [PubMed] [Google Scholar]
- Rochelle PA, Blatchley ER III, Chan PS, Scheible OK, Shen C, 2010. Challenge organisms for inactivation of viruses by ultraviolet treatment. Final Report, Project #3105 Water Research Foundation, Denver CO. [Google Scholar]
- Ryu H, Cashdollar J, Fout S, Schrantz K, Hayes S, 2015. Applicability of integrated cell culture quantitative PCR (ICCqPCR) for UV inactivation of human adenovirus. J. Environ. Sci. Health: Part A e Toxic/Hazardous Substances & Environmental Engineering J. Environ. Sci. Health, Part A Toxic/Hazardous Subst. Environ. Eng 50 (8), 777–787. [DOI] [PubMed] [Google Scholar]
- Schmid FX, 2001. Biological Macromolecules: UV-visible Spectrophotometry. Encyclopedia of Life Sciences Macmillan Publishers Ltd. [Google Scholar]
- Sensor Electronic Technology, Inc. (SETi), 2012. Sensor Electronic Technology, Inc. Reaches Milestone UVC LED Efficiencies of over 10%, Bringing Consumer Disinfection Markets within Reach [cited August 2016] http://www.s-et.com/news/2012-04-23.pdf.
- Setlow P, 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals.” J. Appl. Microbiol 101 (3), 514–525. [DOI] [PubMed] [Google Scholar]
- Sharpless CM, Linden KG, 2005. Interpreting collimated beam ultraviolet photolysis rate data in terms of electrical efficiency of treatment. J. Environ. Eng. Sci 4, S19–S26. [Google Scholar]
- Shin G, Linden KG, Sobsey MD, 2005. Low pressure ultraviolet inactivation of pathogenic enteric viruses and bacteriophages. J. Environ. Eng. Sci 4, S7–S11. [Google Scholar]
- Song K, Mohseni M, Taghipour F, 2016. Application of ultraviolet light-emitting diodes (UV-LEDs) for water disinfection: a review. Water Res 94, 341–349. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Sinsheimer RL, 1963. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J. Mol. Biol 7, 43–54. [DOI] [PubMed] [Google Scholar]
- Thompson SS, Jackson JL, Suva-Castillo M, Yanko WA, El Jack Z, Kuo J, Chen CL, Williams FP, Schnurr DP, 2003. Detection of infectious human adenoviruses in tertiary-treated and ultraviolet-disinfected wastewater. Water Environ. Res 75 (2), 163–170. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (USEPA), 2001. Method 1601: Male-specific (F) and Somatic Coliphage in Water by Two-step Enrichment Procedure Washington, DC. [Google Scholar]
- U.S. Environmental Protection Agency (USEPA), 2006. Ultraviolet Disinfection Guidance Manual for the Final Long Term 2 Enhanced Surface Water Treatment Rule US Environmental Protection Agency, Washington, D.C. [Google Scholar]
- Vilhunen SMS, 2010. Recent developments in photochemical and chemical AOPs in water treatment: a mini-review. Rev. Environ. Sci. Biotechnol 9, 323–330. [Google Scholar]
- Wright HB, Mackey ED, Gaithuma D, Fonseca C, Baumberger L, Drzurny T, Clancy JL, Hargy TM, Fallon K, Cabaj A, Schmalweisser A, Bierman A, Gribbin C, 2007. Optimization of UV Disinfection Water Research Foundation, Denver, CO. [Google Scholar]
- Wurtele MA, Kolbe T, Lipisz M, Kulberg A, Weyers M, Kneissl M, Jekel M, 2011. Application of GaN-based ultraviolet-C light emitting diodes e UV LEDs - for water disinfection. Wat. Res 45, 1481–1489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


