Abstract
Background
Entrainment to the environmental light cycle is an essential property of the circadian clock. Although the compound eye is known to be the major photoreceptor necessary for entrainment in many insects, the molecular mechanisms of photic entrainment remain to be explored.
Results
We found that cryptochromes (crys) and c-fos mediate photic entrainment of the circadian clock in a hemimetabolous insect, the cricket Gryllus bimaculatus. We examined the effects of RNA interference (RNAi)-mediated knockdown of the cry genes, Gb’cry1 and Gb’cry2, on photic entrainment, and light-induced resetting of the circadian locomotor rhythm. Gb’cry2 RNAi accelerated entrainment for delay shifts, while Gb’cry1/ Gb’cry2 double RNAi resulted in significant lengthening of transient cycles in both advance and delay shifts, and even in entrainment failure in some crickets. Double RNAi also strongly suppressed light induced resetting. The Gb’cry-mediated phase shift or resetting of the rhythm was preceded by light-induced Gb’c-fosB expression. We also found that Gb’c-fosB, Gb’cry2 and Gb’period (Gb’per) were likely co-expressed in some optic lobe neurons.
Conclusion
Based on these results, we propose a novel model for photic entrainment of the insect circadian clock, which relies on the light information perceived by the compound eye.
Electronic supplementary material
The online version of this article (10.1186/s40851-018-0109-8) contains supplementary material, which is available to authorized users.
Keywords: C-fos, Circadian clock, Clock gene, Cryptochrome, Insect, Photic entrainment
Background
The circadian clock is an endogenous, highly conserved timing mechanism in animals that is used to anticipate and adapt to daily environmental changes [1]. The oscillatory mechanism of insect clocks consists of interlinked transcriptional and translational feedback loops [2–4]. The major players in the loops are so called ‘clock genes’, including period (per), timeless (tim), Clock (Clk), and cycle (cyc). It is generally thought that the products of Clk and cyc genes heterodimerize to form a CLOCK (CLK)/CYCLE (CYC) complex, which activates transcription of per and tim in the late day to early night, and PERIOD (PER) and TIMELESS (TIM) proteins form a heterodimer that then inhibits CLK/CYC transcriptional activity later at night [2, 3]. This negative feedback is thought to produce an approximately 24 h rhythm. There is an additional loop producing rhythmic expression of either Clk or cyc [4]. This oscillatory mechanism includes vrille (vri) and Par domain protein 1 (Pdp1) for Clk [5, 6], and ecdysone induced protein 75 (E75) and hormone receptor 3 (HR3) for cyc [7].
An essential property of the clock is the ability to synchronize with daily environmental cycles, with sunlight as the most important time cue. The mechanism for this synchronization, or entrainment, is best understood in Drosophila, which uses CRYPTOCHROME (dCRY or CRY1) as a photoreceptor molecule. CRY1 is a flavin-based blue light receptor, and is known to be expressed in a limited number of clock neurons [8], where it leads to TIM degradation in a light dependent manner [9, 10], and resets the clock [11, 12]. However, many insects possess another type of CRY, CRY2 [13, 14], which is more similar to mammalian CRYs. The role of mammalian CRYs is not completely understood. Certain lines of evidence suggest they are involved in the core oscillatory mechanism, working together with PER to repress transcriptional activity of CLK and BRAIN AND MUSCLE ARNT LIKE 1 (BMAL1, the mammalian homologue of CYC) complex [15, 16]. However, some studies have shown that mammalian CRY also plays a role as a photoreceptor, and is involved in photic entrainment of the suprachiasmatic nucleus (SCN) or peripheral clocks [17, 18]. In insects, CRY2 is believed to be involved in the core clock oscillatory mechanism [14], mainly based on assays using cultured cell systems. The role of CRY2 is yet to be explored in vivo.
In the present study, we investigated the role of cry1 and cry2 genes in photic entrainment of the circadian clock in the cricket, Gryllus bimaculatus. In this cricket, photic entrainment solely depends on the compound eye [19, 20], and the major circadian photoreceptor molecule in the compound eye is opsin-Long Wavelength (Gb’OpLW) [21, 22]. We have previously shown that resetting of the clock by the extension of the light phase during the early subjective night includes transcriptional regulation of clock genes, with Pdp1 as the first responder to light [22]. However, the reset mechanism in other situations, e.g. in free-running conditions or during the night, remains unknown. We have recently shown that the cricket genome includes two cry genes, which are involved in the oscillatory machinery of its internal clock [23]. Unlike other insects, Gb’cry2 has several transcriptional variants that form a feedback loop in a specific combination with the isoforms and Gb’cry1 [23]. Our RNAi experiments reveal for the first time that re-entrainment to shifted light cycles was rather accelerated by reduced expression of Gb’cry2, but severely disrupted by Gb’cry1 and Gb’cry2 double knock-down. We also show that Gb’c-fosB is involved in the photic entrainment pathway. Based on these results, we propose a novel model of photic entrainment of the insect circadian clock, which furthers our understanding of the insect circadian system.
Materials and methods
Experimental animals
Eighth instar nymphs and adult males of the cricket, Gryllus bimaculatus, were used. They were purchased or obtained from a laboratory colony maintained under standard environmental conditions, with a lighting regimen of alternating 12 h light and 12 h darkness (LD 12:12; light: 0600–1800; Japan standard time, JST) and at a constant temperature of 25 ± 0.5 °C. They were fed laboratory chow and water.
Measurement of mRNA levels
The mRNA levels of Gb’cry1 (GenBank/EMBL/DDBJ Accession No. LC202047), Gb’cry2 (LC202053), Gb’c-fosA (fra-A, LC215243), Gb’c-fosB (fra-B, LC215244), and Gb’Pdp1 were measured by quantitative real-time polymerase chain reaction (qPCR) [22]. Total RNA was extracted and purified from six adult male optic lobes with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and treated with DNase I to remove contaminating genomic DNA. About 500 ng total RNA from each sample was reverse transcribed with random 6mers using PrimeScript RT reagent kit (TaKaRa, Shiga, Japan). qPCR was performed in the Mx3000P real-time PCR system (Stratagene, La Jolla, CA, USA) using FastStart Universal SYBR Green Master (Roche, Tokyo, Japan) including SYBR Green, with primers listed in Additional file 1: Table S1. We used Gb’rpl18a (GenBank/EMBL/DDBJ Accession No. DC448653) as an internal reference gene. Quantification was based on a standard curve obtained with known amounts of template DNA. The results were analyzed using the instrument vendor-associated software. The values were normalized with those of Gb’rpl18a at each time point. Results of 3–8 independent experiments were used to calculate the mean ± SEM.
RNAi
Double stranded RNA (dsRNA) for Gb’cry1, Gb’cry2, Gb’c-fosA, Gb’c-fos (for targeting both Gb’c-fosA and Gb’c-fosB), Gb’opsin-long wavelength (Gb’opLW) (GenBank/EMBL/DDBJ accession No. LC004297), Gb’opsin-blue (Gb’opBlue) (LC004296), and DsRed2 derived from a coral species (Discosoma sp.), were synthesized using MEGAscript High Yield Transcription Kit (Ambion, Austin, TX, USA). For Gb’cry1, Gb’cry2, Gb’c-fos, Gb’c-fosA, Gb’opLW, and Gb’opBlue, template cDNA fragments for in vitro transcription were amplified by PCR from the cricket brain cDNA library using ExTaq DNA polymerase (TaKaRa). Primers tagged with T7 or T3 promoter sequences were used for PCR amplification (primer sequences are listed in Additional file 1: Table S1). For DsRed2 dsRNA, a DsRed2 cDNA fragment was amplified from pDsRed2-N1 (Clontech, Mountain View, CA, USA) with primers listed in Additional file 1: Table S1. Amplified fragments were purified with phenol/chloroform and precipitated with ethanol. RNA was synthesized from each of these cDNA fragments using T7 or T3 RNA polymerase. Synthesized RNA was extracted with phenol/chloroform, and suspended in 50 μl TE buffer after isopropanol precipitation. The yield and quality of RNA was assessed by spectrophotometer (Genequant Pro, Amersham Bioscience, Piscataway, NJ, USA), and equal amounts of sense and antisense RNAs were then mixed. The RNA mixture was denatured for 5 min at 100 °C and annealed by a gradual cooling to room temperature (25 °C). After ethanol precipitation, the obtained dsRNA was suspended in UltraPure DNase/RNase-Free Distilled Water (Invitrogen) and adjusted to a final concentration of 20 μM. The dsRNA solution was stored at − 80 °C until use. 760 nl of dsRNA solution was injected with Nanoliter Injector (WPI, Sarasota, FL, USA) into the abdomen of 8th instar nymphs or adult crickets anesthetized with CO2.
In situ hybridization
The adult male heads were collected at Zeitgeber time 18 (ZT18: ZT 0 and ZT 12 correspond to light-on and light-off, respectively), fixed for 24 h at 4 °C with PFA solution (4% paraformaldehyde in phosphate buffered saline), dehydrated by a series of butyl alcohol and ethanol, and embedded in paraffin. Tissues were sectioned at 6 μm and mounted on MAS-GP type A coated slides (Matsunami Glass, Osaka, Japan). In situ hybridization (ISH) was performed using ViewRNA ISH Tissue Assay (Affymetrix, Santa Clara, CA) following the manufacturer’s protocol. In brief, tissue sections were subjected to xylene deparaffinization followed by ethanol dehydration. To unmask the RNA targets, deparaffinized sections were incubated in pretreatment buffer at 90–95 °C for 10 min and digested with protease (1:100 dilution) at 40 °C for 10 min, followed by fixation with 10% neutral buffered formalin at room temperature for 5 min. Unmasked tissue sections were subsequently hybridized with the ViewRNA probe set (1:50 dilution) for 2 h at 40 °C, followed by series of post-hybridization washes. The ViewRNA probes used for detecting Gb’per (Accession No. AB375516) and Gb’cry2 were designed and synthesized by Affymetrix, covering 1027–2016 and 1548–2669 base region, respectively. A non-probe sample was utilized as a negative control. Signal amplification was achieved via a series of sequential hybridizations and washes according to the manufacture’s protocol. Signals for Gb’per and Gb’cry2 were detected with Fast Red or Fast Blue substrate, respectively. Slides were post-fixed in 10% neutral buffered formalin, mounted in Dako Ultramount mounting medium (Dako, Carpinteria, CA), observed and photographed using light microscopy (BZ-X700, KEYENCE, Osaka, Japan).
In situ RT-PCR
Heads were collected at ZT21 from adult males that were exposed to light for 1 h from ZT20. They were fixed in 4% PFA solution for 24 h at 4 °C, dehydrated by a series of butyl alcohol and ethanol, and embedded in paraffin. Tissues were sectioned at 6 μm and mounted on MAS-GP type A coated slides. The sections were pretreated with 1 U/μl DNase I (TaKaRa) in DNase buffer containing 2 U/μl RNase inhibitor (TaKaRa) at 37 °C overnight. Following the DNase I treatment, the sections were washed with RNase-free PBS and RNase-free water. One-step in situ RT-PCR was performed using RT-PCR Quick Master Mix (TOYOBO, Osaka, Japan) in a thermal cycler (Mastercycler, Eppendorf) with in situ Adapter. Final concentration of the reaction mixture was as follows: 1× RT-PCR Quick Master Mix, 2.5 mM Mn(OAc)2, 0.2 μM of forward and reverse primers for Gb’c-fosB (described in Additional file 1: Table S1), 40 U/μl RNase inhibitor (TaKaRa), 1.2 μl/100 μl of 1 mM digoxigenin (DIG)-11-dUTP (Roche) and relevant amount of RNase free water. The cDNA was synthesized at 60 °C for 30 min. PCR amplification consisted of an initial denaturation step of 94 °C for 1 min, followed by 20 reaction cycles of denaturation (94 °C, 30 s), annealing (60 °C, 30 s), and extension (72 °C, 1 min), then by termination with a final extension reaction at 72 °C for 7 min. The sections were fixed with 4% PFA solution for 10 min at 4 °C and washed in 0.1× standard saline citrate and washing buffer. After a blocking step, the sections were incubated with alkaline-phosphatase-conjugated sheep anti-digoxigenin Fab antibody (Roche). DIG-labeled PCR products were detected with 4-nitro blue tetrazolium chloride (Roche) and 5-bromo-4-chloro-3-indolyl phosphate (Roche) by incubating the sections for the relevant length of time. The sections were mounted, observed and photographed using light microscopy (BZ-X700, KEYENCE).
Behavioral analysis
Locomotor activities were recorded according to Moriyama et al. [24]. Briefly, the final instar nymphs or adult crickets were individually housed in a transparent plastic box (18 × 9 × 4.5 cm) with a rocking substratum. The number of substratum rocks was recorded every 6 min by a computerized system. Food and water were provided ad libitum. The actographs were placed in an incubator (MIR-153, Sanyo Biomedica, Osaka, Japan) with constant temperature at 25 ± 0.5 °C, and light was introduced by a cool white fluorescent lamp connected to an electric timer. The light intensity was 600–1000 lx at the animal’s level, varying with proximity to the lamp. The raw data were displayed as conventional double-plotted actograms to judge activity patterns, and statistically analyzed by the chi-square periodogram [25] with Actogram J (http://actogramj.neurofly.de/) [26]. If a peak of the periodogram appeared above the 0.05 confidence level, the power value (height of the peak above the confidence level) was greater than or equal to 10, and the width of the peak was greater than or equal to 2, the period for the peak was designated as statistically significant [27].
The magnitude of phase shifts caused by a light pulse or a light phase extension was estimated by fitting a regression line to daily activity onsets at steady-state free-running in constant darkness (DD) after light treatments. The phase of the free-running rhythm was determined on the day of light treatment by extrapolating the regression line. The same value was obtained for the control crickets, which received the same treatments but were transferred to DD without light treatment. The magnitude of phase shift of an animal caused by light treatments was estimated by subtracting the average value of the control group from the value of each light-treated animal.
Statistical analysis
One-way analysis of variance (ANOVA) followed by a post-hoc Tukey-Kramer test was used to compare differences in the mean mRNA levels between the different time points, or in the means of magnitudes of phase shifts between groups with various treatments. To compare the means of two groups, t-test was used. In all statistical tests, the significance level was set at α = 0.05.
Results
Gb’cry1 and Gb’cry2 play an important role in photic entrainment
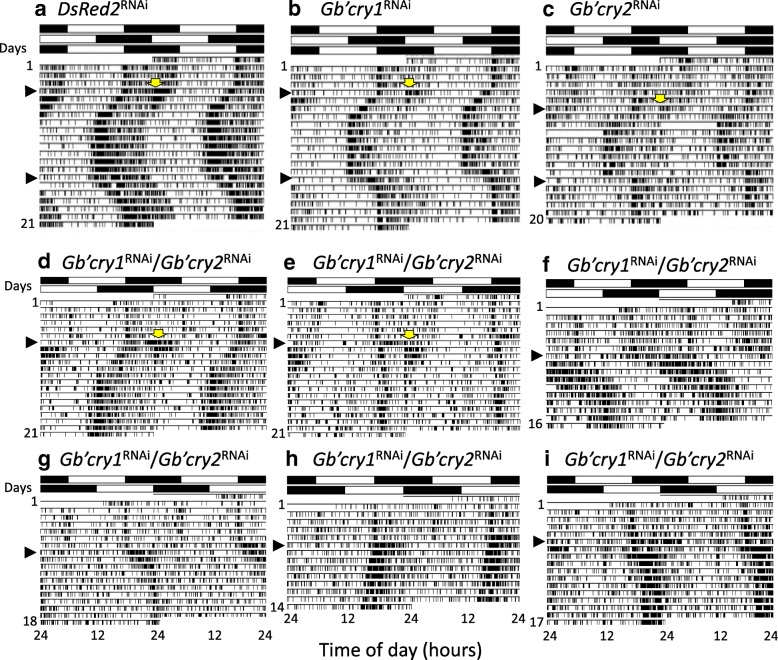
We examined the effects of Gb’cry1RNAi and Gb’cry2RNAi on light entrainment of the locomotor rhythm (Fig. 1). We first confirmed that the crickets were entrained to LD12:12. The onset of their nocturnal activity occurred slightly before light-off for both Gb’cry1RNAi and Gb’cry2RNAi, and their Ψ values were similar to that of DsRed2RNAi control crickets (Table 1). When the light cycle was advanced or delayed by 6 h, control crickets treated with dsDsRed2 re-synchronized to the shifted LD, with transient cycles of approximately four days for both shift directions (Table 2). Although Gb’cry1RNAi and Gb’cry2RNAi crickets also re-synchronized to the shifted LD cycles, the transients tended to be a little shorter than those of the control crickets; however, only the delay shifts of Gb’cry2RNAi crickets were statistically significant (Table 2). Although some of the Gb’cry2RNAi crickets showed instantaneous entrainment to a delayed LD (Fig. 1c), we have to examine whether this is true entrainment or a result of strong negative masking of light. An intense bout of activity occurred at light-on during the transient cycles in advance shifts (Fig. 1a-c). Treatment with dsRNA of Gb’cry2 significantly reduced the light-induced responses (Fig. 1c, Additional file 2: Figure S1). The Ψ after the shifts was again close to that of the DsRed2RNAi controls (Table 1).
Fig. 1.
Entrainment of the locomotor rhythm to LD12:12 in DsRed2RNAi (a), Gb’cry1RNAi (b), Gb’cry2RNAi (c), and Gb’cry1RNAi/Gb’cry2RNAi crickets (Gryllus bimaculatus) (d-i). Light cycles were advanced or delayed by 6 h on the day indicated by an arrowhead on the left side of the actograms. White and black bars above the actograms indicate light (white) and dark (black) cycles. Yellow arrows indicate the light induced activity at light-on after a 6 h phase advance of the light cycle. Transient cycles were shorter in Gb’cry1RNAi and Gb’cry2RNAi crickets than in the control (DsRed2RNAi) crickets, but were longer in Gb’cry1RNAi/Gb’cry2RNAi crickets. Some of the Gb’cry1RNAi/Gb’cry2RNAi crickets (e, h, i) apparently lost entrainability
Table 1.
Effects of Gb’cry1 and Gb’cry2 RNAi on the phase relationship between light-off and activity onset. Different lower case letters indicate that values are significantly different (ANOVA followed by Tukey-test, P < 0.05). Ψadvance and Ψdelay indicate reestablished phase relationship after 6 h phase advance or delay of LD cycles, respectively. Numbers in parenthesis indicate number of animals used
| Treatment | Ψoriginal | Ψadvance | Ψdelay |
|---|---|---|---|
| dsDsRed2 | 0.51 ± 0.46 h (28) | 0.68 ± 0.56a h (18) | 0.92 ± 0.82ab h (13) |
| dsGb’cry1 | 0.60 ± 0.51 h (35) | 0.69 ± 0.60a h (28) | 0.73 ± 0.66ab h (21) |
| dsGb’cry2 | 0.52 ± 0.78 h (15) | 0.56 ± 0.74a h (16) | 0.30 ± 0.28a h (14) |
| dsGb’cry1/dsGb’cry2 | 0.87 ± 0.84 h (45) | 1.69 ± 1.60b h (14) | 1.02 ± 0.76b h (18) |
Table 2.
Effects of Gb’cry1 and Gb’cry2 RNAi on re-entrainment of the circadian locomotor rhythms in Gryllus bimaculatus. Different lower case letters indicate that values are significantly different (ANOVA followed by Tukey-test, P < 0.05)
| Treatment | N | Re-entrained animals (%) | Transient cycles (Mean ± SD; days) |
|---|---|---|---|
| 6 h advance | |||
| dsDsRed2 | 22 | 100 | 4.0 ± 0.84a |
| dsGb’cry1 | 30 | 100 | 3.57 ± 0.72a |
| dsGb’cry2 | 16 | 100 | 3.27 ± 1.22a |
| dsGb’cry1/dsGb’cry2 | 16 | 94 | 6.8 ± 1.52b |
| 6 h delay | |||
| dsDsRed2 | 14 | 100 | 4.0 ± 0.39a |
| dsGb’cry1 | 16 | 100 | 3.25 ± 1.0a |
| dsGb’cry2 | 13 | 100 | 1.15 ± 0.38b |
| dsGb’cry1/dsGb’cry2 | 18 | 89 | 5.56 ± 1.59c |
We then tested the effect of Gb’cry1 and Gb’cry2 double knock-down on photic entrainment (Fig. 1d-i). For both advance and delay shifts, some fraction of the treated crickets could not synchronize within 14 days, as exemplified in Fig. 1e, h, and i. The loss of entrainment was observed in 6% (1/16) and 11% (2/18) of crickets, for advance and delay shifts, respectively. Even in the re-entrained crickets, the transient cycles were significantly greater than in control crickets, with cycles of 6.8 ± 1.52 days and 5.56 ± 1.59 days for advance and delay shifts, respectively (Table 2), and the time course and direction of re-entrainment were variable, with some crickets responding to a 6 h advance shift by synchronizing with delay shifts (Fig. 1f). This variability may be related to the wide variation of the free-running period in the double RNAi crickets [23]. Interestingly, some crickets showed a rhythm splitting, where a component re-synchronized with a gradual advance while the other component did not respond and stayed almost at the same phase (Fig. 1e). The established final phase-relationship with LD was often abnormal in the double RNAi crickets, leading to greater Ψ values (Table 1), although the difference was significant only after advance shifts. Similar to Gb’cry2RNAi crickets, the light-induced activity during advance shifts was significantly reduced than that of the control crickets (Additional file 2: Figure S1).
Effects of Gb’cry1RNAi and Gb’cry2RNAi on phase responses to a light pulse
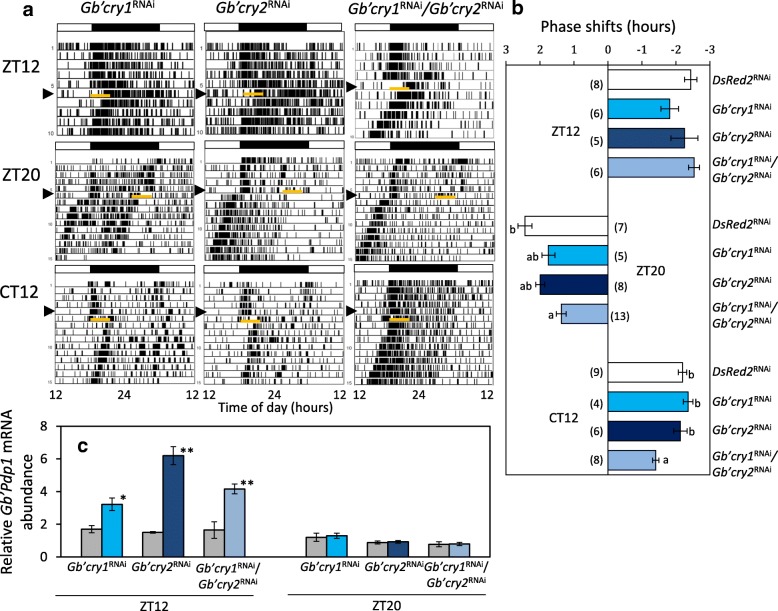
We examined the effects of Gb’cry1RNAi and Gb’cry2RNAi treatment on phase-shifts of the locomotor rhythm caused by a 3 h light pulse given at late night (ZT20) or early subjective night (Circadian time (CT) 12: CT 0 and CT 12 correspond to subjective dawn and subjective sunset, respectively) after a transfer to DD. A 3 h light exposure given at ZT20 caused a phase advance by 2.44 ± 0.54 h (n = 7) in DsRed2RNAi crickets. The Gb’cry1RNAi and Gb’cry2RNAi crickets showed advance shifts (1.75 ± 0.47 h, n = 5; 1.99 ± 0.37 h, n = 8, respectively, Fig. 2) with the magnitude slightly less than that of the control, but the difference was not significant. In Gb’cry1/Gb’cry2 double RNAi crickets, the shift was 1.37 ± 0.53 h (n = 13), and was significantly smaller than that of the control (Fig. 2). A 3 h light pulse given at CT12 induced delay shifts in Gb’cry1RNAi and Gb’cry2RNAi crickets, with the magnitude similar to that of DsRed2RNAi control crickets, while in Gb’cry1RNAi/Gb’cry2RNAi crickets, the magnitude was − 1.41 ± 0.26 h (n = 8), which was significantly smaller than that of the control (− 2.20 ± 0.37 h, n = 9) (Fig. 2a, b).
Fig. 2.
Effects of RNAi of Gb’cry genes on light-induced phase shifts of circadian locomotor rhythm (a, b) and on light-induced Gb’Pdp1 expression (c) in the cricket Gryllus bimaculatus. a Phase shifts of locomotor rhythm caused by a 3 h light phase extension at ZT12 and a 3 h light exposure at ZT20 or CT12. Orange bars indicate a 3 h light phase extension or a light pulse. Black and white bars indicate dark and light phases, respectively. Arrow heads indicate the day of transfer to constant darkness. b Average phase shifts caused by a light phase extension at ZT12 or by a 3 h light pulse given at ZT20 or CT12. Error bars indicate SEM. Number in parenthesis indicates the number of crickets used. Different lower case letters indicate that values are significantly different (ANOVA followed by Tukey-Krammer test, P < 0.05). c Effects of light exposure on Gb’Pdp1 mRNA levels at ZT12 or ZT20. Colored and grey bars indicate the results with and without 60 min light exposure, respectively. Light phase extension at ZT12 induced significant upregulation of Gb’Pdp1 expression (t-test, P < 0.05) in all Gb’cryRNAi crickets, while light exposure at ZT20 had no effect
We also examined the effects of a 3 h extension of light phase (ZT12–15) on the phase of free-running locomotor rhythms in the ensuing DD. There were no significant effects of single or double dsRNA treatment of Gb’cry genes on the magnitude of the phase shifts (Fig. 2a, b).
Effects of Gb’cry1 and Gb’cry2 knock-down on photic responses of Gb’Pdp1
We next examined the effects of Gb’cry1RNAi and Gb’cry2RNAi treatment on light-induced Gb’Pdp1 upregulation, which is the first responder to light phase extension at early subjective night [22]. We measured the mRNA levels of Gb’Pdp1 1 h after light phase extension starting at ZT12, or 1 h after light exposure starting at ZT20.
In all RNAi crickets, Gb’Pdp1 was upregulated 1 h after light-on at ZT12 in comparison to the control without light exposure, while no significant changes were observed in Gb’Pdp1 levels when a light pulse was given at ZT20 (Fig. 2c). The results were similar to those observed in untreated crickets [22], suggesting that Gb’crys are not upstream components of the transcription-dependent entrainment pathway.
Gb’c-fosB is involved upstream of Gb’crys
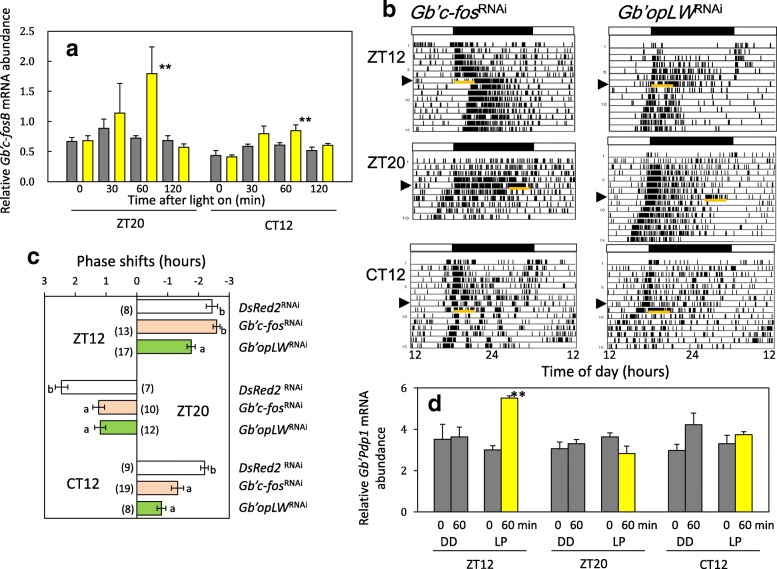
Since a bZip transcription factor gene, c-fos, is known to be up-regulated by light exposure in mammalian circadian clocks [28], we examined whether it also responded to light in crickets. G. bimaculatus has two isoforms of c-fos: Gb’c-fosA (fra-A, LC215243) and Gb’c-fosB (fra-B, LC215244), which arise from alternative promoter usage or alternative splicing from a single locus. We thus tested the effects of a 3 h light exposure at ZT20 and CT12 on their expression levels. Gb’c-fosB was significantly upregulated following light exposure, increasing from 30 min after light-exposure, but increasing significantly after 60 min (Fig. 3a), and then decreasing to basal levels after 120 min.
Fig. 3.
Changes of Gb’c-fosB mRNA levels caused by light exposure (a), effects of RNAi of Gb’c-fos and Gb’opLW genes on the light-induced phase shifts of locomotor rhythms (b, c), and effects of Gb’c-fosRNAi on the light induced Gb’Pdp1 expression (d) in the cricket Gryllus bimaculatus. a Gb’c-fosB was upregulated by light exposure at ZT20 and CT12, and a statistically significant difference was evident after 60 min of exposure (**P < 0.01, t-test). The effect was greater in the late night (ZT20). Yellow and grey bars indicate the results with and without light exposure, respectively. b Phase shifts of locomotor rhythm caused by a 3 h light phase extension at ZT12 or 3 h light exposure at ZT20 or CT12 (orange bars). Black and white bars indicate dark and light phases, respectively. Arrow heads indicate the day of transfer to constant darkness. c Average phase shifts caused by a light phase extension at ZT12 or by a 3 h light pulse given at ZT20 or CT12. Error bars indicate SEM. Different lower-case letters indicate that values are significantly different (ANOVA followed by Tukey-Kramer test, P < 0.05). Gb’c-fosRNAi suppressed the light-induced phase shifts at ZT20 and CT12 but not at ZT12, and Gb’opLWRNAi strongly suppressed them in all cases (ANOVA followed by Tukey-Kramer test, P < 0.01). d Effects of light exposure on Gb’Pdp1 mRNA levels at ZT12, ZT20, and CT12 in Gb’c-fosRNAi crickets. Yellow and grey bars indicate the results with and without 60 min light exposure, respectively. Light phase extension at ZT12 induced significant upregulation of Gb’Pdp1 expression (**P < 0.01, t-test) in Gb’c-fosRNAi crickets, while no apparent effect was observed following light exposure at ZT20 and CT12
We then examined the effect of Gb’c-fosRNAi on the light-induced resetting of the circadian locomotor rhythm. Gb’c-fosA and Gb’c-fosB were both targeted because their nucleotide sequences are mostly identical, and we could not make a specific knock-down of Gb’c-fosB. The dsRNA treatment effectively knocked down the Gb’c-fosB mRNA levels to approximately 25% of that of the DsRed2RNAi controls (Additional file 3: Figure S2). The Gb’c-fosRNAi had no significant effect on the locomotor rhythm; the treated crickets showed a rhythm synchronized to LD cycles and free-running in DD (Fig. 3b). There was no significant difference in free-running period in DD between DsRed2RNAi crickets (23.70 ± 0.38 [mean ± SD] h, n = 9) and Gb’c-fosRNAi crickets (23.81 ± 0.30 h, n = 8) which were transferred directly to DD. A 3 h light pulse given at CT12 or ZT20 delayed or advanced the rhythm in Gb’c-fosRNAi by − 1.32 ± 0.86 h (n = 19) and 1.24 ± 0.59 h (n = 10), respectively, but the magnitude was significantly smaller than that of DsRed2RNAi control crickets (Fig. 3b, c). We also used dsRNA specific for Gb’c-fosA, and found no significant effects on advance shifts caused by a 3 h light pulse at ZT20 (Additional file 4: Figure S3). The suppression of phase-shifts by Gb’c-fosRNAi was therefore apparently caused by knockdown of Gb’c-fosB. However, no suppression was observed when light phase was extended by 3 h at ZT12 (Fig. 3b, c).
Following RNAi of Gb’opLW, light induced phase shifts were significantly reduced in all light treatments (Fig. 3b, c), being consistent with previous observations [21, 22]. We further examined the effects of Gb’c-fos knock-down on Gb’Pdp1 levels in response to light phase extension. In Gb’c-fosRNAi crickets, Gb’Pdp1 mRNA levels were up-regulated when the light phase was extended at ZT12, while no significant changes were observed when light was given at ZT20 or CT12 (Fig. 3d), similar to control crickets [22], suggesting that the Pdp1-dependent entrainment pathway is independent of the c-fos pathway.
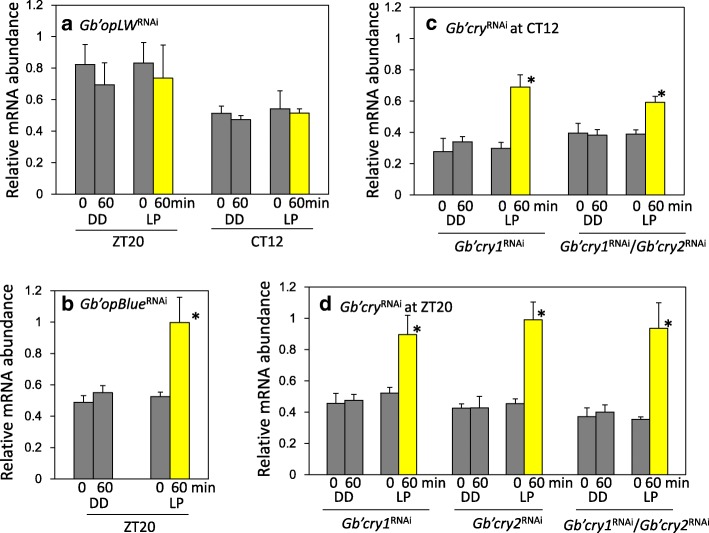
We then examined the effects of Gb’opLWRNAi on Gb’c-fosB levels, since Gb’OpLW is the major photoreceptor for photic entrainment in this cricket [21, 22]. We observed no significant changes in Gb’c-fosB levels following light exposure at CT12 and ZT20 in Gb’opLWRNAi crickets (Fig. 4a), while Gb’c-fosB was significantly upregulated in Gb’opBlueRNAi crickets 1 h after light exposure at ZT20 (Fig.4b). This suggests that the signal from Gb’OpLW is required for the Gb’c-fosB dependent entrainment pathway.
Fig. 4.
Effects of RNAi of Gb’opLW (a), Gb’opBlue (b), and Gb’cry genes (c, d) on light induced Gb’c-fosB mRNA expression in the cricket Gryllus bimaculatus. Gray and yellow columns indicate samples kept in darkness or exposed to light for 60 min, respectively. DD, kept in darkness; LP, exposed to light. Bars indicate SEM. N = 3 or 4. a Gb’opLWRNAi suppressed light-induced upregulation of Gb’c-fosB. b-d RNAi of Gb’opBlue or Gb’cry genes had no effects on the light-induced upregulation of Gb’c-fosB at ZT20 and CT12. *P < 0.05, t-test
We then examined the effects of Gb’cry knock-down on Gb’c-fosB mRNA levels. We found that Gb’cry1 or Gb’cry2 single knock-down or double knock-down did not affect light-induced upregulation of Gb’c-fosB at CT12 or ZT20 (Fig. 4c, d), suggesting that Gb’crys are downstream of Gb’c-fosB.
Gb’cry2, Gb’per, and Gb’c-fosB are expressed in the optic lobe
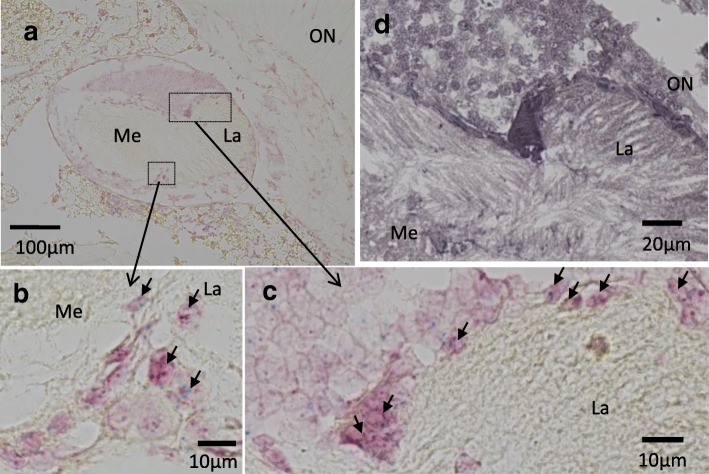
To determine whether Gb’cry2, Gb’per, and Gb’c-fosB are expressed in the clock neurons, we examined their expression in the optic lobe by in situ hybridization or in situ RT-PCR. Although Gb’per and Gb’cry2 were expressed in many cells in the optic lobe at ZT18, they co-localized in some neurons that are located near the outer chiasma between the lamina and medulla and the outer limb of lamina (Fig. 5). By in situ RT-PCR Gb’c-fosB was found to be expressed by 1 h light exposure at ZT20 in cells closely located near the area in which Gb’per and Gb’cry2 are expressed. These results suggest that Gb’per, Gb’cry2 and Gb’c-fosB are co-expressed, at least in some clock neurons.
Fig. 5.
Expression of Gb’per, Gb’cry2, and Gb’c-fosB in the optic lobe of the cricket Gryllus bimaculatus. a-c In situ hybridization of Gb’per (red) and Gb’cry2 (blue) in the optic lobe sampled at ZT18. b and c shows magnification of areas near outer chiasma indicated in a. Short arrows indicate the cells coexpressing Gb’per and Gb’cry2. d In situ PCR of Gb’c-fosB in the optic lobe after 1 h light exposure at ZT20. Gb’c-fosB was strongly expressed in the cells near outer chiasma and along the outer surface of lamina. For further explanations see text
Discussion
Gb’cry1 and Gb’cry2 play separate roles in photic entrainment
In insects, cry1 and cry2 are thought to encode a photoreceptor involved in photic entrainment and a transcriptional repressor in the clock oscillatory mechanism, respectively [14, 29–31]. However, we have recently shown that both Gb’cry1 and Gb’cry2 form an oscillatory loop that can operate independently of the Gb’per/Gb’tim loop in G. bimaculatus [23]. In the present study, we have clearly shown that Gb’cry1 and Gb’cry2 are both involved in the photic entrainment pathway of the cricket’s circadian clock in a manner different from that of Drosophila [11]. Double knock-down of Gb’cry1 and Gb’cry2 severely disrupted re-entrainment to shifted light cycles, with some crickets losing entrainability and others attaining longer transient cycles (Fig. 1, Table 2). The Gb’cry-dependent entrainment pathway is apparently different from the transcription-dependent entrainment pathway, or Gb’Pdp1 pathway, which includes Gb’Pdp1 as the first responder and works only when the light phase is extended [22]. The two Gb’cry genes most likely play different roles, since the effect of double RNAi was not a simple combination of the single knock-down effect for each of these genes.
The Gb’cry2 knock-down accelerated re-synchronization of the locomotor rhythm to delayed LDs (Fig. 1c). Although we still need to examine whether this is true entrainment, this fact is reminiscent of the report that cry knock-out mice have significantly larger phase delays than wild-type mice [32]. Because Gb’cry2 is an essential component of the cry-oscillatory loop [23], its knock-down results in a severe impairment of the loop. The acceleration of light resetting may be caused by the Gb’Pdp1 pathway, which is still functional even in this condition. However, entrainment through the Gb’Pdp1 pathway apparently incomplete, because double RNAi of Gb’cry genes prevents photic entrainment. This may also partly explain the large Ψ of the rhythm after phase shifts in Gb’cry double RNAi crickets (Table 1), which is similar to cry-deficient mice [32]. In contrast, Gb’cry1RNAi only has a small effect on entrainment (Fig. 1b), because the cry-loop can be operated by Gb’cry2 variants [23].
The Gb’c-fosB pathway is involved in photic entrainment
Although the molecular mechanism of Gb’cry-dependent light resetting of the cricket’s clock still remains to be explored, Gb’c-fosB apparently plays a regulatory role upstream of Gb’crys since Gb’c-fosB expression was upregulated by light even after knocking-down of Gb’crys (Fig. 4). This hypothesis is strongly supported by the finding that Gb’c-fosB expression detected by in situ RT-PCR overlapped with that of cells co-expressing Gb’per and Gb’cry2. c-fos has been used as a marker for light input in vertebrate circadian clocks because of its rapid induction after light exposure [33]. Its product protein forms a complex, activating protein 1 (AP-1), with other transcription factor genes, such as jun gene family members, and activates transcription of target genes [34]. However, its role in resetting of the circadian clock remains mostly unclear. Our results revealed for the first time that c-fos plays a major role in resetting the clock in insects, since Gb’c-fosRNAi strongly prevents light induced phase shifts of the circadian rhythm. This also suggests that Gb’crys are downstream of Gb’c-fosB. At present, although the connection between Gb’c-fosB and Gb’crys is unclear, F-box and leucin rich repeat proteins (FBXL) and Bromodomain and WD repeat domain containing 3 (BRWD3) may be mediating this connection, as they are known to regulate CRY degradation by ubiquitination of CRY [35–37]. Light input to the Gb’c-fosB pathway is through Gb’OpLW, because the light-induced upregulation of Gb’c-fosB is eliminated by Gb’opLWRNAi. Based on the present findings and those of previous studies, the most likely hypothesis appears to be that light information is supplied to the clock neurons by neurotransmission through the OpLW pathway, causing induction of Gb’c-fosB, followed by a functional modulation of FBXL or BRWD3, which may ubiquitinate CRYs and reset the oscillatory mechanism including CRYs (Fig. 6). This hypothesis should be tested in future studies.
Fig. 6.
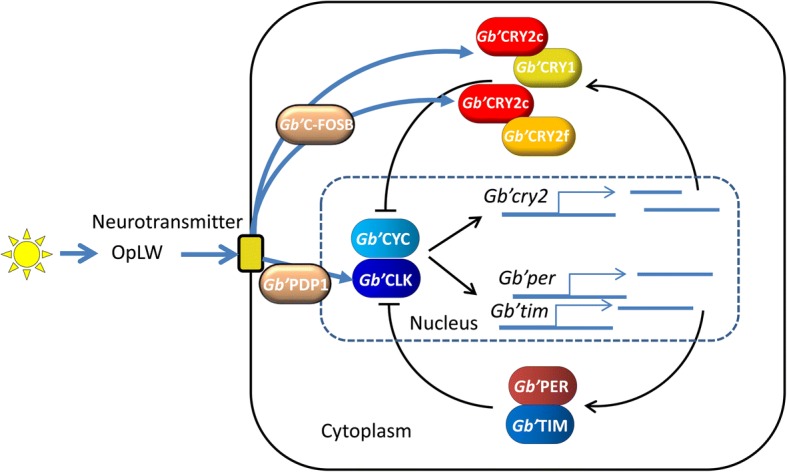
A model for the light entrainment mechanism of the cricket circadian clock. The clock includes two major oscillatory loops, one for Gb’per and Gb’tim, like in Drosophila, and the other for Gb’cry2. The latter is comprised of Gb’cry1 and two Gb’cry2 isoforms, Gb’cry2c and Gb’cry2f, and their product proteins form a complex that suppresses the transcription mediated by Gb’CLK/Gb’CYC complex [23]. Light is perceived by the retinular cells in the compound eye expressing Gb’opLW, and the information is transmitted to the clock neuron in the optic lobe through neurotransmitters. This neurotransmission causes upregulation of Gb’c-fosB in the clock neurons, which finally affects the Gb’cry2 oscillatory loop. The change in the Gb’cry2 loop may reset the whole clock system since the Gb’cry2 loop interact with the Gb’per/Gb’tim loop by influencing Gb’CLK/Gb’CYC. In addition to the Gb’cry2 pathway, the Gb’Pdp1 pathway resets the clock by upregulating Gb’Clk expression when light off was delayed [22]
Besides the Gb’c-fosB mediated pathway, Gb’crys might also be regulated by light through other mechanisms. Our previous study showed that light phase extension at early night induced Gb’cry2 upregulation with an increase of Gb’Pdp1 and Gb’Clk in nymphal crickets [22], suggesting the E-box mediated transcription of Gb’cry2 by Gb’Clk. Another possibility is through a D-box mediated regulation known for zebrafish clocks, in which light induces cry1a expression through a D-box mediated mechanism including PAR bZip factors, PAR and TEF-1 [38]. Because D-boxes are found in the cis-regulatory region of Gb’cry1 and Gb’cry2, a similar mechanism might be involved in the cricket clock. These issues should be addressed in future studies.
Heterogeneous nature of clock cells
In the cricket’s clock, there must be at least two sets of clock neurons since some fraction of the Gb’cry1/Gb’cry2 double RNAi crickets showed a rhythm dissociation into two components when the light cycle was shifted by 6 h: one component still retained photic entrainability, while it was lost in the other (Fig. 1e). The heterogenous cellular organization of the clock is quite similar to that observed in Drosophila and mammals [39, 40], where entrainability to light is different between the cerebral or SCN clock neurons [41–43].
The heterogeneous cellular nature of the clock may explain the effect of Gb’cry2RNAi on the free-running locomotor rhythm. We have previously shown that Gb’cry2RNAi crickets have a wide variety of free-running periods in DD [23], which may reflect the underlying oscillatory neurons. The lack of Gb’CRY2 following RNAi may destroy the stable free-running of the clock neuron, hence causing dissociation among the cells. This is reminiscent of the mammalian SCN clock, where CRY plays a role as a coupling factor of clock cells that have variable free-running periods [39, 44–46].
Comparison with other insect clocks
In the present study, we found for the first time that both Gb’cry1 and Gb’cry2 play important roles in photic entrainment of the circadian clock in crickets. In hemimetabolous insects, the photoreceptor for entrainment is believed to reside in the compound eye [19, 47, 48], and this assumption has been confirmed at the molecular level in the cricket G. bimaculatus [21]. Therefore, Gb’cry1 and Gb’cry2 are both active in the clock resetting mechanism downstream of the neurotransmission from the retinal photoreceptor. Although the exact role of Gb’cry genes should be examined in future studies, the cricket’s Gb’CRY1 likely works together with Gb’CRY2 to form an oscillatory feedback loop, which can operate independently of the Gb’per/Gb’tim loop [23]. As for insect cry2, it is believed to be involved in the clock machinery as a clock component [14]. However, we have shown for the first time that Gb’cry2 also plays an important role in the photic entrainment of the clock. Our results shed light on the insect clock mechanism and provide deeper understanding of its photic entrainment and diversification.
Conclusions
Photic entrainment is an essential property of the animal circadian clock that sets the appropriate timing of behavioral and physiological events in a 24 h cycle. Although most insects use compound eyes as photoreceptors for entrainment, the molecular mechanisms underlying entrainment remain largely unknown. Here we elucidated for the first time the molecular entrainment pathway using a hemimetabolous insect, the cricket Gryllus bimaculatus. Our results suggest that neural signals mediated by green-sensitive opsins in the compound eye first up-regulate an isoform of the bZip transcription factor gene, Gb’c-fos, which subsequently resets the clock through Gb’cry1 and Gb’cry2. These findings contribute to understanding of the photic entrainment mechanism of insect clocks.
Additional files
Table S1. PCR primers used for quantitative RT-PCR and dsRNA synthesis. The primers tagged with T7 or T3 promoter sequences were used for PCR amplification for dsRNA synthesis. T7 and T3 sequences are underlined. (DOCX 18 kb)
Figure S1. Effect of RNAi of cry genes on locomotor activity during the first 3 h after light-on in the cricket Gryllus bimaculatus. The activity was measured on the first day after 6 h phase advance of light-on. Error bars indicate SEM. Numbers in parenthesis indicate the number of animals used. Gb’cry2RNAi and Gb’cry1RNAi/Gb’cry2RNAi significantly reduced the light-induced locomotor activity compared to DsRed2RNAi treatment (*P < 0.05, **P < 0.01, Dunnett’s test). (PDF 58 kb)
Figure S2. Gb’c-fosRNAi significantly down-regulated both Gb’c-fosA and Gb’c-fosB mRNA levels in the optic lobe of the cricket Gryllus bimaculatus (**P < 0.01, t-test). The optic lobes were collected at ZT20 seven days after dsRNA injection. mRNA levels were measured by qPCR and are shown relative to those of Gb’rpl18a. The values shown are mean ± SEM of six samples. (PDF 49 kb)
Figure S3. A: Gb’c-fosARNAi had no significant effects on the light induced phase advance in the cricket Gryllus bimaculatus. A 3 h light pulse was given at ZT20 on the day of transfer to DD, which was seven days after dsRNA injection. Numbers in the parenthesis indicate the number of animals used. B and C: Gb’c-fosARNAi significantly knocked down Gb’c-fosA mRNA levels (*P < 0.05, t-test), but had no significant effect on Gb’c-fosB mRNA levels. mRNA levels were measured by qPCR and are shown relative to those of Gb’rpl18a. The values shown are mean ± SEM of four samples. (PDF 69 kb)
Acknowledgments
We thank Taishi Yoshii for helpful discussions and Tsugumichi Shinohara for his assistance in manuscript preparation.
Authors’ contributions
YK and KT designed the experiments. YK, AT, MN, YT, YM performed the experiments. TB and TW performed promoter analysis of Gb’cry genes and molecular analysis of Gb’c-fos gene, respectively. YK and KT analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported in part by JSPS KAKENHI Grant Number JP15H0440017 and JP18H02480.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuki Kutaragi, Email: pu473j6u@s.okayama-u.ac.jp.
Atsushi Tokuoka, Email: pan27q5m@s.okayama-u.ac.jp.
Yasuaki Tomiyama, Email: paf49f1v@s.okayama-u.ac.jp.
Motoki Nose, Email: pd2w2z46@s.okayama-u.ac.jp.
Takayuki Watanabe, Email: taka226@gmail.com.
Tetsuya Bando, Email: tbando@cc.okayama-u.ac.jp.
Yoshiyuki Moriyama, Email: y.moriyama@med.kawasaki-m.ac.jp.
Kenji Tomioka, Phone: +81-86-251-8498, Email: tomioka@cc.okayama-u.ac.jp.
References
- 1.Saunders D.S., Steel C.G.H., Vafopoulou X., Lewis R.D. Insect Clocks. 2002. Other Types of Insect Clocks; pp. 449–472. [Google Scholar]
- 2.Hardin P. Molecular mechanisms of circadian timekeeping in Drosophila. Sleep Biol Rhythms. 2009;7:235–242. doi: 10.1111/j.1479-8425.2009.00412.x. [DOI] [Google Scholar]
- 3.Tataroglu O, Emery P. The molecular ticks of the Drosophila circadian clock. Curr Opin Insect Sci. 2015;7:51–57. doi: 10.1016/j.cois.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomioka K, Matsumoto A. Circadian molecular clockworks in non-model insects. Curr Opin Insect Sci. 2015;7:58–64. doi: 10.1016/j.cois.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/S0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 6.Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, et al. vrille, Pdp1 and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/S0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 7.Kamae Y, Uryu O, Miki T, Tomioka K. The nuclear receptor genes HR3 and E75 are required for the circadian rhythm in a primitive insect. PLoS One. 2014;9:e114899. doi: 10.1371/journal.pone.0114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila's clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 9.Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, et al. Light-dependent sequentation of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 10.Lin F-J, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol Cell Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/S0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa Tomoko, Matsumoto Akira, Kato Jr Tomohisa, Togashi Shin, Ryo Haruko, Ikenaga Mituo, Todo Takeshi, Ueda Ryu, Tanimura Teiichi. DCRY is aDrosophilaphotoreceptor protein implicated in light entrainment of circadian rhythm. Genes to Cells. 1999;4(1):57–65. doi: 10.1046/j.1365-2443.1999.00237.x. [DOI] [PubMed] [Google Scholar]
- 13.Ingram KK, Kutowoi A, Wurm Y, Shoemaker D, Meier R, Bloch G. The molecular clockwork of the fire ant Solenopsis invicta. PLoS One. 2012;7:e45715. doi: 10.1371/journal.pone.0045715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 15.Langmesser S, Tallone T, Bordon A, Rusconi S, Albrecht U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol Biol. 2008;9:41. doi: 10.1186/1471-2199-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye R, Selby CP, Chiou Y-Y, Ozkan-Dagliyan I, Gaddameedhi S, Sancar A. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and period proteins in the mammalian circadian clock. Genes Dev. 2016;28:1989–1998. doi: 10.1101/gad.249417.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selby CP, Thompson C, Schmitz TM, Van Gelder RN, Sancar A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proc Natl Acad Sci. 2000;97:14697–14702. doi: 10.1073/pnas.260498597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathalie H, Schleicher E, Kacprzak S, Bouly J-P, Picot M, Wu W, et al. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 2008;6:e160. doi: 10.1371/journal.pbio.0060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomioka K, Chiba Y. Effects of nymphal stage optic nerve severance or optic lobe removal on the circadian locomotor rhythm of the cricket. Zool Sci. 1984;1:375–382. [Google Scholar]
- 20.Yukizane M, Tomioka K. Neural pathways involved in mutual interactions between optic lobe circadian pacemakers in the cricket Gryllus bimaculatus. J Comp Physiol A. 1995;176:601–610. doi: 10.1007/BF01021580. [DOI] [Google Scholar]
- 21.Komada S, Kamae Y, Koyanagi M, Tatewaki K, Hassaneen E, Saifullah A, et al. Green-sensitive opsin is the photoreceptor for photic entrainment of an insect circadian clock. Zoological Letters. 2015;1:11. doi: 10.1186/s40851-015-0011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutaragi Y, Miki T, Bando T, Tomioka K. Transcriptional and non-transcriptional events are involved in photic entrainment of the circadian clock in the cricket Gryllus bimaculatus. Physiol Entomol. 2016;41:358–368. doi: 10.1111/phen.12162. [DOI] [Google Scholar]
- 23.Tokuoka A, Itoh TQ, Hori S, Uryu O, Danbara Y, Nose M, et al. cryptochrome genes form an oscillatory loop independent of the per/tim loop in the circadian clockwork of the cricket Gryllus bimaculatus. Zoological Letters. 2017;3:5. doi: 10.1186/s40851-017-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriyama Y, Sakamoto T, Karpova SG, Matsumoto A, Noji S, Tomioka K. RNA interference of the clock gene period disrupts circadian rhythms in the cricket Gryllus bimaculatus. J Biol Rhythm. 2008;23:308–318. doi: 10.1177/0748730408320486. [DOI] [PubMed] [Google Scholar]
- 25.Sokolove PG, Bushell WN. The chi square periodogram: its utility for analysis of circadian rhythm. J Theor Biol. 1978;72:131–160. doi: 10.1016/0022-5193(78)90022-X. [DOI] [PubMed] [Google Scholar]
- 26.Schmid B, Helfrich-Förster C, Yoshii T. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J Biol Rhythm. 2011;26:464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol. 2000;43:207–233. doi: 10.1002/(SICI)1097-4695(20000605)43:3<207::AID-NEU1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS. Photic and circadian regulation of c-fos gene expression in hamster suprachiasmatic nucleus. Neuron. 1990;5:127–134. doi: 10.1016/0896-6273(90)90303-W. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Lear BC, Seluzicki A, Allada R. The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr Biol. 2009;19:2050–2055. doi: 10.1016/j.cub.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Yuan Q, Briscoe AD, Froy O, Casselman A, Reppert SM. The two CRYs of the butterfly. Curr Biol. 2005;15:R953–R954. doi: 10.1016/j.cub.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 31.Michae AK, Fribourgh JL, Gelder RNV, Partch CL. Animal cryptochromes: divergent roles in light perception, circadian timekeeping and beyond. Photochem Photobiol. 2017;93:128–140. doi: 10.1111/php.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spoelstra K, Albrecht U, GTJvd H, Brauer V, Daan S. Phase responses to light pulses in mice lacking functional Per or Cry genes. J Biol Rhythm. 2004;19:518–529. doi: 10.1177/0748730404268122. [DOI] [PubMed] [Google Scholar]
- 33.Moore HAM, Whitmore D. Circadian rhythmicity and light sensitivity of the zebrafish brain. PLoS One. 2014;9:e86176. doi: 10.1371/journal.pone.0086176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guillaumond F, Sage D, Deprez P, Bosler O, Becquet D, François-Bellan AM. Circadian binding activity of AP-1, a regulator of the arylalkylamine N-acetyltransferase gene in the rat pineal gland, depends on circadian Fra-2, c-Jun, and Jun-D expression and is regulated by the clock's zeitgebers. J Neurochem. 2000;75:1398–1407. doi: 10.1046/j.1471-4159.2000.0751398.x. [DOI] [PubMed] [Google Scholar]
- 35.Yoo S-H, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong H-K, et al. Competing E3 ubiquitin ligase govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of Cryptochromes. Cell. 2013;152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 37.Ozturk N, VanVickle-Chavez SJ, Akileswaran L, Van Gelder RN, Sancar A. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1-CUL4-ROC1 E3 ligase complex. Proc Natl Acad Sci. 2013;110:4980–4985. doi: 10.1073/pnas.1303234110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mracek P, Santoriello C, Idda ML, Pagano C, Ben-Moshe Z, Gothilf Y, et al. Regulation of per and cry genes reveals a central role for the D-box enhancer in light-dependent gene expression. PLoS One. 2012;7:e51278. doi: 10.1371/journal.pone.0051278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(SICI)1096-9861(20000619)422:1<66::AID-CNE5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Miyasako Y, Umezaki Y, Tomioka K. Separate sets of cerebral clock neurons are responsible for light and temperature entrainment of Drosophila circadian locomotor rhythms. J Biol Rhythm. 2007;22:115–126. doi: 10.1177/0748730407299344. [DOI] [PubMed] [Google Scholar]
- 42.Yoshii T, Hermann-Luibl C, Kistenpfennig C, Schmid B, Tomioka K, Helfrich-Förster C. Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J Neurosci. 2015;35:6131–6141. doi: 10.1523/JNEUROSCI.0070-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Reviews. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 44.Ono D, Honma S, K-i H. Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Commun. 2013;4:1666. doi: 10.1038/ncomms2670. [DOI] [PubMed] [Google Scholar]
- 45.Evans JA, Pan H, Liu AC, Welsh DK. Cry1−/− circadian rhythmicity depends on SCN intercellular coupling. J Biol Rhythm. 2012;27:443–452. doi: 10.1177/0748730412461246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K-i. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishiitsutsuji-Uwo J, Pittendrigh CS. Central nervous system control of circadian rhythmicity in the cockroach. II. The pathway of light signals that entrain the rhythm. Z vergl Physiol. 1968;58:1–13. doi: 10.1007/BF00302433. [DOI] [Google Scholar]
- 48.Page TL. Interaction between bilaterally paired components of the cockroach circadian system. J Comp Physiol. 1978;124:225–236. doi: 10.1007/BF00657054. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PCR primers used for quantitative RT-PCR and dsRNA synthesis. The primers tagged with T7 or T3 promoter sequences were used for PCR amplification for dsRNA synthesis. T7 and T3 sequences are underlined. (DOCX 18 kb)
Figure S1. Effect of RNAi of cry genes on locomotor activity during the first 3 h after light-on in the cricket Gryllus bimaculatus. The activity was measured on the first day after 6 h phase advance of light-on. Error bars indicate SEM. Numbers in parenthesis indicate the number of animals used. Gb’cry2RNAi and Gb’cry1RNAi/Gb’cry2RNAi significantly reduced the light-induced locomotor activity compared to DsRed2RNAi treatment (*P < 0.05, **P < 0.01, Dunnett’s test). (PDF 58 kb)
Figure S2. Gb’c-fosRNAi significantly down-regulated both Gb’c-fosA and Gb’c-fosB mRNA levels in the optic lobe of the cricket Gryllus bimaculatus (**P < 0.01, t-test). The optic lobes were collected at ZT20 seven days after dsRNA injection. mRNA levels were measured by qPCR and are shown relative to those of Gb’rpl18a. The values shown are mean ± SEM of six samples. (PDF 49 kb)
Figure S3. A: Gb’c-fosARNAi had no significant effects on the light induced phase advance in the cricket Gryllus bimaculatus. A 3 h light pulse was given at ZT20 on the day of transfer to DD, which was seven days after dsRNA injection. Numbers in the parenthesis indicate the number of animals used. B and C: Gb’c-fosARNAi significantly knocked down Gb’c-fosA mRNA levels (*P < 0.05, t-test), but had no significant effect on Gb’c-fosB mRNA levels. mRNA levels were measured by qPCR and are shown relative to those of Gb’rpl18a. The values shown are mean ± SEM of four samples. (PDF 69 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.