Abstract
To counteract host immunity, Cryptosporidium parvum has evolved multiple strategies to suppress host antimicrobial defense. One such strategy is to reduce the production of the antimicrobial peptide beta-defensin 1 (DEFB1) by host epithelial cells but the underlying mechanisms remain unclear. Recent studies demonstrate that a panel of parasite RNA transcripts of low protein-coding potential are delivered into infected host cells and may modulate host gene transcription. Using in vitro models of intestinal cryptosporidiosis, in this study we analyzed the expression profile of host beta-defensin genes in host cells following infection. We found that C. parvum infection caused a significant downregulation of the DEFB1 gene. Interestingly, downregulation of DEFB1 gene was associated with host delivery of Cdg7_FLc_1000 RNA transcript, a C. parvum RNA that has previously demonstrated to be delivered into the nuclei of infected host cells. Knockdown of Cdg7_FLc_1000 in host cells could attenuate the trans-suppression of host DEFB1 gene and decreased the parasite burden. Therefore, our data suggest that trans-suppression of DEFB1 Gene in intestinal epithelial cells following C. parvum infection involves host delivery of parasite Cdg7_FLc_1000 RNA, a process that may be relevant to the epithelial defense evasion by C. parvum at the early stage of infection.
Keywords: Cryptosporidium, intestinal epithelium, DEFB, DEFB1, gene transcription, epithelial defense
Introduction
Cryptosporidium parvum, a zoonotic protozoan parasite, has been recognized as the leading cause of waterborne disease outbreaks worldwide (Checkley et al. 2015). This phylum Apicomplexan parasite is resistant to standard disinfection applied to drinking water and is a potential bioterror pathogen (OʼDonoghue 1995). C. parvum infects human gastrointestinal epithelium and causes an acute, self-limited diarrheal disease in immunocompetent individuals but potentially life-threatening syndromes in immunocompromised patients, young children and elders (Kotloff et al. 2013; Checkley et al. 2015). Humans are infected by ingesting Cryptosporidium oocysts. Once ingested, oocysts excyst in the gastrointestinal tract, releasing infective sporozoites. The sporozoite attaches to the apical membrane of host epithelial cells and forms an intracellular but extracytoplasmic parasitophorous vacuole in which the organism resides. The internalized parasite then develops for its life cycle resulting in autoinfection and release of mature oocysts (Chen et al. 2002). Despite the magnitude and severity of cryptosporidial infection, there is currently no fully effective therapy (Striepen 2013).
Immunity against cryptosporidiosis involves parts of the innate and adaptive host immune responses. Mucosal epithelial cells recognize and respond to C. parvum infection through Toll-like receptors and initial host defense mechanisms, including release of inflammatory cytokines/chemokines and infiltration of immune effector cells at infection sites (Chen et al. 2002). Upon C. parvum infection, epithelial cells quickly initiate a series of innate immune reactions including production and secretion of various cytokines and chemokines, prostaglandin E2 (which stimulates mucin production), and antimicrobial peptides (defensins and cathelicidins) and nitric oxide, which can kill C. parvum or inhibit parasite growth (Laurent et al. 1998; Chen et al. 2005; Rogers et al. 2006). These chemokines/cytokines can mobilize and activate immune effector cells (e.g., lymphocytes, macrophages and neutrophils) to the infection sites (Chen et al. 2002; Rogers et al. 2006). These responses provide the front line of anti-C. parvum defense. However, complete elimination of infection requires the adaptive immune responses, particularly those mediated by CD4+ T cells (Schmidt et al. 2001; Chen et al. 2002). Interferon-γ, mainly released by activated CD4+ T cells (Schmidt et al. 2001), is also critical in the control of cryptosporidiosis (Chen et al. 2002). Eradication of C. parvum infection requires cell-mediated adaptive immunity in which CD4+ T cells play a key role (Chen et al. 2002). Such cell-mediated immune responses become effective about three weeks after initial infection (Schmidt et al. 2001). Therefore, it appears that C. parvum can survive host innate immune attack during the early stage of infection (OʼDonoghue 1995; Chen et al. 2002; Checkley et al. 2015). How C. parvum may evade host innate immune defense at the early stage of infection is still unclear. One immune evasion strategy developed by the parasite is to reduce the production of the antimicrobial peptide beta-defensin 1 (DEFB1) by host epithelial cells via unknown mechanisms (Zaalouk et al. 2004).
We previously demonstrated that several C. parvum RNA transcripts of low protein-coding potential are selectively delivered into the nuclei of host cells through heat shock protein 70-mediated nuclear importing mechanisms during host-parasite interactions and may modulate gene transcription in infected epithelial cells (Puiu et al. 2004; Yamagishi et al. 2011; Wang et al. 2017a). Specifically, delivery of parasite Cdg7_FLc_0990 RNA (GenBank ID: FX115678.1) (Puiu et al. 2004; Yamagishi et al. 2011) into infected intestinal epithelial cells causes trans-suppression of the LDL receptor related protein 5 (LRP5), solute carrier family 7 member 8 (SLC7A8), and interleukin 33 (IL33) genes through histone modification-mediated epigenetic mechanisms (Wang et al. 2017a, b). Delivery of the parasite Cdg7_FLc_1000 transcript (GenBank ID: FX115830.1) (Puiu et al. 2004; Yamagishi et al. 2011) causes trans-suppression of host sphingomyelin phosphodiesterase 3 (SMPD3) gene (Ming et al. 2017). Using a non-malignant human intestinal epithelial cell line for C. parvum infection in vitro, here, we report that downregulation of the DEFB1 gene in intestinal epithelial cells following C. parvum infection involves the host delivery of Cdg7_FLc_1000 RNA transcript, a process relevant to the epithelial defense evasion by C. parvum at the early stage of infection.
2. Materials and methods
2.1. C. parvum, cell lines, and infection models
C. parvum oocysts of the Iowa strain were purchased from a commercial source (Bunch Grass Farm, Deary, ID). INT cells (FHs 74 Int, CCL-241™) and HCT-8 (CCL-244™) were purchased from ATCC (Manassas, VA). The murine intestinal epithelial cell line (IEC4.1) was a kind gift from Dr. Pingchang Yang (McMaster University, Hamilton, Canada) and cultured in Dulbeccoʼs modified Eagleʼs Medium (DMEM-Mediatech Inc., Manassas, VA) supplemented with 5% fetal bovine serum (Hyclone), 100 U/ml penicillin, 100 µg/ml streptomycin, 50 µg/ml gentamicin and 1 ng/ml epidermal growth factor. Models of intestinal cryptosporidiosis using cultured cell lines were employed as previously described (Zhou et al. 2012; Wang et al. 2017a). Briefly, before infecting cells, C. parvum oocysts were treated with 1% sodium hypochlorite on ice for 20 min followed by extensive washing with DMEM-F12 medium and confirmed negative for the Limulus Amebocyte Lysate test (Bio-Whittaker). Real-time PCR and immunofluorescence microscopy were used to assay C. parvum infection as previously reported (Zhou et al. 2012; Wang et al. 2017a).
2.2. siRNAs and plasmids
Custom-designed siRNA oligos against Cdg7_FLc_1000 and a scrambled siRNA were synthesized by Integrated DNA Technologies (Coralville, Iowa) and transfected into cells with Lipofectamine RNAimax (Invitrogen) (Ming et al. 2017). siRNA sequence for Cdg7_FLc_1000: sense, 5ʼ-GAGAUAACUAACGCCACCUU-3ʼ and antisense, 5ʼ-AGGUGGCGUUAGUUAUCUCUU-3ʼ; non-specific control: sense, 5ʼ-UUCUCCGAACGUGUCACGUUU-3ʼ and antisense, 5ʼ-ACGUGACACGUUCGGAGAAUU-3ʼ. The plasmids for Full-Cdg7_FLc_1000 and Full-Cdg7_FLc_0990 were generated by RT-PCR amplification of the corresponding cDNA (Ming et al. 2017; Wang et al. 2017a, b), using RNA from C. parvum sporozoites (Iowa strain) and cloned into the pcDNA3.1(+) vector according to the manufacturerʼs protocol (Invitrogen). Full-Cdg7_FLc_1000 or Full-Cdg7_FLc_0990 were transfected into cells with lipofectamine 2000 (Invitrogen); pcDNA3.1(+) Empty vector transfected as control. The primer sequences for plasmid generation are listed as following: Cdg7_FLc_1000: forward (Nhe I), 5ʼ-CGGCTAGCAGTTTTTACATTTTGTATCTCAGTT-3ʼ and reverse (KpnI), 5ʼ-GGGGTACCTGAGCGAAATTAGAGTAGTCTGA-3ʼ; Cdg7_FLc_0990: forward (Xba1), 5ʼ-GCTCTAGAAGATTTATTAAGGTTTTATTTT-3ʼ and reverse (KpnI), 5ʼ-GGGGTACCAAAAATACAAGAAGGAGTTATG-3ʼ.
2.3. Agilent microarray analysis
The Agilent SurePrint G3 Human Gene Expression Microarray and the LC Sciences service to process the samples were applied to genome-wide analysis. Briefly, INT cells were grown to 80% confluence and then exposed to C. parvum infection or transfected with Full-Cdg7_FLc_1000 or Full-Cdg7_FLc_0990. Total RNA of harvested cells was isolated with the RNeasy Mini kit (Qiagen). A mixture of equal amounts of total RNAs from each group was used as the reference pool. A total of 2 µg RNA from each sample was then labeled with the Agilent Gene Expression Hybridization Kit (Agilent). After hybridization, the slides were scanned with the Agilent Microarray Scanner (Agilent). The Feature Extraction software (version10.7.1.1, Agilent Technologies) was used to analyze array images to get raw data and Genesrping software was employed to finish the basic analysis with the raw data. To begin with, the raw data was normalized with the quantile algorithm. The probes that at least 1 out of all samples have flags as “detected” were treated as positive signals and chosen for further data analysis. Quantified positive signals were then extracted and analyzed by the LC Sciences, in accordance with MIAME guidelines.
2.4. Whole cell extracts, nuclear extracts, and quantitative real-time PCR
Whole cell extracts and nuclear extracts were obtained using the standard approach as previously described (Wang et al. 2017a). For quantitative analysis of mRNA and C. parvum RNA expression, comparative real-time PCR was performed as previous reported (Wang et al. 2017a; Ming et al. 2017) using the SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA). Briefly, RNA was extracted using TRI-reagent, treated with DNA-free Kit (Ambion) to remove any remaining DNA. Quantified 500 ng RNA was reverse-transcribed using T100 thermal cyclers (Bio-Rad). Real-time PCR was then performed using 25 ng of template cDNA for each RNA gene of interest. Each sample was run in triplicate. The relative abundance of each RNA was calculated using the ΔΔCt method and normalized to GAPDH (total mRNA). The sequences for all the primers are as follows: DEFB1, forward, 5ʼ-GCCTCAGGTGGTAACTTTCTC-3ʼ and reverse, 5ʼ-CGTCATTTCTTCTGGTCACTC-3ʼ; GAPDH, forward, 5ʼ-TGCACCACCAACTGCTTAGC-3ʼ and reverse, 5ʼ-GGCATGGACTGTGGTCATGAG-3ʼ; C-X-C motif chemokine ligand 2 (CXCL2), forward, 5ʼ-AAAGGGGTTCGCCGTTCT-3ʼ and reverse, 5ʼ-TGGCAGCGCAGTTCAGTG-3ʼ; NFKB inhibitor zeta (NFKBIZ), forward, 5ʼ-ACTCGGAACTTGGAGAACG-3ʼ and reverse, 5ʼ-CAGGACTGCCTAACTGACAG-3ʼ; CXCL8, forward, 5ʼ-TGCAGCTCTGTGTGAAGGTG-3ʼ and reverse, 5ʼ-ACAACCCTCTGCACCCAGTT-3ʼ; LRP5, forward, 5ʼ-GGCCAGTGTGTCCTCATCAAA-3ʼ and reverse, 5ʼ-AGACACCACCCATGACGAAGA-3ʼ; SLC7A8, 5ʼ-TTGCCCTGTCCACATTTGG-3ʼ and reverse, TGGAGATGCATGTGAAGAGCA-3ʼ; U2, forward, 5ʼ-CTGATACGTCCTCTATCCGAGG-3ʼ and reverse, TGCAATACCAGGTCGATGCGT-3ʼ; Defb1, forward, 5ʼ-CAGGTGTTGGCATTCTCA-3ʼ and reverse, 5ʼ-CCATCGCTCGTCCTTTAT-3ʼ; Gapdh, forward, 5ʼ-AACAGGGTGGTGGACCTCAT-3ʼ and reverse, 5ʼ-AGTTGGGATAGGGCCTCTCTT-3ʼ; Cdg7_FLc_1000, forward, 5ʼ-CTTCAAAAGGGACAGACAAACGGC-3ʼ and reverse, 5ʼ-CTTGATCTCGCCCATAGCCACTT-3ʼ; 18S, forward, 5ʼ-TAGAGATTGGAGGTTGTTCCT-3ʼ and reverse, 5ʼ-CTCCACCAACTAAGAACGGCC-3ʼ.
2.5. Statistical Analysis
All values are given as mean ± standard error of the mean (SEM). Means of groups were from at least three independent experiments and compared with Studentʼs t test (two-tailed unpaired) or the ANOVA test when appropriate. The array data were analyzed and compared statistically by LC Sciences using the Agilent Feature Extraction Software, in accordance with MIAME guidelines. p values < 0.05 were considered statistically significant.
3. Results
3.1. Expression levels of host DEFB genes in cultured INT cells following C. parvum infection revealed by genome-wide array analysis
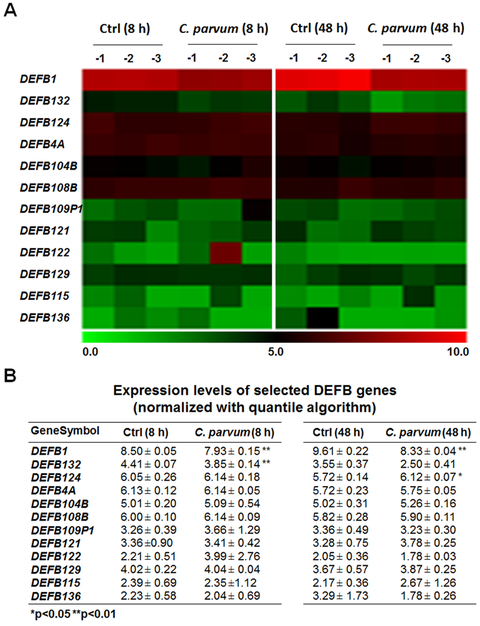
To explore the expression profile of DEFB genes in intestinal epithelial cells following C. parvum infection, we analyzed our previous genome-wide array data on non-malignant human intestinal epithelial cells (INT cells) following C. parvum infection for 8 and 48 h (the GEO database with the accession number: GSE87047) (Wang et al. 2017a). Out of a total of 35 human DEFB genes available in the array chip, signals for 12 DEFB genes were passed the threshold set by the array chip internal control in either the un-infected control or the infected cells and thus, they treated as the detectable DEFB genes in INT cells (Fig. 1A, B). Expression levels for other DEFB genes were then treated as undetectable in the un-infected control or C. parvum infected INT cells. Out of these detectable DEFB genes, DEFB1 was downregulated in cells following infection at 8 h and 48 h. Expression level for DEFB132 was downregulated in INT cells following infection for 8 h but no significant difference was found between the infected cells following infection for 48 h and the control (Fig. 1A, B). Expression level for DEFB124 was upregulated in INT cells following infection for 48 h but no significant difference was found comparing the infected cells following infection for 8 h with the control (Fig. 1A, B). Consistent with results from previous studies (Wang et al. 2017a; Ming et al. 2017; Deng et al. 2004; Yang et al. 2009), upregulation of several epithelial cell defense genes, such as interleukin-8, C-X-C motif chemokine ligand 5, and nitric oxide synthase 2, was detected in cells following C. parvum infection (data not shown).
Fig. 1. Genome-wide array analysis of DEFB genes in INT cells following C. parvum infection.
Alterations in DEFB gene expression profile from genome-wide transcriptome analysis in INT cells following C. parvum infection. INT cells were exposed to C. parvum infection for 8h and 48 h, followed by the microarray analysis; non-infected cells were used as the control. The mean value of the expression level for selected DEFB genes, which passed the filtering criteria variation across the samples and normalized with quantile algorithm using the Genesrping software, was presented as the heatmap (A) and listed in the table (B). *P<0.05 and **P<0.01 versus non-infected cells.
3.2. Expression levels of host DEFB genes in INT cells expressing C. parvum Cdg7_FLc_1000 RNA revealed by genome-wide array analysis
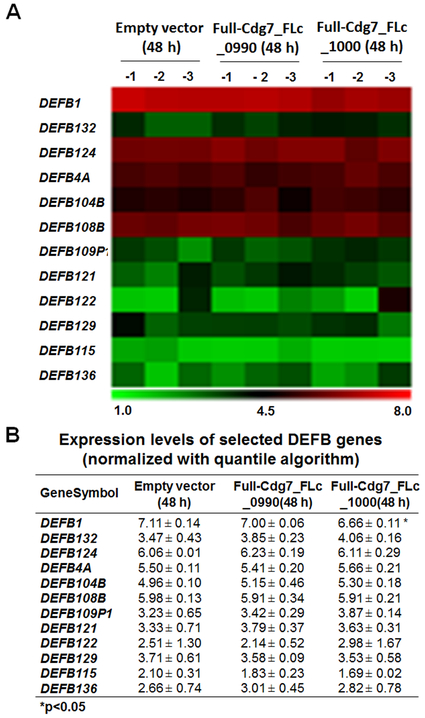
We previously demonstrated that nuclear delivery of C. parvum RNA transcripts of low protein-coding potential, such as Cdg7_FLc_0990 and Cdg7_FLc_1000, is involved in the trans-suppression of specific host genes in C. parvum-infected intestinal epithelial cells (Wang et al. 2017a, b; Ming et al. 2017). We then explored the expression levels of DEFB genes in INT cells expressing Cdg7_FLc_0990 or Cdg7_FLc_1000. We analyzed the previous genome-wide data using the same array chip as described above on INT cells following transfection of the Full-Cdg7_FLc_0990 or Full-Cdg7_FLc_1000 for 48 h (the GEO database with the accession number: GSE87047) (Wang et al. 2017a, b; Ming et al. 2017). No significant difference in the expression levels of DEFB genes was detected in cells transfected with the Full-Cdg7_FLc_0990, compared with cells transfected with the empty vector as the control (Fig. 2A, B). In contrast, downregulation of DEFB1 gene was observed in cells transfected with the Full-Cdg7_FLc_1000 (Fig. 2A, B). The expression level of DEFB104B showed a tendency to increase (with a P value close to 0.05) in INT cells after Full-Cdg7_FLc_1000 transfection (Fig. 2A, B). Notably, although the expression levels of the DEFB104B gene showed a tendency to increase in cells following infection, there was no statistic difference between cells of the non-infected control and cells following C. parvum infection for 8 and 48 h (Fig. 1A, B).
Fig. 2. Genome-wide array analysis of DEFB genes in INT cells expressing specific parasite RNA transcripts.
Alterations in DEFB gene expression profile from genome-wide transcriptome analysis in INT cells expressing specific RNA transcripts. INT cells were transfected with the Full-Cdg7_FLc_1000 or Full-Cdg7_FLc_0990 for 48h, followed by the microarray analysis; cells transfected with the empty vector were used as the control. The mean value of the expression level for these DEFB genes was presented as the heatmap (A) and listed in the table (B). *P<0.05 t test versus cells transfected with the empty vector.
3.3. Trans-suppression of host DEFB1 gene following C. parvum infection involves host delivery of Cdg7_FLc_1000 RNA
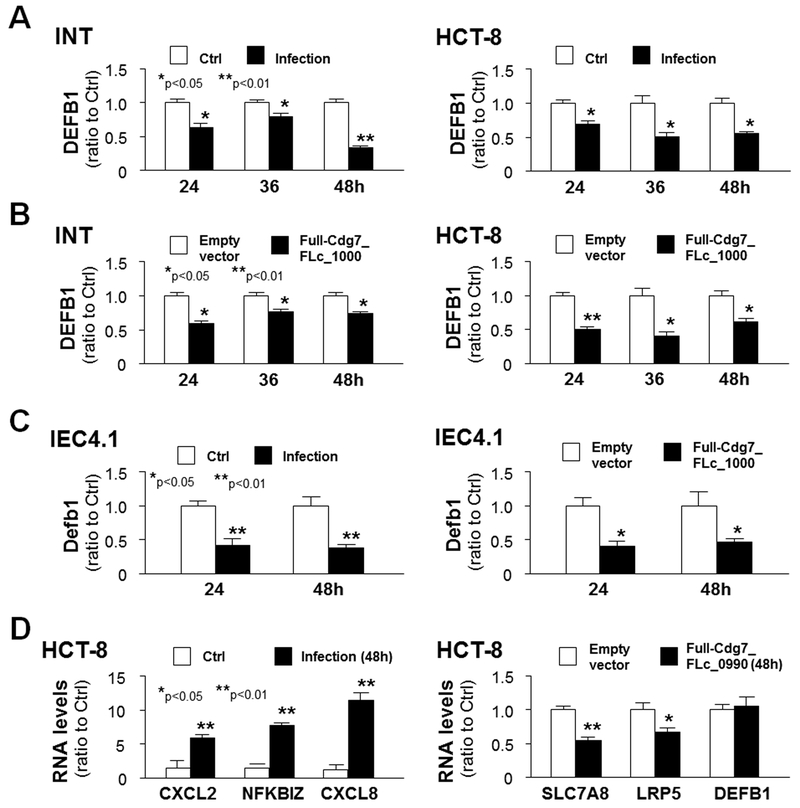
Given the downregulation of DEFB1 gene in cells expressing Cdg7-FLc_1000, we then questioned whether trans-suppression of host DEFB1 gene following infection involves host delivery of Cdg7-FLc_1000 RNA. We first measured the expression levels of DEFB1 gene at the RNA level in INT and HCT-8 cells following C. parvum infection for various periods of time using real-time PCR. As shown in Fig. 3A, expression levels of DEFB1 RNA in INT and HCT-8 cells following infection for 24, 36, and 48 h were significantly lower than that in the un-infected control. Accordingly, suppression of DEFB1 gene was also detected in INT and HCT-8 cells following transfection of the Full-Cdg7-FLc_1000 for 24, 36, and 48 h (Fig. 3B). Moreover, suppression of DEFB1 gene was further confirmed in murine intestinal epithelial (IEC4.1) cells following C. parvum infection or after transfection of the Full-Cdg7_FLc_1000 (Fig. 3C). For control, induction of the CXCL2, NFKBIZ and CXCL8 genes in HCT-8 cells following infection was detected (Fig. 3D). Consistent with results of previous reports (Wang et al. 2017a, b), suppression of SLC7A8 and LRP5 genes was detected in cells transfected with the Full-Cdg7-FLc_0990; in contrast, no changes were detected in the expression level of DEFB1 in the transfected cells (Fig. 3D), further supporting the specificity of trans-suppression of DEFB1 by Cdg7-FLc-1000.
Fig. 3. Trans-suppression of DEFB1 gene in cultured human and murine intestinal epithelial cells following C. parvum infection measured with real-time PCR.
(A) Downregulation of the DEFB1 gene in INT and HCT-8 cells following C. parvum infection. INT and HCT-8 cells were exposed to C. parvum infection for 24, 36, and 48 h, followed by real-time PCR analysis of DEFB1 RNA levels. (B) Downregulation of the DEFB1 gene in INT and HCT-8 cells following C. parvum infection or transfection with the Full-Cdg7_FLc_1000. INT and HCT-8 cells were exposed to C. parvum infection or transfection with the Full-Cdg7_FLc_1000 for 24, 36, and 48 h, followed by real-time PCR analysis of DEFB1 RNA levels. (C) Murine intestinal epithelial (IEC4.1) cells were exposed to C. parvum infection or transfection with the Full-Cdg7_FLc_1000, followed by real-time PCR analysis of Defb1 RNA levels. (D) Expression levels of selected genes induced by C. parvum infection or suppressed by Cdg7_FLc_0990 transfection in HCT-8 cells. HCT-8 cells were exposed to C. parvum infection for 48 h or transfection with the Full-Cdg7_FLc_0990 for 48 h, followed by real-time PCR analysis of selected genes. **P<0.05 and **P<0.01 t test versus non-infected cells or cells transfected with the empty vector.
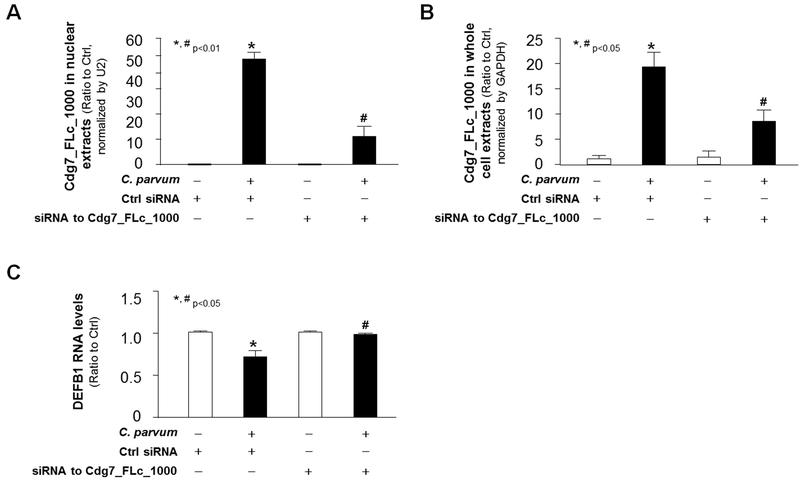
We then measured the effects of Cdg7_FLc_1000 knockdown on DEFB1 expression in cells following infection. Because conventional genetic tools are very difficult, if not impossible, to modify C. parvum genes (Striepen 2013; Vinayak et al. 2015), we designed an siRNA to Cdg7_FLc_1000 and transfected host cells for 12 h, followed by the exposure to C. parvum infection. A non-specific scrambled siRNA was used for control. The increase of Cdg7_FLc_1000 RNA levels in the nuclear extracts (Fig. 4A) and whole cell extracts (Fig. 4B) of HCT-8 cells induced by C. parvum infection was partially suppressed by pre-treatment of the siRNA to Cdg7_FLc_1000. Accordingly, suppression of DEFB1 RNA expression induced by C. parvum infection was significantly attenuated through pretreatment of the siRNA to Cdg7_FLc_1000 in HCT-8 cells (Fig. 4C). Taken together, these data suggest that suppression of DEFB1 gene in host cells induced by C. parvum infection may be mediated through host delivery of Cdg7_FLc_1000.
Fig. 4. Inhibition of Cdg7_FLc_1000 in host cells by the siRNA treatment attenuated the downregulation of DEFB1 following C. parvum infection.
HCT-8 cells were treated with an siRNA to Cdg7_FLc_1000 for 12 h and then exposed to C. parvum infection for additional 24 h. Content of Cdg7_FLc_1000 in the nuclear extracts (A) and whole cell extracts (B) from cells following infection, as well as the expression level of DEFB1 RNA in the infected cells (C), was quantified by real-time PCR. Data represent three independent experiments. *P<0.05 ANOVA versus non-infected cells treated with the control siRNA; # P<0.05 ANOVA versus infected cells treated with the control siRNA.
3.4. Inhibition of Cdg7_FLc_1000 delivery decreases C. parvum infection burden in cultured human intestinal epithelial cells
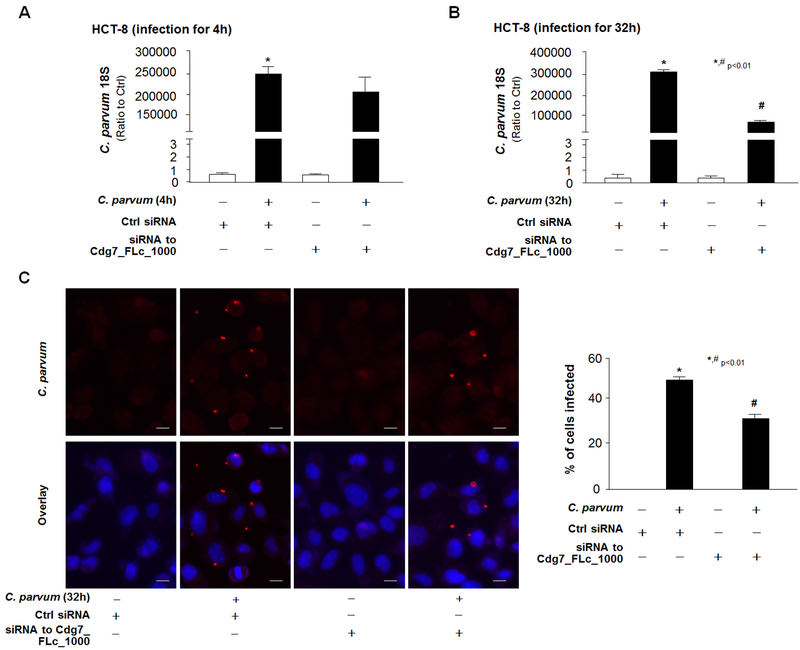
To test whether Cdg7_FLc_1000-mediated trans-suppression is involved in intestinal epithelial defense responses against C. parvum infection, we assessed the effects of Cdg7_FLc_1000 knockdown on the parasite infection burden in cultured cells. HCT-8 cells were transfected with the specific siRNA to Cdg7_FLc_1000, and then exposed to a constant number of C. parvum oocysts for 4 h to allow sufficient host-cell attachment and cellular invasion (Zhou et al. 2012; Chen and LaRusso 2000; Zhou et al. 2009). After extensive washing with culture medium to remove non-attached and non-internalized parasites, cells were either collected to detect the parasite attachment/invasion or cultured for an additional 28 h for the measurement of infection burden at intracellular development stage. Parasite burden was assessed in the samples using a real-time PCR approach as we previously reported (Ming et al. 2017; Zhou et al. 2012). The parasite burden in cells after exposure to C. parvum for 4 h, reflecting parasite attachment/invasion, was similar between cells transfected with the control siRNA and cells transfected with the siRNA to Cdg7_FLc_1000 (Fig. 5A), confirming that the siRNA treatments do not affect initial parasite host cell attachment and cellular invasion. Cells transfected with the siRNA to Cdg7_FLc_1000 displayed a decreased parasite burden at 32 h after infection as compared to cells treated with the non-specific scrambled siRNA (Fig. 5B). Similar results were obtained with INT cells (data not shown). Decrease of parasite burden at 32 h after initial infection in HCT-8 cells treated with the siRNA to Cdg7_FLc_1000 was further assessed and confirmed by immunofluorescent microscopy (Fig. 5C).
Fig. 5. Inhibition of Cdg7_FLc_1000 in host cells by the siRNA treatment decreased parasite infection burden in HCT-8 cells.
Cells were treated with an siRNA to Cdg7_FLc_1000 for 12 h and then exposed to C. parvum infection for 4 h or 32 h. Parasite infection burden was measured by using real-time PCR for the C. parvum 18s (A and B) or by immunofluorescent staining (C). Data represent three independent experiments. *P<0.01 ANOVA versus non-infected cells treated with the control siRNA; # P<0.01 ANOVA versus infected cells treated with the control siRNA. Bar = 10 µm.
4. Discussion
Increasing evidence suggests that RNA molecules may play important regulatory roles in diverse biological processes, including regulation of gene transcription (Prasanth and Spector 2007; Ulitsky and Bartel 2013). Recent genomic research has revealed the expression of novel non-protein coding RNA genes in the protozoan group of parasites, such as P. falciparum and C. parvum (Abrahamsen et al. 2004; Puiu et al. 2004; Yamagishi et al. 2011; Liao et al. 2014; Vembar et al. 2014). The interactions between Cryptosporidium and intestinal epithelial cells involves exchanges of distinct effector molecules from both sides of the host cell and the parasite at the host-parasite interface (Sibley 2004). Several C. parvum proteins have been demonstrated to be delivered into host epithelial cells at the host-parasite interface and are involved in parasite intracellular development (Sibley 2004; O’Connor et al. 2007). Our recent observation of selective delivery of C. parvum RNA transcripts of low protein-coding potential into infected host epithelial cells (Wang et al. 2017a) and consequent trans-suppression of host genes through distinct mechanisms (Ming et al. 2017; Wang et al. 2017b) support the notion that cryptosporidial infection causes transcriptional gene suppression with pathological significance in infected cells through nuclear transfer of specific parasite RNAs. In this study, our data suggest that trans-suppression of host DEFB1 gene in cultured human intestinal epithelial cells following C. parvum infection involves nuclear delivery of parasite Cdg7_FLc_1000 RNA, a process that may be relevant to the epithelial defense evasion by C. parvum at the early stage of infection.
One of the interesting findings of this study is the downregulation of DEFB1 genes in cells following C. parvum infection in vitro. Out of the 12 DEFB genes detectable with the microarray approach in INT cells, DEFB1 was persistently downregulated in cells following infection. Our finding is consistent with data from a previous report on the reduced production of the antimicrobial peptide of DEFB1 by murine and human intestinal epithelial cells following C. parvum infection in vitro and in vivo (Zaalouk et al. 2004). Downregulation of DEFB132 was detected in infected cells but only in cells following infection for 8 h. Downregulation of DEFB1 gene was further confirmed using real-time PCR analysis in cultured INT and HCT-8 cells following infection. In contrast, only DEFB124 was found to be upregulated in INT cells following infection for 48 h. Expression level of DEFB104B (also known as HBD-4), one of the most common inducible DEFB genes (Semple and Dorin 2012; Jarczak et al. 2013), showed a tendency to increase but without statistical significance. Of note, upregulation of DEFB104B induced by C. parvum has previously been demonstrated in human biliary epithelial cells and malignant human intestinal epithelial cell lines following infection in vitro (Zaalouk et al. 2004; Chen et al. 2005; Hu et al. 2013). Many effector molecules released from immune cells, such as IFN-γ from NK cells and many inflammatory cytokines from macrophages and lymphocytes, can regulate the expression of defense genes in the intestinal epithelial cells during microbial infection in vivo (Lantier et al. 2013; McDonald et al. 2013). Coupled with the fact that multiple DEFB genes are expressed in the intestine, future studies should characterize the DEFB gene expression profiles in epithelial cells using intestinal tissues from patients with intestinal cryptosporidiosis and elucidate the role of each DEFB gene in intestinal defense against cryptosporidium infection in vivo using conditional knock-out mice.
Several pieces of evidence implicate that nuclear delivery of the parasite Cdg7_FLc_1000 RNA transcript into infected cells modulates transcription of the DEFB1 gene. First, downregulation of the DEFB1 gene was observed in cells following infection. Intriguingly, genome-wide analysis of the gene expression profile revealed significant suppression of DEFB1 gene in cultured human intestinal epithelial cells overexpressing the Cdg7_FLc_1000 RNA. Second, using a specific siRNA to knockdown Cdg7_FLc_1000 in host cells during C. parvum infection attenuated the downregulation of the DEFB1 gene. Moreover, the association between Cdg7_Flc_1000 delivery and trans-suppression of DEFB1 appears to be specific, as transfection of another nuclear delivery parasite Cdg7_FLc_0990 RNA (Wang et al. 2017a, b), failed to downregulate the expression level of DEFB1. How nuclear delivery of Cdg7_FLc_1000 RNA may suppress DEFB1 transcription in infected cells remains unclear.
Despite significant induction of antimicrobial peptides, release of inflammatory chemokines/cytokines and local inflammatory reactions at infection sites, C. parvum can survive host immune attack at the early stage of infection. How C. parvum evades host innate immune defense at the early stage of infection is still unclear. Intracellular pathogens, including protozoan parasites, have evolved strategies to evade host immune attack. The suicide of host cells following invasion by intracellular pathogens is an ancient defense mechanism (Carmen and Sinai 2007). Many intracellular pathogens have developed strategies to inhibit the induction of host cell death (Carmen et al. 2006; Petersen et al. 2006). Indeed, C. parvum infection induces apoptotic resistance in infected epithelial cells (Chen et al. 2001; Liu et al. 2009). Our data from the current study suggest a new mechanism by which C. parvum may attenuate epithelial cell antimicrobial defense, i.e., trans-suppression of host defense DEFB1 gene through delivery of the parasite Cdg7_FLc_1000 RNA transcript into infected cells. Nuclear transfer of Cdg7_FLc_1000 RNA into infected host cells has also been demonstrated to attenuate intestinal epithelial cell migration via trans-suppression of host cell SMPD3 gene (Ming et al. 2017). Therefore, host delivery of parasite RNAs, such as Cdg7_FLc_1000, may provide several benefits to C. parvum for its intracellular development within intestinal epithelial cells after internalization. Targeting host delivery of specific parasite RNA transcripts may be of therapeutic value and merits further investigation.
Acknowledgments
We thank Dr. Guanghui Zhao (College of Veterinary Medicine, Northwest A&F University) for helpful and stimulating discussions, and Barbara L. Bittner (Creighton University) for her assistance in writing the manuscript. This work was supported by funding from the National Institutes of Health (AI116323 and AI136877) and the Nebraska Stem Cell Research Program (LB606), and by revenue from Nebraskaʼs excise tax on cigarettes awarded to Creighton University through the Nebraska Department of Health & Human Services (DHHS) (LB595). Dr. Zhenping Ming was a visiting scholar supported by the China Scholarship Council and the National Natural Science Foundation of China (NSFC No. 31372194). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the State of Nebraska, DHHS or NSFC.
Footnotes
Disclosures
The authors disclose no conflict of interest.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304: 441–445 [DOI] [PubMed] [Google Scholar]
- Carmen JC, Hardi L, Sinai AP (2006) titleToxoplasma gondii inhibits ultraviolet light-induced apoptosis through multiple interactions with the mitochondrion-dependent programmed cell death pathway. Cell Microbiol 8: 301–315 [DOI] [PubMed] [Google Scholar]
- Carmen JC, Sinai AP (2007) Suicide prevention: disruption of apoptotic pathways by protozoan parasites. Mol Microbiol 64: 904–916 [DOI] [PubMed] [Google Scholar]
- Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER (2015) A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, Keithly JS, Paya CV, LaRusso NF (2002) Cryptosporidiosis. N Engl J Med 346:1723–1731 [DOI] [PubMed] [Google Scholar]
- Chen XM, LaRusso NF (2000) Mechanisms of attachment and internalization of Cryptosporidium parvum to biliary and intestinal epithelial cells. Gastroenterology 118: 368–379 [DOI] [PubMed] [Google Scholar]
- Chen XM, Levine SA, Splinter PL, Tietz PS, Ganong AL, Jobin C, Gores GJ, Paya CV, LaRusso NF (2001). Cryptosporidium parvum activates nuclear factor kappaB in biliary epithelia preventing epithelial cell apoptosis. Gastroenterology 120: 1774–1783 [DOI] [PubMed] [Google Scholar]
- Chen XM OʼHara SP, Nelson JB, Splinter PL, Small AJ, Tietz PS, Limper AH, LaRusso NF(2005) Multiple Toll-like Receptors are expressed in human cholangiocytes and mediate host epithelial responses to C. parvum via activation of NF-kappaB. J Immunol 175: 7447–7456 [DOI] [PubMed] [Google Scholar]
- Deng M, Lancto CA, Abrahamsen MS (2004) Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol. 34: 73–82 [DOI] [PubMed] [Google Scholar]
- Hu G, Gong AY, Roth AL, Huang BQ, Ward HD, Zhu G, Larusso NF, Hanson ND, Chen XM (2013) Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog 9: e1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarczak J, Kościuczuk EM, Lisowski P, Strzałkowska N, Jóźwik A, Horbańczuk J, Krzyżewski J, Zwierzchowski L, Bagnicka E (2013) Defensins: natural component of human innate immunity. Hum Immunol 74: 1069–1079 [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O’Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382: 209–222 [DOI] [PubMed] [Google Scholar]
- Lantier L, Lacroix-Lamandé S, Potiron L, Metton C, Drouet F, Guesdon W, Gnahoui-David A, Le Vern Y, Deriaud E, Fenis A, Rabot S, Descamps A, Werts C, Laurent F (2013) Intestinal CD103+ dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PLoS Pathog 9: e1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent F, Kagnoff MF, Savidge TC, Naciri M, Eckmann L (1998) Human intestinal epithelial cells respond to Cryptosporidium parvum infection with increased prostaglandin H synthase 2 expression and prostaglandin E2 and F2alpha production. Infect Immun 66: 1787–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Shen J, Liu J, Sun X, Zhao G, Chang Y, Xu L, Li X, Zhao Y, Zheng H, Zhao Y, Wu Z (2014) Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol Res 113: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Liu J, Deng M, Lancto CA, Abrahamsen MS, Rutherford MS, Enomoto S (2009) Biphasic modulation of apoptotic pathways in C. parvum-infected intestinal epithelial cells. Infect Immun 77: 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald V, Korbel DS, Barakat FM, Choudhry N, Petry F (2013) Innate immune responses against Cryptosporidium parvum infection. Parasite Immunol 35: 55–64 [DOI] [PubMed] [Google Scholar]
- Ming ZP, Gong AY, Wang Y, Zhang XT, Li M, Mathy NW, Strauss-Soukup JK, Chen XM (2018) Involvement of Cryptosporidium parvum Cdg7_FLc_1000 RNA in the attenuation of intestinal epithelial cell migration via trans-suppression of host cell SMPD3 gene. J Infect Dis 217: 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RM, Wanyiri JW, Wojczyk BS, Kim K, Ward H (2007) Stable expression of Cryptosporidium parvum glycoprotein gp40/15 in Toxoplasma gondii. Mol Biochem Parasitol 152: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OʼDonoghue PJ (1995) Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol 25: 139–195 [DOI] [PubMed] [Google Scholar]
- Petersen CA, Krumholz KA, Carmen J, Sinai AP, Burleigh BA (2006) Trypanosoma cruzi infection and nuclear factor kappa B activation prevent apoptosis in cardiac cells. Infect Immun 74: 1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the genome complexityʼ conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Puiu D, Enomoto S, Buck GA, Abrahamsen MS, Kissinger JC (2004) CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res 32: 329–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KA, Rogers AB, Leav BA, Sanchez A, Vannier E, Uematsu S, Akira S, Golenbock D, Ward HD (2006) MyD88-dependent pathways mediate resistance to Cryptosporidium parvum infection in mice. Infect Immun 74: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Wahnschaffe U, Schäfer M, Zippel T, Arvand M, Meyerhans A, Riecken EO, Ullrich R (2001) Rapid increase of mucosal CD4 T cells followed by clearance of intestinal cryptosporidiosis in an AIDS patient receiving highly active antiretroviral therapy. Gastroenterology 120: 984–987 [DOI] [PubMed] [Google Scholar]
- Semple F, Dorin JR (2012) β-Defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun 4: 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD (2004) Intracellular parasite invasion strategies. Science 304: 248–253 [DOI] [PubMed] [Google Scholar]
- Striepen B (2013) Parasitic infections: Time to tackle cryptosporidiosis. Nature 503: 189–191 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Scherf A, Siegel TN (2014) Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr Opin Microbiol 20: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B (2015) Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523: 477–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong AY, Ma S, Chen X, Li Y, Su CJ, Norall D, Chen J, Strauss-Soukup JK, Chen XM (2017a) Delivery of parasite RNA transcripts into infected epithelial cells during Cryptosporidium infection and its potential impact on host gene transcription. J Infect Dis 215: 636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong AY, Ma S, Chen X, Strauss-Soukup JK, Chen XM (2017b) Delivery of parasite Cdg7_Flc_0990 RNA transcript into intestinal epithelial cells during Cryptosporidium parvum infection suppresses host cell gene transcription through epigenetic mechanisms. Cell Microbiol 10.1111/cmi.12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi J, Wakaguri H, Sugano S, Kawano S, Fujisaki K, Sugimoto C, Watanabe J, Suzuki Y, Kimata I, Xuan X (2011) Construction and analysis of full-length cDNA library of Cryptosporidium parvum. Parasitol Int 60: 199–202 [DOI] [PubMed] [Google Scholar]
- Yang YL, Serrano MG, Sheoran AS, Manque PA, Buck GA, Widmer G (2009) Over-expression and localization of a host protein on the membrane of Cryptosporidium parvum infected epithelial cells. Mol Biochem Parasitol 168: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaalouk TK, Bajaj-Elliott M, George JT, McDonald V (2004) Differential regulation of beta-defensin gene expression during Cryptosporidium parvum infection. Infect Immun 72: 2772–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Gong AY, Eischeid AN, Chen XM (2012) miR-27b targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum infection. PLoS Pathog 8: e1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM (2009) NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog 5: e1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]