Abstract
Accumulating evidence suggests that soluble oligomers are more toxic than final fibrils of amyloid aggregations. Among the mixture of inter-converting intermediates with continuous distribution of sizes and secondary structures, oligomers in the β-barrel conformation – a common class of protein folds with a closed β-sheet – have been postulated as the toxic species with well-defined three-dimensional structures to perform pathological functions. A common mechanism for amyloid toxicity, therefore, implies that all amyloid peptides should be able to form β-barrel oligomers as the aggregation intermediates. Here, we applied all-atom discrete molecular dynamics (DMD) simulations to evaluate the formation of β-barrel oligomers and characterize their structures and dynamics in the aggregation of a seven-residue amyloid peptide, corresponding to the amyloid core of amyloid-β with a sequence of 16KLVFFAE22 (Aβ16-22). We carried out aggregation simulations with various numbers of peptides to study the size dependence of aggregation dynamics and assembly structures. Consistent with previous computational studies, we observed the formation of β-barrel oligomers in all-atom DMD simulations. Using a network-based approach to automatically identify β-barrel conformations, we systematically characterized β-barrels of various sizes. Our simulations revealed the conformational inter-conversion between β-barrels and double-layer β-sheets due to increased structural strains upon forming a closed β-barrel while maximizing backbone hydrogen bonds. The potential of mean force analysis further characterized the free energy barriers between these two states. The obtained structural and dynamic insights of β-barrel oligomers may help better understand the molecular mechanism of oligomer toxicities and design novel therapeutics targeting the toxic β-barrel oligomers.
Introduction
Amyloid depositions of protein aggregation are associated with more than 25 degenerative diseases, including Alzheimer’s disease [1, 2], Parkinson’s disease [3, 4], prion conditions [5] and type-2 diabetes [6–8]. Despite differences in sequence and length of aggregating proteins, different amyloid proteins show similar all-or-none sigmoidal aggregation kinetics and common fibrillar morphology with the characteristic cross-β core of their final aggregates. These similarities together with the shared symptoms of different amyloid diseases suggest a potentially common mechanism for amyloid cytotoxicity [9–11]. Although mature fibrils were long suspected to be the toxic disease agents, more and more experimental studies with different amyloid proteins suggested that the small soluble oligomer intermediates populated during aggregation are more toxic [12–14]. These oligomer intermediates correspond to the mixture of many small aggregates with continuous distributions in sizes and secondary structure contents, not all of which are cytotoxic. In addition, several non-fibril intermediates have been shown important and applicable to develop small molecules that are able to regulate Aβ aggregation and cytotoxicity [15–18]. As the aggregation intermediates, these oligomers are highly heterogeneous, polymorphic and transient in nature, imposing major experimental challenges for pinpointing and characterizing the exact toxic species of amyloid aggregation.
Based on the general structure-function principle in biology [19–21], toxic oligomers are expected to have well-defined three-dimensional structures to carry out their pathological functions. A common amyloid toxicity mechanism also suggests that toxic oligomers of different amyloid proteins may have similar structures. For example, experimental studies using atomic force miscopy (AFM) showed that various amyloid peptides could form pore-like structures in the membrane, causing membrane leakage and disruption [22–24]. Using an 11-residue peptide fragment from a slow-aggregating αB crystalline (with the sequence of 90KVKVLGDVIEV100, named K11V), Laganowsky et al. discovered that the short peptides could form a stable toxic hexamer [12]. Using x-ray crystallography, the hexamer was found to feature a β-barrel or β-cylindrin structure. β-barrel is a common protein topology, which is adopted by both soluble and membrane proteins. In addition, Do et al. combined experimental characterizations with computational modeling to show that the C-terminal fragments of amyloid-β (Aβ) with a length of 11 residues might form similar β-barrel oligomers [20]. The potential formation of β-barrel oligomers during the aggregation of hIAPP8-20 has been supported by a recent mass spectra and ion mobility study [25]. Moreover, the β-barrel structure as a model for small Aβ40 oligomers was also supported by hydrogen exchange mass spectrometry experiments [20]. Other experimental studies of Aβ-membrane interactions suggested the insertion of Aβ into the membrane and formation of an ion channel like configuration, causing an abnormal flux of ions into the cell [26–28]. Hence, these β-barrel oligomers with the capability to be incorporated into the membrane and thus compatible to the “amyloid pore” hypothesis [22–24] have been postulated as the early aggregation intermediates exerting toxic effects on cells [12].
The formation of β-barrel oligomers was also observed in previous computational studies of several short peptides [29–32]. Using Monte Carlo [33] simulations, Irback & Mitternacht found that Aβ16-22 (i.e., the amyloid core of Amyloid-β with a sequence of 16KLVFFAE22) could form β-barrels. Via enhanced sampling with replica exchange molecular dynamics (REMD), β-barrel conformations have been observed in all-atom aqueous simulations of NHVTLSQ of the beta-2 microglobulin protein [30] and KLVFFAE of Aβ peptide [29]. However, the detailed structure and dynamics of β-barrel oligomers – including size dependence, structural characterization, and conversion to protofibrils or fibrils – remain unknown. Recently, Zhang et al. investigated the fibril-barrel transitions of K11V using a structure-based constraints to model both barrels and fibrils [31], however the independent insights from unconstrained simulations remain to be seen. Molecular insights of the structures and dynamics of β-barrel oligomers as aggregation intermediates may help understand the origin of amyloid toxicity and design novel therapeutics targeting these toxic species.
Recently, all-atom discrete molecular dynamics (DMD) simulations – a predictive and computationally efficient molecular dynamics approach [34–37] – have been used to capture the assembly dynamics of Aβ16-22 from monomers to oligomers and to fibril-like cross-β structures [38], where the formation of β-barrel oligomers was not the focus. Motivated by recent experimental and computational studies, we used the same approach to evaluate whether Aβ16-22 could also form β-barrel oligomers and to investigate the detailed structures and dynamics of these well-structured oligomers. Indeed, the 7-residue peptide could form β-barrel oligomers as the aggregation intermediates. We proposed a network-based approach to identify β-barrels in simulations and characterized β-barrels with various sizes. The formation of β-barrels was driven by minimizing the exposure of hydrophobic residues to solvent along with maximizing the number of backbone hydrogen bonds, although the closure of a β-sheet into a β-barrel also resulted into conformational strains in the structure. We also observed the conformational inter-conversion between β-barrels and double-layer β-sheets, suggesting their co-existence with comparable free energies. The potential of mean force (PMF) analysis of DMD simulations further characterized the free energy barrier between these two states. With double-layer β-sheets resembling the cross-beta core of fibrils, our results suggested the β-barrel oligomers as “off-pathway” aggregates towards fibrillization in solution. Future aggregation simulations with full-length amyloid peptides and/or in the membrane environment are necessary to decipher the general roles of β-barrel oligomers in amyloid toxicity.
Results and Discussion
In order to investigate the assembly dynamics and aggregate structures of Aβ16-22, twelve molecular systems with the number of peptides varying from 1 to 12 were studied. In all simulations, the same peptide concentration (~15 mM) was maintained by adjusting the dimensions of simulation boxes. To obtain sufficient sampling and avoid potential biases from the starting configurations, we performed fifty independent simulations for all molecular systems, each of which started with different initial configurations and lasted 200 ns (details of simulations see Methods).
Aβ16-22 shows a high propensity to form β-sheet aggregates
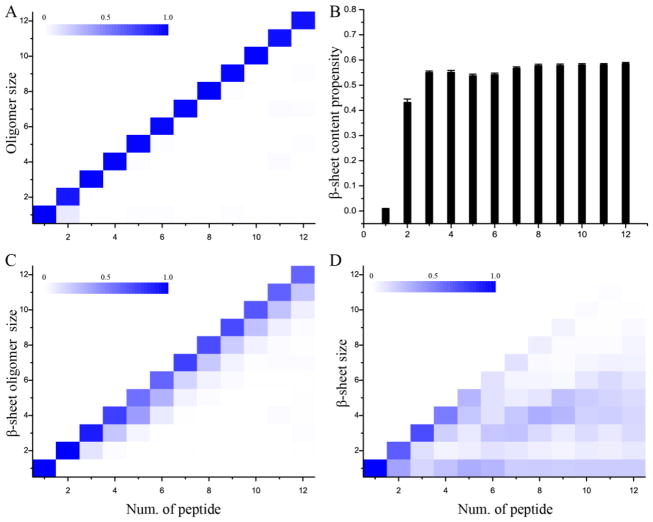
We estimated the aggregation thermodynamics using the last 100 ns trajectories of independent simulations, which reached their corresponding steady states in terms of energetics and secondary structure contents (data not shown). By computing the number of atomic contacts between peptides for each snapshot, we were able to group peptides into connected clusters (i.e., oligomers) and analyze the distribution of oligomer size defined as the number of peptides forming an oligomer (Fig. 1A). The oligomer size was nearly equal to the number of peptides within the corresponding molecular systems, suggesting the high self-association propensity of Aβ16-22 [39, 40].
Figure 1.
Oligomerization and β-sheet formation in DMD simulations with different number of peptides. (A) Probabilities of observing oligomers of various sizes, (B) the total β-sheet content, and probabilities of observing (C) β-sheet rich oligomers formed by various number of β-strands and (D) β-sheets comprised of different numbers of β-strands were calculated for simulations with a given number of peptides. All analyses were averaged over all 50 independent simulations and only the last half of each trajectory was used. In panels A, C & D, the probability of each aggregate shown in the heat map was weighted according to its size (i.e., the number of composite peptides). Error bars in panel B denote the standard error of means (S.E.M).
We also computed the average contents of various secondary structures, and found the aggregates were dominated by β-sheets (Fig. 1B). It was noticeable that even in simulations with only two peptides, the dimer already had ~45% of β-sheet conformations. To characterize the formation of β-sheet rich aggregates, we further investigated the formation of β-sheet oligomers, which were defined as aggregates comprised of β-sheets and whose sizes were determined to be the number of peptides adopting β-strand conformations (details of analysis in Methods). The probability distribution of observing β-sheet oligomers with various sizes as the function of total peptides in a molecular system further suggested Aβ16-22 peptides could readily associate with each together and form β-sheet rich aggregates (Fig. 1C). Since a β-sheet rich aggregate could be comprised of multiple β-sheets, we also evaluated the distribution of individual β-sheet size defined as the number of β-strands in a β-sheet (Fig. 1D). We found that small systems would form a single β-sheet with the size of β-sheet close to the number of total peptides in simulations, while the aggregates of larger systems could end up as double or multi-layer β-sheet structures. Our simulation results are in agreement with previous experimental and computational studies, where Aβ16-22 were observed having a high propensity to self-assemble into aggregates rich in β-strand content, with a preference for the antiparallel arrangement [39].
Size dependence of Aβ16-22 aggregation structures
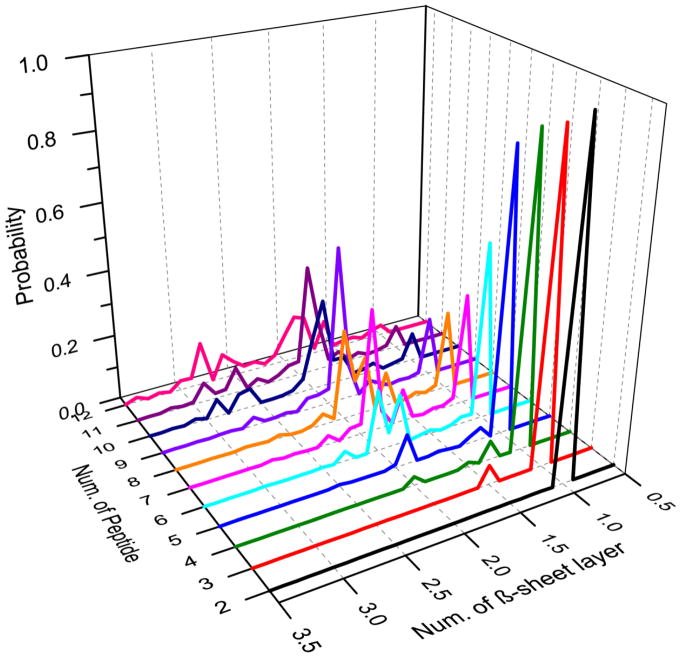
We estimated the number of β-sheets in a β-sheet oligomer by dividing its size with the mass-weighted average β-sheet size (Methods) and computed the distribution of β-sheet numbers in β-sheet oligomers of each molecular system (Fig. 2). As expected from the β-sheet size distribution in Fig. 1D, small molecular systems (with the number of peptides less than 5) mainly aggregated into a single β-sheet structure. Six Aβ16-22 peptides had a probability of ~57% to form a single β-sheet and ~40% to form double-layer β-sheet aggregates. With eight peptides, the systems had approximately equal probability to end up in either single- or double-layer structures. The double-layer structure became the dominated species and the triple-layer structure started to emerge after the system size reached nine and larger. These results suggested that an isolated Aβ16-22 β-sheet aggregate might be stable with up to five peptides, while multi-layer aggregates became more favorable when more peptides were introduced into the system. This size dependence of Aβ16-22 aggregate structures likely resulted from the fact that residues at both surfaces of an Aβ16-22 β-sheet are hydrophobic – i.e., L17, F19, A21 on one side and V18, F20 on the other side. By forming the multi-layer structures, a large number of Aβ16-22 peptides could bury hydrophobic residues to minimize the hydrophobic solvent accessible surface area. These results are consistent with our previous study [38] and also the experimental report that Aβ16-22 peptides could self-assemble into the ribbon-like aggregate [41], indicating the formation of multi-layer β-sheet structures.
Figure 2.
The probabilities of observing β-sheet rich aggregates with different number of β-sheets were computed for simulations with a give number of peptides. For each β-sheet aggregate, the number of β-sheet layers was estimated according to the mass-weighted β-sheet size averaged over all compositing β-sheets.
The dynamics of Aβ16-22 oligomerization and identification of β-barrel structures
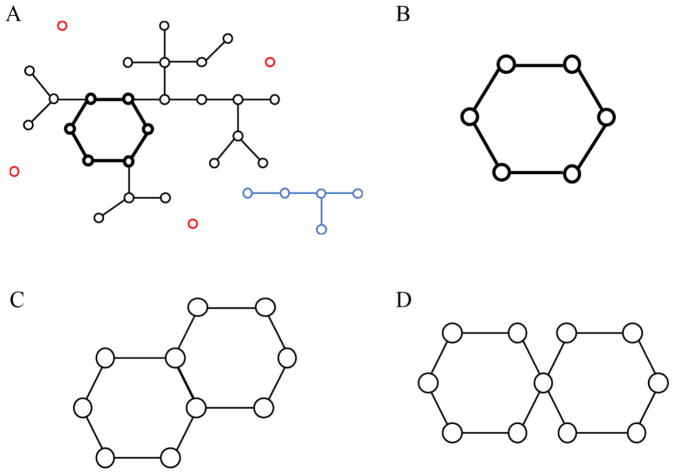
We next examined the dynamics of peptide oligomerization and β-sheet formation by monitoring various structural properties along DMD simulation trajectories, including the sizes of the largest oligomer, the largest β-sheet oligomer, the largest β-sheet, the mass-weighted β-sheet, and the β-barrel. For a given snapshot, a simple method was used to identify the β-barrel structure (Fig. 3), which corresponds to a closed β-sheet. We constructed a network by connecting any two peptides forming a β-sheet with multiple consecutive backbone-bone hydrogen bonds. For each inter-connect cluster, we iteratively eliminated all terminal elements (peptides with only one β-sheet neighbor) until there were no further dangling terminal elements. The clusters left were those with closed circuits of multiple elements (peptides), and thus, determined as β-barrels (Fig. 3B). It is noticeable that the elements inside such a closed circuit could have more than two neighbors, and thus the whole structure may form multiple loops. However, this scenario could only take place when a long peptide had more than two β-sheet neighbors (Fig. 3C, D), which was not observed for a short 7-residue peptide in our simulations.
Figure 3.
The network-based approach to identify β-barrel structures. (A) The schematic β-sheet contact network, where each element represents a peptide and any two peptides forming adjacent β-strands in a β-sheet is connected. The network is composed of two clusters with more than one connected elements (color in black and blue respectively) and isolated elements (colored in red). The circular structure highlighted by thick links represent a β-barrel structure. (B) By iteratively removing dangling elements (i.e., having only one contacted neighbors), the only structure left in addition to isolated elements (removed) is the circular β-barrel. (C, D) Schematics of possible multi-barrel structures where a peptide could form more than two β-sheet neighbors. In our simulations of short peptides, such cases were not observed.
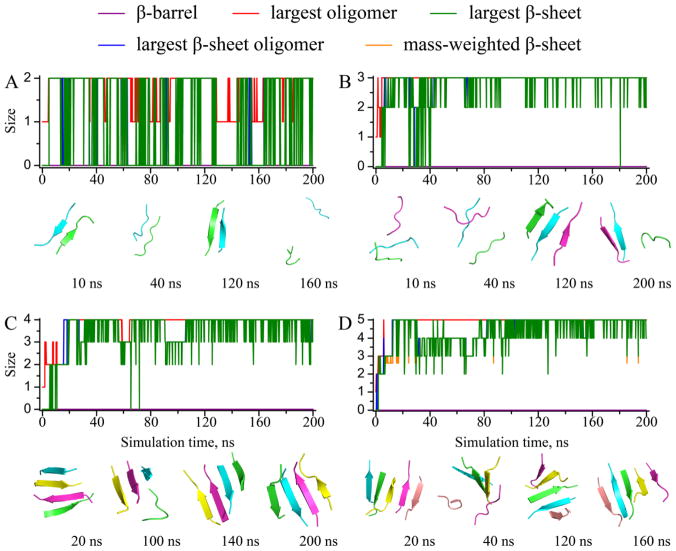
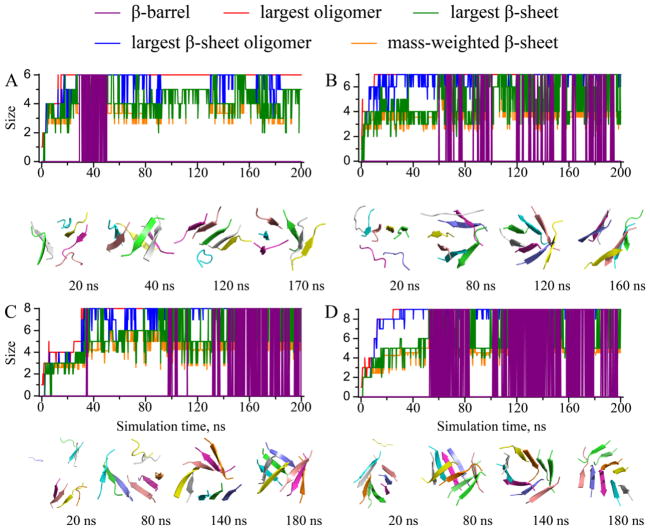
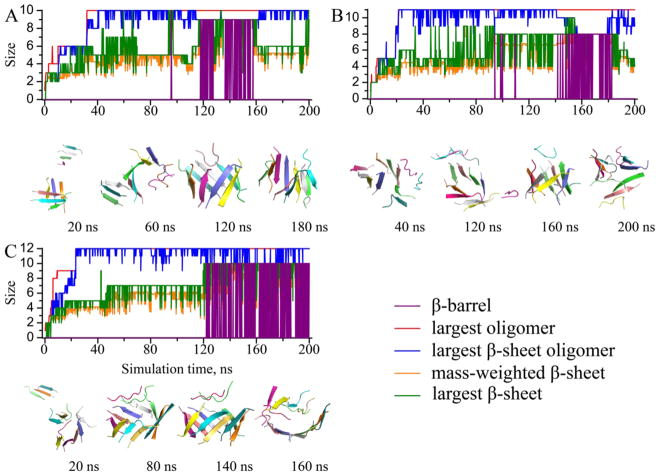
The monitored structural properties were computed along each simulation trajectory, with typical ones shown to illustrate the aggregation process of Aβ16-22 (Figs. 4, 5, 6). In all simulations, we observed rapid peptide association, as the size of the largest oligomer (red) rapidly reached its maximum and equaled the number of total peptides. The time evolution of the largest β-sheet oligomer size (blue) displayed similar trend with only a short “lag time” during which the peptides inside the oligomers underwent conformational changes to adopt β-rich structures. Size of the largest β-sheet (green) displayed high fluctuations, and the mass-weighted β-sheet size (orange) ended up with approximately five in relatively large systems, confirming the observation that the size limit for an isolated Aβ16-22 β-sheet was approximately five (Fig. 1). Remarkably, we observed β-barrel structures (purple) in all simulations with six or more peptides. The formation of β-barrel oligomers appeared rather dynamic. A β-barrel could occur during the early stage of a simulation and then unfold (Fig. 5A), or undergo unfolding/refolding transitions (Fig. 6A). From our simulations, we determined the minimum number of Aβ16-22 peptide required to form a β-barrel was six, slightly above the upper limit of a stable isolated Aβ16-22 β-sheet. In the case of six Aβ16-22 peptides (Fig. 5A), we observed conversions of β-barrel to a single and then a double-layer β-sheet aggregates. A β-barrel could be considered as a unique single-layer β-sheet, where the β-sheet was large enough to curve and form a closed loop to maximize the number of backbone-backbone hydrogen bonds and minimize the hydrophobic solvent accessible surface area by allowing peptides at edges to make contacts and form hydrogen bonds. Similarly, a small double-layer β-sheet with aligned β-strands could have peptides at both ends forming inter-peptide hydrogen bonds to form a closed β-barrel. Since it was rare for peptides in both ends to close simultaneously, the latter process could be described stepwise as double-to-single and then sheet-to-barrel transitions.
Figure 4.
The dynamics of oligomerization and aggregation. Sizes of the largest oligomer, the largest β-sheet oligomer, the largest β-sheet, the mass-weighted β-sheet, and the β-barrels were plotted as functions of the simulation time for typical simulations trajectories with (A) 2, (B) 3, (C) 4, and (D) 5 peptides. Snapshot structures at different times with peptides shown as cartoon with different colors were used to illustrate the aggregate structures and their conformational dynamics.
Figure 5.
The dynamics of oligomerization and aggregation for simulations with (A) 6, (B) 7, (C) 8, and (D) 9 peptides. The same presentation style as Fig. 4 is used.
Figure 6.
The dynamics of oligomerization and aggregation for simulations with (A) 10, (B) 11, and (C) 12 peptides. The same presentation style as Fig. 4 is used.
β-barrel conformations of various sizes
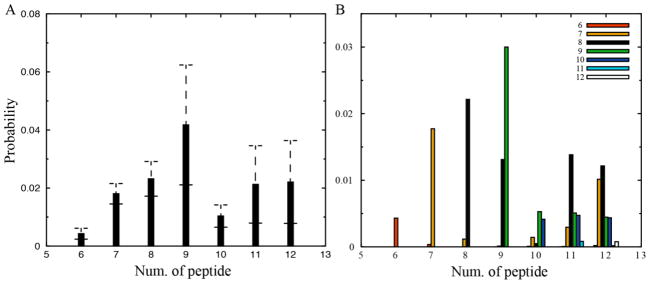
We further focused on the identified β-barrel structures from simulations with six or more peptides. As discussed above, we did not observe multi-barrel structures in our simulations (Fig. 3C, D), which might occur in longer peptides. The probability of finding a peptide in β-barrel oligomers at any given time was computed for each molecular system and averaged over fifty independent simulations (Fig. 7A). The probability increased initially with the increasing number of peptides until reaching the maximum (~4%) with nine peptides, and dropped to an approximately saturated value of ~1–2% in our simulated systems up to 12 peptides. We also obtained the size distribution of β-barrels in different molecular systems, and found that barrels in small systems with less than eight peptides were likely to have the same sizes as the corresponding system sizes. In simulations with more than eight peptides, however, peptides could form β-barrels with different sizes (Fig. 7B) and the octameric β-barrel was the most populated species.
Figure 7.
Probabilities of observing a peptide in (A) a β-barrel structure and (B) in a β-barrel of various sizes were computed for each simulation system with a given number of peptides. Error bars denote S.E.M.
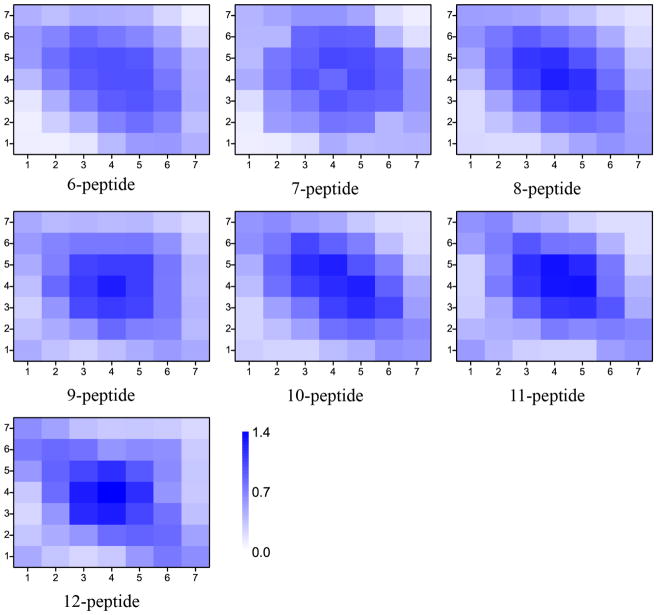
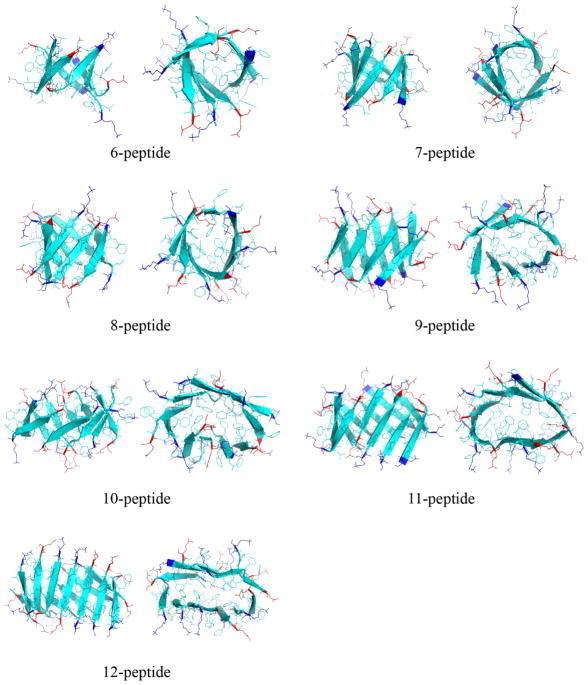
To characterize the 3D structures of β-barrels, we computed the residue-wise contact frequency maps based on inter-peptide main-chain interactions (Fig. 8). β-barrels of various sizes shared similar features. For instance, a common binding pattern perpendicular to the diagonal suggesting anti-parallel alignments of β-strands and similar “hot-spot” regions indicating the same inter-peptide interactions. Typical snapshots of β-barrels with various sizes were selected to illustrate the structural features (both side and top views were provided in Fig. 9). Indeed, β-barrels had mostly anti-parallel alignments of strands with hydrophobic side-chains pointing towards the interior in agreement with previous computational studies [29]. The anti-parallel alignments arose from electrostatic interactions as the peptide contained a positively charged N-terminal residue K16 and a negatively charged C-terminal residue E22. More importantly, the cross-sections of small β-barrels (from size 6 to 8) displayed a round shape, while larger ones changed into more elliptical shapes. The size-dependent shape of either cylindrical or cylindroid β-barrels was mainly determined by hydrophobic interactions. An ideal cylindrical barrel with the number of peptides larger than eight might result into holes in the center, while a cylindroid could reduce the void. As the number of peptide increases, structural strains associated with large curvatures near the two ends of major axis of the cylindroid eventually led to the breakage of hydrogen bonds, limiting the observed β-barrel sizes of Aβ16-22 in our DMD simulations. However, in a lipid membrane environment larger β-barrels with hydrophilic residues pointing inward could be stable and form the pore.
Figure 8.
Inter-chain contact maps of β-barrel with various sizes. By constructing the structural ensemble of all β-barrel with a given size from simulations, the contact frequencies between any two residues were computed based on inter main-chain interactions between different peptides forming a β-barrel.
Figure 9.
Representative structures of β-barrels with various sizes are shown in both side and top views. Each peptide is shown in cartoon and all residues are shown as sticks. Positively-charged lysine residues in the N-terminal are colored blue and negatively-charged glutamate residues in the C-terminal are colored red, while the rest hydrophobic residues are colored cyan.
The β-barrel oligomers might be “off-pathway” intermediates towards fibrillization
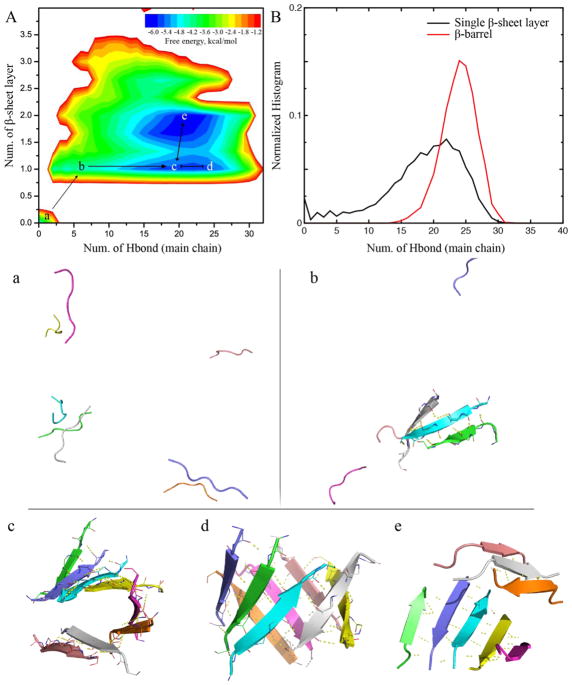
To better understand the aggregation process, we computed the potential of mean force (PMF, i.e., the effective free energy) as a function of the number of hydrogen bonds between main-chains and the number of β-sheet layer for simulations of eight peptides (Fig. 10A). All the 200ns trajectories from 50 independent simulations were included to capture the early aggregation process. This particular molecular system was chosen because of approximately equal probabilities to observe both single- and double-layer β-sheet aggregates (Fig. 2). Indeed, two major free energy basins could be observed corresponding to the single- and double-layer β-sheet aggregates, respectively. β-barrels are unique single-layer β-sheets with more main-chain hydrogen bonds than extended sheets, and thus the normalized histogram of the total number of main-chain hydrogen bonds for β-barrels had peaks at larger numbers than that of all single-layer β-sheets (Fig. 10B). In another word, β-barrels were enriched in the right end of the single-layer aggregate basin in the 2D-PMF.
Figure 10.
The aggregation free energy landscape estimated from simulations with 8 peptides. (A) The PMF was estimated according to the probability observing an aggregate with a total number of hydrogen bonds (inter main-chain) and the number of β-sheet layer in the structure. Basins and saddles were labeled (a–e) and connected to demonstrate the aggregation pathway. Structural snapshots were selected as the representative structures of each corresponding labels in the free energy landscape in panel A. Each peptide is shown in cartoon and hydrogen bonds are highlighted as dashed yellow lines. (B) Normalized histograms of the number of hydrogen bonds for both single-layer β-sheets and β-barrels.
Taken together, we could postulate the formation of β-barrels along aggregation pathway of Aβ16-22. Starting as isolated peptides in our simulations (Fig. 10a), peptides started to associate with each other and rapidly form inter-peptide hydrogen bonds as single-layer β-sheets (Fig. 10b). An anti-parallel alignment of the β-sheet was preferred due to charged K16 and E22. When the β-sheet was larger than five, the hydrophobic interaction drove the sheet into a curved conformation (Fig. 10c) and further into a closed β-barrel (Fig. 10d). Compared to a double-layer β-sheet conformation (Fig. 10e), a β-barrel conformation features a higher number of hydrogen bonds but also with higher structural-strains in curved sheets. Hence, the two types conformations – i.e., β-barrel and double-layer β-sheet – had comparable free energies and thus could inter-convert with each other (Fig. 10d, e). On the other hand, a β-barrel had to break hydrogen bonds along two ends in order to form the double-layer β-sheet with a curved single-layer β-sheet as the intermediate. With the double-layer β-sheet en route toward the final fibrils, the β-barrel therefore could be considered as “off-pathway” intermediates towards the fibrillization of Aβ16-22.
The relevance of β-barrel oligomers with the membrane-associated amyloid toxicity
While the relationship between amyloid deposition and cellular toxicity is still unknown, many studies suggest that Aβ oligomers are polymorphic and transient toward peptide fibrillization [42, 43], and may have different mechanisms of toxicity [44, 45]. For instance, the deposition of intermediate to large Aβ oligomers is believed to induce a general increase in the membrane permeability [46] which eventually leads to the cellular dysfunction [47–50]. Aβ40 or its fragments are found capable of forming pore-like or barrel-like oligomers in neuronal plasma membranes, which can function as cation-selective channels and induce ion fluxes as what had been postulated in a two-step mechanism of membrane disruption [27, 51–56]. Other soluble Aβ oligomers denoted as Aβ-derived diffusible ligands (ADDL), on the other hand, are linked to neuron toxicity via binding with protein receptors [49]. The AMF images of Aβ barrel-like structures revealed an outer diameter of 8–12 nm and an inner diameter of the cavity pore as 1–2 nm [57]. Computational studies about the polymorphism of Aβ barrel-like structures had also been carried out and revealed the linkage of Aβ barrels and amyloid channels in cell membranes [58, 59]. Moreover, the comparison of modeled channels of different amyloid peptides showed shared structural features, suggesting the formation of β-barrel structure is generic mechanism inducing toxicity among amyloid peptides.
Our simulation results suggested the 7-residue Aβ fragment 16KLVFFAE22 could spontaneously form the β-barrel oligomers in solution. With the experimental evidence of the insertion of Aβ into cell membrane, it’s possible for Aβ peptides to form pore-like barrel structures in similar way. Although large beta-barrel oligomers with big pores were unstable in our solution simulations (e.g., Fig. 9), the hydrophobic environment in the membrane could stabilize those oligomers. The formation β-barrel structures with large pores in the membrane could then result in the membrane leakage and abnormal flux of ions into the cell, causing cell death.
Conclusions
In summary, we performed all-atom DMD simulations of Aβ16-22 aggregation with different number of peptides. Consistent with previous studies [60], Aβ16-22 shows a high propensity to form β-sheet aggregates. While monomers were mostly coils, even two peptides could form dimers with extensive β-sheet content. With the system size smaller than six, the peptides mainly formed single-layer β-sheets. Only when the number of peptides reached six and larger, the β-barrel oligomers could be observed. The observation of β-barrel oligomers was consistent with previous all-atom aqueous REMD and Monte Carlo studies [29, 33]. Using the proposed β-barrel detection algorithm, our systematic analysis of β-barrels indicated they were mostly formed by six to nine peptides. High-order β-barrels of larger sizes could be observed but with significantly smaller probabilities (Fig. 7B). The β-barrels of six to eight peptides were mostly cylindrical (Fig. 9), and become more cylindroid with increasing sizes. These barrels were stabilized by maximizing the number of backbone hydrogen bonds and burying the hydrophobic residues, but the closing of a single β-sheet also imposed structural strains. As the result, we observed the dynamic inter-conversion from β-barrels to double-layer β-sheets via an intermediate state of highly-curved single-layer sheets, where the breaking of hydrogen bonds likely happened at β-strands with high strains (or large curvature). Calculation of the contact frequency maps of various β-barrels indicated that adjacent β-strands tent to be in anti-parallel alignment, similar to the experimentally solved K11V β-barrel hexamer [12]. We also performed PMF analysis of the aggregation of eight peptides, where both single- and double-layers β-sheets were populated with similar probability (Fig. 2). Our computational analysis suggested that β-barrels were “off-pathway” intermediates toward fibrillization. Taken together, our unconstrained all-atom DMD simulations offered detailed structural and dynamic insights to the formation of β-barrel intermediates. Although our simulations were performed in a membrane free environment, the spontaneous formation of β-barrels in our simulations may indicate an intrinsic propensity of the amyloid-core fragment of Aβ to form the pore-like structures. How the amyloid peptides form oligomers after their insertion into membrane or whether pre-formed β-barrels could be incorporated into the membrane still need to be established. In addition, our observed β-barrels are formed by the Aβ16-22 fragment, questions like whether and how full length amyloid peptides form the β-barrel structures remain to be answered. Further studies such as the formation of these β-barrels in membrane environments and experimental validation and characterization of these toxic species are necessary to better understand the mechanism of amyloid toxicity and design therapeutic strategies targeting these novel toxic species.
Material and Methods
Simulation setup
In all simulations, we used peptide segment Aβ16-22 (KLVFFAE) to construct our molecular system. Twelve molecular systems were modeled, with the number of peptides varied from 1 to 12 and the peptide concentration maintained the same as that of a single peptide in a cubic box with the dimension of 48Å (~15 mM). The peptides were set fully extend and randomly placed with inter-chain minimum distance no less than 1.5 nm. Periodic boundary conditions were also applied. For each molecular system, we performed 50 independent simulations with each started from different initial configurations (e.g., randomized velocities, intermolecular distances and orientations), and lasted 200ns to ensure sufficient sampling and avoid potential bias.
DMD simulations
Our simulations were carried out by using the discrete molecular dynamics (DMD) algorithm, a special approach of molecular dynamics in which discrete step functions instead of continuous functions are used to mimic the constraints [34]. DMD has been widely used by our group and others in studying protein folding, protein aggregation, protein-nanoparticle interactions and protein-small molecule interactions [34, 61–63] In our simulations, the step function potentials are adapted from the continuous Medusa force field, with non-bounded inter-atomic interactions include van der Waals (VDW), solvation, hydrogen bond and electrostatic terms [63]. The VDW parameters were adopted from the CHARMM force field and the EEF1 implicit solvent model was used to model the solvation term [64, 65]. The hydrogen bond interactions were implicitly modeled with a reaction-like approach[66]. The Debye—Hückel approximation is applied to model screened electrostatic interactions, with the screening length set to 10Å. We also use Anderson’s thermostat to fix the simulation as 300K [67].
Analysis methods
We used the dictionary secondary structure of protein (DSSP) method in our analysis of the system secondary structure [68], and the cutoff of 3.5Å for inter-atomic distance to determine the formation of hydrogen bond. We also applied the same criteria in our previous works to determine the formation of β-sheet structure [32, 60]. If two peptides are connected by at least two backbone hydrogen bonds and each chain contains at least two consecutive residues adopting the β-strand structure, they are determined as forming a β-sheet. In addition, if two peptides are connected by at least one inter-molecular heavy atom contact (with the cutoff 5.5Å), they are determined belonging to the same oligomer, where the size of it is the number of peptide contained. We further defined the β-sheet oligomer as the multiple β-sheets connected by at least one inter-molecular heavy atom contact, and the size of the mass-weighted β-sheet was computed as
where nβ denotes the number of β-sheet and ni is the size of the ith β-sheet in a simulation structure. Calculation of the two-dimensional PMF (or effective free energy) was according to the equation
where KB is the Boltzmann constant, T corresponds to the simulation temperature 300K, and P(NH–bond, Nβ–layer) is the probability of finding a molecular system containing eight Aβ16-22 forming a structure with NH–bond main-chain hydrogen bond as well as Nβ–layer layer of β-sheet at the time, where Nβ–layer was estimated by dividing the size of β-sheet oligomer with the size of mass-weighted β-sheet.
Acknowledgments
The work is supported in part by NSF CAREER CBET-1553945 (Ding) and NIH MIRA R35GM119691 (Ding). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH and NSF.
Footnotes
Conflict of Interest
No conflict of interest is declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Xi W, Wei G. Atomic-level study of the effects of O4 molecules on the structural properties of protofibrillar Abeta trimer: beta-sheet stabilization, salt bridge protection, and binding mechanism. J Phys Chem B. 2015;119:2786–2794. doi: 10.1021/jp508122t. [DOI] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 4.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 6.Anguiano M, Nowak RJ, Lansbury PT. Protofibrillar islet amyloid polypeptide permeabilizes synthetic vesicles by a pore-like mechanism that may be relevant to type II diabetes. Biochemistry-Us. 2002;41:11338–11343. doi: 10.1021/bi020314u. [DOI] [PubMed] [Google Scholar]
- 7.Ma Z, Westermark GT, Sakagashira S, Sanke T, Gustavsson A, Sakamoto H, Engstrom U, Nanjo K, Westermark P. Enhanced in vitro production of amyloid-like fibrils from mutant (S20G) islet amyloid polypeptide. Amyloid. 2001;8:242–249. doi: 10.3109/13506120108993820. [DOI] [PubMed] [Google Scholar]
- 8.Mo YX, Lei JT, Sun YX, Zhang QW, Wei GH. Conformational Ensemble of hIAPP Dimer: Insight into the Molecular Mechanism by which a Green Tea Extract inhibits hIAPP Aggregation. Sci Rep-Uk. 2016;6 doi: 10.1038/srep33076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Tycko R. Solid-State NMR Studies of Amyloid Fibril Structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Abeta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic view of a toxic amyloid small oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson ME, Lesne SE. Soluble A ss oligomer production and toxicity. J Neurochem. 2012;120:125–139. doi: 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieschke J, Herbst M, Wiglenda T, Friedrich RP, Boeddrich A, Schiele F, Kleckers D, del Amo JML, Gruning BA, Wang QW, Schmidt MR, Lurz R, Anwyl R, Schnoegl S, Fandrich M, Frank RF, Reif B, Gunther S, Walsh DM, Wanker EE. Small-molecule conversion of toxic oligomers to nontoxic beta-sheet-rich amyloid fibrils. Nat Chem Biol. 2012;8:93–101. doi: 10.1038/nchembio.719. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Zheng XY, Krishnamoorthy J, Savelieff MG, Park HM, Brender JR, Kim JH, Derrick JS, Kochi A, Lee HJ, Kim C, Ramamoorthy A, Bowers MT, Lim MH. Rational Design of a Structural Framework with Potential Use to Develop Chemical Reagents That Target and Modulate Multiple Facets of Alzheimer’s Disease. Journal of the American Chemical Society. 2014;136:299–310. doi: 10.1021/ja409801p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindo SS, Mancino AM, Braymer JJ, Liu YH, Vivekanandan S, Ramamoorthy A, Lim MH. Small Molecule Modulators of Copper-Induced A beta Aggregation. Journal of the American Chemical Society. 2009;131:16663–16665. doi: 10.1021/ja907045h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi JS, Braymer JJ, Nanga RPR, Ramamoorthy A, Lim MH. Design of small molecules that target metal-A beta species and regulate metal-induced A beta aggregation and neurotoxicity. P Natl Acad Sci USA. 2010;107:21990–21995. doi: 10.1073/pnas.1006091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyung SJ, DeToma AS, Brender JR, Lee S, Vivekanandan S, Kochi A, Choi JS, Ramamoorthy A, Ruotolo BT, Lim MH. Insights into antiamyloidogenic properties of the green tea extract (−)-epigallocatechin-3-gallate toward metal-associated amyloid-beta species. P Natl Acad Sci USA. 2013;110:3743–3748. doi: 10.1073/pnas.1220326110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weibel ER, Taylor CR, Hoppeler H. The Concept of Symmorphosis - a Testable Hypothesis of Structure-Function Relationship. P Natl Acad Sci USA. 1991;88:10357–10361. doi: 10.1073/pnas.88.22.10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Do TD, LaPointe NE, Nelson R, Krotee P, Hayden EY, Ulrich B, Quan S, Feinstein SC, Teplow DB, Eisenberg D, Shea JE, Bowers MT. Amyloid beta-Protein C-Terminal Fragments: Formation of Cylindrins and beta-Barrels. J Am Chem Soc. 2016;138:549–557. doi: 10.1021/jacs.5b09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussinov R, Tsai CJ. Allostery without a conformational change? Revisiting the paradigm. Curr Opin Struc Biol. 2015;30:17–24. doi: 10.1016/j.sbi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Jang H, Ma B, Lal R, Nussinov R. Models of Toxic beta-Sheet Channels of Protegrin-1 Suggest a Common Subunit Organization Motif Shared with Toxic Alzheimer beta-Amyloid Ion Channels. Biophys J. 2008;95:4631–4642. doi: 10.1529/biophysj.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stckl MT, Zijlstra N, Subramaniam V. alpha-Synuclein Oligomers: an Amyloid Pore? Mol Neurobiol. 2013;47:613–621. doi: 10.1007/s12035-012-8331-4. [DOI] [PubMed] [Google Scholar]
- 24.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT. Neurodegenerative disease - Amyloid pores from pathogenic mutations. Nature. 2002;418:291–291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Ilitchev AI, Giammona MJ, Li F, Buratto SK, Bowers MT. Human Islet Amyloid Polypeptide Assembly: The Key Role of the 8–20 Fragment. Journal of Physical Chemistry B. 2016;120:11905–11911. doi: 10.1021/acs.jpcb.6b09475. [DOI] [PubMed] [Google Scholar]
- 26.Arispe N, Diaz JC, Simakova O. Abeta ion channels. Prospects for treating Alzheimer’s disease with Abeta channel blockers. Biochimica et biophysica acta. 2007;1768:1952–1965. doi: 10.1016/j.bbamem.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Lal R, Lin H, Quist AP. Amyloid beta ion channel: 3D structure and relevance to amyloid channel paradigm. Biochimica et biophysica acta. 2007;1768:1966–1975. doi: 10.1016/j.bbamem.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams TL, Serpell LC. Membrane and surface interactions of Alzheimer’s A beta peptide - insights into the mechanism of cytotoxicity. Febs J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 29.Xie LG, Luo Y, Wei GH. A beta(16-22) Peptides Can Assemble into Ordered beta-Barrels and Bilayer beta-Sheets, while Substitution of Phenylalanine 19 by Tryptophan Increases the Population of Disordered Aggregates. Journal of Physical Chemistry B. 2013;117:10149–10160. doi: 10.1021/jp405869a. [DOI] [PubMed] [Google Scholar]
- 30.De Simone A, Derreumaux P. Low molecular weight oligomers of amyloid peptides display beta-barrel conformations: A replica exchange molecular dynamics study in explicit solvent. J Chem Phys. 2010;132 doi: 10.1063/1.3385470. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Xi W, Hansmann UHE, Wei Y. Fibril-Barrel Transitions in Cylindrin Amyloids. J Chem Theory Comput. 2017 doi: 10.1021/acs.jctc.7b00383. [DOI] [PubMed] [Google Scholar]
- 32.Sun YX, Qian ZY, Guo C, Wei GH. Amphiphilic Peptides A(6)K and V6K Display Distinct Oligomeric Structures and Self-Assembly Dynamics: A Combined All-Atom and Coarse-Grained Simulation Study. Biomacromolecules. 2015;16:2940–2949. doi: 10.1021/acs.biomac.5b00850. [DOI] [PubMed] [Google Scholar]
- 33.Irback A, Mitternacht S. Spontaneous beta-barrel formation: An all-atom Monte Carlo study of A beta(16-22) oligomerization. Proteins. 2008;71:207–214. doi: 10.1002/prot.21682. [DOI] [PubMed] [Google Scholar]
- 34.Ding F, Tsao D, Nie HF, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun SJ, Urbanc B, Cruz L, Bitan G, Teplow D, Stanley HE. Role of electrostatic interactions in amyloid beta-protein (Abeta) oligomer formation: A discrete molecular dynamics study. Biophys J. 2007:195a–195a. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodie NI, Popov KI, Petrotchenko EV, Dokholyan NV, Borchers CH. Solving protein structures using short-distance cross-linking constraints as a guide for discrete molecular dynamics simulations. Sci Adv. 2017;3 doi: 10.1126/sciadv.1700479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing YT, Pilkington EH, Wang MY, Nowell CJ, Kakinen A, Sun YX, Wang B, Davis TP, Ding F, Ke PC. Lysophosphatidylcholine modulates the aggregation of human islet amyloid polypeptide. Phys Chem Chem Phys. 2017;19:30627–30635. doi: 10.1039/c7cp06670h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Wang B, Ge X, Ding F. Distinct oligomerization and fibrillization dynamics of amyloid core sequences of amyloid-beta and islet amyloid polypeptide. Phys Chem Chem Phys. 2017;19:28414–28423. doi: 10.1039/c7cp05695h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Favrin G, Irback A, Mohanty S. Oligomerization of amyloid A beta(16-22) peptides using hydrogen bonds and hydrophobicity forces (vol 87, pg 3657, 2004) Biophys J. 2005;89:754–754. doi: 10.1529/biophysj.104.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petty SA, Decatur SM. Experimental evidence for the reorganization of beta-strands within aggregates of the a beta(16-22) peptide. J Am Chem Soc. 2005;127:13488–13489. doi: 10.1021/ja054663y. [DOI] [PubMed] [Google Scholar]
- 41.Lu K, Jacob J, Thiyagarajan P, Conticello VP, Lynn DG. Exploiting amyloid fibril lamination for nanotube self-assembly. J Am Chem Soc. 2003;125:6391–6393. doi: 10.1021/ja0341642. [DOI] [PubMed] [Google Scholar]
- 42.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. A partially folded structure of amyloid-beta(1-40) in an aqueous environment. Biochem Bioph Res Co. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnamoorthy J, Brender JR, Vivekanandan S, Jahr N, Ramamoorthy A. Side-Chain Dynamics Reveals Transient Association of A beta(1-40) Monomers with Amyloid Fibers. Journal of Physical Chemistry B. 2012;116:13618–13623. doi: 10.1021/jp305279w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang HB, Zheng J, Lal R, Nussinov R. New structures help the modeling of toxic amyloid beta ion channels. Trends Biochem Sci. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Miller Y, Ma B, Nussinov R. Polymorphism in Alzheimer A beta Amyloid Organization Reflects Conformational Selection in a Rugged Energy Landscape. Chem Rev. 2010;110:4820–4838. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J Biol Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 47.Etcheberrigaray R, Ito E, Oka K, Tofelgrehl B, Gibson GE, Alkon DL. Potassium Channel Dysfunction in Fibroblasts Identifies Patients with Alzheimer-Disease. P Natl Acad Sci USA. 1993;90:8209–8213. doi: 10.1073/pnas.90.17.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss JH, Pike CJ, Cotman CW. Ca2+ Channel Blockers Attenuate Beta-Amyloid Peptide Toxicity to Cortical-Neurons in Culture. J Neurochem. 1994;62:372–375. doi: 10.1046/j.1471-4159.1994.62010372.x. [DOI] [PubMed] [Google Scholar]
- 49.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacManus A, Ramsden M, Murray M, Henderson Z, Pearson HA, Campbell VA. Enhancement of (45)Ca(2+) influx and voltage-dependent Ca(2+) channel activity by beta-amyloid-(1-40) in rat cortical synaptosomes and cultured cortical neurons. Modulation by the proinflammatory cytokine interleukin-1beta. J Biol Chem. 2000;275:4713–4718. doi: 10.1074/jbc.275.7.4713. [DOI] [PubMed] [Google Scholar]
- 51.Arispe N, Pollard HB, Rojas E. beta-Amyloid Ca(2+)-channel hypothesis for neuronal death in Alzheimer disease. Molecular and cellular biochemistry. 1994;140:119–125. doi: 10.1007/BF00926750. [DOI] [PubMed] [Google Scholar]
- 52.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 53.Lin H, Bhatia R, Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 54.Kagan BL, Hirakura Y, Azimov R, Azimova R, Lin MC. The channel hypothesis of Alzheimer’s disease: current status. Peptides. 2002;23:1311–1315. doi: 10.1016/s0196-9781(02)00067-0. [DOI] [PubMed] [Google Scholar]
- 55.Sciacca MFM, Kotler SA, Brender JR, Chen J, Lee DK, Ramamoorthy A. Two-Step Mechanism of Membrane Disruption by A beta through Membrane Fragmentation and Pore Formation. Biophys J. 2012;103:702–710. doi: 10.1016/j.bpj.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotler SA, Walsh P, Brender JR, Ramamoorthy A. Differences between amyloid-beta aggregation in solution and on the membrane: insights into elucidation of the mechanistic details of Alzheimer’s disease. Chemical Society reviews. 2014;43:6692–6700. doi: 10.1039/c3cs60431d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chimon S, Ishii Y. Capturing intermediate structures of Alzheimer’s beta-amyloid, Abeta(1-40), by solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:13472–13473. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
- 58.Yu X, Zheng J. Polymorphic structures of Alzheimer’s beta-amyloid globulomers. PloS one. 2011;6:e20575. doi: 10.1371/journal.pone.0020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang MZ, Ren BP, Chen H, Sun Y, Ma J, Jiang BB, Zheng J. Molecular Simulations of Amyloid Structures, Toxicity, and Inhibition. Isr J Chem. 2017;57:586–601. [Google Scholar]
- 60.Sun YX, Wang B, Ge XW, Ding F. Distinct oligomerization and fibrillization dynamics of amyloid core sequences of amyloid-beta and islet amyloid polypeptide. Phys Chem Chem Phys. 2017;19:28414–28423. doi: 10.1039/c7cp05695h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radic S, Davis TP, Ke PC, Ding F. Contrasting effects of nanoparticle-protein attraction on amyloid aggregation. RSC Adv. 2015;5:105498. doi: 10.1039/C5RA20182A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurzov EN, Wang B, Pilkington EH, Chen P, Kakinen A, Stanley WJ, Litwak SA, Hanssen EG, Davis TP, Ding F, Ke PC. Inhibition of hIAPP Amyloid Aggregation and Pancreatic beta-Cell Toxicity by OH-Terminated PAMAM Dendrimer. Small. 2016;12:1615–1626. doi: 10.1002/smll.201502317. [DOI] [PubMed] [Google Scholar]
- 63.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. Plos Comput Biol. 2006;2:725–733. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 65.Lazaridis T, Karplus M. Effective energy functions for protein structure prediction. Curr Opin Struc Biol. 2000;10:139–145. doi: 10.1016/s0959-440x(00)00063-4. [DOI] [PubMed] [Google Scholar]
- 66.Ding F, Borreguero JM, Buldyrey SV, Stanley HE, Dokholyan NV. Mechanism for the alpha-helix to beta-hairpin transition. Proteins-Structure Function and Genetics. 2003;53:220–228. doi: 10.1002/prot.10468. [DOI] [PubMed] [Google Scholar]
- 67.Andersen HC. Molecular-Dynamics Simulations at Constant Pressure and-or Temperature. J Chem Phys. 1980;72:2384–2393. [Google Scholar]
- 68.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins-Structure Function and Genetics. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]