Abstract
The presence or absence of minimal residual disease (MRD) in patients with multiple myeloma (MM) has emerged as a useful marker to determine the depth of remission. MRD negativity as an endpoint has been shown to be associated with improved progression-free survival in many studies. MRD detection is therefore part of numerous clinical trial protocols for MM. At the present time, two methodologies are most widely accepted for MRD detection: i) multi-color flow cytometry, and ii) next-generation sequencing (NGS) -based clonotype detection. While both of those methodologies enable accurate quantification of MRD in the bone marrow (BM), with sensitivity as low as 10−5 to 10−6, there are several limitations to these methods. First, these approaches reveal the presence or absence of MRD but provide limited molecular information about MM. More comprehensive characterization of MM cells at the MRD stage may identify molecular mechanisms of drug-resistance. Second, MRD detection in the BM is typically performed at one time point only, but more frequent detection may define the duration of the MRD status and thus refine its prognostic value. Third, less-invasive approaches that avoid the discomfort and risk associated with BM biopsy would be highly desirable, especially in elderly or frail patients. “Liquid biopsy” for the detection and characterization of circulating MM cells may address these issues. While MRD detection in the peripheral blood at the same sensitivity as in the BM may be challenging, the identification of patients who do not achieve MRD negativity might reduce the need for BM biopsies. Here, we give an overview of approaches that have been described to detect and characterize MM cells when they occur at very low frequencies in the peripheral blood or in the BM, emphasizing recently described NGS approaches for more comprehensive characterization of circulating MM cells.
Keywords: circulating multiple myeloma cells, MRD, clonal evolution, next generation sequencing
Rationale for assessment of circulating tumor cells in myeloma
Despite tremendous achievements following the introduction of proteasome inhibitors [1–3], immunomodulatory agents [4] and most recently daratumumab [5,6] and elotuzumab [7,8], multiple myeloma (MM) remains incurable. Novel agents have led to unprecedented depth of response, but residual drug-resistant MM cells lead to inevitable relapse in nearly all patients. Over the past decade, substantial progress has been made in the understanding of MM biology and constant clonal evolution and genetic heterogeneity have been proposed as key drivers for the development of drug-resistant disease [9,10]. The clonal composition of MM is not only diverse, but also highly dynamic and changes repeatedly over time [11]. Although some targetable defects are already known in MM [12,13] serial assessment of the disease may reveal many more mechanisms of drug resistance that can be exploited therapeutically. Serial assessment by BM biopsies is limited by sampling errors, as biopsies are routinely performed from a single anatomic site (iliac crest) only, but MM is heterogeneously spread throughout the entire BM, or extramedullary sites. This represents a particular challenge for extramedullary MM. From a clinician´s point of view, monitoring of the disease by repeated BM biopsies remains inaccessible for the vast majority of MM patients as the procedure is painful, inconvenient and is accompanied by risks. This poses a particular challenge to elderly and frail patients, who might benefit from accurate tracking of the disease to balance treatment efficacy and treatment-related toxicity. In younger patients constant monitoring of the disease may be essential to detect potentially targetable vulnerabilities before the disease burden has reached a critical “point of no return” at which patients are too frail to receive adequate treatment.
Facing the entirety of these social, clinical, diagnostic and basic biological facets, liquid biopsy has recently gained a broad interest to be adapted and standardized in MM. Various tools have been introduced with various objectives, including i) longitudinal quantification of disease burden, ii) monitoring of clonal evolution, ii) determining mechanisms of drug resistance and iv) monitoring progression from pre-malignant conditions to overt MM.
This review focuses on currently existing techniques that may be used to interrogate circulating MM cells (CMMCs) as a tool for longitudinal tracking of MM. Detection of residual CMMCs in patients with low disease burden remains challenging as it aims at the detection of very low levels of disease. In the BM, sensitivities of ≤1: 105 to ≤1:106 MM cells in normal cells are currently pursued [14]. An ideal diagnostic approach for monitoring MM would i) be non-invasive, ii) provide a quick readout and iii) provide enough information for detailed molecular profiling even at low disease burden. Methods for molecular profiling of residual MM cells in the blood are rapidly emerging and today´s isolation techniques range from immunophenotypic approaches such as high-speed multiparameter flow cytometry (MFC) to molecular procedures including real-time quantitative PCR (RQ-PCR), fluorescence-in-situ hybridization (FISH), next-generation sequencing (NGS) and approaches that combine several of these techniques [11,15–21].
Identification and enumeration of CMMCs by flow cytometry
MFC offers fast and conclusive read-outs of CMMCs [19,22,23]. Prior 4-colour MFC panels have gradually been expanded to panels with greater numbers of parameters. For example, an 8-color 2-tube EuroFlow panel has been demonstrated to detect residual MM in the BM at high sensitivity in a standardized manner [24]. Similar sensitivities were reported in a 10-color single-tube approach [25]. Immunophenotypic detection of residual MM cells from the peripheral blood is more challenging as the fraction of MM cells is typically lower than in the BM [19]. Microfluidic cell capture methods for plasma cell antigens such as CD138 have been used to increase the sensitivity of MFC to quantify CMMCs from the peripheral blood reaching capture efficiencies of ~40–70% and detecting 20–24 CD138+ cells/mL and 45–184 CD138+ cells/mL in MM patients in remission and with active disease, respectively [26].
In the context of residual disease detection, the presence of CMMCs detected by MFC represents a marker of unfavorable prognosis. A recent study on n=647 patients with previously treated MM reported a worse survival of 12 vs. 33 months in patients with relapsed disease and ≥100 CMMCs vs. <100 CMMCs per 150,000 gated peripheral blood mononuclear cells (PB-MNCs). Of note, CMMCs in patients at complete remission remained entirely absent in this study [27].
Applications of MFC may not only be limited to MRD. Recent analyses reported on the ability of MFC approaches to also capture CMMCs in smoldering multiple myeloma (SMM) and monoclonal gammopathy of undetermined significance (MGUS) [28,29], potentially depicting a prognostic biomarker for patients with aberrant plasma cells prior to developing symptomatic disease. Paiva and colleagues have recently utilized MFC to identify distinct subpopulations of CMMCs characterized by the downregulation of integrins (CD11a/CD11c/CD29/CD49d/CD49e), adhesion molecules (CD33/CD56/CD117/CD138), and lymphostimulatory molecules (CD28/CD38/CD81) [19]. Circadian fluctuation of CMMCs and negative correlation to chemotactic modulators (CXCL12, CXCR4) in this study was postulated to reflect BM egression of a quiescent subpopulation of plasma cells possibly driving MM dissemination throughout the body [19]. Despite its merits, MFC itself is limited by the small number of parameters for molecular characterization. Approaches to improve the information density obtained from CMMCs include combination of MFC with high-throughput DNA- and RNA-seq methods. Such approaches will also have to take into account the potential interference of diagnostic antibodies directed at the same epitope as therapeutic antibodies (daratumumab, elotuzumab), e.g. by utilizing antibodies prone to different epitopes on the same target molecule [30]. Mass cytometry has recently been developed as a variation of flow cytometry that detects heavy metal-ion tagged antibodies using time-offlight mass spectrometry. Such techniques allow for the detection of up to 38 simultaneous epitopes on a single cell [31].
CMMC detection and isolation by immunomagnetic methods and other approaches
Circulating tumor cells (CTCs) in non-hematologic malignancies have been described as a biomarker for monitoring cancer without the need for invasive diagnostic procedures. A large number of rare cell enrichment technologies has been described. Most of these assays aim to identify CTCs based on three different strategies: i) positive selection, ii) negative selection, and iii) selection-free isolation. Positive selection enriches for cells with CTC-like signatures not exhibited by other blood cell components, including physical (e.g. size, density, deformability, surface charge) and phenotypic properties (expression of EpCAM, cytokeratins) [32–36]. Negative selection selects for and then discards cell subsets that have physiological blood cell properties (e.g. expression of CD45, CD66b, CD34, CD11b, CD14) [37–39]. Selection-free approaches provide high-throughput techniques which do not require positive or negative selection for the detection of CTCs [40,41].
Isolation techniques range from high speed flow cytometric cell sorting to microfluidic cell isolation devices, including some approaches which may warrant further exploration in the analysis of CMMCs. CTC-iChip is an antigen-independent microfluidic technology which uses lateral displacement and magnetophoresis [34]. Another commercial platform provides imaging-based selection of immunofluorescently labeled CTCs that adhere to slides with proprietary coating [41]. Microfluidic devices employ curvilinear microchannels that allow for fast, label-free enrichment of CTCs. Other systems consist of a microfluidic device of microchannels and capture chambers for isolation of CTCs based on their size and deformability [39]. As most microfluidic devices do not require cell labeling, such approaches may be highly valuable for subsequent immunophenotypic analyses. While some of these technologies may be suitable for detection or isolation of CMMCs, they are dependent on distinct properties of MM cells and normal white blood cells, which may be subtle. The automated Cellsearch system represents the only FDA-approved rare cell enrichment platform for clinical use in solid cancer at the moment [37,38]. This platform combines immunomagnetic selection and surface marker expression to purify CTCs. MM patients with CMMC counts ≥100 at remission showed reduced survival as compared to patients with CMMC counts <100, suggesting the eligibility of such assays as prognostic tools in MM [20].
Quantification of CMMCs by PCR
Due to its sensitivity of ≤1:105, allele-specific oligonucleotide real-time quantitative PCR (ASO-RQ-PCR) has become a common tool to investigate MRD from the BM of MM patients [42]. Major improvements in isolating CMMCs have recently driven additional interest to adopt ASO-RQ-PCR for the detection of residual plasma cells in the peripheral blood. Novel approaches that utilize patient-specific primers designed from the complementarity-determining region 3 (CDR3) sequence of the immunoglobulin genes may detect MRD in >90% patients despite somatic hypermutations in MM [43]. In a prior analysis of MM patients, positive IgH-RQ-PCR of CMMC proved prognostically relevant and preceded clinical relapse after HD-ASCT [18]. In the same study however, MRD levels in the peripheral blood were 40-fold lower than in the BM, thereby again stressing the challenges to apply RQ-PCR to liquid biopsy samples in order to detect myeloma MRD [18].
Characterization of CMMCs by FISH
Interphase fluorescence in situ hybridization (FISH) is a technique widely used in routine diagnostics for multiple myeloma to detect cytogenetic aberrations by labeling DNA on chromosomes using a hybridizing complementary strand of nucleic acids [44]. Probes are designed to target common abnormalities in MM [45]. As CMMCs are rare, particularly in MRD, FISH techniques require immunomagnetic enrichment or commercially available cell enrichment platforms to allow for plasma cell isolation from the peripheral blood. Several approaches have recently provided evidence of the feasibility for FISH in MMMRD [20,46]. Despite being less standardized than MFC and NGS, FISH-based methods may unravel cytogenetic data while at the same time being less costly than NGS. Prior analyses of CMMCs and paired BM samples have demonstrated a high degree of concordance in cytogenetic aberrations including deletion del(13q14) [46]. Other investigators have utilized FISH to detect distinct cytogenetic aberrations in CMMCs as compared to paired BM and showed a higher clonogenic potency of CMMCs [19,47]. Future investigations are needed to determine the genetic relationship between CMMCs and spatially heterogeneous MM within the BM [48].
Characterization of CMMCs by NGS
NGS allows detailed molecular characterization of CMMCs with great resolution [17,21]. We recently developed an approach to combine CD138+CD45−cell enrichment by serial dilution and single-cell micromanipulation using fluorescence microscopy with DNA- and RNA-sequencing of single CMMCs [21,49]. This technology currently enables isolation of MM cells based on established marker profiles at a frequency of less than one cell per 106 [21]. Using this approach, we have identified CMMCs in 24/24 randomly selected patient samples, and successfully performed DNA-sequencing of single MM cells, isolated from the blood and BM. Our methodology reproduced all mutations that were previously identified by CLIA-certified genotyping from bulk BM, and was also able to identify canonical MM mutations when the CLIA test from BM failed. In some cases, particular mutations could only be detected in the blood but not in BM, demonstrating that DNA-sequencing of single CMMCs and single BM MM cells is feasible, and suggesting that CMMCs can provide genomic information distinct from BM MM cells. In another study, whole-exome sequencing with additional targeted sequencing of eight matched flow-sorted BM and CMMC bulk samples reported a similar level of concordance between clonal mutations in both MM compartments [17]. RNA-sequencing of single MM cells from blood and BM has also been employed and may allow to define subsets of MM by inferring the presence of chromosomal translocations that result in overexpression of key drivers of MM [21] as well as to establish lineage identity (Figure 1). The ability to comprehensively characterize single MM cells by DNA- and RNA- sequencing with a sensitivity of 10−6 or better offers the chance to obtain molecular information about MRD from BM or from the peripheral blood. This may reveal resistant disease before a clinically apparent relapse occurs, which may help to increase the efficacy of treatments and identify mechanisms of drug resistance, particularly with emerging immunological treatment approaches (Figure 2). Besides being much more convenient for the patient, molecular characterization of minimal disease from blood may allow tracking of the kinetics of MRD loss, which cannot be achieved by BM biopsies. However, the sensitivity of myeloma-derived CMMCs for detection of MRD and its genomic characterization has not been defined. Now, studies are needed to define the exact sensitivity of CMMCs for MRD detection and determine their prognostic and predictive impact.
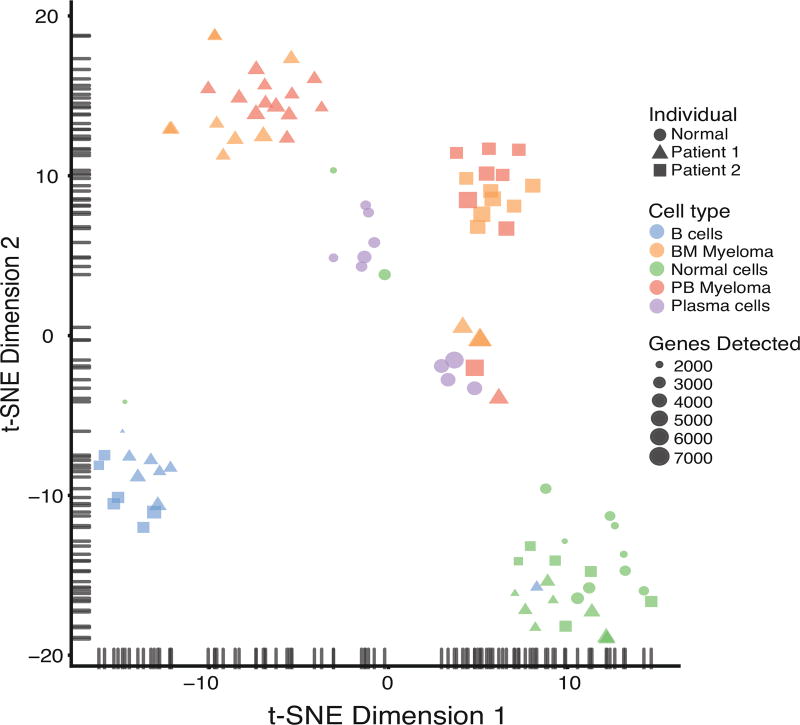
Figure 1. Distinguishing single CMMCs and BM MM cells from other cell types by single-cell RNA-sequencing.
Single CD138+CD45− MM cells were isolated either from the peripheral blood (PB Myeloma) or from the bone marrow (BM myeloma), and single CD19+ B lymphocytes (B cells) or CD45+ normal white blood cells (Normal cells) were isolated from the peripheral blood of two MM patients. CD138+CD45+ plasma cells (Plasma cells) were isolated from a healthy blood donor. Single cell whole transcriptome RNA sequencing was performed, and the gene expression of all single cells was visualized as two-dimensional t-SNE scatter plot. The symbol size represents the number of genes detected in each cell.
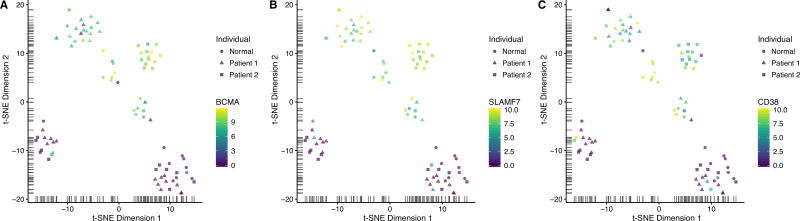
Figure 2. Expression of typical plasma cell markers in single CMMCs, BM MM cells and normal white blood cells.
Single cell whole transcriptome RNA sequencing was performed and visualized as t-SNE scatter plot as in Figure 1. The relative gene expression level of three different myeloma markers was color-coded, demonstrating high expression of typical plasma cell markers only in MM cells from two individual patients, and plasma cells from the blood of a healthy blood donor: (A) BCMA, (B) SLAMF7 and (C) CD38.
Complementary tools for minimal residual disease assessment
Several complementary approaches have been described with the potential to support the detection and characterization of MRD. Imaging techniques, such as whole-body 18F-FDG-PET/CT or contrast-enhanced MRI, may be particularly useful for detecting localized, extramedullary disease [50,51]. MM-derived cell-free DNA (cfDNA) in the blood has been described in MM, but its detection at MRD is limited by the total amount of cfDNA that is present in the blood, creating a challenge for achieving a detection sensitivity better than ~10−4 [16,52,53]. However, deep sequencing of cfDNA from plasma has recently been demonstrated to recapitulate mutational profiles of matched BM aspirates [16]. In this study, Kis and colleagues performed hybrid capture of all exons of KRAS, NRAS, BRAF, EGFR and PIK3CA in 64 cfDNA specimens from 53 myeloma patients and performed DNA-sequencing to >20,000× median coverage. This sequencing approach allowed for the detection of cfDNA-only mutations that were not identifiable in BM aspirates but persisted in serial cfDNA assessment of some patients, consistent with the clinical course of each patient [16]. In another study, mutation enrichment screening of cfDNA in relapsed/refractory (RR) MM patients detected MM key driver gene mutations that at the same time point remained undetectable in paired BM controls (27.2% of all RRMM cases). Interestingly, such peripheral blood-only mutations could only be detected in 6.6% of newly diagnosed MM patients. These observations point towards spatial and genetic heterogeneity over the course of myeloma disease that can be detected through querying cfDNA [52]. DNA-sequencing for clonotypic V(D)J rearrangements in cfDNA from 27 myeloma patients recapitulated the persistence of MM-cfDNA in 91% of non-responders vs. 41% of responders [53]. At the same time, M-protein persisted in the majority of the patients, suggesting that cfDNA provides information about tumor burden in MM patients after treatment that is distinct from M-protein.
Summary and future directions
Liquid biopsy approaches hold great promise for the tracking of MM in the blood. Utilizing CMMCs rather than BM MM cells to gain insight into MM biology may i) allow for frequent molecular analyses over time and may thus be crucial to capture genomic evolution, ii) more accurately reflect multifocal disease than a BM biopsy due to a more representative sampling, and iii) be substantially more convenient for patient and provider.
Several methodologies have been described that allow characterization of MM cells that are present at low frequencies, either in the blood or in the BM. MFC is well-established in clinical routine, but only provides information about a relatively small number of parameters. This allows for accurate quantification of CMMCs, but is typically insufficient for detailed molecular or functional characterization. Mass cytometry approaches may further increase the number of parameters that can be queried [31].
Highly sensitive technologies have been developed to isolate MM cells either from the blood or from the BM, including flow sorting, immunomagnetic enrichment, microscopy-based isolation approaches, or combination approaches [19,20,26,27,31,42,43,46,47,54]. Pairing single-cell or small input NGS with these technologies allows for unprecedented granularity of the obtained information [22,26]. This enables deep insight into the genome or transcriptome of MM from just a handful of cells and may have great utility for following the dynamics of clonal evolution and determining molecular mechanisms of drug resistance [21]. A sensitivity of 10−6 or better for the detection of MM cells is feasible with some of these methods, and they can be applied to BM and blood equivalently. This enables high content characterization of MRD in the BM, rather than being limited to simply quantification of MRD. While the frequency of MM cells in the circulation appears to be about two orders of magnitude lower than in the BM in many cases, larger volumes of blood may partially compensate for the difference in sensitivity. For instance, 40ml of blood can be obtained relatively easily. In contrast, obtaining the same volume of BM typically requires multiple individual biopsies, to avoid contamination with peripheral blood. Multiple BM biopsies are impractical, associated with greater risk and inconvenience, and are therefore rarely performed. More studies are needed to further define the sensitivity of CMMCs for MRD detection, and more importantly their prognostic impact on response, progression free survival and overall survival. The incorporation of comprehensive characterization of minimal disease either from peripheral blood or from BM into future clinical trials will be crucial for more comprehensive functional and molecular characterization of MM and will provide much deeper insight into MM biology than has been previously possible.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
All authors declare no relevant conflicts of interests.
References
- 1.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–6. [PubMed] [Google Scholar]
- 2.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–17. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 3.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352:2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 4.Singhal S, Mehta J, Desikan R, Ayers D, Roberson P, Eddlemon P, et al. Antitumor activity of thalidomide in refractory multiple myeloma. N Engl J Med. 1999;341:1565–71. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- 5.de Weers M, Tai Y-T, van der Veer MS, Bakker JM, Vink T, Jacobs DCH, et al. Daratumumab, a novel therapeutic human CD38 monoclonal antibody, induces killing of multiple myeloma and other hematological tumors. J Immunol Baltim Md 1950. 2011;186:1840–8. doi: 10.4049/jimmunol.1003032. [DOI] [PubMed] [Google Scholar]
- 6.Lokhorst HM, Plesner T, Laubach JP, Nahi H, Gimsing P, Hansson M, et al. Targeting CD38 with Daratumumab Monotherapy in Multiple Myeloma. N Engl J Med. 2015;373:1207–19. doi: 10.1056/NEJMoa1506348. [DOI] [PubMed] [Google Scholar]
- 7.Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:2775–84. doi: 10.1158/1078-0432.CCR-07-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lonial S, Dimopoulos M, Palumbo A, White D, Grosicki S, Spicka I, et al. Elotuzumab Therapy for Relapsed or Refractory Multiple Myeloma. N Engl J Med. 2015;373:621–31. doi: 10.1056/NEJMoa1505654. [DOI] [PubMed] [Google Scholar]
- 9.Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–76. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrulis M, Lehners N, Capper D, Penzel R, Heining C, Huellein J, et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov. 2013;3:862–9. doi: 10.1158/2159-8290.CD-13-0014. [DOI] [PubMed] [Google Scholar]
- 13.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J Clin Oncol Off J Am Soc Clin Oncol. 2015;33:3911–20. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson KC, Auclair D, Kelloff GJ, Sigman CC, Avet-Loiseau H, Farrell AT, et al. The Role of Minimal Residual Disease Testing in Myeloma Treatment Selection and Drug Development: Current Value and Future Applications. Clin Cancer Res Off J Am Assoc Cancer Res. 2017;23:3980–93. doi: 10.1158/1078-0432.CCR-16-2895. [DOI] [PubMed] [Google Scholar]
- 15.Gonsalves WI, Rajkumar SV, Dispenzieri A, Dingli D, Timm MM, Morice WG, et al. Quantification of circulating clonal plasma cells via multiparametric flow cytometry identifies patients with smoldering multiple myeloma at high risk of progression. Leukemia. 2017;31:130–5. doi: 10.1038/leu.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kis O, Kaedbey R, Chow S, Danesh A, Dowar M, Li T, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017;8:15086. doi: 10.1038/ncomms15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishima Y, Paiva B, Shi J, Park J, Manier S, Takagi S, et al. The Mutational Landscape of Circulating Tumor Cells in Multiple Myeloma. Cell Rep. 2017;19:218–24. doi: 10.1016/j.celrep.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korthals M, Sehnke N, Kronenwett R, Schroeder T, Strapatsas T, Kobbe G, et al. Molecular monitoring of minimal residual disease in the peripheral blood of patients with multiple myeloma. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transplant. 2013;19:1109–15. doi: 10.1016/j.bbmt.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 19.Paiva B, Paino T, Sayagues J-M, Garayoa M, San-Segundo L, Martín M, et al. Detailed characterization of multiple myeloma circulating tumor cells shows unique phenotypic, cytogenetic, functional, and circadian distribution profile. Blood. 2013;122:3591–8. doi: 10.1182/blood-2013-06-510453. [DOI] [PubMed] [Google Scholar]
- 20.Foulk B, Schaffer M, Gross S, Rao C, Smirnov D, Connelly MC, et al. Enumeration and characterization of circulating multiple myeloma cells in patients with plasma cell disorders. Br J Haematol. 2018;180:71–81. doi: 10.1111/bjh.15003. [DOI] [PubMed] [Google Scholar]
- 21.Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ, et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med. 2016;8:363ra147. doi: 10.1126/scitranslmed.aac7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paiva B, Cedena M-T, Puig N, Arana P, Vidriales M-B, Cordon L, et al. Minimal residual disease monitoring and immune profiling in multiple myeloma in elderly patients. Blood. 2016;127:3165–74. doi: 10.1182/blood-2016-03-705319. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Lopez J, Lahuerta JJ, Pepin F, González M, Barrio S, Ayala R, et al. Prognostic value of deep sequencing method for minimal residual disease detection in multiple myeloma. Blood. 2014;123:3073–9. doi: 10.1182/blood-2014-01-550020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores-Montero J, Sanoja-Flores L, Paiva B, Puig N, García-Sánchez O, Böttcher S, et al. Next Generation Flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia. 2017;31:2094–103. doi: 10.1038/leu.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roshal M, Flores-Montero JA, Gao Q, Koeber M, Wardrope J, Durie BGM, et al. MRD detection in multiple myeloma: comparison between MSKCC 10-color single-tube and EuroFlow 8-color 2-tube methods. Blood Adv. 2017;1:728–32. doi: 10.1182/bloodadvances.2016003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qasaimeh MA, Wu YC, Bose S, Menachery A, Talluri S, Gonzalez G, et al. Isolation of Circulating Plasma Cells in Multiple Myeloma Using CD138 Antibody-Based Capture in a Microfluidic Device. Sci Rep. 2017;7:45681. doi: 10.1038/srep45681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonsalves WI, Morice WG, Rajkumar V, Gupta V, Timm MM, Dispenzieri A, et al. Quantification of clonal circulating plasma cells in relapsed multiple myeloma. Br J Haematol. 2014;167:500–5. doi: 10.1111/bjh.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Rajkumar SV, Kyle RA, Lacy MQ, Dispenzieri A, Fonseca R, et al. Prognostic value of circulating plasma cells in monoclonal gammopathy of undetermined significance. J Clin Oncol Off J Am Soc Clin Oncol. 2005;23:5668–74. doi: 10.1200/JCO.2005.03.159. [DOI] [PubMed] [Google Scholar]
- 29.Bianchi G, Kyle RA, Larson DR, Witzig TE, Kumar S, Dispenzieri A, et al. High levels of peripheral blood circulating plasma cells as a specific risk factor for progression of smoldering multiple myeloma. Leukemia. 2013;27:680–5. doi: 10.1038/leu.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberle A, Brandt A, Alawi M, Langebrake C, Janjetovic S, Wolschke C, et al. Long-term CD38 saturation by daratumumab interferes with diagnostic myeloma cell detection. Haematologica. 2017;102:e368–70. doi: 10.3324/haematol.2017.169235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baughn LB, Sachs Z, Noble-Orcutt KE, Mitra A, Van Ness BG, Linden MA. Phenotypic and functional characterization of a bortezomib-resistant multiple myeloma cell line by flow and mass cytometry. Leuk Lymphoma. 2017;58:1931–40. doi: 10.1080/10428194.2016.1266621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park J-M, et al. SSA-MOA: a novel CTC isolation platform using selective size amplification (SSA) and a multi-obstacle architecture (MOA) filter. Lab Chip. 2012;12:2874–80. doi: 10.1039/c2lc40065k. [DOI] [PubMed] [Google Scholar]
- 33.Warkiani ME, Guan G, Luan KB, Lee WC, Bhagat AAS, Chaudhuri PK, et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip. 2014;14:128–37. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 34.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J-F, Ho H, Lichterman J, Lu Y-T, Zhang Y, Garcia MA, et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015;121:3240–51. doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Todenhöfer T, Hennenlotter J, Feyerabend S, Aufderklamm S, Mischinger J, Kühs U, et al. Preliminary experience on the use of the Adnatest® system for detection of circulating tumor cells in prostate cancer patients. Anticancer Res. 2012;32:3507–13. [PubMed] [Google Scholar]
- 37.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 38.Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 39.Gogoi P, Sepehri S, Zhou Y, Gorin MA, Paolillo C, Capoluongo E, et al. Development of an Automated and Sensitive Microfluidic Device for Capturing and Characterizing Circulating Tumor Cells (CTCs) from Clinical Blood Samples. PloS One. 2016;11:e0147400. doi: 10.1371/journal.pone.0147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campton DE, Ramirez AB, Nordberg JJ, Drovetto N, Clein AC, Varshavskaya P, et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer. 2015;15:360. doi: 10.1186/s12885-015-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SB, et al. Analytical Validation and Capabilities of the Epic CTC Platform: Enrichment-Free Circulating Tumour Cell Detection and Characterization. J Circ Biomark. 2015;4:3. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig N, Sarasquete ME, Balanzategui A, Martínez J, Paiva B, García H, et al. Critical evaluation of ASO RQ-PCR for minimal residual disease evaluation in multiple myeloma. A comparative analysis with flow cytometry. Leukemia. 2014;28:391–7. doi: 10.1038/leu.2013.217. [DOI] [PubMed] [Google Scholar]
- 43.Bai Y, Wong KY, Fung TK, Chim CS. High applicability of ASO-RQPCR for detection of minimal residual disease in multiple myeloma by entirely patient-specific primers/probes. J Hematol OncolJ Hematol Oncol. 2016;9:107. doi: 10.1186/s13045-016-0336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaughnessy J, Tian E, Sawyer J, Bumm K, Landes R, Badros A, et al. High incidence of chromosome 13 deletion in multiple myeloma detected by multiprobe interphase FISH. Blood. 2000;96:1505–11. [PubMed] [Google Scholar]
- 45.Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau J-L, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol Off J Am Soc Clin Oncol. 2009;27:4585–90. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamande JW, Lindell MAM, Witek MA, Voorhees PM, Soper SA. Isolation of circulating plasma cells from blood of patients diagnosed with clonal plasma cell disorders using cell selection microfluidics. Integr Biol Quant Biosci Nano Macro. 2018 doi: 10.1039/c7ib00183e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paíno T, Paiva B, Sayagués JM, Mota I, Carvalheiro T, Corchete LA, et al. Phenotypic identification of subclones in multiple myeloma with different chemoresistant, cytogenetic and clonogenic potential. Leukemia. 2015;29:1186–94. doi: 10.1038/leu.2014.321. [DOI] [PubMed] [Google Scholar]
- 48.Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8:268. doi: 10.1038/s41467-017-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–84. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavo M, Terpos E, Nanni C, Moreau P, Lentzsch S, Zweegman S, et al. Role of 18F-FDG PET/CT in the diagnosis and management of multiple myeloma and other plasma cell disorders: a consensus statement by the International Myeloma Working Group. Lancet Oncol. 2017;18:e206–17. doi: 10.1016/S1470-2045(17)30189-4. [DOI] [PubMed] [Google Scholar]
- 51.Hillengass J, Ritsch J, Merz M, Wagner B, Kunz C, Hielscher T, et al. Increased microcirculation detected by dynamic contrast-enhanced magnetic resonance imaging is of prognostic significance in asymptomatic myeloma. Br J Haematol. 2016;174:127–35. doi: 10.1111/bjh.14038. [DOI] [PubMed] [Google Scholar]
- 52.Mithraprabhu S, Khong T, Ramachandran M, Chow A, Klarica D, Mai L, et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia. 2017;31:1695–705. doi: 10.1038/leu.2016.366. [DOI] [PubMed] [Google Scholar]
- 53.Oberle A, Brandt A, Voigtlaender M, Thiele B, Radloff J, Schulenkorf A, et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica. 2017;102:1105–11. doi: 10.3324/haematol.2016.161414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowakowski GS, Witzig TE, Dingli D, Tracz MJ, Gertz MA, Lacy MQ, et al. Circulating plasma cells detected by flow cytometry as a predictor of survival in 302 patients with newly diagnosed multiple myeloma. Blood. 2005;106:2276–9. doi: 10.1182/blood-2005-05-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]