Abstract
BRCA1 deficiencies cause breast, ovarian, prostate and other cancers, and render tumours hypersensitive to PARP inhibitors. To understand resistance mechanisms, we conducted whole-genome CRISPR-Cas9 synthetic-viability/resistance screens in BRCA1-deficient breast cancer cells treated with PARP inhibitors. We identified two previously uncharacterized proteins, C20orf196 and FAM35A, whose inactivation confers strong PARP-inhibitor resistance. Mechanistically, we show C20orf196 and FAM35A form a complex, “Shieldin” (SHLD1/2), with FAM35A interacting with single-stranded DNA via its C-terminal OB fold region. We establish that Shieldin acts as the downstream effector of 53BP1/RIF1/MAD2L2 to promote DNA double-strand break (DSB) end-joining through restricting DSB resection and counteract homologous recombination by antagonising BRCA2/RAD51 loading in BRCA1-deficient cells. Notably, Shieldin inactivation further sensitises BRCA1-deficient cells to cisplatin, suggesting how defining the SHLD1/2 status of BRCA1-deficient tumours might aid patient stratification and yield new treatment opportunities. Highlighting this potential, we document reduced SHLD1/2 expression in human breast cancers displaying intrinsic or acquired PARP-inhibitor resistance.
Introduction
DNA double-strand breaks (DSBs) are highly cytotoxic cellular lesions that must be effectively and accurately repaired to maintain genome stability and prevent premature aging, neurodegeneration, immunodeficiency, cancer and other diseases1–3. In response to DSB detection, the apical kinases ATM, ATR and PRKDC (DNA-PKcs) become activated and phosphorylate numerous substrates to initiate the cellular DNA damage response (DDR)4. The ensuing cascade of molecular DDR events, which are promoted by various post-translational modifications including protein phosphorylation, ubiquitylation, sumoylation and poly (ADP-ribosyl)ation, impacts on a myriad of cellular components, amongst other things leading to assembly of DDR factors at DNA-damage sites, arrest or slowing of cell-cycle progression, and activation of DNA repair mechanisms4, 5. The two main types of DSB-repair pathway are non-homologous end-joining (NHEJ) which is active throughout the cell cycle, and homologous recombination (HR), which normally requires a sister chromatid as a template and hence only operates in S and G2 phases of the cell cycle. DSB-repair pathway choice is partly determined by functional antagonism between the HR-promoting factor BRCA1 and NHEJ-promoting proteins such as TP53BP1 (53BP1), RIF1 and MAD2L2 (REV7)6–13.
Inherited or acquired mutations in the BRCA1 or BRCA2 genes that result in protein loss or a mutant BRCA1/2 protein cause breast, ovarian, prostate and other cancers, and render tumours hypersensitive to PARP-inhibitor drugs such as olaparib14–17. Unfortunately, intrinsic or acquired PARP-inhibitor resistance frequently leads to lack-of-response or to patient relapse and tumour regrowth15, 18. In the clinic, the most common PARP-inhibitor resistance mechanisms reported to date are restoration of BRCA1/2 expression or function. Notably, 53BP1 expression is lost in various triple-negative breast cancers7, which may account for certain clinically relevant examples of PARP-inhibitor resistance. Nevertheless, the mechanisms driving PARP-inhibitor resistance in a large proportion of BRCA1/2-deficient tumours remain unexplained18, 19.
To systematically survey for genetic mechanisms of PARP-inhibitor resistance, we conducted whole-genome CRISPR-Cas9 synthetic-viability/resistance screens in human BRCA1-deficient breast cancer cells treated with PARP inhibitors. In addition to identifying known resistance factors such as 53BP1, RIF1 and MAD2L2 loss6–13, we identified two previously uncharacterized proteins, C20orf196 and FAM35A, whose inactivation confers PARP-inhibitor resistance to BRCA1-deficient cells. Our ensuing work lead us to define the “Shieldin” (SHLD1C20orf196/SHLD2FAM35A) complex that promotes NHEJ by serving as the downstream effector of 53BP1, RIF1 and MAD2L2, restricts DSB resection, and counteracts HR in BRCA1-deficient cells by antagonising replacement of replication protein A (RPA) with BRCA2 and RAD51 on resected single-stranded DNA (ssDNA). Finally, we report that SHLD1C20orf196/SHLD2FAM35A loss confers hypersensitivity to the DNA-crosslinking agent cisplatin, and that reduced SHLD1C20orf196 or SHLD2FAM35A expression is associated with evolution of PARP-inhibitor resistance in a patient-derived BRCA1-deficient breast cancer xenograft model and in BRCA1-mutant cancers displaying intrinsic PARP-inhibitor resistance.
Results
FAM35A or C20orf196 loss suppresses PARP-inhibitor sensitivity of BRCA1-mutant cells
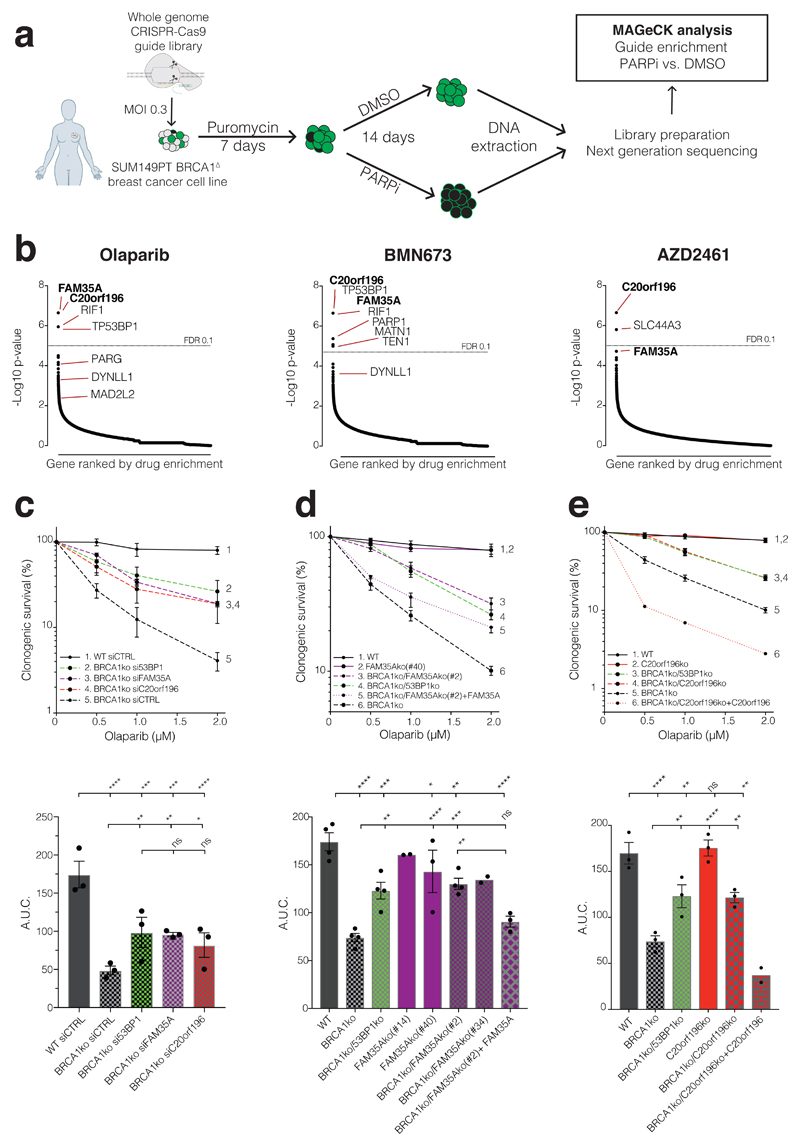
To systematically explore genetic mechanisms imparting PARP-inhibitor resistance, we carried out genome-wide CRISPR-Cas9 gene-inactivation screens with the GeCKO library20 in the BRCA1-mutant breast cancer cell line SUM149PT treated in parallel with the PARP inhibitors olaparib, talazoparib (BMN673) or AZD2461 (Fig 1a, b, Supplementary Fig 1a-c). In addition to identifying the known resistance genes TP53BP1, RIF1 and MAD2L2 whose products form a complex21, we identified several new suppressor candidates (Supplementary Table 1, Supplementary Fig 1d-e). These included DYNLL1, a known 53BP1 interaction partner22, and TEN1, a component of the CST telomere-capping complex (CTC1/STN1/TEN1) that also promotes telomere DNA replication23. In our ensuing studies, however, we focused on the uncharacterised proteins FAM35A and C20orf196 that collectively received the highest scores from our screens (Fig 1b and Supplementary Table 1). Thus, by carrying out short-interfering RNA (siRNA) mediated mRNA silencing in non-transformed, hTERT immortalized human RPE1 cells (Supplementary Fig 1f), we established that, as for 53BP1 loss7, depletion of FAM35A or C20orf196 markedly suppressed PARP-inhibitor sensitivity caused by BRCA1 inactivation while having no discernible effect in BRCA1-proficient cells (Fig 1c, Supplementary Fig 1g). This conclusion was independently confirmed by de novo CRISPR-Cas9 gene editing, with FAM35A or C20orf196 inactivation alleviating the olaparib hypersensitivity of BRCA1-deficient cells in a manner counteracted by re-introduction of wild-type FAM35A or C20orf196 (Fig 1d-e; Supplementary Fig 1h; as shown in supplementary Fig 1i, these effects did not reflect altered cell-cycle profiles).
Figure 1. CRISPR-Cas9 screens identify suppressors of PARP-inhibitor sensitivity in BRCA1-mutant cells.
a, Schematic of screen procedure. b, MAGeCK analysis of guide enrichments following specified drug treatments; false discovery rate (FDR) of 0.1 indicated by dotted line; n=3 technical replicates per drug treatment. c, siRNA mediated verification of hits in clonogenic survival assays; lower panels show area under the curve (AUC); n=3 independent experiments d, De novo Cas9 mediated knockout (ko) verification and complementation for FAM35A in clonogenic survival assays (multiple ko clones are shown in AUC); n=4 independent experiments except FAM35Ako(#14) (n=2), FAM35Ako(#40) (n=3), BRCA1ko/FAM35Ako(#34) (n=2), and BRCA1ko/FAM35Ako(#2) +FAM35A (n=3). e, As (d) but for C20orf196; n=3 independent experiments except BRCA1ko/C20orf196ko + C20orf196 (n=2). c-e Bars represent mean ± SEM, one-way Anova; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05). Individual data points are plotted over bars, and statistical source data including the precise p values can be found in Supplementary Table 5.
The FAM35A/C20orf196 complex interacts with and acts downstream of 53BP1/RIF1/MAD2L2
Sequence analyses indicated that FAM35A and C20orf196 are well conserved in vertebrates. Moreover, structure prediction modelling (RaptorX; http://raptorx.uchicago.edu/) revealed that FAM35A harbours a disordered N-terminus and an ordered C-terminal region containing three OB folds, with the last C-terminal OB fold/FAM domain containing a CXXC-type zinc finger motif (Fig 2a). Notably, this organization is highly similar to those of the RPA1 subunit of ssDNA binding protein RPA and the CTC1 subunit of the CST complex that also binds ssDNA23. In this regard, we noted that while the C20orf196 N-terminus (residues 1-70) is predicted to be intrinsically disordered, its C-terminal part is more structured and may harbour one- or two-winged helix (WH) domains (Fig 2a) similar to those in the yeast CST subunit Stn123, 24, suggesting that C20orf196 and Stn1 might play analogous or complementary roles.
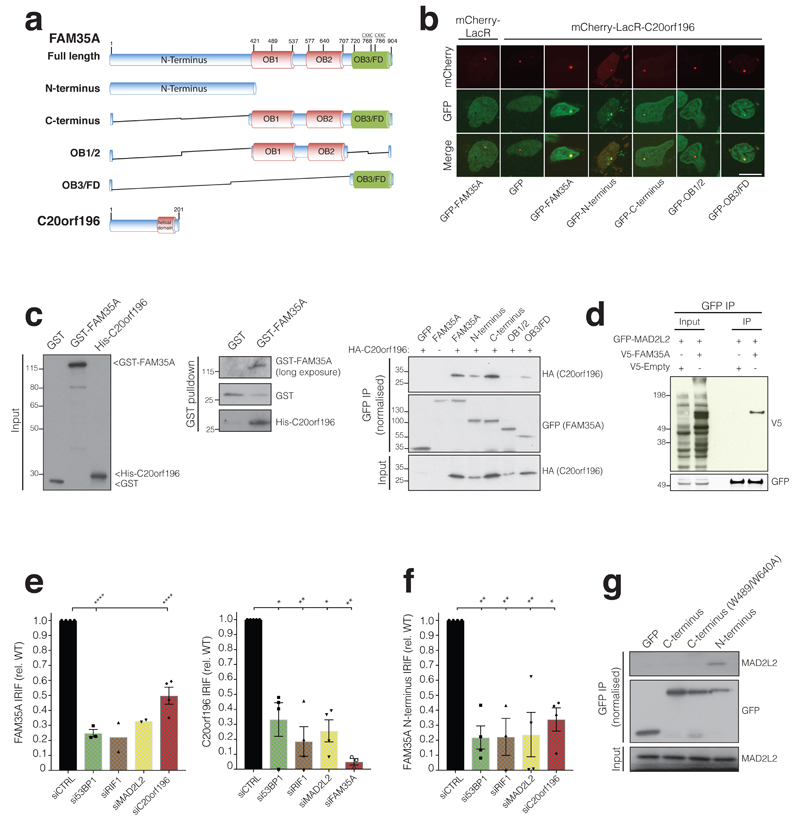
Figure 2. FAM35A and C20orf196 domains, interactions and IRIF formation.
a, FAM35A and C20orf196 predicted domains and variants used, OB fold (OB), FAM domain (OB3/FD). b, Recruitment of FAM35A/derivatives GFP-fusions to a chromosomal Lac-operator array via mCherry-LacR-C20orf196. Data shown represent 3 experiments with quantifications shown in Supplementary Fig 2a. Scale bar 10µm. c, (left and middle panel) Purified recombinant GST-FAM35A directly interacts with recombinant His-C20orf196. c, (right panel) Cell extracts expressing GFP-FAM35A/derivatives and HA-C20orf196 analysed by co-immunoprecipitation and immunoblotting. d, V5-FAM35A co-immunoprecipitates with GFP-MAD2L2; interaction with C20orf196 shown in Supplementary Fig 2c. e, Quantification of inducible GFP-FAM35A (left panel) and GFP-C20orf196 (right panel) IRIF in γH2AX positive cells 5 h after IR (5Gy) treated with indicated siRNAs. N=4 independent experiments except (left panel) si53BP1 (n=3), siRIF1 and siMAD2L2 (n=2); and (right panel) siCTRL(n=5), siRIF1(n=3), siFAM35A(n=3). f, As in (e) but for inducible GFP-FAM35A N-terminus; n=4 independent experiments except siRIF1 (n=3). g, Endogenous MAD2L2 co-immunoprecipitates with GFP-FAM35A N-terminus. e-f, Bars represent mean ± SEM, one-way Anova; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05); individual data points plotted over bars. Statistical source data including the precise p values are shown in Supplementary Table 5. All immunoblots are representative of two independent experiments; unprocessed scans of immunoblots are shown in Supplementary Fig 8.
Through combining cellular co-localization and co-immunoprecipitation experiments, we established that FAM35A and C20orf196 directly interact in a manner that is mainly, but not exclusively, mediated by the FAM35A OB3/FAM domain (Fig 2b-c, Supplementary Fig 2a-b). Because loss of FAM35A or C20orf196 had similar effects to loss of 53BP1/RIF1/MAD2L2 in BRCA1-deficient cells, we tested for possible interactions between these factors. Thus, via co-immunoprecipitation and mass spectrometry (MS) studies, we found that both C20orf196 and FAM35A interact with MAD2L2, the most distal factor of the 53BP1/RIF1/MAD2L2 axis mediating PARP-inhibitor sensitivity in BRCA1-deficient cells6–13 (Fig 2d, Supplementary Fig 2c).
Many DDR proteins accumulate at DSB sites within ionizing radiation (IR)-induced nuclear foci (IRIF)5. We established that both FAM35A and C20orf196 formed IRIF, and by live-cell imaging studies found that the proteins were also recruited to localised DNA-damage sites induced by laser micro-irradiation (Supplementary Fig 2d). Furthermore, we determined by confocal and super-resolution microscopy that FAM35A co-localised with the established DSB markers phosphorylated histone H2AFX25 (γH2AX) and 53BP126 (Fig 2e-f, Supplementary Fig 2e). Notably, siRNA/shRNA-depletion experiments established that while 53BP1 IRIF and MAD2L2 levels and IRIF were not significantly impaired by FAM35A or C20orf196 depletion (Supplementary Fig 2f-h), IRIF formation by FAM35A and C20orf196 required 53BP1, RIF1 and MAD2L2 but not PTIP (Fig 2e and Supplementary Fig 3a-c; note that total levels of GFP-tagged FAM35A/C20orf196 were minimally affected by 53BP1/RIF1/MAD2L2 depletion). We also established that C20orf196 IRIF were almost totally abrogated by FAM35A depletion, while C20orf196 depletion reduced but did not abolish FAM35A IRIF (Fig 2e). In addition, FAM35A formed nuclear foci when cells were treated with the DNA topoisomerase I inhibitor camptothecin (CPT; Supplementary Fig 3d). Significantly, the FAM35A N-terminus was necessary and sufficient for its IRIF formation, these IRIF depended on 53BP1, RIF1, MAD2L2 and C20orf196, and this region could be co-immunoprecipitated with MAD2L2 (Fig 2f-g, Supplementary Fig 3e-f). Collectively, these findings indicated that FAM35A and C20orf196 act as downstream components of the 53BP1/RIF1/MAD2L2 molecular assembly21 at DSB sites.
FAM35A and C20orf196 promote NHEJ
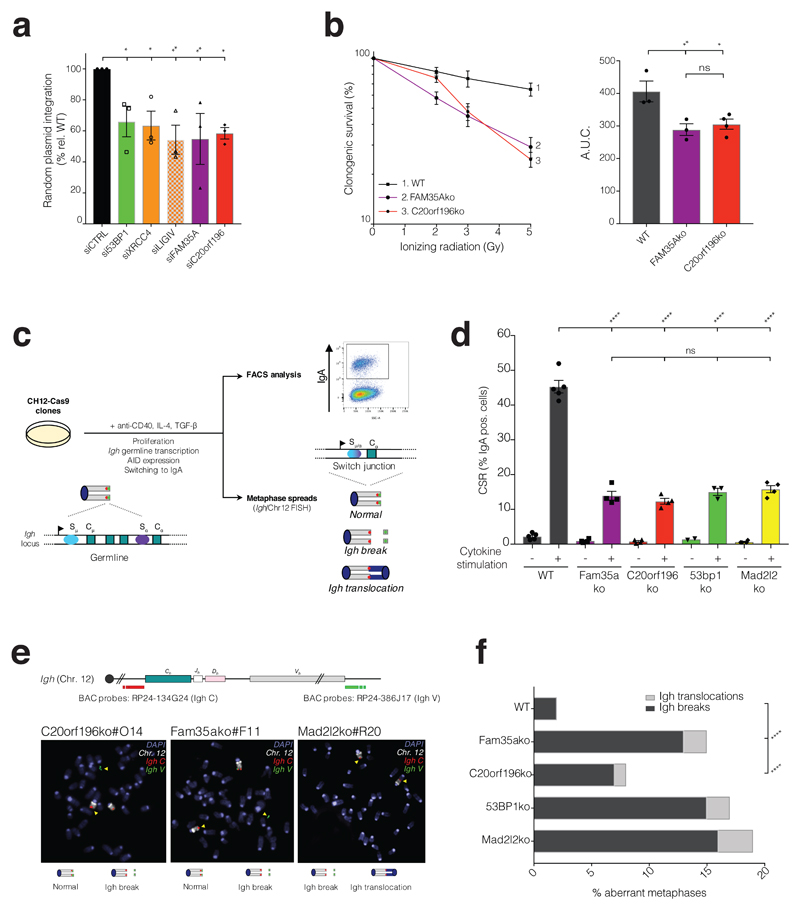
Since 53BP1, RIF1 and MAD2L2 promote NHEJ6–13, we tested whether FAM35A and C20orf196 fulfil a similar role. Indeed, as for depletion of the NHEJ factor XRCC4, siRNA depletion of 53BP1, FAM35A or C20orf196 impaired NHEJ as measured by random integration of plasmid DNA into chromosomes27 (Fig 3a). In addition, FAM35A or C20orf196 depletion conferred IR hypersensitivity to both human and mouse cells (Fig 3b and Supplementary Fig 4a). 53BP1 and associated factors promote NHEJ-mediated class-switch recombination (CSR) at the immunoglobulin heavy-chain locus, a process that allows B-lymphocytes to change antibody production from one type to another28. By CRISPR-Cas9 gene editing in mouse CH12F3 (CH12) B-lymphocytes29 we established that, as for 53BP1/RIF1/MAD2L2 inactivation6, 8, 10–12, 30, 31, loss of FAM35A or C20orf196 markedly reduced CSR (Fig 3c-d and Supplementary Fig 4b-d). Furthermore, analysis of metaphase chromosomal spreads of such cells revealed that FAM35A or C20orf196 inactivation led to chromosomal breaks and translocations symptomatic of aberrant CSR32 (Fig 3e-f; note from Supplementary Fig 4e-f that CSR effects were not associated with defects in cell proliferation, or in Aid or germ-line Sα switch region transcription).
Figure 3. FAM35A and C20orf196 promote NHEJ and immunoglobulin class-switch recombination.
a, Random plasmid integration assay. b, FAM35Ako and C20orf196ko cells were treated with IR and analysed for clonogenic survival, right panel shows AUC. a-b, Bars represent mean ± SEM, one-way Anova; n=3 independent experiments, except C20orf196ko in b (n=4), with individual data points plotted over bars; statistical source data can be found in Supplementary Table 5. c, Schematic representation of class-switch recombination and chromosomal instability in murine IgM+ B cells (germline configuration with Cμ transcription) induced to express AID and undergo CSR to IgA (switch configuration with Cα transcription) upon addition of anti-CD40, IL4 and TGF-β. CSR levels are measured as the % of IgA positive cells after 72 h cytokine stimulation, and DNA fluorescence in situ hybridization (FISH) is performed using a chromosome 12-specific paint (grey chromosome) and Igh locus specific probes (red and green spots) for the measurement of chromosomal instability at the Igh locus upon induction of CSR. d, CSR levels in Fam35Ako and C20orf196ko CH12-Cas9 cells are reduced compared with wild-type (WT) CH12-Cas9 cells. Bars represent mean ± SEM, one-way Anova. N=4 independent experiments of 3 clones except 53BP1ko +cytokine where n=3 of 2 clones, and 53BP1ko -cytokine where n=2 of 2 clones; with individual data points plotted over bars. e, Representative images of Igh translocation and breaks in aberrant metaphases, quantified in f. f, Quantification of Igh breaks and translocations in metaphases of the indicated CH12-Cas9 cells. Horizontal bars represent means, Fisher’s Exact test; n=2 independent experiments except Fam35ako and C20orf196ko where n=3. For a, b, d and f, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05); statistical source data including the precise p values for these panels can be found in Supplementary Table 5.
FAM35A and C20orf196 antagonise DNA-end resection
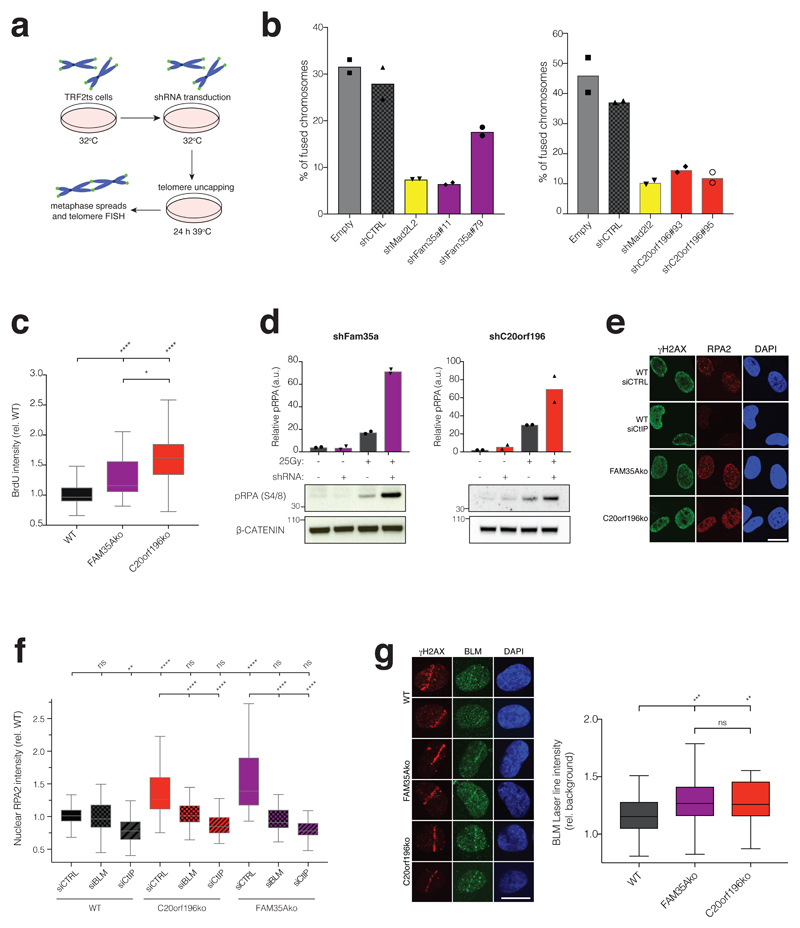
To explore FAM35A and C20orf196 function further, we carried out assays in mouse cells harbouring a temperature-sensitive allele of the telomere-associated factor TRF2 (TRF2ts). TRF2ts inactivation at higher temperatures results in de-protected chromosome ends and causes NHEJ-mediated telomere fusions (Fig 4a)6, 33. Strikingly, through use of short-hairpin RNA (shRNA) mediated mRNA silencing, we found that such chromosome fusions were diminished by FAM35A or C20orf196 depletion like that elicited by MAD2L2 depletion (Fig 4b, Supplementary Fig 5a-b).
Figure 4. FAM35A and C20orf196 promote telomere-mediated fusions and limit DNA-end resection.
a, Schematic of TRF2ts experimental setup. b, shRNA depletion of FAM35A (left panel) or C20orf196 (right panel) reduces un-capped telomere-mediated chromosome fusions. Bars represent means. The experiments were performed twice with ≥1300 chromosomes counted per condition, and individual data points plotted over bars; source data can be found in Supplementary Table 5. c, FAM35Ako and C20orf196ko RPE1 cells labelled with BrdU (10μM) for 48 h then treated with 1µM camptothecin (CPT) for 1 h, pre-extracted, fixed and stained for BrdU under non-denaturing conditions to visualise ssDNA. Box and whisker plot with centre line at median, box limits at 25th/75th centiles and whiskers ±1.5xIQR; one-way Anova; n=3 independent experiments. d, IR-induced pRPA(S4/8) is enhanced in MEFs due to Fam35a or C20orf196 silencing. Bars represent means. The experiments were performed twice with individual data points plotted over bars; source data can be found in Supplementary Table 5. e, RPE1-FAM35Ako or -C20orf196ko cells display hyper DNA-end resection (cells treated with 1μM camptothecin for 1h). Representative images from 3 independent experiments. Scale bar 10µm. f, RPE1-FAM35Ako or -C20orf196ko cells display BLM and CtIP dependent markers of excessive DNA-end resection. Box and whisker plot with centre line at median, box limits at 25th/75th centiles and whiskers ±1.5xIQR; one-way Anova; n=3 independent experiments. g, Enhanced BLM accrual in FAM35Ako and C20orf196ko compared with wild-type (WT) RPE1 cells fixed and stained 2 h after laser micro-irradiation. Representative images shown in left panel and quantification in right panel. Scale bar 10µm. Box and whisker plot with centre line at median, box limits at 25th/75th centiles and whiskers ±1.5xIQR; one-way Anova; n=3 independent experiments. For c, f and g, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05); statistical source data including the precise p values can be found in Supplementary Table 5.
The impacts of 53BP1, RIF1 or MAD2L2 depletion in the TRF2ts system are connected to these factors counteracting DSB resection6–13, 34. We thus explored whether FAM35A and C20orf196 might also have this function. Indeed, as for 53BP1/RIF1/MAD2L2 inactivation6–13, 35, loss of FAM35A or C20orf196 in human cells enhanced DSB-resection as measured by RPA and ssDNA staining intensity in pre-extracted nuclei after treatment with camptothecin (Fig 4c-e; RPA1 kinetics at DNA damage sites induced by laser micro-irradiation were not altered by FAM35A or C20orf196 depletion, Supplementary Fig 5c). In line with this resection being mediated by canonical pathways, it was diminished by depletion of the resection promoting factors RBBP8 (CtIP) or BLM (Fig 4f). Furthermore, we established that recruitment of BLM to sites of laser micro-irradiation was enhanced by FAM35A or C20orf196 depletion (Fig 4g). Similarly, as shown for 53BP1 depletion in mouse cells6, FAM35A or C20orf196 depletion in such cells led to higher levels of the DNA-end resection marker, Ser4/8 phosphorylated RPA2, after IR treatment (Fig 4d). Together with our other findings, these data established FAM35A and C20orf196 as crucial components of 53BP1/RIF1/MAD2L2-mediated chromosomal NHEJ, and suggested that their pro-NHEJ function is connected to limiting DSB resection.
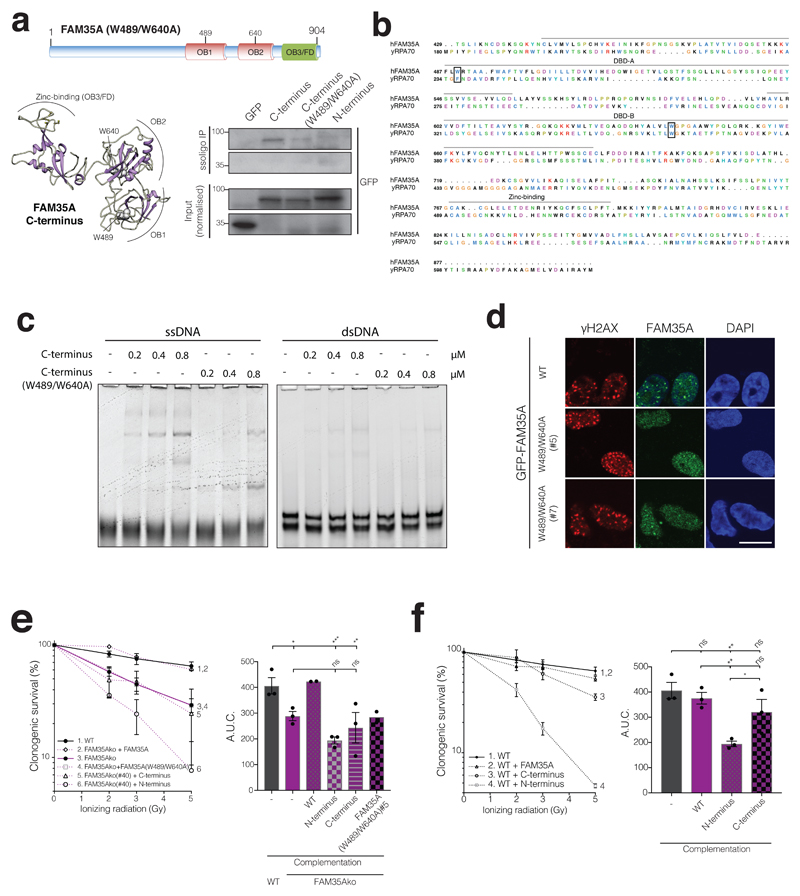
The FAM35A OB fold region interacts with ssDNA and promotes IR survival
Consistent with our prediction of structural similarity between FAM35A and RPA1, the FAM35A C-terminus could be retrieved from cell extracts via interaction with a ssDNA oligonucleotide (Fig 5a). Sequence alignment to RPA1 and structural modelling of FAM35A identified two Trp (W) residues predicted to be at the protein-ssDNA interface, based on analogous residues critical for RPA binding to ssDNA (Fig 5b, Supplementary Fig 5d). In accord with this prediction, we found via electrophoretic gel-mobility shift assays (EMSAs) that the bacterially-expressed, purified FAM35A C-terminal region bound preferentially to ssDNA rather than double-stranded DNA (Fig 5c, Supplementary Fig 5e), and ssDNA binding was reduced when the two Trp residues were mutated to Ala (W489/W640A; Fig 5c). Furthermore, while full-length FAM35A bearing these mutations (FAM35AW489/W640A) still interacted with C20orf196 (Supplementary Fig 5f) and formed IRIF in cells, these IRIF were consistently less pronounced than those of the wild-type FAM35A protein (Fig 5d). This suggested that following IRIF recruitment via its N-terminal region, the FAM35A C-terminal ssDNA binding region may allow further FAM35A recruitment, retention and/or stabilization. In addition, unlike the wild-type protein, FAM35AW489/W640A did not confer significant IR resistance when reintroduced into FAM35A null cells (Fig 5e). In parallel studies, expression of the FAM35A C-terminus did not complement the IR hypersensitivity of FAM35A null cells. Moreover, expression of the FAM35A N-terminus rendered cells IR hypersensitive irrespective of whether they expressed endogenous FAM35A, implying that the N-terminal IRIF-forming domain of FAM35A may have a dominant-negative effect on NHEJ (Fig 5f; overexpression of these FAM35A derivatives did not affect olaparib sensitivity in a wild-type background, Supplementary Fig 5g).
Figure 5. FAM35A OB folds mediate ssDNA interaction and is required for IR resistance.
a, Schematic of FAM35A with residues W489/W640 mutated to A (top panel). Predicted 3D structure of wild-type FAM35A with W489 and W640 positions (lower left panel). FAM35A W489/W640 promote efficient ssDNA binding in cellular extracts (lower right panel). b, Alignment of yRPA1 with FAM35A C-terminus; amino acids critical for yRPA1 ssDNA binding and the corresponding amino acid residues in FAM35A are boxed. c, EMSAs on native (non-denaturing) gels with 10nM ssDNA or dsDNA, and the indicated purified, bacterially expressed FAM35A C-terminus or W489/W640A mutant in µM. d, Inducible GFP-FAM35A W489/W640A fails to efficiently form IRIF (12 h after 5Gy of IR). Scale bar 10µm. Representative images from 3 independent experiments. e, FAM35Ako RPE1 cells complemented with FAM35A derivatives in clonogenic survival assays; right panel shows AUC. f, Overexpression of FAM35A N-terminus but not C-terminus or full-length FAM35A sensitises wild-type cells to IR in clonogenic assays; right panel shows AUC. e-f, Bars represent mean ± SEM, one-way Anova; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05); n=3 independent experiments except group 2 and 4 in e (n=2), with individual data points plotted over bars; statistical source data including the precise p values can be found in Supplementary Table 5. All immunoblots are representative of two independent experiments; unprocessed scans of immunoblots are shown in Supplementary Fig 8.
FAM35A and C20orf196 antagonise HR in BRCA1-deficient cells
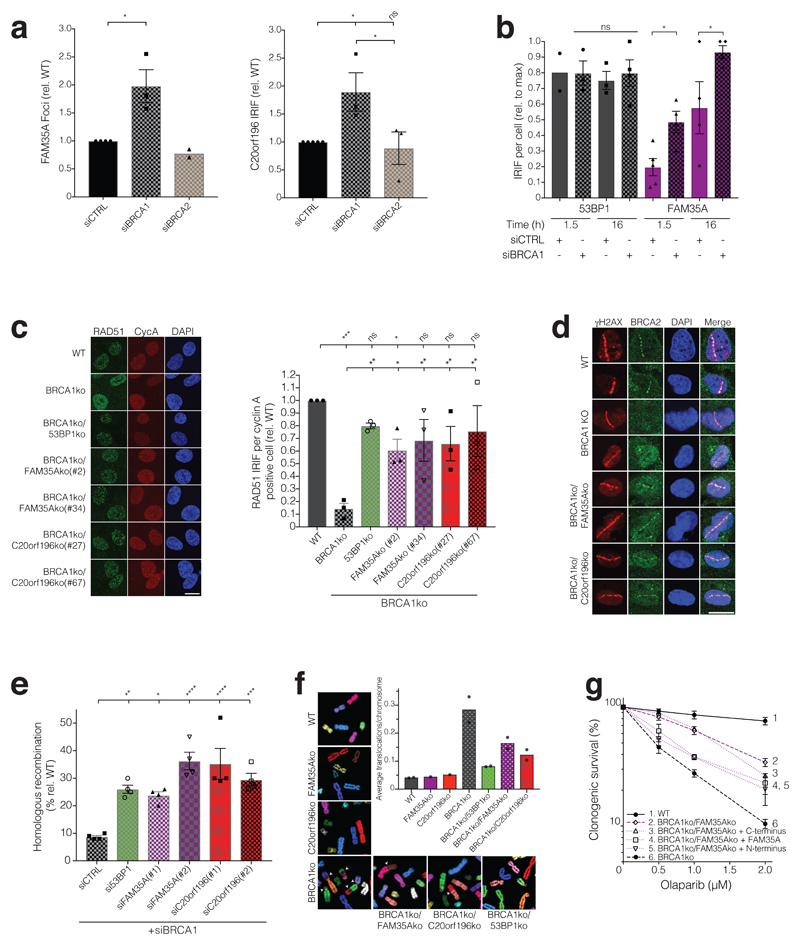
PARP inhibitors generate replication-associated DNA lesions that require BRCA1-mediated HR for their effective repair19, and loss of 53BP1/RIF1/MAD2L2 partly restores the ability of BRCA1-deficient cells to repair such lesions6–13. This has led to a model in which BRCA1 and 53BP1/RIF1/MAD2L2 play antagonistic roles in channelling DSBs towards HR or NHEJ, respectively. We thus speculated that BRCA1 might antagonise FAM35A/C20orf196 action. Accordingly, both FAM35A and C20orf196 IRIF, but not 53BP1 IRIF, were significantly enhanced in number and intensity upon BRCA1 but not BRCA2 depletion (Fig 6a-b, Supplementary Fig 6a-b).
Figure 6. FAM35A or C20orf196 loss restores HR in BRCA1-deficient cells.
a, Quantification of GFP-FAM35A (left panel) and GFP-C20orf196 (right panel) IRIF in U2OS cells after BRCA1 or BRCA2 depletion (5 h after 5Gy). Bars represent mean ± SEM, one-way Anova; n=3 independent experiments, except FAM35A siCTRL (n=4), FAM35A siBRCA2 (n=2), and C20orf196 siCTRL (n=5); with individual data points plotted over bars. b, Quantification of 53BP1 and inducible GFP-FAM35A IRIF in U2OS cells with or without BRCA1 depletion (5Gy, indicated time points). Bars represent mean ± SEM, one-way Anova; n=4 independent experiments, except 53BP1 1.5h siCTRL (n=2), 53BP1 1.5h siBRCA1 and 53BP1 16h siCTRL (n=3), FAM35A 1.5h siCTRL (n=5); with individual data points plotted over bars. c, Representative images (left panel) and quantification (right panel) of RAD51 IRIF (5.5 h after 5Gy) in Cyclin A (CycA) positive RPE1ko cell lines as indicated. Bars represent mean ± SEM, one-way Anova; n=3 independent experiments, with individual data points plotted over bars. Scale bar 10µm. d, FAM35A/C20orf196 loss restore BRCA2 recruitment 2 h after laser-induced DNA-damage sites in BRCA1-null cells (for quantification see Supplementary Fig 6e). Scale bar 10µm. e, HR assay in U2OS-TLR cells treated with indicated siRNAs (for gating strategy see Supplementary Fig 6f). Bars represent mean ± SEM, one-way Anova; n=4 independent experiments, with individual data points plotted over bars. f, Formation of spontaneous chromosomal aberrations in BRCA1ko cells is alleviated by FAM35A/C20orf196 inactivation. Representative images of metaphase spreads shown, and quantified in graph; bars represent means, n=2 independent experiments except FAM35Ako and C20orf196ko (n=1), with individual data points plotted over bars. g, Olaparib clonogenic survival assay with indicated RPE1ko and complemented cell lines. Bars represent mean ± SEM, one-way Anova; n=4 independent experiments, except group 4 and 5 (n=3) and group 3 (n=2); AUC is shown in Supplementary Fig 6g. For a-c and e, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05); statistical source data including the precise p values can be found in Supplementary Table 5.
Collectively, our results suggested that FAM35A/C20orf196 act at the interface between the opposing functions of BRCA1 and 53BP1/RIF1/MAD2L2 to regulate DSB-repair pathway choice. While this action could operate at least in part through control of DSB resection, which is misregulated and of slower kinetics in BRCA1-deficient cells11, 36, we reasoned that FAM35A/C20orf196 might also contribute to the severe defect in BRCA2-mediated RAD51 loading at DNA-damage sites in BRCA1-deficient cells11, 35, 37. Indeed, as for 53BP1 inactivation, loss of FAM35A or C20orf196 restored RAD51 IRIF formation in BRCA1-null cells (Fig 6c). While exploring the mechanism for this effect, we found elevated resection levels in FAM35A and C20orf196 knockout cells, as measured by RPA recruitment at DNA-damage sites, were still maintained in the absence of BRCA1 (Supplementary Fig 6c-d). Furthermore, FAM35A/C20orf196 depletion also alleviated the impaired recruitment of BRCA2 to DNA-damage sites in BRCA1-deficient cells (Fig 6d, Supplementary Fig 6e). Accordingly, studies with a cell-based chromosomal traffic light reporter (TLR) HR system 38, 39 established that FAM35A or C20orf196 depletion in BRCA1-deficient cells restored HR to levels similar to those acquired upon 53BP1 depletion in this setting (Fig 6e, Supplementary 6f). In addition, removing FAM35A or C20orf196 rescued the spontaneous genomic instability phenotype of BRCA1 knockout cells (Fig 6f). Building on our findings that the FAM35A N-terminal region largely mediates its localisation to IRIF (Fig 2f, Supplementary Fig 3e-f), introducing the FAM35A N-terminus, but not the C-terminus, enhanced PARP-inhibitor sensitivity of BRCA1/FAM35A null cells (Fig 6g, Supplementary Fig 6g). Furthermore, FAM35A inactivation was epistatic to 53BP1 inactivation in relation to conferring PARP-inhibitor resistance in BRCA1 knockout cells (Supplementary Fig 6h). Considering our findings together, we propose that C20orf196 and FAM35A be named SHLD1 and SHLD2, respectively, or collectively as the “Shieldin complex” because it shields DSBs from inappropriate activities and promotes appropriate modes of DSB repair.
FAM35A/C20orf196 loss correlates with PARP inhibitor resistance in cancers
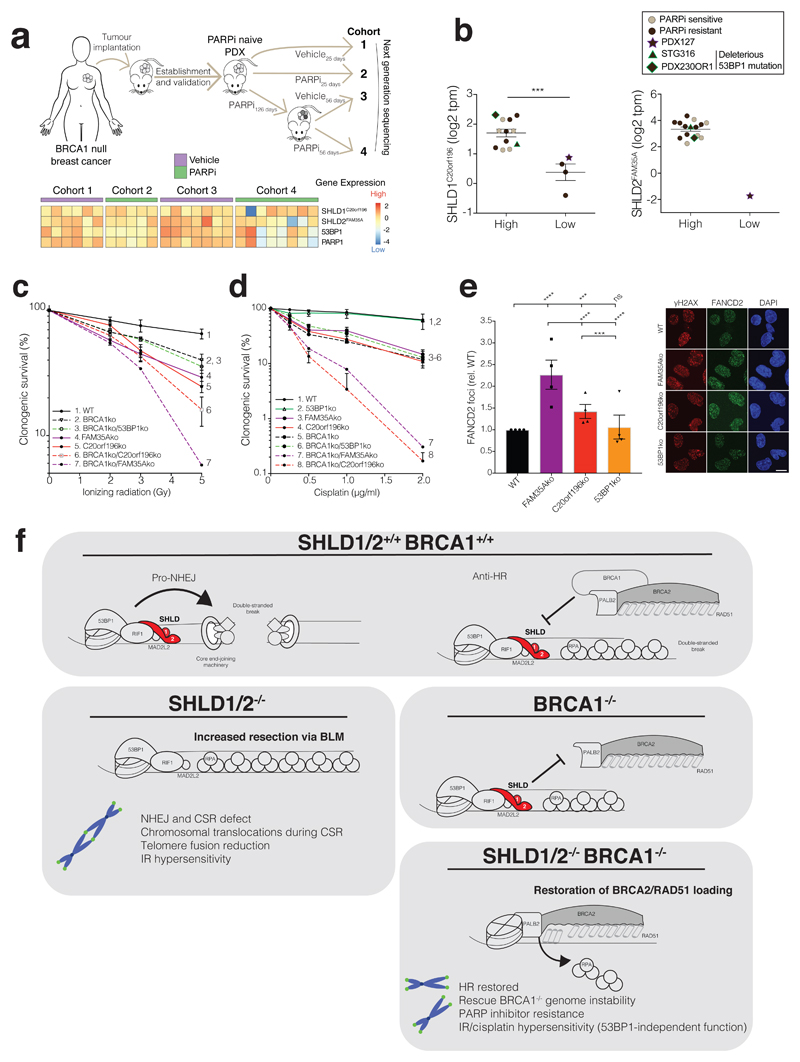
Having identified SHLD1C20orf196 and SHLD2FAM35A as mediating the PARP-inhibitor sensitivity of a BRCA1-deficient breast cancer cell line, we speculated that this might also apply in more physiological settings. Consequently, we employed a patient-derived xenograft (PDX) model of BRCA1-deficient breast cancer propagated in mice in the presence (cohort 2) or absence (cohort 1) of olaparib (Fig 7a). The resistant tumour was further serially passaged into new hosts that were treated in the presence (cohort 4) or absence (cohort 3) of olaparib to confirm and sustain drug resistance (Fig 7a; see also Supplementary Fig 7a). The tumours were then harvested and subjected to whole-genome RNA-sequencing. Notably, our analyses revealed that in contrast to the other cohorts, nearly all resistant tumours after chronic olaparib treatment (cohort 4) correlated with reduced mRNA expression of SHLD1C20orf196, SHLD2FAM35A, 53BP1 and/or PARP1 (Fig 7a; each heatmap column represents one tumour/mouse sample). Because this tumour model is polyclonal40, our data suggested that olaparib resistance mechanisms might arise through parallel evolutionary trajectories converging on loss of Shieldin activity. Furthermore, when we stratified a cohort of BRCA1-deficient PDX tumours by SHLD1/2 expression, ensuing analyses indicated that low SHLD1C20orf196 transcript levels correlated with intrinsic olaparib resistance (Fig 7b). One of the olaparib resistant models (PDX127) demonstrated concomitant loss of both SHLD1C20orf196 and SHLD2FAM35A, while two other resistant models exhibiting normal SHLD1/2 transcript levels harboured deleterious 53BP1 mutations. Notably, several of these resistant PDX models also display BRCA1 nuclear foci41, suggesting the presence of multiple mechanisms of resistance, due to tumour heterogeneity and/or mechanistic cooperation.
Figure 7. FAM35A or C20orf196 loss correlates with PARP inhibitor resistance in cancers.
a, Schematic of in vivo PDX study (top panel). Heat map generated from mRNA-sequencing showing scaled expression levels of indicted genes from corresponding PDX samples (lower panel); n=6, 5, 7, 8 mice for cohorts 1-4 respectively. b, Expression of C20orf196/FAM35A in breast and ovarian cancer PDXs derived from BRCA1-deficient tumours. y-axis: log2 transcript per million. Lines represent mean ± SEM; n=12, 4, 15, 1 for SHLD1-high, SHLD1-low, SHLD2-high, SHLD2-low groups respectively; two-tailed unpaired student t-test; ***p=0.0003. Statistical source data for PDXs can be found in Supplementary Table 5 and methods. c-d, Clonogenic survival assay after IR (c) or cisplatin treatment (d) in the indicated RPE1ko cell lines with AUC shown in Supplementary Fig 7b and 7c, respectively. Data shown represent mean ± SEM (n = 3 independent experiments except for group 7 in c and group 7 in d where n = 2) e, Loss of FAM35A/C20orf196 leads to increased cisplatin-induced FANCD2 foci. Bars represent mean ± SEM, one-way Anova; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant (p≥0.05); n=4 independent experiments, with individual data points plotted over bars; statistical source data can be found in Supplementary Table 5. Scale bar 10µm. f, Proposed model for the action of SHLD1/2 in DSB repair in the presence or absence of functional BRCA1.
Finally, we found that in contrast to 53BP1 deficiency42, SHLD1C20orf196 or SHLD2FAM35A loss increased the sensitivity of BRCA1-proficient and BRCA1-null cells to IR, and even more markedly enhanced their sensitivity to the DNA crosslinking agent cisplatin (Fig 7c-d, Supplementary Fig 7b-c). Furthermore, enhanced cisplatin sensitivity upon SHLD1C20orf196 or SHLD2FAM35A inactivation was associated with increased DNA-damage focus formation by the FANCD2 protein that is involved in detection and repair of DNA crosslinks (Fig 7e, Supplementary Fig 7d). These findings therefore suggested that, if loss/reduced expression of SHLD1/2 occurs in patients, it may provide collateral therapeutic vulnerabilities that could be exploited clinically.
Discussion
Over the past two decades, it has become evident that eukaryotic cells have evolved multiple mechanisms of DNA DSB repair that are regulated in complex and sophisticated ways to optimise genome stability. In particular, much attention has focused on how cells strategically employ the two prime modes of DSB repair – NHEJ and HR – which antagonise one another, operate optimally in different contexts and whose relative usage is regulated by factors such as chromatin structure and cell-cycle stage. In addition to being of academic interest, work on such subjects is also of clinical relevance, particularly in cancer therapy where DSB-inducing chemotherapeutic agents are frequently used, and molecularly-targeted drugs such as PARP inhibitors are being increasingly employed in specific settings. Intrinsic or arising PARP-inhibitor resistance in patients with BRCA1/2 mutations is nevertheless an increasing clinical problem. Using whole-genome CRISPR-Cas9 synthetic-viability/resistance screens, we have uncovered two, previously uncharacterised proteins – SHLD1C20orf196 and SHLD2FAM35A – whose loss mediates PARP-inhibitor resistance and which we have shown act as the most distal factors of the 53BP1/RIF1/MAD2L2 molecular axis to promote NHEJ and restrict HR in BRCA1-deficient settings. Our screens have also identified additional candidate PARP-inhibitor resistance factors that await validation in future studies.
Mechanistically, we have shown that SHLD1C20orf196 and SHLD2FAM35A form a complex, termed Shieldin, with SHLD1C20orf196 recruitment to DNA-damage sites via its interactions with SHLD2FAM35A and other factors, and SHLD2FAM35A interacting with single-stranded DNA via its C-terminal OB fold region. Moreover, we have established that SHLD1C20orf196 and SHLD2FAM35A promote NHEJ in a manner that may be mediated via their effects on restricting DNA-end resection, and serve as a barrier to HR by antagonising the replacement of RPA with BRCA2/RAD51 on resected ssDNA in a manner counteracted by BRCA1. Our work is in line with a recent report43, that independently identified SHLD1C20orf196 and SHLD2FAM35A as NHEJ-promoting factors and antagonists of HR in BRCA1-defective cells. This study also identified a third component, RINN1/SHLD3CTC-534A2.2 that is proposed to serve as a molecular bridge from RIF1 to MAD2L2 and SHLD1/2.
While it seems possible that Shieldin loss contributes to HR restoration in BRCA1-deficient cells through effects on both resection and on BRCA2/RAD51 loading, the relative importance of these mechanisms needs further investigation. We note that more extensive and possibly faster resection in the absence of Shieldin might enhance BRCA2/RAD51 loading. Alternatively, or in addition, Shieldin might serve as a physical barrier to BRCA2/RAD51 loading at dsDNA/ssDNA junctions in BRCA1-deficient cells – perhaps through it being tethered to DSB-flanking chromatin via the 53BP1/RIF1/MAD2L2 complex allowing interactions between the C-terminus of distal SHLD2FAM35A and ssDNA (see Fig 7f for a model, and Supplementary Fig 7e for SHLD2FAM35A domain function summary). Nevertheless, we found that overexpression of the SHLD2FAM35A N- but not the C-terminus confers olaparib hypersensitivity to BRCA1/FAM35A double knockout cells, suggesting that at least in this context, chromatin binding by SHLD2FAM35A plays a dominant role in restricting HR. By contrast, we found that both the SHLD2FAM35A N- and C-termini are important for IR resistance (in BRCA1-proficient cells). As IR sensitivity in Shieldin-deficient cells likely reflects impaired NHEJ, we speculate that Shieldin potentiates NHEJ by restricting DSB resection as well as by assembling with other NHEJ-promoting factors to tether DSB ends together to facilitate their juxtaposition and repair.
Notably, we have found that SHLD1C20orf196 or SHLD2FAM35A inactivation confers enhanced cisplatin sensitivity to BRCA1-null or BRCA1-proficient cells. Such sensitivity probably does not reflect the roles for Shieldin in promoting NHEJ, restricting DNA-end resection or antagonising BRCA1-mediated BRCA2/RAD51 loading because, in our hands, 53BP1 loss does not have pronounced effects on cisplatin sensitivity. Intra-strand DNA crosslinks (ICLs) generated by cisplatin and other compounds are detected and repaired by the Fanconi anemia (FA) pathway, with a key FA protein being FANCD2, which forms foci at sites of these lesions44. We have observed that following cisplatin treatment, FANCD2 foci were more pronounced in cells in which SHLD1C20orf196 or SHLD2FAM35A was inactivated. It will thus be of interest to determine if Shieldin – like MAD2L2, which (with REV3L) functions as a regulatory subunit of the trans-lesion DNA synthesis (TLS) polymerase Pol zeta, and whose biallelic inactivation has been associated with FA45–47 – might also promote ICL repair via TLS mechanisms.
Finally, based on our findings, it will be interesting to evaluate SHLD1/2 expression in tumour biopsies from patients, establish whether this information can be used in patient stratification for PARP-inhibitor therapies, and determine whether SHLD1/2 expression changes arise in patients whose BRCA1-deficient cancers develop resistance after PARP-inhibitor treatment. In this regard, we note that if down-regulation of Shieldin components in BRCA1-deficient cancers does confer clinical resistance, this might allow alternative treatments, such as ones based on platinum compounds.
Methods
CRISPR-Cas9 screen
Performed using genome-scale (GeCKO) v2.020. SUM149PT cells were transduced at multiplicity of infection (MOI) of 0.3 and 250-fold coverage of the library. Cells were then selected with puromycin for 7 days prior to treatment with 3 different PARPi for a further 14 days. IC’s used were; Olaparib IC95-2 μM, BMN673 IC95-5 nM, AZD2461 IC70-4 µM. Surviving clones from each condition were collected, genomic DNA (gDNA) isolated (Blood & Cell Culture DNA Midi Kit, Qiagen) and subjected to PCR with Illumina-compatible primers, followed by Illumina sequencing. Genes enriched or depleted in the inhibitor-treated samples were determined by the software package MAGeCK version 0.5.5 (see commands in the section of “Code availability”).
Cell culture
U2OS, U2OS-derived, HEK293, HEK293T-LentiX cells were cultured as in39. RPE1 p53 null FRT48 and RPE1 p53 null FRT-derived cells were cultured in F-12 (Ham’s F-12; Sigma-Aldrich) supplemented with 17 ml NaHCO3 7.5% per 500ml (Sigma-Aldrich). All media was supplemented with 10% (v/v) foetal bovine serum (FBS; BioSera), 100 U/ml penicillin, 100 µg/ml streptomycin (Sigma-Aldrich) and 2 mM L-glutamine. SUM149PT cells were cultured in Ham's F12 Nutrient Mixture (ThermoFisher) supplemented with 5% (v/v) FBS (BioSera), 10 mM HEPES, 1 μg/ml hydrocortisone, 5 μg/ml insulin, antibiotic as described above. For maintenance and selection of RPE1 FRT p53 null or U20S Trex cells stably expressing GFP or GFP-tagged constructs, 2 μg/ml blasticidin (Sigma-Aldrich) and 0.5 mg/ml G418 (Invitrogen) were used. U2OS-TLR were cultured as in39. In addition to RPE1 p53 null FRT-derived cells, U20S Trex cells stably expressing inducible constructs were cultured with 1 µg/ml doxycycline (Sigma-Aldrich) for 24-48h to induce expression of GFP constructs. All cells were originally obtained from the ATCC cell repository and routinely tested to be mycoplasma free. The U2OS and RPE1 cell lines were recently authenticated using Affymetrix SNP6 copy number analysis. Trf2-/-;p53-/-;TRF2(Ile468Ala) MEFs (TRF2ts MEFs) as described previously33, 49. CH12F3 (CH12)29 and CH12-Cas9 cell lines were cultured in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 50µM 2-mercaptoethanol, 1xMEM non-essential amino acids, 1mM sodium pyruvate and 10mM HEPES.
Generation of human stable cell lines and knockouts
U2OS Trex or RPE1 p53 null FRT-derived cells stably expressing inducible GFP-tagged constructs, were generated by transfection of pcDNA5/FRT/TO-neo containing the GFP-tagged construct and pOG44 (1:4 respectively). Selection began at 48 h using 0.5 mg/ml G418 (Invitrogen). Knockouts were generated in RPE1 p53 null cells by transfecting an ‘All-in-one’ plasmid48. Single-cell sorting by GFP expression was done using MoFlo (Beckman Coulter). Single clones were expanded, genomic DNA extracted and screened by PCR, TOPO-cloning and sequencing. Validated mouse Embryonic Stem Cell (mESC) knockouts of Fam35a and C20orf196 were obtained from Haplobank (www.haplobank.at).
Plasmids and cloning
siRNA and plasmid transfection
siRNAs were obtained from MWG or IDT and transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Plasmid transfections were carried out using TransIT-LT1 (Mirus Bio) according to the manufacturer’s protocol. For siRNA and DNA co-transfections, plasmids were transfected 8h after siRNA treatment. See Supplementary Table 4.
Random plasmid integration assay
Performed as previously described27.
DNA-damage induction using chemical agents, ionizing radiation and laser micro-irradiation
Performed as previously described39.
FRAP and association kinetics
Performed as previously described50.
TLR assays
The Traffic Light Reporter (TLR) assay and the constructs used herein have been described in detail previously38, 39.
Cell-cycle profiling
Performed as previously described39.
Clonogenic survival assays
Whole cell extracts and immunoblotting
Were performed as previously described39. For detection of phospho-RPA (pS4/S8-RPA2), lysates were prepared by scraping cells in 2xSDS buffer followed by SDS-PAGE using 4-12% Bis-Tris gel (Invitrogen), and immunoblotting was done using SuperSignal West Pico PLUS (Thermo Scientific). IRDye800CW- and IRDye680-labelled secondary antibody was used for detection on the Odyssey Infrared imager (LI-COR). Quantification of blots was performed using ImageJ. All protein concentrations were determined using a BCA assay (Pierce). All antibodies are listed in Supplementary Table 2.
Immunoprecipitation
All immunoprecipitation procedures performed twice as previously described39. For co-immunoprecipitation shown in Fig 2d (FAM35A), 293T cells were co-transfected with pMSCV-blas-eGFP-MAD2L2 and either pLX304-blast-V5-Empty or pLX304-V5-FAM35A. 72h post-transfection cells were exposed to 25Gy IR followed by 3h recovery. GFP-Trap_MA beads (ChromoTek) were used, and immunoprecipitation was performed according to the manufacturer’s protocol. For C20orf196 (Supplementary Fig 2c), 293T cells were co-transfected with pMSCV-blas-3xFlag-hMAD2L2 and either pcDNA5.1-GFP or pcDNA5.1-GFP-C20orf196. 72h post-transfection, cells were exposed to 25Gy IR followed by 3h recovery. After washing with cold PBS, cells were lysed in 1 ml lysis buffer (50mM Tris HCl pH7.4; 150mM NaCl; 1mM EDTA; 1% Triton X-100) supplemented with the same inhibitors as above. After 30min incubation on ice followed by centrifugation (16,000g), anti-Flag M2 Magnetic Beads (M8823, Sigma-Aldrich) pre-washed with TBS (50mM Tris HCl, 150 mM NaCl pH7.4), were added to the lysate and rotated over-night at 4°C. Immune-complexes were eluted by 5min boiling.
DNA pulldown experiments
Procedures were described in detail in39 using oligos with the sequence:
5’BiosG/ATCGCATTGGCATTGGCAATGCGATACGACTGATCGAGGGTACTCAGCTAGCTGATTCCGATCGGCTTATTCCGTGTACATACATCGGAT-3’ (IDT)
In vitro GST pull-down
Gluathione sepharose beads (GE Healthcare) were washed with ice-cold PBS and blocked for 30min with PBS supplemented with 10% bacterial lysate (non-induced BL21 cells, lysed using PBS/lysozyme) then resuspended in binding buffer (10mM Tris pH7.5, 150 mM NaCl, 0.5% NP40, 0.5 mM EDTA, 0.5 % BSA). Purified GST (bacterial expression), GST-FAM35A (Novus Biologicals), and His-C20orf196 (Creative BioMart) were added to the beads at 2 pmol and incubated for 30min at 4°C. Beads were washed 5x with 10 mM Tris, pH 7.5, 250 mM NaCl, 0.5 % NP40, 0.5 mM EDTA and eluted with 100 mM Tris pH 8, 20 mM reduced glutathione, 120 mM NaCl for 15min rotating at 4°C. The eluates were boiled for 5min, loaded on 4-12 % Bis-Tris gel (Invitrogen) and subjected to western blotting. The blots were probed with the indicated antibodies.
Recombinant protein purifications and Electrophoretic Mobility Shift Assays (EMSAs)
Wild-type and mutant FAM35A C-terminal domains were purified using the same method. Harvested cells were lysed by sonication in 50 mM Tris pH 8.0, 5% glycerol, 150 mM NaCl, 2 mM β-mercaptoethanol, 10mM imidazole, protease inhibitor (Roche) and 40 g/ml deoxyribonuclease I (Sigma). After centrifugation at 30,000g for 30min, supernatant was loaded onto a gravity column containing Ni-NTA affinity resin (Qiagen) pre-equilibrated with 50 mM Tris pH 8.0, 5% glycerol, 150 mM NaCl, 2 mM β-mercaptoethanol and 10 mM imidazole. After washing beads with the same buffer for 10x column volume, protein was eluted using 50 mM Tris pH 8.0, 5% glycerol, 150 mM NaCl, 2 mM β-mercaptoethanol and 100 mM imidazole. The eluate was dialysed with Q column (GE healthcare) buffer A (20 mM Tris pH 8.0, 50 mM NaCl, 5% glycerol and 2 mM β-mercaptoethanol) and loaded onto a 5 ml Q column. Protein was eluted in a gradient against buffer B (20 mM Tris pH 8.0, 1 M NaCl, 5% glycerol and 2mM β-mercaptoethanol). Fractions containing FAM35A protein were collected and further purified by running through Superdex 200 10/300 column (GE Healthcare) equilibrated in buffer GF (20 mM Tris pH 8, 150 mM NaCl, 5% glycerol and 5 mM DTT). Protein samples during each step of purification were analysed on 4-12% Bis-Tris gels (Invitrogen). Final purified samples were concentrated and stored at -80°C. Both forward and reverse 90-bases DNA oligos (IDT) (F: 6-FAM (6-carboxyfluorescein)-ATCGCATTGGCATTGGCAATGCGATACGACTGATCGAGGGTACTCAGCTAGCTGATTCCGATCGGCTTATTCCGTGTACATACATCGGAT; R:6-FAM-ATCCGATGTATGTACACGGAATAAGCCGATCGGAATCAGCTAGCTGAGTACCCTCGATCAGTCGTATCGCATTGCCAATGCCAATGCGAT) were dissolved in annealing buffer (10 mM Tris pH 8.0, 50 mM NaCl and 1 mM EDTA) to a final concentration of 100 μM. DNA oligo F was used as ssDNA for EMSA. Equal volumes of DNA oligo F and R were mixed and annealed (heated to 95°C for 2min and cooled to 25°C over 45min) to generate dsDNA. Each 20 μl of EMSA reaction contained 10 nM of ssDNA/dsDNA incubated with different concentrations of proteins in 20 mM Tris-HCl pH 7.5, 50 mM KCl, 5% (v/v) glycerol, 100 μM DTT, 10 μg/ml BSA. Samples were incubated at 37°C for 15min and applied onto a 5% polyacrylamide native gel in 0.5xTBE buffer for electrophoresis at 4°C. DNA was visualized by Typhoon 9000 (GE Healthcare).
GFP-Trap pull down for mass spectrometry
HEK293T cells were cultured in SILAC media containing either L-arginine and L-lysine, or L-arginine [13C6, 15N4] and L-lysine [13C6, 15N2] (Cambridge Isotope Laboratories) as described previously51. Cells were lysed 48h post-transfection in modified RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% sodium deoxycholate) supplemented with protease, phosphatase inhibitors and N-ethylmaleimide. Lysates were cleared by centrifugation at 16,000×g for 15min at 4°C and protein concentrations were estimated using QuickStart Bradford Protein assay (BioRad). Per SILAC condition, 20 µl of pre-equilibrated GFP-Trap-A beads (ChromoTek) were added to 2 mg of lysate and incubated 1h at 4°C rotating, followed by 3x washes with modified RIPA buffer. Bound proteins were eluted in NuPAGE® LDS Sample Buffer (Life Technologies) supplemented with 1 mM dithiothreitol, heated at 70 ºC for 10min and alkylated with 5.5 mM chloroacetamide at RT. Samples were loaded onto 4-12% gradient SDS-PAGE gels, proteins were stained using the Colloidal Blue Staining Kit (Life Technologies) and digested in-gel using trypsin. Peptides were extracted from the gel and desalted on reversed phase C18 StageTips52.
Mass spectrometry analysis
Peptide fractions were analyzed on a quadrupole Orbitrap mass spectrometer (Q Exactive Plus, Thermo Scientific) equipped with an EASY-nLC 1000 (Thermo Scientific) as described53. Peptide samples were loaded onto C18 reversed phase columns and eluted with a linear gradient from 8 to 40% acetonitrile containing 0.1% formic acid for 2h. The mass spectrometer was operated in data dependent mode, automatically switching between MS and MS2 acquisition. Survey full-scan MS spectra (m/z 300–1650) were acquired in the Orbitrap. The ten most intense ions were sequentially isolated and fragmented by HCD54. Fragment spectra were acquired in the Orbitrap mass analyzer. Raw data files were analyzed using MaxQuant (development version 1.5.2.8)55. Parent ion and MS2 spectra were searched against a database containing 92,578 human protein sequences obtained from the UniProtKB released in December 2016 using Andromeda search engine56. Spectra were searched with a mass tolerance of 6 ppm in MS mode, 20 ppm in HCD MS2 mode, strict trypsin specificity and allowing up to three miscleavages. Cysteine carbamidomethylation was searched as a fixed modification, whereas protein N-terminal acetylation, methionine oxidation, n-ethylmaleimide modification of cysteines were searched as variable modifications. The dataset was filtered based on posterior error probability to arrive at a false discovery rate below 1% estimated using a target-decoy approach57.
Immunofluorescence and microscopy imaging
Confocal imaging for γH2AX, RAD51, RPA, ssDNA (BrdU), BLM, BRCA2, FANCD2, Cyclin A and GFP (FAM35A and C20orf196) was performed as described in39, for RAD51 and Cyclin A the pre-extraction step was omitted and cells were permeabilised for 15min in 0.2% Triton X-100 (Sigma) in PBS after fixation. Super-resolution images were acquired using a Deltavision OMX 3D-SIM System V3 BLAZE (Applied Precision, a GE Healthcare company) equipped with 3 sCMOS cameras, 405, 488, 592.5nm diode laser illumination, an Olympus Plan Apo N 60x 1.42NA oil objective, and standard excitation and emission filter sets. Imaging of each channel was done sequentially using three angles and five phase shifts of the illumination pattern as described58. Sections were acquired at 0.125 μm z steps. Raw OMX data was reconstructed and channel registered in SoftWoRx software version 6.5.2 (Applied Precision, a GE Healthcare company). Voxelwise nearest-neighbour distances were measured for GFP-FAM35A signal relative to 53BP1 signal using a custom script (Butler R) for Fiji (https://github.com/gurdon-institute/OMX-Spatial-Analysis). The script maps signal volumes using Kapur's maximum entropy thresholding method59 and measures distances using the exact signed 3D Euclidean distance transform with internal distances set to zero for display on the histogram. For all images, scale bars = 10μm.
Multiplex fluorescence in situ hybridisation (M-FISH)
Human 24-colour multiplex FISH (M-FISH) probe preparation and slides treatments followed60. For each human cell sample, 10-30 metaphases were karyotyped based on the M-FISH classification and DAPI-banding pattern. FISH on metaphases spreads using BAC probes was performed as previously described61 and counted manually. For class-switch recombination (CSR) assays, DNA FISH on metaphases spreads was performed as previously described61 and counted manually. At least 470 metaphases were evaluated per genotype, using at least 2 independent clones for each condition. For telomere uncapping, cell harvesting, preparation of metaphase spreads and telomere FISH with an Alexa488-(CCCTAA repeat) peptide nucleic acid custom probe (PN-TC060-005, Panagene/Eurogentec), metaphase chromosome analysis was done as described previously6. These data represent 2 independent experiments, ≥1300 chromosomes for each condition, counted manually after blinding the genotypes.
Telomere fusion assays: MEFs viral transduction
Cells were transduced as before49 with pLKO-puro shRNA lentiviruses obtained from the MISSION shRNA library (Sigma), against mouse genes as described or left untransduced (‘empty’). Assessment of telomere NHEJ. TRF2ts MEFs were grown for 24h at the non-permissive temperature of 39°C to inactivate TRF2 and induce NHEJ-dependent chromosome end-to-end fusions because of telomere uncapping.
Class-switch recombination assays: CH12 and CH12-Cas9 cell lines
The CH12-Cas9 cell line was generated by transducing CH12-Cas9 cells made using spin-infect with lentivirus particles packaged in HEK293T. Plasmids: pKLV2-EF1aBsd2Acas9-W, pxPAX2 (Addgene #12260), VSV-G and pMD2.G (Addgene #12259). Blasticidin selection (10 μg/ml) started 48h after transduction for one week. sgRNA expression plasmids for class-switch recombination assays. sgRNAs were used to target Fam35a, C20orf196 and Mad2l2/Rev7 mouse genes (2 sgRNAs per target gene, sequences listed in Supplementary Table 4. sgRNAs were cloned into pKLV-flipped U6gRNA_CCDB_PB_BbsI_PGKpuro2ABFP vector62. Generation of wild-type and knockout CH12-Cas9 cell clones. 53bp1 null CH12 cell clones (gift from Fred Alt) were as previously described63. 12 million CH12-Cas9 cells were nucleofected with 2.2 μg of each sgRNA-1 and sgRNA-2 and 0.6 μg of piggyBac transposase expression vector64, using an Amaxa Nucleofector, Nucleofector® Kit V solution (Lonza) and program X-001. Two days later, BFP-positive/puromycin-resistant CH12-Cas9 cells were selected with 3 μg/ml puromycin for one week. Cells were then single cell diluted into 96-well plates, further cultured and screened by PCR and Sanger sequenced using PCR primers listed in Supplementary Table 4. Class-switch recombination and cell proliferation assays. CH12 cells were plated at 50,000 cells/ml in complete RPMI supplemented with anti-CD40 antibody (1 μg/ml, Miltenyi), IL-4 (20 ng/ml, Miltenyi) and TGF-β (1 ng/ml, R&D Biotech) to induce IgM to IgA switching. After 3 days, cells were assayed for class-switching by flow cytometry using an IgA-PE antibody (eBiosciences) and a Canto II analyser (BD Biosciences). Viable cells were counted using a Casy cell counter (Roche). CSR and proliferation assays were done on: 3xwild-type (WT), 3xFam35a knockout (Fam35a), 3xC20orf196 knockout (C20orf196), 2x53bp1 knockout (53bp1) and 3xMad2l2 knockout (Mad2l2) in three independent experiments. RT-PCR analysis. Igh, α germ-line transcripts (αGLT) and Aid mRNA were quantified as previously described12. Primers are listed in Supplementary Table 4.
Patient-derived tumour xenografts
PDXs were generated and established from consented breast or ovarian cancer patients’ samples as previously described40. The research was done with the appropriate approval by the National Research Ethics Service, Cambridgeshire 2 REC (REC reference number: 08/H0308/178), and by the Vall d’Hebron Hospital Clinical Investigation Ethical Committee (PR(AG)183/2012). STG201, the PDX model used in this study, is a BRCA-null model featuring BRCA1 promoter methylation, loss of BRCA1 mRNA and protein expression. We have previously shown its sensitivity in vivo and in PDX derived cells to PARP inhibitors, including olaparib. STG201 is also linked to deep molecular and drug sensitivity annotation40 and http://caldaslab.cruk.cam.ac.uk/bcape/. All other PDXs were derived from breast or ovarian tumours from BRCA1-mutation carriers or BRCA1 epigenetic silencing due to promoter hypermethylation41. PDX127 did not show any co-expression of BRCA1 but it was low in both FAM35A and C20orf196 expression. None of the 5 PARPi-sensitive PDXs exhibited low levels of C20orf196, FAM35A or 53BP1 loss nor BRCA1 hypomorphs. The study was compliant with all relevant ethical regulations regarding research involving animal use and human participants.
Generation of acquired drug resistance in vivo
AZD2281 (Olaparib/Lynparza) as a PARP inhibitor (50mg/kg, 5IW) was administered to immunocompromised tumour bearing mice upon randomization as previously described (50mg/kg, 5 days/week)40. To classify the response of the subcutaneous implants we modified the RECIST criteria to be based on the % tumour volume change following continuous olaparib treatment: complete response (CR), best response≤-95%; partial response (PR), -95%<best response≤-30%; stable disease (SD), -30%<best response≤+20%; progressive disease (PD), % tumour volume change at day 21 of treatment >+20%. PARPi-resistant PDXs exhibited PD while PARPi-sensitive models exhibited SD, PR or CR. For STG201, time matched vehicle and olaparib treated samples were collected 25 days after treatment (PARPi naïve PDX) and processed for RNA extractions and sequencing. A couple of mice in the study were left with continued exposure to olaparib until tumour regrowth. One of these resistant tumours was serially passaged 126 days after treatment into new host mice (PARPi resistant PDX) and treated with further vehicle or olaparib. 58 days after treatment the resistance phenotype was confirmed and samples were collected and processed for RNA-sequencing as described below. Growth curves show average and standard deviation of at least 5 independent tumour volumes per trial arm. All experimental procedures were approved by the University of Cambridge Animal Welfare and Ethical Review Committee and by the Vall d’Hebron Hospital Clinical Investigation Ethical Committee and Animal Use Committee. RNA-sequencing. RNA was extracted from all samples using the Qiagen miRNeasy or RNeasy Mini kit (Cat ID, 217004 or 74104) as per manufacturer’s instructions. Libraries for Illumina sequencing were prepared using TruSeq Stranded mRNA HT kit or Total RNA Library Prep kit with Ribo-Zero Gold (Cat ID, RS-122-2103 or RS-122-2301, Illumina). 500ng of total RNA with RNA Integrity Numbers (RINs) above 8 was used for library preparation. Samples were processed following manufacturer’s HS (High-Sample) instructions (part# 15031048 Rev. E, Illumina) with 12 or 15 cycles of PCR used at the Enrichment of DNA Fragments step. All libraries were quantified using KAPA Library Quantification Kit Illumina ROX Low (Cat ID, KK4873, KAPA Biosystems) and normalised. Libraries were pooled in equal volumes and pools were used for clustering on HiSeq4000 sequencing flow cell following manufacturer’s instructions. Sequencing was performed using 150bp or 100bp paired-end run type for dual-indexed libraries. Prior to alignment, sequencing quality of the reads was enforced using Trim Galore! (v0.4.2) http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Then, as described in Callari et al. or Ahdesmaki et al65 reads were aligned to a combined human (hg19) and mouse (mm10) reference genome using STAR (v2.5.2b)66, 67. Counts were assigned to genome features using featureCounts (v1.5.2), whereby the alignment score is used to discern accurately between reads sourced from human and mouse68. Counts from multiple sequencing runs were merged and then normalised using the edgeR package69, 70.
Code availability
A custom FIJI script used in OMX analysis can be found at https://github.com/gurdon-institute/OMX-Spatial-Analysis.
The MAGeCK commands used for CRISPR-Cas9 screens were:
mageck test -k counts.csv -c DMSO -t WC_2461 -n WC_2461
mageck test -k counts.csv -c DMSO -t WC-673 -n WC-673
mageck test -k counts.csv -c DMSO -t WC-2281 -n WC-2281
Statistics and Reproducibility
Unless stated otherwise Prism v7.0b (GraphPad Software) was used to generate graphs, perform statistical tests and calculate p values. Error bars, statistical tests and number of independent repeats (n) are indicated in figure legends with statistical source data including the precise p values provided in Supplementary Table 5. Statistical tests included two-tailed Student t-tests, Fisher’s Exact test, and one-way analysis of variance (ANOVA), the latter all being corrected as recommended for multiple comparisons. Microscopy image analyses were performed using ImageJ/FIJI or Volocity 6.3 (Perkin-Elmer). CRISPR screens were performed with three clones per drug treatment. Mass spectrometry of GFP-FAM35A and GFP-C20orf196 was performed in two independent experiments. RNA-sequencing was performed as three replicates for each trial arm, due to sequencing across multiple lanes (which were merged prior to any further analysis). This was performed for the following number of independent biological samples: six PDXs in cohort 1, five PDXs in cohort 2, seven PDXs in cohort 3 and eight PDXs in cohort 4. For the SHLD1 high and low expression cohorts 12 and 4 independent PDXs were evaluated respectively. All immunofluorescence assay quantification data represent means ±SEM’s of 3 independent biological repeats and n≥30 cells per condition unless otherwise specified. All immunoblots are representative of two independent experiments with unprocessed scans of immunoblots shown in Supplementary Fig 8.
Supplementary Material
Acknowledgements
We thank all SPJ laboratory members for support and advice, and our Cambridge colleagues N. Lawrence for OMX super-resolution microscopy support and R. Butler for help with computational image analyses and programming. We thank S. Selivanova and S. Hough for help with plasmid amplification, sample preparation and tissue culture maintenance, K. Dry for extensive editorial assistance, F. Muñoz-Martinez for assistance with CRISPR-Cas9 knockout generation, L. Radu for assistance with protein purification, C. Lord (Institute of Cancer Research, London) for SUM149PT cells, D. Durocher (University of Toronto, Canada) for U2OS LacSceIII cells, F. Alt (Harvard University, USA) for CH12F3 cells and 53bp1 knockout CH12F3 cell clones, T. Honjo (Kyoto University, Japan) for permission to use the CH12F3 cell line, and J. Serrat in the Jacobs lab for technical assistance. The SPJ lab is largely funded by a Cancer Research UK (CRUK) Program Grant, C6/A18796, and a Wellcome Trust (WT) Investigator Award, 206388/Z/17/Z. Core infrastructure funding was provided by CRUK Grant C6946/A24843 and WT Grant WT203144. SPJ receives salary from the University of Cambridge. HD is funded by WT Clinical Fellowship 206721/Z/17/Z. TWC was supported by a Cambridge International Scholarship. D.P. is funded by Cancer Research UK studentship C6/A21454. The PB lab is supported by the Emmy Noether Program (BE 5342/1-1) from the German Research Foundation and the Marie Curie Career Integration Grant from the European Commission (630763). The LP lab is funded by the WT (investigator award 104641/Z/14/Z) and the Medical Research Council (project grant MR/N000161/1). The CC lab was supported with funding from CRUK. The JJ lab was supported by the European Research Council Grant ERC-StG 311565, The Dutch Cancer Society (KWF) grant KWF 10999, and the Netherlands Organization for Scientific Research (NWO) as part of the National Roadmap Large-scale Research Facilities of the Netherlands, Proteins@Work (project number 184.032.201 to the Proteomics Facility of the Netherlands Cancer Institute). The LD lab is funded by the Institut Pasteur, the Institut National du Cancer (# PLBIO16-181) and the European Research Council under the ERC (starting grant agreement #310917). WW is part of the Pasteur - Paris University (PPU) International PhD program and this project received funding from the CNBG company, China. QW is funded by the Wellcome Trust (200814/Z/16/Z to T.L.B). The VS lab work was funded by the Instituto de Salud Carlos III (ISCIII), an initiative of the Spanish Ministry of Economy and Innovation partially supported by European Regional Development FEDER Funds (PI17-01080 to VS), the European Research Area-NET, Transcan-2 (AC15/00063), a non-commercial research agreement with AstraZeneca UK Ltd, and structural funds from the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR, 2017 SGR 540) and the Orozco Family. VS received salary and travel support to CC’s lab from ISCIII (CP14/00228, MV15/00041) and the FERO Foundation.
Footnotes
Data availability
The raw data files for the whole genome CRISPR-Cas9 screen in SUM149 cells is available on NIH Sequence Read Archive (SRA) via the accession number PRJNA471892. Raw data files for the PDX RNA sequencing is available on NIH Sequence Read Archive (SRA) via the accession number PRJNA473981. Raw data files for the mass spectrometry are available via the ProteomeXchange Consortium on the PRIDE partner repository with the dataset identifier PXD009830. Source data for figures can be found in Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Author contributions
TWC and SPJ conceived the project and TWC initiated the project by performing the CRISPR-Cas9 screens, with MH doing the bioinformatic analyses. LD supervised, and CL and WW performed class switch recombination and Igh locus instability experiments. JJ supervised, and IK performed recombinant MAD2L2 co-IPs, MAD2L2 IRIF/IB, pS4/8-RPA2 IB and telomere fusion experiments. MO, PB performed mass-spectrometry. JC performed and analysed clonogenic survival experiments and random plasmid integration assay; MD and MS generated human knockout cell lines and MS performed in vitro pulldown experiments; JL carried out oligonucleotide interaction studies; DP performed end-resection assays; TWC and RB generated RPE1 p53ko, RPE1 p53ko/BRCAko and p53ko/BRCA1ko/53BP1ko cell lines; YG and SPJ supervised the above. GB performed IR survivals in mouse cells, mouse sgRNA cloning, and FISH with FY and BF. LP performed structural analysis and FAM35A modelling. QW performed purification of bacterially expressed recombinant FAM35A proteins and EMSA. AM, AS, AB and CC performed patient-derived xenograft experiments on PARPi-induced resistance. VS, MO’C and ZL established, performed, analysed and characterised PDXs in the experiments on intrinsic PARPi resistance. HD assisted with many of the above, and devised and performed all other experiments. HD, YG and SPJ wrote the manuscript with input from all others. LD, JJ, YG and SPJ supervised the project.
Competing financial interests
SPJ receives some research funding from AstraZeneca and is a named inventor on patents describing the use of PARP inhibitors in cancer therapy. VS’s laboratory receives research funding support from AstraZeneca. MJO and ZL, are employees and shareholders of AstraZeneca. The other authors declare no competing financial interests.
References
- 1.Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 4.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Boersma V, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5' end resection. Nature. 2015;521:537–540. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daley JM, Sung P. RIF1 in DNA break repair pathway choice. Mol Cell. 2013;49:840–841. doi: 10.1016/j.molcel.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5' end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capoluongo E, et al. Guidance Statement On BRCA1/2 Tumor Testing in Ovarian Cancer Patients. Semin Oncol. 2017;44:187–197. doi: 10.1053/j.seminoncol.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 17.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 18.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 19.Basu B, Sandhu SK, de Bono JS. PARP inhibitors: mechanism of action and their potential role in the prevention and treatment of cancer. Drugs. 2012;72:1579–1590. doi: 10.2165/11635510-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonetta M, et al. H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle. 2018;17(1):124–136. doi: 10.1080/15384101.2017.1404210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo KW, et al. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem. 2005;280:8172–8179. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- 23.Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Sun J, et al. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scully R, Xie A. Double strand break repair functions of histone H2AX. Mutat Res. 2013;750:5–14. doi: 10.1016/j.mrfmmm.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87 doi: 10.1259/bjr.20130685. 20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman S, et al. 53BP1 Contributes to Igh Locus Chromatin Topology during Class Switch Recombination. J Immunol. 2017;198:2434–2444. doi: 10.4049/jimmunol.1601947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 30.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 31.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. 2016;16:387–398. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochs F, et al. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol. 2016;23:714–721. doi: 10.1038/nsmb.3251. [DOI] [PubMed] [Google Scholar]
- 36.Cruz-Garcia A, Lopez-Saavedra A, Huertas P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014;9:451–459. doi: 10.1016/j.celrep.2014.08.076. [DOI] [PubMed] [Google Scholar]
- 37.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 38.Certo MT, et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt CK, et al. Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat Cell Biol. 2015;17:1458–1470. doi: 10.1038/ncb3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruna A, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167:260–274 e222. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruz C, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA mutated breast cancer. Ann Oncol. 2018 doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunting SF, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta R, et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell. 2018;173:972–988 e923. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 45.Makarova AV, Burgers PM. Eukaryotic DNA polymerase zeta. DNA Repair (Amst) 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomida J, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase zeta. Nucleic Acids Res. 2015;43:1000–1011. doi: 10.1093/nar/gku1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bluteau D, et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest. 2016;126:3580–3584. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang TW, le Sage C, Larrieu D, Demir M, Jackson SP. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci Rep. 2016;6:24356. doi: 10.1038/srep24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peuscher MH, Jacobs JJ. DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nat Cell Biol. 2011;13:1139–1145. doi: 10.1038/ncb2326. [DOI] [PubMed] [Google Scholar]
- 50.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 52.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 53.Michalski A, et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. M111 011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen JV, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 55.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 56.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 57.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 58.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kapur JN, Sahoo PK, Wong AKC. A New Method for Gray-Level Picture Thresholding Using the Entropy of the Histogram. Comput Vision Graph. 1985;29:273–285. [Google Scholar]
- 60.Agu CA, et al. Successful Generation of Human Induced Pluripotent Stem Cell Lines from Blood Samples Held at Room Temperature for up to 48 hr. Stem Cell Reports. 2015;5:660–671. doi: 10.1016/j.stemcr.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lescale C, et al. RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair. Nat Commun. 2016;7:10529. doi: 10.1038/ncomms10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metzakopian E, et al. Enhancing the genome editing toolbox: genome wide CRISPR arrayed libraries. Sci Rep. 2017;7:2244. doi: 10.1038/s41598-017-01766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panchakshari RA, et al. DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc Natl Acad Sci U S A. 2018;115:762–767. doi: 10.1073/pnas.1719988115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahdesmaki MJ, Gray SR, Johnson JH, Lai Z. Disambiguate: An open-source application for disambiguating two species in next generation sequencing data from grafted samples. F1000Res. 2016;5:2741. doi: 10.12688/f1000research.10082.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Callari M, et al. Computational approach to discriminate human and mouse sequences in patient-derived tumour xenografts. BMC Genomics. 2018;19:19. doi: 10.1186/s12864-017-4414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casper J, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 69.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.