Abstract
Hunchback is a bifunctional transcription factor that can activate and repress gene expression in Drosophila development. We investigated the regulatory DNA sequence features that control Hunchback function by perturbing enhancers for one of its target genes, even-skipped (eve). While Hunchback directly represses the eve stripe 3+7 enhancer, we found that in the eve stripe 2+7 enhancer, Hunchback repression is prevented by nearby sequences—this phenomenon is called counter-repression. We also found evidence that Caudal binding sites are responsible for counter-repression, and that this interaction may be a conserved feature of eve stripe 2 enhancers. Our results alter the textbook view of eve stripe 2 regulation wherein Hb is described as a direct activator. Instead, to generate stripe 2, Hunchback repression must be counteracted. We discuss how counter-repression may influence eve stripe 2 regulation and evolution.
Author summary
During development, animals express genes in distinct patterns to specify different cell types that are important for function. Enhancers are DNA sequences that generate these patterns by binding proteins that activate gene expression, repress gene expression, or both. We have learned how enhancers generate patterns by carefully studying particular DNA sequences, including the even-skipped stripe 2 enhancer, which generates expression in a single stripe in fruit fly embryos; this enhancer has been an important case study for the field. Although the protein Hunchback has been thought to activate this enhancer, we find that it actually functions as a repressor, and this repression is counteracted by nearby sequences in the enhancer. Our results change how we think the even-skipped stripe 2 enhancer works and evolves over time.
Introduction
Bifunctional transcription factors (TFs) that can activate or repress their target genes are critical in animal development [1,2] and are associated with human disease [3,4]. The function of these TFs can depend on the context of the enhancer sequences they bind [5–9]. To infer accurate regulatory networks from genome sequence, we must define the sequence features that control the activity of bifunctional TFs [10].
Hunchback (Hb) is a bifunctional TF that patterns the Drosophila melanogaster embryo [11]; the Hunchback homolog Ikaros is critical in human hematopoesis [12]. hb is a gap gene with many targets throughout Drosophila development including other gap genes [13], pair-rule genes [14–17], homeotic genes [18–20] and neuronal genes [21]. Its bifunctional role in regulating the pair-rule gene even-skipped (eve) has been particularly well-studied. Classic experiments using minimized enhancer constructs found evidence that Hb directly activates the minimal eve stripe 2 enhancer (eve2min) [14,22,23]. Other studies examined endogenous eve expression by misexpressing hb along the ventral surface of embryos (sna::hb embryos) and found that Hb represses eve stripes 3 and 7 [24]. Qualitative measurements of mutated versions of the eve stripe 3+7 enhancer (eve3+7) driving reporter gene expression suggested that Hb repression of eve3+7 is direct [15,17,25,26].
Even after decades of study, the DNA sequence features that control Hb bifunctionality are not known. One hypothesis is that DNA-bound Hb dimers act as repressors while Hb monomers act as activators (dimerization hypothesis). The dimerization hypothesis is supported by computational work that predicts expression of eve3+7 and the gap gene Krüppel [27,28], as well as in vitro experiments identifying zinc finger domains in Hb and Ikaros that allow for dimerization [29]. In another hypothesis, binding of a different TF converts Hb from a repressor into an activator (co-activation hypothesis). In vivo measurements of synthetic binding site arrays for Bicoid (Bcd) and Hb support the co-activation hypothesis [30], and Hb co-activation by Bcd has been incorporated into computational models of eve enhancer function [31–33]. Recent models have also incorporated Hb co-activation by the TF Caudal (Cad) [34]. Cad activates gap and pair-rule gene expression in the posterior of both long and short germ-band insects [35–38], and Cad homologs are critical in vertebrate development and human disease [39,40]. Though both hypotheses have been included in computational models of enhancer function [28,34], neither has been experimentally tested.
Here, we experimentally test the co-activation hypothesis by perturbing two enhancers that are active in the same cells but respond differently to Hb. eve stripe 7 is generated by two shadow enhancers; Hb represses eve3+7 [24,26] and activates eve2+7, an extended version of eve2min that also generates stripe 7 (Fig 1A and 1B) [41]. Because both enhancers are active in the same nuclei, Hb function must be partially controlled by enhancer sequence. In this system, we can test the co-activation hypothesis by measuring the effects of perturbing both enhancers quantitatively and at cellular resolution (Fig 1) [42–44].
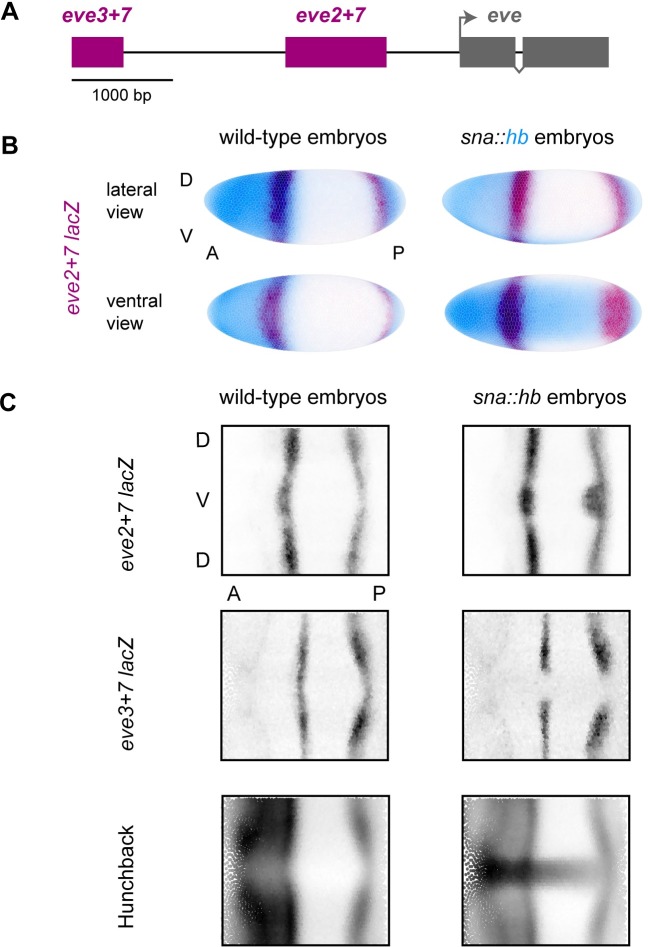
Fig 1. Quantitative gene expression data in wild-type and sna::hb embryos.
(A) Diagram of the upstream half of the even-skipped locus. Enhancers are indicated in maroon, and coding sequence is indicated in grey. (B) We created transgenic lines containing lacZ reporter constructs for eve enhancers and measured gene expression in wild-type embryos and embryos misexpressing ventral hb (sna::hb embryos). Here, we show visual renderings [74] of gene expression atlas data that average measurements from multiple embryos in timepoint 4 (25–50% membrane invagination) [43]. Dorsal (D) and ventral (V) surfaces are indicated for the lateral view, as are anterior (A) and posterior (P) positions. Left: eve2+7 lacZ expression (maroon) in wild-type embryos; Right: eve2+7 lacZ in sna::hb embryos. Hb protein is shown in blue. Individual nuclei are outlined, and darker coloring indicates higher relative expression level. (C) To help visualize all relevant nuclei, we show 2-dimensional projections of expression data throughout this manuscript. Positions of individual nuclei along the dorsal-ventral (DV) axis are plotted as a function of position along the anterior-posterior (AP) axis. Darker color indicates higher expression for each nucleus. Relative expression values are normalized to the maximum value and range from 0 to 1.
We find that Hb directly represses eve3+7 and indirectly activates eve2+7. Indirect Hb activation occurs because nearby sequences corresponding to predicted Cad binding sites prevent Hb repression in eve2+7. We also find that the number of predicted Cad and Hb sites appears correlated in orthologous eve stripe 2 enhancers. Our results suggest that the textbook description of eve2min, where Hb is described as a direct activator that synergizes with Bcd [45], is incomplete. Instead, Hb represses eve stripe 2, but this activity is inhibited by additional factors.
Results
Hb directly represses eve3+7
Previous work indicated that Hb directly represses eve3+7, but did not decipher how Hb regulates expression of each stripe. For example, mutating Hb binding sites in eve3+7 showed that Hb defines the anterior border of stripe 3, but the effect of Hb on stripe 7 was unclear [26]. Furthermore, the stripe 7 pattern driven by eve3+7 retreats in response to ectopic Hb, but this effect could be indirect [41]. Here, we combined perturbations in both enhancer sequence and Hb protein localization and measured the effects on expression of each stripe quantitatively.
We removed predicted Hb binding sites from eve3+7 (eve3+7mutHb) using the most current position weight matrices (PWMs) available; PWMs allow us to predict TF binding at a variety of different stringencies (see Materials and Methods). Some predicted low affinity Hb sites remain because they overlap with sites for other factors (Fig 2A, S1 Appendix). We incorporated eve3+7mutHb into a reporter construct, integrated it into the Drosophila melanogaster genome and measured expression using quantitative in situ hybridization in wild-type and sna::hb embryos, where Hb protein is produced along the ventral side of the embryo. We compared expression driven by this construct to expression driven by a wild-type version (Fig 2). We also measured expression driven by a mutant construct from [26] (S1 Fig). We display our quantitative gene expression data in multiple ways: a line trace of expression level versus anterior-posterior (AP) position along the lateral surfaces of the embryo (Fig 2C and 2E); a 2-dimensional rendering of the expression level in every cell in a single embryo (Fig 2C and 2D) or gene expression atlas (Fig 2B); or a plot of the differences in peak expression level between a lateral and a ventral line trace in individual sna::hb embryos to quantify the effect of hb misexpression (Fig 2D and 2F). Means and standard errors for our quantitative measurements can be found in S1 and S2 Tables. All enhancer sequences in this study were incorporated into the same reporter backbone and integrated into the same genomic location (see Materials and Methods). Annotated eve3+7 and eve2+7 enhancer sequences from this study are included in S1 Appendix.
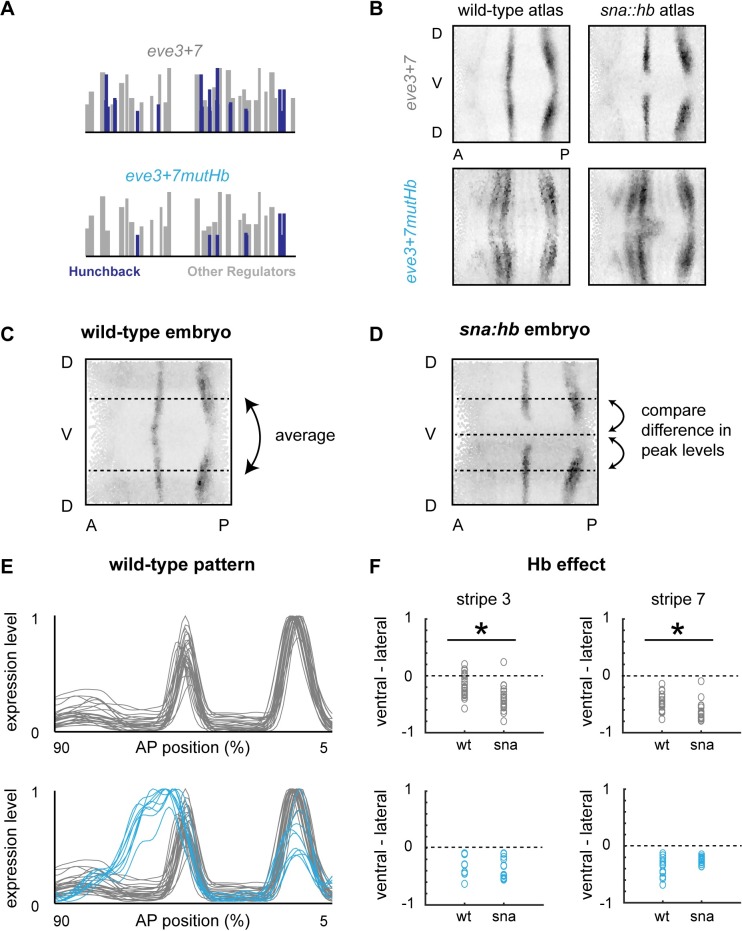
Fig 2. Hunchback directly represses eve3+7.
(A) Predicted Hb binding sites in wild-type and mutant versions of eve3+7 are depicted as vertical bars along the sequence where height is proportional to PATSER score [68]. Hb sites are indicated in blue; other regulators are in grey. (B) 2D projections of atlas data for reporter constructs expressed in wild-type or sna::hb embryos. Data is taken from timepoint 4. (C) 2D projection of a representative wild-type embryo expressing eve3+7 lacZ. We plot gene expression as a function of anterior-posterior (AP) position by averaging measurements from lateral strips in individual embryos and normalizing them to their maximum value (see E). (D) 2D projection of a representative sna::hb embryo expressing eve3+7 lacZ. To quantify the effect of ventral Hb in individual embryos, we take average measurements from lateral strips, extract the local maxima corresponding to stripes 3 and 7, and subtract those values from the corresponding peaks of the ventral strip. We perform the same analysis in wild-type embryos to account for modulation along the DV axis. A decrease in the ventral/lateral difference between wild-type and sna::hb embryos indicates Hb repression, while an increase indicates Hb activation (see F). (E) Lateral line traces from individual wild-type embryos containing eve3+7 reporter constructs (eve3+7: grey, n = 26; eve3+7mutHb: blue, n = 9). Embryos are from all six timepoints in stage 5. (F) Differences between ventral and lateral stripe peaks are plotted for individual wild-type and sna::hb embryos in all 6 timepoints in stage 5. Top: wild-type eve3+7 (wt: n = 26; sna::hb: n = 19); Bottom: eve3+7mutHb (wt: n = 9; sna::hb: n = 13). Asterisks indicate significant differences between wild-type and sna::hb embryos (p-value < 0.001, Mann-Whitney U test). Differences between wild-type and sna::hb embryos containing eve3+7mutHb were not significant (p-value > 0.4 for both stripes, Mann-Whitney U test).
Consistent with previous results [26], eve3+7mutHb drove an expression pattern where stripe 3 expanded anteriorly, though we detected no effect on the boundaries of stripe 7 (Fig 2B and 2E). In addition, the stripe 7 expression pattern driven by eve3+7mutHb did not retreat from ventral Hb in sna::hb embryos (Fig 2B and 2F). By combining perturbations to enhancer sequence and Hb protein localization, these results confirm that Hb directly represses eve3+7.
Hb indirectly activates stripe 7 in eve2+7
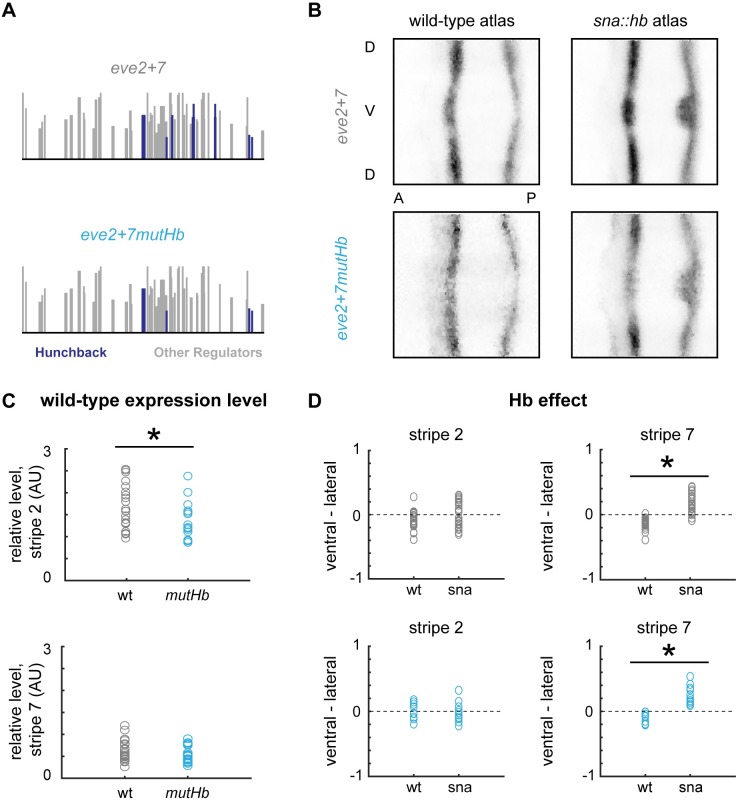
Previous experimental and computational work suggested that Hb directly activates eve stripe 2 [23,31]. We previously found that the stripe 7 pattern driven by eve2+7 bulges in sna::hb embryos, and thus hypothesized that Hb directly activates eve2+7 [41]. Here, we tested whether Hb directly activates stripe 7 in eve2+7 by mutating predicted Hb binding sites in eve2+7 (eve2+7mutHb, Fig 3A) and measuring gene expression in wild-type and sna::hb embryos.
Fig 3. Hunchback indirectly activates eve2+7.
(A) Predicted Hb binding sites in eve2+7 and eve2+7mutHb. Sites are displayed as in Fig 2. (B) 2D projections of atlas data for reporter constructs expressed in wild-type or sna::hb embryos. Data is taken from timepoint 4. (C) Peak stripe expression levels for individual embryos from timepoints 2–4 (4–50% membrane invagination) containing eve2+7 (grey, n = 23) or eve2+7mutHb (blue, n = 20) were measured by normalizing lacZ expression levels using a huckebein co-stain [44] and extracting local maxima from lateral line traces. Asterisks indicate significant differences (p-value < 0.05, Mann-Whitney U test). (D) Differences between ventral and lateral stripe peaks are plotted for individual wild-type and sna::hb embryos in all 6 timepoints in stage 5. Top: wild-type eve2+7 (wt: n = 22; sna::hb: n = 26); Bottom: eve2+7mutHb (wt: n = 9; sna::hb: n = 11). Asterisks indicate significant differences between wild-type and sna::hb embryos (p-value < 0.001, Mann-Whitney U test).
We were surprised to find that mutating Hb binding sites had little effect on the spatial pattern driven by eve2+7 in either genetic background. Compared to eve2+7, eve2+7mutHb drives low level expression anterior to stripe 2 (Fig 3B; line traces for individual embryos are shown in S2 Fig), but other features of the expression pattern were unaffected, including the stripe 7 bulge in sna::hb embryos. We therefore tested for quantitative effects of removing Hb binding sites by measuring expression levels using a co-stain method [44]. We found that eve2+7mutHb drove slightly lower expression levels in stripe 2 (p-value = 0.022, Mann-Whitney U test), but stripe 7 levels were indistinguishable between eve2+7 and eve2+7mutHb (p-value = 0.304, Mann-Whitney U test) (Fig 3C).
Given the subtle effects that Hb binding site mutations had in eve2+7, we examined the behavior of other eve stripe 2 enhancer fragments in the literature (see S2 Appendix for annotated enhancer sequences). The eve stripe 2 fragment used in comparative work is smaller than eve2+7 by 100 base pairs on either side, but contains all three binding sites that we mutated in eve2+7mutHb [46]. When we mutated these binding sites in this fragment, we observed a significant decrease in stripe 2 expression level (S3 Fig). The minimal eve stripe 2 enhancer (eve2min) is smaller still and contains only one of the sites mutated in eve2+7mutHb. Previous work using qualitative methods has suggested that mutating this binding site decreases expression driven by eve2min [23,47], which has been central to cartoon models of eve stripe 2 in textbooks [45] and computational models of enhancer function in the field [31,34]. However, using our quantitative methods, we observed no significant decrease in expression level as a result of mutating this Hb binding site in eve2min (S3 Fig). These data indicate that Hb binding site mutations in eve stripe 2 enhancers have small, context-dependent effects on gene expression. Taken together, our results further suggest that while Hb may be a weak activator of stripe 2, it does not influence stripe 7 expression at endogenous Hb levels in either eve3+7 or eve2+7.
Mutations in predicted Caudal binding sites prevent repression of eve2+7 by Hb
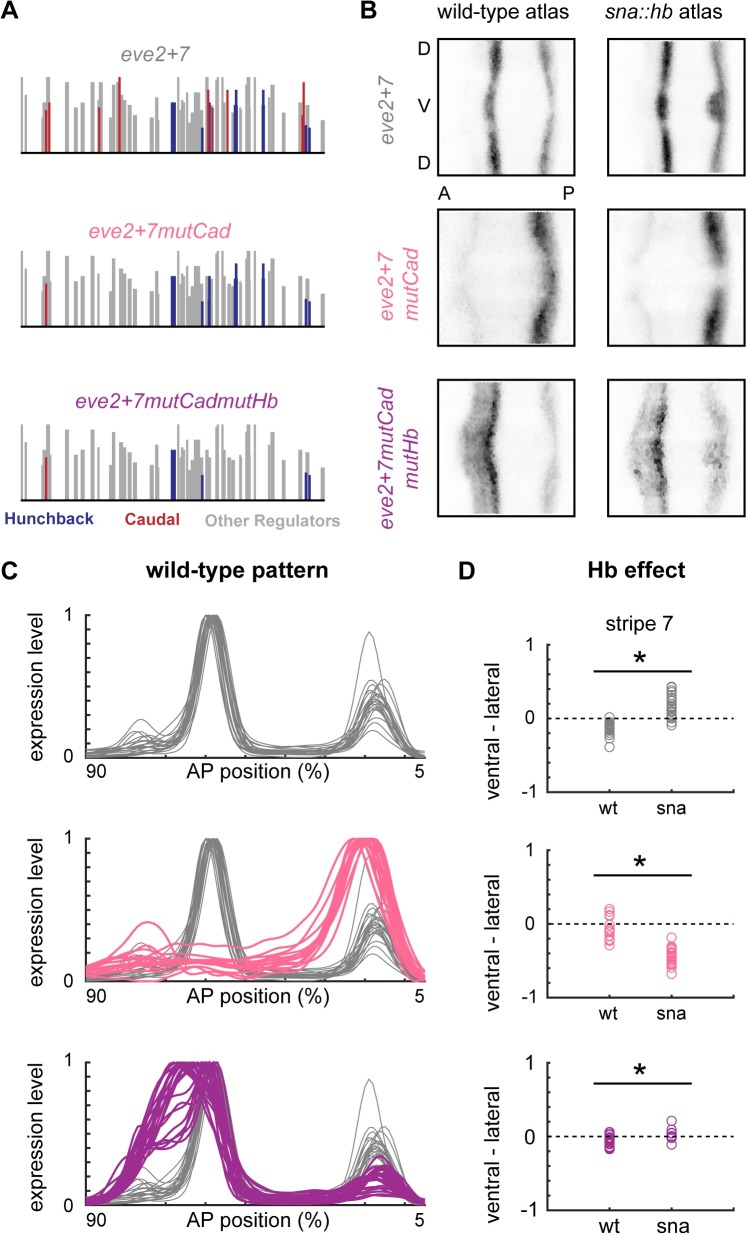
Our data indicate that Hb directly represses eve3+7 but has little effect in eve2+7. One possible explanation for this result is that Hb functions as a weak activator in eve2+7. If this is the case, then the hypothesis that Hb acts as an activator by interacting with Cad (the co-activation hypothesis) makes a strong prediction: mutating Cad sites in eve2+7 should convert Hb from a weak activator to a repressor of that sequence. We tested this hypothesis by mutating predicted Cad binding sites in eve2+7 (eve2+7mutCad, Fig 4A), and measuring gene expression in wild-type and sna::hb embryos. These mutations abolished stripe 2 expression completely and caused an anterior expansion of stripe 7 (Fig 4B and 4C). We hypothesize that this anterior expansion of stripe 7 was caused by unintended mutations in binding sites for the repressor Giant (Gt), whose predicted binding sites overlap with Cad (S4 Fig). In sna::hb embryos, removing Cad binding sites caused the stripe 7 pattern to retreat (Fig 4B and 4D). This result suggests that without Cad, Hb behaves as a repressor in this sequence. To confirm that this effect was due to direct repression by Hb, we mutated both Hb and Cad binding sites in eve2+7 (eve2+7mutCadmutHb, Fig 4A). We found that removing predicted Cad and Hb binding sites restored the stripe 2 expression that was lost following the removal of predicted Cad sites alone (Fig 4B and 4C). We also observed anterior expansions of stripe 2 and stripe 7; we attribute this effect to the unintended loss of a binding site for Gt, as mentioned above (see Discussion). Most importantly, these additional Hb mutations abolished repression by ventral Hb (Fig 4B and 4D). These results therefore support a variation of the co-activation hypothesis where Cad prevents Hb from directly repressing eve2+7.
Fig 4. Hunchback is counter-repressed in eve2+7.
(A) Predicted Cad (red) and Hb (blue) binding sites in wild-type and mutant versions of eve2+7 are shown as in Fig 2. (B) 2D projections of atlas data for reporter constructs expressed in wild-type or sna::hb embryos. Data is taken from timepoint 4. (C) Lateral line traces from individual wild-type embryos containing eve2+7 reporter constructs. Embryos are from all six timepoints in stage 5. (D) Differences between ventral and lateral stripe peaks are plotted for individual wild-type and sna::hb embryos in all 6 timepoints in stage 5. Top: wild-type eve2+7 (wt: n = 22; sna: n = 26); Middle: eve2+7mutCad (wt: n = 15; sna: n = 24); Bottom: eve2+7mutCadmutHb (wt: n = 22; sna: n = 9). Asterisks indicate significant differences between wild-type and sna::hb embryos (p-value < 0.001, Mann-Whitney U test).
If Cad binding to eve2+7 prevents Hb repression, removing Cad from the embryo should result in the loss of eve stripe 2 expression. However, previous work examining endogenous eve expression in Cad mutant embryos did not describe any effect on stripe 2 [38]. We therefore attempted to knock-down cad expression in embryos by expressing short hairpin RNAs targeting cad (cad RNAi embryos). However, we observed no qualitative changes in endogenous eve expression in cad RNAi embryos (S5 Fig), suggesting a low knock-down efficiency of our RNAi method. As cad is both maternally and zygotically expressed [35,36,48,49], these results are consistent with previous observations that this method is effective at depleting maternally deposited transcripts, but not transcripts that are zygotically expressed in the early embryo [50]. We therefore cannot rule out the possibility that some sequence feature other than Cad binding sites controls Hb activity (see Discussion).
Predicted Hunchback and Caudal binding sites co-evolve in orthologous eve stripe 2 enhancers
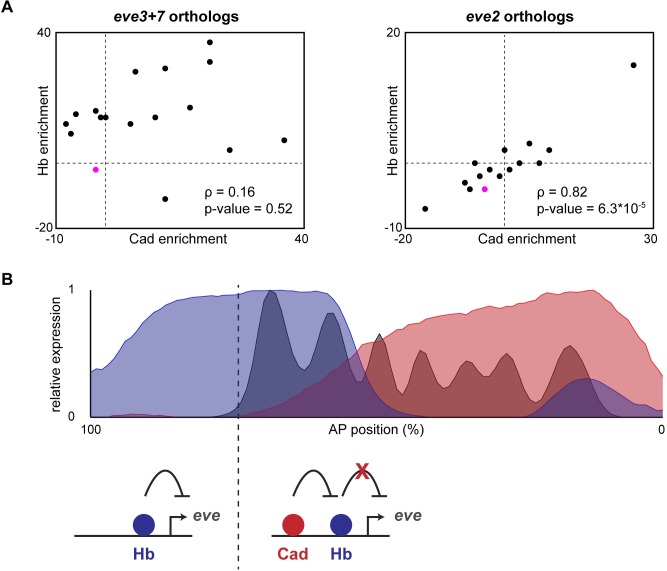
Our results suggest that predicted Cad sites in eve2+7 prevent Hb from repressing eve stripe 2. We therefore hypothesized that the balance between Hb and Cad binding sites is under selective constraint, which would result in Hb and Cad binding sites co-evolving in eve stripe 2 orthologs. To test this hypothesis, we predicted Cad and Hb binding sites in orthologous eve stripe 2 enhancers previously identified in Drosophilid and Sepsid genomes [51], and calculated how much they were enriched compared to other regions of open chromatin during stage 5 of embryogenesis in Drosophila melanogaster [52]. We found that Cad and Hb enrichment scores were significantly correlated in orthologous eve stripe 2 enhancers (Fig 5A). In contrast, we did not observe any correlation between Hb and Cad binding sites in orthologous eve3+7 enhancers when using the same PATSER score threshold used for predicted binding sites (Fig 5A). This result is notable since mutating Cad binding sites in eve3+7 decreased expression of both stripes (S6 Fig), which suggests that Cad also directly activates eve3+7. However, we note that the correlation we observe in predicted sites depends on the PATSER score threshold used for identifying functional sites– at higher values for this threshold, we also observe a correlation in eve3+7 orthologs (S7 Fig). Finally, we used the same method to test for a correlation in Hb and Bcd enrichment scores for eve2 and eve3+7 orthologs, but we observed no significant correlation in either case (S8 Fig). Taken together, these results suggest that counter-repression of Hb by Cad is a conserved feature of eve stripe 2 enhancers, although we cannot rule out the possibility that a similar interaction also occurs in eve3+7 (see Discussion).
Fig 5. Caudal and Hunchback binding sites co-evolve in orthologous eve stripe 2 enhancers.
(A) Enrichment scores for predicted Hb and Cad binding sites in orthologous eve stripe 2 and eve3+7 sequences from different Drosophilid and Sepsid species [51]. Scores were calculated by comparing the number of predicted sites to an expected value calculated from the genomic background [52]. Scores for Drosophila melanogaster enhancers are indicated in magenta. Spearman correlation and p-value are displayed for each set of enhancers. (B) Summary of current findings. Top: levels of Hb protein (blue), Cad protein (red) and Eve protein (grey) are plotted as a function of anterior-posterior position. Data were taken from the FlyEx database [75]. Bottom: cartoon indicating Hb function in eve2+7 at different locations in the embryo. In the anterior, Cad levels are low, so Hb represses eve2+7. In the trunk, Cad binding to eve2+7 prevents Hb repression.
Discussion
The eve enhancers are often used to teach the fundamental principles of patterning [45,53]. Hunchback (Hb) is a key regulator of eve; it is thought to activate the eve stripe 2 enhancer in concert with Bicoid (Bcd), while it acts as a repressor at other enhancers, such as eve3+7 [24,26,54]. Here, we coupled quantitative imaging and systematic perturbations of regulatory DNA and TF expression to uncover the DNA sequence features that control Hb activity. We suggest that Hb activity is controlled by a second TF, Caudal (Cad), and discuss the implications for eve stripe 2 regulation and evolution.
Revising our picture of eve stripe 2 regulation
In the textbook picture of eve stripe 2 [45], Bcd and Hb directly activate the enhancer in the anterior of the embryo, while Gt and Kr directly repress it to carve out a single stripe of expression [14,22,23]. However, attempts to reconstitute the minimal eve stripe 2 enhancer from its component binding sites have failed, which suggests that there is still more to learn about its regulatory logic [55]. For example, these interactions alone cannot explain how eve stripe 2 is repressed in the anterior tip of the embryo. Two other mechanisms have been proposed to account for this discrepancy: direct repression by Slp1 and downregulation of Bcd activity by Torso [56]. Our results suggest an additional mechanism: Hb may repress eve2+7 in the anterior, possibly due to the absence of Cad (Fig 5B). In our data, eve2+7mutHb generates low level expression anterior to stripe 2 (Fig 3B, S2 Fig), which suggests that Hb may function with other factors in defining the anterior boundary of the stripe [22]. However, these results may also be due to mutations in binding sites for other factors, including Gt [46]. Indeed, we observe that mutating Hb and Cad binding sites in eve2+7 causes a dramatic anterior expansion of both stripes, which we hypothesize is partially due to unintended mutations in Gt binding sites that overlap predicted Cad sites (see below).
Bcd and Hb have been proposed to synergistically activate eve stripe 2 [14,30], but our results do not support this hypothesis. Computational models that include Bcd/Hb synergy predict that Hb binding site mutations cause large decreases in expression [31]. However, our data suggest only modest effects of Hb binding site mutations in accordance with qualitative data in the literature [23,47]. In addition, our results do not support the previous model for Bcd/Hb synergy in eve2+7; without predicted Cad sites, Hb represses eve2+7 even though Bcd binding sites are the same.
Hb has long been considered a critical activator of eve stripe 2, but our results suggest that its function is more complicated. First, mutating a footprinted Hb site in the context of the minimal eve stripe 2 enhancer did not affect expression level (S3 Fig), which directly contradicts previous qualitative data in the literature [23]. Mutating the same site and two others in the context of eve2+7 causes a slight decrease in the expression of stripe 2 but not stripe 7 (Fig 3C). From these data, we see that three Hb binding sites in eve2+7 may perform two functions with quantitatively small effects: weak activation of eve stripe 2 and repression of low-level expression anterior to eve stripe 2. Other Hb binding sites in eve2+7 of lower predicted affinity may also contribute to these functions, but we did not mutate these sites to test this hypothesis. Because our findings rely on reporter constructs, it remains to be seen which quantitative features of eve stripe expression are necessary in the endogenous context for development to proceed properly. Thus, we cannot rigorously address whether the presence of Hb binding sites in eve2+7 confers a selective advantage, but this question would be interesting to examine in future work.
Discrepancies between Caudal perturbations in cis and trans
Developmental enhancers are complex, and dissecting their regulatory logic with either cis or trans perturbations carries caveats. Making sequence perturbations in cis can disrupt nearby or overlapping sites for known or unknown regulators. In addition, mutating or misexpressing regulators in trans can affect the location, level and function of other genes in the network [57]. These caveats are important to consider in this study, since endogenous eve stripe 2 is expressed in cad mutant embryos [38], which conflicts with our result that mutations in Cad binding sites in eve2+7 abolish stripe 2 expression. While our binding site mutations may have disrupted other sequence features that are necessary for counter-repression, this discrepancy may be explained in other ways. First, hb expression may be affected in a cad mutant background. While hb expression is detectable in this background by qualitative in situ hybridization [38], hb expression levels may be lower, which may affect its capacity to repress stripe 2. Second, the piece of DNA that we tested in reporters may not contain all relevant DNA that contributes to eve stripe 2 expression. Indeed, in the endogenous eve locus, the intervening DNA between eve2+7 and eve3+7 contains binding sites for many eve regulators, but is notably devoid of predicted Hb binding sites [41]. Therefore, sequence adjacent to eve2+7 may not be affected by Hb repression in the absence of cad. Finally, the placement of the eve2+7 enhancer adjacent to the promoter in our reporter constructs may allow Hb to exert a stronger repressive effect than in the endogenous locus.
In this study, we combined cis perturbations to enhancer sequence and trans perturbation of upstream regulators to mitigate the drawbacks of each method. In our previous work, discrepancies between the eve3+7 reporter pattern and the endogenous stripes revealed the influence of eve2+7 as a stripe 7 shadow enhancer [41]; further dissection of stripe behavior in cad mutant embryos, using both reporter constructs and perturbations to the endogenous locus, may yield additional insights into eve regulatory logic.
Counter-repression may influence regulatory evolution
Our results indicate that Hb binding sites in eve2+7 can repress stripe 2. Previous work has shown that the absence of eve stripe 2 is lethal in Drosophila melanogaster [58]. It follows that selective mechanisms should operate to prevent repression of stripe 2 by Hb. One such mechanism could be selection against Hb sites in eve2. Indeed, we find that Hb binding sites are underrepresented in the eve stripe 2 enhancer sequence from Drosophila melanogaster (Fig 5A) [51]. Similarly, most eve3+7 and eve4+6 orthologs are depleted in binding sites for the repressor Kr, which overlaps stripes 3 and 4, while all eve stripe 2 enhancers are enriched in Kr binding sites, which are thought to define the posterior border of the stripe 2 pattern (S3 Table). These results suggest that selection may generally operate against binding sites for repressors that spatially overlap a developmental expression pattern.
However, our findings suggest that counter-repression may provide an additional mechanism that prevents repression of stripe 2 by Hb. We speculate that counter-repression may be useful, in that it allows for the intrusion of Hb sites over evolutionary time. Hb sites may arise because they play important functional roles in patterning the blastoderm embryo, or at other stages of development (e.g.[59]).
The correlation we observe between enrichment scores for predicted Cad and Hb sites in orthologous eve stripe 2 enhancers may reflect counter-repression in that enhancer specifically. However, this bioinformatic signature is not enough to rule out the possibility that Cad also counter-represses Hb in eve3+7. Sensitivity analyses indicate that the correlation of Hb and Cad binding sites in eve stripe 2 orthologs is highest when sites with lower predicted affinity (i.e. lower PATSER scores) are included, while in eve3+7 orthologs we find that correlation values are highest when only sites of high predicted affinity are included (S7 Fig). These results may reflect differences in the constraints on the regulatory logic of eve2+7 and eve3+7: for example, if Hb repression in eve2+7 must be prevented or counteracted by Cad to allow for stripe 2 expression, whereas Hb repression in eve3+7 is necessary for correct positioning of the stripes but may be modulated by Cad binding. Nonetheless, the co-evolutionary signature we observe in eve stripe 2 orthologs suggests that a functional interaction between Cad and Hb sites may be a conserved feature of eve stripe 2 regulation.
Hb ‘bifunctionality’ is due to counter-repression
Our previous work indicated that Hb activates eve2+7 and represses eve3+7. Because Hb was “known” to be an activator of eve2min and repressor of eve3+7, we hypothesized that these interactions were direct. Here, we find that Hb activation of eve stripe 7 is indirect. We now hypothesize that the sna::hb perturbation causes retreat of Gt and subsequent anterior expansion of the stripe 7 pattern [41]. Knirps forms the anterior boundary of stripe 7 through the eve3+7 enhancer [26], while Gt has been proposed to form the anterior boundary of stripe 7 through eve2+7 [31]. Indeed, we believe that the anterior stripe 7 expansion we observe after mutating predicted Cad sites in eve2+7 is caused by unintended mutations in Gt binding sites. Predicted Gt sites overlap with predicted Cad sites in this sequence, and our mutations disrupt a Cad site and a predicted Gt site that has been confirmed by footprinting in eve2min (S4 Fig) [60]. It is challenging to mutate Cad sites without impacting Gt sites because measured binding preferences for Gt vary greatly between methods and do not exactly correspond to footprinted sites in the literature [22,61]. While these challenges make it difficult to determine whether Cad directly activates eve2+7, they do not affect our conclusions concerning Hb counter-repression. Indeed, the anterior expansion of stripe 7 makes the repressive influence of ventral Hb even more striking (Fig 4B and 4D).
Which TFs activate eve stripe 7 expression is less clear, though Cad and Zelda are obvious candidates shared by eve2+7 and eve3+7. Overall, the conclusion of our previous work—that eve stripe 7 shadow enhancers use different regulatory logic—still holds [41]. Hb represses eve3+7 but has little effect on eve2+7 because of counter-repression. Furthermore, we hypothesize that the two enhancers use different repressors to set the anterior border, though this remains to be tested directly. We also previously showed that Hb activates and represses separate Kr shadow enhancers using the same sna::hb misexpression assay [52]; it is thus possible that Cad also counter-represses Hb in the proximal Kr enhancer.
Cad may counter-repress Hb through any number of molecular mechanisms. For example, Cad may interfere with Hb via direct protein-protein interactions. The simplest idea is that when Cad is present, Hb does not bind. This hypothesis is not supported by genome-wide chromatin immunoprecipitation (ChIP) data for both Hb and Cad, as both are bound to eve2+7 [62,63]. We note two caveats of this data. First, it is not spatially resolved, so it remains possible that Hb binding is affected in a subset of cells. Second, Cad may disrupt Hb binding quantitatively, which may be enough to affect function but not enough to detect changes by ChIP. Another possibility is that direct protein-protein interactions between Cad and Hb interfere with Hb protein domains that execute its repressive function [21]. Finally, interactions could occur indirectly through co-regulators or mechanisms analogous to the effects of short-range repressors on nearby activators [64]. Importantly, other counter-repressors have been identified in Drosophila melanogaster development. Stat92E functions as a counter-repressor in the formation of the posterior spiracle, but its target repressor remains unknown [65]. Different counter repressors may exert their effects through different molecular mechanisms.
A long-term goal of studying gene regulation is predicting the output of a specific enhancer [10]. Context-dependent TF function [e.g. [66]] presents a challenge for meeting this goal. Here, we further elucidate counter-repression, one type of context dependence, and offer some strategies for uncovering it in regulatory DNA. First, single mutations of annotated activators or bifunctional TFs may be misleading. Analyzing the effects of single and double mutations together are necessary to classify activators, repressors, counter-repressors and the targets of counter-repression. For example, let’s say that you mutate binding sites for TF #1 and expression decreases. TF #1 may either be an activator or a counter-repressor. You now combine mutations in TF #1 with mutations in TF #2, and expression is restored. TF #2 may be a repressor or the target of counter-repression. Finally, you examine the effect of mutating TF #2 alone. If TF #2 is a repressor, expression will increase, but if TF #2 is the target of counter-repression, there will be no effect. All three experiments are necessary to correctly classify both TF #1 and TF #2. High-throughput mutational studies both in reporters [9] and in vivo [67] may be able to systematically gather this type of data on combinatorial effects.
Conclusion
By combining targeted genetic perturbations with quantitative expression measurements, we uncovered counter-repression of Hb as a key feature of eve stripe 2 regulation. These results emphasize the complexity of animal developmental enhancers, and argue that the current textbook model of eve stripe 2 regulation is incomplete. We suggest that counter-repression increases the flexibility in the regulatory logic of eve stripe 2, which may impact its plasticity over evolutionary time.
Materials and methods
Binding site predictions and construct design
We used PATSER [68] to predict binding sites in eve3+7 and eve2+7 for blastoderm TFs. We used position weight matrices (PWMs) derived from bacterial 1-hybrid experiments for the following factors: Bicoid, Caudal, Dichaete, Stat92E, Hunchback, Krüppel, and Nubbin [69]. We used other published PWMs for Giant, Knirps, and Tailless [70]. Finally, we used a Zelda PWM from a personal communication with Christine Rushlow. Count matrices were converted into frequency matrices for use in PATSER using a pseudocount of 1. In designing binding site mutations, we predicted the effects on all predicted sites above a p-value cutoff of 0.003. In an effort to minimize impacts on other binding sites, we chose to leave some low affinity sites unmutated in both eve3+7 and eve2+7. Annotated sequences of wild-type and mutated versions of enhancers can be found in S1 Appendix (eve3+7 and eve2+7 enhancers) and S2 Appendix (eve stripe 2 enhancers). We have posted csv files containing binding site predictions for all enhancers on Figshare, along with the frequency matrices we used for designing mutations (https://figshare.com/articles/Quantitative_Data_and_Analyses_for_Vincent_et_al_2018/6233600).
Fly work
All reporter constructs were cloned into the NotI/BglII insertion site in the pBΦY reporter plasmid [51] and integrated into the attP2 landing site using injection services provided by BestGene Inc [71]. Successful transformants were homozygosed using the mini-white marker. We generated sna::hb embryos as described previously [24] by crossing virgin females containing a male germline-specific β2-tubulin–FLP transgene with males containing two copies of the sna::hb transgene flanked by FRT sites, both generous gifts from Steve Small. F1 males were then crossed to virgin females homozygous for the enhancer reporter construct of interest. We generated cad RNAi embryos as described previously [50] by crossing virgin females containing a mat-tub-GAL4 driver with males containing short hairpin RNAs for cad driven by an upstream activating sequence (Bloomington Stock #34702). F1 virgin females were then crossed to males containing eve2+7 reporter constructs to generate cad RNAi embryos.
In situ hybridization and data analysis
Embryos were collected, fixed in heptane and paraformaldehyde, stained using quantitative in situ hybridization, staged into 6 timepoints using membrane invagination as a morphological marker, imaged using 2-photon confocal microscopy and processed to extract cellular resolution expression measurements as previously described [42,72]. For each embryo, we measure fushi-tarazu mRNA using an anti-digoxigenin horseradish peroxidase (HRP) antibody (Roche) and a coumarin-tyramide color reaction (PerkinElmer), as well as lacZ mRNA using an anti-2,4-dinitrophenyl HRP antibody (PerkinElmer) and a Cy3-tyramide color reaction (PerkinElmer). The sna::hb crossing scheme generates wild-type and mutant embryos in approximately equal proportions; we identified embryos misexpressing hb using the ftz expression pattern. For individual genotypes, we use a previously described computational pipeline to average measurements from multiple embryos together to generate gene expression atlases [43], which we display as 2-dimensional projections in this study. For atlas building, we used the coarse registration method for all genotypes because we lacked a ftz expression template for sna::hb embryos. To quantify the effect of ventral Hb in individual embryos, we normalized cellular expression measurements by the maximum value and subtracted the peak stripe expression value along the lateral side from the peak value along the ventral side. For experiments involving a hkb co-stain, we stained embryos in the same batch, discarded outliers, and ensured that hkb levels and lacZ levels were significantly correlated as described previously [44,52]. Means and standard errors for our quantitative measurements can be found in S1 and S2 Tables. We have deposited the processed expression data for all embryos, which we call pointclouds, along with the MATLAB code used to generate all data figures in this study on Figshare (see link above).
Binding site enrichment analysis
Binding site enrichment was calculated as described previously [52]. In summary, we used PWMs from FlyFactorSurvey [61] to predict binding sites in orthologous eve enhancers, and compared that result with a null expectation for an enhancer of identical size that we generated by predicting sites in accessible areas of the genome during state 5 of embryogenesis [73]. This method depends on the PATSER score threshold used to identifying functional binding sites, which is difficult to define rigorously. We therefore calculated enrichment scores at all integer values for this threshold between 1 and the maximum score for the PWM; this method amounts to a sensitivity analysis for the PATSER score threshold. We have deposited our binding site predictions for all orthologous eve enhancers in csv format along with the MATLAB code used to calculate enrichment scores on Figshare (see link above).
Supporting information
(A) Predicted Hb binding sites (blue) in eve3+7 and eve3+7mutHb are plotted as in Fig 2. eve3+7mutHb sequence was taken from [26]. (B) 2D projections of atlas data for reporter constructs expressed in WT or sna::hb embryos. Data is taken from timepoint 4 (25–50% membrane invagination). Low-level anterior and posterior expression is due to an unused hkb co-stain. (C) Lateral line traces from individual wild-type embryos containing eve3+7 reporter constructs (WT: grey, n = 26; mutHb: blue, n = 11). Each trace is normalized to its maximum value. Embryos are from all six timepoints in stage 5. (D) Differences in the maximum values of ventral and lateral line traces are plotted for individual wild-type and sna::hb embryos containing eve3+7mutHb in all 6 timepoints in stage 5. wt: n = 11; sna::hb: n = 12. Differences between wild-type and sna::hb embryos were not significant (Mann-Whitney U test, p-value > 0.1 for both stripes).
(TIF)
Wild-type: grey, n = 47; mutHb: cerulean, n = 42; mutCad: salmon, n = 35; mutCadmutHb: mauve, n = 32. Each trace is normalized to its maximum value. Embryos are from all six timepoints in stage 5. Expression anterior to 90% and posterior to 5% AP length is not shown and may be partially due to an unused huckebein co-stain in some embryos.
(TIF)
(A) Predicted binding sites for Hb (blue) and other eve2+7 regulators (grey) in different eve stripe 2 enhancer constructs (46,47). (B) Peak stripe 2 expression levels for individual embryos from timepoints 2–4 (4–50% membrane invagination) were measured using a hkb co-stain method (44). Asterisks indicate p-values < 0.05 (Mann-Whitney U test). Note that because each experiment was performed in separate hybridizations, comparisons can only be made between wild-type and mutated versions of the same enhancer, not between different stripe 2 enhancers.
(TIF)
Predicted binding for Cad (red) and Gt (lilac) are shown in eve2+7 and eve2+7mutCad. Many predicted Giant binding sites are near predicted Cad sites. One Cad binding site mutation in eve2+7mutCad (grey box) disrupts a predicted Gt binding site that also overlaps an annotated Gt binding site [47].
(TIF)
2D projections of eve expression data from representative wild-type (left) and cadRNAi embryos. Both embryos were from timepoint 3 (15% membrane invagination).
(TIF)
(A) Predicted Cad binding sites in eve3+7 and eve3+7mutCad. Sites were predicted and displayed as described in previous figures. (B) Lateral line traces from individual wild-type embryos containing reporter constructs for eve3+7 (grey, n = 11) and eve3+7mutCad (red, n = 7). Traces were normalized using a co-stain method [44]; embryos are from timepoints 3 and 4 (9–50% membrane invagination). (C) Individual stripe peaks were found by taking local maxima from line traces in B. Asterisks indicate significant differences in stripe level (p-values < 0.001, Mann-Whitney U test).
(TIF)
(A) Spearman correlation values (rho) of Cad and Hb binding site enrichment scores are plotted as a function of binding site threshold for eve2 and eve3+7 orthologs. Binding site threshold refers to the minimum PATSER score for a predicted site to be counted in the analysis. Higher PATSER scores are assumed to reflect higher affinity sites. (B) Hb and Cad enrichment values are plotted for individual eve2 and eve3+7 orthologs at a binding site threshold of 7.2 –the threshold that maximizes rho in eve3+7 orthologs.
(TIF)
Spearman correlation values (rho) of Bcd and Hb binding site enrichment scores are plotted as a function of binding site threshold for eve2 (left) and eve3+7 (right) orthologs. Enrichment scores for Bcd and Hb sites are not significantly correlated at any binding site threshold in either eve2 or eve3+7 orthologs.
(TIF)
Here, we list the sequences for the eve3+7 and eve2+7 reporter constructs used in the study. Predicted Hb binding sites are indicated in blue, and predicted Cad binding sites are indicated in red. We have indicated any base pairs that are shared by both a Hb binding site and a Cad binding site in purple. As detailed in the Materials and Methods, we did not mutate all predicted sites for each factor because we tried to minimize the impact on previously annotated or predicted sites for other eve regulators. PATSER scores for all predicted sites in each enhancer can be found on Figshare (see link above).
(DOCX)
Here, we have listed the sequences for eve stripe 2 reporter constructs included in our study. Previously identified Hb binding sites are indicated in blue [46].
(DOCX)
We quantify the effect of ventral Hb by comparing the difference in ventral and lateral peak expression levels between wild-type and sna::hb embryos (see Fig 2 and Materials and Methods).
(XLSX)
We compare relative stripe levels between embryos expressing different reporter constructs by normalizing lacZ expression using a huckebein co-stain [44,52]. Using this method, only comparisons between embryos stained in the same hybridization batch are meaningful. Samples contained in the same batch are grouped together in the table.
(XLSX)
We calculated Kr enrichment scores for orthologs using two different PATSER score thresholds. Positive values indicate an overrepresentation in Kr binding sites compared to a null model, while negative values indicate underrepresentation. In general, Kr binding sites are enriched in eve2 orthologs and poorly enriched or depleted in eve3+7 and eve4+6 orthologs. Kr is a repressor that is thought to define the posterior border of stripe 2 [14,47], but overlaps the expression patterns driven by eve3+7 and eve4+6 [15,16].
(XLSX)
Acknowledgments
The authors gratefully acknowledge Steve Small for the sna::hb flies, as well as all members of the DePace Lab for helpful comments on the manuscript. Clarissa Scholes deserves particular recognition for her incisive and thorough commentary. Finally, we would like to acknowledge our editor and two anonymous reviewers, whose comments and suggestions greatly improved the clarity and rigor of the manuscript.
Data Availability
Raw data for this paper is available on Figshare.com (https://figshare.com/articles/Quantitative_Data_and_Analyses_for_Vincent_et_al_2018/6233600) along with the code used to generate the figures.
Funding Statement
This work was generously supported by the Albert J. Ryan Foundation (to BJV), the Harvard Herchel Smith Graduate Student Fellowship (to MVS), the Research Scholar Initiative (to FL-R), the Jane Coffin Childs Memorial Fund for Medical Research (to ZW), the Novartis Fellows Fund (to JE), the Giovanni Armenise Foundation (to AHD), the National Institutes of Health (U01GM103804, R21HD072481, and R01GM122928 to AHD, K99/R00 HD73191 to ZW, and F32GM128310 to TTH), and the National Science Foundation (CAREER-IOS 1452557 and Collaborative Research MCB 1715184 to AHD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hong J-W, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci U S A. 2008;105: 20072–20076. 10.1073/pnas.0806476105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1: 81–84. 10.4161/trns.1.2.12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aradhya S, Nelson DL. NF-κB signaling and human disease. Curr Opin Genet Dev. 2001;11: 300–306. [DOI] [PubMed] [Google Scholar]
- 4.Hui C-C, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27: 513–537. 10.1146/annurev-cellbio-092910-154048 [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Rushlow CA, Zhou Q, Small S, Levine M. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 1992;11: 3147–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72: 741–752. [DOI] [PubMed] [Google Scholar]
- 7.Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, et al. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997;11: 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324: 407–410. 10.1126/science.1164265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White MA, Kwasnieski JC, Myers CA, Shen SQ, Corbo JC, Cohen BA. A Simple Grammar Defines Activating and Repressing cis-Regulatory Elements in Photoreceptors. Cell Rep. 2016;17: 1247–1254. 10.1016/j.celrep.2016.09.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HD, Shay T, O’Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325: 429–432. 10.1126/science.1171347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287: 795–801. [DOI] [PubMed] [Google Scholar]
- 12.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48: 1272–1278. 10.1016/j.molimm.2011.03.006 [DOI] [PubMed] [Google Scholar]
- 13.Jaeger J. The gap gene network Cell Mol Life Sci. SP Birkhäuser Verlag; Basel; 2011;68: 243–274. 10.1007/s00018-010-0536-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5: 827–839. [DOI] [PubMed] [Google Scholar]
- 15.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175: 314–324. 10.1006/dbio.1996.0117 [DOI] [PubMed] [Google Scholar]
- 16.Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126: 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palsson A, Wesolowska N, Reynisdóttir S, Ludwig MZ, Kreitman M. Naturally occurring deletions of hunchback binding sites in the even-skipped stripe 3+7 enhancer. PLoS One. Public Library of Science; 2014;9: e91924 10.1371/journal.pone.0091924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White RA, Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986;47: 311–321. [DOI] [PubMed] [Google Scholar]
- 19.Zhang CC, Bienz M. Segmental determination in Drosophila conferred by hunchback (hb), a repressor of the homeotic gene Ultrabithorax (Ubx). Proc Natl Acad Sci U S A. 1992;89: 7511–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Vasisht V, Kosman D, Reinitz J, Small S. Thoracic patterning by the Drosophila gap gene hunchback. Dev Biol. 2001;237: 79–92. 10.1006/dbio.2001.0355 [DOI] [PubMed] [Google Scholar]
- 21.Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137: 1421–1430. 10.1242/dev.048678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254: 1385–1387. [DOI] [PubMed] [Google Scholar]
- 23.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122: 205–214. [DOI] [PubMed] [Google Scholar]
- 24.Clyde DE, Corado MSG, Wu X, Pare A, Papatsenko D, Small S. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. Nature Publishing Group; 2003;426: 849–853. 10.1038/nature02189 [DOI] [PubMed] [Google Scholar]
- 25.Stanojević D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. Nature Publishing Group; 1989;341: 331–335. 10.1038/341331a0 [DOI] [PubMed] [Google Scholar]
- 26.Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, Small S. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development. The Company of Biologists Limited; 2011;138: 4291–4299. 10.1242/dev.065987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papatsenko D, Levine MS. Dual regulation by the Hunchback gradient in the Drosophila embryo. Proc Natl Acad Sci U S A. National Acad Sciences; 2008;105: 2901–2906. 10.1073/pnas.0711941105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilsley GR, Fisher J, Apweiler R, De Pace AH, Luscombe NM. Cellular resolution models for even skipped regulation in the entire Drosophila embryo. Elife. 2013;2: e00522 10.7554/eLife.00522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarty AS, Kleiger G, Eisenberg D, Smale ST. Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell. 2003;11: 459–470. [DOI] [PubMed] [Google Scholar]
- 30.Simpson-Brose M, Treisman J, Desplan C. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78: 855–865. [DOI] [PubMed] [Google Scholar]
- 31.Janssens H, Hou S, Jaeger J, Kim A-R, Myasnikova E, Sharp D, et al. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet. Nature Publishing Group; 2006;38: 1159–1165. 10.1038/ng1886 [DOI] [PubMed] [Google Scholar]
- 32.He X, Samee MAH, Blatti C, Sinha S. Thermodynamics-based models of transcriptional regulation by enhancers: the roles of synergistic activation, cooperative binding and short-range repression. Ohler U, editor. PLoS Comput Biol. 2010;6: e1000935 10.1371/journal.pcbi.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samee MAH, Sinha S. Quantitative modeling of a gene’s expression from its intergenic sequence. Tanay A, editor. PLoS Comput Biol. 2014;10: e1003467 10.1371/journal.pcbi.1003467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim A-R, Martinez C, Ionides J, Ramos AF, Ludwig MZ, Ogawa N, et al. Rearrangements of 2.5 kilobases of noncoding DNA from the Drosophila even-skipped locus define predictive rules of genomic cis-regulatory logic. Levine M, editor. PLoS Genet. 2013;9: e1003243 10.1371/journal.pgen.1003243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivera-Pomar R, Lu X, Perrimon N, Taubert H, Jäckle H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376: 253–256. 10.1038/376253a0 [DOI] [PubMed] [Google Scholar]
- 36.Häder T, La Rosée A, Ziebold U, Busch M, Taubert H, Jäckle H, et al. Activation of posterior pair-rule stripe expression in response to maternal caudal and zygotic knirps activities. Mech Dev. 1998;71: 177–186. [DOI] [PubMed] [Google Scholar]
- 37.Copf T, Schröder R, Averof M. Ancestral role of caudal genes in axis elongation and segmentation. Proc Natl Acad Sci U S A. National Acad Sciences; 2004;101: 17711–17715. 10.1073/pnas.0407327102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, et al. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development. 2006;133: 3973–3982. 10.1242/dev.02576 [DOI] [PubMed] [Google Scholar]
- 39.Guo R-J, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3: 593–601. [DOI] [PubMed] [Google Scholar]
- 40.Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci U S A. 2004;101: 7641–7645. 10.1073/pnas.0401654101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staller MV, Vincent BJ, Bragdon MDJ, Lydiard-Martin T, Wunderlich Z, Estrada J, et al. Shadow enhancers enable Hunchback bifunctionality in the Drosophila embryo. Proc Natl Acad Sci U S A. National Acad Sciences; 2015;112: 785–790. 10.1073/pnas.1413877112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luengo Hendriks CL, Keränen SVE, Fowlkes CC, Simirenko L, Weber GH, DePace AH, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol. BioMed Central Ltd; 2006;7: R123 10.1186/gb-2006-7-12-r123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fowlkes CC, Hendriks CLL, Keränen SVE, Weber GH, Rübel O, Huang M-Y, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133: 364–374. 10.1016/j.cell.2008.01.053 [DOI] [PubMed] [Google Scholar]
- 44.Wunderlich Z, Bragdon MD, DePace AH. Comparing mRNA levels using in situ hybridization of a target gene and co-stain. Methods. 2014;68: 233–241. 10.1016/j.ymeth.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, et al. Molecular Biology of the Cell, Sixth Edition Garland Science; 2014. [Google Scholar]
- 46.Ludwig MZ, Patel NH, Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998;125: 949–958. [DOI] [PubMed] [Google Scholar]
- 47.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11: 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macdonald PM, Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986;324: 537–545. 10.1038/324537a0 [DOI] [PubMed] [Google Scholar]
- 49.Schulz C, Tautz D. Zygotic caudal regulation by hunchback and its role in abdominal segment formation of the Drosophila embryo. Development. 1995;121: 1023–1028. [DOI] [PubMed] [Google Scholar]
- 50.Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, et al. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. Genetics Society of America; 2013;193: 51–61. 10.1534/genetics.112.144915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. Perrimon N, editor. PLoS Genet. 2008;4: e1000106 10.1371/journal.pgen.1000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wunderlich Z, Bragdon MDJ, Vincent BJ, White JA, Estrada J, DePace AH. Krüppel Expression Levels Are Maintained through Compensatory Evolution of Shadow Enhancers. Cell Rep. 2015; 10.1016/j.celrep.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilbert SF. Developmental Biology. Sinauer Associates Incorporated; 2013. [Google Scholar]
- 54.Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57: 413–422. [DOI] [PubMed] [Google Scholar]
- 55.Vincent BJ, Estrada J, DePace AH. The appeasement of Doug: a synthetic approach to enhancer biology. Integr Biol. Royal Society of Chemistry; 2016; 10.1039/C5IB00321K [DOI] [PubMed] [Google Scholar]
- 56.Andrioli LPM, Vasisht V, Theodosopoulou E, Oberstein A, Small S. Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development. 2002;129: 4931–4940. [DOI] [PubMed] [Google Scholar]
- 57.Staller MV, Fowlkes CC, Bragdon MDJ, Wunderlich Z, Estrada J, DePace AH. A gene expression atlas of a bicoid-depleted Drosophila embryo reveals early canalization of cell fate. Development. The Company of Biologists Limited; 2015;142: 587–596. 10.1242/dev.117796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3: e93 10.1371/journal.pbio.0030093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Preger-Ben Noon E, Sabarís G, Ortiz DM, Sager J, Liebowitz A, Stern DL, et al. Comprehensive Analysis of a cis-Regulatory Region Reveals Pleiotropy in Enhancer Function. Cell Rep. 2018;22: 3021–3031. 10.1016/j.celrep.2018.02.073 [DOI] [PubMed] [Google Scholar]
- 60.Gallo SM, Gerrard DT, Miner D, Simich M, Des Soye B, Bergman CM, et al. REDfly v3. 0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. Oxford Univ Press; 2011;39: D118–D123. 10.1093/nar/gkq999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, Basciotta MD, et al. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 2011;39: D111–7. 10.1093/nar/gkq858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradley RK, Li X-Y, Trapnell C, Davidson S, Pachter L, Chu HC, et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 2010;8: e1000343 10.1371/journal.pbio.1000343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paris M, Kaplan T, Li X-Y, Villalta JE, Lott SE, Eisen MB. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. Wittkopp P, editor. PLoS Genet. 2013;9: e1003748 10.1371/journal.pgen.1003748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gray S, Cai H, Barolo S, Levine M. Transcriptional repression in the Drosophila embryo. Philos Trans R Soc Lond B Biol Sci. 1995;349: 257–262. 10.1098/rstb.1995.0111 [DOI] [PubMed] [Google Scholar]
- 65.Pinto PB, Espinosa-Vázquez JM, Rivas ML, Hombría JC-G. JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade. PLoS Genet. 2015;11: e1005412 10.1371/journal.pgen.1005412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stampfel G, Kazmar T, Frank O, Wienerroither S, Reiter F, Stark A. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015;528: 147–151. 10.1038/nature15545 [DOI] [PubMed] [Google Scholar]
- 67.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527: 192–197. 10.1038/nature15521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15: 563–577. [DOI] [PubMed] [Google Scholar]
- 69.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. Oxford University Press; 2008;36: 2547–2560. 10.1093/nar/gkn048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schroeder MD, Greer C, Gaul U. How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development. The Company of Biologists Limited; 2011;138: 3067–3078. 10.1242/dev.062141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keränen SVE, Fowlkes CC, Luengo Hendriks CL, Sudar D, Knowles DW, Malik J, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution II: dynamics. Genome Biol. 2006;7: R124 10.1186/gb-2006-7-12-r124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas S, Li X-Y, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12: R43 10.1186/gb-2011-12-5-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rübel O, Weber GH, Keränen SVE, Fowlkes CC, Hendriks CLL, Simirenko L, et al. PointCloudXplore: Visual Analysis of 3D Gene Expression Data Using Physical Views and Parallel Coordinates. EuroVis. 2006. pp. 203–210. [Google Scholar]
- 75.Pisarev A, Poustelnikova E, Samsonova M, Reinitz J. FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution. Nucleic Acids Res. Oxford University Press; 2009;37: D560–6. 10.1093/nar/gkn717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Predicted Hb binding sites (blue) in eve3+7 and eve3+7mutHb are plotted as in Fig 2. eve3+7mutHb sequence was taken from [26]. (B) 2D projections of atlas data for reporter constructs expressed in WT or sna::hb embryos. Data is taken from timepoint 4 (25–50% membrane invagination). Low-level anterior and posterior expression is due to an unused hkb co-stain. (C) Lateral line traces from individual wild-type embryos containing eve3+7 reporter constructs (WT: grey, n = 26; mutHb: blue, n = 11). Each trace is normalized to its maximum value. Embryos are from all six timepoints in stage 5. (D) Differences in the maximum values of ventral and lateral line traces are plotted for individual wild-type and sna::hb embryos containing eve3+7mutHb in all 6 timepoints in stage 5. wt: n = 11; sna::hb: n = 12. Differences between wild-type and sna::hb embryos were not significant (Mann-Whitney U test, p-value > 0.1 for both stripes).
(TIF)
Wild-type: grey, n = 47; mutHb: cerulean, n = 42; mutCad: salmon, n = 35; mutCadmutHb: mauve, n = 32. Each trace is normalized to its maximum value. Embryos are from all six timepoints in stage 5. Expression anterior to 90% and posterior to 5% AP length is not shown and may be partially due to an unused huckebein co-stain in some embryos.
(TIF)
(A) Predicted binding sites for Hb (blue) and other eve2+7 regulators (grey) in different eve stripe 2 enhancer constructs (46,47). (B) Peak stripe 2 expression levels for individual embryos from timepoints 2–4 (4–50% membrane invagination) were measured using a hkb co-stain method (44). Asterisks indicate p-values < 0.05 (Mann-Whitney U test). Note that because each experiment was performed in separate hybridizations, comparisons can only be made between wild-type and mutated versions of the same enhancer, not between different stripe 2 enhancers.
(TIF)
Predicted binding for Cad (red) and Gt (lilac) are shown in eve2+7 and eve2+7mutCad. Many predicted Giant binding sites are near predicted Cad sites. One Cad binding site mutation in eve2+7mutCad (grey box) disrupts a predicted Gt binding site that also overlaps an annotated Gt binding site [47].
(TIF)
2D projections of eve expression data from representative wild-type (left) and cadRNAi embryos. Both embryos were from timepoint 3 (15% membrane invagination).
(TIF)
(A) Predicted Cad binding sites in eve3+7 and eve3+7mutCad. Sites were predicted and displayed as described in previous figures. (B) Lateral line traces from individual wild-type embryos containing reporter constructs for eve3+7 (grey, n = 11) and eve3+7mutCad (red, n = 7). Traces were normalized using a co-stain method [44]; embryos are from timepoints 3 and 4 (9–50% membrane invagination). (C) Individual stripe peaks were found by taking local maxima from line traces in B. Asterisks indicate significant differences in stripe level (p-values < 0.001, Mann-Whitney U test).
(TIF)
(A) Spearman correlation values (rho) of Cad and Hb binding site enrichment scores are plotted as a function of binding site threshold for eve2 and eve3+7 orthologs. Binding site threshold refers to the minimum PATSER score for a predicted site to be counted in the analysis. Higher PATSER scores are assumed to reflect higher affinity sites. (B) Hb and Cad enrichment values are plotted for individual eve2 and eve3+7 orthologs at a binding site threshold of 7.2 –the threshold that maximizes rho in eve3+7 orthologs.
(TIF)
Spearman correlation values (rho) of Bcd and Hb binding site enrichment scores are plotted as a function of binding site threshold for eve2 (left) and eve3+7 (right) orthologs. Enrichment scores for Bcd and Hb sites are not significantly correlated at any binding site threshold in either eve2 or eve3+7 orthologs.
(TIF)
Here, we list the sequences for the eve3+7 and eve2+7 reporter constructs used in the study. Predicted Hb binding sites are indicated in blue, and predicted Cad binding sites are indicated in red. We have indicated any base pairs that are shared by both a Hb binding site and a Cad binding site in purple. As detailed in the Materials and Methods, we did not mutate all predicted sites for each factor because we tried to minimize the impact on previously annotated or predicted sites for other eve regulators. PATSER scores for all predicted sites in each enhancer can be found on Figshare (see link above).
(DOCX)
Here, we have listed the sequences for eve stripe 2 reporter constructs included in our study. Previously identified Hb binding sites are indicated in blue [46].
(DOCX)
We quantify the effect of ventral Hb by comparing the difference in ventral and lateral peak expression levels between wild-type and sna::hb embryos (see Fig 2 and Materials and Methods).
(XLSX)
We compare relative stripe levels between embryos expressing different reporter constructs by normalizing lacZ expression using a huckebein co-stain [44,52]. Using this method, only comparisons between embryos stained in the same hybridization batch are meaningful. Samples contained in the same batch are grouped together in the table.
(XLSX)
We calculated Kr enrichment scores for orthologs using two different PATSER score thresholds. Positive values indicate an overrepresentation in Kr binding sites compared to a null model, while negative values indicate underrepresentation. In general, Kr binding sites are enriched in eve2 orthologs and poorly enriched or depleted in eve3+7 and eve4+6 orthologs. Kr is a repressor that is thought to define the posterior border of stripe 2 [14,47], but overlaps the expression patterns driven by eve3+7 and eve4+6 [15,16].
(XLSX)
Data Availability Statement
Raw data for this paper is available on Figshare.com (https://figshare.com/articles/Quantitative_Data_and_Analyses_for_Vincent_et_al_2018/6233600) along with the code used to generate the figures.