Abstract
Reliable separation and isolation of target single cells from bodily fluids with high purity is of great significance for an accurate and quantitative understanding of the cellular heterogeneity. Here, we describe a fully integrated single-blood-cell analysis platform capable of size-selective cell separation from a population containing a wide distribution of sizes such as diluted blood sample and highly efficient entrapment of single monocytes. The spiked single U937 cells (human monocyte cell line) are separated in sequence by two different-sized microfilters for removing large cell clumps, white blood cells, and red blood cells and then discriminated by dielectrophoretic force and isolated individually by downstream single-cell trapping arrays. When 2% hematocrit blood cells with a final ratio of 1:1000 U937 cells were introduced under the flow rate of 0.2 ml/h, 400 U937 cells were trapped sequentially and deterministically within 40 s with single-cell occupancy of up to 85%. As a proof-of-concept, we also demonstrated single monocyte isolation from diluted blood using the integrated microfluidic device. This size-selective, label-free, and live-cell enrichment microfluidic single blood-cell isolation platform for the processing of cancer and blood cells has a myriad of applications in areas such as single-cell genetic analysis, stem cell biology, point-of-care diagnostics, and cancer diagnostics.
INTRODUCTION
Understanding intra-sample genomic heterogeneity may hold valuable clues about detailed insight into the origins of human disease pathways and gene expression kinetics that is of great interest in clinical and biomedical communities.1,2 For example, measurement of gene expression by counting single biomolecules from clinical bio-samples such as human tumor tissues3,4 and stem cells5 contributes to the treatment and prevention of major ailments. Additionally, abnormal gene expression of distinct mRNAs can be taken as a good indicator of cellular irregularity. Many analytical cell-based assays, including reverse-transcription quantitative PCR (RT-qPCR), western blot, immunocytochemistry, and enzyme-linked immunosorbent assay (ELISA), measure only the average response from cell population. But the averaging in these measurements masks the intrinsic intra-sample heterogeneity at the single-cell level within cell communities.6,7 This intra-sample heterogeneity provides valuable clues for designing therapeutic administrations and designating treatments for different conditions according to the variability between the responses of patients, which could not be inferred from traditional bulk cell analyses.8–10 Therefore, accurate single-cell phenotyping technologies including isolating, monitoring, and extracting of biomolecules are required to explore the intra-sample heterogeneity caused by stochastic fluctuations in external responses.11,12
For an accurate and quantitative understanding of the cellular heterogeneity, it is important to separate and isolate targeted single-cell populations from the unwanted and contaminated cells and then collect the isolated cells with high purity. Isolation of single cells using microfluidics is becoming an essential tool for the selection and identification of target cells within the array of available biological fluids toward clinical practicality.13 Specifically, the capture and analysis of single monocytes could provide information about the immune system such as phagocytizing and degrading foreign microorganisms in the body.14 As monocytes in blood are rare (5% in whole blood), isolation of target monocytes of interest from the background of erythrocytes and other leukocytes is therefore important to profile expression levels in individual monocytes.15 Powerful approaches for the separation of monocytes from human blood have been reported;16,17 however, many existing devices still needed a time-consuming labeling procedure and have yielded low sample purities, causing challenges in downstream analysis. The inherent heterogeneity of extremely low frequency monocytes dictates the need for an effective analysis method at the single-cell level but approaches for label-free isolation of single monocytes using microfluidic devices have not been fully developed.
Microwell arrays, miniaturized replicas of 96-well plates, allow cells to be localized and monitored at the single-cell level.18–21 Several well-established single-cell isolation technologies based on dielectrophoresis, magnetism, and acoustic and mechanical valves have been utilized to isolate single cells in the miniaturized trapping arrays with high efficiency and accuracy. However, these techniques require external sources and complicated operations and therefore have significant hurdles such as the maintenance of cell viability due to an excessive localized electric field gradient, integration with other microfluidic components, and device parallelization for larger-scale sample processing. Hydrodynamic passive trapping with careful design of microwells that use gravity or fluid flow enables up to 70% single-cell capture without compromising cell viability. However, this approach has not been applied to target cells from a mixture of different-sized cells/particles because the microwell arrays were designed to isolate microparticles of a specific size.20 There are a number of methods that have been adapted to isolate single cells microfluidically in a hydrodynamic manner, but the microfluidic separation module is usually completely separated from the microwell arrays. Kim et al. have reported a cell bandpass filter integrated with a microfluidic single-cell array to separate and isolate single cells with polydisperse distributions.22 They used pinched flow fractionation to continuously separate cells with different sizes by utilizing multiple bypass microchannels; however, these bypass channels resulted in complicated microchannel networks that limited the number of cell traps (<20). Also, Kim et al. developed a trapping-and-sorting microfluidic device that can allocate particles to different trap zones by size.23 They employed additional side channels to isolate and decouple fluid flow between each trap zone. This device, however, has limitations in the separation of high-concentrated samples and is low throughput.
In this paper, we aim to develop a fully-integrated single-blood-cell analysis platform that is capable of size-selective cell separation from diluted blood samples and highly efficient entrapment of single cells in a single device. Two different-sized microfilters are designed in sequence for removing large cell clumps, white blood cells (WBCs), and red blood cells (RBCs) so that continuous separation of a targeted cells population with an intermediate size is achieved. Most cancer cell lines such as MCF-7, SK-BR-3 (human breast), HeLa (human carcinoma), and K562 (leukemia) can also be retained in the filter array due to their intermediate sizes. We spiked U937 cells (human monocyte cell line) as a model cell line to mimic macrophage behavior. The separated targeted U937 cells can then be discriminated by dielectrophoretic force and isolated individually by downstream trapping arrays.24 This straightforward approach enables efficient separation and recovery of medium-sized cells and facilitates the individual trapping of target cells without moving parts or external forces. As a proof-of-concept, we also demonstrated the isolation of 200 single monocytes using the microfilters and dielectrophoretic (DEP) separation from diluted blood.
WORKING PRINCIPLES
Microfluidic device design
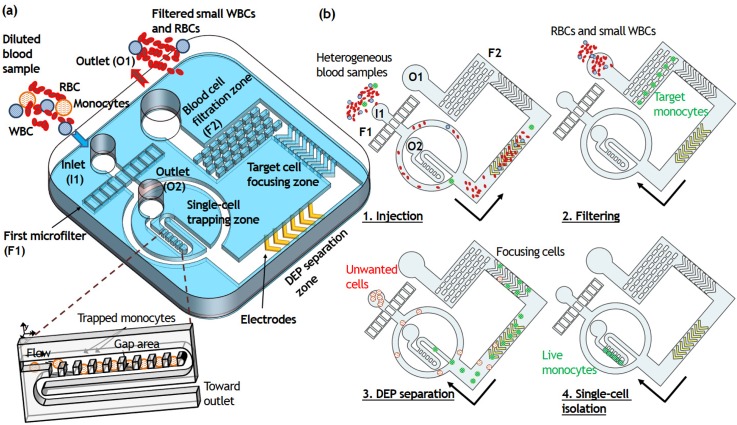
Figure 1(a) shows a conceptual representation of the presented device. The prototype is composed of four different microfluidic modules—a hydrodynamic microfilter, a sheathless cell focuser, a DEP separator, and a single-cell microwell array—connected sequentially from the inlet. The first microfilter F1 is designed for large cell and clumps trapping (target cell diameter >18 μm), and the second microfilter F2 is for medium-size cell separation and recovery (target cell diameter >10 μm) [Fig. 1(b)]. The basic mechanism of cell trapping and size-based sorting is similar to that of the sieve. Cells smaller than the pore size pass through the filter (e.g., RBCs and smaller WBCs except monocytes), whereas cells larger than the pore size such as U937 cells are trapped geometrically. When a U937 cell-spiked blood sample is introduced to the inlet I1, the U937 cell aggregates are filtered through F1 while RBCs, smaller WBCs, and U937 cells are transported to F2 along the main channel. The height of the F2 microfilter and main channel is set to 19 μm. Then, to collect captured single U937 cells, we reverse the flow by adding PBS at the outlet O1 after the removal of RBCs and smaller WBCs at O1 using a syringe pump with the withdrawal mode. Consequently, single U937 cells ranging from 10 to 18 μm are completely separated from RBCs and smaller WBCs and transferred to the downstream isolation area. The overall design is presented in Fig. S1(a) in the supplementary material.
FIG. 1.
A microfluidic platform for the integrated microfluidic platform for size-selective single-cell trapping. (a) Schematic illustration of the microfluidic platform comprising the hydrodynamic filters, obstacle array, DEP electrode, and the array of single-cell traps. (b) Schematic of the process of size-selective single-cell trapping. Two different-sized microfilters are designed in sequence for removing large cell clumps (target cell diameter >18 μm), smaller WBCs and RBCs (target cell diameter <10 μm) so that continuous separation of a targeted U937 cells and monocytes population with an intermediate size is achieved. The separated targeted cancer cells can then be discriminated by dielectrophoretic force and isolated individually by downstream trapping arrays.
The cell focusing channel comprises a series of ridges having the slanted geometry (40 μm in height) that directs cells to either side of the microchannel tightly based on hydrophoresis without the assistance of sheath fluid. Hydrophoresis handles particle displacement laterally by generating helical streamlines at the slanted grooves in microchannels.25 The local anisotropy of the grooves constructs rotational flows that are transverse to the main flow direction. If the groove gap satisfies the design criteria for particle ordering by hydrophoresis,26,27 the cells can be displaced from the original streamline toward the channel wall by steric interactions. To ensure hydrophoretic ordering of the U937 cells ranged from 10 to 18 μm in diameter, we set hg = 18 μm and ht = 22 μm. While the U937 cells were unfocused in the non-grooved channel, most of the U937 cells were successfully focused into the side of the microchannel along the slanted obstacle arrays, achieving a narrow focusing stream [Fig. 1(b)]. The tightly focused U937 cells were transported to the DEP separation zone, where viable U937 cells were separated from non-viable ones by dielectrophoresis under the non-uniform electric field. The chevron-shaped DEP electrodes consist of 25 nm thick chromium, which act as an adhesion layer for the 250 nm thick conductive gold electrodes. The viable U937 cells were deflected toward the microchannel center by positive DEP and transported to the single-cell microwell array, whereas the non-viable U937 cells maintained their trajectory and were removed via outlet O2. Finally, the separated viable U937 cells were delivered to the single-cell trapping zone and captured passively and sequentially in the 18 × 18 × 16 μm3 sized microwells.
In order to trap single cells passively, we placed an array of densely packed traps, previously reported for high-density cell trapping.24,28,33 The continuous single-cell trapping process is based on the combination of perpendicular deformation and horizontal delivery flow. The microfluidic trapping region involves highly packed 400 single-cell traps which enables observation and identification of every single U937 cell flowing through the microfluidic channel. Each trapping unit has a smaller height of the trap (hT) than the height of the main delivery channel (H), resulting in a gap area (hG = H − hT). Therefore, DEP-separated viable U937 cells follow the bulk continuous fluid and are pushed into the traps by the vertical flow through the gap area.
DEP electrode design
DEP is a phenomenon in which a force is exerted on a dielectric particle when it is subjected to a non-uniform electric field. More specifically, this force is induced by the interaction of the particle's dipole and the spatial gradient of the surround electric field. The magnitude and direction of the force is strongly dependent on the particle's physical and dielectric properties, as well as the characteristics of the electric field. For a uniform spherical particle, the time-averaged DEP force can be expressed as follows:
where εm is the permittivity of the medium, R is the radius of the cell, E is the strength of the applied electric field, β is the Clausius–Mossotti (CM) factor, and ∇ is the gradient operator. The complex permittivity (ε*) is a function of frequency (ω), which indicates that the value for the term inside the parenthesis in the equation is a function of the applied electrical field frequency. It defines both the magnitude and directions of the force with respect to the frequency of the applied electric field, and the permittivity and conductivity of the particle and its surrounding medium. For a single shell model, in case of Re(β) < 0 or Re(β) > 0, cells will be migrated away (negative DEP) or toward (positive DEP) the direction of increasing electric field. Furthermore, the direction of DEP mobility at a particular frequency is also sensitive to the cell size/shape and the protein density on the cell membrane and cell wall, which vary widely for different species, and is distinct for live/dead and diseased/healthy cells. As a result, this phenomenon is being used in many research applications as a label-free method to sort and manipulate microparticles, nanoparticles, and biological cells.29,30
The main key to the separation of viable cells is a DEP operation regime, in which Re(β) on the two cell populations are different. Based on a single shell model, viable cells exposed to several kHz with a larger Re(β) experience stronger DEP force resulting in a larger lateral deflection than those with a smaller Re(β) (non-viable cells) because of the different polarization of impaired dead cell membrane under the applied electric field.31 Song et al. reported an oblique interdigitated electrodes array that is capable of continuously separating human mesenchymal stem cells and their differentiation progenies when controlled by an AC electric field with alternating on/off control.32 By modifying the electrode configuration into a V-shaped interdigitated electrode array, this subsystem was then incorporated into our single-cell analysis platform to separate live and dead cells continuously. The COMSOL electrostatic simulation in Figure S1(b) in the supplementary material simulates the electric field converging on the surface of the electrodes when an electric potential is applied between them; hence, at frequencies where the CM factor is positive, the cells will be pulled laterally along the electrode due to DEP. Also, there is a hydrodynamic force induced by fluid flow pushing the cell along the microchannel. Finally, the deflected viable cells based on the combined forces acting on them can be collected at the center outlet and the non-viable cells at the side outlet.
MATERIALS AND METHODS
Cell culture
U937 (monoblast cell line, average diameter ∼14 μm) cells were purchased from American Type Culture Collection and cultured in RPMI1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco). Cells were passaged every 2–3 days following standard protocols and cultured in a humidified incubator at 37 °C with 5% CO2.
Blood samples
The human blood samples were collected, with informed consent, from healthy donors in UCI General Clinical Research Center with Institutional Review Board approval. Vacutainer tubes (BD Bioscience) containing EDTA as an anticoagulant were used for collection.
Device fabrication
We used standard multi-layer microfabrication techniques to fabricate the silicon master of microfluidic channels and to produce the microfluidic device. A 3-in. silicon wafer is first patterned using standard multi-layer photolithography and the patterned silicon wafer was treated with trichloro(1H,1H,2H,2H-perfluorooctyl)silane for 1 h to avoid the adhesion of poly-dimethylsiloxane (PDMS). Then, PDMS prepolymer is mixed in a 10:1 (wt/wt) ratio with a curing agent and poured onto the silanized wafer. After baking at 65 °C for 3 h, the cured PDMS is peeled from the mold, and access holes (1.5 mm) for fluidic inlet and outlets are punched by using a Uni-Core puncher (Ted Pella Inc., Redding, CA, USA). The PDMS device is irreversibly bond to the glass slide upon oxygen plasma treatment.
RESULTS AND DISCUSSION
Separation of single live U937 cells from diluted blood samples
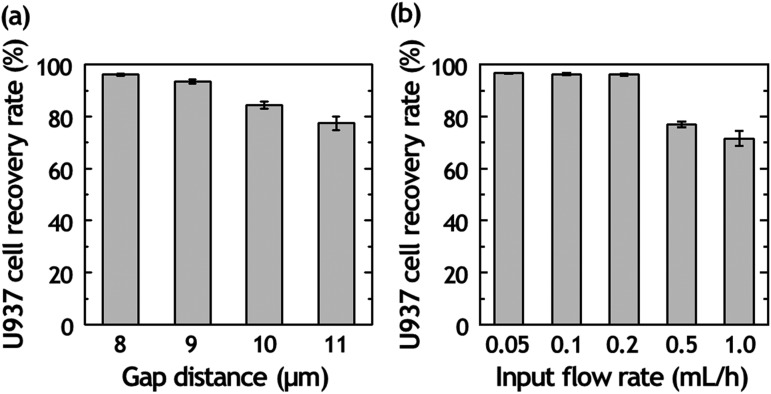
The separation of U937 cells from diluted blood is performed using a matrix of micrometer-sized PDMS structures consisting of 10 rows and 250 columns. The pore size of the filter is kept constant along each row from 8 to 12 μm. The ability to separate U937 cells from diluted blood was initially tested by doping U937 cells into diluted blood (2% hematocrit). We conducted experiments at a doping ratio of one U937 cell to 1000 WBCs to characterize the parameters relevant to the isolation of single U937 cells. When diluted blood is introduced into the bottom-left corner of the matrix, fluid flow transports cells in a diagonal path from the bottom-left to the top-right of the matrix. While RBCs and WBCs flowed to the top of the matrix and exited through the outlet channel because of their smaller diameter than the filter gap, the U937 cells are captured at the hydrodynamic filter due to their size. The capture rate of U937 cells can be tuned by adjusting the filter gap and the flow rates applied at the inlet. We examined the capture rate of U937 cells with diluted whole blood samples of 2% hematocrit to investigate the effect of gap distance on U937 cell capture in the matrix [Fig. 2(a)]. The results explained that the U937 cell loss became greater with increasing gap distance. With the increased gap distance, more RBCs and WBCs attempted to pass through the space between the filter gaps while also pushing against the previously captured U937 cells, resulting in an increase in U937 cell loss and a decrease in the sorting efficiency. However, a further decrease in the gap distance below 8 μm facilitates capturing RBCs and induced a lower purity of U937 cell separation.
FIG. 2.
Separation efficiency of cancer cells from diluted blood. (a) Plot of U937 cell recovery rate according to a gap distance of hydrodynamic filter (F2) array from 8 to 11 μm under the input flow rate of 0.2 ml/h. (b) Plot of U937 cell recovery rate according to input flow rate with 8-μm-gap hydrodynamic filter array.
We also tested the U937 cell capture rate of the device under various input flow rate from 0.05 to 1.0 ml/h [Fig. 2(b)]. When the flow rate increased for achieving higher throughput, the percentage of captured U937 cells in the matrix was decreased; this means that the high flow rate induces the undesirable shear stress-induced cell deformation. Under the input flow rate of 0.2 ml/h, the U937 cell capture rate was about 97%, higher than other flow conditions. The presented approach for U937 cell sorting has a trade-off between the separation efficiency and throughput. If the flow rate is increased for achieving higher throughput, the pushing forces imposed on trapped U937 cells accelerate deformation and the cells can be released into the fluid stream from the matrix without capturing. The higher number of released U937 cells under the high pressure also affects the U937 cell recovery rate. Thus, we chose to use 2% hematocrit under 0.2 ml/h input flow rate for optimizing both the throughput and the separation performance.
Validation of sheathless cell focusing and dielectrophoretic live cell separation
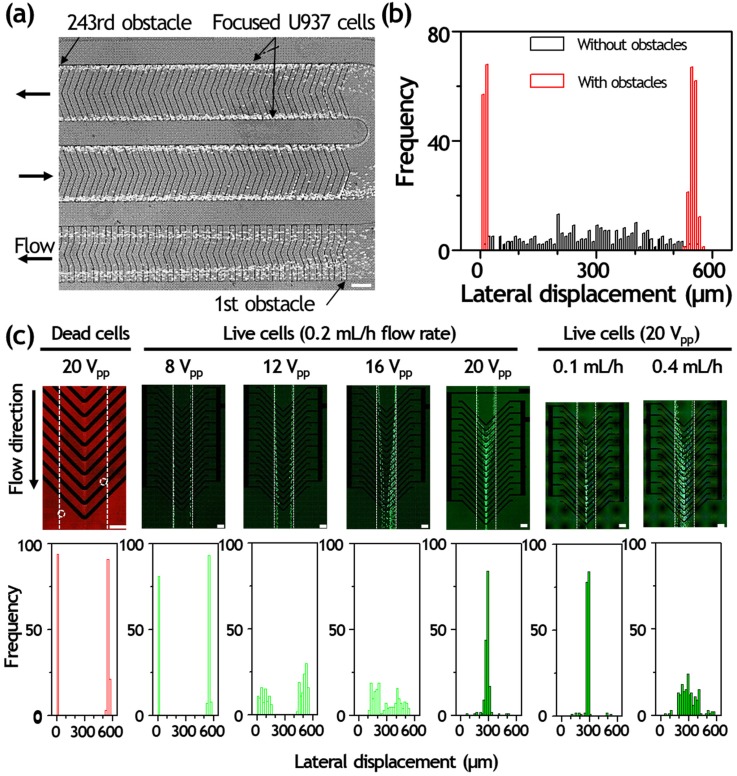
The focusing capability of the device with U937 cells was first tested under the input flow rate of 0.2 ml/h. We determined focusing variation for all experiments by measuring the positions of individual particles in the observation region (562 μm in width). All U937 cells were successfully focused on the side wall after 243 obstacles, achieving two narrow focusing streams [Fig. 3(a)]. At such circumstances, they were driven to either the right and left sidewalls by the obstacles. The focusing positions were 9.94 ± 3.68 and 549.14 ± 8.37 μm for U937 cells. We also investigated the lateral positions of U937 cells passed through the microchannel without obstacles, and their lateral positions were 287.24 ± 140.50 μm [Fig. 3(b)]. The input flow rate affects the cell focusing efficiency. If we increase the flow rate higher than 0.6 ml/h, U937 cells could not keep their focusing widths because of the inertial effects.
FIG. 3.
Dielectrophoretic live cell sorting of hydrodynamically separated cancer cells. (a) Fluorescence streak images of U937 cells in the grooved microchannel. The cells are successfully guided by locally patterned slanted grooves and focusing efficiency is improved with increasing number of obstacles. Input flow rate is 0.2 ml/h. (b) Cell measurement according to the existence of obstacle array was performed at the end of focusing zone (width of 562 μm) under the input flow rate of 0.2 ml/h and plotted in the focusing histograms. Each focusing histogram was obtained from the measurement of more than 180 cells. (c) Continuous sorting of dead and live U937 cells under the different voltage (8 to 20 Vpp) and flow rate conditions (0.1 to 0.4 ml/h). Each focusing histogram was obtained from the measurement of more than 180 cells. The channel wall is outlined with dotted lines for clarity. Scale bars, 200 μm.
At certain high concentration, particle–particle interaction and nonspecific agglomeration of cells and particles tend to influence the separation resolution and purity by deforming the pressure fields and colliding with each other. Thus, a microfluidic separation typically requires multiple rounds of separation to reduce the exact operational limit of particle concentration and particle–particle interaction phenomena. However, our microfluidic platform achieved U937 cell separation purity of >85% in a single round of separation, which is the reason why the smaller blood cells were separated completely at the microfilter F2 and the reduced cell volume did not disrupt the focusing profile.
After studying the particle trajectories of U937 cells in the integrated separation microfluidic device, the DEP separation was investigated and the results are shown in Fig. 3. The cells were introduced into the DEP separation zone with a flow rate of 0.2 ml/h. When the electric field was off, both live and dead U937 cells flowed straightly through the microchannel without any lateral displacement and exited the side channel via the inlet I1 due to the hydrodynamic flow [Fig. 3(c)]. When the electric field was applied with an AC field of 15 V peak-to-peak at a frequency of 0.5 MHz, most of the pre-focused viable U937 cells (in green) exerting strong positive DEP force were deflected laterally toward the center of chevron-shaped DEP electrodes, whereas most of the dead U937 cells (in red) remained a straight trajectory due to weaker DEP force and exited the side channel via the inlet I1. The deflected viable U937 cells were then transferred to the single-cell trapping zone near outlet O2.
To optimize the sorting efficiency, the voltage- and flow rate-dependent DEP cell deflection was also investigated [Fig. 3(c)]. The applied flow rate was fixed as 0.2 ml/h and the applied voltage was adjusted from 8 to 20 Vpp at a frequency of 0.5 MHz. After entering the DEP sorting region, U937 cells were deflected from the left and right sidewalls to the center of DEP electrodes. At the voltage of 20 Vpp, most of the live U937 cells could be focused toward the center; however, the cells were unable to successfully deflect at relatively low voltages of 8 and 12 Vpp. The lateral positions of dead U937 cells were 12.12 ± 3.61 and 552.31 ± 8.54 μm at 20 Vpp. The lateral positions of live U937 cells were 9.85 ± 3.79 and 550.11 ± 7.86 μm at 8 Vpp, 73.71 ± 43.52 μm and 494.96 ± 34.70 μm at 12 Vpp, 294.40 ± 121.45 μm at 16 Vpp, and 284.37 ± 38.79 μm at 20 Vpp. We avoided the high voltages above 20 Vpp to maximize the viability of U937 cells. As increasing flow rate will reduce the cell sorting time and increase the throughput, we tested the DEP deflection of live U937 cells under various input flow rates ranging from 0.1 to 0.4 ml/h. If the flow rate is increased to 0.4 ml/h for achieving higher throughput, the number of introduced U937 cells was increased; however, the percentage of deflected single U937 cells was dramatically decreased. This means that the high flow rate causes undesirable passing through the DEP sorting region due to higher hydrodynamic force. The non-deflected live cell would be transported to outlet O1 and induce low cell purity. Under a total flow rate of 0.2 ml/h, the percentage of deflected single U937 cells was about 98%, higher than other flow conditions. Also, the deflected cells were trapped around the electrode instead of flowing through the microchannel under the application of above 20 Vpp. This results in a trade-off between the DEP sorting efficiency and throughput. In addition, at higher flow rates, the hydrodynamic pressure imposed on single U937 cells trapped in downstream trapping arrays (near outlet O2) accelerates deformation and the trapped single cells can be released into the fluid stream from the single-cell trap without capturing.
On-chip trapping of U937 cells and viability assay
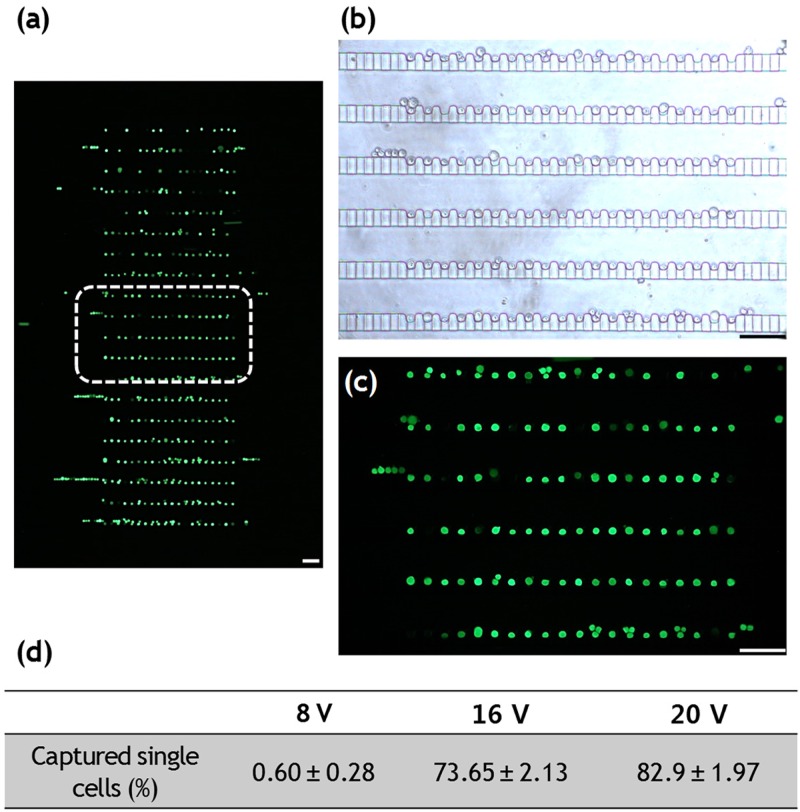
As a proof-of-concept, we demonstrated the trapping of DEP-sorted U937 cells to investigate device trapping efficiency as well as the feasibility of single-cell monitoring and analysis for trapped target cells [Fig. 4(a)]. The high-throughput microfluidic trapping array contains highly packed 400 single-cell traps, designed with small dimensions to fit within a microscopic field of view and can be filled within 3 min, enabling the observation and identification of every single U937 cell flowing through the channel [Figs. 4(b) and 4(c)]. We designed the dummy traps at each end of the arrays to make cells experience similar flow rates near each trap. The dummy traps make cells move closer to the traps after passing the serpentine channel. Therefore, cells were mainly trapped in the middle of each row of the arrays instead of in the dummy traps.24 The trapping efficiency of DEP-sorted U937 cells within the downstream single-cell array (near outlet O2) was also quantified. The percentage of captured U937 cells within the 400 single-cell trapping arrays under the voltage application of 8, 16, and 20 Vpp were 0.60 ± 0.28%, 73.65 ± 2.13%, and 82.9 ± 1.97%, respectively [Fig. 4(d)].
FIG. 4.
Single-cell entrapment of DEP separated live U937 cells. (a) Fluorescent micrograph of high-density 400 single-cell trapping arrays. (b) Bright-field and (c) fluorescent image of stained live U937 cells separated by dielectrophoretic force. Scale bars, 100 μm.
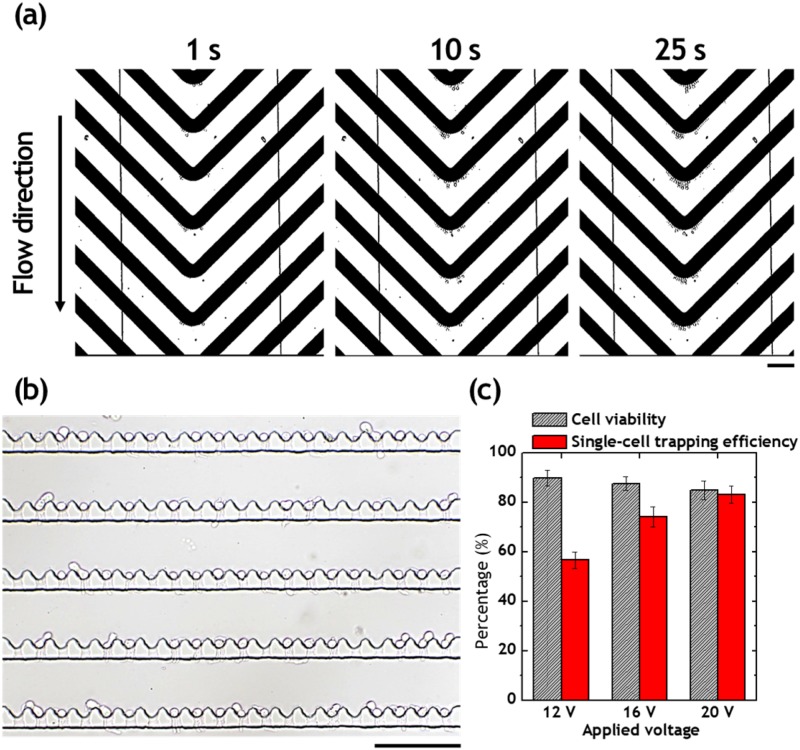
Isolation of single monocytes from blood
Figure 5(a) shows the continuous separation of single monocytes at the chevron-shaped DEP electrodes. After 25 s positive DEP exposure, the single monocytes were successfully deflected toward the center of chevron-shaped electrodes. The deflection of monocytes allows the single-cell isolation at the downstream microwell arrays after turning off the voltage. A cell viability assay was performed to see if the monocytes are actually live in the microwell array [Fig. 5(c)]. After single-cell isolation, we introduced the Calcein AM staining solution to stain the single monocytes for 15 min. We also quantified monocyte viability in microwell arrays under the different voltage applications. The viabilities of monocytes separated under 12, 16, and 20 Vpp were 89.8%, 87.6%, and 84.8%, respectively. Despite the exposure of electric field under the positive DEP regime, the percentage viabilities of monocytes were stable at ∼84%. This was because PBS was exchanged for DEP buffer at the microwell array after DEP separation, resulting in the maintenance of cell viability within the microwell arrays. The viability of cells within the microwell arrays could be further improved by enlargement of channel height over 100 μm, which allow on-chip cell culture. The single-cell capturing efficiency was also quantified according to the different voltage applications. The capturing efficiencies of monocytes separated under the 12, 16, and 20 Vpp were 56.6%, 74.0%, and 83.2%, respectively. The lower voltage interrupts the isolation of single monocytes because of the lower number of deflected cells by the pDEP force. As the number of monocytes is very low in the diluted blood, the appropriate higher voltage would be necessary to isolate single monocytes in the microfluidic device.
FIG. 5.
Single-cell entrapment of DEP separated live U937 cells. (a) DEP separation of live monocytes from diluted blood. (b) Bright-field image of isolated single monocytes separated by DEP force under the application of 20 Vpp and 0.5 MHz. (c) Percentage of live monocytes and single-monocyte capturing efficiency after the DEP separation and isolation. Scale bars, 100 μm.
DISCUSSION
The applicability of this size-selective single-cell trapping platform for monocyte isolation was demonstrated by integrating different microfluidic components: the hydrodynamic separation filter, the DEP electrodes, and the single-cell trapping. The presented system enables single-cell analysis with a large degree of flexibility, which are envisioned to open up a variety of attractive applications, such as (i) increased separation and isolation throughput of the single monocytes by integrating multiple replicates of microfluidic devices; (ii) maintenance of the viability of isolated cells for further analysis; and (iii) the simplicity of single-cell trapping design which enables the integration of active microfluidic components. However, we encountered a challenge during the cell separation that the current platform can only process up to 2% diluted blood. As the hematocrit increases, the separation performance decreases due to the unwanted clogging of RBCs around the filter due to their cell-to-cell interaction. The device can deal with a higher hematocrit level sample if the device is massively parallelized. We determined that the presented device could be arrayed with 10 arranged channels in parallel with a single inlet and 12 outlets integrated into the 3-in. PDMS device that can deal with large volumes of blood samples. We simply calculated that when 2% diluted blood is tested at 0.2 ml/h, 10 000 single monocytes can be isolated in 5 min. The simple and highly scalable channel design facilitates the single-cell isolation throughput to be greatly enhanced by stacking multiple sorting layers without compromising performance parameters such as purity and recovery.
To enhance user-friendliness, microfluidic chips with standard interconnecting inlet ports could prove beneficial. For example, if the microfluidic devices were designed to be compatible with pipettes such that the inlets are directly connected to a pipette tip, constant flow rates can be generated manually with back-and-forth movement. This would eliminate the need for complex fluid interconnections and bulky precision pumps. The combination of the pipette and the proposed microfluidic chip offers the following advantages: (i) stable monocyte separation and isolation with the simple, manual operation of a pipette; (ii) continuous monocyte separation without clogging; and (iii) applicable for a wider community of end-users to be familiar with its use so that non-experts can easily carry out the procedures. On the other end of the spectrum, this platform could be adapted to be compatible with automated liquid dispensing systems for high throughput sample processing.
The presented platform offered the capability to investigate the cellular heterogeneity of normal monocytes after the continuous separation under the DEP regime. One of the main challenges in the proposed platform is that the monocytes can be manipulated only under positive DEP operating conditions, in which cells are trapped at constrictions where electric field maxima may be causing cell attachment to the electrodes. This could lead to the cell damage and deteriorates the separation performance.
While in our proof-of-concept demonstration, we showed high-throughput, size-selective isolation of single U937 cell spiked in blood, the proposed method would be amenable to other applications in which single-cell screening is required. The large-scale microwell arrays can be adapted to high-throughput single-cell analyzers,33 label-free detection via fluorescence lifetime imaging,34 and lensless observation,35 which enable analyzing a large population of cells with high statistical precision in a practical period of time. Such applications include imaging and detection of single cells of interest in stem-cell biology, medicine, pharmaceutics, and environmental science.
CONCLUSION
In this paper, we presented a continuous flow, integrated microfluidic device to isolate single monocytes from the diluted blood in microfluidic arrays in a size-selective manner. The hydrodynamic filtration enables the separation of U937 cells from the diluted blood based on their different sizes. The device also includes the utilization of DEP force in an array of oblique interdigitated electrodes and the AC field control to realize continuous separation of viable monocytes. Significant technical findings are summarized as follows:
-
(1)
It was found that two different-sized microfilters allow for separation and recovery of medium-sized monocytes from the diluted blood cell suspension with heterogeneous size distribution. Extensive experiments demonstrated the notable separation of monocytes and blood cells in terms of various flow conditions and hematocrits. The application of back-flow by switching the flow direction allows the transfer of only monocytes to the DEP electrodes, while the filtered blood cells are removed from the outlet reservoirs.
-
(2)
The separation efficiency and purity for normal U937 cells were quantified, exhibiting consistent performance when the flow rate is fixed as 0.2 ml/h. Separation efficiency up to 96.4% can be obtained for normal U937 cells at the center outlet connected to the microwell arrays, with purity up to 84%.
-
(3)
The separated normal U937 cells were successfully isolated individually in the 400-microwell arrays downstream. Up to 98% single-cell occupancy of the entire traps was achieved, with average single-cell occupancy of 94 ± 4%. With a U937 cell concentration of 1 × 106 cells per ml, and a flow rate of 0.2 ml/h, 400 U937 cells were trapped sequentially and deterministically within 40 s.
The presented size-selective, label-free, and non-destructive microfluidic single-cell analysis platform with the DEP sorting of monocytes would have a myriad of applications in single-cell genetic analysis, stem cell biology, point-of-care diagnostics, and cancer diagnostics.
SUPPLEMENTARY MATERIAL
See supplementary material for the overall microchannel design and the COMSOL electrostatic simulation of the electric field on the surface of the electrodes.
ACKNOWLEDGMENTS
The authors would like to acknowledge the National Science Foundation [Partnerships for Innovation: Accelerating Innovation Research-Research Alliance (PFI:AIR-RA)] and W. M. Keck Foundation (Grant No. KF-52356) for funding support.
References
- 1.Diercks A., Kostner H., and Ozinsky A., PLoS One 4, e6326 (2009). 10.1371/journal.pone.0006326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren L., Bryder D., Weissman I. L., and Quake S. R., Proc. Natl. Acad. Sci. U.S.A. 103, 17807–17812 (2006). 10.1073/pnas.0608512103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powell A. A., Talasaz A. H., Zhang H., Coram M. A., Reddy A., Deng G., Telli M. L., Advani R. H., Carlson R. W., and Mollick J. A., PLoS One 7, e33788 (2012). 10.1371/journal.pone.0033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strati A., Markou A., Parisi C., Politaki E., Mavroudis D., Georgoulias V., and Lianidou E., BMC Cancer 11, 1 (2011). 10.1186/1471-2407-11-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narsinh K. H., Sun N., Sanchez-Freire V., Lee A. S., Almeida P., Hu S., Jan T., Wilson K. D., Leong D., and Rosenberg J., J. Clin. Invest. 121, 1217–1221 (2011). 10.1172/JCI44635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuang Y. and Walt D. R., Anal. Biochem. 345, 320–325 (2005). 10.1016/j.ab.2005.06.028 [DOI] [PubMed] [Google Scholar]
- 7.Brock A., Chang H., and Huang S., Nat. Rev. Genet. 10, 336–342 (2009). 10.1038/nrg2556 [DOI] [PubMed] [Google Scholar]
- 8.Elowitz M. B., Levine A. J., Siggia E. D., and Swain P. S., Science 297, 1183–1186 (2002). 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- 9.Wills A. and Obermair A., Gynecol. Oncol. 131, 467–479 (2013). 10.1016/j.ygyno.2013.07.082 [DOI] [PubMed] [Google Scholar]
- 10.Foulkes W. D., Reis-Filho J. S., and Narod S. A., Nat. Rev. Clin. Oncol. 7, 348–353 (2010). 10.1038/nrclinonc.2010.39 [DOI] [PubMed] [Google Scholar]
- 11.Kærn M., Elston T. C., Blake W. J., and Collins J. J., Nat. Rev. Genet. 6, 451–464 (2005). 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- 12.Acar M., Mettetal J. T., and van Oudenaarden A., Nat. Genet. 40, 471–475 (2008). 10.1038/ng.110 [DOI] [PubMed] [Google Scholar]
- 13.Huang L., Bian S., Cheng Y., Shi G., Liu P., Ye X., and Wang W., Biomicrofluidics 11, 011501 (2017). 10.1063/1.4975666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanberg P. R., Park D.-H., Kuzmin-Nichols N., Cruz E., Hossne N. A., Jr, Buffolo E., and Willing A. E., J. Cell Mol. Med. 14, 553–563 (2010). 10.1111/j.1582-4934.2009.00903.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li P., Mao Z., Peng Z., Zhou L., Chen Y., Huang P.-H., Truica C. I., Drabick J. J., El-Deiry W. S., Dao M., Suresh S., and Huang T. J., Proc. Natl. Acad. Sci. U.S.A. 112, 4970–4975 (2015). 10.1073/pnas.1504484112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis J. A., Inglis D. W., Morton K. J., Lawrance D. A., Huang L. R., Chou S. Y., Strum J. C., and Austin R. H., Proc. Natl. Acad. Sci. U.S.A. 103, 14779–14784 (2006). 10.1073/pnas.0605967103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grenvall C., Magnusson C., Lilja H., and Laurell T., Anal. Chem. 87, 5596–5604 (2015). 10.1021/acs.analchem.5b00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo T., Hou J., Chen S., Chow Y.-T., Wang R., Ma D., Zhu R., and Sun D., Biomicrofluidics 11, 054103 (2017). 10.1063/1.5000917 [DOI] [Google Scholar]
- 19.Schneider T., Yen G. S., Thompson A. M., Burnham D. R., and Chiu D. T., Anal. Chem. 85, 10417–10423 (2013). 10.1021/ac402383n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dura B., Dougan S. K., Barisa M., Hoehl M. M., Lo C. T., Ploegh H. L., and Voldman J., Nat. Commun. 6, 97–103 (2015). 10.1038/ncomms6940 [DOI] [PubMed] [Google Scholar]
- 21.Yesilkoy F., Ueno R., Desbiolles B. X. E., Grisi M., Sakai Y., Kim B. J., and Brugger J., Biomicrofluidics 10, 014120 (2016). 10.1063/1.4942457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H., Lee S., Lee J.-H., and Kim J., Lab Chip 15, 4128–4132 (2015). 10.1039/C5LC00904A [DOI] [PubMed] [Google Scholar]
- 23.Kim J., Erath J., Rodriguez A., and Yang C., Lab Chip 14, 2480–2490 (2014). 10.1039/C4LC00219A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung K., Rivet C. A., Kemp M. L., and Lu H., Anal. Chem. 83, 7044–7052 (2011). 10.1021/ac2011153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi S. and Park J.-K., Lab Chip 7, 890–897 (2007). 10.1039/b701227f [DOI] [PubMed] [Google Scholar]
- 26.Song S. and Choi S., J. Chromatogr. A 1302, 191–196 (2013). 10.1016/j.chroma.2013.06.030 [DOI] [PubMed] [Google Scholar]
- 27.Yan S., Zhang J., Chen H., Alici G., Du H., Zhu Y., and Li W., Biomicrofluidics 8, 064115 (2014). 10.1063/1.4903761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Tao Y., Lee D.-H., Wickramasinghe H. K., and Lee A., Lab Chip 17, 1635–1644 (2017). 10.1039/C7LC00133A [DOI] [PubMed] [Google Scholar]
- 29.Chan J. Y., Kayani A. B. A., Ali M. A. M., Kok C. K., Majiis B. Y., Hoe S. L. L., Marzuki M., Koo A. S.-B., Ostrikov K., Rahaman M. A., and Sriram S., Biomicrofluidics 12, 011503 (2018). 10.1063/1.5010158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang H., Lee D.-H., Choi W., and Park J.-K., Biomicrofluidics 3, 014103 (2009). 10.1063/1.3086600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yildizhan Y., Erdem N., Islam M., Martinez-Durate R., and Elitas M., Sensors 17, 2691 (2017). 10.3390/s17112691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song H., Rosano J. M., Wang Y., Garson C. J., Prabhakarpandian B., Pant K., Klarmann G. J., Perantoni A., Alvarez L. M., and Lai E., Lab Chip 15, 1320–1328 (2015). 10.1039/C4LC01253D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellogg R. A., Gómez-Sjöberg R., Leyrat A. A., and Tay S., Nat. Protoc. 9, 1713–1726 (2014). 10.1038/nprot.2014.120 [DOI] [PubMed] [Google Scholar]
- 34.Lee D.-H., Li X., Ma N., Digman M. A., and Lee A. P., Lab Chip 18, 1349–1358 (2018). 10.1039/C7LC01301A [DOI] [PubMed] [Google Scholar]
- 35.Greenbaum A., Luo W., Su T.-W., Göröcs Z., Xue L., Isikman S. O., Coskun A. F., Mudanyali O., and Ozcan A., Nat. Methods 9, 889–895 (2012). 10.1038/nmeth.2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the overall microchannel design and the COMSOL electrostatic simulation of the electric field on the surface of the electrodes.