Abstract
Auxin plays critical roles in many developmental processes of plants. The auxin signaling pathway is a series of plant responses to auxin stimuli. However, the functions of many genes in this pathway are still obscure. As auxin receptors, TIR/AFB family genes encode F-Box proteins that directly bind auxin and then transduce the stimulus through the signaling pathway. In this paper, we generated an overexpression line of Auxin-signaling F-Box 6 (OsAFB6) in rice, which largely delayed heading, greatly increased spikelets per panicle and primary branch number and ultimately enhanced grain yield by 50%. OsAFB6 is preferentially expressed in young tissues with stronger meristem activities and suppresses flowering by upregulating OsRR1 and downregulating Ehd1 expression levels. Overexpression of OsAFB6 delayed heading, increased cytokinin (CK) by suppressing the expression level of Gn1a and simultaneously decreased the IAA concentration in the young panicle, which promoted inflorescence meristem development and resulted in large panicles with more spikelets per panicle, primary branches and increased grain yield. It would be a beneficial strategy to generate lines with varied expression levels of OsAFB6 to breed high-yielding cultivars for specific regions that can fully utilize the local sunlight and temperature resources.
Introduction
Auxin regulates many aspects of plant growth and development, including embryogenesis1, the architecture of the root system2, gravitropism3, phototropism4, initiation and radial positioning of plant lateral organs and cell elongation5,6. Plants sense auxin by receptors and then transduce its signal to elicit a series of responses. The intracellular receptors of auxin are the Transport Inhibitor Response 1/Auxin-signaling F-Box (TIR1/AFB) gene family7–9. TIR1/AFB genes encode F-box proteins that have an amino-terminal F-box motif for mediating interactions with SKP1 and form part of the SCF (Skp-Cullin-F-Box containing protein) complex, a carboxy-terminal protein-protein interaction domain for targeting specific substrates Auxin/Indole-3-Acetic Acid (Aux/IAA), and a leucine-rich repeat (LRR) domain10. TIR1 is the first auxin receptor identified in this family7,8. Other AFB members have also been demonstrated to work as auxin receptors11,12 but with diverse functions. For example, OsTIR1 and OsAFB2 are targets of microRNAs. Overexpression of OsmiR393 downregulates both genes and results in more tillers, early flowering and less tolerance to salt and drought stress13. Both OsTIR1 and OsAFB2 regulate the auxin response but differ in their relative contributions to seedling development9. The AFBs contribute to auxin response, with diverse intensities of interactions with Aux/IAAs9. AFB4 and AFB5 in Arabidopsis have been confirmed to respond specifically to synthetic auxin picloram. Mutants for both genes are resistant to picloram with no elongation of hypocotyls and are suitable for the analysis of resistance to auxin-like herbicides12.
The TIR1/AFB family members mediate the auxin signaling pathway, which is centered on ubiquitin-dependent SCFTIR1/AFBs-Aux/IAAs-ARFs flow14. With low auxin concentration, Aux/IAAs repress the transcriptional activities of auxin response factor genes (ARFs) by recruiting the corepressor TOPLESS. As auxin levels increase, the auxin receptor TIR1/AFB, a subunit of an E3 ubiquitin-protein ligase SCF complex, binds to auxin directly and recognizes the specific target Aux/IAAs, which are then degraded through ubiquitination15. Subsequently, the repressed ARFs are released and function as either activators or repressors to regulate downstream genes in the auxin signaling pathway16. There are six TIR1/AFBs in rice9 that degrade 31 Aux/IAAs17, which in turn repress 25 ARFs18, resulting in the regulation of other downstream genes. The number of possible combinations of TIR/AFB-Aux/IAA-ARF is large and results in a complex auxin-dependent regulation network. The functions of some combinations have been well characterized19,20.
Similar to auxin, cytokinin (CK) is another important plant hormone regulating many aspects of plant growth21,22. CK plays positive roles in the regulation of shoot apical meristem (SAM) activity23. The LOG (LONELY GUY) gene encodes a novel cytokinin-activating enzyme that works in the final step of bioactive cytokinin synthesis and shows specific expression at the top of shoot meristems where stem cells reside and CK accumulates24. Auxin and CK act either synergistically or antagonistically to play roles in several significant developmental progresses25. Maintenance of the optimum cellular concentration of auxin and CK is rather important and can be regulated in multiple processes, such as biosynthesis, transport, perception, signaling and degradation26.
Auxin and cytokinin can regulate grain yield. For example, PINs, encoding auxin efflux transporters, with the pin-formed mutant phenotype, are regulated by an auxin-dependent Aux/IAA-ARF signaling pathway27. A recent study demonstrated that the auxin transporter PIN5b gene could modulate rice plant architecture and yield28. CK also makes a great contribution to regulating spikelets per panicle, which directly associates with grain yield29.
Rice heading date is a growth-adaptive trait that is related to grain yield and is controlled by multiple genes. A gene family comprising 41 members encoding CCT (CO, CO-LIKE and TOC1) domain proteins plays important roles in regulating heading date. The major heading date genes, including Hd130, Ghd7 and Ghd7.1, belong to the CCT family31,32. In addition to these three genes, nine additional CCT genes were confirmed to control flowering in rice33. OsHAP, another flowering-related gene family containing Ghd834,35, is also a pleiotropic gene regulating heading date, plant height and spikelets per panicle, similar to Ghd7.
In the process of overexpressing CCT05, we generated a late-flowering line. However, this phenotype change was not associated with CCT05 overexpression. In this study, we confirmed that overexpression of the auxin signaling F-box gene OsAFB6 resulted in late flowering. It was also noted that plants with OsAFB6 overexpression exhibited more primary branches, more spikelets per panicle, and a higher grain yield. We demonstrated that OsAFB6 regulated key genes involved in flowering pathways and panicle formation-related hormone pathways.
Results
Cloning and functional characterization of OsAFB6
To test the function of CCT05 (LOC_Os02g08150), CCT05 RNA interference plants and CCT05-overexpressing plants were generated. Transcript levels of CCT05 in the RNAi plants were knocked down to 10–30%, but no differences in phenotypes were observed between the positive and control plants33. Interestingly, among the three CCT05 overexpression lines, only one exhibited variability in heading date, with a segregation ratio of 3:1; late flowering was dominant over early flowering (Fig. 1A). However, the transcript level of the CCT05 gene in this family was not detectibly overexpressed (Fig. S1). Therefore, we considered the late-flowering transgenic plant as a new mutant, and the heading date variation was not caused by CCT05 overexpression but by other gene alterations during the transformation event.
Figure 1.
Performance of OXOsAFB6 mutant plants.The whole plants. (A), main culms (B), panicles (C) and seedlings (D) for the control plant (left) and OsAFB6 overexpression mutant (right); the grain length (E) and grain width (F) for the control plants (up) and OsAFB6 overexpression plants (bottom). Scale bars, 20 cm (A) and (B), 5 cm (C) and (D), and 1 cm (E) and (F).
Southern blot hybridization confirmed a single copy insertion in the mutant (Fig. S2A), consistent with the single gene segregation ratio of 3:1 in the T1 progeny. Thermal asymmetric interlaced PCR (TAIL-PCR) analysis showed that the vector was inserted at 0.56kb upstream of the gene LOC_03g08850 (Fig. S2B), the auxin-signaling F-Box (AFB) gene OsAFB6. To confirm that OsAFB6 was the only gene affected by the insertion, the expression levels of all the genes near the insertion site were investigated (Fig. S2C–E). While adjacent genes showed comparable transcript levels to the control plants, OsAFB6 was the only gene that had significantly increased expression levels in the mutant at the seedling stage; moreover, the increased amount under SD conditions (5 to 20-fold, Fig. S2G) was significantly lower than under LD conditions (15 to 35-fold, Fig. S2E). In addition, OsAFB6 expression exhibited a circadian rhythmic pattern, with high expression during daytime that decreased dramatically at night (Fig. S2F–H). These results highly suggest that OsAFB6 is a strong candidate gene for the mutant phenotype.
To further test this idea, an F2 population of 40 plants was developed by selfing the hybrid of the mutant and wild type (ZH11). Higher OsAFB6 expressions were detected in all 28 late-flowering plants, whereas levels similar to those in the control plants were detected in the 12 early-flowering plants (Fig. S3). This cosegregation test indicated that OsAFB6 was the gene responsible for late heading in the mutant. Hence, the mutant, hereafter named OXOsAFB6, was the result of OsAFB6 overexpression. The seeds of OXOsAFB6 were deposited in the China Center for Type Culture Collection (CCTCC) with accession number P201709.
Overexpression of OsAFB6 can delay heading and increase grain yield
In addition to the heading date variation, we found that the mutant was also photoperiod-sensitive, as flowering was delayed by ~22 days under LD conditions and by ~12 days under SD conditions (Table 1). In addition, the mutant had fewer tillers (Fig. 1A), one more internode in the main culm, longer panicles with more primary and secondary branches in total, more spikelets per panicle (Fig. 1B,C and Table 1), and longer roots at the seedling stage (Fig. 1D). Although the grain size was smaller (Fig. 1E,F), the grain yield per plant was significantly increased by approximately 50% under both LD and SD conditions (Table 1).
Table 1.
Comparison of yield related phenotypes between OsAFB6 overexpression mutants and its control plants under LD and SD conditions.
| Conditions | Traits | Control plants | OXOsAFB6 mutants | P value |
|---|---|---|---|---|
| LD (Wuhan) | Heading date (d) | 73.6 ± 0.6 | 95.6 ± 1.1 | 1.1E-10 |
| Spikelets per panicle | 125.6 ± 9.3 | 175.3 ± 14.8 | 5.7E-05 | |
| Grain yield per plant (g) | 27.0 ± 5.1 | 44.3 ± 8.0 | 9.9E-03 | |
| panicle length (cm) | 20.9 ± 0.9 | 24.7 ± 1.5 | 5.1E-10 | |
| grain length (mm) | 6.8 ± 7E-2 | 6.5 ± 0.1 | 7.0E-05 | |
| grain width (mm) | 2.9 ± 5E-2 | 2.8 ± 3E-2 | 2.9E-03 | |
| kilo-grain weight (g) | 23.2 ± 1.4 | 20.5 ± 1.0 | 9.9E-04 | |
| primary branch number | 8.8 ± 0.9 | 10.3 ± 1.0 | 7.8E-04 | |
| secondary branch number | 25.1 ± 3.9 | 34.5 ± 6.5 | 2.4E-04 | |
| SD (Hainan) | Heading date (d) | 61.6 ± 0.6 | 73.8 ± 3.1 | 1.3E-05 |
| Spikelets per panicle | 61.9 ± 3.9 | 98.4 ± 13.0 | 8.7E-04 | |
| Grain yield per plant (g) | 18.1 ± 6.5 | 28.1 ± 12 | 1.7E-03 | |
| panicle length (cm) | 17.7 ± 1.3 | 20 ± 0.6 | 9.7E-03 | |
| grain length (mm) | 6.7 ± 0.1 | 6.6 ± 0.1 | 3.7E-02 | |
| grain width (mm) | 2.8 ± 0.1 | 2.7 ± 0.1 | 2.5E-02 | |
| kilo-grain weight (g) | 23.5 ± 0.5 | 22.2 ± 0.9 | 5.9E-03 | |
| primary branch number | 7.3 ± 0.5 | 8.6 ± 0.8 | 6.3E-04 | |
| secondary branch number | 13.4 ± 2.7 | 22.7 ± 4.6 | 1.8E-04 | |
| Root length (cm) | 7.2 ± 1.1 | 8.6 ± 1.6 | 1.0E-02 |
Plants for root length measurement were cultivated on MS media for 15 days under short-day condition. Data was represented by mean ± standard deviation.
To further verify whether the various phenotypes were caused by the upregulation of OsAFB6, OsAFB6 was overexpressed in ZH11 under the control of the 35S promoter. We generated 18 transgenic positive plants. T1 lines from all 18 plants segregated with early- and late-flowering phenotypes in both Hainan (winter, 2015) and Wuhan (summer, 2016). OsAFB6 was overexpressed in all late-flowering phenotype plants but not in early-flowering plants.
In addition, we took T2 lines from four single-copy inserted transgenic T0 individuals with different expression levels as examples. Line 1 (OsAFB6 overexpressed approximately 9-fold) delayed flowering by approximately 5 days under SD and 7 days under LD, with an increase in spikelets per panicle and yield but no significant changes in primary branch number. Line 2 (OsAFB6 overexpressed approximately 19-fold) showed a large delay in heading date (17 days under SD and 19 days under LD) with increased primary branch number, spikelets per panicle and yield; lines 3 and 4 (overexpressed approximately 40 and 45-fold) flowered even later (18 and 19 days under SD and 25 and 26 days under LD, respectively) but with comparable or slightly decreased primary branch number, spikelets per panicle and yield (Table 2). These findings confirmed that the overexpression of OsAFB6 would result in a later heading date and increased grain yield.
Table 2.
Performance of OsAFB6 overexpression lines under LD and SD conditions.
| Conditions | Genotype | Heading date (d) | primary branch number | Spikelets per panicle | Grain yield per plant (g) |
|---|---|---|---|---|---|
| LD (Wuhan) | Control | 76.3 ± 2.2 | 9.8 ± 0.8 | 142.5 ± 9.7 | 22.7 ± 4.7 |
| Line 1 | 83.7 ± 3.2** | 10.3 ± 1.0 | 178.3 ± 10.5** | 33.4 ± 5.5** | |
| Line 2 | 95.3 ± 3.6** | 11.8 ± 0.6* | 183.6 ± 10.9** | 37.9 ± 6.2** | |
| Line 3 | 101.4 ± 2.7** | 10.2 ± 0.8 | 148.5 ± 11.7 | 19.8 ± 3.2* | |
| Line 4 | 102.7 ± 3.6** | 9.5 ± 0.7 | 135.7 ± 8.8** | 18.9 ± 3.7* | |
| SD (Hainan) | Control | 70.6 ± 2.9 | 8.7 ± 0.6 | 95.3 ± 1.8 | 16.4 ± 4.3 |
| Line 1 | 75.6 ± 2.9* | 9.2 ± 1.0 | 111.7 ± 11.4** | 21.3 ± 6.8** | |
| Line 2 | 87.0 ± 3.0** | 9.9 ± 0.5* | 118.7 ± 10.1** | 24.6 ± 6.4** | |
| Line 3 | 88.8 ± 1.6** | 8.4 ± 0.8 | 95.1 ± 13.8 | 14.2 ± 5.6 | |
| Line 4 | 90.0 ± 3.4** | 7.6 ± 0.9 | 81.6 ± 9.5** | 13.9 ± 3.7* |
*, ** Significant difference at 5% and 1% level as compared to control lines. Data was represented by mean ± standard deviation.
However, the dsRNA-mediated gene silencing transgenic plants and CRISPR plants of OsAFB6 in the ZH11 background showed no phenotypic changes (Fig. S4). It is likely that gene redundancy exists among six members in the TIR1/AFB family.
Expression of OsAFB6 in the meristem might be associated with increased primary branch number in the OXOsAFB6 mutant
Sequence analysis showed that OsAFB6 covered a 2020-bp region of the genome, consisting of three exons and two introns, with a coding sequence of 1812 bp encoding a protein of 603 amino acids. One F-box domain located in its N-terminal region, described as a recognizer for ubiquitination targets, and two AMN1 domains containing many LRR repeats located in the middle and C terminus of the OsAFB6 protein (Fig. S5). Many cis-acting elements were identified in the promoter (Table S1), including common elements related to transcription activity, elements that respond to light, auxin and other hormones such as ABA and GA, elements involved in the stress response and others.
The expression profile of OsAFB6 was extracted from online microarray data (http://crep.ncpgr.cn/crep-cgi/home.pl). OsAFB6 was highly expressed in tissues such as plumule, shoot apical meristem, young inflorescence and reproductive organs (stigma and ovary) compared to mature leaves and seedlings (Fig. 2A). The OsAFB6 gene expression level gradually decreased with the growth of the inflorescence. Samples of ovary, inflorescences and stigma were collected from exclusive organs. However, for the young plumule, the samples are collected from the whole plumule (from basal to tip). To test whether the expression of OsAFB6 probably exhibited gradient changes in plumula, a construct with the GUS gene driven by the 2-kb OsAFB6 native promoter was introduced into the wild-type plant ZH11. GUS staining showed that the site near the embryo where the plumule came out displayed a stronger signal than the other parts, showing an expression gradient that decreased from the basal part to the tip (Fig. 2B,C). All these results illustrated that from germination to the heading stage, OsAFB6 is preferentially expressed in tissues where meristem cells with more activities are located.
Figure 2.
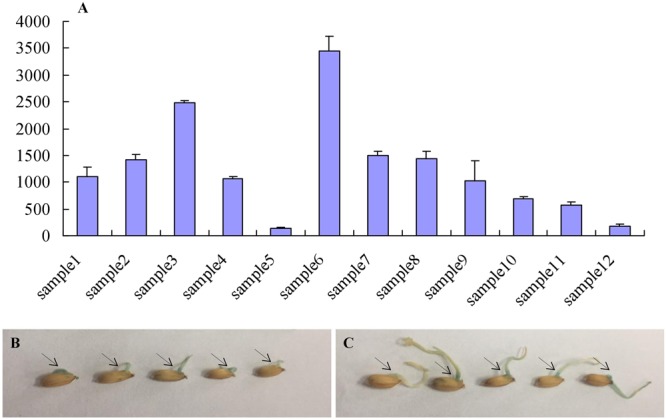
Expression pattern of OsAFB6. (A) Expression of OsAFB6 in different tissues and different stages (data from http://crep.ncpgr.cn/crep-cgi/home.pl). Sample1: plumule; sample2: stigma; sample3: ovary; sample4: 7-day-old seedling; sample5: mature leaf; sample6: SAM sample7: young inflorescence (up to 3 cm); sample8: inflorescence (3–5 cm); sample9: inflorescence (5–10 cm); sample10: inflorescence (10–15 cm); sample11: inflorescence (15–22 cm); sample12: inflorescence (22–30 cm). (B,C) Pictures of the OsAFB6 promoter-driven GUS transgenic material: (B) one day after germination and (C) two days after germination. These materials were soaked in GUS reagent for another 24 hours.
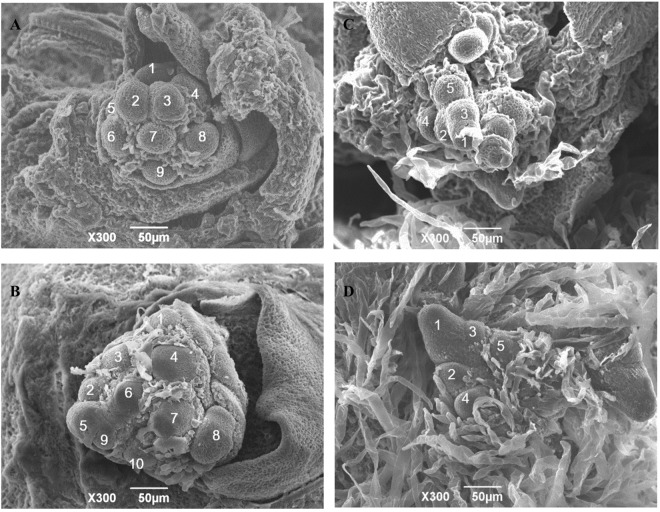
In addition, we performed scanning electron microscopy to identify any cellular changes in tissues with high OsAFB6 expression. The primary and secondary branches of the young panicles before bract hair initiation were observed in OXOsAFB6 mutants. The primary branch number was 8.8 in control plants and 10.3 in OXOsAFB6 mutants, whereas the secondary branch numbers on each primary branch were comparable (approximately 5; Fig. 3).
Figure 3.
Scanning Electronic Microscope pictures for branch primordia of the OXOsAFB6 mutants and the control plants. Primary branch primordia of the control plant (A) and OXOsAFB6 mutant (B), secondary branch primordia of the control plant (C) and (D) OXOsAFB6 mutant. The numbers on the picture indicate the number of branch primordium.
OsAFB6 increased the CK concentration and decreased the IAA concentration in young panicles
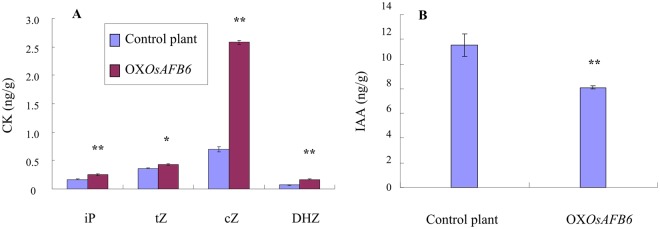
To uncover the function of OsAFB6 in the hormone signaling pathway, we measured endogenous cytokinin and IAA concentrations in young panicles of the OXOsAFB6 mutants and the control plants. N6-(Δ2-isopentenyl) adenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DHZ) were four active CK forms36. Concentrations of all four active CK forms were significantly higher in the OXOsAFB6 mutant than in the wild type (Fig. 4A), especially cZ with more than a three times increase. However, the IAA concentration was significantly lower in the OXOsAFB6 mutant plants (OXOsAFB6 mutant plants: 8.1 ± 0.1 ng/g, control plants: 11.5 ± 0.9 ng/g) (Fig. 4B). In summary, the overexpression mutant of OsAFB6 increased the CK concentration and decreased the IAA concentration in young panicles.
Figure 4.
Cytokinin and IAA concentration of young panicles. iP: N6-(Δ2-isopentenyl) adenine. tZ: trans-zeatin. cZ: cis-zeatin. DHZ: Dihydrozeatin. There were four active forms of CK. OXOsAFB6 and control plants represent the OsAFB6 overexpression mutant and the Zhonghua 11 plant as the negative control, respectively. Error bars, standard deviation. *, ** Indicates significance at the level of P < 0.05 and 0.01, respectively.
OsAFB6 delays heading date by repressing the key flowering gene Ehd1
As shown above, the mutant OXOsAFB6 significantly delayed the heading date and showed strong photoperiod sensitivity (Table 1). To elucidate the associated mechanism, seven key genes involved in the photoperiod flowering pathway were transcriptionally examined by qRT-PCR under both LD and SD conditions. Expression level analysis showed that Ehd1, as well as the florigen genes Hd3a and RFT1, was greatly suppressed in OXOsAFB6 plants (Fig. 5). The expression levels of other genes, such as OsGI, Ghd7, Ghd7.1 and Hd1, were comparable to those of the control plants (Fig. S6). Hence, OsAFB6 overexpression dramatically decreased the expression level of florigen genes by repressing Ehd1, which ultimately resulted in late flowering.
Figure 5.
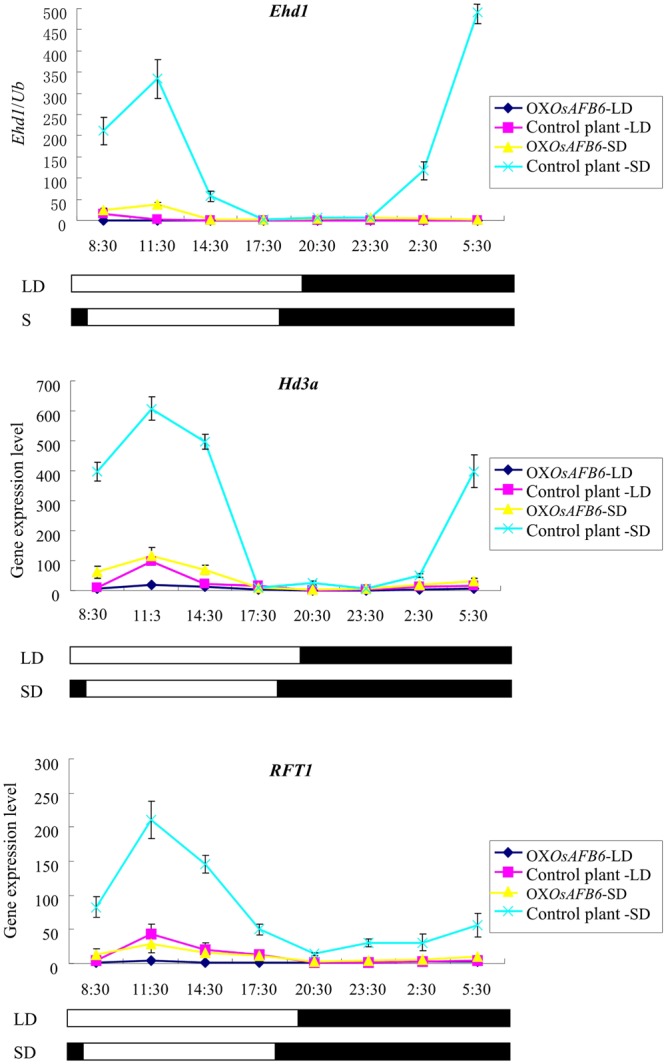
Expression levels of key flowering genes from the photoperiod flowering pathway in the OXOsAFB6 mutant and control plant under LD and SD conditions. OXOsAFB6-LD and control plant-LD represent the genotypes of OsAFB6 for the expression mutant and the negative the control plant, respectively, under long-day conditions. OXOsAFB-SD and control plant-SD represent the genotypes under short day conditions. The white bars indicate the light period, and the black bars indicate the dark period. The numbers below the x-axis indicate hours of the days. Error bars, standard deviation.
Global regulation by OsAFB6 involved in flowering and hormone pathways
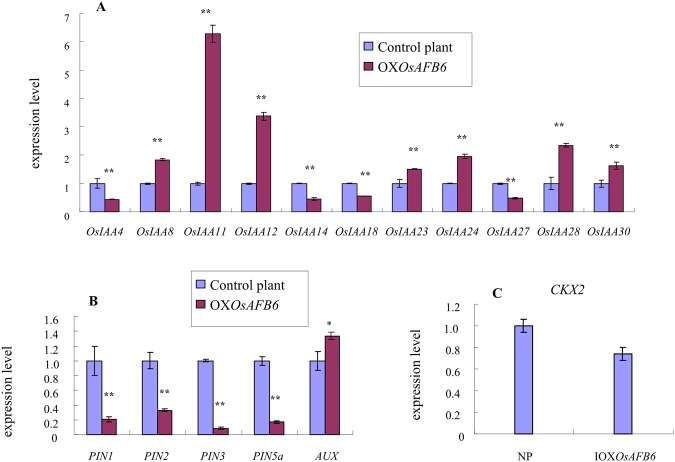
An RNA-seq experiment was also launched to define the global regulation of OsAFB6 in the OXOsAFB6 mutant. A total of 287 differentially expressed genes were identified in young panicles. Among these genes, 58 were upregulated, while the remaining 229 were downregulated (Fig. S7; Excel named RNA sequencing result). GO analysis showed that the up- or down-regulated genes were involved in photosystem, which suggests they might be sensitive to light; in flowering pathway, and in hormone pathways (including auxin and other hormones sensitive, biosynthetic, catabolic processes and signaling pathways). For instance, two flowering-related genes, OsMADS34/PAP2 (LOC_Os03g54170) and OsHAP2F (LOC_Os12g42400), were upregulated, and OsHAP3F (LOC_Os07g41580) was downregulated. Five genes in auxin signaling, degradation and transport pathways were downregulated, including one of the Aux/IAA family genes, OsIAA27 (LOC_Os11g11410); the auxin-rapid response gene OsGH3.3/OsJAR2 (LOC_Os01g12160), which also functioned as a light and JA-signaling gene; the dioxygenase for auxin oxidation gene DAO (LOC_Os04g39980); a gene that promoted auxin polar transport, HOX1 (LOC_Os10g41230); and an auxin efflux transporter gene, OsPIN5b (LOC_Os08g41720). Genes in other hormone pathways, such as the cytokinin-signaling gene OsRR11 (LOC_Os02g42060) and WOX3 (LOC_Os05g02730), which is a gene that might integrate auxin and cytokinin signaling pathways, were upregulated. Four genes in cytokinin pathways were downregulated: cytokinin dehydrogenase CKX5 (LOC_Os01g56810); D14 (LOC_Os03g10620), which functions in the strigolactone (SL) signaling pathway; OsNAC2 (LOC_Os04g38720), which participates in GA signaling; and RAF1 (LOC_Os10g30870), which is responsive to ethylene. These data suggest that OsAFB6 plays an important role in the flowering pathway, the auxin signaling pathway and crosstalk with other hormone signaling pathways.
Further experiments were carried out to confirm the expression levels of genes involved in signaling pathways identified through RNA-seq. In rice, there are 31 genes encoding Aux/IAAs, which are short-lived nuclear proteins, the substrates of the SCF complex17. OsIAA27, a member of the AUX/IAA family, was downregulated in the OXOsAFB6 mutant RNA-seq results. Thus, a total of 31 genes in the family were analyzed by qRT-PCR. Four of them, OsIAA4, 14, 18 and 27, were downregulated, whereas OsIAA8, 11, 12, 23, 24, 28 and 30 were upregulated in the mutant (Fig. 6A). An auxin efflux carrier-like gene OsPIN5b (LOC_Os08g41720) was downregulated in the OXOsAFB6 mutant RNA-seq results. Therefore, the expression of the other 5 auxin transporter genes, including the influx transporter gene AUX1 and efflux transporter genes PIN1, 2, 3, and 5a, was also examined (Fig. 6B). The influx transporter gene AUX1 was upregulated, while all the efflux transporter genes were downregulated. In addition, CKX5, a cytokinin dehydrogenase from the CKX gene family, was found to be downregulated in the OXOsAFB6 mutant RNA-seq results. Interestingly, we found that another well-known gene in this family, CKX2 (Gn1a), which regulates rice grain production29, was also downregulated in the mutant (Fig. 6C). Taken together, our RNA-seq and further experiments show that OsAFB6 plays a critical role in global flowering and hormone regulatory systems and contributes to the phenotype change of the OXOsAFB6 mutant in late flowering and grain yield increases.
Figure 6.
Differentially expressed genes in young panicles by real-time PCR. (A) The expression level of OsIAA/AUX family genes. (B) The expression level of auxin transporter genes. (C) CKX2 expression level. OXOsAFB6 and control plants represent the OsAFB6 overexpression mutant and Zhonghua 11 plant as the negative control, respectively. The expression levels of each gene in the control plants were marked as 1. Error bars, standard deviation. *, ** Indicates significance at the level of P < 0.05 and 0.01, respectively.
Discussion
OsAFB6 overexpression delays heading date by repressing the expression of Ehd1
Rice is a short-day plant that flowers early under short-day conditions. The OsAFB6 overexpression mutant delayed heading under LD and SD conditions but with different effects. This character of OsAFB6 is unique among known flowering genes. Hd1 promotes flowering under SD conditions but delays flowering under LD conditions30. Ghd7, Ghd8, Ghd7.1 and OsMADS56 are specific repressors under LD conditions31,32,34,37. CO3 negatively regulates flowering under SD conditions38. COL4, COL10, COL13 and OsPhyB are constitutive repressors under both SD and LD conditions39–42, while OsMADS50 can promote flowering under both conditions43. Ehd1, a well-known flowering gene, is highly homologous to the B-type response regulator (RR) family genes in the CK signaling pathway44. A recent study revealed that homodimerization of Ehd1 was crucial for its activity, which could be inhibited by OsRR1, an A-type RR member. OsRR1 physically interacted with Ehd1 to form a heterodimer, which was an inactive complex for regulating flowering45. In OXOsAFB6, OsRR1 was upregulated, and more OsRR1 proteins were potentially synthesized. With a lower Ehd1 expression level, more Ehd1 was involved in forming the inactive heterodimer OsRR1-Ehd1; this greatly weakened Ehd1 flowering promotion. OsAFB6 was a light-responsive gene harboring light response cis-elements in the promoter region, which probably explained the fact that its transcription could be induced rapidly after moving from dark to light conditions and decreased dramatically under the inverse conditions (Fig. S2F–H), as well as the fact that the expression levels of OsAFB6 in the OXOsAFB6 mutant were 5–20-fold and 15–35-fold higher compared with the control plants under SD and LD conditions, respectively. Therefore, the flowering difference was dependent on the OsAFB6 expression level, which was regulated by photoperiod.
OsAFB6 increased grain yield through CK pathways
The plant hormone cytokinin is another key regulator of growth and development in plants, such as the formation of SAM23. The CK concentration remained balanced by synthesis and metabolism. The WOX3 gene was reported to downregulate the expression level of the YAB3 gene, which encodes a repressor of the KNOX genes OSH1 and OSH346. KNOX proteins function as positive regulators of CK production through induction of the CK biosynthesis enzyme47. In this study, overexpression of OsAFB6 upregulated the WOX3 gene, which resulted in more CK by alleviating the repression of the CK biosynthesis enzyme by the YAB3 gene. In contrast, Gn1a (OsCKX2), which encodes cytokinin oxidase/dehydrogenase, an enzyme that degrades the phytohormone cytokinin29, was confirmed to be downregulated in the young panicle of the OXOsAFB6 mutant. This would prevent the degradation of CK and result in CK accumulation, which was reported to regulate rice branch and flower numbers48. Therefore, an increased CK level was expected in OXOsAFB6. IAA repressed CK biosynthesis49 and inhibited apical meristem outgrowth50. In fact, the increased endogenous active CK concentration in the mutant OXOsAFB6 agreed with the expression changes of CK-related genes. Taken together, higher CK and lower IAA concentrations in the panicle promoted inflorescence meristem development and resulted in large panicles in the OXOsAFB6 mutant. In addition, the differentiation of both the sieve tubes and the vessels of vascular tissues was induced by auxin and CK51, which was also in accordance with the observation that there were more vascular bundles in OXOsAFB6. The advanced vascular system enhanced assimilate transport capability and ensured a comparable seed setting rate. Finally, the grain yield increased by 50% in the OXOsAFB6 line compared to the control plants (Table 1).
Potential of OsAFB6 for high-yield breeding in different regions
There is a large planting zone for rice worldwide; some regions have a long day length and some have a neutral day length or short day length during the rice-growing season. Therefore, the varieties grown in different regions require distinct photoperiod sensitivity33. In the OXOsAFB6 mutant, grain yield was greatly increased by extending the vegetative growth phase due to delayed heading (Table 1) and an increased CK concentration in young panicles (Fig. 4). Plants with suitable overexpressed levels of OsAFB6 also exhibited late flowering, more primary branches and more grain yield (Table 2). Hence, it is encouraged to generate several overexpression transgenic lines with different expression levels of OsAFB6. Then, all transgenic lines should be recorded for heading date and grain yield in regions with diverse photoperiod and temperature resources. Finally, the most appropriate lines for a region should be determined with an optimal heading date, which will allow the plants to fully utilize the local sunlight and temperature resources to produce high yield potential.
Materials and Methods
Generation of transgenic plants
A 1246-bp genomic fragment of CCT05 from Minghui63 (MH63) and a 1812-bp coding sequence region of OsAFB6 from Zhonghua 11 (ZH11) were separately cloned into the pCAMBIA1301 vector under the 35S promoter. For the OsAFB6-RNAi construct, a 472-bp fragment was amplified from ZH11 and cloned into the pDS1301 vector. For the OsAFB6-CRISPR construct, the target site was located in the first exon. For promoter-GUS assays, a 2-kb promoter fragment of OsAFB6 was cloned into the DX2181 vector. Constructs were transformed into ZH11, a japonica rice variety (O. sativa L. ssp. japonica). For each transformation, we generated transgenic-negative plants from calli that did not integrate any exogenous DNA. They experienced the same tissue culture processes and were used as controls (hereafter named control plants) by Agrobacterium tumefaciens-mediated transformation methods52. The primer sequences for vector construction are listed in Table S2.
Field experiments and trait measurements
In the normal rice growing season, rice plants were grown in the experimental field of Huazhong Agricultural University, Wuhan, China. Each year, seeds were planted in seedling beds in mid-May and were transplanted to the field approximately 25 days later. Ten plants were transplanted in each row as 16.5 × 26 cm within and between rows. Field management followed normal agricultural practices. Heading date (HD) was recorded as the number of days from sowing to the appearance of the first panicle. After ripening, the middle eight of ten plants in a row (the middle five out of seven plants for day length treatment) were measured for plant height (PH) from the paddy field surface to the highest panicle tip. At the harvest stage, the plants were harvested individually to score the number of panicles, the primary and secondary branch number (PBN; SBN), and the lengths of five panicles for each plant. Then, the numbers of full-filled grains and empty grains, as well as the full-filled grain length, width and weight, were obtained. Finally, spikelet number per panicle (SPP), 1000-grain weight (KGW) and grain yield per plant were calculated.
Long-day (LD) and short-day (SD) condition treatments
In the field, the OsAFB6 overexpression mutant (hereafter named OXOsAFB6) and its control plants were grown under natural long-day conditions (LD) in Wuhan (more than 13.5 hours of day length) and natural short-day conditions (SD) in Hainan (less than 12 hours of day length) for recording heading date and yield-related traits. OXOsAFB6 and its control plants were planted in nutrient solution (Table S3) under LD (daytime from 6:00 AM to 8:00 PM) and SD (daytime from 8:00 AM to 6:00 PM) conditions at 28 °C set in growth chambers. The leaves from three plants of each genotype at the same time point were pooled as a single biological sample, and three biological samples were used for checking the expression level of flowering genes by qRT-PCR.
DNA extraction and Southern blotting
Leaves from seedlings were collected for DNA extraction following a previously described method53. A 150-bp fragment of the hygromycin gene was amplified by PCR to use as a probe and was labeled with DIG-High Prime from a DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany). DNA concentrations were measured using a NanoDrop 2000 (Thermo, MA, USA), and 3 µg of DNA was digested with Hind III (Takara, Otsu, Japan) for Southern blotting.
RNA extraction and qRT-PCR
Total RNA was isolated using TransZol reagent (TransGen Biotech, Beijing, China). Then, 3 μg of RNA was reverse-transcribed using oligo (dT) primers and MLV reverse transcriptase (Invitrogen, CA, USA) with the rice Actin1 gene as a control for concentration consistency tests. The ABI7500 real-time PCR detection system was applied (Applied Biosystems, CA, USA). The rice Ubq gene (LOC_Os03g13170) was used as an internal control. Three technical repeats were performed for all analyses. All the primers used are listed in Table S2.
Thermal asymmetric interlaced PCR (TAIL-PCR)
Three specific primers, SP1, SP2 and SP3, were designed according to the T-border sequence of the pCAMBIA vectors, along with six short arbitrary degenerate primers with low annealing temperatures (Table S2). The products from tertiary PCR with clear bands were digested using exonuclease. PCR systems and procedures were performed as previously described54.
Bioinformatic analysis of OsAFB6
We searched the Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE) to understand the gene structure. We used the conserved domain database on NCBI for conserved domain prediction, (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Promoter analysis was performed on PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The expression pattern of OsAFB6 was obtained from the Collection of Rice Expression Profiles on NCPGR (http://crep.ncpgr.cn/crep-cgi/home.pl) through a rice multiplatform microarray search55.
Scanning electron microscopy observation of young panicles
Young panicles at the primary and secondary branch initiation stages were observed using a scanning electron microscope (JSM-6390LV, JEOL, Akishima-shi, Japan), as described previously56.
RNA sequencing and qRT-PCR analysis
Young panicles (<1 mm) were dissected from the main culms of 80 OXOsAFB6 mutant plants and 80 negative control plants in the summer of 2015 and then immediately placed into a 1.5 ml RNase-free tube (Invitrogen, CA, USA) floating in liquid nitrogen. Samples for each group were divided into two halves as 2 biological replicates for RNA extraction. RNA was extracted and sequenced by a biotechnology company (Novogene, Beijing, China). The RNA sequencing data were deposited in The National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database57 under the accession number GSE106954. Several key differentially expressed genes identified from RNA sequencing were later examined by qRT-PCR with RNA samples from young panicles (<1 mm) of 30 OXOsAFB6 mutant plants and 30 negative control plants in the summer of 2016.
Measurement of cytokinin and IAA concentrations
Young panicles (<1 mm) of 40 OXOsAFB6 mutant plants and 40 control plants were dissected in the summer of 2017. Samples for each group were divided into 3 biological replicates. Samples in liquid nitrogen were delivered to Greensword Creation Technology Company (Wuhan, China). Measurements of IAA and four active forms of cytokinin were made using the methods described by Liu et al.58 and Chen et al.59. Three technical repeats were performed for all measurements.
Electronic supplementary material
Acknowledgements
This work was partly supported by the National Special Program for Research of Transgenic Plant of China (2016ZX08009001-002), the National Natural Science Foundation of China (91535301) and the Natural Science Foundation of Hubei province, China (2015CFA006).
Author Contributions
Y.X. conceived and designed the experiments. Q.H. performed the experiments. Q.H. and Y.X. analyzed the data, L.Y., W.H. and J.Z. contributed reagents/materials/analysis tools, Q.H. and Y.X. wrote the paper.
Data Accessibility
The data were deposited into the NCBI GEO database under the accession number GSE106954.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32450-x.
References
- 1.Moller B, Weijers D. Auxin control of embryo patterning. C S H Perspect Biol. 2009;1:a001545. doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benkova E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 3.Rashotte AM, et al. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 2005;139:559–565. doi: 10.1104/pp.104.900172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS. Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 2004;134:28–31. doi: 10.1104/pp.103.031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christian M, et al. How does auxin enhance cell elongation? Roles of auxin-binding proteins and potassium channels in growth control. Plant Biol. 2006;8:346–352. doi: 10.1055/s-2006-923965. [DOI] [PubMed] [Google Scholar]
- 7.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 8.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 9.Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Nati Acad Sci USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napier RM. TIRs of joy: new receptors for auxin. Bioessays. 2005;27:1213–1217. doi: 10.1002/bies.20329. [DOI] [PubMed] [Google Scholar]
- 11.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F- box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Prigge MJ, et al. The Arabidopsis auxin receptor F-Box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3-Genes Genom Genet. 2016;6:1383–1390. doi: 10.1534/g3.115.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia KF, et al. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PloS One. 2012;7:e30039. doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leyser O. Molecular genetics of auxin signaling. Annu Rev Plant Biol. 2002;53:377–98. doi: 10.1146/annurev.arplant.53.100301.135227. [DOI] [PubMed] [Google Scholar]
- 15.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 16.Lee YS, et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010;63:18–30. doi: 10.1111/j.1365-313X.2010.04226.x. [DOI] [PubMed] [Google Scholar]
- 17.Jain M, et al. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Funct Integr Genomics. 2006;6:47–59. doi: 10.1007/s10142-005-0005-0. [DOI] [PubMed] [Google Scholar]
- 18.Shen CJ, et al. Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L) J Exp Bot. 2010;61:3971–3981. doi: 10.1093/jxb/erq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weijers D, et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. Embo J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos T R Soc B. 2012;367:1461–1468. doi: 10.1098/rstb.2011.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarkowski P, et al. Cytokinins in the perianth, carpels, and developing fruit of Helleborus niger L. J Exp Bot. 2006;57:2237–2247. doi: 10.1093/jxb/erj190. [DOI] [PubMed] [Google Scholar]
- 22.Werner T, et al. Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J Exp Bot. 2008;59:2659–2672. doi: 10.1093/jxb/ern134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Nati Acad Sci USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 25.Moubayidin L, Di Mambro R, Sabatini S. Cytokinin–auxin crosstalk. Trends Plant Sci. 2009;14:557–562. doi: 10.1016/j.tplants.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Su YH, Liu YB, Zhang XS. Auxin-cytokinin interaction regulates meristem development. Mol Plant. 2011;4:616–625. doi: 10.1093/mp/ssr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer M, et al. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006;20:2902–2911. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu GW, et al. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015;83:913–925. doi: 10.1111/tpj.12939. [DOI] [PubMed] [Google Scholar]
- 29.Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 30.Yano M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue WY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 32.Yan WH, et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23:969–971. doi: 10.1038/cr.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci Rep. 2015;5:7663–7674. doi: 10.1038/srep07663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan WH, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- 35.Li QP, et al. Duplication of OsHAP family genes and their association with heading date in rice. J Exp Bot. 2016;67:1759–1768. doi: 10.1093/jxb/erv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 37.Ryu CH, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009;32:1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim SK, et al. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta. 2008;228:355–365. doi: 10.1007/s00425-008-0742-0. [DOI] [PubMed] [Google Scholar]
- 39.Lee DJ, Park JW, Lee HW, Kim J. Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J Exp Bot. 2009;60:3935–3957. doi: 10.1093/jxb/erp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa R, et al. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- 41.Sheng PK, et al. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol Biol. 2016;92:209–222. doi: 10.1007/s11103-016-0506-3. [DOI] [PubMed] [Google Scholar]
- 42.Tan JJ, et al. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in Rice. Plant Cell Physiol. 2016;57:798–812. doi: 10.1093/pcp/pcw025. [DOI] [PubMed] [Google Scholar]
- 43.Bian XF, et al. Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating. Ehd1. Plant Cell Rep. 2011;30:2243–2254. doi: 10.1007/s00299-011-1129-4. [DOI] [PubMed] [Google Scholar]
- 44.Doi K, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho LH, Yoon JY, Pasriga R, An G. Homodimerization of Ehd1 is required to induce floering in rice. Plant Physiol. 2016;170:2159–2171. doi: 10.1104/pp.15.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai MQ, Hu YF, Zhao Y, Liu HF, Zhou DX. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007;144:380–390. doi: 10.1104/pp.107.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 48.Barazesh S, McSteen P. Hormonal control of grass inflorescence development. Trends Plant Sci. 2008;13:656–662. doi: 10.1016/j.tplants.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006;45:1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x. [DOI] [PubMed] [Google Scholar]
- 50.Nordstrom A, Tarkowski P, Tarkowska D. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA. 2004;101:8039–8044. doi: 10.1073/pnas.0402504101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aloni R. The induction of vascular tissues by auxin and cytokinin. Plant Hormones 531–546 (1995).
- 52.Supartana P, et al. Development of simple and efficient in Planta transformation method for rice (Oryza sativa L) using Agrobacterium tumefaciens. J Biosci Bioeng. 2005;100:391–397. doi: 10.1263/jbb.100.391. [DOI] [PubMed] [Google Scholar]
- 53.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313X.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 56.Li YB, et al. Chalk5 encodes a vacuolar H+ -translocating pyrophosphatase influencing grain chalkiness in rice. Nat Genet. 2014;46:398–404. doi: 10.1038/ng.2923. [DOI] [PubMed] [Google Scholar]
- 57.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z, Wei F, Feng YQ. Determination of cytokinins in plant samples by polymer monolith microextraction coupled with hydrophilic interaction chromatography-tandem mass spectrometry. Anal Methods-UK. 2010;2:1676–1685. doi: 10.1039/c0ay00334d. [DOI] [Google Scholar]
- 59.Chen ML, et al. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC–ESI-Q-TOF-MS analysis. J Chomatogr B. 2012;905:67–74. doi: 10.1016/j.jchromb.2012.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data were deposited into the NCBI GEO database under the accession number GSE106954.