Abstract
In the last decade, ancient DNA research has grown rapidly and started to overcome several of its earlier limitations through Next-Generation-Sequencing (NGS). Among other advances, NGS allows direct estimation of sample contamination from modern DNA sources. First NGS-based approaches of estimating contamination measured heterozygosity. These measurements, however, could only be performed on haploid genomic regions, i.e. the mitochondrial genome or male X chromosomes, but provided no measures of contamination in the nuclear genome of females with their two X chromosomes. Instead, female nuclear contamination is routinely extrapolated from mitochondrial contamination estimates, but it remains unclear if this extrapolation is reliable and to what degree variation in mitochondrial to nuclear DNA ratios affects this extrapolation. We therefore analyzed ancient DNA from 317 samples of different skeletal elements from multiple sites, spanning a temporal range from 7,000 BP to 386 AD. We found that the mitochondrial to nuclear DNA (mt/nc) ratio negatively correlates with an increase in endogenous DNA content and strongly influenced mitochondrial and nuclear contamination estimates in males. The ratio of mt to nc contamination estimates remained stable for overall mt/nc ratios below 200, as found particularly often in petrous bones but less in other skeletal elements and became more variable above that ratio.
Introduction
The emergence of Next-Generation-Sequencing (NGS) technologies has substantially advanced the field of ancient DNA (aDNA) research1. Besides its high throughput, NGS gave rise to analyses of ancient DNA-specific DNA damage to reveal patterns that authenticate ancient DNA. Post-mortem DNA continuously accumulates characteristic modifications, in particular deamination at the DNA fragment ends2–4. Deamination frequency thus increases over time and can therefore reveal the ancient origin of an aDNA sample3. As a second step required for authenticating ancient human DNA, NGS data allow estimating contamination levels of human DNA directly1. Earlier estimates of contamination levels in NGS data from early modern humans were based on so-called diagnostic positions on the mitochondrial DNA (mtDNA), i.e. nucleotide positions that differ between the sample and a comparative dataset of complete modern human mtDNA sequences from world-wide populations5. Reads exhibiting a different allele at these positions than the majority of reads likely constitute contamination. Today, Bayesian approaches allow even more precise estimates by identifying the proportion of sequencing reads that were considered to be authentic aDNA from the studied individual6.
While quantifications of contamination levels in the haploid and maternally inherited mtDNA are achieved by measuring levels of heterozygosity1, contamination estimates for the diploid nuclear genome can only be obtained for the haploid sex chromosomes in male individuals, especially for the larger X chromosome7. For female individuals, most current studies therefore restrict analyses to nuclear DNA sequences with typical aDNA damage patterns and thus sequences of trusted ancient origin, often reducing datasets by an order of magnitude8. Alternatively, female mtDNA contamination estimates are extrapolated to the nuclear level and only female samples with low mtDNA contamination estimates are used for population genetic analysis on the nuclear genome. Whether this extrapolation is reliable, however, remains untested, in particular given that mitochondrial to nuclear DNA ratios can substantially vary between and even within bone samples9, potentially affecting the extrapolation of mtDNA to nuclear contamination levels.
The reliability of this extrapolation may also depend on the chosen skeletal element given differences in endogenous DNA content and relative amounts of mitochondrial and nuclear DNA. Recent work has identified the petrous part of the temporal bone as a particularly rich source of endogenous DNA and thus an ideal candidate for aDNA studies10,11. Endogenous DNA portions in this skeletal element exceed that in other parts of the skeleton by up to a factor of 40012. This reduces the cost of shotgun sequencing for ancient human genomes to sufficient coverage10 and allows obtaining sequences from geographic regions with typically poor DNA preservation, such as the Near East, Remote Oceania and Africa13–17. The exceptionally good DNA preservation in petrous bones has been linked to the high bone density of the bony labyrinth10, which reaches adult size during gestation18 and shows reduced bone remodeling compared to the surrounding tissue19. These conditions seem to be ideal for DNA preservation. Initial studies suggest low mt/nc ratios for petrous bones20. We hypothesize that these low ratios allow reliable estimates of contamination with human DNA using mtDNA, especially for female individuals.
This is the first study to explore systematic differences in mt/nc ratios between skeletal elements and classically used negative controls, and the relationship between mt/nc ratios and contamination estimates. We compare mt/nc ratios and overall DNA preservation in petrous bones, teeth and other skeletal elements in newly produced and publically available aDNA datasets. Our data show that DNA contamination estimates in males strikingly vary with mt/nc ratios, and argue that the typically low mt/nc ratios in petrous bones make them ideal for making reliable contamination estimates using mtDNA.
Results
Differences in the mt/nc ratio between sites
For each of the three tested skeletal elements we investigated samples from different sites (Table 1), labelled P1 through P7 for petrous bones, T1 through T4 for teeth, and B1 through B4 for other bones. The subsample pairs T3/B1, T4/B2 and P1/T1 originate from the same studies, respectively. Given that any systematic differences between sample origins in, for example, general preservation, laboratory protocols, soil type, climatic conditions and taphonomic alterations can introduce between-sample variation, we first assessed the degree of origin-related variation in mt/nc ratios within each skeletal element (Fig. 1). Indeed, we detected significant variation between sample origins in all skeletal elements (Petrous bones ANOVA F6,50 = 20.15, p < 0.001; Teeth ANOVA F3,181 = 25.25, p < 0.001; Bones ANOVA F3,71 = 9.26, p < 0.001), with at least one sample differing from all remaining ones in each skeletal element (Tukey-HSD tests, Fig. 1). In all the following statistical models comparing differences in mt/nc ratios between tissues, we therefore routinely – and conservatively – accounted for between-origin variation while maintaining the paired nature of samples from the same source by integrating study ID and site ID as random factors.
Table 1.
Samples from different studies used in this analysis.
| Site | Dating | Samples | Reference | ||
|---|---|---|---|---|---|
| Petrous bones | Teeth | Diverse bones | |||
| Oberbipp, Switzerland | approx. 5500 BP | P1 | T1 | This study | |
| Spreitenbach, Switzerland | approx. 4500 BP | T2 | This study | ||
| Europe and Central Asia | 3400 BP–600 AD | T3 | B1 | 38 | |
| Abusir el-Meleq, Egypt | 1300 BP–386 AD | T4 | B2 | 39 | |
| Hungary | 5060–1830 BP | P2 | 10 | ||
| Atapuerca, Spain | 5500–3500 BP | B3 | 40 | ||
| Zagros, Iran | 10000–7000 BP | P3 | B4 | 13 | |
| Zagros, Iran | 10000–9700 BP | 14 | |||
| Muttenz, Switzerland | Neolithic | P4 | This study | ||
| Wartau, Switzerland | Neolithic | P5 | This study | ||
| Seengen, Switzerland | Neolithic | P6 | This study | ||
| Bad Zurzach, Switzerland | Neolithic/Bronze Age | P7 | This study | ||
Figure 1.
Variation in mt/nc ratios between sample sources. Mitochondrial to nuclear DNA ratios (log-transformed) are shown for petrous bones (red), teeth (blue), other bones (green), and controls (grey). Box plots show the raw data median (thick line), interquartile range IQR (box), data within 1.5*IQR (flags), and extreme values (dots). Different lower case letters indicate pairwise differences revealed by Tukey-HSD post-hoc tests.
Differences in the mt/nc ratio between skeletal elements
To assess the mt/nc ratio of potential sources for contamination of archaeological samples, we also measured the mt/nc ratio of human contamination originating from plant extracts21 and laboratory negative controls. If mt/nc ratios of modern human contamination exceeded those of the ancient human DNA, the relative contamination impact would be stronger on the mtDNA than on the nDNA, and vice versa. We found no significant difference in the loge[mt/nc ratio] between both controls (ANOVA F1,21 = 2.33, p = 0.14), therefore combining them for all further analyses.
The mt/nc ratios showed significant variation (Fig. 2) between the three skeletal elements (petrous bones, teeth and bones) and controls (Chi² = 9.54, df = 3, p = 0.023). Petrous bones had significantly lower mt/nc ratios than teeth and showed a trend towards lower mt/nc ratios than other bone samples, while teeth and other bones showed similarly high ratios (Tukey HSD post hoc tests as indicated in Fig. 2). The mt/nc ratios of the combined control group were intermediate between the ratios derived from teeth and bones and those derived from petrous bones. In addition to these differences in mean mt/nc ratios, skeletal elements also showed striking differences in mt/nc ratio variation (Levene’s test F3,336 = 15.01, p < 0.001). Variation was again similar between teeth and other bones (Tukey HSD post hoc P = 0.75) but significantly larger in these two than in petrous bones (both P < 0.0001).
Figure 2.
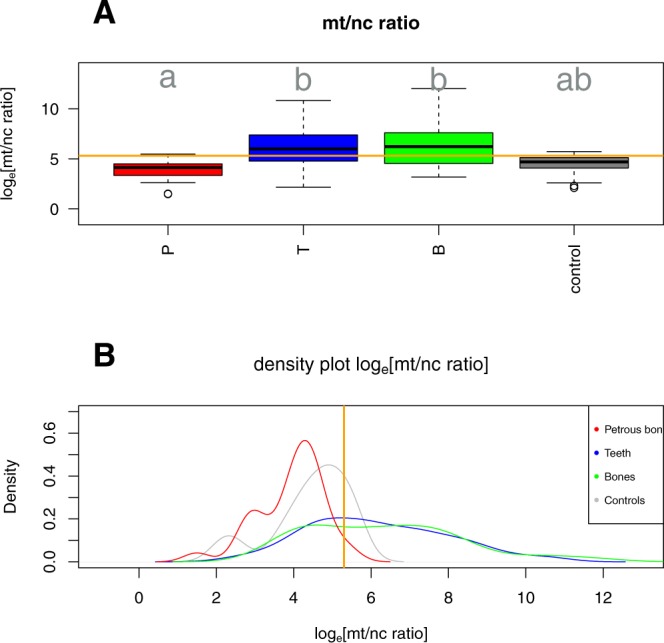
Variation in the mt/nc ratios between skeletal elements. (A) Box plots of the mitochondrial to nuclear DNA ratio (log-transformed) of human DNA in petrous bones (P), teeth (T), other bones (B) and controls. Panel (B) shows density plots of the same data across elements. Orange lines indicate the suggested threshold of the mitochondrial to nuclear ratio of 200 (loge(200) = 5.3).
Factors influencing the mt/nc ratio
To assess the degree to which mt/nc ratios vary with sample characteristics within sample origins, we tested relationships with endogenous DNA content, age, and mapping stringency. Despite much between-sample variation, mt/nc ratios almost consistently decreased with increasing endogenous DNA content across skeletal elements and sampling origins (Fig. 3, mixed model endogenous DNA-effect Chi² = 6.57, df = 1, P = 0.010, overall regression slope = −0.224 ± 0.082, mean ± SE).
Figure 3.
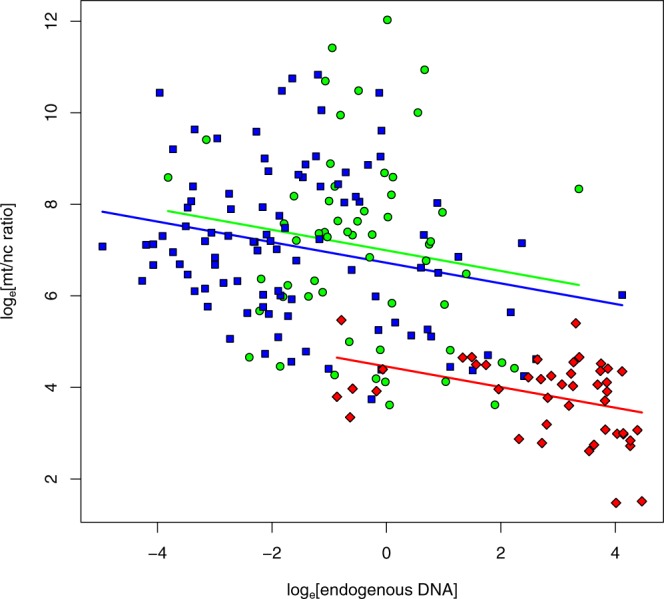
Relationship between the mt/nc ratio and the percentage of endogenous DNA in petrous bones (red), teeth (blue) and other bones (green). Despite overall differences in average mt/nc ratios between skeletal elements and subsamples, these ratios declined consistently when samples contained more endogens DNA.
In contrast, we detected no significant relationship between radio carbon dates of the P2, P3, T3 and B2 samples and their loge[mt/nc ratio] (mixed model sample age-effect Chi² = 0.63, df = 1, P = 0.426). Changes in the mapping stringency in terms of mismatches allowed per read (BWA parameter –n, with tested values 0.001, 0.01, 0.1, 0.2 and 0.8 corresponding to 5, 3.7, 2, 1.7 and 0 mismatches for the average fragment size of T1 and T2 of 55 bp) showed some fluctuation in the mt/nc ratio in T1 and T2 teeth, but no directional effect on mean mt/nc ratios (linear regression F1,125 = 2.049, P = 0.155, R² = 0.016).
Effect of changes in mt/nc ratio on contamination estimates
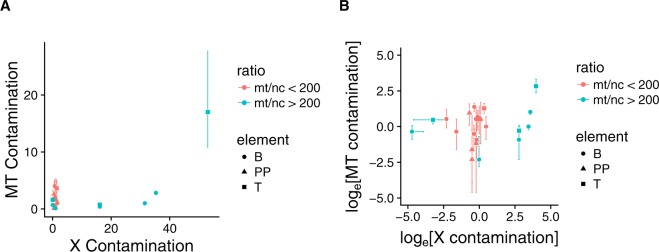
To evaluate whether mitochondrial and nuclear contamination estimates vary with sample mt/nc ratios, we analysed male individuals where sufficient NGS coverage allowed reliable estimates of mtDNA and nuclear contamination levels. Mitochondrial and X-chromosomal contamination rates were by far most strongly biased towards mt-contamination in samples with high mt/nc ratios (Fig. 4). In contrast, where mt/nc ratios were small, nuclear contamination estimates were close to mtDNA contamination rates with an average ratio of 1.47 ± 0.44 SE. In addition, contamination ratios remained stable across loge[mt/nc ratio] between 0 and loge(200) = 5.3 (mixed model loge[mt/nc ratio]-effect Chi2 = 0.80, df = 1, p = 0.37), but strikingly increased with loge[mt/nc ratio] at values exceeding loge(200) = 5.3 (Chi² = 5.95, df = 1, p = 0.015). This stability is reflected in the fact that samples with a ratio below loge(200) = 5.3 are located on the bisecting line when plotting MT contamination against X contamination (Fig. 4).
Figure 4.
Relationship between male mitochondrial and X-chromosome contamination rates in un-transformed (A) and loge transformed (B) data. Different colours indicate samples with low (<loge(200) = 5.3 red) and high (>loge(200) = 5.3 blue) mt/nc ratios.
Discussion
In this work, we show that variation in the mt/nc ratio of ancient DNA samples strongly influences the estimates of contamination by human DNA, which is one of the most critical steps when analysinig ancient human DNA. Besides the characteristic age related misincorporation2, estimates of contamination rates are used for authentication of ancient human DNA. Currently available methods for contamination estimates of the mtDNA and nDNA levels measure heterozygosity in haploid regions of the mtDNA, or the X chromosome in males6,22,23. However, at present it is not possible to estimate contamination levels in the nDNA of female individuals. Recent studies of ancient human nDNA either restrict their analysis to male individuals with known nDNA contamination levels or extrapolate contamination levels of mtDNA to nDNA in the case of females.
Our results show that contamination levels extrapolated from the mtDNA to the nDNA might systematically underestimate the actual level of nDNA contamination. This appears particularly likely for teeth and other bones with a high mt/nc ratio, where such extrapolations (as often done for female specimens) may be highly misleading. For example, three “other bone” samples in the current study exhibited less than 3% contamination on the mtDNA, contrasting to nDNA-contamination between 16 and 30% measured on the X-chromosomes. All three samples were characterized by high mt/nc ratios spanning 870 to >56.000. For these samples, it is not possible to determine from the distribution of reads on the X and Y chromosome with certainty if they are males with a contaminated X chromosome or if they are females with male contamination. Either way, the contamination on the nuclear level is too high for further population genetic analysis and even in absence of Y chromosomal reads as an indicator of contamination, the extrapolation from the mtDNA contamination level could still underestimate the actual nuclear contamination level in case of high mt/nc ratios. Therefore, we advise to take the mt/nc ratio into consideration if only the mtDNA contamination estimate is used in order to choose female samples to be included in population genetic analysis. An effect of mt/nc ratio on the contamination estimates of mtDNA and nDNA as described above was already suggested in the context of analysing Neandertal nuclear and mtDNA9,24 and receives further support by our statistical analysis.
In contrast to samples with a high mt/nc ratio, samples with a ratio below 200 (loge[200] = 5.3) seem to provide a rather reliable estimate of contamination on the mitochondrial level (Fig. 4). We included two types of possible contamination in our study: modern human DNA contamination in archaeological samples measured from aDNA plant extracts from 6,000 year old barley21 and contamination introduced during lab work measured from the negative controls carried along in the entire laboratory workflow. The mt/nc ratios of these potential contamination sources were intermediate to those detected in the three tested skeletal elements (Fig. 2). Therefore, these types of contamination contribute more to the contamination level of mtDNA than nDNA in samples showing lower ratios than these controls. Our systematic comparison of a large number of teeth, diverse bones and petrous bones indicates that the petrous bones display relatively low mt/nc ratios and hence have a high chance to show even lower contamination rates on the nDNA than extrapolated from their mtDNA. In addition, we found that the mt/nc ratio in petrous bones not only tended to be smaller compared to teeth and other bones but also exhibited significantly less variation (Fig. 2). The lower mt/nc ratios in petrous bones increase the chance to obtain reliable extrapolations of nDNA contamination rates from estimated mtDNA contamination. Furthermore, the extraordinarily good preservation of endogenous DNA in petrous bones makes it possible to overcome the limited preservation of aDNA in most environments12. Hansen and colleagues20 show that teeth can have even higher contents of endogenous DNA than petrous bones. In our dataset, however, the majority of petrous bones displayed clearly higher endogenous DNA contents than teeth, leading us to maintain petrous bones as the prime source for highly concentrated and little contaminated endogenous DNA.
From these influences of the mt/nc ratio on the contamination estimates, we conclude that the mt/nc ratio should be considered if mtDNA contamination estimates are used to select female samples for nDNA studies. Reasons for the observed variation in this ratio have already been subject of different studies and no clear reason for the elevation of the mt/nc ratio in some aDNA samples compared to modern DNA has been identified. One possibility could be a bias of the mt/nc ratio originating from bacteria, since mtDNA contains features of a bacterial chromosome and bacterial reads from the environmental background might map better to the mtDNA elevating the mt/nc ratio. However, mapping with different stringencies (different numbers of allowed mismatches per read, BWA parameter -n) did not reveal a particular trend. Allowing more mismatches (-n 0.001, -n 0.01 and -n 0.1) resulted in similar values in the ratio than more stringent mapping (-n 0.2 and -n 0.8). We could therefore not detect any effect of the bacterial background in aDNA extracts on the correlation between high mt/nc ratios and lower percentages of endogenous DNA.
It has been suggested that the higher ratios in aDNA might originate from better protection of DNA in the mitochondrion due to its double membrane25. In addition, Schwarz and colleagues25 and Allentoft and colleagues26 conclude from their observation of longer mtDNA fragments compared to nDNA that mtDNA decays more slowly than nDNA. This would result in increasing ratios through time. No such trend is detectable in our data for petrous bones (P2, P3) and teeth (T3), and only a weak trend in other bones (B2). Therefore, we found no indication that different decay rates cause the observed variation in the mt/nc ratio.
Another reason for differences in mt/nc ratios could be higher metabolic activity in particular skeletal elements, as suggested by Higgins and colleagues27 in their study of different parts of teeth. Their observation indicates that higher mt/nc ratios in dentine may originate from its higher metabolic activity as compared to enamel and cementum. This assumption, of different metabolic activity resulting in different mt/nc ratios could also be applied to other skeletal elements, the petrous bone in particular. In the cortical part of the petrous bone, bone remodelling is suppressed compared to surrounding tissue by a specific signalling-pathway, and the number of metabolic active osteoclasts and osteoblasts is reduced28. Furthermore, the more compact bone regions contain a higher number of osteocytes, which have lower numbers of mitochondria29. The number of mitochondria in osteocytes decreases from periosteal and endosteal surfaces towards the inner and denser bone parts30. Consequently, sampling the hardest part of the petrous bone results in low mt/nc ratios due low mt/nc ratio in this area ante-mortem. Within the densest parts of the cortical bone displaying low mt/nc ratios, the endogenous DNA is also better protected against the environmental background of bacteria, fungi and other microbes resulting in high percentages of endogenous DNA.
In conclusion, we showed that the mt/nc ratio is an important value in aDNA authentication. It strongly influences the accuracy of extrapolating nDNA contamination levels from mtDNA contamination estimates. This approach should be used with particular caution if mt/nc ratios exceed 200. Lower mt/nc ratios are often associated with high percentages of endogenous DNA, as typically found in the densest parts of the petrous bones. In this case, low mt/nc ratios are most likely derived from a low mtDNA concentration in this bone region ante-mortem, which is caused by a low metabolic activity.
Temporal bones, including the petrous bones have already proven to be valuable in physical anthropology. The compact structure of the petrous bone is ideal for preservation in archaeological contexts, therefore, it can be used to estimate the minimal number of individuals31. While age can only be roughly estimated depending on developmental stages32. In addition, the discovery of the higher percentage of endogenous DNA in petrous bone compared to other skeletal elements10,11 constitutes a substantial improvement for the field of aDNA. Our study confirms those findings and shows that mitochondrial to nuclear ratios provide a further argument to extract ancient DNA from petrous bones providing reliable human contamination assessment from both sexes.
Material and Methods
Dataset
We combined shotgun data from 76 new ancient DNA samples from the late Neolithic dolmen burial site of Oberbipp in Switzerland33,34, the late Neolithic multiple burial of Muttenz35, the final Neolithic sites in Switzerland Spreitenbach36, Seengen35, Zurzach35 and cave finds from Wartau37 (detailed description of archaeological sites and laboratory workflow SI section 1 and 2) with published datasets (Table 1). In total, DNA from 317 ancient individuals and 23 controls was analysed for its mtDNA to nDNA (mt/nc) ratio. For each of the three investigated skeletal elements, samples originated from three sources. Different skeletal elements do not originate from the same individuals. Samples for petrous bones (n = 57) included 40 newly processed samples from the previous mentioned Swiss burials (P1, P4, P5, P6, P7), 13 from Gamba and colleagues10 (P2) and four from Broushaki and colleagues13 and Gallego-Llorente and colleagues14 (P3). Teeth samples (n = 185) included 36 from the Swiss burials (T1, T2), 88 from Allentoft and colleagues38 (T3) and 61 from Schuenemann and colleagues39 (T4). P1 and T1 from this study originate from the same multiple burial in Oberbipp, Switzerland but due to the comingled nature of the remains an assignment of the teeth to the petrous bones is not possible. Furthermore, the endogenous DNA in the teeth is too low to allow kinship analysis to identify identical individuals. Therefore, it is possible that teeth and petrous bones are from the same individuals. Finally, diverse bone samples from compact bone parts (n = 75) included 13 from Allentoft and colleagues38 (B1), 52 from Schuenemann and colleagues39 (B2) and eight from Günther and colleagues40 (B3) and two from Broushaki and colleagues13 (B3). Fastq files were downloaded from the European Nucleotide Archive. Radio carbon dates were used from Gamba and colleagues10, Broushaki and colleagues13, Gallego-Llorente and colleagues14 and Schuenemann and colleagues39.
Bioinformatic processing
All data were processed with the EAGER pipeline41. If necessary, adapters were removed and paired-end data was merged using Clip&Merge41. Mapping was performed with BWA with the mismatch parameter set to 0.01 and a seed length of 1,000. If necessary, PCR duplicates were removed using DeDup41. Mitochondrial to nuclear ratios were calculated by dividing the mean coverage of the mitochondrial genome by the mean coverage of the nuclear genome to take into account the different length of nuclear and mitochondrial genome. Samples with no reads on the mitochondrial chromosome resulting in a ratio of zero as found in some negative controls, plant extracts, and extremely bad preserved teeth samples were excluded.
Reads mapping to the mitochondrial genome were extracted from the BAM files and the mitochondrial genome was reconstructed using the software schmutzi22. Mitochondrial contamination was then estimated with a Bayesian approach as described in Fu and colleagues6. Sex determination was performed after Skoglund and colleagues42. X chromosomal contamination in males with more than 0.5-fold coverage on the X chromosome was estimated using ANGSD23.
Statistical analysis
Statistical analysis was performed in R version 3.4.3 (R Core Team 2017). We first used one-way ANOVA to assess differences in loge[mt/nc ratio] (i.e., the raw mt/nc-ratios loge-transformed to approach normality) between the sub-samples within each of the three different skeletal elements and controls, followed by Tukey HSD post hoc tests. Second, to assess overall differences in loge[mt/nc ratio] between skeletal elements and controls, we performed linear mixed models as implemented in the lme4-package for R43. Given substantial variation in loge[mt/nc ratio] between different sample origins and studies detected above, this model contained the source study as a random slope factor, allowing between-study variation in the main effect while maintaining the paired nature of samples from the same source. Pairwise Tukey HSD post hoc tests were calculated from this model using the lsmeans package for R44. Differences in the variance of the loge[mt/nc ratio] between skeletal elements and control were tested using Levene’s test, followed by Tukey HSD post hoc tests.
To assess how loge[mt/nc ratio] varies with DNA-content (log-transformed to approach normality) we constructed a linear mixed model as above with loge[mt/nc ratio] as the response variable, loge[endogenous DNA] as the predictor, and subsample as random slope factor allowing differential regression slopes between subsamples. In addition, we added skeletal element and its interaction with loge[endogenous DNA] as fixed factor to assess slope consistency between skeletal elements. Given that this interaction was clearly insignificant (Chi² = 0.18, df = 2, P = 0.91) we removed it from the final reported model. Information about endogenous DNA content was only available for all Swiss and Egyptian samples. An identical model structure was used to assess the relationship between sample age and loge[mt/nc ratio], now with sample age as the main predictor.
Differences in loge[mt/nc ratio] between different mapping stringencies were assessed using linear regression. Information about loge[mt/nc ratio] at different mapping stringencies was obtained for the teeth of the Swiss burials sites Oberbipp and Spreitenbach (T1) since they have low endogenous content and a high portion of bacterial background.
Finally, to assess how the ratio of mitochondrial over X-chromosomal contamination varies with loge[mt/nc ratio] we constructed linear mixed models as above, but separate for samples with low (<5.3) and high (>5.3) loge[mt/nc ratio]. Contamination ratio served as the response variable, loge[mt/nc ratio] as the predictor, and skeletal element as a random slope factor allowing differential responses to loge[mt/nc ratio].
Electronic supplementary material
Acknowledgements
We would like to thank the archaeological services of the Cantons of Aargau, Baselland, Bern, and Sankt Gallen for providing access to their collections as well as Anne Gibbson for proof reading the manuscript.
Author Contributions
J.K. conceived and designed the general research. A.F., E.R. and G.U.N. performed DNA-extraction and the preparation of sequencing libraries. I.S., N.S., S.L. and A.H. provided samples. I.S. performed morphological analysis of the Swiss skeletons from Oberbipp, Wartau and Zurzach. A.F. and N.A. performed bioinformatic and statistical analyses. A.F., N.A., V.J.S. and J.K. wrote the manuscript and S.L. contributed to it. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anja Furtwängler, Email: anja.furtwaengler@uni-tuebingen.de.
Johannes Krause, Email: krause@shh.mpg.de.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32083-0.
References
- 1.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. NatRev Genet. 2011;12:603–14. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 2.Briggs AW, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104.37:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawyer S, et al. Temporal Patterns of Nucleotide Misincorporations and DNA Fragmentation in Ancient DNA. PLoS ONE. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dabney J, Meyer M, Paabo S. Ancient DNA Damage. Cold Spring Harbor Perspectives in Biology. 2013;5:a012567. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause J, et al. A Complete mtDNA Genome of an Early Modern Human from Kostenki, Russia. Current Biology. 2010;3:231–236. doi: 10.1016/j.cub.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 6.Fu Q, et al. A Revised Timescale for Human Evolution Based on Ancient Mitochondrial Genomes. Current Biology. 2013;7:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–62. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, Q. et al. The genetic history of Ice Age Europe. Nature, 243 (2016). [DOI] [PMC free article] [PubMed]
- 9.Green RE, et al. The Neandertal genome and ancient DNA authenticity. EMBO J. 2009;28:2494–502. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamba, C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Comms, 5257 (2014). [DOI] [PMC free article] [PubMed]
- 11.Rasmussen M, et al. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–29. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinhasi R, et al. Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PLoS ONE. 2015;l0:e0129102. doi: 10.1371/journal.pone.0129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broushaki F, et al. Early Neolithic genomes from the eastern Fertile Crescent. Science. 2016;6298:499–503. doi: 10.1126/science.aaf7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallego-Llorente, M. et al. The genetics of an early Neolithic pastoralist from the Zagros, Iran. Sci. Rep., 31326 (2016). [DOI] [PMC free article] [PubMed]
- 15.Llorente MG, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science. 2015;350:820–22. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 16.Skoglund P, et al. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016;538:510–13. doi: 10.1038/nature19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Loosdrecht, M. et al. Pleistocene North African genomes link Near Eastern and sub-Daharan African human populations Science, eaar8380 (2018). [DOI] [PubMed]
- 18.Richard C, et al. New insight into the bony labyrinth: A microcomputed tomography study. AurisNasus Larynx. 2010;37:155–61. doi: 10.1016/j.anl.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Sorensen MS. Temporal bone dynamics, the hard way. Formation, growth, modeling, repair and quantum type bone remodeling in the otic capsule. Acta Otolaryngol Suppl. 1994;512:1–22. [PubMed] [Google Scholar]
- 20.Hansen HB, et al. Comparing Ancient DNA Preservation in Petrous Bone and Tooth Cementum. PloS ONE. 2017;12.1:e0170940. doi: 10.1371/journal.pone.0170940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mascher M, et al. Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley. Nat Genet. 2016;48:1089–93. doi: 10.1038/ng.3611. [DOI] [PubMed] [Google Scholar]
- 22.Renaud G, Slon V, Duggan AT, Kelso J. Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome biology. 2015;16.1:1. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:6052:94–8. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green RE, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;7117:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz C, et al. New insights from old bones: DNA preservation and degradation in permafrost preserved mammoth remains. Nucleic Acids Research. 2009;37:3215–29. doi: 10.1093/nar/gkp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allentoft ME, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proceedings of the Royal Society B: Biological Sciences. 2012;1748:4724–4733. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins D, et al. Differential Nuclear and Mitochondrial DNA Preservation in Post-Mortem Teeth with Implications for Forensic and Ancient DNA Studies. PLoS ONE. 2015;5:e0126935. doi: 10.1371/journal.pone.0126935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bloch SL, Kristensen SL, Sørensen MS. The Viability of Perilabyrinthine Osteocytes: A Quantitative Study Using Bulk-Stained Undecalcified Human Temporal Bones. Anat Rec. 2012;7:1101–1108. doi: 10.1002/ar.22492. [DOI] [PubMed] [Google Scholar]
- 29.Dudley HR. The Fine Structure Of Bone Cells. The Journal of Cell Biology. 1961;3:627–649. doi: 10.1083/jcb.11.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frikha-Benayed Dorra, Basta-Pljakic Jelena, Majeska Robert J., Schaffler Mitchell B. Regional differences in oxidative metabolism and mitochondrial activity among cortical bone osteocytes. Bone. 2016;90:15–22. doi: 10.1016/j.bone.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterholtz Anna J., Baustian Kathryn M., Martin Debra L., Potts Daniel T. Commingled and Disarticulated Human Remains. New York, NY: Springer New York; 2013. Commingled Human Skeletal Assemblages: Integrative Techniques in Determination of the MNI/MNE; pp. 35–50. [Google Scholar]
- 32.Cunningham Craig, Scheuer Louise, Black Sue. Developmental Juvenile Osteology. 2016. The Skull; pp. 43–148. [Google Scholar]
- 33.Ramstein M, Schimmelpfennig D, Lösch S. Ein neolithischer Dolmen an der Steingasse in Oberbipp. Archäologie Schweiz. 2014;37(3):4–15. [Google Scholar]
- 34.Siebke, I., Furtwängler, A., Hafner, A., Krause, J. & Lösch, S. An Interdisciplinary Project on the Neolithic Population of Modern Switzerland. 86th Annual Meeting of the American Association of Physical Anthropologists. American Journal of Physical Anthropology. p.358 (2017)
- 35.Bleuer, E., Doppler, T & Fetz, H. Gräber im näheren und weiteren Umfeld von Spreitenbach in Spreitenbach-Moosweg (Aargau, Schweiz): Ein Kollektivgrab um 2500 v Chr (ed. Doppler, T.) 233–266 (Basel 2012).
- 36.Doppler, T. Spreitenbach-Moosweg: ein Kollektivgrab um 2500 v. Chr. Archäologie Schweiz, Vol. 51 (2012).
- 37.Stehrenberger T. Obere Höhle bei der “Procha Burg” Höhlenpost148 (2016).
- 38.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;7555:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 39.Schuenemann VJ, et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat Comms. 2017;8:15694. doi: 10.1038/ncomms15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Günther T, et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci USA. 2015;38:11917–11922. doi: 10.1073/pnas.1509851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peltzer A, et al. EAGER: efficient ancient genome reconstruction. Genome biology. 2016;17.1:1. doi: 10.1186/s13059-016-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoglund P, Stora J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. Journal of Archaeological Science. 2013;40.12:4477–4482. doi: 10.1016/j.jas.2013.07.004. [DOI] [Google Scholar]
- 43.Bates D, Maechler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67(1):1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 44.Lenth RV. Least-Squares Means: The R Package lsmeans. Journal of Statistical Software. 2016;69(1):1–33. doi: 10.18637/jss.v069.i01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.