Abstract
ADAMTS proteins are a superfamily of 26 secreted molecules comprising two related, but distinct families. ADAMTS proteases are zinc metalloendopeptidases, most of whose substrates are extracellular matrix (ECM) components, whereas ADAMTS-like proteins lack a metalloprotease domain, reside in the ECM and have regulatory roles vis-à-vis ECM assembly and/or ADAMTS activity. Evolutionary conservation and expansion of ADAMTS proteins in mammals is suggestive of crucial embryologic or physiological roles in humans. Indeed, Mendelian disorders or birth defects resulting from naturally occurring ADAMTS2, ADAMTS3, ADAMTS10, ADAMTS13, ADAMTS17, ADAMTS20, ADAMTSL2 and ADAMTSL4 mutations as well as numerous phenotypes identified in genetically engineered mice have revealed ADAMTS participation in major biological pathways. Important roles have been identified in a few acquired conditions. ADAMTS5 is unequivocally implicated in pathogenesis of osteoarthritis via degradation of aggrecan, a major structural proteoglycan in cartilage. ADAMTS7 is strongly associated with coronary artery disease and promotes atherosclerosis. Autoantibodies to ADAMTS13 lead to a platelet coagulopathy, thrombotic thrombocytopenic purpura, which is similar to that resulting from ADAMTS13 mutations. ADAMTS proteins have numerous potential connections to other human disorders that were identified by genome-wide association studies. Here, we review inherited and acquired human disorders in which ADAMTS proteins participate, and discuss progress and prospects in therapeutics.
Keywords: Osteoarthritis, atherosclerosis, genome-wide association studies (GWAS), metalloprotease, protease, extracellular matrix
1. INTRODUCTION
Most reviews on secreted proteases view them as destructive entities and drug targets, but in fact, their evolutionary expansion uncovered by genome sequencing suggests otherwise. Indeed, they have evolved to serve essential roles in morphogenesis, tissue remodeling and immunity [1]. This may be particularly true for the ADAMTS proteases, which do not appear to be capable of indiscriminate destruction by virtue of having an apparently high catalytic specificity, and whose mutations both in humans and mice suggest participation in distinct pathways [2]. The few instances where ADAMTS proteases are excessively active in disease are overwhelmingly outnumbered by disorders arising from loss of function, typically by gene mutations. Genetic disorders of ADAMTS proteases were reviewed in detail recently [2]. We provide an update on those here and focus the review on ADAMTS proteins in acquired human conditions. Many acquired disorders have a small, but significant genetic component that can be revealed, with inherent limitations, using genome-wide association studies (GWAS). Indeed, GWAS made an initial association of ADAMTS7 with atherosclerosis, which now appears to be widely accepted. On the other hand, several published ADAMTS associations with disease were made solely using ELISA assays or transcriptome analysis to link higher or lower levels of ADAMTS proteins and genes with disease states, but without further analysis. These associations are not discussed here. Several ADAMTS proteases are implicated as anti-cancer or pro-tumorigenic molecules and undoubtedly have a role in the biology of many cancers. However, there are no instances where ADAMTS proteins are unequivocally identified as causal (i.e., primary oncogenes), or substantially protective (i.e., primary tumor suppressors) in specific cancer types. The reader is referred to a recent review of ADAMTS proteases in cancer for coverage of this complex topic [3]. Here, we offer a curated discussion of the most robust established disease roles and emerging disease associations.
1.1. ADAMTS proteases and ADAMTS-like proteins
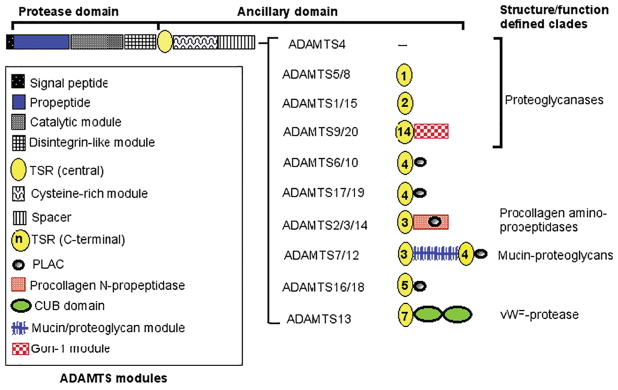
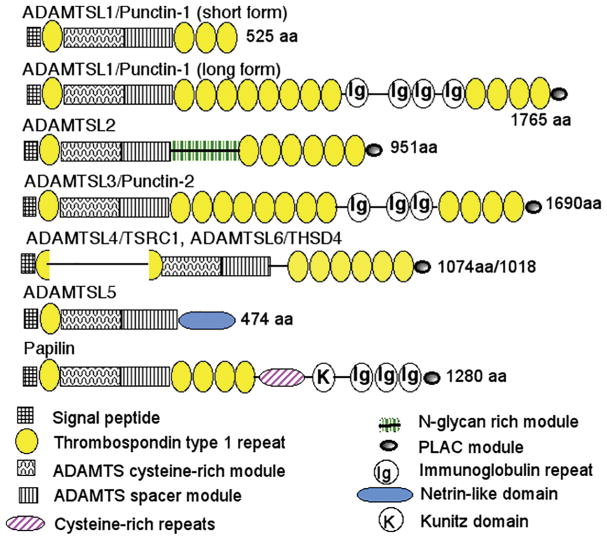
Of the 26 ADAMTS superfamily genes known in humans and mice, 19 encode zinc metalloproteinases, constituting the ADAMTS protease family (Figure 1). Although their numbering extends from ADAMTS1 to ADAMTS20, the symbols ADAMTS5 and ADAMTS11 described the same protease, and ADAMTS11 is no longer used. The discovery of genes encoding 7 ADAMTS-like proteins (ADAMTSL1 through ADAMTSL6 and PAPLN) (Figure 2) completed genetic cataloguing of the superfamily. ADAMTSLs are the products of distinct genes and none arise from alternative splicing of ADAMTS protease genes. They lack the zinc metalloproteinase domain as well as the propeptide and disintegrin-like domain present in all ADAMTS proteases. The latter two domains may therefore be required for regulation of ADAMTS catalytic activity, and there is evidence that they do so. Specifically, the ADAMTS propeptide may function as an intramolecular chaperone for correct folding of the catalytic domain, e.g. as shown for ADAMTS9 [4], and akin to matrix metalloproteinase propeptides, it may also have a role in inhibiting catalytic activity [5, 6]. High-resolution structural analysis has suggested that the ADAMTS4 and ADAMTS5 disintegrin-like module may extend the substrate-binding region and thus contribute to catalytic specificity [7, 8]. ADAMTSLs retain all other ADAMTS modules, i.e., those comprising the C-terminal ancillary domain of ADAMTS proteases, namely the thrombospondin type 1 repeats (TSRs), cysteine-rich and spacer modules and protease and lacunin (PLAC) domain in specific configurations (Figure 2). However, as is evident from Figure 1 and Figure 2, none of the ADAMTSLs have the precise composition of any of the ADAMTS ancillary domains. Since ADAMTS protease ancillary domains have a substantial role in substrate recognition, and therefore in ADAMTS specificity, it is likely that ADAMTSLs also have specific extracellular ligands. Current evidence suggests that several are ECM-binding proteins that function at the cell-matrix interface [9–11]. Fibrillin microfibrils are known to bind ADAMTSL2, ADAMTSL4, ADAMTSL5 and ADAMTSL6 [12–15] and members of the lysyl oxidase family were also recently identified to bind ADAMTSL2 and ADAMTS10 [16]. Additionally, ADAMTSL2 binds latent TGFβ-binding protein 1 and is thought to regulate TGFβ activity via its interaction with this molecule and fibrillin-1 [12, 17]. Thus, ADAMTSLs could be viewed as matricellular proteins, which are dynamically expressed non-structural proteins that reside in ECM and have regulatory roles.
Figure 1. Mammalian ADAMTS proteases.
The domain backbone shared by each ADAMTS protease is shown at the top and modules present in each ADAMTS are shown in the box at left. The modular organization of specific ADAMTS homologous pairs or groups is indicated on the right and the key to these modules is located at the bottom of the figure. The homologous pairs are named according to structural or functional characteristics that best defines them. Domain structures are based on reference sequences obtained from GenBank.
The figure was originally published in the Journal of Biological Chemistry: Apte, S.S. The ADAMTS Superfamily- Functions, Structure and Mechanisms. J. Biol. Chem. 2009; 284(46): 31493–31497. © The American Society for Biochemistry and Molecular Biology.
Figure 2. Mammalian ADAMTSLs.
The domain structure of each ADAMTSL is shown according to the key at the bottom. The short and long forms of ADAMTSL1 are splice variants and the long form comprises a homologous pair with ADAMTSL3. ADAMTSL4 and ADAMTSL6 comprise a homologous pair in which TSR1 is split by an insertion. Domain structures are based on reference sequences obtained from GenBank.
The figure was originally published in the Journal of Biological Chemistry: Apte, S.S. The ADAMTS Superfamily- Functions, Structure and Mechanisms. J. Biol. Chem. 2009; 284(46): 31493–31497. © The American Society for Biochemistry and Molecular Biology.
Although the structural similarities of ADAMTS proteases and ADAMTSLs intuitively suggest a functional relationship of ADAMTSLs vis-à-vis ADAMTS proteases, direct evidence of biochemical or physiological interactions is limited. For example, Drosophila papilin was shown to regulate activity of bovine ADAMTS2 in procollagen processing and to bind fibrillin-1, which interacts with several ADAMTS proteases as well as ADAMTSL2 [18, 19]. The physiological relevance of these interactions is unknown. However, a putative relationship is supported by phenotypically similar disorders arising from mutations of ADAMTS proteases (i.e., ADAMTS10, ADAMTS17) and ADAMTSLs (ADAMTSL2, ADAMTSL4) [20, 21]. Systematic testing of interactions between individual ADAMTS proteases and ADAMTS-like proteins remains to be undertaken and will be relevant to human disorders.
Altogether, evolutionary conservation and expansion of ADAMTS proteins suggests that they are crucial for mammalian physiology, which is supported by strong phenotypes compromising morphogenesis, mobility or reproduction observed in many ADAMTS gene knockouts in mice (see Table 2 in [2]). Because of inherent challenges in expressing and purifying ADAMTS proteases, their enzymology and substrate repertoire are relatively poorly characterized. In contrast, modern genetic tools are not similarly constrained and have provided a wealth of insights on ADAMTS mendelian disorders in humans and other animals and complex multifactorial acquired conditions in humans.
1.2. Relevance of ADAMTS homologous pairing
Several ADAMTS proteases (e.g., ADAMTS9 and ADAMTS20, ADAMTS7 and ADAMTS12) as well as ADAMTSLs (e.g. ADAMTSL1 and ADAMTSL3, ADAMTSL4 and ADAMTSL6) form highly homologous pairs reflecting their evolutionary origin by duplication of ancestral genes and suggesting overlapping function (sub-functionalization) (Figure 1 and 2). Spatial and temporal co-expression of homologous ADAMTS proteins, as well as overlapping activity [22–24] is common, which may ensure over-engineering of key biological pathways and morphogenetic processes. This should be anticipated as a primary consideration in analysis of human disorders and their treatment. Many of the ADAMTS mouse knockouts have more severe phenotypes when combined with knockout of their homologs [22–24]. Thus, the effects of single ADAMTS gene mutations may be masked or modified in human Mendelian conditions by concurrent or compensating expression and functional overlap of a homolog, which then constitutes a key modifier gene. Homologous pairing is also relevant to development of drugs intended to block specific ADAMTS proteases, since cross-inhibition of closely related homologs could cause more severe side effects than may be expected from blockade of a single protease. Active consideration of homologs is therefore vital in thinking about the function and targeting of ADAMTS proteins in disease.
2. ADAMTS gene mutations and genetic associations
2.1. Update on Mendelian disorders resulting from ADAMTS mutations
Table 1 provides an update on human Mendelian disorders discussed in detail in a previous review in this journal, which also described outcomes of inactivation of ADAMTS protease genes in mice [2]. Befitting enzymes and/or non-structural molecules, most ADAMTS mutations act recessively, i.e., requiring two mutant alleles to be manifest. ADAMTS2 mutations lead to Ehlers-Danlos syndrome (dermatosparactic type or VIIc), whose major manifestation is severe skin fragility, although other anomalies, including abnormal dentition have been described [25, 26]. ADAMTS10, ADAMTS17 and ADAMTSL2 mutations result in acromelic dysplasias, syndromes characterized by short stature and disproportionate distal limb shortening, each accompanied by other characteristic anomalies. These conditions have been extensively reviewed [20, 21, 27, 28]. ADAMTSL4 mutations lead to ectopia lentis and ectopia lentis et pupillae, which arise from defects in the ocular zonule, an acellular structure comprising fibrillin microfibrils that centers the lens in the optic path and in the iris, respectively [21, 29].
Table 1.
Mendelian disorders resulting from mutations in human ADAMTS genes
| Mendelian condition | MIM number | Gene name, chromosomal locus | Inheritance |
|---|---|---|---|
| Ehlers-Danlos syndrome (EDS), dermatosparaxis type or type VIIC | 225410 | ADAMTS2, 5q35.3 [26] | Autosomal recessive |
| Hennekam lymphangiectasia-Lymphedema syndrome 3 | None available | ADAMTS3, 4q13.3 [30] | Autosomal recessive |
| Weill-Marchesani Syndrome 1/Weill-Marchesani syndrome, Autosomal recessive/Mesodermal Dysmorphodystrophy, Congenital | 277600 | ADAMTS10/19p13.2 [103] | Autosomal recessive |
| Thrombotic thrombocytopenic purpura, congenital/Upshaw-Schulman Syndrome | 274150 | ADAMTS13, 9q34.2 [91] | Autosomal recessive |
| Weill-Marchesani-like Syndrome | 613195 | ADAMTS17, 15q26.3 [104] | Autosomal recessive |
| Microcornea, Myopic Chorioretinal atrophy and Telecanthus (MMCAT) | 615458 | ADAMTS18, 16q23.1 [105] | Autosomal recessive |
| Geleophysic dysplasia 1 | 231050 | ADAMTSL2, 9q34.2 [17] | Autosomal recessive |
| Ectopia lentis et pupillae | 225200 | ADAMTSL4, 1q21.2 [29] | Autosomal recessive |
| Ectopia lentis, isolated, autosomal recessive | 225100 | ADAMTSL4, 1q21.2 [106] | Autosomal recessive |
2.1.1. ADAMTS3 and primary lymphedema
The most recent incrimination of an ADAMTS protease in a Mendelian disorder occurred via identification of recessive ADAMTS3 mutations in Hennekam lymphangiectasia-lymphedema syndrome 3 [30]. Primary lymphedema, which typically affects lower extremities, results from abnormal lymphatic morphogenesis or function. Mutations in several different genes result in this condition, many affecting VEGF-C to VEGFR3 signaling, a central pathway in lymphangiogenesis [31]. VEGF-C is synthesized as a precursor with restricted ability to activate VEGFR3, and its N- and C-terminal propeptides require proteolytic excision for maximal receptor activation by the central VEGF homology domain. ADAMTS3 was implicated as the protease responsible for proteolytic activation of VEGF-C via its interaction with the N-terminal domain of collagen and calcium-binding EGF domain 1 (CCBE1), a secreted, ECM- and VEGF-C-binding molecule. CCBE1 is also essential for lymphangiogenesis, and is mutated in a different form of Hennekam syndrome [32]. VEGF-C is located both at the cell surface and sequestered in the ECM; these localizations contribute in specific ways to proliferation of lymphatic endothelium and formation of organized lymphatic networks, respectively [33]. The C-terminal domains of VEGF-C, ADAMTS3 and CCBE1 each bind to cell-surface heparan-sulfate proteoglycans (HSPGs), forming an obligate activation complex localized to the cell-surface, where VEGF-C activation occurs [33]. Adamts3-deficient mouse embryos do not survive past 15 days of gestation owing to severe lymphedema resulting from lack of lymphangiogenesis [34]. Interestingly, ADAMTS3 is closely related to the procollagen I amininopropeptidase ADAMTS2, which is mutated in Ehlers-Danlos syndrome dermatosparaxis type (also designated as VIIc) [26]. Indeed, ADAMTS3 was previously shown to cleave the N-propeptide of procollagen II, a major component of cartilage ECM where it is strongly expressed during development [35, 36]. However, the observed human and mouse lymphatic phenotypes argue that VEGF-C activation is a definitive ADAMTS3 function whereas its predicted role in procollagen maturation and cartilage health is yet to be similarly validated.
2.1.2. ADAMTS disease associations arising from genome-wide analysis
The challenge of unequivocally establishing causality in acquired disorders is a daunting one requiring a heavy burden of proof, i.e., consistent disease association in independent studies, convincing statistical significance, support from animal models, and precise mechanisms of causality. A number of ADAMTS proteins were connected to disease via GWAS, which are large population-based analyses with significant statistical power to elicit multi-gene contributions to complex common disorders (Table 2). In contrast to fully penetrant Mendelian conditions, the caveats of GWAS caution against immediate presumption of causality. First, GWAS generally links the vicinity of gene loci to traits and only a few variants map to protein coding sequences. Second, variants occurring in inter-gene regions are frequently identified by the nearest known locus, but a presumption that the most proximate locus is associated with the disease may be in error. Third, directionality of an observed variant (whether it is protective or deleterious) is unclear unless specific mechanistic testing is undertaken. Fourth, multiple ethnic groups need to be analyzed to determine effect size in distinct populations, i.e., variants causally involved in one ethnic group may not be relevant to another. Finally, many acquired disorders are truly multifactorial and individual genes that emerge from GWAS analysis probably have very small effects that may not justify therapeutic targeting. The most compelling disease associations are those supported by multiple studies over an extended period of time that utilize well-characterized study populations and demonstrate high experimental rigor. This benchmark has been achieved in only a few specific ADAMTS-disease connections, which are discussed in detail (sections on ADAMTS7 and ADAMTS13 below).
Table 2.
GWAS-identified ADAMTS locus associations with human and animal phenotypic traits and complex disorders
| ADAMTS1 | Degenerative intervertebral disc disease [47] |
| ADAMTS2 | Syndromic, common myopia [53]; cerebral aneurysm [49]; pediatric stroke [48] |
| ADAMTS3 | Bronchodilator response [107]; lipoprotein subclasses and triglyceride measurement [108]; height [109] |
| ADAMTS5 | Degenerative intervertebral disc disease [46, 47] |
| ADAMTS6 | Inguinal hernia [110] |
| ADAMTS7 | Coronary atherosclerosis; peripheral arterial disease [38, 39, 70, 71, 75, 76, 82, 111] |
| ADAMTS8 | Pulse pressure and mean arterial pressure [112] |
| ADAMTS9 | Diabetes mellitus type 2/insulin resistance [113–115]; obesity/waist-hip ratio and other anthropomorphic traits [116, 117]; asthma [118]; psoriatic arthritis [119]; smoking and coronary artery calcification [120]; age at menopause [121]; age-related macular degeneration [43, 122–124]; cognitive aging [125]; altitude adaptation (pig) [126]; |
| ADAMTS12 | Cerebral vascular aneurysm [49]; pediatric stroke [49] |
| ADAMTS13 | Ischemic stroke [127]; pediatric stroke [49]; prolonged gestation [128] |
| ADAMTS14 | Suicidal behavior [129] |
| ADAMTS16 | Urgency urinary incontinence in women [130]; functional impairment in schizophrenia [131]; hypertension (rat) [132] |
| ADAMTS17 | Primary open angle glaucoma (dog) [50]; height [51]; pediatric stroke [49] |
| ADAMTS18 | Syndromic, common myopia [53]; Bone mass/bone mineral density [54, 133] |
| ADAMTS19 | Premature ovarian failure [134, 135] |
| ADAMTS20 | Cryptorchidism (dog) [136]; cleft palate (dog and human) [52] |
| ADAMTSL1 | Systemic lupus erythematosus [137] |
| ADAMTSL3 | Lean body mass [138]; height (human) [139, 140]; birth, weanling and yearling weight (cow) [141]; schizophrenia [139] |
| ADAMTSL4 | Syndromic common myopia [53]; coronary artery disease protection [142] |
| PAPLN | Suicidal ideation [143] |
Table 2 lists diverse phenotypic traits and complex disorders with which ADAMTS gene loci have been associated, with the caveats listed above applying to nearly all of them. Nevertheless, this metadata provides specific insights that may be instructive. It is notable that the initial GWAS linkage of ADAMTS7 to atherosclerosis and coronary artery disease was validated in a mouse model [37] and supported independently by other studies, suggesting that subsequently observed associations with peripheral vascular disease [38] and vascular calcific disease [39] are worthy of detailed investigation. ADAMTS9 is associated with diverse traits and phenotypes, perhaps because it is widely expressed, for example, by capillary endothelial cells of nearly all organs [40], or cleaves a widely distributed ECM component, versican [41] or because the associated genome variants may be highly polymorphic. Nevertheless, its associations with metabolism, i.e., obesity, diabetes and associated traits appear to be consistent in several ethnic groups (Table 2). Because efficient ADAMTS9 secretion depends on the activity of one of its modifying enzymes B3GLCT (also known as B3GALTL) [42], it is intriguing that B3GLCT and ADAMTS9 are strongly expressed in the eye [42] and vascular endothelial cells [40] and each is associated with age-related macular degeneration [43]. Since ADAMTS5 is indisputably causal in cartilage aggrecan destruction in osteoarthritis (OA) [44, 45], association of the conjoint locus for ADAMTS5 and another aggrecanase, ADAMTS1 in degenerative disk disease [46, 47] is intriguing, since the intervertebral disk is similar in its composition to cartilage. ADAMTS12 is highly homologous to ADAMTS7 [24], and it may have a similar role in smooth muscle cells in association with vascular/hemostatic disorders [48, 49]. ADAMTS17 mutations result in a Mendelian condition characterized by skeletal and eye anomalies, in support of its independent association by GWAS in canine open angle glaucoma and variation in human height [50, 51]. Reinforcing possible ADAMTS20 variant association with cleft lip and palate in humans, a canine ADAMTS20 mutation was identified to cause cleft lip and palate and syndactyly in Nova Scotia Duck Tolling Retrievers [52]. Indeed, cleft palate was previously characterized in detail in combined mouse mutants of Adamts20 and Adamts9 and syndactyly was identified in combined mutants of Adamts20 with Adamts9 or Adamts5 [22, 23]. An association of ADAMTSL4 and ADAMTS2 loci with syndromic myopia has credence arising from their association with connective tissue assembly, providing a potential role in axial growth of the eye globe, whose outermost layer, the sclera, is a dense connective tissue [53]. Association of the ADAMTS18 locus with myopia and bone density [53, 54] suggests a role in collagen metabolism, since collagen I mutations leading to osteogenesis imperfecta esult in “blue” sclera and collagen I is the major component of the sclera.
3. ADAMTS proteins in acquired disorders
3.1. ADAMTS proteases as primary aggrecanases in osteoarthritis
Osteoarthritis (OA) arises from degenerative loss of articular cartilage in synovial joints with presumed secondary changes in bone and synovium, and is a common condition in the population. It has a multi-factorial etiology, with a strong genetic contribution in some instances that leads to structurally defective cartilage, but more frequently results from enzymatic breakdown of cartilage ECM initiated by unknown factors. Of these, prior injuries, altered joint biomechanics or increased loading are probably the most significant. One of the major structural components of articular cartilage is the chondroitin sulfate (CS) proteoglycan aggrecan, which forms giant aggregates with the glycosaminoglycan hyaluronan (HA). The CS chains of aggrecan, which carry a net negative charge, absorb large quantities of water, leading to swelling of the HA-aggrecan aggregates. The swollen aggregates are constrained by entrapment within networks of collagen II, another major cartilage component, and this composite structure gives cartilage ECM its compressive strength and endows articular cartilage with shock-absorbing properties. Several ADAMTS proteases, but not matrix metalloproteases (MMPs), cleave the aggrecan core protein at the Glu373-Ala374 peptide bond between the G1 and G2 domains (known as the interglobular region) [55, 56], releasing the entire CS-bearing region [57]. This compromises the mechanical properties of cartilage and exposes other structural molecules such as collagen II to proteolysis [58]. A key study generated transgenic mice with a mutation of this ADAMTS cleavage site and found reduced aggrecan loss and cartilage erosion in a surgical model of OA [59]. This model not only demonstrated the significance of ADAMTS-mediated aggrecan proteolysis, but also appeared to show an anabolic effect in the wake of acute inflammation [59]. Notably, the pro-inflammatory activity of cleaved ECM fragments such as from fibronectin and aggrecan is known to amplify disease pathology [60, 61]. Both ADAMTS4 and ADAMTS5 are thought to contribute to aggrecan cleavage, but ADAMTS5 was implicated as the major culprit by demonstration of favorable enzymatic properties against specific aggrecanase cleavage sites in aggrecan, its association with OA, lack of susceptibility to surgically-induced OA in Adamts5- but not Adamts4-deficient mice and protection against aggrecan breakdown by ADAMTS5-specific antibody blockade [62, 63]. The availability of highly specific ADAMTS5 blocking antibodies [63–65] is a major breakthrough, since small molecule active-site inhibitors, despite the advantage of oral bioavailability, generally lack exquisite specificity and have broad side-effects, as noted with MMP inhibitors developed for cancer therapy [66]. Nevertheless, concerns about side effects of ADAMTS5 inhibitors are valid, mostly arising from observed roles of ADAMTS5 in embryogenesis, as well as potentially in ECM turnover in the adult cardiovascular system [67–69]. A possible solution to bypassing a systemic toxicity is intra-articular administration of the blocking antibodies, but it has not been explored. Another challenge presented by OA is its long sub-clinical period, such that ADAMTS5 inhibition may be most effective early in the disease process. Early treatment necessitates not only improvement of biomarkers for early OA diagnosis, but due consideration to long-term side-effects, or complications that may only become apparent decades after treatment initiation.
3.2. ADAMTS7 and cardiovascular disease
3.2.1. Novel locus for coronary atherosclerosis and mechanisms of action in vascular disease
GWAS of coronary artery disease (CAD) identified highly associated ADAMTS7 single nucleotide polymorphisms (SNPs). Although the lead SNPs in two of the GWAS, i.e., rs1994016 [70] and rs4380028 [71] are located in intron 8 and 7.6 kb upstream of ADAMTS7, respectively, rs3825807 represents an A to G coding SNP, resulting in replacement of Ser214 by Pro [72]. Each SNP is in linkage disequilibrium with ADAMTS7, with rs3825807 possibly affecting protein function. Moreover, rs3825807 is significantly associated with coronary artery calcification [39, 73] and independently predicts the severity and survival of angiographically documented CAD [74, 75]. Individuals with the GG variant (Pro substitution) showed approximately three times fewer cardiovascular events in relation to the AA (Ser) genotype [74]. The G allele is also associated with lower obstructive CAD and angiographic severity [76]. Expression of ADAMTS7 constructs carrying the Pro (G) substitution [77] demonstrated reduced furin-mediated ADAMTS7 propeptide excision, which occurs at the cell-surface [78] and is presumably an activating step. Consistent with this, cells expressing the GG variant of ADAMTS7 had lower migratory activity than cells of the AA genotype [77], possibly owing to reduced proteolysis of a reported ADAMTS7 substrate COMP, which is strongly associated with vascular smooth muscle cell migration and neointima formation.
Several subsequent studies undertook functional analysis to test the genetic association and together they strongly suggest that ADAMTS7 contributes to atherogenesis. Analysis of an intragenic lacZ reporter mouse allele identified Adamts7-expressing vascular smooth muscle cells in early atheroma lesions (4 weeks) formed in Adamts7−/−ApoE−/− mice fed a western diet [37]. Despite lack of expression in older atheromatous lesions (10 weeks), Adamts7−/−ApoE−/− mice formed smaller atheromatous plaques than ApoE−/− littermates. These mouse studies suggested that ADAMTS7 acted early in atheroma development in this mouse model of atherosclerosis, possibly in response to TNFα in the inflammatory milieu [37]. A corresponding analysis of human atheroma lesions demonstrated ADAMTS7 localization to smooth muscle cells, but not macrophages in the lesion, and pinpointed its localization to their cell surface and filopodia [37], consistent with the demonstrated effect of ADAMTS7 in facilitating SMC migration in vitro [79]. Another analysis suggested that ADAMTS7 content in atherosclerotic plaques from symptomatic patients was increased over that of asymptomatic patients and was associated with a vulnerable plaque phenotype [80]. ADAMTS7 was co-localized with macrophages and smooth muscle cells in coronary and carotid atherosclerotic plaque with staining present throughout the plaque, including the shoulder, cap and core [77, 80].
Plasma ADAMTS7 was elevated in patients with severe obstructive CAD [81]. Indeed, the level of ADAMTS7 increased the risk for future cardiovascular events in the symptomatic patients, but not the asymptomatic patients [80]. rs7177699, which is in strong linkage disequilibrium with coronary artery-associated SNP rs3825807, is associated with postoperative cardiovascular death [80]. In a further study, SNP rs3825807, which impairs ADAMTS7 function, is associated with reduced CAD burden [76]. Specifically, the SNP is associated with a smaller fibrous cap and percentage area of smooth muscle cells in the intima [76]. The ADAMTS7 SNPs, rs1994016 and rs3825807 were further shown to be over-represented at the mRNA and protein level in peripheral artery disease, which like CAD, is most commonly caused by atherosclerosis [38]. CAD has a complex etiology and several lifestyle factors are contributory. For example, in a further GWAS analysis, the allelic variant rs7178051 was associated with reduced ADAMTS7 expression, which conferred stronger CAD protection in never-smokers than in ever-smokers [82]. Therefore, ADAMTS7 expression has a specific correlation to and may contribute to loss of CAD protection in smokers.
3.2.2. Ventricular remodeling
Acute myocardial infarction (AMI) is associated with high morbidity and mortality. Following AMI, myocardial ECM is degraded, which causes dilation and thinning of the infarcted zone with catastrophic consequences. Plasma levels of ADAMTS7 were elevated in patients with diminished left ventricular ejection fraction, an indicator of poor ventricular contraction [83, 84]. Therefore, elevated ADAMTS7 levels may contribute to deleterious ventricular remodeling after AMI.
3.2.3. Response to arterial injury
Wire injury of the carotid artery in Adamts7−/− mice promoted greater re-endothelialization than in wild-type mice [85]. ADAMTS7 inhibits both endothelial cell proliferation and migration; these effects appear to be independent of COMP, one of the reported ADAMTS7 substrates [85]. Instead, thrombospondin appears to be the relevant substrate for this effect, since the impact of Adamts7 deficiency on re-endothelialization was not seen in Tsp1−/− mice [85].
3.2.4. ADAMTS7 as a therapeutic target in vascular disease
ADAMTS7 is an attractive drug target for vascular disease, based on the cumulative evidence from population genetics, animal models and in vitro analysis. Its pro-atherogenic and anti-endothelial roles could conceivably be countered using systemic or local administration (such as via drug eluting stents) of ADAMTS7 blocking agents, respectively. The limited analysis of Adamts7−/− mice to date has suggested that they lack major phenotypes, promising a low-risk of side effects from the use of anti-ADAMTS7 inhibitors or antibodies [24, 37]. Whereas the development of such targeting agents is necessary, there is also an urgent need for a better understanding of the substrates targeted by this protease, its cooperative roles and regulatory relationships with its homolog ADAMTS12 [24], and potential side-effects related to its physiological functions, which remain to be fully elucidated. For example, musculoskeletal tissues from Adamts7−/− mice were shown recently to have upregulation of Adamts12 [24], whose roles in cardiovascular disease remain unexplored, and Adamts7 is constitutively expressed in smooth muscle cells of the pulmonary arterial tree, where its functions are unknown [37]. Notably, consistent with the introductory emphasis on ADAMTS homologous pairs and functional overlap, ADAMTS7 and ADAMTS12 constitute a distinct homologous pair of metalloprotease-proteoglycans that shows high evolutionary conservation [24], suggesting vital (for supporting mammalian evolution) and protected functions. Moreover, a recent study identified tendon, meniscal and ligamentous heterotopic ossification in Adamts7−/−Adamts12−/− mice, and significantly lower ADAMTS7 mRNA levels were seen in human tendons having degenerative morphology, including heterotopic ossification [24]. Thus, one requirement for specific inhibitors is that they should not cross-inhibit ADAMTS12. It remains to be determined whether pharmacologic inhibition of ADAMTS7 activity might result in ADAMTS12 upregulation, and what impact that could have on cells. Nothing is known about ADAMTS12 in cardiovascular disease, and none of the work on ADAMTS7 in cardiovascular disease has considered a potential role for ADAMTS12. A salutary historical lesson was provided by clinical trials of MMP inhibitors in cancer, which were found to have potential toxicity related to connective tissue turnover as well as to be unsuccessful in cancer therapy [86, 87]. Recently, remarkably specific ADAMTS5 inhibitory antibodies with a high level of target engagement have been generated, but their clinical application is proceeding cautiously owing to implication of ADAMTS5 in a number of morphogenetic events in the embryo, as well as potential contribution of reduced ADAMTS5 to atherosclerosis and ascending aortic aneurysms [67, 68]. Ideally, development of ADAMTS7 inhibitors should proceed alongside an equal emphasis on elucidating ADAMTS7 and ADAMTS12 regulation, molecular structure, life cycle, interacting partners, expression profile and in vivo functions at the sites of expression, which remain woefully understudied.
3.3. ADAMTS13 in platelet coagulopathy
3.3.1. Clinical characteristics and mechanisms
ADAMTS13 is primarily synthesized by stellate cells of the liver and vascular endothelial cells and proteolyzes ultra-large von Willebrand factor (vWF) multimers (ULvWF) at the Tyr1605-Met1606 bond in the A2 domain [88]. This site is cryptic until exposed to the action of ADAMTS13 by shear-mediated extension vWF. When ADAMTS13 levels are reduced, pro-hemostatic ULvWF accumulates in plasma, promoting platelet aggregation and resulting in microthrombi that imperil the circulation of vital organs. ADAMTS13 cleavage of ULvWF into smaller fragments mitigates the procoagulative function (reviewed in [89]). ADAMTS13 deficiency can have two origins. Acquired thrombotic thrombocytopenic purpura (aTTP), the more common form, is an autoimmune disorder caused by antibodies to ADAMTS13 [90], whereas, congenital TTP (cTTP) is much rarer and caused by ADAMTS13 mutations [91]. TTP is a blood coagulation disease characterized by the presence of VWF and platelet-rich microthrombi in the microvasculature of many organs [92]. cTTP has been associated with over eighty mutations in ADAMTS13 that cause severe ADAMTS13 deficiency. An imbalance of ADAMTS13 and VWF has also been noted in other acquired cardiovascular disorders as well as preeclampsia, sepsis, inflammatory bowel disease and liver cirrhosis [93–98].
3.3.2. ADAMTS13 replacement for treatment of thrombotic thrombocytopenic purpura
TTP treatment requires replacement of ADAMTS13, and this has been done to date by fresh frozen plasma infusion for cTTP and plasma exchange for aTTP [92]. aTTP additionally requires some measure of immunosuppression to combat its autoimmune basis. These treatments have largely been successful, reducing the mortality of an acute TTP episode from 100% to ~20% [92]. Plasma infusion is relatively straightforward and inexpensive, and plasma exchange not only replaces ADAMTS13, but also removes anti-ADAMTS13 antibodies and supplies other coagulation factors that may have been consumed in the acute episode. However, it is a prolonged infusion with attendant risks of volume overload, hyperviscosity, allergic reactions ranging from mild to severe and pathogen transmission. Furthermore, catheter-related complications can occur in patients undergoing long-term infusion. Given these risks, the possibility of preventing cTTP or treating aTTP with recombinant ADAMTS13 has been an attractive one, since the effective levels of activity needed to prevent platelet coagulation have a broad range, the infused volume can be small, and there is reduced risk of allergic reactions or pathogen transmission [99, 100]. High vWF levels seen in stroke and cardiovascular disease could also potentially be mitigated by recombinant ADAMTS13 more safely than by plasma transfusion.
Baxalta, a biopharmaceutical company, generated a recombinant ADAMTS13, BAX390, in genetically engineered Chinese hamster ovary cells cultured in a chemically defined medium. The pharmacokinetics of rADAMTS13 was characterized in knockout mice, rats and monkeys, whose vWF was effectively cleaved, and efficacy was demonstrated in Adamts13 knockout mice [101]. BAX390 was recently used in a prospective Phase I study of 15 cTTP patients ranging from 12–65 years of age using dosages of 5–40 units/kg [100]. It was well tolerated in all patients without allergic reactions and without development of an immune response to the recombinant ADAMTS13, and the appropriate response of ULvWF and cleaved vWF fragments was observed.
4. Future needs and anticipated challenges
At the present rate of progress, associations of ADAMTS proteases with disease will continue to emerge. They will need to be validated systematically in a sufficiently large population using diverse approaches that include rigorously validated antibodies, specific functional assays and animal models, otherwise the findings will remain preliminary and unconvincing. ADAMTS knockout mice have yielded a plethora of knowledge and a blueprint for anticipated human disease connections. At the same time, our fundamental understanding of ADAMTS proteins is in its infancy. ADAMTS5 and ADAMTS13 offer superb case studies on inhibitor targeting and enzyme replacement, respectively, for the field. Each case study has a fascinating history and translational development was based on a comprehensive fundamental understanding. Whereas these two enzymes have been intensively studied, there are a number of family members about which virtually nothing is known. It is vital to “fill in the blanks” because they will help to elucidate general principles of ADAMTS proteins and determine similarities and contrasts between the family members. A detailed dissection of ADAMTS domain structure, key residues, molecular dynamics, post-translational modification, and life cycle should therefore be made a priority, since it will be necessary for understanding their involvement in disease and seeking therapeutic strategies. One of the major steps yet to be taken by the field is the widespread use of proteomics for identification of ADAMTS substrates [102], so that assays can be developed for inhibitor evaluation, the protease’s specificity bandwidth is known, and the pathways in which the proteases act can be defined. With this information in hand, drug development will be better informed about potential side-effects and benefit-to-risk ratio. Continued fundamental research on the ADAMTS molecules should therefore continue hand in hand with drug development.
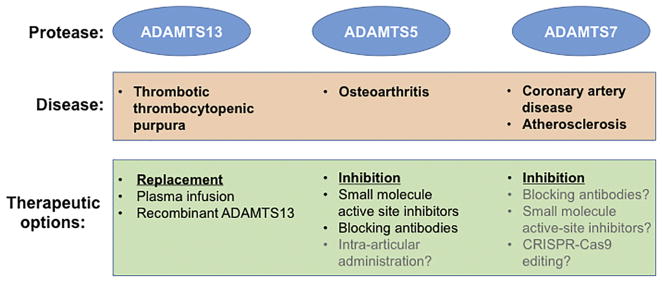
Figure 3.
Current translational targets in the ADAMTS family. The figure shows three targets with strong associations with disease, ADAMTS13, ADAMTS5 and ADAMTS7 which are at various stages of development of clinical therapeutics, the specific approach (replacement versus inhibition) and additional specifics. The text in gray indicates untested/undeveloped possibilities.
Highlights.
The ADAMTS superfamily comprises an important group of extracellular matrix modifying proteases and non-structural proteins.
ADAMTS gene mutations lead to diverse genetic disorders and ADAMTS variants are associated with several complex acquired diseases.
Inhibition of ADAMTS protease activity has emerged as a translational possibility in treatment of osteoarthritis and atherosclerosis.
Enzymatic replacement with recombinant ADAMTS13 is under development for thrombotic thrombocytopenic purpura.
Acknowledgments
Support for this work was provided by the National Institutes of Health award EY024943 and by the Allen Distinguished Investigator Program, through support made by The Paul G. Allen Frontiers Group and the American Heart Association.
Footnotes
Author conflicts: The authors have no relevant conflicts to declare
References
- 1.Apte SS, Parks WC. Metalloproteinases: A parade of functions in matrix biology and an outlook for the future. Matrix Biol. 2015;44–46:1–6. doi: 10.1016/j.matbio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Dubail J, Apte SS. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 2015;44–46:24–37. doi: 10.1016/j.matbio.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Cal S, Lopez-Otin C. ADAMTS proteases and cancer. Matrix Biol. 2015;44–46:77–85. doi: 10.1016/j.matbio.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Koo BH, Longpre JM, Somerville RP, Alexander JP, Leduc R, Apte SS. Regulation of ADAMTS9 secretion and enzymatic activity by its propeptide. J Biol Chem. 2007;282:16146–16154. doi: 10.1074/jbc.M610161200. [DOI] [PubMed] [Google Scholar]
- 5.Longpre JM, McCulloch DR, Koo BH, Alexander JP, Apte SS, Leduc R. Characterization of proADAMTS5 processing by proprotein convertases. Int J Biochem Cell Biol. 2009;41(5):1116–26. doi: 10.1016/j.biocel.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Tortorella MD, Arner EC, Hills R, Gormley J, Fok K, Pegg L, Munie G, Malfait AM. ADAMTS-4 (aggrecanase-1): N-terminal activation mechanisms. Arch Biochem Biophys. 2005;444(1):34–44. doi: 10.1016/j.abb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, Hargreaves D, Ting A, Pauptit RA, Parker AE, et al. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373(4):891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 8.Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, Mackie S, Olland S, Lin L, Zhong X, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17(1):16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirohata S, Wang LW, Miyagi M, Yan L, Seldin MF, Keene DR, Crabb JW, Apte SS. Punctin, a novel ADAMTS-like molecule (ADAMTSL-1) in extracellular matrix. J Biol Chem. 2002;22:22. doi: 10.1074/jbc.M109665200. [DOI] [PubMed] [Google Scholar]
- 10.Hall NG, Klenotic P, Anand-Apte B, Apte SS. ADAMTSL-3/punctin-2, a novel glycoprotein in extracellular matrix related to the ADAMTS family of metalloproteases. Matrix Biol. 2003;22(6):501–10. doi: 10.1016/s0945-053x(03)00075-1. [DOI] [PubMed] [Google Scholar]
- 11.Koo BH, Le Goff C, Jungers KA, Vasanji A, O’Flaherty J, Weyman CM, Apte SS. ADAMTS-like 2 (ADAMTSL2) is a secreted glycoprotein that is widely expressed during mouse embryogenesis and is regulated during skeletal myogenesis. Matrix Biol. 2007;26(6):431–41. doi: 10.1016/j.matbio.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Le Goff C, Mahaut C, Wang LW, Allali S, Abhyankar A, Jensen S, Zylberberg L, Collod-Beroud G, Bonnet D, Alanay Y, et al. Mutations in the TGFbeta binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am J Hum Genet. 2011;89(1):7–14. doi: 10.1016/j.ajhg.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel LA, Wang LW, Bader H, Ho JC, Majors AK, Hollyfield JG, Traboulsi EI, Apte SS. ADAMTSL4, a secreted glycoprotein widely distributed in the eye, binds fibrillin-1 microfibrils and accelerates microfibril biogenesis. Invest Ophthalmol Vis Sci. 2012;53(1):461–469. doi: 10.1167/iovs.10-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bader HL, Wang LW, Ho JC, Tran T, Holden P, Fitzgerald J, Atit RP, Reinhardt DP, Apte SS. A disintegrin-like and metalloprotease domain containing thrombospondin type 1 motif-like 5 (ADAMTSL5) is a novel fibrillin-1-, fibrillin-2-, and heparin-binding member of the ADAMTS superfamily containing a netrin-like module. Matrix Biol. 2012;31(7–8):398–411. doi: 10.1016/j.matbio.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsutsui K, Manabe R, Yamada T, Nakano I, Oguri Y, Keene DR, Sengle G, Sakai LY, Sekiguchi K. ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J Biol Chem. 2010;285(7):4870–82. doi: 10.1074/jbc.M109.076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aviram R, Zaffryar-Eilot S, Hubmacher D, Grunwald H, Maki JM, Myllyharju J, Apte SS, Hasson P. Interactions between lysyl oxidases and ADAMTS proteins suggest a novel crosstalk between two extracellular matrix families. Matrix Biol. 2018 doi: 10.1016/j.matbio.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Goff C, Morice-Picard F, Dagoneau N, Wang LW, Perrot C, Crow YJ, Bauer F, Flori E, Prost-Squarcioni C, Krakow D, et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet. 2008;40(9):1119–23. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramerova IA, Kawaguchi N, Fessler LI, Nelson RE, Chen Y, Kramerov AA, Kusche-Gullberg M, Kramer JM, Ackley BD, Sieron AL, et al. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development. 2000;127(24):5475–85. doi: 10.1242/dev.127.24.5475. [DOI] [PubMed] [Google Scholar]
- 19.Sengle G, Tsutsui K, Keene DR, Tufa SF, Carlson EJ, Charbonneau NL, Ono RN, Sasaki T, Wirtz MK, Samples JR, et al. Microenvironmental regulation by fibrillin-1. PLoS Genet. 2012;8(1):e1002425. doi: 10.1371/journal.pgen.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubmacher D, Apte SS. Genetic and functional linkage between ADAMTS superfamily proteins and fibrillin-1: a novel mechanism influencing microfibril assembly and function. Cell Mol Life Sci. 2011;68(19):3137–48. doi: 10.1007/s00018-011-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubmacher D, Apte SS. ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 2015;47:34–43. doi: 10.1016/j.matbio.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enomoto H, Nelson C, Somerville RPT, Mielke K, Dixon L, Powell K, Apte SS. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 2010;137:4029–4038. doi: 10.1242/dev.050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCulloch DR, Nelson CM, Dixon LJ, Silver DL, Wylie JD, Lindner V, Sasaki T, Cooley MA, Argraves WS, Apte SS. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell. 2009;17(5):687–98. doi: 10.1016/j.devcel.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mead TJ, McCulloch DR, Ho JC, Du Y, Adams SM, Birk DE, Apte SS. The metalloproteinase-proteoglycans ADAMTS7 and ADAMTS12 provide an innate, tendon-specific protective mechanism against heterotopic ossification. JCI Insight. 2018;3(7):e92941. doi: 10.1172/jci.insight.92941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bekhouche M, Colige A. The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol. 2015;44–46:46–53. doi: 10.1016/j.matbio.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Colige A, Sieron AL, Li SW, Schwarze U, Petty E, Wertelecki W, Wilcox W, Krakow D, Cohn DH, Reardon W, et al. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am J Hum Genet. 1999;65(2):308–17. doi: 10.1086/302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Goff C, Cormier-Daire V. Genetic and molecular aspects of acromelic dysplasia. Pediatr Endocrinol Rev. 2009;6(3):418–23. [PubMed] [Google Scholar]
- 28.Le Goff C, Cormier-Daire V. The ADAMTS(L) family and human genetic disorders. Hum Mol Genet. 2011;20(R2):R163–7. doi: 10.1093/hmg/ddr361. [DOI] [PubMed] [Google Scholar]
- 29.Ahram D, Sato TS, Kohilan A, Tayeh M, Chen S, Leal S, Al-Salem M, El-Shanti H. A homozygous mutation in ADAMTSL4 causes autosomal-recessive isolated ectopia lentis. Am J Hum Genet. 2009;84(2):274–8. doi: 10.1016/j.ajhg.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brouillard P, Dupont L, Helaers R, Coulie R, Tiller GE, Peeden J, Colige A, Vikkula M. Loss of ADAMTS3 activity causes Hennekam lymphangiectasia-lymphedema syndrome 3. Hum Mol Genet. 2017;26(21):4095–4104. doi: 10.1093/hmg/ddx297. [DOI] [PubMed] [Google Scholar]
- 31.Brouillard P, Boon L, Vikkula M. Genetics of lymphatic anomalies. J Clin Invest. 2014;124(3):898–904. doi: 10.1172/JCI71614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeltsch M, Jha SK, Tvorogov D, Anisimov A, Leppanen VM, Holopainen T, Kivela R, Ortega S, Karpanen T, Alitalo K. CCBE1 enhances lymphangiogenesis via A disintegrin and metalloprotease with thrombospondin motifs-3-mediated vascular endothelial growth factor-C activation. Circulation. 2014;129(19):1962–71. doi: 10.1161/CIRCULATIONAHA.113.002779. [DOI] [PubMed] [Google Scholar]
- 33.Jha SK, Rauniyar K, Karpanen T, Leppanen VM, Brouillard P, Vikkula M, Alitalo K, Jeltsch M. Efficient activation of the lymphangiogenic growth factor VEGF-C requires the C-terminal domain of VEGF-C and the N-terminal domain of CCBE1. Sci Rep. 2017;7(1):4916. doi: 10.1038/s41598-017-04982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janssen L, Dupont L, Bekhouche M, Noel A, Leduc C, Voz M, Peers B, Cataldo D, Apte SS, Dubail J, et al. ADAMTS3 activity is mandatory for embryonic lymphangiogenesis and regulates placental angiogenesis. Angiogenesis. 2016;19(1):53–65. doi: 10.1007/s10456-015-9488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandes RJ, Hirohata S, Engle JM, Colige A, Cohn DH, Eyre DR, Apte SS. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J Biol Chem. 2001;276(34):31502–9. doi: 10.1074/jbc.M103466200. [DOI] [PubMed] [Google Scholar]
- 36.Le Goff C, Somerville RP, Kesteloot F, Powell K, Birk DE, Colige AC, Apte SS. Regulation of procollagen amino-propeptide processing during mouse embryogenesis by specialization of homologous ADAMTS proteases: insights on collagen biosynthesis and dermatosparaxis. Development. 2006;133(8):1587–96. doi: 10.1242/dev.02308. [DOI] [PubMed] [Google Scholar]
- 37.Bauer RC, Tohyama J, Cui J, Cheng L, Yang J, Zhang X, Ou K, Paschos GK, Zheng XL, Parmacek MS, et al. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation. 2015;131(13):1202–1213. doi: 10.1161/CIRCULATIONAHA.114.012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayoglu B, Arslan C, Tel C, Ulutin T, Dirican A, Deser SB, Cengiz M. Genetic variants rs1994016 and rs3825807 in ADAMTS7 affect its mRNA expression in atherosclerotic occlusive peripheral arterial disease. J Clin Lab Anal. 2018;32(1):e22174. doi: 10.1002/jcla.22174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Setten J, Isgum I, Smolonska J, Ripke S, de Jong PA, Oudkerk M, de Koning H, Lammers JW, Zanen P, Groen HJ, et al. Genome-wide association study of coronary and aortic calcification implicates risk loci for coronary artery disease and myocardial infarction. Atherosclerosis. 2013;228(2):400–5. doi: 10.1016/j.atherosclerosis.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 40.Koo BH, Coe DM, Dixon LJ, Somerville RP, Nelson CM, Wang LW, Young ME, Lindner DJ, Apte SS. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am J Pathol. 2010;176(3):1494–504. doi: 10.2353/ajpath.2010.090655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somerville RP, Longpre JM, Jungers KA, Engle JM, Ross M, Evanko S, Wight TN, Leduc R, Apte SS. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J Biol Chem. 2003;278(11):9503–13. doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- 42.Dubail J, Vasudevan D, Wang LW, Earp SE, Jenkins MW, Haltiwanger RS, Apte SS. Impaired ADAMTS9 secretion: A potential mechanism for eye defects in Peters Plus Syndrome. Sci Rep. 2016;6:33974. doi: 10.1038/srep33974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–9. 439e1–2. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434(7033):648–52. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 45.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434(7033):644–8. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 46.Rajasekaran S, Kanna RM, Senthil N, Raveendran M, Ranjani V, Cheung KM, Chan D, Kao PY, Yee A, Shetty AP. Genetic susceptibility of lumbar degenerative disc disease in young Indian adults. Eur Spine J. 2015;24(9):1969–75. doi: 10.1007/s00586-014-3687-y. [DOI] [PubMed] [Google Scholar]
- 47.Virtanen IM, Noponen N, Barral S, Karppinen J, Li H, Vuoristo M, Niinimaki J, Ott J, Ala-Kokko L, Mannikko M. Putative susceptibility locus on chromosome 21q for lumbar disc disease (LDD) in the Finnish population. J Bone Miner Res. 2007;22(5):701–7. doi: 10.1359/jbmr.070123. [DOI] [PubMed] [Google Scholar]
- 48.Arning A, Hiersche M, Witten A, Kurlemann G, Kurnik K, Manner D, Stoll M, Nowak-Gottl U. A genome-wide association study identifies a gene network of ADAMTS genes in the predisposition to pediatric stroke. Blood. 2012;120(26):5231–6. doi: 10.1182/blood-2012-07-442038. [DOI] [PubMed] [Google Scholar]
- 49.Arning A, Jeibmann A, Kohnemann S, Brokinkel B, Ewelt C, Berger K, Wellmann J, Nowak-Gottl U, Stummer W, Stoll M, et al. ADAMTS genes and the risk of cerebral aneurysm. J Neurosurg. 2016;125(2):269–74. doi: 10.3171/2015.7.JNS154. [DOI] [PubMed] [Google Scholar]
- 50.Forman OP, Pettitt L, Komaromy AM, Bedford P, Mellersh C. A Novel Genome-Wide Association Study Approach Using Genotyping by Exome Sequencing Leads to the Identification of a Primary Open Angle Glaucoma Associated Inversion Disrupting ADAMTS17. PLoS One. 2015;10(12):e0143546. doi: 10.1371/journal.pone.0143546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Duyvenvoorde HA, Lui JC, Kant SG, Oostdijk W, Gijsbers AC, Hoffer MJ, Karperien M, Walenkamp MJ, Noordam C, Voorhoeve PG, et al. Copy number variants in patients with short stature. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, Cox TC, McHenry T, Narayan N, Feingold E, et al. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 2015;11(3):e1005059. doi: 10.1371/journal.pgen.1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flitcroft DI, Loughman J, Wildsoet CF, Williams C, Guggenheim JA Cream Consortium for the. Novel Myopia Genes and Pathways Identified From Syndromic Forms of Myopia. Invest Ophthalmol Vis Sci. 2018;59(1):338–348. doi: 10.1167/iovs.17-22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koller DL, Ichikawa S, Lai D, Padgett LR, Doheny KF, Pugh E, Paschall J, Hui SL, Edenberg HJ, Xuei X, et al. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J Clin Endocrinol Metab. 2010;95(4):1802–9. doi: 10.1210/jc.2009-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arner EC, Pratta MA, Trzaskos JM, Decicco CP, Tortorella MD. Generation and characterization of aggrecanase. A soluble, cartilage-derived aggrecan-degrading activity. J Biol Chem. 1999;274(10):6594–601. doi: 10.1074/jbc.274.10.6594. [DOI] [PubMed] [Google Scholar]
- 56.Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, Burn T, Arner E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4) J Biol Chem. 2000;275(24):18566–73. doi: 10.1074/jbc.M909383199. [DOI] [PubMed] [Google Scholar]
- 57.Sandy JD, Flannery CR, Neame PJ, Lohmander LS. The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu 373-Ala 374 bond of the interglobular domain. J Clin Invest. 1992;89(5):1512–6. doi: 10.1172/JCI115742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pratta MA, Yao W, Decicco C, Tortorella MD, Liu RQ, Copeland RA, Magolda R, Newton RC, Trzaskos JM, Arner EC. Aggrecan protects cartilage collagen from proteolytic cleavage. J Biol Chem. 2003;278(46):45539–45. doi: 10.1074/jbc.M303737200. [DOI] [PubMed] [Google Scholar]
- 59.Little CB, Meeker CT, Golub SB, Lawlor KE, Farmer PJ, Smith SM, Fosang AJ. Blocking aggrecanase cleavage in the aggrecan interglobular domain abrogates cartilage erosion and promotes cartilage repair. J Clin Invest. 2007;117(6):1627–36. doi: 10.1172/JCI30765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Homandberg GA. Cartilage damage by matrix degradation products: fibronectin fragments. Clin Orthop. 2001;(391 Suppl):S100–7. doi: 10.1097/00003086-200110001-00010. [DOI] [PubMed] [Google Scholar]
- 61.Miller RE, Ishihara S, Tran PB, Golub SB, Last K, Miller RJ, Fosang AJ, Malfait AM. An aggrecan fragment drives osteoarthritis pain through Toll-like receptor 2. JCI Insight. 2018;3(6) doi: 10.1172/jci.insight.95704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fosang AJ, Little CB. Drug insight: aggrecanases as therapeutic targets for osteoarthritis. Nat Clin Pract Rheumatol. 2008;4(8):420–7. doi: 10.1038/ncprheum0841. [DOI] [PubMed] [Google Scholar]
- 63.Larkin J, Lohr TA, Elefante L, Shearin J, Matico R, Su JL, Xue Y, Liu F, Genell C, Miller RE, et al. Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification. Osteoarthritis Cartilage. 2015;23(8):1254–66. doi: 10.1016/j.joca.2015.02.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiusaroli R, Visentini M, Galimberti C, Casseler C, Mennuni L, Covaceuszach S, Lanza M, Ugolini G, Caselli G, Rovati LC, et al. Targeting of ADAMTS5’s ancillary domain with the recombinant mAb CRB0017 ameliorates disease progression in a spontaneous murine model of osteoarthritis. Osteoarthritis Cartilage. 2013;21(11):1807–10. doi: 10.1016/j.joca.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 65.Santamaria S, Yamamoto K, Botkjaer K, Tape C, Dyson MR, McCafferty J, Murphy G, Nagase H. Antibody-based exosite inhibitors of ADAMTS-5 (aggrecanase-2) Biochem J. 2015;471(3):391–401. doi: 10.1042/BJ20150758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–92. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 67.Cikach FS, Koch CD, Mead TJ, Galatioto J, Willard BB, Emerton KB, Eagleton MJ, Blackstone EH, Ramirez F, Roselli EE, et al. Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight. 2018;3(5):e97167. doi: 10.1172/jci.insight.97167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Didangelos A, Mayr U, Monaco C, Mayr M. Novel role of ADAMTS-5 protein in proteoglycan turnover and lipoprotein retention in atherosclerosis. J Biol Chem. 2012;287(23):19341–5. doi: 10.1074/jbc.C112.350785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fava M, Barallobre-Barreiro J, Mayr U, Lu R, Didangelos A, Baig F, Lynch M, Catibog N, Joshi A, Barwari T, et al. Role of ADAMTS (A Disintegrin and Metalloproteinase With Thrombospondin Motifs)-5 in Aortic Dilatation and Extracellular Matrix Remodeling. Arterioscler Thromb Vasc Biol. 2018 doi: 10.1161/ATVBAHA.117.310562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377(9763):383–92. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43(4):339–44. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 72.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, Sun YV, Province MA, Aspelund T, Dehghan A, et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124(25):2855–64. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pereira A, Palma Dos Reis R, Rodrigues R, Sousa AC, Gomes S, Borges S, Ornelas I, Freitas AI, Guerra G, Henriques E, et al. Association of ADAMTS7 gene polymorphism with cardiovascular survival in coronary artery disease. Physiol Genomics. 2016;48(11):810–815. doi: 10.1152/physiolgenomics.00059.2016. [DOI] [PubMed] [Google Scholar]
- 75.You L, Tan L, Liu L, Shen R, Chaugai S, Wang DW, Cui W. ADAMTS7 locus confers high cross-race risk for development of coronary atheromatous plaque. Mol Genet Genomics. 2016;291(1):121–8. doi: 10.1007/s00438-015-1092-9. [DOI] [PubMed] [Google Scholar]
- 76.Chan K, Pu X, Sandesara P, Poston RN, Simpson IA, Quyyumi AA, Ye S, Patel RS. Genetic Variation at the ADAMTS7 Locus is Associated With Reduced Severity of Coronary Artery Disease. J Am Heart Assoc. 2017;6(11) doi: 10.1161/JAHA.117.006928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pu X, Xiao Q, Kiechl S, Chan K, Ng FL, Gor S, Poston RN, Fang C, Patel A, Senver EC, et al. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am J Hum Genet. 2013;92(3):366–74. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Somerville RP, Longpre JM, Apel ED, Lewis RM, Wang LW, Sanes JR, Leduc R, Apte SS. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J Biol Chem. 2004;279(34):35159–75. doi: 10.1074/jbc.M402380200. [DOI] [PubMed] [Google Scholar]
- 79.Wang L, Zheng J, Bai X, Liu B, Liu CJ, Xu Q, Zhu Y, Wang N, Kong W, Wang X. ADAMTS-7 mediates vascular smooth muscle cell migration and neointima formation in balloon-injured rat arteries. Circ Res. 2009;104(5):688–98. doi: 10.1161/CIRCRESAHA.108.188425. [DOI] [PubMed] [Google Scholar]
- 80.Bengtsson E, Hultman K, Duner P, Asciutto G, Almgren P, Orho-Melander M, Melander O, Nilsson J, Hultgardh-Nilsson A, Goncalves I. ADAMTS-7 is associated with a high-risk plaque phenotype in human atherosclerosis. Sci Rep. 2017;7(1):3753. doi: 10.1038/s41598-017-03573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu J, Zhou B, Yu H, Han J, Cui M, Zhang F, Wang G, Guo L, Gao W. Association between plasma ADAMTS-7 levels and severity of disease in patients with stable obstructive coronary artery disease. Medicine (Baltimore) 2016;95(48):e5523. doi: 10.1097/MD.0000000000005523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saleheen D, Zhao W, Young R, Nelson CP, Ho W, Ferguson JF, Rasheed A, Ou K, Nurnberg ST, Bauer RC, et al. Loss of Cardioprotective Effects at the ADAMTS7 Locus as a Result of Gene-Smoking Interactions. Circulation. 2017;135(24):2336–2353. doi: 10.1161/CIRCULATIONAHA.116.022069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu W, Zhou Y, Li Y, Li J, Ke Y, Wang Y, Zheng J. Association between plasma ADAMTS-7 levels and ventricular remodeling in patients with acute myocardial infarction. Eur J Med Res. 2015;20:27. doi: 10.1186/s40001-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu W, Wang H, Yu C, Li J, Gao Y, Ke Y, Wang Y, Zhou Y, Zheng J. Association of ADAMTS-7 Levels with Cardiac Function in a Rat Model of Acute Myocardial Infarction. Cell Physiol Biochem. 2016;38(3):950–8. doi: 10.1159/000443047. [DOI] [PubMed] [Google Scholar]
- 85.Kessler T, Zhang L, Liu Z, Yin X, Huang Y, Wang Y, Fu Y, Mayr M, Ge Q, Xu Q, et al. ADAMTS-7 inhibits re-endothelialization of injured arteries and promotes vascular remodeling through cleavage of thrombospondin-1. Circulation. 2015;131(13):1191–201. doi: 10.1161/CIRCULATIONAHA.114.014072. [DOI] [PubMed] [Google Scholar]
- 86.Drummond AH, Beckett P, Brown PD, Bone EA, Davidson AH, Galloway WA, Gearing AJ, Huxley P, Laber D, McCourt M, et al. Preclinical and clinical studies of MMP inhibitors in cancer. Ann N Y Acad Sci. 1999;878:228–35. doi: 10.1111/j.1749-6632.1999.tb07688.x. [DOI] [PubMed] [Google Scholar]
- 87.Krzeski P, Buckland-Wright C, Balint G, Cline GA, Stoner K, Lyon R, Beary J, Aronstein WS, Spector TD. Development of musculoskeletal toxicity without clear benefit after administration of PG-116800, a matrix metalloproteinase inhibitor, to patients with knee osteoarthritis: a randomized, 12-month, double-blind, placebo-controlled study. Arthritis Res Ther. 2007;9(5):R109. doi: 10.1186/ar2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng X, Majerus EM, Sadler JE. ADAMTS13 and TTP. Curr Opin Hematol. 2002;9(5):389–94. doi: 10.1097/00062752-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 89.Zheng XL. ADAMTS13 and von Willebrand Factor in Thrombotic Thrombocytopenic Purpura. Annu Rev Med. 2015;66:211–25. doi: 10.1146/annurev-med-061813-013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thomas MR, de Groot R, Scully MA, Crawley JT. Pathogenicity of Anti-ADAMTS13 Autoantibodies in Acquired Thrombotic Thrombocytopenic Purpura. E Bio Medicine. 2015;2(8):942–52. doi: 10.1016/j.ebiom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, McGee BM, Yang AY, Siemieniak DR, Stark KR, Gruppo R, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488–94. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 92.Crawley JT, Scully MA. Thrombotic thrombocytopenic purpura: basic pathophysiology and therapeutic strategies. Hematology Am Soc Hematol Educ Program. 2013;2013:292–9. doi: 10.1182/asheducation-2013.1.292. [DOI] [PubMed] [Google Scholar]
- 93.Akyol O, Akyol S, Chen CH. Update on ADAMTS13 and VWF in cardiovascular and hematological disorders. Clin Chim Acta. 2016;463:109–118. doi: 10.1016/j.cca.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 94.Bongers TN, de Bruijne EL, Dippel DW, de Jong AJ, Deckers JW, Poldermans D, de Maat MP, Leebeek FW. Lower levels of ADAMTS13 are associated with cardiovascular disease in young patients. Atherosclerosis. 2009;207(1):250–4. doi: 10.1016/j.atherosclerosis.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 95.Bongers TN, Emonts M, de Maat MP, de Groot R, Lisman T, Hazelzet JA, Leebeek FW. Reduced ADAMTS13 in children with severe meningococcal sepsis is associated with severity and outcome. Thromb Haemost. 2010;103(6):1181–7. doi: 10.1160/TH09-06-0376. [DOI] [PubMed] [Google Scholar]
- 96.Feys HB, Canciani MT, Peyvandi F, Deckmyn H, Vanhoorelbeke K, Mannucci PM. ADAMTS13 activity to antigen ratio in physiological and pathological conditions associated with an increased risk of thrombosis. Br J Haematol. 2007;138(4):534–40. doi: 10.1111/j.1365-2141.2007.06688.x. [DOI] [PubMed] [Google Scholar]
- 97.Martin K, Borgel D, Lerolle N, Feys HB, Trinquart L, Vanhoorelbeke K, Deckmyn H, Legendre P, Diehl JL, Baruch D. Decreased ADAMTS-13 (A disintegrin-like and metalloprotease with thrombospondin type 1 repeats) is associated with a poor prognosis in sepsis-induced organ failure. Crit Care Med. 2007;35(10):2375–82. doi: 10.1097/01.ccm.0000284508.05247.b3. [DOI] [PubMed] [Google Scholar]
- 98.Molvarec A, Rigo J, Jr, Boze T, Derzsy Z, Cervenak L, Mako V, Gombos T, Udvardy ML, Harsfalvi J, Prohaszka Z. Increased plasma von Willebrand factor antigen levels but normal von Willebrand factor cleaving protease (ADAMTS13) activity in preeclampsia. Thromb Haemost. 2009;101(2):305–11. [PubMed] [Google Scholar]
- 99.Scully M, Hibbard C, Ewenstein B. Recombinant ADAMTS 13 in thrombotic thrombocytopenic purpura. Oncoscience. 2017;4(11–12):160–161. doi: 10.18632/oncoscience.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scully M, Knobl P, Kentouche K, Rice L, Windyga J, Schneppenheim R, Kremer Hovinga JA, Kajiwara M, Fujimura Y, Maggiore C, et al. Recombinant ADAMTS-13: first-in-human pharmacokinetics and safety in congenital thrombotic thrombocytopenic purpura. Blood. 2017;130(19):2055–2063. doi: 10.1182/blood-2017-06-788026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kopic A, Benamara K, Piskernik C, Plaimauer B, Horling F, Hobarth G, Ruthsatz T, Dietrich B, Muchitsch EM, Scheiflinger F, et al. Preclinical assessment of a new recombinant ADAMTS-13 drug product (BAX930) for the treatment of thrombotic thrombocytopenic purpura. J Thromb Haemost. 2016;14(7):1410–9. doi: 10.1111/jth.13341. [DOI] [PubMed] [Google Scholar]
- 102.Bekhouche M, Leduc C, Dupont L, Janssen L, Delolme F, Vadon-Le Goff S, Smargiasso N, Baiwir D, Mazzucchelli G, Zanella-Cleon I, et al. Determination of the substrate repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and TGF-beta signaling as primary targets. FASEB J. 2016;30(5):1741–56. doi: 10.1096/fj.15-279869. [DOI] [PubMed] [Google Scholar]
- 103.Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, Dollfus H, Alembik Y, Munnich A, Legeai-Mallet L, et al. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am J Hum Genet. 2004;75(5):801–6. doi: 10.1086/425231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morales J, Al-Sharif L, Khalil DS, Shinwari JM, Bavi P, Al-Mahrouqi RA, Al-Rajhi A, Alkuraya FS, Meyer BF, Al Tassan N. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85(5):558–68. doi: 10.1016/j.ajhg.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aldahmesh MA, Alshammari MJ, Khan AO, Mohamed JY, Alhabib FA, Alkuraya FS. The syndrome of microcornea, myopic chorioretinal atrophy, and telecanthus (MMCAT) is caused by mutations in ADAMTS18. Hum Mutat. 2013;34(9):1195–9. doi: 10.1002/humu.22374. [DOI] [PubMed] [Google Scholar]
- 106.Christensen AE, Fiskerstrand T, Knappskog PM, Boman H, Rodahl E. A novel ADAMTSL4 mutation in autosomal recessive ectopia lentis et pupillae. Invest Ophthalmol Vis Sci. 2010;51(12):6369–73. doi: 10.1167/iovs.10-5597. [DOI] [PubMed] [Google Scholar]
- 107.Mak AC, White MJ, Eckalbar WL, Szpiech ZA, Oh SS, Pino-Yanes M, Hu D, Goddard P, Huntsman S, Galanter J, et al. Whole Genome Sequencing of Pharmacogenetic Drug Response in Racially Diverse Children with Asthma. Am J Respir Crit Care Med. 2018 doi: 10.1164/rccm.201712-2529OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davis JP, Huyghe JR, Locke AE, Jackson AU, Sim X, Stringham HM, Teslovich TM, Welch RP, Fuchsberger C, Narisu N, et al. Common, low-frequency, and rare genetic variants associated with lipoprotein subclasses and triglyceride measures in Finnish men from the METSIM study. PLoS Genet. 2017;13(10):e1007079. doi: 10.1371/journal.pgen.1007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542(7640):186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jorgenson E, Makki N, Shen L, Chen DC, Tian C, Eckalbar WL, Hinds D, Ahituv N, Avins A. A genome-wide association study identifies four novel susceptibility loci underlying inguinal hernia. Nat Commun. 2015;6:10130. doi: 10.1038/ncomms10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vargas JD, Manichaikul A, Wang XQ, Rich SS, Rotter JI, Post WS, Polak JF, Budoff MJ, Bluemke DA. Common genetic variants and subclinical atherosclerosis: The Multi-Ethnic Study of Atherosclerosis (MESA) Atherosclerosis. 2016;245:230–6. doi: 10.1016/j.atherosclerosis.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43(10):1005–11. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boesgaard TW, Gjesing AP, Grarup N, Rutanen J, Jansson PA, Hribal ML, Sesti G, Fritsche A, Stefan N, Staiger H, et al. Variant near ADAMTS9 known to associate with type 2 diabetes is related to insulin resistance in offspring of type 2 diabetes patients--EUGENE2 study. PLoS One. 2009;4(9):e7236. doi: 10.1371/journal.pone.0007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grarup N, Andersen G, Krarup NT, Albrechtsen A, Schmitz O, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57(9):2534–40. doi: 10.2337/db08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trombetta M, Bonetti S, Boselli ML, Miccoli R, Trabetti E, Malerba G, Pignatti PF, Bonora E, Del Prato S, Bonadonna RC. PPARG2 Pro12Ala and ADAMTS9 rs4607103 as “insulin resistance loci” and “insulin secretion loci” in Italian individuals. The GENFIEV study and the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 4. Acta Diabetol. 2013;50(3):401–8. doi: 10.1007/s00592-012-0443-9. [DOI] [PubMed] [Google Scholar]
- 116.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Randall JC, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL, Kilpelainen TO, Esko T, Magi R, Li S, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9(6):e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barreto-Luis A, Pino-Yanes M, Corrales A, Campo P, Callero A, Acosta-Herrera M, Cumplido J, Ma SF, Martinez-Tadeo J, Villar J, et al. Genome-wide association study in Spanish identifies ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9), as a novel asthma susceptibility gene. J Allergy Clin Immunol. 2016;137(3):964–6. doi: 10.1016/j.jaci.2015.09.051. [DOI] [PubMed] [Google Scholar]
- 119.Julia A, Pinto JA, Gratacos J, Queiro R, Ferrandiz C, Fonseca E, Montilla C, Torre-Alonso JC, Puig L, Perez Venegas JJ, et al. A deletion at ADAMTS9-MAGI1 locus is associated with psoriatic arthritis risk. Ann Rheum Dis. 2015;74(10):1875–81. doi: 10.1136/annrheumdis-2014-207190. [DOI] [PubMed] [Google Scholar]
- 120.Polfus LM, Smith JA, Shimmin LC, Bielak LF, Morrison AC, Kardia SL, Peyser PA, Hixson JE. Genome-wide association study of gene by smoking interactions in coronary artery calcification. PLoS One. 2013;8(10):e74642. doi: 10.1371/journal.pone.0074642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pyun JA, Kim S, Cho NH, Koh I, Lee JY, Shin C, Kwack K. Genome-wide association studies and epistasis analyses of candidate genes related to age at menarche and age at natural menopause in a Korean population. Menopause. 2014;21(5):522–9. doi: 10.1097/GME.0b013e3182a433f7. [DOI] [PubMed] [Google Scholar]
- 122.Fan Q, Cheung CMG, Chen LJ, Yamashiro K, Ahn J, Laude A, Mathur R, Mun CC, Yeo IY, Lim TH, et al. Shared genetic variants for polypoidal choroidal vasculopathy and typical neovascular age-related macular degeneration in East Asians. J Hum Genet. 2017;62(12):1049–1055. doi: 10.1038/jhg.2017.83. [DOI] [PubMed] [Google Scholar]
- 123.Helisalmi S, Immonen I, Losonczy G, Resch MD, Benedek S, Balogh I, Papp A, Berta A, Uusitupa M, Hiltunen M, et al. ADAMTS9 locus associates with increased risk of wet AMD. Acta Ophthalmol. 2014;92(5):e410. doi: 10.1111/aos.12341. [DOI] [PubMed] [Google Scholar]
- 124.Whitmore SS, Braun TA, Skeie JM, Haas CM, Sohn EH, Stone EM, Scheetz TE, Mullins RF. Altered gene expression in dry age-related macular degeneration suggests early loss of choroidal endothelial cells. Mol Vis. 2013;19:2274–97. [PMC free article] [PubMed] [Google Scholar]
- 125.Lin E, Tsai SJ, Kuo PH, Liu YL, Yang AC, Kao CF, Yang CH. The ADAMTS9 gene is associated with cognitive aging in the elderly in a Taiwanese population. PLoS One. 2017;12(2):e0172440. doi: 10.1371/journal.pone.0172440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dong K, Yao N, Pu Y, He X, Zhao Q, Luan Y, Guan W, Rao S, Ma Y. Genomic scan reveals loci under altitude adaptation in Tibetan and Dahe pigs. PLoS One. 2014;9(10):e110520. doi: 10.1371/journal.pone.0110520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hanson E, Jood K, Nilsson S, Blomstrand C, Jern C. Association between genetic variation at the ADAMTS13 locus and ischemic stroke. J Thromb Haemost. 2009;7(12):2147–8. doi: 10.1111/j.1538-7836.2009.03617.x. [DOI] [PubMed] [Google Scholar]
- 128.Schierding W, Antony J, Karhunen V, Vaarasmaki M, Franks S, Elliott P, Kajantie E, Sebert S, Blakemore A, Horsfield JA, et al. GWAS on prolonged gestation (post-term birth): analysis of successive Finnish birth cohorts. J Med Genet. 2018;55(1):55–63. doi: 10.1136/jmedgenet-2017-104880. [DOI] [PubMed] [Google Scholar]
- 129.Galfalvy H, Haghighi F, Hodgkinson C, Goldman D, Oquendo MA, Burke A, Huang YY, Giegling I, Rujescu D, Bureau A, et al. A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2015;168(7):557–63. doi: 10.1002/ajmg.b.32330. [DOI] [PubMed] [Google Scholar]
- 130.Richter HE, Whitehead N, Arya L, Ridgeway B, Allen-Brady K, Norton P, Sung V, Shepherd JP, Komesu Y, Gaddis N, et al. Genetic contributions to urgency urinary incontinence in women. J Urol. 2015;193(6):2020–7. doi: 10.1016/j.juro.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McGrath LM, Cornelis MC, Lee PH, Robinson EB, Duncan LE, Barnett JH, Huang J, Gerber G, Sklar P, Sullivan P, et al. Genetic predictors of risk and resilience in psychiatric disorders: a cross-disorder genome-wide association study of functional impairment in major depressive disorder, bipolar disorder, and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(8):779–88. doi: 10.1002/ajmg.b.32190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Joe B, Saad Y, Dhindaw S, Lee NH, Frank BC, Achinike OH, Luu TV, Gopalakrishnan K, Toland EJ, Farms P, et al. Positional identification of variants of Adamts16 linked to inherited hypertension. Hum Mol Genet. 2009;18(15):2825–38. doi: 10.1093/hmg/ddp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L, Sha BY, Tang ZH, Pan F, Yang TL, Chen XD, et al. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84(3):388–98. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Knauff EA, Franke L, van Es MA, van den Berg LH, van der Schouw YT, Laven JS, Lambalk CB, Hoek A, Goverde AJ, Christin-Maitre S, et al. Genome-wide association study in premature ovarian failure patients suggests ADAMTS19 as a possible candidate gene. Hum Reprod. 2009;24(9):2372–8. doi: 10.1093/humrep/dep197. [DOI] [PubMed] [Google Scholar]
- 135.Pyun JA, Kim S, Kwack K. Epistasis between polymorphisms in ACVR2B and ADAMTS19 is associated with premature ovarian failure. Menopause. 2015;22(2):212–6. doi: 10.1097/GME.0000000000000285. [DOI] [PubMed] [Google Scholar]
- 136.Zhao X, Onteru S, Saatchi M, Garrick D, Rothschild M. A genome-wide association study for canine cryptorchidism in Siberian Huskies. J Anim Breed Genet. 2014;131(3):202–9. doi: 10.1111/jbg.12064. [DOI] [PubMed] [Google Scholar]
- 137.Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, Adler AJ, Li H, Rasmussen A, Williams AH, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet. 2012;90(4):648–60. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zillikens MC, Demissie S, Hsu YH, Yerges-Armstrong LM, Chou WC, Stolk L, Livshits G, Broer L, Johnson T, Koller DL, et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat Commun. 2017;8(1):80. doi: 10.1038/s41467-017-00031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dow DJ, Huxley-Jones J, Hall JM, Francks C, Maycox PR, Kew JN, Gloger IS, Mehta NA, Kelly FM, Muglia P, et al. ADAMTSL3 as a candidate gene for schizophrenia: gene sequencing and ultra-high density association analysis by imputation. Schizophr Res. 2011;127(1–3):28–34. doi: 10.1016/j.schres.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 140.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martinez R, Bejarano D, Gomez Y, Dasoneville R, Jimenez A, Even G, Solkner J, Meszaros G. Genome-wide association study for birth, weaning and yearling weight in Colombian Brahman cattle. Genet Mol Biol. 2017;40(2):453–459. doi: 10.1590/1678-4685-GMB-2016-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Abramowitz Y, Roth A, Keren G, Isakov O, Shomron N, Laitman Y, Weissglas-Volkov D, Arbel Y, Banai S, Finkelstein A, et al. Whole-exome sequencing in individuals with multiple cardiovascular risk factors and normal coronary arteries. Coron Artery Dis. 2016;27(4):257–66. doi: 10.1097/MCA.0000000000000357. [DOI] [PubMed] [Google Scholar]
- 143.Laje G, Allen AS, Akula N, Manji H, John Rush A, McMahon FJ. Genome-wide association study of suicidal ideation emerging during citalopram treatment of depressed outpatients. Pharmacogenet Genomics. 2009;19(9):666–74. doi: 10.1097/FPC.0b013e32832e4bcd. [DOI] [PMC free article] [PubMed] [Google Scholar]