Abstract
The intestinal epithelial barrier is important to mucosal immunity, although how it is maintained after damage is unclear. Here, we show that G protein-coupled receptor 109A (GPR109A) supports barrier integrity and decreases mortality in a mouse cecum ligation and puncture (CLP) sepsis model. Data from 16S RNA sequencing showed that the intestinal microbiota of WT and Gpr109a−/− mice clustered differently and their compositions were disrupted after CLP surgery. GPR109A-deficient mice showed increased mortality, intestinal permeability, altered inflammation, and lower tight junction gene expression. After eliminating the intestinal flora with antibiotics, all experimental mice died within 48 h of CLP surgery. This demonstrates the critical role of the gut microbiota in CLP-induced sepsis. Importantly, mortality and other pathologies in the model were decreased after Gpr109a−/− mice received WT gut microbiota. These findings indicate that GPR109A regulates the gut microbiota, contributing to intestinal epithelial barrier integrity and decreased mortality in CLP-induced sepsis.
Keywords: GPR109A, gut microbiota, intestinal epithelial barrier, CLP, sepsis
Introduction
Sepsis is a frequent cause of mortality in intensive care units and is one of the 10 most common causes of death worldwide (1–3). However, sepsis is not a single disease but a collection of several related syndromes that often occur after bacterial, viral, or fungal infection (4). Sepsis can also arise after sterile tissue injuries, such as pancreatitis, ischemia reperfusion injury, cancer, or other disorders (1). The intestine has been hypothesized to play a central role in the pathogenesis of sepsis and been referred to as the “motor” of the systemic inflammatory response (5–7). Sepsis is also often associated with impairment of the intestinal epithelial barrier. This increases intestinal permeability, causing the contents of intestinal lumen, including the microbiota, to leak out of the intestine, resulting in multiple organ failure or even death in the most extreme cases (8–10). The protective effects of the intestinal epithelial barrier are therefore essential for the prevention and treatment of sepsis.
In 2003, G protein-coupled receptor 109A (GPR109A) was identified as a receptor for niacin or nicotinic acid (NA) and was found to mediate the antilipolytic effects of NA (11–13). GPR109A belongs to the large G protein-coupled receptor family and is activated by 3-hydroxy-butyrate. Except for lymphocytes, it is expressed on most cells, including adipocytes, monocytes, macrophages, and dermal dendritic cells (14). GPR109A activation has been shown to reduce inflammation during atherosclerosis, sepsis, obesity, diabetic retinopathy, and renal disease (15–19). In recent years, GPR109A was thought to be associated with regulation of the microbiotametabolites, suppressing colonic inflammation, and maintaining intestinal barrier integrity (20, 21). However, dietary fiber has also been shown to regulate the composition of the intestinal microbiota (22–24). As the receptor for butyrate, previous studies have shown that microbiota-derived butyrate and NA inhibit inflammation of the intestinal tract by activating GPR109A in intestinal epithelial cells, dendritic cells, and macrophages (25). However, whether these microbiota-associated metabolites are the only associations that exist between the GPR109A receptor and the gut microbiota is unknown, as is the exact role that GPR109A plays in regulating the gut microbiota.
To investigate the function of GPR109A in more detail, our study used cecum ligation and puncture surgery (CLP), described as the “gold standard” model for sepsis(26), to examine the effects of Gpr109a deletion. We performed CLP surgery using WT and Gpr109a−/− mice with a C57BL/6 background to investigate the protective effects of GPR109A receptor in this sepsis model. We also examined the association between GPR109A and the gut microbiota, in particular the role that the microbiota may play in controlling the pathology of sepsis. These experiments revealed that GPR109A regulates the intestinal microbiota, leading to the preservation of intestinal epithelial barrier integrity. This demonstrates the value of investigating the links between host and microbiota and may lead to improvements in sepsis therapy and control.
Materials and methods
Animals
Gpr109a−/−and WT mice were on a C57BL/6 background and maintained under specific pathogen-free (SPF) conditions. All mice were provided with food and water ad libitum and housed under a strict 12 h light-dark cycle. Six-to-eight week old female mice were used for all experiments. The Gpr109a−/− was generous gift form Dr. Martin Sager (Zentrale Einrichtung für Tierforschung und Tierschutzaufgabender Heinrich-Heine Universität Düsseldorf, Germany). The WT mice were provided by the Centre of Experimental Animalsof the Baiqiuen Medical College of Jilin University (Changchun, China). Studies were performed in accordance with guidelines established by the Jilin University Institutional Animal Care and Use Committee (Permit number: 20170401).
CLP model
CLP surgerywas performed as descried previously(27). Briefly, prior to CLP surgery, autoclaved the scissors and forceps, and anesthetized the mice with pentobarbital sodium (45 mg/kg). About 1 cm incision is made to the left ventral surface of the abdomen, and the cecum is exposed. The cecum is partially ligated (about 1 cm) at its base with a 3-0 silk suture, then the ligated cecum was punctured with a 21-gauge needle. The cecum was returned to the peritoneal cavity and the incision was sutured with 3–0 silk suture. The mice would recover in about 1 h.
Body weight and temperature
Before the CLP surgery, measured the body weight using an electronic scale and recorded W-0, examined the rectal temperature with a thermometer and recorded T-0, after CLP surgery, measured the body weight and temperature at 6, 12, 24 and 48 h, and recorded W-6, W-12, W-24, W-48, and T-0, T-12, T-24, T-48 respectively. Weight loss-n = W-0 - W-n, Temperature change-n = T-0–T-n.
Clinical score
During the experiment, roachback or emaciation, colon thickening and pellet morphology were recorded. The clinical score was calculated based on the scoring system as shown in Table 1.
Table 1.
The scoring system of clinical score.
| Roachback or Emaciation | Colon thickening | Pellet morphology | |
|---|---|---|---|
| 0 | None | None | Normal |
| 1 | Yes | Slight | Soft stool |
| 2 | Moderate | Diarrhea | |
| 3 | Severe | Bloody stool |
Antibiotic treatment
Mice were given a combination of vancomycin (1 g/L), kanamycin (1 g/L), ampicillin (1 g/L), and metronidazole (1 g/L) in their drinking water following a previously published regimen (28, 29). After two months, colon samples were collected and incubated in lysogeny broth (LB) solid medium for 12 h to assess whether the gut microbiota had been completely removed by the antibiotics. All antibiotics used in this experiment were purchased from Sigma-Aldrich, St. Louis, MO, USA.
Intestinal permeability assay
Intestinal permeability was assessed using fluorescein isothiocyanate (FITC)-dextran with an average molecular weight of 3,000–5,000 (Sigma-Aldrich), as previously described (30). Briefly, 2 d after CLP surgery, mice were deprived of food for 4 h and then gavaged with FITC-dextran (0.6 mg/g body weight at a concentration of 125 mg/mL). Four hours later, blood was collected from the eyes of micecentrifuged at 12,000 g for 3 min. The FITC-dextran content of the serum was determined using a microplate reader with an excitation wavelength of 490 nm and emission detection at 525 nm. Each sample was measured in triplicate.
Fecal microbial transplantation
Between three to four pellets were collected from adult female mice, re-suspended in 2 mL sterile PBS/glycerol (80:20 v:v), and homogenized. Next, 200 μL of the homogenized fecal samples were introduced by gavage with a flexible plastic tube into the stomachs of experimental mice (WT intestinal microbiota transplanted into Gpr109a−/− mice, Gpr109a−/− intestinal microbiota transplanted into WT mice). The fecal suspension was administered three times, 7 d apart. The recipient mice were analyzed 14 d after the third microbiota transfer, as described previously (31).
Gut microbiota profiling
Fresh colon samples were collected and immediately frozen using liquid nitrogen. These were then sent to Beijing Allwegene Gene Technology Co., Ltd., Beijing, China. Bacterial DNA was extracted using a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA), following the manufacturer's instructions. The purity and quality of the genomic DNA were checked using 0.8% agarose gels. Bacterial DNA was PCR amplified with barcoded universal bacterial primers targeting the V3 and V4 variable regions of the 16S rRNA gene. Three PCR products per sample were pooled to mitigate reaction-level PCR biases. PCR products were purified using a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), quantified using Real Time PCR, and sequenced at Auwigene, Beijing, China. Deep sequencing was performed using the Miseq platform at Auwigene. After sequencing, image analysis, base calling, and error estimation were performed using Illumina Analysis Pipeline Version 2.6 (Illumina, San Diego, CA, USA). The raw data were first screened and sequences removed if they were shorter than 200 bp, had a low-quality score (≤ 20), contained ambiguous bases, or did not exactly match the primer sequences and barcode tags. The remaining high-quality reads were separated using sample-specific barcode sequences and trimmed with Illumina Analysis Pipeline Version 2.6 for analysis using QIIME. The sequences were clustered into operational taxonomic units (OTUs) at a similarity level of 97% to generate rarefaction curves and to calculate richness and diversity indices. The Ribosomal Database Project (RDP) Classifier tool (https://rdp.cme.msu.edu/classifier/) was used to classify all sequences into different taxonomic groups. To examine the similarities between different samples, clustering analyses and PCoA were performed based on the OTUs from each sample using R. The evolutionary distances between microbial communities from each sample were calculated using thetayc coefficients and represented as an unweighted pair group method with arithmetic mean (UPGMA) clustering tree that described the dissimilarity (1-similarity) between multiple samples. These data were used to generate a Newick-formatted tree file. To compare group memberships and the structures of the communities in different samples, heat maps were generated using the 20 most abundant OTUs and mothur's Bayesian classifier.
Hematoxylin and eosin (H&E)-staining and immunofluorescence
After CLP surgery 48 h later, mice were sacrificed and the colonic and ileac segments 2 to 3 cm in length were excised, washed in phosphate buffered saline (PBS), fixed in 4% formaldehyde, embedded in paraffin, and sectioned (5-μm thick). A portion of the paraffin sections were stained with hematoxylin and eosin (H&E) and the rest was used for immunofluorescence using procedures detailed previously (32). Briefly, antigens were unmasked by boiling under pressure in sodium citrate buffer. Tissue sections were cooled to room temperature (about 20°C) naturally and then washed with PBS three times (5 min each wash). The sections were incubated with a blocking buffer (5% donkey serum in PBS) for 60 min against the species of the secondary antibody. Primary antibodies (Mucin 2 H-300, rabbit polycional lgG, Santa Cruz, USA) were incubated at overnight 4°C and then the sections were washed three times with PBS. Alexa Fluor® 488 and 594, donkey anti-rabbit lgG (H+L; Life, USA) were diluted 1:1000 and incubated for 1 h at RT and washed three times in PBS. Nuclei were counterstained using DAPI. The tissue sections were then photographed with Nikon Eclipse Ti-U microscope and Nikon Intensilight C-HGFI (Japan).
Quantitative real-time PCR (qRT-PCR)
After CLP surgery 48 h later, mice were sacrificed. Fresh colon and ileum tissues were collected and frozen using liquid nitrogen. These were then stored in a −80 °C ultra-low temperature freezer until use. The samples were pulverized with a cell/tissue grinder and total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. cDNA was then generated using a commercial RT-PCR Kit (TaKaRa, Kyoto, Japan) and amplified with a SYBR Green QuantiTect RT-PCR Kit (Roche, Basel, Switzerland) and the primers of IL-6, IL-1β, Cldn1, Cldn2, Cldn3, Ocln, Zo-1, Zo2, and β-actin to evaluate the mRNA levels of various genes. Each of the samples was analyzed in triplicate using procedures referenced in our previous work (33). The sequences of primers used in this investigation are shown below.
| Gene | Forward primer | Reverse primer |
| IL-6 | 5′-AGCCACTGCCTTCCCTAC-3′ | 5′-TTGCCATTGCACAACTCTT-3′ |
|---|---|---|
| IL-1β | 5′-TGTGATGTTCCCATTAGAC-3′ | 5′-AATACCACTTGTTGGCTTA-3′ |
| Cldn1 | 5′-AGGTCTGGCGACATTAGTGG-3′ | 5′-CGTGGTGTTGGGTAAGAGGT-3′ |
| Cldn2 | 5′-AGTGGCTGTAGTGGGTGGAG-3′ | 5′-AAAGGATGACTCCGGCTACC-3′ |
| Cldn3 | 5′-GAGATGGGAGCTGGGTTGTA-3′ | GTAGTCCTTGCGGTCGTAGG |
| Zo-1 | 5′-GACCTTGATTTGCATGACGA-3′ | 5′-AGGACCGTGTAATGGCAGAC-3′ |
| Zo-2 | 5′-CAGTCCCTATGCCTGAGAGC-3′ | 5′-TTGGAACCGCATAGATGTCA-3′ |
| Ocln | 5′-ACACTTGCTTGGGACAGAGG-3′ | 5′-AAGGAAGCGATGAAGCAGAA-3′ |
| β-actin | 5′-GTCAGGTCATCACTATCGGCAAT-3′ | 5′-AGAGGTCTTTACGGATGTCAACGT-3′ |
Statistics
Data are presented as means ± SEM. Data were analyzed using the statistical software package SPSS 12.0 (SPSS Inc., Chicago, IL, USA). Groups were compared by one-way analysis of variance (ANOVA) followed by the least significant difference test. *P < 0.05 was considered significant, and **P < 0.01 was considered markedly significant.
Results
GPR109A-deficient mice show severe inflammation and increased mortality in a murine CLP-induced sepsis model
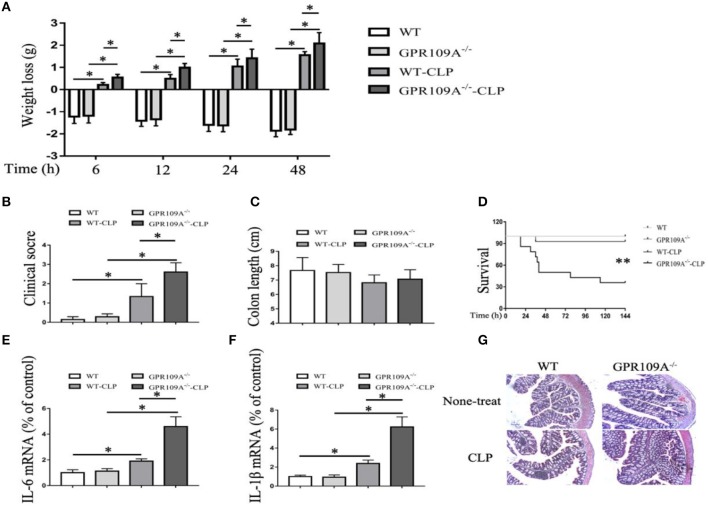
Previous studies have reported that GPR109A inhibits inflammation during inflammatory bowel disease (IBD) (20, 21). To determine whether GPR109A functions in a similar manner during sepsis, we compared WT and Gpr109a−/−C57BL/6 female mice in a CLP-induced sepsis model. This showed that the pathogenic manifestations in Gpr109a−/− mice were more serious than those in WT mice. These included more rapid loss of body weight, and more severe clinical scores, but there were no effects on colon length (Figures 1A–C). In particular, the Gpr109a−/− mice had significantly decreased survival rate (Figure 1D), suggesting that GPR109A has a protective effect against CLP-induced sepsis in the model. These effects were also observed in colon tissue homogenate pro-inflammatory cytokine release (Figures 1E,F), indicating that GPR109A inhibited the expression of these pro-inflammatory markers during sepsis. We next examined the colon and ileum using hematoxylin and eosin (HE)-stained specimens (Figure 1G), revealing that GPR109A offered a protection against tissue damage during CLP-induced sepsis in the intestinal epithelium. All of these data indicate that GPR109A protects against intestinal inflammation in CLP-induced sepsis.
Figure 1.
GPR109A-Deficient mice show an altered inflammatory response and increased mortality in a CLP-induced sepsis model. all measurements were performed using 6 to 8 week-old female mice, *p < 0.05. (A) Body weight loss in none-treat WT and Gpr109a−/−mice, and CLP-treat WT and Gpr109a−/− mice(n = 7 ~ 9), *p < 0.05. (B) Clinical score of none-treat WT and Gpr109a−/−mice, and CLP-treat WT and Gpr109a−/− mice (n = 5 ~ 9), *p < 0.05. (C) The colon lengths of WT and Gpr109a−/− mice, and CLP-treat WT and Gpr109a−/− mice (n = 5 ~ 9). (D) The survival rates of none-treat WT and Gpr109a−/−, and CLP-treat mice (n = 14) mice, **p < 0.01. (E,F) The expression levels of IL-6 and IL-1β mRNA in colon tissue homogenate from none- and CLP-treated WT and Gpr109a−/− mice (n = 3, three determinations per mouse). The results shown are means ± SEM, *p < 0.05. (G) Hematoxylin and eosin (H&E)-stained colon and ileum samples from none- and CLP-treated WT and Gpr109a−/− mice (n = 3) mice. Magnification shown is 10×. The scale bar represents 500 μm.
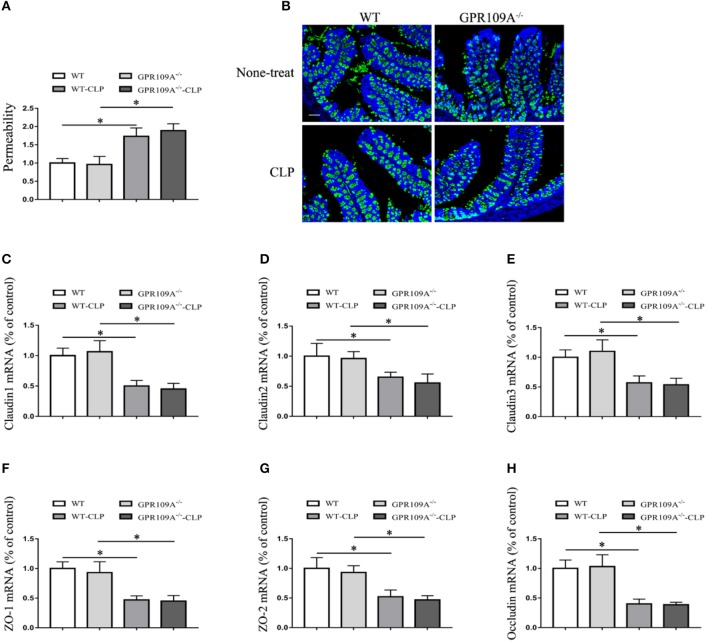
GPR109A protects mice from CLP-induced intestinal permeability and a decrease in tight junction gene and mucin-2 protein expression
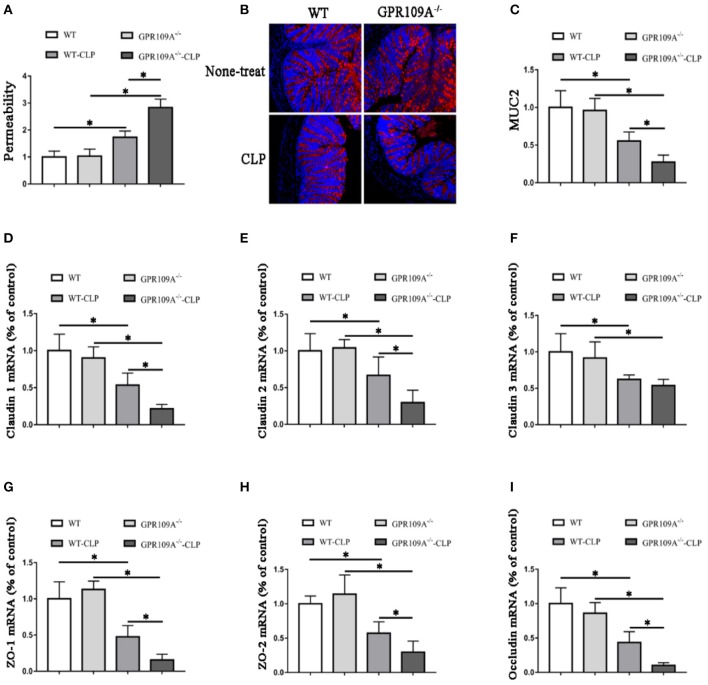
Although previous studies have shown that GPR109A protects against intestinal inflammation in IBD (20, 21), there was no investigation of the functions of GPR109A in maintaining intestinal barrier integrity in sepsis. To study intestinal leakage in WT and Gpr109a−/− mice after CLP surgery, intestinal permeability was tested by orally administrating fluorescently-labeled FITC-dextran to the experimental mice. This showed decreased intestinal permeability in Gpr109a−/− mice relative to that in WT, suggesting that GPR109A is involved in regulating permeability (Figure 2A). To further explore the mechanisms that underlie the effects of GPR109A on decreasing intestinal permeability, the expression of various tight junction gene transcripts in the colon was evaluated. This showed that the expression of claudin-1 (Cldn1), claudin-2 (Cldn2), Zo1, Zo2, and occludin (Ocln) was lower in Gpr109a−/− mice relative to WT mice, although there was no difference in claudin-3 (Cldn3) expression (Figures 2D–I). We next examined the expression of mucin-2 (MUC2) protein using immunofluorescence. MUC2 is one of the main components of the mucosa and is typically expressed by goblet cells. We found that Gpr109a−/− mice had less MUC2 protein expression than WT, indicating that GPR109A is involved in maintaining natural levels of MUC2 in WT mice (Figures 2B,C). We also measured the expression of tight junction genes and MUC2 in the ileum, demonstrating that these were consistent with the colon data (Figures S1A–G). All of these results indicate that GPR109A maintains the expression of tight junction genes and MUC2 protein, and has a role in controlling the permeability of the intestine in CLP-induced sepsis in mice.
Figure 2.
GPR109A Protects mice from CLP-induced intestinal permeability by regulating the expression of tight junction genes and Mucin-2 protein. (A) The intestinal of permeability of none- and CLP-treated WT and Gpr109a−/− mice (n = 3, three determinations per mouse). The results shown are means ± SEM, *p < 0.05. (B) Sections were prepared from the colons of none- and CLP-treated WT and Gpr109a−/− mice (n = 3), and observed by fluorescence microscopy. Nuclear staining with DAPI is shown in blue. MUC2 staining is indicated in red. Magnification shown is 10×. The scale bar represents 500 μm. (C) Representative thequantization of MUC2 in B. (D–I) Representative the expression levels of claudin-1 (Cldn1), claudin-2 (Cldn2), claudin-3 (Cldn3), Zo1, Zo2, and occludin of colon tissue homogenate from none- and CLP-treated WT and Gpr109a−/− mice (n = 3, three determinations per mouse). The results shown are means ± SEM, *p < 0.05.
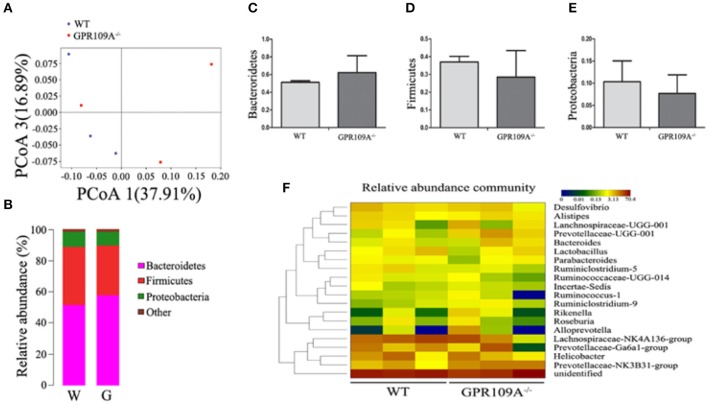
GPR109A regulates the gut microbiota and CLP-induced sepsis affects the composition of the intestinal microbiota
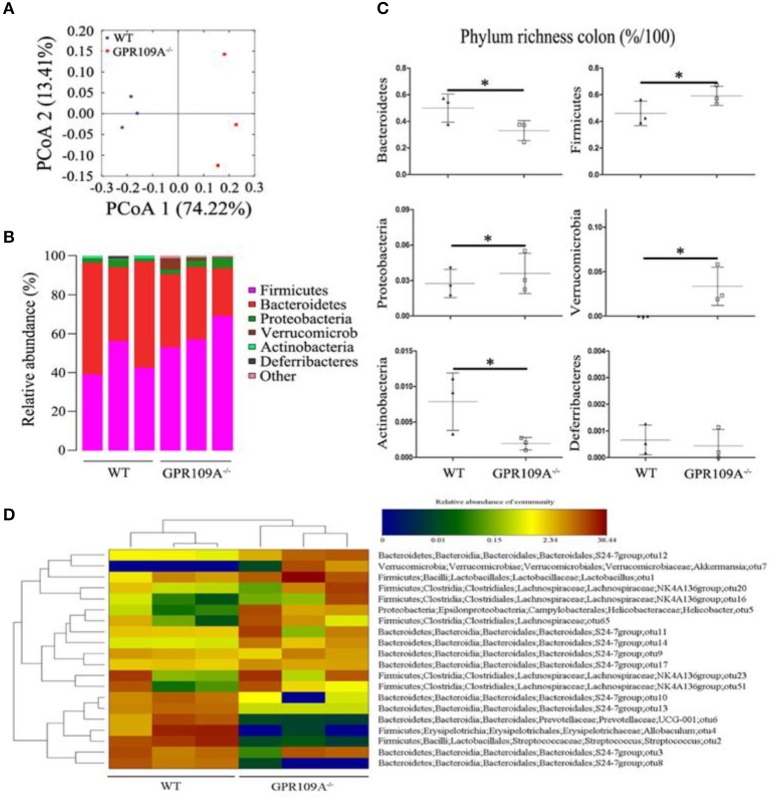
Bacteria play a critical role in the pathogenesis of IBD and changes to the composition and abundances of the species in the gut microbiota can accompany the disease (34, 35). However, there are no reports on whether there is any alteration of the gut microbiota during sepsis. To investigate this, we performed 16S rRNA sequencing to assess the composition of the microbiota in colon samples from both WT and Gpr109a−/− mice. A principal coordinates analysis (PCoA) using unweighted UniFrac distances showed that there were major differences between the gut microbiota of the two groups of mice (Figure 3A). As expected, the Firmicutes and Bacteroidetes were the most abundant phyla in samples (Figure 3B). However, the proportion that were Bacteroidetes or Actinobacteria was higher in the colons of WT mice compared to that in Gpr109a−/− mice. Conversely, the Firmicutes, Proteobacteria, and Verrucomicrobia showed an opposite effect and were more abundant in the Gpr109a−/− mice. There was no difference in the proportion that were Deferribacteres between both groups (Figure 3C). We next assessed if there was a difference in the abundances of operational taxonomic units (OTUs) at the phylum level in the Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia. This analysis revealed that the OTUs present within the WTorGpr109a−/− mice clustered together, but the OTUs were different betweenWT and Gpr109a−/−mice (Figure 3D). We also compared the heterozygous Gpr109a+/– and Gpr109a–/– mice, and the results showed that there is obvious difference between the mice (Figure S2A). The Shannon, Simpson, PCoA and gut microbiota gene function and pathway analysis results showed that there was obvious difference on the intestinal microbiota composition of the Gpr109a–/– and Gpr109a+/–, WT mice. The intestinal microbiota structure of Gpr109a+/– is more similar than Gpr109a–/– with WT mice (Figures S2B–E). All the results demonstrate that GPR109A regulates the structure of the gut microbiota in mice.
Figure 3.
GPR109A regulates the gut microbiota of mice. (A) Principal coordinates analysis (PCoA) based on unweighted UniFrac distances of OTUs. Each symbol represents a single WT or Gpr109a−/− colon sample (n = 3 for both). (B) Comparison of phylum-level proportional OTU abundances in WT or Gpr109a−/− colon samples (n = 3 for both). (C) Representative proportions of OTUs classified at the phylum level (n = 3 mice for each). *p < 0.05. (D) Heatmap depicting the OTU abundances at the phylum level within the Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia. OTUs are shown as Phylum, Class, Order, Family, Genus, and Species.
In order to understand the effects of CLP and sepsis on the gut microbiota, fecal samples were collected two days after surgery. Further 16S rRNA gene sequencing was used to analyze the microbiota and a PCoA showed that the individual differences in the sepsis mice were larger than in untreated mice (Figures S3A). This indicates that the compositions of gut microbiota were dramatically altered after CLP surgery. Notably, the proportions of Bacteroidetes increased by 25.87 and 56.57% in WT and Gpr109a−/− mice compared to that in untreated mice, respectively. However, the proportions that were Firmicutes was reduced by 36.68 and 53.60% in WT and Gpr109a−/− mice, respectively (Figures S3B,C). Changes to other phyla were also observed (Figure S2C). Compared to that in the untreated mice, OTU composition at the phylum level in the Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia was also dramatically changed. Additionally, OTUs could not be clustered within the WT and Gpr109a−/−-CLP mice as before (Figure S3D). These data indicate that CLP-induced sepsis affects the structure of the gut microbiota in the mice colon.
The gut microbiota protects against the effects of CLP-induced sepsis in the mouse model
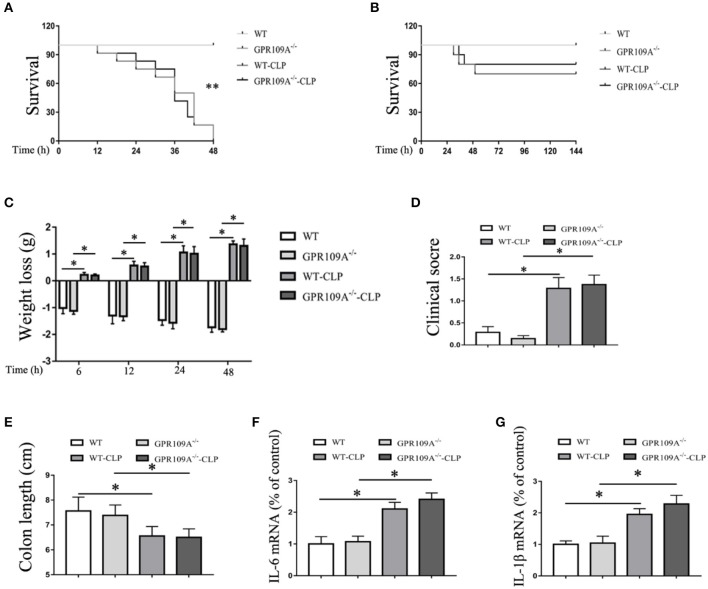
In order to assess the role of the gut microbiota during CLP-induced sepsis, we used orally administered antibiotics and gut flora transplantation to either remove or replace the microbiota. WT and Gpr109a−/− mice were administered a combination of vancomycin (1 g/L), kanamycin (1 g/L), ampicillin (1 g/L), and metronidazole (1 g/L) in their drinking water for two months using a validated regimen (28, 29) (Figure S4A). Colon samples were collected and incubated in lysogeny broth (LB) solid medium for 12 h. Intact colonies were observed in the WT-Sham (W-Sham) and Gpr109a−/−-Sham (G-Sham) groups. However, the antibiotic treated groups displayed negligible colony growth (Figure S4B), demonstrating that the gut microbiota had been removed by the antibiotics. We then examined the effects of antibiotic treatment and CLP-induced sepsis. During this experiment, all of the mice died within 48 h (n ≥ 30) of CLP surgery (Figure 4A). This demonstrates that the gut microbiota plays a critical role in protection against CLP-induced sepsis in the mouse model.
Figure 4.
The gut microbiota plays a protective role during CLP-induced sepsis in mice. (A) The survival of none-treat WT and Gpr109a−/– mice, andantibiotic treated WTand Gpr109a−/– mice which undergo the CLP surgery (n ≥ 30)), **p < 0.01. (B) Overall survival rate of none-treat WT and Gpr109a−/– mice, and microbiota-transferred WT and Gpr109a−/– mice which undergo the CLP surgery (n = 14). (C) Weight loss of none- and CLP-treated WT and Gpr109a−/− mice (n = 7 ~ 9). (D) Clinical score of none- and CLP-treated WT and Gpr109a−/− mice (n = 7 ~ 9), *p < 0.05. (E) Colon lengths of different treatment mice (n = 7 ~ 9). (F,G) The expression of IL-6 and IL-1β mRNA in colon tissue homogenate from different treatment mice (n = 3).
To validate this hypothesis, we transplanted the intestinal flora of WT mice into Gpr109a−/− mice via fecal transfer. After sequencing the gut microbiota of the microbiota-transplanted mice, the PCoAresultsshowed large individual differences (Figure 5A), but the relative abundances of phyla were almostidentical in the Bactericides, Firmicutes, and Proteobacteria (Figures 5B–E). We then examined the 20 most abundant genera in the gut microbiota, revealing that there were no differences betweenthe WTmice (receive the intestinal microbiota of Gpr109a−/− mice) and Gpr109a−/−mice (receive the intestinal of WT mice; Figure 5F). This indicated that the gut flora had been successfully transferred between the WT and Gpr109a−/− mice. CLP surgery was then performed in these gut microbiota transplanted mice, revealing that there were no differences between WT and Gpr109a−/− mice when assessed using various pathological measures, including lossof body weight, change in body temperature, clinical score, colon length, and overall survival (Figures 4B–E). The expression of colon epithelium pro-inflammatory cytokines was also similar between the WT and Gpr109a−/−mice (Figures 4F,G). The results demonstrated that there is no pathological differences between WT mice and Gpr109a−/−mice with a WT microbiota.
Figure 5.
16S rRNA Sequencing of colon samples from microbiota transferred mice. (A) Principal coordinates analysis (PCoA) based on the unweighted UniFrac distances of OTUs. Each symbol represents a single colon sample from WT (n = 3) or Gpr109a−/– (n = 3) mice. (B) Representative comparisons of the average phylum-level proportional abundances in colon samples from WT (n = 3) and Gpr109a−/– (n = 3) mice. (C–E) Representative proportions of OTUs classified at the phylum level (n = 3 for each). (F) Heatmap depicting the top 20 genera in the gut microbiota of colon samples from WT (n = 3) and Gpr109a−/– (n = 3) mice.
To further investigate the involvement of host microbiota, we examined the permeability of microbiota-transferred mice. This showed that there was no difference between WT and Gpr109a−/− mice after transplantations of intestinal microbiota (Figure 6A). We next measured MUC2 protein expression using immunofluorescence and the expression of tight junctions genes by qRT-PCR. This again showed that there were no differences between the two groups after transfer of a WT microbiota into Gpr109a−/− mice and Gpr109a−/−microbiota into WT mice (Figures 6B–H). We also measured tight junction gene expression and MUC2 protein in ileum samples, demonstrating that they were similar to those in the colon samples (Figures S5A–G). Together, these results indicate that the gut microbiota has a protective effect against CLP-induced sepsis in the mouse model.
Figure 6.
The gut microbiota affects intestinal permeability and promotes the natural expression of MUC2 protein and tight junction genes in the CLP-induced Sepsis Model. (A) The intestinal of permeability of different treatment mice (n = 3). The results are shown as means ± SEM, *p < 0.05. (B) Tissue sections were prepared from the colons of none-treat WT and Gpr109a−/– mice, and microbiota-transferred WT and Gpr109a−/– mice which undergo the CLP surgery (n = 3) and observed by fluorescence microscopy. The nucleus was stained with DAPI (blue) and MUC2 was stained green. Magnification shown is 10×. The scale bar represents 500 μm. (C–H) Representative expression levels of claudin-1 (cldn1), claudin-2 (cldn2), claudin-3 (cldn3), ZO-1, ZO-2, and occludin (ocln) in colon tissue homogenates from different treatment mice (n = 3). The results shown are means ± SEM, *p < 0.05.
Discussion
The intestinal epithelium is a critical barrier that protects against potentially harmful microbiota and pathogens (36). Defective tight junctions in the intestinal epithelial barrier can lead to an increase in intestinal permeability, contributing to the development of intestinal inflammation (37, 38, 39). After organisms and viruses from the microbiota cross the intestinal epithelium and enter the blood, they can result in multi-organ dysfunction and even death (8–10). The protective effects of the intestinal epithelium barrier are therefore central to the prevention and treatment of sepsis.GPR109A have been demonstrated plays an important role in maintaining intestinal epithelium barrierand inhibiting inflammationby us and others (21, 40).
In the present study, we have examined the differences in survival, inflammation, and epithelial barrier integrity in WT and Gpr109a−/− mice using a CLP-induced model of sepsis. This has demonstrated that Gpr109a−/−mice showed overall lower survival, altered inflammation, and disruption of epithelial barrier integrity. This is consistent with previous studies that indicate that microbial-derived butyrate protects against inflammation through GPR109A signaling during IBD (20, 21). Butyrate is one of the most important microbiota metabolites involved in the activation of the GPR109A receptor. Activation by butyrate and NA in intestinal epithelial cells, dendritic cells, and macrophages, can lead to inhibition of inflammation in the intestinal tract (25). However, there have been no studies investigating the direct influence of host GPR109A on the intestinal microbiota, although recent studies have revealed that dietary fiber can help to maintain the stability of the intestinal flora (22–24).
To investigate the relationship between GPR109A and the microbiota, we performed 16S rRNA sequencing using colon fecal samples from WT and Gpr109a−/− mice, demonstrating that the GPR109A receptor regulated the structure of the gut microbiota and that the microbial populations were different between WT and Gpr109a−/−mice. Bacteroidetes and Firmicutes, two major phyla of gut microbiota in humans, which are involved in the regulation of lipid and bile acid metabolism in the gut(41). Proteobacteria is phylum of Gram-negativebacteria includes many pathogenic and lipopolysaccharide-containing bacteria, such as Escherichia. Jacqueline M. reported a relative high abundance of Proteobacteria was observed in critically ill patients (42). In particular, the proportions of Firmicutes, Actinobacteria, Bacteroidetes, Proteobacteria, and Verrucomicrobia we observed differed between the two groups of mice (Figure 3C). It's remarkable that the abundance of Proteobacteria is higher in Gpr109a−/−mice than control. The OTU abundances at the phylum level within the Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia were also different (Figure 3D). Similar research has revealed that mice deficient in the intestinal inflammatory regulator NLRP12 possess a dysbiotic microbiome and exacerbated colitis (43). Additionally, SPF mice have been found to display different inflammatory responses in the gut depending on the composition of their intestinal microbiota in a DSS-induced model of colitis (44). These data, combined with our results, indicate that that GPR109A and the microbiota interact to maintain homeostasis in the host epithelial barrier.
Our data also showed that the gut microbiota in the WT and Gpr109a−/− groups changed dramatically after CLP-induced sepsis relative to that in untreated mice. This indicated that CLP-induced sepsis severely disrupted the composition of the gut microbiota, leading to dysbiosis. The ratio of Firmicutes to Bacteroidetes (F/B ratio) was thought as amicrobiota marker in critically ill patients which represent decreased microbial diversity and severe inflammatory response. Ojima M have reported that extreme changes in the F/B ratio were observed in almost all of the sepsis patients with a poor prognosis (45). In the present study, Gpr109a−/−mice showedextraordinary decrease in F/B ratio after CLP (data was not shown) and eventually lead to an increase in mortality.Previous work using TNFdeltaARE mice has also shown that inflammation associates with the development of dysbiosis (32). Several other diseases have also been shown to associate with imbalances in the composition of the gut microbiota, although whether dysbiosis contributes directly to the development of these diseases, or is a reflection of the altered physiological state of the host, is still debated (46–48). Bacteroidetes and Firmicutes are the predominant bacterial phyla colonizing the healthy human large intestine that both ferment dietary fiber, they are the main producer of butyrate which have been authenticated contribute to intestinal health. To investigate the potential role of the microbiota in the sepsis model, we examined the effects of CLP surgery in mice with their microbiota removed via antibiotics. Surprisingly, all the experimental mice died within 48 h, suggesting that the gut microbiota diversity is crucial for survival in the CLP-induced sepsis model. Accordinglymicrobiota deletion exacerbate the severity of CLP induced sepsis. To further investigate the role of the gut microbiota in the CLP-induced sepsis model, we transplanted the gut microbiota of WTandGpr109a−/−into each other and performed CLP surgery. This revealed increased survival in the Gpr109a−/− mice that received the WT microbiota compared to that in control mice, and there was no significant change after the WT mice received the microbiota from Gpr109a−/− mice. There were also changes in the expression of inflammatory markers and improved epithelium barrier integrity in the CLP-induced sepsis model. This is supported by prior work demonstrating that the gut microbiota from wild mice can be banked and maintained in a genetically similar laboratory mouse colony. Transfer of the more natural gut microbiota from the wild mice improved outcomes during viral infection and in tumorigenesis models (49). Another study also showed attenuation of colonic inflammation in Nlrp12−/− mice using fecal transplants (43).
In summary, our results indicate that the GPR109A receptor contributes to improved survival, altered inflammatory marker expression, and increased gut epithelium barrier integrity in a CLP-induced sepsis model. We have also demonstrated that GPR109A is involved in regulating the gut microbiota of mice, although the underlying mechanisms remain unclear. Despite this, our work demonstrates the important role that the microbiota and GPR109A play during sepsis and may lead to improved treatment and diagnosis for patients.
Author contributions
GC, LX, JL, and WW designed the experiments. GC, BH, and SF performed the experiments and analyzed data. BL did the H&E staining. XR, DH, LJ, YL, and BdL participated in the CLP surgery. BdL from Xie Lab at Guangdong Institute of Microbiology carried out 16S rRNA sequencing analysis. GC, LX, and WW collected the data and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Martin Sager (Zentrale Einrichtung für Tierforschung und Tierschutzaufgaben der Heinrich-Heine Universität Düsseldorf, Germany) for generously providing the Gpr109a−/− mice, Liang Zhao for assistance with the 16S rRNA sequencing.
Footnotes
Funding. This research was supported by funding from the National Nature Science Foundation of China (31572479, 31872442, 31702211, 31602020), Graduate Innovation Fund of Jilin University (2017168) and GDAS Project of Science and Technology Development (2018GDASCX-0806).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02079/full#supplementary-material
References
- 1.Deutschman CS, Tracey KJ. Sepsis: current dogma and new perspectives. Immunity (2014) 40:463–75. 10.1016/j.immuni.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. (2013) 13:862–74. 10.1038/nri3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leentjens J, Kox M, van der Hoeven JG, Netea MG, Pickkers P. Immunotherapy for the adjunctive treatment of sepsis: from immunosuppression to immunostimulation. Time for a paradigm change? Am J Respir Crit Care Med. (2013) 187:1287–93. 10.1164/rccm.201301-0036CP [DOI] [PubMed] [Google Scholar]
- 4.Vincent JL, Opal S M, Marshall JC, Tracey KJ. Sepsis definitions: time for change. Lancet (2013) 381:774–5. 10.1016/S0140-6736(12)61815-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrico CJ, Meakins JL, Marshall JC, Fry D, Maier RV. Multiple-organ-failure syndrome. Arch Surg. (1986) 121:196–208. [DOI] [PubMed] [Google Scholar]
- 6.Hassoun HT, Kone BC, Mercer DW, Moody FG, Weisbrodt NW, Moore FA. Post-injury multiple organ failure: the role of the gut. Shock (2001) 15:1–10. [DOI] [PubMed] [Google Scholar]
- 7.Clark JA, Coopersmith CM. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock (2007) 28:384–93. 10.1097/shk.0b013e31805569df [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark JA, Gan H, Samocha AJ, Fox AC, Buchman TG, Coopersmith CM. Enterocyte-specific epidermal growth factor prevents barrier dysfunction and improves mortality in murine peritonitis. Am J Physiol Gastrointest Liver Physiol. (2009) 297:G471–9. 10.1152/ajpgi.00012.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark JA, Clark AT, Hotchkiss RS, Buchman TG, Coopersmith CM. Epidermal growth factor treatment decreases mortality and is associated with improved gut integrity in sepsis. Shock (2008) 30:36–42. 10.1097/shk.0b013e31815D0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandenbroucke RE, Dejonckheere E, Van Hauwermeiren F, Lodens S, De Rycke R, Van Wonterghem E, et al. Matrix metalloproteinase 13 modulates intestinal epithelial barrier integrity in inflammatory diseases by activating TNF. EMBO Mol Med. (2013) 5:1000–16. 10.1002/emmm.201202100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soga T, Kamohara M, Takasaki J, Matsumoto S, Saito T, Ohishi T, Hiyama H, et al. Molecular identification of nicotinic acid receptor. Biochem Biophys Res Commun. (2003) 303:364–9. 10.1016/S0006-291X(03)00342-5 [DOI] [PubMed] [Google Scholar]
- 12.Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, et al. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. (2003) 9:352–5. 10.1038/nm824 [DOI] [PubMed] [Google Scholar]
- 13.Wise A, Foord SM, Fraser NJ, Barnes AA, Elshourbagy N, Eilert M, et al. Molecular identification of high and low affinity receptors for nicotinic acid. J Biol Chem. (2003) 278:9869–74. 10.1074/jbc.M210695200 [DOI] [PubMed] [Google Scholar]
- 14.Taggart AK, Kero J, Gan X, Cai TQ, Cheng K, Ippolito M, et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. (2005) 280:26649–52. 10.1074/jbc.C500213200 [DOI] [PubMed] [Google Scholar]
- 15.Lukasova M, Malaval C, Gille A, Kero J, Offermanns S. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor GPR109A expressed by immune cells. J Clin Invest. (2011) 121:1163–73. 10.1172/JCI41651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanders D, Graff EC, White BD, Judd RL. Niacin increases adiponectin and decreases adipose tissue inflammation in high fat diet-fed mice. PLoS ONE (2013) 8:e71285. 10.1371/journal.pone.0071285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambhir D, Ananth S, Veeranan-Karmegam R, Elangovan S, Hester S, Jennings E, et al. GPR109A as an anti-inflammatory receptor in retinal pigment epithelial cells and its relevance to diabetic retinopathy. Invest Ophthalmol Vis Sci. (2012) 53:2208–17. 10.1167/iovs.11-8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho KH, Kim HJ, Rodriguez-Iturbe B, Vaziri ND. Niacin ameliorates oxidative stress, inflammation, proteinuria, and hypertension in rats with chronic renal failure. Am J Physiol Renal Physiol. (2009) 297:F106–13. 10.1152/ajprenal.00126.2009 [DOI] [PubMed] [Google Scholar]
- 19.Kwon WY, Suh GJ, Kim KS, Kwak YH. Niacin attenuates lung inflammation and improves survival during sepsis by downregulating the nuclear factor-kappaB pathway. Crit Care Med. (2011) 39:328–34. 10.1097/CCM.0b013e3181feeae4 [DOI] [PubMed] [Google Scholar]
- 20.Macia L, Tan J, Vieira A T, Leach K, Stanley D, Luong S, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. (2015) 6:6734. 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- 21.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity (2014) 40:128–39. 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng W, Lu J, Li B, Lin W, Zhang Z, Wei X, et al. Effect of functional oligosaccharides and ordinary dietary fiber on intestinal microbiota diversity. Front Microbiol. (2017) 8:1750. 10.3389/fmicb.2017.01750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrario C, Statello R, Carnevali L, Mancabelli L, Milani C, Mangifesta M, et al. How to feed the mammalian gut microbiota: bacterial and metabolic modulation by dietary fibers. Front Microbiol. (2017) 8:1749. 10.3389/fmicb.2017.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell (2016) 167:1339–3.e21. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jobin C. GPR109a: the missing link between microbiome and good health? Immunity (2014) 40:8–10. 10.1016/j.immuni.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dejager L, Pinheiro I, Dejonckheere E, Libert C. Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol. (2011) 19:198–208. 10.1016/j.tim.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 27.Wen H. Sepsis induced by cecal ligation and puncture. Methods Mol Biol. (2013) 1031:117–24. 10.1007/978-1-62703-481-4_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell (2014) 159:514–29. 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- 29.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell (2016) 166:1231–46.e13. 10.1016/j.cell.2016.07.043 [DOI] [PubMed] [Google Scholar]
- 30.Jeong EM, Son YH, Choi Y, Kim JH, Lee JH, Cho SY, et al. Transglutaminase 2 is dispensable but required for the survival of mice in dextran sulfate sodium-induced colitis. Exp Mol Med. (2016) 48:e267. 10.1038/emm.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigoni R, Fontana E, Guglielmetti S, Fosso B, D'Erchia AM, Maina V, et al. Intestinal microbiota sustains inflammation and autoimmunity induced by hypomorphic RAG defects. J Exp Med. (2016) 213:355–75. 10.1084/jem.20151116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, et al. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut (2016) 65:225–37. 10.1136/gutjnl-2015-309333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Fu S, Feng W, Huang B, Xu S, Wang W, et al. AMP010014A09 in sus scrofa encodes an analog of g protein-coupled receptor 109a, which mediates the anti-inflammatory effects of beta-hydroxybutyric acid. Cell Physiol Biochem. (2017) 42:1420–30. 10.1159/000479206 [DOI] [PubMed] [Google Scholar]
- 34.West CE, Renz H, Jenmalm MC, Kozyrskyj AL, Allen KJ, Vuillermin P, et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. J Allergy Clin Immunol. (2015) 135:3–13. 10.1016/j.jaci.2014.11.012 [DOI] [PubMed] [Google Scholar]
- 35.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology (2011) 140:1720–28. 10.1053/j.gastro.2011.01.054 [DOI] [PubMed] [Google Scholar]
- 36.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. (2014) 14:141–53. 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- 37.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. (2009) 9:799–809. 10.1038/nri2653 [DOI] [PubMed] [Google Scholar]
- 38.Arrieta MC, Madsen K, Doyle J, Meddings J. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut (2009) 58:41–8. 10.1136/gut.2008.150888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyatt J, Vogelsang H, Hubl W, Waldhoer T, Lochs H. Intestinal permeability and the prediction of relapse in Crohn's disease. Lancet (1993) 341:1437–9. 10.1016/0140-6736(93)90882-H [DOI] [PubMed] [Google Scholar]
- 40.Chen G, Ran X, Li B, Li Y, He D, Huang B, et al. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBio Med. (2018) 30:317–25. 10.1016/j.ebiom.2018.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 42.Lankelma JM, van Vught LA, Belzer C, Schultz MJ, van der Poll T, de Vos WM, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. (2017) 43:59–68. 10.1007/s00134-016-4613-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Wilson JE, Koenigsknecht MJ, Chou WC, Montgomery SA, Truax AD, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol. (2017) 18:541–51. 10.1038/ni.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy U, Galvez EJC, Iljazovic A, Lesker TR, Blazejewski AJ, Pils MC, et al. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep. (2017) 21:994–1008. 10.1016/j.celrep.2017.09.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ojima M, Motooka D, Shimizu K, Gotoh K, Shintani A, Yoshiya K, et al. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci. (2016) 61:1628–34. 10.1007/s10620-015-4011-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. (2013) 13:321–35. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- 47.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. (2005) 102:11070–5. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe (2008) 3:213–23. 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell (2017) 171:1015–28.e13. 10.1016/j.cell.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.