Abstract
While long non-coding RNAs (lncRNAs) may play important roles in cellular function and biological process, we still know little about them. Growing evidences indicate that subcellular localization of lncRNAs may provide clues to their functionality. To facilitate researchers functionally characterize thousands of lncRNAs, we developed a database-driven application, lncSLdb, which stores and manages user-collected qualitative and quantitative subcellular localization information of lncRNAs from literature mining. The current release contains >11 000 transcripts from three species. Based on the accumulated region of lncRNAs, we classify transcripts into three basic localization types (nucleus, cytoplasm and nucleus/cytoplasm). In some conditions, the nucleus and cytoplasm types can be divided into three more accurate subtypes (chromosome, nucleoplasm and ribosome). Besides browsing and downloading data in lncSLdb, our system provides a set of comprehensive tools to search by gene symbols, genome coordinates or sequence similarity. We hope that lncSLdb will provide a convenient platform for researchers to investigate the functions and the molecular mechanisms of lncRNAs in the view of subcellular localization.
Introduction
Long non-coding RNAs (lncRNAs) are non-coding transcripts whose lengths are >200 nucleotides (1, 2). In recent years, with the development of biological technique, especially the broad application of high-throughput RNA sequencing (RNA-Seq) (3, 4), more and more novel lncRNAs have been identified and annotated in genomes (5–7). Growing evidences suggest that lncRNAs have important function in various aspects of cellular function and biological process (8–10). However, the function of most lncRNAs is still unclear (10).
Unlike mRNAs, which are transported to cytoplasm and translated into proteins on ribosomes, lncRNAs have little coding potential. Similar to proteins, the function of lncRNAs heavily depends on their subcellular localization (10, 11). The accumulated lncRNAs in nucleus may take part in the nuclear organization or regulate the gene expression before transcription (11, 12), whereas the accumulated lncRNAs in cytoplasm have important roles in the post-transcriptional regulation and post-translational modification (11, 12). For example, lncRNA Airn, accumulated in nucleus, is involved in silencing Igf2r by overlapping with its promoter (13); Neat1 is an essential component to form paraspeckles and related with the nuclear retention of structured or edited mRNAs (14). Cytoplasmic lncRNA NKILA can influence NF-κB activation via inhibiting IKK-induced IκBα phosphorylation (15); TUG1 and CTB-89H12.4 can regulate the PTEN expression by acting as the sponge regulators to complete the microRNA with PTEN transcripts (16).
Therefore, the subcellular localization of lncRNAs is a very important property to understand the function of lncRNAs. Nowadays, researchers have investigated the subcellular localization of a set of lncRNAs. There is a great need for integrated platforms to manage, search and analyse these data. Amaral et al. (17) published the lncRNAdb, which contains subcellular localization information of ∼80 lncRNAs gene. Zhang et al. (18) has developed a database, RNALocate, to collect the subcellular localization of all kinds of RNA, which contains >1700 lncRNAs genes from 10 different species. Mas Ponte et al. (19) publish the LncATLAS, which collects the subcellular localization of 7267 human lncRNAs genes in 15 cell lines and define the RCI (Relative concentration index) for measuring the localization types. However, these systems usually focus on the lncRNA genes instead of lncRNA transcripts and only cover a small fraction of available lncRNAs in different species. We also note that these systems only provide limited support for qualitative and/or quantitative experimental results, such as photos or expression levels in different cell compartments. More details are shown in Table 1.
Table 1.
Statistics comparison between lncSLdb and other lncRNA subcellular localization databases
| #LncRNA gene | #LncRNA transcript | #Localization entry | #Species | Data type | Data source | #Paper | |
|---|---|---|---|---|---|---|---|
| lncSLdb | 9494a | 11 698b | 14 973 | 7 | Figure, expression ratio, description | Text Mining | 99 |
| lncATLAS | 7267 | - | 30 580 | 1 | Expression ratio | ENCODE | 1 |
| RNALocate | 1792 | - | 2383 | 10 | Description | Text Mining | 192 |
| lncRNAdb | ∼80 | - | 91 | ∼2 | Description | Text Mining | - |
a5581 with annotation, 3913 without annotation
b5356 with official names, 6342 without official names
#refers to “The number of”
We develop an lncRNA subcellular localization system (lncSLdb), which collects qualitative and quantitative subcellular localization information of lncRNAs by manually curating the literatures. The current release contains subcellular location information of >11 000 lncRNA transcripts from 9494 genes and three main species (human, mouse and fruit fly), classified into three basic subcellular localization types (nucleus, cytoplasm and nucleus/cytoplasm) and three subtypes (ribosome, chromosome and nucleoplasm), all of which are supported by biological experiments. Our aim is to provide a comprehensive platform to help researchers investigate the subcellular localization of lncRNAs and further for function and potential molecular mechanism. lncSLdb collects a set of information of lncRNAs, including gene IDs/symbols, transcript IDs, genome coordinates, gene/transcript biotype, subcellular localization and relative expression ratio or experimental pictures. The data set used by our system can be downloaded freely. Furthermore, researchers can submit new subcellular localization of lncRNAs to lncSLdb.
Data collection and implementation
We searched published papers in the PubMed Central (PMC) database by using ‘long non coding RNA subcellular localization’ and ‘lncRNA subcellular localization’ as keywords, which leads to >3000 papers. All papers are filtered manually to find if they are related to lncRNA subcellular localization. Papers that are not included in the result set but cited by some paper in the result set are also considered. The current release includes ∼100 papers, filtered from the first 1000 search results and their reference (Figure 1). We also collected the gene/transcript genome information from other database such as FlyBase (20), Ensembl (21), UCSC (22), MGD (23), GenBank (24) and Gencode (25).
Figure 1.
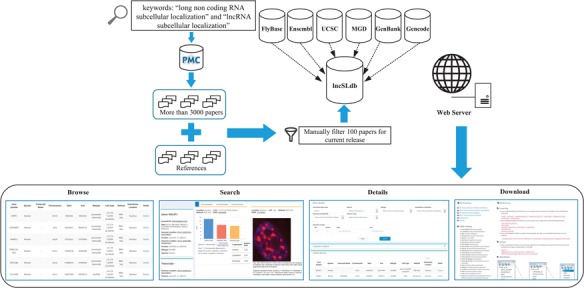
Workflow and construction process of lncSLdb.
lncSLdb is developed with HTML/JSP and Java languages using MySQL (http://www.mysql.com/) as the database manage system. The web interface is based on the Bootstrap (http://getbootstrap.com/2.3.2/) and AdminLTE (https://www.almsaeedstudio.com/) frameworks, and JavaScript scripts developed to support user interaction.
Database structure and content
For every localization item in lncSLdb, we consider three aspects, including transcript information, gene information and subcellular localization information. All information contained in lncSLdb are listed in the Table 2.
Table 2.
The information collected in lncSLdb
| Transcript information | |
|---|---|
| Transcript ID | The transcript id of the transcript |
| Chromosome | The chromosome of the transcript |
| Start | The transcript start position of the transcript |
| End | The transcript end position of the transcript |
| Strand | The strand of the transcript |
| Biotype | The biotype of the transcript |
| Sequence source | The source of transcript sequences |
| Gene information | |
| Gene symbol | The official symbol of the gene |
| ensembl id | The ensembl id of the gene |
| alias | The alias of the gene |
| chromosome | The chromosome of the gene |
| start | The transcript start position of the gene |
| end | The transcript start position of the gene |
| strand | The strand of the gene |
| biotype | The biotype of the gene |
| species | The species of the gene |
| version | The reference version of the genomic information |
| Subcellular Localization Information | |
| cell | The cell line or tissue used for experiments |
| method | The method used for experiments |
| localization | The subcellular localization of the transcript in this experiment |
| pmid | The pmid of this experiment |
| title | The article title of this experiment |
| source | The qualitative or quantitative results of this experiment |
Transcript information records the basic information of transcripts, including transcript ID, genomic coordinates and biotype. Since novel lncRNAs are being identified daily, many of these transcripts may still have no official names. We add the genomic coordinates, including transcript start site position, transcript end site position, chromosome and strand, as an identifier for every transcript. We fetch the genomic coordinates from Ensembl (21), UCSC (22), MGD (23), GenBank (24) and FlyBase (20), according to their transcript IDs. For transcripts without official IDs, we use the genomic coordinates described in corresponding articles. GRCh37 and GRCh38 are used as the reference genome for human, while GRCm38 for mouse and BDGP6 for fruit fly, respectively. We also get the transcript biotype from Ensembl database for those with Ensembl IDs. For the transcripts with accession number in GenBank, we use FEELnc (26), a tool for lncRNA annotation, to classify transcript into different biotype by comparing the genome location of transcripts with that of Gencode (25) transcripts. The biotype of other transcripts is obtained based on the description in corresponding papers or marked as ‘lncRNA’ if no description.
Gene information consists of gene symbol, Ensembl ID, alias and genomic coordinates and gene biotype. Since an lncRNA gene may have plenty of isoforms, which may have different subcellular localization types, we gather all transcripts belonging to the same gene to show its localization type. For intronic lncRNAs, information of host genes is used as gene information. In order to avoid the mismatch due to alias names, we convert all names to Ensembl ID and get gene symbol from Ensembl database. All other names are thought to be alias. For genes that cannot be found in Ensembl database, the Ensembl ID field will be unknown, while the known gene names are used as gene symbol. For some transcripts that do not belong to any genes, the genes are marked as unknown.
Subcellular localization information collects the experiment condition and the results, which mainly contain the cell line or tissue used, experiment method, experiment conclusion and specific experiment results. lncRNA subcellular localization is typically obtained from two types of experiments: one is based on in situ hybridization, for example ISH (27) and RNA-FISH (fluorescence in situ hybridization) (28, 29). The other combines nuclear-cytoplasm fraction with an expression assay using either microarrays (30) or RNA-Seq technologies (31). The first-type method will produce images showing subcellular localization of a certain lncRNA, while the second method will provide specific expression levels in different cellular compartments. In lncSLdb, we show the photos of in situ hybridization methods collected from papers or public databases, like Fly-Fish (32). For sequence results, we show bar plots about the expression level in different cell compartments and compute the relative ratio for every compartment with following formula:
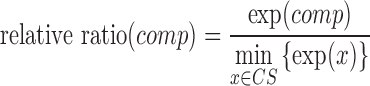 |
where 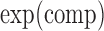 is the transcript expression in the chosen cellular compartment (comp),
is the transcript expression in the chosen cellular compartment (comp), 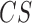 is the cellular compartment set of corresponding experiments,
is the cellular compartment set of corresponding experiments, 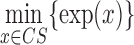 is the minimal expression value in all cell compartments. For example, for transcript ENST00000400436 in Clark et al. (31), the experiment separates cells into two compartments, nucleus and cytoplasm, of which we can compute the relative ratio. Here,
is the minimal expression value in all cell compartments. For example, for transcript ENST00000400436 in Clark et al. (31), the experiment separates cells into two compartments, nucleus and cytoplasm, of which we can compute the relative ratio. Here,  and the relative ratio in nucleus and in cytoplasm respectively is
and the relative ratio in nucleus and in cytoplasm respectively is
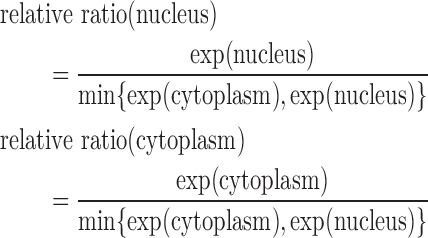 |
We think there are three basic types of subcellular localization in a cell, accumulated in nucleus, accumulated in cytoplasm and accumulated in both (nucleus/cytoplasm). In some condition, where the location region is more accurate, our system includes the most specific sub regions in nucleus or cytoplasm. According to the data we collect, we indicate that some lncRNAs are accumulated in chromosome or nucleoplasm in nucleus and some lncRNAs are accumulated in ribosome in cytoplasm. The type of the lncRNA subcellular localization is fetched directly from the papers. If authors did not state the type explicitly, we provide the reference types by considering the transcripts are nuclear accumulated if the nuclear expression level is more than 2-fold of the cytoplasm expression and cytoplasm accumulated if cytoplasm expression level is >2-fold of the nuclear expression and accumulated in both in other situations, similar with the definition in (30).
The current release contains >11 000 transcripts from ∼100 papers, mainly involving three species. Specifically, there are 9003 transcripts for human, 2630 for mouse, 59 for fruit fly and 6 for other species. In total, we collect >14 000 subcellular localization information. The distribution of localization types is shown in Figure 2.
Figure 2.
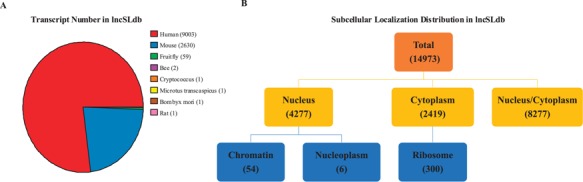
Statistics of species and types in lncSLdb. (A) The number of lncRNA transcript of different species in lncSLdb. lncSLdb collects more than 11 000 lncRNAs, mainly from three species (human,mouse and fruit fly). (B) The distribution of subcellular localization types in lncSLdb. lncSLdb collects >14 000 subcellular localization items, classified into three basic types and three subtypes.
Querying the database
lncSLdb is available online at: http://bioinformatics.xidian.edu.cn/lncSLdb. Users can browse, query and download data through the web interface.
In the browse page, all items are listed, which can be filtered by certain subtypes, including species, localization and transcript biotype. Every item has a detail page about the transcripts and localization, including transcript ID, transcript genome coordinates, subtype, method and cell used for experiment, reference article and its PMID, localization conclusion and the specific result. Transcripts belonging to the same gene are listed in the same detail page, where the gene information is shown in the beginning.
In the search page, we provide a comprehensive query tool. Users can query the lncRNA localization by using the gene name or transcript name as the keywords, selecting the specific species, biotype and subcellular localization type. We also offer a tool to search transcripts in a genome region in order to find novel transcripts without official names. In addition, there is a tool for searching the location type of homologous transcript via supplying the sequences in the fasta format.
All data can be downloaded from the download page with txt format or Microsoft Excel format. We also open the SQL interface to allow users to develop their program to access our database.
Researchers can submit new subcellular localization to lncSLdb online. More details can be found on the submission and help page.
Discussion and future prospects
Increasing evidence has proven that lncRNAs play important roles in cell activities. But we still have little knowledge about their basic properties, such as the subcellular localization. The study in the protein subcellular localization helps researchers understand the function of protein. We hope the effort in lncRNA subcellular localization can provide another view to explain their function and biogenesis (11). Although some researchers have developed some databases containing lncRNA subcellular localization (17–19), they only cover a small fraction of available lncRNAs in different species. Here, we developed lncSLdb, an lncRNA subcellular localization database, collecting the qualitative and quantitative localization information of >10 000 of lncRNAs subcellular localization information from published articles from three species, classified into three basic subcellular localization types and three subtypes. To our knowledge, this is the most complete database for lncRNA subcellular localization up to now. We hope that lncSLdb can provide researchers an integrated platform for studying the basic property and subcellular localization of lncRNAs, and further for figuring out if lncRNAs share the same or similar exportation mechanism with mRNAs and other potential molecular roles. We are interested in mining the features of transcripts in different cellular compartments and predicting the distribution of lncRNAs in different cell compartments. We will continue to update an improve lncSLdb in the future.
Acknowledgements
We thank Xiaofei Yang for the help of development of lncSLdb. We thank Peizhuo Wang and Ran Duan for discussion about the design of web server. We are grateful to Hao Lin and Quan Zou for suggestion about the manuscript.
Funding
National Natural Science Foundation of China (61532014, 61672407, 61432010 and 91530113). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest. None declared.
Database URL: http://bioinformatics.xidian.edu.cn/lncSLdb
References
- 1. Kapranov P., Cheng J., Dike S. et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- 2. Mattick J.S. and Rinn J.L. (2015) Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol., 22, 5–7. [DOI] [PubMed] [Google Scholar]
- 3. Mortazavi A., Williams B.A., McCue K. et al. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- 4. Wang Z., Gerstein M. and Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet., 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pauli A., Valen E., Lin M.F. et al. (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res., 22, 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iyer M.K., Niknafs Y.S., Malik R. et al. (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet., 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. White N.M., Cabanski C.R., Silva-Fisher J.M. et al. (2014) Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol., 15, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mercer T.R., Dinger M.E. and Mattick J.S. (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet, 10, 155–159. [DOI] [PubMed] [Google Scholar]
- 9. Zhang B., Gunawardane L., Niazi F. et al. (2014) A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA. Mol. Cell. Biol., 34, 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cabili M.N., Dunagin M.C., McClanahan P.D. et al. (2015) Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol., 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L.L. (2016) Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci., 41, 761–772. [DOI] [PubMed] [Google Scholar]
- 12. Batista P.J. and Chang H.Y. (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell, 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Latos P.A., Pauler F.M., Koerner M.V. et al. (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science, 338, 1469–1472. [DOI] [PubMed] [Google Scholar]
- 14. Chen L.L. and Carmichael G.G. (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell., 35, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu B., Sun L., Liu Q. et al. (2015) A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell., 27, 370–381. [DOI] [PubMed] [Google Scholar]
- 16. Du Z., Sun T., Hacisuleyman E. et al. (2016) Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun., 7, 10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amaral P.P., Clark M.B., Gascoigne D.K. et al. (2011) lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res., 39, D146–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang T., Tan P., Wang L. et al. (2017) RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Res., 45, D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mas-Ponte D., Carlevaro-Fita J., Palumbo E. et al. (2017) LncATLAS database for subcellular localization of long noncoding RNAs. RNA, 23, 1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tweedie S., Ashburner M., Falls K. et al. (2009) FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res., 37, D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yates A., Akanni W., Amode M.R. et al. (2016) Ensembl 2016. Nucleic Acids Res., 44, D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tyner C., Barber G.P., Casper J. et al. (2017) The UCSC Genome Browser database: 2017 update. Nucleic Acids Res., 45, D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blake J.A., Eppig J.T., Kadin J.A. et al. (2017) Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res., 45, D723–D729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benson D.A., Cavanaugh M., Clark K. et al. (2017) GenBank. Nucleic Acids Res, 45, D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrow J., Frankish A., Gonzalez J.M. et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res., 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wucher V., Legeai F., Hedan B. et al. (2017) FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res., 45, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mercer T.R., Dinger M.E., Sunkin S.M. et al. (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A., 105, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raj A., Bogaard P., Rifkin S.A. et al. (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods, 5, 877–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raj A., Rifkin S.A., Andersen E. et al. (2010) Variability in gene expression underlies incomplete penetrance. Nature, 463, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clark M.B., Johnston R.L., Inostroza-Ponta M. et al. (2012) Genome-wide analysis of long noncoding RNA stability. Genome Res., 22, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Derrien T., Johnson R., Bussotti G. et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res., 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilk R., Hu J., Blotsky D. et al. (2016) Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev., 30, 594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]


