Abstract
Introduction
Since 2005 the S7B and E14 guidances from ICH and FDA have been in place to assess a potential drug candidate’s ability to cause long QT syndrome. To refine these guidelines, the FDA proposed the Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative, where the assessment of drug effects on cardiac repolarization was one subject of investigation. Within the myocyte validation study, effects of pharmaceutical compounds on human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) were assessed and this article will focus on the evaluation of the proarrhythmic potential of 23 blinded drugs in four hiPSC-CM cell lines.
Methods
Experiments were performed on the CardioExcyte 96 at different sites. A combined readout of contractility (via impedance) and electrophysiology endpoints (field potentials) was performed.
Results
Our data demonstrates that hERG blockers such as dofetilide and further high risk categorized compounds prolong the field potential duration. Arrhythmia were detected in both impedance as well as field potential recordings. Intermediate risk compounds induced arrhythmia in almost all cases at the highest dose. In the case of low risk compounds, either a decrease in FPDmax was observed, or not a significant change from pre-addition control values.
Discussion
With exceptions, hiPSC-CMs are sensitive and exhibit at least 10% delayed or shortened repolarization from pre-addition values and arrhythmia after drug application and thus can provide predictive cardiac electrophysiology data. The baseline electrophysiological parameters vary between iPS cells from different sources, therefore positive and negative control recordings are recommended.
Keywords: Arrhythmia, EAD, CardioExcyte 96, Cardiomyocytes, CiPA, Contractility, Extracellular Field Potential, hiPSC, Impedance, Safety Pharmacology
1. Introduction
In order to revise existing cardiac safety guidelines S7B and E14, human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) were investigated in a blinded pilot (phase I) and validation (phase II) study as part of the Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative. The CiPA initiative represents an international collaborative effort to reevaluate safety assessment procedures (Colatsky et al., 2016; Stockbridge, Morganroth, Shah, & Garnett, 2013). Results are being used to confirm in silico reconstructions of the electrophysiological effects of drugs within a parallel CiPA study. One objective of the myocyte validation study was to discover proarrhythmic mechanisms that the ion channels and in silico assays did not discover, such as trafficking or disruption of ion channel function. Human iPSC-CMs are currently being evaluated as an alternative to acutely isolated cardiomyocytes due to their recapitulation of native behavior, long-term stability in culture and routine availability. Additionally, they are ready-to-use on high throughput screening (HTS) platforms and have been studied successfully on automated patch clamp systems (Becker et al., 2013; Friedrichs, Malan, Voss, & Sasse, 2015; Goversen et al., 2018; Obergrussberger et al., 2015; Obergrussberger, Brüggemann, et al., 2016; Obergrussberger, Haarmann, et al., 2017; Obergrussberger et al., 2018; Scheel et al., 2014; Stoelzle, Haythornthwaite, et al., 2011; Stoelzle, Obergrussberger, et al., 2011), MEA platforms (Clements, Millar, Williams, & Kalinka, 2015; Clements & Thomas, 2014; Gilchrist, Lewis, Gay, Sellgren, & Grego, 2015; Guo et al., 2011; Navarrete et al., 2013) and impedance devices (Abassi et al., 2012; Clements & Thomas, 2014; Doerr et al., 2015; Guo et al., 2011; Jonsson, Wang, & Becker, 2011; Kho et al., 2015; Lamore, Scott, & Peters, 2015; Nguemo, Šaric, Pfannkuche, Reppel, & Hescheler, 2012; Obergrussberger, Juhasz, et al., 2016; Obergrussberger, Thomas, et al., 2017; Peters, Lamore, Guo, Scott, & Kolaja, 2015; Xi et al., 2011), amongst others. Within the CiPA myocyte pilot study, coordinated by the Health and Environmental Sciences Institute (HESI), MEA systems and optical measurement systems have been used to evaluate monolayers of a number of different commercially available hiPS-CMs. Changes in the extracellular field potentials of spontaneously active hiPS-CMs using microelectrode array (MEA) readouts (Millard et al., 2018), or changes in the action potential configuration recorded optically using voltage-sensitive dyes (VSD) or integral membrane proteins were investigated in the presence of 8 (Millard et al., 2018) or more different reference drugs (Blinova et al., 2016; Kanda, Yamazaki, Kurokawa, Inutsuka, & Sekino, 2016; Zhang et al., 2016). Understanding the necessity to investigate this further, the FDA awarded a Grant to the HESI Myocyte Subteam to extend the pilot study to a larger set of 28 compounds. Based on clinical findings the compounds are categorized according to high, intermediate, and low/no risk of proarrhythmia. Commercially available hiPSC-CMs were utilized primarily as monolayers on MEA, VSD technology, impedance or other platforms. Data generated on novel instruments, such as described in this manuscript, were part of the validation study. It is expected that such an extended data set of beating networks of hiPSC-CMs, in combination with innovative technologies, would reveal potential gaps in cellular electrophysiological effects on single ion channels heterologously expressed, and thus may impact future decisions of TdP risk assessment regulations. In addition, technologies allowing multi-parametric readouts as deployed in the myocyte validation study, and as described here, allow comprehensive and robust assessments in the drug discovery process. This article will focus on an extensive evaluation of the proarrhythmic potential of 23 blinded drugs on 4 commercially available hiPSC-CM cell lines as investigated at 4 different sites. Experimental readout combined endpoints defining the electrophysiologic behavior as well as contractile behavior of the cells, via extracellular field potential and impedance readout, respectively. The goal of this study was to evaluate the consistency of results from hiPSC-CMs using a standardized protocol across multiple myocyte types and experimental sites, by using the same experimental platform, the CardioExcyte 96 (Nanion Technologies GmbH). Multi-parameter datasets encompassing electrophysiology, contractility and viability of beating networks of monolayers of cells were collected, and a cross-site and cross-cell comparison was performed.
2. Methods
2.1 Sensor plate preparation and cell seeding
Four different types of hiPSC-CMs were used for this study: CDI (iCell Cardiomyocytes2, Cellular Dynamics International), AXG and PLR (Cor.4U® and Pluricyte® Cardiomyocytes, Ncardia) and CLS (Cellartis® Cardiomyocytes, Takara Bio Clontech). The four sites consisted of four independent labs, located at Roche in Switzerland, Novartis in Switzerland, Nanion Technologies Inc. in the USA and Nanion GmbH in Germany.
Cells were stored in liquid nitrogen until thawed and cultured according to manufacturer instructions or were received pre-plated on the CardioExcyte 96 sensor plates. Two different types of CardioExcyte 96 sensor plates (NSP-96) were used among participating sites, either a 0.6 mm or a 2 mm central (recording) electrode. No significant difference was observed in the statistical assessment of field potential duration, impedance/EFP amplitude or beat rate when comparing baseline data from the two electrode types (Suppl. Fig 2). All cell types were plated and maintained according to cell supplier recommendations, including extracellular matrix coating procedures, cell density, media composition and volume per well, and time in culture required to mature cells prior to compound application. Briefly, the wells to be used were incubated in 10 μg/ml fibronectin (Sigma Aldrich, St. Louis, MO, Product: FIBRP-RO 1mg solved in 1ml PBS) in PBS (including Ca2+ and Mg2+; Thermofisher/Gibco 14040 – 083 DPBS) for 2 hours at 37°C or overnight at 4°C. The fibronectin was removed shortly before cell seeding. After the cells were seeded, the NSP-96 was left in the sterile cabinet for 30 min without moving it, to allow the cells to settle evenly. Medium exchange was performed according to cell supplier recommendations.
2.2 CardioExcyte 96 monitoring of hiPSC–CM attachment, viability, contractility and extracellular field potential
The CardioExcyte 96 uses impedance measurements to study pharmacological effects on contractility and coordinated ion channel activity through extracellular field potential (EFP) recordings (Doerr et al., 2015; Obergrussberger, Juhasz, et al., 2016; Obergrussberger, Thomas, et al., 2017). All hiPSC-CMs used in this study were monitored during spontaneous beating. Cell attachment, viability, contractility and field potential durations were recorded in combination using the CardioExcyte Control software (Nanion Technologies GmbH). Prior to compound application, all hiPSC-CM cells formed syncytial monolayers suitable for electrophysiological and phenotypical assessments via extracellular field potential and impedance. Impedance has been used in the past as an indirect measure of contractility (Zhang et al., 2016). An indirect measure of contractility can be assessed by looking at impedance amplitude, in correlation with time-to-peak, upstroke or downstroke velocity. For this manuscript, data was assessed for impedance amplitude only. A typical example of cell monolayer formation in a recording well is shown in Fig. 1A. A key parameter for estimation of cell viability and monolayer formation over time is the base impedance (or the absolute impedance recorded from each monolayer). A representative base impedance recording over 5 days from CDI cells is depicted in Fig. 1B. An exemplary long term chronic effect of nifedipine on the cell viability is shown in Fig. 1C. Nifedipine is a Ca2+ channel blocker with negative inotropic and chronotropic effects which can be observed as acute effects which was one goal of this work (see nifedipine effects in table 3), but also has toxic effects which impacts the cells’ viability. The base impedance is altered by adhesion, spreading and proliferation of cells, a decrease of this parameter being a surrogate for reduced viability.
Figure 1.
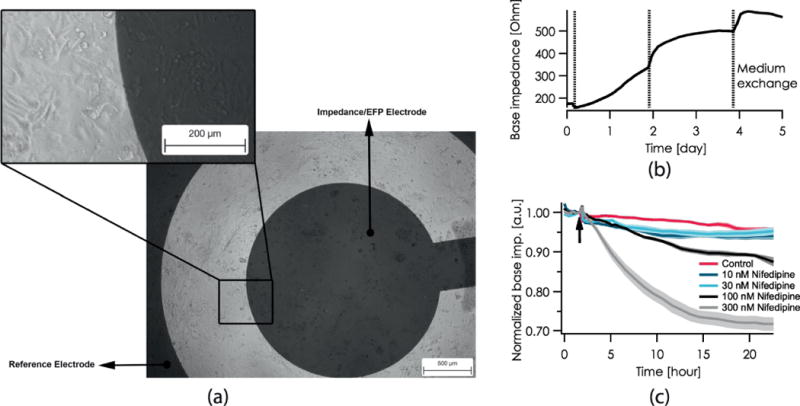
A, Phase contrast images of AXG cardiomyocytes cultured for 21 days on a CardioExcyte 96 NSP-96 transparent sensor plate. B, Representative base impedance recording over 5 days from CDI cells. C, Chronic effect of nifedipine as recorded over 24 h on AXG cardiomyocytes. A time and concentration-dependent reduction of cell viability was observed. The arrow marks the timepoint of compound addition; a.u. stands for arbitrary unit.
Table 3.
Summary of % FPDmax changes at 30 min post compound addition for all compounds, all cell types and across all participating sites.
| Test article concentrations (μM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | Site/Cell type | Trend | ETPC-Free (uM) | Dose 1 | Dose 2 |
type |
Dose 3 |
type |
Dose 4 |
type |
| HIGH RISK | ||||||||||
| Disopyramide | Site 1, CDI |  |
0.7 | 0.1 | 1 | – | 10 | † | 100 | †† |
| Disopyramide | Site 2, CDI |  |
0.7 | 0.1 | 1 | – | 10 | † | 100 | †† |
| Disopyramide | Site 3, CDI |  |
0.7 | 0.1 | 1 | – | 10 | † | 100 | †† |
| Disopyramide | Site 1, AXG |  |
0.7 | 0.1 | 1 | – | 10 | † | 100 | †† |
| Disopyramide | Site 2, AXG |  |
0.7 | 0.1 | 1 | – | 10 | – | 100 | † |
| Disopyramide | Site 4, AXG | 0.7 | 0.1 | 1 | – | 10 | – | not tested | ||
| Disopyramide | Site 2, CLS | 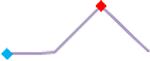 |
0.7 | 0.1 | 1 | – | 10 | – | 100 | †† |
| Disopyramide | Site 4, CLS |  |
0.7 | 0.1 | 1 | – | 10 | † | not tested | |
| Disopyramide | Site 2, PLR | 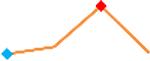 |
0.7 | 0.1 | 1 | – | 10 | † | 100 | †† |
| Quinidine | Site 1, CDI |  |
3 | 0.95 | 3 | †† | 9.49 | †† | 30 | quiescence |
| Quinidine | Site 2, CDI | 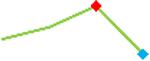 |
3 | 0.95 | 3 | † | 9.49 | † | 30 | † |
| Quinidine | Site 3, CDI | 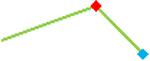 |
3 | 0.95 | 3 | †† | 9.49 | †† | 30 | quiescence |
| Quinidine | Site 1, AXG | 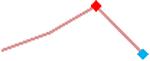 |
3 | 0.95 | 3 | † | 9.49 | †† | 30 | quiescence |
| Quinidine | Site 2, AXG |  |
3 | 0.95 | 3 | – | 9.49 | † | 30 | † |
| Quinidine | Site 4, AXG |  |
3 | 0.95 | 3 | – | 9.49 | † | 30 | †† |
| Quinidine | Site 2, CLS | 3 | 0.95 | 3 | – | 9.49 | quiescence | 30 | quiescence | |
| Quinidine | Site 4, CLS |  |
3 | 0.95 | 3 | †† | 9.49 | quiescence | 30 | quiescence |
| Quinidine | Site 2, PLR | 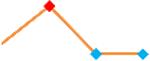 |
3 | 0.95 | 3 | † | 9.49 | † | 30 | † |
| D,I Sotalol | Site 1, CDI |  |
15 | 0.1 | 1 | – | 10 | †† | 100 | †† |
| D,l Sotalol | Site 2, CDI |  |
15 | 0.1 | 1 | – | 10 | †† | 100 | †† |
| D,l Sotalol | Site 3, CDI |  |
15 | 0.1 | 1 | – | 10 | †† | 100 | †† |
| D,l Sotalol | Site 1, AXG |  |
15 | 0.1 | 1 | – | 10 | – | 100 | †† |
| D,l Sotalol | Site 2, AXG |  |
15 | 0.1 | 1 | – | 10 | – | 100 | †† |
| D,l Sotalol | Site 4, AXG | 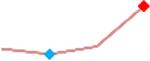 |
15 | 0.1 | 1 | – | 10 | – | 100 | †† |
| D,l Sotalol | Site 2, CLS | 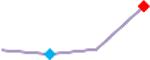 |
15 | 0.1 | 1 | – | 10 | – | 100 | †† |
| D,l Sotalol | Site 4, CLS | 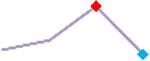 |
15 | 0.1 | 1 | – | 10 | – | 100 | †† |
| D,l Sotalol | Site 2, PLR |  |
15 | 0.1 | 1 | – | 10 | †† | 100 | †† |
| Vandetanib | Site 1, CDI | 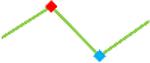 |
0.3 | 0.01 | 0.1 | – | 1.00 | † | 10 | †† |
| Vandetanib | Site 2, CDI | 0.3 | 0.01 | 0.1 | 1.00 | – | 10 | quiescence | ||
| Vandetanib | Site 3, CDI | 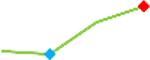 |
0.3 | 0.01 | 0.1 | – | 1.00 | † | 10 | †† |
| Vandetanib | Site 1, AXG | 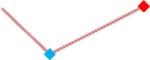 |
0.3 | 0.01 | 0.1 | – | 1.00 | – | 10 | †† |
| Vandetanib | Site 2, AXG |  |
0.3 | 0.01 | 0.1 | – | 1.00 | – | 10 | †† |
| Vandetanib | Site 4, AXG |  |
0.3 | 0.01 | 0.1 | – | 1.00 | – | 10 | †† |
| Vandetanib | Site 2, CLS |  |
0.3 | 0.01 | 0.1 | – | 1.00 | – | 10 | †† |
| Vandetanib | Site 4, CLS | 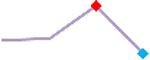 |
0.3 | 0.01 | 0.1 | – | 1.00 | – | 10 | †† |
| Vandetanib | Site 2, PLR |  |
0.3 | 0.01 | 0.1 | – | 1.00 | † | 10 | † |
| INTERMEDIATE RISK: | ||||||||||
| Astemizole | Site 1, CDI |  |
0.0003 | 0.0001 | 0.001 | – | 0.01 | †† | 0.1 | †† |
| Astemizole | Site 3, CDI | 0.0003 | 0.0001 | 0.001 | – | 0.01 | † | 0.1 | †† | |
| Astemizole | Site 1, AXG |  |
0.0003 | 0.0001 | 0.001 | – | 0.01 | †† | 0.1 | †† |
| Chlorpromazine | Site 1, CDI | 0.03 | 0.10 | 0.30 | – | 0.95 | – | 3.00 | – | |
| Chlorpromazine | Site 1, AXG | 0.03 | 0.10 | 0.30 | – | 0.95 | – | 3.00 | – | |
| Cisapride | Site 1, CDI |  |
0.003 | 0.003 | 0.01 | – | 0.03 | † | 0.1 | †† |
| Cisapride | Site 1, AXG | 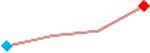 |
0.003 | 0.003 | 0.01 | – | 0.03 | †† | 0.1 | †† |
| Clarithromycin | Site 1, CDI | 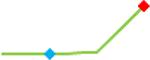 |
1.21 | 0.1 | 1 | – | 10 | †† | 100 | †† |
| Clarithromycin | Site 1, AXG |  |
1.21 | 0.1 | 1 | – | 10 | †† | 100 | †† |
| Domperidone | Site 2, CDI | 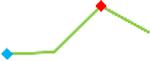 |
0.02 | 0.003 | 0.03 | – | 0.3 | †† | 3 | †† |
| Domperidone | Site 2, AXG |  |
0.02 | 0.003 | 0.03 | – | 0.3 | †† | 3 | †† |
| Domperidone | Site 2, CLS |  |
0.02 | 0.003 | 0.03 | – | 0.3 | †† | 3 | †† |
| Domperidone | Site 2, PLR |  |
0.02 | 0.003 | 0.03 | – | 0.3 | †† | 3 | †† |
| Droperidol | Site 2, CDI |  |
0.02 | 0.03 | 0.1 | – | 0.32 | † | 1 | † |
| Droperidol | Site 2, AXG |  |
0.02 | 0.03 | 0.1 | – | 0.32 | – | 1 | †† |
| Droperidol | Site 2, CLS | 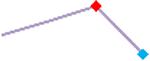 |
0.02 | 0.03 | 0.1 | – | 0.32 | † | 1 | †† |
| Droperidol | Site 2, PLR |  |
0.02 | 0.03 | 0.1 | – | 0.32 | †† | 1 | †† |
| Ondansetron | Site 3, CDI |  |
0.37 | 0.03 | 0.3 | – | 3.00 | † | 30.00 | † |
| Ondansetron | Site 4, AXG | 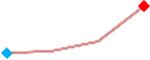 |
0.37 | 0.03 | 0.3 | – | 3.00 | – | 30.00 | †† |
| Ondansetron | Site 4, CLS |  |
0.37 | 0.03 | 0.3 | – | 3.00 | †† | 30.00 | quiescence |
| Pimozide | Site 2, CDI | 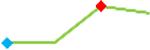 |
0.0004 | 0.001 | 0.003 | – | 0.01 | † | 0.03 | † |
| Pimozide | Site 3, CDI | 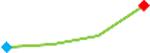 |
0.0004 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | † |
| Pimozide | Site 2, AXG | 0.0004 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | †† | |
| Pimozide | Site 4, AXG | 0.0004 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | – | |
| Pimozide | Site 2, CLS | 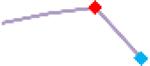 |
0.0004 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | †† |
| Pimozide | Site 4, CLS |  |
0.0004 | 0.001 | 0.003 | – | 0.01 | †† | 0.03 | †† |
| Pimozide | Site 2, PLR | 0.0004 | 0.001 | 0.003 | – | 0.01 | †† | 0.03 | †† | |
| Risperidone | Site 3, CDI | 0.002 | 0.003 | 0.01 | – | 0.03 | – | 0.10 | – | |
| Risperidone | Site 4, AXG | 0.002 | 0.003 | 0.01 | – | 0.03 | no sg ch | 0.10 | – | |
| Risperidone | Site 4, CLS | 0.002 | 0.003 | 0.01 | – | 0.03 | no sg ch | 0.10 | – | |
| Terfenadine | Site 2, CDI | 0.0003 | 0.001 | 0.01 | – | 0.1 | – | 1 | quiescence | |
| Terfenadine | Site 2, AXG | 0.0003 | 0.001 | 0.01 | – | 0.1 | – | 1 | quiescence | |
| Terfenadine | Site 4, AXG | 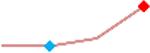 |
0.0003 | 0.001 | 0.01 | – | 0.1 | – | 1 | †† |
| Terfenadine | Site 2, CLS | 0.0003 | 0.001 | 0.01 | – | 0.1 | – | 1 | quiescence | |
| Terfenadine | Site 4, CLS | 0.0003 | 0.001 | 0.01 | – | 0.1 | – | 1 | †† | |
| Terfenadine | Site 2, PLR | 0.0003 | 0.001 | 0.01 | – | 0.1 | quiescence | 1 | quiescence | |
| LOW RISK | ||||||||||
| Diltiazem | Site 1, CDI | 0.13 | 0.01 | 0.1 | – | 1.00 | – | 10.00 | – | |
| Diltiazem | Site 1, AXG | 0.13 | 0.01 | 0.1 | – | 1.00 | quiescence | 10.00 | quiescence | |
| Diltiazem | Site 4, AXG | 0.13 | 0.01 | 0.1 | – | 1.00 | – | 10.00 | quiescence | |
| Diltiazem | Site 4, CLS | 0.13 | 0.01 | 0.1 | – | 1.00 | quiescence | 10.00 | quiescence | |
| Loratadine | Site 1, CDI | 0.0005 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | – | |
| Loratadine | Site 1, AXG | 0.0005 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | – | |
| Loratadine | Site 4, AXG | 0.0005 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | – | |
| Loratadine | Site 4, CLS | 0.0005 | 0.001 | 0.003 | – | 0.01 | – | 0.03 | – | |
| Metoprolol | Site 4, AXG | 1.80 | 3.17 | 10.01 | – | 31.65 | no sg ch | 100 | – | |
| Metoprolol | Site 4, CLS | 1.80 | 3.17 | 10.01 | – | 31.65 | – | 100 | †† | |
| Mexiletine | Site 3, CDI | 2.50 | 0.1 | 1 | – | 10 | no sg ch | 100 | – | |
| Mexiletine | Site 4, AXG | 2.50 | 0.1 | 1 | – | 10 | – | 100 | – | |
| Mexiletine | Site 4, CLS | 2.50 | 0.1 | 1 | – | 10 | – | 100 | †† | |
| Nifedipine | Site 2, CDI | 0.01 | 0.001 | 0.01 | – | 0.1 | – | 1 | – | |
| Nifedipine | Site 2, AXG | 0.01 | 0.001 | 0.01 | – | 0.1 | – | 1 | – | |
| Nifedipine | Site 2, CLS | 0.01 | 0.001 | 0.01 | – | 0.1 | quiescence | 1 | quiescence | |
| Nifedipine | Site 2, PLR | 0.01 | 0.001 | 0.01 | – | 0.1 | – | 1 | – | |
| Nitrendipine | Site 2, CDI | 0.003 | 0.01 | 0.03 | – | 0.09 | – | 0.3 | – | |
| Nitrendipine | Site 2, AXG | 0.003 | 0.01 | 0.03 | – | 0.09 | – | 0.3 | – | |
| Nitrendipine | Site 2, CLS | 0.003 | 0.01 | 0.03 | quiescence | 0.09 | quiescence | 0.3 | quiescence | |
| Nitrendipine | Site 2, PLR | 0.003 | 0.01 | 0.03 | – | 0.09 | – | 0.3 | – | |
| Ranolazine | Site 2, CDI | 1.95 | 0.1 | 1 | – | 10 | – | 100 | quiescence | |
| Ranolazine | Site 3, CDI | 1.95 | 0.1 | 1 | – | 10 | – | 100 | †† | |
| Ranolazine | Site 2, AXG | 1.95 | 0.1 | 1 | – | 10 | – | 100 | quiescence | |
| Ranolazine | Site 2, CLS | 1.95 | 0.1 | 1 | – | 10 | – | 100 | †† | |
| Ranolazine | Site 2, PLR | 1.95 | 0.1 | 1 | – | 10 | – | 100 | †† | |
| Tamoxifen | Site 1, CDI | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Tamoxifen | Site 2, CDI | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Tamoxifen | Site 3, CDI | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Tamoxifen | Site 1, AXG | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Tamoxifen | Site 2, AXG | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Tamoxifen | Site 2, CLS | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Tamoxifen | Site 2, PLR | 0.02 | 0.1 | 0.3 | – | 0.95 | – | 3 | – | |
| Verapamil | Site 1, CDI | 0.05 | 0.001 | 0.01 | – | 0.1 | – | 1 | quiescence | |
| Verapamil | Site 3, CDI | 0.05 | 0.001 | 0.01 | – | 0.1 | – | 1 | – | |
| Verapamil | Site 1, AXG | 0.05 | 0.001 | 0.01 | – | 0.1 | – | 1 | quiescence | |
“Trend” lines represent average data per compound, n= 4 or 5 replicates each, with min (blue dot) and max (red dot) values.
represents type A arrhythmia as specified in the HESI Myocyte Phase II Validation Study Protocol. All other types of/combinations of arrhythmia observed are marked with
(type B, C, D arrhythmias listed Myocyte Phase II Validation Study Protocol). Colour shades represent the first dose where FPDmax was > or <10% change (or >< 30%, in case compounds were more potent).
2.3 Chemical reagents and solutions
Stem cell culture media was cell type specific and was obtained from the stem cell providers. On the day of compound applications, culture media was completely removed from the wells and fresh media was added to ensure an exact volume per well. Cells were allowed to re-equilibrate for 2-3 hours, and online parameters were monitored to ensure a stable baseline. The blinded chemical reagents were provided by the Chemotherapeutic Agents Repository of the National Cancer Institute and consisted of a random subset of CiPA compounds, dedicated to this study. All sites received compounds from each risk category, high risk compounds were common across all sites, difference quantity resulted in 23 individual compounds in total. Chemical stock solutions at 1000-fold of the target concentrations were prepared under sterile conditions in DMSO and stored at −20°C, according to HESI Myocyte Phase II Validation Study Protocol instructions. The serial diluted chemicals were further prepared in DMSO immediately, prior to compound addition. The 2-fold or 10-fold final dilutions of the chemicals were prepared with culture medium, for single time use only. Cells were exposed to concentrations of drug, under sterile conditions in single point additions (i.e. one concentration per well) in five replicates for each dose, following the dilution scheme provided by the HESI Myocyte Phase II Validation Study Protocol. Vehicle control was 0.1 % DMSO, and different doses of dofetilide were used among participating sites as positive control (Fig. 3B). After application of compound, measurements of impedance and EFP were taken every 5 minutes for 10-30 s for a period of 2 hours. Site 2 used serum-free media for compound applications across all cell types tested; all other sites used serum-containing media. The effects on cardiomyocyte contractility and electrophysiology of 23 compounds ranked as low, medium and high risk were tested at 4 different sites on 4 different commercially available hiPSC-CM types. All compounds were part of the HESI myocyte validation study (Table 2). Cells were treated with test compounds 7 – 14 days after thawing, according to the recommendations from cell providers.
Figure 3. FPDmax values in vehicle and positive control across all sites and cell types.
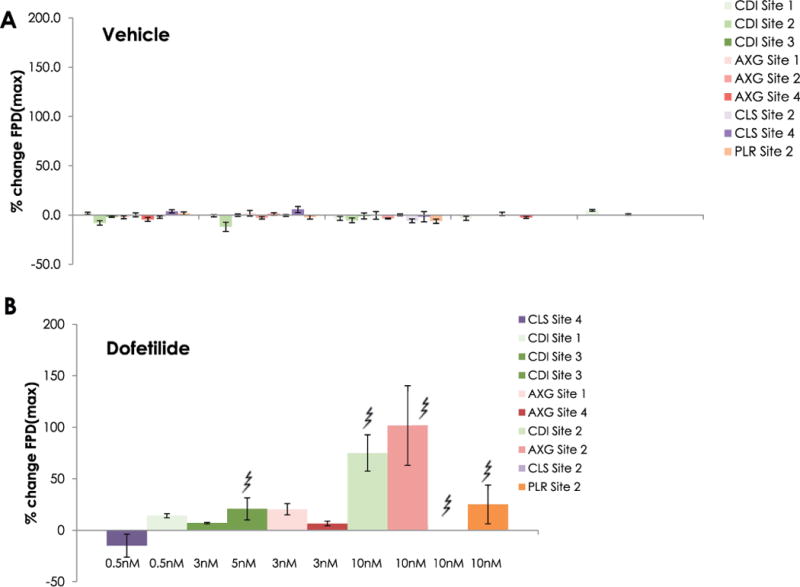
Percent change in FPDmax following administration of DMSO (A) and dofetilide (B) across all sites and cell types. Markers depict arrhythmic events.
Table 2.
All cell types and all compounds tested at each participating site.
| Compound | Cell Type | Site | |
| High Risk | Disopyramide | AXG, CDI, PLR, CLS | 1,2,3,4 |
| Quinidine | AXG, CDI, PLR, CLS | 1,2,3,4 | |
| D,l Sotalol | AXG, CDI, PLR, CLS | 1,2,3,4 | |
| Vandetanib | AXG, CDI, PLR, CLS | 1,2,3,4 | |
| Intermediate Risk | Astemizole | AXG, CDI | 1,3 |
| Chlorpromazine | AXG, CDI | 1 | |
| Cisapride | AXG, CDI | 1 | |
| Clarithromycin | AXG, CDI | 1 | |
| Droperidol | AXG, CDI, PLR, CLS | 2 | |
| Domperidone | AXG, CDI, PLR, CLS | 2 | |
| Ondansetron | AXG, CDI, CLS | 3,4 | |
| Pimozide | AXG, CDI, PLR, CLS | 2,3,4 | |
| Risperidone | AXG, CDI, CLS | 3,4 | |
| Terfenadine | AXG, CDI, PLR, CLS | 2,4 | |
| Low risk | Diltiazem | AXG, CDI, CLS | 1,4 |
| Loratadine | AXG, CDI, CLS | 1,4 | |
| Metoprolol | AXG, CLS | 4 | |
| Mexiletine | AXG, CDI, CLS | 3,4 | |
| Nifedipine | AXG, CDI, PLR, CLS | 2 | |
| Nitrendipine | AXG, CDI, PLR, CLS | 2 | |
| Ranolazine | AXG, CDI, PLR, CLS | 2,3 | |
| Tamoxifen | AXG, CDI, PLR, CLS | 1,2,3 | |
| Verapamil | AXG, CDI, CLS | 1,3 |
Compounds are grouped by risk of manifesting Human TdP, and are ranked as high, intermediate and low risk.
2.4 Data acquisition and analysis
Impedance data of spontaneous beating cells was recorded with a sampling rate of 1 ms (1 kHz), and EFP data were collected at 0.1 ms (10 kHz). Data acquisition was controlled by CardioExcyte 96 Control software which operates the CardioExcyte 96 hardware. CardioExcyte 96 Control software is used to define the experimental setup such as timing of sweeps, sweep interval and online analysis parameters, for details see Doerr et al., 2015; Obergrussberger, Juhasz, et al., 2016; Obergrussberger, Thomas, et al., 2017. Briefly, a mean beat was generated in impedance (IMP) and EFP modes by aligning individual beats and calculating mean beat and standard deviation. A selection of parameters including primary and secondary (corresponding to arrhythmic events) beat differentiation for IMP (Base Impedance, Amplitude, Beat Rate (BR), BRCoV, Pulsewidth at 30%, Falling time, #primary, #secondary) and, for EFP (Amplitude, BR, BRCoV, FPDc(max), FPDc(zero)) were automatically detected. BrCoV – Beat rate (beat period) coefficient of variation is defined as the ratio of the standard deviation to the mean. An alongside calculated shape regularity index (data not shown) allows to monitor signal shapes and thus enables the user to identify any drug induced shape change like arrhythmia or other shape changes (due to differentiation or maturation of cells) that are captured and are manifested in any of the above listed parameters. Data was analyzed using DataControl 96 (Nanion Technologies GmbH). Grand average statistics are used for parameter values (for every sweep, a mean beat is calculated and further averaged across replicates, see Doerr et al., 2015). Statistical information of compound results is provided in summary Table 3, where column “Trend” provides visualization of concentration responses for average FPDmax for each compound tested at each site. Data points on trendlines represent averages of 4 or 5 replicates for each concentration tested. If replicates did not pass inclusion criteria indicated in the study protocol, they were excluded. Where trendline goes to zero, a FPDmax calculation could not be performed due to arrhythmia being present. Additional statistical information for cross-correlations is provided in Supplemental Figures 2–4.
3. Results
Impedance technology was recently introduced in electrophysiology assays as a non-invasive, multi-parameter methodology to investigate compound-related toxicity (Peters et al., 2015). Impedance effects are mediated by various effects on action potentials, calcium flux or mechanical contractions (Sahu, 2017). Impedance signals could be monophasic or biphasic (Supplemental Figure 1 C), and both types of signals were observed in the cells participating in the study. Occurrence of the biphasic profile in impedance traces varies among cell lines, with cell seeding density and with age of the monolayer in culture. Example calculation of automated beat detection (beat rate and amplitude) for each type of impedance signal is depicted in Supplemental Figure 1 C.
A rapid, concerted activation of voltage-gated Na+ channels results in a depolarization of the cell monolayer and is marked as a depolarization spike. As the monolayer undergoes repolarization, an influx of calcium occurs (ICa-L, mainly by activation of voltage-gated L-type Ca2+ channels), followed by an efflux of potassium (IKr (rapid), IKs (slow) with several other underlying currents of delayed rectification). The repolarization phase is characterized by a repolarization peak (or an analogous of the in vivo T-wave from a clinical electrocardiogram (ECG)). The interval between the depolarization spike and maximum value of the repolarization feature represents the field potential duration (FPDmax) and is analogous to the QT interval of a clinical ECG. Automated and semi-automated detection of FPDmax is performed in CardioExcyte Control, as per Supplemental Fig. 1A and B.
Four different cell types (CDI, AXG, CLS and PLR) were investigated at four sites deploying a combined contractility and field potential readout. Cell attachment on the sensor plate as well as viability during maturation time or compound treatment was observed via monitoring of base impedance (Fig. 1B, C), amongst different other endpoints. The primary endpoint in drug-free baseline conditions, FPDmax, is shown across the different cell types in Fig. 2. AXG cells had a faster spontaneous beat rate by approximatively 50% and shorter FPDmax as compared to the other hiPSC-CMs tested (Table 1). Graphs in Fig. 2 show the number of wells across all sites plotted against FPDmax, and values in the table represent the average FPD ± standard deviation.
Figure 2. Baseline FPDmax endpoints.
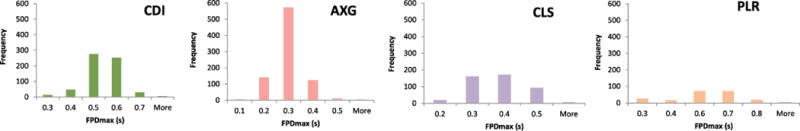
Graphs show the number of wells across all sites and cells plotted against the FPDmax.
Table 1.
Average baseline statistics for two assay endpoints.
| CDI | AXG | CLS | PLR | |
|---|---|---|---|---|
| Average FPDmax (ms) | 468 ±76 (16%) | 246 ± 41 (17%) | 333 ± 98 (29%) | 548 ± 163 (29%) |
| Average Beat Rate (bpm) | 34 ±3 (9%) | 70 ± 10 (14%) | 47 ± 6 (12%) | 38 ± 4 (10%) |
FPDmax +/− standard deviation and beat rate baseline values of all cells types.
Most hiPSC-CMs reproduce expected responses to positive and vehicle controls
Figure 3 shows FPDmax changes of all cells at all sites in the presence of vehicle control (Fig. 3A) and positive control, dofetilide (Fig. 3B) at 0.5 nM, 3 nM, 5 nM and 10 nM. Vehicle control did not produce a discernible effect across cell types or sites (Fig. 3A). The IKr inhibitor dofetilide elicited a prolongation of FPD and arrhythmia in wells treated with 5 nM and 10 nM. CLS hiPSC-CMs did not exhibit an expected FPD prolongation upon dofetilide application. The other cell types showed an FPD prolongation ranging from approx. 10 % for 0.5 nM dofetilide to 100 % FPD prolongation for 10 nM dofetilide, due to arrhythmia presence and a high dose being administered.
3.1 Cross-site comparisons across cell types indicate consistency of the impedance/field potential assay
A common protocol was utilized across four hiPSC-CM cell types across four independent sites. Four high risk compounds were common across all sites (disopyramide, quinidine, vandetanib, sotalol), one intermediate and one low risk compound (pimozide and tamoxifen, respectively) across three sites and the remaining compounds being investigated at one or two sites. A detailed list including all cell types and all compounds tested at each participating site is presented in Table 2. Compounds are grouped by risk of manifesting Human TdP, and are ranked as high, intermediate and low risk. A representative compound for each risk category was selected to discuss multi ion-channel effects.
Disopyramide is part of the high risk category and is an anti-arrhythmic agent that inhibits the fast sodium channel conduction. In addition to that, it has a negative inotropic effect on ventricular myocardium, significantly decreasing the contractility (Levites & Anderson, 1979). In Fig. 4A, example raw impedance and field potential waveforms (AXG) are presented before (red) and after dosing (shades of blue) with increasing concentrations of disopyramide at 0.1, 1, 100 μM. Fig. 4B shows endpoints across all cell types. Primary endpoints are quantified across replicates at one site (n=4 or 5 replicates). Average % change in impedance amplitude is decreased as concentration of disopyramide increased and showed variability at 10 and 100 μM due to presence of arrhythmia, with exception of CDI cells. Average % change in EFP amplitude decreased with increasing concentrations of disopyramide, while FPDmax increased. The presence of EADs was always associated with increased variability in the beat period as illustrated by the larger error bars at higher doses. Arrhythmia was detected and marked, for 10 and 100 μM disopyramide. The most prominent effects of disopyramide were decreased contractility (as suggested by Levites et al., see (Levites & Anderson, 1979)). Furthermore, we observed a reduction in EFP spike amplitude across all cell types. EADs were present in various patterns of occurrence, either in an isolated beat or multiple times in consecutive beats, or in an alternans pattern.
Figure. 4 Disopyramide demonstrates expected electrophysiological responses.
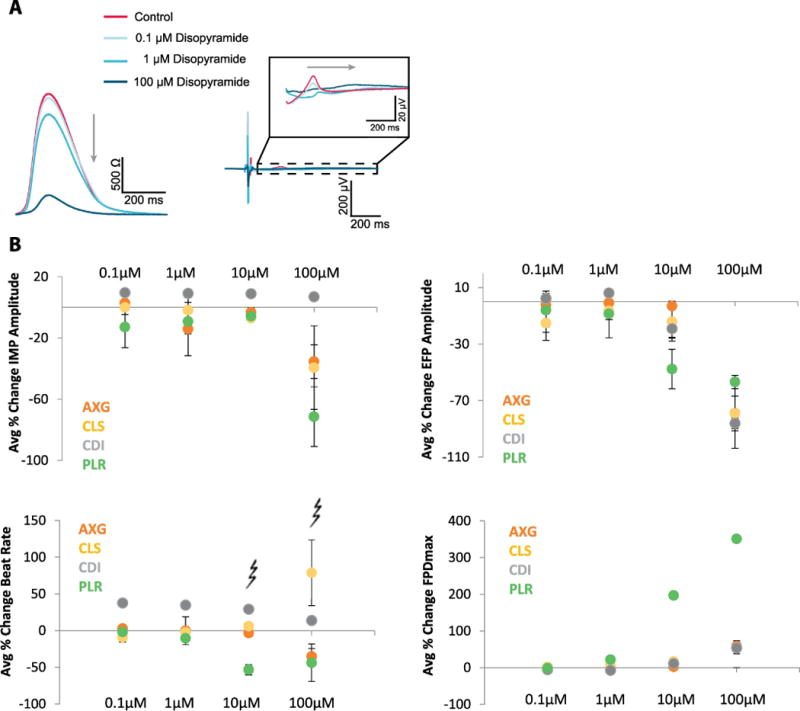
A, Example raw mean beat impedance (AXG) and field potential waveforms (AXG) (left) are presented before (red) and after (shades of blue) dosing with increasing concentrations of disopyramide at 0.1, 1, 10, 100 μM. B, Primary endpoints are quantified across replicates at one site (n=4 or 5 replicates). Error bars indicate +/− standard error across the replicates, and arrhythmia is marked at 10, 100 μM.
Domperidone is part of the intermediate risk category and is a potent inhibitor of IKr. It prolongs the cardiac repolarization at clinically relevant drug concentrations (Benoit, Guy, Pascal, René, & Jacques, 2000), and ventricular tachyarrhythmias have been reported after the use of domperidone (Joss, Goldhirsch, Brunner, & Galeazzi, 1982). In Fig. 5A, example raw impedance and field potential waveforms are presented for CLS cardiomyocytes before (red) and after dosing (shades of blue) with increasing concentrations of domperidone at 0.003, 0.03, 0.3 and 3 μM. Fig. 5B illustrates primary endpoints which are quantified across replicates at one site (n=4 or 5 replicates). Error bars indicate +/− standard error across the replicates. Average % change in impedance amplitude showed variability at 3 μM due to presence of arrhythmia. Average % change in EFP amplitude decreased with increasing concentrations of domperidone, while FPDmax increased. Additionally, the peak of the impedance transient is decreasing with increasing domperidone concentrations for CLS and PLR cells, while CLS and AXG cells show the opposite trend at the highest dose. This effect is due to different types of arrhythmia experienced in the cell types.
Figure 5. Domperidone demonstrates expected electrophysiological response.
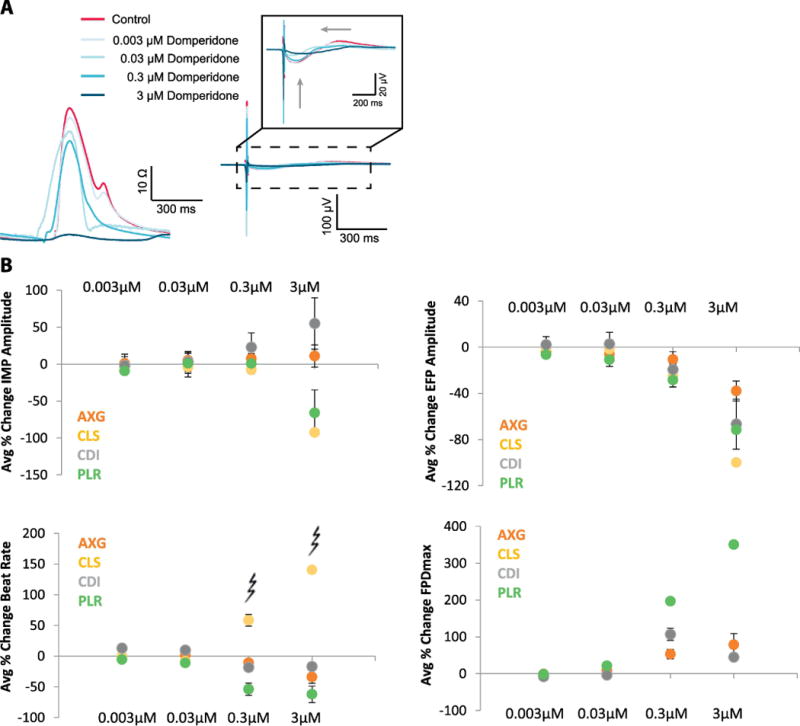
A, Example raw mean beat impedance and field potential waveforms from CLS hiPSC-CMs (left) are presented before (red) and after dosing (shades of blue) with increasing concentrations of domperidone at 0.003, 0.03, 0.3 and 3 μM. B, Primary endpoints are quantified across replicates at one sites (n= 4 or 5 replicates). Error bars indicate +/− standard error across the replicates and arrhythmia is marked at 0.3, 3 μM.
Tamoxifen is an antioxidant used in the treatment of breast cancer and exhibits antiarrhythmic capabilities by modulation of ionic transport pathways (Diez et al., 2013). In Fig. 6A, example raw impedance and field potential waveforms are presented for CDI cardiomyocytes before (red) and after dosing (shades of blue) with increasing concentrations of tamoxifen at 0.3, 0.9 and 3 μM. No significant changes in average impedance signals were observed, and tamoxifen demonstrated expected electrophysiological responses. Primary endpoints are quantified across replicates at one site (n= 4 or 5 replicates), Fig. 6B. Error bars indicate +/− standard error across the replicates. An increase of ~40% in beat rate was observed for CDI, at 3 μM, while average % change in EFP amplitude and FPDmax decreased by ~40% at the same dose.
Figure 6. Tamoxifen demonstrates expected electrophysiological responses.

A, Example raw mean beat impedance and field potential waveforms from CDI (left) are presented before (red) and after dosing (shades of blue) with increasing concentrations of tamoxifen at 0.3, 0.9 and 3 μM. 0.09 μM was left out due to visibility reasons. B, Primary endpoints are quantified across replicates at one site (n= 4 or 5 replicates). Error bars indicate +/− standard error across the replicates.
3.2 Arrhythmia occurrence and identification of early afterdepolarization (EAD) events
Arrhythmic events were observed in both impedance and EFP modes (Fig. 7), from each arrhythmia category specified in the HESI Myocyte Phase II Validation Study Protocol, supplied by the organizational team. In this study protocol, as well as in our manuscript, the types are defined as early after depolarization (EAD; type A), rolling EAD (type B), ectopic beat (type C), tachyarrhythmia (type D) and irregular beating (Type E). Arrhythmia occurrence was automatically quantified by an algorithm in CardioExcyte Control that allows calculation of total number of arrhythmic events in each recording, for details see Doerr et al., 2015; Obergrussberger, Juhasz, et al., 2016; Obergrussberger, Thomas, et al., 2017 and Supplemental Fig. 1A. In addition to that, arrhythmia occurrence and identification of arrhythmic types were identified manually through inspection of raw traces and marked in summary Table 3. EAD Definitions and fibrillation-like arrhythmia encountered in both impedance and EFP modes are depicted in Fig. 7.
Figure 7. Arrhythmia types.
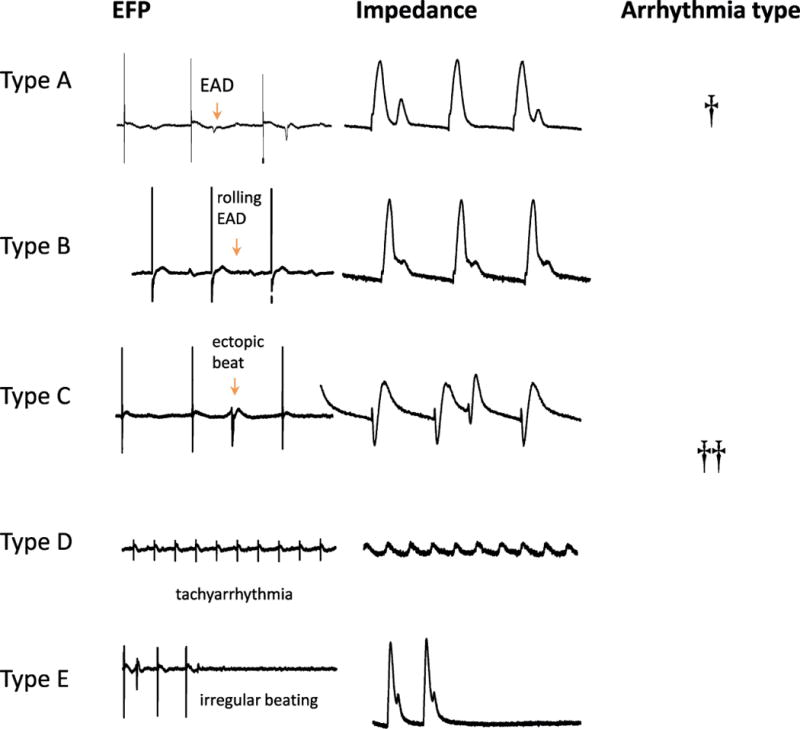
EAD Definitions and fibrillation-like arrhythmia encountered in both impedance and EFP modes.
In control experiments, both baseline pre-addition and time-matched vehicle control conditions, the beat period coefficient of variation (BRCoV) is ~0.01 – 0.02 (1-2%) (left panel, Fig. 8). This value is well under the elimination criteria recommended in the HESI Myocyte Phase II Validation Study Protocol (a value of 5% or lower is accepted, pre-compound treatment).
Figure 8. Droperidol effects on PLR cells.
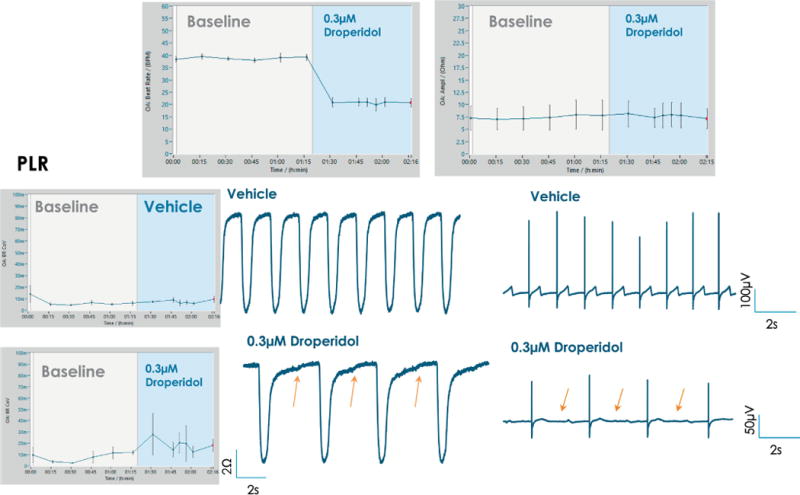
FPDmax prolongation and subtle, rolling EAD-like events are observed in the case of PLS cardiomyocytes treated with 0.3 μM droperidol (orange arrows). A decrease in beat rate is observed post addition of 0.3 μM droperidol, while impedance amplitude remains unchanged (top panel). Beat rate coefficient of variation shows an increase post addition of droperidol, indicative of arrhythmia.
Droperidol is a potent dopamine receptor antagonist, with intermediate risk of QT prolongation. In vitro QT-prolongation for droperidol is recapitulated in hiPSC-CMs; an example is listed in Fig.8 in the case of PLR cardiomyocytes. Addition of 0.3 μM droperidol produced an increase in FPDmax and rolling EAD-like events were observed (Fig.8, orange arrows point to arrhythmic events). A decrease in beat rate is observed post addition of 0.3 μM droperidol, while impedance amplitude remains unchanged (top panel). Grand average plots for BRCoV, beat rate or impedance amplitude are directly exported from DataControl 96 software, error bars represent standard error.
In the case of d, l sotalol, multiple types of arrhythmia are present (Fig. 9). Sotalol is part of the high risk category, as a potent hERG blocker and non-selective competitive beta-adrenergic receptor blocker. As a representative example, effects post addition of 10 μM d, l sotalol are shown in Fig. 9, where it produced EAD-like events and ectopic beats in CDI cardiomyocytes (orange arrows). A decrease in beat rate was observed post addition of 10 μM d, l sotalol, while impedance amplitude remains unchanged (top panel). BRCoV shows a large increase post addition of d, l sotalol, indicative of arrhythmia. In control experiments, BRCoV remains low, at ~0.01 (~1%) (left panel, Fig. 9). Grand average plots for BRCoV, beat rate or impedance amplitude are directly exported from DataControl 96 software, error bars represent standard error.
Figure 9. Sotalol effects on CDI cells.
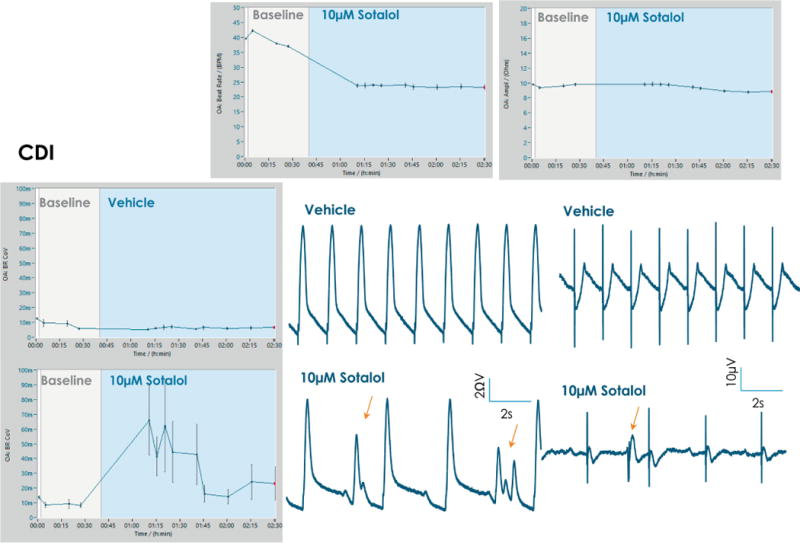
Addition of 10 μM sotalol produced EAD-like events and ectopic beats in CDI cardiomyocytes (orange arrows). A decrease in beat rate is observed post addition of 10 μM sotalol, while impedance amplitude remains unchanged (top panel). Beat rate coefficient of variation shows a large increase post addition of sotalol, indicative of arrhythmia.
3.3 Compound results across sites and cell types indicate consistency of the field potential assay
A summary of % FPDmax changes at 30 min post compound addition compared to baseline is summarized for all compounds tested, all cell types and across all participating sites (Table 3). Compounds are ranked as per HESI Myocyte Phase II Validation Study Protocol instructions, under each risk category. The “Trend” column displays actual average data per compound, n= 4 or 5 replicates each, with min (blue dot) and max (red dot) values pointed out. Changes in FPDmax are concentration dependent, as illustrated by column “Trend”. Arrhythmia classification was simplified by marking the occurrence of EAD-like events with † (type A arrhythmia listed under arrhythmia category specified in the HESI Myocyte Phase II Validation Study Protocol), next to each test article concentration. All other types of/combinations of arrhythmia observed are marked with †† (type B, C, D arrhythmias listed HESI Myocyte Phase II Validation Study Protocol). Where signals from hiPSC-CMs were arrested, “quiescence” was marked in Table 3. Exemplary raw traces for each arrhythmia category are listed in Fig.7. All high risk compounds showed a significant prolongation of FPDmax, with arrhythmia detection starting at dose 2 (quinidine) or dose 3 (disopyramide, d, l sotalol, vandetanib). Incidence of FPDmax>10% or > 30% started at dose 1 in the case of quinidine and was prevalent starting with dose 3 in almost all high risk compounds. In the case of intermediate risk compounds, incidence of FPDmax >10% or > 30% was present in almost all compounds at dose 3. Astemizole, cisapride, clarithromycin, domperidone, droperidol, ondansetron and pimozide produce arrhythmia in most cases at dose 3, and almost all cases at dose 4. In the case of low risk compounds, either a decrease in FPDmax was observed (diltiazem, nifedipine, nitrendipine, verapamil), or not a significant change (loratadine) when compared to pre-addition values. A summary of acute effects of 23 drugs on mean %change FPDmax investigated in four commercially available hiPSC-CM cell lines is presented in Supplemental Fig. 3. Two intermediate range doses (dose 2 and dose 3) are shown, as investigated at four different sites; vehicle effects subtracted. An increase in average % FPDmax is captured in high risk compounds and most of intermediate risk compounds, while low risk compounds illustrate the opposite effect. Furthermore, a positive correlation of all common compounds tested at all sites for AXG cells is presented in Supplemental Fig. 4.
4. Discussion
A subset of experiments allowed a direct comparison across four commercial hiPSC-CM cell types with the same compound. A high correlation for the high risk compounds was found for all cell types, arrhythmogenic properties of the compounds were observed from dose 2 (quinidine) or from dose 3 (disopyramide, d, l sotalol, vandetanib) in 33 from 36 individual experiments. In two cases (quinidine, vandetanib) signals from hiPSC-CMs were arrested at dose 3 (quinidine, site 2, CLS and site 4, CLS) and dose 4 (quinidine, site 1, CDI; site 3, CDI; site 1, AXG and vandetanib, site 2, CDI). Intermediate risk compounds induced arrhythmia from dose 3 in 19 from 36 experiments, chlorpromazine did not induce arrhythmic events independent of the cell type or site. Pimozide at site 4, AXG cells, was not arrhythmogenic but a FDP prolongation was observed. Terfenadine induced arrhythmia in 2 experiments (site 4, AXG and site 4, CLS) and quiescence in 4 other experiments. Risperidone induced no significant change in the FPD in AXG and CLS cells and >10% in CDI cells. In the case of low risk compounds, either a decrease in FPDmax was observed (diltiazem, nifedipine, nitrendipine, verapamil), or not a significant change (loratadine), arrhythmogenesis only at highest dose and in 5 from 36 experiments. The summary presented in Supplemental Fig. 3 illustrates a clean separation between low risk vs high risk compounds, based on %change in FPDmax. The positive correlation of all common compounds tested at all sites for AXG cells (Supplemental Fig. 4) illustrates a high degree of site-to-site reproducibility of this assay. Present results demonstrate that the baseline electrophysiological characteristics measured differ between the four cell lines, the baseline beat rate of AXG cardiomyocytes being near physiological values in humans (Nes, Janszky, Wisloff, Stoylen, & Karlsen, 2013). The observed difference could potentially be explained by different expression levels of ion channels and by the donor’s specific genetic profile which varies between different iPS cell lines. Additionally, it is also known for hiPSC-CMs that the action potential morphology and some ion channel currents vary with cell ages (differentiation times).
With CLS cells mostly behaving as an exception, hiPSC-CMs are sensitive and exhibit at least 10% delayed or shortened repolarization from pre-addition values and arrhythmia after drug application and thus can provide predictive cardiac electrophysiology data. The minimum effective concentration was different between cell types, but not between sites.
This indicates that the technology and test protocol appear to be robust between the different test sites and laboratories. Cell specific sensitivities to detect a lengthening of FPD and arrhythmia following acute test substance application may be related to differing FPD in baseline conditions between the cell types. The measurement of impedance in addition to the field potential added value to the characterization of arrhythmia types. Even though use of the impedance signal as a marker for cellular contractility was not the primary focus of the study, compounds like nifedipine, verapamil, disopyramide, domperidone and others could be identified to have a negative inotropic effect on hiPSC-CMs.
4.1 Future direction
Pacing of hiPSC-CMs during the assay, whereby the beat rate is controlled through electrical or optogenetic stimulation (Rehnelt et al., 2017), would eliminate variability due to differences in spontaneous beating across cell types. A comparative analysis of contractility assessments of native cardiomyocytes with hiPSC-CM would reinforce utility of an impedance screen, as it is highly sensitive at providing information about arrhythmic properties of compounds.
4.2 Conclusion
In the present study, we investigated the effects of 23 drugs on FPDmax recorded from hiPSC-CM with the CardioExcyte 96 system. Impedance data confirms arrhythmia detected in EFP modes. Arrhythmia occurrence was consistent with previous literature findings, and hiPSC-CMs tested exhibited prolongation of FPDmax in agreement with mechanism of action reported for compounds under testing. Altogether, these results demonstrate the potential of predicting torsadogenic risk with hiPSC-CMs using the CardioExcyte 96 system.
Supplementary Material
Supplementary Figure 1. EFP and Impedance endpoint detection. Secondary beat detection allows quantification of number of arrhythmic events for each recorded sweep (A). Automated detection of T-wave in an interval specified by user (B). Automated detection of beat period, impedance amplitude in case of monophasic (C, left figure) and biphasic (C, middle) impedance traces. Field potential duration maximum (FPDmax) is measured from initial spike depolarization to the maximum value of the repolarization (C, right).
Supplementary Figure 2. FPD correlation to beat rate for Cor.4U and CDI cells. The scatter plot illustrating the relation between beat rate and FPDmax for Cor.4U and CDI cells on different recording plates under baseline conditions. The different marker types represent the different plates and each marker represents a single well. The comparison of baseline information of Cor.4U and CDI cells on plate type A (green, 2 mm electrode size) and plate type B (purple, 0.6 mm electrode size) showed no impact on the spontaneous beating of the cells. The same was true for different culturing conditions (serum-free (site 2) versus serum containing medium (all other sites)). A prominent source of the variance appeared to be the site-to-site heterogeneity which can be explained by the different operators at the different sites and the cells’ batch-to-batch variance.
Supplementary Figure 3. Summary of acute effects of 23 drugs on mean %change FPDmax investigated in all cells and all sites; vehicle effects subtracted. Compound incubation time was 30 min. An increase in average % FPDmax is captured in high risk compounds and most of intermediate risk compounds, while low risk compounds illustrate the opposite effect.
Supplementary Figure 4. Cor.4U FPDmax results of dose 2 of high risk (disopyramide, quinidine, sotalol, vandetanib) medium risk (pimozide, terfenadine) and low risk compounds (diltiazem, loratadine, tamoxifen) are correlated across different sites. The FPDmax (% change) results from different sites are compared. Each data point represents the mean across replicates for a given compound and concentration evaluated at a different site. A good cross site correlation was observed based on linear regression calculations. The calculated R2 values are 0.91 for site 2 - site 4, 0.8 for site 1 - site 4, and 0.79 for site 1- site 4. Note the pattern cloud for low risk compounds staying in the low FPDmax changes vs. the high/intermediate risk compounds which separate themselves in the higher FPDmax change range.
Acknowledgments
We thank Cellular Dynamics International, a FujiFilm Company, for providing us with cardiomyocytes (iCell cardiomyocytes2) and Ncardia, for providing us with cardiomyocytes (Cor.4U®, Pluricyte®). We thank Takara Bio Europe AB for providing Cellartis® hiPS-CMs. The authors would also like to thank the Drug Synthesis & Chemistry Branch or Toxicology & Pharmacology Branch at the Division of Cancer Treatment and Diagnosis (DCTD), National Cancer Institute (NCI) for managerial support on compound supply; NCI-Chemotherapeutic Agents Repository for compound preparation and distribution. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E and R24 HL117756. The study was also funded in part by Eureka’s EUROSTARS grant FKZ 01QE1502 (CardioPredict).
Abbreviations
- AXG
Cor.4U®, hiPSC-CMs, Ncardia
- BrCoV
Beat rate (beat period) coefficient of variation
- CDI
iCell Cardiomyocytes2 hiPSC-CMs, Cellular Dynamics International, a FujiFilm Company
- CLS
Cellartis®, hiPSC-CMs, Takara Bio Clontech
- EAD
early after depolarization
- EFP
extracellular field potential
- FPDmax
field potential duration at maximum repolarization
- HESI
Health and Environmental Sciences Institute
- hiPSC-CMs
human induced pluripotent stem cell-derived cardiomyocytes
- PLR
Pluricyte®, hiPSC-CMs, Ncardia
- TdP
torsade de pointes
- VSD
voltage-sensing dyes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
CTB, SSF, KJ are employed by Nanion Technologies GmbH, the provider of the CardioExcyte 96.
No competing interests are declared.
References
- Abassi YA, Xi B, Li N, Ouyang W, Seiler A, Watzele M, Xu X. Dynamic monitoring of beating periodicity of stem cell-derived cardiomyocytes as a predictive tool for preclinical safety assessment. British Journal of Pharmacology. 2012;165(5):1424–1441. doi: 10.1111/j.1476-5381.2011.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker N, Stoelzle S, Göpel S, Guinot D, Mumm P, Haarmann C, Brüggemann A. Minimized cell usage for stem cell-derived and primary cells on an automated patch clamp system. Journal of Pharmacological and Toxicological Methods. 2013;68(1):82–87. doi: 10.1016/j.vascn.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Benoit D, Guy R, Pascal D, René C, Jacques T. Domperidone Should Not Be Considered a No-Risk Alternative to Cisapride in the Treatment of Gastrointestinal Motility Disorders. Circulation. 2000;102:1883–1885. doi: 10.1161/01.cir.102.16.1883. [DOI] [PubMed] [Google Scholar]
- Blinova K, Stohlman J, Vicente J, Chan D, Hortigon-vinagre MP, Zamora V, Strauss DG. Comprehensive Translational Assessment of Human- Induced Pluripotent Stem Cell Derived Cardiomyocytes for Evaluating Drug-Induced Arrhythmias. Toxicological Sciences. 2016;155(1):234–247. doi: 10.1093/toxsci/kfw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M, Millar V, Williams A, Kalinka S. Bridging Functional and Structural Cardiotoxicity Assays using Human Embryonic Stem- Cell Derived Cardiomyocytes for a more Comprehensive Risk Assessment. Toxicological Sciences. 2015;148(1):241–260. doi: 10.1093/toxsci/kfv180. [DOI] [PubMed] [Google Scholar]
- Clements M, Thomas N. High-Throughput Multi-Parameter Profiling of Electrophysiological Drug Effects in Human Embryonic Stem Cell Derived Cardiomyocytes Using Multi-Electrode Arrays. Toxicological Sciences. 2014;140(2):445–461. doi: 10.1093/toxsci/kfu084. [DOI] [PubMed] [Google Scholar]
- Colatsky T, Fermini B, Gintant G, Pierson JB, Sager P, Sekino Y, Stockbridge N. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative — Update on progress. Journal of Pharmacological and Toxicological Methods. 2016;81:15–20. doi: 10.1016/j.vascn.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Diez ER, Prado NJ, Carrión AM, Petrich ER, Ponce Zumino AZ, Miatello RM. Electrophysiological Effects of Tamoxifen: Mechanism of Protection Against Reperfusion Arrhythmias in Isolated Rat Hearts. Journal of Cardiovascular Pharmacology. 2013;62(2):184–191. doi: 10.1097/FJC.0b013e318295b611. [DOI] [PubMed] [Google Scholar]
- Doerr L, Thomas U, Guinot DR, Bot CT, Stoelzle-Feix S, Beckler M, Fertig N. New Easy-to-Use Hybrid System for Extracellular Potential and Impedance Recordings. Journal of Laboratory Automation. 2015;20(2):175–188. doi: 10.1177/2211068214562832. [DOI] [PubMed] [Google Scholar]
- Friedrichs S, Malan D, Voss Y, Sasse P. Scalable Electrophysiological Investigation of iPS Cell-Derived Cardiomyocytes Obtained by a Lentiviral Purification Strategy. Journal of Clinical Medicine. 2015;4(1):102–123. doi: 10.3390/jcm4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist KH, Lewis GF, Gay EA, Sellgren KL, Grego S. High-throughput cardiac safety evaluation and multi-parameter arrhythmia profiling of cardiomyocytes using microelectrode arrays. Toxicology and Applied Pharmacology. 2015;288(2):249–257. doi: 10.1016/j.taap.2015.07.024. [DOI] [PubMed] [Google Scholar]
- Goversen B, Becker N, Stoelzle-Feix S, Obergrussberger A, Vos MA, van Veen TAB, de Boer TP. A hybrid model for safety pharmacology on an automated patch clamp platform: Using dynamic clamp to join iPSC-derived cardiomyocytes and simulations of Ik1 ion channels in real-time. Frontiers in Physiology. 2018;8:1094. doi: 10.3389/fphys.2017.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Abrams R, Babiarz JE, Cohen JD, Kameoka S, Sanders MJ, Kolaja KL. Estimating the Risk of Drug-induced Proarrhythmia using Human Induced Pluripotent Stem Cell Derived Cardiomyocytes. Toxicological Sciences : An Official Journal of the Society of Toxicology. 2011;123(1):281–289. doi: 10.1093/toxsci/kfr158. [DOI] [PubMed] [Google Scholar]
- Jonsson MKB, Wang QD, Becker B. Impedance-based detection of beating rhythm and proarrhythmic effects of compounds on stem cell-derived cardiomyocytes. ASSAY and Drug Development Technologies. 2011;9(6):589–599. doi: 10.1089/adt.2011.0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joss RA, Goldhirsch A, Brunner KW, Galeazzi RL. Sudden death in a cancer patient on high dose domperidone. Lancet. 1982;1(8297):1019. doi: 10.1016/s0140-6736(82)92016-5. [DOI] [PubMed] [Google Scholar]
- Kanda Y, Yamazaki D, Kurokawa J, Inutsuka T, Sekino Y. Points to consider for a validation study of iPS cell-derived cardiomyocytes using a multi-electrode array system. Journal of Pharmacological and Toxicological Methods. 2016;81(2016):196–200. doi: 10.1016/j.vascn.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Kho D, MacDonald C, Johnson R, Unsworth CP, O’Carroll SJ, du Mez E, Graham ES. Application of xCELLigence RTCA Biosensor Technology for Revealing the Profile and Window of Drug Responsiveness in Real Time. Biosensors. 2015;5(2):199–222. doi: 10.3390/bios5020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamore SD, Scott CW, Peters MF. Cardiomyocyte Impedance Assays. In: Sittampalam G, Coussens N, Nelson H, Arkin M, Auld D, Austin C, Weidner J, editors. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Transational Sciences; 2015. [PubMed] [Google Scholar]
- Levites R, Anderson GJ. Electrophysiological effects of disopyramide phosphate during experimental myocardial ischemia. American Heart Journal. 1979;98(3):339–344. doi: 10.1016/0002-8703(79)90046-2. [DOI] [PubMed] [Google Scholar]
- Millard D, Dang Q, Shi H, Zhang X, Strock C, Kraushaar U, Gintant G. Cross-Site Reliability of Human Induced Pluripotent Stem-Cell Derived Cardiomyocyte Based Safety Assays using Microelectrode Arrays: Results from a Blinded CiPA Pilot Study. Toxicological Sciences. 2018 doi: 10.1093/toxsci/kfy110. EPub ahead of Print. [DOI] [PMC free article] [PubMed]
- Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, Wu JC. Screening Drug-Induced Arrhythmia Using Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes and Low-Impedance Microelectrode Arrays. Circulation. 2013;128(11_suppl_1):S3–S13. doi: 10.1161/CIRCULATIONAHA.112.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes B, Janszky I, Wisloff U, Stoylen A, Karlsen T. Age‐ predicted maximal heart rate in healthy subjects: The HUNT Fitness Study. Scandinavian Journal of Medicine & Science in Sports. 2013;23(6):697–704. doi: 10.1111/j.1600-0838.2012.01445.x. http://doi.org/doi:10.1111/j.1600-0838.2012.01445.x. [DOI] [PubMed] [Google Scholar]
- Nguemo F, Šaric T, Pfannkuche K, Reppel M, Hescheler J. In vitro Model for Assessing Arrhythmogenic Properties of Drugs Based on High-resolution Impedance Measurements. Cellular Physiology and Biochemistry. 2012;29:819–832. doi: 10.1159/000188069. [DOI] [PubMed] [Google Scholar]
- Obergrussberger A, Brüggemann A, Goetze TA, Rapedius M, Haarmann C, Rinke I, Fertig N. Automated Patch Clamp Meets High-Throughput Screening: 384 Cells Recorded in Parallel on a Planar Patch Clamp Module. Journal of Laboratory Automation. 2016;21(6):779–793. doi: 10.1177/2211068215623209. [DOI] [PubMed] [Google Scholar]
- Obergrussberger A, Goetze TA, Brinkwirth N, Becker N, Friis S, Rapedius M, Fertig N. An update on the advancing high-throughput screening techniques for patch clamp-based ion channel screens: implications for drug discovery. Expert Opinion on Drug Discovery. 2018;13(3):269–277. doi: 10.1080/17460441.2018.1428555. [DOI] [PubMed] [Google Scholar]
- Obergrussberger A, Haarmann C, Stölzle-Feix S, Becker N, Ohtsuki A, Brüggemann A, Fertig N. Automated patch clamp recordings of human stem cell-derived cardiomyocytes. In: Clements M, Roquemore L, editors. Stem cell-derived models in Toxicity. 1st. New York: Springer Science+Business Media New York; 2017. pp. 57–82. [DOI] [Google Scholar]
- Obergrussberger A, Juhasz K, Thomas U, Stölzle-Feix S, Becker N, Dörr L, Fertig N. Safety pharmacology studies using EFP and impedance. Journal of Pharmacological and Toxicological Methods. 2016;81:223–232. doi: 10.1016/j.vascn.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Obergrussberger A, Stölzle-Feix S, Becker N, Brüggemann A, Fertig N, Möller C. Novel screening techniques for ion channel targeting drugs. Channels. 2015;9(6):367–375. doi: 10.1080/19336950.2015.1079675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obergrussberger A, Thomas U, Stoelzle-Feix S, Becker N, Dörr L, Beckler M, Fertig N. Combined Impedance and extracellular field potential recordings from human stem cell-derived cardiomyocytes. In: Clements M, Roquemore L, editors. Stem cell-derived models in Toxicity. 1st. New York: Springer Science+Business Media New York; 2017. pp. 191–209. [DOI] [Google Scholar]
- Peters MF, Lamore SD, Guo L, Scott CW, Kolaja KL. Human Stem Cell-Derived Cardiomyocytes in Cellular Impedance Assays: Bringing Cardiotoxicity Screening to the Front Line. Cardiovascular Toxicology. 2015;15(2):127–139. doi: 10.1007/s12012-014-9268-9. [DOI] [PubMed] [Google Scholar]
- Rehnelt S, Malan D, Juhasz K, Wolters B, Doerr L, Beckler M, Bruegmann T. Frequency-Dependent Multi-Well Cardiotoxicity Screening Enabled by Optogenetic Stimulation. International Journal of Molecular Sciences. 2017;18(12):2634. doi: 10.3390/ijms18122634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu S, editor. Stem cells in Toxicology and Medicine. John Wiley & Sons, Inc; 2017. [Google Scholar]
- Scheel O, Frech S, Amuzescu B, Eisfeld J, Lin KH, Knott T. Action potential characterization of human induced pluripotent stem cell-derived cardiomyocytes using automated patch-clamp technology. Assay and Drug Development Technologies. 2014;12(8):457–69. doi: 10.1089/adt.2014.601. [DOI] [PubMed] [Google Scholar]
- Stockbridge N, Morganroth J, Shah RR, Garnett C. Dealing with global safety issues: Was the response to QT-liability of non-cardiac drugs well coordinated? Drug Safety. 2013;36(3):167–182. doi: 10.1007/s40264-013-0016-z. [DOI] [PubMed] [Google Scholar]
- Stoelzle S, Haythornthwaite A, Kettenhofen R, Kolossov E, Bohlen H, George M, Fertig N. Automated Patch Clamp on mESC-Derived Cardiomyocytes for Cardiotoxicity Prediction. Journal of Biomolecular Screening : The Official Journal of the Society for Biomolecular Screening. 2011;16(8):910–916. doi: 10.1177/1087057111413924. [DOI] [PubMed] [Google Scholar]
- Stoelzle S, Obergrussberger A, Brüggemann A, Haarmann C, George M, Kettenhofen R, Fertig N. State-of-the-art automated patch clamp devices: Heat activation, action potentials, and high throughput in ion channel screening. Frontiers in Pharmacology. 2011;2:76. doi: 10.3389/fphar.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi B, Wang T, Li N, Ouyang W, Zhang W, Wu J, Abassi YA. Functional Cardiotoxicity Profiling and Screening Using the xCELLigence RTCA Cardio System. Journal of Laboratory Automation. 2011;16(6):415–421. doi: 10.1016/j.jala.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo L, Zeng H, White S, Furniss M, Balasubramanian B, Abassi Y. Multi-parametric assessment of cardiomyocyte excitation-contraction coupling using impedance and field potential recording: A tool for cardiac safety assessment. Journal of Pharmacological and Toxicological Methods. 2016;81:201–216. doi: 10.1016/j.vascn.2016.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. EFP and Impedance endpoint detection. Secondary beat detection allows quantification of number of arrhythmic events for each recorded sweep (A). Automated detection of T-wave in an interval specified by user (B). Automated detection of beat period, impedance amplitude in case of monophasic (C, left figure) and biphasic (C, middle) impedance traces. Field potential duration maximum (FPDmax) is measured from initial spike depolarization to the maximum value of the repolarization (C, right).
Supplementary Figure 2. FPD correlation to beat rate for Cor.4U and CDI cells. The scatter plot illustrating the relation between beat rate and FPDmax for Cor.4U and CDI cells on different recording plates under baseline conditions. The different marker types represent the different plates and each marker represents a single well. The comparison of baseline information of Cor.4U and CDI cells on plate type A (green, 2 mm electrode size) and plate type B (purple, 0.6 mm electrode size) showed no impact on the spontaneous beating of the cells. The same was true for different culturing conditions (serum-free (site 2) versus serum containing medium (all other sites)). A prominent source of the variance appeared to be the site-to-site heterogeneity which can be explained by the different operators at the different sites and the cells’ batch-to-batch variance.
Supplementary Figure 3. Summary of acute effects of 23 drugs on mean %change FPDmax investigated in all cells and all sites; vehicle effects subtracted. Compound incubation time was 30 min. An increase in average % FPDmax is captured in high risk compounds and most of intermediate risk compounds, while low risk compounds illustrate the opposite effect.
Supplementary Figure 4. Cor.4U FPDmax results of dose 2 of high risk (disopyramide, quinidine, sotalol, vandetanib) medium risk (pimozide, terfenadine) and low risk compounds (diltiazem, loratadine, tamoxifen) are correlated across different sites. The FPDmax (% change) results from different sites are compared. Each data point represents the mean across replicates for a given compound and concentration evaluated at a different site. A good cross site correlation was observed based on linear regression calculations. The calculated R2 values are 0.91 for site 2 - site 4, 0.8 for site 1 - site 4, and 0.79 for site 1- site 4. Note the pattern cloud for low risk compounds staying in the low FPDmax changes vs. the high/intermediate risk compounds which separate themselves in the higher FPDmax change range.


