Abstract
Gram-negative bacteria have become the main pathogens and cause serious clinical problems with increased morbidity and mortality. However, the slow discovery of new antimicrobial agents is unable to meet the need for the treatment of bacterial infections caused by drug-resistant strains. The interaction of L12 and L10 is essential for ribosomal function and protein synthesis. In this study, a yeast two-hybrid system was established to successfully detect the interaction between L12 and L10 proteins from gram-negative bacteria Escherichia coli, which allows us to screen compounds that specifically disrupt this interaction. With this system, we identified two compounds IMB-84 and IMB-87 that block L12−L10 interaction and show bactericidal activity against E. coli. We used glutathione-S-transferase (GST) pull-down and surface plasmon resonance (SPR) assays to demonstrate that these compounds disrupt L12−L10 interaction in vitro and the target of compounds was further confirmed by the overexpression of target proteins. Moreover, protein synthesis and elongation factor G-dependent GTPase activities are inhibited by two compounds. Therefore, we have identified two antibacterial agents that disrupt L12−L10 interaction by using yeast two-hybrid system.
KEY WORDS: Yeast two-hybrid, Escherichia coli, Ribosome, L12/L10, Antimicrobial agents
Graphical abstract
In this study, a yeast two-hybrid system was established to detect the interaction between L12 and L10 proteins from E. coli. With this system, we identified two compounds IMB-84 and IMB-87 that block L12–L10 interaction and show bactericidal activity against E. coli.
1. Introduction
Gram-negative bacteria have become the main pathogens in clinic, and one main reason is the generation of drug-resistant bacterial strains1, 2, 3, 4. Unfortunately, very few drugs are available in clinical treatment of drug-resistant gram negative strains. Also, few new agents are under development to keep pace with the emerging of drug-resistant bacteria5, 6, 7. Therefore, it is imminent to identify new antimicrobial agents with novel targets to deal with the emergence of drug-resistant bacteria.
Proteins carry out all cellular functions in each organism and the ribosome is the factory where protein synthesis occurs. The difference between ribosome structures in bacteria and human cells allows some antibiotics to kill bacteria specifically8. For example, gentamicin binds to the A site of 16S ribosomal RNA to prevent protein synthesis in bacteria9. Tetracycline inhibits protein synthesis in bacteria because it prevents aminoacyl-tRNA from entering the A site during translation10. Now, protein–protein interaction inhibitors provide new opportunities for drug discovery. The ribosome is a multi-protein complex and the protein–protein interactions in a ribosome are attractive targets for new antibiotics because of their central role in protein synthesis and cellular functions.
In bacteria, ribosomal proteins L12 (encoded by the rplL gene) and L10 (encoded by the rplJ gene) are part of the stalk, which belongs to the large ribosomal subunit (50S). It has been shown that the elongation factors EF-G and EF-Tu are recruited to the stalk by the L12 C-terminal domain to enhance the GTPase activity11, 12. Consistently, a ribosomal stalk lacking L12 is unable to interact with elongation factors13. The C-terminal α-helix of L10 anchors two or three L12 dimers by associating with the N-terminal domains of L12, and disruption of L12–L10 interaction prevents the binding of EF-G and EF-Tu to the stalk and causes the loss of ribosomal GTPase activity14. A yeast two-hybrid system has been established to identify small molecules that block the interaction between L12 and L10 proteins from Mycobacterium tuberculosis. L12–L10 interaction appears to be a potential target for anti-tuberculosis agents15, 16. In addition, L12–L10 interaction is highly conserved in Gram-negative bacteria, but these two proteins show low homology with corresponding human proteins. Therefore, the L12−L10 interaction could be used to screen new agents that kill Gram-negative bacteria.
In this study, we developed a screening system based on the interaction between Escherichia coli ribosomal proteins L12 and L10. Firstly, L12–L10 interaction is confirmed by yeast two-hybrid (Y2H) system, with which we can identify the compounds that specifically inhibit this interaction. After screening, two compounds IMB-84 and IMB-87 were selected. These compounds block L12–L10 interaction and inhibit the growth of E. coli with certain toxicity to mammalian cells. Surface plasmon resonance (SPR) and glutathione-S-transferase (GST) pull-down were further used to confirm that IMB-84 and IMB-87 inhibit L12–L10 interaction in vitro. Consistently, both EF-G-dependent GTPase activity and ribosome-mediated protein synthesis are inhibited by these two compounds in vitro. In addition, overexpression of either L12 or L10 in E. coli increases the minimum inhibitory concentration (MIC) of these two compounds, indicating that L12 and L10 are likely the targets in vivo. In summary, using the Y2H assay, we successfully identified new antibacterial agents targeting the L12–L10 interaction in Gram-negative bacteria.
2. Materials and methods
2.1. Regents
The yeast two-hybrid system was purchased from Clontech (Dalian, China). The expression vectors pET-16b, pET-30a and pGEX-4T-1 were obtained from Novagen (Shanghai, China). The compound library is a combination of synthetic (from Enamine Ltd., Ukraine and the Institute of Medicinal Biotechnology, CAMS, China). All the compounds were suspended in DMSO and stored at 4 °C. The purity of compounds is higher than 90%. Monoclonal antibodies (anti-HA, anti-c-Myc and anti-His) were supplied by Cwbio (Beijing, China). Glutathione sepharose 4B was purchased from GE Healthcare (Uppsala, Sweden). T4 DNA ligase, restriction endonucleases and DNA polymerase were purchased from TaKaRa (Dalian, China). All other chemicals were from Sigma. The small molecule synthesized by MedPharma Partners (Boston, MA, USA). Compounds IMB-84 and IMB-87 were synthesized by the Institute of Medicinal Biotechnology and were suspended in DMSO at 10 mg/mL concentration. Further dilutions for use in the SPR assays and cell culture were done in buffer or culture medium.
2.2. Plasmid construction
The DNA fragments of rplJ and rplL genes that encode L10 and L12 proteins, respectively, were amplified by PCR from E. coli ATCC 25922 genomic DNA. The primer pairs were designed as follows: rplJ forward primer, 5′-CTCATATGGCTTTAAATCTTCAAGAC-3′, rplJ reverse primer, 5′-ATGGATCCTTAAGCAGCTTCTTT-3′; rplL forward primer, 5′-CTCATATGTCTATCACTAAAGATCAAAT-3′, rplL reverse primer, 5′-ATGGATCCTTATTTAACTTCAACTT-3′. After digestion with NdeI and BamHI, the PCR fragments were inserted into pGADT7 (DNA activation domain, AD) to generate plasmids pAD-L10 and pAD-L12, in such way that the DNA fragments of L10 and L12 were fused in frame with Gal4 transcription activating domain. Similarly, the DNA fragments for L10 and L12 proteins were inserted into pGBKT7 (DNA binding domain, BD) to generate pBD-L10 and pBD-L12, in which the L10 and L12 fragments were fused in frame with Gal4 DNA binding domain. The control plasmids pAD-T, pBD-53 and pBD-lam were obtained from Clontech.
For the construction of the His-fusion plasmids, the gene for L10 protein was amplified by PCR with primer pairs: forward primer, 5′-GGAATTCCATATGGCTTTAAATCTTCAAGAC-3′, reverse primer, 5′-CGCGGATCCAAAGCAGCTTCTTTCGCAT-3′. After digestion with NdeI and BamHI, the fragment was inserted into pET30a vector; the resulting recombinant plasmid pET30a-L10 will express L10 protein with 6× His-tag at the C terminal. After digestion with NdeI and BamHI, the DNA fragment for L12 protein from pAD-L12 was ligated with pET16b to generate recombinant plasmid pET16b-L12, which has 6× His-tag at the N terminal of L12 protein.
For the construction of the GST-fusion plasmids, the gene for L12 protein was amplified by PCR with primer pairs: forward primer, 5′-CGCGGATCCATGTCTATCACTAAAGATCAAATCA-3′, reverse primer, 5′-CCGCTCGAGTTTAACTTCAACTTCAGCGCCAGCTTCT-3′. This PCR fragment was inserted into the pGEX-4T-1 expression vector after digestion with the BamHI and XhoI. All constructs were sequenced for verification.
2.3. Detection of L12–L10 interaction using the yeast two-hybrid system
The Gal4 yeast two-hybrid system was used to detect L10–L12 interaction. The pAD-L12 and pBD-L10 plasmids were co-transformed into AH109 yeast cells using LiAC method17. The transformants were selected by growing the cells on SD/–Leu–Trp plates at 30 °C for 3 days. The growing colonies should be AH109 with pAD-L12 and pBD-L10. AH109 (pAD-L10+pBD-L12), AH109 (pAD+pBD-L12), AH109 (pAD-L10+pBD), (pAD-T+pBD-53) and AH109 (pAD-T+pBD-lam) were obtained using the same method. Strains AH109 (pAD-T+pBD-53) and AH109 (pAD-T+pBD-lam) were used as positive and negative control, respectively. All transformants were grown on SD/–Leu–Trp–Ade–His plates at 30 °C for 3–4 days. The yeast cells are expected to grow on the quadruple dropout plates. If the two fusion proteins interact with each other, the reporter genes will be activated. The L12–L10 interaction in the positive clones was further assessed by β-galactosidase (β-gal) activity assay.
β-Gal activity was measured as described previously15. Yeast cells from fresh cultures in SD/–Leu–Trp dropout medium were transferred onto filter paper, and the cells were lysed by freeze/thaw treatment. Five mL of Z buffer (0.1 mol/L Na2HPO4, 35 mmol/L NaH2PO4, 10 mmol/L KCl, and 1 mmol/L MgSO4, pH 7.0), 83.5 μL 20 mg/mL X-gal and 0.27% β-mercaptoethanol were added into the lysates. The filters were incubated at 30 °C, and examined periodically until color change was observed.
To quantify β-gal activity, 4.5 mL yeast cultures in mid-log phase (OD600=0.5–0.8) were harvested by centrifugation and then resuspended in 300 μL Z buffer. A total of 0.1 mL of cell suspension was lysed by freeze/thaw treatment. Then, 700 μL Z buffer with 0.27% β-mercaptoethanol and 160 μL o-nitrophenyl β-d-galactopyranoside (ONPG, 4 mg/mL in Z buffer) was added to the reaction tube successively, and incubated at 30 °C until color change was observed. The reaction was stopped by addition of 0.4 mL of 1 mol/L Na2CO3 and the reaction tube was centrifuged at 14,000×g for 10 min. A420 absorption of the supernatant was measured and the β-gal activity was quantified using Eq. (1):
| (1) |
where t is the incubation time (min) and V is the volume of the cell cultures used for the assay (mL). The experiments were repeated three times.
The expression of the two fusion proteins in yeast AH109 was verified by Western blotting (Tanon 5200, Shanghai, China). Yeast cells were crushed using vortex with glass beads, and protein extracts were obtained. Protein expression in yeast cells was verified by SDS-PAGE and followed with Western blotting using anti-Myc and anti-HA monoclonal antibodies.
2.4. Compound library screening
Yeast cells AH109 (pAD-L12+pBD-L10), AH109 (pAD-T+pBD-53) and AH109 were used for screening. The screening assays were performed in 96-well plates in a final volume of 200 μL. Fresh yeast cells (OD600=0.8) of AH109 (pAD-L12+pBD-L10) and AH109 (pAD-T+pBD-53) were diluted 100-fold in SD/–Leu–Trp–Ade–His dropout medium, but AH109 cells were diluted 100 times in YPD rich medium. We added 198 μL of diluted culture and 2 μL of compounds into each well and the final concentration of compound is 25 μg/mL with 1% DMSO. The yeast cells were incubated at 30 °C for 3 days to assess the growth inhibition.
2.5. Expression and purification of recombinant proteins
E. coli BL21 (DE3) was used to express His-tagged L10 protein. The E. coli cells with the pET30a-L10 plasmid were grown in LB media containing 50 μg/mL kanamycin at 37 °C. The expression of L10 was induced by addition of 0.1 mmol/L isopropyl β-d-1-thiogalactopyranoside (IPTG) for 10 h at 37 °C. Cells were then resuspended in binding buffer (20 mmol/L sodium phosphate, 500 mmol/L NaCl, 40 mmol/L imidazole, pH 7.4) and disrupted by high pressure systems. After centrifugation at 12,000×g for 60 min to remove debris, the supernatant was loaded onto a column of Ni2+ His-Trap HP (GE Healthcare), and attached His-tagged L10 proteins were then eluted using a linear imidazole gradient in elution buffer (20 mmol/L sodium phosphate, 500 mmol/L NaCl, 100–500 mmol/L imidazole, pH 7.4). Protein level were determined by 15% SDS-PAGE followed by coomassie blue staining. For the purified His-tagged L10, the concentration was measured by BCA method and confirmed by western blotting using anti-His antibody. We also used E. coli BL21 (DE3) to express His-tagged L12 protein. The recombinant strain was grown in LB media containing 100 μg/mL ampicillin at 37 °C. His-tagged L12 protein was expressed and purified using the same method.
The E. coli Rosetta (DE3) cells containing pGEX-4T-1-L12 were grown in LB, and the expression of L12 was induced by auto-inducible ZYM-5052 media at 20 °C overnight18. GST-tagged L12 was purified with GST-Trap HP (GE Healthcare). The binding buffer contained 140 mmol/L NaCl, 2.7 mmol/L KCl, 10 mmol/L Na2HPO4 and 1.8 mmol/L KH2PO4 (pH 7.3) and the elution buffer comprised 50 mmol/L Tris and 10 mmol/L reduced glutathione (pH 8.0). The purified proteins were confirmed by Western blotting using anti-GST antibody. For GST protein, the plasmid pGEX-4T-1 was transformed into E. coli Rosetta (DE3) cells, and then GST protein was expressed and purified using the same method.
2.6. GST pull-down assay
GST pull-down was used to determine if IMB-84 and IMB-87 inhibit L12–L10 interaction in vitro. After washing three times with working buffer (150 mmol/L NaCl, 50 mmol/L Tris, 1% Triton X-100, pH 7.5), 30 µL glutathione sepharose beads (GE Healthcare) were incubated with 4 µg/mL GST-tagged L12 in 500 µL working buffer for 2 h at 4 °C. Beads were collected by centrifugation and resuspended in 500 µL working buffer with 4 µg/mL His-tagged L10 and various concentrations of IMB-84 or IMB-87 from 0 to 25 μg/mL. The reaction mixtures were incubated at 4 °C for 4 h and the beads were washed three times with working buffer. Finally, proteins were solubilized and separated by SDS-PAGE and probed with anti-His and anti-GST primary antibodies. After blotting with horseradish peroxidase (HRP)-conjugated secondary antibody and exposure, the protein levels were shown and quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Results were analyzed by the Student's t-test. P values of less than 0.05 were considered statistically significant. Reactions were treated with 1% DMSO as the positive control and GST-tagged L12 was replaced by GST protein for the negative control.
2.7. SPR assay
The SPR assay were performed using a Biacore T100 system (GE Healthcare) at 25 °C in a HBS-P+ running buffer (10 mmol/L HEPES, 150 mmol/L NaCl, 0.05% surfactant P20, pH 7.4 and 5% DMSO). A Ni2+-nitrilotriacetic acid (NTA) sensor chip sensor chip was primed and loaded with Ni2+, and then purified His-tagged L12 (2 μg/mL) was captured on the sensor chip by chelation of Ni2+ in HBS-P+ buffer, the ligand density is ~1700 response units (RU). To determine the binding affinity of His-tagged L12 and the compounds, solutions of compounds IMB-84 and IMB-87 (5% DMSO) at a given concentration were injected into the L12 immobilized chambers. The surface was regenerated with 350 mmol/L EDTA and 250 mmol/L imidazole. Before a new assay, the sensor chip surface was reconstructed with fresh L12 proteins. The concentrations of compounds were ranging from 3.125 to 50 μmol/L. On the other hand, purified His-tagged L10 (3 μg/mL) was immobilized onto the NTA sensor chip, with a ligand density of ~1500 RU. For the binding affinity assays, the concentrations of compounds were ranging from 1.5625 to 25 μmol/L.
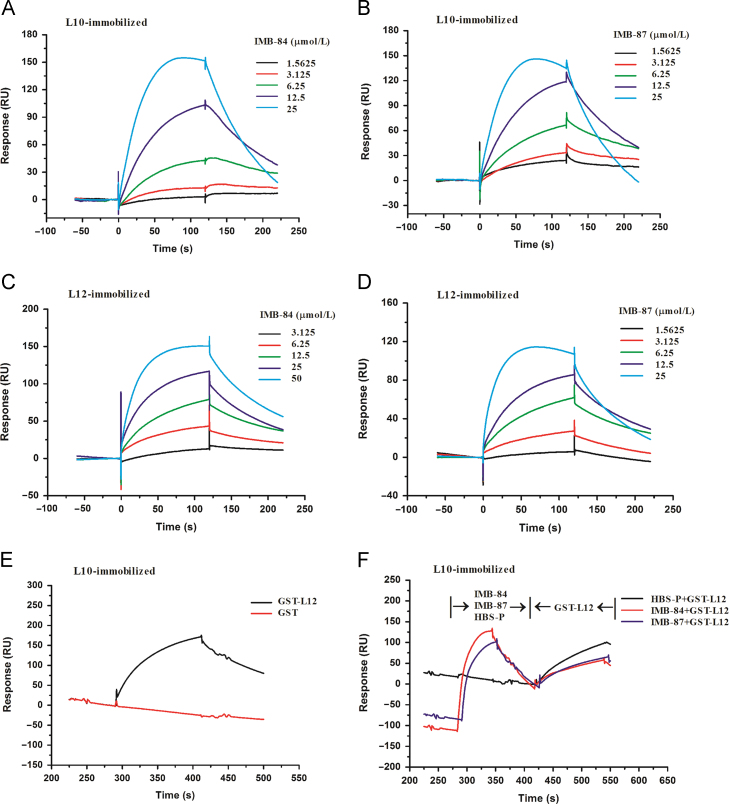
To examine the block of L12–L10 interaction by compounds IBM-84 and IBM-87, GST, GST-tagged L12 and His-tagged L10 were used. The interaction between GST-tagged L12 and His-tagged L10 was first determined and the possibility of non-specific interactions excluded. For this purpose, 0.1 μmol/L GST or GST-tagged L12 was injected to ligand L10 (~500 RU) that is immobilized on sensor surface. The interaction between L12 or GST and L10 was represented by RU. To examine the inhibition of L12–L10 interaction by IMB-84 or IMB-87, 25 μmol/L compounds was injected to saturate ligand L10 (~400 RU) that is immobilized on sensor surface. Then GST-tagged L12 (0.05 μmol/L) was injected. In the control assay, GST-tagged L12 was injected after the injection of HBS-P+ running buffer.
2.8. The assay for the inhibition of GTPase activity
The GTP hydrolysis activity of ribosomes was assessed as previously described19, 20. To determine ribosome-stimulated GTP hydrolysis by EF-G, 0.2 µmol/L ribosome extract and 0.4 µmol/L EF-G were added to the reaction buffer, which contains 50 mmol/L Tris–HCl (pH 7.5), 70 mmol/L NH4Cl, 30 mmol/L KCl, and 7 mmol/L MgCl2. The reaction was initiated by the addition of 1 mmol/L GTP and incubated at 37 °C for 30 min. The reaction was terminated by the addition of 100 μL malachite green and developed for 30 min at room temperature and the absorbance was then measured at 620 nm. To detect the inhibition of GTPase activity by IMB-84 and IMB-87, various concentrations of compounds (0.16–640 μg/mL) were added into the reaction mixtures and incubated at 37 °C for 15 min and the absorbance was then measured.
2.9. In vitro protein synthesis inhibition assay
An in vitro cell-free translation system and a luciferase reporter were used to assess protein synthesis inhibition by compound IMB-84 and IMB-87. The E. coli S30 ribosome extract or rabbit reticulocyte, nucleotide triphosphates mixture, amino acids, and a reporter plasmid with luciferase gene were included in the translation system according to manufacturer׳s instruction (Promega). Light emission was recorded with a luminescence counter (PerkinElmer, Wellesley, MA, USA). The final concentrations of compounds range from 0.08 to 625 μg/mL. The IC50 was determined on the basis of the ratio of light emission units to the concentration of compounds (log plots) that fits to a variable-slope dose–response equation.
2.10. Determination of the MIC for E. coli BL21 (DE3) overexpressing L12 or L10
Recombinant plasmids pET16b-L12 and pET30a-L10 were transformed into E. coli BL21 (DE3) and then the expression of recombinant proteins was verified by Western blotting with anti-His antibody. The MICs of compounds IMB-84 and IMB-87 against E. coli BL21 (DE3) overexpressing L12 or L10 were determined as described previously21, 22. In short, cells were grown in LB with 100 μg/mL ampicillin or 50 μg/mL Kanamycin at 37 °C (OD600=0.5) and 0.5 mmol/L IPTG was added to the culture. After 6 h, cultures were diluted with LB/ampicillin or kanamycin/IPTG medium (OD600=0.08) and used for MIC assay as described above. The BL21 strains with control vectors (pET16b, pET30a) and expression vector with non-related gene (pET16b-L11, pET30a-DHQS) were used as a negative control.
2.11. Determination of anti-E. coli activity
The anti-E. coli activities of compounds IMB-84 and IMB-87 were measured using broth microdilution assay23. The final concentrations of compounds ranged from 1 to 64 μg/mL. All the E. coli cells, including ATCC 25922 and clinical isolated strains were grown to mid-log phase in Mueller–Hinton broth and then diluted with the same medium to the density of 5×105 CFU/mL. Then the cells with gradient concentrations of compounds were dispensed at 0.2 mL/well in sterile 96-well microplates. After incubation at 37 °C for 20 h, the MIC was recorded. The first-line of anti-Gram-negative drugs, cefoxitin, meropenem and colistin, were used as reference.
2.12. Mode of action: bacteriostatic vs. bactericidal
E. coli ATCC 25922 was grown in LB medium to mid-log phase. The cultures were diluted to 5×105 CFU/mL in fresh medium and compound concentrations ranged from 0 to 8× MIC. The bacteria were collected every hour, serially diluted and spread onto plates. The colonies were counted after grown in 37 °C for 12 h.
3. Cytotoxicity assay
Human embryonic kidney 293 cells were used to detect the cytotoxic effect of IMB-84 and IMB-87. The cells were seeded in triplicate in 96 well plates and the cell density was 5×103 cells per well. Cells were incubated for 24 h at 37 °C until reached to log-phase. Then, the cells were incubated with compounds at gradient concentrations ranging from 3.125 to 100 μg/mL in the DEME medium without fetal bovine serum. After incubation for 48 h, the medium was removed and fresh medium were added. Cells were incubated for 24 h at 37 °C, MTT dye reagent was then added and incubation for 4 h. After added 50 μL of DMSO, the absorbance was measured at 570 nm. The IC50 values were calculated on a concentration–response curve.
4. Result
4.1. Detection of the interaction between E. coli ribosomal proteins L12 and L10 by yeast two-hybrid assay
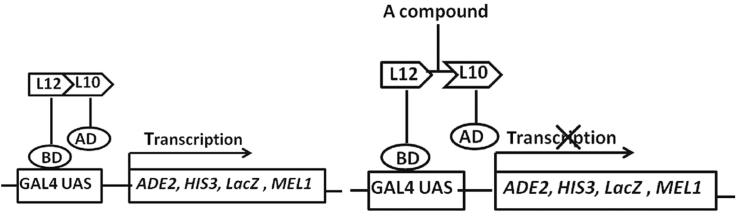
Ribosomal proteins L12 and L10 interact with each other24, 25, and this interaction has been further verified by the crystal structure of the ribosome stalk26, 27. To identify compounds that disrupt L12–L10 interaction, we first constructed the yeast two-hybrid system to detect this interaction. For this purpose, the plasmids pAD-L12, pAD-L10, pBD-L12 and pBD-L10 were constructed. A pair of plasmids of pAD-L12 and pBD-L10 or pAD-L10 and pBD-L12 was introduced into AH109 yeast strain28. The interaction between L12 and L10 is expected to activate the transcription of three reporter genes ADE2, HIS3 and LacZ in AH109 cells. Thus, the L12–L10 interaction can be determined based on the growth of yeast cells on the plates lacking adenine and histidine, as well as LacZ-dependent color change after incubation in the presence of LacZ substrates (Fig. 1A).
Figure 1.
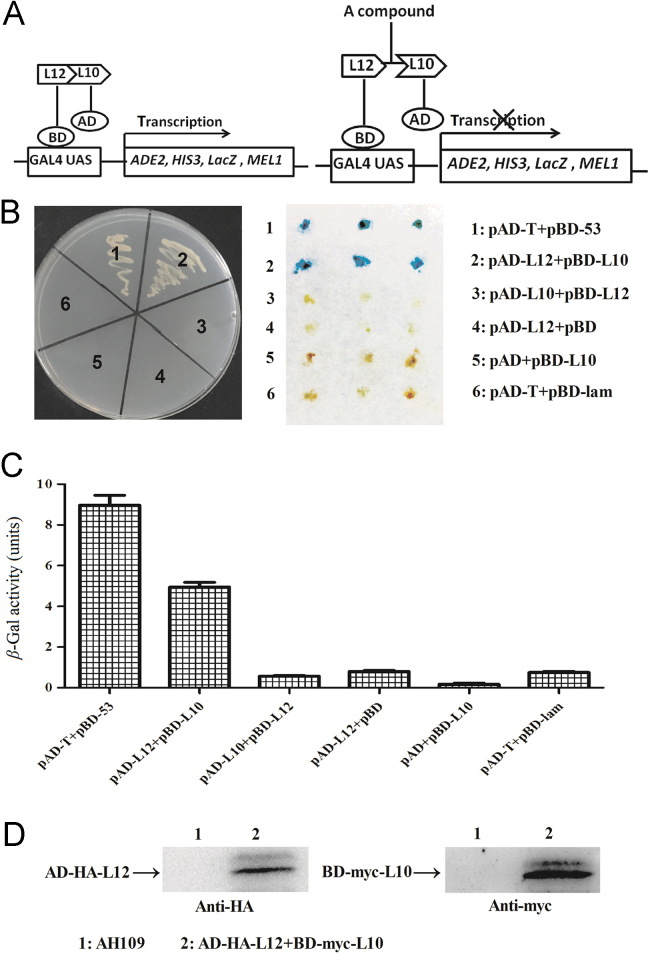
Detection of L10–L12 interaction using yeast two-hybrid assay. (A) The rationale for high throughput screen using yeast two-hybrid system. L10–L12 interaction reconstitutes the function of the Gal4 protein and enables the expression of reporter genes, ADE2, HIS3, and LacZ. The disruption L12–L10 interaction by a compound inhibits yeast cell growth in SD/Ade/His dropout medium and β-gal will not be produced. (B) The growth and β-gal activity of yeast cells expressing various combinations of BD and AD fusions. The combination of the different plasmids in AH109 strains is shown on the right. Among them, strains 1 and 6 are the positive and negative controls, respectively. The left panel shows the growth of yeast cells with indicated plasmids on a SD/Leu/Trp/Ade/His dropout plate. The right panel shows the β-gal activity of indicated strains. (C) Quantification of β-gal activity in yeast cells containing various combinations of plasmids. The results are the average from triplicate assays. (D) The expression of L12 and L10 proteins in yeast cells. L12 and L10 fusion proteins were expressed in yeast cells, and the expression was detected using anti-HA and anti-Myc antibodies.
As the positive control, AH109 yeast cells with plasmids pAD-T and pBD-53 grew well on a SD/Ade–His dropout plate and showed strong β-gal activity because of the interaction of expressed proteins p53 and SV40-T, but AH109 cells with pAD-T and pBD-lam (human lamin C) did not grow on SD/-Ade–His dropout plates, which was used as a negative control29 (Fig. 1B and C). AH109 cells with plasmids pAD-L12 and pBD-L10 grew well on a SD/-Ade–His plate and were positive for β-gal activity, indicating the interaction between L12 and L10. Surprisingly, AH109 with pAD-L10 and pBD-L12 plasmids neither grew on the dropout plates nor showed β-gal activity. One possible explanation is that the fusion of the Gal4 DNA binding domain at the N terminus of L12 protein affects its interaction with L10 as described previously15. Self-activation was excluded, because yeast cells harboring either pAD-L12 or pBD-L10 alone did not show growth and were negative for β-gal activity (Fig. 1B and C). We also used Western blotting to validate the expression of L12 and L10 proteins in yeast cells (Fig. 1 D). Therefore, we established the yeast two-hybrid system to detect the interaction between E.coli L12 and L10, which could be used for drug screening.
4.2. Screening for the compounds blocking L12–L10 interaction
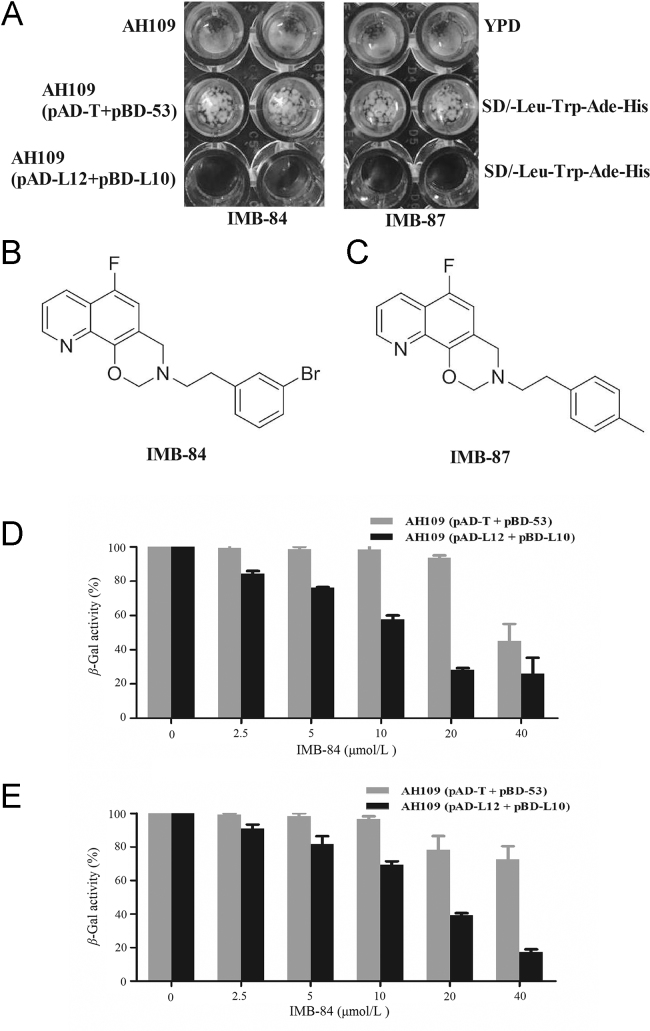
To screen for the compounds that can block the interaction between E. coli L12 and L10 proteins, the growth of AH109 (pAD-L12+pBD-L10) was assessed in 96-well plates in the presence of compounds at 25 μg/mL. If a compound disrupts the Gal4 expression system or shows antifungal activity, the growth of AH109 (pAD-L12+pBD-L10) will be inhibited by this compound. However, this inhibition will be non-specific and the growth of AH109 (pAD-T+pBD-53) cells will also be inhibited by this compound. To exclude possibilities, the growth of AH109 (pAD-T+pBD-53) and AH109 were assessed as well. The compounds that show specific growth inhibition for AH109 (pAD-L12+pBD-L10), but not AH109 (pAD-T+pBD-53) or AH109 were selected (Fig. 2A). With this method, six compounds were identified.
Figure 2.
Identification of compounds that block L12–L10 interaction. (A) Growth inhibition of yeast cells by specific inhibitors. Yeast strains with indicated plasmids were inoculated into SD/Leu/Trp/Ade/His dropout medium in 96-well plates, AH109 cells were inoculated into YPD medium. The compound was added into one of the two wells for each strain at the concentration of 25 μg/mL and the growth was examined after 48 h at 30 °C. (B) and (C) The structure of compounds IMB-84 and IMB-87. (D) and (E) The inhibition of β-gal activity of AH109 (pAD-L12 + pBD-L10) cells by IMB-84 (D) and IMB-87 (E) at various concentrations. Strain AH109 (pAD-T+pBD-53) was used as a control. Values represent the β-gal activity ratio of cells treated with the compounds over that in untreated cells. The results are the average of triplicate assays.
To further confirm that the identified compounds selectively block L12–L10 interaction, we performed a quantitative β-gal assay with different concentrations of the selected compounds. Two of them, IMB-84 {6-fluoro-3-(3-bromophenethyl)-3,4-dihydro-2H-[1,3]oxazine[5,6-h]quinoline} and IMB-87{6-fluoro-3-(4-methylphenethyl)-3,4-dihydro-2H-[1,3]oxazine[5,6-h]quinoline}, inhibited the β-gal activity of AH109 (pAD-L12 + pBD-L10) in a dose-dependent manner, while they showed no effect on β-gal activity of strain AH109 (pAD-T + pBD-53) (Fig. 2D and E). Therefore, IMB-84 and IMB-87 were selected for further study and their structures are shown in Fig. 2B and C.
4.3. Compounds IMB-84 and IMB-87 disrupt L12–L10 interaction in vitro
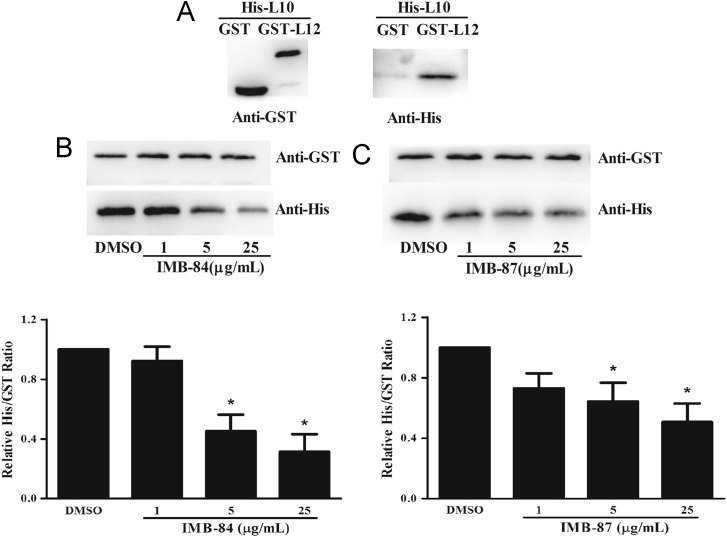
The disruption of L12–L10 interaction by IMB-84 and IMB-87 was further examined using GST pull-down assays, which is widely used to determine protein–protein interactions30. For this purpose, we first expressed 6×His-tagged L10, GST-tagged L12 and GST proteins in E. coli and the purified proteins were used to examine their interaction (Supplementary Information Fig. S1). A specific protein band (Anti-His) was observed in the resulting pull-down products when GST-L12 protein (bait) but not GST protein was incubated with His-L10 protein, which confirmed L12–L10 interaction in vitro (Fig. 3A). To determine whether compounds IMB-84 and IMB-87 disrupt L12–L10 interaction, GST-L12 protein (bait) and His-tagged L10 were incubated with these compounds and then the level of the His-tagged L10 protein in the resulting pull-down products was detected by Western blotting using the anti-His antibody. The product treated with DMSO was used as a positive control. Strikingly, the level of the His-tagged L10 protein from the compound-treated pull-down products decreased significantly in a dose-dependent manner (Fig. 3B and C). These data collectively demonstrate that compounds IMB-84 and IMB-87 disrupt L12–L10 interaction in vitro.
Figure 3.
Disruption of L12–L10 protein interaction by IMB-84 and IMB-87. (A) Demonstration of L10–L12 interaction by GST pull-down assay. Purified GST-L12 or GST proteins were bound to glutathione-sepharose beads, and then incubated with purified His-tagged L10. Here show protein bands of resulting pull-down products after western blotting with anti-GST and anti-His antibodies. (B) and (C) Compounds IMB-84 and IMB-87 block L12–L10 interaction. Compound IMB-84 or IMB-87 was incubated with GST-L12 proteins bound to glutathione-sepharose beads, and then His-tagged L10 was added. Equal volume of DMSO was added as control. Protein bands of resulting pull-down products are shown after Western blotting with anti-GST and anti-His antibodies. Data are shown as the mean±SD, n=3. *P<0.05 vs control group.
Next, we assessed the binding of compounds IMB-84 and IMB-87 to L12 and L10 proteins as well as the effect of these compounds on L12–L10 interaction by SPR, which has been widely used to measure the interaction between small molecules and proteins in real time. 6×His-tagged L10 and 6×His-tagged L12 proteins were expressed and purified successfully (Fig. S1). Both IMB-84 and IMB-87 bound to L10 as indicated by the changes in response units with the KD of 20 μmol/L for IMB-84 and 6.7 μmol/L for IMB-87 (Fig. 4A and B). Moreover, these two compounds were able to bind to L12 with the KD of 13.8 μmol/L for IMB-84 and 16 μmol/L for IMB-87 (Fig. 4C and D). In addition, the two compounds decreased the interaction between L12 and L10 when GST-L12 proteins flowed through the L10-coated sensor chip (Fig. 4E and F).
Figure 4.
SPR analysis for the binding of compounds IMB-84 and IMB-87 to proteins L12 and L10 and their effect on L12–L10 protein interaction. (A) and (B) Compounds IMB-84 and IMB-87 bind to L10 protein. Solutions with series concentrations of IMB-84 (A) or IMB-87 (B) were injected into the L10-coated sensor chip chamber. The change of response units is shown. (C) and (D) Compounds IMB-84 and IMB-87 bind to L12 protein. Solutions with series concentrations of IMB-84 (C) or IMB-87 (D) were injected into the L12-coated sensor chip chamber. The change of response units is shown. (E) Binding of the GST and GST-L12 to L10 protein. Solutions of GST or GST-L12 were injected into the L10-coated sensor chip chamber. The change of response units is shown. (F) Compounds IMB-84 and IMB-87 block L12–L10 interaction. IMB-84 or IMB-87 (25 μmol/L) was injected into the L12-coated sensor chip chamber. Then purified L10 protein was injected. As a control, HBS-P+ was injected at first and followed by the injection of L10. The change of response units was measured.
4.4. IMB-84 and IMB-87 inhibit ribosomal GTPase activity and ribosome-dependent protein synthesis
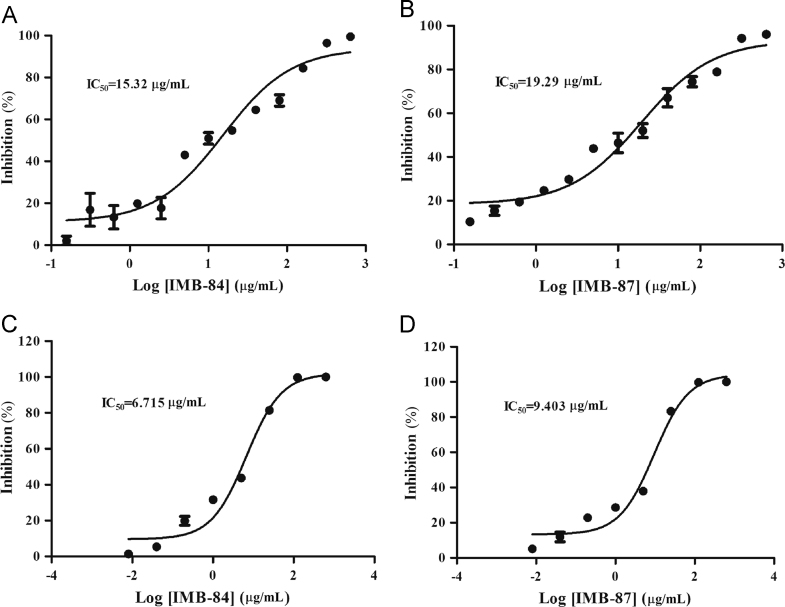
The disruption of L12–L10 interaction disables the binding of EF-G to the stalk and causes the loss of ribosomal GTPase activity12. Because IMB-84 and IMB-87 block L12–L10 interaction in vitro, we speculate that these compounds will decrease the ribosome mediated GTPase activity of EF-G. To test this, an in vitro system was used to detect the GTPase activity in E. coli. In this assay, GTPase activity was measured on the basis of the release of Pi from ribosome-bound EF-G·GTP, and the free Pi subsequently forms complex with malachite green, which allows the detection of enzyme activity. As expected, both IMB-84 and IMB-87 showed inhibition on the GTPase activity of E. coli in a dose-dependent manner, and the IC50 values were 15.32 and 19.29 μg/mL, respectively (Fig. 5A and B).
Figure 5.
In vitro inhibition of GTPase activity and protein translation by IMB-84 and IMB-87. (A) and (B) Compounds IMB-84 and IMB-87 reduce ribosome GTPase activity. The GTPase activity was assessed by measuring free Pi from GTP using malachite green solution. (C) and (D) Inhibition of protein translation by IMB-84 and IMB-87. Inhibition was determined using cell-free translation system containing E. coli or rabbit reticulocyte, and luciferase was used as a reporter.
The GTPase activity is essential for ribosome-dependent protein synthesis. Therefore, we further used an in vitro transcription–translation system to determine whether IMB-84 and IMB-87 inhibit ribosome-mediated protein synthesis in E. coli and rabbit reticulocyte. In this assay, compounds were added to E. coli or rabbit reticulocyte cell-free transcription–translation system with a luciferase reporter. The results showed that IMB-84 and IMB-87 showed an inhibitory effect on protein translation for E. coli at IC50 of 6.715 and 9.403 μg/mL, respectively. Also, the inhibition is dose-dependent (Fig. 5C and D). But for rabbit reticulocyte, we did not detect any inhibitory effect of these compounds until their concentration reached as high as 125 μg/mL, indicating the specific inhibition of protein synthesis in E. coli by these compounds.
4.5. The anti-E. coli activity of IMB-84 and IMB-87
Because the L12–L10 interaction is essential for ribosome function, we speculate that disruption of this interaction will inhibit the growth of E. coli cells. Indeed, both IMB-84 and IMB-87 showed a MIC at 2 and 4 μg/mL against E. coli ATCC 25922, respectively (Table 1). For some clinical drug-resistant E. coli strains, IMB-84 and IMB-87 showed growth inhibition with MICs ranging from 1 to 64 μg/mL (Table 1). Since L12 and L10 proteins exhibit low homology with the corresponding human proteins, we also examined the growth inhibition of IMB-84 and IMB-87 to human cells. We showed that the IC50s of IMB-84 and IMB-87 for human embryonic kidney 293 cells were 15 and 24 μg/mL, respectively, indicating that human cells are less sensitive to these compounds.
Table 1.
MICs of IMB-84 and IMB-87 against E. coli strains.
| Strains | MIC (μg/mL) |
||||
|---|---|---|---|---|---|
| IMB-84 | IMB-87 | Cefoxitin | Meropenem | Polymyxin | |
| 259922 | 2 | 4 | 2 | <0.5 | <0.5 |
| ZG2016M78-1 | 64 | 64 | 2 | <0.5 | 256 |
| ZP2016MH3-1 | 16 | 8 | 512 | 64 | 4 |
| ZP2016M106-1 | 16 | 8 | >512 | <0.5 | 2 |
| ZP2016CH6-2 | 16 | 16 | 512 | 32 | 4 |
| ZP2016CH5-1 | 16 | 16 | 512 | 64 | 1 |
| ZP2016CH8-1 | 16 | 16 | 512 | 128 | 4 |
| ZP2016CH9-1 | 32 | 16 | >512 | 64 | 2 |
| ZP2016CH6-1 | 16 | 8 | 512 | 32 | 8 |
ZG2016M78-1, ZP2016MH3-1, ZP2016M106-1, ZP2016CH6-2, ZP2016CH5-1, ZP2016CH8-1, ZP2016CH9-1 and ZP2016CH6-1 are all clinical drug-resistant E. coli strains. 25922: ATCC25922 E. coli.
4.6. Higher MICs for strains overexpressing L12 and L10
If L12–L10 interaction is the target of IMB-84 and IMB-87 for their antibacterial activity, high level of L10 or L12 protein expression will alleviate the antibacterial activity of these compounds. Therefore, we constructed expression plasmids containing genes encoding L12 or L10 protein. We transformed the expression plasmids pET16b-L12 and pET30a-L10 into E. coli BL21 (DE3). Expression of the recombinant proteins was confirmed by western blotting with anti-His antibody (Supplementary Information Fig. S2). For IMB-84, the MICs of strains overexpressing L12 or L10 were 25 and 12.5 µg/mL, which was 4-fold or 2-fold of the strains with the control vector. The MICs of IMB-87 for strains overexpressing L12 or L10 were both 12.5 µg/mL, which is 2-fold of the control strain (Table 2).
Table 2.
Antibacterial activity of compounds IMB-84 and IMB-87 against E. coli strains with a control vector or plasmids overexpressing L12, L10, L11 and DHQS.
| Strains | MIC (μg/mL) |
|
|---|---|---|
| IMB-84 | IMB-87 | |
| Vector control (pET16b) | 6.25 | 6.25 |
| L12 overexpression | 25 | 12.5 |
| L11 overexpression | 6.25 | 6.25 |
| Vector control (pET30a) | 6.25 | 6.25 |
| L10 overexpression | 12.5 | 12.5 |
| DHQS overexpression | 6.25 | 6.25 |
4.7. The mode of growth inhibition of E. coli by compounds IMB-84 and IMB-87
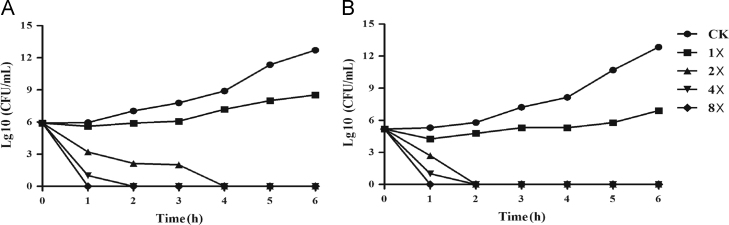
IMB-84 and IMB-87 may inhibit the growth of E. coli through bacteriostatic or bactericidal effect. We examined the viability of E. coli 25922 over time in the presence of various concentrations of IMB-84 and IMB-87. In the presence of 1-fold of MIC of IMB-84 and IMB-87, the two compounds showed bacteriostatic effect and the number of colonies increased very slowly during the time course for 5 h. But these compounds showed bactericidal activity against this strain at 2-fold of MIC and the activity was increased significantly when the concentrations were at 4- and 8-fold of MIC (Fig. 6). After 1 h of incubation, almost all of the bacteria were killed in the presence of compounds at 8-fold MIC, indicating a strong bactericidal activity of these compounds against E. coli.
Figure 6.
Dose-dependent bactericidal activity of compounds IMB-84 and IMB-87 against E. coli 25922. The MICs of IMB-84 and IMB-87 for E. coli 25922 is 2 and 4 μg/mL, respectively. The viability of E. coli 25922 was determined after treatment with IMB-84 (A) and IMB-87 (B) at 1- to 8-fold of MIC.
5. Discussion
The interaction between L12 and L10 proteins is critical for ribosome function, therefore, the abolishment of L12–L10 interaction will disrupt the ribosome function and block protein synthesis. Several compounds were found to inhibit the interaction between L12 and L10 proteins from M. tuberculosis. Their anti-TB activities indicate that L12–L10 interaction could be a target for anti-TB agents15, 16. The development of drug resistant Gram-negative bacteria is a serious clinical problem that causes increased morbidity and mortality. The purpose of this study is to identify some compounds that target L12–L10 interaction in Gram-negative bacteria, such as E. coli, using yeast two-hybrid system. With this system, two compounds IMB-84 and IMB-87 were identified, and we further demonstrated that they block L12–L10 interaction by a series of assays including protein synthesis inhibition, GTPase activity inhibition, SPR and GST pull-down. More importantly, these two compounds showed potent inhibitory activity against E. coli ATCC 25922, and exhibited antibacterial activity against some drug-resistant strains as well. In summary, we have identified two anti-E. coli compounds that likely target L12–L10 interaction.
The clinical isolates used in this study showed some extent resistance to compounds IMB-84 and IMB-87 as indicated by increased MIC value. Compared with the corresponding control drugs, the increase of MIC is less significant, which suggests that the antibacterial mechanism of these two compounds is likely different from that of the existing antibiotics. For some clinical multidrug resistant E. coli strains, the mechanism of drug resistance could be reduced drug permeability or enhanced drug efflux, in addition to the mutation of drug targets. Therefore, more experiments are needed to clarify if the resistance to compounds IMB-84 and IMB-87 is due to reduced permeability or increased efflux.
In general, the inhibitory activity of a compound to the target should correlate with its anti-bacterial activity. We noticed that the IC50 of IMB-84 and IMB-87 for both GTPase activity and cell-free protein synthesis are higher than their MIC for E. coli ATCC 25922. One possible explanation is the difference between the extracellular and intracellular environments, which affect the activities of these compounds. Another possibility is that IMB-84 and IMB-87 are multi-target compounds, and these compounds may show lower MICs toward unidentified targets.
Compounds IMB-84 and IMB-87 inhibit the GTPase activity of E. coli cell ribosomal extracts, and the protein sequences between the E. coli elongation factor G and human homologue share 39% similarity31, 32 Moreover, these compounds inhibit the growth of human embryonic kidney 293 cells with IC50 of 15 and 24 μg/mL. The GTPase activity of the elongation factor G is essential for protein translation, so an open question is whether the cytotoxicity is attributed to the inhibition of the GTPase activity by these compounds. IMB-84 and IMB-87 inhibit protein translation in E. coli ribosomal extract at IC50 of 6.715 and 9.403 μg/mL, respectively. However, these compounds do not inhibit protein translation in the cell ribosomal extract of rabbit reticulocyte when their concentration is as high as 125 μg/mL. The selective inhibition of protein translation in bacteria by these two compounds favors the possibility that they inhibit L12–L10 interaction, which subsequently blocks EF-G recruitment onto the ribosome. Compare to the elongation factor, The L12 and L10 proteins are much less conserved between bacteria and mammalian cells33. Therefore, we speculate the compounds may inhibit the growth of mammalian cells through other targets but not the GTPase or L12–L10 interaction. Our future work is to use these compounds as a lead to synthesize more efficient antibacterial molecules with low toxicity and high selectivity by structural modification. Furthermore, the selective inhibition of protein synthesis by IMB-84 and IMB-87 confirms that L12–L10 interaction could be used for the identification of anti-Gram-negative bacteria agents.
Our SPR data showed the RU values for compounds IMB-84 and IMB-87 that bind to a protein-coated surface, but the RU values appear high for such small molecules. These two compounds may bind to the chip or the protein non-specifically. For the SPR experiment, the binding of these two compounds to the chip without protein coating was examined and the value was used as a negative control. Therefore, the RU values shown in the figure is the difference between RUprotein and RUcontrol. By doing so, we exclude the possibility that the high RU value is a result of non-specific binding to the chip. The interaction of IMB-84 or IMB-87 with other irrelevant proteins LptA and BamB were also examined, but no interaction was detected, indicating that the interaction of IMB-84 or IMB-87 with L12 and L10 should be specific. However, further studies are needed to determine the detailed binding mode between the two compounds and L12, L10 proteins.
In summary, our research work demonstrates that L12–L10 interaction is likely a feasible drug target for Gram-negative bacteria, such as E. coli. Moreover, we identified two compounds IMB-84 and IMB-87 that inhibit L12–L10 interaction and show antibacterial activity. Although the anti-bacteria activity and safety of these compounds are not ideal, it is promising to find more potent anti-E. coli drug candidates after structural modification of these compounds. Our work also demonstrates that the combination of yeast-two-hybrid system and GST pull-down assay will be a useful tool to discover compounds that disrupt a specific protein–protein interaction.
Acknowledgements
We thank Prof. Juan Li and Dr. Min Yuan from the Chinese Center for Disease Control and Prevention (CCDC) for helping us test the anti-E. coli activity of compounds. We would like to thank Prof. Xuefu You at Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College for his team to help us to test the MIC of compounds against clinical resistant E. coli stains. This work was supported by the National Natural Science Foundation of China (Grant nos. 81370089, 81529003, 81621064 and 81361138020) and the Foundation for Innovative Research Groups and the Funds for International Cooperation and Exchange between China–Sweden and CAMS Initiative for Innovative Medicine (2016-12M-3-014).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.apsb.2018.07.006.
Contributor Information
Yan Li, Email: 13391513480@163.com.
Shuyi Si, Email: sisyimb@hotmail.com.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
References
- 1.Vincent J.L., Rello J., Marshall J., Silva E., Anzueto A., Martin C.D. International study of the prevalence and outcomes of infection in intensive care units. J Am Med Assoc. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q., Lambert G., Liao D., Kim H., Robin K., Tung C.K. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 3.Wellington E.M., Boxall A.B., Cross P., Feil E.J., Gaze W.H., Hawkey P.M. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhary A.S. A review of global initiatives to fight antibiotic resistance and recent antibiotics׳ discovery. Acta Pharm Sin B. 2016;6:552–556. doi: 10.1016/j.apsb.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown E.D., Wright G.D. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 6.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 7.Nambiar S., Laessig K., Toerner J., Farley J., Cox E. Antibacterial drug development: challenges, recent developments, and future considerations. Clin Pharmacol Ther. 2014;96:147–149. doi: 10.1038/clpt.2014.116. [DOI] [PubMed] [Google Scholar]
- 8.Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 9.Fourmy D., Recht M.I., Blanchard S.C., Puglisi J.D. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 10.Aleksandrov A., Simonson T. Molecular dynamics simulations of the 30S ribosomal subunit reveal a preferred tetracycline binding site. J Am Chem Soc. 2008;130:1114–1115. doi: 10.1021/ja0741933. [DOI] [PubMed] [Google Scholar]
- 11.Oleinikov A.V., Perroud B., Wang B., Traut R.R. Structural and functional domains of Escherichia coli ribosomal protein L7/L12. The hinge region is required for activity. J Biol Chem. 1993;268:917–922. [PubMed] [Google Scholar]
- 12.Savelsbergh A., Mohr D., Wilden B., Wintermeyer W., Rodnina M.V. Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J Biol Chem. 2000;275:890–894. doi: 10.1074/jbc.275.2.890. [DOI] [PubMed] [Google Scholar]
- 13.Burma D.P. Conformational change of 23S RNA in 50s ribosome is responsible for translocation in protein-synthesis. J Biosci. 1984;6:419–430. [Google Scholar]
- 14.Hagiya A., Naganuma T., Maki Y., Ohta J., Tohkairin Y., Shimizu T. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J Biol Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y., Li Y., Zhu Y., Zhang J., Li Y., Liu X. Identification of antituberculosis agents that target ribosomal protein interactions using a yeast two-hybrid system. Proc Nat Acad Sci U S A. 2012;109:17412–17417. doi: 10.1073/pnas.1110271109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y., Li Y., Zhu N., Han Y., Jiang W., Wang Y. The antituberculosis antibiotic capreomycin inhibits protein synthesis by disrupting interaction between ribosomal proteins L12 and L10. Antimicrob Agents Chemother. 2014;58:2038–2044. doi: 10.1128/AAC.02394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gietz R.D., Schiestl R.H., Willems A.R., Woods R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 18.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Ash M.R., Maher M.J., Guss J.M., Jormakka M. The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure. PLoS ONE. 2011;6:e23355. doi: 10.1371/journal.pone.0023355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savelsbergh A., Mohr D., Kothe U., Wintermeyer W., Rodnina M.V. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baum E.Z., Crespo-Carbone S.M., Foleno B.D., Simon L.D., Guillemont J., Macielag M. MurF inhibitors with antibacterial activity: effect on muropeptide levels. Antimicrob Agents Chemother. 2009;53:3240–3247. doi: 10.1128/AAC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yogiara Kim D., Hwang J.K., Pan J.G. Escherichia coli ASKA clone library harboring tRNA-specific adenosine deaminase (tadA) reveals resistance towards xanthorrhizol. Molecules. 2015;20:16290–16305. doi: 10.3390/molecules200916290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng C.J., Sohn M.J., Kim W.G. Vinaxanthone, a new FabI inhibitor from Penicillium sp. J Antimicrob Chemother. 2009;63:949–953. doi: 10.1093/jac/dkp058. [DOI] [PubMed] [Google Scholar]
- 24.Pettersson I. Studies on the RNA and protein binding sites of the E. coli ribosomal protein L10. Nucleic Acids Res. 1979;6:2637–2646. doi: 10.1093/nar/6.7.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson D.N., Nierhaus K.H. Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol. 2005;40:243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 26.Mitra K., Schaffitzel C., Fabiola F., Chapman M.S., Ban N., Frank J. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol Cell. 2006;22:533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff L., Berninghausen O., Beckmann R. Molecular basis for the ribosome functioning as an l-tryptophan sensor. Cell Rep. 2014;9:469–475. doi: 10.1016/j.celrep.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 28.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B., Fields S. Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- 30.Vikis H.G., Guan K.L. Glutathione-S-transferase-fusion based assays for studying protein–protein interactions. Methods Mol Biol. 2004;261:175–186. doi: 10.1385/1-59259-762-9:175. [DOI] [PubMed] [Google Scholar]
- 31.Zengel J.M., Archer R.H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984;12:2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanes J., Freudenstein J., Rapp G., Scheit K.H. Construction of a plasmid containing the complete coding region of human elongation factor 2. Biol Chem Hoppe Seyler. 1992;373:201–204. doi: 10.1515/bchm3.1992.373.1.201. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalo P., Reboud J.P. The puzzling lateral flexible stalk of the ribosome. Biol Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.