Abstract
Long time-lapse super-resolution imaging in live cells requires a labeling strategy that combines a bright, photostable fluorophore with a high-density localization probe. Lipids are ideal high-density localization probes, as they are >100 times more abundant than most membrane-bound proteins and simultaneously demark the boundaries of cellular organelles. Here, we describe Cer-SiR, a two-component, high-density lipid probe that is exceptionally photostable. Cer-SiR is generated in cells via a bioorthogonal reaction of two components: a ceramide lipid tagged with trans-cyclooctene (Cer-TCO) and a reactive, photostable Si-rhodamine dye (SiR-Tz). These components assemble within the Golgi apparatus of live cells to form Cer-SiR. Cer-SiR is benign to cellular function, localizes within the Golgi at a high density, and is sufficiently photostable to enable visualization of Golgi structure and dynamics by 3D confocal or long time-lapse STED microscopy.
Keywords: Bioorthogonal chemistry, Click chemistry, Fluorophores, Membranes, Super-resolution microscopy, Inverse electron demand Diels-Alder reaction
1. Introduction
1.1. Visualizing the Boundaries of Cellular Organelles
Super-resolution microscopy can visualize cellular structure and dynamics at resolutions that simply cannot be attained using conventional microscopes [1–6]. However, increased spatial resolution extracts a price, as the concomitant high sampling rate and photon flux places enormous demands on both the photophysical properties of the fluorophore and the structural properties of the targeting probes. Nearly all super-resolution techniques [1–6] rely on fluorophores that cycle between ON and OFF states [1–7], either stochastically (as in PALM and STORM) [8, 9] or using point spread function engineering (as in STED) [10, 11]. In STED, cycling is achieved with highly powerful depletion and excitation lasers that switch the dye ON and OFF multiple times [1]. Fluorophore photostability is therefore paramount, with a clear preference for far red, bright, photostable dyes such as Atto647N [12] and SiR [13–15]. The structural properties of the targeting probes are also restrictive, as they must be simultaneously cell- permeable, benign to cell function, and possess the ability to localize at high density to achieve bright labeling.
Although proteins can now be tagged easily with fluorophores [16,17], either by the expression of fusion proteins [18] or variants containing orthogonally reactive unnatural amino acids [19, 20], the labeling density achieved with most tagged proteins is generally insufficient for long time-lapse super-resolution imaging. The exceptions are protein polymers such as tubulin, actin, and clathrin - structures that are inherently dense and considered gold standards fore super-resolution imaging. By contrast, lipids represent ideal high-density probes, as they represent the primary constituent of an organelle membrane and are present at densities that are at least one hundred fold higher than the membrane-bound proteins they harbor [21–27]. Perhaps more importantly, it is the lipid—not the proteins held within—that defines the molecular outline of the organelle [28]. Although many lipid-dye conjugates, such as BOD- IPY FL C5 and NBD C6 ceramide have long been commercially available [29, 30], these molecules lack the requisite photostability to survive the powerful lasers required for super-resolution methods such as STED [1–5].
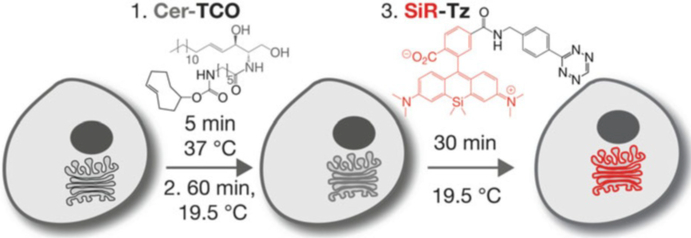
1.2. Labeling the Golgi with Cer-TCO and Sir-Tz
Herein, we describe a general approach to label organelle lipids at high density using fluorophores that are compatible with long time-lapse super-resolution imaging, and apply this approach to visualize the Golgi apparatus [31]. This approach relies on the well-characterized selective partitioning of lipids among cellular organelles [32, 33]; in the case of the Golgi apparatus, the enriched lipids are derived from ceramide [30, 32,33]. We synthesized a ceramide derivative tagged with trans-cyclooctene (Cer-TCO), localized it to the Golgi apparatus in live cells, and reacted it in situ with a tetrazine-modified version of a photostable siliconrhodamine dye (SiR-Tz) [13, 34–38] (Fig. 1). The resulting product, Cer-SiR, is not cytotoxic and does not perturb protein trafficking within and through the Golgi: it is indeed benign to organelle function [31]. The two-component Cer-TCO/SiR-Tz labeling system we describe is suitable for STED imaging of the Golgi for over 900 frames and allows the visualization of vesicles budding and exiting the Golgi [31]. The one challenge is that the Golgi is a convoluted three dimensional object and typically STED only enhances the resolution in the xy axis, however new PSF engineering isotropic 3D STED microscopes, such as 4Pi STED should circumvent this challenge [39].
Fig. 1.
Labeling of the Golgi with Cer-SiR: a two-component, photostable, high-density lipid probe for live cells. Hela cells are incubated with 2 μM Cer-TCO for 5 min, washed, and incubated at 19.5 °C for 60 min to localize the lipid to the Golgi. The cells are then incubated with 2 μM SiR-Tz for 30 min at 19.5 °C, washed, and imaged
2. Materials
2.1. Microscopy Systems
2.1.1. STED Imaging (Commercial Microscope)
STED imaging was carried out on a commercial Leica TCS STED microscope. A picosecond pulsed laser diode (PicoQuant, LDH-P- F-640B) was used for excitation while depletion was achieved with a femtosecond pulsed mode-locked Ti:Sapphire laser (Spectra Physics, Mai Tai) tuned to 755 nm with the output pulses stretched to approximately 200 ps via propagation through a 120 m single mode polarization maintaining fiber. The excitation and depletion beams were directed into a 100 × 1.4 NA oil immersion objective (Leica) which also collected fluorescence from the sample. Fluorescence was filtered from excitation light with a bandpass filter (Semrock, FF01–685/40) and detected with an avalanche photodiode.
2.1.2. STED Imaging (Custom Microscope)
Additional STED imaging was performed on a custom-built system with faster scanning capabilities compared to the Leica TCS STED microscope (Bewersdorf lab, Yale University). The system is centered around an 80 MHz mode-locked Ti:Sapphhire laser (Chameleon Ultra II, Coherent) tuned to 755 nm as the STED depletion beam. The output pulses from the depletion laser were stretched to several hundred picoseconds by first passing through a glass block made from a high dispersion material (SF6, Schott) and then coupled into a 100 m polarization-maintaining single mode optical fiber. After leaving the optical fiber, the depletion beam was collimated and directed onto an SLM, conjugate to the objective back pupil plane, where a 2π phase ramp was displayed. Excitation for fluorescence was accomplished with a 640 nm pulsed diode laser (PicoQuant) electronically synchronized to the depletion beam with an additional computer controlled electronic delay (Colby Instruments). The STED depletion and fluorescence excitation beams were combined with a dichroic mirror and directed into a 100 × 1.4 NA oil immersion objective lens (UPLSAPO 100XO/ PSF, Olympus) where they were focused at the sample. A 16 kHz resonance scanning mirror, in combination with a galvanometer mirror, allowed imaging via beam scanning. The two mirrors were imaged into the objective pupil plane and allowed the beams to scan through the sample at a rate of 16 kHz along the fast axis. Fluorescence from the sample was collected by the objective lens, de- scanned by the scan mirrors, and separated from the excitation and depletion light using dichroic mirrors and band pass filters (FF01–685/40, Semrock). The fluorescence was then focused into a 105 μm core (~0.7 Airy units) multimode fiber connected to a single photon counting avalanche photodiode (ARQ-13-FC, Perkin Elmer). Counts from the APD were collected using an FPGA based data acquisition card (PCIe-7852R, National Instruments) and custom acquisition software (LabVIEW, National Instruments). Data collection was synchronized with the resonance mirror for uni-directional collection during the two-thirds of the halfperiod where the motion ofthe mirror is most linear. Recorded pixel values were therefore linearized (on the DAQ card) to account for the sinusoidal velocity profile of the resonant mirror and normalized according to the pixel dwell times such that the center pixel was divided by unity.
2.1.3. Spinning Disk Confocal Microscopy
Spinning-disk confocal microscopy was performed using an Improvision UltraVIEW VoX system (Perkin-Elmer) built around a Nikon Ti-E inverted microscope, equipped with PlanApo objectives (60 × 1.45-NA) and controlled by the Volocity software (Improvision). SiR was excited with a 640 nm laser and for the detection a 705 ± 45 nm filter was used. The microscope stage was surrounded by a box constantly held at 37 °C.
2.2. Synthesis of Cer-TCO
2.2.1. Equipment
3 and 20 mL scintillation vials.
Erlenmeyer flasks.
Round-bottom flasks.
Filter funnels and filter paper.
Separatory funnel.
Chromatography column (see Note 1).
Spatulas.
Magnetic stir bar.
Rotary evaporator (see Note 2).
TLC developing chamber.
Fume hood.
Temperature controlled magnetic stir plate.
TLC plates.
2.2.2. Chemicals
TOC-PNB ester (((4E)-cyclooct-4-en-1-yl) (4-nitrophenyl) carbonate).
6-aminohexanoic acid.
Sphingosine ((2S,3R)-2-aminooctadec-4-ene-1,3-diol).
DMF (N,N-Dimethylformamide).
Dichloromethane.
Methanol.
1 M HCl.
Saturated aqueous NaHCO3.
MgSO4 (anhydrous).
HBTU (N,N,N′,N′-Tetramethyl-O-(1H-benzotriazol-1-yl) uronium hexafluorophosphate).
iPr2NEt (N,N-Diisopropylethylamine).
Silica gel.
Iodine chamber: Add 50 g of silica gel to a jar and add a few crystals of iodine.
KMnO4 TLC stain: dissolve 3 g KMnO4, 20 g K2CO3, 5 mL 5% NaOH (aq.) in 300 mL of deionized water (see Note 3).
2.3. Preparing Solutions for Cell Labeling
Prepare Cer-TCO working solution (2 μM): Add 1 μL of the 2 mM stock solution to 20 μL of 10% puronic F127 and mix thoroughly by drawing and releasing the solution into/from the pipet tip. Add 1 mL of 1% casein in PBS and mix again (see Notes 4 and 5).
Prepare SiR-Tz working solution (2 μM): Add 1 μL of the 2 mM stock solution to 1 mL of 1% casein in PBS and mix by pipetting up and down (see Note 5).
2.4. Imaging with Cer-TCO and SiR-Tz
All the solutions were prepared using ultrapure water (deionized water further purified to a sensitivity of 18 MΩ cm at 25 °C) and analytical grade reagents. All reagents and solutions were stored at —20 °C unless noted otherwise. HeLa cells were cultured in T75 flasks in Dulbecco’s Modified Eagle Medium with phenol red (DMEM(ph+)) supplemented with 10% FBS and 1% Penicil- lin/Streptomycin solution (10,000 U/mL).
2 mM SiR-Tz in DMSO (see Notes 6 and 7).
2 mM Cer-TCO in DMSO (see Notes 6 and 8).
Dulbecco’s Modified Eagle Medium without phenol red, supplemented with 10% FBS (DMEM (ph-)) (store at 4 °C).
10% Pluronic F127 in H2O (see Note 9).
1% casein hydrolysate in 1x PBS (sterile filtered; store at 4 °C).
35 mm glass-bottom dish (glass no. 1.5).
Parafilm™.
Waterbath with thermostat (see Note 10).
3. Methods
This section describes our protocols for the synthesis of Cer-TCO, maintenance of the cells to be labeled, preparation of all working solutions, labeling of the cells using Cer-TCO and SiR-Tz, as well as the imaging of labeled cells by 3D confocal and STED microscopy.
3.1. Synthesis of Cer-TCO
All the chemical steps should be performed in a well-vented fume hood, and appropriate personal protective equipment (googles, gloves, lab coat) should be worn throughout all the procedures. All waste should be disposed of in accord with appropriate regulations. None of the reactions described requires special measures to prevent contact with air or moisture.
3.1.1. Synthesis of TCO- C6 (rel-(1R-4E-pR)- Cyclooct-4-ene-1-yl-N- hexanoic Acid Carbamate)
Add 100 mg (343 μmol, 1.0 eq) ofTCO-PNB ester to a 20 mL scintillation vial (see Note 11).
Add 135 mg of 6-aminohexanoic acid (1.03 mmol, 3.0 eq) to the vial.
Add a 5 mm magnetic stir bar and 3 mL of DMF.
Add 360 μL of ipr2Tr2NEt (2.06 mmol, 6.0 eq) and close the vial with its cap (see Notes 12 and 13).
Place an oil bath on a magnetic stir plate and set the temperature to 65 °C.
Mount the vial into a clamp and lower the closed vial into the oil bath until the level of the oil bath matches the level of the suspension in the vial.
Cover the reaction setup with aluminum foil to exclude light.
Stir the reaction for 3 days at 65 °C. Switch off the oil bath heater and remove the vial from the clamp.
Concentrate the reaction mixture under vacuum (see Note 2).
Transfer the reaction mixture to a separatory funnel together with 20 mL of dichloromethane.
Wash the organic layer 3 × with 5 mL of 1 M aqueous HCl and combine the aqueous layers.
Extract the combined aqueous layers 2x with 5 mL of dichloromethane.
Combine the organic layers of step 12 with the organic layer of step 11.
Dry combined organic layers over MgSO4.
Concentrate the dried organic layers by rotary evaporation.
Prepare a chromatography column of silica gel (25 g) packed in dichloromethane (see Note 1).
Load the column with the crude product obtained in step 15 and elute with a gradient from 100% dichloromethane to 5% methanol in dichloromethane (400 mL) and further elute with 5% methanol in dichloromethane (150 mL).
Identify the fractions containing the desired product by running silica gel TLC plates with 5% methanol in dichloromethane. The product can be visualized by developing the TLC place in an iodine chamber for 3 min. The product appears as a brown/yellow spot with a retention factor of 0.35 (see Note 14).
Combine the fractions containing the product and concentrate them by rotary evaporation to obtain the product as a pale yellow oil with a yield of approximately 40% (see Note 15).
Analyze the product by NMR and ESI-HRMS. The following analytical data is anticipated.
1H NMR (400 MHz, MeOD-d3) δ/ppm = 5.66–5.54 (m, 1H), 5.47 (ddd, J = 16.0, 10.7, 3.5 Hz, 1H), 4.39–4.25 (m, 1H), 3.06 (t, J = 7.0 Hz, 2H), 2.37–2.31 (m, 3H), 2.28 (t, J = 7.4 Hz, 2H), 2.04–1.86 (m, 4H), 1.79–1.65 (m, 2H), 1.65–1.54 (m, 3H), 1.54–1.44 (m, 2H), 1.39–1.27 (m, 2H).
13C NMR (101 MHz, MeOD-d3) δ 177.4, 158.6, 136.0, 133.7, 81.5, 42.2, 41.5, 39.6, 35.2, 34.8, 33.5, 32.1, 30.6, 27.3, 25.7.
HRMS (ESI): m/z for C15H26NO4+: 284.1856.
3.1.2. Synthesis of Cer-TCO
Solution A: Dissolve 25.0 mg of sphingosine (83.5 μmol, 1.3 eq)in2 mL ofDMF.
Solution B: Dissolve 40.1 mg of HBTU (77.1 μmol, 1.2 eq) in 0.8 mL of DMF and add 33.7 μL of iPr2NEt (193 pmol, 3.0 eq).
Add 18.2 mg of TCO-C6 (from Subheading 3.2, step 1) (64.2 μmol, 1.0 eq) to solution A, add a magnetic stir bar, and stir for 2 min.
Combine solutions A and B and stir for 2.5 h at room temperature (see Note 16).
Concentrate the reaction mixture under reduced pressure to obtain a brownish-yellowish residue (see Note 2).
Dissolve the residue in 50 mL of dichloromethane and transfer into a separatory funnel.
Wash the organic layer 3× with 10 mL of sat. aqueous NaHCO3 and wash 3× with 10 mL of 1 M aqueous HCl (see Note 17).
Combine the aqueous NaHCO3 layers and wash 1× with 10 mL of dichloromethane.
Combine the aqueous HCl layers and wash 1 × with 10 mL of dichloromethane.
Combine the organic layers from steps 7 to 9, dry over MgSO4, and concentrate by rotary evaporation.
Prepare a chromatography column of silica gel (12 g) packed in dichloromethane (see Note 1).
Load the column with the crude product obtained in step 10 and elute with a gradient from 100% dichloromethane to 5% methanol in dichloromethane (200 mL) and further elute with 5% methanol in dichloromethane (100 mL).
Identify the fractions containing the product using silica gel TLC plates eluted with 5% methanol in dichloromethane. The product can be visualized by developing the TLC place in an iodine chamber for 3 min. It appears as a brown/yellow spot with a retention factor of 0.35. Product purity can be assessed by staining the TLC plate with KMnO4. Impurities in the reaction mixture will appear as additional yellow spots on the TLC plate once the KMnO4-stained plate is heated with a heat gun (see Note 18).
Combine those fractions that lack impurities as judged by TLC and concentrate them by rotary evaporation to obtain the product as a colorless wax with a yield of approximately 70% (see Note 15).
Analyze the product by NMR and ESI-HRMS. The following analytical data is anticipated.
1H NMR (500 MHz, MeOD-d3): δ/ppm = 5.72 (dt, J = 14.3, 6.7 Hz, 1H), 5.68–5.56 (m, 1H), 5.54–5.44 (m, 2H), 4.42–4.27 (m, 1H), 4.09 (t, J = 7.2 Hz, 1H), 3.89 (q, J = 5.8 Hz, 1H), 3.71 (d, J = 5.0 Hz, 2H), 3.08 (t, J= 6.8 Hz,2H),2.42–2.29 (m, 3H),2.23 (t, J= 7.5 Hz, 2H), 2.06 (q, J = 7.1 Hz, 2H), 2.03–1.89 (m, 4H), 1.81–1.68 (m, 2H), 1.68– 1.56 (m, 3H), 1.50 (p, J = 7.3 Hz, 2H), 1.31 (s, 23H), 0.92 (t, J = 6.7 Hz, 3H).
13C NMR (126 MHz, MeOD-d3): δ/ppm = 176.5, 159.1, 136.5, 135.1, 134.2, 131.6, 81.9, 74.1, 62.7, 57.2, 42.7, 41.9.40.1, 37.6, 35.6, 33.9, 33.8, 33.5, 32.6, 31.2, 31.2, 31.1, 30.9, 30.8, 30.8, 27.9, 27.1, 24.2, 14.9.
HRMS (ESI): m/z calc. For C33H61N2O5+: 565.4575.
3.2. Preparing Cells and Their Labeling with Cer-TCO and SiR-Tz
Seed 160,000 HeLa cells on a 35 mm glass-bottom dish in DMEM(ph+)and incubate for 24 h at 37 °C (see Note 19).
Wash the attached cells in the 35 mm glass-bottom dish 3× with 2 mL of DMEM (ph—).
Add 500 yL of the Cer-TCO working solution (2 μΜ) to the dish and incubate for 5 min at 37 °C.
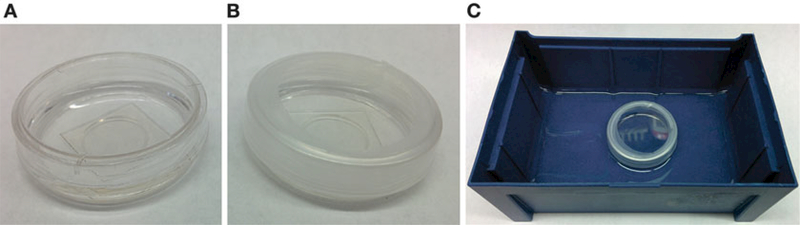
Wash the cells 3× with 2 mL of DMEM (ph—), add 2 mL of DMEM (ph—), and seal the dish with Parafilm™ (see Fig. 2).
Incubate the dish for 60 min in a water bath held at 19.5 °C (see Notes 10 and 20; Fig. 2).
Wash the cells 3 × with 2 mL of DMEM (ph—) and add 500 yL of the SiR-Tz working solution.
Seal the dish again with Parafilm™ and incubate at 19.5 °C for 30 min (seeNote 10).
Wash the cells 3 × with 2 mL of DMEM (ph—) and add 2 mL of DMEM (ph—) (see Note 21).
Fig. 2.
Preparing cells for temperature block. (a) Hela cells in 35 mm glass bottom dish after incubation with Cer-TCO; (b) Glass bottom dish sealed with parafilm to prevent water from leaking into the dish; (c) Sealed glass bottom dish placed in a water bath which can be cooled to 19.5 °C
3.3. Imaging Cells by 3D Confocal Microscopy
Place the dish on the stage of the microscope (see Note 22).
Find cells using the bright field mode of the microscope.
Turn on the 640 nm laser.
Adjust the exposure time to <300 ms and increase the camera sensitivity settings in order to decrease the laser intensity as low as possible while still obtaining an image with a good signal to noise ratio.
Find cells to image and adjust settings from step 4 if necessary.
Set the z-slice distance to 0.5 ym and define the upper and lower z-boundary for imaging in the microscope settings (see Note 23).
Start imaging at the maximum rate (see Note 24).
3.4. Imaging Cells by STED Microscopy
Place the dish on the stage of the microscope (see Note 22).
Find cells using the eyepiece of the microscope.
Turn on the 640 nm laser.
Find cells to image.
Set field of view to 512 × 512 pixels with a pixel size of 30–40 nm.
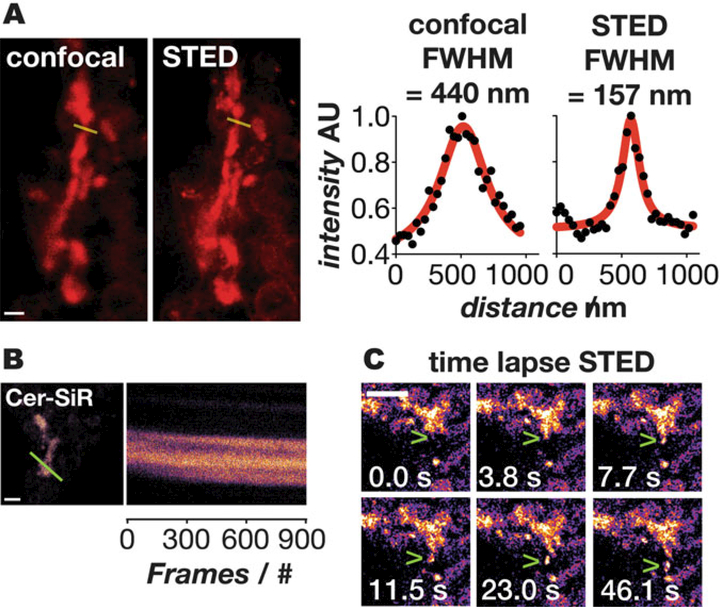
Image with STED laser set to 755 nm with 60–120 accumulations per line (see Note 25, Fig. 3).
Fig. 3.
Imaging of the Golgi with Cer-SiR. (a) Comparison of images obtained in confocal (left) and STED (right) mode; scale bar: 800 nm. The line profiles through the Golgi (yellow) illustrate the improvement in resolution. (b) Kymograph along the yellow lines illustrating the photostability of the probe; scale bar: 2 μm. (c) Timelapse STED images of vesicles budding and exiting the Golgi; scale bar: 1 μm
4. Notes
Alternatively, separation can be achieved with an automated chromatography system (such as a Teledyne Isco CombiFlash Rfwith columns prepacked with RediSep Rf Silica (40–60 μm) or RediSep Rf Gold Silica (20–40 μm, spherical)). Use of the CombiFlash allows for better gradient control and better separation.
Rotary evaporation of DMF from the reaction mixture requires careful heating with a heat gun or use of a high vacuum pump to assist the rotary evaporator. Alternatively, a small-volume evaporation system can be used (e.g., Biotage V-10 evaporator).
The KMnO4 solution should be stored in the dark. To minimize light-induced decomposition and contamination with silica particles from previously stained plates, a jar should be filled with only the amount of solution required to develop the TLC plates and the remainder of the solution should be stored separately.
The order of the addition is important. Dissolving the lipid in Pluronic F127 prevents the lipid from aggregating.
This solution should be prepared right before its use to avoid degradation.
Aliquots of the 2 mM stock solutions of Cer-TCO and SiR-Tz in DMSO can be stored at —20 °C for up to 9 and 18 months, respectively.
Stock solutions of SiR-Tz in DMSO are blue in its frozen from and become colorless to slightly pink when thawed.
Newly synthesized TCO-Cer should be stored as solid under argon/nitrogen at —20 °C, as the solid form is more stable than the stock solution. After more than 12 months ofstorage, Cer-TCO purity should be evaluated by both TLC and NMR. If impurities are detected, repeat purification steps 11-13 (Subheading 3.1.2).
Pluronic F-127 dissolves very slowly. The dissolution rate can be increased by shaking the mixture in a thermoshaker at 37 ° C for 2 h. The solution must be stored at RT (not 4 °C) to avoid precipitation.
Alternatively, the dish can also be incubated at 19.5 °C in an incubator with cooling capacities. The dish temperature must remain <20 °C to ensure that Cer-TCO accumulates in the Golgi. Variations of 1–2 °C below 19.5 °C are acceptable.
Alternatively, the reaction can be carried out at half the scale.
After the addition of ipr2NEt the reaction mixture might start to turn yellowish to deep yellow after 3 days.
The reaction mixture will not fully dissolve, even after 3 days.
If two spots are evident by TLC then the material should be chromatographed again using a shallower gradient to obtain pure material. Alternatively, the impure product can be used in the next synthetic step as the impurity does not interfere and can be removed during the workup of the next reaction.
Examples including images of the 1H and 13C NMR spectra of TCO-C6 and Cer-TCO can be found in the supporting information of reference [31].
The reaction can be stirred for up to 24 h without negatively impacting purity or yield.
The aqueous layer of the final wash with sat. NaHCO3 (aq) should be (almost) colorless. If it is not, then continue to wash with sat. NaHCO3 (aq) until the aqueous layer is clear.
If multiple yellow spots are visible on the TLC after staining with KMnO4 the chromatography step should be repeated with a shallower gradient.
The confluency of the cells affects the labeling results. Ideal results are obtained with cells at 60–85% confluency.
The incubation time can be extended up to 2.5 h and will lead to brighter labeling of the Golgi. However, the morphology of the Gsolgi will become more compact over an extended incubation time.
Fixing the cells with 10% PFA is possible after three additional washing steps with PBS. However, the cells should still be imaged within one day since Golgi signal will blur over longer storage time, potentially as the probe is non- or weakly fixable.
For optimal cell health an incubator chamber around the microscope stage is useful.
Imaging of the whole Golgi usually requires 18–25 z-slices.
If the experiment requires imaging over a prolonged time (30 min and more), slower imaging speeds are recommended to minimize bleaching and phototoxicity.
Note that the depletion wavelength differs from the depletion wavelength commonly used to deplete the fluorescence of SiR attached to proteins.
Acknowledgments
This study was supported by the Wellcome Trust Foundation and by the National Institutes of Health (GM83257 to A.S.). R S.E. was supported by an Advanced Postdoc. Mobility fellowship from the Swiss National Science Foundation.
References
- 1.Hell SW (2007) Far-field optical nanoscopy. Science 316:1153–1158 [DOI] [PubMed] [Google Scholar]
- 2.Huang B, Bates M, Zhuang XW (2009) Superresolution fluorescence microscopy. Annu Rev Biochem 78:993–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toomre D, Bewersdorf J (2010) Anew wave of cellular imaging. Annu Rev Cell Dev Biol 26:285–314 [DOI] [PubMed] [Google Scholar]
- 4.Van De Linde S, Heilemann M, Sauer M (2012) Live-cell super-resolution imaging with synthetic fluorophores. Annu Rev Phys Chem 63:519–540 [DOI] [PubMed] [Google Scholar]
- 5.Fornasiero EF, Opazo F (2015) Superresolution imaging for cell biologists. BioEssays 37:436–451 [DOI] [PubMed] [Google Scholar]
- 6.Uno SN, Tiwari DK, Kamiya M et al. (2015) A guide to use photocontrollable fluorescent proteins and synthetic smart fluorophores for nanoscopy. Microscopy 64:263–277 [DOI] [PubMed] [Google Scholar]
- 7.Chozinski TJ, Gagnon LA, Vaughan JC (2014) Twinkle, twinkle little star: photoswitchable fluorophores for super-resolution imaging. FEBS Lett 588:3603–3612 [DOI] [PubMed] [Google Scholar]
- 8.Betzig E, Patterson GH, Sougrat R et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645 [DOI] [PubMed] [Google Scholar]
- 9.Rust MJ, Bates M, Zhuang XW (2006) Subdiffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3:793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hell SW, Wichmann J (1994) Breaking the diffraction resolution limit by stimulated- emission - stimulated-emission-depletion fluorescence microscopy. Opt Lett 19:780–782 [DOI] [PubMed] [Google Scholar]
- 11.Vicidomini G, Moneron G, Han KY et al. (2011) Sharper low-power STED nanoscopy by time gating. Nat Methods 8:571–U575 [DOI] [PubMed] [Google Scholar]
- 12.Westphal V, Rizzoli SO, Lauterbach MA et al. (2008) Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science 320:246–249 [DOI] [PubMed] [Google Scholar]
- 13.Lukinavicius G, Umezawa K, Olivier N et al. (2013) A near-infrared fluorophore for livecell super-resolution microscopy of cellular proteins. Nat Chem 5:132–139 [DOI] [PubMed] [Google Scholar]
- 14.Bottanelli F, Kromann EB, Allgeyer ES et al. (2016) Two-colour live-cell nanoscale imaging of intracellular targets. Nat Commun 7:10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bottanelli F, Kilian N, Ernst AM et al. (2017) A novel physiological role for ARF1 in the formation of bi-directional tubules from the Golgi. Mol Biol Cell 28(12):1676–1687. doi: 10.1091/mbc.E16-12-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Suarez M, Ting AY (2008) Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol 9:929–943 [DOI] [PubMed] [Google Scholar]
- 17.Heilemann M, Van De Linde S, Mukherjee A et al. (2009) Super-resolution imaging with small organic fluorophores. Angew Chem Int Ed 48:6903–6908 [DOI] [PubMed] [Google Scholar]
- 18.Hinner MJ, Johnsson K(2010) How to obtain labeled proteins and what to do with them. Curr Opin Biotechnol 21:766–776 [DOI] [PubMed] [Google Scholar]
- 19.Nikic I, Plass T, Schraidt O et al. (2014) Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew Chem Int Ed 53:2245–2249 [DOI] [PubMed] [Google Scholar]
- 20.Uttamapinant C, Howe JD, Lang K et al. (2015) Genetic code expansion enables livecell and super-resolution imaging of site- specifically labeled cellular proteins. J Am Chem Soc 137:4602–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guidotti G (1972) Membrane proteins. Annu Rev Biochem 41:731–752 [DOI] [PubMed] [Google Scholar]
- 22.Quinn P, Griffiths G, Warren G (1984) Density of newly synthesized plasma-membrane proteins in intracellular membranes. 2. Biochemical-studies. J Cell Biol 98:2142–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neef AB, Schultz C (2009) Selective fluorescence labeling of lipids in living cells. Angew Chem Int Ed 48:1498–1500 [DOI] [PubMed] [Google Scholar]
- 24.Jao CY, Roth M, Welti R et al. (2009) Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc Natl Acad Sci U S A 106:15332–15337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Seckute J, Cole CM et al. (2012) Livecell imaging of cyclopropene tags with fluoro- genic tetrazine cycloadditions. Angew Chem Int Ed 51:7476–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hang HC, Wilson JP, Charron G (2011) Bioorthogonal chemical reporters for analyzing protein lipidation and lipid trafficking. Acc Chem Res 44:699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiele C, Papan C, Hoelper D et al. (2012) tracing fatty acid metabolism by click chemistry. ACS Chem Biol 7:2004–2011 [DOI] [PubMed] [Google Scholar]
- 28.Shroff H, Galbraith CG, Galbraith JA et al. (2008) Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods 5:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagano RE, Martin OC, Kang HC et al. (1991) a novel fluorescent ceramide analog for studying membrane traffic in animal-cells - accumulation at the golgi-apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol 113:1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks DL, Bittman R, Pagano RE (2008) Use of bodipy-labeled sphingolipid and cholesterol analogs to examine membrane microdomains in cells. Histochem Cell Biol 130:819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erdmann RS, Takakura H, Thompson AD et al. (2014) Super-resolution imaging of the Golgi in live cells with a bioorthogonal ceramide probe. Angew Chem Int Ed 53:10242–10246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Meer G, De Kroon AIPM (2011) Lipid map of the mammalian cell. J Cell Sci 124:5–8 [DOI] [PubMed] [Google Scholar]
- 34.Blackman ML, Royzen M, Fox JM (2008) Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc 130:13518–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devaraj NK, Hilderbrand S, Upadhyay R et al. (2010) Bioorthogonal turn-on probes for imaging small molecules inside living cells. Angew Chem Int Ed 49:2869–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karver MR, Weissleder R, Hilderbrand SA (2011) Synthesis and evaluation of a series of 1,2,4,5-tetrazines for bioorthogonal conjugation. Bioconjug Chem 22:2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Karver MR, Li WL et al. (2012) Metal- catalyzed one-pot synthesis of tetrazines directly from aliphatic nitriles and hydrazine. Angew Chem Int Ed 51:5222–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson JCT, Meimetis LG, Hilderbrand SA et al. (2013) BODIPY-tetrazine derivatives as superbright bioorthogonal turn-on probes. Angew Chem Int Ed 52:6917–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dyba M, Keller J, Hell SW (2005) Phase filter enhanced STED-4Pi fluorescence microscopy: theory and experiment. New J Phys 7:134 [Google Scholar]