Abstract
Changes in the abundance and activity of long non-coding RNAs (lncRNAs) have an important impact on the development of cancer. The nuclear paraspeckle assembly transcript 1 (NEAT1) has been reported to be overexpressed in many types of cancer since its discovery. However, inconsistencies exist as NEAT1 can also function as a tumor suppressor in certain types of cancer, such as acute promyelocytic leukemia. Here we systematically describe our current understanding of NEAT1 in tumor initiation and progression. First, we analyzed the expression patterns of NEAT1 in various normal tissues and malignant cancers using data from public data portals, the Genotype-Tissue Expression Project (GTEx) and the Cancer Genome Atlas (TCGA), together with recent progress in the study of NEAT1 in various types of cancer. Second, we discussed the functions and mechanisms of NEAT1 in modulating tumor activity. Then, the upstream transcription factors and downstream microRNA targets of NEAT1 in the transcription cascade of cancers were also summarized. These data highlight the emerging role of NEAT1 in tumorigenesis, and present promising targetable pathways and clinical opportunities for tumor prevention and classifications.
Keywords: Cancer, Long non-coding RNA, NEAT1, Pan-cancer analysis, Regulation
Introduction
Long non-coding RNAs (lncRNAs) have emerged as an important element in modulating cell growth, differentiation, and apoptosis in the development of normal and malignant tissues.1, 2 Over the past decades, accumulating evidence has demonstrated that numerous lncRNAs are aberrantly regulated and play critical oncogenic or tumor-suppressive roles in many types of cancers.2, 3 Elucidating the functions and mechanisms of these lncRNAs would be of great help to understand the complex transcriptional machineries in tumors.
Nuclear paraspeckle assembly transcript 1 (NEAT1), located on chromosome 11q13.1, is a lncRNA that encodes two transcriptional variants, namely NEAT1-1 (3.7 kb) and NEAT1-2 (23 kb). Previous studies have revealed that NEAT1 functions as an essential structural component of a nuclear domain called paraspeckle, which participates in gene regulation mainly by nuclear retention of proteins and RNAs.4 Cumulatively, numerous studies have identified the aberrant expression and prognostic value of NEAT1 in various types of tumors, the majority of which suggest NEAT1 as an oncogene that is overexpressed in tumors compared to their respective normal tissues and promotes tumor cell progression.5 However, in hematological malignancies such as acute promyelocytic leukemia (APL), NEAT1 can function as a tumor suppressor by inducing the differentiation of APL cells.6 This prompted us to systematically analyze the role of NEAT1 from a pan-cancer perspective.
Here we first identified the expression profiles of NEAT1 in various normal tissues and certain types of cancer utilizing data from the Genotype-Tissue Expression (GTEx) and the Cancer Genome Atlas (TCGA) data portals. Then, a literature search was conducted to further examine the expression patterns and potential clinical significance of NEAT1 in distinct tumors. The discrepancy in NEAT1 expression patterns in specific tumors between TCGA data and previous publications were also discussed. Finally, we summarized the functions and potential upstream and downstream factors of NEAT1 in tumor initiation and progression.
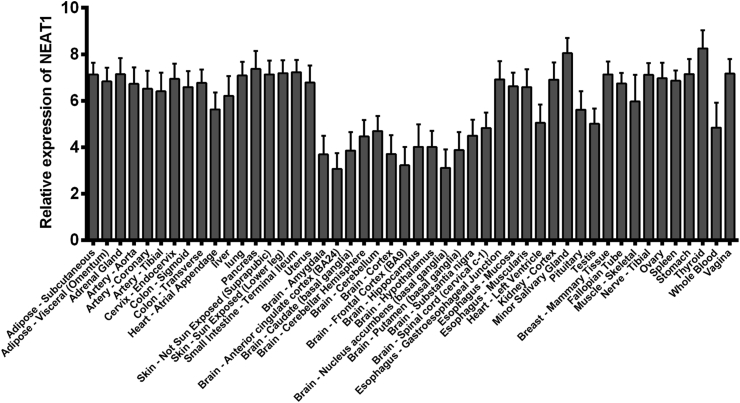
NEAT1 expression in different normal tissues
The GTEx project establishes a database and a tissue bank that analyzes gene expression in different human tissues. To explore the expression levels of NEAT1 in different tissues, we extracted data from the GTEx portal (https://www.gtexportal.org/home/, updated to June 20, 2017) and analyzed 48 types of tissues from 7862 samples. NEAT1 demonstrated a similar expression level among different tissues, with relatively low expression in the brain, heart and whole blood compared with other tissues (Fig. 1). This indicated that NEAT1 was widely expressed in various tissues and may exert potential effects in the maintenance of tissue function.
Figure 1.
NEAT1 expression in different normal human tissues (data from GTEx).
Dysregulation of NEAT1 in distinct types of cancers leveraging TCGA portals
TCGA, containing molecular features of over 30 types of cancer, provides a platform to conveniently and effectively understand genes using multi-layered genome profiles. Reanalyzing these large public datasets may help us draw a comprehensive picture of a gene in abnormal cancerous situations, compared to normal physiological conditions.
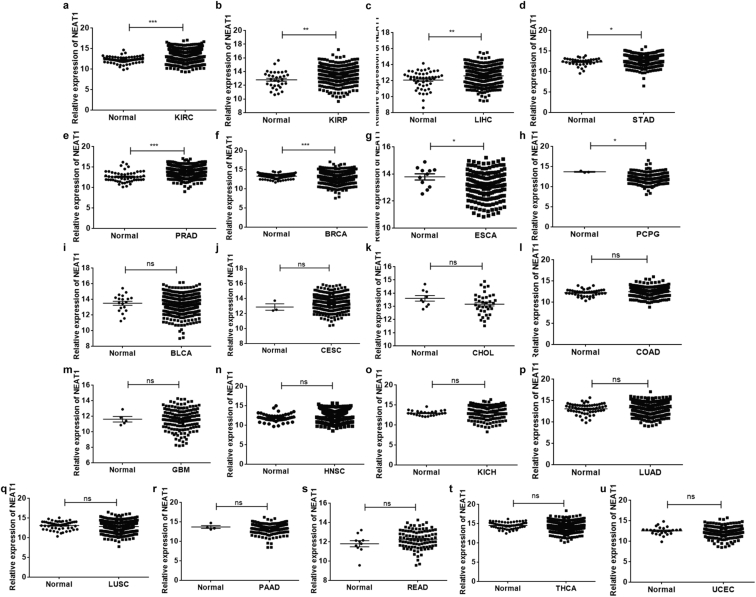
NEAT1 has been reported to be an oncogene highly expressed in multiple types of cancer and promotes tumor progression.1, 7, 8 To investigate the function of NEAT1 from a pan-cancer perspective, we analyzed NEAT1 expression using RNA-sequence (RNA-seq) data downloaded from the TCGA Pan-Cancer (PANCAN) of UCSC Xena Network (http://xena.ucsc.edu, updated to June 16, 2017). Only the cancers with data from both primary tumors and corresponding normal tissues (n ≥ 3) were chosen for further analysis. In all, data of 21 types of cancer with 722 normal tissues (N) and 7536 primary tumors (T) were collected and analyzed, which included bladder urothelial carcinoma (BLCA, N = 19, T = 407), breast invasive carcinoma (BRCA, N = 114, T = 1097), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, N = 3, T = 303), cholangiocarcinoma (CHOL, N = 9, T = 36), colon carcinoma (COAD, N = 41, T = 286), esophageal carcinoma (ESCA, N = 11, T = 184), glioblastoma multiforme (GBM, N = 5, T = 154), heck & neck squamous cell carcinoma (HNSC, N = 44, T = 520), kidney clear cell carcinoma (KIRC, N = 72, T = 533), kidney chromophore (KICH, N = 25, T = 123), kidney papillary cell carcinoma (KIRP, N = 32, T = 290), liver hepatocellular carcinoma (LIHC, N = 50, T = 371), lung adenocarcinoma (LUAD, N = 59, T = 515), lung squamous cell carcinoma (LUSC, N = 51, T = 502), pancreatic adenocarcinoma (PAAD, N = 4, T = 178), pheochromocytoma & paraganglioma (PCPG, N = 3, T = 179), prostate adenocarcinoma (PRAD, N = 52, T = 497), rectal cancer (READ, N = 10, T = 94), stomach adenocarcinoma (STAD, N = 35, T = 538), thyroid carcinoma (THCA, N = 59, T = 553), and uterine corpus endometrioid (UCEC, N = 24, T = 176).
Overexpression of NEAT1 in prostate, stomach, liver, kidney clear cell carcinoma and papillary cell carcinoma
Specifically, the expression of NEAT1 in prostate adenocarcinoma, stomach adenocarcinoma, liver hepatocellular carcinoma, kidney papillary cell carcinoma and kidney clear cell carcinoma were relatively high compared to their corresponding normal tissues (Fig. 2a–e). The upregulation of NEAT1 in primary tumors may be involved in their complex transcription networks and contribute to tumorigenesis, indicating the potential oncogenic function of NEAT1 in specific cancers.
Figure 2.
The expression of NEAT1 in different tumors leveraging RNA-seq data from TCGA. Relative gene expression levels are presented in the form of mean ± standard deviation (SD). Comparisons between groups were examined by the Student's t-test (two-tailed). A p-value of less than 0.05 was considered statistically significant. BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: cholangiocarcinoma; COAD: colon carcinoma; ESCA: esophageal carcinoma; GBM: glioblastoma multiforme; HNSC: heck & neck squamous cell carcinoma; KICH: kidney chromophore; KIRC: kidney clear cell carcinoma; KIRP: kidney papillary cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma & paraganglioma; PRAD: prostate adenocarcinoma; READ: rectal cancer; STAD: stomach adenocarcinoma; THCA: thyroid carcinoma; UCEC: uterine corpus endometrioid.
Downregulation of NEAT1 in breast invasive cancer, esophageal carcinomas and pheochromocytoma & paraganglioma
Surprisingly, the expression of NEAT1 in breast invasive carcinoma, esophageal carcinoma and pheochromocytoma & paraganglioma displayed relatively low expression with respect to their corresponding normal tissues (Fig. 2f–h). The down-expression of NEAT1 in these tumors indicates that NEAT1 may function as a tumor suppressor and not solely an oncogene, to some extent.
No significant difference in NEAT1 expression between cancer and normal groups in most types of our studied cancer
However, in the majority (14/21) of these types of cancer, i.e. BLCA, CESC, CHOL, COAD, GBM, HNSC, KICH, LUAD, LUSC, PAAD, READ, THCA, and UCEC, the expression of NEAT1 remained unchanged in primary tumors as compared with their expression in corresponding normal tissues (Fig. 2i–u).
Dysregulation of NEAT1 in distinct types of cancers according to literature
To further examine the dysregulation and potential clinical significance of NEAT1 in distinct tumors, a literature search (last search updated to Sep. 13, 2017) was conducted in PubMed using the following search terms: (NEAT1 OR Nuclear paraspeckle assembly transcript 1) AND (cancer OR tumor OR neoplasm). In all, 93 articles (including 20 types of cancers) were obtained and carefully analyzed.
Overexpression of NEAT1 in the majority of solid tumors
As demonstrated by Table 1, NEAT1 is overexpressed in most of the solid tumors, including breast cancer, cholangiocarcinoma, clear cell renal cell carcinoma, colorectal cancer, endometrioid adenocarcinoma, esophageal squamous cell carcinoma, gastric cancer, glioma, hepatocellular carcinoma, laryngeal squamous cell cancer, lung cancer, nasopharyngeal carcinoma, ovarian cancer, pancreatic cancer, prostate cancer, and thyroid carcinoma. The function of NEAT1 in tumorigenesis ranges from regulation of cell proliferation, apoptosis, invasion, migration, metastasis to the adjustment of epithelial-to-mesenchymal transition (EMT), blood-tumor barrier permeability and chemo-sensitivity.9, 10, 11, 12 For example, in breast cancer, overexpression of NEAT1 promotes EMT and invasion in vitro as well as dissemination and metastasis in vivo.13 Exogenous modulation of NEAT1 modulates gemcitabine sensitivity in cholangiocarcinoma.14 Although inconsistencies exist, NEAT1 is a good diagnostic and prognostic biomarker and high expression of NEAT1 indicates unfavorable prognosis in colorectal cancer.15 Moreover, in breast cancer, hepatocellular carcinoma, and papillary kidney cancer, whole-genome analysis indicates NEAT1 carries specific mutations that affect their expression levels.16, 17 rs512715 of NEAT1 is also associated with an increased risk of cervical cancer.18 Together, these results indicate that NEAT1 is overexpressed and functions as an oncogene in most of the solid tumors.
Table 1.
The expression patterns and functions of NEAT1 in different tumors.
| Tumors | NEAT1 expression | Function | Reference (PMID) |
|---|---|---|---|
| Breast cancer | overexpressed mutated | Promote proliferation, EMT, invasion and inhibit apoptosis in vitro Promote dissemination and metastasis in vivo |
28805661, 28658208, 28338194, 28034643, 27556296, 27147820, 25417700 |
| Cervical cancer | Polymorphism | Indicate an increased risk of cervical cancer (rs512715) | 28423658 |
| Cholangiocarcinoma | Overexpressed | Promote growth and metastasis Modulate chemo-sensitivity |
28810932, 28122578 |
| Clear cell renal cell carcinoma | Overexpressed | Promote cell proliferation, invasion, migration, and EMT Inhibit apoptosis |
28269753 |
| Colorectal cancer | Overexpressed | Promote proliferation, invasion and migration Inhibit apoptosis Indicate poor outcome and recurrence |
28013491, 26552600, 26314847 |
| Endometrioid adenocarcinoma | Overexpressed | Accelerate cell growth, invasion and metastasis Reduce apoptosis |
27664948 |
| Esophageal squamous cell carcinoma | Overexpressed | Promote cell proliferation, invasion and migration | 26609486 |
| Gastric cancer | Overexpressed | Promote cell proliferation, EMT and invasion inhibit apoptosis Accelerate chemotherapy resistance |
28401449, 27095450, 26911892 |
| Glioma | Overexpressed | Promote permeability of the blood-tumor barrier Facilitate stem-like properties Promote cell proliferation, invasion and migration Inhibit apoptosis |
28185956, 27878295, 26582084, 26242266,27556696 |
| Hepatocellular carcinoma | Overexpressed mutated | Promote cell proliferation and invasion | 28783584, 28732670, 28526689, 27064257, 26191242 |
| Laryngeal squamous cell cancer | Overexpressed | Promote cell proliferation Inhibit apoptosis and cell cycle arrest |
26822763 |
| Leukemia | Downregulated | Accelerate multidrug resistance Interfere with p53-dependent DNA damage response (in chronic lymphocytic leukemia and lymphoma) Impairs myeloid differentiation (in acute promyelocytic leukemia) |
27446393, 25971364, 25245097 |
| Non-small cell lung cancer | Overexpressed | Promote cell proliferation, invasion and migration in vitro Accelerate tumor growth in vivo Modulate chemo-sensitivity |
28762332, 28615056, 28239820, 27351135, 27270317, 25854373 |
| Multiple myeloma | Downregulated | Correlate with cytogenetic aberrations | 28543758 |
| Nasopharyngeal carcinoma | Overexpressed | Promote EMT and radio-resistance | 27020592 |
| Ovarian cancer | Overexpressed | Indicate poor prognosis | 27608895, 20969748 |
| Pancreatic cancer | Overexpressed | Promote cell proliferation inhibit apoptosis |
27888106, 27822425 |
| Papillary kidney cancer | Mutated | Be associated with NEAT1 expression and indicate poor outcome | 28358873 |
| Prostate cancer | Overexpressed | Promote tumorigenesis, chemotherapy, and tumor progression in vivo | 25415230 |
| Thyroid carcinoma | Overexpressed | Accelerate cell growth and metastasis in vitro increase tumor size in vivo | 28000845 |
Down-expression of NEAT1 in hematological malignancies
However, in hematological malignancies such as leukemia and multiple myeloma, NEAT1 is significantly downregulated.6, 19, 20 The down-expression of NEAT1 accelerates multidrug resistance in leukemia, and interferes with p53-dependent DNA damage response machinery in chronic lymphocytic leukemia and lymphoma.19 In acute promyelocytic leukemia, NEAT1 expression is inhibited by the fusion protein PML/RARα and reduced expression of NEAT1 impairs myeloid differentiation.6 In addition, NEAT1 expression is closely correlated with cytogenetic aberrations of multiple myeloma. These reports uncover the tumor-suppressive properties of NEAT1 in hematological malignancies.
Comparison of NEAT1 expression between TCGA data and literature
As the largest publicly available repository of multi-layered genome and clinical profiles, TCGA undoubtedly presents a comprehensive resource of tumor patients. The data from TCGA and literature can verify and complement with each other. Firstly, the numbers of samples would be enlarged greatly in many types of cancer leveraging TCGA. Secondly, the tumor types from TCGA are a little bit different from those discussed in literature studies, which largely expand the tumor spectrum of our research from the pan-cancer perspective. Lastly, as some types of cancer, such as hematological malignancies, have missed the data of normal tissues, the reports from literature are of significant importance. Thus, integrating and analyzing these two aspects of data would tremendously enrich our current understanding of NEAT1 in tumor development.
As to the expression of NEAT1, the data from TCGA and literature demonstrate the same tendency in most cancers. In some cases, the number of normal tissues in TCGA data is too small to get a significant difference in the expression level of NEAT1 between cancers and normal groups. For instance, there is only 3 control samples in cervical squamous cell carcinoma and endocervical adenocarcinoma. Nevertheless, NEAT1 do present a rising trend in some specific cancers, such as cervical squamous cell carcinoma and endocervical adenocarcinoma, colon carcinoma, glioblastoma multiforme, and rectal cancer. In other cases, the data from TCGA indicates the expression of NEAT1 shows no significant difference between cancer and corresponding normal tissues in certain types of cancer, i.e. cholangiocarcinoma, non-small cell lung cancer, pancreatic adenocarcinoma, and thyroid carcinoma. However, several studies have shown overexpression of NEAT1 in these types of cancer. This discrepancy may be resulted from differences in patient characteristics, including the numbers, racial, and clinical subtypes of patients, or analysis methods and procedures adopted by different literature. For example, the results from RNA-seq data of TCGA patients may be different from the results based on microarray or tumor cell lines. Studies with a larger number of cases and stricter classification and analysis procedures are required to extensively explore this problem.
In addition, TCGA data showed that NEAT1 was down-expressed in breast invasive cancer, esophageal carcinomas and pheochromocytoma & paraganglioma, compared with their corresponding normal tissues. This is somehow surprising since many studies have demonstrated NEAT1 overexpressed and facilitated breast cancer cell growth and invasion via microRNA-related signaling pathways.21, 22 However, a recent study indicated that in breast invasive cancer, from TCGA datasets, NEAT1 was focally deleted in ∼8% of breast cancers and its promoters carried various mutations, and three out of the four mutations they validated reproducibly decreased NEAT1 expression compared with the wild type sequence.16 Therefore, the expression pattern of NEAT1 in breast cancer needs to be extensively studied according to their clinical and pathological type. As for esophageal carcinoma, although Chen et al showed that NEAT1 was highly expressed in esophageal squamous cell carcinoma and promoted esophageal tumor cell proliferation,23 Xiong et al concluded that NEAT1 was down-regulated in esophageal carcinoma through summarizing previous published lncRNA datasets and literature.7 Given that downregulation of NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia,20 NEAT1 may play a tumor-suppressive role, to some extent, in specific tumors. Further studies investigating the tumor-suppressive property and mechanisms of NEAT1 are imperative.
NEAT1 emerges as an important participant of the complex transcription network in cancer development
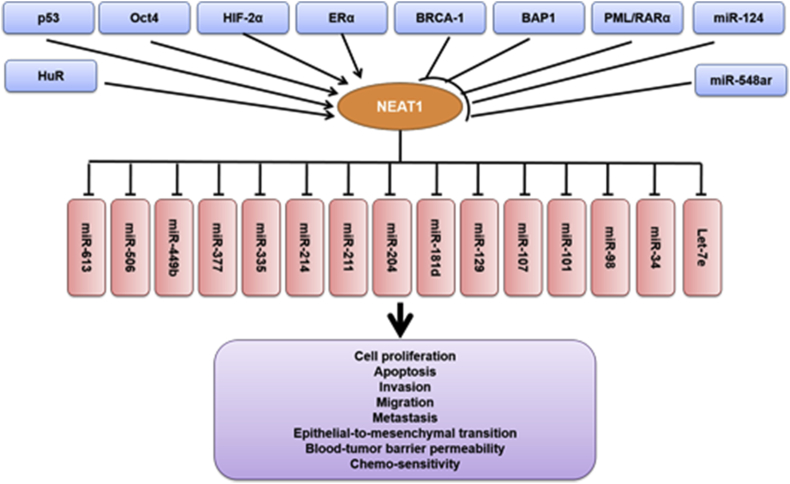
Upstream transcription factors that regulate NEAT1 expression
To explain the distinct expression patterns of NEAT1 in different cancers, we further summarized the reported upstream regulators of NEAT1. NEAT1 expression can be regulated by various transcription factors (Fig. 3). p53, Oct4, HIF-2α, estrogen receptor alpha (ERα), and Runx1 can induce the upregulation of NEAT1,19, 24, 25, 26, 27, 28 while BRCA1, BRCA-1 associated protein-1 (BAP1) and PML/RARα inhibit the expression of NEAT1.6, 14, 29 The expression of these transcription factors varies among different tumor backgrounds, which may lead to the distinct expression patterns of NEAT1 in different types of cancer.
Figure 3.
The upstream factors and downstream microRNA targets of NEAT1 in the transcription cascade of tumorigenesis.
Of the transcription factors demonstrated to regulate NEAT1 expression, p53 is the most reported one in different types of cancer. Indeed, NEAT1 is involved in the versatile roles of p53 in tumor regulation, including transformation suppression and chemo-sensitivity.19, 24, 25, 30 It is also closely correlated with p53-dependent DNA damage response in chronic lymphocytic leukemia and lymphoma.19 The ERα-regulated NEAT1 is a critical modulator of prostate cancer.27 In lung cancer, NEAT1 is up-regulated by the direct binding of Oct4, since knockdown of NEAT1 abolishes Oct4-mediated lung cancer cell growth and motility.26 In breast cancer, NEAT1 is inversely regulated by BRCA1 and miR-548ar-3p.29, 31 BRCA1 deficiency in both human normal/cancerous breast cells leads to NEAT1 overexpression and stimulates in vitro and in vivo breast tumorigenicity.29 On the other hand, Runx1 binds to the promoter of NEAT1 and positively regulates the expression of NEAT1.32 The ERα-NEAT1-FOXN3/NEAT1/SIN3A-GATA3 axis is also reported to be implicated in the progression of hormonally responsive breast cancer.13 In addition, tumor hypoxia induces NEAT1-associated nuclear paraspeckle formation through HIF-2α and accelerates breast cancer cell proliferation but reduces apoptosis.28 In cholangiocarcinoma, BAP1 dependent expression of NEAT1 contributes to drug sensitivity.14 In ovarian cancer, NEAT1 is stabilized by an RNA-binding protein HuR, but suppressed by miR-124-3p.33 In acute promyelocytic leukemia, NEAT1 expression is repressed by oncogenic PML/RARα and the reduction of NEAT1 by small interfering RNAs could block all trans retinoic acid-induced cell differentiation.6 Moreover, infection with Japanese encephalitis, rabies and HIV are reported to induce NEAT1 expression. Given to the close association between virus infection and cancer development, the overexpression of NEAT1 in specific tumors may be linked to viral infection.4 Taken together, diverse transcription factors either regulate the expression of NEAT1 positively or negatively. The aberrant expression of NEAT1 in certain types of cancer may be resulted from the cooperation and antagonism between different transcription factors.
Downstream microRNA targets of NEAT1 in diverse tumors
The mechanisms underlying tumor regulation are mostly reliant on the physical and functional interaction of NEAT1 with various microRNAs. We next summarized the target genes of NEAT1 through analyzing literature. A myriad of microRNAs have been reported to be regulated by NEAT1 (either through altered expression or sponging effect) and exerted tumor regulation function, i.e. let-7e, miR-34, miR-98, miR-101, miR-107, miR-129, miR-181d, miR-204, miR-214, miR-335, miR-377, miR-449b, miR-506, and miR-6139, 10, 11, 12, 21, 22, 34, 35, 36, 37, 38 (Fig. 3). Through influencing their target genes and associated signaling pathways, these microRNAs can regulate tumor development and progression. For example, the NEAT1/miR-335-5p/c-met axis plays a pivotal role in pancreatic cancer in vivo and in vitro.9 In laryngeal squamous cell cancer, high expression of NEAT1 decreases miR-107 expression, thus regulating CDK6 level and exerting an oncogenic effect.39 In hepatocellular carcinoma, overexpression of NEAT1 inhibits the expression of miR-129-5p and promotes tumor cell proliferation.10 In breast cancer, NEAT1 suppresses tumor growth through miR-101 dependent EZH2 regulation.22 NEAT1 promotes the malignant progression of glioma stem cells by downregulating let-7e expression.37 Also, miR-181d-5p can directly bind to NEAT1 and regulates permeability of the blood-tumor barrier through its downstream targets ZO-1, occludin, and claudin-5.12 In addition, NEAT1 functions as a miR-449b-5p sponge and promotes glioma pathogenesis.40 And NEAT1 can upregulate the green tea polyphenol (EGCG)-induced CTR1 to enhance cisplatin sensitivity in lung cancer by sponging hsa-mir-98-5p.41 Through interacting with various microRNAs, NEAT1 creates a complex transcription network that contributes to tumor modulation.
NEAT1 may exert its tumor modulation function through other approaches
Apart from sponging related microRNAs, NEAT1 may mediate tumor progression via other approaches, such as, paraspeckle-related function regulation. Paraspeckles are dynamic nuclear structures that sequester transcriptionally active proteins as well as RNA transcripts. It has been reported that NEAT1 could regulate transcription through protein sequestration into paraspeckles.42 Actually, tumor hypoxia induces the formation of NEAT1-containing paraspeckles and accelerates tumor cell growth and clonogenic survival.28 Carmen Adriaens et al also demonstrated that NEAT1-containing paraspeckles are upregulated by p53 and modulate tumor chemo-sensitivity.24 In addition, NEAT1 are also reported to be located in numerous non-paraspeckle foci, indicating potential paraspeckle-independent functions.43
Conclusions and future directions
Emerging evidence has demonstrated that NEAT1 plays critical roles in tumorigenesis of many types of cancer.5, 7, 8 Here, we systematically analyzed the expression patterns of NEAT1 in various normal and malignant tissues using public data portals, together with the recent progress in the study of NEAT1 in various types of tumor. Although discrepancy exists, NEAT1 expression is deregulated in certain types of tumor and its dysregulation is closely related with tumorigenesis and tumor progression through regulating tumor cell proliferation, invasion, EMT, etc. The expression of NEAT1 is directly induced or inhibited by different transcription factors, such as p53 or PML/RARα. NEAT1, in turn, can regulate a myriad of microRNAs and their target genes and thus modulating tumorigenesis. This suggests that NEAT1 is an important participant of the complex transcription network in cancer development.
Recently, several studies have revealed the mutations and polymorphisms of NEAT1 are also closely correlated with the prognosis of cancers,16, 18 which has added complexity to NEAT1-associated signaling and function. Moreover, since the two major isoforms of NEAT1 produced from alternative transcription have been reported to have different cellular location and tumor regulation properties,43, 44 it is necessary to distinguish the exact expression and function of these two isoforms. Apart from the reported microRNAs or transcription factors listed above, further studies illustrating the mechanisms of NEAT1 in modulating tumorigenesis are needed. More importantly, considering the distinct expression patterns of NEAT1 in different cancers (especially discrepancy in solid tumors and hematological malignancies), the diagnostic and prognostic value of NEAT1 as an independent biomarker in certain types of cancer should be more extensively confirmed for clinical application.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported in part by National Natural Science Foundation Grants of China (81530003) and China Postdoctoral Science Foundation (2017M611580).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Forrest M.E., Khalil A.M. Review: regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett. 2017;407:106–112. doi: 10.1016/j.canlet.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 2.Paralkar V.R., Weiss M.J. Long noncoding RNAs in biology and hematopoiesis. Blood. 2013;121(24):4842–4846. doi: 10.1182/blood-2013-03-456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 4.Bond C.S., Fox A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186(5):637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang J., Qiao F., Tu J. High expression of long non-coding RNA NEAT1 indicates poor prognosis of human cancer. Oncotarget. 2017;8(28):45918–45927. doi: 10.18632/oncotarget.17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng C., Xu Y., Xu L. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Canc. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong D.D., Feng Z.B., Cen W.L. The clinical value of lncRNA NEAT1 in digestive system malignancies: a comprehensive investigation based on 57 microarray and RNA-seq datasets. Oncotarget. 2017;8(11):17665–17683. doi: 10.18632/oncotarget.14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X., Li Z., Zheng H., Chan M.T., Wu W.K. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2) doi: 10.1111/cpr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J., Zhang Y., Yang J. NEAT1 regulates pancreatic cancer cell growth, invasion and migration though mircroRNA-335-5p/c-met axis. Am J Canc Res. 2016;6(10):2361–2374. [PMC free article] [PubMed] [Google Scholar]
- 10.Fang L., Sun J., Pan Z. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IkappaB. Am J Physiol Gastrointest Liver Physiol. 2017;313(2):G150–G156. doi: 10.1152/ajpgi.00426.2016. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y., Li T., Wei G. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2016;37(9):11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 12.Guo J., Cai H., Zheng J. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim Biophys Acta. 2017;1863(9):2240–2254. doi: 10.1016/j.bbadis.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Li W., Zhang Z., Liu X. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest. 2017;127(9):3421–3440. doi: 10.1172/JCI94233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parasramka M., Yan I.K., Wang X. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Canc. 2017;16(1):22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Li Y., Chen W. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641–27650. doi: 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rheinbay E., Parasuraman P., Grimsby J. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547(7661):55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Shuch B.M., Gerstein M.B. Whole-genome analysis of papillary kidney cancer finds significant noncoding alterations. PLoS Genet. 2017;13(3):e1006685. doi: 10.1371/journal.pgen.1006685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J., Li Y., Zhang J. Correlation between polymorphisms in microRNA-regulated genes and cervical cancer susceptibility in a Xinjiang Uygur population. Oncotarget. 2017;8(19):31758–31764. doi: 10.18632/oncotarget.15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blume C.J., Hotz-Wagenblatt A., Hullein J. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29(10):2015–2023. doi: 10.1038/leu.2015.119. [DOI] [PubMed] [Google Scholar]
- 20.Sedlarikova L., Gromesova B., Kubaczkova V. Deregulated expression of long non-coding RNA UCA1 in multiple myeloma. Eur J Haematol. 2017;99(3):223–233. doi: 10.1111/ejh.12908. [DOI] [PubMed] [Google Scholar]
- 21.Li X., Wang S., Li Z. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105(Pt 1):346–353. doi: 10.1016/j.ijbiomac.2017.07.053. [DOI] [PubMed] [Google Scholar]
- 22.Qian K., Liu G., Tang Z. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Archives Biochem Biophys. 2017;615:1–9. doi: 10.1016/j.abb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Chen X., Kong J., Ma Z., Gao S., Feng X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Canc Res. 2015;5(9):2808–2815. [PMC free article] [PubMed] [Google Scholar]
- 24.Adriaens C., Standaert L., Barra J. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 25.Mello S.S., Sinow C., Raj N. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31(11):1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jen J., Tang Y.A., Lu Y.H., Lin C.C., Lai W.W., Wang Y.C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Canc. 2017;16(1):104. doi: 10.1186/s12943-017-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakravarty D., Sboner A., Nair S.S. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhry H., Albukhari A., Morotti M. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34(34):4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo P.K., Zhang Y., Wolfson B. Dysregulation of the BRCA1/long non-coding RNA NEAT1 signaling axis contributes to breast tumorigenesis. Oncotarget. 2016;7(40):65067–65089. doi: 10.18632/oncotarget.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idogawa M., Ohashi T., Sasaki Y., Nakase H., Tokino T. Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int J Canc. 2017;140(12):2785–2791. doi: 10.1002/ijc.30689. [DOI] [PubMed] [Google Scholar]
- 31.Ke H., Zhao L., Feng X. NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Biol. 2016;10(suppl 1):11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barutcu A.R., Hong D., Lajoie B.R. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta. 2016;1859(11):1389–1397. doi: 10.1016/j.bbagrm.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai Y., Liu J., Zhang Z., Liu L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Canc Med. 2016;5(7):1588–1598. doi: 10.1002/cam4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B., Liu C., Wu Q. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophysl Res Commun. 2017;482(4):828–834. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 35.Yang X., Xiao Z., Du X., Huang L., Du G. Silencing of the long non-coding RNA NEAT1 suppresses glioma stem-like properties through modulation of the miR-107/CDK6 pathway. Oncol Rep. 2017;37(1):555–562. doi: 10.3892/or.2016.5266. [DOI] [PubMed] [Google Scholar]
- 36.Sun C., Li S., Zhang F. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7(32):51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong W., Zheng J., Liu X., Ma J., Liu Y., Xue Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget. 2016;7(38):62208–62223. doi: 10.18632/oncotarget.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Zou Q., Song M., Chen J. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed Pharmacother Biomed Pharmacother. 2017;94:612–618. doi: 10.1016/j.biopha.2017.07.111. [DOI] [PubMed] [Google Scholar]
- 39.Wang P., Wu T., Zhou H. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Canc Res CR. 2016;35:22. doi: 10.1186/s13046-016-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhen L., Yun-Hui L., Hong-Yu D., Jun M., Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2016;37(1):673–683. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 41.Jiang P., Wu X., Wang X., Huang W., Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget. 2016;7(28):43337–43351. doi: 10.18632/oncotarget.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirose T., Virnicchi G., Tanigawa A. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25(1):169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R., Harvey A.R., Hodgetts S.I., Fox A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. Rna. 2017;23(6):872–881. doi: 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Yang L., Zhao J. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Canc. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]