Abstract
Acetolactate synthase (ALS) catalyzes the initial step in the biosynthesis of branched-chain amino acids, and is highly conserved from bacteria to higher plants. ALS is encoded by a single copy gene in rice genome and is a target enzyme of several classes of herbicides. Although ALS mutations conferring herbicide-resistance property to plants are well documented, effect of Imazamox (IMZ) on rice and the mutations in ALS correlated with IMZ tolerance were unclear. In this article, the effect of IMZ on rice calli and seedlings in tissue culture conditions were evaluated. Also, the ALSA96V mutation was confirmed to improve IMZ tolerance of rice calli. Based on these results, ALS-assisted multiplex targeted base editing in rice was demonstrated in combination with Target-AID, a CRISPR/Cas9-cytidine deaminase fusion system [1], [2].
Specifications table
| Subject area | Biotechnology |
| More specific subject area | Plant Biotechnology, Plant genome editing and Plant breeding technique |
| Type of data | Tables and Figures |
| How data was acquired | Agrobacterium-mediated transformation, Plant tissue culture technique, Sanger sequencing and Fluorescence microscopic analysis |
| Data format | Analyzed data |
| Experimental factors | Not applicable |
| Experimental features | Multiplex targeted base editing using the Target-AID system [1] and rice callus lines carrying switch-mEGFP[2] |
| Data source location | Kobe University, Kobe, Japan |
| Data accessibility | Data are provided with this article |
| Related research article | Z. Shimatani, U. Fujikura, H. Ishii, Y. Matsui, M. Suzuki, Y. Ueke, K. Taoka, R. Terada, K. Nishida and A. Kondo Inheritance of co-edited genes by CRISPR-based targeted nucleotide substitutions in rice. Plant Physiol Biochem. 131 (2018), pp. 78-83. |
Value of the data
-
●
Effective IMZ concentrations were determined to suppress rice callus proliferation and seedling growth in tissue culture conditions.
-
●
The data demonstrated that ALS A96V mutation confers IMZ tolerance to rice calli and is thus it is applicable as an endogenous selectable marker indicating the activity of Target-AID system.
-
●
Simultaneous engineering of multiplex traits of rice calli was successfully demonstrated by Target-AID in combination with ALS-assisted selection. This will contribute to more efficient selection of the prospective cells carrying desired mutations as IMZ tolerance provide a useful index of Target-AID activity.
1. Data
This article shows the optimization of an ALS-assisted screening strategy that facilitates more efficient targeted nucleotide substitutions in rice using Target-AID system. Optimal IMZ concentrations to inhibit rice callus proliferation and plant growth in tissue culture conditions were determined (Fig. 1B and C). The conferring of IMZ tolerance to rice calli by introducing A96V mutation in ALS was confirmed (Table 1). On the basis of these results, simultaneous multiplex gene editing with ALS-assisted Target-AID syrategy was demonstrated to introduce A96V mutation to endogenous ALS as well as restoration of EGFP (Fig. 2 B–D, Table 2).
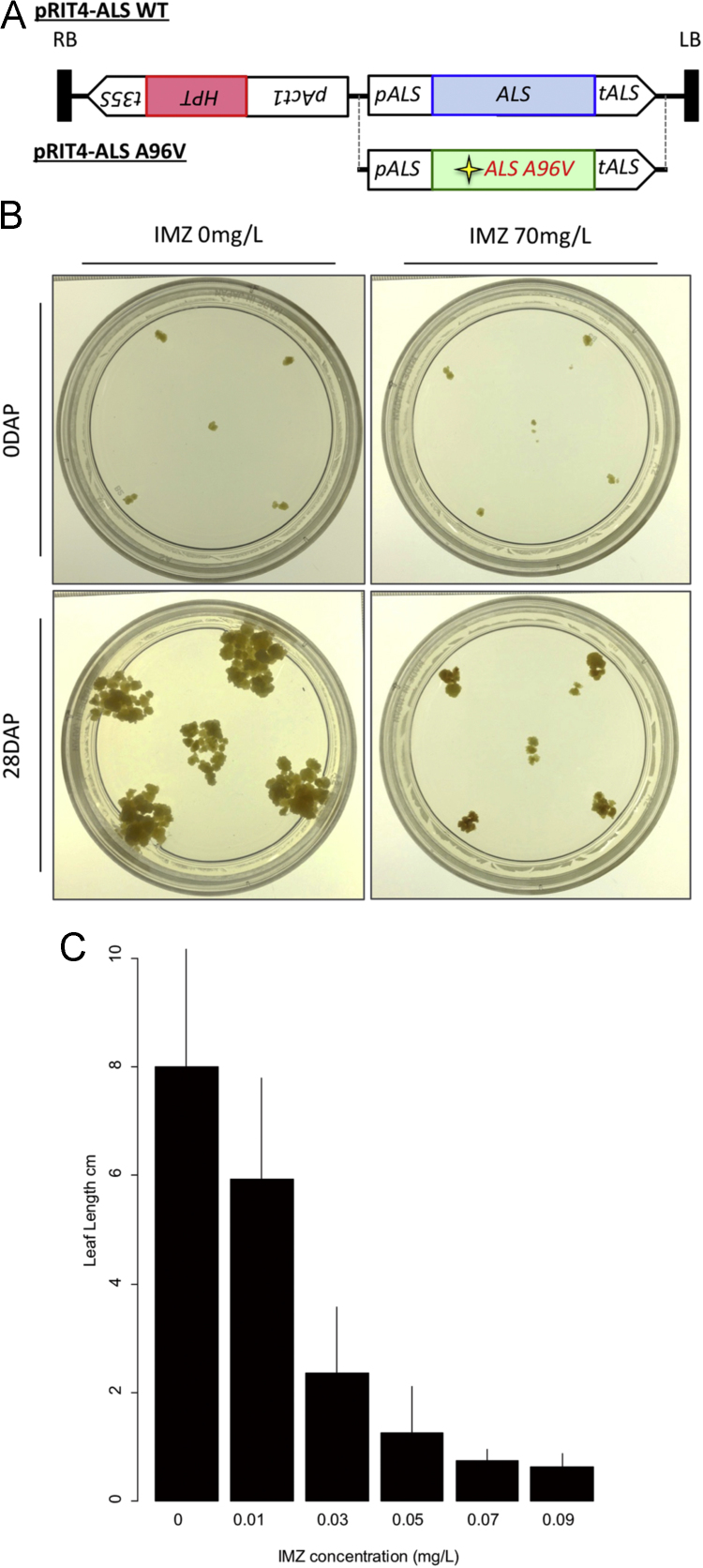
Fig. 1.
Imazamox tolerance-assay of rice calli and seedlings. (A) The T-DNA structures of the binary vectors used for Imazamox (IMZ) tolerance assay. The rice acetolactate synthase (ALS) gene was cloned and integrated into pRIT4 with its authentic promoter and terminator. The four-pointed star indicates the artificially induced nucleotide substitution leading to A96V mutation. pAct1, rice Actin 1 promoter with its intron 1; HPT, hygromycin phosphotransferase; t35S, cauliflower mosaic virus 35S terminator; RB, right border; LB, left border. (B) Effect of IMZ on proliferation of rice callus. Rice calli were cultured for 28 days on N6D medium supplemented with 0 and 70 mg/L IMZ. (C) Effect of IMZ on growth of rice seedlings. Rice seeds were germinated and grown for 10 days on 1/2MS medium supplemented with 0–0.09 mg/L IMZ under continuous light conditions. Mean lengths of leaf were indicated (n > 8). Bar = SD.
Table 1.
The efficiency of Imazamox selection of rice calli with ALS A96V mutation.
| Vector | Number of callus lines |
||
|---|---|---|---|
| Hygromycin resistant | Imazamox tolerant | Frequency (%) | |
| pRIT4-ALS WT | 169 | 6 | 3.6 |
| pRIT4-ALS A96V | 263 | 261 | 99.2 |
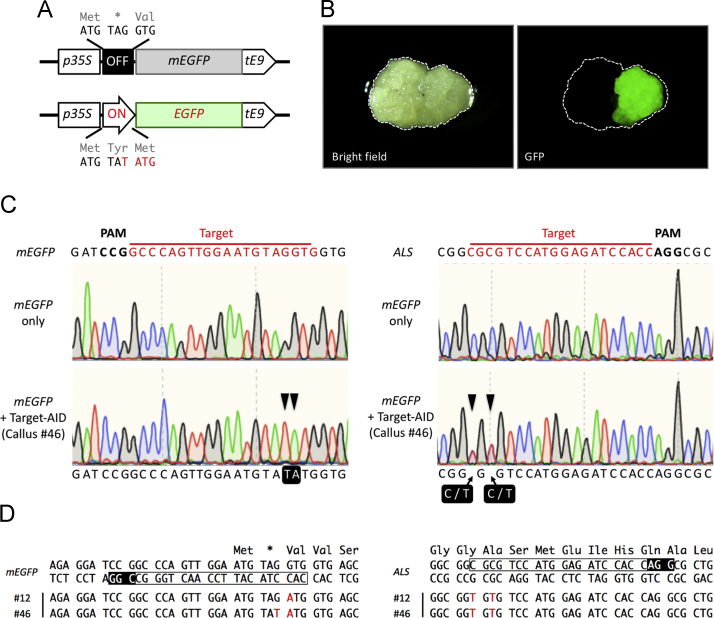
Fig. 2.
Simultaneous multiplex target editing by ALS-assisted Target-AID. (A) Schematic illustration of EGFP reporter assay. In switch-mEGFP reporter vector, single stop codon inserted immediately downstream of the initiation codon (Top). The switching module, TAG was altered to TAT by Target-AID to express EGFP (Bottom). (B) Fluorescence microscopic images of the rice callus. Expression of EGFP reporter was detected among double transformed callus lines carrying mEGFP and Target-AID. (C) Sequencing chromatograms showing the nucleotide substitutions by Target-AID in switch-mEGFP and ALS genes of calli carrying switch-mEGFP only (Top) and exhibiting EGFP expression and IMZ tolerance after introduced Target-AID vector (Bottom), respectively. Arrowheads with highlighted character indicate the mutation. (D) Sequence alignment of the mutations at mEGFP and ALS loci. Red letters indicate substituted nucleotides.
Table 2.
Number of rice calli carrying multiplex edited genes by Target-AID.
| Phenotype analysis | Sequencing analysis | ||
|---|---|---|---|
| Analyzed | GFP-positive and Imazamox tolerant | ALS A96V | Recovered EGFP |
| 124 | 3 | 2 | 2 |
2. Experimental design, materials, and methods
2.1. Evaluation of Imazamox tolerance of rice in tissue culture conditions
Effects of IMZ concentration in N6D media [3] on rice callus were evaluated. N6D and N6DSE-IMZ medium containing IMZ at gradual concentrations (30, 50, 70 mg/L) were used in this assay. The proliferation of rice callus was strictly inhibited by 70 mg/L IMZ (Fig. 1B).
Wild type rice ALS gene was cloned from genomic DNA by PCR amplification using the appropriate primers numbered as 1 and 2 in Table 3. The ALS A96V gene was synthesized via overlapping PCR procedure using the primers 1–6. The DNA sequence of the clones carrying ALS genes were confirmed using the primers 7–24. The cloned genes were installed to pRIT4, a derivative of binary vector pRIT3 [2] harboring a modified HPT gene [4]. The resultant vectors, pRIT4-ALS WT and pRIT4-ALS A96V (Fig. 1A) were introduced to rice calli by Agrobacterium-mediated transformation according to a previous report [5] using the plant media shown in Table 4. Transformed calli were selected on N6DSE-H40 medium over 3 weeks, then subcultured on N6DSE-H40IMZ70 medium to evaluate IMZ tolerance. After the selection over 2 months, 99.2% of calli introduced ALS A96V exhibited IMZ tolerance and proliferated on the media, whereas almost all the calli were sensitive to IMZ when introduced WT ALS (Table 1).
Table 3.
List of oligonucleotides used in this study.
| Serial number | Name | Sequence (5′--->3′) | Note |
|---|---|---|---|
| pRIT4-ALS A96V | |||
| 1 | ALS cloning-F | AGTCCCTGCAGGTTAATTAACTTGCGCTGCGTTTGTGCGGGTGCG | Construction of pRIT4-ALS vectors |
| 2 | ALS cloning-R | TGACGGTACCACTAGTTAGTAGTACCCAATAAGATCGACCGAAGAGA | |
| 3 | ALSA96V-F | CGGGCGGCGTGTCCATGGAGATCCACCAGGCGCTG | Generating ALS A96V variant by overlapping-PCR |
| 4 | ALSA96V-F2 | GGCGTCAGCGACGTGTTCGCCTACCCGGGCGGCGTGTCCATGGAGATCCACCAGGCGCTG | |
| 5 | ALSA96V-R2 | GAGCGCGTCAGCGCCTGGTGGATCTCCATGGACACGCCGCCCGGGTAGGCGAACACGTCG | |
| 6 | ALSA96V-R | TCCATGGACACGCCGCCCGGGTAGGCGAACACGTC | |
| ALS | |||
| 7 | pALS F-1 | CATCCAATCGACTGACACGCGGGCCCAGAT | PCR and sequencing analysis of rice ALS gene |
| 8 | pALS R-1 | GGTTTCTGGGTTTGGGCGAGAGGGAGAGAG | |
| 9 | pALS R-2 | ATCTGGGCCCGCGTGTCAGTCGATTGGATG | |
| 10 | ALS F-1 | CCGTAAGAACCACCAGCGACACCACGTCCT | |
| 11 | ALS F-2 | GGAGACGCCCATAGTCGAGGTCACCCGCTC | |
| 12 | ALS F-3 | CAGGGCCAAGATTGTGCACATTGACATTGA | |
| 13 | ALS F-4 | CTTGGGCAACCCGGAATGTGAGAGCGAGAT | |
| 14 | ALS F-5 | GGTGCTTCTGTGGCTAACCCAGGTGTCACA | |
| 15 | ALS R-1 | TTAATACACAGTCCTGCCATCACCATCCAG | |
| 16 | ALS R-2 | GTGTAATATTGTGCCGCCCACATCTGGTGC | |
| 17 | ALS R-3 | CCAACCAGACGCAAGACCTGCTCAAGCAAT | |
| 18 | ALS R-4 | TCGCCCTGCTCGTGGCGGAAGAGGTGGTTG | |
| 19 | ALS R-5 | ATGTCCGCGCCCTTGCGGGGCTCGGCCGGC | |
| 20 | ALS R-6 | GAGCGGGTGACCTCGACTATGGGCGTCTCC | |
| 21 | tALS F-1 | GGCAAAGCACCAGCCCGGCCTATGTTTGAC | |
| 22 | tALS F-2 | TCTATGCAATAGCTCTGAGTTAAGTGTTTC | |
| 23 | tALS R-1 | GGAGAGTACTTCGTGTGATGACAGTTGAGC | |
| 24 | tALS R-2 | CACATACAAACATCATAGGCATACCACTCT | |
| switch-mEGFP | |||
| 25 | SbfI-p35S-F | ATGCATCCTGCAGGCTCTAGAGGATCCCCCCTCAG | PCR and sequencing analysis of mEGFP gene on pRIT3-mEGFP |
| 26 | EGFP-NotI-R | AGCCGGGCGGCCGCTTTACTTGTACAGCTCGTCCA | |
| 27 | p35SF-1 | CGCACAATCCCACTATCCTTCGCAAGACCC |
Table 4.
Media composition for plant tissue culture in this study.
| Medium | N6D | N6DSE-H40 | N6DSE-H40P50 | N6DSE-IMZ | N6DSE-H40IMZ70 | 1/2MS | 1/2MS-IMZ | |
|---|---|---|---|---|---|---|---|---|
| Application | Callus proliferation | Selection of transgenic calli | Selection of ALS A96V calli | Germination | Selection of ALS A96V Plants | |||
| Basal medium | N6D | N6D | N6D | N6D | 1/2MS | |||
| Selective agents | Hygromycin | – | 40 mg/L | 40 mg/L | – | 40 mg/L | – | – |
| Paromomycin | – | – | 50 mg/L | – | – | – | – | |
| Imazamox | – | – | – | 30, 50, 70 mg/L | 70 mg/L | – | 0.01–30 mg/L | |
| Gelling agents | Gelrite | 4 g/L | 4 g/L | – | 4 g/L | 4 g/L | 4 g/L | 4 g/L |
| Agarose | – | – | 8 g/L | – | – | – | – | |
The minimum effective concentration of IMZ on rice seedlings in aseptic conditions were determined as follows. Wild-type rice seeds were germinated and grown on 1/2MS media containing IMZ at concentration of 0.01, 0.03, 0.05, 0.07, 0.09, 0.1 and 0.25 mg/L (Table 4). The growth of the seedlings was analyzed by measuring their shoot length at 7 days after planting. As a result, seedling growth was remarkably suppressed by IMZ at 0.07 mg/L or higher concentration (Fig. 1C).
2.2. Multiplex editing of endogenous genes by Target-AID
To demonstrate the multiplex gene editing by Target-AID, the callus lines harboring pRIT3-mEGFP [2] were used in this experiment. Such calli were confirmed to carry dysfunctional EGFP (switch-mEGFP) containing a premature stop codon right after the initiation codon (Fig. 2A). A vector for Target-AID system expressing nCas9(D10A)-PmCDA1 with gRNAs corresponding to endogenous ALS and mEGFP was introduced by Agrobacterium-mediated transformation. After selection on N6DSE-H40P50 media, the double transformants were subcultured on N6DSE-IMZ70 medium over 2 months. As a result, 3 callus lines exhibiting IMZ tolerance and EGFP expression were obtained from 124 double transformants (Table 2, Fig. 2B). The targeted nucleotide substitutions were confirmed by direct DNA sequencing analysis using primers 10, 24 for endogenous ALS and 25–27 for switch-mEGFP (Table 3, Table 2 calli were found to harbor both of the desired mutations leading to ALS A96V and functional recovery of EGFP gene (Fig. 2A, C, D).
Acknowledgments
We thank Y. Ueke, M. Suzuki and A. Miyabe for their technical assistance. We also thank Dr. C. Vavricka for helpful discussions. This work was supported by Cross-ministerial Strategic Innovation Promotion Program (SIP); ‘Technologies for Creating Next-generation Agriculture, Forestry and Fisheries.’ This work was also partly supported by a Special Coordination Fund for Promoting Science and Technology, Creation of Innovative Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) from the Ministry of Education, Culture, Sports and Technology (MEXT) of Japan, the commission for Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterial from the Ministry of Economy, Trade and Industry (METI), of Japan and by JSPS KAKENHI Grant numbers 26119710 and 16K14654.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.dib.2018.08.124.
Transparency document. Supporting information
Supporting information
References
- 1.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y., Shimatani Z., Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 2.Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., Teramura H., Yamamoto T., Komatsu H., Miura K., Ezura H., Nishida K., Ariizumi T., Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 3.Toki S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Report. 1997;15:16–21. [Google Scholar]
- 4.Bilang R., Iida S., Peterhans A., Potrykus I., Paszkowski J. The 3′-terminal region of the hygromycin-B-resistance gene is important for its activity in Escherichia coli and Nicotiana tabacum. Gene. 1991;100:247–250. doi: 10.1016/0378-1119(91)90375-l. [DOI] [PubMed] [Google Scholar]
- 5.Terada R., Johzuka-Hisatomi Y., Saitoh M., Asao H., Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 2007;144:846–856. doi: 10.1104/pp.107.095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information