Abstract
Reactive oxygen species (ROS), which are a byproduct of oxidative metabolism, serve as signaling molecules in a number of physiological settings. However, if their levels are not tightly maintained, excess ROS lead to potentially cytotoxic oxidative stress. Accordingly, several transcriptional regulatory networks have evolved to include components that are highly ROS-responsive. Depending on the context, these regulatory networks can leverage ROS to respond to nutrient conditions, metabolism, or other physiological signals, or to respond to oxidative stress. However, ROS signaling is complex, so regulatory interactions between various ROS-responsive transcription factors are still being mapped out. Here we show that the transcription factor NRF2, a key regulator of the adaptive response to oxidative stress, directly regulates expression of HIF1A, which encodes HIF1α, a key transcriptional regulator of the adaptive response to hypoxia. We used an integrative genomics approach to identify HIF1A as a ROS-responsive transcript and we found an NRF2-bound antioxidant response element (ARE) approximately 30 kilobases upstream of HIF1A. This ARE sequence is deeply conserved, and we verified that it is directly bound and activated by NRF2. In addition, we found that HIF1A is upregulated in breast and bladder tumors with high NRF2 activity. Taken together, our results demonstrate that NRF2 targets a functional ARE at the HIF1A locus, and reveal a direct regulatory connection between two important oxygen responsive transcription factors.
Highlights
-
•
HIF1A transcription is induced by oxidative stress.
-
•
NRF2 binds to an antioxidant response element (ARE) ~ 32 kilobases upstream of HIF1A.
-
•
Regulatory output at the ARE upstream of HIF1A is increased by oxidative stress.
-
•
NRF2 activation is associated with HIF1A upregulation in select cancer types.
Graphical abstract
1. Introduction
The relationship between the cellular response to oxygen and the detoxification of reactive oxygen species (ROS) is an ancient one [1]. Oxygen is used for the conversion of sugar into ATP via mitochondrial oxidative phosphorylation, creating endogenous ROS as a byproduct. ROS have the potential to react with and damage all classes of macromolecules, so they are often viewed as cytotoxic. Consistent with this, decades of toxicity studies have reliably shown that damaging levels of ROS are generated when cells are exposed to a range of environmental xenobiotics. However, despite their cytotoxic potential, ROS are also required for proper cell signaling and development. Many properties of ROS make them ideal signaling molecules, including their potential for rapid modification of proteins and close ties to cellular metabolism [2], [3]. In fact, many proteins that regulate angiogenesis, cell cycle progression, stem cell self-renewal, and differentiation are activated upon being oxidized by ROS [4], [5], [6], [7]. Thus, the role of cellular ROS is somewhat paradoxical: ROS can cause damage with potentially dire consequences, yet ROS are required as signaling molecules for normal development. Fortunately, cells have evolved many antioxidant mechanisms that can directly combat the harmful effects of ROS, while keeping ROS levels tightly balanced in order to prevent disruption of redox homeostasis.
Much of the response to ROS is regulated at the transcriptional level, and vertebrates have multiple redox-responsive transcription factors (TFs). This includes TFs commonly viewed as stress-responsive like NRF2 (encoded by the gene NFE2L2), p53 (encoded by the gene TP53), HIF1α (encoded by the gene HIF1A), as well as immunomodulatory TFs such as NF-κB, STAT, and the AP-1 complex (Fos/Jun), and more [8], [9], [10], [11], [12], [13], [14], [15]. The range of processes regulated by these TFs is broad: factors like NRF2 and p53 regulate extensive gene networks responsible for mitigating ROS-induced damage, whereas HIF1α regulates the adaptive response to hypoxia [9], [16], [17]. Considering the number of redox-responsive TFs encoded within metazoan genomes, there is significant potential for crosstalk between these TF regulatory networks during the fine-tuning of gene expression changes in response to changing levels of oxygen or ROS.
NRF2, a key regulator of the transcriptional response to oxidative stress, and HIF1α, a key regulator of the transcriptional response to hypoxic stress, represent an interesting pair of TFs with underappreciated potential for functional connectivity. Both TFs regulate oxygen-associated processes but, considering the environments to which they are most responsive, are often viewed to be on opposite ends of spectrum. Nevertheless, the activities of both NRF2 and HIF1α are regulated by ROS. NRF2 activity is largely regulated by KEAP1, an inhibitory protein that binds cytoplasmic NRF2 and targets it for proteosomal degradation, and KEAP1 contains over two dozen cysteine residues that can be modified by ROS [18]. ROS modification of KEAP1's cysteine residues decreases its ability to target NRF2 for ubiquitination so, as a consequence, NRF2 activity is increased by ROS. HIF1α is regulated via a similar mechanism. In general, HIF1α activity is low during normoxia – this is regulated by prolyl hydroxylase (PHD) proteins that hydroxylate HIF1α and target it for proteasomal degradation [19], [20]. However, hypoxia triggers a paradoxical increase in ROS generation at the mitochondria, these hypoxia-induced ROS inhibit PHD activity and, therefore, increase HIF1α stability and activity [21], [22], [23], [24], [25]. Thus, although they regulate two very different adaptive responses, the stability and activity of both NRF2 and HIF1α are regulated by ROS.
Because ROS can modify the activity of many TF networks, an unbiased genome-wide view of the transcriptional response to ROS has the potential to identify novel connections between redox-responsive TFs. With this in mind, we integrated publicly available transcriptomic data that measured the dynamics of gene expression in response of human hepatoma cells (HepG2) to multiple ROS inducers [26] with TF binding data (genome-wide chromatin immunoprecipitation, or ChIP-seq) from multiple sources [27], [28], [29], [30]. Several unique gene expression clusters were identified, including a prominent gene set induced within hours of ROS exposure that was also enriched for NRF2 ChIP-seq targets. Notably, network analysis revealed HIF1A, the gene that encodes HIF1α, as a central member of this gene set. We confirmed these genomic findings in multiple cell lines, we identified a functional NRF2 binding site (i.e., an antioxidant response element, or ARE) approximately 32 kilobases upstream of HIF1A, and we found that oncogenic NRF2 mutations are associated with high HIF1A expression in multiple cancer types. These findings are consistent with recent work suggesting NRF2 plays a role in HIF1A expression [31], [32], and our work lends further support to a model in which NRF2 is a direct regulator of HIF1A transcription.
2. Results and discussion
2.1. A genome-wide view of the transcriptional response to oxidative stress
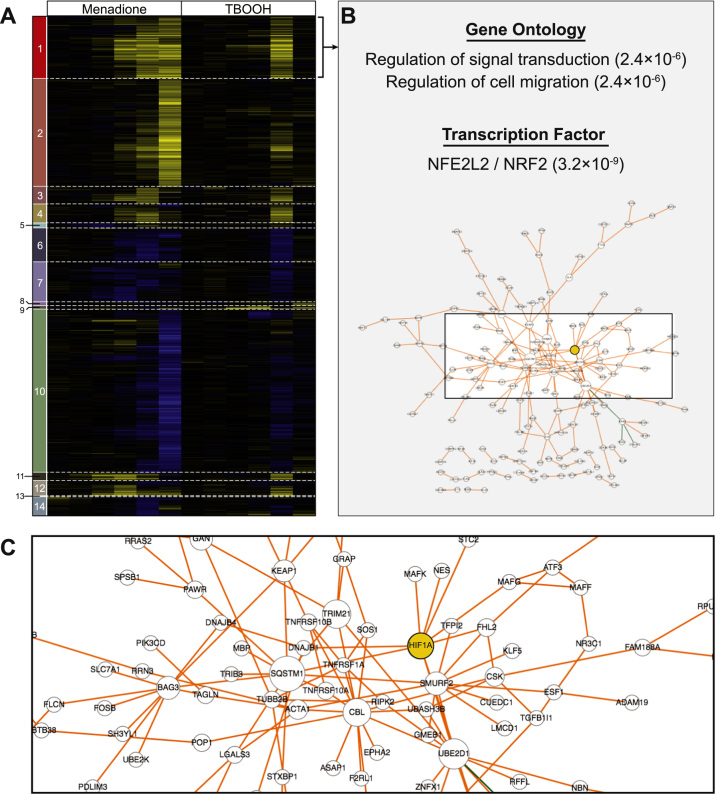
To get a global view of the transcriptional response to ROS we made use of a well-designed, previously published expression profiling dataset [26]. This time course microarray study monitored gene expression in HepG2 cells treated with menadione, tert-butyl hydroperoxide, or hydrogen peroxide; we focused on the 0.5, 1, 2, 4, 8, and 24 h time points following exposure to menadione or tert-butyl hydroperoxide (TBOOH), as these samples had the most robust ROS-mediated gene expression changes. From these data, we generated a list of differentially expressed genes consisting of any gene that was induced or repressed at least 1.5-fold at any time point post-ROS induction, and used hierarchical clustering to identify gene sets with similar expression patterns (Fig. 1A). The responses to menadione and TBOOH were generally concordant, although transcriptional changes in response to menadione were apparent at earlier time points and often continued until 24 h post-treatment, whereas the transcriptional response to TBOOH peaked after 8 h. Nevertheless, despite this difference, the gene expression changes associated with these distinct oxidants allowed for identification of genes consistently up- or downregulated by ROS. Several interesting gene expression clusters emerged from this analysis (Fig. 1A; Supplementary Table 1). For example, Cluster 1 consists of genes induced within the first 8 h after oxidative stress, whereas Cluster 2 consists of genes that required a full 24 h of menadione treatment before they were robustly induced. Similarly, genes in Clusters 6 and 7 were repressed within the first 8 h after oxidative stress, and genes in Cluster 10 required 24 h of menadione treatment before they were strongly repressed.
Fig. 1.
A cluster of putative NRF2-dependent, ROS-responsive genes includes HIF1A. (A) Hierarchical clustering of gene expression changes after exposure of human hepatoma cell line (HepG2) to the ROS inducers menadione or tert-butyl hydroperoxide (TBOOH) for 0.5, 1, 2, 4, 8, and 24 h. Data are from Deferme et al. Genes that were induced or repressed at least 1.5-fold in any of these conditions are included; yellow represents induction by ROS, blue represents repression by ROS, and black represents no change. (B) Cluster 1 is enriched for genes involved in signal transduction and cell motility, and enriched for genes called as NRF2 targets based on genome-wide chromatin immunoprecipitation (ChIP-seq) data. P-values (Fisher's exact test) are as calculated by Enrichr [30]. Network analysis [38] based on protein-protein interactions highlighted HIF1A (yellow) as central to Cluster 1 (see also Supplementary Table 2). (C) The same network described in (B), zoomed in to highlight HIF1A.
Considering each expression cluster consists of genes following the same expression pattern, we next asked whether the genes in each cluster were likely to be regulated by the same transcription factors (TFs). We used Enrichr [30], which has compiled hundreds of publicly available ChIP-seq datasets, to scan for TFs that bind near a significant fraction of the genes in each cluster (see Methods) (Supplementary Table 2). Nine of the fourteen clusters were significantly associated with at least one TF. Cluster 1, for example, is most significantly associated with loci bound by NRF2, one of the established ROS-responsive TFs [33], [34]. NRF2 is also one of the top three TFs associated with Cluster 2, but this cluster is most enriched for RUNX1-bound genes. Interestingly, although RUNX1 has not previously been implicated with gene induction in response to ROS, RNT-1, a nematode ortholog of RUNX1, regulates oxidative stress responsive gene expression in Caenorhabditis elegans [35]; this connection may indicate a previously unrecognized redox-responsive role for human RUNX1. Repressed gene sets are associated with TFs like the cell cycle regulator E2F4 (Clusters 6, 10, 14) [36] and hepatocyte specification factor HNF4A (Cluster 10) [37], suggesting that prolonged ROS exposure represses both proliferation and hepatocyte identity regulatory networks. Overall, these clusters of co-expressed genes and their associated TFs represent a valuable platform for identifying potential connections between the regulatory networks that respond to ROS.
2.2. Network analysis highlights HIF1A as a ROS-induced gene
We next chose to focus on Cluster 1, which was interesting for multiple reasons. First, of the two largest clusters upregulated by ROS (Clusters 1 and 2), genes in Cluster 1 responded much faster and were generally induced within the first 8 h after oxidant exposure. Second, this cluster was enriched for NRF2 target genes based on overlap with ChIP-seq data, and the cluster contains many expected NRF2 targets, including HMOX1, GCLC, GCLM, SLC7A11, SQSTM1, KEAP1, and EPHA2. Combined, these trends suggest this is a rapidly-responding and functionally relevant gene set. However, Cluster 1 consists of 381 genes, so it is made up of more than just canonical antioxidant response genes; this is also evident based on the top gene ontology enrichment categories for this cluster (regulation of cell migration [GO:0030334]; regulation of signal transduction [GO:0009966]) neither of which are the clearly linked to the antioxidant response (Fig. 1B). Therefore, we used network analysis [38] to find potentially interesting genes within Cluster 1 (Fig. 1B–C). Network analysis links genes based on known physical or genetic interactions, and highly linked genes in a network are often functionally important [39], [40]. The five most central members of the Cluster 1 network (based on betweenness centrality) were SQSTM1, UBE2D1, CBL, TRIM21, and HIF1A (Supplementary Table 3). SQSTM1 is involved in multiple protein degradation pathways, UBE2D1 is a ubiquitin-conjugating enzyme, and CBL and TRIM21 are both ubiquitin ligases – although all four are likely to play an important role in the response to ROS, and SQSTM1 is a well-characterized NRF2 target gene, the network centrality of these proteins can be explained by the broad protein-protein interaction spectrum associated with ubiquitin mediated degradation [41], [42], [43], [44]. However, HIF1A, which encodes HIF1α, is typically associated with hypoxic stress and, therefore, an unanticipated member of this gene set. Nevertheless, HIF1A's central presence in Cluster 1 suggests it is subject to ROS-responsive transcriptional regulation, and implies a potential role for NRF2, so we set out to explore HIF1A regulation in more detail.
The HIF1A expression pattern described above was based on microarray data from HepG2 cells. To verify that HIF1A is a ROS-responsive transcript in multiple contexts, we monitored its expression in HepG2 and two breast cancer cell lines, MCF7 and MDA-MB-231, after exposure to menadione or TBOOH. Breast cancer cells, particularly the triple-negative cell line MDA-MB-231, were chosen because high HIF1α expression in these cells is associated with multiple negative characteristics (chemoresistance, pluripotency, etc.) [45], [46]. To ensure we were measuring a response to ROS and avoid measuring gene expression changes associated with cell death, gene expression was monitored after eight hours of exposure, based on menadione or TBOOH concentrations that left at least 80% viable cells after 24 h for each cell line. Indeed, HIF1A expression was significantly induced by both menadione and TBOOH in all three cell lines (Fig. 2 and Supplementary Fig. 1).
Fig. 2.
Menadione treatment increases HIF1A mRNA expression in multiple cell types. (A) HIF1A expression in HepG2 cells treated for 8 h with vehicle control (ethanol, EtOH) or menadione. Gene expression values were measured by quantitative reverse transcription PCR, and normalized relative to ACTB expression. (B) Same as (A) only for MCF7 breast cancer cell. (C) Same as (A) only for MDA-MB-231 breast cancer cells. Gene expression changes were measured after 8 h of menadione exposure and, to avoid confounding cell death gene expression signatures, menadione treatments for each cell line were based on a concentration that resulted in 80% cell viability after 24 h (24 h EC80). The 24 h EC80 for HepG2 was 17 μM, for MCF7 was 6.3 μM, and for MDA-MB-231 was 10 μM. Asterisks represent p-values for menadione versus vehicle for each cell type (***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05, Welch's t-test).
The work described above is not the first to demonstrate that HIF1A is a ROS-responsive transcript, however. Although much research effort has focused on the post-translational regulation of HIF1α levels, largely because the rapid increase in HIF1α protein stability is central to the adaptive response to hypoxia because [47], [48], several groups have recognized that regulation of HIF1α expression is also important [45], [49], [50], [51], [52]. And, importantly, HIF1A expression has been linked to ROS in multiple settings: HIF1A mRNA is upregulated by ROS-inducing mutations in mouse lung carcinoma cells, by multiple sources of ROS in pulmonary artery smooth muscle cells, by chemotherapeutic-induced ROS, and by the ROS-inducer arsenite [32], [45], [53], [54]. Overall, in combination with this previously published work, these results demonstrate that ROS can activate HIF1A expression in multiple cellular contexts.
2.3. An NRF2-bound antioxidant response element (ARE) at the HIF1A locus
The gene expression cluster that includes HIF1A (Cluster 1, Fig. 1A) is significantly enriched for genes called as NRF2 targets based on ChIP-seq data. Overall, this cluster contained 53 putative NRF2 target genes (Supplementary Table 4) – HIF1A is not on this list. However, the program we used to identify significant TFs for each cluster, Enrichr, is conservative in its identification of a TF's putative target genes because it selects for TF binding sites that are close to transcription start sites (TSS) [30], [55]. This conservative approach is suitable for unbiased scanning for enriched TFs across gene lists, but it is likely to miss many TF target genes because TSS-distal binding events are just as likely to regulate gene expression [56]. Therefore, we used published NRF2 ChIP-seq data [33], [34] to manually search for NRF2 binding sites near HIF1A; we focused on data from human lymphoblastoid cell lines treated with sulforaphane, an NRF2 activator, because this dataset is robust and highly concordant with NRF2 ChIP-seq data from additional conditions and cell lines [33], [34], [57]. These data revealed a strong NRF2 binding site approximately 32.5 kb upstream of the HIF1A TSS (Fig. 3A) (NRF2 ChIP peak coordinates: chromosome 14, 62,129,227 – 62,129,613; GRCh37/hg19 human genome assembly). This region was also called as a strong NRF2 ChIP-seq peak in HepG2 cells, K562 leukemia cells, A549 lung cancer cells, and BEAS-2B bronchial epithelial cells (not shown). Although this NRF2 binding site is > 30 kb from HIF1A, functional NRF2 targeted AREs are often distal to gene promoters [58]. This binding site also falls within a topogically associated domain with HIF1A (Supplementary Fig. 2A), so it is structurally associated with the HIF1A locus and, therefore, just as likely to affect HIF1A expression as more proximal cis-regulatory regions [59], [60], [61]. In addition, this NRF2 binding site falls within the most prominent enhancer region upstream of HIF1A based on compiled chromatin state data from 127 human tissues and cell types (Supplementary Fig. 2B) [62]. Finally, this NRF2 binding event is centered on a perfect match to the ARE consensus (Fig. 3A) and, importantly, this perfect ARE sequence is strongly conserved across mammals (Fig. 3B).
Fig. 3.
An NRF2 binding site at the HIF1A locus. (A) An NRF2 ChIP-seq peak and antioxidant response element (ARE) sequence at the human HIF1A locus. ChIP-seq data are from lymphoblastoid cell lines treated with sulforaphane [33], [34], [57], and the same ChIP-seq peak is seen in NRF2 data from four additional human cell lines (see main text). Only protein coding genes are represented; see Fig. S2 for non-coding RNAs at this locus. (B) The ARE highlighted in (A) is highly conserved. (C) NRF2-MAFG electrophoretic mobility shift assay (EMSA) using the ARE from the NQO1 promoter as a labeled probe. Lane 1 contains the labeled NQO1 ARE probe with no protein, lane 2 contains the NQO1 probe with purified MAFG only, lane 3 contains the NQO1 probe with purified NRF2 only, and lane 4 contains the NQO1 probe with NRF2 and MAFG. Binding is only seen when both proteins are present in the reaction. Lanes 5–8: NRF2 and MAFG are present in each lane. Competition reactions included addition of excess unlabeled competitor probes containing the wild-type (WT) HIF1A ARE, a mutated (mut) HIF1A ARE, the wild-type NQO1 ARE, or a mutated HIF1A ARE, as indicated.
Based on the ChIP-seq data, NRF2 binding at the ARE-containing HIF1A enhancer is significantly stronger in cells treated with the NRF2-inducer sulforaphane in comparison cells treated with vehicle control (Supplementary Fig. 3A–B). In addition, electrophoretic mobility shift assays (EMSAs) performed with nuclear lysates from either vehicle- or menadione-treated MCF7 cells revealed a ROS-responsive protein binding signal at the HIF1A ARE is nearly identical to ROS-responsive binding at the NQO1 ARE, a well-studied NRF2 binding site (Supplementary Fig. 3C–D). These results further support the idea that NRF2 can bind ARE sequence within the ChIP-seq peak upstream of HIF1A, so we then used EMSAs with affinity purified proteins to determine whether NRF2, in complex with its DNA binding cofactor MAFG, can directly bind the HIF1A ARE. Specifically, we performed competition-based EMSAs using a labeled DNA probe based on the well-studied ARE at the NQO1 locus – this probe was bound by NRF2-MAFG heterodimers (Fig. 3C). As expected, an unlabeled version of the NQO1 probe competes for binding with the labeled probe, and mutation of the NQO1 ARE sequence eliminates competition for binding; thus, this interaction is specific and ARE-dependent. Importantly, a similar pattern was seen with the HIF1A ARE sequence, which also competed for binding with the labeled NQO1 probe in an ARE-dependent manner. Thus, NRF2, in complex with its DNA binding cofactor MAFG, directly binds the HIF1A ARE sequence.
To determine whether the HIF1A ARE has cis-regulatory activity, we generated plasmid constructs in which this ARE-containing region drives expression of the luciferase reporter gene. Both wild type (HIF1AWT) and ARE mutant (HIF1AMut) constructs were generated, and reporter assays were performed in HepG2, MCF7, and MDA-MB-231 cells, with or without menadione treatment. Reporter assays performed with TBOOH treatment were unsuccessful because all luciferase activity, even for positive controls, was eliminated by TBOOH (not shown); menadione did not have this effect, so it was our ROS inducer of choice for these assays. As seen in Fig. 4, menadione treatment led to a significant increase in HIF1AWT reporter activity in all three cell types, and this induction was eliminated by mutation of the ARE sequence (HIF1AMut). Interestingly, the ARE is required for both basal (non-stress) and inducible activity of this region in MCF7 and HepG2 cells, while it is only required for inducible activity in MDA-MB-231 cells. This suggests that additional, non-ARE binding TFs might also act on this DNA region to promote basal expression in MDA-MB-231 cells. Despite these cell-specific differences, the HIF1A ARE is clearly responsible for menadione-driven increases in the regulatory output of this DNA region. Taken together, the results described in this section indicate that this deeply conserved NRF2 binding site is a functional ARE that falls within an important HIF1A-associated enhancer region.
Fig. 4.
Menadione treatment increases HIF1A ARE activity in multiple cell types. (A) Reporter assays in which the region surrounding the HIF1A ARE (HIF1AWT), or a construct in which the ARE was mutated (HIF1AMut), were cloned upstream of a luciferase reporter gene. HepG2 cells were transfected with each plasmid and treated with either vehicle control (EtOH) or menadione for 24 h. Cells were then assessed for luciferase activity, and all values were normalized to a control reporter plasmid driven by the LDHA promoter region. (B) Same as (A) only for MCF7 breast cancer cell. (C) Same as (A) only for MDA-MB-231 breast cancer cells. P-values for menadione versus vehicle for each reporter construct are represented with asterisks (***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05, Welch's t-test).
2.4. A role for NRF2 in HIF1A expression
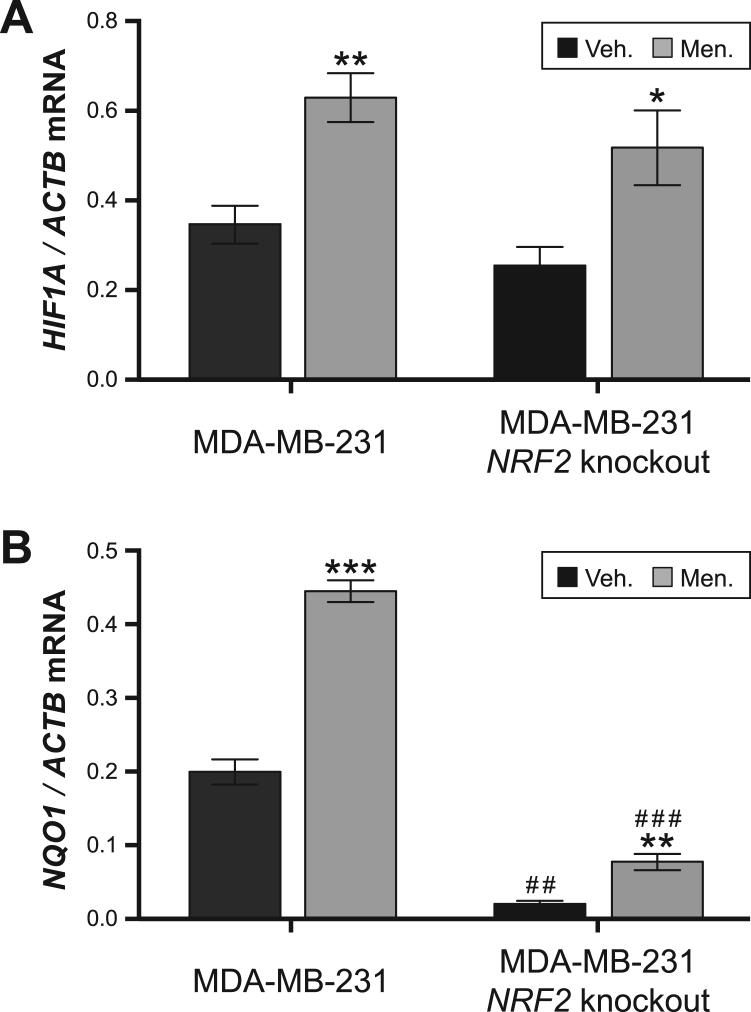
The above findings indicate that (1) HIF1A gene expression is ROS-responsive and (2) the ROS-responsive TF NRF2 can bind and transactivate an ARE upstream of HIF1A. Both points suggest NRF2 is a transcriptional regulator of HIF1A, and this is further supported by previous work implicating NRF2 in regulation of HIF1α. For example, NRF2 knockdown impairs hypoxia-mediated induction of HIF1α in both glioblastoma and colon cancer cells, and NRF2 overexpression drives HIF1α induction in multiple cancer cell lines [63], [64], [65]. NRF2 is also required for HIF1α mRNA and protein induction during induced pluripotent stem cell reprogramming [31]. And, importantly, NRF2 is required for arsenite-mediated upregulation of HIF1α in HepG2 hepatoma cells [32]. To build on these data, we used genome-edited NRF2 knockout MDA-MB-231 cells to test whether NRF2 plays a role in menadione-driven HIF1A induction in this cell line. Consistent with the results from Fig. 2, HIF1A was induced by menadione in control MDA-MB-231 cells (Fig. 5A). However, HIF1A was also induced by menadione in NRF2 knockout cells, but this induction was not as robust as that of control MDA-MB-231 cells (Fig. 5A). For context, we also tested expression of NQO1, a well characterized NRF2 target gene [66], in these conditions. Consistent with its role as a canonical NRF2 target, NQO1 expression was largely NRF2-dependent under basal conditions and after treatment with menadione; however, it was still upregulated by menadione, suggesting additional TFs play a small role in inducible NQO1 expression in this cell type. Thus, although much work suggests that HIF1A expression is regulated by NRF2, it is not overwhelmingly NRF2-dependent like NQO1, at least in MDA-MB-231 breast cancer cells [32], [63], [64], [65]. Instead, this experiment implies NRF2 plays a role in HIF1A expression, but additional TFs also regulate HIF1A in certain cellular contexts. Consistent with this model, ROS-mediated activation of HIF1α is regulated in part by the phosphatidylinositol 3-Kinase/AKT signaling pathway and the TF NFκB [51], [53], [54], [67], so NRF2 is just one of multiple ROS-responsive factors regulating HIF1α levels.
Fig. 5.
Menadione-driven upregulation of HIF1A is reduced in NRF2 knockout cells cells. (A) HIF1A expression in control MDA-MB-231 breast cancer cells and NRF2 knockout MDA-MB231 cells treated with vehicle control (Veh.) or menadione (Men.) for 8 h. Gene expression values were measured by quantitative reverse transcription PCR and normalized relative to ACTB expression. (B) Same as (A), only for NQO1 expression. P-values for menadione versus vehicle within a genotype are represented with asterisks (***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05, Welch's t-test) and p-values for NRF2 knockout versus the control MDA-MB-231 within a treatment are represented with hashes (# # #p ≤ 0.001; # #p ≤ 0.01; #p ≤ 0.05, Welch's t-test).
Data from The Cancer Genome Atlas (TCGA) provide an additional avenue for exploring the relationship between NRF2 activity and expression of its downstream genes. The TCGA project has generated genome-wide profiling data (genotype and gene expression) for thousands of clinical tumor samples across more than 30 cancer types. This is useful because mutations that disrupt the interaction between NRF2 and its inhibitor KEAP1 are common in multiple cancer types – these mutations, which can occur in either NRF2 or KEAP1, drive constitutive NRF2 activity and provide cells with multiple oncogenic advantages [68], [69], [70]. To test the impact of oncogenic NRF2 activity on downstream expression of HIF1A (and, for comparison, NQO1) we looked at the expression of both genes in the three cancer types where mutations in NRF2 or KEAP1 are most prevalent: bladder carcinoma (BLCA), lung squamous cell carcinoma (LUSC), and lung squamous cell carcinoma (LUAD) [71], [72], [73], [74]. For each cancer, we separated individual tumors into those with no mutation in NRF2 or KEAP1 (NRF2WT; KEAP1WT), those with a mutation in NRF2 (NRF2mut), and those with a mutation in KEAP1 (KEAP1mut); we then compared the expression of HIF1A and NQO1 across the three tumor classes for each cancer type (Fig. 6A). NQO1 upregulation in NRF2mut tumors was significant across all three cancer types, consistent with the idea that cancer-associated NRF2 mutations often lead to NRF2 hyperactivation; a similar, though less robust, pattern was seen for NQO1 expression in KEAP1mut tumors. The situation was more complex with HIF1A expression. HIF1A was generally unaffected in KEAP1mut tumors and in LUAD tumors, but NRF2mut tumors were consistently associated with higher than average HIF1A expression in bladder and lung carcinoma (BLCA and LUSC). Thus, whereas NQO1 is almost always induced in tumors with presumed NRF2 hyperactivating mutations, NRF2-associated induction of HIF1A is limited to select cancer types (BLCA and LUSC).
Fig. 6.

NRF2 target gene induction in tumors with high NRF2 activity. (A) Gene expression values for NQO1 and HIF1A in bladder carcinoma (BLCA), lung squamous cell carcinoma (LUSC), and lung adenocarcinoma (LUAD). For each cancer, tumors with mutated NRF2 (NRF2mut) or mutated KEAP1 (KEAP1mut) were separated from tumors with no mutations in either gene (NRF2WT; KEAP1WT). Expression values are based on pan-cancer normalized RNA-seq data from The Cancer Genome Atlas (TCGA); each dot represents expression levels from a single tumor, and horizontal lines represent the mean expression for the cancer/genotype population. Significant changes in NRF2mut or KEAP1mut relative to NRF2WT; KEAP1WT are indicated (Wilcoxon rank-sum test; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05) (B) Gene expression values for NQO1 and HIF1A in breast cancer (BRCA) tumors. Tumors with high inferred NRF2 activity were separated from normal tumors based on a 32 gene NRF2 expression signature (see Methods). As in (A), expression values are based on pan-cancer normalized RNA-seq data from TCGA; each dot represents expression levels from a single tumor, and horizontal lines represent the mean expression for each population. Expression in tumors with high inferred NRF2 activity was compared to expression in tumors with normal inferred NRF2 activity, and significant differences are indicated (Wilcoxon rank-sum test; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05).
HIF1A expression is also important in breast cancer cells, where high HIF1α levels are associated with cancer stem cell enrichment and chemoresistance [45], [46]. Although oncogenic NRF2 mutations are rare in breast cancer, NRF2 can also be indirectly hyperactivated in many cancer types [75], [76]. Thus, we used TCGA data to identify breast cancer (BRCA) tumors with high inferred NRF2 activity based on a 32 gene NRF2 expression signature (Levings et al. [87]) (see Methods) [77]. Consistent with our finding that HIF1A is ROS-responsive in breast cancer cell lines, HIF1A is also high in breast cancer tumors with high inferred NRF2 activity (Fig. 6B). Overall, the gene expression patterns in clinical tumor samples are consistent with the regulatory strategies implied by the NRF2 knockout results: activation of NQO1 by oncogenic NRF2 was consistent across all four cancer types, whereas NRF2-associated activation of HIF1A was dependent on cancer type, and most robust in bladder and breast cancer. Together, these results support a model in which HIF1A expression is the result of input from multiple TFs, and one of these TFs is NRF2.
3. Conclusion
One of the benefits of genomics-based approaches is that they can highlight unexpected or underappreciated connections between regulatory networks. Here, we leveraged multiple sources of publicly available functional genomics data to identify a direct regulatory connection between the redox-responsive TFs NRF2 and HIF1α. We identified a robust in situ NRF2 binding event upstream of HIF1A. This NRF2 binding site is centered on a strongly conserved ARE that is directly bound by NRF2 in vitro and drives ROS-responsive gene expression in luciferase reporter assays. In addition, this ARE falls within the most prominent enhancer region upstream of HIF1A based on chromatin state data, suggesting it falls within an important cis-regulatory region. Additionally, we find that HIF1A expression is enhanced by oncogenic NRF2 mutations in certain cancer types. In combination with previously published studies implying a role for NRF2 in HIF1A expression [31], [32], [63], [64], [65], this work suggests NRF2 is a direct regulator of HIF1A.
The exact setting(s) where direct regulation of HIF1A transcription by NRF2 is most relevant remain subject to speculation, with both hypoxic and non-hypoxic contexts as possibilities. Hypoxia triggers a paradoxical increase in ROS, largely via the mitochondria, that is thought to inhibit PHD-mediated degradation of HIF1α and increase HIF1α activity [78], [79]. ROS-dependent transcriptional induction of HIF1A, via NRF2 and other TFs, would provide a parallel mechanism for increasing HIF1α activity during hypoxia. NRF2-mediated induction of HIF1A might also be important in cancer settings, where we found NRF2-associated HIF1A increases in bladder carcinoma, lung carcinoma, and breast cancer. NRF2 can be directly activated in cancer via mutation in NRF2 or KEAP1, indirectly activated via additional oncogenic pathways, and possibly activated by ROS generated during hypoxia, which is common in solid tumors – downstream activation of HIF1A could be important in any of these tumorigenic conditions. An additional cancer related link is that arsenite-mediated induction of HIF1α via NRF2 is growth promoting, and may explain part of arsenite's carcinogenicity [32]. Further, in triple negative breast cancer cells, ROS induction by chemotherapeutic drugs leads to an increase HIF1α (mRNA and protein), which then promotes cancer stem cell formation and chemoresistance [45]. In this context, it is also worth mentioning that NRF2 and HIF1α both directly regulate the antioxidant genes GCLM and SLC7A11, which have also been linked to chemoresistance and cancer stem cell formation [45], [46], [80], [81]. Overall, there are several links between ROS, NRF2 activity, and HIF1α levels in cancer, and the connections between NRF2 and HIF1α likely extend to shared target genes regulated by both TFs.
Beyond hypoxia and cancer, a third setting where NRF2-driven induction of HIF1A could be important is during induced pluripotent stem cell (iPSC) formation [31]. Early iPSC reprogramming is associated with a burst of oxidative phosphorylation, elevated ROS, and increased NRF2 activity. Shortly thereafter, HIF1α mRNA and protein levels increase in an NRF2-dependent manner, and HIF1α drives a shift toward glycolytic metabolism that is required for efficient iPSC reprogramming. The exact mechanism by which NRF2 upregulates HIF1α during iPSC generation is yet unknown, but our results suggest that direct transcriptional regulation is a likely mechanism.
Regulation of HIF1α levels by NRF2 also extends beyond direct control of its transcription; the protein products of two prominent NRF2 target genes, NQO1 and TXN (also known as TRX1), also interact with HIF1α and increase its stability [82], [83], [84]. NRF2 activation, therefore, has the potential to increase both HIF1A expression and HIF1α stability. Ultimately, additional work will be required to find the settings where NRF2's induction of HIF1A transcription is most critical. Regardless, the fact that HIF1α is both part of the NRF2 regulatory network (directly targeted by NRF2) and stabilized by members of the NRF2 regulatory network (TXN, NQO1), suggests that the connection between these two redox-responsive TFs is an important one.
4. Materials and methods
4.1. Chemicals and materials
FuGENE HD transfection reagent was purchased from Promega (Madison, WI). Menadione was purchased from MP Biomedicals (Solon, OH), and Luperox (tert-butyl hydroperoxide solution; TBOOH) was purchased from Sigma-Aldrich Chemical Company (St. Louis, MO). Stock solutions of all test compounds were prepared in 100% Ethanol. MISSION LightSwitch Luciferase Assay Reagent was purchased from Sigma-Aldrich. All oligonucleotides used for gel mobility shift assays and cloning were purchased from Integrated DNA Technologies (Coralville, IA).
4.2. Cells lines and cell culture
The human hepatocellular carcinoma HepG2 cells and the human breast cancer MCF7 and MDA-MB-231 cells were purchased from the American Type Culture Collection (ATCC). Genome edited NFE2L2/NRF2 knockout MDA-MB-231 cells were generated by GenScript (Piscataway, NJ) using GenCRISPR gene editing technology. Targeted disruption of NRF2's final exon resulted in a biallelic 1b deletion upstream of the basic leucine zipper domain, which was confirmed by Sanger sequencing; the deleted base coordinate is chromosome 2, position 178095987 (GRCh37/hg19 genome assembly). HepG2 cells were grown and maintained in EMEM culture media (ATCC), supplemented with 10% fetal bovine serum purchased from Atlanta Biologicals (Norcross, GA), and 1% (v/v) penicillin/streptomycin purchased from Invitrogen-Life technologies (Carlsbad, CA). MCF7 and MDA-MB-231 cells were grown and maintained in IMEM culture media (Gibco), supplemented with 5% fetal bovine serum purchased from (Atlanta Biologicals), 1% (v/v) penicillin/streptomycin (Invitrogen-Life technologies), 11.25 nM bovine insulin from Sigma-Aldrich, and 2.5 μg/L plasomicin prophylactic (InvivoGen, San Diego, CA). All cell lines were maintained at 37 °C in humidified 5% CO2 air.
4.3. Microarray gene expression profiling and ChIP-seq data
Gene expression microarray data as described by Deferme et al. (GEO series accession number GSE39291). To calculate log2 fold change values, average expression values from the 0.5, 1, 2, 4, 8, and 24 h time points for each treatment were compared to the average expression value from the corresponding control sample at each time point. Genes with |log2 fold change| > 0.58 in at least one of the twelve comparisons were then grouped into clusters of similar expression pattern using the hierarchical clustering tool within TM4's MeV Stand-Alone Client (http://mev.tm4.org). Individual clusters were then analyzed using Enrichr [30] to identify putative transcription factors regulating genes within the cluster; we called a TF (from the ‘ENCODE and ChEA Consensus TFs from ChIP-X’ category) as enriched for binding genes in a given cluster if its adjusted enrichment p-value was < 0.001 (Fisher's exact test) and combined Enrichr score was > 10 (see Supplementary Table 1). NRF2 ChIP-seq data are from lymphoblastoid cell lines treated with sulforaphane, as described previously – the sequencing data for this ChIP-seq experiment can be accessed under GEO Accession Number GSE37589 [33], [34], [57].
4.4. Electrophoretic mobility shift assays
NRF2 was purified from BL21 bacteria transfected with pPROHEX-HC-Flag3-NRF2 as His-tagged fusions using Ni-agarose beads as described previously [85], [86]. Purified his-tagged NRF2 (28.1 nM), MAFG (28.1 nM), and IRDye-700 labeled double stranded DNA were incubated for 30 min at room temperature with poly(dI-dC) in the binding buffer (20 mM HEPES, 4 mM MgCl2, 100 μg/ml BSA, 4% glycerol, 20 mM KCl, 5 mM DTT, and 1 mM EDTA). Orange loading dye (LI-COR, Lincoln, NE) was added and samples were electrophoresed through a native acrylamide gel in 1X TBE. Gels were imaged using the Odyssey® infrared imaging system (LI-COR, Lincoln, NE). The EMSA in Supplementary Fig. 3A was performed with MCF7 nuclear lysates isolated using the Active Motif Nuclear Extract kit (Carlsbad, CA). The double stranded oligonucleotides containing the following sequences were used for the electrophoretic mobility shift assay (EMSA): IRDye-700 5’-AATCCGCAGTCACAGTGACTCAGCAGAATCTGAGCCTAG-3’ (NQO1 ARE, Labeled), 5'-AATCCGCAGTCACAGTGACTCAGCAGAATCTGAGCCTAG-3’(NQO1 cold competitor), 5'-AATCCGCAGTCACAGACTCCTACGAGAATCTGAGCCTAG-3’ (Mutant NQO1 cold competitor), 5’-TCAGTGACCCATCTTGCTGAGTCATGCTAAACCTGTTGA-3’ (HIF1A cold competitor; Labeled version used in Supplementary Fig. 3), 5’-TCAGTGACCCATCTTGCTGAAAAATGCTAAACCTGTTGA-3’ (Mutant HIF1A cold competitor).
4.5. Reporter assays
All reporter plasmids are based on the Switchgear Genomics (Carlsbad, CA) LightSwitch optimized luciferase reporter vector system. The LDHA (lactate dehydrogenase A) control reporter construct was purchased directly from Switchgear Genomics (Product ID: S721613). To generate the HIF1A ARE constructs (both wild type and ARE mutant), we annealed oligonucleotides containing the ARE enhancer sequences into the pLightSwitch-LR Reporter Vector (SwitchGear Genomics, catalog number 32024). The pLightSwitch vector was cut with BglII and MluI purchased from New England Biolabs (Ipswich, MA), and the annealed oligonucleotides contained overhanging sequences compatible with a BglII and MluI insertion site. Annealing the following two oligonucleotides generated the HIF1AWT insert: 5’-CGCGGGTGTTCAGTGACCCATCTTGCTGAGTCATGCTAAACCTGTTGAGCATG-3’ and 5’- GATCCATGCTCAACAGGTTTAGCATGACTCAGCAAGATGGGTCACTGAACACC-3’ (ARE region is bold, and overhang cloning sequences are italicized). An equivalent oligonucleotide pair generated the HIF1AMut insert: 5’-CGCGGGTGTTCAGTGACCCATCTTGCTGAAAAATGCTAAACCTGTTGAGCATG-3’ and 5’-GATCCATGCTCAACAGGTTTAGCATTTTTCAGCAAGATGGGTCACTGAACACC-3’ (ARE region is bold, mutated sequences within the ARE region are underlined, and overhang cloning sequences are italicized). For reporter assays, cells were seeded and transfected in Opti-MEM (Invitrogen) supplemented with 10% FBS. 96 well plates were seeded with 15,000 cells/well and simultaneously transfected using the SwitchGear Genomic High-throughput transfection protocol. Each transfection included 0.15 μl of FuGENE HD transfection reagent and approximately 50 ng/well of reporter plasmid. Each construct was transfected in at least four replicates, and plates were incubated at 37 °C. The transfected cells were treated with 10 μM menadione 24 h post transfection. Then the cells were incubated for an additional 24 h at 37 °C before being frozen at −80 °C. Plates were removed from the freezer and allowed to reach room temperature. 100 μl of MISSION LightSwitch Luciferase Assay Reagent was added to each well, and plates were incubated at room temperature for 30 min and read on a SpectraMax M3 (Molecular Devices, Sunnyvale, CA) according to the manufacturer's instructions. Statistical analysis (Welch's t-test) was performed using Graphpad Prism (La Jolla, CA).
4.6. Reverse transcription and quantitative PCR
Cells were seeded at 125,000 cells/ml in a 6-well plate and grown at 37 °C for 24 h, then treated with either vehicle (0.1% ETOH), or the experimentally determined EC80 (the concentration required to maintain a live cell population of 80%) of the two ROS inducers- TBOOH or menadione. The EC80 for TBOOH and menadione was determined individually for each cell type using the CellTiter-Fluor kit from Promega (Madison, WI). The EC80 concentrations used to induce ROS in the cell lines were as follows: HEPG2, 300 μM TBOOH, 17 μM menadione; MCF7, 80 μM TBOOH, 6.3 μM menadione; MDA-MB-231, 100 μM TBOOH, 10 μM menadione. Each cell type was treated in a minimum of three replicates, and plates were incubated at 37 °C. EtOH concentrations remained at a constant concentration of 0.1% across all treatment groups. Following drug treatment for 8 h, total RNA was isolated using the Qiagen RNeasy kit. Reverse transcriptase PCR was performed according to manufacturer's instructions (Omniscript RT kit, Qiagen) using 1.0 μg of total RNA to generate cDNA. Primers for HIF1A (FAM-hydrolysis probe, Hs.PT.58.534274), SQSTM1 (FAM-hydrolysis probe, Hs.PT.58.39829257), NQO1 (FAM-hydrolysis probe, Hs.PT.58.2697277), and ACTB (HEX-hydrolysis probe, Hs.PT.39a.22214847) were purchased from the predesigned PrimeTime qPCR Assays (Integrated DNA Technologies). RT-PCR reactions were performed using a LightCycler 480 Multiwell Plate 96 containing 5 μM of each primer set, 1× LightCycler 480 Probes Master (Roche Diagnostics, Bazel, Switzerland), and 2 μl of cDNA template, in a final reaction volume of 10 μl. The RT-PCR was performed using the following cycle parameters: initial enzyme activation at 95 °C for 10 min; followed by 45 cycles of 95 °C for 10 s, 60 °C for 20 s, and 65 °C for 30 s. Following the amplification phase, a cooling step was performed at 40 °C for 10 s (ramp rate of 1.5 °C/s). Acquisition of the fluorescence signal was performed using the Dual Hydrolysis Probe setting for FAM (465–510 nm) and HEX/Yellow555 (533–580) dyes following the 65 °C extension phase of each cycle. Relative expression of selected genes was evaluated using qPCR by calculating the ratio of the specific gene to β-Actin (ACTB). Statistical analysis (Welch's t-test) was performed using Graphpad Prism (La Jolla, CA).
4.7. Analysis of TCGA gene expression data
To assess if HIF1A expression is induced in cancers with NRF2 hyperactivation, we used data from The Cancer Genome Atlas (TCGA). First, using XenaPython we downloaded two sets of data for all samples from the cancer types BLCA, HNSC, LUSC and UCEC in TCGA: 1) pan-cancer normalized gene expression values from the UCSC TOIL recompute for NQO1 and HIF1A, and 2) all non-silent mutations in NFE2L2 for these samples. We then used R to generate dotplots depicting the spread of the gene expression values, with means for the two genes listed above in NRF2WT and NRF2mut tumors from each cancer. P-values were calculated using the non-parametric Mann-Whitney-Wilcox test in R. N's for each group are as follows (each gene has the same N for each cancer/NRF2 status combination): BLCA – NRF2WT/KEAP1WT = 359, NRF2mut = 23, KEAP1mut = 8; LUSC – NRF2WT/KEAP1WT = 128, NRF2mut = 27, KEAP1mut = 22; LUAD – NRF2WT/KEAP1WT = 385, NRF2mut = 12, KEAP1mut = 88.
For analysis of BRCA, where NRF2 mutations are rare, we used a 32 gene NRF2 signature to infer NRF2 activity in individual tumors. The 32 gene signature is based on direct NRF2 target genes that are reliably upregulated by oncogenic NRF2 (Levings et al. [87]) and consists of the following genes: ABCB6, ABCC3, AKR1C3, ANXA10, ASF1A, DNAJB4, EPHX1, FECH, FTH1, GCLC, GCLM, GSR, GSTM3, KEAP1, MAFG, ME1, NAMPT, NECAB2, NQO1, PANX2, PIR, PRDX1, SLC3A2, SLC7A11, SRXN1, TKT, TLK1, TMTC3, TRIM16L, TXN, TXNRD1, ZNF746. We calculated a mean expression value across all 32 NRF cancer signature genes for each BRCA sample, then computed the value for the 95th percentile of this mean expression across all samples. Any sample with a mean expression above this 95th percentile was assigned to the high NRF2 group (N = 49), and any at or below this value was assigned normal NRF2 (N = 928).
Acknowledgements
The authors wish to acknowledge Juan Palacios Moreno for helpful discussions. We are also indebted to the researchers who generated the genome-wide data used in this study. This work was supported by funding from the National Institute of General Medical Sciences (R35-GM-119553 to M.S.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.08.014.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Briehl M.M. Oxygen in human health from life to death–an approach to teaching redox biology and signaling to graduate and medical students. Redox Biol. 2015;5:124–139. doi: 10.1016/j.redox.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorlach A., Dimova E.Y., Petry A., Martinez-Ruiz A., Hernansanz-Agustin P., Rolo A.P., Palmeira C.M., Kietzmann T. Reactive oxygen species, nutrition, hypoxia and diseases: problems solved? Redox Biol. 2015;6:372–385. doi: 10.1016/j.redox.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samoylenko A., Hossain J.A., Mennerich D., Kellokumpu S., Hiltunen J.K., Kietzmann T. Nutritional countermeasures targeting reactive oxygen species in cancer: from mechanisms to biomarkers and clinical evidence. Antioxid. Redox Signal. 2013;19:2157–2196. doi: 10.1089/ars.2012.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu J., Dawes I.W. Redox control of cell proliferation. Trends Cell Biol. 2012;22:592–601. doi: 10.1016/j.tcb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y.W., Byzova T.V. Oxidative stress in angiogenesis and vascular disease. Blood. 2014;123:625–631. doi: 10.1182/blood-2013-09-512749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K., Zhang T., Dong Q., Nice E.C., Huang C., Wei Y. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013;4:e537. doi: 10.1038/cddis.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martindale J.L., Holbrook N.J. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 9.Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sanchez-Perez P., Cadenas S., Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kietzmann T., Gorlach A. Reactive oxygen species in the control of hypoxia-inducible factor-mediated gene expression. Semin. Cell Dev. Biol. 2005;16:474–486. doi: 10.1016/j.semcdb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Hahn M.E., McArthur A.G., Karchner S.I., Franks D.G., Jenny M.J., Timme-Laragy A.R., Stegeman J.J., Woodin B.R., Cipriano M.J., Linney E. The transcriptional response to oxidative stress during vertebrate development: effects of tert-butylhydroquinone and 2,3,7,8-tetrachlorodibenzo-p-dioxin. PLoS One. 2014;9:e113158. doi: 10.1371/journal.pone.0113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linher-Melville K., Haftchenary S., Gunning P., Singh G. Signal transducer and activator of transcription 3 and 5 regulate system Xc- and redox balance in human breast cancer cells. Mol. Cell. Biochem. 2015;405:205–221. doi: 10.1007/s11010-015-2412-4. [DOI] [PubMed] [Google Scholar]
- 14.Linher-Melville K., Singh G. The complex roles of STAT3 and STAT5 in maintaining redox balance: lessons from STAT-mediated xCT expression in cancer cells. Mol. Cell. Endocrinol. 2017;451:40–52. doi: 10.1016/j.mce.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Jennings P., Limonciel A., Felice L., Leonard M.O. An overview of transcriptional regulation in response to toxicological insult. Arch. Toxicol. 2013;87:49–72. doi: 10.1007/s00204-012-0919-y. [DOI] [PubMed] [Google Scholar]
- 16.Cadenas S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic. Biol. Med. 2018;117:76–89. doi: 10.1016/j.freeradbiomed.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Kipp A.P., Deubel S., Arner E.S.J., Johansson K. Time- and cell-resolved dynamics of redox-sensitive Nrf2, HIF and NF-kappaB activities in 3D spheroids enriched for cancer stem cells. Redox Biol. 2017;12:403–409. doi: 10.1016/j.redox.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sihvola V., Levonen A.L. Keap1 as the redox sensor of the antioxidant response. Arch. Biochem. Biophys. 2017;617:94–100. doi: 10.1016/j.abb.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Epstein A.C., Gleadle J.M., McNeill L.A., Hewitson K.S., O'Rourke J., Mole D.R., Mukherji M., Metzen E., Wilson M.I., Dhanda A., Tian Y.M., Masson N., Hamilton D.L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P.H., Pugh C.W., Schofield C.J., Ratcliffe P.J. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 20.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandel N.S., Maltepe E., Goldwasser E., Mathieu C.E., Simon M.C., Schumacker P.T. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandel N.S., McClintock D.S., Feliciano C.E., Wood T.M., Melendez J.A., Rodriguez A.M., Schumacker P.T. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 23.Kaelin W.G., Jr. ROS: really involved in oxygen sensing. Cell Metab. 2005;1:357–358. doi: 10.1016/j.cmet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Wang G.L., Jiang B.H., Semenza G.L. Effect of altered redox states on expression and DNA-binding activity of hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 1995;212:550–556. doi: 10.1006/bbrc.1995.2005. [DOI] [PubMed] [Google Scholar]
- 25.Guzy R.D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K.D., Simon M.C., Hammerling U., Schumacker P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Deferme L., Briede J.J., Claessen S.M., Jennen D.G., Cavill R., Kleinjans J.C. Time series analysis of oxidative stress response patterns in HepG2: a toxicogenomics approach. Toxicology. 2013;306:24–34. doi: 10.1016/j.tox.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle A.P., Araya C.L., Brdlik C., Cayting P., Cheng C., Cheng Y., Gardner K., Hillier L.W., Janette J., Jiang L., Kasper D., Kawli T., Kheradpour P., Kundaje A., Li J.J., Ma L., Niu W., Rehm E.J., Rozowsky J., Slattery M., Spokony R., Terrell R., Vafeados D., Wang D., Weisdepp P., Wu Y.C., Xie D., Yan K.K., Feingold E.A., Good P.J., Pazin M.J., Huang H., Bickel P.J., Brenner S.E., Reinke V., Waterston R.H., Gerstein M., White K.P., Kellis M., Snyder M. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512:453–456. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T., Rowe L.D., Dreszer T.R., Roe G., Podduturi N.R., Tanaka F., Hong E.L., Cherry J.M. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44:D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw377. W90-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins K.E., Joy S., Delhove J.M., Kotiadis V.N., Fernandez E., Fitzpatrick L.M., Whiteford J.R., King P.J., Bolanos J.P., Duchen M.R., Waddington S.N., McKay T.R. NRF2 orchestrates the metabolic shift during induced pluripotent stem cell reprogramming. Cell Rep. 2016;14:1883–1891. doi: 10.1016/j.celrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Taleb Z., Petry A., Chi T.F., Mennerich D., Gorlach A., Dimova E.Y., Kietzmann T. Differential transcriptional regulation of hypoxia-inducible factor-1alpha by arsenite under normoxia and hypoxia: involvement of Nrf2. J. Mol. Med. 2016;94:1153–1166. doi: 10.1007/s00109-016-1439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacher S.E., Lee J.S., Wang X., Campbell M.R., Bell D.A., Slattery M. Beyond antioxidant genes in the ancient Nrf2 regulatory network. Free Radic. Biol. Med. 2015;88:452–465. doi: 10.1016/j.freeradbiomed.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K., Shim J., Bae J., Kim Y.J., Lee J. Stabilization of RNT-1 protein, runt-related transcription factor (RUNX) protein homolog of Caenorhabditis elegans, by oxidative stress through mitogen-activated protein kinase pathway. J. Biol. Chem. 2012;287:10444–10452. doi: 10.1074/jbc.M111.314146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B.K., Bhinge A.A., Iyer V.R. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011;39:3558–3573. doi: 10.1093/nar/gkq1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLaForest A., Nagaoka M., Si-Tayeb K., Noto F.K., Konopka G., Battle M.A., Duncan S.A. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138:4143–4153. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bean D.M., Heimbach J., Ficorella L., Micklem G., Oliver S.G., Favrin G. esyN: network building, sharing and publishing. PLoS One. 2014;9:e106035. doi: 10.1371/journal.pone.0106035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khurana E., Fu Y., Chen J., Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput. Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carter H., Hofree M., Ideker T. Genotype to phenotype via network analysis. Curr. Opin. Genet. Dev. 2013;23:611–621. doi: 10.1016/j.gde.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thien C.B., Langdon W.Y. C-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem. J. 2005;391:153–166. doi: 10.1042/BJ20050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan J.A., Sun Y., Jiang Y.P., Bott A.J., Jaber N., Dou Z., Yang B., Chen J.S., Catanzaro J.M., Du C., Ding W.X., Diaz-Meco M.T., Moscat J., Ozato K., Lin R.Z., Zong W.X. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol. Cell. 2016;61:720–733. doi: 10.1016/j.molcel.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J., Xu Y., Lin S., Guo Y., Deng W., Zhang Y., Guo A., Xue Y. iUUCD 2.0: an update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res. 2018;46:D447–D453. doi: 10.1093/nar/gkx1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei L., Bandola-Simon J., Roche P.A. Ubiquitin-conjugating enzyme E2 D1 (Ube2D1) mediates lysine-independent ubiquitination of the E3 ubiquitin ligase March-I. J. Biol. Chem. 2018;293:3904–3912. doi: 10.1074/jbc.RA117.001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samanta D., Gilkes D.M., Chaturvedi P., Xiang L., Semenza G.L. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:E5429–E5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Lu H., Samanta D., Xiang L., Zhang H., Hu H., Chen I., Bullen J.W., Semenza G.L. Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc. Natl. Acad. Sci. USA. 2015;112:E4600–E4609. doi: 10.1073/pnas.1513433112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lendahl U., Lee K.L., Yang H., Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat. Rev. Genet. 2009;10:821–832. doi: 10.1038/nrg2665. [DOI] [PubMed] [Google Scholar]
- 48.Dengler V.L., Galbraith M., Espinosa J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014;49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page E.L., Robitaille G.A., Pouyssegur J., Richard D.E. Induction of hypoxia-inducible factor-1alpha by transcriptional and translational mechanisms. J. Biol. Chem. 2002;277:48403–48409. doi: 10.1074/jbc.M209114200. [DOI] [PubMed] [Google Scholar]
- 50.Blouin C.C., Page E.L., Soucy G.M., Richard D.E. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 51.Belaiba R.S., Bonello S., Zahringer C., Schmidt S., Hess J., Kietzmann T., Gorlach A. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol. Biol. Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iommarini L., Porcelli A.M., Gasparre G., Kurelac I. Non-canonical mechanisms regulating hypoxia-inducible factor 1 alpha in cancer. Front. Oncol. 2017;7:286. doi: 10.3389/fonc.2017.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koshikawa N., Hayashi J., Nakagawara A., Takenaga K. Reactive oxygen species-generating mitochondrial DNA mutation up-regulates hypoxia-inducible factor-1alpha gene transcription via phosphatidylinositol 3-kinase-Akt/protein kinase C/histone deacetylase pathway. J. Biol. Chem. 2009;284:33185–33194. doi: 10.1074/jbc.M109.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonello S., Zahringer C., BelAiba R.S., Djordjevic T., Hess J., Michiels C., Kietzmann T., Gorlach A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler. Thromb. Vasc. Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 55.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordan R., Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Campbell M.R., Lacher S.E., Cho H.Y., Wan M., Crowl C.L., Chorley B.N., Bond G.L., Kleeberger S.R., Slattery M., Bell D.A. A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of parkinsonian disorders. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shenvi S.V., Smith E.J., Hagen T.M. Transcriptional regulation of rat gamma-glutamate cysteine ligase catalytic subunit gene is mediated through a distal antioxidant response element. Pharmacol. Res. 2009;60:229–236. doi: 10.1016/j.phrs.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R., Santos-Simarro F., Gilbert-Dussardier B., Wittler L., Borschiwer M., Haas S.A., Osterwalder M., Franke M., Timmermann B., Hecht J., Spielmann M., Visel A., Mundlos S. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez-Sandoval A., Gasser S.M. On TADs and LADs: spatial control over gene expression. Trends Genet.: TIG. 2016;32:485–495. doi: 10.1016/j.tig.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 61.Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., Aiden E.L. Juicebox Provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Amin V., Whitaker J.W., Schultz M.D., Ward L.D., Sarkar A., Quon G., Sandstrom R.S., Eaton M.L., Wu Y.C., Pfenning A.R., Wang X., Claussnitzer M., Liu Y., Coarfa C., Harris R.A., Shoresh N., Epstein C.B., Gjoneska E., Leung D., Xie W., Hawkins R.D., Lister R., Hong C., Gascard P., Mungall A.J., Moore R., Chuah E., Tam A., Canfield T.K., Hansen R.S., Kaul R., Sabo P.J., Bansal M.S., Carles A., Dixon J.R., Farh K.H., Feizi S., Karlic R., Kim A.R., Kulkarni A., Li D., Lowdon R., Elliott G., Mercer T.R., Neph S.J., Onuchic V., Polak P., Rajagopal N., Ray P., Sallari R.C., Siebenthall K.T., Sinnott-Armstrong N.A., Stevens M., Thurman R.E., Wu J., Zhang B., Zhou X., Beaudet A.E., Boyer L.A., De Jager P.L., Farnham P.J., Fisher S.J., Haussler D., Jones S.J., Li W., Marra M.A., McManus M.T., Sunyaev S., Thomson J.A., Tlsty T.D., Tsai L.H., Wang W., Waterland R.A., Zhang M.Q., Chadwick L.H., Bernstein B.E., Costello J.F., Ecker J.R., Hirst M., Meissner A., Milosavljevic A., Ren B., Stamatoyannopoulos J.A., Wang T., Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim T.H., Hur E.G., Kang S.J., Kim J.A., Thapa D., Lee Y.M., Ku S.K., Jung Y., Kwak M.K. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Cancer Res. 2011;71:2260–2275. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- 64.Ji X., Wang H., Zhu J., Zhu L., Pan H., Li W., Zhou Y., Cong Z., Yan F., Chen S. Knockdown of Nrf2 suppresses glioblastoma angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Int. J. Cancer. 2014;135:574–584. doi: 10.1002/ijc.28699. [DOI] [PubMed] [Google Scholar]
- 65.Wang H., Liu X., Long M., Huang Y., Zhang L., Zhang R., Zheng Y., Liao X., Wang Y., Liao Q., Liu W., Tang Z., Tong Q., Wang X., Fang F., Rojo de la Vega M., Ouyang Q., Zhang D.D., Yu S., Zheng H. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aad6095. (334ra351) [DOI] [PubMed] [Google Scholar]
- 66.Venugopal R., Jaiswal A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao N., Ding M., Zheng J.Z., Zhang Z., Leonard S.S., Liu K.J., Shi X., Jiang B.H. Vanadate-induced expression of hypoxia-inducible factor 1 alpha and vascular endothelial growth factor through phosphatidylinositol 3-kinase/Akt pathway and reactive oxygen species. J. Biol. Chem. 2002;277:31963–31971. doi: 10.1074/jbc.M200082200. [DOI] [PubMed] [Google Scholar]
- 68.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leinonen H.M., Kansanen E., Polonen P., Heinaniemi M., Levonen A.L. Dysregulation of the Keap1-Nrf2 pathway in cancer. Biochem. Soc. Trans. 2015;43:645–649. doi: 10.1042/BST20150048. [DOI] [PubMed] [Google Scholar]
- 70.Lacher S.E., Slattery M. Gene regulatory effects of disease-associated variation in the NRF2 network. Curr. Opin. Toxicol. 2016;1:71–79. doi: 10.1016/j.cotox.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Tamborero D., Schroeder M.P., Jene-Sanz A., Santos A., Lopez-Bigas N. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods. 2013;10:1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., Scrimieri F., Winter J.M., Hruban R.H., Iacobuzio-Donahue C., Kern S.E., Blair I.A., Tuveson D.A. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menegon S., Columbano A., Giordano S. The dual roles of NRF2 in cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gorlach A., Kietzmann T. Superoxide and derived reactive oxygen species in the regulation of hypoxia-inducible factors. Methods Enzymol. 2007;435:421–446. doi: 10.1016/S0076-6879(07)35022-2. [DOI] [PubMed] [Google Scholar]
- 79.Sabharwal S.S., Schumacker P.T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat. Rev. Cancer. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erickson A.M., Nevarea Z., Gipp J.J., Mulcahy R.T. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. Revision of the ARE consensus sequence. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- 81.Habib E., Linher-Melville K., Lin H.X., Singh G. Expression of xCT and activity of system xc(-) are regulated by NRF2 in human breast cancer cells in response to oxidative stress. Redox Biol. 2015;5:33–42. doi: 10.1016/j.redox.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Welsh S.J., Bellamy W.T., Briehl M.M., Powis G. The redox protein thioredoxin-1 (Trx-1) increases hypoxia-inducible factor 1alpha protein expression: Trx-1 overexpression results in increased vascular endothelial growth factor production and enhanced tumor angiogenesis. Cancer Res. 2002;62:5089–5095. [PubMed] [Google Scholar]
- 83.Malec V., Gottschald O.R., Li S., Rose F., Seeger W., Hanze J. HIF-1 alpha signaling is augmented during intermittent hypoxia by induction of the Nrf2 pathway in NOX1-expressing adenocarcinoma A549 cells. Free Radic. Biol. Med. 2010;48:1626–1635. doi: 10.1016/j.freeradbiomed.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Oh E.T., Kim J.W., Kim J.M., Kim S.J., Lee J.S., Hong S.S., Goodwin J., Ruthenborg R.J., Jung M.G., Lee H.J., Lee C.H., Park E.S., Kim C., Park H.J. NQO1 inhibits proteasome-mediated degradation of HIF-1alpha. Nat. Commun. 2016;7:13593. doi: 10.1038/ncomms13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gebelein B., Culi J., Ryoo H.D., Zhang W., Mann R.S. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev. Cell. 2002;3:487–498. doi: 10.1016/s1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- 86.Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P., Dror I., Zhou T., Rohs R., Honig B., Bussemaker H.J., Mann R.S. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Levings D.C., Wang X., Kohlhase D., Bell D.A., Slattery M. A distinct class of antioxidant response elements is consistently activated in tumors with NRF2 mutations. Redox Biol. 2018;19:235–249. doi: 10.1016/j.redox.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material