Abstract
The Botryosphaeriaceae is a species-rich family that includes pathogens of a wide variety of plants, including species of Eucalyptus. Recently, during disease surveys in China, diseased samples associated with species of Botryosphaeriaceae were collected from plantation Eucalyptus and other plants, including Cunninghamina lanceolata, Dimocarpus longan, Melastoma sanguineum and Phoenix hanceana, which were growing adjacent to Eucalyptus. In addition, few samples from Araucaria cunninghamii and Cedrus deodara in two gardens were also included in this study. Disease symptoms observed mainly included stem canker, shoot and twig blight. In this study, 105 isolates of Botryosphaeriaceae were collected from six provinces, of which 81 isolates were from Eucalyptus trees. These isolates were identified based on comparisons of the DNA sequences of the internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS), and partial translation elongation factor 1-alpha (tef1), β-tubulin (tub), DNA-directed RNA polymerase II subunit (rpb2) and calmodulin (cmdA) genes, the nuclear ribosomal large subunit (LSU) and the nuclear ribosomal small subunit (SSU), and combined with their morphological characteristics. Results showed that these isolates represent 12 species of Botryosphaeriaceae, including Botryosphaeria fusispora, Cophinforma atrovirens, Lasiodiplodia brasiliense, L. pseudotheobromae, L. theobromae and Neofusicoccum parvum, and six previously undescribed species of Botryosphaeria and Neofusicoccum, namely B. pseudoramosa sp. nov., B. qingyuanensis sp. nov., B. wangensis sp. nov., N. hongkongense sp. nov., N. microconidium sp. nov. and N. sinoeucalypti sp. nov. Aside from B. wangensis, C. atrovirens and N. hongkongense, the other nine Botryosphaeriaceae species were isolated from Eucalyptus trees in South China. Botryosphaeria fusispora (26 % of the isolates from Eucalyptus) is the dominant species, followed by L. pseudotheobromae (23 % of the isolates from Eucalyptus). In addition to species found on Eucalyptus trees, we also found B. pseudoramosa on M. sanguineum; B. wangensis on C. deodara; C. atrovirens on D. longan; L. theobromae on C. lanceolata, D. longan and P. hanceana; and N. hongkongense on A. cunninghamii. Pathogenicity tests showed that the 12 species of Botryosphaeriaceae are pathogenic to three Eucalyptus clones and that Lasiodiplodia species are the most aggressive. The results of our study suggest that many more species of the Botryosphaeriaceae remain to be discovered in China. This study also provides confirmation for the wide host range of Botryosphaeriaceae species on different plants.
Keywords: Botryosphaeria, Cophinforma, Lasiodiplodia, Neofusicoccum, pathogenicity, plant pathogen
INTRODUCTION
The Botryosphaeriaceae includes a range of phylogenetically and morphologically diverse fungi with a broad host range and geographic distribution globally (Punithalingam 1980, Slippers & Wingfield 2007, Liu et al. 2012, Phillips et al. 2013). These fungi occur primarily on woody plants including both economically important crops and native trees (Slippers & Wingfield 2007). Many species of Botryosphaeriaceae are well-known pathogens that can cause stem canker, shoot blight and dieback on woody plants; however, some species of Botryosphaeriaceae have been described as latent pathogens or endophytes that cause disease when the plant is under stress conditions (Slippers & Wingfield 2007).
Species of Eucalyptus are widely planted in more than 100 countries, and because of the rapid growth of some Eucalyptus trees, they represent one of the most widely planted genera for commercial forestry worldwide, with approximately 20 million hectares (Mha) established in plantations (Iglesias-Trabad et al. 2009). In China, Eucalyptus plantations have expanded substantially during the past 30 years, with more than 4.5 Mha of Eucalyptus established in South China by the end of 2013 (Chen & Chen 2013). Industrial Eucalyptus plantations in China are typically single species or hybrid plantings, often from a few clones that share a common parentage (Wei 2005, Turnbull 2007, Zhou & Wingfield 2011). The model of large-scale plantations with few clones greatly increases the threat from pests and diseases (Wingfield 2003, Wingfield et al. 2008). In recent years, the sustainable development of Eucalyptus plantations in China has been increasingly threatened by pathogens and pests (Zhou & Wingfield 2011). The important diseases in Chinese Eucalyptus plantations include stem canker/wilt caused by species of Botryosphaeriaceae (Chen et al. 2011c), Ceratocystis (Chen et al. 2013, Liu et al. 2015), Chrysoporthe (Chen et al. 2010) and Teratosphaeria (Chen et al. 2011a); leaf blight/spot caused by species of Teratosphaeriaceae (Burgess et al. 2006), Mycosphaerellaceae (Burgess et al. 2007), Calonectria (Lombard et al. 2010, Chen et al. 2011b) and Quambalaria (Zhou et al. 2007); and bacterial wilt associated with Ralstonia solanacearum (Cao 1982, Old et al. 2003).
Relatively little research has been conducted on diseases caused by Botryosphaeriaceae on Eucalyptus trees in China (Chen et al. 2011c, Li et al. 2015a). Based on DNA sequence comparisons and morphological features, five species of Botryosphaeriaceae have been identified from Eucalyptus in China to date, including Botryosphaeria fabicerciana from FuJian, GuangXi and HaiNan Provinces, Lasiodiplodia pseudotheobromae from GuangXi Province, L. theobromae from GuangDong and GuangXi Provinces, Neofusicoccum parvum from FuJian and GuangXi Provinces and N. ribis s.lat. from FuJian Province (Chen et al. 2011c, Li et al. 2015a). These species were collected from cankered stems and blighted branches or twigs, and pathogenicity tests showed that all five species could produce lesions on Eucalyptus seedlings or trees (Chen et al. 2011c, Li et al. 2015a).
In China, species of Botryosphaeriaceae also have been isolated from a number of other woody and horticultural plants, including Acacia confusa (Zhao et al. 2010), Actinidia chinensis (Zhou et al. 2015), Bougainvillea spectabilis, Polyscias balfouriana (Li et al. 2015a), Juglans regia (Li et al. 2015b, Yu et al. 2015), Malus domestica (Tang et al. 2012, Xu et al. 2015a), Rosa rugosa (Chen et al. 2016), Vitis vinifera (Yan et al. 2012, 2013) and Vaccinium corymbosum (Xu et al. 2015b). Botryosphaeriaceae species identified from these plants resided in Botryosphaeria, Lasiodiplodia and Neofusicoccum. These Botryosphaeriaceae were all isolated from diseased tissue of the respective plant hosts.
From 2013–2014, surveys were conducted on Eucalyptus in plantations and some plants adjacent to Eucalyptus, and diseases with symptoms typical of those caused by Botryosphaeriaceae were observed. Diseased samples were collected and the putative Botryosphaeriaceae fungi (based on microscopic morphology) were isolated. In addition, few samples previously collected from Araucaria cunninghamii and Cedrus deodara were also included in this study. The aims of this study are to:
– identify these species of Botryosphaeriaceae based on phylogenetic analyses and morphological characteristics;
– clarify the geographic distribution of these Botryosphaeriaceae species; and
– evaluate pathogenicity of the identified Botryosphaeriaceae species on different Eucalyptus clones.
MATERIALS AND METHODS
Disease symptoms, sample collection and fungal isolation
Disease surveys were mainly conducted on species of Eucalyptus in plantations distributed in FuJian, GuangDong, GuangXi and HaiNan Provinces. Disease symptoms typically caused by Botryosphaeriaceae include tree dieback, stem canker, branch canker and twig blight (Fig. 1). Other plants, including Cunninghamina lanceolata, Dimocarpus longan, Melastoma sanguineum and Phoenix hanceana, which were growing in close proximity to Eucalyptus trees, were also randomly surveyed in this study. These surveys were conducted during 2013–2014. Samples of diseased materials, including stems, branches and twigs that showed typical symptoms of Botryosphaeriaceae infection, were collected and taken to the laboratory for fungal isolation. Diseased branches of C. deodara in HeNan Province and A. cunninghamii in Hong Kong Region with similar symptoms typical of Botryosphaeriaceae collected previously, were also added in this study (Fig. 1).
Fig. 1.

Disease symptoms on Eucalyptus trees caused by Botryosphaeriaceae. a. Typical dieback of a Eucalyptus grandis clone in FunJian Province; b. dieback of Eucalyptus globulus; c–e. stem cankers and lesions on main stems of different Eucalyptus clones/genotypes; f. branch and twig blight of a Eucalyptus grandis clone; g. fruiting structures with abundant mature dark conidia on a Eucalyptus branch; h. new branches germinated after main stem infection.
Fungi were isolated from diseased stems, branches and twigs, as well as from pycnidia produced on diseased tissues of Eucalyptus and other plants. When pycnidia formed on the surface of diseased tissue, the pycnidia were scratched lightly with a sterile scalpel and transferred with a sterile steel needle to 2 % malt extract agar (MEA) media containing 20 g of malt extract powder (Beijing Shuangxuan Microbial Culture Medium Products Factory, Beijing, China) and 20 g of agar per litre of water (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) under a stereomicroscope (Carl Zeiss Ltd., Munchen, Germany). For diseased tissues that did not produce pycnidia, small tissue pieces (approximately 0.25 cm2) were cut from inner wood and transferred to 2 % MEA. Pieces of pycnidia and wood were incubated at room temperature for 2–5 d until colonies formed. Colonies with morphological characteristics typical of Botryosphaeriaceae were transferred to fresh 2 % MEA plates. Pure cultures were obtained by transferring single hyphal tips from colonies to 2 % MEA. Cultures were deposited in the culture collection of the China Eucalypt Research Centre (CERC), Chinese Academy of Forestry (CAF), ZhanJiang, GuangDong Province, China. Isolates linked to type specimens of the fungal species were deposited in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China. The specimens were deposited in the Collection of Central South Forestry Fungi of China (CSFF), GuangDong Province, China.
DNA extraction, PCR amplification and sequencing
DNA extractions and sequence comparisons were conducted on selected isolates collected from different trees and different regions (Table 1). For the selected isolates, mycelia were scraped from 7-d-old cultures using sterile scalpels and transferred to 2 mL Eppendorf tubes. A CTAB-based protocol, ‘Method 5’ described by Van Burik et al. (1998), was used to extract the DNA samples. The resulting DNA was checked for purity and concentration using a NanoDrop 2000 Spectrometer (Thermo Fisher Scientific Inc. Waltham, MA, USA). Prior to PCR amplification, each DNA sample was diluted to approximately 100 ng/μL with DNase/RNase-free ddH2O (Sangon Biotech Co., Ltd., Shanghai, China). The internal transcribed spacer (ITS) region was amplified using the primers ITS1/ITS4 (White et al. 1990), a part of the translation elongation factor 1-alpha (tef1) gene was amplified using the primers EF1-728F/EF1-986R (Carbone & Kohn 1999) or EF1F/EF2R (Jacobs et al. 2004), a part of the β-tubulin (tub) gene was amplified using the primers BT-2a/BT-2b (Glass & Donaldson 1995), a part of DNA-directed RNA polymerase II subunit (rpb2) gene was amplified using the primers fRPB2-5F/fRPB2-7cR for Botryosphaeria and Cophinforma (Liu et al. 1999), rpb2-LasF/rpb2-LasR for Lasiodiplodia (Cruywagen et al. 2017) and RPB2bot6F/RPB2bot7R for Neofusicoccum (Pavlic et al. 2009a, Sakalidis et al. 2011), the nuclear ribosomal large subunit (LSU) region was amplified using the primers LR0R/LR5 (Vilgalys & Hester 1990, Cubeta et al. 1991), the nuclear ribosomal small subunit (SSU) region was amplified using the primers NS1/NS4 (White et al. 1990). For the isolates of Lasiodiplodia, a portion of the calmodulin (cmdA) gene was amplified using the primers CAL-228F/CAL-737R (Carbone & Kohn 1999). All primers were synthesised by Life Technologies (Thermo Fisher Scientific Inc., Shanghai, China). The PCR mixtures to amplify the ITS, tef1, tub, rpb2, cmdA, LSU, SSU regions used the TopTaq™ Master Mix Kit (Qiagen Inc., Hilden, Germany). All amplification reactions consisted of 25 μL TopTaq™ Master Mix (contain 1.25 U TopTaq™ DNA Polymerase, 200 μM of each dNTP and 1.5 mM MgCl2), 0.2 mM of each primer and 50 ng template DNA (made up to a total volume of 50 μL with RNase-free water). The amplification conditions consisted of an initial denaturation step at 94 °C for 3 min, 35 cycles of 94 °C for 1 min, 55 °C (except 45 °C for SSU) for 1 min, and 72 °C for 1 min, followed by a final elongation step at 72 °C for 10 min.
Table 1.
Isolates sequenced and used for phylogenetic analyses, morphological studies and pathogenicity tests in this study.
| Species1 | Isolate No.2,3 | Genotype4 | Host | Location | GPS information | Collector | GenBank accession No.5 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub | rpb2 | cmdA | LSU | SSU | |||||||
| Botryosphaeria fusispora | CERC1997 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277967 | KX278072 | KX278177 | MF410116 | N/A | MF410007 | MF410205 |
| CERC2273 | AAAA-AA | Eucalyptus hybrid | FuZhou Region, FuJian Province, China | N26°13′39″ E119°10′51″ | S.F. Chen & G.Q. Li | KX277968 | KX278073 | KX278178 | MF410117 | N/A | MF410008 | MF410206 | |
| CERC22746,7 | AAAA-AA | Eucalyptus hybrid | FuZhou Region, FuJian Province, China | N26°13′39″ E119°10′51″ | S.F. Chen & G.Q. Li | KX277969 | KX278074 | KX278179 | MF410118 | N/A | MF410009 | MF410207 | |
| CERC2910 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | Unknown | S.F. Chen & G.Q. Li | KX277970 | KX278075 | KX278180 | MF410119 | N/A | MF410010 | MF410208 | |
| CERC2912 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | Unknown | S.F. Chen & G.Q. Li | KX277971 | KX278076 | KX278181 | MF410120 | N/A | MF410011 | MF410209 | |
| CERC2913 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | Unknown | S.F. Chen & G.Q. Li | KX277972 | KX278077 | KX278182 | MF410121 | N/A | MF410012 | MF410210 | |
| CERC34416 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277974 | KX278079 | KX278184 | MF410123 | N/A | MF410014 | MF410212 | |
| CERC3469 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277975 | KX278080 | KX278185 | MF410124 | N/A | MF410015 | MF410213 | |
| CERC3474 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277976 | KX278081 | KX278186 | MF410125 | N/A | MF410016 | MF410214 | |
| CERC3426 | AAAA-AB | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX277973 | KX278078 | KX278183 | MF410122 | N/A | MF410013 | MF410211 | |
| CERC1998 7 | ABAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277977 | KX278082 | KX278187 | MF410126 | N/A | MF410017 | MF410215 | |
| CERC2006 | ABAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N21°15′26″ E110°07′00″ | S.F. Chen & G.Q. Li | KX277978 | KX278083 | KX278188 | MF410127 | N/A | MF410018 | MF410216 | |
| CERC29116 | ABAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | Unknown | S.F. Chen & G.Q. Li | KX277979 | KX278084 | KX278189 | MF410128 | N/A | MF410019 | MF410217 | |
| CERC29186 | ABAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277980 | KX278085 | KX278190 | MF410129 | N/A | MF410020 | MF410218 | |
| CERC2921 | ABAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277981 | KX278086 | KX278191 | MF410130 | N/A | MF410021 | MF410219 | |
| CERC2925 | ABAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277982 | KX278087 | KX278192 | MF410131 | N/A | MF410022 | MF410220 | |
| CERC2948 | ABAA-AA | Eucalyptus hybrid | QingYuan Region, GuangDong Province, China | N23°51′44″ E113°10′58″ | S.F. Chen & G.Q. Li | KX277983 | KX278088 | KX278193 | MF410132 | N/A | MF410023 | MF410221 | |
| CERC2949 | ABAA-AA | Eucalyptus hybrid | QingYuan Region, GuangDong Province, China | N23°51′44″ E113°10′58″ | S.F. Chen & G.Q. Li | KX277984 | KX278089 | KX278194 | MF410133 | N/A | MF410024 | MF410222 | |
| CERC2954 | ABAA-AA | Eucalyptus hybrid | QingYuan Region, GuangDong Province, China | N23°51′44″ E113°10′58″ | S.F. Chen & G.Q. Li | KX277985 | KX278090 | KX278195 | MF410134 | N/A | MF410025 | MF410223 | |
| CERC3446 7 | ABAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277986 | KX278091 | MF409964 | MF410135 | N/A | MF410026 | MF410224 | |
| CERC29307 | ACAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277987 | KX278092 | KX278196 | MF410136 | N/A | MF410027 | MF410225 | |
| B. pseudoramosa | CERC19996 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277988 | KX278093 | KX278197 | MF410139 | N/A | MF410030 | MF410228 |
| CERC2001 = CGMCC3.187396,7,8,9 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277989 | KX278094 | KX278198 | MF410140 | N/A | MF410031 | MF410229 | |
| CERC20049 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′41″ E109°43′01″ | S.F. Chen & G.Q. Li | KX277990 | KX278095 | KX278199 | MF410141 | N/A | MF410032 | MF410230 | |
| CERC2019 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | Unknown | S.F. Chen & G.Q. Li | KX277991 | KX278096 | KX278200 | MF410142 | N/A | MF410033 | MF410231 | |
| CERC2983 = CGMCC3.187406 | AAAA-AA | Melastoma sanguineum | ZhanJiang Region, GuangDong Province, China | N21°13′24″ E110°24′04″ | S.F. Chen | KX277992 | KX278097 | KX278201 | MF410143 | N/A | MF410034 | MF410232 | |
| CERC2985 | AAAA-AA | M. sanguineum | ZhanJiang Region, GuangDong Province, China | N21°13′24″ E110°24′04″ | S.F. Chen | KX277993 | KX278098 | KX278202 | MF410144 | N/A | MF410035 | MF410233 | |
| CERC29876,9 | AAAA-AA | M. sanguineum | ZhanJiang Region, GuangDong Province, China | N21°13′24″ E110°24′04″ | S.F. Chen | KX277994 | KX278099 | KX278203 | MF410145 | N/A | MF410036 | MF410234 | |
| CERC29886 | AAAA-AA | M. sanguineum | ZhanJiang Region, GuangDong Province, China | N21°13′24″ E110°24′04″ | S.F. Chen | KX277995 | KX278100 | KX278204 | MF410146 | N/A | MF410037 | MF410235 | |
| CERC34527 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277996 | KX278101 | KX278205 | MF410147 | N/A | MF410038 | MF410236 | |
| CERC3455 = CGMCC3.187416 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277997 | KX278102 | KX278206 | MF410148 | N/A | MF410039 | MF410237 | |
| CERC3462 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277998 | KX278103 | KX278207 | MF410149 | N/A | MF410040 | MF410238 | |
| CERC3472 | AAAA-AA | Eucalyptus hybrid | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX277999 | KX278104 | KX278208 | MF410150 | N/A | MF410041 | MF410239 | |
| B. qingyuanensis | CERC2946 = CGMCC3.187426,7,8,9 | AAAA-AA | Eucalyptus hybrid | QingYuan Region, GuangDong Province, China | N23°44′30″ E112°48′49″ | S.F. Chen & G.Q. Li | KX278000 | KX278105 | KX278209 | MF410151 | N/A | MF410042 | MF410240 |
| CERC2947 = CGMCC3.187437,9 | AAAA-AA | Eucalyptus hybrid | QingYuan Region, GuangDong Province, China | N23°44′30″ E112°48′49″ | S.F. Chen & G.Q. Li | KX278001 | KX278106 | KX278210 | MF410152 | N/A | MF410043 | MF410241 | |
| B. wangensis | CERC2298 = CGMCC3.187446,7,8,9 | AAAA-AA | C. deodara | XinZhuang, MangChuan, RuZhou Region, HeNan Province, China | N34°04′09.8″ E112°49′00.7″ | S.F. Chen | KX278002 | KX278107 | KX278211 | MF410153 | N/A | MF410044 | MF410242 |
| CERC2299 = CGMCC3.187456,7 | AAAA-AA | C. deodara | XinZhuang, MangChuan, RuZhou Region, HeNan Province, China | N34°04′09.8″ E112°49′00.7″ | S.F. Chen | KX278003 | KX278108 | KX278212 | MF410154 | N/A | MF410045 | MF410243 | |
| CERC2300 = CGMCC3.187466,9 | AAAA-AA | C. deodara | XinZhuang, MangChuan, RuZhou Region, HeNan Province, China | N34°04′09.8″ E112°49′00.7″ | S.F. Chen | KX278004 | KX278109 | KX278213 | MF410155 | N/A | MF410046 | MF410244 | |
| Cophinforma atrovirens | CERC3481 | AAAA-AA | Dimocarpus longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278005 | KX278110 | KX278214 | MF410156 | N/A | MF410047 | MF410245 |
| CERC3482 | AAAA-AA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278006 | KX278111 | KX278215 | MF410157 | N/A | MF410048 | MF410246 | |
| CERC34847 | AAAA-AA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278007 | KX278112 | KX278216 | MF410158 | N/A | MF410049 | MF410247 | |
| CERC34897 | BAAA-AA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278008 | KX278113 | KX278217 | MF410159 | N/A | MF410050 | MF410248 | |
| CERC3490 | BAAA-AA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278009 | KX278114 | KX278218 | MF410160 | N/A | MF410051 | MF410249 | |
| Lasiodiplodia brasiliense | CERC22846,7 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278010 | KX278115 | KX278219 | MF410163 | MF409967 | MF410054 | MF410252 |
| L. pseudotheobromae | CERC2262 | AAAAAAA | Eucalyptus hybrid | YuLin Region, GuangXi Province, China | N22°09′12″ E110°12′08″ | S.F. Chen & G.Q. Li | KX278011 | KX278116 | KX278220 | MF410164 | MF409968 | MF410055 | MF410253 |
| CERC2280 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278012 | KX278117 | KX278221 | MF410165 | MF409969 | MF410056 | MF410254 | |
| CERC2281 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278013 | KX278118 | KX278222 | MF410166 | MF409970 | MF410057 | MF410255 | |
| CERC2282 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278014 | KX278119 | KX278223 | MF410167 | MF409971 | MF410058 | MF410256 | |
| CERC2283 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278015 | KX278120 | KX278224 | MF410168 | MF409972 | MF410059 | MF410257 | |
| CERC22866,7 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278016 | KX278121 | KX278225 | MF410169 | MF409973 | MF410060 | MF410258 | |
| CERC22876 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278017 | KX278122 | KX278226 | MF410170 | MF409974 | MF410061 | MF410259 | |
| CERC2288 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278018 | KX278123 | KX278227 | MF410171 | MF409975 | MF410062 | MF410260 | |
| CERC2289 | AAAAAAA | Eucalyptus hybrid | ZhangZhou Region, FuJian Province, China | N24°46′06″ E117°51′02″ | S.F. Chen & G.Q. Li | KX278019 | KX278124 | KX278228 | MF410172 | MF409976 | MF410063 | MF410261 | |
| CERC2960 | AAAAAAA | Eucalyptus hybrid | YunFu Region, GuangDong Province, China | N23°15′12″ E111°41′51″ | S.F. Chen & G.Q. Li | KX278020 | KX278125 | KX278229 | MF410173 | MF409977 | MF410064 | MF410262 | |
| CERC2961 | AAAAAAA | Eucalyptus hybrid | YunFu Region, GuangDong Province, China | N23°15′12″ E111°41′51″ | S.F. Chen & G.Q. Li | KX278021 | KX278126 | KX278230 | MF410174 | MF409978 | MF410065 | MF410263 | |
| CERC34177 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278023 | KX278128 | KX278232 | MF410176 | MF409980 | MF410067 | MF410265 | |
| CERC34326 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278024 | KX278129 | KX278233 | MF410177 | MF409981 | MF410068 | MF410266 | |
| CERC3434 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278025 | KX278130 | KX278234 | MF410178 | MF409982 | MF410069 | MF410267 | |
| CERC3438 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278026 | KX278131 | KX278235 | MF410179 | MF409983 | MF410070 | MF410268 | |
| CERC3475 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278027 | KX278132 | KX278236 | MF410180 | MF409984 | MF410071 | MF410269 | |
| CERC34957 | AAAAAAA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278028 | KX278133 | KX278237 | MF410181 | MF409985 | MF410072 | MF410270 | |
| CERC3496 | AAAAAAA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278029 | KX278134 | KX278238 | MF410182 | MF409986 | MF410073 | MF410271 | |
| CERC2962 | AAAAABA | Eucalyptus hybrid | YunFu Region, GuangDong Province, China | N23°15′12″ E111°41′51″ | S.F. Chen & G.Q. Li | KX278022 | KX278127 | KX278231 | MF410175 | MF409979 | MF410066 | MF410264 | |
| L. theobromae | CERC20246 | AAAAAAA | Phoenix hanceana | ZhanJiang Region, GuangDong Province, China | N21°15′26″ E110°07′01″ | S.F. Chen & G.Q. Li | KX278030 | KX278135 | KX278239 | MF410183 | MF409987 | MF410074 | MF410272 |
| CERC34206,7 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278031 | KX278136 | KX278240 | MF410184 | MF409988 | MF410075 | MF410273 | |
| CERC34246 | AAAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278032 | KX278137 | KX278241 | MF410185 | MF409989 | MF410076 | MF410274 | |
| CERC2025 | ABAAAAA | P. hanceana | ZhanJiang Region, GuangDong Province, China | N21°15′26″ E110°07′01″ | S.F. Chen & G.Q. Li | KX278033 | KX278138 | KX278242 | MF410186 | MF409990 | MF410077 | MF410275 | |
| CERC2264 | ABAAAAA | E. urophylla × E. grandis | YuLin Region, GuangXi Province, China | N22°09′12″ E110°12′08″ | S.F. Chen & G.Q. Li | KX278034 | KX278139 | KX278243 | MF410187 | MF409991 | MF410078 | MF410276 | |
| CERC2275 | ABAAAAA | E. urophylla × E. grandis | YongAn Region, FuJian Province, China | N26°01′40″ E117°27′11″ | S.F. Chen & G.Q. Li | KX278035 | KX278140 | KX278244 | MF410188 | MF409992 | MF410079 | MF410277 | |
| CERC2934 | ABAAAAA | Eucalyptus hybrid | DingAn County, HaiNan Province, China | N19°36′41″ E110°17′16″ | S.F. Chen & G.Q. Li | KX278036 | KX278141 | KX278245 | MF410189 | MF409993 | MF410080 | MF410278 | |
| CERC2957 | ABAAAAA | Cunninghamina lanceolata | ShaoGuan Region, GuangDong Province, China | N24°31′32″ E113°37′40″ | S.F. Chen & G.Q. Li | KX278037 | KX278142 | KX278246 | MF410190 | MF409994 | MF410081 | MF410279 | |
| CERC2958 | ABAAAAA | C. lanceolata | ShaoGuan Region, GuangDong Province, China | N24°31′32″ E113°37′40″ | S.F. Chen & G.Q. Li | KX278038 | KX278143 | KX278247 | MF410191 | MF409995 | MF410082 | MF410280 | |
| CERC2963 | ABAAAAA | Eucalyptus hybrid | YunFu Region, GuangDong Province, China | N23°15′12″ E111°41′51″ | S.F. Chen & G.Q. Li | KX278039 | KX278144 | KX278248 | MF410192 | MF409996 | MF410083 | MF410281 | |
| CERC3418 | ABAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278040 | KX278145 | KX278249 | MF410193 | MF409997 | MF410084 | MF410282 | |
| CERC3422 | ABAAAAA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278041 | KX278146 | KX278250 | MF410194 | MF409998 | MF410085 | MF410283 | |
| CERC3485 | ABAAAAA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278042 | KX278147 | KX278251 | MF410195 | MF409999 | MF410086 | MF410284 | |
| CERC3486 | ABAAAAA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278043 | KX278148 | KX278252 | MF410196 | MF410000 | MF410087 | MF410285 | |
| CERC3487 | ABAAAAA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278044 | KX278149 | KX278253 | MF410197 | MF410001 | MF410088 | MF410286 | |
| CERC3491 | ABAAAAA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278045 | KX278150 | KX278254 | MF410198 | MF410002 | MF410089 | MF410287 | |
| CERC3493 | ABAAAAA | D. longan | ZhanJiang Region, GuangDong Province, China | Unknown | S.F. Chen | KX278046 | KX278151 | KX278255 | MF410199 | MF410003 | MF410090 | MF410288 | |
| CERC35136,7 | ABAAAAA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278047 | KX278152 | KX278256 | MF410200 | MF410004 | MF410091 | MF410289 | |
| CERC3514 | ABAAAAA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278048 | KX278153 | KX278257 | MF410201 | MF410005 | MF410092 | MF410290 | |
| CERC35167 | ABAAAAA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278049 | KX278154 | KX278258 | MF410202 | MF410006 | MF410093 | MF410291 | |
| Neofusicoccum hongkongense | CERC2967= CGMCC3.18747 | AAAA-AA | Araucaria cunninghamii | Hong Kong, China | Unknown | S.F. Chen | KX278050 | KX278155 | KX278259 | KX278281 | N/A | MF410094 | MF410292 |
| CERC2968 = CGMCC3.187486,7,9 | AABA-AA | A. cunninghamii | Hong Kong, China | Unknown | S.F. Chen | KX278051 | KX278156 | KX278260 | KX278282 | N/A | MF410095 | MF410293 | |
| CERC2973 = CGMCC3.187496,7,8,9 | AABA-AA | A. cunninghamii | Hong Kong, China | Unknown | S.F. Chen | KX278052 | KX278157 | KX278261 | KX278283 | N/A | MF410096 | MF410294 | |
| N. microconidium | CERC3497 = CGMCC3.187506,7,8,9 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278053 | KX278158 | KX278262 | MF410203 | N/A | MF410097 | MF410295 |
| CERC3498 = CGMCC3.187516,7,9 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278054 | KX278159 | KX278263 | MF410204 | N/A | MF410098 | MF410296 | |
| N. parvum | CERC29517 | AAAA-AA | E. urophylla × E. grandis | QingYuan Region, GuangDong Province, China | N23°51′44″ E113°10′58″ | S.F. Chen & G.Q. Li | KX278055 | KX278160 | KX278264 | KX278284 | N/A | MF410099 | MF410297 |
| CERC3508 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278056 | KX278161 | KX278265 | KX278285 | N/A | MF410100 | MF410298 | |
| CERC3509 7 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278057 | KX278162 | KX278266 | KX278286 | N/A | MF410101 | MF410299 | |
| CERC3502 | ABAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278058 | KX278163 | KX278267 | KX278287 | N/A | MF410102 | MF410300 | |
| CERC35036 | ABAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278059 | KX278164 | KX278268 | KX278288 | N/A | MF410103 | MF410301 | |
| CERC35046,7 | ABAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278060 | KX278165 | KX278269 | KX278289 | N/A | MF410104 | MF410302 | |
| N. sinoeucalypti | CERC2005 = CGMCC3.187526,7,8,9 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°15′26″ E110°07′00″ | S.F. Chen & G.Q. Li | KX278061 | KX278166 | KX278270 | KX278290 | N/A | MF410105 | MF410303 |
| CERC34156 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278063 | KX278168 | KX278272 | KX278292 | N/A | MF410107 | MF410305 | |
| CERC3416 = CGMCC3.187546 | AAAA-AA | Eucalyptus hybrid | BeiHai Region, GuangXi Province, China | N21°35′49″ E109°43′49″ | S.F. Chen & G.Q. Li | KX278064 | KX278169 | KX278273 | KX278293 | N/A | MF410108 | MF410306 | |
| CERC3457 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX278066 | KX278171 | KX278275 | KX278295 | N/A | MF410110 | MF410308 | |
| CERC3458 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX278067 | KX278172 | KX278276 | KX278296 | N/A | MF410111 | MF410309 | |
| CERC34637 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX278068 | KX278173 | KX278277 | KX278297 | N/A | MF410112 | MF410310 | |
| CERC3464 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX278069 | KX278174 | KX278278 | KX278298 | N/A | MF410113 | MF410311 | |
| CERC3467 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX278070 | KX278175 | KX278279 | KX278299 | N/A | MF410114 | MF410312 | |
| CERC3517 | AAAA-AA | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N21°13′31″ E110°23′47″ | S.F. Chen & G.Q. Li | KX278071 | KX278176 | KX278280 | KX278300 | N/A | MF410115 | MF410313 | |
| CERC2265 = CGMCC3.187536,9 | AAAA-AB | E. urophylla × E. grandis | YuLin Region, GuangXi Province, China | N22°08′55″ E110°12′00″ | S.F. Chen & G.Q. Li | KX278062 | KX278167 | KX278271 | KX278291 | N/A | MF410106 | MF410304 | |
| CERC3451 | AAAA-AB | E. urophylla × E. grandis | ZhanJiang Region, GuangDong Province, China | N20°41′20″ E110°01′17″ | S.F. Chen & G.Q. Li | KX278065 | KX278170 | KX278274 | KX278294 | N/A | MF410109 | MF410307 | |
1 Species names in bold are novel species described in this study.
2 Isolates in bold are in the phylogenetic trees.
3 CERC: Culture Collection of China Eucalypt Research Centre, Chinese Academy of Forestry, ZhanJiang, GuangDong Province, China; CGMCC: China General Microbiological Culture Collection Center, Beijing, China.
4 Genotype within each identified species, determined by ITS, tef1, tub, rpb2, cmdA, LSU and SSU regions; ‘–’ means not available.
5 ITS, internal transcribed spacer region and intervening 5.8S nrRNA gene; tef1, translation elongation factor 1-alpha; tub, β-tubulin; rpb2, DNA-directed RNA polymerase II subunit; cmdA, calmodulin; LSU, nuclear ribosomal large subunit; SSU, nuclear ribosomal small subunit; N/A = not available.
6 Isolates used for morphological studies.
7 Isolates used for pathogenicity tests on three Eucalyptus clones.
8 Isolates represent ex-type.
9 Isolates used for culture growth studies.
PCR amplifications were carried out in a thermocycler (Bio-Rad Laboratories, Inc., Berkeley, California, USA). The PCR products were separated by electrophoresis in 1.5 % agarose gels with SYBR Safe DNA Gel Stain (Thermo Fisher Scientific Inc., USA) in 1× Tris-acetate-EDTA (TAE) buffer at a constant voltage (80 V) for 30 min. All PCR products were sequenced in both directions using the primers specified above by Beijing Genomics Institution, Guangzhou, GuangDong Province, China. The nucleotide sequences were edited with MEGA v. 6.0.5 software (Tamura et al. 2013). Sequences obtained in this study were all deposited in GenBank (http://www.ncbi.nlm.nih.gov) (Table 1).
Phylogenetic analyses
The preliminary identities of the isolates sequenced in this study were obtained by conducting a standard nucleotide BLAST search using the ITS, tef1, tub, rpb2, cmdA, LSU, SSU sequences. The sequences of the ex-type strains that were closely related to the Botryosphaeriaceae isolates sequenced in this study were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/) and used for polygenetic analyses (Table 2). Sequences were aligned using MAFFT online v. 7 (http://mafft.cbrc.jp/alignment/server/) (Katoh & Standley 2013), with the iterative refinement method (FFT-NS-i setting). The alignments were further edited manually with MEGA v. 6.0.5 software (Tamura et al. 2013). Resulting alignments and phylogenetic trees for all the datasets were deposited in TreeBASE (http://treebase.org).
Table 2.
Isolates from other studies used in the phylogenetic analyses for this study.
| Species | Isolate numbers1 | Host | Location | Collector | GenBank accession numbers2 | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub | rpb2 | cmdA | LSU | SSU | ||||||
| Botryosphaeria agaves | MFLUCC 11-0125 = CBS 1339923 | Agave sp. | Thailand | R. Phookamsak | JX646791 | JX646856 | JX646841 | N/A | N/A | JX646808 | JX646825 | Liu et al. (2012) |
| MFLUCC 10-0051 | Agave sp. | Thailand | P. Chomnunti | JX646790 | JX646855 | JX646840 | N/A | N/A | JX646807 | JX646824 | Liu et al. (2012) | |
| B. auasmontanum | CMW 25413 = CBS 1217693 | Acacia mellifera | Namibia | F.J.J. van der Walt & J. Roux | EU101303 | EU101348 | N/A | N/A | N/A | KF766332 | KF766252 | Slippers et al. (2013, 2014) |
| B. corticis | CBS 1190473 | Vaccinium corymbosum | USA | P.V. Oudemans | DQ299245 | EU017539 | EU673107 | N/A | N/A | EU673244 | EU673175 | Phillips et al. (2006, 2008), Lazzizera et al. (2008) |
| ATCC 22927 | Vaccinium sp. | USA | R.D. Millholland | DQ299247 | EU673291 | EU673108 | N/A | N/A | EU673245 | EU673176 | Phillips et al. (2006, 2008) | |
| B. dothidea | CBS 115476 = CMW 80003 | Prunus sp. | Switzerland | B. Slippers | AY236949 | AY236898 | AY236927 | EU339577 | N/A | AY928047 | EU673173 | Slippers et al. (2004a), Phillips et al. (2008) |
| CBS 110302 | Vitis vinifera | Portugal | A.J.L. Phillips | AY259092 | AY573218 | EU673106 | N/A | N/A | EU673243 | EU673174 | Alves et al. (2004), Phillips et al. (2008) | |
| B. fabicerciana | CMW 27094 = CBS 1271933 | Eucalyptus sp. | China | M.J. Wingfield | HQ332197 | HQ332213 | KF779068 | MF410137 | N/A | MF410028 | MF410226 | Chen et al. (2011c), This study |
| CMW 27121 = CBS 127194 | Eucalyptus sp. | China | M.J. Wingfield | HQ332198 | HQ332214 | KF779069 | MF410138 | N/A | MF410029 | MF410227 | Chen et al. (2011c), This study | |
| B. fusispora | MFLUCC 10-00983 | Entada sp. | Thailand | S. Boonmee | JX646789 | JX646854 | JX646839 | N/A | N/A | JX646806 | JX646823 | Liu et al. (2012) |
| MFLUCC 11-0507 | Entada sp. | Thailand | R. Cheewangkoon | JX646788 | JX646853 | JX646838 | N/A | N/A | JX646805 | JX646822 | Liu et al. (2012) | |
| B. kuwatsukai | CBS 135219 = PG 23 | Malus domestica | China | C.S. Wang | KJ433388 | KJ433410 | N/A | N/A | N/A | N/A | N/A | Xu et al. (2015a) |
| LSP 5 | Pyrus sp. | China | C.S. Wang | KJ433395 | KJ433417 | N/A | N/A | N/A | N/A | N/A | Xu et al. (2015a) | |
| B. minutispermatia | GZCC 16-00133 | Dead wood | Guizhou, China | H.A. Ariyawansa | KX447675 | KX447678 | N/A | N/A | N/A | N/A | N/A | Ariyawansa et al. (2016) |
| GZCC 16-0014 | Dead wood | Guizhou, China | H.A. Ariyawansa | KX447676 | KX447679 | N/A | N/A | N/A | N/A | N/A | Ariyawansa et al. (2016) | |
| B. ramosa | CBS 122069 = CMW 261673 | Eucalyptus camaldulensis | Australia | T.I. Burgess | EU144055 | EU144070 | KF766132 | N/A | N/A | KF766333 | KF766253 | Pavlic et al. (2008), Slippers et al. (2013) |
| B. rosaceae | CGMCC3.180073 | Malus sp. | Shandong, China | Y. Zhang & J.Q. Zhang | KX197074 | KX197094 | KX197101 | N/A | N/A | KX197083 | N/A | Zhou et al. (2017) |
| CGMCC3.18008 | Amygdalus sp. | Shandong, China | Y. Zhang, J.Q. Zhang & Z.P. Dou | KX197075 | KX197095 | KX197102 | N/A | N/A | KX197084 | N/A | Zhou et al. (2017) | |
| B. scharifii | IRAN 1529C = CBS 1247033 | Mangifera indica | Iran | J. Abdollahzadeh | JQ772020 | JQ772057 | N/A | N/A | N/A | N/A | N/A | Abdollahzadeh et al. (2013) |
| IRAN 1543C = CBS 124702 | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | JQ772019 | JQ772056 | N/A | N/A | N/A | N/A | N/A | Abdollahzadeh et al. (2013) | |
| B. sinensia | CGMCC3.17723 | Morus sp. | Henan, China | Z.P. Dou | KT343254 | KU221233 | KX197107 | N/A | N/A | KX197090 | N/A | Zhou et al. (2016, 2017) |
| CGMCC3.17724 | Juglans regia | Henan, China | Z.P. Dou | KT343256 | KU221234 | KX197108 | N/A | N/A | N/A | N/A | Zhou et al. (2016, 2017) | |
| Cophinforma atrovirens | CBS 124934 = CMW 226743 | Pterocarpus angolensis | South Africa | J. Mehl & J. Roux | FJ888473 | FJ888456 | N/A | N/A | N/A | N/A | N/A | Mehl et al. (2011) |
| CBS 124935 = CMW 22682 | Pterocarpus angolensis | South Africa | J. Mehl & J. Roux | FJ888476 | FJ888457 | N/A | N/A | N/A | N/A | N/A | Mehl et al. (2011) | |
| CBS 117445 = CMW 13425 | Acacia mangium | Venezuela | S. Mohali | EF118046 | GU134939 | N/A | N/A | N/A | N/A | N/A | Mohali et al. (2007) | |
| CBS 117446 = CMW 13429 | Eucalyptus hybrid | Venezuela | S. Mohali | EF118048 | GU134940 | N/A | N/A | N/A | N/A | N/A | Mohali et al. (2007) | |
| Lasiodiplodia avicenniae | CMW 414673 | Avicennia marina | South Africa | J.A. Osorio & J. Roux | KP860835 | KP860680 | KP860758 | KU587878 | N/A | N/A | N/A | Osorio et al. (2017) |
| LAS 199 (DNA) | Avicennia marina | South Africa | J.A. Osorio & J. Roux | KU587957 | KU587947 | KU587868 | KU587880 | N/A | N/A | N/A | Osorio et al. (2017) | |
| L. americana | CERC1961 = CFCC500653 | Pistachia vera | Arizona, USA | T.J. Michailides | KP217059 | KP217067 | KP217075 | MF410161 | MF409965 | MF410052 | MF410250 | Chen et al. (2015), This study |
| CERC1960 = CFCC50064 | Pistachia vera | Arizona, USA | T.J. Michailides | KP217058 | KP217066 | KP217074 | MF410162 | MF409966 | MF410053 | MF410251 | Chen et al. (2015), This study | |
| L. brasiliense | CMM 40153 | Mangifera indica | Brazil | M.W. Marques | JX464063 | JX464049 | N/A | N/A | N/A | N/A | N/A | Netto et al. (2014) |
| CMW 35884 | Adansonia madagascariensis | Madagascar | KU887094 | KU886972 | KU887466 | KU696345 | KU886755 | N/A | N/A | Cruywagen et al. (2017) | ||
| L. bruguierae | CMW 414703 | Bruguiera gymnorrhiza | South Africa | J.A. Osorio & J. Roux | KP860833 | KP860678 | KP860756 | KU587875 | N/A | N/A | N/A | Osorio et al. (2017) |
| CMW 41614 | Bruguiera gymnorrhiza | South Africa | J.A. Osorio & J. Roux | KP860834 | KP860679 | KP860757 | KU587877 | N/A | N/A | N/A | Osorio et al. (2017) | |
| L. caatinguensis | CMM 13253 | Citrus sinensis | Itarema, Ceará, Brazil | I.B.L. Coutinho & J.S. Lima | KT154760 | KT008006 | KT154767 | N/A | N/A | N/A | N/A | Coutinho et al. (2017) |
| IBL 40 | Spondias mombin | Itarema, Ceará, Brazil | J.S. Lima & J.E. Cardoso | KT154762 | KT154755 | KT154769 | N/A | N/A | N/A | N/A | Coutinho et al. (2017) | |
| L. chinensis | CGMCC3.180613 | Unknown | China | W. He & Z.P. Dou | KX499889 | KX499927 | KX500002 | KX499965 | N/A | N/A | N/A | Dou et al. (2017a) |
| CGMCC3.18066 | Hevea brasiliensis | China | Y. Zhang & Y.P. Zhou | KX499899 | KX499937 | KX500012 | KX499974 | N/A | N/A | N/A | Dou et al. (2017a) | |
| L. citricola | CBS 124707 = IRAN 1522C3 | Citrus sp. | Iran | J. Abdollahzadeh & A. Javadi | GU945354 | GU945340 | KU887505 | KU696351 | KU886760 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) |
| CBS 124706 = IRAN 1521C | Citrus sp. | Iran | A. Shekari | GU945353 | GU945339 | KU887504 | KU696350 | KU886759 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) | |
| L. crassispora | CBS 118741 = WAC125333 | Santalum album | Kununurra, Australia | T.I. Burgess & B. Dell | DQ103550 | EU673303 | KU887506 | KU696353 | KU886761 | DQ377901 | N/A | Burgess et al. (2006), Phillips et al. (2008), Cruywagen et al. (2017) |
| CBS 110492 | Unknown | Unknown | Unknown | EF622086 | EF622066 | EU673134 | N/A | N/A | EU673251 | N/A | Alves et al. (2008), Phillips et al. (2008) | |
| L. euphorbicola | CMM 36093 | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234543 | KF226689 | KF254926 | N/A | N/A | N/A | N/A | Machado et al. (2014) |
| CMW 33350 | Adansonia digitata | Botswana | KU887149 | KU887026 | KU887455 | KU696346 | KU886754 | N/A | N/A | Cruywagen et al. (2017) | ||
| L. exigua | CBS 1377853 | Retama raetam | Tunisia | B.T. Linaldeddu | KJ638317 | KJ638336 | KU887509 | KU696355 | KU886764 | N/A | N/A | Linaldeddu et al. (2015), Cruywagen et al. (2017) |
| BL 184 | Retama raetam | Tunisia | B.T. Linaldeddu | KJ638318 | KJ638337 | N/A | N/A | N/A | N/A | N/A | Linaldeddu et al. (2015) | |
| L. gilanensis | CBS 124704 = IRAN 1523C3 | Unknown | Iran | J. Abdollahzadeh & A. Javadi | GU945351 | GU945342 | KU887511 | KU696357 | KU886765 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) |
| CBS 124705 = IRAN 1501C | Unknown | Iran | J. Abdollahzadeh & A. Javadi | GU945352 | GU945341 | KU887510 | KU696356 | KU886766 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) | |
| L. gonubiensis | CBS 115812 = CMW 140773 | Syzygium cordatum | South Africa | D. Pavlic | AY639595 | DQ103566 | DQ458860 | KU696359 | KU886768 | DQ377902 | EU673193 | Pavlic et al. (2004), Burgess et al. (2006), Phillips et al. (2008), Cruywagen et al. (2017) |
| CBS 116355 = CMW 14078 | Syzygium cordatum | South Africa | D. Pavlic | AY639594 | DQ103567 | EU673126 | KU696358 | KU886767 | EU673252 | EU673194 | Pavlic et al. (2004), Burgess et al. (2006), Phillips et al. (2008), Cruywagen et al. (2017) | |
| L. gravistriata | CMM 45643 | Anacardium humile | Brazil | M.S.B. Netto | KT250949 | KT250950 | N/A | N/A | N/A | N/A | N/A | Netto et al. (2017) |
| CMM 4565 | Anacardium humile | Brazil | M.S.B. Netto | KT250947 | KT266812 | N/A | N/A | N/A | N/A | N/A | Netto et al. (2017) | |
| L. hormozganensis | CBS 124709 = IRAN 1500C3 | Olea sp. | Iran | J. Abdollahzadeh & A. Javadi | GU945355 | GU945343 | KU887515 | KU696361 | KU886770 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) |
| CBS 124708 = IRAN 1498C | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | GU945356 | GU945344 | KU887514 | KU696360 | KU886769 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) | |
| L. hyalina | CGMCC3.179753 | Acacia confusa | China | Y. Zhang & Y.P. Zhou | KX499879 | KX499917 | KX499992 | KX499955 | N/A | N/A | N/A | Dou et al. (2017b) |
| CGMCC3.18383 = B 6180 | Unknown tree | China | Z.P. Dou & Z.C. Liu | KY767661 | KY751302 | KY751299 | KY751296 | N/A | N/A | N/A | Dou et al. (2017b) | |
| L. indica | IBP 013 | Angiospermous tree | India | I.B. Prasher & G. Singh | KM376151 | N/A | N/A | N/A | N/A | N/A | N/A | Prasher & Singh (2014) |
| L. iraniensis | IRAN 1520C3 | Salvadora persica | Iran | J. Abdollahzadeh & A. Javadi | GU945348 | GU945336 | KU887516 | KU696363 | KU886771 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) |
| IRAN 1502C | Juglans sp. | Iran | A. Javadi | GU945347 | GU945335 | KU887517 | KU696362 | KU886772 | N/A | N/A | Abdollahzadeh et al. (2010), Cruywagen et al. (2017) | |
| L. laeliocattleyae | CBS 167.283 | Laeliocattleya | Italy | C. Sibilia | KU507487 | KU507454 | N/A | N/A | N/A | DQ377892 | N/A | Crous et al. (2006), Rodríguez-Gálvez et al. (2017) |
| LAREP1 | Mangifera indica | Repartidor, Peru | P. Guerrero | KU507484 | KU507451 | N/A | N/A | N/A | N/A | N/A | Rodríguez-Gálvez et al. (2017) | |
| L. lignicola | MFLUCC 11-0435 = CBS1341123 | Unknown | Thailand | A.D. Ariyawansa | JX646797 | KU887003 | JX646845 | KU696364 | N/A | JX646814 | JX646830 | Liu et al. (2012), Cruywagen et al. (2017) |
| L. macrospora | CMM 38333 | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234557 | KF226718 | KF254941 | N/A | N/A | N/A | N/A | Machado et al. (2014) |
| L. mahajangana | CBS 124925 = CMW 278013 | Terminalia catappa | Madagascar | J. Roux | FJ900595 | FJ900641 | FJ900630 | KU696365 | KU886773 | N/A | N/A | Begoude et al. (2010), Cruywagen et al. (2017) |
| CBS 124926 = CMW 27820 | Terminalia catappa | Madagascar | J. Roux | FJ900596 | FJ900642 | KU887519 | KU696366 | KU886774 | N/A | N/A | Begoude et al. (2010), Cruywagen et al. (2017) | |
| L. margaritacea | CBS 122519 = CMW 261623 | Adansonia gibbosa | WA, Tunnel Creek Gorge | T.I. Burgess | EU144050 | EU144065 | KU887520 | KU696367 | KU886775 | KX464354 | N/A | Pavlic et al. (2008), Cruywagen et al. (2017) |
| L. mediterranea | CBS 1377833 | Quercus ilex | Italy | B.T. Linaldeddu | KJ638312 | KJ638331 | KU887521 | KU696368 | KU886776 | N/A | N/A | Linaldeddu et al. (2015) |
| CBS 137784 | Vitis vinifera | Italy | S. Serra | KJ638311 | KJ638330 | KU887522 | KU696369 | KU886777 | N/A | N/A | Linaldeddu et al. (2015) | |
| L. missouriana | CBS 128311 = UCD2193MO3 | Vitis sp. × Vitis labruscana | Missouri, USA | K. Striegler & G.M. Leavitt | HQ288225 | HQ288267 | HQ288304 | KU696370 | KU886778 | N/A | N/A | Úrbez-Torres et al. (2012), Cruywagen et al. (2017) |
| CBS 128312 = UCD2199MO | Vitis sp. × Vitis labruscana | Missouri, USA | K. Striegler & G.M. Leavitt | HQ288226 | HQ288268 | HQ288305 | KU696371 | KU886779 | N/A | N/A | Úrbez-Torres et al. (2012), Cruywagen et al. (2017) | |
| L. parva | CBS 456.783 | Cassava-field soil | Colombia | O. Rangel | EF622083 | EF622063 | KU887523 | KU696372 | KU886780 | KF766362 | N/A | Alves et al. (2008), Cruywagen et al. (2017) |
| CBS 494.78 | Cassava-field soil | Colombia | O. Rangel | EF622084 | EF622064 | EU673114 | KU696373 | KU886781 | EU673258 | EU673201 | Alves et al. (2008), Phillips et al. (2008), | |
| Cruywagen et al. (2017) | ||||||||||||
| L. plurivora | CBS 1208323 | Prunus salicina | Stellenbosch, Western Cape, South Africa | U. Damm | EF445362 | EF445395 | KU887524 | KU696374 | KU886782 | KX464356 | N/A | Damm et al. (2007), Cruywagen et al. (2017) |
| CBS 121103 | Vitis vinifera | South Africa | F. Halleen | AY343482 | EF445396 | KU887525 | KU696375 | KU886783 | KX464357 | N/A | Damm et al. (2007), Cruywagen et al. (2017) | |
| L. pontae | CMM 12773 | Spondias purpurea | Pio-IX/Piauí/Brazil | J.S. Lima & F.C.O. Freire | KT151794 | KT151791 | KT151797 | N/A | N/A | N/A | N/A | Coutinho et al. (2017) |
| L. pseudotheobromae | CBS 1164593 | Gmelina arborea | Costa Rica | J. Carranza & Velásquez | EF622077 | EF622057 | EU673111 | KU696376 | KU886784 | EU673256 | EU673199 | Alves et al. (2008), Phillips et al. (2008), Cruywagen et al. (2017) |
| CMM 3887 | Jatropha curcas | Brazil | A. R. Machado | KF234559 | KF226722 | KF254943 | N/A | N/A | N/A | N/A | Machado et al. (2014) | |
| L. pyriformis | CBS 121770 = CMW 254143 | Acacia mellifera | Dordabis, Namibia | F.J.J. van der Walt & J. Roux | EU101307 | EU101352 | KU887527 | KU696378 | KU886786 | N/A | N/A | Slippers et al. (2014), Cruywagen et al. (2017) |
| CBS 121771 = CMW 25415 | Acacia mellifera | Dordabis, Namibia | F.J.J. van der Walt & J. Roux | EU101308 | EU101353 | KU887528 | KU696379 | KU886787 | N/A | N/A | Slippers et al. (2014), Cruywagen et al. (2017) | |
| L. rubropurpurea | CBS 118740 = CMW 14700 = WAC 125353 | Eucalyptus grandis | Tully, Queensland | T.I. Burgess & G. Pegg | DQ103553 | DQ103571 | EU673136 | KU696380 | KU886788 | DQ377903 | EU673191 | Burgess et al. (2006), Phillips et al. (2008), Cruywagen et al. (2017) |
| WAC 12536 = CMW 15207 | Eucalyptus grandis | Tully, Queensland | T.I. Burgess & G. Pegg | DQ103554 | DQ103572 | KU887530 | KU696381 | N/A | N/A | N/A | Burgess et al. (2006), Cruywagen et al. (2017) | |
| L. sterculiae | CBS 342.783 | Sterculia oblonga | Germany | S. Bruhn | KX464140 | KX464634 | KX464908 | KX463989 | N/A | JX681073 | N/A | Yang et al. (2017) |
| L. subglobosa | CMM 38723 | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234558 | KF226721 | KF254942 | N/A | N/A | N/A | N/A | Machado et al. (2014) |
| CMM 4046 | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234560 | KF226723 | KF254944 | N/A | N/A | N/A | N/A | Machado et al. (2014) | |
| L. thailandica | CPC 227953 | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193637 | KJ193681 | N/A | N/A | N/A | N/A | N/A | Trakunyingcharoen et al. (2015) |
| CPC 22755 | Phyllanthus acidus | Thailand | T. Trakunyingcharoen | KM006433 | KM006464 | N/A | N/A | N/A | N/A | N/A | Trakunyingcharoen et al. (2015) | |
| L. theobromae | CBS 164.963 | Fruit along coral reef coast | New Guinea | A. Aptroot | AY640255 | AY640258 | KU887532 | KU696383 | KU886789 | EU673253 | EU673196 | Alves et al. (2008), Phillips et al. (2008), Cruywagen et al. (2017) |
| CBS 111530 | Unknown | Unknown | Unknown | EF622074 | EF622054 | KU887531 | KU696382 | KU886790 | N/A | N/A | Alves et al. (2008), Cruywagen et al. (2017) | |
| L. venezuelensis | CBS 118739 = CMW 13511 = WAC 125393 | Acacia mangium | Acarigua, Venezuela | S. Mohali | DQ103547 | DQ103568 | KU887533 | KU696384 | KU886791 | DQ377904 | EU673192 | Burgess et al. (2006), Phillips et al. (2008), Cruywagen et al. (2017) |
| CMW 13512 = WAC 12540 | Acacia mangium | Acarigua, Venezuela | S. Mohali | DQ103548 | DQ103569 | KU887534 | N/A | KU886792 | N/A | N/A | Burgess et al. (2006), Cruywagen et al. (2017) | |
| L. viticola | CBS 128313 = UCD 2553AR3 | Vitis vinifera | USA | K. Striegler & G.M. Leavitt | HQ288227 | HQ288269 | HQ288306 | KU696385 | KU886793 | N/A | N/A | Úrbez-Torres et al. (2012), Cruywagen et al. (2017) |
| CBS 128315 = UCD 2604MO | Vitis vinifera | USA | K. Striegler & G.M. Leavitt | HQ288228 | HQ288270 | HQ288307 | KU696386 | KU886794 | N/A | N/A | Úrbez-Torres et al. (2012), Cruywagen et al. (2017) | |
| L. vitis | CBS 1240603 | Vitis vinifera | Italy | S. Burruano | KX464148 | KX464642 | KX464917 | KX463994 | N/A | KX464367 | N/A | Yang et al. (2017) |
| Neofusicoccum algeriense | CBS 137504= ALG 13 | Vitis vinifera | Algeria | A. Berraf-Tebbal | KJ657702 | KJ657715 | KX505915 | N/A | N/A | N/A | N/A | Berraf-Tebbal et al. (2014), Lopes et al. (2017) |
| CAA 322 | Malus domestica | Portugal | KX505906 | KX505894 | KX505916 | N/A | N/A | N/A | N/A | Lopes et al. (2017) | ||
| N. andinum | CBS 117453 = CMW 134553 | Eucalyptus sp. | Me’ rida state, Venezuela | S. Mohali | AY693976 | AY693977 | KX464923 | KX464002 | N/A | KX464373 | N/A | Mohali et al. (2006), Yang et al. (2017) |
| CBS 117452 = CMW 13446 | Eucalyptus sp. | Me’ rida state, Venezuela | S. Mohali | DQ306263 | DQ306264 | KX464922 | KX464001 | N/A | DQ377914 | N/A | Mohali et al. (2006), Yang et al. (2017) | |
| N. arbuti | CBS 1161313 | Arbutus menziesii | Washington, USA | M. Elliott | AY819720 | KF531792 | KF531793 | KX464003 | N/A | DQ377915 | KF531814 | Farr et al. (2005), Crous et al. (2006), Phillips et al. (2013), Yang et al. (2017) |
| CBS 117090 | Arbutus menziesii | California, USA | M. Elliott | AY819724 | KF531791 | KF531794 | N/A | N/A | DQ377919 | KF531813 | Farr et al. (2005), Crous et al. (2006), Phillips et al. (2013) | |
| N. australe | CMW 68373 | Acacia sp. | Batemans Bay, Australia | M.J. Wingfield | AY339262 | AY339270 | AY339254 | EU339573 | N/A | KF766367 | N/A | Slippers et al. (2004b, 2013), Burgess et al. (2007) |
| CBS 110865 = CPC 4599 | Vitis vinifera | South Africa | F. Halleen | AY343408 | KX464661 | KX464937 | KX464005 | N/A | KX464385 | N/A | Van Niekerk et al. (2004), Yang et al. (2017) | |
| N. batangarum | CBS 124924 = CMW 283633 | Terminalia catappa | Cameroon | D. Begoude & J. Roux | FJ900607 | FJ900653 | FJ900634 | FJ900615 | N/A | KX464401 | N/A | Begoude et al. (2010), Yang et al. (2017) |
| CBS 124923 = CMW 28320 | Terminalia catappa | Cameroon | D. Begoude & J. Roux | FJ900608 | FJ900654 | FJ900635 | FJ900616 | N/A | KX464400 | N/A | Begoude et al. (2010), Yang et al. (2017) | |
| N. brasiliense | CMM 13383 | Mangifera indica | Brazil | M.W. Marques | JX513630 | JX513610 | KC794031 | N/A | N/A | N/A | N/A | Marques et al. (2013) |
| CMM 1285 | Mangifera indica | Brazil | M.W. Marques | JX513628 | JX513608 | KC794030 | N/A | N/A | N/A | N/A | Marques et al. (2013) | |
| N. buxi | CBS 116.753 | Buxus sempervirens | France | H.A. van der Aa | KX464165 | KX464678 | N/A | KX464010 | N/A | KX464406 | N/A | Yang et al. (2017) |
| CBS 113714 | Buxus sempervirens | Sweden | O. Constantinescu | KX464164 | KX464677 | KX464954 | KX464009 | N/A | KX464405 | N/A | Yang et al. (2017) | |
| N. cordaticola | CBS 123634 = CMW 139923 | Syzigium cordatum | South Africa | D. Pavlic | EU821898 | EU821868 | EU821838 | EU821928 | N/A | KX464409 | N/A | Pavlic et al. (2009b), Yang et al. (2017) |
| CBS 123635 = CMW 14056 | Syzigium cordatum | South Africa | D. Pavlic | EU821903 | EU821873 | EU821843 | EU821933 | N/A | KX464410 | N/A | Pavlic et al. (2009b), Yang et al. (2017) | |
| N. cryptoaustrale | CMW 23785 = CBS 1228133 | Eucalyptus trees | South Africa | H.M. Maleme | FJ752742 | FJ752713 | FJ752756 | KX464014 | N/A | KX464416 | N/A | Crous et al. (2013), Yang et al. (2017) |
| N. eucalypticola | CBS 115679 = CMW 65393 | Eucalyptus grandis | Orbost, Victoria, Australia | M.J. Wingfield | AY615141 | AY615133 | AY615125 | N/A | N/A | KF766368 | KF766288 | Slippers et al. (2004c, 2013) |
| CBS 115766 = CMW 6217 | Eucalyptus rossii | Tidbinbilla, NSW, Australia | M.J. Wingfield | AY615143 | AY615135 | AY615127 | N/A | N/A | N/A | N/A | Slippers et al. (2004c, 2013) | |
| N. eucalyptorum | CBS 115791 = CMW 101253 | Eucalyptus grandis | South Africa | H. Smith | AF283686 | AY236891 | AY236920 | N/A | N/A | N/A | N/A | Smith et al. (2001), Slippers et al. (2004b) |
| CMW 10126 | Eucalyptus grandis | South Africa | H. Smith | AF283687 | AY236892 | AY236921 | N/A | N/A | N/A | N/A | Smith et al. (2001), Slippers et al. (2004b) | |
| N. grevilleae | CBS 129518 = CPC 169993 | Grevillea aurea | Australia | P.W. Crous & R.G. Shivas | JF951137 | N/A | N/A | N/A | N/A | JF951157 | N/A | Crous et al. (2011) |
| N. hellenicum | CERC1947 = CFCC500673 | Pistachia vera | Thessaloniki, Greece | T.J. Michailides | KP217053 | KP217061 | KP217069 | N/A | N/A | N/A | N/A | Chen et al. (2015) |
| CERC1948 = CFCC50068 | Pistachia vera | Aghios Mamas, Chalkidiki, Greece | T.J. Michailides | KP217054 | KP217062 | KP217070 | N/A | N/A | N/A | N/A | Chen et al. (2015) | |
| N. illicii | CGMCC3.183103 | Illicium verum | Guangxi, China | L. Wang | KY350149 | N/A | KY350155 | N/A | N/A | N/A | N/A | Zhang et al. (2017) |
| CGMCC3.18311 | Illicium verum | Guangxi, China | L. Wang | KY350150 | KY817756 | KY350156 | N/A | N/A | N/A | N/A | Zhang et al. (2017) | |
| N. kwambonambiense | CBS 123639 = CMW 140233 | Syzigium cordatum | South Africa | D. Pavlic | EU821900 | EU821870 | EU821840 | EU821930 | N/A | KX464422 | N/A | Pavlic et al. (2009b), Yang et al. (2017) |
| CBS 123641 = CMW 14140 | Syzigium cordatum | South Africa | D. Pavlic | EU821919 | EU821889 | EU821859 | EU821949 | N/A | KX464424 | N/A | Pavlic et al. (2009b), Yang et al. (2017) | |
| N. lumnitzerae | CMW 414693 | Lumnitzera racemosa | South Africa | J.A. Osorio & J. Roux | KP860881 | KP860724 | KP860801 | KU587925 | N/A | N/A | N/A | Osorio et al. (2017) |
| CMW 41228 | Lumnitzera racemosa | South Africa | J.A. Osorio & J. Roux | KP860882 | KP860725 | KP860803 | KU587926 | N/A | N/A | N/A | Osorio et al. (2017) | |
| N. luteum | CBS 562.92 = ATCC 581933 | Actinidia deliciosa, lesion on ripe fruit | New Zealand | S.R. Pennycook | KX464170 | KX464690 | KX464968 | KX464020 | N/A | KX464430 | N/A | Yang et al. (2017) |
| N. macroclavatum | CBS 118223 = WAC 124443 | Eucalyptus globulus | Western Australia | T. Burgess | DQ093196 | DQ093217 | DQ093206 | KX464022 | N/A | KX464436 | N/A | Burgess et al. (2005), Yang et al. (2017) |
| N. mangiferae | CBS 118531 = CMW 70243 | Mangifera indica | Australia | G.I. Johnson | AY615185 | DQ093221 | AY615172 | N/A | N/A | DQ377920 | EU673153 | Slippers et al. (2005), Phillips et al. (2008) |
| CBS 118532 = CMW 7797 | Mangifera indica | Australia | G.I. Johnson | AY615186 | DQ093220 | AY615173 | KX464023 | N/A | DQ377921 | EU673154 | Slippers et al. (2005), Phillips et al. (2008), Yang et al. (2017) | |
| N. mangroviorum | CMW 413653 | Avicennia marina | South Africa | J.A. Osorio & J. Roux | KP860859 | KP860702 | KP860779 | KU587905 | N/A | N/A | N/A | Osorio et al. (2017) |
| CMW 42481 | Bruguiera gymnorrhiza | South Africa | J.A. Osorio & J. Roux | KP860848 | KP860692 | KP860770 | KU587895 | N/A | N/A | N/A | Osorio et al. (2017) | |
| N. mediterraneum | CBS 121718 = CPC 131373 | Eucalyptus sp. | Greece | P.W. Crous, M.J. Wingfield & A.J.L. Phillips | GU251176 | GU251308 | GU251836 | KX464024 | N/A | N/A | N/A | Crous et al. (2007), Yang et al. (2017) |
| N. nonquaesitum | CBS 126655 = PD 4843 | Umbellularia californica | USA | F.P. Trouillas | GU251163 | GU251295 | GU251823 | KX464025 | N/A | KX464437 | N/A | Inderbitzin et al. (2010), Yang et al. (2017) |
| PD 301 | Vaccinum corymbosum cv. Elliot | Chile | E.X. Bricenõ, J.G. Espinoza, B.A. Latorre & J.G. Espinoza | GU251164 | GU251296 | GU251824 | N/A | N/A | N/A | N/A | Inderbitzin et al. (2010) | |
| N. occulatum | CBS 128008 = MUCC 2273 | Eucalyptus grandis hybrid | Australia | T.I. Burgess | EU301030 | EU339509 | EU339472 | EU339558 | N/A | KX464438 | N/A | Sakalidis et al. (2011), Yang et al. (2017) |
| MUCC 286 = WAC 12395 | Eucalyptus pellita | Australia | T.I. Burgess | EU736947 | EU339511 | EU339474 | EU339560 | N/A | N/A | N/A | Sakalidis et al. (2011) | |
| N. parvum | ATCC 58191 = CMW 90813 | Populus nigra | New Zealand | G.J. Samuels | AY236943 | AY236888 | AY236917 | EU821963 | N/A | AY928045 | EU673151 | Slippers et al. (2004a), Alves et al. (2005), Phillips et al. (2008), Pavlic et al. (2009b) |
| CMW 9080 = ICMP 8002 | Populus nigra | New Zealand | G.J. Samuels | AY236942 | AY236887 | AY236916 | EU821962 | N/A | N/A | N/A | Slippers et al. (2004a), Pavlic et al. (2009b) | |
| N. pennatisporum | WAC 13153 = MUCC 5103 | Allocasuarina fraseriana | Western Australia | K.M. Taylor | EF591925 | EF591976 | EF591959 | N/A | N/A | EF591942 | N/A | Taylor et al. (2009) |
| N. pistaciae | CBS 595.763 | Pistacia vera | Greece | D.G. Zachos | KX464163 | KX464676 | KX464953 | KX464008 | N/A | KX464404 | N/A | Yang et al. (2017) |
| N. pistaciarum | CBS 113083 = CPC 52633 | Pistacia vera | USA | T.J. Michailides | KX464186 | KX464712 | KX464998 | KX464027 | N/A | KX464465 | N/A | Yang et al. (2017) |
| CBS 113084 = CPC 5284 | Redwood | USA | T.J. Michailides | KX464187 | KX464713 | KX464999 | KX464028 | N/A | KX464466 | N/A | Yang et al. (2017) | |
| N. protearum | CBS 114176 = STE-U 17753 | Leucadendron salignum | South Africa | S. Denman | AF452539 | KX464720 | KX465006 | KX464029 | N/A | JX556245 | N/A | Denman et al. (2003), Yang et al. (2017) |
| CBS 111200 = CPC 1357 | Leucadendron sp. | South Africa | P.W. Crous | KX464193 | KX464719 | KX465005 | N/A | N/A | KX464472 | N/A | Yang et al. (2017) | |
| N. ribis | CBS 115475 = CMW 77723 | Ribes sp. | USA | B. Slippers & G. Hudler | AY236935 | AY236877 | AY236906 | EU821958 | N/A | AY928044 | KF766292 | Slippers et al. (2004a, 2013), Alves et al. (2005), Pavlic et al. (2009b) |
| CBS 121.26 = CMW 7054 | Ribes rubrum | USA | N.E. Stevens | AF241177 | AY236879 | AY236908 | EU821960 | N/A | KX464473 | N/A | Slippers et al. (2004a), Pavlic et al. (2009b), Yang et al. (2017) | |
| N. sinense | CGMCC3.183153 | Unknown woody plant | Guizhou, China | J.J. Gan | KY350148 | KY817755 | KY350154 | N/A | N/A | N/A | N/A | Zhang et al. (2017) |
| N. stellenboschiana | CBS 110864 = CPC 45983 | Vitis vinifera | South Africa | F. Halleen | AY343407 | AY343348 | KX465047 | KX464042 | N/A | KX464513 | N/A | Van Niekerk et al. (2004), Yang et al. (2017) |
| N. terminaliae | CBS 125263 = CMW 266793 | Terminalia sericea | South Africa | D. Begoude & J. Roux | GQ471802 | GQ471780 | KX465052 | KX464045 | N/A | KX464518 | N/A | Begoude (2010), Yang et al. (2017) |
| CBS 125264 = CMW 26683 | Terminalia sericea | South Africa | D. Begoude & J. Roux | GQ471804 | GQ471782 | KX465053 | KX464046 | N/A | KX464519 | N/A | Begoude (2010), Yang et al. (2017) | |
| N. umdonicola | CBS 123645 = CMW 140583 | Syzigium cordatum | South Africa | D. Pavlic | EU821904 | EU821874 | EU821844 | EU821934 | N/A | KX464522 | N/A | Pavlic et al. (2009b), Yang et al. (2017) |
| CBS 123646 = CMW 14060 | Syzigium cordatum | South Africa | D. Pavlic | EU821905 | EU821875 | EU821845 | EU821935 | N/A | KX464523 | N/A | Pavlic et al. (2009b), Yang et al. (2017) | |
| N. ursorum | CMW 24480 = CBS 1228113 | Eucalyptus trees | South Africa | H.M. Maleme | FJ752746 | FJ752709 | KX465056 | KX464047 | N/A | N/A | N/A | Crous et al. (2013), Yang et al. (2017) |
| CMW 23790 | Eucalyptus trees | South Africa | H.M. Maleme | FJ752745 | FJ752708 | KX465057 | N/A | N/A | N/A | N/A | Crous et al. (2013), Yang et al. (2017) | |
| N. viticlavatum | CBS 112878 = STE-U 50443 | Vitis vinifera | South Africa | F. Halleen | AY343381 | AY343342 | KX465058 | KX464048 | N/A | KX464527 | N/A | Phillips et al. (2013), Yang et al. (2017) |
| CBS 112977 = STE-U 5041 | Vitis vinifera | South Africa | F. Halleen | AY343380 | AY343341 | KX465059 | N/A | N/A | KX464528 | N/A | Phillips et al. (2013), Yang et al. (2017) | |
| N. vitifusiforme | CBS 110887 = STE-U 52523 | Vitis vinifera | South Africa | J.M. van Niekerk | AY343383 | AY343343 | KX465061 | KX464049 | N/A | KX464530 | N/A | Van Niekerk et al. (2004), Yang et al. (2017) |
| CBS 110880 = STE-U 5050 | Vitis vinifera | South Africa | J.M. van Niekerk | AY343382 | AY343344 | KX465008 | N/A | N/A | KX464475 | N/A | Van Niekerk et al. (2004), Yang et al. (2017) | |
1 ALG: Personal culture collection A. Berraf-Tebbal; ATCC: American Type Culture Collection, Virginia, USA; BL: Personal number of B.T. Linaldeddu; CAA: Personal culture collection Artur Alves, Universidade de Aveiro, Portugal; CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; CERC: Culture collection of China Eucalypt Research Centre, Chinese Academy of Forestry, ZhanJiang, GuangDong, China; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Microbiological Culture Collection Center, Beijing, China; CMM: Culture Collection of Phytopathogenic Fungi ‘Prof. Maria Menezes’, Universidade Federal Rural de Pernambuco, Recife, Brazil; CMW: Tree Pathology Co-operative Program, Forestry and Agricultural Biotechnology Institute, University of Pretoria, South Africa; CPC: Working collection of P.W. Crous, housed at CBS; GZCC: Guizhou Academy of Agricultural Sciences Culture Collection, GuiZhou, China; IBL: Personal culture collection, I.B.L. Coutinho; IBP: Personal culture collection, I.B. Prasher; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; IRAN: Iranian Fungal Culture Collection, Iranian Research Institute of Plant Protection, Iran; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUCC: Culture collection of Murdoch University, Perth, Australia; STE-U: Culture collection of the Department of Plant Pathology, University of Stellenbosch, South Africa; UCD: University of California, Davis, Plant Pathology Department Culture Collection; WAC: Department of Agriculture, Western Australia Plant Pathogen Collection, South Perth, Western Australia.
2 ITS, internal transcribed spacer region and intervening 5.8S nrRNA gene; tef1, translation elongation factor 1-alpha; tub, β-tubulin; rpb2, DNA-directed RNA polymerase II subunit; cmdA, calmodulin; LSU, nuclear ribosomal large subunit; SSU, nuclear ribosomal small subunit; N/A = not available.
3 Isolates represent ex-type or are from samples that have been linked morphologically to type materials of the species.
The BLAST results showed that the isolates collected in this study were grouped in the genera Botryosphaeria, Cophinforma, Lasiodiplodia and Neofusicoccum. Phylogenetic analyses were conducted for each of the ITS, tef1, tub, rpb2, cmdA, LSU and SSU datasets for genera Botryosphaeria/Cophinforma, Lasiodiplodia and Neofusicoccum, respectively. As the cmdA sequences are only available for Lasiodiplodia, and not for Botryosphaeria, Cophinforma and Neofusicoccum, the analyses for cmdA sequences were only conducted for the genus Lasiodiplodia.
Phylogenetic analyses were also conducted for combined datasets, as the LSU and SSU sequences are not available for some of the previously described species of Botryosphaeria, Cophinforma, Lasiodiplodia and Neofusicoccum, and the rpb2 sequences are not available for some species of Botryosphaeria. The ITS, tef1 and tub sequences were combined for phylogenetic analyses of Botryosphaeria/Cophinforma isolates, the ITS, tef1, tub, rpb2 and cmdA sequences were combined for Lasiodiplodia isolates, and ITS, tef1, tub and rpb2 sequences were combined for Neofusicoccum isolates.
Two phylogenetic analysis methods were used: PAUP v. 4.0b10 (Swofford 2003) for the maximum parsimony (MP) analyses and PhyML v. 3.0 (Guindon et al. 2010) for maximum likelihood (ML) tests. For MP analyses, gaps are treated as a fifth character and the characters are unordered and of equal weight with 1 000 random addition replicates. The equally most parsimonious trees were obtained using the heuristic search function and tree bisection and reconstruction (TBR) as the branch swapping algorithms. MAXTREES were limited to 5 000, and branch lengths of zero were collapsed. A bootstrap analysis (50 % majority rule, 1 000 replicates) was performed to determine the confidence levels of the tree-branching points (Felsenstein 1985). Tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were used to evaluate the trees (Hillis & Huelsenbeck 1992).
For ML analyses of each dataset, the best models of nucleotide substitution were determined using jModelTest v. 2.1.5 (Darriba et al. 2012). Additional ML parameters in PhyML include the retention of the maximum number of 1 000 trees and the determination of nodal support by non-parametric bootstrapping with 1 000 replicates. All phylogenetic trees were viewed using MEGA v. 6.0.5 (Tamura et al. 2013). Neofusicoccum parvum (ATCC 58191) was used as the outgroup taxon for analyses of Botryosphaeria and Cophinforma; Botryosphaeria dothidea (CBS 115476) was used as the outgroup taxon for analyses of Lasiodiplodia and Neofusicoccum (Table 2).
Morphology
Representative isolates for each genotype of Botryosphaeriaceae species identified by DNA sequence comparisons were selected for morphological study. To induce sporulation, selected isolates were transferred to 2 % water agar (WA) media (20 g of agar per litre of water) with double-sterilised pine needles placed on the surface of the media (Smith et al. 1996). These cultures were incubated at 25 °C under near-ultraviolet light for 4–6 wk. Conidia in the pycnidia were mounted in one drop of 80 % lactic acid on glass slides and examined under a stereomicroscope (Carl Zeiss Ltd., Munchen, Germany). Conidia and other structures were examined and recorded using a Zeiss Axio Imager A1 microscope and a Zeiss AxioCam MRc digital camera with Zeiss Axio Vision v. 4.8 software (Carl Zeiss Ltd.). Measurements of conidiomata, conidiophores and conidiogenous cells were made to determine the smallest and the largest values. For the isolates selected as a holotype, the lengths and widths of 100 conidia per isolate were measured, as well as 25 measurements of the remaining isolates of each taxon. Average (mean), standard deviation (SD), minimum (min) and maximum (max) measurements are presented as (min–)(mean–SD)–(mean+SD)(–max). The average length/average width ratio (L/W) of the conidial measurements was calculated.
Colony morphology was characterised by cultures grown on 2 % MEA for 7 d and colony colour was determined using the colour charts of Rayner (1970). For growth studies, a 5-mm-diam plug from the growing margin of 7-d-old colonies of each representative isolate was placed in the centre of 90-mm-diam Petri plates containing 2 % MEA. These cultures were incubated in the dark at 5 °C intervals from 5–40 °C. Five replicate plates of each isolate at each temperature were conducted. Two diameter measurements, orthogonally, were recorded daily until the fastest growing culture reached the edge of the Petri plate. The experiment was repeated once and the average for each of the eight temperatures was calculated.
Pathogenicity tests
To determine the pathogenicity of the identified species on Eucalyptus seedlings, representative isolates of all Botryosphaeriaceae species identified in this study were selected to inoculate on Eucalyptus seedlings. Three Eucalyptus clones, CEPT-11 (Eucalyptus urophylla × E. grandis), CEPT-12 (E. urophylla) and CEPT-13 (E. urophylla × E. tereticornis), were used for inoculations. The Eucalyptus seedlings were 1-yr-old, approximately 1.7 m in height, and had a 2.0 cm diam at the root collar. For each clone, 10 seedlings were inoculated with each isolate. On each inoculated seedling, a 5-mm-diam wound was made on the tree stem using a cork borer to remove the bark and expose the xylem. The wounds are located approximately 30 cm above the root collar. For inoculation, 5-mm-diam plugs of mycelia from the margins of colonies grown on 2 % MEA for 7 d in the dark were taken and placed into the wounds with the mycelia facing the cambium. Inoculated wounds were encased with masking tape to prevent contamination and desiccation. Ten seedlings of each Eucalyptus clone were inoculated with sterile MEA plugs to serve as negative controls. One month after inoculation, the bark of inoculated seedlings was removed and the internal lesion/wound length on the cambium was measured. The inoculated fungi were re-isolated by cutting small pieces of wood from the edges of the lesions and cultivating them in 2 % MEA at 25 °C. Re-isolations were made from the seedlings inoculated by mycelium plugs and MEA plugs. The data were analysed by one-way analyses of variance (ANOVA) using SAS v. 9.3 (SAS Institute Inc. 2011).
RESULTS
Fungal isolation
In this study, 105 isolates from Eucalyptus and other plants that show typical morphology of Botryosphaeriaceae were isolated. Eighty-one isolates were collected from Eucalyptus trees: 12 from FuJian Province, 39 from GuangDong Province, 29 from GuangXi Province and one from HaiNan Province. Eighteen isolates with typical characteristics of Botryosphaeriaceae were collected from other plants which were growing in close proximity to Eucalyptus: two from C. lanceolata, 10 from D. longan, four from M. sanguineum, and two from P. hanceana. In addition, three isolates were collected from A. cunninghamii and C. deodara, respectively (Table 1).
Phylogenetic analyses
For all the 105 isolates in this study, ITS, tef1, tub, rpb2, cmdA, LSU and SSU sequence data were generated and deposited in GenBank (Table 1). The PCR fragments are approximately 520 bps for the ITS region, 280 bps for the tef1 region, 430 bps for the tub region, 610 bps for the rpb2 region, 850 bps for the LSU region and 1040 bps for the SSU region. The genotype for each isolate was determined by the ITS, tef1, tub, rpb2, LSU, SSU sequences for isolates in the genera Botryosphaeria, Cophinforma and Neofusicoccum, and by ITS, tef1, tub, rpb2, cmdA, LSU, SSU sequences for isolates in the genus Lasiodiplodia (Table 1). The preliminary identities of the isolates were determined from conducting a standard nucleotide BLAST with the sequences of ITS, tef1, tub, rpb2, cmdA, LSU and SSU, the results consistently showed that the isolates sequenced in this study resided in Botryosphaeria, Cophinforma, Lasiodiplodia or Neofusicoccum. One to two isolates of each genotype were selected and used for phylogenetic analyses, depending on the number of isolates of each genotype (Table 1). Based on the comparisons for six to seven region sequences generated in this study and published sequences from ex-type strains of Botryosphaeriaceae downloaded from NCBI, sequences of Botryosphaeria, Cophinforma, Lasiodiplodia or Neofusicoccum related to species emerging from this study were used for analyses (Table 2). The aligned sequences of each region of ITS, tef1, tub, rpb2, cmdA, LSU, SSU, as well as the combined sequences of three to five (Botryosphaeria/Cophinforma: three; Lasiodiplodia: five; Neofusicoccum: four) regions were deposited in TreeBASE (No. 21430). These datasets for genera Botryosphaeria/Cophinforma, Lasiodiplodia and Neofusicoccum, as well as statistical values for the trees for the MP analyses and parameters for the best-fit substitution models of ML analyses, are provided in Table 3.
Table 3.
Datasets used and statistics resulting from phylogenetic analyses.
| Genus | Dataset | No. of taxa | No. of bp1 | Maximum parsimony |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIC2 | No. of trees | Tree length | CI3 | RI4 | RC5 | HI6 | ||||||
| Botryosphaeria/Cophinforma | ITS | 45 | 543 | 42 | 18 | 67 | 0.8209 | 0.9506 | 0.7804 | 0.1791 | ||
| tef1 | 45 | 356 | 147 | 875 | 206 | 0.8932 | 0.9677 | 0.8643 | 0.1068 | |||
| tub | 34 | 415 | 43 | 8 | 58 | 0.9138 | 0.9688 | 0.8852 | 0.0862 | |||
| rpb2 | 22 | 718 | 92 | 2 | 116 | 0.9397 | 0.9809 | 0.9217 | 0.0603 | |||
| LSU | 34 | 847 | 21 | 3 | 24 | 0.9583 | 0.9865 | 0.9454 | 0.0417 | |||
| SSU | 31 | 1024 | 9 | 342 | 9 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |||
| ITS/tef1/tub | 45 | 1314 | 232 | 5000 | 337 | 0.8665 | 0.9585 | 0.8305 | 0.1335 | |||
| Lasiodiplodia | ITS | 76 | 526 | 48 | 5000 | 87 | 0.6897 | 0.8945 | 0.6169 | 0.3103 | ||
| tef1 | 75 | 332 | 142 | 852 | 402 | 0.6219 | 0.9067 | 0.5639 | 0.3781 | |||
| tub | 67 | 409 | 41 | 5000 | 60 | 0.7667 | 0.9352 | 0.7170 | 0.2333 | |||
| rpb2 | 57 | 532 | 104 | 861 | 190 | 0.6421 | 0.8761 | 0.5626 | 0.3579 | |||
| cmdA | 45 | 521 | 74 | 496 | 91 | 0.9121 | 0.9776 | 0.8916 | 0.0879 | |||
| LSU | 28 | 835 | 20 | 3 | 27 | 0.7407 | 0.9247 | 0.6850 | 0.2593 | |||
| SSU | 19 | 1020 | 17 | 2 | 23 | 0.8261 | 0.9048 | 0.7474 | 0.1739 | |||
| ITS/tef1/tub/rpb2/cmdA | 76 | 2320 | 409 | 5000 | 948 | 0.5918 | 0.8713 | 0.5156 | 0.4082 | |||
| Neofusicoccum | ITS | 77 | 532 | 81 | 1404 | 187 | 0.5615 | 0.8707 | 0.4889 | 0.4385 | ||
| tef1 | 75 | 307 | 147 | 5000 | 303 | 0.7426 | 0.9321 | 0.6922 | 0.2574 | |||
| tub | 75 | 424 | 71 | 1430 | 141 | 0.6170 | 0.8784 | 0.5420 | 0.3830 | |||
| rpb2 | 55 | 607 | 114 | 652 | 191 | 0.7173 | 0.9069 | 0.6505 | 0.2827 | |||
| LSU | 55 | 841 | 33 | 3260 | 70 | 0.5714 | 0.8324 | 0.4757 | 0.4286 | |||
| SSU | 21 | 1027 | 3 | 1 | 3 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |||
| ITS/tef1/tub/rpb2 | 77 | 1870 | 413 | 336 | 884 | 0.6267 | 0.8824 | 0.5530 | 0.3733 | |||
| Genus | Dataset | Maximum likelihood |
||||||||||
| Subst. model7 | NST8 | Rate matrix | Ti/Tv ratio9 | p-inv | Gamma | Rates | ||||||
| Botryosphaeria/Cophinforma | ITS | TrN+I | 6 | 1.0000 | 1.4207 | 1.0000 | 1.0000 | 8.3891 | – | 0.8010 | – | Equal |
| tef1 | TPM2uf+G | 6 | 1.7386 | 4.6965 | 1.7386 | 1.0000 | 4.6965 | – | – | 0.4470 | Gamma | |
| tub | HKY+G | 2 | – | – | – | – | – | 4.1414 | – | 0.0220 | Gamma | |
| rpb2 | TrN+G | 6 | 1.0000 | 3.1864 | 1.0000 | 1.0000 | 10.4238 | – | – | 0.2610 | Gamma | |
| LSU | TrN+I | 6 | 1.0000 | 5.1213 | 1.0000 | 1.0000 | 14.2608 | – | 0.8440 | – | Equal | |
| SSU | TIM2 | 6 | 0.2353 | 0.4397 | 0.2353 | 1.0000 | 3.3084 | – | – | – | Equal | |
| ITS/tef1/tub | TrN+G | 6 | 1.0000 | 3.2977 | 1.0000 | 1.0000 | 5.9498 | – | – | 0.0970 | Gamma | |
| Lasiodiplodia | ITS | TPM1uf+I+G | 6 | 1.0000 | 8.3075 | 3.1185 | 3.1185 | 8.3075 | – | 0.6760 | 0.7400 | Gamma |
| tef1 | TrN+G | 6 | 1.0000 | 3.6014 | 1.0000 | 1.0000 | 5.5149 | – | – | 0.3870 | Gamma | |
| tub | TIM3+G | 6 | 2.6761 | 3.8909 | 1.0000 | 2.6761 | 10.7362 | – | – | 0.4190 | Gamma | |
| rpb2 | TrN+G | 6 | 1.0000 | 4.8566 | 1.0000 | 1.0000 | 13.8753 | – | – | 0.3530 | Gamma | |
| cmdA | HKY+I | 2 | – | – | – | – | – | 2.7918 | 0.5470 | – | Equal | |
| LSU | TrN+I | 6 | 1.0000 | 7.7385 | 1.0000 | 1.0000 | 16.1138 | – | 0.7970 | – | Equal | |
| SSU | TIM2+I | 6 | 3.1234 | 3.8646 | 3.1234 | 1.0000 | 15.5731 | – | 0.9220 | – | Equal | |
| ITS/tef1/tub/rpb2/cmdA | TIM3+I+G | 6 | 0.6986 | 3.2898 | 1.0000 | 0.6986 | 5.4586 | – | 0.5380 | 0.6730 | Gamma | |
| Neofusicoccum | ITS | TIM1+I+G | 6 | 1.0000 | 11.6895 | 3.1944 | 3.1944 | 22.131 | – | 0.5510 | 0.6030 | Gamma |
| tef1 | HKY+G | 2 | – | – | – | – | – | 2.8135 | 0.0740 | 0.6810 | Gamma | |
| tub | TrN+G | 6 | 1.0000 | 4.0352 | 1.0000 | 1.0000 | 7.8348 | – | – | 0.1980 | Gamma | |
| rpb2 | TIM3+G | 6 | 1.9463 | 7.0524 | 1.0000 | 1.9463 | 19.4804 | – | – | 0.2840 | Gamma | |
| LSU | TrN+I | 6 | 1.0000 | 6.0800 | 1.0000 | 1.0000 | 25.6908 | – | 0.9010 | – | Equal | |
| SSU | TrN | 6 | 1.0000 | 0.9096 | 1.0000 | 1.0000 | 7.9272 | – | – | – | Equal | |
| ITS/tef1/tub/rpb2 | TrN+I+G | 6 | 1.0000 | 4.8874 | 1.0000 | 1.0000 | 9.1711 | – | 0.4320 | 0.7150 | Gamma | |
1 bp = base pairs.
2 PIC = number of parsimony informative characters.
3 CI = consistency index.
4 RI = retention index.
5 RC = rescaled consistency index.
6 HI = homoplasy index.
7 Subst. model = best fit substitution model.
8 NST = number of substitution rate categories.
9 Ti/Tv ratio = transition/transversion ratio.
Species residing in Botryosphaeria
For the isolates grouping in the genus Botryosphaeria, isolates clustered into four phylogenetic groups (Groups A–D) for each of the ITS, tef1, tub, rpb2 and ITS/tef1/tub datasets (Fig. 2a–d, g). For each of the LSU and SSU datasets, Groups A, C and D clustered together (Fig. 2e–f). The ITS sequences of Botryosphaeria fabicerciana, B. fusispora, B. kuwatsukai, B. rosaceae and the six Chinese isolates (CERC2274, CERC2911, CERC2918, CERC2930, CERC3426 and CERC3441) in Group A are consistent, and all of them grouped into one phylogenetic clade (Fig. 2a). For the tef1 sequence analyses, the isolates in Group A clustered closely to B. fabicerciana and B. fusispora (Fig. 2b). For the tub sequences, the isolates in Group A resided in the same phylogenetic clade with B. fusispora (Fig. 2c). For the rpb2, LSU and SSU sequences, the isolates in Group A clustered to the same clade with B. fabicerciana and B. fusispora (rpb2 is not available to B. fusispora) (Fig. 2d–f). The phylogenetic analyses for ITS, tef1, tub, rpb2, LSU and SSU sequences showed that the six Chinese isolates in Group A are most closely related to B. fabicerciana and B. fusispora (Fig. 2a–f). The analyses of the combination of ITS, tef1 and tub sequences indicated that the six isolates are not forming a well-resolved clade, but are phylogenetically more closely related to B. fusispora than to B. fabicerciana (Fig. 2g). Based on the phylogenetic analyses for ITS, tef1, tub, rpb2, LSU, SSU and the combination of the ITS, tef1 and tub sequences, the six isolates were identified as B. fusispora.
Fig. 2.
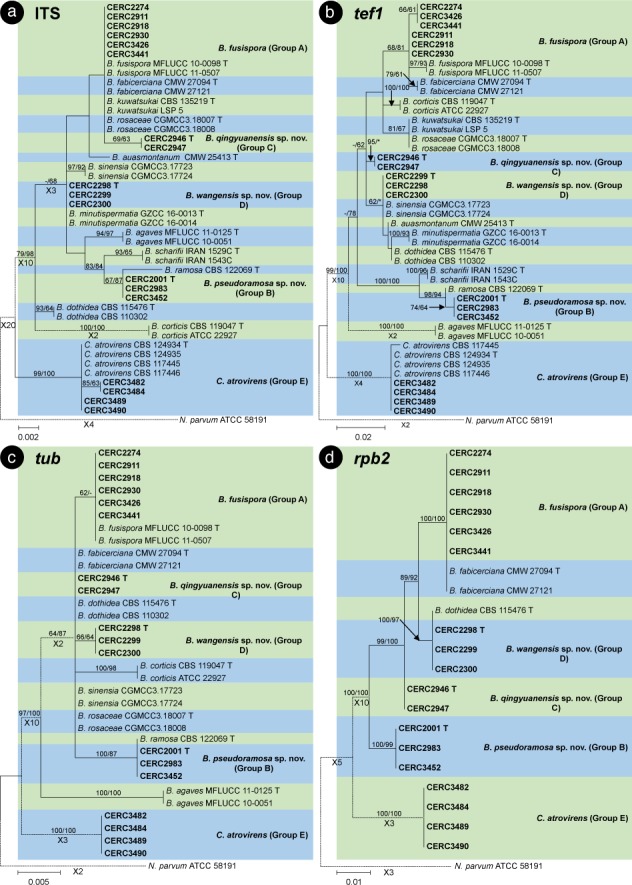
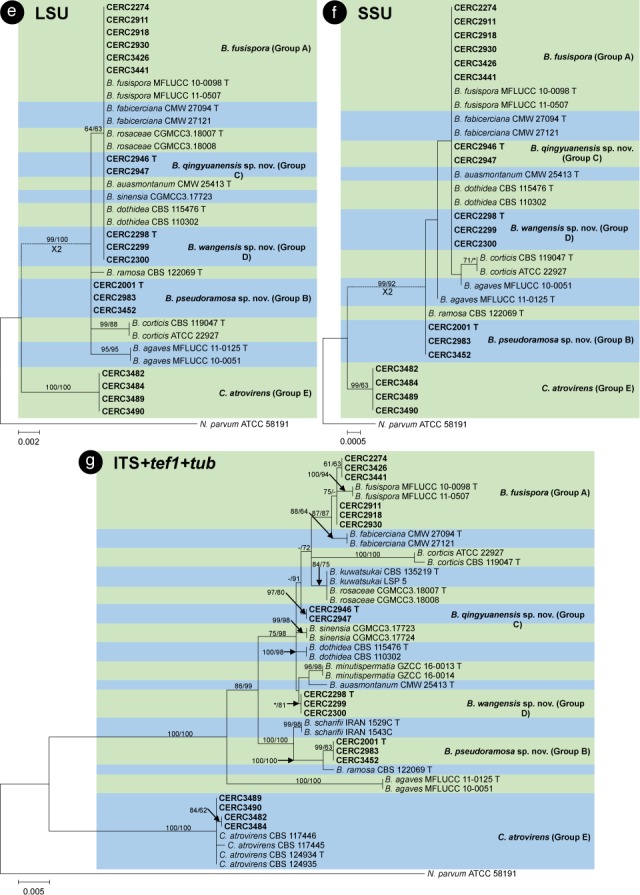
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Botryosphaeria and Cophinforma. a. ITS region; b. tef1 gene region; c. tub gene region; d. rpb2 gene region; e. LSU region; f. SSU region; g. combination of ITS, tef1 and tub regions. Isolates sequenced in this study are in bold. Bootstrap support values ≥ 60 % for ML and MP are presented above branches as follows: ML/MP, bootstrap support values < 60 % are marked with ‘–’, and absent (< 50 %) are marked with ‘*’. Isolates representing ex-type sequences are marked with ‘T’. Neofusicoccum parvum (ATCC 58191) was used as the outgroup taxon.
Isolates in Group B (CERC2001, CERC2983 and CERC3452) and Group C (CERC2946 and CERC2947) were found to be consistently distinct from other known phylogenetically related species of Botryosphaeria by congruent distinction in the multiple datasets (Group B: ITS, tef1, rpb2 and LSU datasets; Group C: ITS, tef1 and rpb2 datasets) (Fig. 2a–f). The analyses of the combination of ITS, tef1 and tub sequences indicated that the isolates in Group B and Group C form two well-resolved clades supported by relatively high bootstrap values (Fig. 2g). Isolates in Groups B and C represent two previously undescribed species of Botryosphaeria.
The phylogenetic analyses based on ITS, tef1, tub, rpb2, LSU and SSU sequences consistently showed that three isolates (CERC2298, CERC2299 and CERC2300) in Group D were phylogenetically most closely related to B. auasmontanum, B. dothidea, B. minutispermatia and B. sinensia (Fig. 2a–f). The analyses of combined ITS, tef1 and tub sequences showed that isolates in Group D form one well-resolved clade (Fig. 2g). Isolates in Group D were identified as a new species of Botryosphaeria.
Species residing in Cophinforma
The BLAST results for ITS sequences show that isolates CERC3482, CERC3484, CERC3489 and CERC3490 (Group E) are related to the genus Cophinforma. Only two species of Cophinforma have previously been described, Cophinforma atrovirens (Mehl et al. 2011, Phillips et al. 2013) and C. mamane (Gardner 1997, Phillips et al. 2013). The two species of Cophinforma are morphologically very similar, but can be distinguished based on ITS sequence data. BLAST results of the ITS sequences indicate that the four Chinese isolates are more closely related to C. atrovirens than to C. mamane. A BLAST search of the tef1 sequences show that the Chinese isolates and the ex-type isolate of C. atrovirens (CBS 124934) are identical. Cophinforma mamane does not have a tef1 sequence and cultures are not available (Phillips et al. 2013). Based on the sequence comparisons of the ITS and tef1 regions (tub gene sequences are not available for species of Cophinforma), isolates in Group E were identified as C. atrovirens (Fig. 2a–b, g).
Species residing in Lasiodiplodia
The isolates in our study that clustered in the genus Lasiodiplodia grouped into three phylogenetic groups for the tef1 dataset (Group F: CERC2284; Group G: CERC2024, CERC3420, CERC3513, CERC3516; Group H: CERC2286, CERC2962, CERC3495) (Fig. 3b), and two clades (Group F = Group G; Group H) for the ITS, tub, rpb2, cmdA, LSU and SSU datasets (Fig. 3a, c–g). For Group F, the sequence analyses of the ITS, tef1, tub, rpb2, cmdA datasets showed that the Chinese isolates clustered into the same (ITS, tef1, rpb2 and cmdA) clade or close (tub) to L. brasiliense (LSU and SSU sequences are not available to L. brasiliense) (Fig. 3a–g). The analyses indicated that isolates in Group G and Group H clustered into the same (ITS, tub, rpb2, cmdA, LSU and SSU) clade or close (tef1) to L. theobromae and L. pseudotheobromae, respectively (Fig. 3a–g). The analyses of the combination of the ITS, tef1, tub, rpb2 and cmdA sequences indicated that the isolates in Groups F, G and H are phylogenetically most closely related to L. brasiliense, L. theobromae and L. pseudotheobromae, respectively (Fig. 3h). Altogether, the results of these phylogenetic analyses identified isolates in Groups F, G and H as L. brasiliense, L. theobromae and L. pseudotheobromae, respectively.
Fig. 3.
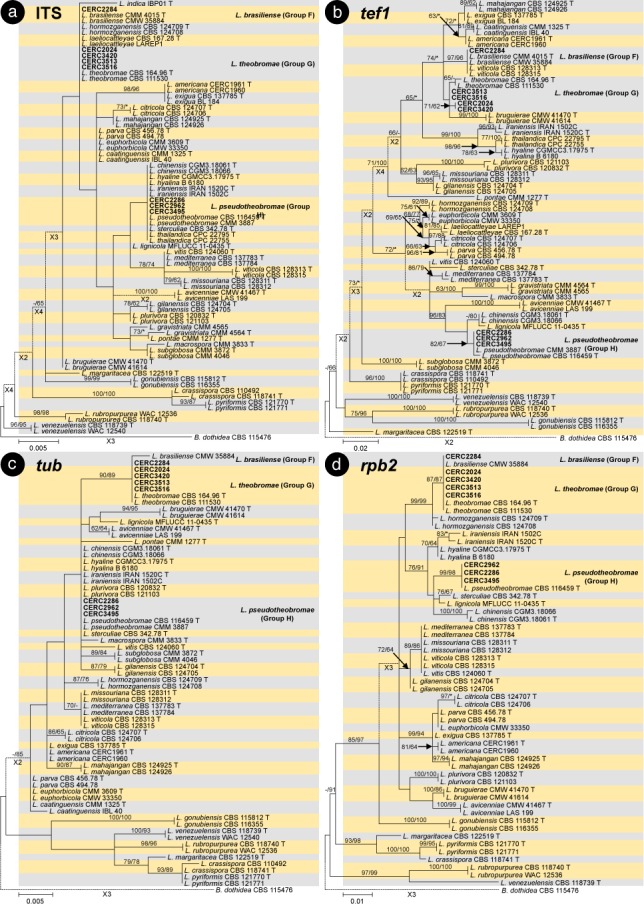
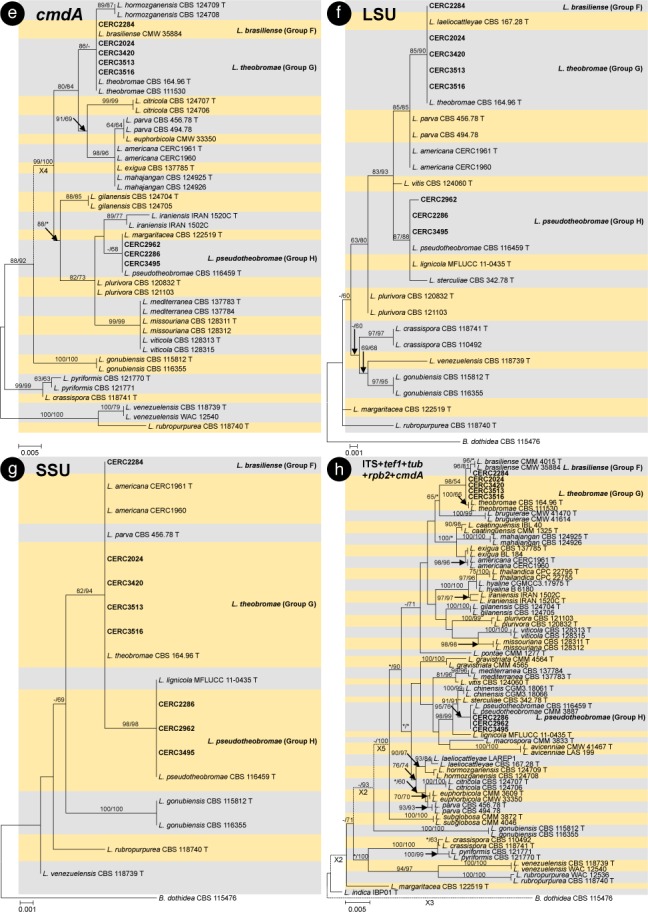
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Lasiodiplodia. a. ITS region; b. tef1 gene region; c. tub gene region; d. rpb2 gene region; e. cmdA gene region; f. LSU region; g. SSU region; h. combination of ITS, tef1, tub, rpb2 and cmdA regions. Isolates sequenced in this study are in bold. Bootstrap support values ≥ 60 % for ML and MP are presented above branches as follows: ML/MP, bootstrap values < 60 % are marked with ‘–’, and absent (< 50 %) are marked with ‘*’. Isolates representing ex-type sequences are marked with ‘T’. Botryosphaeria dothidea (CBS 115476) was used as the outgroup taxon.
Species residing in Neofusicoccum
For the Chinese isolates that grouped in the genus Neofusicoccum, the isolates in this study clustered into four phylogenetic groups for the ITS, tef1, tub and rpb2 datasets (Group I: CERC3497, CERC3498; Group J: CERC2967, CERC2968, CERC2973; Group K: CERC2005, CERC2265, CERC3416, CERC3451; Group L: CERC2951, CERC3503, CERC3504, CERC3508) (Fig. 4a–d). The Chinese Neofusicoccum isolates clustered into three groups (Group I, Group J = Group K, Group L) for the LSU dataset, and two groups (Group I, Group J = Group K = Group L) for the SSU dataset (Fig. 4e–f).
Fig. 4.

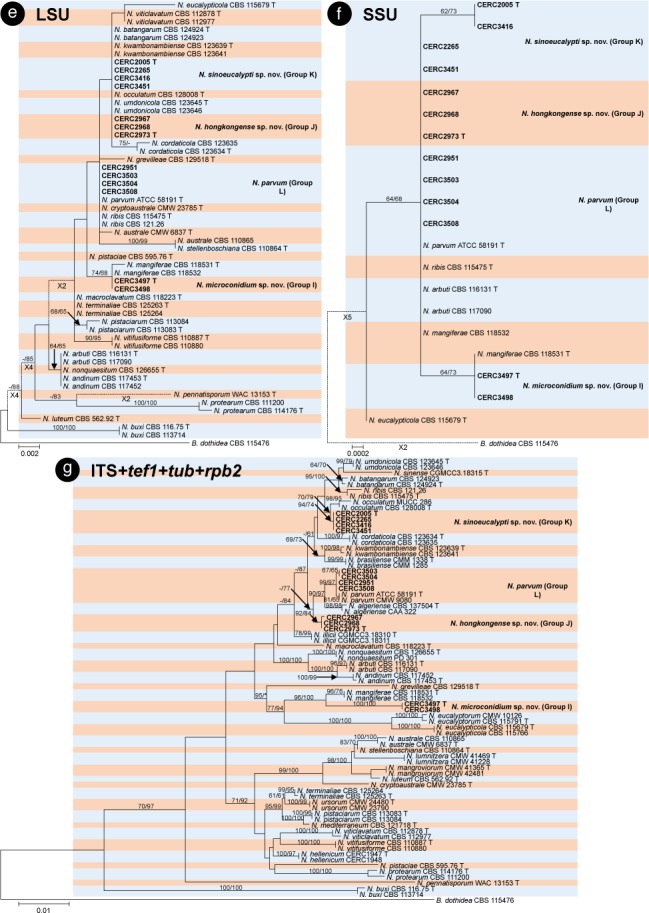
Phylogenetic trees based on maximum likelihood (ML) analyses for species in Neofusicoccum. a. ITS region; b. tef1 gene region; c. tub gene region; d. rpb2 gene region; e. LSU region; f. SSU region; g. combination of ITS, tef1, tub and rpb2 regions. Isolates sequenced in this study are in bold. Bootstrap support values ≥ 60 % for ML and MP are presented above branches as follows: ML/MP, bootstrap support values < 60 % are marked with ‘–’, and absent (< 50 %) are marked with ‘*’. Isolates representing ex-type sequences are marked with ‘T’. Botryosphaeria dothidea (CBS 115476) was used as the outgroup taxon.
Previous studies have shown that phylogenetic analyses of the ITS, tef1, tub and rpb2 sequences, especially a combination of the four gene sequences, is an efficient method for species identification in Neofusicoccum (Pavlic et al. 2009a, Sakalidis et al. 2011, Osorio et al. 2017). The isolates in each of Group I and J were found to be consistently distinct from other known phylogenetically closely related species of Neofusicoccum by congruent distinction in all the ITS, tef1, tub and rpb2 datasets (Fig. 4a–d). Isolates in Group K formed a single independent clade that was distinct from any known Neofusicoccum species in the tef1 and rpb2 datasets (Fig. 4b, d). The analyses of the combination of the ITS, tef1, tub and rpb2 sequences indicated that isolates in each of Groups I, J and K formed a well-resolved clade that was distinct from any described Neofusicoccum species which are supported by high bootstrap values (Fig. 4g). Therefore, we considered isolates in Groups I, J and K to represent three undescribed species of Neofusicoccum.
The Chinese isolates in Group L grouped in the same clade as N. parvum based on the ITS, tub, rpb2, LSU and SSU sequence analyses (Fig. 4a, c–f), and close to N. parvum on the tef1 sequence analysis (Fig. 4b). In the phylogenetic analyses combining four gene regions, isolates in Group L were identified as N. parvum (Fig. 4g).
Morphology and taxonomy
Representative isolates (Table 1, 4) selected for morphological studies produced asexual fruiting structures on pine needles on WA media within 4–6 wk. No sexual structures were observed during the same period of time. For the 12 phylogenetic groups of Botryosphaeriaceae which were distinguished by DNA sequences, morphological studies, including culture and conidia characteristics, show that isolates in each of Group A, E, F, G, H and L were morphologically similar to the type specimens linked to it via sequence data, especially in the morphological characterisation of conidia (Table 4), namely B. fusispora, C. atrovirens, L. brasiliense, L. theobromae, L. pseudotheobromae and N. parvum, respectively. For phylogenetic groups B, C, D, I, J and K, morphological differences were observed compared to the phylogenetically most closely related species based on sequence data, and consequently each of the six groups were considered as new species. Based on the phylogenetic analyses and the morphological characteristics, the fungi collected from Eucalyptus and other plants in this study represent 12 species of Botryosphaeriaceae, including six previously undescribed species. These new species are described as follows.
Table 4.
Conidial measurements of Botryosphaeriaceae species examined in this study and comparison with measurements described in previous studies.
| Species1 | Conidial size (μm) (L × W)2 | Mean (μm) (L × W)3 | L/W4 | Reference |
|---|---|---|---|---|
| Botryosphaeria auasmontanum | (8.1–)8.8–11.3(–13) × (2.5–)2.9–3.9(–5) | 10.1 × 3.4 | 3.0 | Slippers et al. (2014) |
| B. corticis | (20.5–)23.5–32.5(–34.5) × (5.0–)5.5–7(–7.5) | 28.9 × 6.4 | 4.5 | Phillips et al. (2006) |
| B. dothidea | (20–)23–27(–30) × 4–5(–6) | 26.2 × 5.4 | 4.9 | Slippers et al. (2004a) |
| B. fabicerciana | (16.5–)19.5–24.5(–26) × (4.5–)5–6.5(–7.5) | 22.0 × 5.8 | 3.8 | Chen et al. (2011c) |
| B. fusispora | (16.5–)19–23.5(–28.5) × 5–6(–8) | 21.2 × 5.6 | 3.8 | This study |
| B. fusispora | 16–22 × 4–5.5 | 20.0 × 5.0 | 4.0 | Liu et al. (2012) |
| B. kuwatsukai | (18.5–)20–24.5(–26) × 5–7(–8) | 22.3 × 6.2 | 3.6 | Xu et al. (2015a) |
| B. minutispermatia | 8–14 × 3–4 | 13.0 × 3.5 | 3.7 | Ariyawansa et al. (2016) |
| B. pseudoramosa5 | (8–)10–13(–16) × (4–)4.5–5(–6) | 11.5 × 4.6 | 2.5 | This study |
| B. qingyuanensis5 | (15–)19.5–24.5(–28.5) × (5–)6–6.5(–7.5) | 22.0 × 6.2 | 3.5 | This study |
| B. ramosa | (11–)12–15(–16) × (4.7–)5–6(–7) | 13.5 × 5.5 | 2.3 | Pavlic et al. (2008) |
| B. rosaceae | 20–31 × 6–8 | 26.2 × 6.7 | 3.9 | Zhou et al. (2017) |
| B. scharifii | (11.5–)13–17(–19) × 4–6.5 | 15.4 × 5.2 | 2.7 | Abdollahzadeh et al. (2013) |
| B. sinensia | (15–)19–29 × 5–7 | 24.3 × 5.9 | 4.1 | Zhou et al. (2016) |
| B. wangensis5 | (20.5–)22–26(–29) × (4.5–)5.5–6.5(–7.5) | 23.8 × 6.0 | 3.9 | This study |
| Lasiodiplodia brasiliense | 22.7–29.2 × 11.7–17 | 26.0 × 14.6 | 1.8 | Netto et al. (2014) |
| L. brasiliense | (22–)25–27(–28) × (12–)13.5–15(–15.5) | 26.0 × 14.4 | 1.8 | This study |
| L. pseudotheobromae | (22.5–)23.5–32(–33) × (13.5–)14–18(–20) | 28.0 × 16.0 | 1.7 | Alves et al. (2008) |
| L. pseudotheobromae | (22.5–)24.5–28.5(–31.5) × (12–)13–15(–16) | 26.5 × 13.8 | 1.9 | This study |
| L. theobromae | (19–)21–31(–32.5) × (12–)13–15.5(–18.5) | 26.2 × 14.2 | 1.9 | Alves et al. (2008) |
| L. theobromae | (21–)24–26.5(–29.5) × (11–)12.5–14(–16) | 25.3 × 13.1 | 1.9 | This study |
| Neofusicoccum algeriense | (14.5–)17–18(–21) × (4.5–)5.5–5.7(–6.5) | 17.6 × 5.6 | 3.1 | Berraf-Tebbal et al. (2014) |
| N. batangarum | (12–)14–17.5(–20) × (4–)4.5–6(–6.5) | 15.5 × 5.5 | 2.9 | Begoude et al. (2010) |
| N. cordaticola | 18–28 × 4.5–7 | 23.3 × 5.3 | 4.3 | Pavlic et al. (2009b) |
| N. hongkongense5 | (11.5–)13–15.5(–17.5) × (4–)4.5–5(–5.5) | 14.1 × 4.7 | 3.0 | This study |
| N. kwambonambiense | 16–28 × 5–8 | 22.3 × 6.3 | 3.6 | Pavlic et al. (2009b) |
| N. microconidium5 | (10–)11.5–13(–14.5) × (4–)4.5–5.5(–6) | 12.3 × 5.0 | 2.5 | This study |
| N. mangiferae | (11–)12–15(–17.5) × 5–6.6 | 13.6 × 5.4 | 2.0–2.5 | Slippers et al. (2005) |
| N. occulatum | 14–22 × 3.5–7.5 | 18.3 × 5.2 | 3.5 | Sakalidis et al. (2011) |
| N. parvum | (12–)13.5–21(–24) × 4–6(–10) | 17.1 × 5.5 | 3.2 | Phillips et al. (2013) |
| N. parvum | (15.5–)16.5–19(–21) × (4.5–)5–6(–6.5) | 17.9 × 5.5 | 3.3 | This study |
| N. ribis | (16–)19–23(–24) × 5–6(–7) | 20.8 × 5.5 | 3.8 | Slippers et al. (2004a) |
| N. sinense | (15.2–)17.6–20.4(–23) × (6.9–)7.4–8(–9) | 18.7 × 7.7 | 2.4 | Zhang et al. (2017) |
| N. sinoeucalypti5 | (13–)15–20.5(–25.5) × (4–)5–5.5(–6.5) | 17.7 × 5.2 | 3.4 | This study |
| N. umdonicola | 15–23.5 × 4.5–6.5 | 19.4 × 5.5 | 3.5 | Pavlic et al. (2009b) |
1 Isolates and measurements in bold were examined in this study.
2 Minimum–(average – standard deviation)–(average + standard deviation)–maximum or minimum–maximum, L × W = length × width.
3 L × W = average length × average width.
4 L/W = average length/average width.
5 Novel species described in this study.
TAXONOMY
Botryosphaeria pseudoramosa G.Q. Li & S.F. Chen, sp. nov. — MycoBank MB822323; Fig. 5
Fig. 5.
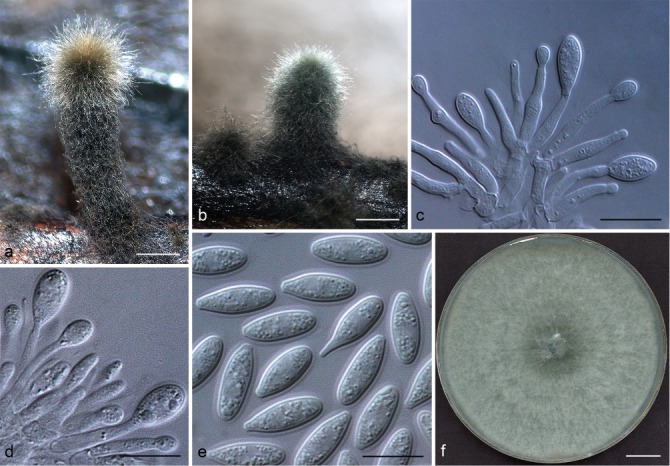
Botryosphaeria pseudoramosa. a–b. Conidiomata formed on pine needle culture; c–d. conidiogenous cells and developing conidia; e. conidia; f. living culture after 10 d on 2 % MEA (front). — Scale bars: a–b = 500 μm; c–e = 10 μm; f = 1 cm.
Etymology. Named for its phylogenetic resemblance to B. ramosa.
Sexual morph unknown. Conidiomata pycnidial, produced on pine needles on WA within 2–4 wk, globose to ovoid, dark brown to black, up to 698 μm wide, sometimes with a neck up to 1 660 μm long, arising from the substrate, covered by hyphal hairs, embedded in needle tissue, semi-immersed to superficial, unilocular, with a central ostiole. Conidiophores reduced to conidiogenous cells. Conidiogenous cells holoblastic, discrete, hyaline, cylindrical to lageniform, phialidic with periclinal thickening, (10–)11–16(–22.5) × (1–)2–3.5(–4) μm. Paraphyses not seen. Conidia hyaline, thin-walled, smooth with granular contents, unicellular, aseptate ellipsoid to fusoid, base subtruncate to bluntly rounded, (8–)10–13(–16) × (4–)4.5–5(–6) μm (av. = 11.5 × 4.6 μm, n = 100; L/W = 2.5) (Table 4).
Culture characteristics — Colonies on MEA have fluffy mycelia with an uneven margin and a few cottony aerial mycelia reaching to the lid of the Petri plate, with an appressed mycelial mat that is sparse to moderately dense. Colony mycelia initially white, becoming smoke grey (21’’’’f) to pale mouse grey (15’’’’’d) at the surface and olivaceous (21’’k) to iron grey (23’’’’’k) at the reverse within 10–14 d. Optimal growth temperature is 30 °C, covering the 90 mm plates after 5 d. No growth at 5 °C. After 5 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C, 35 °C and 40 °C reached 17 mm, 20 mm, 64 mm, 80 mm, 87 mm, 33 mm and 8 mm, respectively.
Specimens examined. China, GuangXi, from twigs of one Eucalyptus tree, fruiting structures induced on needles of Pinus sp. on water agar, 24 May 2014, S.F. Chen & G.Q. Li (holotype CSFF2025, culture ex-type CERC2001 = CGMCC3.18739); GuangDong, from twigs of one Eucalyptus tree, 24 May 2014, S.F. Chen & G.Q. Li (CSFF2026, culture CERC3455 = CGMCC3.18741); GuangDong, from twigs of one Melastoma sanguineum plant, 17 Mar. 2014, S.F. Chen (CSFF2027, culture CERC2983 = CGMCC3.18740).
Notes — Botryosphaeria pseudoramosa is phylogenetically closely related to B. ramosa and B. scharifii. Botryosphaeria pseudoramosa can be distinguished from B. ramosa and B. scharifii based on the morphology of their conidia. Conidia of B. pseudoramosa (av. 11.5 × 4.6; L/W = 2.5) are smaller than B. ramosa (av. 13.5 × 5.5; L/W = 2.3) (Pavlic et al. 2008) and B. scharifii (av. 15.4 × 5.2; L/W = 2.7) (Abdollahzadeh et al. 2013) (Table 4).
Botryosphaeria qingyuanensis G.Q. Li & S.F. Chen, sp. nov. — MycoBank MB822324; Fig. 6
Fig. 6.
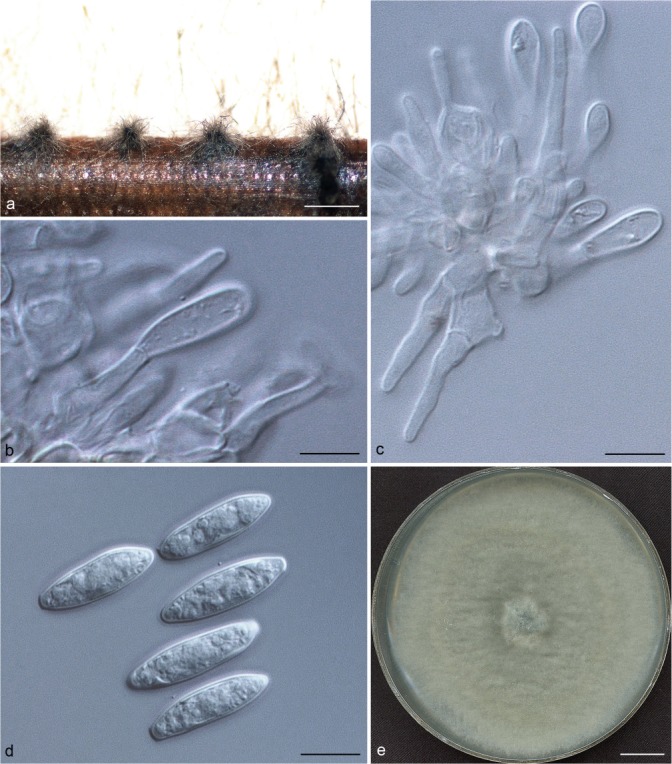
Botryosphaeria qingyuanensis. a. Conidiomata formed on pine needle culture; b–c. conidiogenous cells and developing conidia; d. conidia; e. living culture after 10 d on 2 % MEA (front). — Scale bars: a = 500 μm; b–d = 10 μm; e = 1 cm.
Etymology. Named for the QingYuan Region where the fungus was isolated for the first time.
Sexual morph unknown. Conidiomata pycnidial, produced on pine needles on WA within 2–4 wk, solitary, globose to ovoid, dark brown to black, up to 317 μm wide, 229 μm high, embedded in needle tissue, semi-immersed to superficial, unilocular, with a central ostiole. Conidiophores absent. Conidiogenous cells holoblastic, discrete, hyaline, cylindrical to lageniform, phialidic with periclinal thickening, (7–)7.5–12(–14.5) × (2–)2.5–3.5 μm. Paraphyses not seen. Conidia hyaline, thin-walled, smooth with granular contents, unicellular, aseptate narrowly fusiform, base subtruncate to bluntly rounded, (15–)19.5–24.5(–28.5) × (5–)6–6.5(–7.5) μm (av. = 22 × 6.2 μm, n = 100; L/W = 3.5) (Table 4).
Culture characteristics — Colonies on MEA have fluffy mycelia with an uneven margin and few cottony aerial mycelia reaching to the lid of the Petri plate, with an appressed mycelial mat that is sparse to moderately dense. Colony mycelia initially white, becoming smoke grey (21’’’’f) to pale mouse grey (15’’’’’d) at the surface and smoke grey (21’’’’f) to iron grey (23’’’’’k) at the reverse within 10–14 d. Optimal growth temperature is (25–)30 °C, reaching the edge of the 90 mm plates after 5 d. No growth is observed at 5 °C and 40 °C. After 5 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C and 35 °C reach 14 mm, 22 mm, 52 mm, 73 mm, 74 mm and 12 mm, respectively.
Specimens examined. China, GuangDong, from twigs of one Eucalyptus tree, fruiting structures induced on needles of Pinus sp. on water agar, 4 Dec. 2013, S.F. Chen & G.Q. Li (holotype CSFF2028, culture ex-type CERC2946 = CGMCC3.18742); GuangDong, from twigs of one Eucalyptus hybrid tree, fruiting structures induced on needles of Pinus sp. on water agar, 4 Dec. 2013, S.F. Chen & G.Q. Li (CSFF2029, culture CERC2947 = CGMCC3.18743).
Notes — Botryosphaeria qingyuanensis is phylogenetically closely related to B. corticis, B. fabicerciana, B. fusispora, B. kuwatsukai and B. rosaceae, but can be distinguished from these species based on morphological or growth characteristics. Conidia of B. qingyuanensis (av. 22 × 6.2; L/W = 3.5) are wider than these of B. fabicerciana (av. 22 × 5.8; L/W = 3.8) and the optimal growth temperature of B. qingyuanensis ((25–)30 °C) is different from that of B. fabicerciana (25(–30) °C) (Chen et al. 2011c). Conidia of B. qingyuanensis are longer and wider than B. fusispora (av. 20 × 5; L/W = 4) (Liu et al. 2012). Conidia of B. qingyuanensis are smaller than B. corticis (av. 28.9 × 6.4; L/W = 4.5) (Phillips et al. 2006) and B. rosaceae (av. 26.2 × 6.7; L/W = 3.9) (Zhou et al. 2017). Conidia of B. qingyuanensis are slightly shorter than B. kuwatsukai (av. 22.3 × 6.2; L/W = 3.6) (Xu et al. 2015a) and no conidia or microconidia are observed for B. qingyuanensis, but conidia with 1–3 septa before germination and microconidia (3–8 × 1–2 μm) have been found for B. kuwatsukai (Xu et al. 2015a) (Table 4).
Botryosphaeria wangensis G.Q. Li & S.F. Chen, sp. nov. — MycoBank MB822325; Fig. 7
Fig. 7.
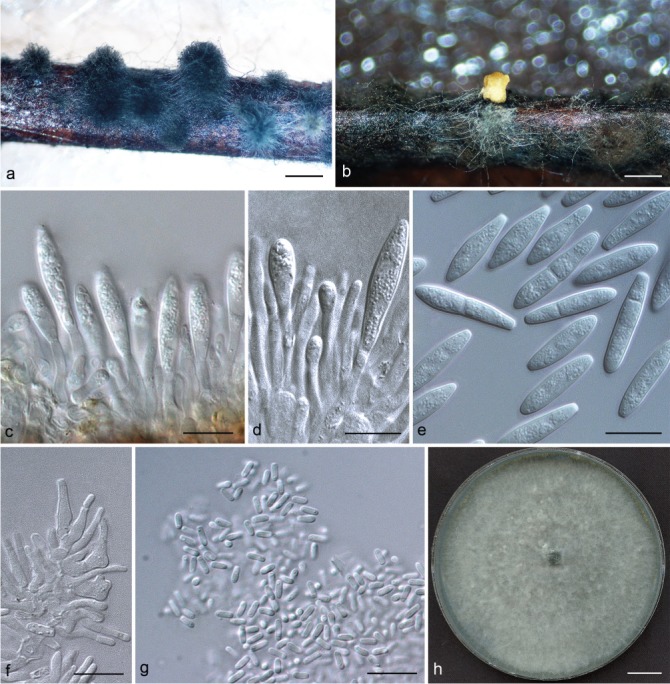
Botryosphaeria wangensis. a–b. Conidiomata on pine needle culture; c–d. conidiogenous cells and developing conidia; e. conidia with 1 septum; f. spermatogenous cells; g. spermatia; h. living culture after 10 d on 2 % MEA (front). — Scale bars: a–b = 500 μm; c–g = 10 μm; h = 1 cm.
Etymology. Named after the Wang village where the fungus was isolated for the first time.
Sexual morph unknown. Conidiomata pycnidial, produced on pine needles on WA within 2–4 wk, solitary, globose to ovoid, dark brown to black, up to 698 μm wide, 484 μm high, embedded in needle tissue, semi-immersed to superficial, unilocular, with a central ostiole, exuding conidia in a yellow mucoid mass. Conidiophores absent. Conidiogenous cells holoblastic, discrete, hyaline, cylindrical to lageniform, phialidic with periclinal thickening, (6–)8.5–13.5(–15) × 2–3(–3.5) μm. Paraphyses not seen. Conidia hyaline, thin-walled, smooth with granular contents, unicellular, aseptate, becoming 1-septate before germination, narrowly fusiform, base subtruncate to bluntly rounded, (20.5–)22–26(–29) × (4.5–)5.5–6.5(–7.5) μm (av. = 23.8 × 6 μm, n = 100; L/W = 3.9) (Table 4). Spermatophores hyaline, smooth, branched, cylindrical to subcylindrical (Fig. 7f). Spermatogenous cells discrete or integrated, hyaline, smooth, cylindrical, producing spermatia on their tips, holoblastic or proliferating via phialides with periclinal thickenings, 6.5–16 × 1.5–2.5 μm. Spermatia unicellular, aseptate, hyaline, thin-walled, allantoid to rod-shaped, 3.5–4.5 × 1–1.5 μm, L/W = 2.9.
Culture characteristics — Colonies on MEA have fluffy mycelia with an uneven margin and a few cottony aerial mycelia reaching to the lid of the Petri plate, with an appressed mycelial mat that is sparse to moderately dense. Colony mycelia initially white, becoming smoke grey (21’’’’f) to mouse grey (13’’’’’i) at the surface and olivaceous grey (21’’’’’i) to iron grey (23’’’’’k) at the reverse within 10–14 d. Optimal growth temperature is 30 °C, covering the 90 mm plates after 5 d. No growth at 5 °C and 40 °C. After 5 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C and 35 °C reach 15 mm, 21 mm, 50 mm, 69 mm, 89 mm and 24 mm, respectively.
Specimens examined. China, HeNan, from twigs of one Cedrus deodara tree, fruiting structures induced on needles of Pinus sp. on water agar, 26 Nov. 2013, S.F. Chen (holotype CSFF2030, culture ex-type CERC2298 = CGMCC3.18744); HeNan, from twigs of one Cedrus deodara tree, 26 Nov. 2013, S.F. Chen (CSFF2031, culture CERC2300 = CGMCC3.18746).
Notes — Botryosphaeria wangensis is phylogenetically closely related to B. auasmontanum, B. dothidea, B. minutispermatia and B. sinensia. Botryosphaeria wangensis can be distinguished from its phylogenetically closely related species by the size of their conidia. Conidia of B. wangensis (av. 23.8 × 6; L/W = 3.9) are longer and wider than those of B. auasmontanum (av. 10.1 × 3.4; L/W = 3) (Slippers et al. 2014) and B. minutispermatia (av. 13 × 3.5; L/W = 3.7) (Ariyawansa et al. 2016) and shorter and wider than those of B. dothidea (av. 26.2 × 5.4; L/W = 4.9) (Slippers et al. 2004a) and B. sinensia (av. 24.3 × 5.9; L/W = 4.1) (Zhou et al. 2016) (Table 4).
Neofusicoccum hongkongense G.Q. Li & S.F. Chen, sp. nov. — MycoBank MB822328; Fig. 8
Fig. 8.
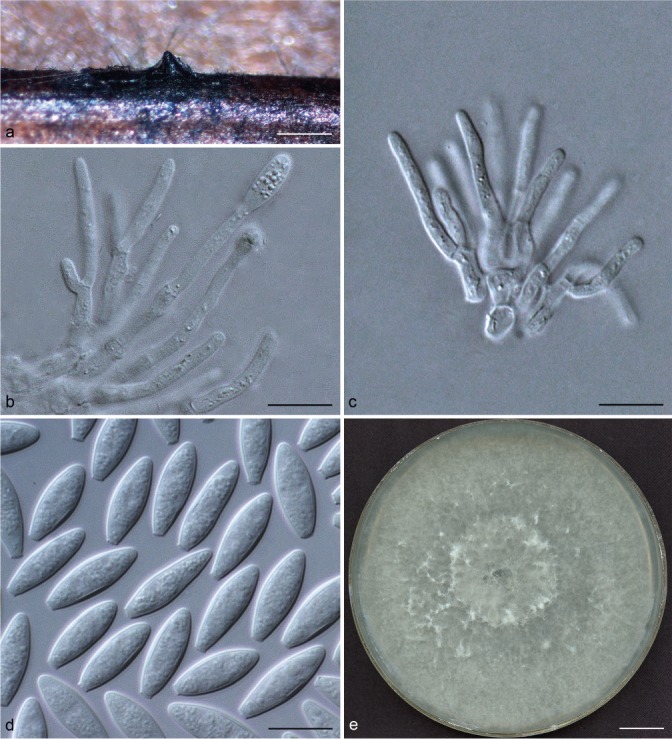
Neofusicoccum hongkongense. a. Conidiomata formed on pine needle culture; b. conidiogenous cells and developing conidia; c. conidiogenous cells; d. conidia; e. living culture after 10 d on 2 % MEA (front). — Scale bars: a = 500 μm; b–d = 10 μm; e = 1 cm.
Etymology. Named after the Hong Kong Region where it was isolated for the first time.
Sexual morph unknown. Conidiomata pycnidial, produced on pine needles on WA within 2–4 wk, solitary, globose to ovoid, dark brown to black, up to 694 μm wide, up to 776 μm high, embedded in needle tissue, semi-immersed to superficial, unilocular, with a central ostiole. Conidiophores reduced to conidiogenous cells. Conidiogenous cells holoblastic, discrete, hyaline, cylindrical, phialidic with periclinal thickening, (9.5–)12–18.5(–22) × (1.5–)2–2.5(–3) μm. Paraphyses not seen. Conidia hyaline, thin-walled, smooth with granular contents, unicellular, aseptate narrowly fusiform, base subtruncate to bluntly rounded, (11.5–)13–15.5(–17.5) × (4–)4.5–5(–5.5) μm (av. = 14.1 × 4.7 μm, n = 100; L/W = 3) (Table 4).
Culture characteristics — Colonies on MEA have fluffy mycelia with an uneven margin and a few cottony aerial mycelia reaching to the lid of the Petri plate, with an appressed mycelial mat that is sparse to moderately dense. Colony mycelia initially white, becoming smoke grey (21’’’’f) to grey olivaceous (21’’’’b) at the surface and grey olivaceous (21’’’’b) to olivaceous grey (21’’’’’i) at the reverse within 10–14 d. Optimal growth temperature is 25 °C, covering the 90 mm plates after 5 d. No growth at 5 °C or 40 °C. After 5 d, colonies grown at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C and 35 °C reach 25 mm, 41 mm, 66 mm, 90 mm, 84 mm and 9 mm, respectively.
Specimens examined. China, Hong Kong, from twigs of Araucaria cunninghamii, fruiting structures induced on needles of Pinus sp. on water agar, 11 Mar. 2014, S.F. Chen (holotype CSFF2034, culture ex-type CERC2973 = CGMCC3.18749); Hong Kong, from twigs of Araucaria cunninghamii, 11 Mar. 2014, S.F. Chen (CSFF2035, culture CERC2968 = CGMCC3.18748).
Notes — Based on phylogenetic analyses, N. hongkongense phylogenetically clustered in the N. parvum/N. ribis species complex. Neofusicoccum hongkongense can be distinguished from other species in the N. parvum/N. ribis complex by the size and shape of their conidia. The conidia of N. hongkongense (av. 14.1 × 4.7; L/W = 3) are shorter and narrower than those of N. algeriense (av. 17.6 × 5.6; L/W = 3.1) (Berraf-Tebbal et al. 2014), N. batangarum (av. 15.5 × 5.5; L/W = 2.9) (Begoude et al. 2010), N. cordaticola (av. 23.3 × 5.3; L/W = 4.3) (Pavlic et al. 2009b), N. kwambonambiense (av. 22.3 × 6.3; L/W = 3.6) (Pavlic et al. 2009b), N. occulatum (av. 18.3 × 5.2; L/W = 3.5) (Sakalidis et al. 2011), N. parvum (av. 17.1 × 5.5; L/W = 3.2) (Phillips et al. 2013), N. ribis (av. 20.8 × 5.5; L/W = 3.8) (Slippers et al. 2004a), N. sinense (av. 18.7 × 7.7; L/W = 2.4) (Zhang et al. 2017), N. sinoeucalypti (av. 17.7 × 5.2; L/W = 3.4) (this study) and N. umdonicola (av. 19.4 × 5.5; L/W = 3.5) (Pavlic et al. 2009b). The conidial size of N. brasiliense remains unknown (Marques et al. 2013) (Table 4).
Neofusicoccum microconidium G.Q. Li & S.F. Chen, sp. nov. — MycoBank MB822326; Fig. 9
Fig. 9.
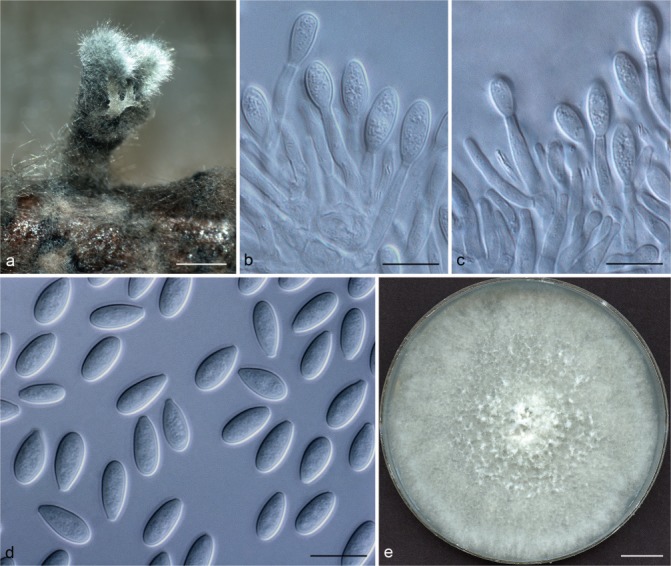
Neofusicoccum microconidium. a. Conidiomata formed on pine needle culture; b–c. conidiogenous cells and developing conidia; d. conidia; e. living culture after 10 d on 2 % MEA (front). — Scale bars: a = 500 μm; b–d = 10 μm; e = 1 cm.
Etymology. Named for the small conidia of this fungus.
Sexual morph unknown. Conidiomata pycnidial, produced on pine needles on WA within 2–4 wk, solitary, globose to ovoid, dark brown to black, up to 895 μm wide, 1 729 μm high, embedded in needle tissue, semi-immersed to superficial, unilocular, with a central ostiole, exuding conidia in a white mucoid mass. Conidiophores reduced to conidiogenous cells. Conidiogenous cells holoblastic, discrete, hyaline, cylindrical, phialidic with periclinal thickening, (10.5–)12.5–18(–20.5) × (2–)2.5–3(–3.5) μm. Paraphyses not seen. Conidia hyaline, thin-walled, smooth with granular contents, unicellular, aseptate narrowly fusiform, base subtruncate to bluntly rounded, (10–)11.5–13(–14.5) × (4–)4.5–5.5(–6) μm (av. = 12.3 × 5 μm, n = 100; L/W = 2.5) (Table 4).
Culture characteristics — Colonies on MEA have fluffy mycelia with an uneven margin and a few cottony aerial mycelia reaching to the lid of the Petri plate, with an appressed mycelial mat that is sparse to moderately dense. Colony mycelia initially white, becoming pale mouse grey (15’’’’’d) to olivaceous grey (21’’’’’i) at the surface and olivaceous grey (21’’’’’i) to iron grey (23’’’’’k) at the reverse within 10–14 d. Optimal growth temperature is 30 °C, reaching the edge of the 90 mm plates after 5 d. No growth at 5 °C. After 5 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C, 35 °C and 40 °C reach 24 mm, 34 mm, 66 mm, 74 mm, 86 mm, 36 mm and 8 mm, respectively.
Specimens examined. China, GuangDong, from twigs of E. urophylla × E. grandis, fruiting structures induced on needles of Pinus sp. on water agar, 22 July 2014, S.F. Chen & G.Q. Li (holotype CSFF2032, culture ex-type CERC3497 = CGMCC3.18750); GuangDong, from twigs of E. urophylla × E. grandis, 22 July 2014, S.F. Chen & G.Q. Li (CSFF2033, culture CERC3498 = CGMCC3.18751).
Notes — Neofusicoccum microconidium is phylogenetically closely related to N. mangiferae. The two species can be distinguished from each other based on conidial morphology. Conidia of N. microconidium (av. 12.3 × 5; L/W = 2.5) are smaller than those of N. mangiferae (av. 13.6 × 5.4; L/W = 2–2.5) (Slippers et al. 2005) (Table 4).
Neofusicoccum sinoeucalypti G.Q. Li & S.F. Chen, sp. nov. — MycoBank MB822327; Fig. 10
Fig. 10.
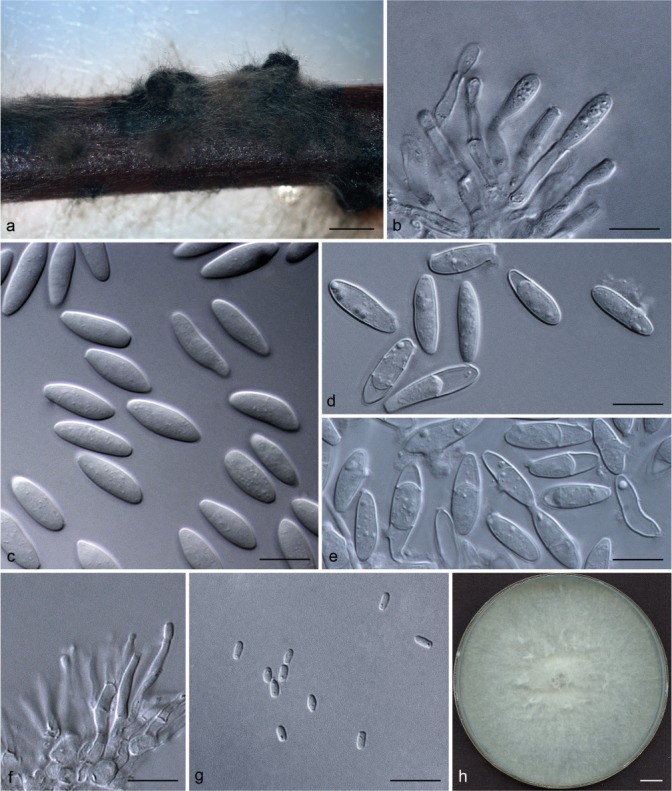
Neofusicoccum sinoeucalypti. a. Conidiomata formed on pine needle culture; b. conidiogenous cells and developing conidia; c. immature conidia; d–e. mature conidia with 1–2 septa; f. spermatogenous cells; g. spermatia; h. living culture after 10 d on 2 % MEA (front). — Scale bars: a = 500 μm; b–g = 10 μm; h = 1 cm.
Etymology. Named after the host genus Eucalyptus from which it was isolated for the first time.
Sexual morph solitary, globose to ovoid, dark brown to black, up to 1 007 μm wide, 685 μm high, embedded in needle tissue, semi-immersed to superficial, unilocular, with a central ostiole. Conidiophores absent. Conidiogenous cells holoblastic, discrete, hyaline, cylindrical to lageniform, phialidic with periclinal thickening, (10–)10.5–11 × 2–3 μm. Paraphyses not seen. Conidia hyaline, thin-walled, smooth with granular contents, unicellular, aseptate, narrowly fusiform, base subtruncate to bluntly rounded, (13–)15–20.5(–25.5) × (4–)5–5.5(–6.5) μm (av. = 17.7 × 5.2 μm, n = 100; L/W = 3.4). Spermatophores hyaline, smooth, cylindrical to subcylindrical. Spermatogenous cells discrete or integrated, hyaline, smooth, cylindrical, producing spermatia on their tips, holoblastic or proliferating via phialides with periclinal thickenings, 8.5–15.5 × 1.5–2 μm. Spermatia unicellular, aseptate, hyaline, thin-walled, allantoid to rod-shaped, 2.5–4.5 × 1.5 μm, L/W = 2.1.
Culture characteristics — Colonies on MEA have fluffy mycelia with an uneven margin and a few cottony aerial mycelia that reach to the lid of the Petri plate, with an appressed mycelial mat that is sparse to moderately dense. Colony mycelia are initially white, becoming pale mouse grey (15’’’’’d) to mouse grey (13’’’’’i) at the surface and olivaceous buff (21’’’d) to iron grey (23’’’’’k) at the reverse within 10–14 d. Optimal growth temperature is 30 °C, reaching the edge of the 90 mm plates after 5 d. No growth at 5 °C or 40 °C. After 5 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C, 30 °C and 35 °C reach 25 mm, 31 mm, 53 mm, 78 mm, 90 mm and 11 mm, respectively.
Specimens examined. China, GuangDong, from twigs of E. urophylla × E. grandis, fruiting structures induced on needles of Pinus sp. on water agar, 30 July 2013, S.F. Chen & G.Q. Li (holotype CSFF2036, culture ex-type CERC2005 = CGMCC3.18752); GuangXi, from twigs of E. urophylla × E. grandis, 25 Oct. 2013, S.F. Chen & G.Q. Li (CSFF2037, culture CERC2265 = CGMCC3.18753); GuangXi, from twigs of Eucalyptus hybrid, 22 May 2014, S.F. Chen & G.Q. Li (CSFF2038, culture CERC3416 = CGMCC3.18754).
Notes — Neofusicoccum sinoeucalypti clustered in the N. parvum/N. ribis species complex. Other species in this complex include N. algeriense, N. batangarum, N. brasiliense, N. cordaticola, N. hongkongense (this study), N. kwambonambiense, N. occulatum, N. parvum, N. ribis and N. umdonicola. For these species, except N. brasiliense (morphological data not available) (Marques et al. 2013), spermatia have been reported only in N. sinoeucalypti and are allantoid to rod-shaped. Conidia of N. sinoeucalypti (av. 17.7 × 5.2; L/W = 3.4) are longer and wider than those of N. hongkongense (av. 14.1 × 4.7; L/W = 3), longer and narrower than those of N. batangarum (av. 15.5 × 5.5; L/W = 2.9) (Begoude et al. 2010) and N. parvum (av. 17.1 × 5.5; L/W = 3.2) (Phillips et al. 2013), shorter and narrower than those of N. cordaticola (av. 23.3 × 5.3; L/W = 4.3) (Pavlic et al. 2009b), N. kwambonambiense (av. 22.3 × 6.3; L/W = 3.6) (Pavlic et al. 2009b), N. ribis (av. 20.8 × 5.5; L/W = 3.8) (Slippers et al. 2004a), N. sinense (av. 18.7 × 7.7; L/W = 2.4) (Zhang et al. 2017) and N. umdonicola (av. 19.4 × 5.5; L/W = 3.5) (Pavlic et al. 2009b), shorter than those of N. occulatum (av. 18.3 × 5.2; L/W = 3.5) (Sakalidis et al. 2011), and narrower than those of N. algeriense (av. 17.6 × 5.6; L/W = 3.1) (Berraf-Tebbal et al. 2014). The optimal growth temperature of N. sinoeucalypti (30 °C) is different compared to N. algeriense (25 °C) (Berraf-Tebbal et al. 2014), N. batangarum (25 °C) (Begoude et al. 2010), N. brasiliense (27.7 °C) (Marques et al. 2013), N. hongkongense (25 °C) (this study), N. occulatum (25 °C) (Sakalidis et al. 2011) and N. ribis (25 °C) (Slippers et al. 2004a) (Table 4).
Distribution of Botryosphaeriaceae
According to the phylogenetic and morphological analyses of the 105 isolates collected in this study, twelve species of Botryosphaeriaceae were identified from seven hosts in the FuJian, GuangDong, GuangXi, HaiNan and HeNan Provinces and the Hong Kong Region of China (Fig. 11). These species include B. fusispora (21 isolates: all from Eucalyptus hybrids), B. pseudoramosa (12 isolates: 8 from Eucalyptus hybrids, 4 from M. sanguineum), B. qingyuanensis (2 isolates: both from one Eucalyptus hybrid), B. wangensis (3 isolates: all from C. deodara), C. atrovirens (5 isolates: all from D. longan), L. brasiliense (1 isolate: from a Eucalyptus hybrid), L. pseudotheobromae (19 isolates: 17 from unknown Eucalyptus hybrids, two from E. urophylla × E. grandis), L. theobromae (20 isolates: six from unknown Eucalyptus hybrids, five from E. urophylla × E. grandis, 2 from C. lanceolata, 5 from D. longan, 2 from P. hanceana), N. hongkongense (3 isolates: all from A. cunninghamii), N. microconidium (2 isolates: both from E. urophylla × E. grandis), N. parvum (6 isolates: all from E. urophylla × E. grandis) and N. sinoeucalypti (11 isolates: nine from E. urophylla × E. grandis, two from Eucalyptus hybrids) (Table 1, Fig. 11). The 81 isolates collected from Eucalyptus trees include nine species (except for B. wangensis, C. atrovirens and N. hongkongense) of Botryosphaeriaceae. Of these nine species from Eucalyptus, B. fusispora (26 % of the isolates), L. pseudotheobromae (23 % of the isolates) and L. theobromae (14 % of the isolates) are dominant and are distributed throughout the surveyed Provinces of South China. Of the 12 species of Botryosphaeriaceae, L. theobromae (isolated from C. lanceolata, D. longan, a Eucalyptus hybrid and P. hanceana) and B. pseudoramosa (isolated from a Eucalyptus hybrid and M. sanguineum) were collected from more than one plant host (Fig. 11).
Fig. 11.
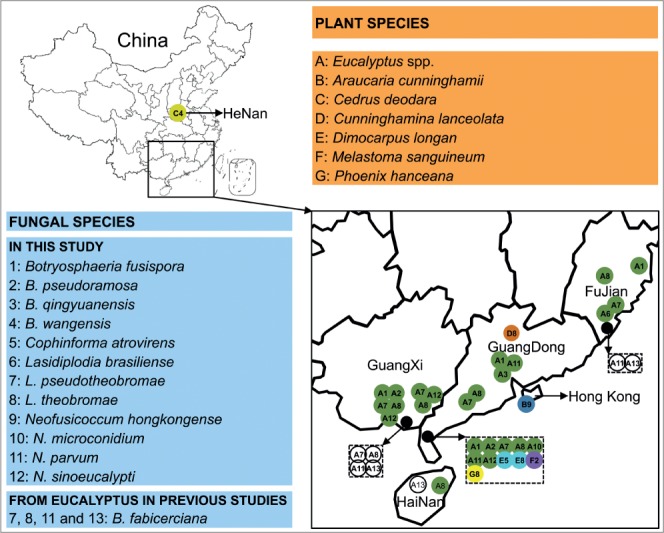
Map showing the 12 species of Botryosphaeriaceae detected from different regions and plant hosts. The different Botryosphaeriaceae species are indicated as numbers 1 to 12; the plant hosts are shown as letters A to G. For example, A8 indicates L. theobromae (number 8 of fungal species) isolated from Eucalyptus spp. (letter A of plant species) in HaiNan Province. The pies in colours indicate Botryosphaeriaceae isolated from different plant hosts in this study, the pies without colour indicate Botryosphaeriaceae species reported from Eucalyptus in previous studies (Chen et al. 2011c, Li et al. 2015a).
Pathogenicity tests
Twenty-eight isolates representing the 12 species of Botryosphaeriaceae identified in this study were used for inoculations on three different Eucalyptus clones (different parents) (Table 1, 5). Pathogenicity tests indicate that all of the Botryosphaeriaceae isolates tested produce lesions on stems of the three Eucalyptus clones, while MEA unclonised plugs produced only wounds. Overall, isolates in species of Lasiodiplodia produce relatively longer lesions than that of Botryosphaeria, Cophinforma and Neofusicoccum. For all three tested Eucalyptus clones, the lesions produced by Lasiodiplodia isolates are all significantly longer than the wounds caused by negative controls, except isolate CERC3420 (L. theobromae) on CEPT-11 and CEPT-13 (P < 0.05) (Table 5). For isolates in the genera of Botryosphaeria, Cophinforma and Neofusicoccum, isolates CERC3497 (N. microconidium) and CERC2005 (N. sinoeucalypti) also produce significantly longer lesions on CEPT-11 and CEPT-13 (P < 0.05) (Table 5). Analysis of variance shows significant differences in the susceptibility of the three Eucalyptus clones to some of the isolates we tested. For example, the lesions produced by isolate CERC2284 (L. brasiliense) on three Eucalyptus clones are significantly different from each other (P < 0.05) (Table 5). Analysis of results also show that not all the isolates of the same species of Botryosphaeriaceae react in the same manner to the Eucalyptus clones. For example, lesions produced by isolate CERC3420 (L. theobromae) on clone CEPT-12 are significantly longer than those on CEPT-13, whereas lesions produced by isolate CERC3513 (L. theobromae) on CEPT-12 are significantly shorter than those on CEPT-13 (P < 0.05) (Table 5). In addition, based on the lesions caused by all Botryosphaeriaceae isolates in this study, CEPT-11 (average lesion length: 33.0 ± 2.4 mm) is more tolerant than CEPT-12 (average lesion length: 44.2 ± 3.2 mm) and CEPT-13 (average lesion length: 42.0 ± 3.4 mm). All 12 species of Botryosphaeriaceae were re-isolated successfully from the lesions, and no Botryosphaeriaceae were isolated from the negative controls, thus fulfilling Koch’s postulates.
Table 5.
Average lesion length (mm) on seedlings of three Eucalyptus clones inoculated with Botryosphaeriaceae.
| Species | Isolates |
Eucalyptus clones |
||
|---|---|---|---|---|
| CEPT-11 | CEPT-12 | CEPT-13 | ||
| Botryosphaeria fusispora | CERC1998 | 17.6 ± 1.8 m-p1 | 27.1 ± 9.3 k-p | 10.6 ± 0.7 op |
| CERC2274 | 10.3 ± 0.4 op | 10.7 ± 0.4 op | 9.2 ± 0.2 op | |
| CERC2930 | 12.3 ± 1.2 op | 13.9 ± 2.4 m-p | 14.5 ± 1.6 m-p | |
| CERC3446 | 12.0 ± 0.6 op | 17.5 ± 4.5 m-p | 15.5 ± 2.8 m-p | |
| B. pseudoramosa | CERC2001 | 13.5 ± 0.8 n-p | 15.5 ± 4.0 m-p | 11.1 ± 0.5 op |
| CERC3452 | 16.8 ± 2.0 m-p | 26.9 ± 7.3 k-p | 13.9 ± 1.9 m-p | |
| B. qingyuanensis | CERC2946 | 10.4 ± 0.5 op | 16.2 ± 5.6 m-p | 11.1 ± 0.8 op |
| CERC2947 | 11.0 ± 0.5 op | 18.2 ± 5.2 l-p | 10.4 ± 0.5 op | |
| B. wangensis | CERC2298 | 10.6 ± 0.5 op | 12.3 ± 0.8 op | 10.0 ± 0.2 op |
| CERC2299 | 9.3 ± 1.1 op | 11.1 ± 0.3 op | 9.8 ± 0.1 op | |
| Cophinforma atrovirens | CERC3484 | 11.0 ± 1.5 op | 10.0 ± 1.5 op | 8.8 ± 1.1 op |
| CERC3489 | 12.2 ± 0.7 op | 9.2 ± 0.2 op | 9.4 ± 0.3 op | |
| Lasiodiplodia brasiliense | CERC2284 | 41.7 ± 5.6 j-m | 139.7 ± 27.3 cd | 95.8 ± 15.7 f |
| L. pseudotheobromae | CERC2286 | 90.7 ± 16.9 fg | 84.5 ± 12.6 f-h | 100.1 ± 21.8 ef |
| CERC3417 | 78.4 ± 9.7 fg | 121.0 ± 12.6 de | 128.4 ± 12.2 d | |
| CERC3495 | 120.9 ± 11.5 de | 138.1 ± 12.8 cd | 85.9 ± 9.5 fg | |
| L. theobromae | CERC3420 | 26.7 ± 2.3 k-p | 46.6 ± 7.5 i-l | 26.8 ± 5.6 k-p |
| CERC3513 | 123.9 ± 16.0 d | 150.7 ± 21.9 b | 219.5 ± 19.8 a | |
| CERC3516 | 126.3 ± 13.1 d | 142.0 ± 21.6 bc | 173.5 ± 18.7 b | |
| Neofusicoccum hongkongense | CERC2968 | 17.7 ± 1.2 m-p | 12.9 ± 2.2 op | 14.6 ± 1.2 m-p |
| CERC2973 | 21.6 ± 1.2 k-p | 31.8 ± 5.6 k-p | 18.7 ± 1.3 m-p | |
| N. microconidium | CERC3497 | 32.7 ± 2.2 k-p | 47.7 ± 7.5 i-k | 40.8 ± 6.8 j-n |
| CERC3498 | 16.6 ± 1.2 m-p | 17.2 ± 1.7 m-p | 20.8 ± 4.3 l-p | |
| N. parvum | CERC2951 | 10.3 ± 0.5 op | 11.9 ± 0.9 op | 10.3 ± 0.3 op |
| CERC3504 | 17.3 ± 0.7 m-p | 30.1 ± 5.2 k-p | 22.1 ± 3.2 k-p | |
| CERC3509 | 16.0 ± 1.5 m-p | 17.5 ± 4.0 k-p | 15.3 ± 2.4 m-p | |
| N. sinoeucalypti | CERC2005 | 27.5 ± 4.7 k-p | 68.0 ± 9.0 g-i | 62.3 ± 8.7 h-j |
| CERC3463 | 30.9 ± 4.8 k-p | 24.0 ± 4.3 k-p | 39.3 ± 7.7 j-o | |
| Control | 10.5 ± 0.6 op | 10.0 ± 0.2 op | 9.4 ± 0.3 op | |
1 Mean ± SE followed by different lowercase letters indicates treatments that are significantly different (P < 0.05); Mean = average lesion length; SE = standard error of mean.
DISCUSSION
In this study, disease samples from symptomatic trees with stem cankers, shoot and twig blight were collected mainly from Eucalyptus and six other plant hosts in China. Botryosphaeriaceae was isolated from these diseased samples. Based on phylogenetic analyses and morphological characteristics, 12 species of Botryosphaeriaceae were isolated from these samples and the genera Botryosphaeria, Cophinforma, Lasiodiplodia and Neofusicoccum were identified from among a relatively large collection of isolates. These species include Botryosphaeria fusispora, Cophinforma atrovirens, Lasiodiplodia brasilience, L. pseudotheobromae, L. theobromae, Neofusicoccum parvum and each of three previously undescribed species of Botryosphaeria and Neofusicoccum, namely B. pseudoramosa sp. nov., B. qingyuanensis sp. nov., B. wangensis sp. nov., N. hongkongense sp. nov., N. microconidium sp. nov. and N. sinoeucalypti sp. nov.
In this study, ITS, tef1, tub, rpb2, cmdA, LSU and SSU sequences were generated to distinguish and describe new species of Botryosphaeria, Cophinforma, Lasiodiplodia and Neofusicoccum. For the six to seven regions used for analyses of Botryosphaeria, Lasiodiplodia and Neofusicoccum, phylogenetic analyses based on sequence comparisons show that polymorphic nucleotides exist between some isolates collected in this study and other closely related species. Sequences of the ITS, tef1 and tub regions are widely used to distinguish and describe new species of Botryosphaeria, Lasiodiplodia and Neofusicoccum of Botryosphaeriaceae (Phillips et al. 2013, Chen et al. 2015, Linaldeddu et al. 2015, Coutinho et al. 2017), except ITS, tef1 and tub, rpb2 genes are also used for the species identification of Neofusicoccum (Pavlic et al. 2009a, Sakalidis et al. 2011, Osorio et al. 2017, Yang et al. 2017) and rpb2 and cmdA are also used for Lasiodiplodia (Cruywagen et al. 2017, Dou et al. 2017a, b, Osorio et al. 2017). The phylogenetic analyses based on a combination of the three to five regions (Botryosphaeria: ITS, tef1 and tub; Lasiodiplodia: ITS, tef1, tub, rpb2 and cmdA; Neofusicoccum: ITS, tef1, tub and rpb2) indicated that these isolates form independent phylogenetic clades supported by high bootstrap values, which are identified and described as six new species. In the other Chinese isolates, the differences we did find occurred only in one of the two (Cophinforma), six (Botryosphaeria and Neofusicoccum) or seven (Lasiodiplodia) regions, these isolates reside in the same clade to previously identified species or form independent phylogenetic clades but not supported by high bootstrap values, and they were identified as B. fusispora, C. atrovirens, L. brasiliense, L. pseudotheobromae, L. theobroma and N. parvum.
The identification of 12 Botryosphaeriaceae species is also supported by morphological and/or biological characteristics. For each of the six species that have been described previously, their culture morphology and conidial characteristics are very similar to that of the type specimens. For the six newly described species in this study, morphological differences exist among them and other phylogenetically closely related species, especially in terms of the size and shape of conidia, as well as conidium septum characteristics. We also observed biological differences, for example optimal growth temperatures, among some of the species. For the six new species, B. pseudoramosa, B. wangensis, N. hongkongense and N. microconidium are easily distinguished from other phylogenetically close species based on conidial morphology. Although some overlap in conidial shape and size is observed among some species, such as B. fabicerciana, B. kuwatsukai and B. qingyuanensis, these species can be distinguished from each other by the presence of a conidial septum (older conidia) and microconidia, as well as the optimal growth temperature. The newly described species N. sinoeucalypti can be distinguished from other species with similar conidia in the N. parvum/N. ribis complex by conidial morphology and the optimal growth temperature.
Except for B. wangensis, C. atrovirens and N. hongkongense, the other nine species were isolated from Eucalyptus trees in South China. Of the Botryosphaeriaceae species isolated from Eucalyptus, B. fusispora, L. pseudotheobromae and L. theobromae are dominant and distributed in the GuangDong, GuangXi and HaiNan Provinces; L. pseudotheobromae and L. theobromae have also been found in previous studies (Chen et al. 2011c, Li et al. 2015a), suggesting that they may be widely distributed on Eucalyptus trees in other areas in South China. Four new species, B. pseudoramosa, B. qingyuanensis, N. microconidium and N. sinoeucalypti, were isolated from Eucalyptus in China. This study also presents the first report of L. brasiliense on Eucalyptus in the world. Species of Botryosphaeriaceae are distributed in all the areas surveyed where Eucalyptus is planted. The results of our study suggest that the species diversity of Botryosphaeriaceae on Eucalyptus in China may be higher than what was previously expected (Chen et al. 2011c).
In addition to Botryosphaeriaceae species identified on Eucalyptus, we also identified B. pseudoramosa from Melastoma sanguineum, B. wangensis from C. deodara, C. atrovirens from D. longan, L. theobromae from C. lanceolata, D. longan and P. hanceana, and N. hongkongense from A. cunninghamii. Aside from L. theobromae from P. hanceana (Lu et al. 2000), which has been reported previously, these Botryosphaeriaceae species are reported from their respective plant hosts for the first time. Disease materials were collected randomly from limited areas, including the areas which were adjacent to Eucalyptus plantations, and further work is needed to better understand the biodiversity and distribution of Botryosphaeriaceae on their hosts.
Based on sequence comparisons of the seven gene regions, the same genotype of L. theobromae was shared by species of Eucalyptus in all the surveyed provinces in South China, and C. lanceolata, D. longan and P. hanceana planted in GuangDong Province (Table 1). We isolated the newly described species B. pseudoramosa from both Eucalyptus trees and M. sanguineum, and isolates from different hosts in geographically close areas do share the same genotype (Table 1). These results provide confirmation for the wide host range of L. theobromae and B. pseudoramosa on different plants. Previous studies used genetic diversity and geographic distribution comparisons to show the wide host range of N. mediterraneum on different crop trees in California (Chen et al. 2014a, b). The results of our current study further show that some Botryosphaeriaceae have wide geographic and host ranges.
Inoculation experiments revealed that all species of Botryosphaeriaceae identified in this study are pathogenic to the tested Eucalyptus clones, which is consistent with previous work showing that Botryosphaeriaceae species include important pathogens of Eucalyptus (Pavlic et al. 2007, Mohali et al. 2009, Rodas et al. 2009, Chen et al. 2011c). Pathogenicity tests in this study showed that species of Lasiodiplodia are more aggressive than Botryosphaeria and Neofusicoccum on three Eucalyptus clones, including one clone of E. urophylla × E. grandis, which is consistent with results in previous studies (Chen et al. 2011c). Results in Mohali et al. (2009) showed that some species of Neofusicoccum were more aggressive than Lasiodiplodia on clones of E. urophylla × E. grandis, which indicated that resistance of different genotypes of E. urophylla × E. grandis can be significantly different. Therefore, the identification of commercially available Eucalyptus genotypes resistant to Botryosphaeriaceae will promote the selection of resistant materials for wide-scale planting.
Of the fungal species we found, L. theobromae and L. pseudotheobromae are the most aggressive and are also widely distributed on Eucalyptus trees in different regions; it is essential that these pathogens be monitored carefully to help make decisions regarding disease management. Except for species of Lasiodiplodia, other fungi of the genera Botryosphaeria, Cophinforma and Neofusicoccum also produce lesions on inoculated seedlings; although these species are not highly virulent to Eucalyptus and are not widespread, these fungi still need to be monitored carefully because some of them may be highly aggressive to their original hosts or may spread and act as important pathogens in a suitable environment.
Our results in this study indicate that some species of Botryosphaeriaceae are widely distributed in different geographic regions on different hosts. These fungal species have significant potential to cause diseases of Eucalyptus. Management of the diseases on Eucalyptus reported in this study will need to rely on sound breeding programs to select Eucalyptus genotypes to match climatic and edaphic factors and silvicultural practices (spacing and thinning) as part of an integrated management strategy (Old et al. 2003). Further study is needed to better understand the genetic diversity of the species at the population level and to understand the biological and epidemiological characteristics of these species to help with long-term disease management.
Acknowledgments
This study was supported by the National Key R&D Program of China (project no. 2017YFD0600103), the National Natural Science Foundation of China (NSFC) (project nos 31622019 and 31400546). We thank Ms CaiYun Xu and Mr ShengLong Zhang for their assistance in collecting disease samples in GuangDong and GuangXi Provinces. We thank WILEY for its linguistic assistance during the preparation of this manuscript.
REFERENCES
- Abdollahzadeh J, Javadi J, Mohammadi Goltapeh E, et al. 2010. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 25: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahzadeh J, Zare R, Phillips AJL. 2013. Phylogeny and taxonomy of Botryosphaeria and Neofusicoccum species in Iran, with description of Botryosphaeria scharifii sp. nov. Mycologia 105: 210–220. [DOI] [PubMed] [Google Scholar]
- Alves A, Correis A, Luque J, et al. 2004. Botryosphaeria corticola sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia 96: 598–613. [PubMed] [Google Scholar]
- Alves A, Crous PW, Correis A, et al. 2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13. [Google Scholar]
- Alves A, Phillips AJL, Henriques I, et al. 2005. Evaluation of amplified ribosomal DNA restriction analysis as a method for the identification of Botryosphaeria species. FEMS Microbiology Letters 245: 221–229. [DOI] [PubMed] [Google Scholar]
- Ariyawansa HA, Hyde KD, Liu JK, et al. 2016. Additions to Karst Fungi 1: Botryosphaeria minutispermatia sp. nov., from Guizhou Province, China. Phytotaxa 275: 35–44. [Google Scholar]
- Begoude BAD. 2010. Characterization of Botryosphaeriaceae and Cryphonectriaceae associated with terminalia spp. in Africa. PhD thesis, Department of Microbiology and Plant Pathology, Forestry and Agricultural Biotechnology Institute, University of Pretoria, South Africa. [Google Scholar]
- Begoude BAD, Slippers B, Wingfield MJ, et al. 2010. Botryosphaeriaceae associated with Terminalia catappa in Cameroon, South Africa and Madagascar. Mycological Progress 9: 101–123. [Google Scholar]
- Berraf-Tebbal A, Guereiro MA, Phillips AJL. 2014. Phylogeny of Neofusicoccum species associated with grapevine trunk diseases in Algeria, with description of Neofusicoccum algeriense sp. nov. Phytopathologia Mediterranea 53: 416–427. [Google Scholar]
- Burgess TI, Andjic V, Hardy GEStJ, et al. 2006. First report of Phaeophleospora destructans in China. Journal of Tropical Forest Science 18: 144–146. [Google Scholar]
- Burgess TI, Barber PA, Hardy GEStJ. 2005. Botryosphaeria spp. associated with eucalypts in Western Australia, including the description of Fusicoccum macroclavatum sp. nov. Australasian Plant Pathology 34: 557–567. [Google Scholar]
- Burgess TI, Barber PA, Sufaati S, et al. 2007. Mycosphaerella spp. on Eucalyptus in Asia: new species, new hosts and new records. Fungal Diversity 24: 135–157. [Google Scholar]
- Cao JD. 1982. Investigation of bacterial wilt in Eucalyptus saligna and E. grandis introduced from Brazil. Guangxi Forestry Science and Technology 4: 30–31. [In Chinese.] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Chen SF, Barnes I, Chungu D, et al. 2011a. High population diversity and increasing importance of the Eucalyptus stem canker pathogen, Teratosphaeria zuluensis, in South China. Australasian Plant Pathology 40: 407–415. [Google Scholar]
- Chen SF, Gryzenhout M, Roux J, et al. 2010. Identification and pathogenicity of Chrysoporthe cubensis on Eucalyptus and Syzygium spp. in South China. Plant Disease 94: 1143–1150. [DOI] [PubMed] [Google Scholar]
- Chen SF, Li GQ, Liu FF, et al. 2015. Novel species of Botryosphaeriaceae associated with shoot blight of pistachio. Mycologia 107: 780–792. [DOI] [PubMed] [Google Scholar]
- Chen SF, Li GQ, Liu QL, et al. 2016. Characteristics of Lasiodiplodia theobromae from Rosa rugosa in South China. Crop Protection 79: 51–55. [Google Scholar]
- Chen SF, Lombard L, Roux J, et al. 2011b. Novel species of Calonectria associated with Eucalyptus leaf blight in Southeast China. Persoonia 26: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Morgan DP, Hasey JK, et al. 2014a. Phylogeny, morphology, distribution, and pathogenicity of Botryosphaeriaceae and Diaporthaceae from English walnut in California. Plant Disease 98: 636–652. [DOI] [PubMed] [Google Scholar]
- Chen SF, Morgan DP, Michailides TJ. 2014b. Botryosphaeriaceae and Diaporthaceae associated with panicle and shoot blight of pistachio in California, USA. Fungal Diversity 67: 157–179. [Google Scholar]
- Chen SF, Pavlic D, Roux J, et al. 2011c. Characterization of Botryosphaeriaceae from plantation-grown Eucalyptus species in South China. Plant Pathology 60: 739–751. [Google Scholar]
- Chen SF, Van Wyk M, Roux J, et al. 2013. Taxonomy and pathogenicity of Ceratocystis species on Eucalyptus trees in South China, including C. chinaeucensis sp. nov. Fungal Diversity 58: 267–279. [Google Scholar]
- Chen SX, Chen XF. 2013. Technical problems and thinking on Eucalypt plantation management in China. Eucalypt Science & Technology 30: 52–59. [In Chinese.] [Google Scholar]
- Coutinho IBL, Freire FCO, Lima CS, et al. 2017. Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in northeastern Brazil. Plant Pathology 66: 90–104. [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, et al. 2011. Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, et al. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, et al. 2013. Fungal Planet description sheets: 154–213. Persoonia 31: 188–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Phillips AJL. 2007. Neofusicoccum mediterraneum. Fungal Planet 19. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- Cruywagen EM, Slippers B, Roux J, et al. 2017. Phylogenetic species recognition and hybridisation in Lasiodiplodia: a case study on species from baobabs. Fungal Biology 121: 420–436. [DOI] [PubMed] [Google Scholar]
- Cubeta MA, Echandi E, Abernethy T, et al. 1991. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81: 1395–1400. [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2007. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia 99: 664–680. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman S, Crous PW, Groenewald JZ, et al. 2003. Circumscription of Botryosphaeria species associated with Proteaceae based on morphology and DNA sequence data. Mycologia 95: 294–307. [PubMed] [Google Scholar]
- Dou ZP, He W, Zhang Y. 2017a. Lasiodiplodia chinensis, a new holomorphic species from China. Mycosphere 8: 521–532. [Google Scholar]
- Dou ZP, He W, Zhang Y. 2017b. Does morphology matter in taxonomy of Lasiodiplodia? An answer from Lasiodiplodia hyalina sp. nov. Mycosphere 8: 1014–1027. [Google Scholar]
- Farr DF, Elliott M, Rossman AY, et al. 2005. Fusicoccum arbuti sp. nov. causing cankers on Pacific madrone in western North America with notes on Fusicoccum dimidiatum, the correct name for Scytalidium dimidiatum and Nattrassia mangiferae. Mycologia 97: 730–741. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gardner DE. 1997. Botryosphaeria mamane sp. nov. associated with witches’-brooms on the endemic forest tree Sophora chrysophylla in Hawaii. Mycologia 89: 298–303. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Huelsenbeck JP. 1992. Signal, noise and reliability in molecular phylogenetic analyses. Journal of Heredity 83: 189–195. [DOI] [PubMed] [Google Scholar]
- Iglesias-Trabad G, Carbaeira-Tenreiro R, Folgueiia-Lozano J. 2009. Eucalyptus universalis. Global cultivated eucalypt forest map Version 1.2 In: GIT Forestry Consulting’s EUCALYPTOLOGICS: Information resources on Eucalyptus cultivation worldwide. Retrieved from http://www.git-forestry.com [accessed: 19 Oct. 2009].
- Inderbitzin P, Bostock RM, Trouillas FP, et al. 2010. A six locus phylogeny reveals high species diversity in Botryosphaeriaceae from California almond. Mycologia 102: 1350–1368. [DOI] [PubMed] [Google Scholar]
- Jacobs K, Bergdahl DR, Wingfield MJ, et al. 2004. Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycological Research 108: 411–418. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzizera C, Frisullo S, Alves A, et al. 2008. Morphology, phylogeny and pathogenicity of Botryosphaeria and Neofusicoccum species associated with drupe rot of olives in southern Italy. Plant Pathology 57: 948–956. [Google Scholar]
- Li GQ, Arnold RJ, Liu FF, et al. 2015a. Identification and pathogenicity of Lasiodiplodia species from Eucalyptus urophylla × grandis, Polyscias balfouriana and Bougainvillea spectabilis in Southern China. Journal of Phytopathology 163: 956–967. [Google Scholar]
- Li GQ, Liu FF, Li JQ, et al. 2015b. Characterization of Botryosphaeria dothidea and Lasiodiplodia pseudotheobromae from English walnut in China. Journal of Phytopathology 164: 348–353. [Google Scholar]
- Linaldeddu BT, Dedda A, Scanu B, et al. 2015. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Diversity 71: 201–214. [Google Scholar]
- Liu FF, Mbenoun M, Barnes I, et al. 2015. New Ceratocystis species from Eucalyptus and Cunninghamia in South China. Antonie van Leeuwenhoek 107: 1451–1473. [DOI] [PubMed] [Google Scholar]
- Liu JK, Phookamsak R, Doilom M, et al. 2012. Towards a natural classification of Botryosphaeriales. Fungal Diversity 57: 149–210. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lombard L, Zhou XD, Crous PW, et al. 2010. Calonectria species associated with cutting rot of Eucalyptus. Persoonia 24: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes A, Phillips AJL, Alves A. 2017. Mating type genes in the genus Neofusicoccum: Mating strategies and usefulness in species delimitation. Fungal Biology 121: 394–404. [DOI] [PubMed] [Google Scholar]
- Lu B, Hyde KD, Ho WH, et al. 2000. Checklist of Hong Kong fungi. Fungal Diversity Press, Hong Kong. [Google Scholar]
- Machado AR, Pinho DB, Pereira OL. 2014. Phylogeny, identification and pathogenicity of the Botryosphaeriaceae associated with collar and root rot of the biofuel plant Jatropha curcas in Brazil, with a description of new species of Lasiodiplodia. Fungal Diversity 67: 231–247. [Google Scholar]
- Marques MW, Lima NB, De Morais MA, Jr, et al. 2013. Botryosphaeria, Neofusicoccum, Neoscytalidium and Pseudofusicoccum species associated with mango in Brazil. Fungal Diversity 61: 195–208. [Google Scholar]
- Mehl JWM, Slippers B, Roux J, et al. 2011. Botryosphaeriaceae associated with Pterocarpus angolensis (kiaat) in South Africa. Mycologia 103: 534–553. [DOI] [PubMed] [Google Scholar]
- Mohali S, Slippers B, Wingfield MJ. 2006. Two new Fusicoccum species from Acacia and Eucalyptus in Venezuela, based on morphology and DNA sequence data. Mycological Research 110: 405–413. [DOI] [PubMed] [Google Scholar]
- Mohali SR, Slippers B, Wingfield MJ. 2007. Identification of Botryosphaeriaceae from Eucalyptus, Acacia and Pinus in Venezuela. Fungal Diversity 25: 103–125. [Google Scholar]
- Mohali SR, Slippers B, Wingfield MJ. 2009. Pathogenicity of seven species of the Botryosphaeriaceae on Eucalyptus clones in Venezuela. Australasian Plant Pathology 38: 135–140. [Google Scholar]
- Netto MSB, Assunção IP, Lima GSA, et al. 2014. Species of Lasiodiplodia associated with papaya stem-end rot in Brazil. Fungal Diversity 67: 127–141. [Google Scholar]
- Netto MSB, Lima GSA, Correia KC, et al. 2017. Analysis of phylogeny, distribution, and pathogenicity of Botryosphaeriaceae species associated with gummosis of Anacardium in Brazil, with a new species of Lasiodiplodia. Fungal Biology 121: 437–451. [DOI] [PubMed] [Google Scholar]
- Old KM, Wingfield MJ, Yuan ZQ. 2003. A manual of diseases of Eucalypts in South-East Asia. Centre for International Forestry Research, Indonesia. [Google Scholar]
- Osorio JA, Crous CJ, De Beer ZW, et al. 2017. Endophytic Botryosphaeriaceae, including five new species, associated with mangrove trees in South Africa. Fungal Biology 121: 361–393. [DOI] [PubMed] [Google Scholar]
- Pavlic D, Slippers B, Coutinho TA, et al. 2007. Botryosphaeriaceae occurring on native Syzygium cordatum in South Africa and their potential threat to Eucalyptus. Plant Pathology 56: 624–636. [Google Scholar]
- Pavlic D, Slippers B, Coutinho TA, et al. 2009a. Multiple gene genealogies and phenotypic data reveal cryptic species of the Botryosphaeriaceae: A case study on the Neofusicoccum parvum/N. ribis complex. Molecular Phylogenetics and Evolution 51: 259–268. [DOI] [PubMed] [Google Scholar]
- Pavlic D, Slippers B, Coutinho TA, et al. 2009b. Molecular and phenotypic characterization of three phylogenetic species discovered within the Neofusicoccum parvum/N. ribis complex. Mycologia 101: 636–647. [DOI] [PubMed] [Google Scholar]
- Pavlic D, Slippers B, Teresa A, et al. 2004. Lasiodiplodia gonubiensis sp. nov., a new Botryosphaeria anamorph from native Syzygium cordatum in South Africa. Studies in Mycology 50: 313–322. [Google Scholar]
- Pavlic D, Wingfield MJ, Barber P, et al. 2008. Seven new species of the Botryosphaeriaceae from baobab and other native trees in Western Australia. Mycologia 100: 851–866. [DOI] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Abdollahzadeh J, et al. 2013. The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology 76: 51–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Pennycook SR, et al. 2008. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21: 29–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJL, Oudemans PV, Correia A, et al. 2006. Characterisation and epitypification of Botryosphaeria corticis, the cause of blueberry cane canker. Fungal Diversity 21: 141–155. [Google Scholar]
- Prasher IB, Singh G. 2014. Lasiodiplodia indica – A new species of coelomycetous mitosporic fungus from India. Kavaka 43: 64–69. [Google Scholar]
- Punithalingam E. 1980. Plant diseases attributed to Botryodiplodia theobromae Pat. Cramer, Vaduz, Liechtenstein. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society, Kew, Surrey, UK. [Google Scholar]
- Rodas CA, Slippers B, Gryzenhout M, et al. 2009. Botryosphaeriaceae associated with Eucalyptus canker diseases in Colombia Forest Pathology 39: 110–123. [Google Scholar]
- Rodríguez-Gálvez E, Guerrero P, Barradas C, et al. 2017. Phylogeny and pathogenicity of Lasiodiplodia species associated with dieback of mango in Peru. Fungal Biology 121: 452–465. [DOI] [PubMed] [Google Scholar]
- Sakalidis ML, Hardy GEStJ, Burgess TI. 2011. Use of the Genealogical Sorting Index (GSI) to delineate species boundaries in the Neofusicoccum parvum–Neofusicoccum ribis species complex. Molecular Phylogenetics and Evolution 60: 333–344. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. 2011. SAS® 9.3 System Options: Reference, 2nd edn. Cary, North California, USA. [Google Scholar]
- Slippers B, Boissin E, Phillips AJL, et al. 2013. Phylogenetic lineages in the Botryosphaeriales: a systematic and evolutionary framework. Studies in Mycology 76: 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slippers B, Crous PW, Denman S, et al. 2004a. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96: 82–101. [PubMed] [Google Scholar]
- Slippers B, Fourie G, Crous PW, et al. 2004b. Multiple gene sequences delimit Botryosphaeria australis sp. nov. from B. lutea. Mycologia 96: 1030–1041. [PubMed] [Google Scholar]
- Slippers B, Fourie G, Crous PW, et al. 2004c. Speciation and distribution of Botryosphaeria spp. on native and introduced Eucalyptus trees in Australia and South Africa. Studies in Mycology 50: 343–358. [Google Scholar]
- Slippers B, Johnson GI, Crous PW, et al. 2005. Phylogenetic and morphological re-evaluation of the Botryosphaeria species causing diseases of Mangifera indica. Mycologia 97: 99–110. [DOI] [PubMed] [Google Scholar]
- Slippers B, Roux J, Wingfield MJ, et al. 2014. Confronting the constraints of morphological taxonomy in the Botryosphaeriales. Persoonia 33: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slippers B, Wingfield MJ. 2007. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biology Reviews 21: 90–106. [Google Scholar]
- Smith H, Crous PW, Wingfield MJ, et al. 2001. Botryosphaeria eucalyptorum sp. nov., a new species in the B. dothidea-complex on Eucalyptus in South Africa. Mycologia 93: 277–285. [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, et al. 1996. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ding Z, Zhou ZQ, et al. 2012. Phylogenetic and pathogenic analyses show that the causal agent of apple ring rot in China is Botryosphaeria dothidea. Plant Disease 96: 486–496. [DOI] [PubMed] [Google Scholar]
- Taylor K, Barber PA, Hardy GEStJ, et al. 2009. Botryosphaeriaceae from tuart (Eucalyptus gomphocephala) woodland, including descriptions of four new species. Mycological Research 113: 337–353. [DOI] [PubMed] [Google Scholar]
- Trakunyingcharoen T, Lombard L, Groenewald JZ, et al. 2015. Caulicolous Botryosphaeriales from Thailand. Persoonia 34: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull JW. 2007. Development of sustainable forestry plantations in China: a review. ACIAR Impact Assessment Series Report No. 45. Australian Centre for International Agricultural Research, Canberra. [Google Scholar]
- Úrbez-Torres JR, Peduto F, Striegler RK, et al. 2012. Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Diversity 52: 169–189. [Google Scholar]
- Van Burik JAH, Schreckhise RW, White TC, et al. 1998. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Medical Mycology 36: 299–303. [PubMed] [Google Scholar]
- Van Niekerk JM, Crous PW, Groenewald JZ, et al. 2004. DNA phylogeny, morphology and pathogenicity of Botryosphaeria species on grapevines. Mycologia 96: 781–798. [PubMed] [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RP. 2005. Genetic diversity and sustainable productivity of Eucalypt plantations in China. In: Wang H. (ed), Changing patterns: tree introduction and phytogeography: 19–27. China Forestry Publishing House, Beijing, China. [In Chinese.] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR Protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, CA, USA. [Google Scholar]
- Wingfield MJ. 2003. Increasing threat of diseases to exotic plantation forests in the Southern Hemisphere: lessons from Cryphonectria canker. Australasian Plant Pathology 32: 133–139. [Google Scholar]
- Wingfield MJ, Slippers B, Hurley BP, et al. 2008. Eucalypt pests and diseases: growing threats to plantation productivity. Southern Forests 70: 139–144. [Google Scholar]
- Xu C, Wang CS, Ju LL, et al. 2015a. Multiple locus genealogies and phenotypic characters reappraise the causal agents of apple ring rot in China. Fungal Diversity 71: 215–231. [Google Scholar]
- Xu C, Zhang H, Zhou Z, et al. 2015b. Identification and distribution of Botryosphaeriaceae species associated with blueberry stem blight in China. Europe Journal of Plant Pathology 143: 737–752. [Google Scholar]
- Yan JY, Xie Y, Yao SW, et al. 2012. Characterization of Botryosphaeria dothidea, the causal agent of grapevine canker in China. Australasian Plant Pathology 41: 351–357. [Google Scholar]
- Yan JY, Xie Y, Zhang W, et al. 2013. Species of Botryosphaeriaceae involved in grapevine dieback in China. Fungal Diversity 61: 221–236. [Google Scholar]
- Yang T, Groenewald JZ, Cheewangkoon R, et al. 2017. Families, genera and species of Botryosphaeriales. Fungal Biology 121: 322–346. [DOI] [PubMed] [Google Scholar]
- Yu ZD, Tang GH, Peng SB, et al. 2015. Neofusicoccum parvum causing canker of seedlings of Juglans regia in China. Journal of Forestry Research 26: 1019–1024. [Google Scholar]
- Zhang M, Lin S, He W, et al. 2017. Three species of Neofusicoccum (Botryosphaeriaceae, Botryosphaeriales) associated with woody plants from southern China. Mycosphere 8: 797–808. [Google Scholar]
- Zhao JP, Lu Q, Liang J, et al. 2010. Lasiodiplodia pseudotheobromae, a new record of pathogenic fungus from some subtropical and tropical trees in southern China. Cryptogamie Mycologie 31: 431–439. [Google Scholar]
- Zhou XD, De Beer ZW, Xie YJ, et al. 2007. DNA-based identification of Quambalaria pitereka causing severe leaf blight of Corymbia citriodora in China. Fungal Diversity 25: 245–254. [Google Scholar]
- Zhou XD, Wingfield MJ. 2011. Eucalypt diseases and their management in China. Australasian Plant Pathology 40: 339–345. [Google Scholar]
- Zhou Y, Gong GS, Cui YL, et al. 2015. Identification of Botryosphaeriaceae species causing kiwifruit rot in Sichuan Province, China. Plant Disease 99: 699–708. [DOI] [PubMed] [Google Scholar]
- Zhou YP, Dou ZP, He W, et al. 2016. Botryosphaeria sinensia sp. nov., a new species from China. Phytotaxa 245: 43–50. [Google Scholar]
- Zhou YP, Zhang M, Dou ZP, et al. 2017. Botryosphaeria rosaceae sp. nov. and B. ramosa, new botryosphaeriaceous taxa from China. Mycosphere 8: 162–171. [Google Scholar]


