Abstract
The native ‘ōhi’a lehua (Metrosideros polymorpha) has cultural, biological and ecological significance to Hawai’i, but it is seriously threatened by a disease commonly referred to as rapid ‘ōhi’a death (ROD). Preliminary investigations showed that a Ceratocystis species similar to C. fimbriata s.lat. was the cause of the disease. In this study, we used a combination of the phylogenetic, morphological and biological species concepts, as well as pathogenicity tests and microsatellite analyses, to characterise isolates collected from diseased ‘ōhi’a trees across Hawai’i Island. Two distinct lineages, representing new species of Ceratocystis, were evident based on multigene phylogenetic analyses. These are described here as C. lukuohia and C. huliohia. Ceratocystis lukuohia forms part of the Latin American clade (LAC) and was most closely associated with isolates from Syngonium and Xanthosoma from the Caribbean and elsewhere, including Hawai’i, and C. platani, which is native to eastern USA. Ceratocystis huliohia resides in the Asian-Australian clade (AAC) and is most closely related to C. uchidae, C. changhui and C. cercfabiensis, which are thought to be native to Asia. Morphology and interfertility tests support the delineation of these two new species and pathogenicity tests show that both species are aggressive pathogens on seedlings of M. polymorpha. Characterisation of isolates using microsatellite markers suggest that both species are clonal and likely represent recently-introduced strains. Intensive research is underway to develop rapid screening protocols for early detection of the pathogens and management strategies in an attempt to prevent the spread of the pathogens to the other islands of Hawai’i, which are currently disease free.
Keywords: Ceratocystidaceae, fungal barcoding genes, invasive species, ITS types, new taxa
INTRODUCTION
The Ascomycete genus Ceratocystis (Sordariomycetes, Hypocreomycetidae, Microascales) is one of 11 clearly defined genera in the Ceratocystidaceae (De Beer et al. 2014, 2017, Mayers et al. 2015, Nel et al. 2017). Ceratocystis, as currently recognized, includes 36 species (Marin-Felix et al. 2017, Liu et al. 2018) that are characterized by their mostly black, globose ascomatal bases with long, elongated necks terminating in an ostiole, through which sticky hat-shaped ascospores exude (Upadhyay 1981, Seifert et al. 1993). Based on phylogenetic inference, the species reside in four broadly defined geographic clades. These include the Latin American clade (LAC) (Harrington 2000, Engelbrecht & Harrington 2005), the North American clade (NAC) (Johnson et al. 2005), the African clade (AFC) (Heath et al. 2009, Mbenoun et al. 2014) and the Asian-Australian clade (AAC) (Johnson et al. 2005, Thorpe et al. 2005, Li et al. 2017).
Diseases caused by Ceratocystis spp. include vascular wilts, cankers, as well as rot of various root crops (Kile 1993, Roux & Wingfield 2009). Of particular concern in recent years has been the emerging epidemics caused by aggressive Ceratocystis spp., particularly those residing in the LAC. For example, C. manginecans, that was introduced into Oman and Pakistan (Al Adawi et al. 2014) has caused devastating losses to the mango industry (Al Adawi et al. 2006, Van Wyk et al. 2007) and resulted in apparent host jumps to native Prosopis cineraria and Dalbergia sissoo, causing branch and stem cankers (Al Adawi et al. 2013). A pathogen of the same name has emerged in South East Asia, causing a severe canker and wilt disease of plantation grown Acacia mangium (Tarigan et al. 2011, Thu et al. 2014, Brawner et al. 2015). Ceratocystis platani, also residing in the LAC, is the causal agent of a devastating canker and wilt disease of Platanus orientalis in Europe, where it was introduced from the USA in the early 1940s (Engelbrecht et al. 2004, Tsopelas et al. 2017). A strain of C. fimbriata, the type species of the genus, is well known as the causal agent of post-harvest black rot of sweet potato (Ipomoea batatas) (Li et al. 2016, Scruggs et al. 2017).
A new disease that is rapidly assuming crisis status in Hawai’i has colloquially been termed rapid ‘ōhi’a death (ROD). Hundreds of thousands of native M. polymorpha have recently died on the Big Island of Hawai’i (Keith et al. 2015, Mortenson et al. 2016). Dying trees were first noticed in the Puna district of Hawai’i Island as early as 2010, and the disease has now spread across most of the Big Island. This iconic tree species, more commonly known as ‘ōhi’a lehua, is the most common and widespread native tree species of Hawai’i, occurring from sea level to 2 500 m elevation in both dry and wet forests and on substrates ranging from 50 to 4 million years in age (Friday & Herbert 2006, Loope 2016). It is the most ecologically important native tree in Hawai’i, defining native forest succession and ecosystem function over broad areas, providing critical habitat for rare and endangered native bird and insect species, and exemplifying the strong links between native Hawaiian culture and the islands’ environment (Dawson & Stemmermann 1990). Extensive mortality of ‘ōhi’a associated with abiotic factors, known as ‘ōhi’a dieback, has been noted in the past (Hodges et al. 1986), but the recent mortality appears to be caused by a new, biotic factor. Trees of all ages, across all environmental conditions, in native forests and residential areas, are currently affected, with mortality ranging from 10–90 % in certain areas (F. Hughes, pers. comm.).
A characteristic feature of much of the current mortality is the rapid development of symptoms from the onset of the first expression of the disease. Initially, leaves on individual branches in the crown appear to turn yellowish and become necrotic within days to weeks. The brown leaves typically remain attached to dead or dying branches as the disease symptoms rapidly develop throughout the canopy. Dark-brown to black radial discolouration or staining is observed in the outer woody xylem of affected trees that is typical of wilt symptoms caused by Ceratocystis species (Harrington 2013, Roux & Wingfield 2013).
The pathogen responsible for the wilt symptoms was tentatively identified as C. fimbriata s.lat., and the disease was referred to as Ceratocystis wilt (Keith et al. 2015). This identification was based on a combination of disease symptoms typical of Ceratocystis infections, morphological structures similar to those of C. fimbriata s.lat. and a 98 % similarity blast hit of the ITS (internal transcribed spacer) sequence to an isolate identified as C. fimbriata in GenBank (KC493164) (Keith et al. 2015). This isolate, CBS 115167, was obtained from basal rot symptoms on Syngonium podophyllum (arrowhead plant) in Florida, and its ITS sequence was identical to those of other Syngonium isolates in Florida, as well as Syngonium isolates from Brazil, Australia and Hawai’i (Uchida & Aragaki 1979, Thorpe et al. 2005). Other strains of Ceratocystis from Hawai’i include C. fimbriata from sweet potato, which has been known on this crop plant in Hawai’i since 1925 (Chung 1923, Brown & Matsuura 1941, Li et al. 2016), and strains from Colocasia esculenta (taro), which have been known in Hawai’i since the 1970s (Uchida & Aragaki 1979). These isolates from taro, also previously referred to as C. fimbriata (Thorpe et al. 2005), were recently described as a new species, C. uchidae, which resides in the AAC (Li et al. 2017).
The increasing importance of ROD in Hawai’i demands that the isolates of the Ceratocystis sp. associated with the disease on M. polymorpha be accurately identified. The objective of this study was to identify and describe a collection of isolates associated with ROD based on morphology, the biological species concept and by using a multigene phylogenetic approach. The study included closely related Ceratocystis species and isolates treated as C. fimbriata s.lat. previously reported from Hawai’i. The pathogenicity of the isolates from M. polymorpha were confirmed in inoculation studies and the ability of isolates to cross-infect other hosts was determined. In addition, isolates collected from different regions on the Island of Hawai’i were characterized using microsatellite markers to determine their genetic diversity.
MATERIALS AND METHODS
Sampling and isolations
Surveys were conducted in the Puna, Hilo and Kona districts of Hawai’i Island during 2014 and 2015 for M. polymorpha trees showing typical symptoms of ROD (Fig. 1). Wood samples were collected from M. polymorpha showing brown to black streaking in the woody xylem. Carrot baiting was used to isolate Ceratocystis from discoloured wood (Moller & Devay 1968). Primary isolations were made by transferring ascospore drops from the tips of the ascomata formed on the wood or carrot discs onto 10 % V8 agar. Pure cultures were stored at -80 °C in 15 % glycerol. Single ascospore cultures were generated by transferring single ascospores to 2 % malt extract agar (MEA, Biolab, Midrand, South Africa), supplemented with 100 μg/L thymine (Sigma-Aldrich, Germany). Cultures were incubated at 25 °C.
Fig. 1.
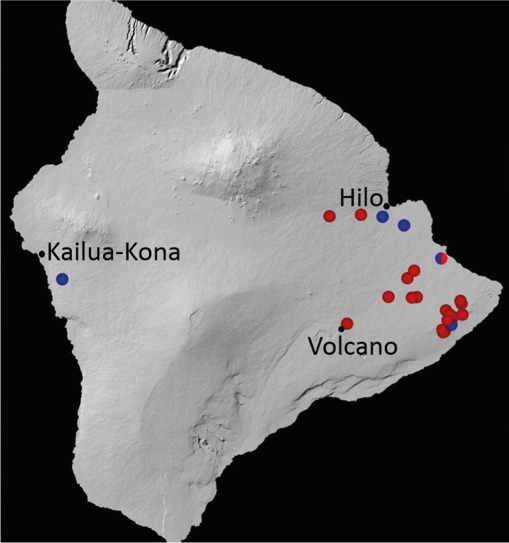
Distribution of samples yielding isolates of Ceratocystis lukuohia (red circles) and C. huliohia (blue circles) on Hawai’i Island during 2014–2015.
Cultures obtained in this study are stored at the USDA Agricultural Research Service, Hilo, HI; the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa; or at Iowa State University (Table 1; Appendix). Representative cultures have been deposited at the Westerdijk Fungal Biodiversity Institute (CBS-KNAW), Utrecht, The Netherlands, Mycothèque de l’Université Catholique de Louvain, Belgium (MUCL) and dried specimens have been deposited in the National Collection of Fungi (PREM), Pretoria, South Africa (Table 1).
Table 1.
Details of the Ceratocystis isolates from the Latin American clade (LAC) and the Asian-Australian clade (AAC) used in the phylogenetic analyses, mating tests and pathogenicity tests in this study.
| Species1 | Isolate no.2,3 | Substrate | Country | Locality | Year collected | GenBank accession numbers |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS-rDNA | bt1 | tef1 | ms204 | rpb2 | ||||||
| C. adelphaT | CBS 115169; CMW 14809; PREM 61152; C1833 | Theobroma cacao | Ecuador | Pichilingue | 2001 | DQ520637 | KJ601509 | KJ601516 | KJ601563 | KJ601599 |
| C. adelpha | CBS 152.62; CMW 15051; C940 | Theobroma cacao | Costa Rica | Atlantic side | 1962 | AY157951 | KJ601510 | KJ601517 | KJ601564 | KJ601600 |
| C. albifundus | CBS 128992; CMW 4068 | Acacia mearnsii | South Africa | KwaZulu-Natal, Bloemendal | 1997 | DQ520638 | EF070429 | EF070400 | KY643987 | KY644041 |
| C. atroxT | CBS 120518; CMW 19385; PREM 59012 | Eucalyptus grandis | Australia | Queensland | 2005 | NR_136981; EF070415 | EF070431 | EF070403 | KY643976 | KY644030 |
| C. atrox | CBS 120517; CMW 19383 | Eucalyptus grandis | Australia | Queensland | 2006 | EF070414 | EF070430 | EF070402 | KY643975 | KY644029 |
| C. cacaofunestaT | CBS 115172; CMW 26375; C1983; BPI 843740 | Theobromae cacao | Brazil | Rondonia | 2002 | AY157953 | KJ601512 | KJ601519 | KJ601566 | KJ601602 |
| C. cacaofunesta | CBS 114722; CMW 14798; C1548; BPI 843730 | Theobromae cacao | Costa Rica | La Lola Experiment Station | 1999 | AY157952 | KJ601511 | KJ601518 | KJ601565 | KJ601601 |
| C. cercfabiensisT | CBS 139654; CMW 43029; CERC 2170; PREM 61229 | Eucalyptus sp. | China | HaiNan Province, LinGao County | 2013 | KP727592 (Group 2, Clone F); KP727593 (Group 2, Clone G); KP727594 (Group 2, Clone K) | KP727618 | KP727643 | KY643963 | KY644016 |
| C. cercfabiensis | CBS 139656; CMW 42795; CERC 2687 | Eucalyptus sp. | China | GuangDong | 2013 | same as KP727583 (Group 2, Clone A) | KP727619 | KP727644 | KY643968 | KY644022 |
| C. cercfabiensis | CMW 42736 | Eucalyptus sp. | China | GuangDong | 2013 | KP727583 (Group 2, Clone A); KP727584 (Group 1, Clone B) | – | – | – | – |
| C. cercfabiensis | CMW 42741 | Eucalyptus sp. | China | GuangDong | 2013 | KP727586 (Group 2, Clone A); KP727588 (Group 1, Clone E) | – | – | – | – |
| C. cercfabiensis | C3301; CERC 2548 | Eucalyptus sp. | China | GuangDong | 2013 | KY306679 | – | – | – | – |
| C. cercfabiensis | C3302; CERC 2549 | Eucalyptus sp. | China | GuangDong | 2013 | KY306680 | – | KP727623 | – | – |
| C. cercfabiensis | CMW 42577; C3305; CERC 2552 | Eucalyptus sp. | China | GuangDong | 2013 | KY306681 | – | KP727624 | – | – |
| C. cercfabiensis | C3356, CERC 2166 | Eucalyptus sp. | China | Hainan | 2013 | – | – | – | – | – |
| C. cercfabiensis | CMW 42489; C3358; CERC 2168 | Eucalyptus sp. | China | Hainan | 2013 | – | – | KP727630 | – | – |
| C. changhuiT | CBS 139797; CMW 43281; CERC 3615; PREM 61241 | Colocasia esculenta | China | YunNan Province, KunMing | 2014 | KY643884 | KY643919 | KY643941 | KY643962 | KY644015 |
| C. changhui | CBS 139798; CMW 43272; CERC 3605; PREM 61242 | Colocasia esculenta | China | YunNan Province, KunMing | 2015 | KY643886 | KY643915 | KY643937 | KY643958 | KY644011 |
| C. collisensisT | CBS 139679; CMW 42552; CERC 2459; PREM 61232 | Cunninghamia lanceolata | China | FuJian Province, ZhangZhou Region | 2013 | KP727578 | KP727614 | KP727639 | KY643970 | KY644024 |
| C. collisensis | CBS 139647; CMW 42554; CERC 2466 | Cunninghamia lanceolata | China | FuJian Province, ZhangZhou Region | 2013 | KP727580 | KP727615 | KP727640 | KY643972 | KY644026 |
| C. colombianaT | CBS 121792; CMW 5751; PREM 59434 | Coffea arabica | Colombia | Valle del Cauca | 2000 | NR_119483; AY177233 | AY177225 | EU241493 | KJ601567 | KJ601603 |
| C. colombiana | CBS 121791; CMW 5761; PREM 59435 | Coffea arabica | Colombia | – | 2000 | AY177234 | AY177224 | EU241492 | KJ601568 | KJ601604 |
| C. colombiana | C1945 | Coffea arabica | Colombia | – | 2002 | |||||
| C. corymbiicolaT | CBS 127215; CMW 29120; PREM 60431 | Corymbia variegata | Australia | Wedding Bells State Forest, New South Wales | 2008 | NR_119830; HM071902 | HM071914 | HQ236453 | KY643983 | KY644037 |
| C. corymbiicola | CBS 127216; CMW 29349; PREM 60433 | Eucalyptus pilularis | Australia | Ingalba State Forest, New South Wales | 2008 | HM071919 | HQ236455 | HM071905 | KY643984 | KY644038 |
| C. curvataT | CBS 122603; CMW 22442; PREM 60151 | Eucalyptus deglupta | Ecuador | Salinas | 2004 | NR_137018; FJ151436 | FJ151448 | FJ151470 | KJ601570 | KJ601606 |
| C. curvata | CBS 122604; CMW 22435 | Eucalyptus deglupta | Ecuador | Salinas | 2004 | FJ151437 | FJ151449 | FJ151471 | KJ601569 | KJ601605 |
| C. diversiconidiaT | CBS 123013; CMW 22445; PREM 60160 | Terminalia ivorensis | Ecuador | Salinas | 2004 | FJ151440 | FJ151452 | FJ151474 | KJ601571 | KJ601607 |
| C. diversiconidia | CBS 122605; CMW 22448; PREM 60162 | Terminalia ivorensis | Ecuador | Salinas | 2004 | FJ151441 | FJ151453 | FJ151475 | KJ601572 | KJ601608 |
| C. ecuadorianaT | CBS 124020; CMW 22092; PREM 60155 | Eucalyptus deglupta | Ecuador | Salinas | 2004 | FJ151432 | FJ151444 | FJ151466 | KJ601573 | KJ601609 |
| C. ecuadoriana | CBS 124022; CMW 22097 | Eucalyptus deglupta | Ecuador | Salinas | 2004 | FJ151434 | FJ151446 | FJ151468 | KJ601574 | KJ601610 |
| C. eucalypticolaT | CBS 124016; CMW 11536; PREM 60168 | Eucalyptus sp. | South Africa | KwaZulu-Natal, KwaMbonambi | 2002 | FJ236723 | FJ236783 | FJ236753 | KJ601576 | KJ601612 |
| C. eucalypticola | CBS 124019; CMW 10000 | Eucalyptus sp. | South Africa | KwaZulu-Natal, KwaMbonambi | 2002 | FJ236722 | FJ236782 | FJ236752 | KJ601575 | KJ601611 |
| C. ficicolaT | CMW 38543; NIAES 20600; C1355; BPI 843724; MAFF 625119 | Ficus carica | Japan | Fukuoka Prefecture | 1990 | NR_119410 | KY685077 | KY316544 | KY685080 | KY685082 |
| C. ficicola | CMW 38544 | Ficus carica | Japan | Fukuoka Prefecture | 1991 | KY685076 | KY685078 | KY685079 | KY685081 | KY685083 |
| C. fimbriata | CBS 114723; CMW 14799; C1421 | Ipomoea batatas | USA | North Carolina | 1998 | KC493160 | KF302689 | KJ631109 | KJ601578 | KJ601614 |
| C. fimbriata | CBS 123010; CMW 1547 | Ipomoea batatas | Papua New Guinea | – | 1984 | AF264904 | EF070443 | EF070395 | KJ601577 | KJ601613 |
| C. fimbriata | CMW 46734; P15-31 | Ipomoea batatas | USA | Hawai’i Island, Hawai’i | 2015 | KY809154 | KY809103 | KY809116 | KY809128 | KY809141 |
| C. fimbriata | C4135 | Ipomoea batatas | USA | Hawai’i Island, Hawai’i | 2016 | same as KC493160 | – | – | – | – |
| C. fimbriatomimaT | CBS 121786; CMW 24174; PREM 59439 | Eucalyptus grandis × E. urophylla | Venezuela | Portuguesa state, Acarigua | 2006 | EF190963 | EF190951 | EF190957 | KJ601579 | KJ601615 |
| C. fimbriatomima | CMW 24377 | Eucalyptus grandis × E. urophylla | Venezuela | Portuguesa state, Acarigua | 2006 | EF190966 | EF190954 | KJ601520 | KJ601581 | KJ601617 |
| C. huliohiaT | CBS 142794; CMW 47149; P15-58; PREM 61769; MUCL 56340 | Metrosideros polymorpha | USA | Hōlualoa, North Kona District, Hawai’i | 2015 | KY809156 | KY809105 | KY809118 | KY809130 | KY809143 |
| C. huliohia | CBS 142795; CMW 47135; P14-10; PREM 61768; MUCL 56341; C4211 | Metrosideros polymorpha | USA | ‘Opihikao, Puna District, Hawai’i | 2014 | KY809155 | KY809104 | KY809117 | KY809129 | KY809142 |
| C. huliohia | CMW 44104; P14-9-1 | Metrosideros polymorpha | USA | ‘Opihikao, Puna District, Hawai’i | 2014 | same as KY809156 | same as KY809105 | same as KY809118 | same as KY809130 | same as KY809143 |
| C. huliohia | CMW 46716; P14-14 | Metrosideros polymorpha | USA | Hawaiian Paradise Park, Puna District, Hawai’i | 2014 | same as KY809156 | same as KY809105 | same as KY809118 | same as KY809130 | same as KY809143 |
| C. huliohia | CMW 46721; P15-3 | Metrosideros polymorpha | USA | Hilo, South Hilo District, Hawai’i | 2014 | same as KY809156 | same as KY809105 | same as KY809118 | same as KY809130 | same as KY809143 |
| C. huliohia | CMW 46738; P15-59; C4215 | Metrosideros polymorpha | USA | Hōlualoa, North Kona District, Hawai’i | 2015 | same as KY809156 | same as KY809105 | same as KY809118 | same as KY809130 | same as KY809143 |
| C. huliohia | CMW 47143; P15-33 | Metrosideros polymorpha | USA | Pana’ewa, South Hilo District, Hawai’i | 2015 | same as KY809156 | same as KY809105 | same as KY809118 | same as KY809130 | same as KY809143 |
| C. huliohia | C4190 | Metrosideros polymorpha | USA | Hawai’i Island, Hawai’i | 2016 | same as KY809156 | – | – | – | – |
| C. huliohia | C4191 | Metrosideros polymorpha | USA | Hawai’i Island, Hawai’i | 2016 | same as KY809156 | – | – | – | – |
| C. huliohia | C4192 | Metrosideros polymorpha | USA | Hawai’i Island, Hawai’i | 2016 | same as KY809156 | – | – | – | – |
| C. lariumT | CBS 122512; CMW 25434; PREM 60193 | Styrax benzoin | Indonesia | North Sumatra | 2007 | NR_137016; EU881906 | EU881894 | EU881900 | KY643981 | KY644035 |
| C. larium | CBS 122606; CMW 25435 | Styrax benzoin | Indonesia | North Sumatra | 2007 | EU881907 | EU881895 | EU881901 | KY643982 | KY644036 |
| C. lukuohiaT | CBS 142792; CMW 44102; P14-1-1; PREM 61770; MUCL 56342; C4212 | Metrosideros polymorpha | USA | Leilani Estates, Puna District, Hawai’i | 2014 | KP203957/KY809157 | KY809106 | KY809119 | KY809131 | KY809144 |
| C. lukuohia | CMW 44103; P14-6 | Metrosideros polymorpha | USA | Orchidlands Estates, | 2014 | same as KY809157 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| Puna District, Hawai’i | ||||||||||
| C. lukuohia | CMW 44105; P14-12 | Metrosideros polymorpha | USA | Hawaiian Acres, Puna District, Hawai’i | 2014 | same as KY809157 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | CMW 44106; P14-13 | Metrosideros polymorpha | USA | Hawaiian Paradise Park, Puna District, Hawai’i | 2014 | same as KY809157 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | CMW 44107; P14-15 | Metrosideros polymorpha | USA | Hawaiian Acres, Puna District, Hawai’i | 2014 | same as KY809157 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | CBS 142793; CMW 46741; P15-64; PREM 61771; MUCL 56343; C4210 | Metrosideros polymorpha | USA | Nānāwale Estates, Puna District, Hawai’i | 2015 | KY809158 | KY809107 | KY809120 | KY809132 | KY809145 |
| C. lukuohia | CMW 47857; P15-11 | Metrosideros polymorpha | USA | Keau’ohana Forest Reserve, Puna District, Hawai’i | 2015 | same as KY809158 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | CBS 142591; CMW 47152; P15-65; PREM 61772; MUCL 56344; C4213 | Metrosideros polymorpha | USA | Waiākea Forest Reserve, South Hilo District, Hawai’i | 2015 | haplotype A same as KY809157; haplotype C same as KY809158 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | CMW 46743; P15-27 | Metrosideros polymorpha | USA | Kopua Farm Lots, Puna District, Hawai’i | 2015 | haplotype A same as KY809157; haplotype C same as KY809158 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | CMW 47139; P15-17 | Metrosideros polymorpha | USA | Mālama Kî Forest Reserve, Puna District, Hawai’i | 2015 | haplotype A same as KY809157; haplotype C same as KY809158 | same as KY809106 | same as KY809119 | same as KY809131 | same as KY809144 |
| C. lukuohia | C4133 | Metrosideros polymorpha | USA | Hawai’i Island, Hawai’i | 2016 | haplotype A same as KY809157 | – | – | – | – |
| C. lukuohia | C4185 | Metrosideros polymorpha | USA | Hawai’i Island, Hawai’i | 2016 | haplotype A same as KY809157 | – | – | – | – |
| C. lukuohia | C4186 | Metrosideros polymorpha | USA | Hawai’i Island, Hawai’i | 2016 | haplotype A same as KY809157 | – | – | – | – |
| C. mangicolaT | CBS 114721; CMW 14797; C1688; PREM 60182 | Mangifera indica | Brazil | São Paulo State | 2000 | AY953382 | EF433307 | EF433316 | KJ601582 | KJ601618 |
| C. mangicola | CMW 28907; PREM 60184 | Mangifera indica | Brazil | São Paulo State | 2008 | FJ200257 | FJ200270 | FJ200283 | KJ601583 | KJ601619 |
| C. manginecansT | CBS 121659; CMW 13851; PREM 59612 | Mangifera indica | Oman | – | 2002 | NR_119532; AY953383 | EF433308 | EF433317 | KJ601584 | KJ601620 |
| C. manginecans | CBS 121660; CMW 13852; PREM 59613 | Mangifera indica | Oman | – | 2002 | AY953384 | EF433309 | EF433318 | KJ601585 | KJ601621 |
| C. mangivoraT | CBS 128340; CMW 27305; PREM 60570 | Mangifera indica | Brazil | São Paulo State, Campinas | 2001 | FJ200262 | FJ200275 | FJ200288 | KJ601587 | KJ601623 |
| C. mangivora | CBS 600.70; CMW 15052; PREM 60188 | Mangifera indica | Brazil | São Paulo State | 1970 | EF433298 | EF433306 | EF433315 | KJ601586 | KJ601622 |
| C. neglectaT | CBS 121789; CMW 17808; PREM 59616 | Eucalyptus grandis | Colombia | – | 2004 | NR_137552; EF127990 | EU881898 | EU881904 | KJ601588 | KJ601624 |
| C. neglecta | CMW 18194; PREM 59617 | Eucalyptus grandis | Colombia | – | 2004 | EF127991 | EU881899 | EU881905 | KJ601589 | KJ601625 |
| C. obpyriformisT | CBS 122511; CMW 23808; PREM 59796 | Acacia mearnsii | South Africa | Mpumalanga, Piet Retief | 2006 | EU245003 | EU244975 | EU244935 | KY643978 | KY644032 |
| C. obpyriformis | CBS 122608; CMW 23807; PREM 59797 | Acacia mearnsii | South Africa | Mpumalanga, Piet Retief | 2007 | EU245004 | EU244976 | EU244936 | KY643977 | KY644031 |
| C. papillataT | CBS 121793; CMW 8856; PREM 59438 | Citrus × Tangelo hybrid | Colombia | Caldas | 2001 | NR_119486; AY233867 | AY233874 | EU241484 | KJ601590 | KJ601626 |
| C. papillata | CMW 10844; PREM 60172 | Coffea arabica | Colombia | Antioquia | 1998 | AY177238 | AY177229 | EU241481 | KJ601591 | KJ601627 |
| C. pirilliformisT | CBS 118128; CMW 6579; PREM 57323 | Eucalyptus nitens | Australia | Australian Capital Territory, Uriarra | 2000 | NR_119452; AF427105 | DQ371653 | AY528983 | KJ601594 | KJ601630 |
| C. pirilliformis | CMW 6569; PREM 57322 | Eucalyptus nitens | Australia | Australian Capital Territory, Uriarra | 2000 | AF427104 | DQ371652 | AY528982 | KY643985 | KY644039 |
| C. plataniPT | CBS 115162; CMW 14802; C1317 | Platanus occidentalis | USA | Janet Day Plantation, North Carolina | 1998 | DQ520630 | EF070425 | EF070396 | KJ601592 | KJ601628 |
| C. platani | CMW 23450 | Platanus orientalis | Greece | Peloponese | 2006 | KJ631107 | KJ601513 | KJ601521 | KJ601593 | KJ601629 |
| C. platani | CMW 1896 | Platanus sp. | Switzerland | – | – | AF395681 | KY809113 | KY809126 | KY809138 | KY809151 |
| C. platani | CMW 2219 | Platanus sp. | France | Saint-Maurice | 1991 | AF395679 | KY809114 | AY528975 | KY809139 | KY809152 |
| C. platani | CMW 9499 | Platanus sp. | Italy | – | – | KY809159 | KY809115 | KY809127 | KY809140 | KY809153 |
| C. platani | C1329 | Platanus occidentalis | USA | Bertie, North Carolina | 1998 | – | – | – | – | – |
| C. platani | C1339 | Platanus occidentalis | USA | Sussex, Virginia | 1998 | same as AY157958 | – | – | – | – |
| C. platani | C1342 | Platanus occidentalis | USA | Bertie, North Carolina | 1998 | – | – | – | – | – |
| C. polychromaT | CBS 115778; CMW 11424; PREM 57818 | Syzygium aromaticum | Indonesia | Sulawesi, Touliang Oki | 2002 | AY528970 | AY528966 | AY528978 | KY643973 | KY644027 |
| C. polychroma | CBS 115777; CMW 11436; PREM 57819 | Syzygium aromaticum | Indonesia | Sulawesi, Touliang Oki | 2003 | AY528971 | AY528967 | AY528979 | KY643974 | KY644028 |
| C. polychroma | CBS 115775; CMW 11449; C2240; PREM 57821 | Syzygium aromaticum | Indonesia | Sulawesi, Touliang Oki | 2003 | AY528972 | AY528968 | KY316543 | Pending | Pending |
| C. polyconidiaT | CBS 122289; CMW 23809; PREM 59788 | Acacia mearnsii | South Africa | Mpumalanga, Piet Retief | 2006 | EU245006 | EU244978 | EU244938 | KY643979 | KY644033 |
| C. polyconidia | CBS 122290; CMW 23818; PREM 59789 | Acacia mearnsii | South Africa | Mpumalanga, Piet Retief | 2007 | EU245007 | EU244979 | EU244939 | KY643980 | KY644034 |
| Ceratocystis sp. | CBS 115167; CMW 48508; C1809 | Syngonium podophyllum | USA | Florida | 2001 | AY526295 | KY809108 | KY809121 | KY809133 | KY809146 |
| Ceratocystis sp. | CBS 114719; CMW 48500; C1717 | Syngonium sp. | USA | Hawai’i | 1987 | AY526294 | KY809110 | KY809123 | KY809135 | KY809148 |
| Ceratocystis sp. | CMW 50456; P16-1 | Syngonium podophyllum | USA | Hawai’i Island, Hawai’i | 2016 | same as AY526294 | – | – | – | – |
| Ceratocystis sp. | CMW 50457; P16-2 | Syngonium podophyllum | USA | Hawai’i Island, Hawai’i | 2016 | same as AY526294 | – | – | – | – |
| Ceratocystis sp. | C4118 | Syngonium podophyllum | USA | Hawai’i Island, Hawai’i | 2016 | same as AY526294 | – | – | – | – |
| Ceratocystis sp. | C1774 | Syngonium podophyllum | USA | Florida | 2000 | AY526295 | – | – | – | – |
| Ceratocystis sp. | CBS 115165; CMW 14805/48506; C1780 | Xanthosoma sagittifolium | Costa Rica | Cartago | 2001 | AY526297 | KY809111 | KY809124 | KY809136 | KY809149 |
| Ceratocystis sp. | CBS 114718; CMW 14794/48499; C1817 | Xanthosoma sp. or C. esculenta | Cuba | Havana | 2001 | AY526298 | KY809112 | KY809125 | KY809137 | KY809150 |
| C. uchidaeT | CBS 115164; CMW 14804/48505; C1714; BPI 843732 | Colocasia esculenta | USA | O’ahu, Hawai’i | 1991 | AY526306 | KY643899 | KY643921 | KY643943 | KY643996 |
| C. uchidae | CBS 114720; CMW 14796/48501; C1715; | Colocasia esculenta | USA | Kaua’i, Hawai’i | 1988 | AY526307 | KY643898 | KY643920 | KY643942 | KY643995 |
| BPI 843733 | ||||||||||
| C. uchidae | C1931; DAR 58902 | Xanthosoma sagittifolium | Fiji | – | 1987 | AY526308 | – | HM569618 | – | – |
1 T = Extype material of Ceratocystis species used in the phylogeneic studies.
2 BPI = US National Fungus Collections, Beltsville, Maryland, USA; C = Personal collection of Tom Harrington, Iowa State University; CBS = Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CERC = China Eucalypt Research Centre (CERC), Chinese Academy of Forestry (CAF), ZhanJiang, GuangDong Province, China; CMW = Tree Pathology Co-operative Program, Forestry and Agricultural Biotechnology Institute, University of Pretoria, South Africa; MUCL = Mycothèque de l’Université Catholique de Louvain, Belgium; P = USDA Agricultural Research Service, Hilo, HI; PREM = National Collection of Fungi, Agricultural Research Council, Pretoria, South Africa.
DNA extraction, PCR and sequencing
Cultures were grown for 2 wk on 2 % MEA. Mycelium was scraped from the surface of the agar plates, lyophilized and crushed using the Retsch® GmbH MM301 homogenizer (Haan, Germany). DNA was extracted using the phenol/chloroform method described by Goodwin et al. (1992). DNA quantity was measured using a Thermo Scientific NanoDrop® ND-1000 spectrophotometer (Wilmington, DE, USA) and diluted to 30 ng/μL working stock solutions.
For the phylogenetic analyses of Ceratocystis species, partial regions of five loci were amplified (Marin-Felix et al. 2017). These included the Internal Transcribed Spacer (ITS) rDNA regions and the 5.8S rRNA gene using the primers ITS1 and ITS4 (White et al. 1990), Beta-tubulin 1 (bt1) gene using primers Bt1a and Bt1b (Glass & Donaldson 1995), Transcription Elongation Factor-1 alpha (tef1) gene with primers TEF1F and TEF2R (Jacobs et al. 2004), the guanine nucleotide-binding protein subunit beta-like protein (ms204) using primers MS204F.cerato (AAG GGC ACC CTC GAG GGC CAC) and MS204R.cerato (GAT GGT RAC GGT GTT GAT GTA) (Fourie et al. 2015) and second largest subunits of RNA polymerase II (rpb2) using degenerate primers RPB2-5Fb (GAY GAY CGT GAT CAC TTY GG) and RPB2-7Rb (CCC ATR GCY TGY TTR CCC AT) (Fourie et al. 2015).
PCR amplification reactions and conditions, including annealing temperatures and MgCl2 concentrations, for all gene regions were the same as those described by Fourie et al. (2015). When rbp2 was difficult to amplify, a second round of PCR was carried out using the cleaned product of the first PCR reaction as a template. Successful PCR amplicons were determined by gel electrophoresis on 1 % agarose gels and visualized with GelRed™ (Biotium, Hayward, CA, USA) under UV illumination (Gel Doc EZ Imager, BioRad, Johannesburg, South Africa). PCR amplicons were purified through 6 % Sephadex G-50 (Sigma-Aldrich, Steinheim, Germany) using Centri-Sep spin columns (Princeton Separations, Inc., Adelphia, NJ, USA).
PCR amplicons were sequenced in both directions using the BigDye® Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Thermo Fisher, Foster City, CA, USA) cleaned by filtration through 6 % Sephadex G-50 and run on an ABI 3100 Genetic Analyzer (Applied BioSystems, Thermo Fisher). In cases where the ITS region was difficult to sequence due to multiple ITS types (Marin-Felix et al. 2017), cloning and sequencing of the PCR product was carried out using the pGEM®-T Easy Vector System, according to manufacturer’s instructions (Promega, Madison, WI, USA). Forward and reverse sequences were assembled into consensus sequences using CLC MainWorkbench v. 6.6.2 (CLC Bio, www.clcbio.com). All unique sequences generated in this study were submitted to GenBank.
Multi-gene phylogenetic analyses
Initial BLAST analyses of the sequences obtained against the NCBI GenBank database (NCBI; http://www.ncbi.nlm.nih.gov) were performed to identify the species of Ceratocystis most closely related to the isolates from Hawai’i and the biogeographic clade in which they reside. Representative sequences showing the highest similarity matches were downloaded and included in the datasets. Separate ITS datasets were generated using ex-type and ex-paratype sequences representing all described species of Ceratocystis (Li et al. 2017, Marin-Felix et al. 2017, Liu et al. 2018) for LAC and AAC. These datasets also included the different ITS types that have previously been described (Al Adawi et al. 2013, Naidoo et al. 2013, Harrington et al. 2014, Fourie et al. 2015, Liu et al. 2015). A third dataset included the combined sequences of the bt1, ef1, ms204 and rpb2 loci for both the LAC and AAC.
For phylogenetic tree construction, the consensus sequence reads were aligned using the online software MAFFT v. 7 (Katoh & Standley 2013) (http://mafft.cbrc.jp/alignment/server/) and the best alignment strategy for each locus was automatically selected by the software. Alignments were edited and trimmed in MEGA v. 7 (Kumar et al. 2016). Sequence alignments for all three datasets were deposited in TreeBASE (No. S22005) (http://purl.org/phylo/treebase/phylows/study/TB2:S22005).
Aligned sequences were used as input for phylogenetic tree construction using a maximum parsimony (MP) and maximum likelihood (ML) approach. Ceratocystis albifundus, residing in AFC, was used as the outgroup in all tree constructions (Table 1). Maximum parsimony analysis was performed with PAUP v. 4.0b10 (Swofford 2003), and most parsimonious trees were obtained using a heuristic search and TBR branch swopping strategy, based on 1 000 repeats. Gaps were treated as missing except for the LAC ITS dataset, where gaps were treated as a fifth character state. For the ITS datasets, sites 227–234 in the LAC dataset and sites 222–226 in the AAC dataset were excluded from the analyses because they contained repeating thymine nucleotides. Sites 867–880 in the combined dataset, forming part of the ms204 region, were excluded due to poly-C nucleotides and ambiguous sites. Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC) were all calculated. Branch confidence was determined using 1 000 bootstrap replicates.
Maximum likelihood analysis was performed using RAxML (Stamatakis 2014) with standard parameters and the GTRGAMMA setting to determine the optimal nucleotide substitution model. Branch confidence for ML was determined using 1 000 bootstrap replicates. Single gene trees for bt1, ef1, ms204 and rpb2 and a species tree based on the combined sequences of the four genes were generated using both MP and ML analysis. Phylogenetic trees were edited using MEGA v. 7 (Kumar et al. 2016) and Adobe Illustrator CS6.
Culture characteristics and morphology
Isolates used for morphological characterisation were grown on 2 % MEA and incubated for 3 wk at 22 °C with natural day/night light intervals. Colony colour was determined using the colour charts of Rayner (1970). Fungal material was mounted in 80 % lactic acid amended with bromothymol blue on glass slides. Observations of structures were made using the Nikon SMZ 18 stereo microscope and Nikon Eclipse Ni compound microscope with differential interference contrast (DIC) illumination. Measurements were made, and electronic images captured, with a Nikon DS-Ri2 camera and NIS-Elements BR (Nikon Instruments Software-Elements Basic Research) software program.
Up to 50 measurements were taken for each characteristic morphological structure including ascomata, ascospores, phialides and the different conidial forms for isolates representing the holotypes and paratypes for all new species. Measurements are presented as (min–)mean-SD – mean+SD(–max), height × width. All relevant data pertaining to type specimens were deposited in MycoBank (www.MycoBank.org).
Growth in culture
In order to determine growth rate in culture, 4 mm agar plugs covered with mycelium were taken from the outer edges of 10-d-old cultures and placed face down in the centres of 90 mm Petri dishes containing 2 % MEA. Four replica plates were made for all isolates chosen to represent holotypes and paratypes and grown in the dark in incubators at temperatures ranging from 10–30 °C at 5 °C intervals. Measurements of colony diam were taken for each culture every 2nd d for 14 d and averages calculated.
Genotyping of Ceratocystis isolates
Twenty-seven microsatellite markers (Barnes et al. 2001, Steimel et al. 2004, Fourie et al. 2016) were used to determine the genetic diversity of the isolates obtained from Hawai’i. PCR reactions and conditions were the same as those described by Fourie et al. (2015). PCR amplicons were visualised on 2 % agarose gels. Products were pooled into three panels and run on an ABI 3500xl Genetic Analyzer (Thermo Fisher Scientific) and sized using GeneScan software and Liz500 (-250) size standard (Thermo Fisher Scientific). Alleles, based on fragment length, were determined using GeneMapper® v. 4.1 software (Applied Biosystems, Thermo Fisher Scientific). Microsatellite profiles of isolates that were phylogenetically most closely related to those from Hawai’i were also generated and included for comparative purposes.
Interfertility tests
Two series of mating tests were performed on strains of the identified new species and their close relatives as described by Li et al. (2017). Most field isolates of Ceratocystis are MAT2 and self-fertile, but self-sterile sectors of MAT2 isolates that had lost female characteristics (loss of protoperithecium production) are often found in laboratory cultures and can be used as male testers (Harrington & Mcnew 1997, 1998, Ferreira et al. 2010, Oliveira et al. 2015). Such self-sterile MAT2 tester strains were recovered as V-shaped sectors during the cultivation of field isolates on malt yeast extract agar (MYEA; 2 % malt extract, 0.2 % yeast extract, 2 % agar). Female, MAT1 tester strains that produced protoperithecia were obtained by recovering single-ascospore progeny from self-fertile field isolates, which yield both self-fertile (MAT2) and self-sterile (MAT1) strains.
Two putative species of Ceratocystis, referred to as species A and species B, were recovered from symptomatic ‘ōhi’a trees. Suitable male, MAT2 tester strains were derived from three isolates of Ceratocystis sp. A and two isolates of species closely related to it in the phylogenetic analyses. These were a North Carolina isolate of C. platani and a Florida isolate of a Ceratocystis sp. from Syngonium podophyllum (Thorpe et al. 2005). In two replicate experiments, the MAT2 testers were used to spermatize MAT1 testers from eight field isolates of Ceratocystis sp. A, C. platani and Ceratocystis sp. from Syngonium, plus MAT1 testers of C. colombiana, C. cacaofunesta and C. fimbriata of the LAC of Ceratocystis. In the second series of pairings, suitable male, MAT2 testers were derived from two isolates of Ceratocystis sp. B, two isolates of C. uchidae and an isolate of C. cercfabiensis (Li et al. 2017). These MAT2 testers were used to spermatize 11 MAT1 testers derived from field isolates of Ceratocystis sp. B, C. uchidae, C. cercfabiensis and C. polychroma, all of the AAC of Ceratocystis (Li et al. 2017).
The tester strains were grown on MYEA for 7 d at room temperature and lighting before spermatization. Sterile water was added to the male tester, and a suspension of conidia, conidiophores and mycelial fragments was scraped from the agar surface. An aliquot of 1–2 mL of the suspension was dispersed over the colony of the female tester. The plates were incubated at room temperature and lighting for 7 d and examined through a dissecting microscope for the presence of ascomata and ascospore masses that accumulated at the tip of the ascomatal necks. If ascospore masses were evident, the ascospores were examined and rated by making microscope slides of ascospore masses from three ascomata mounted in cotton blue and examining at 1125× with Normarski interference microscopy. Mounts of normal ascospore masses had greater than 90 % uniform ascospores, while abnormal ascospore masses had 10 % or more of the spores misshapen or without cytoplasm, and there was generally debris within the spore mass, assumed to be aborted asci (Li et al. 2017).
Germination of the ascospores from the crosses was evaluated by placing an ascospore mass from atop one perithecium into 25 μL of light oil (Isopar M) on a MYEA plate. Small amounts of the oil suspension were taken with a wire loop (bacterial loop) and thoroughly streaked across each of two additional plates of MYEA. The remaining oil suspension on the original plate was also thoroughly spread with the wire loop. Germination was evaluated on the three plates after 6–8 d incubation at room temperature (Li et al. 2017). If the colonies on the three plates were too numerous to count (TNTC), then the germination percentage was considered high. If fewer than 40 total colonies developed on the three plates, the germination was considered low. If there were 40 or more total colonies on the three plates, but sill countable, then the germination was considered medium. The phenotypes of the colonies were also evaluated after incubating for 1–2 wk to confirm that a cross had occurred, that is, that the progeny colonies were from a genetic recombination between the two parent strains and not from a selfing event. In the case of recombination, the streaks of the ascospore masses resulted in colonies of differing phenotype (the fluffy mycelial phenotype of the male parent and the flat phenotype of the female parent), and some of the colonies produced ascomata and ascospores (self-fertile, MAT2), while others produced no ascomata.
Pathogenicity tests
Metrosideros polymorpha plants were propagated from seeds or cuttings taken from 6–12-mo-old seedlings and grown in 10 cm pots. Platanus × acerifolia ‘Bloodgood’ was imported as 1.5 m bare rooted whips and planted into 38 L pots. Colocasia esculenta ‘Lehua’ and ‘Bun-long’ plants were propagated from corms grown in 15 cm pots, divided and individually transplanted into 15 cm pots. Syngonium podophyllum cv. ‘White Butterfly’ plants were propagated by rooting cuttings from locally purchased stock plants and grown in 10 cm pots. All host plants were grown in Sunshine Mix #4 (Sun Gro Horticulture, Agawam, MA, USA), fertilized every 3 months with Nutricote Total 13-11-11 Type 100 (Arysta LifeScience) and watered twice daily for 5 min with an automatic irrigation system. All plants were grown in a greenhouse in Hilo, HI, with shade ranging from 30 to 72 %, except for C. esculenta, which was grown outdoors in a partially shaded area.
Pathogenicity tests were conducted on 1–2-year-old, ~3–12 mm diam, ~13–96 cm tall M. polymorpha plants; 1-mo-old, ~19–38 mm diam, ~147–177 cm tall Platanus × acerifolia; ~30 cm tall C. esculenta plants with only one fully expanded leaf; and 11-mo-old transplanted S. podophyllum plants. Trial inoculations of M. polymorpha using multiple isolates of Ceratocystis spp. were previously conducted to assess if variation in pathogenicity and/or virulence existed in the population. Since no variation was observed (data not shown), one isolate of Ceratocystis sp. A (P14-1-1), one isolate of Ceratocystis sp. B (P15-59), and two isolates of Ceratocystis from Syngonium (P16-1, P16-2) were used. Three to six plants were inoculated for each isolate and tests were repeated at least once. Plants were inoculated either by a stem injection method or a stem-flap wound method (Uchida & Aragaki 1979, Harrington 2004, Thorpe et al. 2005, Keith et al. 2015). For both methods, an inoculum suspension was produced from 7-d-old cultures grown on 10 % V8 agar at 24 °C under 24 h continuous fluorescent lighting and adjusted to 1×106 spores/mL using a Bright-Line haemocytometer. Meterosideros polymorpha, C. esculenta and S. podophyllum were inoculated with a 31 gauge insulin syringe (BD brand) with ± 1 μL of inoculum injected into the stems, pseudopetioles and pseudostems, respectively. Meterosideros polymorpha and Platanus × acerifolia were also wound inoculated using a sterile scalpel to produce a longitudinal stem flap ~0.5 mm deep × 10–15 mm long and the approximate width of the stem. Inoculum for stem flaps included 5 mm diam filter paper discs soaked in the conidial suspension, plated onto fresh 10 % V8 agar, and grown for 2 d at 24 °C under 24 h continuous fluorescent lighting. Two filter paper discs were inserted into the stem flap of Platanus × acerifolia plants; a single strip of filter paper was trimmed from one of the discs and inserted into the stem of M. polymorpha plants. Control plants were injected with sterile distilled water or inoculated with filter paper discs soaked in sterile distilled water. All stem flaps were wrapped with parafilm. All plants, except for Platanus × acerifolia, were placed in a growth chamber set at 24 °C with 12-h light and ~70 % relative humidity. Platanus × acerifolia plants were placed in a greenhouse with 70 % shade and daily temperatures ranging from 22 °C to 32 °C. All inoculated plants were observed for wilting of leaves, stem discolouration, and plant death every 3–7 d for as little as 2 wk and up to 1 yr depending on the host. Ceratocystis spp. were reisolated using carrot baiting and identified by morphology and PCR amplification and sequencing of the ITS region of rDNA.
RESULTS
Sampling and isolations
Trees exhibiting wilt symptoms of the foliage, including death of the entire canopy (Fig. 2, 3), were sampled by chopping into the trees with a hatchet or axe, while the stems of other trees were dissected with a chain saw. Internal signs of infection included brown to black discolouration of the woody xylem. The symptomatic trees lacked bark cankers, and mycelial mats and/or ascomata were not observed on the surface of intact bark of dead trees. Symptoms were not directly associated with obvious wounding. Tunnels and boring frass of ambrosia beetles were evident in dead trees at some locations, especially in the Puna district, where there was widespread mortality and boring dust/frass could be readily found.
Fig. 2.

Typical symptoms associated with Ceratocystis lukuohia (sp. A). a, b. Affected trees exhibiting rapid, synchronized death of leaves on a major trunk fork or the entire crown; c. dark brown to black vertical streaks of discolouration seen in the woody xylem of an affected tree; d–e. radiating pattern of sapwood discolouration in stem cross-sections of wilting trees; f–g. ascomata of C. lukuohia with long necks supporting sticky masses of ascospores.
Fig. 3.

Typical symptoms associated with Ceratocystis huliohia (sp. B). a. Affected trees exhibiting rapid, synchronized death of leaves on individual branches and spreading to the entire crown; b. brown to grey-black vertical streaks of discolouration in the woody xylem of an affected tree; c–e. blotchy, diffuse black to grey staining of sapwood; and thin black lines of host reaction following the contours of the cambium (arrows); f–g. ascomata of C. huliohia with necks supporting sticky masses of ascospores.
The majority of the sampled symptomatic trees exhibited vertical streaks in the wood that were dark brown-black in colour (Fig. 2). When transverse sections of the trunk were made, the darkest discolouration appeared to follow radially along the ray parenchyma (Fig. 2). In some of the sampled trees, including some completely dead trees and also some trees with partial crown death, a second pattern of wood discolouration was observed in which the vertical streaks were lighter, more diffuse and brown to grey-black in colour. Discolouration in transverse sections of the second pattern was typically diffuse and blotchy and in a few cases, black lines of discolouration appeared in the woody xylem that followed the contours of the cambium (Fig. 3, arrows).
Successful isolations of Ceratocystis spp. were made from both the radial and diffuse patterns of xylem discolouration using the carrot baiting technique. A total of 68 isolates were obtained from the discoloured xylem of wilted trees ranging in age from young saplings to mature trees at 17 different sites across Hawai’i Island (Table 1; Fig. 1; Appendix).
PCR and sequencing
The ITS region was amplified and sequenced for 64 isolates, with an amplicon size of ± 550 bp in length. The PCR products were directly sequenced with the PCR primers, but the electropherograms of the PCR products from the extracted DNA of 35 isolates showed overlapping base calls indicative of intragenomic variation among the repeats of the rDNA operon. These PCR products were cloned, and at least 4–6 cloned products were sequenced per isolate. Three ITS haplotypes were obtained from the cloned PCR products or from direct sequencing: A haplotype ‘A’ was obtained from 18 isolates, a haplotype ‘B’ was obtained from nine isolates, and a haplotype ‘C’ obtained from two isolates (Table 1; Appendix; GenBank accession numbers KY809157, KY809156 and KY809158). Both haplotype ‘A’ and ‘C’ sequences were obtained from the cloned fragments of each of the 35 samples that could not be directly sequenced. When the ITS sequences of haplotypes A (KY809157) and C (KY809158) were aligned with each other, they differed at six base positions: at site 103 in the ITS1 region (T in haplotype A vs C in haplotype C), at site 373 in the ITS2 region (an extra T present in haplotype C), and at sites 517–519 in the ITS2 region where three extra cytosine bases were present in haplotype A that were absent in haplotype C. No recombinants among the three polymorphisms were present in any of the direct sequences or cloned fragments of the A and C haplotypes. The haplotype A and C sequences were most similar to the ITS sequences of members of the LAC. The sequence of the haplotype B was most similar to the ITS sequences of members of the AAC.
Based on the results of the ITS sequencing, isolates representing the three haplotypes, and mixtures of haplotypes A and C from different locations on the island, were chosen for further PCR and sequencing of the bt1, ef1, ms204 and rpb2 loci. These amplified products were ~610 bp, ~780 bp, ~970 bp and ~1 200 bp, respectively.
Multi-gene phylogenetic analyses
The total number of aligned characters used for the phylogenetic analysis of the ITS (LAC) dataset (56 isolates), the ITS (AAC) dataset (48 isolates) and the combined dataset of bt1, ef1, ms204 and rpb2 (82 isolates) were 549, 630 and 3 264, respectively. The number of parsimony-informative characters were 157, 273 and 510, and the number of parsimony-uninformative characters were 66, 36 and 14, respectively. MP analyses for the ITS_LAC loci resulted in 205 most parsimonious trees with TL = 383, CI = 0.736, RI = 0.892 and RC = 0.656. The ITS_AAC loci resulted in 34 most parsimonious trees with TL = 571, CI = 0.758, RI = 0.917 and RC = 0.695. The combined dataset had 15 most parsimonious trees with TL = 763, CI = 0.765, RI = 0.952 and RC = 0.729. The log-likelihood of the most likely tree obtained from ML analyses were -1871.29 (ITS_LAC), -2203.75 (ITS_AAC) and -8552.56.55 (combined). Bootstrap confidence values > 60 % for both MP and ML analyses are presented on a single most parsimonious tree based on MP for each dataset (Fig. 4, 5, 6).
Fig. 4.
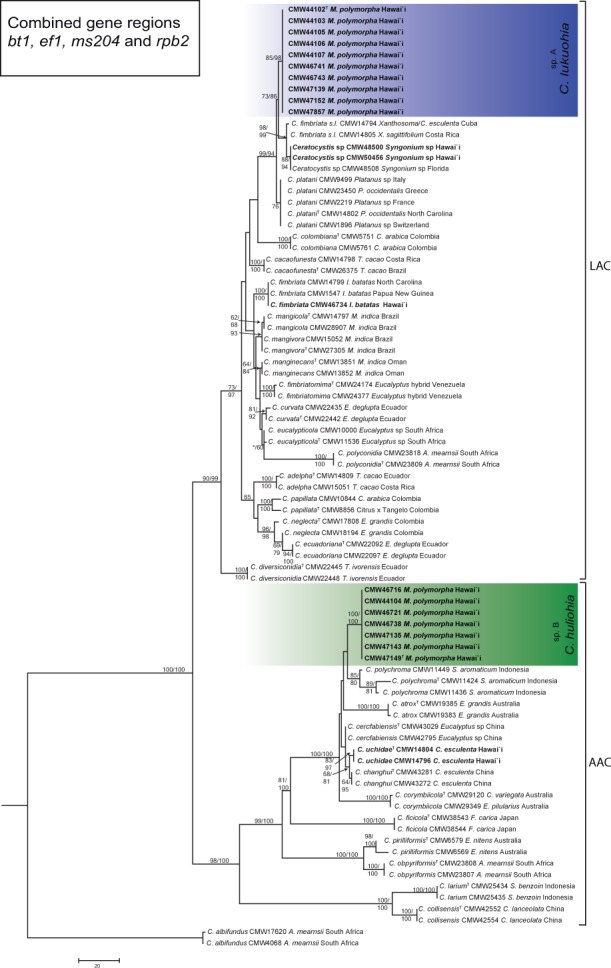
Maximum parsimony tree based on the combined dataset of four gene regions (bt1, tef1, ms204 and rpb2) for species in the Latin American clade (LAC) and the Asian-Australian clade (AAC) of Ceratocystis. Isolates sequenced in this study from M. polymorpha in Hawai’i are highlighted in blue in the LAC, as C. lukuohia (sp. A), and in green in the AAC, as C. huliohia (sp. B). Bootstrap values > 60 % for MP/ML are presented at the branches. Bootstrap values lower than 70 % are indicated with *. Ceratocystis albifundus (CMW 4068) from the African clade was used as the outgroup.
Fig. 5.

Phylogenetic tree based on MP of the ITS-rDNA of Ceratocystis species in the Latin American clade (LAC). Two ITS haplotypes (Group 1 and Group 2) were evident in the isolates of C. lukuohia (sp. A) from M. polymorpha in Hawai’i. These grouped closest to isolates from Xanthosoma sp. from Cuba and Costa Rica and Syngonium sp. from Hawai’i and Florida. Bootstrap values > 70 % for MP/ML are presented at the branches. Ceratocystis albifundus (CMW 4068) from the African clade was used as the outgroup.
Fig. 6.
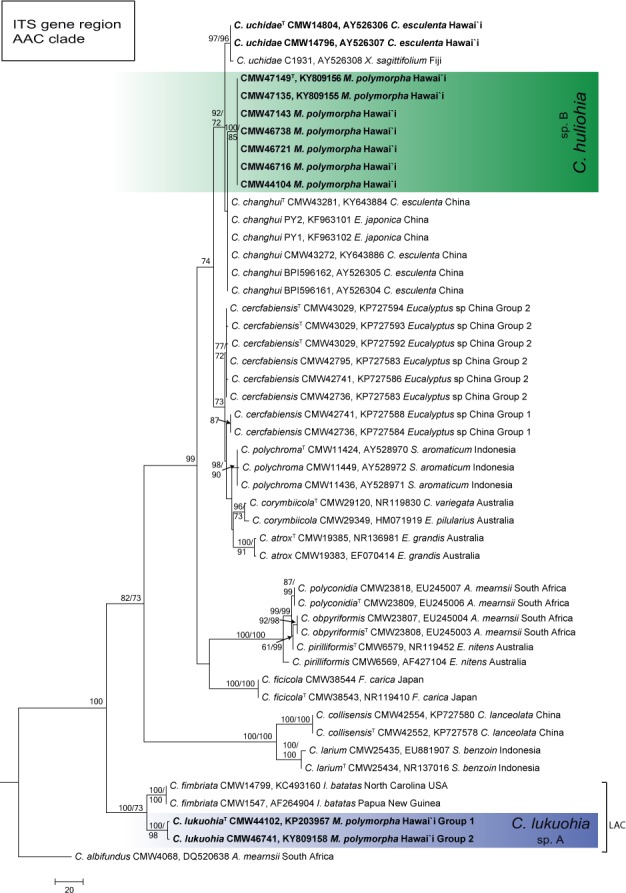
Phylogenetic tree based on MP of the ITS-rDNA of Ceratocystis species in the Asian-Australian clade (AAC). Ceratocystis huliohia (sp. B) forms a monophyletic clade and is closest to species of C. uchidae, also described from Hawai’i and C. changhui. Bootstrap values > 70 % for MP/ML are presented at the branches. Ceratocystis albifundus (CMW 4068) from the African clade was used as the outgroup.
For all loci tested, the isolates from M. polymorpha in Hawai’i separated into two biogeographical clades, viz. LAC and AAC in both the ML and MP analyses (Fig. 4, 5, 6). Although the ITS phylogenies differed in topology with that of the combined phylogeny, the isolates from M. polymorpha formed two monophyletic lineages supported with high bootstrap values and were distinct from all other described Ceratocystis species. These unique lineages represent two new species of Ceratocystis from Hawai’i, hereafter referred to as Ceratocystis sp. A and Ceratocystis sp. B.
In both the ITS_LAC and the combined phylogeny (Fig. 4, 5), one of the emerging new taxa (Ceratocystis sp. A) grouped most closely with isolates collected from Xanthosoma sp. from Cuba and Costa Rica and those from Syngonium sp. from Hawai’i and Florida. There was little or no sequence difference observed between Ceratocystis sp. A and these isolates based on the bt1 and ef1 loci. Ceratocystis sp. A differed, however, from these isolates by up to six and eight fixed polymorphisms, respectively, across all five loci (Table 2). In the combined phylogeny (Fig. 4), Ceratocystis sp. A also grouped closely to C. platani (differing at five fixed nucleotide sites), C. cacaofunesta (differing at 29 sites) and C. colombiana (differing at 35 sites) (Table 2). In the ITS_LAC phylogeny, the two ITS types of Ceratocystis sp. A (haplotypes A and C) were clustered together and were designated as Group 1 and Group 2 in Fig. 5.
Table 2.
Number of fixed nucleotide differences between Ceratocystis lukuohia (sp. A) and close relatives of the Latin American clade, and C. huliohia (sp. B) and close relatives from the Asian-Australian clade, observed at five loci used in the phylogenetic analyses.
| ITS | bt1 | ms204 | rpb2 | ef1 | Total | |
|---|---|---|---|---|---|---|
| C. lukuohia compared with | ||||||
| C. cacaofunesta | 33 | 3 | 19 | 6 | 1 | 61 |
| C. colombiana | 30 | 2 | 18 | 10 | 5 | 69 |
| C. fimbriata (Ipomoea batatas) | 29 | 5 | 18 | 6 | 1 | 55 |
| Ceratocystis sp. from Syngonium | 2 | 0 | 2 | 5 | 1 | 8 |
| Ceratocystis sp. from Xanthosoma | 2 | 0 | 2 | 4 | 0 | 6 |
| C. platani | 13 | 1 | 1 | 2 | 1 | 18 |
| C. huliohia compared with | ||||||
| C. atrox | 36 | 6 | 6 | 6 | 13 | 67 |
| C. cercfabiensis | 23 | 2 | 1 | 4 | 5 | 35 |
| C. changhui | 9 | 0 | 2 | 4 | 5 | 20 |
| C. corymbiicola | 32 | 4 | 20 | 4 | 7 | 67 |
| C. polychroma | 34 | 2 | 0 | 6 | 5 | 47 |
| C. uchidae | 13 | 0 | 1 | 5 | 6 | 25 |
Within the AAC, Ceratocystis sp. B formed a distinct lineage in the C. polychroma cluster and separated from six closely related species (C. uchidae C. changhui, C. polychroma, C. atrox, C. cerfabiensis and C. corymbiicola) with a minimum of 20 fixed nucleotide differences across the five loci (Table 2; Fig. 4, 6). In the ITS_AAC phylogeny (Fig. 6), Ceratocystis sp. B was sister to C. changhui, known only from China, and C. uchidae, known from Hawai’i and Fiji. In the combined analyses, Ceratocystis sp. B was closest to C. polychroma, known from S. aromaticum from Indonesia. In the AAC, the ef1 and the ITS loci showed the greatest differences among species. Ceratocystis sp. B could not be distinguished from C. changhui and C. uchidae using bt1 or from C. polychroma using ms204.
Culture characteristics and morphology
Isolates representing the different ITS types of Ceratocystis sp. A (CMW 44102/P14-1-1 = haplotype A, CMW 46741/P15-64 = haplotype C and CMW 47152/P15-65 = mixture of haplotypes A and C) and the haplotype B of Ceratocystis sp. B (CMW 47149/P15-58 and CMW 47135/P14-10) were used to characterise the culture and morphological characteristics of these fungi (Table 3; Fig. 7, 8, 9, 10).
Table 3.
Morphological measurements (in μm) of isolates of Ceratocystis lukuohia (sp. A) and C. huliohia (sp. B) on 2 % MEA taken after 2 wk of growth.
| C. lukuohia CMW 44102/P14-1-1 (ITS_A) | C. lukuohia CMW 47152/P15-65 (ITS_AC) | C. lukuohia CMW 46741/P15-64 (ITS_C) | C. huliohia CMW 47149/P15-58 | C. huliohia CMW 47135/P14-10 | |
|---|---|---|---|---|---|
| Ascomatal base | (151–)161.5–239(–308) × (146.5–)153.5–241.5(–311) | (157.5–)173.5–242(–271) × (142–)169–229(–250.5) | (134–)163–244.5(–306.5) × (121–)161.5–244.5(–272.5) | (128–)175.5–258.5(–310.5) × (116–)156–234(–287.5) | (169.5–)196.5–279(–331.5) × (161–)182–257(–321.5) |
| Av. ascomatal base | 200 × 198 | 208 × 199 | 204 × 203 | 217 × 195 | 238 × 220 |
| Neck length | (324.5–)534.5–833.5(–971.5) | (560–)727–1022.5(–1244) | (503.5–)690.5–1002.5(–1144.5) | (287.5–)480–824.5(–928.5) | (504–)573.5–781(–871.5) |
| Av. neck length | 684 | 875 | 847 | 652 | 677 |
| Neck width at base | (12–)22–37(–42.5) | (16–)23.5–31.5(–33.5) | (25–)28.5–35.2(–42.5) | (26–)31–40(–43) | (24–)29.5–39(–41.5) |
| Av. neck width at base | 30 | 28 | 32 | 35.5 | 34 |
| Neck width at tip | (9–)15–26(–31.5) | (13–)15–21(–28) | (15–)16–20(–22) | (13.5–)16.5–24(–34) | (14–)16.5–22(–28) |
| Av. neck width at tip | 20 | 18 | 18 | 20 | 19 |
| Ostiolar hyphae length | (40–)56.5–89.5(–111.5) | (38.5–)49–82.5(–98) | (48–)61.5–86.5(–102) | (30–)37.5–56(–70) | (26.5–)29–49.5(–65) |
| Av. ostiolar hyphae length | 73 | 66 | 74 | 47 | 39 |
| Conidiogenous cell length | (28–)42.5–65.5(–81) | (24.5–)38–53.5(–57.5) | (22.5–)29.5–86.5(–178.5) | (48.5–)50–64.5(–72) | (20.5–)26.5–51(–61.5) |
| Av. conidiogenous cell length | 54 | 46 | 58 | 57 | 39 |
| Cylindrical conidia | (10–)13–19(–24.5) × (3.5–)4–5 | (9.5–)12–19.5(–24) × (2–)3–4(–6) | (11–)14–26(–34.5) × 3–4.4(–6) | (12–)16.5–23.5(–28.5) × (2.5–)3–4(–4.5) | (10–)13–23(–39.5) × 3–4(–5) |
| Av. cylindrical conidia length | 16 × 4 | 16 × 4 | 20 × 4 | 20 × 3.5 | 18 × 4 |
| Aleurioconidia | (11–)13–15(–16.5) × (8.5–)9.5–10.5(–11.5) | (11–)12–14(–14.5) × (7.5–)8.5–10.5(–12) | (12–)12.5–15.5(–18) × (8–)9.5–11(–11.5) | (10–)12–14.5(–17) × (8–)9.5–12(–14) | (10–)11.5–13.5(–14.5) × (9.5–)10–12(–13.5) |
| Av. aleurioconidia | 14 × 10 | 13 × 10 | 14 × 10 | 13 × 11 | 13 × 11 |
| Ascospores top view | (4.5–)5–5.5(–6) × (3.5–)4–4.5(–5) | (4–)4.5–5.5 × (3–)3.5–4.5(–5) | (5–)5.5(–6) × 4–4.5 | (4–)4.5–5.5(–6) × 3.5–4(–4.5) | 4.5–5.5(–6) × (3–)3.5–4(–4.5) |
| Av. ascospores top view | 5 × 4 | 5 × 4 | 5 × 4 | 5 × 4 | 5 × 4 |
| Ascospores side view | (4.5–)5–6 × 2.5–3(–3.5) | (4–)4.5–5.5 × (2–)2.5–3(–3.5) | 5–6 × (2–)2.5–3(–3.5) | (3.5–)4.5–5(–5.5) × (1.5–)2–2.5(–3) | (3–)4–5(–6) × 2–3(–3.5) |
| Av. ascospores side view | 5 × 3 | 5 × 3 | 5.5 × 3 | 5 × 2 | 4 × 2 |
Fig. 7.
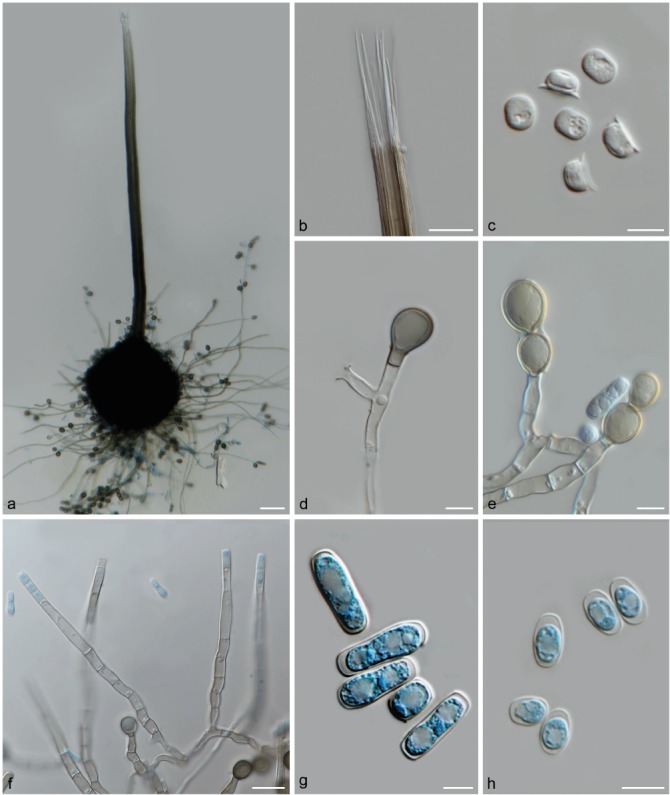
Morphological characteristics of Ceratocystis lukuohia (sp. A). a. Globose ascomata base with long neck; b. straight ostiolar hyphae; c. hat-shaped ascospores from top and side view; d. terminal thick-walled aleurioconidium; e. aleurioconidia in chains; f. flask-shaped conidiophores; g. cylindrical conidia; h. short, barrel-shaped conidia. — Scale bars: a = 50 μm; b, f = 20 μm; c, g–h = 5 μm; d–e = 10 μm.
Fig. 8.
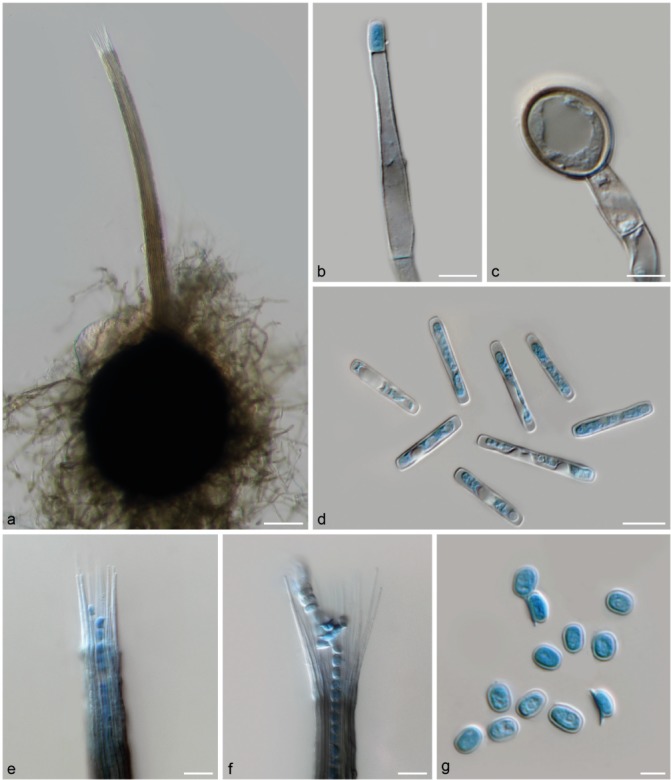
Morphological characteristics of Ceratocystis huliohia (sp. B). a. Ascomata with globose to obpyriform base and short neck; b. conidiophore with emerging, short, barrel-shaped conidium; c. terminal aleurioconidium; d. cylindrical conidia; e. short, straight ostiolar hyphae; f. divergent ostiolar hyphae; g. hat-shaped ascospores in side and top view. — Scale bars: a = 50 μm; b, d–f = 10 μm; c, g = 5 μm.
Fig. 9.
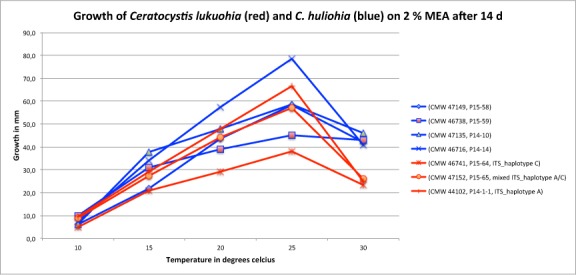
Diameter of colonies in mm of Ceratocystis lukuohia (sp. A) and C. huliohia (sp. B) isolates on 2 % malt extract agar at five different temperatures taken after 14 d growth. Ceratocystis huliohia grows slightly faster than C. lukuohia. Both species have optimal growth at 25 °C.
Fig. 10.
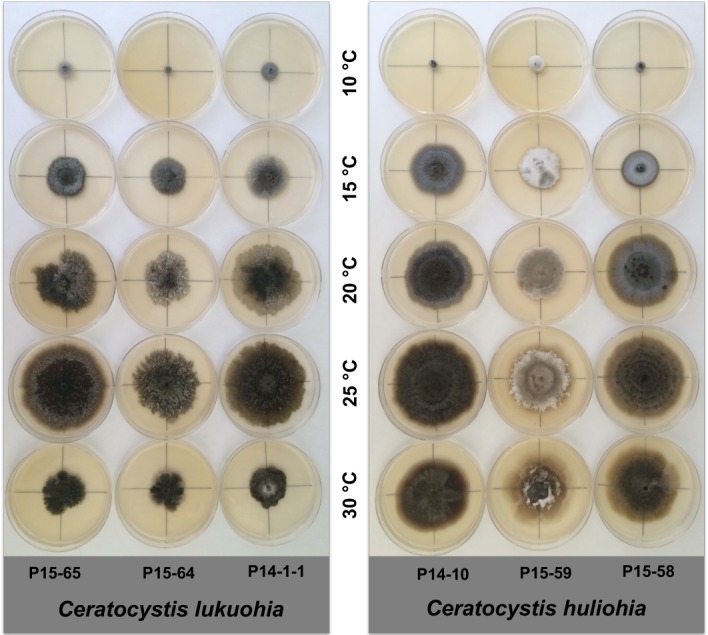
Variation in culture morphology of isolates of Ceratocystis lukuohia (sp. A) and C. huliohia (sp. B) after 14 d growth on 2 % MEA at temperatures 10, 15, 20, 25 and 30 °C.
Ceratocystis sp. A and Ceratocystis sp. B could be distinguished from each other based on culture characteristics and some morphological features. In culture, the colony growth of Ceratocystis sp. A was circular to irregular in form with undulate to lobate colony margins, whereas the colonies of Ceratocystis sp. B were circular with smooth to undulate colony margins (Fig. 10). The cultures of Ceratocystis sp. A were olivaceous green to brown in colour due to the production of excessive quantities of aleurioconidia in long chains in the medium, with grey aerial mycelium. Cultures of Ceratocystis sp. B varied in colour from white to grey to olivaceous green. Sectoring occurred in both taxa. Some isolates of Ceratocystis sp. B produced cultures with concentric rings of black ascomata (Fig. 10), and occasionally, multiple necks were observed on single ascomatal bases, particularly in culture CMW 47135/P14-10. In Ceratocystis sp. A, the light- to dark-brown to black ascomata were generally scattered throughout the colony. The ascomatal bases of Ceratocystis sp. A were generally more globose (av. = 204 × 200 μm) than the more pyriform bases (av. = 227.5 × 207.5 μm) of Ceratocystis sp. B, but the differences were minor.
The most obvious morphological characters distinguishing Ceratocystis sp. A and Ceratocystis sp. B were the average lengths of the necks and ostiolar hyphae. Ceratocystis sp. A had longer necks (av. ranging from 684–875 μm among the three isolates) and ostiolar hyphae (av. ranging from 66–74 μm among the three isolates) compared to the shorter necks (av. of the two isolates = 652 and 677 μm) and ostiolar hyphae (av. of the two isolates = 39 and 47 μm) of Ceratocystis sp. B. Both cylindrical conidia and the thick-walled aleurioconidia were more abundant in cultures of Ceratocystis sp. A, and the aleurioconidia of Ceratocystis sp. A were produced in long chains, whereas they were only produced in short chains in Ceratocystis sp. B.
Growth in culture
For three isolates of Ceratocystis sp. A (CMW 46741, P15-64; CMW 47152, P15-65 and CMW 44102, P14-1-1) and for four isolates of Ceratocystis sp. B (CMW 47149, P15-58; CMW 46716, P14-14; CMW 46738, P15-59 and CMW 47135, P14-10), the optimal temperature for growth was 25 °C (Fig. 9). Overall, Ceratocystis sp. B grew faster than Ceratocystis sp. A at 15, 20, 25 and 30 °C and grew an average of 4.3 mm/d at 25 °C, while Ceratocystis sp. A grew an average of 3.6 mm/d. For both taxa, there was a significant reduction in growth at 10 °C. Although there was a sharp decline in growth at 30 °C as compared to 25 °C for both taxa, Ceratocystis sp. B (43 mm diam) had almost double the growth of Ceratocystis sp. A (25 mm diam) at this temperature after 14 d. For both taxa, ascomatal production was most abundant when grown at 20 and 25 °C, and no ascomata were produced at 10 and 30 °C.
TAXONOMY
Phylogenetic inference based on five gene regions showed conclusively that there are two distinct taxa associated with the death of M. polymorpha trees on the Island of Hawai’i. Ceratocystis sp. A and Ceratocystis sp. B reside in the LAC and AAC biogeographical clades of Ceratocystis s.lat., respectively, and they are phylogenetically distinct from the other described species in those clades. These two putative new taxa could be distinguished from each other based on morphology. These results provide robust evidence to justify describing the two taxa as new species as follows (where Ceratocystis sp. A = Ceratocystis lukuohia and Ceratocystis sp. B = Ceratocystis huliohia):
Ceratocystis lukuohia I. Barnes, T.C. Harr. & L.M. Keith, sp. nov. — MycoBank MB824050; Fig. 2, 7
Etymology. The name lukuohia is derived from the Hawaiian words luku (destruction) and ‘ōhi’a, and reflects the fact that this fungus destroys ‘ōhi’a trees.
Type. USA, Hawai’i, Puna District, Leilani Estates, isolated from diseased Metrosideros polymorpha, 20 Feb. 2014, J.B. Friday (holotype PREM 61770, culture ex-type CMW 44102 = P14-1-1 = CBS 142792 = MUCL 56342). ITS sequence GenBank KY809157.
Ascomata perithecial, developing within 8 d and mature within 12 d, mostly scattered, sometimes gregarious, partially embedded or superficial in agar; bases subglobose to globose (151–)161.5–239(–308) × (146.5–)153.5–241.5(–311) μm, dark brown to black, covered with undifferentiated hyphae and conidiophores with aleurioconidia. Necks straight, occasionally curved, (324.5–)534.5–833.5(–971.5) μm long, smooth, brown to black at base (12–)22–37(–42.5) μm wide and becoming paler towards tapering apex (9–)15–26(–31.5) μm wide. Ostiolar hyphae mostly straight, occasionally divergent, non-septate, (40–)56.5–89.5(–111.5) μm long, hyaline.Asci evanescent, not seen. Ascospores accumulate in white to creamy masses at tips of necks, single-celled, hyaline, ellipsoidal in top view (4.5–)5–5.5(–6) × (3.5–)4–4.5(–5) μm with gelatinous sheath giving hat-shaped appearance in side view (4.5–)5–6 × 2.5–3(–3.5) μm. Conidiophores branched or unbranched, straight or flexuous, hyaline to pale brown, multiseptate, smooth-walled. Conidiogenous cells endophialidic with enteroblastic conidium ontogeny, flask-shaped (lageniform) or tubular (28–)42.5–65.5(–81) μm long. Conidia mostly cylindrical with apices rounded, smooth, non-septate, hyaline, (10–)13–19(–24.5) × (3.5–)4–5 μm, borne in long chains; other conidia barrel-shaped with truncate bases, smooth-walled, non-septate, hyaline, 4–5.5(–6.5) × 3–4.5(–5) μm. Aleurioconidia abundant, pale brown to brown, pyriform to ovoid (11–)13–15(–16.5) × (8.5–)9.5–10.5(–11.5) μm, thick-walled, formed singly or in long chains.
Cultural characteristics — Colonies on 2 % MEA circular to irregular with undulate to lobate colony margins, mycelium submerged to aerial, producing fruity aroma; submerged mycelium greenish olivaceous, mainly due to aleurioconidia; aerial mycelium light grey, frequently sectoring. Optimal temperature for growth 25 °C, with colony diam reaching 66.5 mm after 14 d. Optimal temperature for ascomatal production 20–25 °C.
Substratum — Woody xylem of trunk, stems and branches of M. polymorpha.
Distribution — Hawai’i Island, HI, USA.
Additional specimens. USA, Hawai’i, Puna District, Nānāwale, isolated from diseased M. polymorpha, 3 June 2015, T. Sowards (paratype PREM 61771, culture ex-paratype CMW 46741 = P15-64 = CBS 142793 = MUCL 56343) (ITS haplotype C); South Hilo District, Waiākea Forest Reserve, isolated from diseased M. polymorpha, 27 May 2015, T. Sowards (paratype PREM 61772, culture ex-paratype CMW 47152 = P15-65 = CBS 142591 = MUCL 56344) (ITS haplotype A/C).
Notes — Typical symptoms associated with C. lukuohia include rapid, synchronized death of the entire crown and extensively stained sapwood.
Ceratocystis lukuohia has two different ITS haplotypes (GenBank KY809157 and KY809158), and most isolates appear to have a mixture of the two haplotypes among the tandem repeats of the rDNA operons. It is phylogenetically most closely related to C. platani (Engelbrecht & Harrington 2005) and an undescribed species of Ceratocystis isolated from Xanthosoma sp. in Cuba and Costa Rica and from Syngonium in Hawai’i and Florida (Thorpe et al. 2005). Ceratocystis lukuohia can be distinguished from these Ceratocystis spp. based on ITS, ms204 and rpb2 and from C. platani with ITS, ms204, rpb2, ef1 and bt1 sequence data. Morphologically, C. lukuohia is very similar to these other Ceratocystis spp. and they cannot reliably be distinguished from each other, although isolates from Syngonium and Xanthosoma generally have less aerial mycelium.
Ceratocystis huliohia I. Barnes, T.C. Harr. & L.M. Keith, sp. nov. — MycoBank MB824051; Fig. 3, 8
Etymology. The name huliohia is derived from the Hawaiian words huli (to turn) and ‘ōhi’a and refers to the fact that this fungus changes the natural state of ‘ōhi’a trees.
Type. USA, Hawai’i, North Kona District, Hōlualoa, isolated from diseased M. polymorpha, 10 June 2015, J.B. Friday (holotype PREM 61769, culture ex-type CMW 47149 = P15-58 = CBS 142794 = MUCL 56340). ITS sequence GenBank KY809156.
Ascomata perithecial, developing within 8 d and mature within 12 d, aggregated, partially embedded in agar; bases subglobose to pyriform (128–)175.5–258.5(–310.5) × (116–)156–234(–287.5) μm, dark brown to black, covered with undifferentiated hyphae. Spines and ornamentations absent. Necks straight or occasionally slightly curved and (287.5–)480–824.5(–928.5) μm long, wider at base (26–)31–40(–43) μm and tapering towards apex (13.5–)16.5–24(–34) μm, unbranched, light brown to dark brown or black, smooth to crenulate. Ostiolar hyphae straight to mostly divergent, non-septate, hyaline to light brown, (30–)37.5–56(–70) μm long. Asci evanescent, not seen. Ascospores accumulate in white to creamy masses at tips of necks, single-celled, hyaline, ellipsoidal in top view (4–)4.5–5.5(–6) × 3.5–4(–4.5) μm with gelatinous sheath giving hat-shaped appearance in side view (3.5–)4.5–5(–5.5) × (1.5–)2–2.5(–3) μm. Conidiophores branched or unbranched, straight or flexuous, hyaline to pale brown, multiseptate, smooth-walled. Conidiogenous cells endophialidic, flask-shaped (lageniform) (48.5–)50–64.5(–72) μm in length producing primary conidia in chains by means of enteroblastic conidium ontogeny. Conidia hyaline, non-septate, smooth-walled, cylindrical or bacilliform with rounded apices, mostly (12–)16.5–23.5(–28.5) μm long × (2.5–)3–4(–4.5) μm wide. Aleurioconidia ovoid, thick-walled, hyaline when young, becoming brown when mature, smooth, formed mostly singly (terminal) or in short chains, (10–)12–14.5(–17) × (8–)9.5–12(–14) μm in size.
Culture characteristics — Colonies on 2 % MEA showing circular growth with entire (smooth) to undulate colony margins. Mycelium submerged to aerial, producing fruity aroma; submerged mycelium greenish olivaceous, aerial mycelium light grey, turning dark olivaceous green to brown with age. Ascomata/perithecia either produced in clumps or in a concentric ‘ring-like’ fashion. Optimal temperature for growth 25 °C, with colony diam reaching 58.4 mm after 14 d. Optimal temperature for ascomatal production 20–25 °C.
Substratum — Woody xylem of trunk, stems and branches of M. polymorpha.
Distribution — Hawai’i Island, HI, USA.
Additional specimens. USA, Hawai’i, Puna District, ‘Opihikao, isolated from diseased M. polymorpha, 8 July 2014, T. Sowards (paratype PREM 61768, culture ex-paratype CMW 47135 = P14-10 = CBS 142795 = MUCL 56341).
Notes — Ceratocystis huliohia has been isolated from stained sapwood of trees showing rapid, synchronized death of major branches or death of the entire crown. However, it has also been isolated from trees that have appeared to be dying gradually over a period of years. Phylogenetically, C. huliohia groups in the C. polychroma cluster of the AAC and is closest to C. uchidae, C. changhui, C. cercfabiensis and C. polychroma, which are generally less aggressive plant pathogens than members of the LAC. Morphological features within individuals of the same species can show great variation in the dimensions of measured morphological characteristics and in some cases, these overlap with other species (Li et al. 2017, Liu et al. 2018). However, based on average measurements, C. huliohia can be distinguished from its closest relatives based on ascomatal size, the length of the neck and ostiolar hyphae (Li et al. 2017, Liu et al. 2018). The ascomatal bases of C. huliohia (av. of two isolates = 227.5 × 207.5 μm, Table 3) are somewhat larger than those of C. uchidae (range 95–190 diam), C. changhui (av. = 182 × 180.5 μm) and C. cercfabiensis (av. = 178 × 184 μm). The ascomatal necks of C. huliohia (av. of two isolates = 664.5 μm) are longer than C. uchidae (av. = 422.5 μm) but shorter than C. changhui (av. = 702 μm), C. cercfabiensis (av. = 1 114.5 μm) and C. polychroma (av. = 960 μm). The length of ostiolar hyphae of C. huliohia (av. of two isolates = 43 μm) is shorter than that of C. uchidae (av. = 57.5 μm), C. changhui (av. = 82 μm) and C. cercfabiensis (av. = 59 μm), but slightly longer than C. polychroma (av. = 38 μm).
Ceratocystis lukuohia and C. huliohia can clearly be differentiated from each other based on culture characteristics and morphology. On MEA, C. lukuohia grows more slowly than C. huliohia (Fig. 9) and has smaller, more rounded ascomata that are distributed evenly in olivaceous green to brown cultures. Sectoring is more common in cultures of C. lukuohia than in cultures of C. huliohia, but both species frequently change mycelial phenotype. The culture colour in C. huliohia varies considerably and can be white, to grey, to olivaceous green (Fig. 10). The larger ascomata of C. huliohia are often clumped together (Fig. 3) and in some cases, multiple necks are seen from the same ascomatal base. In both species, optimal growth is at 25 °C (Fig. 9) with ascomata production occurring between 20–25 °C. The most distinguishing morphological features of these two species are the longer necks and ostiolar hyphae in C. lukuohia. Ceratocystis lukuohia also produces prolific amounts of conidia and aleurioconidia in long chains.
Genotyping of Ceratocystis isolates
The 10 C. lukuohia isolates used in the phylogenetic studies for the LAC were clonal at 27 microsatellite loci. The remaining 49 isolates that were screened with a reduced number of markers (10–20) were also all clonal (Appendix). Isolates having the A ITS haplotype (18 isolates), the C ITS haplotype (2 isolates) or mixtures of A and C haplotype (35 isolates) did not differ in their multilocus microsatellite profile. The six Ceratocystis isolates from Xanthosoma and Syngonium, which were phylogenetically closest to C. lukuohia, had ± 20 % alleles in common with this species. Similarly, the clonal lineages of C. platani isolates from continental USA, Greece, Switzerland, France and Italy were identical to C. lukuohia at only 33.3 % of the scorable loci (Appendix). The multilocus genotype (MLG) of the C. fimbriata isolate from sweet potato in Hawai’i was very different to that of C. lukuohia and only shared 7.4 % of the total alleles.
The nine C. huliohia isolates from Hawai’i all shared the same MLG and were, therefore, clonal at all 27 microsatellite loci. Isolates of C. huliohia shared 47.80 % similar alleles with C. polychrome, 52.17 % with C. uchidae, 60 % with C. changhui and 70.83 % with C. cercfabiensis. The two Ceratocystis species causing disease on M. polymorpha only shared a single allele, out of a total of 26. Primer set AF6 did not amplify in either species.
Interfertility tests
In the parings among closely-related species in the LAC or in pairings among closely-related species in the AAC, almost all of the crosses between male, MAT2 and female, MAT1 testers resulted in ascomata with ascospore masses at the tips of their necks. Ascospores germinated when they were streaked onto the surface of MYEA, but the germination percentages were relatively low in crosses between different species (Table 4, 5). When ascospores were produced in either interspecific or intraspecific pairings, progeny arising from the ascospores plated onto MYEA showed some variation in the mycelial phenotype (colony texture and in presence/absence of ascomata), which suggested that sexual recombination had occurred. However, microscopic examination of the ascospores showed substantial variation among interspecific crosses in the abundance and proportion of misshapen ascospores. In most cases of pairings between two strains of the same species, less than 10 % of the ascospores were misshapen or empty, but in most interspecific pairings, greater than 10 % of the ascospores were misshapen (Table 4, 5). In most of the cases, where a pairing of isolates was conducted twice, the identical results were obtained.
Table 4.
Mating reactions (production of normal or abnormal ascospores/with high, medium, low or no germination) in two replicated pairing experiments between MAT1, female tester strains of Ceratocystis spp. in the Latin American clade and MAT2, male testers of Ceratocystis lukuohia, C. platani and Ceratocystis sp. from Syngonium.
| MAT2 males |
||||||
|---|---|---|---|---|---|---|
|
C. lukuohia |
C. platani
|
Ceratocystis sp. from Syngonium |
||||
| MAT1 females | From C4133 | From C4185 | From C4186 | From C1342 | From C1774 | |
| C. lukuohia | From C4185 | Normal/high, Normal/high1 | Normal/high, Normal/high | Normal/high, Normal/high | Abnormal/NT, Abnormal/medium | Abnormal/medium, Normal/medium |
| From C4210 (P15-64) | Abnormal/high, Normal/high | Normal/high, Normal/high | Normal/high, Normal/high | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | |
| From C4212 (P14-1-1) | Abnormal/high, Abnormal/high | Normal/high, Abnormal/high | Normal/high, Abnormal/high | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | |
| From C4213 (P15-65) | Abnormal/high, Normal/high | Normal/high, Normal/high | Normal/high, Normal/high | No ascomata, Abnormal/medium | Abnormal/medium, Abnormal/medium | |
| C. platani | From C1339 | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Normal/high, Normal/high, | Abnormal/low, Abnormal/medium |
| From C1329 | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Normal/high, Normal/high, | Abnormal/medium, Abnormal/medium | |
| Ceratocystis sp. from Syngonium | From C1717 | Abnormal/high, Abnormal/high | Abnormal/medium, Abnormal/low | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Normal/medium, Normal/medium, |
| From C4118 | Abnormal/medium, Abnormal/medium | Abnormal/high, Normal/high | Abnormal/high, Abnormal/medium | Abnormal/medium, Abnormal/medium | Normal/high, Normal/high | |
| C. colombiana | From C1945 | Abnormal/low, Abnormal/medium | Abnormal/low, Abnormal/medium | Abnormal/low, Abnormal/low | Abnormal/medium, Abnormal/medium | Abnormal/low Abnormal/low |
| C. cacaofunesta | From C1833 | Abnormal/low, Abnormal/none | Abnormal/none, Abnormal/none | Abnormal/low, Abnormal/none | Abnormal/none, Abnormal/low | Abnormal/low, Abnormal/medium |
| C. fimbriata | From C4135 | No ascomata, No ascomata | No ascomata, No ascomata | No ascomata, No ascomata | No ascomata, No ascomata | No ascomata, No ascomata |
1 Microscopic examination of abnormal ascospore masses showed more than 10 % of the ascospores misshapen or empty and typically with ascus or ascospore debri. Streaked ascospore mass on malt yeast extract agar: high germination = colonies too numerous to count on three plates; medium germination = 40 or more colonies on three plates; and low germination = fewer than 40 colonies on the three plates. Pairings or ascospore germinations not tested are indicated by NT.
Table 5.
Mating reactions (production of normal or abnormal ascospores/with high, medium, low or no germination) in two replicated pairing experiments between MAT1, female tester strains of Ceratocystis spp. from the Asian-Australian clade and MAT2, male testers of Ceratocystis huliohia, C. uchidae, C. cercfabiensis and C. polychroma.
| MAT2 males |
||||||
|---|---|---|---|---|---|---|
|
C. huliohia |
C. uchidae |
C. cercfabiensis |
||||
| MAT1 females | From C4215 (P15-59) | From C4192 | From C1714 | From C1931 | From C3358 | |
| C. huliohia | From C4191 | Normal/high, Normal/high1 | Normal/high, Normal/high | Abnormal/low, Abnormal/low | Abnormal/medium, Abnormal/medium | Abnormal/medium, NT |
| From C4192 | Normal/high, Normal/high | Normal/high, Normal/high | Abnormal/medium, Abnormal/low | Abnormal/medium, NT | Abnormal/low, Abnormal/medium | |
| From C4211 | Normal/high, Normal/high | Normal/high, Normal/high | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | |
| From C4190 | Normal/high, Normal/high | Normal/NT, Normal/high | Abnormal/low, Abnormal/low | Abnormal/medium, Abnormal/medium | Abnormal/high, Abnormal/medium | |
| C. uchidae | From C1714 | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Abnormal/high, Normal/high | Normal/high, Normal/high | Abnormal/medium, Abnormal/medium |
| From C1715 | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Normal/high, Normal/high | Normal/high, Normal/high | Abnormal/medium, Abnormal/medium | |
| C. cercfabiensis | From C3301 | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/low | Abnormal/low, Abnormal/medium | Normal/medium, Normal/medium | Normal/high, Normal/high |
| From C3356 | Abnormal/medium, Abnormal/low | Abnormal/NT, Abnormal/medium | Abnormal/medium, Abnormal/medium | Abnormal/high, Abnormal/medium | Normal/high, Normal/high | |
| From C3302 | Normal/medium, NT | Normal/medium, NT | Abnormal/medium, NT | Abnormal/medium, NT | Normal/high, NT | |
| From C3305 | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Abnormal/medium, Abnormal/medium | Normal/medium, Normal/medium | Normal/high, Normal/medium | |
| C. polychroma | From C2240 | Sterile ascomata | Sterile ascomata | Abnormal/low, Abnormal/low | Abnormal/low, Abnormal/medium | Abnormal/medium, Abnormal/medium |
1 Microscopic examination of abnormal ascospore masses showed more than 10 % of the ascospores misshapen or empty and typically with ascus or ascospore debri. Streaked ascospore mass on malt yeast extract agar: high germination = colonies too numerous to count on three plates; medium germination = 40 or more colonies on three plates; and low germination = fewer than 40 colonies on the three plates. Pairings or ascospore germinations not tested are indicated by NT.
With few exceptions, the female, MAT1 testers were only fully interfertile (production of normal appearing ascospores and high germination percentage) with male testers of the same species. For C. lukuohia, three of the four female testers produced normal ascospores with high germination percentages with each of the three male testers of C. lukuohia in at least one of the two tests (Table 4). The fourth female tester (from C4212) produced abnormal ascospores in both crosses with the male tester from isolate C4133 of C. lukuohia, but the percentage of germinating ascospores was high in both cases. The four female testers of C. lukuohia produced abnormal ascospores and only medium germination percentages with the male testers of C. platani and the strain isolated from Syngonium. The female tester strains of C. platani produced normal ascospores with high germination percentage only with the male tester of C. platani. The female testers of the Syngonium strain produced normal ascospores with high germination percentages with the male Syngonium tester, but high germination percentages also were obtained for some of the ascospore masses taken from the Syngonium MAT1 testers after spermatization with male testers of C. lukuohia (Table 4). The female tester strains of C. colombiana and C. cacaofunesta produced abnormal ascospores with low or medium germination percentages when crossed with the male testers, and in some cases ascospores taken from female C. cacaofunesta ascomata spermatized with C. lukuohia male testers failed to germinate (Table 4). The female tester of the sweet potato strain of C. fimbriata produced no ascomata with any of the male tester strains.
Similar results were found in pairings among representative tester strains of the AAC (Table 5). The four female testers of C. huliohia produced normal ascospores and high germination percentages only in pairings with the two male testers of C. huliohia; the two female testers of C. uchidae produced normal ascospores with high germination when paired with the two male testers of C. uchidae; and the four female testers of C. cercfabiensis only produced normal ascospores with high germination with the male testers of C. cercfabiensis. Normal ascospores with low to medium germination were found in some crosses between three of the C. cercfabiensis female testers and males of either C. huliohia or C. uchidae (Table 5). The female tester of C. polychroma produced only sterile ascomata (no ascospore masses) when paired with C. huliohia and only abnormal ascospores with medium to low germination percentages with C. uchidae and C. cercfabiensis male tester strains (Table 5).
Pathogenicity tests
Inoculations of M. polymorpha with isolates of C. lukuohia and C. huliohia resulted in wilt symptoms in 2–4 wk. As symptoms progressed, leaves withered, died and remained attached to the plants. Complete plant mortality occurred between 4 wk to 1 yr for 91.7 % of the seedlings inoculated with C. lukuohia and 70 % of the seedlings inoculated with C. huliohia (Table 6). For both species, xylem discolouration was observed in the main stem and the pathogen was re-isolated from all inoculated plants. The reisolated cultures were the same Ceratocystis species inoculated into the plant based on morphology and sequencing the ITS region. All control plants remained healthy. Metrosideros polymorpha inoculated with the Ceratocystis isolates from Syngonium did not produce symptoms (Table 6). Similarly, no wilt symptoms were observed on seedlings of Platanus × acerifolia regardless of whether they had been inoculated with either of the new Ceratocystis spp. or with the isolates from Syngonium (Table 6).
Table 6.
Percent mortality of four host species with isolates of Ceratocystis lukuohia, C. huliohia and Ceratocystis sp. from Syngonium.
| Inoculated host | Source of inoculum1 | Total no. of inoculated plants | Mortality (%) |
|---|---|---|---|
| Metrosideros polymorpha | C. lukuohia from M. polymorpha (CMW 44102, P14-1-1) | 12 | 91.7 |
| C. huliohia from M. polymorpha (CMW 46738, P15-59) | 10 | 70.0 | |
| Ceratocystis sp. from Syngonium (CMW 50456, P16-1) | 8 | 0.0 | |
| Platanus × acerifolia | C. lukuohia from M. polymorpha (CMW 44102, P14-1-1) | 8 | 0.0 |
| C. huliohia from M. polymorpha (CMW 46738, P15-59) | 8 | 0.0 | |
| Ceratocystis sp. from Syngonium (CMW 50456, P16-1) | 8 | 0.0 | |
| Syngonium podophyllum | C. lukuohia from M. polymorpha (CMW 44102, P14-1-1) | 8 | 0.0 |
| C. huliohia from M. polymorpha (CMW 46738, P15-59) | 8 | 0.0 | |
| Ceratocystis sp. from Syngonium (CMW 50456, P16-1 and CMW 50457, P16-2) | 16 | 75.0 | |
| Colocasia esculenta | C. lukuohia from M. polymorpha (CMW 44102, P14-1-1) | 6 | 16.72 |
| C. huliohia from M. polymorpha (CMW 46738, P15-59) | 6 | 16.72 |
1 CMW = Culture collection of the Tree Pathology Co-operative Program, Forestry and Agricultural Biotechnology Institute, University of Pretoria, South Africa; P = culture collection of the USDA Agricultural Research Service, Hilo, HI.
2 One pseudopetiole of one plant.
Syngonium podophyllum inoculated with Ceratocystis isolates from Syngonium resulted in tissue collapse and discolouration of the petiole. Inoculated pseudostems of S. podophyllum died within two weeks. No symptoms were observed on Syngonium inoculated with either of the two new species of Ceratocystis from M. polymorpha (Table 6).One pseudopetiole of Colocasia esculenta inoculated with C. lukuohia and one inoculated with C. huliohia died (Table 6). Mortality was most likely due to natural senescence and not the pathogen since observed discolouration was no more than in the controls.
DISCUSSION
Results of this study have shown that two new Ceratocystis species, C. lukuohia and C. huliohia, are associated with the devastating mortality seen in native M. polymorpha forests, known as rapid ‘ōhi’a death (ROD). Recognition of these species as novel taxa in Ceratocystis was based on a combination of phenotypic, biological, morphological and phylogenetic data. Phylogenetically, C. lukuohia resides in the Latin American clade and C. huliohia in the Asia-Australian clade of Ceratocystis. Neither species was fully interfertile with their closest relatives in those respective clades, and neither appeared to be pathogenic to Syngonium podophylum, Colocasia esculenta or Platanus × acerifolia, the hosts of their nearest known relatives. Microsatellite analyses have shown that they represent two clonal lineages, suggesting that they represent recent introductions into Hawai’i.
Widespread mortality of M. polymorpha has been noted in past decades (Hodges et al. 1986), but in the current epidemic, the trees die more rapidly (Keith et al. 2015). In most ROD cases, a major branch or the whole crown suddenly shows wilt symptoms, and discolouration that follows along the ray parenchyma of the woody xylem is strongly evident. These symptoms are characteristic of wilt or canker-stain diseases caused by those Ceratocystis spp. that reside in the Latin American clade (Harrington 2013, Roux & Wingfield 2013). The initial Ceratocysis sp. associated with M. polymorpha mortality was originally identified as C. fimbriata based on morphology and ITS sequences, which were similar to those of other members of the LAC, including the canker-stain pathogen C. platani (Baker et al. 2003) and a Syngonium strain that had been previously reported in Hawai’i (Thorpe et al. 2005, Keith et al. 2015). Koch’s postulates were completed with small seedlings of M. polymorpha, and the disease was reported as Ceratocystis wilt (Keith et al. 2015). The present study confirms the association of this Ceratocystis sp. with the Ceratocystis wilt epidemic and describes the pathogen as a new species, C. lukuohia. This study also associates a second Ceratocystis sp., C. huliohia, with the M. polymorpha mortality, and seedling inoculations confirm its capacity to cause wilt-like symptoms in ‘ōhi’a.
Minor differences in symptoms and rate of symptom development are associated with the two Ceratocystis species. For example, mortality caused by C. lukuohia typically occurs rapidly and is associated with an extensive staining of the wood that is brown-black in colour, with discolouration developing in a radial pattern. In contrast, C. huliohia is commonly isolated from dying trees that show partial crown death, and the staining of the sapwood is often more diffuse and brown to grey-black in colour. In transverse sections of wood, the staining associated with C. huliohia appears more blotchy and can be seen following the contours of the cambium, perhaps due to host defence responses in sapwood tissues near the cambium. Inoculation of seedlings indicates that C. huliohia is aggressive on M. polymorpha, but field observations on the progression of symptoms in naturally infected trees suggest that it is less aggressive than C. lukuohia. This is consistent with the generalization that species of Ceratocystis in the Asian-Australian clade are less aggressive pathogens than species in the Latin American clade (Harrington 2013, Li et al. 2016). Further work is needed to characterize the disease on M. polymorpha caused by C. huliohia and its role in the currently widespread mortality known as rapid ‘ōhi’a death. In the present study, C. lukuohia was the more commonly isolated pathogen from suddenly wilting trees, and it has been consistently associated with the dramatic mortality found in new outbreak areas on Hawai’i Island.
Thus far, the distribution of the two Ceratocystis pathogens appears to be limited to the Island of Hawai’i. The spread of the pathogens, from the initial discovery in the Puna district on the southeast side of the island in 2010, has been hypothesised to be due to a combination of anthropogenic movement of infected material used for firewood or contaminated tools and aerial dispersal of infected insect boring dust. Some Ceratocystis species are known for their loose association with bark beetles (Scolytinae), ambrosia beetles (Scolytinae and Platypodinae), nitidulid beetles (Coleoptera: Nitidulidae) and flies (Diptera) that act as pathogen vectors (Wingfield et al. 2017). However, members of the LAC have been more commonly associated with wind dispersal of aleurioconidia in the frass of wood-boring ambrosia beetles (Harrington 2013). Non-native ambrosia beetles, such as Xyleborinus saxesenii and various Asian Xyleborus spp., bore into the colonised sapwood of killed M. polymorpha trees (Samuelson 1981), and both species of Ceratocystis have been detected in the frass expelled to the outside of the trees during beetle tunnelling. The frass, contaminated with abundant aleurioconidia, could then be airborne and dispersed in winds. Infections of healthy trees would then take place via natural or induced wounds. However, the modes of transmission and the associated insects involved, require further investigation.
Ceratocystis species are capable of selfing through unidirectional mating type switching, but they can also outcross (Harrington & Mcnew 1997, Witthuhn et al. 2000, Wilken et al. 2014, Wilson et al. 2015). Thus, introduced strains are capable of reproducing clonally through sexual as well as asexual reproduction, and introduced populations typically show only limited genetic variation (Engelbrecht et al. 2004, 2007, Ocasio-Morales et al. 2007, Al Adawi et al. 2014, Oliveira et al. 2015, Li et al. 2016, Scruggs et al. 2017). Isolates of both C. lukuohia and C. huliohia, collected across three different districts of Hawai’i Island, showed no variation in alleles at 23 microsatellite loci. The genetic homogeneity of both pathogens indicates that they are each reproducing clonally and more importantly, this signifies recent introductions of single genotypes of each of the pathogens into Hawai’i. Distinct microsatellite alleles and phylogenetic concordance among the DNA sequences of each of the nuclear genes show that the two genotypes are members of the respective Asian-Australian and Latin American geographic clades, which are only distantly related. Thus, it is unlikely that the two species are two genotypes of a single hybridization event between an Asian species and a Latin American species. However, further genetic studies are needed to confirm intersterility and reproductive isolation between the two species.
The possible origins of C. lukuohia and C. huliohia and their pathways into Hawai’i are unknown, but species of Ceratocystis have been frequently dispersed to new regions in vegetatively propagated plant material (Thorpe et al. 2005, Engelbrecht et al. 2007, Ferreira et al. 2011, Harrington 2013, Oliveira et al. 2015, Li et al. 2016). Phylogenetically, C. lukuohia and C. huliohia are distinct from each other, and they reside in distinctly different geographical clades of Ceratocystis. Ceratocystis lukuohia resides in the LAC, and its closest relatives are C. platani and strains of an undescribed Ceratocystis species on Xanthosoma and Syngonium (Thorpe et al. 2005, Li et al. 2017). Ceratocystis platani is an aggressive canker and wilt pathogen of Platanus spp. that is specific to this host (Panconesi 1999, Engelbrecht et al. 2004). It is considered to be indigenous to the south-eastern USA, but it has been spread to California and several countries in southern Europe (Tsopelas et al. 2017). As demonstrated in the present study, C. lukuohia and C. platani do not infect each others’ hosts, they are only partially interfertile and they differ by more than 66 % in their microsatellite profiles. Phylogenetically, C. lukuohia is very similar to isolates from Xanthosoma and Syngonium, and these strains are believed to be native to the Caribbean region (Thorpe et al. 2005). A strain on the American ornamental Syngonium podophyllum has apparently been moved in infected cuttings to other continents by the commercial nursery trade, and this strain was reported from an ornamental nursery in Hilo, Hawai’i (Uchida & Aragaki 1979, Thorpe et al. 2005), close to the area where Ceratocystis wilt of M. polymorpha was first recognized. However, C. lukuohia is morphologically distinct from the Syngonium strain, it has mostly distinct microsatellite alleles, and the two species are only partially interfertile. In addition, Syngonium isolates did not cause disease in ‘ōhi’a seedlings. In general, isolates from Araceae are not aggressive on dicotyledonous hosts and vice versa (Baker et al. 2003, Thorpe et al. 2005), and in the present study, Ceratocystis isolates from M. polymorpha did not cause disease in the aroids S. podyphyllum or Colocasia esculenta. Nonetheless, it is possible that C. lukuohia, or something closely related, was introduced to Hawai’i in plant propagative material, and it may have hybridized with the Syngonium strain.
Black rot of sweet potato caused by C. fimbriata has been known in Hawai’i since 1925, but it is host specialized and infects only sweet potato (Brown & Matsuura 1941, Li et al. 2016). Ceratocystis lukuohia is clearly distinct from the sweet potato pathogen. A sweet potato isolate from Hawai’i Island was found to have the same combination of microsatellite alleles as other sweet potato isolates from around the world (Li et al. 2016). Although the sweet potato strain resides in the LAC, the present study confirmed that it is not interfertile with C. platani (Engelbrecht & Harrington 2005), and it is not interfertile with C. lukuohia.
Ceratocystis huliohia falls in the C. polychroma cluster in the AAC, where the taxonomic placement and differences among species have recently been well debated and characterised (Li et al. 2017, Liu et al. 2018). The species most closely related to C. huliohia based on ITS sequence are the recently described C. uchidae and C. changhui (Li et al. 2017, Liu et al. 2018). Both of these species are pathogens of root crops in the Araceae. Ceratocystis uchidae was described from Hawai’i and Fiji on C. esculenta and X. sagittifolium, while C. changhui is currently only known from China on Eucalyptus sp. and Colocasia esculenta. Thorpe et al. (2005) hypothesized that C. uchdiae was introduced from Asia to Hawai’i many years ago on corms of Colocasia esculenta. Interestingly, C. huliohia was not pathogenic on Colocasia esculenta, and it is distinct from the other species in the ACC (including C. polychroma and C. cercfabiensis) based on phylogenetic inference, partial interfertility and host range. The origin and introductory pathway of C. huliohia will need further study.
The ITS region is considered the barcoding region of choice for species identification of fungi (Schoch et al. 2012, 2014, Stielow et al. 2015), and it has been used extensively for delimiting species in Ceratocystis (Fourie et al. 2015). However, Harrington et al. (2014) recognised that this region is very variable and heterogeneous and needs to be used with caution for species delimitation in Ceratocystis. Even in the current study, the topology of the species in the ITS phylogeny differed somewhat to the other gene regions used. In addition, the discovery of multiple ITS types within a single spore can complicate species delineation (Al Adawi et al. 2013, Naidoo et al. 2013, Harrington et al. 2014, Li et al. 2017, Liu et al. 2018), and at least two ITS types were found in most isolates of C. lukuohia. However, both of these sequences clustered closely together in the ITS phylogeny and were separated from the Araceae isolates with moderate bootstrap support. Although species concepts in Ceratocystis remain open to debate, it is clear that the two species associated with ROD are phylogenetically distinct from all other described species of Ceratocystis based on multiple gene regions.
Ceratocystis lukuohia and C. huliohia now represent the fourth and fifth species of Ceratocystis to have been introduced into Hawai’i. It is evident that these two species did not arise through sympatric speciation on the same host because they reside in distinct phylogeographic lineages of Ceratocystis. The ROD epidemic provides a sobering example of where introduced tree pathogens can threaten natural ecosystems. The rapid spread of C. lukuohia and C. huliohia in Hawai’i presents a great threat to the diversity, structure and function of M. polymorpha forests and the services they provide. If the disease is not contained, much of the landscape is likely to be replaced by non-native species (Mortenson et al. 2016). These pathogens not only threaten the natural biodiversity of Hawai’i, but also the culture of a people that is intricately tied to this native tree species. The implementation of strict quarantine and biosecurity measures are, therefore, crucial to prevent the spread of the pathogens to other continents and islands, such as New Zealand, that have extensive Meterosideros forests.
Acknowledgments
We thank members of the Tree Protection Co-operative Programme (TPCP), the National Research Foundation (NRF), South Africa, the Hawai’i Invasive Species Council and the U.S. Forest Service for financial support. FeiFei Liu, Andi Wilson, Conrad Trollip, Flint Hughes, James Friday, Travis Sowards, Kyson Dunn, Nathanael Friday and Kelly Hodson provided invaluable technical assistance.
Mention of trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Dept. of Agriculture and does not imply its approval to the exclusion of other products or vendors that also may be suitable.
Appendix.
Multilocus microsatellite profiles of isolates used in this study using primers developed for Ceratocystis sp. (Part 1)
| Species | CMW Culture number | Personal Collection number | Host | ITS haplotype | Country/Locality | Date collected |
Fourie et al. 2016 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF2 | AF3 | AF4 | AF5 | AF6 | AF7 | AF8 | AF9 | AF11 | |||||||
| Latin American Clade (LAC) | |||||||||||||||
| C. fimbriata | 46734 | P15-31 | Ipomoea batatas | - | USA, Hawai’i Island | 2015 | 198 | 215 | 246 | 242 | 285 | 331 | 349 | 415 | 460 |
| Ceratocystis sp. | 48499 | C1817 | Xanthosoma sp. | - | Cuba | 2001 | 206 | 203 | 254 | 262 | 300 | 325 | 334 | 415 | 432 |
| Ceratocystis sp. | 48506 | C1780 | Xanthosoma sagittifolium | - | Costa Rica | 2001 | 209 | 203 | 257 | 262 | 300 | 325 | 334 | 415 | 432 |
| Ceratocystis sp. | 48500 | C1717 | Syngonium sp. | - | USA, Hawai’i | - | 192 | 206 | 252 | 258 | 294 | 325 | 334 | 415 | 432 |
| Ceratocystis sp. | 48508 | C1809 | Syngonium podophyllum | - | USA, Florida | 2001 | 192 | 206 | 252 | 258 | 294 | 325 | 334 | 415 | 432 |
| Ceratocystis sp. | 50456 | P16-1 | Syngonium podophyllum | - | USA, Hawai’i, Hilo, South Hilo District | 2016 | 192 | 206 | 252 | 258 | 294 | 325 | 334 | 415 | 432 |
| Ceratocystis sp. | 50457 | P16-2 | Syngonium podophyllum | - | USA, Hawai’i, Hilo, South Hilo District | 2016 | 192 | 206 | 252 | 258 | 294 | 325 | 334 | 415 | 432 |
| C. lukuohia | 44102 | P14-1-1 | Metrosideros polymorpha | A | USA, Hawai’i, Leilani Estates, Puna District | 2014 Feb 20 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 44105 | P14-12 | Metrosideros polymorpha | A | USA, Hawai’i, Hawaiian Acres, Puna District | 2014 Jul 31 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 44106 | P14-13 | Metrosideros polymorpha | A | USA, Hawai’i, Hawaiian Paradise Park, Puna District | 2014 Jul 8 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 44107 | P14-15 | Metrosideros polymorpha | A | USA, Hawai’i, Hawaiian Acres, Puna District | 2014 Jul 31 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 44103 | P14-6 | Metrosideros polymorpha | A | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 47857 | P15-11 | Metrosideros polymorpha | C | USA, Hawai’i, Keau’ohana Forest Reserve, Puna District | 2015 Jan 6 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 47139 | P15-17 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Mālama Kî Forest Reserve, Puna District | 2015 Feb 2 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 46743 | P15-27 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Kopua Farm Lots, Puna District | 2015 Mar 12 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 46741 | P15-64 | Metrosideros polymorpha | C | USA, Hawai’i, Nānāwale Estates, Puna District | 2015 Jun 3 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 47152 | P15-65 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Waiākea Forest Reserve, South Hilo District | 2015 May 27 | 195 | 215 | 249 | 262 | NR | 316 | 334 | 412 | 432 |
| C. lukuohia | 47153 | P15-67 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Keau’ohana Forest Reserve, Puna District | 2015 Jun 9 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47154 | P15-68 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Volcano, Puna District | 2015 Jul 15 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46702 | P14-1-3 | Metrosideros polymorpha | A | USA, Hawai’i, Leilani Estates, Puna District | 2014 Feb 20 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 46706 | P14-4-3 | Metrosideros polymorpha | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – | |
| C. lukuohia | 46707 | P14-4-4 | Metrosideros polymorpha | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – | |
| C. lukuohia | 46711 | P14-7 | Metrosideros polymorpha | A | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 47856 | P15-10 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Keau’ohana Forest Reserve, Puna District | 2015 Jan 26 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 46725 | P15-15 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Nānāwale Estates, Puna District | 2015 Jan 29 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 47140 | P15-18 | Metrosideros polymorpha | A | USA, Hawai’i, Mālama Kî Forest Reserve, Puna District | 2015 Feb 2 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 47858 | P15-20 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Mālama Kî Forest Reserve, Puna District | 2015 Feb 2 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 47854 | P15-6 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Leilani Estates, Puna District | 2015 Jan 13 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 46722 | P15-7 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Leilani Estates, Puna District | 2015 Jan 15 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 47855 | P15-8 | Metrosideros polymorpha | A | USA, Hawai’i, Leilani Estates, Puna District | 2015 Jan 15 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – |
| C. lukuohia | 46723 | P15-9 | Metrosideros polymorpha | USA, Hawai’i, Leilani Estates, Puna District | 2015 Jan 15 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | – | |
| C. lukuohia | 46710 | P14-6-1 | Metrosideros polymorpha | A | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46714 | P14-11 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hawaiian Acres, Puna District | 2014 Jul 31 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46718 | P14-16 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hawaiian Acres, Puna District | 2014 Jul 31 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46703 | P14-2 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46704 | P14-2-1 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46705 | P14-3 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46712 | P14-7-3 | Metrosideros polymorpha | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 | |
| C. lukuohia | 46713 | P14-8 | Metrosideros polymorpha | A | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46719 | P15-1 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Waiākea Forest Reserve, South Hilo District | 2014 Nov 20 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47138 | P15-13 | Metrosideros polymorpha | A | USA, Hawai’i, Nānāwale Estates, Puna District | 2015 Jan 29 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46724 | P15-14 | Metrosideros polymorpha | A | USA, Hawai’i, Nānāwale Estates, Puna District | 2015 Jan 29 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46726 | P15-16 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Nānāwale Estates, Puna District | 2015 Jan 29 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46727 | P15-19 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Mālama Kî Forest Reserve, Puna District | 2015 Feb 2 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46720 | P15-2 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Waiākea Forest Reserve, South Hilo District | 2014 Nov 20 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46728 | P15-21 | Metrosideros polymorpha | A | USA, Hawai’i, Leilani Estates, Puna District | 2014 Jun 17 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46729 | P15-22 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Leilani Estates, Puna District | 2015 Jan 27 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46730 | P15-24 | Metrosideros polymorpha | A | USA, Hawai’i, ‘Opihikao, Puna District | 2015 Mar 3 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46731 | P15-25 | Metrosideros polymorpha | A | USA, Hawai’i, ‘Opihikao, Puna District | 2015 Mar 3 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46732 | P15-28 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hawaiian Acres, Puna District | 2015 Mar 12 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46733 | P15-29 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Pu’u Kali’u, Puna District | 2015 Mar 6 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47142 | P15-32 | Metrosideros polymorpha | A | USA, Hawai’i, Pu’u Kali’u, Puna District | 2015 Mar 6 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46735 | P15-34 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hawaiian Paradise Park, Puna District | 2015 Mar 12 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47144 | P15-35 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hawaiian Paradise Park, Puna District | 2015 Mar 12 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47145 | P15-36 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Volcano, Puna District | 2015 Mar 3 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47146 | P15-38 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Lava Tree, Puna District | 2015 Mar 3 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47147 | P15-39 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Lava Tree, Puna District | 2015 Mar 3 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47136 | P15-4 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Dec 12 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46708 | P14-5 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Jun 11 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47137 | P15-5 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2014 Dec 12 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46736 | P15-53 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Orchidlands Estates, Puna District | 2015 Mar 31 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47148 | P15-54 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Leilani Estates, Puna District | 2015 May 27 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46737 | P15-55 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hilo Watershed Forest Reserve, South Hilo District | 2015 Jun 8 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 47150 | P15-61 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, Hilo Watershed Forest Reserve, South Hilo District | 2015 Jul 1 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46740 | P15-63 | Metrosideros polymorpha | Mix A/C | USA, Hawai’i, ‘Opihikao, Puna District | 2015 May 27 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. lukuohia | 46742 | P15-66 | Metrosideros polymorpha | A | USA, Hawai’i, Keau’ohana Forest Reserve, Puna District | 2015 Jun 9 | 195 | 215 | 249 | 262 | – | 316 | 334 | 412 | 432 |
| C. platani | 9499 | CF20 | Platanus sp. | - | Italy | - | 203 | 239 | 243 | 262 | NR | 322 | 334 | 418 | 432 |
| C. platani | 1896 | BRENO10 | Platanus sp. | - | Switzerland | - | 203 | 239 | 243 | 262 | NR | 322 | 334 | 418 | 432 |
| C. platani | 2219 | 1012/D0 | Platanus sp. | - | France, Saint-Maurice | 1991 | 203 | 239 | 243 | 262 | NR | 322 | 334 | 418 | 432 |
| C. platani | 23450 | c-1 | Platanus orientalis | - | Greece | 2006 | 203 | 239 | 243 | 262 | NR | 322 | 334 | 418 | 432 |
| C. platani | 14802 | C1317 | Platanus occidentalis | - | USA, North Carolina | 1998 Jul | 201 | 215 | 243 | 262 | – | 322 | 334 | 426 | – |
| C. platani | 9496 | CF34 | Platanus sp. | - | Italy | - | 203 | 239 | 243 | 262 | – | 322 | 334 | 418 | – |
| C. platani | 9000 | CF8 | Platanus sp. | - | Italy | 2002 Feb | 203 | 239 | 243 | 262 | – | 322 | 334 | 418 | – |
| C. platani | 2218 | 1012/D4 | Platanus sp. | - | France, Saint-Maurice | 1991 | 203 | 239 | 243 | 262 | – | 322 | 334 | 418 | – |
| C. platani | 23918 | Platanus sp. | - | Greece | 2006 Jul | 203 | 239 | 243 | 262 | – | 322 | 334 | 418 | – | |
| Asian_Australian Clade (AAC) | |||||||||||||||
| C. huliohia | 47135 | P14-10 | Metrosideros polymorpha | B | USA, Hawai’i, ‘Opihikao, Puna District | 2014 Jul 8 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 46716 | P14-14 | Metrosideros polymorpha | B | USA, Hawai’i, Hawaiian Paradise Park, Puna District | 2014 Jul 8 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 44104 | P14-9-1 | Metrosideros polymorpha | B | USA, Hawai’i, Hawaiian Paradise Park, Puna District | 2015 Feb 04 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 46721 | P15-3 | Metrosideros polymorpha | B | USA, Hawai’i, Hilo, South Hilo District | 2014 Nov 26 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 47143 | P15-33 | Metrosideros polymorpha | B | USA, Hawai’i, Pana’ewa, South Hilo District | 2015 Mar 3 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 47149 | P15-58 | Metrosideros polymorpha | B | USA, Hawai’i, Hōlualoa, North Kona District | 2015 Jun 10 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 46738 | P15-59 | Metrosideros polymorpha | B | USA, Hawai’i, Hōlualoa, North Kona District | 2015 Jun 10 | 212 | 194 | 234 | 226 | NR | 328 | 328 | 401 | 444 |
| C. huliohia | 46739 | P15-60 | Metrosideros polymorpha | B | USA, Hawai’i, Hōlualoa, North Kona District | 2015 Jun 10 | 212 | 194 | 234 | 226 | – | 328 | 328 | 401 | – |
| C. huliohia | 47151 | P15-62 | Metrosideros polymorpha | B | USA, Hawai’i, Hōlualoa, North Kona District | 2015 Jun 10 | 212 | 194 | 234 | 226 | – | 328 | 328 | 401 | – |
| C. uchidae | 14796 | CBS 114720 | Colocasia esculenta | - | USA, Hawai’i, O’ahu | 1991 | 219,68 | 194 | 249 | 226 | 270 | 328 | – | 401 | 444 |
| C. uchidae | 14804 | CBS 115164 | Colocasia esculenta | - | USA, Hawai’i, Kaua’i | 1988 | 219,75 | 194 | 249 | 226 | 270 | 328 | – | 401 | 444 |
| C. cercfabiensis | 43029 | CERC 2170 | Eucalyptus sp. | - | China, HaiNan Province | 2013 | 203 | 194 | 237 | 226 | 294 | 328 | – | 401 | 444 |
| C. cercfabiensis | 42795 | CERC 2687 | Eucalyptus sp. | - | China, GuangDong | 2013 | 206 | 194 | 237 | 226 | 291 | 334 | 328 | 401 | 447 |
| C. changhui | 43272 | CERC 3605 | Colocasia esculenta | - | China, YunNan Province | 2015 | 220 | 194 | 237 | 226 | 270 | 328 | – | 401 | 444 |
| C. changhui | 43281 | CERC 3615 | Colocasia esculenta | - | China, YunNan Province | 2014 | 220 | 194 | 237 | 226 | 270 | 328 | – | 401 | 444 |
| C. changhui | 49317 | C3371 | Eucalyptus sp. | - | China, YunNan Province | 2014 | 232 | 194 | 243 | 226 | 270 | 328 | 328 | 401 | 444 |
| C. changhui | 49318 | C3372 | Eucalyptus sp. | - | China, YunNan Province | 2014 | 232 | 194 | 243 | 226 | 270 | 328 | 328 | 401 | 444 |
| C. changhui | 49319 | C3373 | Eucalyptus sp. | - | China, YunNan Province | 2014 | 232 | 194 | 243 | 226 | 270 | 328 | 328 | 401 | 444 |
| C. changhui | 49320 | C3374 | Eucalyptus sp. | - | China, YunNan Province | 2014 | 232 | 194 | 243 | 226 | 270 | 328 | 328 | 401 | 444 |
| C. changhui | 49321 | C3375 | Eucalyptus sp. | - | China, YunNan Province | 2014 | 232 | 194 | 243 | 226 | 270 | 328 | 328 | 401 | 444 |
| C. changhui | 49322 | C3376 | Eucalyptus sp. | - | China, YunNan Province | 2014 | 232 | 194 | 243 | 226 | 270 | 328 | 328 | 401 | 444 |
| C. polychroma | 11424 | Syzygium aromaticum | - | Indonesia | 2002 | 209 | 194 | 234 | 226 | 294 | 328/334 | – | 401 | 444 | |
| C. polychroma | 11436 | Syzygium aromaticum | Indonesia | 2002 | 209 | 194 | 234 | 226 | 294 | 334 | – | 401 | 444 | ||
MB = multiple bands observed during PCR, did not do fragment analyses; - = no data available; – = PCR was not done; FP = finger print pattern that was difficult to score; NR = no PCR amplification; MA = multiple alleles observed.
(Part 2)
| Species | CMW Culture number | Stiemel et al. 2004 |
Barnes et al. 2001 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAG8 | AAG9 | CAA9 | CAA10 | CAA15 | CAA38 | CAA80 | CAT1 | CAT3K | CAT9X | CAT1200 | CAG5 | GACA650 | AG1/AG2 | AG7/AG8 | CF13/CF14 | CF15/CF16 | CF23/CF24 | ||
| Latin American Clade (LAC) | |||||||||||||||||||
| C. fimbriata | 46734 | 185 | 397 | 206 | 138 | 320 | 150 | 301 | 255 | 322 | 280 | 376 | 319 | 219 | 269 | 284 | 406 | 465 | 155 |
| Ceratocystis sp. | 48499 | 171.37 close to bin171 | 392 | 199.66 | 131 | 314 | 184 | 295 | 263 | MA | 281 | 374 | 328 | 212 | 270 | 302 | 401 | 490 | 153 |
| Ceratocystis sp. | 48506 | 171.49 close to bin171 | 392 | 199.66 | 131 | 314 | 184 | 295 | 263 | MA | 281 | 374 | 328 | 212 | 270 | 304 | 401 | 490 | 174 |
| Ceratocystis sp. | 48500 | 165 | 392 | 254 | 131 | 314 | 250 | 263 | 269 | MA | 281, MA | 374 | 328 | 212 | 270 | 302 | 401 | 466 | 172 |
| Ceratocystis sp. | 48508 | 165 | 392 | 254 | 131 | 314 | 250 | 263 | 269 | MA | 281, MA | 374 | 328 | 212 | 270 | 302 | 401 | 466 | 172 |
| Ceratocystis sp. | 50456 | 165 | 392 | 254 | 131 | 314 | 250 | 263 | 269 | MA | 281, 287 | 374 | 328 | 212 | 270 | 302 | 401 | 466 | 172 |
| Ceratocystis sp. | 50457 | 165 | 392 | 254 | 131 | 314 | 250 | 263 | 269 | MA | 281, 287 | 374 | 328 | 212 | 270 | 302 | 401 | 466 | 172 |
| C. lukuohia | 44102 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 44105 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 44106 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 44107 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 44103 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 47857 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 47139 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 46743 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 46741 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 47152 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | 257 | MA | 284, MA | MB | 328 | 227 | 269 | 293 | FP | 471 | 161 |
| C. lukuohia | 47153 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 47154 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46702 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46706 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46707 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46711 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 47856 | 188 | 392 | 266 | 129 | 314 | 232 | – | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46725 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 47140 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 47858 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 47854 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46722 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 47855 | 188 | 392 | 266 | 129 | 314 | 232 | – | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46723 | 188 | 392 | 266 | 129 | 314 | 232 | 313 | – | – | – | – | 328 | 227 | – | 293 | FP | 471 | 161 |
| C. lukuohia | 46710 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46714 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46718 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46703 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46704 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46705 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46712 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46713 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46719 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47138 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46724 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46726 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46727 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46720 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46728 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46729 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46730 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46731 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46732 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46733 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47142 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46735 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47144 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47145 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47146 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47147 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47136 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46708 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47137 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46736 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47148 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46737 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 47150 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46740 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. lukuohia | 46742 | – | – | 266 | – | – | – | 313 | – | – | – | – | – | – | – | – | – | – | – |
| C. platani | 9499 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | 257 | MA | 281, MA | 393 | 328 | 287 | 271 | 300 | FP | 480 | 161 |
| C. platani | 1896 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | 257 | MA | 281, MA | 393 | 328 | 301 | 271 | 300 | FP | 480 | 161 |
| C. platani | 2219 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | 257 | MA | 281, MA | 393 | 328 | 286.9 | 271 | 300 | FP | 480 | 161 |
| C. platani | 23450 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | 257 | MA | 281, MA | 393 | 328 | 316 | 271 | 300 | FP | 480 | 161 |
| C. platani | 14802 | 174 | 406 | 367 | 129 | 285 | 133 | 295 | – | – | – | – | 328 | - | – | 300 | 406.47 | 482 | 161 |
| C. platani | 9496 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | – | – | – | – | 328 | 287 | – | 300 | FP | 480 | 161 |
| C. platani | 9000 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | – | – | – | – | 328 | 287 | – | 300 | FP | 480 | 161 |
| C. platani | 2218 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | – | – | – | – | 328 | 287 | – | 300 | FP | 480 | 161 |
| C. platani | 23918 | 174 | 406 | 294 | 129 | 314 | 157 | 295 | – | – | – | – | 328 | 287 | – | 300 | FP | 480 | 161 |
| Asian_Australian Clade (AAC) | |||||||||||||||||||
| C. huliohia | 47135 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 46716 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 44104 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 46721 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 47143 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 47149 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 46738 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | 232 | 306 | 270 | 355 | > 500 | 182 | 266 | 278/291 | 417 | 471 | 157 |
| C. huliohia | 46739 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | – | – | – | – | – | 182 | – | 278/291 | 417 | 471 | 157 |
| C. huliohia | 47151 | 174 | 395 | 166 | 126 | 383 | 136 | 304 | – | – | – | – | – | 182 | – | 278/291 | 417 | 471 | 157 |
| C. uchidae | 14796 | 171 | 392 | 147 | 126 | – | 139 | 284 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278 | 446 | – | 157 |
| C. uchidae | 14804 | 171 | 392 | 147 | 126 | – | 139 | 284 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278 | 446 | – | 157 |
| C. cercfabiensis | 43029 | 174 | 392 | 160 | 126 | – | 136 | 304 | 232 | 306 | 270 | 355 | – | 182 | 266 | 278 | 431 | – | 157 |
| C. cercfabiensis | 42795 | 174 | 392 | 160 | 126 | – | 136 | 304 | 232 | 306 | 270 | 355 | – | 182 | 266 | 278 | 431 | – | 157 |
| C. changhui | 43272 | 171 | 392 | 147 | 126 | – | 143 | 304 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278 | 431 | – | 157 |
| C. changhui | 43281 | 171 | 392 | 147 | 126 | – | 143 | 304 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278 | 431 | – | 157 |
| C. changhui | 49317 | 171 | 392 | 147 | 126 | – | 136 | 292 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278/291 | 431 | 472,8 | 157 |
| C. changhui | 49318 | 171 | 392 | 147 | 126 | – | 136 | 292 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278/291 | 431 | 472,8 | 157 |
| C. changhui | 49319 | 171 | 392 | 147 | 126 | – | 136 | 292 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278/291 | 431 | 472,8 | 157 |
| C. changhui | 49320 | 171 | 392 | 147 | 126 | – | 136 | 292 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278/291 | 431 | 472,8 | 157 |
| C. changhui | 49321 | 171 | 392 | 147 | 126 | – | 136 | 292 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278/291 | 431 | 472,8 | 157 |
| C. changhui | 49322 | 171 | 392 | 147 | 126 | – | 136 | 292 | 232 | 306 | 267 | 355 | – | 182 | 266 | 278/291 | 431 | 472,8 | 157 |
| C. polychroma | 11424 | 188 | 389 | 153 | 126 | – | 139 | 307 | 229 | – | 264 | 355 | – | 182 | 266 | 278 | 431 | – | 157 |
| C. polychroma | 11436 | 188 | 389 | 153 | 126 | – | 139 | 307 | 229 | 306 | 264 | 355 | – | 182 | 266 | 278 | 431 | – | 157 |
REFERENCES
- Al Adawi AO, Barnes I, Khan IA, et al. 2013. Ceratocystis manginecans associated with a serious wilt disease of two native legume trees in Oman and Pakistan. Australasian Plant Pathology 42: 179–193. [Google Scholar]
- Al Adawi AO, Barnes I, Khan IA, et al. 2014. Clonal structure of Ceratocystis manginecans populations from mango wilt disease in Oman and Pakistan. Australasian Plant Pathology 43: 393–402. [Google Scholar]
- Al Adawi AO, Deadman ML, Al Rawahi AK, et al. 2006. Aetiology and causal agents of mango sudden decline disease in the Sultanate of Oman. European Journal of Plant Pathology 116: 247–254. [Google Scholar]
- Baker CJ, Harrington TC, Krauss U, et al. 2003. Genetic variability and host specialization in the Latin American clade of Ceratocystis fimbriata. Phytopathology 93: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Barnes I, Gaur A, Burgess T, et al. 2001. Microsatellite markers reflect intra-specific relationships between isolates of the vascular wilt pathogen Ceratocystis fimbriata. Molecular Plant Pathology 2: 319–325. [DOI] [PubMed] [Google Scholar]
- Brawner J, Japarudin Y, Lapammu M, et al. 2015. Evaluating the inheritance of Ceratocystis acaciivora symptom expression in a diverse Acacia mangium breeding population. Southern Forests: a journal of forest science 77: 83–90. [Google Scholar]
- Brown AC, Matsuura M. 1941. Black rot of sweet potato. Agricultural Extension Circular #134, University of Hawai’i. [Google Scholar]
- Chung HL. 1923. The sweet potato in Hawai’i. In: Hawai’i Agricultural Experiment Station Bulletin No. 50.
- Dawson JW, Stemmermann RL. 1990. Metrosideros (Gaud.). In: Wagner WL, Herbst DR, Summer SH. (eds), Manual of flowering plants of Hawai’i: 964–970. University of Hawai’i Press, Honolulu, Hawai’i. [Google Scholar]
- De Beer ZW, Duong TA, Barnes I, et al. 2014. Redefining Ceratocystis and allied genera. Studies in Mycology 79: 187–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Marincowitz S, Duong TA, et al. 2017. Bretziella, a new genus to accommodate the oak wilt fungus, Ceratocystis fagacearum (Microascales, Ascomycota). MycoKeys 27: 1–19. doi: https://doi.org/10.3897/mycokeys.27.20657. [Google Scholar]
- Engelbrecht CJB, Harrington TC. 2005. Intersterility, morphology and taxonomy of Ceratocystis fimbriata on sweet potato, cacao and sycamore. Mycologia 97: 57–69. [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJB, Harrington TC, Alfenas AC, et al. 2007. Genetic variation in populations of the cacao wilt pathogen, Ceratocystis cacaofunesta. Plant Pathology 56: 923–933. [Google Scholar]
- Engelbrecht CJB, Harrington TC, Steimel J, et al. 2004. Genetic variation in eastern North American and putatively introduced populations of Ceratocystis fimbriata f. platani. Molecular Ecology 13: 2995–3005. [DOI] [PubMed] [Google Scholar]
- Ferreira EM, Harrington TC, Thorpe DJ, et al. 2010. Genetic diversity and interfertility among highly differentiated populations of Ceratocystis fimbriata in Brazil. Plant Pathology 59: 721–735. [Google Scholar]
- Ferreira MA, Harrington TC, Alfenas AC, et al. 2011. Movement of genotypes of Ceratocystis fimbriata within and among Eucalyptus plantations in Brazil. Phytopathology 101: 1005–1012. [DOI] [PubMed] [Google Scholar]
- Fourie A, Wingfield MJ, Wingfield BD, et al. 2015. Molecular markers delimit cryptic species in Ceratocystis sensu stricto. Mycological Progress 14: 1–18. [Google Scholar]
- Fourie A, Wingfield MJ, Wingfield BD, et al. 2016. A possible centre of diversity in South East Asia for the tree pathogen, Ceratocystis manginecans. Infection, Genetics and Evolution 41: 73–83. [DOI] [PubMed] [Google Scholar]
- Friday JB, Herbert DA. 2006. Metrosideros polymorpha (‘ōhi’a), ver. 3.2. In: Elevitch CR. (eds), Species profiles for Pacific Island agroforestry. Permanent Agricultural Resources (PAR), Hōlualoa, Hawai’i, http://www.traditionaltree.org. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SB, Drenth A, Fry WE. 1992. Cloning and genetic analyses of two highly polymorphic, moderately repetitive nuclear DNAs from Phytophthora infestans. Current Genetics 22: 107–115. [DOI] [PubMed] [Google Scholar]
- Harrington TC. 2000. Host specialization and speciation in the American wilt pathogen Ceratocystis fimbriata. Fitopatologia Brasileira 25 (Suppl): 262–263. [Google Scholar]
- Harrington TC. 2004. Ceratocystis fimbriata. Crop Protection Compendium CD-ROM. CABI. [Google Scholar]
- Harrington TC. 2013. Ceratocystis diseases. In: Gonthier P, Nicolotti G. (eds), Infectious Forest Diseases: 230–255. CABI. [Google Scholar]
- Harrington TC, Kazmi MR, Al-Sadi AM, et al. 2014. Intraspecific and intragenomic variability of ITS rDNA sequences reveals taxonomic problems in Ceratocystis fimbriata sensu stricto. Mycologia 106: 224–242. [DOI] [PubMed] [Google Scholar]
- Harrington TC, McNew DL. 1997. Self-fertility and uni-directional mating-type switching in Ceratocystis coerulescens, a filamentous ascomycete. Current Genetics 32: 52–59. [DOI] [PubMed] [Google Scholar]
- Harrington TC, McNew DL. 1998. Partial interfertility among the Ceratocystis species on conifers. Fungal Genetics and Biology 25: 44–53. [DOI] [PubMed] [Google Scholar]
- Heath RN, Wingfield MJ, Wingfield BD, et al. 2009. Ceratocystis species on Acacia mearnsii and Eucalyptus spp. in eastern and southern Africa including six new species. Fungal Diversity 34: 41–68. [Google Scholar]
- Hodges CS, Adee KT, Stein JD, et al. 1986. Decline of ‘ōhi‘a (Metrosideros polymorpha) in Hawai’i: a review. In: General Technical Report PSW-86, Berkeley, CA: 22. Pacific Southwest Forest and Range Experiment Station Forest Service, U.S. Department of Agriculture. [Google Scholar]
- Jacobs K, Bergdahl DR, Wingfield MJ, et al. 2004. Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycological Research 108: 411–418. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Harrington TC, Engelbrecht CJB. 2005. Phylogeny and taxonomy of the North American clade of the Ceratocystis fimbriata complex. Mycologia 97: 1067–1092. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith L, Hughes RF, Sugiyama L, et al. 2015. First report of Ceratocystis wilt on ‘ōhi‘a. Plant Disease 99: 1276. [Google Scholar]
- Kile GA. 1993. Plant diseases caused by species of Ceratocystis sensu stricto and Chalara. In: Wingfield MJ, Seifert KA, Webber J. (eds), Ceratocystis and Ophiostoma: taxonomy, ecology and pathogenicity: 173–183. APS Press, St. Paul, Minnesota. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Harrington TC, McNew D, et al. 2016. Genetic bottlenecks for two populations of Ceratocystis fimbriata on sweet potato and pomegranate in China. Plant Disease 100: 2266–2274. [DOI] [PubMed] [Google Scholar]
- Li Q, Harrington TC, McNew D, et al. 2017. Ceratocystis uchidae, a new species on Araceae in Hawai’i and Fiji. MycoScience 58: 398–412. [Google Scholar]
- Liu FF, Barnes I, Roux J, et al. 2018. Molecular phylogenetics and microsatellite analysis reveal a new pathogenic Ceratocystis species in the Asian-Australian clade. Plant Pathology. doi: https://doi.org/10.1111/ppa.12820.
- Liu FF, Mbenoun M, Barnes I, et al. 2015. New Ceratocystis species from Eucalyptus and Cunninghamia in South China. Antonie Van Leeuwenhoek 107: 1451–1473. [DOI] [PubMed] [Google Scholar]
- Loope L. 2016. Guidance document for rapid Ohia death: http://www.cgaps.org/wp-content/uploads/Guidance-Document-for-Rapid-Ohia-Death-Final-L-Loope.pdf (last accessed 25 January 2018).
- Marin-Felix Y, Groenewald JZ, Cai L, et al. 2017. Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology 86: 99–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers CG, McNew DL, Harrington TC, et al. 2015. Three genera in the Ceratocystidaceae are the respective symbionts of three independent lineages of ambrosia beetles with large, complex mycangia. Fungal Biology 119: 1075–1092. [DOI] [PubMed] [Google Scholar]
- Mbenoun M, Wingfield MJ, Begoude Boyogueno AD, et al. 2014. Molecular phylogenetic analyses reveal three new Ceratocystis species and provide evidence for geographic differentiation of the genus in Africa. Mycological Progress 13: 219–240. [Google Scholar]
- Moller W, Devay J. 1968. Carrot as a species-selective isolation medium for Ceratocystis fimbriata. Phytopathology 58: 123–124. [Google Scholar]
- Mortenson LA, Hughes RF, Friday JB, et al. 2016. Assessing spatial distribution, stand impacts and rate of Ceratocystis fimbriata induced ‘ōhi’a (Metrosideros polymorpha) mortality in a tropical wet forest, Hawai’i Island, USA. Forest Ecology and Management 377: 83–92. [Google Scholar]
- Naidoo K, Steenkamp ET, Coetzee MPA, et al. 2013. Concerted evolution in the ribosomal RNA cistron. PLoS ONE 8: e59355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel WJ, Duong TA, Wingfield BD, et al. 2017. A new genus and species for the globally important, multi-host root pathogen Thielaviopsis basicola. Plant Pathology. doi: https://doi.org/10.1111/ppa.12803.
- Ocasio-Morales RG, Tsopelas P, Harrington TC. 2007. Origin of Ceratocystis platani on native Platanus orientalis in Greece and its impact on natural forests. Plant Disease 91: 901–904. [DOI] [PubMed] [Google Scholar]
- Oliveira LSS, Guimarães LMS, Ferreira MA, et al. 2015. Aggressiveness, cultural characteristics and genetic variation of Ceratocystis fimbriata on Eucalyptus spp. Forest Pathology 45: 505–514. [Google Scholar]
- Panconesi A. 1999. Canker stain of plane tree: A serious danger to urban plantings in Europe. Journal of Plant Pathology 81: 3–15. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society. [Google Scholar]
- Roux J, Wingfield MJ. 2009. Ceratocystis species: emerging pathogens of non-native plantation Eucalyptus and Acacia species. Southern Forests 71: 115–120. [Google Scholar]
- Roux J, Wingfield MJ. 2013. Ceratocystis species on the African continent, with particular reference to C. albifundus, and African species in the C. fimbriata sensu lato species complex. In: Seifert KA, De Beer ZW, Wingfield MJ. (eds), The Ophiostomatoid fungi: expanding frontiers: 131–138. CBS, Utrecht, The Netherlands. [Google Scholar]
- Samuelson GA. 1981. A synopsis of Hawaiian Xyleborini (Coleoptera: Scolytidae). Pacific Insects 23: 50–92. [Google Scholar]
- Schoch CL, Robbertse B, Robert V, et al. 2014. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database 2014: 1–21. doi: https://doi.org/10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs AC, Basaiah T, Adams ML, et al. 2017. Genetic diversity, fungicide sensitivity, and host resistance to Ceratocystis fimbriata infecting sweetpotato in North Carolina. Plant Disease 101: 994–1001. [DOI] [PubMed] [Google Scholar]
- Seifert KA, Wingfield MJ, Kendrick WB. 1993. A nomenclator for described species of Ceratocystis, Ophiostoma, Ceratocystiopsis, Ceratostomella and Sphaeronaemella. In: Wingfield MJ, Seifert KA, Webber J. (eds), Ceratocystis and Ophiostoma: Taxonomy, ecology and pathogenicity: 269–287. APS Press, St. Paul, Minnesota. [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9): 1312–1313. doi: https://doi.org/10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimel J, Engelbrecht CJB, Harrington TC. 2004. Development and characterization of microsatellite markers for the fungus Ceratocystis fimbriata. Molecular Ecology Notes 4: 215–218. [Google Scholar]
- Stielow JB, Lévesque CA, Seifert KA, et al. 2015. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35: 242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*: Phylogenetic analysis using parsimony (* and other methods), version 4.0b 10. Sinauer Associates. [Google Scholar]
- Tarigan M, Roux J, Van Wyk M, et al. 2011. A new wilt and die-back disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. South African Journal of Botany 77: 292–304. [Google Scholar]
- Thorpe DJ, Harrington TC, Uchida JY. 2005. Pathogenicity, internal transcribed spacer-rDNA variation, and human dispersal of Ceratocystis fimbriata on the family Araceae. Phytopathology 95: 316–323. [DOI] [PubMed] [Google Scholar]
- Thu PQ, Quynh DN, Fourie A, et al. 2014. Ceratocystis wilt – a new and serious threat to Acacia plantations in Vietnam: taxonomy and pathogenicity. In: IUFRO Working Party 2.08.07: Genetics and silviculture of Acacia, sustaining the future of Acacia plantation forestry, Hue, Vietnam. [Google Scholar]
- Tsopelas P, Santini A, Wingfield MJ, et al. 2017. Canker stain: A lethal disease destroying iconic plane trees. Plant Disease 101: 645–658. [DOI] [PubMed] [Google Scholar]
- Uchida JY, Aragaki M. 1979. Ceratocystis blight of Syngonium podophyllum. Plant Disease Reporter 63: 1053–1056. [Google Scholar]
- Upadhyay H. 1981. A monograph of Ceratocystis and Ceratocystiopsis. University of Georgia Press. [Google Scholar]
- Van Wyk M, Al Adawi AO, Khan IA, et al. 2007. Ceratocystis manginecans sp. nov., causal agent of a destructive mango wilt disease in Oman and Pakistan. Fungal Diversity 27: 213–230. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and application: 315–322. Academic Press, San Diego, USA. [Google Scholar]
- Wilken PM, Steenkamp ET, Wingfield MJ, et al. 2014. DNA loss at the Ceratocystis fimbriata mating locus results in self-sterility. PLoS ONE 9: e92180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AM, Wilken PM, Van der Nest MA, et al. 2015. Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus 6: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield MJ, Barnes I, De Beer ZW, et al. 2017. Novel associations between ophiostomatoid fungi, insects and tree hosts: current status – future prospects. Biological Invasions 19: 3215–3228. [Google Scholar]
- Witthuhn RC, Harrington TC, Wingfield BD, et al. 2000. Deletion of the MAT-2 mating-type gene during uni-directional mating-type switching in Ceratocystis. Current Genetics 38: 48–52. [DOI] [PubMed] [Google Scholar]


