Endogenous retroviruses are harbored by many animals, and their interactions with exogenous retroviral infections have not been widely studied. Feline leukemia virus (FeLV) is a relevant model system to examine this question, as endogenous and exogenous forms of the virus exist. In this analysis of a large domestic cat breeding colony naturally infected with FeLV, we documented that enFeLV copy number was higher in males and inversely related to FeLV viral load and associated with better FeLV disease outcomes. Females had lower enFeLV copy numbers and were more likely to have progressive FeLV disease and FeLV-B subtypes. FFV viral load was correlated with FeLV progression. FFV, FcaGHV-1, and FeLV displayed markedly different patterns of infection with respect to host demographics. This investigation revealed complex coinfection outcomes and viral ecology of chronic infections in a closed population.
KEYWORDS: endogenous, feline foamy virus, feline leukemia virus
ABSTRACT
Exogenous feline leukemia virus (FeLV) is a feline gammaretrovirus that results in a variety of disease outcomes. Endogenous FeLV (enFeLV) is a replication-defective provirus found in species belonging to the Felis genus, which includes the domestic cat (Felis catus). There have been few studies examining interaction between enFeLV genotype and FeLV progression. We examined point-in-time enFeLV and FeLV viral loads, as well as occurrence of FeLV/enFeLV recombinants (FeLV-B), to determine factors relating to clinical disease in a closed breeding colony of cats during a natural infection of FeLV. Coinfections with feline foamy virus (FFV), feline gammaherpesvirus 1 (FcaGHV-1), and feline coronavirus (FCoV) were also documented and analyzed for impact on cat health and FeLV disease. Correlation analysis and structural equation modeling techniques were used to measure interactions among disease parameters. Progressive FeLV disease and FeLV-B presence were associated with higher FeLV proviral and plasma viral loads. Female cats were more likely to have progressive disease and FeLV-B. Conversely, enFeLV copy number was higher in male cats and negatively associated with progressive FeLV disease. Males were more likely to have abortive FeLV disease. FFV proviral load was found to correlate positively with higher FeLV proviral and plasma viral load, detection of FeLV-B, and FCoV status. Male cats were much more likely to be infected with FcaGHV-1 than female cats. This analysis provides insights into the interplay between endogenous and exogenous FeLV during naturally occurring disease and reveals striking variation in the infection patterns among four chronic viral infections of domestic cats.
IMPORTANCE Endogenous retroviruses are harbored by many animals, and their interactions with exogenous retroviral infections have not been widely studied. Feline leukemia virus (FeLV) is a relevant model system to examine this question, as endogenous and exogenous forms of the virus exist. In this analysis of a large domestic cat breeding colony naturally infected with FeLV, we documented that enFeLV copy number was higher in males and inversely related to FeLV viral load and associated with better FeLV disease outcomes. Females had lower enFeLV copy numbers and were more likely to have progressive FeLV disease and FeLV-B subtypes. FFV viral load was correlated with FeLV progression. FFV, FcaGHV-1, and FeLV displayed markedly different patterns of infection with respect to host demographics. This investigation revealed complex coinfection outcomes and viral ecology of chronic infections in a closed population.
INTRODUCTION
Feline leukemia virus (FeLV), feline foamy virus (FFV), feline gammaherpesvirus 1 (FcaGHV-1), and feline coronavirus (FeCoV) have been the topics of many studies of feline viral pathogenesis and disease; however, few studies have systematically examined the impacts of these infections on disease outcomes or interactions between these infections and the endogenous FeLV (enFeLV) genotype, especially in natural systems. Inferences from population-level analysis are relevant to formulating and corroborating in vitro and natural infections and documenting biological relevance of less complex systems. It was with this consideration that this study was undertaken to conduct a multipathogen analysis in a closed breeding colony naturally infected with all four of these pathogens.
Feline leukemia virus is a gammaretrovirus that infects a wide range of felids (1, 2). Exogenous FeLV (exFeLV) exists as a replication-competent, horizontally transmissible form (1, 2). Endogenous FeLV is a replication-defective provirus and is found in species belonging to the Felis genus, including the domestic cat (Felis catus) (3–8). Endogenous retroviruses have been hypothesized to play a role in exogenous retroviral infection either through protection against exogenous retrovirus infection or, conversely, by enhancing the exogenous retrovirus infection (9–11). FeLV subtype A (FeLV-A) is the most commonly described subtype of FeLV (2, 12). During FeLV-A infection in domestic cats, recombination often occurs between host-encoded enFeLV and FeLV-A in the env region, resulting in the production of the FeLV-B subtype. There are other described subtypes which result from FeLV-A recombination events, but these are less common (13).
FeLV-A is thought to be transmitted through contact with saliva during mutual grooming and sharing of food dishes, as well as through blood, feces, and mother's milk (1, 14–16). Studies have indicated that FeLV-B primarily arises through de novo recombination in domestic cats infected with FeLV-A, and it is thought to require FeLV-A as a helper virus for transmission (17–20). FeLV-A infection alone is considered to be minimally pathogenic (21, 22), but it may result in immunosuppression and development of cytopenia, anemia, and myelodysplastic syndromes (22–24). Emergence of FeLV-B following FeLV-A infection is considered to result in higher morbidity and mortality and has been associated with development of leukemia and lymphoma (2, 12). Diagnosis of FeLV in cats is typically achieved by detection of FeLV antigenemia (25), though modern molecular approaches afford opportunities to evaluate disease outcomes with more precision (26).
FFV and FcaGHV-1 are common chronic infections of domestic cats that to date are considered to represent apathogenic infections (27–33). FCoV is a prevalent chronic infection of domestic cats that predominantly occurs in an apathogenic enteric form but can emerge as a highly virulent pathogen, resulting in feline infectious peritonitis (FIP) in certain circumstances (34). While these viruses often exist simultaneously in multicat households, synergistic interactions among these infections have not been studied. Similarly, little work has been undertaken to investigate outcomes of FeLV natural infections using molecular techniques to characterize FeLV dynamics.
To further document viral pathogenicity and coinfection interactions that occur during natural FeLV infection, we evaluated the dynamics of an FeLV outbreak with high morbidity in a large colony of domestic cats living in a closed environment. We evaluated individual host factors, including age, sex, enFeLV copy number, FeLV subtype and viral load, and coinfections with FFV, FcaGHV-1, and FCoV to establish impacts of enFeLV and chronic viral coinfections on FeLV disease progression. The findings presented here document association of lower enFeLV copy number with male cats and better disease outcome. An interesting range of coinfection parameters highlight variations in disease transmission and expression among chronic pathogens.
RESULTS
Sixty-five cats were enrolled in this analysis.
Blood samples from one time point were obtained from 65 cats that originated from leopard cat/domestic cat hybrid matings that had been backcrossed to domestic cats for at least 8 generations. Animals ranged in age from 8 weeks to 9 years, with 30 males (8 neutered and 22 intact), 32 females (3 spayed, 27 intact, and 2 pregnant), and 3 animals of unidentified sex.
Cats harbored FeLV, FFV, FCoV, and FcaGHV-1 infections but were FIV negative.
The prevalence of four feline viral infections is summarized in Table 1. Thirty-two cats (49%) were positive for FeLV antigenemia by enzyme-linked immunosorbent assay (ELISA) SNAP test. All cats with antigenemia detected by ELISA SNAP had detectable plasma viremia by quantitative PCR (qPCR), as anticipated. Thirty-four of 65 cats (52%) tested positive for FeLV plasma viremia by qPCR, and the median viral load of FeLV plasma viremia was 1.9 × 108 copies per ml of plasma (Table 1). Two cats that were ELISA antigen negative but had detectable plasma viremia by qPCR had lower levels of viremia. Forty-three cats (66%) were FeLV provirus positive by qPCR, and the median proviral load of FeLV provirus was 8.4 × 106 copies per million cells (Table 1). Twenty-two (68%) of FeLV ELISA SNAP-positive cats were positive for FeLV-B by PCR. All cats were positive for enFeLV by qPCR. The median enFeLV copy number was estimated to be 34.5 copies per cell (range, 19.5 to 57.7 copies per cell) (Table 1). Thirty-nine cats (60%) were FFV provirus qPCR positive, and the median copy number of FFV was 3.3 × 103 copies per million cells (Table 1). While 20 cats (30%) were seropositive for FCoV antibodies, only 1 cat (1%) had detectable FCoV by qPCR (confirmed by Sanger sequencing). Eight cats (12%) were qPCR positive for FcaGHV-1, and 7 out of 8 (88%) FcaGHV-1-infected cats were male. No cats were positive for FIV antibody (Table 1).
TABLE 1.
Infection of cats with multiple viral pathogensa
| Virus and/or parameter | Assay | No. of cats positive/total | Total % positive | Median copy load (copies/million cells) |
|---|---|---|---|---|
| FeLV p27 antigen | ELISA SNAP test | 32/65 | 49 | NA |
| FeLV provirus | qPCR | 43/65 | 66 | 8.4 × 106 |
| FeLV viremia | qPCR | 34/65 | 52 | 1.9 × 108 copies/ml of plasma |
| FeLV-B | Conventional PCR | 22/32 | 68 | NA |
| enFeLV copy no. | qPCR | 65/65 | 100 | 34.5 copies/cell |
| FFV provirus | qPCR | 39/65 | 60 | 3.3 × 103 |
| FCoV serology | ELISA | 20/65 | 30 | NA |
| FCoV | qPCR | 1/65 | 1 | NA |
| FcaGHV-1 | qPCR | 8/65 | 12 | 1.9 × 103 |
| FIV antibody | ELISA SNAP test | 0/65 | 0 | NA |
Plasma and serum were used to detect FeLV p27 antigen and FIV antibody by ELISA SNAP test and in FCoV serology testing. RNA was extracted from plasma to test for FeLV viremia and FCoV by qPCR. All other tests utilized DNA extracted from PBMCs or whole blood. NA, not available.
A high proportion of cats were FeLV progressors.
Cats were classified into four different FeLV disease groups (progressive, latent, regressive, and regressive/abortive/uninfected) based on ELISA SNAP test, FeLV proviral load, and FeLV plasma viremia load using previously developed definitions (35, 36) (Table 2). The minimum FeLV viral load to result in a positive ELISA SNAP test result was determined to be 1 × 103 copies per ml of plasma (Table 2). Nearly half of all cats were classified as progressively infected (49%), with high FeLV proviral and plasma viremia loads (Table 2). Two cats (3%) were classified as latently infected. These cats had, on average, proviral loads and plasma viremia that were 3 and 6 orders of magnitude lower, respectively, than those of progressors, and they did not have detectable antigenemia by ELISA SNAP test. Nine cats (14%) had detectable proviral loads at very low levels but were negative for plasma viremia by ELISA SNAP test and qPCR and were classified as regressively infected. Twenty-two cats (34%) were negative for FeLV by qPCR and ELISA SNAP test and were classified as regressive/abortive/uninfected. As anticipated, high FeLV proviral load (greater than 4 × 105 copies per million cells) strongly correlated with high FeLV plasma viremia load (Spearman correlation P < 0.001) (Table 3; Fig. 1).
TABLE 2.
Diagnosis of 49% of cats in the colony as FeLV progressors with high mean proviral loads and plasma viremia
| Group classification | No. of cats/total | % of cats | SNAP ELISA result | Provirus status | Median proviral load (copies/million cells) | Range proviral load (copies/million cells) | Plasma viremia status | Median viremia load (copies/ml of plasma) | Range viremia load (copies/ml of plasma) |
|---|---|---|---|---|---|---|---|---|---|
| Progressive | 32/65 | 49 | Positive | Positive | 5 × 106 | 5 × 105–5 × 107 | Positive | 4 × 108 | 1 × 103–3 × 109 |
| Latent | 2/65 | 3 | Negative | Positive | 4 × 103 | 3 × 103–4 × 103 | Positive | 6 × 102 | 1 × 102–1 × 103 |
| Regressive | 9/65 | 14 | Negative | Positive | 3 × 102 | 2 × 102–6 × 102 | Negative | NA | NA |
| Regressive/abortive/uninfected | 22/65 | 34 | Negative | Negative | NA | NA | Negative | NA | NA |
TABLE 3.
Spearman correlation matrix of interrelationships among viral variablesa
| Virus and/or parameter | Value for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| FeLV status | Log10 FeLV proviral load | Log10 FeLV viremia load | Log10 enFeLV proviral load | FeLV-B status | Log10 FFV proviral load | FcaGHV1 status | FCoV ELISA status | |
| FeLV status | <0.001 | <0.001 | 0.702 | <0.001 | 0.001 | 0.215 | 0.498 | |
| Log10 FeLV proviral load | 0.876 | <0.001 | 0.410 | <0.001 | 0.004 | 0.850 | 0.673 | |
| Log10 FeLV viremia load | 0.898 | 0.904 | 0.214 | <0.001 | 0.001 | 0.498 | 0.364 | |
| Log10 enFeLV proviral load | −0.060 | −0.129 | −0.194 | 0.186 | 0.446 | 0.455 | 0.697 | |
| FeLV-B status | 0.718 | 0.778 | 0.777 | −0.206 | 0.001 | 0.273 | 0.914 | |
| Log10 FFV proviral load | 0.469 | 0.433 | 0.474 | −0.119 | 0.480 | 0.215 | 0.020 | |
| FcaGHV1 status | 0.193 | 0.030 | 0.106 | 0.117 | 0.171 | 0.193 | 0.606 | |
| FCoV ELISA status | 0.106 | 0.066 | 0.142 | 0.061 | 0.017 | 0.354 | −0.081 | |
The lower triangle represents the Spearman ρ values. The upper triangle shows associated P values. Significant results are in bold.
FIG 1.
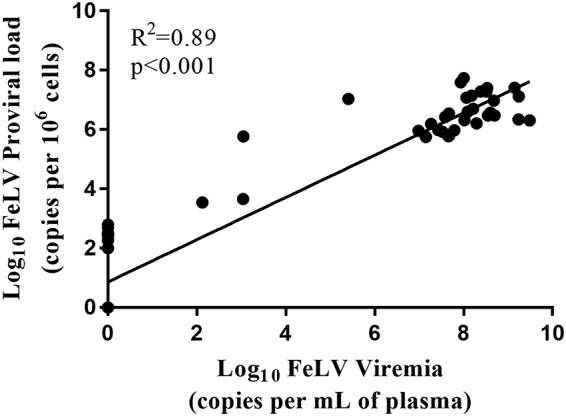
FeLV proviral load strongly correlated with plasma viremia as determined by Spearman correlation.
Combining demographic factors with pathogen associations revealed sex, but not age, effects.
We undertook a Spearman correlation analysis to evaluate patterns of coinfection among FeLV, FFV, FCoV, and FcaGHV-1. FeLV status, proviral load and viremia, FeLV-B status, and FFV were positively associated (Table 3). FCoV ELISA status was found to be positively associated with FFV proviral load (Table 3).
In order to simultaneously account for effects of cat demographic factors (sex and age) and associations between pathogens, we employed a structural equation modeling (SEM) approach (37). FeLV variables and FFV were included in this analysis based upon FeLV associations detected with correlation analysis (Table 3). We modeled sex and age as causal predictors of pathogen status/load and covariance among FeLV variables and FFV (Fig. 2). This analysis preserved the covariance relationships among FeLV variables and FFV determined by correlation analysis (Table 3). SEM revealed that (i) pathogen variables were not significantly predicted by cat age, (ii) enFeLV copy number was higher in male than female cats, and (iii) FeLV viremia and FeLV-B status were higher in female than male cats (Fig. 2). Consequently, enFeLV loads, which were higher in male cats, were inversely associated with FeLV viremia, a finding that was corroborated by multinomial logistic regression analysis (see below).
FIG 2.
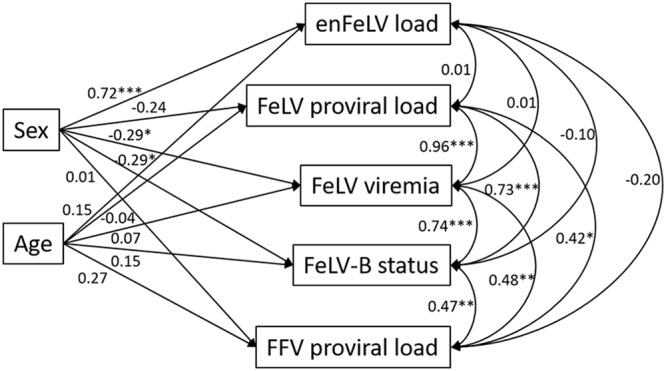
Structural equation model reveals FeLV associations with demographic factors and coinfection. FeLV variables covary with one another (curved double-headed arrows) and FFV proviral load (FFV is the only pathogen to covary with FeLV variables; see Table 3) and are predicted (single-headed arrows) by cat sex and age. Values represent standardized coefficients and are thus comparable in their relative-effect sizes (importance). Positive values associated with sex indicate higher response variable values for males, and negative coefficients indicate higher response variable values for females. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All FeLV and FFV variables are log10 transformed, except FeLV-B status. Values for variation (r2) are as follows: enFeLV load, 0.55; FeLV proviral load, 0.06; FeLV viremia, 0.09; FeLV status, 0.10; FFV proviral load, 0.07.
FeLV disease classifications, proviral load, and viremia were associated with enFeLV.
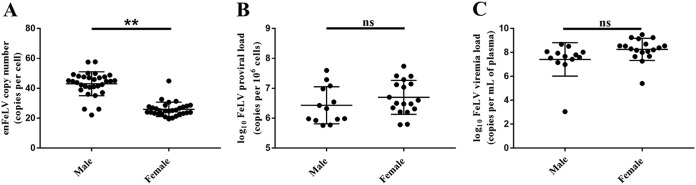
enFeLV copy numbers were lower in cats with progressive FeLV infections (P = 0.02) (Table 4; Fig. 3). Male cats were more likely to have an abortive infection and females were more likely to have a progressive infection (multinomial logistic regression, P = 0.045) (Table 5). Male cats had significantly higher enFeLV copy numbers than female cats (analysis of variance [ANOVA], P < 0.005) (Fig. 2 and 4A), and female cats had higher mean FeLV proviral loads (ANOVA, P = 0.116) (Fig. 2 and 4B) and higher FeLV viremia (ANOVA, P = 0.09) (Fig. 2 and 4C). These associations between enFeLV copy number, FeLV viral loads, and sex were supported by SEM (positive coefficient indicates greater infection in males).
TABLE 4.
Relationship of FeLV variables to FeLV disease classificationsa
| Variable and group classification | Coefficient | SE | t | P |
|---|---|---|---|---|
| enFeLV load | ||||
| Latent | −0.139 | 0.096 | −1.445 | 0.154 |
| Progressive | −0.084 | 0.036 | −2.317 | 0.024 |
| Regressive | −0.088 | 0.052 | −1.693 | 0.096 |
| FeLV proviral load | ||||
| Latent | 3.604 | 0.316 | 11.401 | <0.001 |
| Progressive | 6.586 | 0.119 | 55.556 | <0.001 |
| Regressive | 2.513 | 0.169 | 14.838 | <0.001 |
| FeLV viremia | ||||
| Latent | 2.584 | 0.629 | 4.108 | <0.001 |
| Progressive | 7.925 | 0.236 | 33.597 | <0.001 |
| Regressive | 0.000 | 0.337 | <0.001 | >0.999 |
| FeLV-B status | ||||
| Latent | <0.001 | 7,942.365 | <0.001 | >0.999 |
| Progressive | 20.355 | 2,292.763 | 0.009 | 0.993 |
| Regressive | <0.001 | 4,255.189 | <0.001 | >0.999 |
All values are relative to regressive/abortive/uninfected FeLV disease classification. All FeLV variables are log10 transformed, except FeLV-B status. All linear models based on Gaussian error distribution, except FeLV-B status, which has a binomial error distribution. Significant results are in bold.
FIG 3.
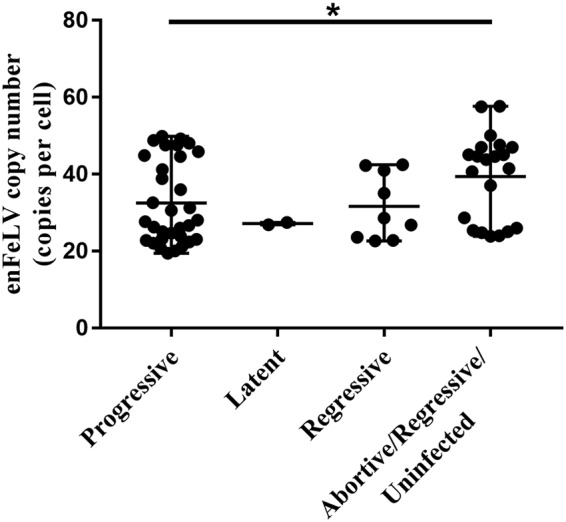
EnFeLV loads were significantly lower in cats with progressive disease than in those with regressive/abortive/uninfected disease (P = 0.02, Spearman correlation). There were 32 cats in the progressive group, the latent group had 2 cats, the regressive group had 9 cats, and the regressive/abortive/uninfected group had 22 cats. ns, not significant (a P value of <0.05 was considered statistically significant). *, P ≤ 0.05.
TABLE 5.
Relationship of FeLV disease classification to sexa
| Group classification | No. of cats |
|
|---|---|---|
| Female | Male | |
| Progressive | 18 | 13 |
| Latent | 2 | 0 |
| Regressive | 3 | 5 |
| Regressive/abortive/uninfected | 6 | 15 |
Males had proportionately more regressive/abortive/uninfected infections than females (χ2 = 6.93; P = 0.045), and females had proportionately more progressive infections than males. Three cats did not have gender recorded at the time of sample collection and were excluded from this analysis. Chi-square P value was simulated using Monte Carlo simulations, owing to expected values of <5 in latent and regressive disease classifications.
FIG 4.
EnFeLV and FeLV are correlated to sex. Male cats had significantly higher enFeLV proviral loads than female cats (P < 0.005) (A), while female cats had higher mean FeLV proviral loads than male cats (P = 0.116) (B) and higher FeLV plasma viremia loads than male cats (P = 0.09) (C) that approached statistical significance, respectively. P values were determined by using ANOVA. ns, not significant (a P value of <0.05 was considered statistically significant). **, P ≤ 0.005.
The presence of FeLV-B is associated with high proviral load and plasma viremia.
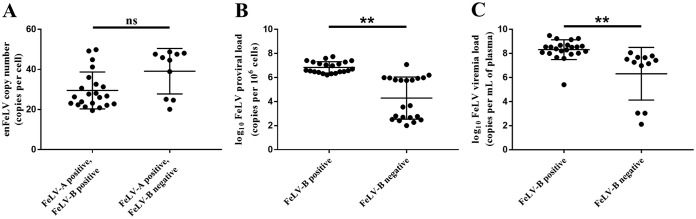
EnFeLV and FeLV-B status had a trend to be negatively correlated (Spearman correlation, P = 0.06) (Table 3; Fig. 5A). FeLV-B-positive cats had higher FeLV proviral loads (Spearman correlation, P < 0.001) (Table 3; Fig. 5B) and higher FeLV plasma viremia loads (Spearman correlation, P < 0.001) (Table 3; Fig. 5C). Female cats were more likely to be FeLV-B positive (ANOVA, P = 0.05). EnFeLV copy numbers were not significantly different in FeLV-B-positive and -negative cats (Table 3; Fig. 5A).
FIG 5.
FeLV-B status is associated with higher levels of exFeLV viremia and tends to be associated with higher enFeLV copy number. (A) Cats with exFeLV infection that were PCR positive for FeLV-B tended to have lower enFeLV copy numbers (P = 0.06). (B) Cats with higher FeLV proviral loads (P < 0.001) (B) and higher FeLV plasma viremia loads (P < 0.001) (C) were more likely to develop FeLV-B. P values were determined by using Spearman correlation. ns, not significant (a P value of <0.05 was considered statistically significant). **, P ≤ 0.005.
FFV-FeLV coinfection was frequently detected, and FFV proviral load was positively associated with progression of FeLV infection or disease.
Twenty-one of 65 cats (32%) were coinfected with both FeLV and FFV. FFV proviral load was found to covary with FeLV proviral load (Spearman correlation, P = 0.004) (Table 3; Fig. 2), FeLV plasma viremia load (Spearman correlation, P = 0.001) (Table 3; Fig. 2), and FeLV-B status (Spearman correlation, P = 0.001) (Table 3; Fig. 2).
DISCUSSION
This study documented relationships between FeLV infection and enFeLV, as well as coinfection with chronic apathogenic viruses, FFV, FCoV, and FcaGHV-1, in a large closed colony of cats. We documented several new observations regarding viral interactions previously unreported in a natural infection. Notably, enFeLV copy number was higher in males, and FeLV progressive disease status was inversely correlated with enFeLV copy numbers. FFV proviral load was found to correlate with FeLV proviral load, FeLV plasma viremia load, and FeLV-B status. Interestingly, three distinct patterns of viral infection emerged among FeLV, FFV, and FcaGHV-1, suggesting that different susceptibilities and infection rates exist for these three viruses. Three of our findings related to FeLV confound previously reported experimental or observational studies. Our study found higher enFeLV copy numbers than in previous studies. We also documented a higher proportion of animals in this study that progressed to FeLV disease, as well as a higher incidence of FeLV-B infection. Overall, our analysis indicates that lower enFeLV copy number correlates with subsequent exogenous viral disease progression, a finding with interesting implications relating to endogenous retrovirus (ERV) function in cats and other species.
enFeLV as measured by LTR sequences: high copy number and relationship to sex.
The average enFeLV copy number per cell as measured by qPCR in this colony was higher than reported in previous studies (34.5 copies/cell; range, 19.5 to 57.7 copies per cell versus 6 to 24 copies per diploid genome) (3, 7, 10, 38–41). The qPCR assay used to quantify enFeLV copy number detected long terminal repeat (LTR) sequences, which would consequently detect enFeLV sequences ranging from full-length endogenous retroviruses to solo LTRs, which are estimated to occur at high frequency in the cat genome (7, 38). This study used as a reference the feline CCR5 gene for quantification of enFeLV copies, while others have used the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as a reference (10). Multiple copies of GAPDH pseudogene sequences have been found in the feline genome, indicating that GAPDH may not be the best reference gene for estimating cell copy number (42); in contrast, CCR5 is present as a diploid gene (2 copies/cell [43]). These differences likely account for the higher number of enFeLV copies than that estimated by fluorescent in situ hybridization (FISH) (9) or qPCR detecting the U3 region in the enFeLV LTR (the same region as detected in this study) and the enFeLV env gene (10).
We found several associations between enFeLV copy number, gender, and naturally occurring infections. Male cats had higher enFeLV proviral loads than female cats, and FeLV viremia loads tended to be lower in male cats. Studies have suggested that male cats have higher enFeLV proviral loads due to increased copy numbers on the Y chromosome (10, 38, 44). Since the Y chromosome does not recombine with other chromosomes, sequences are rarely lost due to recombination, which underlies enFeLV copy number preservation in males (35, 44–46). Further analysis of the ratio of full-length enFeLV genomes to solo LTRs would further clarify this observation (7, 38, 47, 48).
This colony originated as a hybrid between leopard cats (Felis bengalensis) and domestic cats (Felis catus) approximately 10 years prior to sampling. Leopard cats do not harbor enFeLV, potentially leading to a dilution of enFeLV copy number relative to that in domestic cats. Backcross to domestic cats had occurred for multiple generations. Since F1 species hybrid individuals of heterogametic sex are typically sterile, leopard cat genotypes remaining in this colony would be mostly derived from female ancestry, since F1 males are likely to be infertile (49). This may contribute to the high level of progressive FeLV disease and/or the differences in enFeLV copy number between males and females and/or the susceptibility to FeLV-B noted in this colony. The relationship between F. bengalensis genomic elements and FeLV susceptibility is deserving of additional study to further elaborate this potential association.
FeLV progressive disease, demographics, and viral kinetics.
Our analysis recapitulated four infection outcomes for FeLV infections: regressive/abortive/uninfected, regressive, latent, and progressive. The findings from this study investigating a naturally infected group of cats largely corroborate results from studies of experimentally FeLV-infected cats (35, 36). Progressively infected cats are classified by high plasma viremia loads and high proviral loads. Latently infected cats have limited viral replication, resulting in low proviral and plasma viremia loads. Regressively infected cats possess low number of provirally infected cells and no detectable plasma viremia. Abortively infected cats have no actively replicating virus and no detectable provirus. Thirty-two cats (49%) in this colony had progressive FeLV disease as classified by active viral replication and high proviral loads (greater than 5 × 105 copies per million cells) and high plasma viremia loads (greater than 1 × 103 copies per ml of plasma). This frequency is higher than noted in most previous studies, which have reported that 30 to 40% of cats experimentally exposed to virulent FeLV developed progressive disease (1, 13, 50–52). The high level of progressive FeLV disease in this colony may be influenced by environmental factors, such as the close living proximity of animals in this colony, density of animals, and/or exposure and transmission of FeLV at an early age. As previously reported, we noted a strong correlation between FeLV proviral and plasma viral loads, suggesting that high proviral integration rates reflect active viral replication and persistent antigenemia. We could not conclusively differentiate cats that had not been exposed to FeLV and those that were exposed that cleared the infection (35). Analysis of anti-FeLV antibodies would be a useful adjunct to future studies to assist in relating host immune response to disease outcome. Additionally, this point-in-time collection is not predictive of future disease states.
Relationship between enFeLV and FeLV.
We documented an inverse relationship between enFeLV copy number and FeLV proviral load in cats that had a progressive infection, the first documentation of such a finding. Additionally, we found that cats with high enFeLV copy numbers were less likely to have progressive FeLV infections and tended to be less likely to develop FeLV-B than cats with low enFeLV copy numbers. These findings suggest a protective role of enFeLV.
Previous studies have investigated the relationship between endogenous retroviruses and related exogenous retroviruses. Tandon et al. sampled cats originating from five different specific-pathogen-free (SPF) catteries, privately owned cats from Switzerland, and European wildcats (Felis silvestris silvestris) (10). These cats were tested for enFeLV using assays specific for the U3 region of the LTR and the env gene. FeLV was detected in this cohort using real-time PCR targeting the unique U3 region for FeLV and FeLV p27 ELISA. Tandon et al. concluded that higher enFeLV copy numbers were found in cats that were FeLV provirus positive than in provirus-negative cats (10), contradicting findings reported here.
A follow-up study by Tandon et al. exposed cats with high or low enFeLV copy number with FeLV-A/Glasglow-1 (11, 53). FeLV and enFeLV viral loads and FeLV antigen levels were measured for 15 weeks (11). EnFeLV quantification was conducted via qPCR assays detecting two unique regions in the U3 region of the LTR and a third assay amplifying enFeLV env (10, 11). No significant differences were observed between cats with high and low enFeLV copy numbers or FeLV infection outcomes (regressive and progressive) (11). Persistently antigenemic cats with high enFeLV copy numbers were found to have higher levels of FeLV provirus, FeLV p27 antigen, and FeLV plasma viral RNA during early time points in the study (11). However, contradicting many of the conclusions from their paper, the authors noted that cats with high enFeLV loads were less likely to become persistently antigenemic than cats with low enFeLV loads over time (11). In the present study, we found that cats with higher enFeLV loads were also less likely to be progressively FeLV infected than those with lower enFeLV loads during a natural infection. The potential conclusions from this study strengthen the hypothesis that enFeLV may have a protective mechanism to prevent cats from developing progressive FeLV disease, though enFeLV does not seem to protect against FeLV infection. These studies suggest that enFeLV copy number could be a genetic predictor of susceptibility to FeLV infection and provide an avenue for further investigations determining endogenous and exogenous FeLV interactions.
Previous studies have indicated that some endogenous retroviruses may provide protection against homologous exogenous retroviral infection through receptor interference. Evidence of endogenous production of related envelope surface proteins could block receptors utilized by exogenous retroviruses, which has been documented for Friend murine leukemia virus and FeLV (9, 54–59). Experiments conducted by McDougall et al. demonstrated that infection of permissive fibroblasts with FeLV-B was prevented in cell culture medium containing an enFeLV-produced protein thought to be the envelope protein (58). A different endogenous retrovirus protection mechanism has been seen in sheep with the Jaagsiekte sheep retrovirus (JSRV). Endogenous production of a defective Gag antigen was shown to interfere with late stages of the viral replication cycle, resulting in accumulation of endogenous JSRV and exogenous JSRV particles in the cell cytoplasm (60). Thus, endogenous retroviruses could have multiple points of interference against a related exogenous retrovirus, resulting in truncation of exogenous viral replication.
Conversely, evidence has been found to suggest that endogenous retroviruses may enhance exogenous retroviral infection. Conservation of viral epitopes between endogenous and exogenous retroviruses may prevent the production of antibodies capable of neutralization of the virus, as has been seen in Friend murine leukemia virus infections (61) and avian leukosis virus infections (62, 63). Overall, further investigation of endogenous and exogenous interactions is required to determine the influence that endogenous retroviruses have over exogenous retroviral infections. The FeLV system provides a unique opportunity for these follow-up studies.
Emergence of FeLV-B: risk factors and disease associations.
FeLV-B was detected in 68% of FeLV-A-infected cats. The presence of FeLV-B correlated with higher FeLV proviral and viremia loads and was more likely to be detected in female cats. FeLV-B incidence in this colony is higher than in previous studies, in which FeLV-B was estimated to arise in 30 to 60% of FeLV-A infections (17, 18, 22, 64, 65). FeLV-B infection has been associated with leukemia, lymphoma, and increased pathogenesis (17). FeLV-B is nearly always identified in the presence of FeLV-A and is not typically horizontally transmissible in the absence of FeLV-A (17–20), although there has been in vitro evidence that FeLV-B can be isolated without FeLV-A being present (66). FeLV-A has been determined to use the thiamine transport protein (feTHTR1) as a cell entry receptor, whereas FeLV-B uses inorganic phosphate transporters Pit1 and Pit2 as receptors to gain cell entry (67). This difference in receptor specificity results in different cell tropisms for the two strains (67). Further studies will determine what proportion of FeLV infection can be attributed to FeLV-A in light of FeLV-B coinfection, whether all FeLV-B infections represent de novo recombination events, and if FeLV-B may be transmitted to a new host as a coinfection with FeLV-A.
Diagnosis of progressive FeLV.
Our study demonstrated that FeLV antigenemia as diagnosed by ELISA SNAP test is consistent with high plasma viremia and high proviral load. A plasma viremia load of 1 × 103 copies per ml of plasma or greater was consistent with a positive ELISA SNAP test. Two animals diagnosed as FeLV negative by antigen ELISA SNAP test had detectable plasma viremia by qPCR. Additional testing of these animals could be helpful in determining whether plasma qPCR is more sensitive in determining FeLV progressors or if a viral load of 103 particles/ml (equivalent to antigen ELISA SNAP positive) is more specific in determining FeLV progressor status than plasma qPCR.
FFV dynamics in a closed colony.
Feline foamy virus (FFV) is a retrovirus (genus Spumavirus) that is considered apathogenic in domestic cats despite persistent lifelong infection (68), though one prior study has documented renal micropathology in association with experimental disease (32). FFV is found at relatively high prevalence in free-ranging cat populations and is thought to be transmitted by social interactions (33, 69, 70). Two-thirds of cats in this colony were qPCR positive for FFV, and FFV and FeLV coinfection was common (32%).
We documented associations between FFV proviral load and FeLV proviral load, FeLV viremia, FeLV-B status, and FCoV ELISA status. This could indicate some facilitation of FFV load with infection with FeLV and FCoV. Indeed, the covariance relationship between FFV and FeLV persisted when cat sex and age were accounted for in the structural equation model, indicating that the pathogen associations cannot be attributed to host demographic factors alone (37). Further studies examining mechanisms of FFV associations with FeLV and FCoV, and coinfection relationships to disease, would be helpful.
FcaGHV-1 sex bias.
FcaGHV-1 has been recently discovered as a worldwide infection of domestic cats that to date has not been associated with a specific disease association (28–31). FcaGHV-1 was identified in approximately 12% in this colony by qPCR, a prevalence similar to that in other populations (28–31). Serological analysis has indicated that approximately half of cats with FcaGHV-1 exposure (as evidenced by reactive antibodies) are qPCR negative (27), suggesting that the actual incidence in this colony may be higher than 12%. Despite the close proximity of members of this colony, all but one of the FcaGHV-1-positive cats was male. This has also been seen in other populations in the United States, Europe, Brazil, and Australia (28–31, 71) but not Singapore (30). Distribution of FcaGHV-1 among male cats may be linked to male cat behavior, including aggressive encounters or territorial disputes, or to biological factors that influence immune system function, such as sex hormones (28). This very interesting finding suggests unique transmission characteristics for this agent compared to FeLV and FFV.
FCoV viremia is rare despite other cofactors that might promote FIP disease.
FCoV seroprevalence in this colony was approximately 30%. Previously reported prevalence ranges are from 14.6% in Japan to 70% in Austria (34). Risk factors for FCoV infection include multicat households (72); however, this colony had only one cat that had an active infection of FCoV as diagnosed by qPCR of blood cells. The presence of a single FCoV qPCR-positive cat suggests that FeLV, FFV, and FcaGHV-1 infections do not promote FCoV conversion to virulent feline infectious peritonitis (FIP) disease in this setting. However, testing for FCoV in feces was not conducted in this study, and since the fecal-oral transmission route is thought to be the main mode of transmission of this virus, further testing is required (73).
Comparing and contrasting disease dynamics in a multipathogen environment.
One very interesting observation, in this complex single point-in-time analysis of multiple pathogens in a large colony, is the very distinct pattern of infection that occurs with four concurrent viral agents. This colony experienced a virulent FeLV outbreak that was most severe in females and was potentially inversely correlated with enFeLV copy number. It is very likely that most animals in this colony were exposed to FeLV and approximately one-half of them were able to overcome infection. FFV is present in a large number of cats with and without FeLV progressive disease, FFV proviral load increases with age, and FFV infection appears to enhance FeLV and FCoV replication. The fact that FcaGHV-1 is present in a much lower proportion of animals, and predominantly in males, suggests a different route of transmission or susceptibility to this agent than for FFV and FeLV. FCoV typically manifests its more virulent phase as FIP in shelter situations or in association with FeLV (74, 76), and as noted above, it is surprising that we did not detect higher levels of FCoV viremia in more individuals in this colony.
Conclusions.
The association between enFeLV copy number, being male, and progressive FeLV infection in this colony of cats represents an interesting observation about the interaction between endogenous and exogenous retroviruses. This breeding colony represents a closed community of individuals with related genetic backgrounds exposed to similar environmental conditions. Additional studies of disease dynamics and relationship to genotype and demographic characteristics will provide useful insights into mechanisms underlying progressive FeLV disease, impacts of chronic FFV infection, and transmission dynamics underlying FcaGHV-1 propensity to infect males versus females.
MATERIALS AND METHODS
Animals and sampling.
Blood samples were obtained from a private breeding colony consisting of 65 leopard cat/domestic cat hybrids. All procedures were performed by a licensed veterinarian who managed the colony. The original breeding colony consisted of leopard cat/domestic cat hybrids that had been backcrossed to domestic cats for several generations. Four milliliters of blood collected in EDTA and serum was collected for colony health diagnostic purposes, and aliquots were shipped overnight on ice. One-time blood collections were used in this study.
Sample processing.
Blood was fractionated into plasma and peripheral blood mononuclear cells (PBMCs). Ficoll-Histopaque density gradient centrifugation (Histopaque-1077; Sigma Diagnostics, St. Louis, MO) was used to isolate PBMCs from whole blood. DNA was extracted from whole blood using a DNeasy blood and tissue extraction kit (Qiagen, Inc., Valencia, CA). RNA was extracted from plasma samples using an RNeasy minikit (Qiagen, Inc.). DNA and RNA concentrations were determined using a NanoDrop-1000 spectrophometer (Thermo Fisher Scientific, Waltham, MA). cDNA was synthesized from DNase-treated RNA extracts using the Superscript IV first-strand synthesis kit (Thermo Fisher Scientific). All nucleic acid samples were stored at −80°C.
FIV and FCoV serology/FeLV antigen analysis.
Commercially available ELISA SNAP tests (IDEXX Laboratories, Westbrook, ME) were used to determine FeLV and FIV status using plasma and serum samples. FCoV seropositivity was tested using a commercially available ELISA in plasma and serum (IVD Technologies, Santa Ana, CA).
enFeLV and FeLV quantitation.
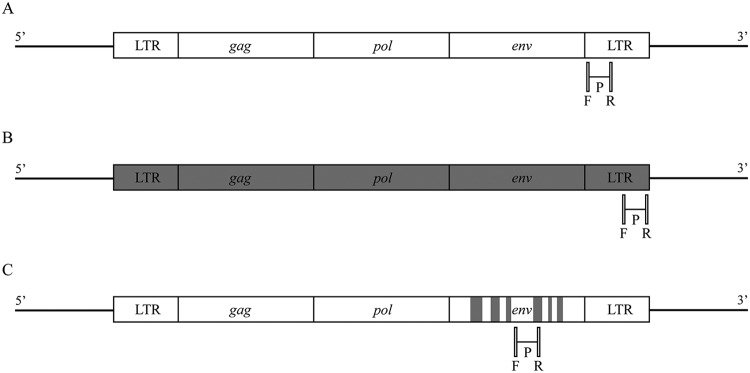
Real-time quantitative PCR (qPCR) was used to quantify enFeLV copy numbers by (forward, 5′-GTCTTATCCTAAGTCCACCGTTTA-3′; reverse, 5′-CTAGGCTCATCTCTAGGGTCTATC-3′; probe, 5′-CCTGGCCCTAAGATGGGAATGGAAA-3′) amplifying enFeLV LTRs. This assay detected both solo LTRs and full-length enFeLV (Fig. 6; Table 6). The probe was labeled with 6-carboxyfluorescein (FAM) reporter dye at the 5′ end, ZEN (Integrated DNA Technologies [IDT], Coralville, IA) internal quencher, and IBFQ (Iowa black fluorescein quencher; IDT). For exFeLV, the proviral and viral loads were quantified by qPCR using previously described primers and probe (36) (Fig. 6; Table 6). This qPCR detects all FeLV subtypes.
FIG 6.
Schematic of FeLV genome (A), enFeLV genome (B), and FeLV-B genome (C) with associated primers (forward and reverse [R]) and probes (P) for specific amplification of these viruses. Recombination of FeLV subtype A with enFeLV in the env gene, resulting in FeLV subtype B, is depicted.
TABLE 6.
Primers and probe sequences, amplification efficiencies, and limits of detection for all PCR assays used in this study
| Type of PCR and primers and probes utilized | Sequence | Amplicon length (bp) | Amplification efficiency (%) | Limit of detection |
|---|---|---|---|---|
| enFeLV provirus LTR real-time qPCR | 137 | 95 | 10 copies/cell | |
| Forward primer | GTCTTATCCTAAGTCCACCGTTTA | |||
| Reverse primer | CTAGGCTCATCTCTAGGGTCTATC | |||
| Probe | CCTGGCCCTAAGATGGGAATGGAAA | |||
| exFeLV provirus and viremia real-time qPCR | 67 | 95 | 10 proviral copies/million cells; 200 copies/reaction; 100 copies/ml of plasma | |
| Forward primer | AGTTCGACCTTCCGCCTCAT | |||
| Reverse primer | AGAAAGCGCGCGTACAGAAG | |||
| Probe | TAAACTAACCAATCCCCATGCCTCTCGC | |||
| Feline CCR5 real-time qPCR | 115 | 95 | 10 copies/million cells | |
| Forward primer | ACGTCTACCTGCTCAACCTGG | |||
| Reverse primer | ACCGTCTTACACATCCCATCCC | |||
| Probe | TCCGACCTGCTCTTCCTCTTCACCCTCC | |||
| FeLV-B screening conventional PCR | 1,932 | NA | NA | |
| Forward primer | CAGATCAGGAACCATTCCCAGG | |||
| Reverse primer | CCTCTAACTTCCTTGTATCTCATGG | |||
| FFV real-time qPCR | 144 | 97 | 100 copies/million cells | |
| Forward primer | GGACGATCTCAACAAGGTCAACTAAA | |||
| Reverse primer | TCCACGAGGAGGTTGCGA | |||
| Probe | AGACCCCCTAGACAACAACAGCAACACT | |||
| FcaGHV-1 real-time qPCR | 113 | 97 | 10 copies/million cells | |
| Forward primer | ACATCTTCACTGGACAACTGG | |||
| Reverse primer | GTGCATTTGATGTCCTGACTG | |||
| Probe | TGAACAGCTGAGTCTCTACAAGTCTCCA | |||
| FCoV real-time qPCR | 171 | 95 | NA | |
| Forward primer | AGCAACTACTGCCACRGGAT | |||
| Reverse primer | GGAAGGTTCATCTCCCCAGT | |||
| Probe | AATGGCCACACAGGGACAACGC |
qPCRs for enFeLV copy numbers, FeLV proviral loads, and FeLV plasma viremia contained 400 nM forward and reverse primers, 80 nM probe, iTaq Universal Probe Supermix (Bio-Rad, Hercules, CA), water, and 2 μl of DNA template for a total reaction volume of 25 μl. Thermal cycling conditions for enFeLV copy number, FeLV provirus, and FeLV plasma viremia assays were 95°C for 3 min, followed by 40 cycles of 95°C for 5 s and 60°C for 15 s.
Standard curves were created as custom synthetic oligonucleotides (gBlocks; IDT) containing a relevant fragment of enFeLV, FeLV-A, and CCR5. Standard dilutions and controls were run in duplicate, and samples were run in triplicate. Starting quantity (SQ) values were averaged for all samples. Samples that did not have at least two out of three reactions cross the threshold prior to cycle 40 were considered to be negative. The limit of detection for the enFeLV and FeLV provirus assays was 10 copies per cell. Controls used included (i) an FeLV-negative feline DNA sample from a specific-pathogen-free (SPF) (free of FeLV and FIV) cat colony, (ii) an FeLV-positive feline DNA sample, and (iii) a no-template control (DNA-free PCR-grade water). Canine DNA was used as a negative control in the feline CCR5 and enFeLV qPCR assays. A standard curve for FeLV plasma viremia was developed from cell culture supernatant from a persistently FeLV-infected Crandall Rees feline kidney cell (CrFK) line. Supernatant RNA was extracted and treated with DNase, cDNA was synthesized, and 10-fold serial dilutions were made from the stock cDNA. The limit of detection for the FeLV viremia assay was determined to be 100 copies per ml of plasma.
Quantification.
The feline CCR5 gene exists as two copies per cell, and previously described CCR5 primers and probe were used to normalize enFeLV and FeLV proviral copy numbers (Table 5) (41). CCR5 reactions and standard curves were run on the same plate with the enFeLV and FeLV reactions and standard curves. The CCR5 SQ values were determined using the standard curve included on every qPCR plate. The SQ value was then averaged and divided by two to find the number of cells in each sample since there are two copies of the CCR5 gene per cell. Next, the number of cells was divided by the SQ values for FeLV or enFeLV, as determined by the specific standard curves for each of these viruses, to give the proviral copies per cell. FeLV plasma viral load per milliliter of plasma was calculated using the appropriate dilution factor to consider original plasma volume, elution volume for RNA synthesis, and cDNA synthesis volume. The prevalence of FeLV-B was detected in samples using PCR with primers designed in the env gene region and LTR region unique to FeLV-B and enFeLV to exclude FeLV-A amplification (forward, 5′-CAGATCAGGAACCATTCCCAGG-3′; reverse, 5′-CCTCTAACTTCCTTGTATCTCATGG-3′) (Fig. 6; Table 6). Each reaction mixture contained 250 nM forward and reverse primers, Kapa HiFi mastermix (Kapa Biosystems), water, and 2 μl of DNA template in a 10-μl reaction. Thermal cycling conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and then one step of 72°C for 5 min. The controls included (i) feline DNA negative for FeLV-A but positive for enFeLV to ensure that the primers were not amplifying enFeLV sequences, (ii) a no-template control (PCR grade water with no DNA), and (iii) a positive control (FeLV-B-positive feline DNA). FeLV-B detection was confirmed by Sanger sequencing of PCR products (Quintarabio, San Francisco, CA). Finally, FIV serologic analysis was performed using a commercially available ELISA SNAP test. As all samples were negative, no further diagnostics were conducted. Feline foamy virus (provirus) was detected using viral DNA extracted from whole blood with a qPCR targeting the FFV gag gene (75) (Table 6). Feline gammaherpesvirus 1 viral load was determined using qPCR targeting glycoprotein B using DNA extracted from whole blood as previously described (28) (Table 6). FCoV antigen was detected using reverse transcriptase PCR (RT-PCR) with viral RNA extracted from plasma and serum. Primers and probe targeted the M and N genes using RNA extracted from plasma as previously described (70) (Table 6). FCoV detection was confirmed by Sanger sequencing of PCR products (Quintarabio).
Statistical analysis.
Data were compiled using Excel (Microsoft, Redmond, WA) and Prism software (GraphPad, San Diego, CA). Statistical analysis was performed using R (v. 3.3.1; R Development Core Team). Statistical differences between enFeLV copy number, FeLV proviral and plasma viremia loads, FeLV-B status, FFV proviral load, FcaGHV-1 status, and FCoV status were determined using Spearman correlation analysis. A structural equation model was used to build on the findings from the Spearman correlations, simultaneously accommodating cat sex and age with covariation among FeLV and FFV factors. Fit of the SEM was assessed as described by Carver et al. (37). Statistical differences of enFeLV copy number, FeLV proviral and plasma viremia loads, and FeLV-B status for FeLV infection outcomes were determined using multinomial logistic regressions. Statistical differences between viral loads using sex as a predictor variable were also determined using repeated-measure analysis of variance (ANOVA). A statistically significant difference was determined using a P value of ≤0.05. FeLV and FFV proviral copy number per million cells and FeLV viremia per milliliter were log transformed prior to analysis.
ACKNOWLEDGMENTS
This material is based in part upon work supported by NSF REU and EID DEB 1413925, NIH 5T32OD012201 and 1F30OD023386, and the Colorado State University Department of Microbiology, Immunology, and Pathology Undergraduate Research Fellows program.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
REFERENCES
- 1.Hoover EA, Mullins JI. 1991. Feline leukemia virus infection and diseases. J Am Vet Med Assoc 199:1287–1297. [PubMed] [Google Scholar]
- 2.Miyazawa T. 2002. Infections of feline leukemia virus and feline immunodeficiency virus. Front Biosci 7:d504–d518. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste RE, Todaro GJ. 1975. Segregation of RD-114 and FeLV-related sequences in crosses between domestic cat and leopard cat. Nature 257:506–508. doi: 10.1038/257506a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O'Brien SJ, Macdonald DW. 2007. The Near Eastern origin of cat domestication. Science 317:519–523. doi: 10.1126/science.1139518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O'Brien SJ. 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- 6.Coffin JM. 2004. Evolution of retroviruses: fossils in our DNA. Proc Am Philos Soc 148:264–280. [PubMed] [Google Scholar]
- 7.Roca AL, Pecon-Slattery J, O'Brien SJ. 2004. Genomically intact endogenous feline leukemia viruses of recent origin. J Virol 78:4370–4375. doi: 10.1128/JVI.78.8.4370-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology 3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen HB. 1997. Interactions between exogenous and endogenous retroviruses. J Biomed Sci 4:1–8. [DOI] [PubMed] [Google Scholar]
- 10.Tandon R, Cattori V, Willi B, Meli ML, Gomes-Keller MA, Lutz H, Hofmann-Lehmann R. 2007. Copy number polymorphism of endogenous feline leukemia virus-like sequences. Mol Cell Probes 21:257–266. doi: 10.1016/j.mcp.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Tandon R, Cattori V, Pepin AC, Riond B, Meli ML, McDonald M, Doherr MG, Lutz H, Hofmann-Lehmann R. 2008. Association between endogenous feline leukemia virus loads and exogenous feline leukemia virus infection in domestic cats. Virus Res 135:136–143. doi: 10.1016/j.virusres.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Jarrett O, Hardy WD, Golder MC, Hay D. 1978. The frequency of occurrence of feline leukaemia virus subgroups in cats. Int J Cancer 21:334–337. doi: 10.1002/ijc.2910210314. [DOI] [PubMed] [Google Scholar]
- 13.Dunham SP, Graham E. 2008. Retroviral infections of small animals. Vet Clin North Am Small Anim Pract 38:879–901. doi: 10.1016/j.cvsm.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Gomes-Keller MA, Gönczi E, Grenacher B, Tandon R, Hofman-Lehmann R, Lutz H. 2009. Fecal shedding of infectious feline leukemia virus and its nucleic acids: a transmission potential. Vet Microbiol 134:208–217. doi: 10.1016/j.vetmic.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Pacitti AM, Jarrett O, Hay D. 1986. Transmission of feline leukaemia virus in the milk of a non-viraemic cat. Vet Rec 118:381–384. doi: 10.1136/vr.118.14.381. [DOI] [PubMed] [Google Scholar]
- 16.Cattori V, Tandon R, Riond B, Pepin AC, Lutz H, Hofmann-Lehmann R. 2009. The kinetics of feline leukaemia virus shedding in experimentally infected cats are associated with infection outcome. Vet Microbiol 133:292–296. doi: 10.1016/j.vetmic.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Overbaugh J, Riedel N, Hoover EA, Mullins JI. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 18.Stewart M, Warnock M, Wheeler A, Wilkie N, Mullins J, Onions D, Neil J. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol 58:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarma PS, Log T. 1973. Subgroup classification of feline leukemia and sarcoma viruses viral interference and neutralization tests. Virology 64:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 20.Jarrett O, Russell PH. 1978. Differential growth and transmission in cats of feline leukaemia viruses of subgroups A and B. Int J Cancer 21:466–472. [DOI] [PubMed] [Google Scholar]
- 21.Donahue PR, Hoover EA, Beltz GA, Riedel N, Hirsch VM, Overbaugh J, Mullins JI. 1988. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J Virol 62:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neil JC, Fulton R, Rigby M, Stewart M. 1991. Feline leukaemia virus: generation of pathogenic and oncogenic variants. Curr Top Microbiol Immunol 171:67–93. [DOI] [PubMed] [Google Scholar]
- 23.Abkowitz JL, Holly RD, Grant CK. 1987. Retrovirus-induced feline pure red cell aplasia. J Clin Invest 80:1056–1063. doi: 10.1172/JCI113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleich S, Hartmann K. 2009. Hematology and serum biochemistry of feline immunodeficiency virus-infected and feline leukemia virus-infected cats. J Vet Intern Med 23:552–558. doi: 10.1111/j.1939-1676.2009.0303.x. [DOI] [PubMed] [Google Scholar]
- 25.Levy J, Crawford C, Hartmann K, Hofmann-Lehmann R, Little S, Sundahl E, Thayer V. 2008. 2008 American Association of Feline Practitioners feline retrovirus management guidelines. American Association of Feline Practitioners, Hillsborough, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu ES, Hoover EA, VandeWoude S. 2018. A retrospective examination of feline leukemia subgroup characterization: viral interference assays to deep sequencing. Viruses 10:29. doi: 10.3390/v10010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stutzman-Rodriguez K, Rovnak J, VandeWoude S, Troyer RM. 2016. Domestic cats seropositive for Felis catus gammaherpesvirus 1 are often qPCR negative. Virology 498:23–30. doi: 10.1016/j.virol.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troyer RM, Beatty JA, Stutzman-Rodriguez KR, Carver S, Lozano CC, Lee JS, Lappin MR, Riley SPD, Serieys LEK, Logan KA, Sweanor LL, Boyce WM, Vickers TW, McBride R, Crooks KR, Lewis JS, Cunningham MW, Rovnak J, Quackenbush SL, VandeWoude S. 2014. Novel gammaherpesviruses in North American domestic cats, bobcats, and pumas: identification, prevalence, and risk factors. J Virol 88:3914–3924. doi: 10.1128/JVI.03405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ertl R, Korb M, Langbein-Detsch I, Klein D. 2015. Prevalence and risk factors of gammaherpesvirus infection in domestic cats in Central Europe. Virol J 12:146. doi: 10.1186/s12985-015-0381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beatty JA, Troyer RM, Carver S, Barrs VR, Espinasse F, Conradi O, Stutzman-Rodriguez K, Chan CC, Tasker S, Lappin MR, VandeWoude S. 2014. Felis catus gammaherpesvirus 1; a widely endemic potential pathogen of domestic cats. Virology 460-461:100–107. [DOI] [PubMed] [Google Scholar]
- 31.McLuckie A, Tasker S, Dhand NK, Spencer S, Beatty JA. 2016. High prevalence of Felis catus gammaherpesvirus 1 infection in haemoplasma-infected cats supports co-transmission. Vet J 214:117–121. doi: 10.1016/j.tvjl.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 32.German AC, Harbour DA, Helps CR, Gruffydd-Jones TJ. 2008. Is feline foamy virus really apathogenic? Vet Immunol Immunopathol 123:114–118. doi: 10.1016/j.vetimm.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 33.Winkler IG, Löchelt M, Flower RLP. 1999. Epidemiology of feline foamy virus and feline immunodeficiency virus infections in domestic and feral cats: a seroepidemiological study. J Clin Microbiol 37:2848–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kummrow M, Meli ML, Haessig M, Goenczi E, Poland A, Pedersen NC, Hofmann-Lehmann R, Lutz H. 2005. Feline coronavirus serotypes 1 and 2: seroprevalence and association with disease in Switzerland. Clin Diagn Lab Immunol 12:1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann-Lehmann R, Cattori V, Tandon R, Boretti FS, Meli ML, Riond B, Pepin AC, Willi B, Ossent P, Lutz H. 2007. Vaccination against the feline leukaemia virus: outcome and response categories and long-term follow-up. Vaccine 25:5531–5539. doi: 10.1016/j.vaccine.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Torres AN, Mathiason CK, Hoover EA. 2005. Re-examination of feline leukemia virus: host relationships using real-time PCR. Virology 332:272–283. doi: 10.1016/j.virol.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 37.Carver S, Beatty JA, Troyer RM, Harris RL, Stutzman-Rodriguez K, Barrs VR, Chan CC, Tasker S, Lappin MR, VandeWoude S. 2015. Closing the gap on causal processes of infection risk from cross-sectional data: structural equation models to understand infection and co-infection. Parasit Vectors 8:658. doi: 10.1186/s13071-015-1274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roca AL, Nash WG, Menninger JC, Murphy WJ, O'Brien SJ. 2005. Insertional polymorphisms of endogenous feline leukemia viruses. J Virol 79:3979–3986. doi: 10.1128/JVI.79.7.3979-3986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koshy R, Gallo RC, Wong-Staal F. 1980. Characterization of the endogenous feline leukemia virus-related DNA sequences in cats and attempts to identify exogenous viral sequences in tissues of virus-negative leukemic animals. Virology 103:434–445. [DOI] [PubMed] [Google Scholar]
- 40.Niman HL, Akhavi M, Gardner MB, Stephenson JR, Roy-Burman P. 1980. Differential expression of two distinct endogenous retrovirus genomes in developing tissues of the domestic cat. J Natl Cancer Inst 64:587–594. [PubMed] [Google Scholar]
- 41.Tandon R, Cattori V, Willi B, Lutz H, Hofmann-Lehmann R. 2008. Quantification of endogenous and exogenous feline leukemia virus sequences by real-time PCR assays. Vet Immunol Immunopathol 123:129–133. doi: 10.1016/j.vetimm.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 42.Helfer-Hungerbuehler AK, Widmer S, Hofmann-Lehmann R. 2013. GAPDH pseudogenes and the quantification of feline genomic DNA equivalents. Mol Biol Int 2013:587680. doi: 10.1155/2013/587680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard KE, Reckling SK, Egan EA, Dean GA. 2010. Acute mucosal pathogenesis of feline immunodeficiency virus is independent of viral dose in vaginally infected cats. Retrovirology 7:2. doi: 10.1186/1742-4690-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kjellman C, Sjögren HO, Widegren B. 1995. The Y chromosome: a graveyard for endogenous retroviruses. Gene 161:163–170. [DOI] [PubMed] [Google Scholar]
- 45.Charlesworth B. 2003. The organization and evolution of the human Y chromosome. Genome Biol 4:226. doi: 10.1186/gb-2003-4-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy WJ, Sun S, Chen ZQ, Pecon-Slattery J, O'Brien SJ. 1999. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res 9:1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casey JW, Roach A, Mullins JI, Burck KB, Nicolson MO, Gardner MB, Davidson N. 1981. The U3 portion of feline leukemia virus DNA identifies horizontally acquired proviruses in leukemic cats. Proc Natl Acad Sci U S A 78:7778–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berry BT, Ghosh AK, Kumar DV, Spodick DA, Roy-Burman P. 1988. Structure and function of endogenous feline leukemia virus long terminal repeats and adjoining regions. J Virol 62:3631–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schilthuizen M, Giesbers MCWG, Beukeboom LW. 2011. Haldane's rule in the 21st century. Heredity (Edinb) 107:95. doi: 10.1038/hdy.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardy WD, McClelland AJ, Zuckerman EE, Hess PW, Essex M, Cotter SM, MacEwen EG, Hayes AA. 1976. Prevention of the contagious spread of feline leukaemia virus and the development of leukaemia in pet cats. Nature 263:326–328. [DOI] [PubMed] [Google Scholar]
- 51.Hoover EA, Rojko JL, Wilson PL, Olsen RG. 1981. Determinants of susceptibility and resistance to feline leukemia virus infection. I. Role of macrophages. J Natl Cancer Inst 67:889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojko JL, Hoover EA, Mathes LE, Olsen RG, Schaller JP. 1979. Pathogenesis of experimental feline leukemia virus infection. J Natl Cancer Inst 63:759–768. [DOI] [PubMed] [Google Scholar]
- 53.Jarrett O, Laird HM, Hay D. 1973. Determinants of the host range of feline leukaemia viruses. J Gen Virol 20:69–75. [DOI] [PubMed] [Google Scholar]
- 54.Buller RS, Wehrly K, Portis JL, Chesebro B. 1990. Host genes conferring resistance to a central nervous system disease induced by a polytropic recombinant Friend murine retrovirus. J Virol 64:493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buller RS, Sitbon M, Portis JL. 1988. The endogenous mink cell focus-forming (MCF) gp70 linked to the Rmcf gene restricts MCF virus replication in vivo and provides partial resistance to erythroleukemia induced by Friend murine leukemia virus. J Exp Med 167:1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gardner MB. 1993. Genetic control of retroviral disease in aging wild mice. Genetica 91:199–209. [DOI] [PubMed] [Google Scholar]
- 57.Gardner MB, Kozak CA, O'Brien SJ. 1991. The Lake Casitas wild mouse: evolving genetic resistance to retroviral disease. Trends Genet 7:22–27. [DOI] [PubMed] [Google Scholar]
- 58.McDougall AS, Terry A, Tzavaras T, Cheney C, Rojko J, Neil JC. 1994. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol 68:2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buller RS, Ahmed A, Portis JL. 1987. Identification of two forms of an endogenous murine retroviral env gene linked to the Rmcf locus. J Virol 61:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 101:11117–11122. doi: 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyazawa M, Fujisawa R. 1994. Physiology and pathology of host immune responses to exogenous and endogenous murine retroviruses—from gene fragments to epitopes. Tohoku J Exp Med 173:91–103. [DOI] [PubMed] [Google Scholar]
- 62.Smith EJ. 1986. Endogenous avian leukemia viruses, p 101–120. In de Boer GF. (ed), Developments in veterinary virology, vol 4 Avian leukosis. Springer, Boston, MA. [Google Scholar]
- 63.Wainberg MA, Halpern MS. 1986. Avian sarcomas: immune responsiveness and pathology, p 131–152. In de Boer GF. (ed), Developments in veterinary virology, vol 4 Avian leukosis. Springer, Boston, MA. [Google Scholar]
- 64.Roy-Burman P. 1995. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes 11:147–161. [DOI] [PubMed] [Google Scholar]
- 65.Phipps AJ, Hayes KA, Al-dubaib M, Roy-Burman P, Mathes LE. 2000. Inhibition of feline leukemia virus subgroup A infection by coinoculation with subgroup B. Virology 277:40–47. doi: 10.1006/viro.2000.0606. [DOI] [PubMed] [Google Scholar]
- 66.Stewart H, Jarrett O, Hosie MJ, Willett BJ. 2013. Complete genome sequences of two feline leukemia virus subgroup B isolates with novel recombination sites. Genome Announc 1:e00036-12. doi: 10.1128/genomeA.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–11057. doi: 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bleiholder A, Mühle M, Hechler T, Bevins S, VandeWoude S, Denner J, Löchelt M. 2011. Pattern of seroreactivity against feline foamy virus proteins in domestic cats from Germany. Vet Immunol Immunopathol 143:292–300. doi: 10.1016/j.vetimm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Nakamura K, Miyazawa T, Ikeda Y, Sato E, Nishimura Y, Nguyen NT, Takahashi E, Mochizuki M, Mikami T. 2000. Contrastive prevalence of feline retrovirus infections between northern and southern Vietnam. J Vet Med Sci 62:921–923. doi: 10.1292/jvms.62.921. [DOI] [PubMed] [Google Scholar]
- 70.Dye C, Helps CR, Siddell SG. 2008. Evaluation of real-time RT-PCR for the quantification of FCoV shedding in the faeces of domestic cats. J Feline Med Surg 10:167–174. doi: 10.1016/j.jfms.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kurissio JK, Rodrigues MV, Taniwaki SA, Zanutto M de S, Filoni C, Galdino MV, Araújo Júnior JP. 15 March 2018. Felis catus gammaherpesvirus 1 (FcaGHV1) and coinfections with feline viral pathogens in domestic cats in Brazil. Cienc Rural doi: 10.1590/0103-8478cr20170480. [DOI] [Google Scholar]
- 72.Foley JE, Poland A, Carlson J, Pedersen NC. 1997. Risk factors for feline infectious peritonitis among cats in multiple-cat environments with endemic feline enteric coronavirus. J Am Vet Med Assoc 210:1313–1318. [PubMed] [Google Scholar]
- 73.Kipar A, Meli ML. 2014. Feline infectious peritonitis: still an enigma? Vet Pathol 51:505–526. doi: 10.1177/0300985814522077. [DOI] [PubMed] [Google Scholar]
- 74.Cornell University College of Veterinary Medicine. 2014. FIP—feline infectious peritonitis. Cornell University College of Veterinary Medicine, Ithaca, NY: https://www2.vet.cornell.edu/departments-centers-and-institutes/cornell-feline-health-center/health-information/feline-health-topics/feline-infectious-peritonitis. [Google Scholar]
- 75.Lee JS, Mackie RS, Harrison T, Shariat B, Kind T, Kehl T, Löchelt M, Boucher C, VandeWoude S. 2017. Targeted enrichment for pathogen detection and characterization in three felid species. J Clin Microbiol 55:1658–1670. doi: 10.1128/JCM.01463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.UC Davis Koret Shelter Medicine Program. 1 September 2015. Feline infectious peritonitis/feline coronavirus (FIP/FCoV). Information sheet. http://www.sheltermedicine.com/library/resources/feline-infectious-peritonitis-feline-coronavirus-fip-fcov.