Influenza A viruses (IAVs) cause a significant burden on human and animal health, and mechanisms for interspecies transmission of IAVs are far from being understood. Findings from this study suggest that an equine-origin A(H3N8) IAV with mutation W222L at its hemagglutinin increased binding to canine-specific receptors with sialyl Lewis X and Neu5Gc motifs and, thereby, may have facilitated viral adaption from horses to dogs. These findings suggest that in addition to the glycosidic linkage (e.g., α2,3-linked and α2,6-linked), the substructure in the receptor saccharides (e.g., sialyl Lewis X and Neu5Gc) could present an interspecies transmission barrier for IAVs and drive viral mutations to overcome such barriers.
KEYWORDS: influenza A virus, equine influenza virus, canine influenza virus, A(H3N8), sialyl Lewis X, Neu5Gc, host tropism, submucosal glands, host adaption
ABSTRACT
An outbreak of respiratory disease caused by the equine-origin influenza A(H3N8) virus was first detected in dogs in 2004 and since then has been enzootic among dogs. Currently, the molecular mechanisms underlying host adaption of this virus from horses to dogs is unknown. Here, we have applied quantitative binding, growth kinetics, and immunofluorescence analyses to elucidate these mechanisms. Our findings suggest that a substitution of W222L in the hemagglutinin of the equine-origin A(H3N8) virus facilitated its host adaption to dogs. This mutation increased binding avidity of the virus specifically to receptor glycans with N-glycolylneuraminic acid (Neu5Gc) and sialyl Lewis X (SLeX) motifs. We have demonstrated these motifs are abundantly located in the submucosal glands of dog trachea. Our findings also suggest that in addition to the type of glycosidic linkage (e.g., α2,3-linkage or α2,6-linkage), the type of sialic acid (Neu5Gc or 5-N-acetyl neuraminic acid) and the glycan substructure (e.g., SLeX) also play an important role in host tropism of influenza A viruses.
IMPORTANCE Influenza A viruses (IAVs) cause a significant burden on human and animal health, and mechanisms for interspecies transmission of IAVs are far from being understood. Findings from this study suggest that an equine-origin A(H3N8) IAV with mutation W222L at its hemagglutinin increased binding to canine-specific receptors with sialyl Lewis X and Neu5Gc motifs and, thereby, may have facilitated viral adaption from horses to dogs. These findings suggest that in addition to the glycosidic linkage (e.g., α2,3-linked and α2,6-linked), the substructure in the receptor saccharides (e.g., sialyl Lewis X and Neu5Gc) could present an interspecies transmission barrier for IAVs and drive viral mutations to overcome such barriers.
INTRODUCTION
Influenza A viruses (IAVs) can infect a wide range of species, including humans, poultry, wild birds, pigs, horses, mink, marine mammals, and dogs (1–3). IAV was first isolated from dogs in the United States in January 2004 during an outbreak of respiratory disease among dogs in Florida. Epidemiologic evidence along with antigenic characterization and genetic analyses of the isolated subtype A(H3N8) IAV suggested that the virus had “jumped” from horses to dogs (3). The virus gained the ability to transmit between dogs and became enzootic among dog populations in the United States.
IAV transmission from birds to humans, or even between different mammalian hosts, is not uncommon, but such transmission is usually transient and causes only sporadic cases or small outbreaks with limited spread. It has been postulated that the adaption of avian IAVs to humans requires an intermediate mammalian host, such as the pig, that harbors both avian-like receptor saccharides containing α2,3-linked sialic acid-galactose (SA2,3Gal) and human-like receptor saccharides containing α2,6-linked sialic acid-galactose (SA2,6Gal) in the respiratory tract (4, 5). However, the glycosidic linkages alone cannot explain interspecies transmission of IAV between mammalian hosts or between mammalian and avian hosts. For example, receptor saccharides containing SA2,3Gal (6–8) are predominant in the respiratory tracts of dogs and horses, but equine IAV (EIV) cannot be transmitted freely between dogs and horses, as evidenced by the fact that the North American-lineage A(H3N8) EIV was only recently introduced into dogs and subsequently became enzootic among them (9).
The adaption of an influenza virus to a new host usually requires mutation(s) at one or multiple virus gene segments (10, 11). It has been suggested that the A(H3N8) EIV and A(H3N8) canine IAV (CIV) have minimal biological differences, and their cross-species transmission and adaption may be mediated by subtle changes in virus biology (12). Sequence analyses suggested that A(H3N8) CIV, compared to its precursor North American lineage A(H3N8) EIV, had mutations N54K, N83S, W222L, I328T, and N483T in the hemagglutinin (HA) (Table 1) and a few additional mutations in neuraminidase and other internal gene segments (13, 14). A difference in receptor binding between the wild-type A(H3N8) EIV (wt-eH3N8) and the wild-type A(H3N8) CIV (wt-cH3N8) has also been suggested (15).
TABLE 1.
Predominant residues at the receptor binding sites of H3N2 (canine and avian) and H3N8 (canine and equine) influenza virusesa
| Virus | Receptor binding site(s) |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| – | 130 loop |
– | – | 190 helix |
220 loop |
|||||||||||||||||||
| 98 | 134 | 135 | 136 | 137 | 138 | 153 | 183 | 188 | 189 | 190 | 191 | 192 | 193 | 194 | 195 | 221 | 222 | 223 | 224 | 225 | 226 | 227 | 228 | |
| Equine (H3N8) | Y | G | R/G | S | G | A | W | H | N/T | Q/K | E | Q | T | K | L | Y | P | W | V | R | G | Q | S | G |
| Canine (H3N8) | Y | G | R | S | G | A | W | H | N | Q/N | E | Q | T | K | L | Y | P | L | V/I | R | G | Q | S | G |
| Avian (H3N2) | Y | G | G | S | G | A | W | H | N | Q | E | Q | T | S | L | Y | P | W | V | R | G | Q | S | G |
| Canine (H3N2) | Y | G | G | S | G | A | W | H | N | Q | E | Q | T | S | L | Y | P | L | V | R | G | Q | S | G |
–, Sites not part of the 130 loop, 190 helix, or 220 loop. Boldfacing (site 222) indicates the key residues of interest in this study.
The molecular mechanism for how A(H3N8) IAV adapts from equine to canine remains unclear, largely because the types of sialosides and their relative distributions and abundance in the respiratory tract of dogs and horses have not been fully elucidated. We aim to gain an understanding of the molecular mechanisms that enabled A(H3N8) IAV to switch from horses to dogs and to provide insight into how this emerging IAV adapted to a new host.
RESULTS
Growth kinetics.
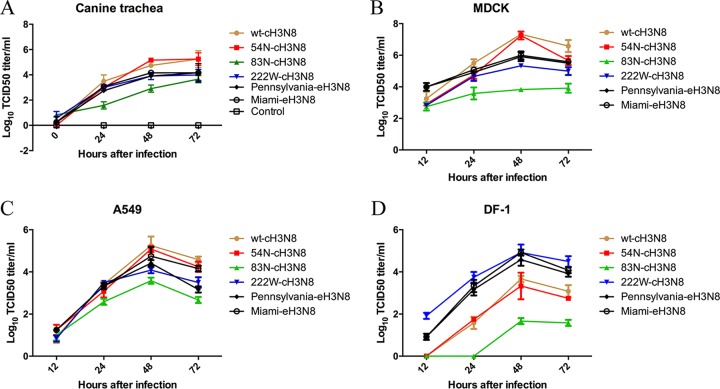
To evaluate the effect of mutations K54N, S83N, and L222W in the HA on the growth properties of A(H3N8) IAV, we compared the growth kinetics in canine trachea explants for the two EIV strains [i.e., one prototype EIV, A/equine/Miami/63(H3N8) (abbreviated as Miami-eH3N8), and a wild-type field strain of EIV, A/equine/Pennsylvania/1/2007(H3N8) (abbreviated as Pennsylvania-eH3N8)], wt-cH3N8, and three wt-cH3N8-derived HA mutants (54N, 83N, and 222W). No significant difference (P > 0.05) was found between the infectivity titers for 54N-cH3N8 and wt-cH3N8 in canine trachea explants (Fig. 1A). In contrast, the growth ability of cH3N8 in the explant tissues was significantly hampered by mutations S83N and L222W. wt-cH3N8 reached an average titer of 105.249 50% tissue culture infective dose (TCID50)/ml at 72 h after infection; this titer was 26.1-fold higher than that for 83N-cH3N8 (P < 0.05) and 21.5-fold higher than that for 222W-cH3N8 (P < 0.05). Of interest, titers for both Miami-eH3N8 and Pennsylvania-eH3N8 in canine trachea explants were similar to those for 222W-cH3N8 (P > 0.05). No virus infectivity was detected in the mock-infected canine trachea explants.
FIG 1.
Growth properties of equine influenza A virus, canine influenza A virus, and mutant viruses derived from canine influenza A viruses in canine trachea explants (A), MDCK cells (B), A549 cells (C), and DF-1 cells (D). Trachea explants from dogs were infected with 400 TCID50 of each virus. Cells were infected in triplicate with each virus at a multiplicity of infection of 0.01 and incubated at 37°C. Supernatant fluids were collected at indicated time points, and the viral titers were determined by TCID50 assay in MDCK cells. Each data point represents the mean virus yield (log10 TCID50/ml) from three individually infected wells ± standard deviation (vertical bars).
We further evaluated the growth properties of wt-cH3N8, three mutants, and wt-eH3N8 on Madin-Darby canine kidney (MDCK) cells, human lung carcinoma (A549) cells, and chicken fibroblast (DF-1) cells. Compared to wt-cH3N8, mutants 83N-cH3N8 and 222W-cH3N8 exhibited poorer growth in MDCK (Fig. 1B) and A549 cells (Fig. 1C), and 83N-cH3N8 had the poorest growth among the three mutants tested. Of interest, similar to the results in canine trachea explants, the replication kinetics for Miami-eH3N8 and Pennsylvania-eH3N8 in MDCK and A549 cells were similar to those for 222W-cH3N8 (P > 0.05) (Fig. 1B and C). In DF-1 cells, wt-cH3N8, 54N-cH3N8, and 83N-cH3N8 showed limited replication ability compared to the other three testing viruses (i.e., 222W-cH3N8, Miami-eH3N8, and Pennsylvania-eH3N8), which reached mean titers of 104.83, 104.58, and 104.91 TCID50/ml, respectively (Fig. 1D).
Receptor binding avidity and specificity.
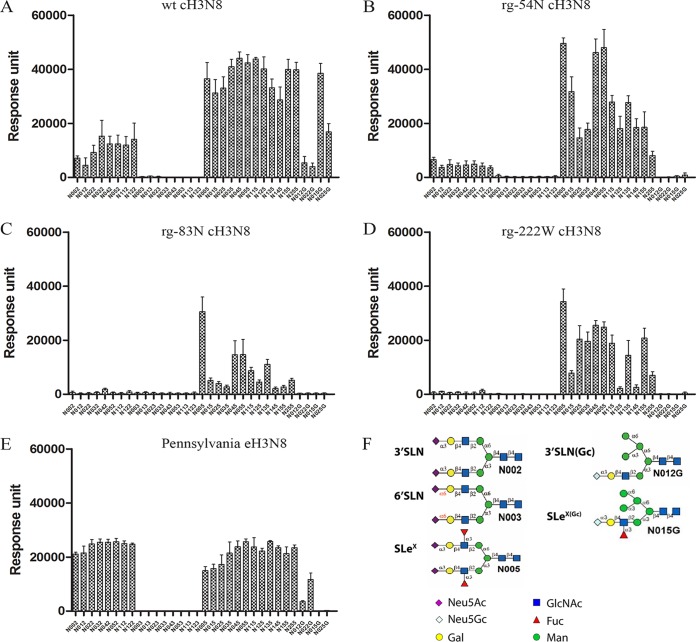
To determine the glycan structures specific for CIV, we generated four reassortant viruses using reverse genetics (rg; rg-wt–cH3N8, rg-54N, rg-83N, and rg-222W), which have HA from cH3N8 (wild-type HA, HA with mutation K54N, HA with mutation S83N, and HA with mutation L222W, respectively), neuraminidase from wt-cH3N8, and six internal genes from A/PR/8/1934(H1N1), and then compared the glycan binding profiles of these four reassortant viruses using an array of 611 glycans from the Consortium for Functional Glycomics (CFG; http://www.functionalglycomics.org) and an array of 83 N-linked glycan isoforms (here referred to as isoform microarray). To compare the glycan binding profiles of the CIVs to those of their precursor EIVs, we also determined the glycan binding profiles of the wild-type EIV field strain Pennsylvania-eH3N8 using the isoform microarray. Glycans on the CFG array have diverse terminal structures and a variety of spacer arms; glycans on the isoform microarray have the same base structures and spacer arms but different terminal structures (see Fig. S1 in the supplemental material).
Results from the CFG array showed that rg-wt–cH3N8 virus and all three mutants preferred binding to glycans containing α2,3-linkage sialic acids over those containing α2,6-linked sialic acids (see Fig. S2A in the supplemental material). The binding specificity of the rg-54N–cH3N8 mutant to some SA2,3GA-like glycans was slightly increased over that of the rg-wt–cH3N8 virus (see Table S1 in the supplemental material).
Consistent with results from the CFG array, results from the isoform microarray showed that all the testing viruses bound to glycans containing branched α2,3-linked sialic acids but not α2,6-linked sialic acids (Fig. 2A to E). Of interest, the data from this isoform microarray demonstrated that rg-wt–cH3N8 virus had stronger binding affinity to the α2,3-linked sialic acids with core fucose than those without (see Table S2 in the supplemental material). For example, the binding affinity of rg-wt–cH3N8 virus to N012 was about 7-fold less than that to N015, which is the fucosylated form of N012; rg-wt–cH3N8 virus showed strong binding signals to both linear (N025G) and branched (N015G) α2,3-linked, fucosylated N-glycolylneuraminic acid (Neu5Gc) but not to linear (N022G) or branched (N012G) α2,3-linked, nonfucosylated Neu5Gc (Fig. 2A; Table S2 in the supplemental material). Compared with the binding signals for rg-wt–cH3N8, the binding signals for all three mutants to the fucosylated α2,3-linked N-acetyl neuraminic acid (Neu5Ac) or Neu5Gc were reduced to different degrees (Fig. 2B to D). In contrast, Pennsylvania-eH3N8 showed moderate binding responses to α2,3-linked sialic acids, and the responses of the Pennsylvania-eH3N8 to α2,3-linked sialic acids with core fucose were similar to those without (Fig. 2E). Of note, the binding responses of Pennsylvania-eH3N8 to α2,3-linked sialic acids with core fucose are much weaker than those of rg-wt–cH3N8. The Pennsylvania-eH3N8 showed moderate or low responses to fucosylated Neu5Gc (N015G and N025G), linear nonfucosylated Neu5Gc (N022G), and branched nonfucosylated Neu5Gc (N012G), which are similar to cH3N8-222W but much weaker than those by rg-wt–cH3N8 (Fig. 2E).
FIG 2.
N-glycan microarray binding profiles of canine influenza A virus (CIV) and CIV-derived mutant viruses to representative linear and branched glycans. (A) wt–cH3N8. (B) rg-54N–cH3N8. (C) rg-83N–cH3N8. (D) rg-222W–cH3N8. (E) Pennsylvania-eH3N8. Viruses are indicated at the top of figure panels. The data are the mean fluorescent binding signal intensities ± the standard deviations (vertical bars). (F) Representative glycan structures on the isoform N-glycan microarray. N002-N122, Neu5Acα2,3 glycans; N003-N123, Neu5Acα2,6 glycans; N005-N255, fucosylated Neu5Acα2,3 glycans; N12G-N22G, Neu5Gcα2,3 glycans; N015G-N025G, fucosylated Neu5Gcα2,3 glycans. Neu5Ac, N-acetyl neuraminic acid; Neu5Gc, N-glycolylneuraminic acid. The structure details of these glycans are listed in Fig. S1 in the supplemental material.
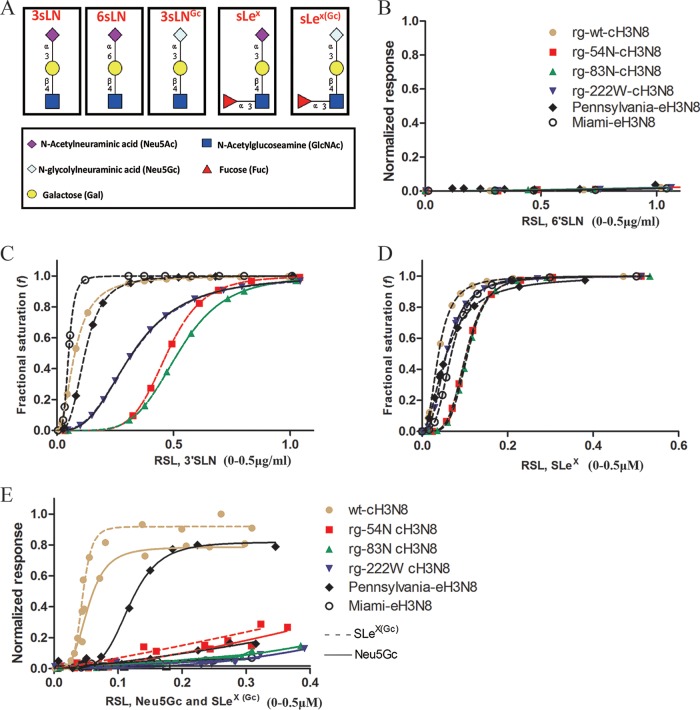
To confirm these results from glycan microarray analyses and further determine the distinct patterns derived from three mutations in CIV, we used biolayer interferometry analyses to quantify the binding avidity and specificity of wt-cH3N8 and the three mutants to five glycan analogs {Neu5Acα2-3Galβ1-4GlcNAcβ (3′SLN), Neu5Acα2-6Galβ1-4GlcNAcβ (6′SLN), Neu5Acα2-3Galβ1-4[Fucα1-3]GlcNAcβ (SLeX), Neu5Gcα2-3Galβ1-4GlcNAcβ [3′SLN(Gc)], and Neu5Gcα2-3Galβ1-4[Fucα1-3]GlcNAcβ (SLeX(Gc))} (Fig. 3A). For data interpretation, wt-eH3N8 was also included as a comparison. In agreement with glycan array analyses, these analyses showed that all tested viruses bound to 3′SLN (Fig. 3C) and SLeX glycan analogs (Fig. 3D), both of which contain α2,3-linked Neu5Ac, but not to 6′SLN (Fig. 3B), which contains α2,6-linked Neu5Ac. The rg-wt–cH3N8 showed strong binding affinity to SLeX(Gc) and 3′SLN(Gc), two glycan analogs with α2,3-linked Neu5Gc (Fig. 3E); Pennsylvania-eH3N8 showed decent binding responses to 3′SLN(Gc) but limited binding responses to SLeX(Gc) (Fig. 3E). Compared to Pennsylvania-eH3N8, the laboratory-adapted prototype CIV Miami-eH3N8 had much weaker binding responses to α2,3-linked Neu5Gc (Fig. 3E).
FIG 3.
Glycan binding specificity of canine influenza A virus (CIV) and CIV-derived mutant viruses to biotinylated α2,6–linked sialic acid (6′SLN), α2,3-linked sialic acid (3′SLN), Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ (SLeX), Neu5Gcα2-3Galβ1-4GlcNAcβ 3′SLN(3′SLN Gc), and Neu5Gcα2-3Galβ1-4(Fucα1-3)GlcNAcβ (SLeX(Gc)) glycan analogs as determined by biolayer interferometry (Pall ForteBio). (A) Structures of the glycan analogs. (B) Normalized response of virus to 6′SLN by dividing the maximum of wild-type (wt) virus response to 3′SLN. (C) Fractional saturation of viruses to 3′SLN. (D) Fractional saturation of viruses to SLeX. (E) Normalized response of viruses to 3′SLN Gc and SLeX(Gc). Values on the y axes represent the response of viruses to glycans after the association step. Normalized response data were determined by dividing the maximum response to each glycan or fractional saturation of the sensor at each relative sugar loading (RSL) at a fixed virus concentration of 100 pM. The RSL0.5 is the relative sugar loading on the streptavidin biosensor when the fractional saturation of the biosensor surface equals to 0.5. Given two testing viruses and a specific glycan, the higher binding response, and the smaller value of RSL0.5, the stronger the binding avidity a virus will have; given two testing glycans and a specific virus, the higher binding response, and the smaller value of RSL0.5, the stronger the binding avidity a virus will have.
To determine distinct patterns of binding among the three mutants (i.e., rg-54N-cH3N8, rg-83N-cH3N8, and rg-222W-cH3N8), rg-wt–cH3N8, and Pennsylvania-eH3N8 against these testing glycan analogs, we quantified 50% relative sugar loading concentration (RSL0.5), which is the relative sugar loading on the streptavidin biosensor when the fractional saturation of the biosensor surface equals to 0.5. Given two testing viruses and a specific glycan, the smaller value of RSL0.5, the stronger binding avidity a virus will have. The results suggested that among three mutants, RSL0.5 of Pennsylvania-eH3N8 to these testing glycan analogs reassemble to those of rg-222W-cH3N8 but not to those of the other two mutants. Specifically, for 3′SLN, rg-wt-cH3N8, rg-54N–cH3N8, and rg-83N–cH3N8 had RSL0.5 values of 0.0771, 0.4726, and 0.5194, respectively, whereas Pennsylvania-eH3N8 and rg-222W–cH3N8 had RSL0.5 values of 0.1207 and 0.3239, respectively; for SLeX, wt-cH3N8, rg-54N–cH3N8, and rg-83N–cH3N8 had RSL0.5 values of 0.0385, 0.1030, and 0.1066, respectively, whereas Pennsylvania-eH3N8, rg-222W–cH3N8 had values of 0.0572 and 0.0573, respectively; for SLeX(Gc), wt-cH3N8, rg-54N–cH3N8, and rg-83N–cH3N8 had RSL0.5 values of 0.1446, 0.3097, and 0.3851, respectively, whereas Pennsylvania-eH3N8 and rg-222W–cH3N8 had values of 1.332 and 1.074, respectively. In summary, rg-222W–cH3N8 and Pennsylvania-eH3N8 had a similar glycan binding pattern, and mutation L222W of HA decreased binding affinities of A(H3N8) CIV to glycans with α2,3-linked, fucosylated Neu5Gc.
SLeX and Neu5Gc glycan distribution.
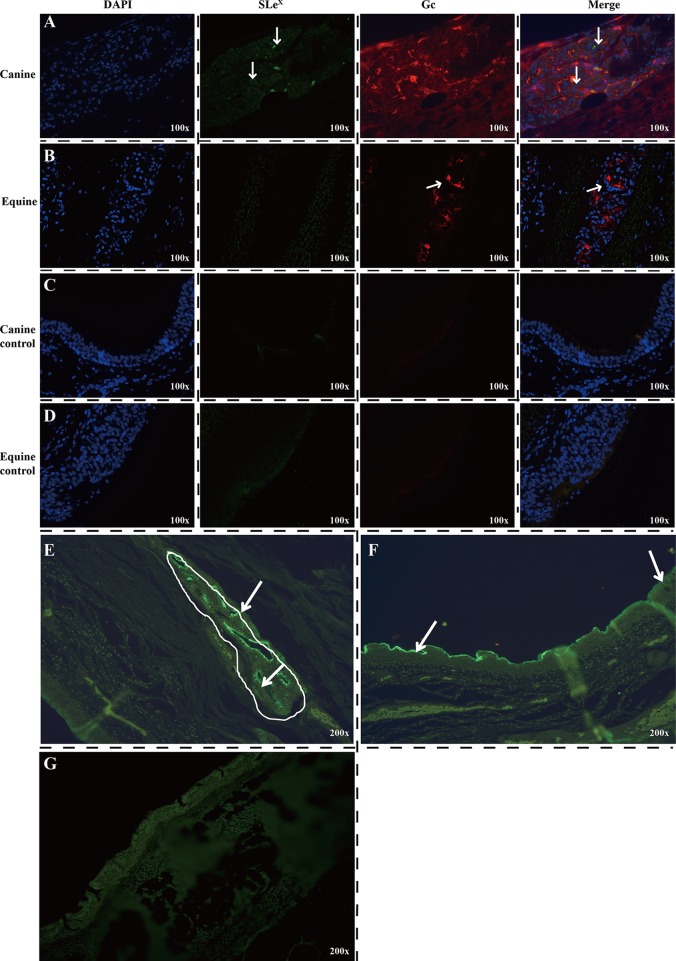
Based on the receptor binding results, we hypothesized that α2,3-linked fucosylated Neu5Gc is the receptor that determines the host tropism of A(H3N8) CIV and that α2,3-linked fucosylated Neu5Gc expression in the dog trachea would differ from expression in horse trachea. To test this hypothesis, we used immunofluorescence to detect SLeX and Neu5Gc in dog and horse tracheas to determine glycan receptor distribution. The canine submucosal glands were extensively and strongly immunopositive, indicating high SLeX glycan expression (Fig. 4A and E). Although the highest level of SLeX glycans was mainly found in the dog submucosal glands, staining was also present along the surface of the respiratory epithelium (Fig. 4F). In contrast, in the horse trachea, no SLeX was detected in the ciliated cells, nonciliated cells, or submucosal glands (Fig. 4B and G). On the other hand, Neu5Gc glycans were widely distributed in both dog and horse tracheas, including the ciliated and nonciliated epithelial cells and submucosal glands (Fig. 4A and B). These results demonstrate high expression of Neu5Gc in horse trachea but not SLeX, whereas dog trachea extensively expresses both SLeX and Neu5Gc.
FIG 4.
Immunofluorescence detection of SLeX (green) and Neu5Gc glycans (red) in dog (A, C, E, and F) and horse (B and D) tracheas. (A and E) SLeX glycans were mainly found in the epithelial cells of submucosal glands in dogs. (B) No SLeX glycans were detected in the ciliated cells, nonciliated cells, or submucosal glands of the horse trachea. Neu5Gc glycans were widely distributed in both dog and horse tracheas, including the ciliated and nonciliated epithelial cells and submucosal glands. (F) Expression of SLeX on the canine surface tracheal respiratory epithelium. (G) Detection of SLeX on the horse tracheal respiratory epithelium and submucosal glands. Original magnifications: ×100 (A, B, C, and D) or ×200 (E, F, and G). The SLeX glycan was detected by using Alexa Fluor 488 anti-human Sialyl Lewis X (dimeric) antibody (green), and the Neu5Gc glycans were detected by using Alexa Fluor 594 anti-Neu5Gc antibody (red). The arrows indicate immunopositive staining.
DISCUSSION
To understand the molecular mechanisms of the host adaption of an emerging influenza virus, it is necessary to characterize the virus receptor binding properties and the types and distributions of host glycan receptors, two key factors determining successful introduction and adaption of an IAV to a new host (16). In this study, we provide evidence supporting that A(H3N8) CIV prefers glycans with a SLeX epitope and that SLeX was differentially distributed in the trachea of the dog versus the horse. Our comparative analyses, combined with our previous observations (17), suggests that mutation W222L of HA increases the binding ability of A(H3N8) CIV to SLeX and Gc epitopes and that gaining the SLeX(Gc) binding ability could have facilitated the adaption of the equine-origin A(H3N8) virus to dogs.
Since the first outbreak of A(H3N8) IAV among dogs in 2004, this virus has become enzootic among the U.S. dog population (14). Prior epidemiologic and genetic studies demonstrated that this emerging A(H3N8) CIV originated in horses and was most closely related to the Florida sublineage strains (3). Interspecies transmission of A(H3N8) EIV from horses to dogs was observed when dogs were put in close contact with experimentally infected horses (18); however, A(H3N8) CIV-infected dogs cannot transmit CIV to horses (19), and A(H3N8) CIV replicates inefficiently in experimentally infected equine respiratory epithelial cells (20). This suggests that CIV and EIV are not exchangeable between equine and canine species and that A(H3N8) CIV has been well adapted in dogs. Nevertheless, A(H3N8) EIV and CIV have similar replication abilities in canine trachea primary cell lines (12) and similar growth patterns in MDCK cells, canine A72 cells, Norden Laboratory feline kidney cells, ferret Mpf cells, human A549 cells, and equine EQKD cells (12). However, differences in glycan binding preferences were observed between A(H3N8) CIV, Eurasia-lineage A(H3N8) EIV, and North America-lineage A(H3N8) EIV. Like Collins et al. (15), we observed that, compared with the Florida sublineage A(H3N8) EIV, A(H3N8) CIV had higher avidity to SLeX, supporting that CIV and EIV are not exchangeable between equine and canine species.
Only three mutations (K54N, S83N, and L222W) were observed between the head of HA protein of A(H3N8) CIV and that of A(H3N8) EIV. It is plausible that one of the mutations could facilitate virus adaption from horse to dogs. von Grotthuss and Rychlewski, who used computational simulation in a prior study (21), suggested that N54K could affect the glycosylation of HA and play a role in host adaption of A(H3N8) CIV. Our data demonstrated that mutations S83N and L222W, but not K54N, have a significant effect on the growth properties and binding properties of A(H3N8) CIV, a finding that does not support the hypothesis of von Grotthuss and Rychlewski (21) (Fig. 1 and 3). It is striking that mutant cH3N8-222W has a growth pattern similar to that of wt-eH3N8. Compared to cH3N8-222W, the mutant cH3N8-83N reduced significantly replication abilities of A(H3N8) CIV not only in canine trachea tissues but also in three cell lines: MDCK, A549, and DF-1 cells. A prior study reported that the infection of enzootic H3N8 CIV could lead to the quasispecies in the position 83 of CIV HA, suggesting that this position was likely to be associated with a viral fitness cost (22). Of interests, further analyses of HA sequences of H3N8 CIVs showed that there are polymorphisms at position 83 of CIV HA (1.4% N; 0.3% T; 0.3% R) (data not shown) and that, in contrast, all H3N8 CIVs have L at position 222 compared to the conserved W at this position for H3N8 EIVs (Table 1). This finding suggests that mutation W222L, but not N83S, could facilitate host adaption of A(H3N8) IAV from horses to dogs. Mutation W222L is located in the 220 loop of the HA receptor binding site, and the importance of residues in the 220 loop in determining the receptor specificity has been reported in other IAV subtypes (23, 24).
Previous studies demonstrated that the substructure of the carbohydrate receptors determine influenza host range and tissue tropisms. The abundance of Neu5Gcα2,3Gal moiety (a type of SAα2,3GA) is important for EIV to replicate in equine respiratory epithelium cells (6), as well as for CIV to replicate in canine respiratory epithelium cells (7, 8). Such results were confirmed by viral binding data from the CFG array, which showed that wt-cH3N8 and wt-eH3N8 prefer SAα2,3GA, including Neu5Gcα2,3Gal. However, such data still cannot explain the potential host factor(s) leading to host adaption of A(H3N8) from horses to dogs. Internal complexity of glycan structures below the sialic acid can influence glycan interaction with HA (25, 26); thus, host adaption cannot be understood without first understanding the glycan substructures of Neu5Gcα2,3Gal. The data from the isoform microarray and biolayer interferometry analyses demonstrated clear patterns of A(H3N8) CIV binding on a set of glycans with fucosylated α2,3-linked glycans (Fig. 2 and 3), and, of interest, a single L222W mutation dismissed that preference so that cH3N8-222W decreased binding to Neu5Gcα2,3Gal with the fucosylated motif (Fig. 3). Thus, mutation W222L seems to have increased the A(H3N8) binding preference from Neu5Gcα2,3Gal to SLeX(Gc), a Neu5Gcα2,3Gal with a fucosylated motif. Of note, a prior study suggested that amino acid residues at positions 222 and 227 of HA could affect IAV binding to glycans with the SLeX motif (27).
Immunofluorescent staining further demonstrated the distribution of SLeX in the tracheas (i.e., virus entry sites) of dogs and the lack of SLeX motifs in the tracheal tissues of horses. These results supported our hypothesis that SLeX motifs could be one of the key factors driving host adaption of A(H3N8) EIV in dogs; specifically, host adaption to dogs could have been driven by the presence of mutation W222L in the virus HA protein. In addition, the distribution of SLeX motifs is not only host specific but also tissue specific. SLeX glycans were primarily expressed in canine tracheal submucosal glands, with some expression noted in ciliated epithelial cells (Fig. 4). As a comparison, we probed SLeX in chicken trachea; the results showed that, unlike in dog trachea, SLeX glycans in chicken trachea were extensively expressed in both the ciliated epithelial cells and the submucosal glands (Fig. 5). The tracheobronchial submucosal glands play multiple roles during infection by producing mucus to inactivate the virus, but may also be infected and thus enhance the infection (28). The receptors for human IAV in ferrets was reported to be O-linked sialylated glycans, which are predominantly distributed in the submucosal glands, and the infection of such cells facilitates the efficient airborne transmission of virus by easily making the virus encapsulated into respiratory droplets (29). The L222W mutation was shown to increase binding of H3N8 CIV to SLeX glycans, which are present in abundance in the submucosal glands, thereby potentially facilitating the airborne transmission of the H3N8 CIV.
FIG 5.
Immunofluorescence assay detection of SLeX glycans in chicken trachea. The nuclei were stained with DAPI (blue). The SLeX glycan was detected by using Alexa Fluor 488 anti-human Sialyl Lewis X (dimeric) antibody (green). The original magnification was ×200.
Neu5Ac and Neu5Gc are two major sialic acids present in mammalian cells; they play important roles in the recognition of influenza virus during the initial step of viral infection. Neu5Gc has been reported to be present in horses (6), dogs (17), pigs (30), and mice (31), but not in humans. Of interest, although it has been reported that horse trachea expresses 90% of the α2,3-linked Gc receptors present in horses (6), our results demonstrate that Pennsylvania-eH3N8 binds moderately to 3′SLN(Gc), a finding that is consistent with that in a previous study (32). However, compared with those of Pennsylvania-eH3N8, the laboratory adapted EIV prototype strain Miami-eH3N8 showed much weaker binding responses to 3′SLN(Gc); such low responses of Miami-eH3N8 to 3′SLN(Gc) might have been caused by the extensive passage of this virus in embryonated chicken eggs, which lack Neu5Gc. Suzuki et al. (6) reported the presence of Gc glycans in equine trachea but not chicken trachea, a finding that was also observed in our study. Our immunofluorescence staining also showed a strong signal for Neu5Gc glycans in dog trachea (Fig. 4). The interaction of Neu5Ac or Neu5Gc glycans to the receptor binding site of viruses could affect the host tropism of those viruses. It has been reported that the large hydrophobic side chain of F75 of two human polyomaviruses (BK polyomavirus and JC polyomavirus) would clash with the glycolyl hydroxyl group and prevent binding of those two viruses to receptors terminating in Neu5Gc (33). Similarly, the L222W mutation introduced a large hydrophobic side chain and possibly caused the loss of binding to Neu5Gc glycans.
In this study, we only characterized the distribution and abundance of two individual glycan motifs, SLeX and Gc, in dog and horse tracheas by using immunofluorescent staining. One potential limitation of this study was that we were unable to test the SLeX(Gc)-like glycans due to the unavailability of SLeX(Gc)-specific antibody; thus, the abundance of this type of glycan substructure in the respiratory tracts of dogs and horses is unknown.
Like H3N8 CIV, avian H3N2 virus has been identified and is enzootic in canine populations in Southeast Asia (34, 35); furthermore, it has been shown that avian H3N2 virus was transmitted to the U.S. dog population (36). A previous study suggested that the W222L mutation of HA facilitated the viral binding affinity of H3N2 CIV to Neu5Gc and improved its replication ability in canine trachea primary cells (17). Interestingly, similar to W222L of HA in H3N8 CIV (Fig. 2), the W222L mutation of HA increased binding affinities of H3N2 CIV to SLeX(Gc) (17, 37).
In summary, these findings suggest that mutation W222L facilitates the host adaption of avian-origin H3N2 and equine-origin A(H3N8) IAVs in dogs by increasing the virus's ability to bind to receptors with Neu5Gc and/or SLeX, which are widely distributed in canine trachea tissues.
MATERIALS AND METHODS
Cells and viruses.
MDCK, A549, and DF-1 cells were purchased from the American Type Culture Collection (Manassas, VA). The cells were maintained at 37°C with 5% CO2 in Dulbecco modified Eagle medium (DMEM; Gibco/BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and penicillin (100 U/ml)–streptomycin (100 μg/ml). Influenza viruses A/canine/Iowa/13628/2005(H3N8), Miami-eH3N8, and Pennsylvania-eH3N8 maintained in our laboratory were used in the study; the viruses were propagated and titrated in the MDCK cells to determine their TCID50 values.
Gene cloning, site-directed mutagenesis, and virus rescue.
All gene segments of A/canine/Iowa/13628/2005(H3N8) virus were cloned into the phw2000 vector, as previously described (38). The QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) was used to create specific mutations in the HA gene by using the following primers: forward primer 5′-TGGGGAAAATATGCAACAACTCATATAGAATTCTAGATGG-3′ and reverse primer 5′-CCATCTAGAATTCTATATGAGTTGTTGCATATTTTCCCCA-3′ were used to generate mutation K54N in HA of A/canine/Iowa/13628/2005 (H3N8) virus; forward primer 5′-GCCCTTCAGTATGAGAATTGGGACCTCTTTATAG-3′ and reverse primer 5′-CTATAAAGAGGTCCCAATTCTCATACTGAAGGGC-3′ were used to generate mutation S83N in HA of A/canine/Iowa/13628/2005 (H3N8) virus; and forward primer 5′-CGAATCTAGACCGTGGGTCAGAGGTCAATC-3′ and reverse primer 5′-GATTGACCTCTGACCCACGGTCTAGATTCG-3′ were used to generate mutation L222W in HA of A/canine/Iowa/13628/2005 (H3N8) virus. In brief, the site-directed mutagenesis PCR amplification mixture contained 38.5 μl of water, 5 μl of 10× reaction buffer, 1 μl of a 2.5 mM deoxynucleoside triphosphate mix, 1.25 μl (100 ng/μl) of each primer, 2 μl (5 ng/μl) of wild-type (wt) HA segments of A/canine/Iowa/13628/2005 (H3N8) plasmid, and 1 μl of PfuUltra HF DNA polymerase (2.5 U/μl). The parameters of the site-directed mutagenesis PCR were as follows: one cycle at 95°C for 30 s, followed by 16 cycles at 95°C for 30 s, 55°C for 1 min, and 68°C for 5 min. The PCR products were then digested with DpnI at 37°C for 1 h, and the PCR product (2 μl) was transfected into XL1-Blue Supercompetent cells (Agilent Technologies). The transformed cells were directly inoculated onto Luria broth agar plates. The plasmids were then extracted, and the mutant viruses were rescued by plasmid-based reverse genetics methods, as described by Hoffmann et al. (39). All of the viruses were sequenced to confirm no unwanted mutations.
Replication kinetics.
To determine multistep growth curves, we infected MDCK, A549, and DF-1 cells with viruses at a multiplicity of infection of 0.01. After 1 h of incubation at 37°C, the cells were washed twice with warm phosphate-buffered saline (PBS) and overlaid with infection medium (Opti-MEM; Gibco/BRL) supplied with 1 μg/ml of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin and penicillin (100 U/ml)–streptomycin (100 μg/ml). Supernatants were collected at 12, 24, 48, and 72 h after infection and stored at −80°C for titration by TCID50 assay in MDCK cells.
Trachea collection and preparation.
Trachea tissues were obtained within 2 h from adult dogs following euthanasia for non-respiratory-related illness. Whole tracheas were transported in prewarmed wash medium consisting of a 1:1 mixture of DMEM and bronchial epithelial cell growth medium (BEGM; Lonza, Walkersville, MD) supplemented with penicillin (1,000 U/ml; Sigma, St. Louis, MO), streptomycin (500 μg/ml; Sigma). Once in the laboratory, tracheas were kept at 37°C in a 5% CO2 humidified incubator. Over 3 h, four to six washes were performed by immersing the tissues in fresh, warm wash medium.
Preparation of the tracheal explants.
After the washing steps, we removed the surrounding connective tissue exterior to the tracheal cartilage, opened the tracheas, and cut them lengthwise into four strips. Each segment was then cut into approximately 5-mm explants by biopsy punch (IntegraMiltex, York, PA). The explant slices were placed on a 24-well transwell plate (Corning, Corning, NY) supplied with 200 μl of explant growth medium consisting of a 1:1 mixture of DMEM and BEGM supplemented with penicillin (200 U/ml), streptomycin (200 μg/ml), amphotericin B (5 μg/ml; Lonza), and a BEGM SingleQuots kit (Lonza; one kit in 500 ml of 1:1 mixture of DMEM and BEGM). Explants were maintained at 37°C in a 5% CO2 humidified incubator; the apical and basal growth media were changed every 4 h to remove any secreted cytokines and inflammatory mediators. Before being infected, the trachea explants were washed two times with 2× sterile PBS. Then, 200 μl of infection medium containing BEGM with 0.5% bovine serum albumin (BSA), penicillin (200 U/ml), streptomycin (200 μg/ml), and amphotericin B (5 μg/ml) was added into the apical chamber, and the tissue was allowed to equilibrate for 30 min in the incubator. After removal of the infection medium, 400 TCID50 each of wt-cH3N8, Pennsylvania-eH3N8, Miami-eH3N8, rg54N-cH3N8, rg83N-cH3N8, and rg222W-cH3N8 mutants was added to the apical chamber of the well and incubated for 1 h at 37°C. After three washes with 1× sterile PBS, 500 μl of fresh growth medium was added to the basal chamber, and 200 μl of infection medium was added to the apical membrane. Infection medium was removed from the apical chamber at different time points, and the viral titers were determined by TCID50 assay in MDCK cells.
CFG glycan array.
Viruses binding to a wide range of glycan analogs were evaluated by Consortium for Functional Glycomics (CFG) glycan array. Viruses were first purified through a sucrose cushion at 100,000 × g for 3 h. The purified viruses were labeled with desiccated Alexa Fluor 488 NHS Ester (succinimidyl ester; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After dialysis with the Slide-A-Lyzer Mini dialysis device (Thermo Scientific, Rockford, IL), the Alexa Fluor 488-labeled viruses were transferred to clean tubes and stored at 4°C until used in glycan microarray hybridizations. In glycan hybridization, the version 5.0 glycan slides (CFG) were used as described previously (40). The binding image was read in a Perkin-Elmer ProScanArray scanner and analyzed using ImaGene 6.0 image analysis software (BioDiscovery, Inc., El Segundo, CA). The relative fluorescence unit (RFU) data were normalized by adjusting the total RFU to the same level across all experiments. A threshold of 2,000 RFU was used to floor the samples; only glycans with at least 2,000 RFU were analyzed statistically. A Wilcoxon signed rank sum test was used to compare the glycan-binding patterns among rg-wt–cH3N8, rg-54N–cH3N8, rg-83N–cH3N8, and rg-222W–cH3N8 mutants.
N-glycan isoform microarray.
The 83 N-glycans (41) were printed on N-hydroxysuccinimide (NHS)-derivatized slides as described previously (42). All glycans were printed in replicates of six in a subarray, and eight subarrays were printed on each slide. All glycans were prepared at a concentration of 100 pM in phosphate buffer (100 mM sodium phosphate buffer [pH 8.5]). The slides were fitted with an eight-chamber adapter to separate the subarray into individual wells for assay. Before the assay, slides were rehydrated for 5 min in TSMW buffer (20 mM Tris-HCl, 150 mM NaCl, 0.2 mM CaCl2, and 0.2 mM MgCl2, 0.05% Tween). Viruses are purified by sucrose density gradient ultracentrifugation and titrated to about 105 hemagglutination units/ml. Then, 15 μl of 1.0 M sodium bicarbonate (pH 9.0) was added to 150 μl of virus, and the virus was incubated with 25 μg of Alexa Fluor 488 NHS Ester (succinimidyl ester; Invitrogen) for 1 h at 25°C. After overnight dialysis to remove excess Alexa 488, viruses HA titer were checked and then bound to the glycan array. Labeled viruses were incubated on the slide at 4°C for 1 h, washed, and centrifuged briefly before being scanned with an InnoScan 1100 AL fluorescence imager (Innopsys, Carbonne, France).
Biolayer interferometry.
Two biotinylated glycan analogs (3′SLN [Neu5Acα2-3Galβ1-4GlcNAcβ] and 6′SLN [Neu5Acα2-6Galβ1-4GlcNAcβ]) linked to 30-kDa polymers containing 20% mol sugar and 5% mol biotin (GlycoTech, Gaithersburg, MD) were used to represent α2,3-linked sialic acid (3′SLN) and α2,6-linked sialic acid (6′SLN). The stock solution (1 mg/ml) was prepared in 1× PBS (vol/vol) according to the manufacturer's instructions. Biotinylated glycans Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ (SLeX), Neu5Gcα2-3Galβ1-4GlcNAcβ 3′SLN (Gc), and Neu5Gcα2-3Galβ1-4(Fucα1-3)GlcNAcβ [SLeX(Gc)] were synthesized. All glycan analogs and viruses were further diluted into the kinetics buffer (pH 7.4; PBS solution containing 0.01% BSA and 0.002% Tween 20) with neuraminidase inhibitors (10 μM zanamivir hydrate and 10 μM oseltamivir phosphate) for binding analysis. The binding affinities of viruses (100 pM) to glycan analogs [ranging from 0 to 0.5 μg/ml for 3′SLN and 6′SLN; 0 to 0.5 μM for SLeX, Gc, and SLeX(Gc)] were determined using an Octet RED96 interferometer equipped with streptavidin biosensors (Pall ForteBio LLC, Fremont, CA). In summary, RSL of the biosensor was calculated at the end of a 5- to 10-min loading, and the binding signal was measured at 25°C in a 20-min association step and 20-min dissociation step, with orbital shaking at 1,000 × rpm. The response of virus binding to a certain glycan loading was recorded at the end of the association step. The normalized response was calculated by dividing the maximum response of each glycan. The fractional saturation of the biosensor surface was calculated by using the Hill equation as reported by Xiong et al. (43). RSL0.5 is the relative sugar loading on the streptavidin biosensor when the fractional saturation of the biosensor surface equals to 0.5. Given two testing viruses and a specific glycan, a higher binding response, and a smaller value of RSL0.5, the stronger the binding avidity of a virus will be; given two testing glycans and a specific virus, a higher binding response, and a smaller value of RSL0.5, the stronger the binding avidity of a virus will be.
Detection of SLeX and Neu5Gc glycans in horse and dog trachea.
Normal horse and dog tracheal tissues were fixed in 10% neutral buffered formalin for 30 h at room temperature. The tissues were paraffin embedded and sectioned at 5 μm and then mounted on 3-aminopropyltrethoxy-silane-coated slides (Sigma). The slides were then deparaffinized in xylene and rehydrated through graded alcohols. Antigen was then retrieved using target retrieval solution (Dako, Carpinteria, CA), after which the slides were blocked by blocking reagent in 5% BSA for 1 h at room temperature and then washed three times with PBST (PBS with 0.05% Tween 20). For detection of sialyloligosaccharides reactive with SLeX antibody, the sections were incubated overnight at 4°C with 250 μl of Alexa Fluor 488 anti-human Sialyl Lewis X (dimeric) antibody (1:100; BioLegend, San Diego, CA). Sections were washed three times each with PBST and PBS; the slides were then counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Thermo Scientific, Rockford, IL). For detection of sialyloligosaccharides reactive with Neu5Gc, slides were incubated overnight at 4°C with anti-Neu5Gc antibody (1:400; BioLegend). The slides were washed three times each in PBST and PBS and then incubated at room temperature for 1 h with goat anti-chicken IgY(H+L) secondary antibody and Alexa Fluor 594 (Thermo Scientific) diluted 1:100 in PBS with 1% BSA. The sections were counterstained with DAPI for 3 min at room temperature. After three washes with PBS, the sections were mounted on coverslips with ProLong Gold Antifade reagent (Thermo Scientific) and evaluated with a fluorescence microscope (Nikon, Tokyo, Japan). Photos were taken with a digital microscope camera (Olympus, Tokyo, Japan).
Sequence alignment and statistical analyses.
HA sequences of H3N8 (equine and canine) and H3N2 (canine and avian) influenza viruses were extracted from the influenza research database (http://www.fludb.org). The H3N2 CIVs included 120 isolates recovered from 2006 to 2018 and the H3N8 CIVs contained 347 isolates from 2003 to 2018. The multiple sequence alignments and the identification of the mutations were identified by using Bioedit software (44). Analysis of variance or unpaired t test was used to compare viral titers and binding to IAV glycan receptors.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the critical comments and insightful discussions from James Stevens and Robert Woods.
This study was supported by the National Institutes of Health (grants 1R15AI107702 and P20GM103646).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01115-18.
REFERENCES
- 1.Morens DM, Taubenberger JK. 2010. Historical thoughts on influenza viral ecosystems, or behold a pale horse, dead dogs, failing fowl, and sick swine. Influenza Other Respir Viruses 4:327–337. doi: 10.1111/j.1750-2659.2010.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3:e00166-00112. doi: 10.1128/mBio.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, Smith C, Hill RC, Ferro P, Pompey J, Bright RA, Medina MJ, Johnson CM, Olsen CW, Cox NJ, Klimov AI, Katz JM, Donis RO. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 4.Webby RJ, Webster RG. 2001. Emergence of influenza A viruses. Philos Trans R Soc Lond B Biol Sci 356:1817–1828. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster RG, Govorkova EA. 2014. Continuing challenges in influenza. Ann N Y Acad Sci 1323:115–139. doi: 10.1111/nyas.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki Y, Ito T, Suzuki T, Holland RE Jr, Chambers TM, Kiso M, Ishida H, Kawaoka Y. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 74:11825–11831. doi: 10.1128/JVI.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ning ZY, Wu XT, Cheng YF, Qi WB, An YF, Wang H, Zhang GH, Li SJ. 2012. Tissue distribution of sialic acid-linked influenza virus receptors in beagle dogs. J Vet Sci 13:219–222. doi: 10.4142/jvs.2012.13.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly JM, Blunden AS, Macrae S, Miller J, Bowman SJ, Kolodziejek J, Nowotny N, Smith KC. 2008. Transmission of equine influenza virus to English foxhounds. Emerg Infect Dis 14:461–464. doi: 10.3201/eid1403.070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbs EP, Anderson TC. 2010. Equine and canine influenza: a review of current events. Anim Health Res Rev 11:43–51. doi: 10.1017/S1466252310000046. [DOI] [PubMed] [Google Scholar]
- 10.Ince WL, Gueye-Mbaye A, Bennink JR, Yewdell JW. 2013. Reassortment complements spontaneous mutation in influenza A virus NP and M1 genes to accelerate adaptation to a new host. J Virol 87:4330–4338. doi: 10.1128/JVI.02749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manz B, Schwemmle M, Brunotte L. 2013. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol 87:7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng KH, Gonzalez G, Deng L, Yu H, Tse VL, Huang L, Huang K, Wasik BR, Zhou B, Wentworth DE, Holmes EC, Chen X, Varki A, Murcia PR, Parrish CR. 2015. Equine and canine influenza H3N8 viruses show minimal biological differences despite phylogenetic divergence. J Virol 89:6860–6873. doi: 10.1128/JVI.00521-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivailler P, Perry IA, Jang Y, Davis CT, Chen LM, Dubovi EJ, Donis RO. 2010. Evolution of canine and equine influenza (H3N8) viruses cocirculating between 2005 and 2008. Virology 408:71–79. doi: 10.1016/j.virol.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Payungporn S, Crawford PC, Kouo TS, Chen LM, Pompey J, Castleman WL, Dubovi EJ, Katz JM, Donis RO. 2008. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg Infect Dis 14:902–908. doi: 10.3201/eid1406.071270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins PJ, Vachieri SG, Haire LF, Ogrodowicz RW, Martin SR, Walker PA, Xiong X, Gamblin SJ, Skehel JJ. 2014. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A 111:11175–11180. doi: 10.1073/pnas.1406606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Graaf M, Fouchier RA. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang G, Li S, Blackmon S, Ye J, Bradley KC, Cooley J, Smith D, Hanson L, Cardona C, Steinhauer DA, Webby R, Liao M, Wan XF. 2013. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol 94:2599–2608. doi: 10.1099/vir.0.054692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka T, Nemoto M, Tsujimura K, Kondo T, Matsumura T. 2009. Interspecies transmission of equine influenza virus (H3N8) to dogs by close contact with experimentally infected horses. Vet Microbiol 139:351–355. doi: 10.1016/j.vetmic.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Yamanaka T, Nemoto M, Bannai H, Tsujimura K, Kondo T, Matsumura T, Muranaka M, Ueno T, Kinoshita Y, Niwa H, Hidari KI, Suzuki T. 2012. No evidence of horizontal infection in horses kept in close contact with dogs experimentally infected with canine influenza A virus (H3N8). Acta Vet Scand 54:25. doi: 10.1186/1751-0147-54-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintana AM, Hussey SB, Burr EC, Pecoraro HL, Annis KM, Rao S, Landolt GA. 2011. Evaluation of infectivity of a canine lineage H3N8 influenza A virus in ponies and in primary equine respiratory epithelial cells. Am J Vet Res 72:1071–1078. doi: 10.2460/ajvr.72.8.1071. [DOI] [PubMed] [Google Scholar]
- 21.von Grotthuss M, Rychlewski L. 2006. Influenza mutation from equine to canine. Science 311:1241–1242. doi: 10.1126/science.311.5765.1241. [DOI] [PubMed] [Google Scholar]
- 22.Hoelzer K, Murcia PR, Baillie GJ, Wood JL, Metzger SM, Osterrieder N, Dubovi EJ, Holmes EC, Parrish CR. 2010. Intrahost evolutionary dynamics of canine influenza virus in naive and partially immune dogs. J Virol 84:5329–5335. doi: 10.1128/JVI.02469-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 24.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 25.Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA, Paulson JC. 2012. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem 51:4860–4863. doi: 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji Y, White YJ, Hadden JA, Grant OC, Woods RJ. 2017. New insights into influenza A specificity: an evolution of paradigms. Curr Opin Struct Biol 44:219–231. doi: 10.1016/j.sbi.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiono T, Okamatsu M, Igarashi M, McBride R, de Vries RP, Peng W, Paulson JC, Sakoda Y, Kida H. 2016. Amino acid residues at positions 222 and 227 of the hemagglutinin together with the neuraminidase determine binding of H5 avian influenza viruses to sialyl Lewis X. Arch Virol 161:307–316. doi: 10.1007/s00705-015-2660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol 171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayaraman A, Chandrasekaran A, Viswanathan K, Raman R, Fox JG, Sasisekharan R. 2012. Decoding the distribution of glycan receptors for human-adapted influenza A viruses in ferret respiratory tract. PLoS One 7:e27517. doi: 10.1371/journal.pone.0027517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, Horiike G, Yamazaki Y, Kawabe K, Masuda H, Miyamoto D, Matsuda M, Nishimura SI, Yamagata T, Ito T, Kida H, Kawaoka Y, Suzuki Y. 1997. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett 404:192–196. doi: 10.1016/S0014-5793(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 31.Hedlund M, Tangvoranuntakul P, Takematsu H, Long JM, Housley GD, Kozutsumi Y, Suzuki A, Wynshaw-Boris A, Ryan AF, Gallo RL, Varki N, Varki A. 2007. N-Glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol 27:4340–4346. doi: 10.1128/MCB.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, Bovin NV, Klenk HD, Matrosovich MN. 2012. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol 86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stehle T, Khan ZM. 2014. Rules and exceptions: sialic acid variants and their role in determining viral tropism. J Virol 88:7696–7699. doi: 10.1128/JVI.03683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song D, Kang B, Lee C, Jung K, Ha G, Kang D, Park S, Park B, Oh J. 2008. Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 14:741–746. doi: 10.3201/eid1405.071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Jia K, Qi W, Zhang M, Sun L, Liang H, Du G, Tan L, Shao Z, Ye J, Sun L, Cao Z, Chen Y, Zhou P, Su S, Li S. 2013. Genetic characterization of avian-origin H3N2 canine influenza viruses isolated from Guangdong during 2006-2012. Virus Genes 46:558–562. doi: 10.1007/s11262-013-0893-3. [DOI] [PubMed] [Google Scholar]
- 36.Voorhees IEH, Glaser AL, Toohey-Kurth K, Newbury S, Dalziel BD, Dubovi EJ, Poulsen K, Leutenegger C, Willgert KJE, Brisbane-Cohen L, Richardson-Lopez J, Holmes EC, Parrish CR. 2017. Spread of canine influenza A(H3N2) virus, United States. Emerg Infect Dis 23:1950–1957. doi: 10.3201/eid2312.170246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pulit-Penaloza JA, Simpson N, Yang H, Creager HM, Jones J, Carney P, Belser JA, Yang G, Chang J, Zeng H, Thor S, Jang Y, Killian ML, Jenkins-Moore M, Janas-Martindale A, Dubovi E, Wentworth DE, Stevens J, Tumpey TM, Davis CT, Maines TR. 2017. Assessment of molecular, antigenic, and pathological features of canine influenza A(H3N2) viruses that emerged in the United States. J Infect Dis 216:S499–S507. doi: 10.1093/infdis/jiw620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley KC, Jones CA, Tompkins SM, Tripp RA, Russell RJ, Gramer MR, Heimburg-Molinaro J, Smith DF, Cummings RD, Steinhauer DA. 2011. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (novel 2009 H1N1). Virology 413:169–182. doi: 10.1016/j.virol.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Liu Y, Ma C, Qu J, Calderon AD, Wu B, Wei N, Wang X, Guo Y, Xiao Z, Song J, Sugiarto G, Li Y, Yu H, Chen X, Wang PG. 2015. Efficient chemoenzymatic synthesis of an N-glycan isomer library. Chem Sci 6:5652–5661. doi: 10.1039/C5SC02025E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen F, Li L, Zhao N, Chiang MJ, Xie H, Cooley J, Webby R, Wang PG, Wan XF. 2017. A Y161F hemagglutinin substitution increases thermostability and improves yields of 2009 H1N1 influenza A virus in cells. J Virol 92:e01621-17. doi: 10.1128/JVI.01621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, Skehel JJ, Gamblin SJ. 2013. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497:392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 44.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.