LMP1 is one of the most divergent sequences in the EBV genome and phylogenetically segregates into seven LMP1 strains. The LMP1 strains differ in geographical distribution and NPC tumor prevalence, but the molecular basis for this potential selection is not clear. While there are signaling motifs conserved in all LMP1 sequences from circulating EBV isolates, phylogenetic studies of NPC also suggest that there may be sequence selection for tumor-associated LMP1 strains and polymorphisms. The present study describes a modified anoikis assay that can distinguish LMP1 strains into two groups by biological properties. The pleiotropic LMP1 signaling properties and sequence diversity may offer a unique opportunity to illuminate the complex mechanisms of metastasis. Although the host genomic landscape is variable between NPC tumors, the present functional-mapping studies on LMP1 support the notion that viral proteins could serve as molecular tool kits and potentially reveal sequence-associated risk factors in NPC metastatic progression.
KEYWORDS: Epstein-Barr virus, anoikis, functional mapping, latent membrane protein 1 strains
ABSTRACT
Nasopharyngeal carcinoma (NPC) is a metastatic Epstein-Barr virus (EBV)-associated cancer that expresses the viral oncogenic protein, latent membrane protein 1 (LMP1). During epithelial metastasis, detached cells must overcome anoikis-induced cell death and gain the ability to reattach and restore growth potential. Anoikis assays have revealed cell survival mechanisms during suspension, but few studies have tracked the fate of cells surviving anoikis-inducing conditions. In this study, a modified anoikis assay was used to examine if the expression of LMP1 confers the recovery of epithelial cells from anoikis. Cells expressing LMP1 mutants and strains were evaluated for distinguishing properties in survival during suspension, reattachment, and outgrowth potential. Expression of LMP1 promoted the outgrowth of the NPC cell line HK1 following anoikis induction that was not attributed to enhanced cell survival in suspension or reattachment. The mechanism of LMP1-induced outgrowth required Akt signaling and the conserved PXQXT motif on LMP1, which activates Akt. Deletion of any of the three LMP1 C-terminal activation regions (CTAR) abrogated anoikis recovery, suggesting that additional LMP1-regulated signaling pathways are likely involved. Of the seven LMP1 strains, only B958, China1, and Med+ promoted HK1 outgrowth from anoikis. This distinguishing biological property segregates LMP1 strains into two categories (anoikis recovering and nonrecovering) and suggests that LMP1 strain-specific sequences may be important in determining metastatic outgrowth potential in NPC tumors.
IMPORTANCE LMP1 is one of the most divergent sequences in the EBV genome and phylogenetically segregates into seven LMP1 strains. The LMP1 strains differ in geographical distribution and NPC tumor prevalence, but the molecular basis for this potential selection is not clear. While there are signaling motifs conserved in all LMP1 sequences from circulating EBV isolates, phylogenetic studies of NPC also suggest that there may be sequence selection for tumor-associated LMP1 strains and polymorphisms. The present study describes a modified anoikis assay that can distinguish LMP1 strains into two groups by biological properties. The pleiotropic LMP1 signaling properties and sequence diversity may offer a unique opportunity to illuminate the complex mechanisms of metastasis. Although the host genomic landscape is variable between NPC tumors, the present functional-mapping studies on LMP1 support the notion that viral proteins could serve as molecular tool kits and potentially reveal sequence-associated risk factors in NPC metastatic progression.
INTRODUCTION
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus that infects greater than 90% of the global population and is associated with several B cell and epithelial cell cancers, including nasopharyngeal carcinoma (NPC) (1, 2). NPC is prevalent in specific geographic regions, such as Southeast Asia and southern China, with an annual incidence reaching approximately 25 to 30 cases per 100,000 (3). Although primary NPC tumors are sensitive to radio- and chemotherapies, NPC tumors are often not diagnosed until advanced stages of the disease, whereupon high rates of local and distal metastasis are observed (4). Recurrent and metastatic NPC tumors can be refractory to conventional therapies and account for the high mortality rates associated with this cancer (4). Early events in NPC metastasis attributed to the expression of the EBV latent membrane protein 1 (LMP1) have begun to be elucidated and are in part attributed to the induction of epithelial-mesenchymal transition (EMT) (5–7). However, the mechanisms underlying later metastatic events that promote the recolonization and outgrowth of distant metastatic tumors are largely undetermined.
In NPC, neoplastic cells are consistently positive for clonal and latent EBV infection, with expression of a restricted set of viral genes termed type II latency that include EBV nuclear antigen 1 (EBNA1), LMP1, LMP2A, and LMP2B, as well as the noncoding EBV-encoded RNAs (EBERs) and BamHI rightward transcripts (BARTs) (1). Expression of LMP1 transcript is detected in more than 90% of NPC tumors, although it is appreciated that there is cellular heterogeneity in the intensity and frequency of LMP1 staining within an NPC tumor (8, 9). LMP1 acts as a constitutively active signaling mimic of the tumor necrosis factor receptor (TNFR) CD40 and is considered the primary viral oncogene, as it is essential for the immortalization of B cells in vitro and promotes the oncogenic potential of epithelial cells (9–12). The LMP1 multipass transmembrane protein localizes to intracellular membranes, interacting with multiple cellular signaling pathways in lipid raft microdomains (1, 13). Sequence polymorphisms and hydrophobic domains have shown that LMP1 contains 6 transmembrane domains and a C-terminal cytoplasmic domain that includes 3 conserved signaling motifs termed C-terminal activation regions 1 (CTAR1), -2, and -3 (1, 8). Through its CTAR domains, LMP1 modulates various cellular signaling pathways, including Akt, NF-κB, and mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) signaling and retinoblastoma (Rb)-regulated cell cycle pathways (13–19). CTAR1 enhances Akt, Rb, and MAPK/ERK signaling by binding TNFR-associated factor 1 (TRAF1), -2, -3, and -5 and activates canonical and noncanonical NF-κB signaling through multiple NF-κB dimers, including p52/p50, p52/p65, and p50/p50 (16). Canonical NF-κB signaling, regulated by the inhibitor of NF-κB alpha (IκBα), is primarily activated by CTAR2 through TRAF2 (20). While the biological properties attributed to CTAR1 and CTAR2 have been extensively analyzed, the potential functions of CTAR3 have only begun to be elucidated. Recently, CTAR3 has been shown to directly interact with the SUMO-conjugating enzyme Ubc9 during latency, which contributes to the oncogenic phenotypes promoted by LMP1 (21). In addition to the conserved CTAR signaling domains, the C terminus also encodes sequence polymorphisms with signature amino acid changes that phylogenetically segregate into distinct LMP1 strains that can be found in circulating lymphocytes and oral secretions of asymptomatic carriers (22). There are at least seven LMP1 strains, denoted B958, China1, China2, Med+, Med−, Alaskan (AL), and North Carolina (NC), with China1 as the most prevalent strain detected in NPC tumors (23, 24). Although notable differences in NF-κB signaling and biological properties have been described, overall, all LMP1 strains confer oncogenic potential and have not been clearly distinguished or grouped by phenotypes (25, 26).
Resistance to anoikis-induced cell death, which is triggered by the detachment of epithelial cells from the extracellular matrix, is a measure of metastatic potential for epithelial cells in vitro (27). There are broadly defined steps in metastasis that culminate in the outgrowth of recolonized cells, which initially involve the detachment of cells from the primary tumor, followed by regional metastasis to lymph nodes, survival during circulation, and finally reattachment to the extracellular matrix with reinitiation of cell growth (28). Circulating single cells and micromets may enter a quiescent state, sometimes termed “dormancy,” in which previously active proliferative programs are suppressed (29). The tumor cells that survive anoikis-induced cell death may remain dormant until a signal reinitiates proliferation, leading to the establishment of a metastatic tumor. One of the latent proteins expressed in NPC, LMP2A, has been shown to resist anoikis and to suppress cell death in suspended acini through the activation of ERK (30, 31). The pleiotropic signaling properties of LMP1 may augment epithelial metastasis by promoting a combination of EMT, prosurvival, focal adhesions, and cell cycle effects (13, 32). While the early events in conferring anoikis resistance have been examined in independent studies for LMP2A, the possible recovery of cells that escape anoikis has not been documented (30, 31).
In this study, the effect of LMP1 expression on metastatic outgrowth was evaluated by a modified anoikis assay that examines three phenotypes in epithelial cells: (i) survival in suspension, (ii) ability to reattach, and (iii) restoration of cell growth. Many established adherent cell lines are resistant to anoikis-induced cell death, including the hTERT-immortalized nasopharyngeal cell line NP460hTERT and the NPC tumor-derived cell lines HONE1 and HNE1 (data not shown), which are suspected to have been contaminated at some point with HeLa cells (33). However, the EBV-negative NPC cell line HK1 was identified as anoikis sensitive. In the modified anoikis assay, expression of LMP1, but not LMP2A, promoted the recovery of HK1 cells following anoikis induction, and this outgrowth property required the three conserved CTAR signaling domains (CTAR1, -2, and -3). Dominant-negative expression and mutational analyses determined that the mechanism involved Akt activation through the PXQXT motif localized to CTAR1. Remarkably, only three (B958, China1, and Med+) of the seven LMP1 strains promoted anoikis recovery, and this is the first clearly distinguishable phenotypic difference that can functionally segregate LMP1 strains. These data indicate that conserved signaling domains, as well as LMP1 strain-specific sequences, both determine the potential for anoikis recovery.
RESULTS
LMP1, but not LMP2A, promotes the outgrowth of HK1 cells after anoikis induction.
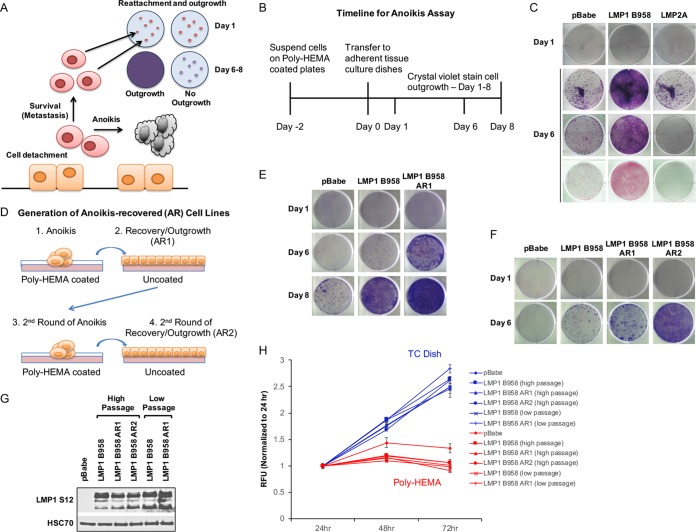
The traditional anoikis assay measures anchorage independence and proliferation during suspension of adherent cells on poly(2-hydroxyethyl methacrylate) (poly-HEMA)-coated culture dishes. In the present study, the anoikis assay was extended to additionally measure the fate of cells that survive anoikis-inducing conditions and are allowed to reattach and grow as adherent cultures. A schematic and a time line of the modified anoikis assay are presented in Fig. 1A and B. Epithelial cell lines were seeded on poly-HEMA-coated dishes for 48 h to induce anoikis. Anoikis-induced cell clumps were mechanically disrupted and transferred to tissue culture-treated dishes to attach overnight. Nonadherent dead cells were washed away, and the remaining adherent surviving cells were cultured and allowed to restore proliferation. Reattachment and outgrowth were assessed by crystal violet staining 1 day postreseeding and after recovery (4 to 8 days).
FIG 1.
LMP1 recovers HK1 cells in a modified anoikis assay. (A) Model for the modified anoikis assay. Cells are detached from the extracellular matrix using 0.05% trypsin and seeded onto poly-HEMA-coated dishes to induce anoikis. The cells that survive can reattach to a tissue culture-treated surface and reinitiate proliferation and outgrowth, which is measured by crystal violet staining after 6 to 8 days. (B) Time line for the modified anoikis assay. (C) Crystal violet staining of recovered HK1 stable cell lines following induction of anoikis after 1 and 6 days. The day 6 images are from three independent experiments. (D) Schematic representation of the generation of AR cell lines. AR1 indicates cell lines recovered after one round of anoikis induction, and AR2 indicates cell lines recovered after two rounds of anoikis induction. (E and F) Crystal violet staining of outgrowth following induction of anoikis for AR1 (E) and AR2 (F) cell lines. To better distinguish differences in the recovery of the AR cell lines, half the proportion of cells seeded in panel C was reseeded and analyzed for up to 8 days for recovery. (G) Immunoblot analysis of LMP1 expression for AR cell lines using LMP1 S12 hybridoma antibody. High-passage cell lines were passaged for >20 passages. Low-passage cell lines were passaged for <20 passages. HSC70 was used as a loading control. (H) Anoikis-induced changes in viability were assessed with the resazurin assay and analyzed after 24, 48, and 72 h on poly-HEMA-coated wells. The relative fluorescence units (RFU) and standard deviations were determined from 3 biological replicates. The blue lines represent tissue culture-treated dishes, and the red lines represent poly-HEMA-coated tissue culture dishes. The error bars indicate standard deviations. TC, tissue culture treated.
The majority of epithelial cell lines are resistant to anoikis-induced cell death. Several cell lines tested in these studies were resistant to anoikis, including the NPC-derived cell lines HONE1 and HNE1; a head and neck cancer cell line, PCI-15A; and an hTERT-immortalized epithelial cell line, NP460hTERT (data not shown). The immortalized breast epithelial cell line MCF10a is sensitive to anoikis and has been used for previous studies of anoikis resistance, but LMP1 expression only modestly promoted recovery from anoikis (reference 31 and data not shown). Therefore, the NPC cell line HK1, which was the most sensitive to induction of anoikis and recovery, was chosen for further evaluation.
In the modified anoikis assay, the effects of LMP1 and LMP2A stable expression were evaluated. Stable HK1 cell lines expressing LMP1 (strain B958) or LMP2A were assessed for outgrowth potential by crystal violet staining and compared to the pBabe vector control. At day 1 postrecovery, all stable cell lines (pBabe, LMP1, and LMP2A) reattached similarly (Fig. 1C, day 1); however, by day 6 postrecovery, only LMP1 efficiently restored proliferation to yield a confluent crystal violet-stained adherent culture (Fig. 1C, day 6). These data indicated that LMP1, but not LMP2A, can promote the outgrowth of cells recovering from anoikis-inducing conditions.
The modified anoikis assay selects for isogenic cell lines.
To further define the modified anoikis assay, cells recovering from anoikis were tested for isogenic properties. Anoikis-recovered (AR) LMP1-expressing cells were subjected to sequential rounds of anoikis induction and recovery (Fig. 1D). Half the proportion of cells were seeded for recovery in the AR cell line experiments to better distinguish differences in recovery over longer periods of up to 8 days postreseeding. After one round of anoikis induction and recovery, LMP1 AR1 cell lines demonstrated enhanced outgrowth compared to the parental LMP1 stable cell line (Fig. 1E). A second round of anoikis induction and recovery (AR2) further enhanced the outgrowth potential of LMP1-expressing cells (Fig. 1F). Although the titration of LMP1 levels is important for balancing LMP1's contrasting oncogenic and cytotoxic properties, the isogenic enrichment could not be explained by selection of LMP1 levels, as similar levels were expressed in the LMP1 parental and isogenic AR lines (Fig. 1G). Additionally, the resazurin cellular-viability assay demonstrated that the AR1 and AR2 cell lines were equivalently arrested and viable under poly-HEMA conditions, which is consistent with the hypothesis that the difference in recovery is attributable to properties postreseeding and not due to differences during anoikis (Fig. 1H). These results indicate that the modified anoikis assay is indeed a functional assay that can enrich for cells with enhanced anoikis recovery potential.
LMP1-induced outgrowth is not due to enhanced anoikis resistance or anchorage independence.
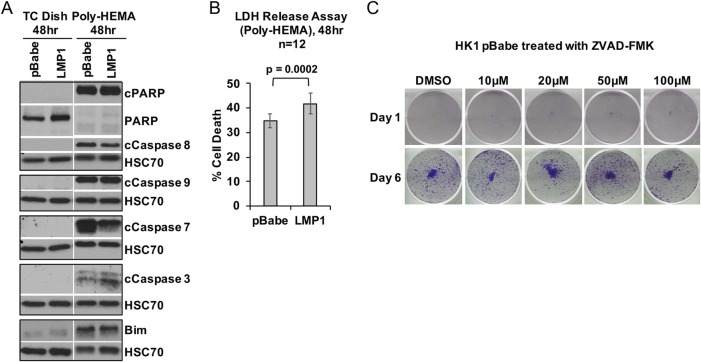
Prior to the reattachment of surviving cells, properties, including enhanced resistance to anoikis-induced cell death and/or gain in anchorage independence during suspension, could contribute to the increased outgrowth observed in the modified anoikis assay. Therefore, the effect of LMP1 expression during cell suspension was examined by immunoblotting for cell death markers and by cytotoxicity assays. Immunoblotting for activated caspases, cleaved poly(ADP-ribose) polymerase (PARP), and expression of the proapoptotic protein Bim was performed on lysates harvested from the pBabe vector control or stably LMP1-expressing cells grown on tissue culture-treated dishes and compared to anoikis-inducing poly-HEMA-coated conditions. Both pBabe and LMP1 cells from tissue culture-treated dishes showed little to no expression of Bim or cleavage of PARP and caspases (Fig. 2A). In contrast, pBabe- and LMP1-expressing cells cultured on poly-HEMA-coated dishes induced the activation of proapoptotic markers, including the cleavage of caspases (caspase 8, caspase 9, caspase 3, and caspase 7) and PARP and expression of Bim (Fig. 2A). However, the levels of caspase and PARP cleavage, as well as Bim expression, were similar between pBabe- and LMP1-expressing cells (Fig. 2A), suggesting that LMP1 does not promote cell survival under anoikis-inducing conditions. Additionally, a lactate dehydrogenase (LDH) release assay demonstrated that LMP1-expressing cells can recover from anoikis despite increased anoikis-induced cytotoxicity (41.8%) compared to pBabe control cells (34.9%) (Fig. 2B) (P = 0.0002) under poly-HEMA conditions, further providing evidence that LMP1 does not promote anoikis resistance under suspension conditions. Furthermore, treatment of HK1 pBabe cells with increasing concentrations of the pan-caspase inhibitor ZVAD-FMK under poly-HEMA-coated conditions did not rescue the outgrowth potential following anoikis induction (Fig. 2C). Therefore, we conclude that LMP1 likely promotes the recovery of cells by affecting a step subsequent to surviving anoikis.
FIG 2.
LMP1 does not enhance anoikis resistance. (A) Immunoblotting analysis of cell death markers using HK1 stable cell lines, grown for 48 h on either tissue culture-treated dishes (TC dish) or poly-HEMA-coated tissue culture dishes. The cell death markers used for these analyses included full-length and cleaved PARP (cPARP), cleaved caspases (caspases 8, 9, 7, and 3), and the proapoptotic protein Bim. HSC70 was used as a loading control. (B) After 48 h on poly-HEMA-coated dishes to induce anoikis, HK1 stable cell lines were analyzed for cytotoxicity by measuring the release of LDH from dying cells. Each data point represents the mean and standard deviation from 12 biological replicates; the P value was determined by two-tailed Student t test. (C) During a 48-h incubation on poly-HEMA-coated dishes, HK1 pBabe cells were treated with increasing concentrations of the pan-caspase inhibitor ZVAD-FMK to inhibit activation of caspases. The cells were transferred to adherent tissue culture dishes and assessed for outgrowth at 1 and 6 days by crystal violet staining. DMSO, dimethyl sulfoxide.
LMP1 outgrowth is attributable to the restoration of growth potential following reattachment to extracellular matrix.
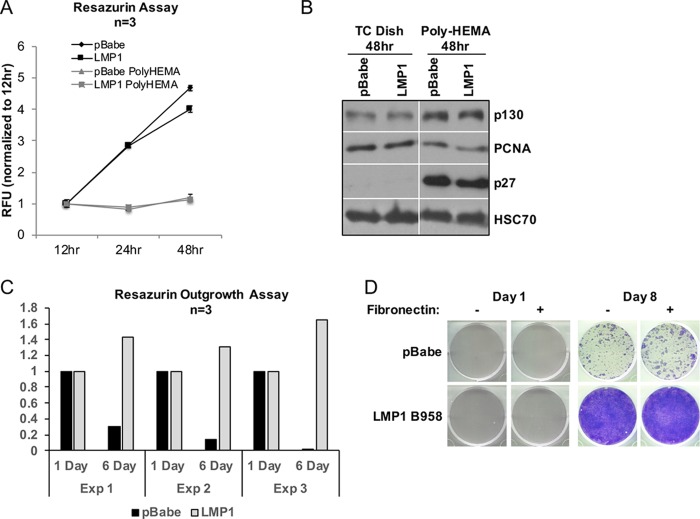
LMP1 induces anchorage-independent cell growth in soft-agar assays and promotes extracellular matrix interactions (5, 6, 15). In the absence of a difference in cell death markers to justify LMP1-induced outgrowth, one mechanism of anoikis resistance is the induction of anchorage-independent cell growth. To determine if LMP1-mediated outgrowth was due to enhanced growth under suspension conditions, a resazurin assay was performed to examine potential differences in proliferation during suspension (Fig. 3A). Resazurin is metabolized by mitochondrial dehydrogenase, and the assay measures proliferation of metabolically active cells. Under normal adherent tissue culture conditions (Fig. 3A, black lines), both pBabe and LMP1 cells show increasing metabolic activity, indicating that the cells are proliferating. However, when seeded onto poly-HEMA-coated dishes neither pBabe nor LMP1 cells have increasing metabolic activity over the 48-h period, demonstrating that the two cell lines are equivalently arrested during suspension (Fig. 3A, gray lines). Additionally, immunoblot analysis of cell cycle markers was performed, including the cell cycle arrest markers p130 and p27 and the S-phase proliferation marker PCNA (Fig. 3B). Both p130 and p27 were upregulated at similar levels in pBabe- and LMP1-expressing cells, with decreased PCNA expression in both cell lines, providing further evidence that LMP1 does not provide a growth advantage during cell suspension that could account for the increased outgrowth following anoikis recovery (Fig. 3C).
FIG 3.
LMP1 outgrowth is attributable to restoration of growth potential following reattachment to extracellular matrix. (A) Anoikis-induced cell cycle arrest was assessed by resazurin assay and analyzed after 12, 24, and 48 h on poly-HEMA-coated wells. The RFU and standard deviations were determined from 3 biological replicates. (B) Immunoblot analysis of cell cycle proteins p130, p27, and PCNA after 48 h incubation of HK1 stable cells on tissue culture-treated dishes (TC dish) or poly-HEMA-coated dishes. HSC70 was used as a loading control. (C) Outgrowth of HK1 pBabe and LMP1 cells following induction of anoikis was examined by resazurin assay at 1 and 6 days postreseeding after 48 h incubation on poly-HEMA-coated dishes. (D) Crystal violet staining of recovered HK1 stable cell lines 1 and 8 days following induction of anoikis. Fibronectin, a component of extracellular matrix, was included to facilitate attachment of HK1 pBabe cells.
LMP1 promotes the spontaneous assembly of focal adhesions and the production of fibronectin, a constituent of extracellular matrix, which could enhance cellular adhesion (7). Therefore, to test the importance of adhesion for outgrowth potential, anoikis-induced HK1 pBabe- and LMP1-expressing cells were transferred to tissue culture plates with or without fibronectin coating. The outgrowth potential for HK1 LMP1-expressing cells was the same regardless of the presence or absence of fibronectin coating after 8 days of recovery (Fig. 3D). Moreover, the outgrowth potential for HK1 pBabe cells did not increase despite the presence of fibronectin to promote cell adhesion (Fig. 3D). These data indicate that LMP1 most likely affects a step subsequent to the induction of anoikis and reattachment of viable cells.
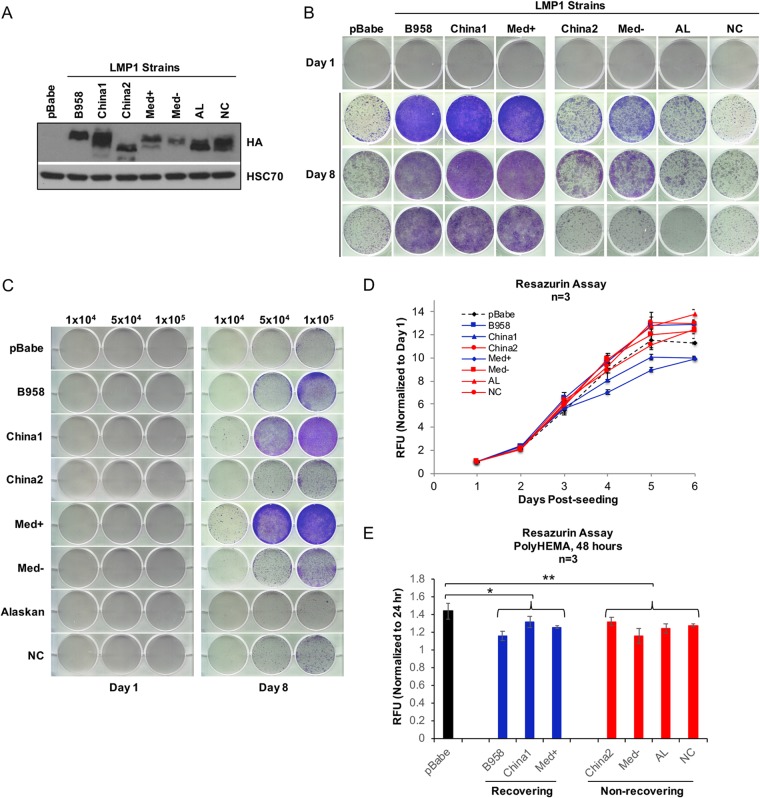
LMP1 strain variants B958, China1, and Med+ enhance outgrowth potential.
Although China1 is the predominant strain associated with NPC, LMP1 strains show similar biological and oncogenic properties in vitro, including transformation of Rat-1 fibroblasts, activation of phosphatidylinositol 3-kinase (PI3K)-Akt signaling, increased cell motility, and induction of cell cycle progression markers (26). To examine for potential LMP1 strain-dependent properties, HK1 stable cell lines expressing the previously analyzed representative isolates of the seven LMP1 strains were generated and tested in the modified anoikis assay (26). LMP1 cytostatic effects can result from high-level expression (34), which is highest in early passages during stable cell line selection. Therefore, to select for nontoxic levels of LMP1 expression, all the stable cells lines were analyzed for expression and biological properties above passage 9. To avoid differing reactivities to LMP1-specific antibodies, expression of the LMP1 strains was evaluated by immunoblotting for the common N-terminal hemagglutinin (HA) tag (Fig. 4A). In anoikis recovery and crystal violet staining, only the B958, China1, and Med+ strains promoted the recovery of HK1 cells, while China2, Med−, AL, and NC were impaired (Fig. 4B). To account for potential differences in viable-cell seeding during anoikis recovery, the anoikis recovery assay was further modified by trypan blue exclusion in which an equivalent number of viable cells were seeded for anoikis recovery, denoted “trypan blue anoikis recovery” experiments (Fig. 4C). Seeding of 1 × 105 viable cells resulted in the increased outgrowth in B958, China1, and Med+ strains, as previously defined in Fig. 4B. Notably, fewer viable cells (5 × 104 cells) were required for outgrowth in the China1 and Med+ strains, suggesting that China1 and Med+ may confer greater recovery potential. Under normal adherent growth conditions, all seven LMP1 strains grew similarly (Fig. 4D), indicating that the difference in outgrowth was indeed due to LMP1-mediated effects during anoikis recovery and not to inherent variability in proliferation rates. An assessment of cell viability during suspension on poly-HEMA by the resazurin assay (Fig. 4E) supported the results of the LDH cytotoxicity assay (Fig. 2A), indicating that pBabe vector control cells do not recover despite increased viability compared to the LMP1 strains and that the recovering LMP1 strains (B958, China1, and Med+) do not significantly differ from the nonrecovering strains (China2, Med−, AL, and NC), suggesting that the difference in recovery is likely attributable to properties postreseeding.
FIG 4.
LMP1 strain variants B958, China1, and Med+ enhance outgrowth potential. (A) Immunoblot analysis of HA-tagged LMP1 strain variants. HSC70 was used as a loading control. (B) Crystal violet staining of outgrowth for HK1 stable cells of LMP1 strain variants at 1 and 8 days. The day 8 images are from three independent experiments. (C) Crystal violet staining of HK1 outgrowth at 8 days. Specific numbers of viable cells (1 × 104, 5 × 104, and 1 × 105) were seeded following induction of anoikis as indicated at the top. (D) Analysis of growth kinetics by resazurin assay for HK1 stable cells on adherent tissue culture dishes. The dashed black line indicates the HK1 pBabe control, the blue lines indicate HK1 LMP1 strain variants that recover following induction of anoikis (B958, China1, and Med+), and the red lines indicate HK1 LMP1 strains that do not recover (China2, Med−, AL, and NC). The RFU and standard deviations were determined from 3 biological replicates. (E) Cell viability was assessed during anoikis induction by resazurin assay and analyzed after 24 and 48 h on poly-HEMA-coated wells. The RFU and standard deviations were determined from 3 biological replicates. The black bar indicates the control cell line (pBabe), the blue bars represent recovering cell lines, and red bars represent nonrecovering cell lines. The P values were calculated using a two-tailed Mann-Whitney test for unpaired, nonparametric samples to compare the control group (pBabe), recovering group, and nonrecovering group. P values of ≤0.05 are displayed. *, P = 0.03; **, P = 0.02.
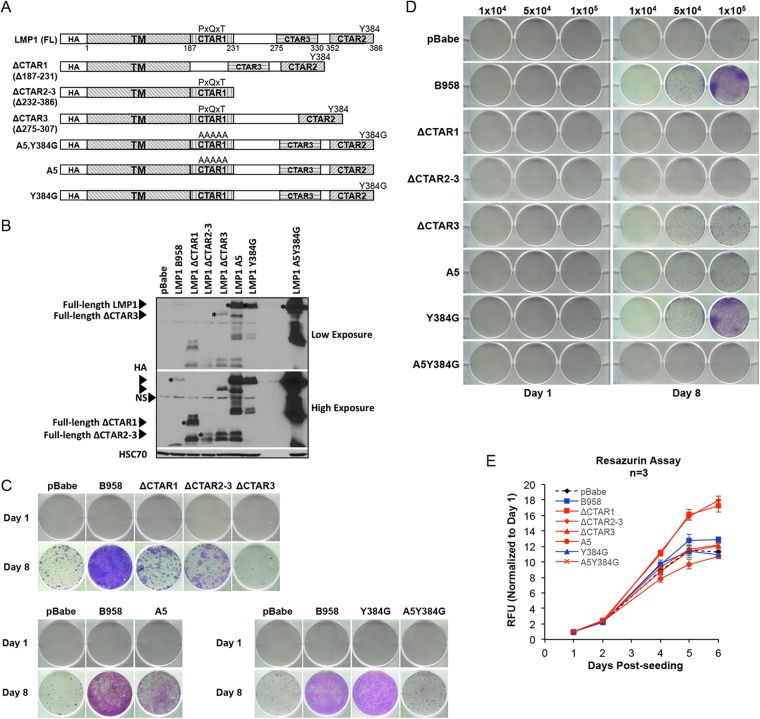
The three conserved signaling domains are important for LMP1-mediated outgrowth.
The C terminus of LMP1 contains three signaling domains (CTAR1, CTAR2, and CTAR3) that interact with cellular signaling molecules, including TRAFs, TRADD/RIP, and the SUMO-conjugating enzyme Ubc9, respectively (13, 15, 20, 21). These conserved signaling domains and motifs were mutated on the LMP1 B958 strain backbone and analyzed by trypan blue anoikis recovery. The LMP1 mutants included the following truncation mutations [ΔCTAR1 (Δ187-231), ΔCTAR2-3 (Δ232-386), and ΔCTAR3 (Δ275-307)] and point mutations at CTAR1 and CTAR2 binding residues (A5 [PXQXT to AAAAA], Y384G, and A5Y384G), as illustrated in Fig. 5A. Stable HK1 cell lines were generated for each mutant, and expression was confirmed by anti-HA immunoblotting (Fig. 5B). Deletion of any one of the three major signaling domains (ΔCTAR1, ΔCTAR2, and ΔCTAR3) resulted in loss of anoikis recovery (Fig. 5C). Mutation of the PXQXT signaling motif in either the single CTAR1 (A5) or double CTAR1 and CTAR2 motif binding mutant (A5Y384G), but not mutation of the CTAR2 (Y384) binding motif, attenuated anoikis recovery, although the A5Y394G double mutant appeared more attenuated than the A5 mutant (Fig. 5C). The attenuated recovery phenotype in the deletion mutants (ΔCTAR1, ΔCTAR2, and ΔCTAR3) and point mutants (A5 and A5Y384G) were consistent with results from viable-cell seeding by trypan blue exclusion (Fig. 5D). These data indicate that signaling through any of the three major signaling domains (CTAR1, CTAR2, and CTAR3) is important for promoting anoikis recovery. However, further dissection of specific signaling motifs within the major signaling domains demonstrated that the CTAR1 PXQXT motif is required for anoikis recovery. Under normal adherent conditions, the proliferation rates of LMP1 signaling mutants were similar to those of the pBabe vector control, with ΔCTAR1 and ΔCTAR2-3 even showing slightly enhanced proliferation kinetics (Fig. 5E). This again lends support to the notion that the impaired outgrowth characteristics in the LMP1 signaling mutants are not due to inherent differences in proliferation rates between the cell lines (Fig. 5E).
FIG 5.
Three conserved signaling domains and the PXQXT motif are critical for outgrowth potential. (A) Schematic representation of LMP1 signaling mutants. (B) Immunoblot analysis of HA-tagged LMP1 signaling mutants. The asterisks indicate the full-length protein for each mutant. NS, nonspecific band produced by HA antibody. HSC70 was used as a loading control. (C) Crystal violet staining to assess the outgrowth of HK1 cell lines expressing LMP1 signaling mutants 1 and 8 days after recovery from anoikis. The day 8 images represent one of three independent experiments. (D) Crystal violet staining at 1 and 8 days of outgrowth for LMP1 signaling mutants following induction of anoikis. Specific numbers of viable cells (1 × 104, 5 × 104, and 1 × 105) were seeded per well for assessment of outgrowth potential as indicated above the wells. (E) Analysis of growth kinetics by resazurin assay for LMP1 mutants on tissue culture-treated dishes. The dashed black line indicates the HK1 pBabe control, the blue lines represent LMP1 mutants that recover following induction of anoikis (B958 and Y384G), and the red lines represent LMP1 mutants that do not recover (ΔCTAR1, ΔCTAR2-3, ΔCTAR3, A5, and A5Y384G). The RFU and standard deviations were determined from 3 biological replicates.
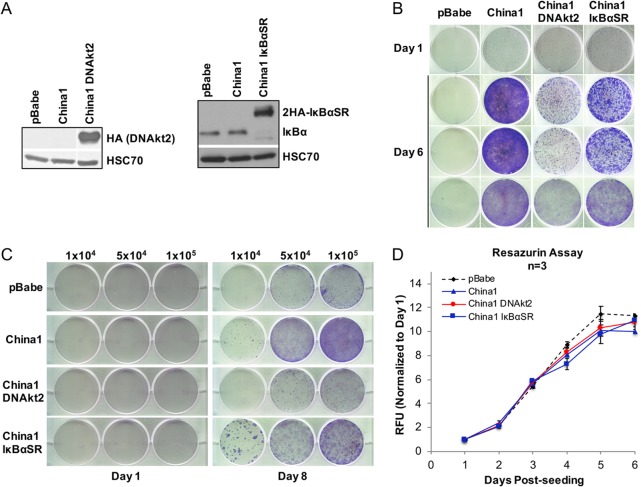
The mechanism of LMP1-induced anoikis recovery is mediated by Akt.
CTAR1 and CTAR2 modulate multiple cellular signaling pathways through interactions with TRAFs and TRADD/RIP, including PI3K/Akt, canonical and noncanonical NF-κB, MAPK/ERK, and JNK/p38 pathways, that result in growth-promoting effects in epithelial cells (13, 15, 20). To determine if Akt or NF-κB signaling contributes to LMP1-mediated restoration of outgrowth after induction of anoikis, dominant-negative constructs for Akt (DNAkt2 [K181M/T309A/S474A] kinase mutant) and a superrepressor of NF-κB (IκBαSR [S32A/S36A]) signaling were stably expressed and analyzed in HK1 LMP1 China1-expressing cells, which had the highest recovery potential. Expression of HA-tagged constructs was confirmed by immunoblotting for the single HA tag on the Akt2 kinase mutant and the 2HA-tagged IκBα superrepressor, which is resolvable from the lower molecular weight of the endogenous IκBα (Fig. 6A). Anoikis recovery was inhibited by expression of the Akt kinase mutant but only partially inhibited by expression of the IκBα superrepressor (Fig. 6B), indicating that activation of Akt signaling is critical for LMP1-mediated outgrowth. These results were further confirmed by trypan blue anoikis recovery, demonstrating gross impairment of LMP1-mediated outgrowth by expression of dominant-negative Akt (Fig. 6C). Proliferation rates under normal growth conditions were similar in all cell lines, indicating that the impairment in outgrowth upon Akt inhibition is not due to intrinsic growth differences between the cell lines (Fig. 6D).
FIG 6.
Akt signaling is critical for outgrowth potential. (A) Immunoblot analysis of HA-tagged DNAkt2 (K181M/T309A/S474A) and 2HA-tagged IκBαSR (S32A/S36A). Anti-HA tag antibody was used for the detection of HA-tagged DNAkt2. Due to a nonspecific band produced by the HA antibody, an anti-IκBα antibody was used for the detection of the size-shifted 2HA-tagged IκBαSR protein. HSC70 was used as a loading control. (B) Crystal violet staining of outgrowth at 1 and 6 days following induction of anoikis. China1 was used as a positive control for outgrowth. The day 6 images are from three independent experiments. (C) Crystal violet staining of outgrowth at 8 days. Specific numbers of viable cells (1 × 104, 5 × 104, and 1 × 105) were seeded per well as indicated at the top. (D) Analysis of growth kinetics by resazurin assay for dominant-negative signaling constructs on tissue culture-treated dishes. The dashed black line indicates the HK1 pBabe control, the blue lines represent cell lines that recover following induction of anoikis (China1, China1, and IκBαSR), and the red line represents a cell line that does not recover (China1 DNAkt2). The RFU and standard deviations were determined from 3 biological replicates.
Targeted mapping of anoikis recovery from LMP1 sequence diversity.
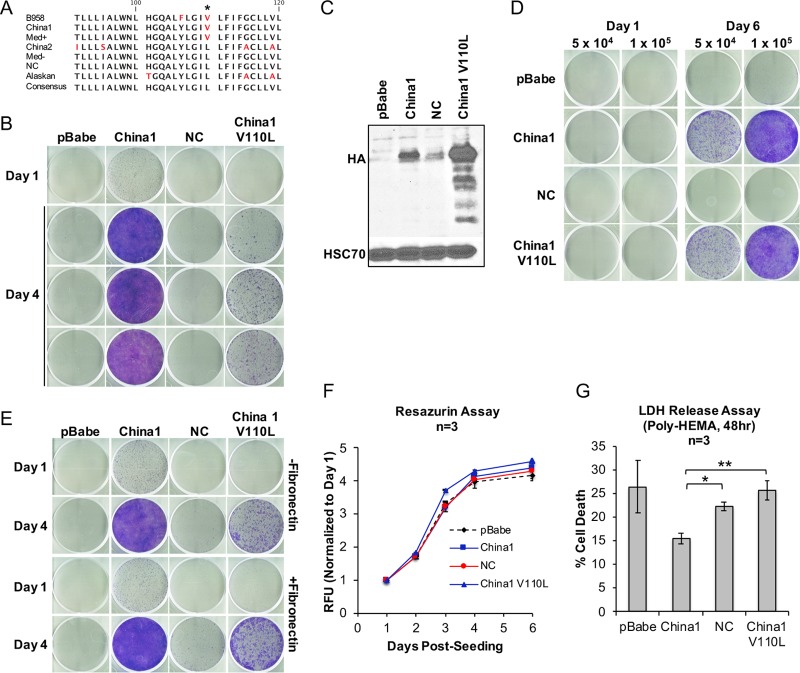
Although there are conserved motifs in the C-terminal signaling domain, LMP1 is one of the most divergent sequences in the EBV genome (35). Polymorphisms can also exist between isolates belonging to the same LMP1 strain (23). A complementary method to systematic mapping of LMP1 is to compare the primary amino acid sequence for potential residues that could distinguish the LMP1 anoikis-recovering strains from the nonrecovering strains. The LMP1 amino acid sequences of the isolates tested in this study were analyzed for residues that were unique to the anoikis-recovering strains (B958, China1, and Med+), but not the nonrecovering strains (China2, Med−, NC, and Alaskan). In the representative strain isolates tested (Fig. 4), there was only one distinguishing amino acid residue at position 110, encoding valine in the anoikis-recovering strains but leucine in the nonrecovering strains (Fig. 7A). We extended the analyses to include publicly deposited LMP1 sequences in the NCBI GenBank database. Genomic LMP1 sequences from 28 NPC biopsy specimens revealed that 25 of the 28 sequences encoded a valine at position 110 while the others encoded leucine, indicating that the polymorphism is well conserved in NPC tumors (36, 37). Of the NPC samples analyzed, only two patients had distant metastasis at the time of diagnosis, and both had a valine at position 110, which would be consistent with observations from this study in conferring anoikis recovery (36). Mutation of V110L on the China1 backbone (China1 V110L) resulted in selection and enhanced expression of LMP1 during selection of outgrowing stable cell lines and attenuation of anoikis recovery (Fig. 7B and C). However, upon normalizing for viable-cell seeding by trypan blue exclusion, the China1 V110L mutant demonstrated recovery potential comparable to that of the parent, China1 (Fig. 7D). Fibronectin coating did not rescue the attenuated recovery of China1 V110L seeded in the traditional anoikis recovery assay (Fig. 7E). Furthermore, the proliferation rates in monolayer culture were similar, indicating that the V110L mutation and higher LMP1 expression levels do not affect cell viability but suggesting that the attenuation could be attributable to enhanced anoikis-mediated cell death without loss of recovery potential (Fig. 7F). This was consistent with results from the LDH release assay showing that the China1 V110L mutant had a higher level of anoikis-induced cell death—26% compared to 15% in China1 (P = 0.005) (Fig. 7G). Anoikis-induced cell death was also higher in the nonrecovering NC LMP1 strain than in the recovering China1 LMP1 strain (P = 0.001) (Fig. 7G). These data indicate that valine at position 110 contributes to survival of anoikis but that additional residues are necessary to confer anoikis recovery.
FIG 7.
Characterization of the China1 V110L mutant for anoikis recovery and survival. (A) Alignment of LMP1 strain amino acid sequences used in this study, ranging from amino acids 91 to 120. Each sequence is from an isolate that phylogenetically segregates into one of seven distinguished LMP1 strain variants. The asterisk denotes the amino acid residue (110) that may distinguish the anoikis-recovering LMP1 strains (B958, China1, and Med+) from the nonrecovering LMP1 strains (Chin 2, Med−, NC, and Alaskan). Sequences were aligned in CLC Main Workbench6, and residues different from the consensus are colored in red. (B) Crystal violet staining of outgrowth at 1 and 4 days following induction of anoikis. The day 4 images are from three independent experiments. (C) Immunoblot analysis of HA-tagged LMP1 strains and the China1 V110L mutant stably expressed in HK1 cells. (D) Crystal violet staining at 1 and 6 days of outgrowth following induction of anoikis, seeding equivalent numbers of viable cells (1 × 104, 5 × 104, and 1 × 105) for assessment of outgrowth potential. (E) Crystal violet staining of HK1 stable cell lines recovered on fibronectin-coated or non-fibronectin-coated tissue culture wells following 1 and 4 days of recovery from anoikis. (F) Analysis of growth kinetics by resazurin assay on tissue culture-treated dishes. The dashed black line represents pBabe, the blue lines represent recovering cell lines, and the red lines represent nonrecovering cell lines. (G) LDH release assay of HK1 stable cells after inducing anoikis for 48 h on poly-HEMA-coated dishes. The P values were calculated using a two-tailed Student t test for unpaired samples with unequal variance (*, P = 0.001; **, P = 0.005). The error bars indicate standard deviations.
DISCUSSION
Up to 40% of NPC tumors relapse with metastases, which are often refractory to treatment with standard chemotherapy and radiation therapy (4). Understanding the underlying risk factors for progressive disease would benefit the assessment of metastatic risk. In this study, a novel LMP1-mediated effect on recovery from anoikis is described in the NPC cell line HK1. Not all of the seven LMP1 strains (China1, China2, B958, Med+, Med−, Alaskan, and NC) were able to confer anoikis recovery. The LMP1 strains have distinct geographic distributions and are characterized by signature amino acid changes in the C terminus (23, 35). Of these strains, China1 and Med+ are consistently detected in NPC tumors in Southeast Asia and northern Africa, respectively (38, 39). These NPC-prevalent LMP1 strains have a characteristic 30-bp deletion in the C terminus, a feature that has been linked to the stabilization and enhanced oncogenic potential of LMP1 (40, 41). Target enrichment of EBV genomes has enabled the high-throughput sequencing of whole EBV genomes from infected cells and biospecimens to reveal genetic diversity, which is greatest in the latent genes (35). There is early evidence supporting the notion that the diversity of EBV genomes within an infected individual is highest during early infection and steadily decreases during establishment of chronic infection and latency (42). Studies of EBV populations circulating in the peripheral blood and oral lavage fluids of asymptomatic carriers also demonstrate that EBV populations are compartmentalized (22). Furthermore, tracking EBV isolates by unique polymorphisms has shown that EBV can circulate between compartments within an individual (22). These studies of clinical specimens illustrate the dynamic nature of EBV transmission but also imply the importance of intrahost EBV sequence variation in disease association.
Historically, the heteroduplex tracking assay showed that China1 was the predominant strain typed in clonally infected NPC tumors (24, 43). The potential selection of the China1 strain was not attributed to superior oncogenic transforming properties, although it is suspected that selection for epitopes that escape immune recognition may be important (24, 26). As more EBV genomes are being sequenced, the potential selection and molecular basis for tumor-associated sequences may become more apparent (32). A comparison of different tumor-derived and laboratory-passaged EBV strains demonstrated that EBV strains could differ in B cell-transforming potential, cell-type-specific infection efficiencies, and infection outcomes (44). Moreover, a single amino acid in EBNA2 can drive superior growth and maintenance of lymphoblastoid cell lines (45). These data indicate that polymorphisms are selected during the establishment of latency and that sequence diversity could be an important risk factor for EBV oncogenic potential. Interestingly, HK1 cells expressing the China1, Med+, or B958 LMP1 strain had the strongest anoikis recovery potential (Fig. 4B and C). Many of the studies of LMP1 have attributed oncogenic properties to the C-terminal signaling domains (8, 13). Findings from this study combined with clinical evidence that only 2 of the possible 25 NPC biopsy specimens with valine encoded at position 110 showed distant metastasis at the time of diagnosis imply that the transmembrane domain may be important for conferring survival in metastasis but that selection of additional residues is likely important for increasing metastatic risk (36, 37). Further fine mapping at position 110 on different LMP1 strain backbones and future epidemiology studies that stratify metadata with the LMP1 strain should help to unravel whether the China1, Med+, and B958 strains are indeed more likely to be associated with metastatic NPC tumors. Examination of the primary sequences of the seven LMP1 strains revealed a conserved residue in anoikis-recovering (V110) versus nonrecovering (L110) strains, which are the two polymorphisms observed in NPC tumors. Mutation of V110 to leucine in the China1 strain decreased anoikis resistance but did not affect the recovery phenotype (Fig. 7), suggesting that primary sequence analysis alone may not be sufficient to identify polymorphisms associated with increased risk for NPC metastasis. As more EBV genomes are being sequenced and deposited in public repositories, future analysis of LMP1 polymorphisms may necessitate comparison of many more naturally derived isolates to overcome the challenge of predicting functional residues simply from the primary sequence. Complementary biochemical methods, such as LMP1 protein-protein interactions, may help to further distinguish potential functionally selected polymorphisms (32). Despite the appreciation for further LMP1 fine mapping to elucidate the complex mechanisms contributing to metastatic outgrowth, the modified anoikis assay does clearly distinguish a phenotypic difference between the LMP1 strains.
The mechanism of anoikis resistance has been intensely studied (27, 46). Several cytoplasmic kinases, including ERK, p38 MAPK, SRC, and FAK, promote anoikis resistance through stabilization of prosurvival proteins and downregulation of proapoptotic proteins, such as Bim-EL (47). Stimulation of signaling through the PI3K/Akt and NF-κB pathways also contributes to anoikis resistance by upregulating expression of prosurvival proteins (47). In this study, the two major signaling pathways that are modulated by LMP1, Akt and NF-κB, were examined using LMP1 signaling mutants and dominant-negative or inhibitory constructs. The results demonstrate that Akt signaling, and not NF-κB, is critical for the recovery of HK1 cells after induction of anoikis (Fig. 6). Additionally, LMP1 did not downregulate expression of Bim during suspension (Fig. 2A), further providing evidence that LMP1 does not affect anoikis resistance. Interestingly, deletion of any of the three CTAR domains abrogated anoikis recovery (Fig. 5), suggesting that while this is a conserved property localized in part to the conserved signaling domains, there are also additional divergent residues that are involved in anoikis recovery (Fig. 4). The loss of anoikis recovery potential after deletion of CTAR3 is notable. CTAR3 does not modulate pathways known to be involved in anoikis resistance but interacts with the SUMO-conjugating enzyme Ubc9, resulting in increased sumoylation of cellular proteins and enhanced cell migration (21). Further study of CTAR3 motifs and targets may aid in the elucidation of additional novel cellular pathways involved in anoikis mechanisms.
Several oncogenic viruses promote anoikis resistance in infected cells. Kaposi's sarcoma-associated herpesvirus (KSHV) enhances anoikis resistance of endothelial cells through stimulation of NF-κB signaling by vFLIP (48). Additionally, KSHV increases anchorage-independent growth, a defining characteristic of anoikis-resistant cells, through repression of the tumor suppressor tropomyosin 1 by microRNAs (49). The E7 protein of bovine papillomavirus type 1 inhibits anoikis, at least partially, through its interaction with p600, a protein that has been implicated in cell survival (50). Although LMP2A did not promote anoikis recovery of HK1 cells in this study, expression of LMP2A in MCF10a and Intestine 407 cells diminishes anoikis activation (30, 31). The current study differs from earlier studies by rescuing cells from anoikis and examining for phenotypes postanoikis, including cell reattachment and outgrowth. Perhaps, the modified anoikis recovery assay described in this study could be extended to the dissection of other viral proteins with suspected metastatic outgrowth properties. These studies highlight the breadth of mechanisms utilized by oncogenic viruses to subvert cell death pathways to promote cancer cell survival, but also suggest that oncogenic viral proteins may be useful molecular mapping tools with which to elucidate complex cellular processes, such as metastasis.
MATERIALS AND METHODS
Cell lines and reagents.
HK1 stable cell lines were generated by transduction with pBabe retroviruses expressing HA-tagged LMP1 strain variants and mutants, including B958, China1, China2, Med+, Med−, AL, NC, ΔCTAR1 (Δ187-231), ΔCTAR2-3 (Δ232-386), ΔCTAR3 (Δ275-307), A5 (PXQXT to AAAAA), Y384G, and the A5-Y384G double mutant, or LMP2A and selected with 500 μg/ml neomycin or 1 μg/ml puromycin (26, 51). Additional stable cell lines expressing LMP1 (strain B958) were generated using two NPC-derived cell lines (HONE1 and HNE1 [NPC AoE Tissue Bank at Hong Kong University]), one primary head and neck squamous cell carcinoma (HNSCC) cell line (PCI-15A) (Robert Ferris, University of Pittsburgh), one immortalized nasopharyngeal epithelial cell line (NP460hTERT) (Sai-Wah Tsao, University of Hong Kong), and one immortalized breast epithelial cell line (MCF10a) (Nancy Raab-Traub, University of North Carolina). For downregulation of Akt and NF-κB signaling pathways, HA-tagged dominant-negative kinase mutants of Akt2 (K181M, T309A, and S474A mutants) and an IκBα superrepressor (S32/36A mutant) were cloned into the pBabe retroviral construct and stably expressed in the HK1 LMP1 China1 cell line. For further dissection of the role of the Akt pathway in outgrowth following induction of anoikis, a constitutively active myristoylated Akt was expressed in the HK1 cell line to rescue outgrowth after anoikis. To suppress caspase activity for potential rescue of outgrowth of HK1 pBabe cells, cells were treated with increasing concentrations of the pan-caspase inhibitor ZVAD-FMK (R&D Systems) during 48 h of incubation on poly-HEMA-coated dishes and induction of anoikis.
Induction of anoikis and outgrowth assays.
Tissue culture dishes (100 mm) were coated (at least 3 times for sufficient coverage) with 10 μg/ml poly-HEMA dissolved in 95% ethanol and allowed to dry under sterile conditions. To induce anoikis, 6 × 106 to 8 × 106 HK1 stable cells were seeded onto poly-HEMA-coated tissue culture plates and incubated for 48 h at 37°C. For each experimental-group comparison, the same number of cells were seeded on poly-HEMA-coated dishes. During suspension, cells float as single cells or clumps, ranging in size from a few cells to >100 cells per clump. In the traditional anoikis recovery experiment, cell clumps were broken up mechanically by repeated pipetting, and equal volumes of disrupted cells were transferred to adherent tissue culture dishes to allow reattachment and assessment of outgrowth potential. Cell-counting experiments using trypan blue exclusion demonstrated >98% viability for reattached cells 1 day after transfer to adherent tissue culture dishes, while cells in suspension demonstrated ∼50% viability. At specific time points, 1 day posttransfer and 4 to 8 days posttransfer, cells were fixed and stained with crystal violet (0.1% crystal violet, 10% ethanol). In fibronectin-facilitated recovery experiments, tissue culture dishes were also coated with fibronectin to facilitate attachment of viable cells after induction of anoikis. To normalize the number of viable cells seeded per well, in trypan blue anoikis recovery experiments, the cell clumps were further broken apart by treatment with 1 mM EDTA for 10 min and passing through a cell strainer. The viable cells were counted using trypan blue exclusion, and increasing numbers of viable cells were seeded per well (5 × 102, 1 × 103, 5 × 103, 1 × 104, 5 × 104, and 1 × 105). Cells were fixed with crystal violet at 1 day postseeding and 4 to 8 days postseeding. Each anoikis recovery experiment was repeated, for a minimum of 3 independent experiments.
Selection of isogenic cell lines.
HK1 stable cell lines expressing LMP1 (strain B958) were seeded onto poly-HEMA-coated dishes to induce anoikis for 48 h. The cells were transferred to adherent 6-well plates to allow outgrowth of surviving cells. After 1 week, the proliferating cells were transferred to 100-mm tissue culture dishes for continued proliferation (AR1 cell line). If the AR1 cells showed no evidence of cell death, the cells were seeded on poly-HEMA-coated dishes for the second round of anoikis induction and recovery. These recovered cell lines were designated AR2 cell lines.
Immunoblotting analysis.
Whole-cell lysates were made as described previously. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA) supplemented with 1 mM phenylmethylsulfonyl, 2 mM activated sodium orthovanadate, and a 1:100 dilution of protease and phosphatase inhibitor cocktails (Sigma). Immunoblotting was performed with the following antibodies: LMP1 S12 (S12 hybridoma tissue culture supernatant; 1:10 dilution); HSC70, p130, PCNA, and IκBα C-21 (Santa Cruz Biotechnology) and cleaved PARP, PARP, cleaved caspase 8, cleaved caspase 9, cleaved caspase 7, cleaved caspase 3, Bim, p27, and anti-HA tag (Cell Signaling Technologies).
Cell death and viability assays.
For the resazurin assay (Cell Signaling Technologies), HK1 stable cell lines were seeded in RPMI 1640 with 10% fetal bovine serum (FBS) at 5 × 103 cells per uncoated well or 5 × 104 cells per poly-HEMA-coated well of a 96-well plate to determine metabolically viable cells according to the manufacturer's protocol. For each resazurin assay, measurements were calculated for triplicate wells. To determine the percentage of dying cells on poly-HEMA-coated plates, HK1 stable cell lines were seeded in RPMI 1640 with 10% FBS at 5 × 104 cells per well of a 96-well plate and incubated at 37°C for 48 h. After the incubation, 100 μl of supernatant was removed for analysis using an LDH cytotoxicity kit (Roche) according to the manufacturer's instructions.
Statistics.
Comparisons between individual cell lines were performed using the two-tailed Student t test for unpaired samples with unequal variance. Group comparisons between the pBabe vector, recovering, or nonrecovering groups were performed using the two-tailed Mann-Whitney test for unpaired, nonparametric samples in PRISM.
ACKNOWLEDGMENTS
This study was supported by the Hillman Foundation, the Samuel and Emma Winters Foundation (project 710580 to K.H.Y.S.), and a National Institutes of Health Mechanisms of Microbial Persistence and Pathogenesis Training Fellowship (NIH T32AI049820-13).
We thank Kristin DePeaux for generation of the HK1 V110L cell line.
REFERENCES
- 1.Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 3.Jia WH, Qin HD. 2012. Non-viral environmental risk factors for nasopharyngeal carcinoma: a systematic review. Semin Cancer Biol 22:117–126. doi: 10.1016/j.semcancer.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Tsang J, Lee VH, Kwong DL. 2014. Novel therapy for nasopharyngeal carcinoma—where are we. Oral Oncol 50:798–801. doi: 10.1016/j.oraloncology.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Shair KH, Schnegg CI, Raab-Traub N. 2008. EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res 68:6997–7005. doi: 10.1158/0008-5472.CAN-08-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shair KH, Schnegg CI, Raab-Traub N. 2009. Epstein-Barr virus latent membrane protein-1 effects on junctional plakoglobin and induction of a cadherin switch. Cancer Res 69:5734–5742. doi: 10.1158/0008-5472.CAN-09-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasil LR, Shair KH. 2015. Epstein-Barr virus LMP1 induces focal adhesions and epithelial cell migration through effects on integrin-alpha5 and N-cadherin. Oncogenesis 4:e171. doi: 10.1038/oncsis.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol 12:431–441. doi: 10.1016/S1044579X0200086X. [DOI] [PubMed] [Google Scholar]
- 9.Tsao SW, Tramoutanis G, Dawson CW, Lo AK, Huang DP. 2002. The significance of LMP1 expression in nasopharyngeal carcinoma. Semin Cancer Biol 12:473–487. doi: 10.1016/S1044579X02000901. [DOI] [PubMed] [Google Scholar]
- 10.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 12.Shair KH, Bendt KM, Edwards RH, Nielsen JN, Moore DT, Raab-Traub N. 2012. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) and LMP2A function cooperatively to promote carcinoma development in a mouse carcinogenesis model. J Virol 86:5352–5365. doi: 10.1128/JVI.07035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson CW, Port RJ, Young LS. 2012. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol 22:144–153. doi: 10.1016/j.semcancer.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Dawson CW, Laverick L, Morris MA, Tramoutanis G, Young LS. 2008. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J Virol 82:3654–3664. doi: 10.1128/JVI.01888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mainou BA, Everly DN Jr, Raab-Traub N. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917–6924. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- 16.Mainou BA, Everly DN Jr, Raab-Traub N. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J Virol 81:9680–9692. doi: 10.1128/JVI.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pratt ZL, Zhang J, Sugden B. 2012. The latent membrane protein 1 (LMP1) oncogene of Epstein-Barr virus can simultaneously induce and inhibit apoptosis in B cells. J Virol 86:4380–4393. doi: 10.1128/JVI.06966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shair KH, Bendt KM, Edwards RH, Bedford EC, Nielsen JN, Raab-Traub N. 2007. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog 3:e166. doi: 10.1371/journal.ppat.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtani N, Brennan P, Gaubatz S, Sanij E, Hertzog P, Wolvetang E, Ghysdael J, Rowe M, Hara E. 2003. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J Cell Biol 162:173–183. doi: 10.1083/jcb.200302085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Nakano H, Wu Z. 2006. The C-terminal activating region 2 of the Epstein-Barr virus-encoded latent membrane protein 1 activates NF-kappaB through TRAF6 and TAK1. J Biol Chem 281:2162–2169. doi: 10.1074/jbc.M505903200. [DOI] [PubMed] [Google Scholar]
- 21.Bentz GL, Whitehurst CB, Pagano JS. 2011. Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J Virol 85:10144–10153. doi: 10.1128/JVI.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitki-Green D, Covington M, Raab-Traub N. 2003. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J Virol 77:1840–1847. doi: 10.1128/JVI.77.3.1840-1847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards RH, Seillier-Moiseiwitsch F, Raab-Traub N. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79–95. doi: 10.1006/viro.1999.9855. [DOI] [PubMed] [Google Scholar]
- 24.Edwards RH, Sitki-Green D, Moore DT, Raab-Traub N. 2004. Potential selection of LMP1 variants in nasopharyngeal carcinoma. J Virol 78:868–881. doi: 10.1128/JVI.78.2.868-881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo AK, Huang DP, Lo KW, Chui YL, Li HM, Pang JC, Tsao SW. 2004. Phenotypic alterations induced by the Hong Kong-prevalent Epstein-Barr virus-encoded LMP1 variant (2117-LMP1) in nasopharyngeal epithelial cells. Int J Cancer 109:919–925. doi: 10.1002/ijc.20051. [DOI] [PubMed] [Google Scholar]
- 26.Mainou BA, Raab-Traub N. 2006. LMP1 strain variants: biological and molecular properties. J Virol 80:6458–6468. doi: 10.1128/JVI.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frisch SM, Screaton RA. 2001. Anoikis mechanisms. Curr Opin Cell Biol 13:555–562. doi: 10.1016/S0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 28.Chaffer CL, Weinberg RA. 2011. A perspective on cancer cell metastasis. Science 331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg RA. 2008. The many faces of tumor dormancy. APMIS 116:548–551. doi: 10.1111/j.1600-0463.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- 30.Iwakiri D, Minamitani T, Samanta M. 2013. Epstein-Barr virus latent membrane protein 2A contributes to anoikis resistance through ERK activation. J Virol 87:8227–8234. doi: 10.1128/JVI.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fotheringham JA, Raab-Traub N. 2013. Epstein-Barr virus latent membrane protein 2 effects on epithelial acinus development reveal distinct requirements for the PY and YEEA motifs. J Virol 87:13803–13815. doi: 10.1128/JVI.02203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shair KHY, Reddy A, Cooper VS. 2018. New insights from elucidating the role of LMP1 in nasopharyngeal carcinoma. Cancers 10:E86. doi: 10.3390/cancers10040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan SY, Choy KW, Tsao SW, Tao Q, Tang T, Chung GT, Lo KW. 2008. Authentication of nasopharyngeal carcinoma tumor lines. Int J Cancer 122:2169–2171. doi: 10.1002/ijc.23374. [DOI] [PubMed] [Google Scholar]
- 34.Lam N, Sandberg ML, Sugden B. 2004. High physiological levels of LMP1 result in phosphorylation of eIF2 alpha in Epstein-Barr virus-infected cells. J Virol 78:1657–1664. doi: 10.1128/JVI.78.4.1657-1664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. 2015. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok H, Wu CW, Palser AL, Kellam P, Sham PC, Kwong DL, Chiang AK. 2014. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol 88:10662–10672. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C, Xiong F, Xiang B, Zhou M, Gong Z, Liao Q, Yu J, He Y, Zhang W, Li X, Li Y, Li G, Xiong W. 2017. Genome-wide analysis of 18 Epstein-Barr viruses isolated from primary nasopharyngeal carcinoma biopsy specimens. J Virol 91:e00301-. doi: 10.1128/JVI.00301-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung NS, Edwards RH, Seillier-Moiseiwitsch F, Perkins AG, Zeng Y, Raab-Traub N. 1998. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int J Cancer 76:207–215. doi:. [DOI] [PubMed] [Google Scholar]
- 39.Dardari R, Khyatti M, Cordeiro P, Odda M, ElGueddari B, Hassar M, Menezes J. 2006. High frequency of latent membrane protein-1 30-bp deletion variant with specific single mutations in Epstein-Barr virus-associated nasopharyngeal carcinoma in Moroccan patients. Int J Cancer 118:1977–1983. doi: 10.1002/ijc.21595. [DOI] [PubMed] [Google Scholar]
- 40.Knecht H, Bachmann E, Brousset P, Sandvej K, Nadal D, Bachmann F, Odermatt BF, Delsol G, Pallesen G. 1993. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkin's disease and identical to those observed in nasopharyngeal carcinoma. Blood 82:2937–2942. [PubMed] [Google Scholar]
- 41.Vallat-Decouvelaere AV, Bretel MA, Vassias I, Laplanche JL, Polivka M, Wassef M, Brunet M, Thiebaut JB, Gosselin B, Morinet F, Mikol J. 2002. High frequency of a 30-bp deletion of Epstein-Barr virus latent membrane protein 1 gene in primary HIV non-Hodgkin's brain lymphomas. Neuropathol Appl Neurobiol 28:471–479. doi: 10.1046/j.1365-2990.2002.t01-1-00418.x. [DOI] [PubMed] [Google Scholar]
- 42.Weiss ER, Lamers SL, Henderson JL, Melnikov A, Somasundaran M, Garber M, Selin L, Nusbaum C, Luzuriaga K. 2018. Early Epstein-Barr virus genomic diversity and convergence toward the b95.8 genome in primary infection. J Virol 92:e01466-. doi: 10.1128/JVI.01466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raab-Traub N, Flynn K. 1986. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 44.Tsai MH, Lin X, Shumilov A, Bernhardt K, Feederle R, Poirey R, Kopp-Schneider A, Pereira B, Almeida R, Delecluse HJ. 2017. The biological properties of different Epstein-Barr virus strains explain their association with various types of cancers. Oncotarget 8:10238–10254. doi: 10.18632/oncotarget.14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzellos S, Correia PB, Karstegl CE, Cancian L, Cano-Flanagan J, McClellan MJ, West MJ, Farrell PJ. 2014. A single amino acid in EBNA-2 determines superior B lymphoblastoid cell line growth maintenance by Epstein-Barr virus type 1 EBNA-2. J Virol 88:8743–8753. doi: 10.1128/JVI.01000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson CD, Anyiwe K, Schimmer AD. 2008. Anoikis resistance and tumor metastasis. Cancer Lett 272:177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 47.Buchheit CL, Weigel KJ, Schafer ZT. 2014. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer 14:632–641. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 48.Efklidou S, Bailey R, Field N, Noursadeghi M, Collins MK. 2008. vFLIP from KSHV inhibits anoikis of primary endothelial cells. J Cell Sci 121:450–457. doi: 10.1242/jcs.022343. [DOI] [PubMed] [Google Scholar]
- 49.Kieffer-Kwon P, Happel C, Uldrick TS, Ramalingam D, Ziegelbauer JM. 2015. KSHV microRNAs repress tropomyosin 1 and increase anchorage-independent growth and endothelial tube formation. PLoS One 10:e0135560. doi: 10.1371/journal.pone.0135560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeMasi J, Chao MC, Kumar AS, Howley PM. 2007. Bovine papillomavirus E7 oncoprotein inhibits anoikis. J Virol 81:9419–9425. doi: 10.1128/JVI.00422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasil LR, Wei L, Chang C, Lan L, Shair KH. 2015. Regulation of DNA damage signaling and cell death responses by Epstein-Barr virus latent membrane protein 1 (LMP1) and LMP2A in nasopharyngeal carcinoma cells. J Virol 89:7612–7624. doi: 10.1128/JVI.00958-15. [DOI] [PMC free article] [PubMed] [Google Scholar]