Investigating evolutionary processes between viruses and nonhuman primates has led to the discovery of a large number of herpesviruses. No study published so far on primate cytomegaloviruses has extensively studied New World monkeys (NWMs) at the subspecies, species, genus, and family levels. The present study sought to identify cytomegalovirus homologues in NWMs and to decipher their evolutionary relationships. This led us to characterize novel viruses from 12 of the 20 primate species tested, which are representative of the three NWM families. The identification of distinct viruses in these primates not only significantly expands our knowledge of the host range of this viral genus but also sheds light on its evolutionary history. Phylogenetic analyses and molecular dating of the sequences obtained support a virus-host coevolution.
KEYWORDS: Cytomegalovirus, CMV, New World monkeys, evolution, phylogeny
ABSTRACT
Over the past few decades, a large number of studies have identified herpesvirus sequences from many mammalian species around the world. Among the different nonhuman primate species tested so far for cytomegaloviruses (CMVs), only a few were from the New World. Seeking to identify CMV homologues in New World monkeys (NWMs), we carried out molecular screening of 244 blood DNA samples from 20 NWM species from Central and South America. Our aim was to reach a better understanding of their evolutionary processes within the Platyrrhini parvorder. Using PCR amplification with degenerate consensus primers targeting highly conserved amino acid motifs encoded by the herpesvirus DNA polymerase gene, we characterized novel viral sequences from 12 species belonging to seven genera representative of the three NWM families. BLAST searches, pairwise nucleotide and amino acid sequence comparisons, and phylogenetic analyses confirmed that they all belonged to the Cytomegalovirus genus. Previously determined host taxa allowed us to demonstrate a good correlation between the distinct monophyletic clades of viruses and those of the infected primates at the genus level. In addition, the evolutionary branching points that separate NWM CMVs were congruent with the divergence dates of their hosts at the genus level. These results significantly expand our knowledge of the host range of this viral genus and strongly support the occurrence of cospeciation between these viruses and their hosts. In this respect, we propose that NWM CMV DNA polymerase gene sequences may serve as reliable molecular markers with which to infer Platyrrhini phylogenetics.
IMPORTANCE Investigating evolutionary processes between viruses and nonhuman primates has led to the discovery of a large number of herpesviruses. No study published so far on primate cytomegaloviruses has extensively studied New World monkeys (NWMs) at the subspecies, species, genus, and family levels. The present study sought to identify cytomegalovirus homologues in NWMs and to decipher their evolutionary relationships. This led us to characterize novel viruses from 12 of the 20 primate species tested, which are representative of the three NWM families. The identification of distinct viruses in these primates not only significantly expands our knowledge of the host range of this viral genus but also sheds light on its evolutionary history. Phylogenetic analyses and molecular dating of the sequences obtained support a virus-host coevolution.
INTRODUCTION
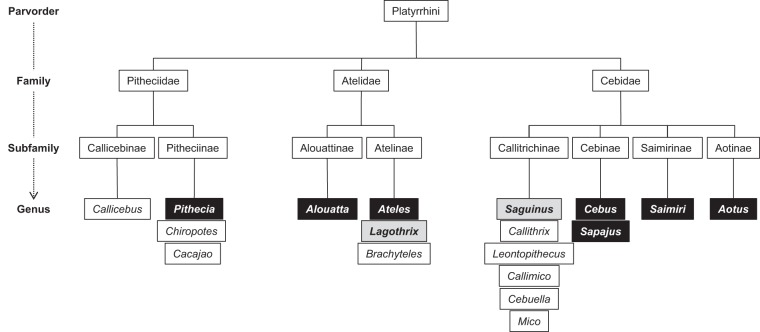
New World monkeys (NWMs) of tropical forests from Central to South America belong to the Platyrrhini parvorder (1). They first appeared in the Neotropics in the late Eocene or early Oligocene and subsequently evolved into broad and diverse families, subfamilies, and genera (Fig. 1) (2, 3). To shed light on their phylogeny and evolution, NWMs have been studied extensively through use of morphological, biogeographical, behavioral, and molecular data (2, 4–17). Over the last few decades, contrasting hypotheses have been proposed, presumably due to different markers and the presence of polymorphisms in the features considered. Agreement on the main clades of NWMs has been reached by use of different approaches, revealing a unique phylogenetic arrangement of Platyrrhini, with three monophyletic families: Pitheciidae, Atelidae, and Cebidae (Table 1; Fig. 1) (4, 5, 11, 12, 14–16). Nevertheless, the relationships between them continue to be debated. Through the analysis of intergeneric and intrageneric relationships, intrafamily relationships have also been studied in depth. By incorporating all the available data, major advances have been made, and many taxonomic controversies have been clarified (6). Therefore, the Pitheciidae family is composed of the genera Callicebus, Pithecia, Chiropotes, and Cacajao, the Atelidae family of Alouatta, Ateles, Brachyteles, and Lagothrix, and the Cebidae family of Cebuella, Mico, Callithrix, Callimico, Saguinus, Leontopithecus, Saimiri, Cebus, Sapajus, and Aotus. However, relationships between or within some subfamilies and/or genera remain under discussion. Among the Cebidae, the phylogenetic position of the Aotinae subfamily remains unclear (15). Indeed, molecular data did not allow determination of whether Aotinae is a sister clade of Callitrichinae or, alternatively, if Aotinae, Saimiriinae, and Cebinae are sisters to Callitrichinae (4, 12, 14–16). Moreover, the number of platyrrhine genera is also still under discussion, such as the division of Cebus into the Sapajus (tufted capuchins) and Cebus (untufted capuchins) genera (4, 17–19). As a result, neither the diversity nor the taxonomy of NWMs is fully known. To appreciate the details of Platyrrhini evolution, much work still needs to be done at various taxonomic levels.
FIG 1.
Diagram representation of Platyrrhini taxa in descending order down to the genus level. Black and gray boxes represent NWM genera tested for CMVs. Black boxes correspond to NWM genera from which CMV sequences have been characterized, while gray boxes represent NWM genera from which no CMV sequence was obtained in the present study. (Adapted from reference 3 with permission of the publisher.)
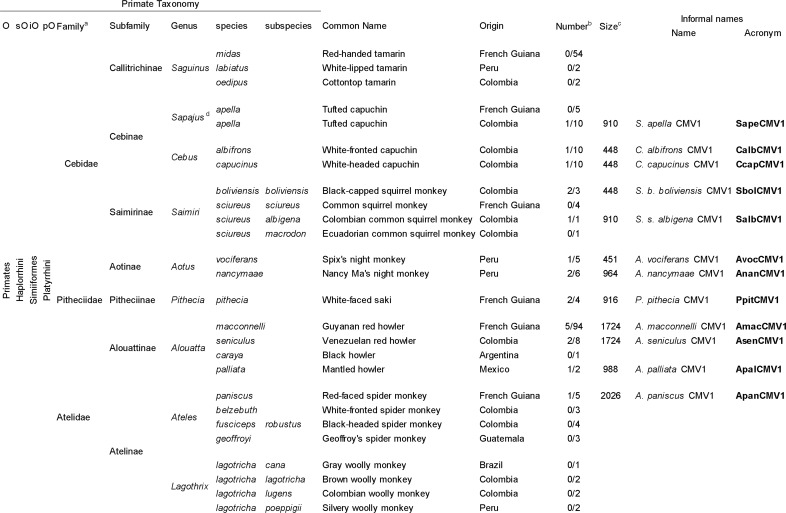
TABLE 1.
New World nonhuman primates tested for cytomegaloviruses by use of molecular methods and survey resultse
According to the work of Perelman et al. (4).
Number of CMV-positive animals (by PCR, cloning, and sequencing)/number of tested animals.
Sizes of the DNA polymerase gene fragments obtained, in base pairs.
According to the work of Alfaro et al. (17).
Abbreviations: O, order; sO, suborder; iO, infraorder; pO, parvorder; CMV, cytomegalovirus.
Viruses of the genus Cytomegalovirus belong to the Betaherpesvirinae subfamily within the Herpesviridae family of the order Herpesvirales (20). Eight cytomegaloviruses (CMVs) are recognized as species by the International Committee on Taxonomy of Viruses (ICTV), according to the latest master species list (MSL 32), released on 12 March 2018 (https://talk.ictvonline.org/files/master-species-lists/m/msl/7185). Human betaherpesvirus 5 (HHV5), commonly referred to in the literature as human cytomegalovirus (HCMV), is the CMV type species. So far, cytomegaloviruses have been characterized only from primates. Since the initial description of a cytomegalovirus in African green monkeys in 1957, whose current species name is Cercopithecine betaherpesvirus 5 (CeHV5), natural infections by such viruses have been described for several Old World monkey (OWM) species, including baboons, macaques, colobuses, chimpanzees, gorillas, and others (21–32). In contrast, cytomegaloviruses of NWMs are represented by only three viral entities, from Aotus trivirgatus (northern owl monkey), Saimiri sciureus (common squirrel monkey), and Cebus sp. (capuchin) animals, despite the wide diversity of platyrrhines (33–35). It is presumed that all primate species harbor CMVs following a cospeciation process, but data supporting this assumption are scarce. The most extensive analyses of primate CMVs conducted, to date, are those of Leendertz et al. (27) and Anoh et al. (31). Using phylogenetic analyses, these studies demonstrated a species-specific distribution of these viruses. This species specificity indicates a long-term coevolution of CMVs with their natural hosts. The identification of two clades, each composed of chimpanzee and gorilla CMVs, suggests that they have coevolved following a horizontal transmission event between these great apes millions of years ago (27). Nevertheless, interspecies transmissions in the wild are rare events (27, 29, 31, 32).
With the exception of the three above-mentioned CMVs of NWMs, there has been little prior organized effort to discover cytomegaloviruses in neotropical primates. The number of NWM species tested, to date, therefore accounts for only a tiny part of their diversity. We thought that additional investigations on a larger number of species were required. We therefore addressed the possible presence of CMVs in different NWM species for which we previously partially characterized Epstein-Barr virus (EBV)-like viruses (36, 37). The purpose was to gain greater insight into the distribution and diversity of CMVs infecting the Platyrrhini primates. Furthermore, based on the coevolution observed between OWMs and their specific CMVs, we wished to determine whether NWM CMV sequences could help to decipher evolutionary relationships of their host species (27, 31). Given that multiple molecular markers of mitochondrial and nuclear DNAs are available, host species can be characterized along with their viruses, allowing progress to be made on their respective patterns of diversification. Here we report the finding of sequences of cytomegaloviruses in different NWM species and achieve a better understanding of the evolutionary processes between these viruses and their Platyrrhini hosts.
RESULTS
To look for the presence of CMV-like viruses in our collection of NWMs, we attempted to amplify a fragment of the highly conserved herpesvirus DNA polymerase gene from the peripheral blood mononuclear cell (PBMC) DNA of each wild-caught primate under previously described PCR conditions (28, 36). A total of 244 samples from 20 different species of the three NWM families were tested (Table 1). DNA samples from 12 species scored positive after nested PCRs (nPCRs) (Table 1). No primate belonging to the Saguinus and Lagothrix genera scored positive. Indeed, no amplification was observed for any samples from the three tamarin species (Saguinus midas, Saguinus labiatus, and Saguinus oedipus) and the four woolly monkey subspecies (Lagothrix lagotricha cana, L. l. lagotricha, L. l. lugens, and L. l. poeppigii). We then used different pairs of consensus-degenerate and specific PCR primers to obtain longer sequences of the DNA polymerase gene from each positive animal (Table 2; Fig. 2). The concatenated nucleotide sequences generated were between 448 and 2,026 bp long, depending on the viral strain (Table 1).
TABLE 2.
Oligonucleotide primers used for cytomegalovirus DNA polymerase gene consensus and specific PCRs
| Oligonucleotide | Orientationa | Locationb | Sequence (5′ → 3′)c | CMV sequence(s) amplified |
|---|---|---|---|---|
| DNA polymerase degenerate primers | ||||
| CMV1F1 | + | 721–749 | GAC AAG AAG TTG ACN ACN TTY GGN TGG TG | AmacCMV1, AsenCMV1, ApanCMV1 |
| CMV1R1 | − | 1534–1559 | ACG CCG GCY TCR TAR TGR AAR TTD AT | AmacCMV1, AsenCMV1, ApanCMV1 |
| CMV1R2 | − | 1480–1504 | CGT CCT GHA CRC ART AYT TNC CNA C | AmacCMV1, AsenCMV1, ApanCMV1 |
| CMV2F1 | + | 1330–1352 | GAY ATG TAY CCN GTS TGY ATG GC | AmacCMV1, AsenCMV1, ApanCMV1, PpitCMV1, SapeCMV1, SalbCMV1 |
| CMV2F2 | + | 1372–1394 | TAC AAR YTV AAY ACB ATG GCS GA | AmacCMV1, AsenCMV1, ApanCMV1, PpitCMV1, SapeCMV1, SalbCMV1 |
| DFASAd | + | 1768–1793 | GTG TTC GAC TTY GCN AGY YTN TAY CC | All |
| CMV3F1 | + | 1795–1817 | TCH ATY ATY ATG GCN CAY AAY CT | All |
| CMV3F2 | + | 2062–2090 | ACG TGC AAT TCT TTY TAY GGB TTY ACN GG | All |
| CMV3R1 | − | 2269–2303 | CGA TAG CAC ACA AAC ACR CTR TCN GTR TCN CCR TA | All |
| CMV3R2 | − | 2125–2147 | CCG ATD CGN GTR ATR CTR GCC GC | All |
| CMV4F1 | + | 2266–2291 | ATC TAY GGK GAC ACS GAY AGY GTS TT | AmacCMV1, AsenCMV1, ApalCMV1, AnanCMV1, ApanCMV1 |
| CMV4R1 | − | 2782–2801 | GCC GCY ARN CGY TTD ATG AC | ApalCMV1, AnanCMV1, ApanCMV1 |
| CMV4R2 | − | 2477–2498 | CGC ACC ARR TCR ACN CCY TTC A | AmacCMV1, AsenCMV1, ApalCMV1, AnanCMV1, ApanCMV1 |
| CMV4R3 | − | 2419–2444 | ATA TAC CGY TTY TTR CAG ATC ATC AT | AmacCMV1, AsenCMV1, ApalCMV1, AnanCMV1, ApanCMV1 |
| DNA polymerase antisense specific primers (in combination with CMV2F1 or CMV2F2)e | ||||
| PitR1 | − | 1958–1978 | TGC GCT GAG CAA CCC ATT TAG | PpitCMV1 |
| PitR2 | − | 1912–1932 | ACG CAC CTC CGA CTT CAC AAA | PpitCMV1 |
| AotR1 | − | 2019–2039 | TTG TCG AGC AGC GTC CTC TTG | AnanCMV1, AvocCMV1 |
| AotR2 | − | 1884–1904 | ACC GTA CCG TTT TCG AAG TTA | AnanCMV1, AvocCMV1 |
| SapR1 | − | 2106–2126 | GCG ACT GGC AAA CAC GGT AAC | SapeCMV1 |
| CapR2 | − | 1994–2016 | GGG ATC TGT GCA ATC TTT CAT GG | CalbCMV1, CcapCMV1 |
| CapR3 | − | 1939–1961 | CGG GTC AAC AAT TCA GAA AGC AC | SapeCMV1, CalbCMV1, CcapCMV1 |
| AteR1 | − | 1994–2016 | CGG ATC TCT GCA ATT TTT CAT GG | ApanCMV1 |
| AteR2 | − | 1935–1957 | TCA GCA GTT CGG ACA ACA CTG AA | ApanCMV1 |
| SaiR1 | − | 1948–1969 | CCA CCC ACT TCG TTA GCA GCT C | SbolCMV1, SalbCMV1 |
| SaiR2 | − | 2155–2175 | ACG CGC GGT GTC TTG TAA CAT | SbolCMV1, SalbCMV1 |
| AloR1 | − | 1959–1979 | TTC CGT TGA GCC ACC CAT TTA | AmacCMV1, AsenCMV1, ApalCMV1 |
| AloR2 | − | 1932–1954 | GCA ATT CCG ATA GCA CTG AGG AA | AmacCMV1, AsenCMV1, ApalCMV1 |
+, sense; −, antisense.
Positions relative to ATG of the DNA polymerase gene of AoHV1 (accession number FJ483970).
Letters at positions of degeneracy follow the International Unit Base codes.
Degenerate oligonucleotide primer described by Rose et al. (46).
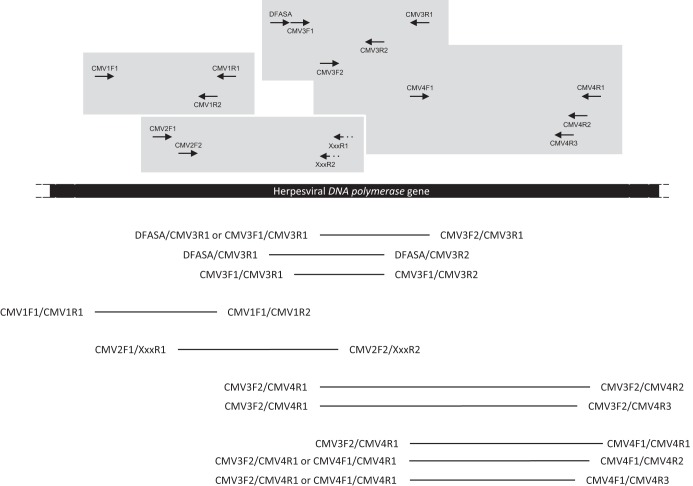
For clarity, all antisense specific primers are indicated as XxxR1 for R1 primers and XxxR2 for R2 primers in Fig. 2.
FIG 2.
Relative positions and orientations of the PCR primers used in this study. The different combinations of primers used in nested or seminested PCRs are represented above the herpesviral DNA polymerase gene sequence, in different gray boxes. Primers XxxR1 and XxxR2, represented by dotted arrows, correspond to the different antisense specific primers used in a degenerate (CMV2F1 or CMV2F2)-nondegenerate nPCR assay (Table 2). Bars below the sequence represent the different nested PCR products expected. The pairs of primers on the left side of the bars indicate those used for the first-round PCR, while those on the right side correspond to those used in the nested PCRs. The sequences of the oligonucleotide primers are given in Table 2.
BLAST searches demonstrated that all sequences identified belonged to the Cytomegalovirus genus and revealed the presence of 12 distinct sequences. Four sequences were identified twice in Saimiri boliviensis boliviensis, Aotus nancymaae, Pithecia pithecia, and Alouatta seniculus animals, while the sequence in Alouatta macconnelli was identified in five individuals (Table 1). Virus names and abbreviations were given to the 12 distinct viruses (Table 1), as follows: the viruses were named after the host species (for Saimiri hosts, after the subspecies), followed by three uppercase letters corresponding to the viral genus (CMV for Cytomegalovirus), to which they were then assigned the Arabic numeral 1, as previously done by us and others (27, 36).
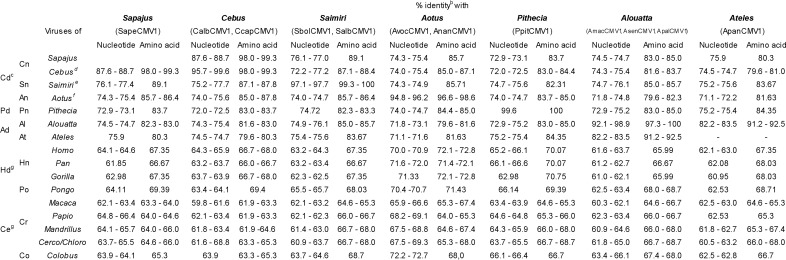
To obtain a full vision of the genetic diversity of these new CMV sequences, pairwise sequence comparisons were made for the 447-bp/149-amino-acid (aa) fragment of the DNA polymerase gene common to all primate CMVs. All 12 sequences obtained differed from each other at the nucleotide level. Sequences that were identified in different specimens of the same primate species, e.g., AnanCMV1, AsenCMV1, AmacCMV1, SbolCMV1, and PpitCMV1, were 100% identical, with the exception of the two PpitCMV1 sequences, which showed 99.6% nucleotide identity (Table 3). For clarity, comparisons of the percentages of identity between the different newly identified NWM CMVs are reported by grouping viral sequences at the host genus level (Table 3). Overall, the new sequences exhibited 71.1% (AnanCMV1 versus ApanCMV1) to 99.6% (CalbCMV1 versus CebHV1 or Cebus sp. herpesvirus) nucleotide identity and 79.6% (CcapCMV1 versus ApanCMV1 and AnanCMV1 versus ApalCMV1) to 100% (AmacCMV1 versus AsenCMV1 as well as SbolCMV1 versus SalbCMV1) amino acid identity among themselves and the other available NWM CMV sequences (Table 3). Viruses infecting NWMs of the same genus presented more than 92% nucleotide and amino acid identities (Table 3). Comparisons between CMVs of different NWM genera ranged from 71.1% (Aotus CMVs versus Ateles CMVs) to 88.7% (Sapajus CMVs versus Cebus CMVs) at the nucleotide level and from 79.6% (Aotus and Alouatta CMVs versus Cebus and Ateles CMVs) to 99.3% (Sapajus CMVs versus Cebus CMVs) at the amino acid level. NWM CMV sequences exhibited 59.8% (Cebus CMVs versus Macaca CMVs) to 72.7% (Aotus CMVs versus Colobus CMVs) nucleotide sequence identities and 61.9 to 68.7% amino acid sequence identities with OWM CMVs. The levels of nucleotide and amino acid sequence identities with CMVs of the Hominidae, except HHV5, ranged from 61 to 72% and from 66 to 72.8%, respectively. Identities with HHV5 ranged from 61.6% (Alouatta CMVs) to 70.9% (Aotus CMVs) at the nucleotide level and from 66% (Alouatta CMVs) to 72.8% (Aotus CMVs) at the amino acid level.
TABLE 3.
Nucleotide and amino acid identities between the novel cytomegaloviruses and all other nonhuman primate cytomegaloviruses and HCMVa
Numbers refer to values obtained in comparison with the 447-bp fragment of the conserved DNA polymerase gene that is available for all viruses.
Sequences identified from specimens from the same primate species showing 100% nucleotide identity, e.g., SbolCMV1, AnanCMV1, AmacCMV1, and AsenCMV1, are not included.
Abbrevations: Cd, Cebidae; Cn, Cebinae; Sn, Saimiriinae; An, Aotinae; Pd, Pitheciidae; Pn, Pitheciinae; Ad, Atelidae; Al, Alouattinae; At, Atelinae; Hd, Hominidae; Hn, Homininae; Po, Pongidae; Ce, Cercopithecidae; Cr, Cercopithecinae; Co, Colobinae.
Nucleotide and amino acid identities of viruses of Cebus rely on the sequences generated in this study as well as on sequences of CebHV1 (accession number JQ264772) and CebusHV (accession number AF292067).
Nucleotide and amino acid identities of viruses of Saimiri rely on the sequences generated in this study as well as on sequences of SaHV4 (accession number FJ483967) and SscHV (accession number AF292065).
Nucleotide and amino acid identities of viruses of Aotus rely on the sequences generated in this study as well as on the sequence of AoHV1 (accession number FJ483970).
Hominidae and Cercopithecidae viral sequences used to calculate nucleotide and amino acid identities correspond to those shown in Fig. 3. Their GenBank accession numbers and associated publications are all reported in the figure and its legend.
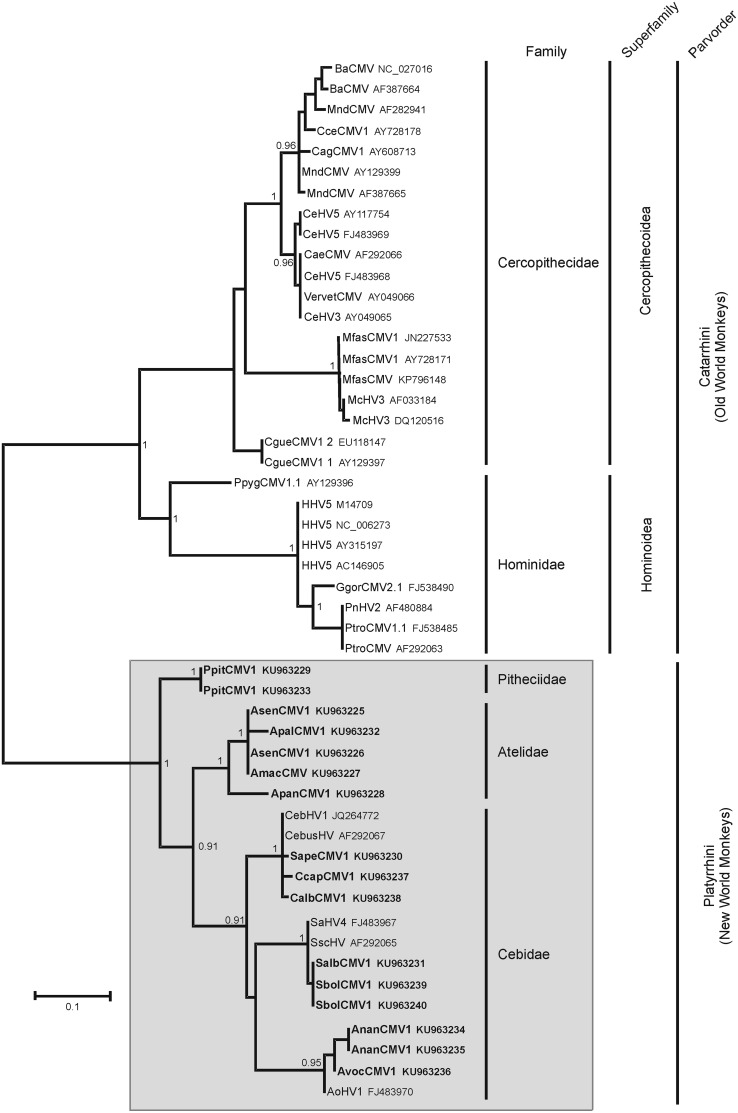
All phylogenetic analyses performed on nucleotide or amino acid sequences of the newly characterized CMV sequences and those of other primate CMVs available in the databases clearly placed the new sequences in a monophyletic lineage of NWM viruses in the Cytomegalovirus genus. The phylogenetic analysis presented in Fig. 3 is based on amino acid sequences. The NWM CMV lineage diverged from the OWM CMV lineage with a posterior probability value of 1. Remarkably, considering the OWM CMV lineage, the phylogenetic tree formed two major monophyletic groups, consisting of Hominoidea and Cercopithecoidea viruses. Within the Cercopithecoidea sequences, the Colobus CMV sequences are the basal taxon, with the formation of additional clades comprising Asian Macaca, Cercopithecus/Chlorocebus, and Papio/Mandrillus/Cercocebus taxa.
FIG 3.
Phylogenetic analysis of primate cytomegalovirus sequences. The phylogenetic tree was derived from the partial amino acid sequences of the DNA polymerase genes (149 aa) of 50 representatives of primate cytomegaloviruses, using the Bayesian method with the JTT+G model of amino acid evolution. Sequences generated in this study are shown in bold. Posterior probabilities of the Bayesian analysis (>0.9) are shown next to the nodes. The scale bar indicates the number of amino acid substitutions per site. The major clades representing Old World and New World primate families, superfamilies (for OWM), and parvorders are labeled on the right side of the figure. The virus names are associated with their accession numbers. Viruses of the Cercopithecidae comprise those of the chacma (BaCMV NC_027016 [21, 59]) and olive (BaCMV AF387664 [22]) baboons, the moustached guenon (CceCMV1 AY728178), the agile mangabey (CagCMV1 AY608713), the drill (MndCMV AF282941 and MndCMV AF387665 [strain OCOM6-2] [22, 28]), the mandrill (MndCMV AY129399), the African green monkey (CeHV5 AY117754, CeHV5 FJ483969 [strain Colburn], CeHV5 FJ483969 [strain 2715], CaeCMV AF292066, VervetCMV AY049066 [strain CSG], and CeHV3 AY049065 [22, 60]), the cynomolgus macaque (MfasCMV1 JN227533 [strain Ottawa], MfasCMV1 AY728171, and MfasCMV KP796148 [strain Mauritius] [61, 62]), the rhesus macaque (McHV3 AF033184 and McHV3 DQ120516 [isolate CMV 180.92] [63, 64]), and the mantled guereza (CgueCMV1.1 AY129397 and CgueCMV1.2 EU118147 [30]). Viruses of the Hominidae comprise those of the Bornean orangutan (PpygCMV1.1 AY129396), the human (HHV5 M14709 [strain AD169], HHV5 NC_006273 [strain Merlin], HHV5 AY315197 [strain Towne], and HHV5 AC146905 [isolate Toledo] [65–68]), the Western gorilla (GgorCMV2.1 FJ538490 [27]), and the common chimpanzee (PnHV2 AF480884 [strain Heberling], PtroCMV1.1 FJ538485, and PtroCMV AF292063 [27, 66]). Regarding viruses of New World monkeys, in addition to those described in the present study, viruses of the Cebidae comprise those of the capuchin monkey (CebHV1 JQ264772 and CebusHV AF292067 [from Cebus spp.]), the common squirrel monkey (SaHV4 FJ483967 and SscHV AF292065), and the three-striped night monkey (AoHV1 FJ483970).
Considering NWM CMVs, analyses demonstrated the existence of five distinct lineages supported by high posterior probability values. The phylogenetic relationships between the different NWM CMVs were correlated with the families and genera to which the infected primates belong. The only exception was the hierarchical branching order of the Aotus/Saimiri/Cebus genera within the Cebidae, which was not supported. Thus, viruses from Aotus spp. (AoHV1, AvocCMV1, and AnanCMV1) all grouped together in a monophyletic clade, as did those from Saimiri (SaHV4, SscHV, SalbCMV1, and SbolCMV1), Cebus/Sapajus (CebHV1, CebusHV, CalbCMV1, CcapCMV1, and SapeCMV1), Alouatta (ApalCMV1, AmacCMV1, and AsenCMV1), and Pithecia (PpitCMV1). Furthermore, viruses from Alouatta spp. were related to ApanCMV1 from Ateles in a monophyletic clade of viruses infecting Atelidae monkeys, with a posterior probability of 1, while those from Saimiri, Aotus, and Cebus/Sapajus belonged to a monophyletic clade of Cebidae, which was supported by a posterior probability of 0.91.
To explore the cospeciation hypothesis, a time calibration analysis was performed on our data set. Cytomegaloviruses identified in NWMs diverged from those of OWMs about 32.45 million years ago (MYA) (95% highest posterior density interval [HPD], 17.76 to 52.33 MYA) (Table 4). In the New World clade, three major groups were identified, corresponding to viruses hosted by members of the (i) Pitheciidae, (ii) Atelidae, and (iii) Cebidae. The Pitheciidae viruses diverged from those of the two other groups 22.33 MYA (95% HPD, 16.25 to 28.21 MYA), and intraspecific divergence of PpitCMV1 occurred 1.15 MYA. The divergence between Atelidae and Cebidae viruses is estimated to have occurred 17.16 MYA (95% HPD, 10.29 to 24.25 MYA). Within the group of Atelidae viruses, the divergence between ApanCMV1, identified in Ateles paniscus, and viruses identified in Alouatta spp. (ApalCMV1, AmacCMV1, and AsenCMV1) is estimated to have occurred 8.29 MYA (95% HPD, 2.57 to 14.82 MYA). Viruses identified in the three different Alouatta species (Alouatta palliata, Alouatta macconnelli, and Alouatta seniculus) diverged 3.11 MYA (95% HPD, 0.47 to 6.82 MYA) (ApalCMV1 versus AmacCMV1/AsenCMV1), while AmacCMV1 diverged from AsenCMV1 1.33 MYA (95% HPD, 0.02 to 3.45 MYA). Within the Cebidae group, the divergence between viruses of the Aotinae, Cebinae, and Saimiriinae occurred around 11.54 MYA (95% HPD, 5.82 to 17.89 MYA). Within the group of cytomegaloviruses hosted by the different Aotus species, AoHV1, identified in A. trivirgatus, diverged from the others about 3.98 MYA (95% HPD, 0.8 to 8.25 MYA), while AnanCMV1 diverged from AvocCMV1 2.15 MYA (95% HPD, 0.2 to 4.88 MYA). Within the group of viruses identified in the Cebinae, CcapCMV1 from Cebus capucinus diverged from the others 3.26 MYA (95% HPD, 0.59 to 6.94 MYA). SapCMV1, identified in Sapajus apella, diverged from the other viruses detected in Cebus albifrons and Cebus spp. 2.09 MYA (0.32 to 4.68 MYA). Finally, within the group of viruses identified in the Saimiri genus, SsciCMV1/SaHV4, detected in S. sciureus, diverged from those hosted by S. boliviensis and Saimiri albigena (SbolCMV1 and SalbCMV1) 2.68 MYA (95% HPD, 0.28 to 6.14 MYA).
TABLE 4.
Estimates of Platyrrhini divergence times based on CMV DNA polymerase gene sequence data and comparison with other estimates
| Node | Divergence time (MYA [95% HPD]) |
||
|---|---|---|---|
| This study | Perelman et al. (4) | Jameson Kiesling et al. (16) | |
| Catarrhini/Platyrrhini | 32.45 (17.76–52.33) | 43.47 (38.55–48.36) | 37.72 (36.04–42.07) |
| Pitheciidae/Atelidae + Cebidae | 22.33 (16.25–28.21) | 24.82 (20.55–29.25) | 25.51 (25.14–26.36) |
| Atelidae/Cebidae | 17.16 (10.29–24.25) | 22.76 (18.14–27.08) | 24.04 (22.6–25.29) |
| Atelinae/Alouattinae | 8.29 (2.57–14.82) | 16.13 (10.52–21.35) | 15.29 (13.29–17.99) |
| Within Alouatta | 3.11 (0.47–6.82) | Not determined | 5.14 (3.65–6.8) |
| Within Cebidae | 11.54 (5.82–17.89) | 19.95 (15.66–24.03) | 20.86 (18.48–22.86) |
| Within Aotus | 3.98 (0.8–8.25) | 5.54 (3.20–7.85) | 4.39 (3.12–5.75) |
| Within Cebus | 3.26 (0.59–6.94) | 6.00 (3.13–9.35) | 5.19 (3.69–6.78) |
| Within Saimiri | 2.68 (0.28–6.14) | 2.24 (1.05–3.73) | 0.97 (0.51–1.45) |
DISCUSSION
This study is the largest conducted, to date, to molecularly characterize CMVs in NWMs in terms of species diversity. It partially characterized 12 cytomegaloviruses from 12 distinct species belonging to seven genera and three NWM families. BLAST searches of the Cytomegalovirus sequences identified further revealed that all but one were new viral sequences close to but distinct from already published CMV sequences from Aotus trivirgatus, Saimiri sciureus, and Cebus spp. The only exception was the viral sequence from C. albifrons, which showed 99.6% identity at the nucleotide level to CebusHV (accession number AF292067) and CebHV1 (accession number JQ264772), both identified from unspecified Cebus spp. These three viral sequences were therefore considered to correspond to the same viral species. In addition, the newly identified viral sequences are completely host specific, with no identification of cross-species transmission in our sample. The observations on sequence comparisons, phylogenetic analysis, and host specificity of the sequences reported in this paper are close to the species demarcation criteria outlined in the 9th ICTV report for formal recognition of new herpesvirus species (38; https://talk.ictvonline.org/ictv-reports/ictv_9th_report/).
By refining the degeneracy of the PCR primers used to screen the sample collection, we were able to specifically target and identify CMV sequences, even though some of the primates tested were coinfected with lymphocryptoviruses (36). Indeed, we formerly identified 17 EBV-related viruses from 15 NWM species belonging to seven genera and three families from the same collection of samples (36, 37). These new combinations of screening primers are therefore good molecular tools to be used for future studies. Nevertheless, among the 20 NWM species tested, we did not characterize any CMV sequences from our collection of Saguinus and Lagothrix samples. Considering the relatively small sampling size for most species belonging to these two genera, with the exception of Saguinus midas, it is conceivable that we missed a CMV-like virus from them. Nevertheless, for the other primate species tested, the sampling size was equivalent or even smaller, and we identified CMV sequences for almost all of them. More strikingly, despite the large sample size of Saguinus midas monkeys screened (54 individuals) and the different PCR approaches used (different combinations of primers with different levels of degeneracy and different PCR cycling conditions), no PCR product was identified. The negativity of the Saguinus and Lagothrix genera for CMV-related viruses can be explained by a lack of primer matching or by a loss of CMVs during evolution within these genera. Likewise, in our former studies of EBV-related sequences, we were unsuccessful in amplifying EBV sequences from individuals of the Aotus and Alouatta genera (36, 37). Taken together, these results highlight the need for more in-depth analyses of representative samples of these and other species of these genera to clarify this point. Moreover, for some of the positive samples, we were unsuccessful in generating longer sequences of the DNA polymerase gene. Whether this is due to the low quality/small amount of the remaining DNA or a low viral load or reflects technical difficulties, i.e., an inadequate level of degeneracy of the primers designed for some of these viruses, is not clear. Nonetheless, the sequence data generated here were sufficient to gain insight into the genetic relationships.
Pairwise nucleotide and amino acid sequence comparisons demonstrated that the viral sequences analyzed present different levels of genetic diversity among them (Table 3); the smallest divergences were detected when viral sequences from primates belonging to the same genus were analyzed. Phylogenetic analyses showed that CMV sequences grouped according to the primate genera from which they were detected. Thereafter, the phylogenetic clustering and diversification followed those proposed for NWM species, corroborating the hypothesis of joint evolution of the viruses with the speciation of their hosts (4, 6). In contrast, analyses of NWM EBV sequences have fallen short of achieving a completely resolved phylogeny (36). While a clear cospeciation can be seen in the terminal branchings within major lineages according to the primate subfamilies, the phylogenetic relationships between them are not concordant with the current interpretations of the host pattern of diversification at the family level. In addition, for OWMs, there is a similar incongruence between the Lymphocryptovirus phylogeny and that of the corresponding host lineages (39, 40). One can therefore argue that, within the Herpesviridae family, DNA polymerase gene sequences from viruses of the Cytomegalovirus genus are better molecular markers than those from viruses of the Lymphocryptovirus genus for testing hypotheses of herpesvirus-primate coevolution. On the basis of the available data, our analysis nevertheless has two limitations regarding viruses of Cebidae that do not perfectly reflect current interpretations of their hosts' diversification pattern. While viral sequences from members of the Cebidae segregate into three well-supported clades, each corresponding to the host genus from which they were identified, i.e., Cebus/Sapajus, Saimiri, and Aotus, the relationships between the three clades are not phylogenetically supported (Fig. 3). The second limitation concerns SapeCMV1, identified from Sapajus apella, which phylogenetically falls within the group of Cebus viruses (Fig. 3). Nevertheless, pairwise sequence comparison of SapeCMV1 with the viral sequences identified from Cebus spp. shows that the nucleotide divergence of SapeCMV1 is over the maximum 8% observed for viral sequences identified from NWM species of the same genus (Table 3). These combined results (on SapeCMV1 and the other Cebus viruses) do not, for the moment, make it possible to confidently separate the Cebus genus into two genera as observed on analyses of Alu elements and by phylogenomics (4, 18). However, our virus results agree quite well with the mitogenomics findings obtained in recent studies, where Sapajus is a taxon within Cebus (19). These limits should be resolved by screening an extensive taxon sampling of the different Sapajus spp. as well as of Callitrichinae for the presence of cytomegaloviruses.
Finally, these data support virus-host coevolution in terms of branching order as well as divergence time. Indeed, for each NWM genus tested, the estimated timing of diversification of viruses is in agreement with host sequence divergence date estimates from previously published studies (Table 4) (4, 11, 16). Nevertheless, dates obtained at superior taxonomic levels are more recent than those based on different types of data sets or models. These discrepancies can be attributed to the fact that a majority of NWM taxa remain to be tested and that no CMV sequence is available for numerous genera. This therefore limits the significance of our estimates for the major primate lineages, for the moment, and emphasizes the need for further studies.
Here we conclusively expand our knowledge of the viral diversity, distribution, and evolutionary relationships of NWM CMVs. Even if the evolutionary history of these viruses is not fully resolved, and despite the limitations mentioned above, these results strongly support the hypothesis of coevolution of these new viruses with their hosts. In light of these data, we propose that CMV DNA polymerase gene sequences may serve as genetic markers to define the evolutionary links of their host species. Indeed, despite the number of studies conducted over the past few decades and the fast-growing number of host DNA sequence data sets, a unifying consensus of the evolutionary hierarchy of NWMs has not fully been reached, partly because not all phylogenies from these data sets agree but also due to a large proportion of missing data for some taxa (2, 4–16). The search for and identification of CMV DNA polymerase gene sequences therefore seem to be an alternative to help solve this issue. Given the high prevalence rates of CMVs in wild primates, their spread through close contact with infectious bodily fluids, and their persistence for the lifetime of the host, cytomegalovirus sequences, if present, should be obtained easily through our PCR approach (21, 22, 41–43). Considering the number of all presently known NWM species, we tested only a fraction of their diversity. This suggests that a great number of cytomegaloviruses remain to be identified in this important group of primates. These results argue for a wider and more systematic sampling and exploration of NWMs to evaluate the presence of CMVs and to confirm the usefulness of those sequences as a new molecular tool to infer the systematics of Platyrrhini.
MATERIALS AND METHODS
Sample collection.
The collection of blood DNA samples has been described in detail elsewhere (Table 1) (36, 37, 44–46). In brief, we tested a total of 244 DNA samples from 20 NWM species (26 subspecies) belonging to the three families and six of the seven subfamilies, according to Schneider and Sampaio (6). All samples were previously genetically identified by phylogenetic analysis of mitochondrial DNA (mtDNA) genes, including the cytochrome c oxidase subunit I (COX1) and/or cytochrome b (CytB) gene (36).
Ethics.
This study is based on samples that were collected several years ago. Biological material from French Guiana was collected in 1994 and 1995, along the Sinnamary River, Petit Saut Hydroelectric Dam, under the supervision of veterinarians of the “Faune Sauvage” team, led by Jean-Christophe Vié (47). Blood sampling from live animals was carried out in accordance with French animal care regulations and the laws of France. The other samples were collected directly from animals killed in the field by indigenous hunters for their own purposes, with the full consent of the hunters and in accordance with the laws of Brazil, Colombia, Mexico, Peru, Guatemala, and Argentina.
Initial screening of samples.
Molecular screening was done by seminested PCR amplification with degenerate consensus primers targeting highly conserved amino acid motifs of the herpesvirus DNA polymerase gene (Table 2). To maximize the chances of amplifying CMV-like sequences, the primers of Rose et al. were refined based on the alignment of all primate cytomegalovirus DNA polymerase gene sequences available in the databases (Table 2) (48). The CMV3F1, CMV3F2, and CMV3R1 primers were designed and used in place of primers QAHNA, VYGA, and GDTD1B, respectively, while the sense primer DFASA was kept as it was. Two different combinations of primers (DFASA/CMV3R1 and CMV3F1/CMV3R1) were used on each DNA sample in separate reaction mixtures for the first-round PCR (Fig. 2). In the second-round PCR, the CMV3F2/CMV3R1 primers were used. PCR analyses were performed at an annealing temperature of 60°C, with an elongation time of 30 s, for 35 cycles. All amplicons of approximately the expected size were purified, cloned by TA cloning, and sent for sequencing to Genewiz, Takeley, United Kingdom.
Partial DNA polymerase gene amplification.
To obtain the nucleotide sequence upstream of the CMV3F2 motif, a degenerate primer (CMV3R2) was derived from the complementary sequences of the small fragments and used in an nPCR amplification with the DFASA or CMV3F1 primer pool, using the initial PCR products as templates (Table 2; Fig. 2). Then, to generate longer segments of the DNA polymerase gene for each newly identified virus, we tried to obtain upstream and downstream sequences by using different sets of consensus degenerate and species-specific primers designed using the primate CMV DNA polymerase gene sequence alignment (Table 2; Fig. 2). Overlapping amplicons were generated, cloned, and sequenced as described above. Each sequence corresponds to at least three independent clones sequenced on both strands. Contig sequences were then assembled using MEGA 5.05 software (49).
Phylogenetic analysis.
Raw sequences were analyzed and edited in MEGA 5.05 (49). Sequences were confirmed to be CMV sequences by homology analysis using the NCBI BLAST search tool (50). Multiple-sequence alignments with all other previously published primate CMV sequences were constructed using ClustalW, and alignments were checked manually. Sequences were translated into amino acids, and both nucleotide and amino acid sequences were checked for irregularities. Hypervariable regions were removed before performing analyses. Sequence identity was calculated using uncorrected P distances. Phylogenetic trees were inferred from the aligned amino acid sequences. The JTT+G model was selected as the best-fitting model of amino acid evolution under the corrected Akaike information criterion (AICc) by use of MEGA 5.05 and was used for the Bayesian approach (51), which was performed with MrBayes 3.2.2 to infer phylogenetic relationships (52). Markov chain Monte Carlo (MCMC) simulations were run for 10,000,000 generations, with four simultaneous chains, using a sample frequency of 500 and a burn-in of 25,000. Majority-rule consensus trees were obtained from the output. Validation of the inference was assessed based on the standard deviation (SD) of split frequencies, which was less than the expected threshold value of 0.01 (calculated value of 0.002).
Time calibration.
Divergence times between clades were calculated using a relaxed Bayesian molecular clock model with an uncorrelated lognormal rate of distribution, as implemented in BEAST, version 1.7.4 (53). A monophyletic constraint was imposed for the nodes used to calibrate evolutionary rates. Two calibration points were applied as normal priors to constrain the ages of the Platyrrhini and Homo-Pan clades: the time of the most recent common ancestor (tMRCA) of Platyrrhini to 23.5 MYA (SD = 3.0) and that of Homo-Pan to 6.5 MYA (SD = 0.8) (54–56). These calibration points are based on fossil dates (57). The amino acid substitution model was the same as that described above. A Yule process of speciation was used as the tree prior. Results were obtained for 10,000,000 generations, with the first 2,500,000 discarded as burn-in and parameter values sampled every 100 generations. The effective sample sizes for parameter estimates and convergence were checked using Tracer, version 1.5.0, software (58). The final tree, with divergence estimates and their 95% HPDs, was computed in TreeAnnotator v1.4.5 (53).
Accession number(s).
The sequences reported in this paper have been deposited in the GenBank database under accession numbers KU963225 to KU963240.
ACKNOWLEDGMENTS
Warm thanks go to Benoît de Thoisy for providing NWM DNA samples from French Guiana.
S.J. was supported by a grant from the Université de la Guyane, École Doctorale 587-Diversités, Santé et Développement en Amazonie, and by a grant from the Collectivité Territoriale de la Guyane. This study was funded by a European commission REGPOT-CT-2011-285837-STRonGer grant within the FP7 and an Investissement d'Avenir grant managed by the Agence Nationale de la Recherche (CEBA) (grant ANR-10-LABX-25-01). It was also supported by grants 1203-09-11239 (Colciencias) and 120108-E0102141 (Fondo para la Acción Ambiental) to M.R.-G.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that we have no competing financial interests.
A.L., M.R.-G., and V.L. contributed to the study design. M.R.-G. collected some of the samples. S.J., D.D., J.-F.P., A.L., and V.L. performed the molecular and genetic analyses. S.J., A.L., and V.L. analyzed the data and wrote the article. All authors participated in the final writing and editing.
REFERENCES
- 1.Groves CP. 2005. Order Primates, p 111–184. In Wilson DE, Reeder DM (ed), Mammal species of the world: a taxonomic and geographic reference, 3rd ed, vol 2 The Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 2.Rylands AB, Mittermeier RA. 2009. The diversity of the New World primates (Platyrrhini): an annotated taxonomy, p 23–54. In Garber PA, Estrada A, Bicca-Marques JC, Heymann EW, Stier KB (ed), South American primates—comparative perspectives in the study of behavior, ecology, and conservation. Springer-Verlag New York, New York, NY. doi: 10.1007/978-0-387-78705-3. [DOI] [Google Scholar]
- 3.Voevodin AF, Marx PA Jr. 2009. Classification of nonhuman primates, p 3–38. In Simian virology. Wiley-Blackwell, Ames, IA. [Google Scholar]
- 4.Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O'Brien SJ, Pecon-Slattery J. 2011. A molecular phylogeny of living primates. PLoS Genet 7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider H, Canavez FC, Sampaio I, Moreira MA, Tagliaro CH, Seuanez HN. 2001. Can molecular data place each neotropical monkey in its own branch? Chromosoma 109:515–523. doi: 10.1007/s004120000106. [DOI] [PubMed] [Google Scholar]
- 6.Schneider H, Sampaio I. 2015. The systematics and evolution of New World primates—a review. Mol Phylogenet Evol 82:348–357. doi: 10.1016/j.ympev.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Schneider H, Sampaio I, Harada ML, Barroso CM, Schneider MP, Czelusniak J, Goodman M. 1996. Molecular phylogeny of the New World monkeys (Platyrrhini, primates) based on two unlinked nuclear genes: IRBP intron 1 and epsilon-globin sequences. Am J Phys Anthropol 100:153–179. doi:. [DOI] [PubMed] [Google Scholar]
- 8.Schneider H, Schneider MP, Sampaio I, Harada ML, Stanhope M, Czelusniak J, Goodman M. 1993. Molecular phylogeny of the New World monkeys (Platyrrhini, primates). Mol Phylogenet Evol 2:225–242. doi: 10.1006/mpev.1993.1022. [DOI] [PubMed] [Google Scholar]
- 9.Rylands AB, Mittermeier RA, Silva JS Jr. 2012. Neotropical primates: taxonomy and recently described species and subspecies. Int Zoo Yearb 46:11–24. doi: 10.1111/j.1748-1090.2011.00152.x. [DOI] [Google Scholar]
- 10.Rylands AB, Schneider H, Langguth A, Mittermeier RA, Groves CP, Rodrigues-Luna E. 2000. An assessment of the diversity of New World primates. Neotrop Primates 8:61–93. [Google Scholar]
- 11.Opazo JC, Wildman DE, Prychitko T, Johnson RM, Goodman M. 2006. Phylogenetic relationships and divergence times among New World monkeys (Platyrrhini, Primates). Mol Phylogenet Evol 40:274–280. doi: 10.1016/j.ympev.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Wildman DE, Jameson NM, Opazo JC, Yi SV. 2009. A fully resolved genus level phylogeny of neotropical primates (Platyrrhini). Mol Phylogenet Evol 53:694–702. doi: 10.1016/j.ympev.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Singer SS, Schmitz J, Schwiegk C, Zischler H. 2003. Molecular cladistic markers in New World monkey phylogeny (Platyrrhini, Primates). Mol Phylogenet Evol 26:490–501. doi: 10.1016/S1055-7903(02)00312-3. [DOI] [PubMed] [Google Scholar]
- 14.Ray DA, Xing J, Hedges DJ, Hall MA, Laborde ME, Anders BA, White BR, Stoilova N, Fowlkes JD, Landry KE, Chemnick LG, Ryder OA, Batzer MA. 2005. Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol 35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 15.Osterholz M, Walter L, Roos C. 2009. Retropositional events consolidate the branching order among New World monkey genera. Mol Phylogenet Evol 50:507–513. doi: 10.1016/j.ympev.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Jameson Kiesling NM, Yi SV, Xu K, Gianluca Sperone F, Wildman DE. 2015. The tempo and mode of New World monkey evolution and biogeography in the context of phylogenomic analysis. Mol Phylogenet Evol 82:386–399. doi: 10.1016/j.ympev.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Alfaro JW, Silva JD Jr, Rylands AB. 2012. How different are robust and gracile capuchin monkeys? An argument for the use of Sapajus and Cebus. Am J Primatol 74:273–286. doi: 10.1002/ajp.22007. [DOI] [PubMed] [Google Scholar]
- 18.Martins AM Jr, Amorim N, Carneiro JC, de Mello Affonso PR, Sampaio I, Schneider H. 2015. Alu elements and the phylogeny of capuchin (Cebus and Sapajus) monkeys. Am J Primatol 77:368–375. doi: 10.1002/ajp.22352. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Garcia M, Castillo MI, Luengas-Villamil K. 2016. It is misleading to use Sapajus (robust capuchins) as a genus? A review of the evolution of the capuchins and suggestions on their systematics, p 209–268. In Ruiz-Garcia M, Shostell JM (ed), Phylogeny, molecular population genetics, evolutionary biology and conservation of the neotropical primates. Nova Science Publisher, Inc, New York, NY. [Google Scholar]
- 20.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blewett EL, White G, Saliki JT, Eberle R. 2001. Isolation and characterization of an endogenous cytomegalovirus (BaCMV) from baboons. Arch Virol 146:1723–1738. doi: 10.1007/s007050170059. [DOI] [PubMed] [Google Scholar]
- 22.Blewett EL, Lewis J, Gadsby EL, Neubauer SR, Eberle R. 2003. Isolation of cytomegalovirus and foamy virus from the drill monkey (Mandrillus leucophaeus) and prevalence of antibodies to these viruses amongst wild-born and captive-bred individuals. Arch Virol 148:423–433. doi: 10.1007/s00705-002-0937-9. [DOI] [PubMed] [Google Scholar]
- 23.Asher DM, Gibbs CJ Jr, Lang DJ, Gajdusek DC, Chanock RM. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc Soc Exp Biol Med 145:794–801. doi: 10.3181/00379727-145-37897. [DOI] [PubMed] [Google Scholar]
- 24.Malherbe H, Harwin R. 1957. Seven viruses isolated from the vervet monkey. Br J Exp Pathol 38:539–541. [PMC free article] [PubMed] [Google Scholar]
- 25.Black PH, Hartley JW, Rowe WP. 1963. Isolation of a cytomegalovirus from African green monkey. Proc Soc Exp Biol Med 112:601–605. doi: 10.3181/00379727-112-28115. [DOI] [PubMed] [Google Scholar]
- 26.Wroblewska Z, Gilden D, Devlin M, Huang ES, Rorke LB, Hamada T, Furukawa T, Cummins L, Kalter S, Koprowski H. 1979. Cytomegalovirus isolation from a chimpanzee with acute demyelinating disease after inoculation of multiple sclerosis brain cells. Infect Immun 25:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leendertz FH, Deckers M, Schempp W, Lankester F, Boesch C, Mugisha L, Dolan A, Gatherer D, McGeoch DJ, Ehlers B. 2009. Novel cytomegaloviruses in free-ranging and captive great apes: phylogenetic evidence for bidirectional horizontal transmission. J Gen Virol 90:2386–2394. doi: 10.1099/vir.0.011866-0. [DOI] [PubMed] [Google Scholar]
- 28.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Rigoulet J, Petit T, Gessain A. 2000. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J Virol 74:11993–11999. doi: 10.1128/JVI.74.24.11993-11999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murthy S, Couacy-Hymann E, Metzger S, Nowak K, De Nys H, Boesch C, Wittig R, Jarvis MA, Leendertz FH, Ehlers B. 2013. Absence of frequent herpesvirus transmission in a nonhuman primate predator-prey system in the wild. J Virol 87:10651–10659. doi: 10.1128/JVI.01104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prepens S, Kreuzer KA, Leendertz F, Nitsche A, Ehlers B. 2007. Discovery of herpesviruses in multi-infected primates using locked nucleic acids (LNA) and a bigenic PCR approach. Virol J 4:84. doi: 10.1186/1743-422X-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anoh AE, Murthy S, Akoua-Koffi C, Couacy-Hymann E, Leendertz FH, Calvignac-Spencer S, Ehlers B. 2017. Cytomegaloviruses in a community of wild nonhuman primates in Tai National Park, Cote D'Ivoire. Viruses 10:E11. doi: 10.3390/v10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seimon TA, Olson SH, Lee KJ, Rosen G, Ondzie A, Cameron K, Reed P, Anthony SJ, Joly DO, Karesh WB, McAloose D, Lipkin WI. 2015. Adenovirus and herpesvirus diversity in free-ranging great apes in the Sangha region of the Republic of Congo. PLoS One 10:e0118543. doi: 10.1371/journal.pone.0118543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel MD, Melendez LV, King NW, Barahona HH, Fraser CE, Garcia FG, Silva D. 1973. Isolation and characterization of a new virus from owl monkeys: herpesvirus aotus type 3. Am J Phys Anthropol 38:497–500. doi: 10.1002/ajpa.1330380254. [DOI] [PubMed] [Google Scholar]
- 34.Daniel MD, Melendez LV, King NW, Fraser CE, Barahona HH, Hunt RD, Garcia FG, Trum BF. 1971. Herpes virus aotus: a latent herpesvirus from owl monkeys (Aotus trivirgatus) isolation and characterization. Proc Soc Exp Biol Med 138:835–845. doi: 10.3181/00379727-138-36002. [DOI] [PubMed] [Google Scholar]
- 35.Rangan SR, Chaiban J. 1980. Isolation and characterization of a cytomegalovirus from the salivary gland of a squirrel monkey (Saimiri sciureus). Lab Anim Sci 30:532–540. [PubMed] [Google Scholar]
- 36.Lavergne A, de Thoisy B, Pouliquen JF, Ruiz-Garcia M, Lacoste V. 2011. Partial molecular characterisation of New World non-human primate lymphocryptoviruses. Infect Genet Evol 11:1782–1789. doi: 10.1016/j.meegid.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 37.de Thoisy B, Pouliquen JF, Lacoste V, Gessain A, Kazanji M. 2003. Novel gamma-1 herpesviruses identified in free-ranging New World monkeys (golden-handed tamarin [Saguinus midas], squirrel monkey [Saimiri sciureus], and white-faced saki [Pithecia pithecia]) in French Guiana. J Virol 77:9099–9105. doi: 10.1128/JVI.77.16.9099-9105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellett PE, Davison AJ, Eberle R, Hayward BEGS, Lacoste V, Minson AC, Nicholas J, Roizman B, Studdert MJ, Wang F. 2011. Order Herpesvirales, p 99–107. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Inc, Oxford, United Kingdom. [Google Scholar]
- 39.Ehlers B, Ochs A, Leendertz F, Goltz M, Boesch C, Matz-Rensing K. 2003. Novel simian homologues of Epstein-Barr virus. J Virol 77:10695–10699. doi: 10.1128/JVI.77.19.10695-10699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehlers B, Spiess K, Leendertz F, Peeters M, Boesch C, Gatherer D, McGeoch DJ. 2010. Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus. J Gen Virol 91:630–642. doi: 10.1099/vir.0.017251-0. [DOI] [PubMed] [Google Scholar]
- 41.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 42.Cannon MJ, Hyde TB, Schmid DS. 2011. Review of cytomegalovirus shedding in bodily fluids and relevance to congenital cytomegalovirus infection. Rev Med Virol 21:240–255. doi: 10.1002/rmv.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves M, Sinclair J. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol 325:297–313. [DOI] [PubMed] [Google Scholar]
- 44.de Thoisy B, Vogel I, Reynes JM, Pouliquen JF, Carme B, Kazanji M, Vie JC. 2001. Health evaluation of translocated free-ranging primates in French Guiana. Am J Primatol 54:1–16. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Garcia M, Castillo MI, Vasquez C, Rodriguez K, Pinedo-Castro M, Shostell J, Leguizamon N. 2010. Molecular phylogenetics and phylogeography of the white-fronted capuchin (Cebus albifrons; Cebidae, Primates) by means of mtCOII gene sequences. Mol Phylogenet Evol 57:1049–1061. doi: 10.1016/j.ympev.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Ruiz-Garcia M, Pinedo-Castro MO. 2010. Molecular systematics and phylogeography of the genus Lagothrix (Atelidae, Primates) by means of the mitochondrial COII gene. Folia Primatol (Basel) 81:109–128. doi: 10.1159/000315070. [DOI] [PubMed] [Google Scholar]
- 47.Vie JC. 1999. Wildlife rescues—the case of the Petit Saut hydroelectric dam in French Guiana. Oryx 33:115–126. [Google Scholar]
- 48.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW Jr, Thouless ME, Tsai CC, Bosch ML. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol 71:4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poux C, Douzery EJ. 2004. Primate phylogeny, evolutionary rate variations, and divergence times: a contribution from the nuclear gene IRBP. Am J Phys Anthropol 124:1–16. doi: 10.1002/ajpa.10322. [DOI] [PubMed] [Google Scholar]
- 55.Kay RF, Fleagle JG, Mitchell TR, Colbert M, Bown T, Powers DW. 2008. The anatomy of Dolichocebus gaimanensis, a stem platyrrhine monkey from Argentina. J Hum Evol 54:323–382. doi: 10.1016/j.jhevol.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Vignaud P, Duringer P, Mackaye HT, Likius A, Blondel C, Boisserie JR, De Bonis L, Eisenmann V, Etienne ME, Geraads D, Guy F, Lehmann T, Lihoreau F, Lopez-Martinez N, Mourer-Chauvire C, Otero O, Rage JC, Schuster M, Viriot L, Zazzo A, Brunet M. 2002. Geology and palaeontology of the Upper Miocene Toros-Menalla hominid locality, Chad. Nature 418:152–155. doi: 10.1038/nature00880. [DOI] [PubMed] [Google Scholar]
- 57.Tavare S, Marshall CR, Will O, Soligo C, Martin RD. 2002. Using the fossil record to estimate the age of the last common ancestor of extant primates. Nature 416:726–729. doi: 10.1038/416726a. [DOI] [PubMed] [Google Scholar]
- 58.Rambaut A, Drummond AJ. 2009. Tracer, version 1.5.0. http://beast.community/tracer.
- 59.Blewett EL, Sherrod CJ, Texier JR, Conrad TM, Dittmer DP. 2015. Complete genome sequences of Mandrillus leucophaeus and Papio ursinus cytomegaloviruses. Genome Announc 3:e00781-15. doi: 10.1128/genomeA.00781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson G, Dick D, Ayers M, Petric M, Tellier R. 2003. Detection and species-level identification of primate herpesviruses with a comprehensive PCR test for human herpesviruses. J Clin Microbiol 41:1256–1258. doi: 10.1128/JCM.41.3.1256-1258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marsh AK, Willer DO, Ambagala AP, Dzamba M, Chan JK, Pilon R, Fournier J, Sandstrom P, Brudno M, MacDonald KS. 2011. Genomic sequencing and characterization of cynomolgus macaque cytomegalovirus. J Virol 85:12995–13009. doi: 10.1128/JVI.05840-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell JN, Marsh AK, Willer DO, Ambagala AP, Dzamba M, Chan JK, Pilon R, Fournier J, Brudno M, Antony JM, Sandstrom P, Evans BJ, MacDonald KS. 2016. A novel strain of cynomolgus macaque cytomegalovirus: implications for host-virus co-evolution. BMC Genomics 17:277. doi: 10.1186/s12864-016-2588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swanson R, Bergquam E, Wong SW. 1998. Characterization of rhesus cytomegalovirus genes associated with anti-viral susceptibility. Virology 240:338–348. doi: 10.1006/viro.1997.8935. [DOI] [PubMed] [Google Scholar]
- 64.Rivailler P, Kaur A, Johnson RP, Wang F. 2006. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J Virol 80:4179–4182. doi: 10.1128/JVI.80.8.4179-4182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kouzarides T, Bankier AT, Satchwell SC, Weston K, Tomlinson P, Barrell BG. 1987. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol 61:125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84:17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 67.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]