We found that stable transduction with lentiviral shRNA was able to nonspecifically inhibit HCV infection by the dysregulation of host miRNAs. Previous studies showed that the overexpression of shRNAs oversaturated the host miRNA pathways to inhibit HCV infection. In contrast, the miRNA machinery was not affected in our study. Knockout studies suggested that the nonspecific effect was independent of the transcription of shRNAs. The lentiviral vector itself and the integration sites in the host genome determined the changes in miRNAs. Stable transduction with lentiviral vectors was able to increase the expression of JUN, which in turn upregulated miR-216a-5p, miR-216b-5p, and miR-217. miR-216a-5p and miR-216b-5p might inhibit HCV by suppressing the host autophagic machinery. Our study suggested a novel nonspecific effect of lentiviral vectors, and this side effect should be considered when transduction with lentiviral vectors is performed for other purposes, especially in therapy.
KEYWORDS: hepatitis C virus, lentiviral vector, microRNA, off-target effect, shRNA
ABSTRACT
RNA interference (RNAi) is widely used in gene knockdown analysis and as a tool to screen host genes involved in viral infection. Owing to the limitations of transducing cells with synthetic small interfering RNAs (siRNAs), lentiviral short hairpin RNA (shRNA) vectors are more widely used. However, we found that stable transduction with lentiviral shRNA vectors inhibited hepatitis C virus (HCV) propagation in human hepatoma cells. We found by microRNA (miRNA) microarray analysis that this inhibition was induced by the alteration of host miRNA expression. In addition to one miRNA (miR-196b-5p) previously reported to be involved in HCV infection, other miRNAs (miR-216a-5p, -216b-5p, 217, and -30b-5p) were found to influence HCV infection in this study. Further studies suggested that this effect was independent of the transcription of shRNAs. The lentiviral vector itself and the integration site of the lentiviral vector might determine the change in miRNA expression. Moreover, the upregulation of JUN contributed to the dysregulation of miR-216a-5p, -216b-5p, and -217 in stably transduced cells. Although the changes in miRNA expression were beneficial for inhibiting HCV infection in our study, this off-target effect should be considered when transduction with lentiviral vectors is performed for other purposes, especially in therapy.
IMPORTANCE We found that stable transduction with lentiviral shRNA was able to nonspecifically inhibit HCV infection by the dysregulation of host miRNAs. Previous studies showed that the overexpression of shRNAs oversaturated the host miRNA pathways to inhibit HCV infection. In contrast, the miRNA machinery was not affected in our study. Knockout studies suggested that the nonspecific effect was independent of the transcription of shRNAs. The lentiviral vector itself and the integration sites in the host genome determined the changes in miRNAs. Stable transduction with lentiviral vectors was able to increase the expression of JUN, which in turn upregulated miR-216a-5p, miR-216b-5p, and miR-217. miR-216a-5p and miR-216b-5p might inhibit HCV by suppressing the host autophagic machinery. Our study suggested a novel nonspecific effect of lentiviral vectors, and this side effect should be considered when transduction with lentiviral vectors is performed for other purposes, especially in therapy.
INTRODUCTION
Hepatitis C virus (HCV) infects more than 170 million people worldwide and is a major cause of chronic liver disease, such as liver cirrhosis, hepatocellular carcinoma, and metabolic diseases (1, 2). Similar to other viruses, HCV is dependent on host cell proteins for viral entry, uncoating, replication, virion assembly, and release (reviewed in references 3 and 4). Knockdown or knockout of related host genes has been essential in studying the interaction between host cells and the virus. Although new technology, such as Cas-9 mediated CRISPR (clustered regularly interspaced short palindromic repeat), has shown great advantages in gene knockout experiments, RNA interference (RNAi) is still widely used.
Many previous studies have attempted to screen host factors involved in HCV infection by using small interfering RNA (siRNA) libraries (5–8). However, the host factors identified in these studies had little overlap, and many well-established factors were not identified owing to the limitations of using synthetic siRNA in the studies. Synthetic-siRNA transfection can be ineffective in many types of mammalian cells, and some cells are even resistant to such transfection. In the cells that can be transfected, synthetic-siRNA transfection evokes only transient gene silencing, and it can have nonspecific or toxic effects on the host cells (9). Lentiviral vector delivery of short hairpin RNAs (shRNAs) can be used to overcome this obstacle; this alternative method has advantages over siRNAs owing to its wide-range tropism and genomic integration properties. Accordingly, it has been applied in the identification of viral-infection-related host factors (10–13).
However, viral shRNA vectors also have their own limitations and can induce off-target effects. In addition to specific off-target effects caused by the limited degree of shRNA complementarity to nontargeted mRNAs, as well as the problem of host toxicity due to the delivery vehicle, the high level of shRNA produced by this method could overwhelm the host RNAi machinery (14, 15). Grimm et al. found that a host factor in the microRNA (miRNA) pathway's exportin 5 was saturated by shRNA transduction (15). It was also reported that high levels of transduced lentiviral shRNA could disturb endogenous miR-122 biogenesis that was able to inhibit HCV replication (16).
In this study, we initially set out to investigate the underlying mechanism by which the host cell-derived charged multivesicular body protein 4B (CHMP4B) gene can regulate HCV assembly, as it has been reported that CHMP4B participates in the release of infectious HCV particles, but not in viral RNA replication (17, 18). During our knockdown studies, we surprisingly discovered that the cell clones stably transduced with both the CHMP4B shRNA and the nontarget control shRNA showed significant decreases in HCV propagation.
However, because the Huh7.5 cell line we used was heterogeneous, consisting of various cell clones with different sensitivities to viral infection, the inhibition of HCV in stably shRNA-transduced cell clones might be clone dependent. A single cell clone (Huh7.5-1) was derived from Huh7.5 and was utilized to repeat the studies. Moreover, to avoid the possible influences of CHMP4B suppression on miRNA expression and to monitor more effectively the expression of the shRNA, we applied a commercial control shRNA targeted to enhanced green fluorescent protein (eGFP), and we found that stable transduction of Huh7.5-1 cells with the eGFP shRNA also impacted HCV infection, indicating that this effect of lentiviral shRNA vectors in disrupting the HCV infection process is a nonspecific and off-target effect. miRNA microarray analyses of stably transduced cell pools and selected cell clones showed that the stable expression of integrated shRNAs caused a change of endogenous miRNA expression. Some of the changed miRNAs were validated to regulate HCV infection.
Knocking out the integrated shRNA sequence in stably transduced cell clones did not eliminate the influence on miRNA expression, which suggested that this off-target effect was independent of the transcription of shRNAs. The results of the identification of lentiviral vector integration sites in the genomes of selected clones suggested that integration sites determined whether the lentiviral vector affected the expression of miRNAs. Further studies revealed that the dysregulation of the particular miRNAs resulted partially from the elevated expression of JUN. Moreover, the effects were also observed when lentiviral vectors were stably transduced into HEK293 or HeLa cells. Although the changes in miRNAs were beneficial for inhibiting HCV infection in our study, this off-target effect should be considered when transduction with lentiviral vectors is performed for other purposes, especially in therapy.
RESULTS
Stable transduction with shRNA vectors exhibited nonspecific off-target effects.
In an effort to understand what host cell factors are involved in helping to assemble the HCV virion in HCV-infected cells, we analyzed the expression of several endosomal sorting complex required for transport (ESCRT) proteins during HCV infection (data not shown). We found that the protein level of CHMP4B, a component of the ESCRT complex machinery, was moderately upregulated (Fig. 1a). To analyze the function of CHMP4B, we deployed lentiviral shRNA vectors (obtained from the commonly used TRC RNAi library [Sigma-Aldrich, Switzerland]) to knock down CHMP4B in HCV-infected Huh7.5 cells. Because high levels of transduced lentiviral shRNA disturbed endogenous miR-122 biogenesis that was able to inhibit HCV replication (16), an optimal dose of lentiviruses should be able to knock down CHMP4B efficiently without a significant influence on host miRNAs. Huh7.5 cells were transiently transduced with shRNA lentiviruses at different multiplicities of infection (MOI), followed by HCV infection, and HCV proteins, HCV RNA, and cellular miR-122-5p were analyzed (Fig. 1b to d). CHMP4B was efficiently knocked down by lentiviruses expressing the CHMP4B shRNA at an MOI of 2, 5, or 10. Transduction with the control shRNA or the CHMP4B shRNA lentiviruses at an MOI of 5 or 10 significantly reduced host miR-122-5p, HCV proteins, and HCV RNA. In contrast, at an MOI of 2, host miR-122 was not significantly changed after transduction with the control shRNA or the CHMP4B shRNA lentiviruses. Therefore, an MOI of 2 might be an optimal dose for the knockdown of CHMP4B. Transient knockdown of CHMP4B with an optimal dose (2 MOI) of the lentiviral shRNA vector did not affect HCV RNA and expression of the HCV proteins (Fig. 1b and c).
FIG 1.
Stable, but not transient, transduction with lentiviral shRNA vectors decreased HCV protein and RNA. (a) Huh7.5 cells were infected with HCV at an MOI of 2. The protein levels of CHMP4B and HCV core were analyzed by Western blotting 48 h after infection. (b) Huh7.5 cells were transiently transduced with different doses of the lentiviral CHMP4B shRNA or a nontarget control, followed by infection with HCV at an MOI of 2 24 h after transduction. The protein levels of CHMP4B, HCV NS5A, and HCV core were assayed 48 h after HCV infection. Actin was used as a loading control. (c) Huh7.5 cells were treated as for panel b. The level of intracellular HCV RNA was assayed by real-time PCR 48 h after infection. (d) Huh7.5 cells were treated as for panel b. The level of cellular miR-122-5p was analyzed by real-time PCR 48 h after infection. (e) Huh7.5 cells were stably transduced with the nontarget control shRNA or the CHMP4B shRNA at an MOI of 2. The pool of stably transduced Huh7.5 cells with the nontarget control (CON) and two clones (S5 and S8) derived from stably CHMP4B-shRNA-transduced Huh7.5 cells were infected with HCV at an MOI of 2. Untreated and puromycin resistance gene plasmid-transfected Huh7.5 cells (Huh7.5-puro) were used as controls. The protein levels of CHMP4B, HCV core, and NS5A were assayed 48 h after infection. Actin was used as a loading control. (f) Cells were treated as for panel e, and the intracellular HCV RNA was assayed by real-time PCR 48 h after infection. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
For the next step of our study, stable transduction with the shRNA lentivectors against CHMP4B at an MOI of 2 was performed on Huh7.5 cells. A highly knocked-down clone (the S8 clone) and a moderately knocked-down clone (the S5 clone) of CHMP4B were selected according to the Western blotting results for intracellular CHMP4B (Fig. 1e). The pools of Huh7.5 stably transduced with the nontarget control (the CON pool) and the pools of Huh7.5 stably transfected with a puromycin plasmid (the Huh7.5-puro pool) were used as controls. Unexpectedly, the expression of HCV NS5A and core proteins was decreased in the CON pool and the S5 and S8 clones compared with the Huh7.5-puro pool and Huh7.5 cells. In contrast to the transient-transduction results, both the control CON pool and the CHMP4B-targeted S5 clone surprisingly exhibited ∼60% and ∼50% reductions, respectively, in intracellular HCV RNA compared with the Huh7.5-puro pool at 48 h after HCV infection; furthermore, the S8 clone exhibited an even greater (3-log-unit) reduction in HCV RNA (Fig. 1f). Notably, there were no differences between Huh7.5 and Huh7.5-puro control cells. Since this shRNA effect was independent of CHMP4B specificity, these results suggested that stable transduction with lentiviral shRNA vectors nonspecifically might reduce intracellular HCV RNA.
Stable transduction with an shRNA targeted to eGFP confirmed the nonspecific off-target effects.
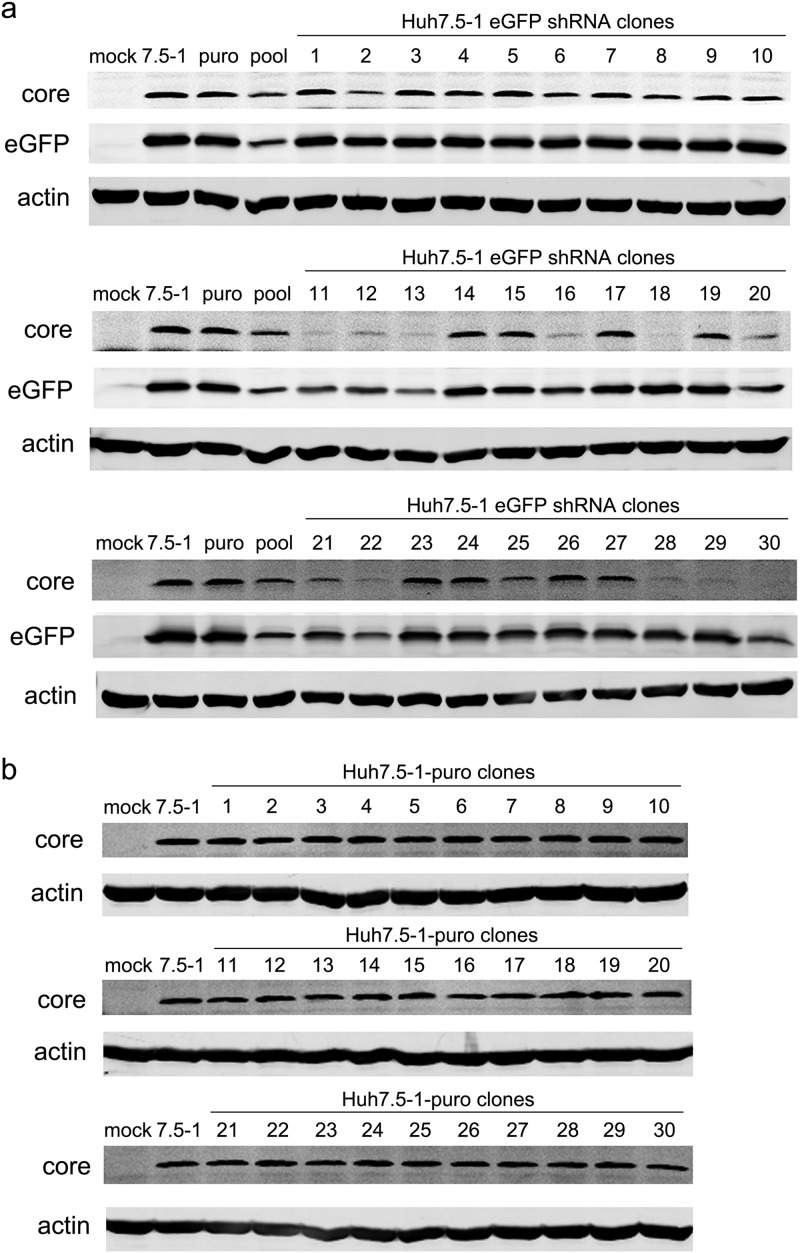
Although the Huh7.5 cell line was a single cell clone derived from Huh7 cells (19), after many passages, the cell line might consist of various cell clones with different sensitivities to virus infection (20). The inhibition of HCV infection in the S5 and S8 clones might result from this heterogeneity. To eliminate the possible influences of the heterogeneity of Huh7.5 cells on HCV infection, we isolated a single cell clone from Huh7.5 (designated Huh7.5-1) and repeated the study. Moreover, to avoid the possible influences of host gene suppression on HCV infection and to allow better monitoring of the expression of shRNAs, a commercial control shRNA targeted to eGFP was applied. Untreated Huh7.5-1 cells and Huh7.5-1 cells stably transfected with a plasmid expressing a puromycin resistance gene were used as controls. Thirty cell clones were isolated from the stably eGFP shRNA-transduced Huh7.5-1 cells, and the expression levels of eGFP and HCV core proteins were examined in these cell clones (Fig. 2a). Of the 30 clones, 15 exhibited a reduction in HCV core. The no. 18 and no. 30 clones showed the most inhibition of HCV core expression and were chosen for further studies. As a control, 30 clones were also isolated from Huh7.5-1 cells transfected with a puromycin plasmid (Fig. 2b). There were no differences in the protein levels of HCV core among these clones, which suggested the inhibition of HCV in the shRNA-transduced cell clones might not have resulted from the heterogeneity of the newly isolated Huh7.5-1 cells but might have been induced by the lentiviral transduction.
FIG 2.
Stable transduction of Huh7.5-1 cells with lentiviral shRNA vectors targeted to eGFP and stable transfection of Huh7.5-1 cells with a plasmid expressing the puromycin resistance gene. (a) Huh7.5-1 cells were stably transduced with lentiviral shRNA vectors targeted to eGFP at an MOI of 2, and 30 clones were isolated. Untreated Huh7.5-1, puromycin plasmid-transfected Huh7.5-1, and a stably transduced pool were used as controls. The indicated cells were infected with HCV and with lentiviruses expressing eGFP simultaneously at an MOI of 2. The protein levels of the HCV core and eGFP were assayed by Western blotting 48 h after infection. Actin was used as a protein-loading control. (b) Huh7.5-1 cells were stably transfected with a plasmid expressing the puromycin resistance gene, and 30 clones were isolated. The indicated cells were infected with HCV at an MOI of 2. The protein level of the HCV core was assayed by Western blotting 48 h after infection. Actin was used as a protein-loading control.
Intracellular HCV RNA was examined in the no. 18 and no. 30 clones after infection with HCV. Both the no. 18 and no. 30 clones exhibited ∼90% reduction in HCV RNA (Fig. 3a). To examine whether this reduction in intracellular HCV RNA came from inhibition of HCV entry into host cells, we performed an entry assay with HCV pseudoparticles (HCVpp). As shown in Fig. 3b, the luciferase activity in HCVpp-infected no. 18 and no. 30 cells was not significantly changed, indicating that stable transduction had no influence on HCV entry. We then proceeded to examine whether these stable transductions inhibited HCV replication. In vitro-transcribed HCV RNA was transfected into the cell clones, and the intracellular HCV RNA was analyzed by real-time PCR 72 h after transfection. As shown in Fig. 3c, the level of HCV RNA decreased significantly in no. 18 and no. 30 cells. The translation of the incoming viral genome was also examined. The activity of the HCV internal ribosome entry site (IRES) was analyzed using a construct that expressed HCV IRES followed by firefly luciferase. In the no. 18 and no. 30 clones, HCV IRES activity showed a significant reduction (Fig. 3d). The intensity of the reduction of HCV IRES activity was less than that of intracellular HCV RNA, which suggested the impaired HCV IRES activity was only partially responsible for the inhibition of HCV infection and that HCV RNA replication might also be inhibited. The intracellular viral titer decreased in no. 18 and no. 30 cells (Fig. 3e). Extracellular viral titers also decreased in these two cell clones (Fig. 3f).
FIG 3.
Stable transduction with lentiviral shRNA vectors exhibits off-target effects. (a) Huh7.5-1, Puro (Huh7.5-1 cells transfected with a plasmid expressing a puromycin resistance gene), no. 18, and no. 30 cells were infected with HCV at an MOI of 2. Intracellular HCV RNA was examined by real-time PCR 48 h after infection. (b) HCVpp was used to infect Huh7.5-1, Puro, no. 18, and no. 30 cells, and firefly luciferase activity was measured to determine HCV entry. (c) Huh7.5-1, Puro, no. 18, and no. 30 cells were transfected with in vitro-synthesized HCV RNA. Intracellular HCV RNA was examined by real-time PCR 48 h after transfection. (d) Huh7.5-1, Puro, no. 18, and no. 30 cells were transfected with a Tk promoter-Renilla luciferase-HCV IRES-firefly luciferase construct, and the relative IRES activity was measured 48 h after transfection. (e and f) Huh7.5-1, Puro, no. 18, and no. 30 cells were infected with HCV at an MOI of 2, and intracellular (e) and extracellular (f) infectivity was measured 48 h after infection. The viral titer was normalized to that of Huh7.5-1 cells. (g and h) Luciferase under the IFN-β promoter (g) or the ISRE promoter (h) was transfected into Huh7.5-1, Puro, no. 18, and no. 30 cells. The cells were infected with HCV 24 h after transfection. The relative luciferase activity was measured at the indicated time points and normalized to that of Huh7.5-1 at 3 h postinfection. Huh7.5-1 cells transfected with the N terminus of RIG-I was used as a positive control. (i) Conditioned culture media from Huh7.5-1, Puro, no. 18, and no. 30 cells were used to treat HCV-infected Huh7.5-1 cells for 12 h. The intracellular HCV RNA was measured by real-time PCR 48 h postinfection. As a positive control, HCV-infected Huh7.5 cells were treated with culture medium from RIG-I-transfected Huh7.5-1 cells. (j) Huh7.5-1, Puro, no. 18, and no. 30 cells were infected with HCV at an MOI of 2. mRNAs of 4 ISG genes, OAS1, IFIT1, MX1, and ISG20, were measured by real-time PCR 48 h after infection. IFN-α-treated Huh7.5-1 was applied as a positive control. Values are presented as means and SD (n = 3). *, P < 0.05; **, P < 0.01 (considered significant) (t test; n = 3).
The off-target effects of the shRNA vectors were independent of type I interferon production.
It is known that RNAi vectors can induce production of antiviral type I interferon (IFN) responses (21). To determine whether the shRNA-mediated nonspecific inhibition of HCV infection (measured by intracellular HCV RNA) in stably transduced cells was due to induction of type I IFN, we examined IFN-β and interferon-stimulated response element (ISRE) promoter activities in Huh7.5-1 cells stably transduced with lentiviral shRNA. As the endogenous retinoic acid-inducible gene I (RIG-I) is defective in Huh7.5 cells (22), we used Huh7.5-1 cells transfected with the type I IFN-inducing N-terminal region of RIG-I (RIG-I N) as a positive control. The IFN-β promoter and the ISRE promoter in no. 18 and no. 30 cells were just slightly activated above those of the negative controls, whereas the positive-control Huh7.5-puro cells transfected with RIG-I N showed high IFN-β promoter and ISRE promoter activation (Fig. 3g and h).
To exclude the possibility that type I IFNs or any other antiviral cytokines or compounds were secreted (and therefore not examined by measuring IFN-β promoter activity), the culture medium was collected from stably transduced Huh7.5-1 cells and used to treat HCV-infected naive Huh7.5-1 cells, and type I IFN-mediated inhibition of viral RNA was measured as a functional readout. As shown in Fig. 3i, the culture medium from the transduced cells had no effect on the HCV RNA abundance, whereas the culture medium from Huh7.5-1 cells transfected with RIG-I, used as a positive control, induced a significant reduction in the intracellular HCV RNA.
Furthermore, the expression of IFN-simulated genes (ISG), including OAS1, IFIT1, MX1, and ISG20, was also measured (Fig. 3j). OAS1 and MX1 were slightly increased in no. 18 cells, but not in no. 30 cells. IFIT1 and ISG20 were unchanged in no. 18 and no. 30 cells. However, the 4 ISG genes were greatly induced in Huh7.5-1 cells treated with IFN-α. These results suggested that type I IFN might not play a role in the reduction of HCV infection in no. 18 and no. 30 cells.
The host genes into which the lentiviral vector integrated did not influence HCV infection.
Another possible reason for the reduction of HCV infection after viral transduction is insertion mutagenesis. After lentiviral infection, the viral vector integrates into the host genome, which may disrupt or enhance the expression of genes surrounding the integration site (23), and those specific genes may be involved in HCV infection. Therefore, the integration sites of lentiviral vectors were determined in no. 18 and no. 30 cells by inverse PCR.
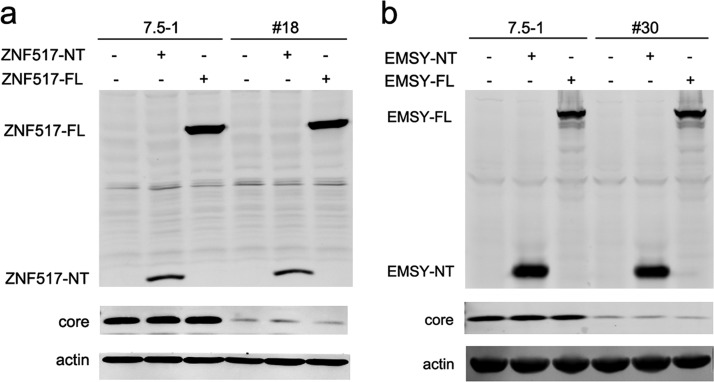
In no. 18 cells, the vector integrated into the 3rd intron of the ZNF517 gene, while in no. 30 cells, the vector integrated into the 9th intron of the EMSY gene. However, both ZNF517 and EMSY were not reported to be associated with HCV infection. Then, we analyzed whether ZNF517 and EMSY had an impact on HCV infection. As shown in Fig. 4, the ZNF517 and EMSY proteins were not detectable in Huh7.5-1 cells or stably shRNA-transduced cells. Overexpression of the full length of ZNF517 (ZNF517-FL) or EMSY (EMSY-FL) protein did not influence the expression of the HCV core. Furthermore, the integration of the lentiviral vector might also result in transcription of truncated mRNA (the exons before the integration site). Thus, the first 3 exons of ZNF517 (ZNF517-NT) or the first 9 exons of EMSY (EMSY-NT) were overexpressed in Huh7.5-1 cells and stably transduced cells, respectively (Fig. 4). Similarly, overexpression of ZNF517-NT or EMSY-NT did not have an impact on the HCV core. These results suggested that the inhibition of HCV infection in no. 18 and no. 30 cells did not result from insertion mutagenesis.
FIG 4.
The host genes into which the lentiviral vector integrated did not influence HCV replication. (a) Huh7.5-1 and no. 18 cells were transfected with the full-length version or an N-terminal (the first 3 exons before the lentiviral vector integration site) fragment of ZNF517, after which they were infected with HCV at an MOI of 2 24 h after transfection. The protein levels of ZNF517 and HCV core were assayed by Western blotting 48 h after infection. (b) Huh7.5-1 and no. 30 cells were transfected with the full-length or an N-terminal (the first 9 exons before the lentiviral vector integration site) fragment of EMSY, after which they were infected with HCV at an MOI of 2 24 h after transfection. The protein levels of EMSY and HCV core were measured by Western blotting 48 h after infection.
Stable transduction with the shRNA vectors altered endogenous miRNA biogenesis.
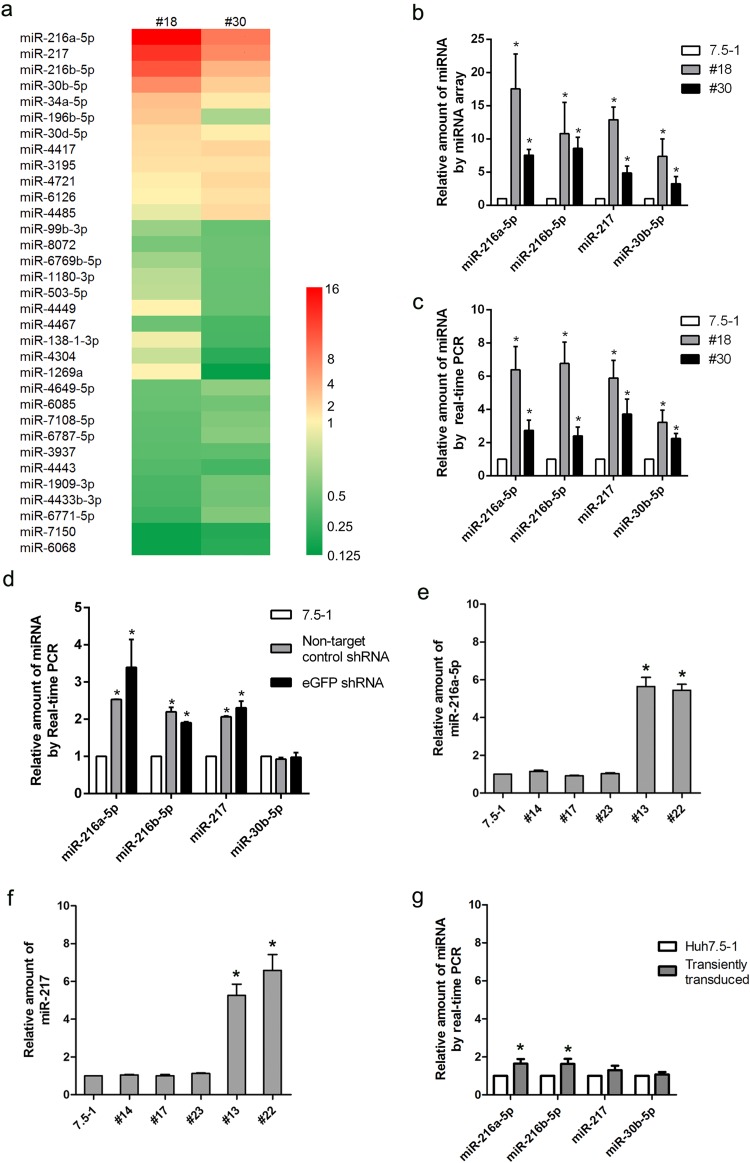
It has been reported that lentiviral shRNA libraries can cause disruption of the host miRNA pathway and thus could potentially influence HCV infection, depending on which miRNAs were affected (16). To determine whether this effect on miRNAs caused the HCV RNA inhibition in our model, we examined the changes in miRNA expression by employing microarray analysis. A total of 22 miRNAs (12 upregulated and 10 downregulated) exhibited >2.0-fold changes in the no. 18 clone (15 miRNAs changed) or the no. 30 clone (17 miRNAs changed) compared with Huh7.5-1 control cells (Fig. 5a). Ten miRNAs (6 upregulated and 4 downregulated) exhibited >2.0-fold changes in both no. 18 and no. 30 cells. Four miRNAs, i.e., miR-216a-5p, miR-217, miR-216b-5p, and miR-30b-5p, showed the greatest changes in both no. 18 and no. 30 cells. Real-time PCR was performed to confirm the upregulation of these four miRNAs, and the real-time PCR results were consistent with the microarray data (Fig. 5b and c).
FIG 5.
The host microRNA expression profile was changed in the cells that were stably transduced with a lentiviral shRNA vector. (a) Hierarchical clustering of differentially expressed miRNAs in no. 18 and no. 30 cells compared with Huh7.5-1 cells by microarray analysis. (b) Quantities of four selected miRNAs in no. 18 and no. 30 cells compared with Huh7.5-1 cells from the microarrays. (c) Real-time quantitative PCR confirmation of the four selected miRNAs. (d) The indicated miRNAs were measured by real-time PCR in the stably control-shRNA-transduced pool and the stably eGFP-shRNA-transduced pool compared with Huh7.5-1 cells. (e and f) miR-216a-5p (e) and miR-217 (f) were measured by real-time PCR in selected stably transduced cell clones. (g) Huh7.5-1 cells were transiently transduced with lentiviral eGFP shRNA at an MOI of 2, and the indicated miRNAs were analyzed by real-time PCR 48 h after transduction. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
As the cell pool stably transduced with the lentiviral shRNA exhibited inhibition of HCV infection (as shown in Fig. 1e and f), we wondered whether the change in the miRNA profile was also present in the pool. Real-time PCR was performed in the pool of nontarget control shRNA or stably eGFP shRNA-transduced Huh7.5-1 cells. miR-216a-5p, miR-217, and miR-216a were significantly upregulated in the pools (Fig. 5d), which suggested that the changes in these particular miRNAs might be general in stably transduced clones. However, miR-30b-5p was not significantly changed in the pools.
To further verify whether the change in miRNA expression was present in other stably transduced clones, the expression levels of miR-216a-5p and miR-217 were determined by real-time PCR in the other 2 clones (no. 13 and no. 22) that exhibited inhibition of HCV and in 3 clones (no. 14, no. 17, and no. 23) that showed no inhibition of the HCV core (Fig. 5e and f). As expected, miR-216a-5p and miR-217 were upregulated in no. 13 and no. 22 cells. In contrast, the two miRNAs were not changed in no. 14, no. 17, or no. 23 cells. These results suggested a possible association between the change in miRNA expression and inhibition of HCV infection.
Then, we proceeded to investigate whether the expression of miRNAs was changed in the cells transiently transduced with lentiviral shRNA vectors. In transiently transduced Huh7.5-1 cells, miR-216a-5p and miR-216b-5p were increased slightly but significantly. However, miR-217 and miR-30b-5p were not significantly changed (Fig. 5g).
We next screened all of the miRNAs that exhibited expression changes in our study to determine whether any miRNAs were known to regulate HCV propagation, and we found that miR-196b-5p had previously been shown to potentially inhibit HCV infection (24). Consistent with this, we found that miR-196b-5p was upregulated in no. 18 cells and the stably transduced pools. Although the expression of miR-196b-5p was downregulated in no. 30 cells, the difference lacked statistical significance.
Differentially expressed miRNAs regulated HCV infection.
To experimentally test whether the miRNAs identified in our microarray screen influence HCV infection, we selected several miRNAs that exhibited large expression changes in both no. 18 and no. 30 cells (Fig. 6a to d). A commercial mimic or inhibitor from Caenorhabditis elegans that had minimal homology with miRNAs from the miRbase database was used as a negative control. The efficiency of transfection was >80%, as indicated by observing the fluorescence of Huh7.5-1 cells transfected with equivalent levels of Cy3-labeled nontarget control siRNA (data not shown).
FIG 6.
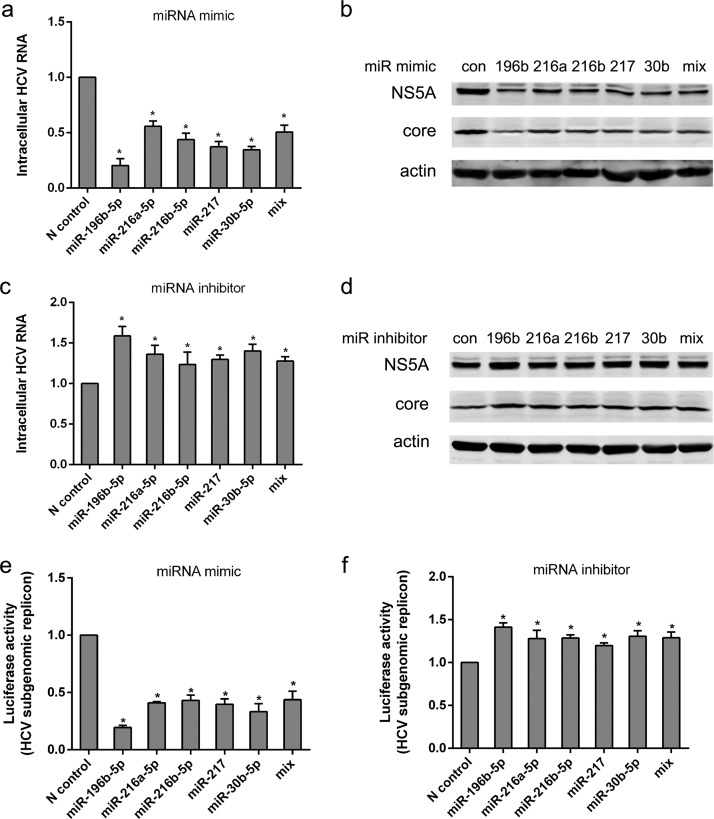
Potential functions of differentially expressed miRNAs in HCV infection. (a) Huh7.5-1 cells were transfected with 50 nM mimics of the indicated miRNAs, followed by HCV infection at 24 h after transfection. The mix included a mixture of the mimics of miR-216a-5p, miR-216b-5p, miR-217, and miR-30b-5p (12.5 nM each). Intracellular HCV RNA was examined by real-time PCR 48 h after infection. (b) Huh7.5-1 cells were treated as for panel a, and the protein levels of HCV NS5A and HCV core were assayed by Western blotting 48 h after HCV infection. (c) No. 18 cells were transfected with 50 nM inhibitors of the indicated miRNAs, followed by HCV infection 24 h after transfection. The mix contained a mixture of the inhibitors of miR-216a-5p, miR-216b-5p, miR-217, and miR-30b-5p (12.5 nM each). Intracellular HCV RNA was examined by real-time PCR 48 h after infection. (d) No. 18 cells were treated as for panel c, and the protein levels of HCV NS5A and HCV core were assayed by Western blotting 48 h after HCV infection. (e) Huh7.5 cells harboring a JFH1 subgenomic replicon (Huh7.5-SGR) that expressed Renilla luciferase were transfected with mimics of the indicated miRNAs as for panel a. Luciferase activity was examined 48 h after transfection. (f) Huh7.5-SGR cells were transfected with inhibitors of the indicated miRNAs as for panel c. Luciferase activity was examined 48 h after transfection. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
As expected from its known role in inhibition of HCV entry, we found that transfection of Huh7.5-1 cells with 50 nM miR-196b-5p mimic reduced intracellular HCV RNA and proteins and that transfection of no. 18 cells with the miR-196b-5p inhibitor increased HCV RNA and proteins. The miR-216a-5p mimic, the miR-216b-5p mimic, the miR-217 mimic, or the miR-30b-5p mimic individually was sufficient to significantly reduce HCV RNA and proteins in Huh7.5-1 cells compared with the negative-control mimic. We also found that transfection of no. 18 cells with inhibitors of miR-216a-5p, miR-216b-5p, miR-217, or miR-30b-5p significantly increased the intracellular HCV RNA abundance and the levels of HCV proteins. However, other mimics or inhibitors of two of the downregulated miRNAs (miR-6068 and miR-7150) we tested exhibited no influence on HCV (data not shown).
In addition, we analyzed whether there was an additive or synergistic effect from concurrent overexpression or inhibition of the four miRNAs (Fig. 6a to d). Because concentrations of mimics or inhibitors higher than 50 nM significantly impaired cell viability, the concentration of each of the mimics or inhibitors was decreased to 12.5 nM in the mixture of mimics or inhibitors, resulting in a final concentration of 50 nM. The mixtures had effects on HCV RNA similar to those of a single mimic or inhibitor (50 nM), which suggested that there might be additive effects, but no synergistic effects.
The effects of these miRNAs on HCV RNA replication was also validated. A Huh7.5 cell line harboring a JFH1 subgenomic replicon that expressed Renilla luciferase was utilized. HCV RNA replication was inhibited by the mimics of miR-196b-5p, miR-216a-5p, miR-216b-5p, miR-217, and miR-30b-5p but potentiated by the inhibitors of these miRNAs (Fig. 6e and f).
Bioinformatic analyses were performed to investigate whether there were potential target sequences of these miRNAs in the HCV genome. However, potential target sites were not identified, which suggested that the miRNAs might not directly target the HCV genome to inhibit viral infection.
miR-216a-5p and miR-216b-5p interfered with host autophagy to inhibit HCV infection.
Both miR-216a-5p and miR-216b-5p were reported to be able to suppress the expression of Beclin-1 and Atg5 genes (25, 26), which are critical genes involved in the autophagy pathway. As a host autophagy mechanism is required for HCV infection (27–29), we set out to determine whether miR-216a-5p and miR-216b-5p suppressed host autophagy to inhibit HCV infection.
As shown in Fig. 7a, the expression of Beclin-1 and Atg5 was decreased in no. 18 and no. 30 cells compared with Huh7.5-1 cells. The autophagy marker LC3-II was also decreased in no. 18 and no. 30 cells. Overexpression of Beclin-1 and/or Atg5 enhanced autophagy and rescued HCV core proteins and intracellular HCV RNA (Fig. 7a and b). Transfection of Huh7.5-1 cells with the miR-216a-5p mimic or the miR-216b-5p mimic inhibited the expression of Beclin-1 and Atg5 (Fig. 7c). HCV core protein and intracellular HCV RNA were also decreased by the miR-216a-5p mimic or the miR-216b-5p mimic (Fig. 7d and e). Similar results were produced from transfection of Huh7.5-1 cells with siRNA of Beclin-1 or Atg5 (Fig. 4d and e). These results suggested that miR-216a-5p and miR-216b-5p might inhibit HCV infection by suppressing host autophagy.
FIG 7.
miR-216a-5p and miR-216b-5p interfered with host autophagy to inhibit HCV infection. (a) eGFP, Beclin-1, or Atg5 was overexpressed in Huh7.5-1, no. 18, and no. 30 cells, followed by HCV infection 24 h after transfection. The expression levels of the indicated proteins were measured by Western blotting 48 h after HCV infection. (b) Huh7.5-1, no. 18, and no. 30 cells were treated as for panel a, and the intracellular HCV RNA was quantified by real-time PCR 48 h after HCV infection. (c) Huh7.5-1 cells were transfected with 50 nM mimics of miR-216a-5p or miR-216b-5p, followed by HCV infection 24 h after transfection. The expression levels of the indicated proteins were measured by Western blotting 48 h after HCV infection. (d) Huh7.5-1 cells were transfected with 50 nM siRNAs for Beclin-1 or Atg5, followed by HCV infection 24 h after transfection. The expression levels of the indicated proteins were measured by Western blotting 48 h after HCV infection. (e) Huh7.5-1 cells were treated as for panels c and d, and the intracellular HCV RNA in transfected cells was determined by real-time PCR 48 h after HCV infection. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
The expressed lentiviral vector itself, but not the shRNA, was responsible for the alteration of host miRNAs.
The above-described studies suggested that the stable transduction of lentiviral shRNA vectors regulated particular miRNA expression, which in turn affected HCV infection. Next, we tried to identify which component of the lentiviral vector caused the change in the miRNA expression profile. The Mission lentiviral packaging system we used was a third-generation packaging system that eliminated most lentiviral genes (Δvpr, Δvif, Δvpu, and Δnef). Only the region between the viral long terminal repeats (LTRs) of the transfer vector is packaged within the virus, as shown in Fig. 8a.
FIG 8.
Knocking out the shRNA sequence in no. 18 and no. 30 cells did not influence HCV replication or miRNA expression. (a) Schematic depiction of the lentiviral shRNA vector used in this study. (b) The shRNA sequence was knocked out by CRISPR in no. 18 and no. 30 cells. Two guide RNAs were designed to target upstream (highlighted in yellow) and downstream (highlighted in green) of the shRNA. The protospacer-adjacent motif (PAM) sequence is also indicated. The no. 18-2 and 18-5 clones were isolated from no. 18 cells, and the no. 30-3 and 30-5 clones were isolated from no. 30 cells. Knockout was confirmed by sequencing. (c and d) The indicated cell clones, Huh7.5-1 cells, no. 18 cells, and no. 30 cells were infected with HCV and lentiviruses expressing eGFP simultaneously at an MOI of 2. The protein levels of HCV core and eGFP were measured by Western blotting 48 h after infection. Actin was used as a loading control. (e) Cells were treated as for panels c and d, and the intracellular HCV RNA levels were measured by real-time PCR 48 h after infection. (f and g) The expression levels of miR-216a-5p and miR-217 were examined by real-time PCR in no. 18-2, no. 18-5, no. 30-3, and no. 30-5 clones compared with Huh7.5-1 cells. (h) Sequences of the eGFP shRNA used in this study and the corresponding region of the lentiviral vector that integrated into no. 18 cells. (i) Huh7.5-1 cells were stably transduced with an empty lentiviral vector without the shRNA or the U6-shRNA cassette, or the eGFP shRNA vector at an MOI of 2, after which the pools were infected with HCV at an MOI of 2. The protein level of the HCV core was measured by Western blotting 48 h after HCV infection. (j) Cells were treated as for panel a, and the intracellular HCV RNA was measured by real-time PCR 48 h after HCV infection. (k) Expression levels of miR-216b-5p and miR-217 were measured by real-time PCR in the indicated stably transduced cells. Untreated Huh7.5-1 cells were used as controls. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
It has been reported that shRNA transduction inducing oversaturation of the host miRNA pathway is the reason for alteration of the miRNA profile (15, 16). Moreover, we noted that inhibition of HCV core proteins was associated with the expression of shRNAs (indicated by the suppression of eGFP) in stably transduced cell clones, excluding the no. 18 clone (Fig. 2a). To clarify whether the expression of shRNAs is responsible for the change in miRNA expression, CRISPR was applied to knock out the shRNA sequence in no. 18 and no. 30 cells. After genome editing, gene sequencing was performed in isolated cell clones for knockout confirmation. Two clones (no. 18-2 and no. 18-5) from no. 18 cells and 2 clones (no. 30-3 and no. 30-5) from no. 30 cells were identified (Fig. 8b). After the shRNA sequence was knocked out, the cells stably transduced with the lentiviral vector still exhibited suppression of HCV core proteins and HCV RNA (Fig. 8c to e). The expression of miR-216a-5p and miR-217 was still upregulated in no. 18-2, no. 18-5, no. 30-3, and no. 30-5 cells (Fig. 8f and g). These results suggested that the alteration of host miRNAs and the inhibition of HCV infection in the cell clones stably transfected with lentiviral shRNA vectors did not result from the expression of shRNAs. It should be noted that eGFP could not be knocked down in no. 18 cells, which suggested shRNA might not be expressed in this cell clone. Indeed, genome sequencing showed that the “shRNA sequence” in no. 18 cells had no homology with the eGFP shRNA used in this study, and the transcribed RNAs could not form correct shRNAs (Fig. 8h). Although we could not find an explanation for the change in the shRNA sequence in no. 18 cells, the finding that the cell clone exhibited changes in miRNAs also supported our conclusion that the alteration of the miRNA profile in our model was not induced by shRNAs.
U6 is a strong RNA polymerase (Pol) III promoter and may compete with endogenous promoters that share the same transcription factors to affect miRNA genesis. We found that the lentiviral vectors lacking U6-shRNA (ΔU6-shRNA) or shRNA (ΔshRNA) exhibited inhibition of the HCV core and RNA similar to that of the intact vector (Fig. 8i and j). The expression of miR-216a-5p and miR-217 was significantly upregulated in cells stably transduced with ΔU6-shRNA (Fig. 9k). These results suggested that the U6 promoter might not be responsible for the change in miRNAs. Because the expression of puroR had no influence on HCV infection in the above-mentioned studies (Fig. 1e and f), the hPKG-puroR cassette might not be responsible for the change in miRNAs. Therefore, it might be the lentiviral vector itself that induced the change in the host miRNA profile and thus inhibited HCV infection.
FIG 9.
Corresponding pri-miRNAs were upregulated in lentiviral-vector-transduced cell clones. (a) Pri-miR-216a-5p, pri-miR-216b-5p, pri-miR-217, and pri-miR-30b-5p were measured in the indicated cell clones by real-time PCR. (b) Schematic depiction of the products of the MIR217HG gene. (c) Expression of the lncRNA MIR217HG was measured in the indicated cells by real-time PCR. (d to g) Expression of the miRNA-processing-related Drosha, exportin 5 (XPO5), Dicer, and Argonaute 2 (AGO2) genes was measured in no. 13, no. 18, no. 22, and no. 30 cells by Western blotting. Corresponding siRNA-transfected Huh7.5-1 cells were used to indicate the specific band for each protein (rightmost lanes). Untreated Huh7.5-1 cells were used as controls (leftmost lanes). Actin was used as a loading control. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
The integration sites determined the expression of the lentiviral vector, which in turn influenced the expression of host miRNAs.
To further validate the above speculation, we set out to determine whether the lentiviral vector existed in the cell clones that showed no inhibition of HCV core proteins. Inverse PCR was performed in no. 14, no. 17, and no. 23 cells that showed no inhibition of the HCV core to identify whether lentiviral vectors integrated into these cell clones. Unexpectedly, the lentiviral shRNA vector was present in the genomes of the three cell clones. However, it was noteworthy that the lentiviral vector integrated in nongene regions in the 3 cell clones, while the vector was present in genes in no. 18 and no. 30 cells. Then, we analyzed two other cell clones (no. 13 and no. 22) that also showed inhibition of the HCV core and alteration of miRNAs (Fig. 5e and f), and we found that the lentiviral vector integrated in the introns of genes. There was good association among the type of the integration sites (in genes or nongene regions), the expression of the vector (as indicated by the inhibition of eGFP, excluding no. 18 cells, in which the shRNA was not correct), and the inhibition of HCV. The results of inverse PCR are summarized in Table 1. These data suggested that the lentiviral vector was expressed only when it integrated into an active region of the host genome and then induced a change of host miRNAs.
TABLE 1.
Summarized results of inverse PCR
| Cell clone | Lentiviral vector integration site |
Inhibition of HCV | Inhibition of eGFP | |||
|---|---|---|---|---|---|---|
| Chromosome | Gene symbol or GenBank accession no. | Sequencea | Direction of integration | |||
| 13 | 19 | MYO9B (2nd intron) | AAACCCCGTC67290TCTACTAAAA | Forward | Yes | Yes |
| 14 | 17 | SMURF2P1b | ACCATTATTG3313TTTTTGAAAA | Reverse | No | No |
| 17 | 1 | AC006500.4c | TCTTAAGTAG4061GGGACCTTCA | Reverse | No | No |
| 18 | 8 | ZNF517 (3rd intron) | CAGAGTCTTC6177TCTAACTACC | Forward | Yes | No |
| 22 | 12 | SARNP (9th intron) | CTGTAGTCCC39477AACTACTCGG | Forward | Yes | Yes |
| 23 | 17 | AC091529.1c | AACTCACTAG134359CTCAAGATCA | Forward | No | No |
| 30 | 11 | EMSY (9th intron) | GCTTTGTCAC54075CCAGGCTGGA | Forward | Yes | Yes |
The nucleotides after which the lentiviral vector integrated are in boldface. The numbers indicate the positions of the nucleotides in the sequences.
SMURF2P1 is a pseudogene.
The genomic fragment does not contain genes.
Corresponding pri-miRNAs were upregulated in lentiviral-vector-transduced cell clones.
To find the mechanism by which these particular miRNAs were regulated, we investigated the expression of corresponding primary miRNAs (pri-miRNAs) and the expression of genes involved in pri-miRNA processing in the cell clones that showed inhibition of HCV and upregulation of miRNAs. Except for pri-miR-30b-5p, which was not changed in no. 13 cells, all the pri-miRNAs analyzed were significantly upregulated in no. 13, no. 18, no. 22, and no. 30 cell clones (Fig. 9a). The data suggested that the upregulation of corresponding pri-miRNAs might result in the elevated expression of these four miRNAs in stably transduced cell clones. The pri-miRNAs were also investigated in the cell clones that showed no influences on HCV and no changes in miRNAs. All the pri-miRNAs were not changed in no. 14, no. 17, and no. 23 cells (Fig. 9a).
When studying the transcription of the pri-miRNAs, we found that miR-216a-5p, miR-216b-5p, and miR-217 were all located in the MIR217HG gene within chromosome 2. MIR217HG is a noncoding gene and contains one transcript, long noncoding RNA (lncRNA) MIR217HG, which contains 3 exons (Fig. 9b). miR-216a-5p, miR-216b-5p, and miR-217 are all processed from the second intron of the same transcript. The transcription of the lncRNA MIR217HG was also analyzed by real-time PCR, and we found that it was also upregulated in the no. 13, no. 18, no. 22, and no. 30 cell clones but not in no. 14, no. 17, and no. 24 cells (Fig. 9c). These findings suggested that the upregulation of the pri-miRNAs and the lncRNA might share the same mechanism and might be induced by the enhanced transcription of the MIR217HG gene.
To find whether the processing of the pri-miRNAs was dysregulated, we analyzed the expression of host proteins involved in the processing of miRNAs. As shown in Fig. 9d to g, there was no difference in the protein levels of Drosha, exportin 5, Dicer, or Argonaute 2 among no. 13, no. 18, no. 22, no. 30, no. 14, no. 17, no. 24, and the parental Huh7.5-1 cells, suggesting the processing of miRNAs might not be dysregulated by the lentiviral vector.
Dysregulated genes identified by mRNA microarray are not reported to be associated with the biogenesis of miRNAs or HCV infection.
To identify potential factors that were consistently dysregulated and might affect the expression of particular miRNAs or the HCV infection process, the gene expression profiles of no. 18 and no. 30 cells were analyzed by mRNA microarray. Multiple probe sets were designed to detect each transcript of the genes included in the microarray. The probe identifiers (IDs) that exhibited changes of more than 2.0-fold in both no. 18 and no. 30 cell clones are shown in the heat map in Fig. 10a, which includes 13 genes: NTS, CTBP2, ESRP1, KRT19, ANO1, GPX2, MARVELD3, DMKN, ANXA3, LITAF, HLA-DMB, CP, and VNN2. All these genes were upregulated in the microarray analysis. However, none of the genes was reported to be associated with the regulation of miRNAs or HCV infection. In the current study, we were not able to investigate the potential effect of each gene on miRNA expression.
FIG 10.
Overexpression of JUN induced upregulation of miR-216a-5p, miR-217, and miR-216b-5p. (a) Gene expression array analysis was performed in Huh7.5-1, no. 18, and no. 30 cells. Hierarchical clustering of differentially expressed mRNAs in both no. 18 and no. 30 cells with changes of >2.0-fold are shown. (b) Relative amounts of JUN mRNA in no. 18 and no. 30 cells compared with Huh7.5-1 cells from the microarrays. (c) mRNA levels of JUN in the stably transduced pool and no. 18 and no. 30 cells was measured by real-time PCR. (d) Protein level of JUN in the stably transduced pool and no. 18 and no. 30 cells was measured by Western blotting. Untreated Huh7.5-1 cells were used as controls. (e) Huh7.5-1 cells or no. 18 cells were transfected with eGFP, JUN, 50 nM control siRNA, or 50 nM JUN siRNA. The protein level of JUN was measured by Western blotting 48 h after transfection. Actin was used as a loading control. (f) The indicated miRNAs were measured in GFP- or JUN-transfected Huh7.5-1 cells by real-time PCR 48 h after transfection. (g) The indicated miRNAs were measured in control siRNA- or JUN siRNA-transfected Huh7.5-1 or no. 18 cells by real-time PCR 48 h after transfection. Values are presented as means and SD (n = 3). *, P < 0.05 (considered significant) (t test; n = 3).
We noted that Beclin-1 and Atg5 were not identified by the microarray, probably because they were only moderately inhibited in no. 18 and no. 30 cells (Fig. 7a), and this downregulation might not have been identified by the microarray.
The overexpression of JUN increased the expression of miR-216a-5p, miR-216b-5p, and miR-217 in stably transduced cells.
As hypothesized above, the upregulation of miR-216a-5p, miR-216b-5p, and miR-217 might be a result of the enhanced transcription of MIR217HG. Predicted enhancers for the MIR217HG gene and corresponding transcription factors were obtained from the GeneHancer database (30). The predicted enhancers with scores of >1.0 were GH02G056008 and GH02G056015. These two enhancers contained binding sites for transcription factors, including CTCF, MXI1, JUN, MAX, RAD21, GATA2, POLR2A, SMC3, FOS, MAFK, RFX1, ZNF654, BHLHE40, MNT, REST, EP300, SMARCA4, ZNF692, and PRDM1. We screened the results of the gene expression microarray to determine whether any transcription factor listed above was dysregulated. The expression levels of these genes, other than JUN, were not significantly changed. JUN (short for Jun proto-oncogene, AP-1 transcription factor subunit), also known as AP-1 or c-Jun, was upregulated in no. 18 and no. 30 cells, but the change was <2.0-fold (Fig. 10b). Moreover, JUN has been reported to inhibit HCV replication (31). The elevated expression of JUN was confirmed in the stably transduced pool and clones by real-time PCR and Western blotting (Fig. 10c and d).
Then, we proceeded to investigate whether JUN was able to enhance the expression of miR-216a-5p, miR-216b-5p, and miR-217. As the predicted enhancers (GH08G134830, GH08G134682, GH08G134777, and GH08G134718) (30) for miR-30b-5p also contained binding sites for JUN, the expression of miR-30b-5p was also analyzed. The overexpression of JUN in Huh7.5-1 cells increased the expression levels of miR-216a-5p, miR-216b-5p, and miR-217, but not miR-30b-5p (Fig. 10e and f). Although the knockdown of JUN in Huh7.5-1 cells did not change the expression of these miRNAs, silencing JUN reduced the levels of miR-216a-5p, miR-216b-5p, and miR-217 in no. 18 cells. However, the expression levels of these miRNAs in no. 18 cells with JUN knocked down were still higher than those in the parental Huh7.5-1 cells (Fig. 10g). These data suggested that the upregulation of JUN only partially accounted for the dysregulation of miRNAs in cells stably transduced with lentiviral vectors.
The effects of lentiviral vectors on the particular miRNAs were also observed in HEK293 and HeLa cells.
To clarify whether the dysregulation of miRNAs observed in Huh7.5 cells was an artifact inherent to that cell line, we transduced HEK293 and HeLa cells with the same lentiviral shRNA vector. A single cell clone derived from HEK293 or HeLa cells was stably transduced. Forty-five stably transduced HEK293 cell clones and 22 stably transduced HeLa cell clones were then isolated, and the transcription of the vector was examined through the knockdown of eGFP (Fig. 11). Four clones (HEK293-8, HEK293-42, HeLa-3, and HeLa-4) in which eGFP was obviously knocked down were selected for further analyses. The expression levels of miR-216a-5p, miR-216b-5p, miR-217, and miR-30b-5p were examined in the four clones and stably transduced pools by real-time PCR. miR-216b-5p and miR-30b-5p were significantly upregulated in the stably transduced HEK293 pool and clones compared with the parental HEK293 cells (Fig. 11c). For HeLa cells, miR-216b-5p was detected in the stably transduced pool and cell clones, but not in the parental HeLa cells (Fig. 11d). miR-30b-5p was upregulated in the pool and cell clones compared with the parental HeLa cells. However, miR-216a-5p and miR-217 were not detected in the parental or transduced HEK293 or HeLa cells.
FIG 11.
Stable transduction of HEK293 or HeLa cells with lentiviral shRNA vectors targeted to eGFP. (a and b) HEK293 or HeLa cells were stably transduced with lentiviral shRNA vectors targeting eGFP at an MOI of 2, after which independent cellular clones were isolated. Untreated HEK293 or HeLa cells were used as controls. The indicated cells were infected with lentiviruses expressing eGFP at an MOI of 2. The protein level of eGFP was measured by Western blotting 48 h after infection. Actin was used as the protein-loading control. (c) Relative amounts of miR-216b-5p and miR-30b-5p were examined in the stably transduced HEK293 pool and cell clones (HEK293-8 and HEK293-42) by real-time PCR. Untreated HEK293 cells were used as controls. (b) Relative amounts of miR-216b-5p and miR-30b-5p were examined in the stably transduced HeLa pool and cell clones (HeLa-3 and HeLa-4) by real-time PCR. Untreated HeLa cells were used as controls. Because miR-216b-5p was not detected in the untreated HeLa cells, the relative amount of miR-216b-5p in the transduced cells is presented as 2−ΔΔCT. (e and f) The protein levels of JUN in stably transduced pools and clones were measured by Western blotting. Actin was used as a loading control. *, P < 0.05 (considered significant) (t test; n = 3).
The expression of JUN was also examined, and we found that JUN was upregulated in the stably lentiviral-vector-transduced HEK293 and HeLa cells (Fig. 11e and f). Taken together, our evidence shows that stable transduction with lentiviral vectors increased the expression of miR-216b-5p, miR-30b-5p, and JUN in HEK293 and HeLa cells, suggesting that the effect was not inherent only to Huh7.5 cells but might be general among different cell lines.
DISCUSSION
In this study, we found that lentiviral shRNA vectors nonspecifically disrupted the HCV life cycle when host cells were stably, but not transiently, transduced with shRNA vectors. Using miRNA microarray analysis, we found that the expression levels of several miRNAs were altered when shRNAs were stably transduced by lentiviral infection. The change in miRNA expression was not induced by oversaturation of the miRNA machinery by shRNAs, as reported previously (15, 16). Some of the miRNAs altered by the shRNA vectors were able to influence HCV propagation. The integration loci of lentiviral vectors determined whether expression of miRNAs was altered. Furthermore, stable transduction increased the expression of the transcription factor JUN, which in turn partially accounted for the dysregulation of the miRNAs.
Our primary goal on entering this study was to evaluate the role of CHMP4B in the HCV life cycle, as two previous studies had revealed that CHMP4B participated in the release of HCV particles, but not in HCV replication (17, 18). We chose to evaluate this role by using a lentiviral shRNA vector system from the commonly used TRC library to avoid the limitations (mentioned above) of the conventional synthetic-siRNA system. Surprisingly, we found that stable transduction with the shRNA vector-targeted CHMP4B reduced HCV RNA abundance, but transient transduction did not. The nontarget shRNA control also exhibited similar inhibitory effects on HCV RNA abundance, strongly suggesting that an off-target effect of shRNA vectors existed; it further suggested that these effects were independent of CHMP4B specificity. Importantly, these off-target effects were not due to the puromycin resistance gene in the vector being used for stable-clone screening because the stable transfection with a puromycin plasmid did not affect intracellular HCV RNA abundance. Because Huh7.5 cells might consist of various cell clones with different sensitivities to virus infections, we could not exclude the possibility that inhibition of HCV propagation in S5 and S8 monoclones resulted from the heterogeneity of Huh7.5 cells. To avoid possible influences from heterogeneity, we isolated a single clone (Huh7.5-1) from Huh7.5 cells to repeat the study. Similar inhibition of HCV was observed by using this single clone, which indicated that the inhibition of HCV infection was not due to the heterogeneity of the host cells.
A well-known side effect of RNAi vectors is to trigger interferon responses (21). However, the inhibition of HCV RNA in our model was independent of type I IFN induction by the lentivirus, because IFN-β promoter activity, ISRE promoter activity, and several ISGs were not significantly activated in the lentivirus-infected cells. Furthermore, the stably transduced cells did not secrete any antiviral cytokines or other compounds. It is known that lentiviral vectors can induce insertion mutagenesis, which might have an impact on HCV infection. However, possible protein products of the host genes in which lentiviral vectors integrated did not influence HCV infection.
It has been reported that transient transduction with high doses of lentiviral shRNA vectors can oversaturate the cellular miRNA pathway to influence HCV infection (16). Thus, to avoid this off-target effect, we used the minimal lentivirus dose for transient transduction that did not inhibit HCV infection. However, the pools of cells stably transduced with the same minimal lentivirus dose also exhibited off-target effects; we then selected cell clones as indicated and determined whether these off-target effects were due to the disruption of the miRNA pathway. Microarray analysis of the stably shRNA-transduced cells showed that 22 miRNAs exhibited >2.0-fold expression level changes, and 1 miRNA known to regulate HCV RNA replication was involved. Stable transduction indeed changed the miRNA expression profile. Further analysis revealed that miRNAs identified by the microarrays were able to inhibit HCV infection.
miR-216a-5p and miR-216b-5p were found to suppress host autophagy, which is required for the replication of HCV (27–29). These results suggested that the dysregulated host miRNAs might inhibit HCV infection through the regulation of host genes. It should be noted that the overexpression of Beclin-1 and Atg5 only partially recued the inhibition of HCV in stably transduced cells, suggesting that miR-216a-5p and miR-216b-5p also utilized other mechanisms to inhibit HCV infection (Fig. 7a and b). The knockdown of Beclin-1 or Atg5 by the siRNAs was much more significant than that of the miRNAs. However, the effects on HCV core protein and RNA were identical (Fig. 4c to e). These results also suggested that the two miRNAs employed mechanisms other than targeting Beclin-1 and Atg5 to inhibit HCV infection. The inhibition of Beclin-1 and Atg5 by the miRNAs only partially contributed to overall effects in HCV.
Aside from host mRNA, lncRNA may also be regulated by miRNAs to influence HCV infection. Experimentally validated lncRNAs that were targeted by the four miRNAs are identified in Table 2. UCA1, targeted by miR-216b-5p, was upregulated during HCV infection but had no impact on HCV infection (32). The association between the other lncRNAs and HCV infection was not reported. The lncRNAs that were predicted to be targeted by these four miRNAs were obtained from LncBase Predicted v.2 (33). The predicted lncRNAs with scores of >0.99 are listed in Table 2. However, none of these predicted lncRNAs was reported to be associated with HCV infection. There were several lncRNAs, including AC017076.14 (34), EGOT (35), BISPR (36), RN7SK (37), and IGF2-AS (37), that were reported to influence HCV infection. However, these lncRNAs were not predicted to be targeted by the four miRNAs. Further studies might be designed to analyze the possibility that the dysregulated miRNAs target lncRNAs and thus affect HCV infection.
TABLE 2.
LncRNAs targeted by miRNAs
Pan et al. and Grimm et al. reported that overexpression of shRNA by lentiviral vectors or adeno-associated virus (AAV) vectors induced saturation of the miRNA pathway and reduced the expression of miR-122 (15, 16), which tightly regulates HCV replication (38). However, the expression of miR-122 was not changed in our microarrays, and most of the miRNAs identified by the microarrays were upregulated. Therefore, the alteration of miRNA expression in our model was not induced by the saturation of endogenous miRNA biogenesis. Knockout experiments showed that the change in miRNAs and the inhibition of HCV RNA were independent of shRNA expression, which further confirmed that the off-target effects appearing in our model were not due to disruption of miRNA biogenesis caused by shRNA overexpression.
We also analyzed whether the U6 promoter within the lentiviral vector influenced the miRNA profile, because it might compete with endogenous promoters that share the same transcription factors to affect miRNA genesis. However, stable transduction with the lentiviral vectors lacking U6-shRNA (ΔU6-shRNA) also significantly upregulated the expression of miR-216a-5p and miR-217, suggesting that the U6 promoter might not be responsible for the change in miRNAs.
Then, we speculated that the cause of the change in miRNAs was the vector itself. Because the inhibition of the HCV core was tightly associated with the suppression of eGFP in stably transduced clones, the vectors should be absent in those clones in which there was no impact on HCV core proteins. Conversely, the results of inverse PCR showed that lentiviral vectors integrated into all of the cell clones and exhibited no inhibition of HCV. By comparing the integration sites of lentiviral vectors between clones with and without inhibition of HCV/eGFP, we found that the vector could induce changes in miRNAs only when it integrated into active region of the host genome (Table 1). When the lentiviral shRNA vector integrated into an inactive region, the lentiviral vector was expressed at a very low level, as indicated by the protein level of eGFP; the puromycin resistance gene was also expressed at a low level but was sufficient for cell survival; the expression level of the viral vector was too low to simulate the change in host miRNAs. Therefore, the alteration of host miRNAs was induced by the viral vector and was determined by whether it integrated into active or inactive regions of the host genome.
If the change in miRNA expression was induced by the highly expressed lentiviral vector, an important question was why transient transduction had no influence on HCV. Since we used minimal doses of lentiviruses to infect Huh7.5-1 cells, transiently transduced cells probably included uninfected cells. Moreover, because integration sites also influenced the outcome, not all transduced cells could exhibit inhibition of HCV. Therefore, the effect was not significant in transiently transduced cells. As shown in Fig. 5g, miRNAs were also changed in transiently transduced Huh7.5-1 cells, but the change was much less significant than in stably transduced cells and might not be sufficient to impact HCV infection.
Next, we tried to determine how the miRNAs were dysregulated by the lentiviral vector transduction. We found that the upregulation of miR-216a-5p, miR-216b-5p, miR-217, and miR-30b-5p resulted from the upregulation of corresponding pri-miRNAs. Furthermore, the host factors that were involved in the processing of pri-miRNAs were analyzed in stably transduced cells. If these host factors were changed, a great number of miRNAs would be dysregulated, but that set would not be limited to these particular miRNAs. As we expected, these factors were not changed. Then, gene expression microarray analysis was performed to identify factors that might be responsible for the dysregulation of miRNAs and the inhibition of HCV infection. However, none of the genes identified by the microarray was known to be associated with miRNA expression or HCV infection. The identified genes might be involved in the regulation of miRNAs. However, we were not able to investigate the function of each gene identified in this study.
The three miRNAs with the greatest change were located in a noncoding gene, MIR217HG, which was upregulated in the lentiviral-vector-transduced cells. The predicted enhancers for this noncoding gene contained binding sites for JUN, which was upregulated in stably transduced cells. Moreover, it was reported that JUN signaling was able to increase the expression of the miRNA let-7g to inhibit HCV replication (31). We then examined whether JUN was able to regulate the miRNAs that were identified by our study, and we found that overexpression of JUN increased the expression of miR-216a-5p, miR-216b-5p, and miR-217, but not miR-30b-5p. Our study provided new mechanisms for JUN-induced inhibition of HCV. However, silencing JUN in no. 18 cells only partially rescued the dysregulation of these miRNAs. These data suggested that mechanisms other than JUN signaling also contributed to the dysregulation of the miRNAs.
Several proteins of human immunodeficiency virus type I (HIV-1) have been shown to activate JUN (39–41). However, the lentiviral vectors we used did not express any HIV-1 proteins. It was interesting that stable integration of Moloney murine leukemia virus (Mo-MuLV) LTR increased the level of JUN mRNA (42). Therefore, the upregulation of JUN in the stably lentiviral-vector-transduced cells might be induced by the LTRs contained in the vectors through similar mechanisms. This hypothesis might be studied in the future.
In conclusion, we found that stable, but not transient, transduction with lentiviral shRNAs inhibited HCV infection in host cells due to the alteration of host endogenous miRNAs. The miRNA expression profile changes were not induced by overexpression of shRNAs but were caused by the expression of the lentiviral vectors and depended on the integration loci. Further studies revealed that the elevated expression of JUN partially accounted for the upregulation of miR-216a-5p, miR-216b-5p, and miR-217 in stably transduced cells. Although our findings revealed that the off-target effect observed can be beneficial in inhibiting HCV infection, concerns should be raised about off-target effects when lentiviral vectors are used to produce stable gene knockdown or perform therapy, as with changes in miRNA expression.
MATERIALS AND METHODS
Cells, reagents, and antibodies.
The human hepatoma Huh7.5 cell line, an Huh7 variant cell line that is highly permissive to HCV RNA replication (19), was kindly provided by Charles M. Rice (Laboratory of Virology and Infectious Disease, Rockefeller University). We isolated a monoclone from Huh7.5 cells without any treatment and named it Huh7.5-1; this is not the same as Huh7.5.1, an interferon-cured Huh7.5 cell line (43). Huh7.5 and Huh7.5-1 were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM nonessential amino acids (complete DMEM). The 293FT cell line was purchased from Thermo Fisher Scientific (Waltham, MA, USA) and maintained in complete DMEM with an additional 2 mM l-glutamate. All the cell culture reagents were from Thermo Fisher Scientific. Recombinant human IFN-α 2a was from Sigma-Aldrich and was reconstituted in complete DMEM and stored in aliquots at −80°C. Anti-HCV core protein (sc-57800), anti-CHMP4B (sc-82557), and anti-eGFP (SC-8334) antibodies were purchased from Santa Cruz (Santa Cruz, CA, USA). Anti-HCV NS5A protein antibody (HCV-7B5) was purchased from BioFront Technologies (Tallahassee, FL, USA). Anti-actin (A3853) antibody was from Sigma-Aldrich. IRdye 680RD-labeled anti-rabbit IgG or 800CW-labeled anti-mouse IgG secondary antibodies were from Li-Cor Biotechnology (Lincoln, NE, USA).
Generation of the highly infectious HCV JFH1 strain and the subgenomic replicon.
The highly infectious HCV JFH1 strain with adaptive mutations (44) was kindly provided by Guangxiang Luo (Department of Microbiology, School of Medicine, University of Alabama at Birmingham). Viral stock was generated as described previously (45). For experiments in this study, Huh7.5 cells were infected (MOI = 2) for 6 h with HCV stock supplemented with 4 μg/ml Polybrene (Sigma-Aldrich, Switzerland). The infected cells were collected for analysis at selected time points. A dicistronic JFH1 subgenomic replicon containing the neomycin resistance gene and the Renilla luciferase gene was created following previously reported methods (46). A stable Huh7.5 cell line harboring this replicon (Huh7.5-SGR) was generated as reported previously (46, 47).
Production of HCV pseudoparticles and entry assay.
HCV pseudoparticles expressing firefly luciferase (HCVpp), vesicular stomatitis virus (VSV) G protein pseudoparticles expressing firefly luciferase (VSVGpp), and no-envelope control viruses were generated in 293T cells as described previously (48) by cotransfection of an envelope-deficient HIV genome (pNL4-3.luc.E-R−; obtained from the NIAID AIDS Research and Reagent Program) and a plasmid expressing HCV JFH1 glycoproteins (cloned from the JFH1 genome), VSV G (obtained from Sigma), or an empty vector (no envelope). The supernatants containing viruses were collected, filtered, supplemented with 20 mM HEPES buffer (pH 7.5), and stored at −80°C as aliquots.
For virus entry assays, cells were seeded at 1 × 104 cells per well on 96-well plates and incubated overnight. On the following day, the cells were spin infected with 100 μl of HCVpp, VSVGpp, or no envelope as a control supplemented with 8 μg/ml of Polybrene for 1 h in a table-top centrifuge (2,500 rpm; 30°C), followed by another 2 h of incubation in a CO2 cell incubator. The cells were lysed 72 h after infection and assayed with a luciferase assay system (Promega. Madison, WI, USA) in a Modulus microplate luminometer (Turner BioSystems). HCVpp relative light unit (RLU) values were normalized to VSVGpp values for each sample. All experiments were performed in triplicate.
In vitro transcription of HCV RNA and transfection of Huh7.5 cells.
HCV RNA was in vitro transcribed from the plasmid pJFH1 (43) using a Mega-script kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. The transcribed HCV RNA was delivered into Huh7.5 cells using the DMRIE-C transfection reagent (Thermo Fisher Scientific) following the manufacturer's protocol.
HCV IRES reporter assay.
The HCV IRES sequence, including a part of the core protein-coding region, of HCV JFH1 (nucleotides 1 to 392) was amplified by PCR from the HCV JFH1 cDNA construct pcDNA6/TR-Tight/JFH1-FL/AR (49), which was provided by Guangxiang Luo (Department of Microbiology, School of Medicine, University of Alabama at Birmingham). The HCV IRES sequence was then inserted into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI, USA). The resulting construct was designated pGL3-IRES.
Transduced Huh7.5 cells were seeded in 24-well plates (1 × 105 cells per well). The next day, the cells were transfected with pGL3-IRES and pRL-SV40. At 12 h after transfection, the cells were harvested and lysed, and the luciferase activity was measured using a Dual-Luciferase reporter assay systems (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Lentivirus packaging, titration, and transduction.
For gene knockdown experiments, the shRNA transfer plasmids for CHMP4B (TRCN0000147769), eGFP (catalog no. SHC005), and a nontarget negative control (catalog no. SHC002) were obtained from the Mission TRC library (Sigma-Aldrich, Switzerland). The nontarget shRNA control can activate RISC and the RNAi pathway but cannot target any human genes, as described in the manufacturer's instructions. All of the shRNAs were predicted not to match the HCV RNA sequence by bioinformatics analysis. For packaging lentiviruses, 6 μg of lentiviral packaging mix (SHP001; Sigma-Aldrich) and 3 μg of the transfer vector mentioned above were cotransfected into 4 × 106 293FT cells with 27 μl of FuGene HD (Roche, Germany) according to the manufacturer's manual. The supernatants containing lentiviruses were collected, filtered, and stored at −80°C as aliquots.
Lentivirus titers were measured with a Lenti-X p24 Rapid Titer kit (632200; Clontech, Palo Alto, CA, USA), following the manufacturer's protocol. The number of transducing units (TU) per milliliter of lentivirus was calculated based upon the concentration of p24 (1 pg of p24 was assumed to be equivalent to 100 TU). Specific shRNA clones and infection MOI were selected according to the target downregulation efficiency and the absence of cytotoxic effects. For transduction, Huh7.5 cells were incubated with viral stocks supplemented with 4 μg/ml Polybrene for 6 h and then supplied with fresh medium.
Selection of cells stably transduced with lentiviral vectors.
Huh7.5, Huh7.5-1, HEK293, or HeLa cells were transduced with a CHMP4B shRNA lentiviral vector, an eGFP shRNA lentiviral vector, a nontarget control vector, or an empty vector without any shRNA sequences. The transduced cells were then supplied with 2 μg/ml puromycin 2 days after transduction to clear the nontransduced cells. Ten days after transduction, isolation of cell clones was performed. The pools of the nontarget control or the stably shRNA vector-transduced cells were selected for 10 days with puromycin and used as controls in the study.
SDS-PAGE and Western blotting.
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) containing a protease inhibitor cocktail (Roche, Germany). The lysates were centrifuged at 16,000 × g at 4°C, and the protein concentration of the supernatant was measured with a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, IL, USA). Equal amounts of total protein were electrophoresed in an SDS-polyacrylamide gel electrophoresis (PAGE) gel and then transferred onto a BioTrace nitrocellulose membrane (Pall, NY, USA). After being blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline, pH 7.4 (PBS), the membrane was incubated with primary antibodies (1 μg/ml diluted in PBS with 5% BSA and 0.1% Tween 20), followed by incubation with the IRdye-labeled secondary antibodies mentioned above. Finally, the membrane was scanned with an Odyssey infrared imaging system (Li-Cor Biotechnology, Lincoln, NE, USA).
Real-time PCR.
Total RNA from cell samples was isolated using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and treated with RNase-free DNase I (Promega, Madison, WI, USA). For analyzing the intracellular HCV RNA, 1 μg of total RNA was reverse transcribed in a 20-μl volume using a reverse transcription kit (Thermo Fisher Scientific) with random primers. The resulting cDNA was used in a real-time quantitative PCR assay with Power SYBR Green PCR master mix (Thermo Fisher Scientific). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control. The primers used in this experiment are listed in Table 3. To analyze pri-miRNAs, we performed real-time PCR with TaqMan Fast Universal master mix (Thermo Fisher Scientific, Waltham, MA, USA) and TaqMan Pri-miRNA assays (Thermo Fisher Scientific) according to the manual without modification. To analyze miRNAs, we purified miRNAs from the indicated cells with a mirVana miRNA isolation kit. cDNA was prepared from the entire pool of miRNAs with a TaqMan advanced miRNA cDNA synthesis kit (Thermo Fisher Scientific) according to the manual without modification. The resulting cDNA was amplified with miR-Amp reagents included in the TaqMan advanced miRNA cDNA synthesis kit to uniformly increase the amount of cDNA for each target. Then, real-time PCR was performed with TaqMan advanced miRNA assays (Thermo Fisher Scientific).
TABLE 3.
Primers used in this study
| Name | Sequence |
|---|---|
| CHMP4B sense | 5′-ATGTCGGTGTTCGGGAAGCT-3′ |
| CHMP4B antisense | 5′-ATCTCTTCCGTGTCCCGCAG-3′ |
| JFH1 sense | 5′-TCTGCGGAACCGGTGAGTA-3′ |
| JFH1 antisense | 5′-TCAGGCAGTACCACAAGGC-3′ |
| JUN sense | 5′-TCCAAGTGCCGAAAAAGGAAG-3′ |
| JUN anti-ense | 5′-CGAGTTCTGAGCTTTCAAGGT-3′ |
| GAPDH sense | 5′-TGGGCTACACTGAGCACCAG-3′ |
| GAPDH antisense | 5′-AAGTGGTCGTTGAGGGCAAT-3′ |
| IP-1 | 5′-GTAAGACCACCGCACA-3′ |
| IP-2 | 5′-TCCTCTGGTTTCCCTTTCGC-3′ |
| IP-3 | 5′-GATCTTCAGACCTGGAGGAG-3′ |
| IP-4 | 5′-CCAGAGTCACACAACAGACG-3′ |
The reaction was run on a 7900HT fast real-time PCR machine (Thermo Fisher Scientific, Waltham, MA, USA), and the results were analyzed using RQ Manager software (Thermo Fisher Scientific).
Dual-luciferase assay.
For IFN-β or ISRE promoter activity assays, transduced Huh7.5 cells were seeded in 24-well plates (1 × 105 cells per well). The next day, the cells were transfected with pGL3-Luc (expressing IFN-β or ISRE promoter firefly luciferase), pRL-SV40 (expressing actin promoter Renilla luciferase), and RIG-I N (expressing the N terminus of RIG-I, used as the positive control) or with an empty vector using Lipofectamine 2000 (Thermo Fisher Scientific). The total amount of DNA was kept consistent. At 24 h after transfection, the transfected cells were infected with HCV at an MOI of 2. The cells were harvested and lysed at the indicated time points.
For HCV replicon assays, Huh7.5-SGR cells were transfected with pGL3-Luc (expressing actin promoter firefly luciferase). The cells were harvested and lysed 24 h after transfection.
The luciferase activity was measured by using dual-luciferase reporter assay systems (Promega, Madison, WI, USA) according to the manufacturer's instructions.
miRNA transfection.
Selected miRNA mimics and inhibitors were synthesized by Ribobio (Guangzhou, China). The negative-control mimic and inhibitor (Ribobio, Guangzhou, China) were from C. elegans and had minimal homology with miRNAs from the miRbase database. These miRNA mimics or inhibitors were transfected into the cells at a final concentration of 50 nM with Lipofectamine RNAiMax (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. All experiments were carried out in triplicate. To confirm the efficiency of transfection, we also transfected an equivalent amount of Cy3-labeled negative control (Ribobio, Guangzhou, China), followed by observation using a fluorescence microscope (TE2000U; Nikon, Japan).
miRNA microarray analysis.
Total RNA was isolated from the indicated cells with TRIzol reagent (Thermo Fisher Scientific). miRNA hybridization was performed by CapitalBio Corp. (Beijing, China) using an Affymetrix GeneChip miRNA 4.0 array (covering 30,424 mature miRNA probes from miRBase V20; 3,770 human, mouse, and rat hairpin pre-miRNAs; and 1,996 snoRNA and scaRNA probes). Two-channel microarray technology was employed. For each sample pair, the experiment was performed with two independent hybridizations (Cy3 and Cy5 labels were interchanged). Raw data were extracted from the TIFF images using Affymetrix GeneChip Command Console 1.1 (Affymetrix, UK). Hierarchical cluster analysis was carried out with Gene Cluster 3.0 (Stanford University, Stanford, CA, USA).
mRNA microarray analysis.
Total RNA was isolated from the indicated cells with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). miRNA hybridization was performed by CapitalBio Corp. (Beijing, China) using an Affymetrix GeneChip Prime View human gene expression array (including 49,395 probe sets, covering more than 20,000 human genes). A two-channel microarray technology was employed. For each sample pair, the experiment was performed with two independent hybridizations (Cy3 and Cy5 labeling was interchanged). Stained microarrays were scanned with an Affymetrix GeneChip scanner 3000 7G. Quality checks and data analyses were carried out using Affymetrix GeneChip operating software (GCOS) and Quality Reporter. Raw data were extracted from the TIFF images using Affymetrix GeneChip Command Console 3.1 (Affymetrix, UK).
Inverse PCR and sequencing.
Inverse PCR was conducted as previously described (50). Briefly, genomic DNA isolated from the indicated cells was digested with EcoRI and MfeI (New England BioLabs, Ipswich, MA, USA). The doubly digested DNA was then ligated by T4 ligase (New England BioLabs). The reaction mixture was purified and used as the template for the first PCR, which used the primers IP-1 and IP-2. The first PCR product was used as the template for the second PCR, which used the primers IP-3 and IP-4. The second PCR product was subjected to agarose gel electrophoresis, and DNA fragments from gel purification were sequenced. The integration site was mapped using bioinformatics tools from NCBI. The sequences of the primers used in inverse PCR are listed in Table 3.
Statistical analysis.
All data are presented as means and standard deviations (SD). A two-tailed Student t test (unpaired) was used to compare differences between groups; P values of <0.05 or <0.01 were considered statistically significant.
ACKNOWLEDGMENTS
The work is sponsored by the CAMS Innovation Fund for Medical Science (CIFMS 2016-12M-1-013 and 2016-12M-1-014), the National Key Plan for Scientific Research and Development of China (2016YFD0500300), the National Natural Science Foundation of China (81301440), and the PUMC Youth Fund/Fundamental Research Funds for the Central Universities (3332014026 and 2017PT31010). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that we have no conflicts of interest in this work.
REFERENCES
- 1.Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. 2009. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol 50:1142–1154. doi: 10.1016/j.jhep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Georgel P, Schuster C, Zeisel MB, Stoll-Keller F, Berg T, Bahram S, Baumert TF. 2010. Virus-host interactions in hepatitis C virus infection: implications for molecular pathogenesis and antiviral strategies. Trends Mol Med 16:277–286. doi: 10.1016/j.molmed.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Bode JG, Brenndorfer ED, Karthe J, Haussinger D. 2009. Interplay between host cell and hepatitis C virus in regulating viral replication. Biol Chem 390:1013–1032. doi: 10.1515/BC.2009.118. [DOI] [PubMed] [Google Scholar]
- 5.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A 104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A 106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, Packer J, Schurdak M, Molla A. 2007. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 9.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran AT, Rahim MN, Ranadheera C, Kroeker A, Cortens JP, Opanubi KJ, Wilkins JA, Coombs KM. 2013. Knockdown of specific host factors protects against influenza virus-induced cell death. Cell Death Dis 4:e769. doi: 10.1038/cddis.2013.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landi A, Vermeire J, Iannucci V, Vanderstraeten H, Naessens E, Bentahir M, Verhasselt B. 2014. Genome-wide shRNA screening identifies host factors involved in early endocytic events for HIV-1-induced CD4 down-regulation. Retrovirology 11:118. doi: 10.1186/s12977-014-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Workenhe ST, Ketela T, Moffat J, Cuddington BP, Mossman KL. 2016. Genome-wide lentiviral shRNA screen identifies serine/arginine-rich splicing factor 2 as a determinant of oncolytic virus activity in breast cancer cells. Oncogene 35:2465–2474. doi: 10.1038/onc.2015.303. [DOI] [PubMed] [Google Scholar]
- 13.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. 2009. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh S, Narang AS, Mahato RI. 2011. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res 28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 15.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 16.Pan Q, de Ruiter PE, von Eije KJ, Smits R, Kwekkeboom J, Tilanus HW, Berkhout B, Janssen HL, van der Laan LJ. 2011. Disturbance of the microRNA pathway by commonly used lentiviral shRNA libraries limits the application for screening host factors involved in hepatitis C virus infection. FEBS Lett 585:1025–1030. doi: 10.1016/j.febslet.2011.02.035. [DOI] [PubMed] [Google Scholar]
- 17.Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. 2011. The ESCRT system is required for hepatitis C virus production. PLoS One 6:e14517. doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corless L, Crump CM, Griffin SD, Harris M. 2010. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol 91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sainz B Jr, Barretto N, Uprichard SL. 2009. Hepatitis C virus infection in phenotypically distinct Huh7 cell lines. PLoS One 4:e6561. doi: 10.1371/journal.pone.0006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 22.Sumpter R Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uren AG, Kool J, Berns A, van Lohuizen M. 2005. Retroviral insertional mutagenesis: past, present and future. Oncogene 24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 24.Hou W, Tian Q, Zheng J, Bonkovsky HL. 2010. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology 51:1494–1504. doi: 10.1002/hep.23401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menghini R, Casagrande V, Marino A, Marchetti V, Cardellini M, Stoehr R, Rizza S, Martelli E, Greco S, Mauriello A, Ippoliti A, Martelli F, Lauro R, Federici M. 2014. MiR-216a: a link between endothelial dysfunction and autophagy. Cell Death Dis 5:e1029. doi: 10.1038/cddis.2013.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Fu Y, Tong J, Fan S, Xu K, Sun H, Liang Y, Yan C, Yuan Z, Ge Y. 2014. MicroRNA-216b/Beclin 1 axis regulates autophagy and apoptosis in human Tenon's capsule fibroblasts upon hydroxycamptothecin exposure. Exp Eye Res 123:43–55. doi: 10.1016/j.exer.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke PY, Chen SS. 2011. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, Lancet D, Cohen D. 1 January 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou WW, Huang CF, Yeh ML, Tsai YS, Hsieh MY, Huang CI, Huang JF, Tsai PC, Hsi E, Juo SH, Tsai WL, Chuang WL, Yu ML, Dai CY. 2016. MicroRNA let-7g cooperates with interferon/ribavirin to repress hepatitis C virus replication. J Mol Med 94:311–320. doi: 10.1007/s00109-015-1348-1. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. 2015. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 6:7899–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, Hatzigeorgiou AG. 2016. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, Valadkhan S. 2014. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 42:10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carnero E, Barriocanal M, Prior C, Pablo Unfried J, Segura V, Guruceaga E, Enguita M, Smerdou C, Gastaminza P, Fortes P. 2016. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 17:1013–1028. doi: 10.15252/embr.201541763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barriocanal M, Carnero E, Segura V, Fortes P. 2014. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol 5:655. doi: 10.3389/fimmu.2014.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Jia M, Yuan J, Zhang C, Zhu Y, Kuang X, Lan L, Wang X. 2015. STAT3 regulated long noncoding RNAs lnc7SK and lncIGF2AS promote hepatitis C virus replication. Mol Med Rep 12:6738–6744. doi: 10.3892/mmr.2015.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jangra RK, Yi M, Lemon SM. 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar A, Manna SK, Dhawan S, Aggarwal BB. 1998. HIV-Tat protein activates c-Jun N-terminal kinase and activator protein-1. J Immunol 161:776–781. [PubMed] [Google Scholar]
- 40.Varin A, Manna SK, Quivy V, Decrion AZ, Van Lint C, Herbein G, Aggarwal BB. 2003. Exogenous Nef protein activates NF-kappa B, AP-1, and c-Jun N-terminal kinase and stimulates HIV transcription in promonocytic cells. Role in AIDS pathogenesis J Biol Chem 278:2219–2227. [DOI] [PubMed] [Google Scholar]
- 41.Varin A, Decrion AZ, Sabbah E, Quivy V, Sire J, Van Lint C, Roques BP, Aggarwal BB, Herbein G. 2005. Synthetic Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-kappaB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages. J Biol Chem 280:42557–42567. doi: 10.1074/jbc.M502211200. [DOI] [PubMed] [Google Scholar]
- 42.Weng H, Choi SY, Faller DV. 1995. The Moloney leukemia retroviral long terminal repeat trans-activates AP-1-inducible genes and AP-1 transcription factor binding. J Biol Chem 270:13637–13644. doi: 10.1074/jbc.270.23.13637. [DOI] [PubMed] [Google Scholar]
- 43.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang J, Luo G. 2012. Cell culture-adaptive mutations promote viral protein-protein interactions and morphogenesis of infectious hepatitis C virus. J Virol 86:8987–8997. doi: 10.1128/JVI.00004-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol 80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao W, Herlihy KJ, Zhang NJ, Fuhrman SA, Doan C, Patick AK, Duggal R. 2007. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob Agents Chemother 51:95–102. doi: 10.1128/AAC.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 48.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A 100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J Virol 79:13963–13973. doi: 10.1128/JVI.79.22.13963-13973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, Gudkov AV, Stark GR. 2009. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci U S A 106:16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]