Plants employ repressive viral genome methylation as an epigenetic defense against geminiviruses. In turn, geminiviruses encode proteins that inhibit methylation by TGS. Previous studies showed that TYLCV V2 can efficiently suppress TGS, but the mechanism remains unknown. We showed that V2 interacted with NbHDA6 but did not inhibit its enzymatic activity. As HDA6 is known to be involved in gene silencing in cooperation with MET1, we explored the relationship between V2, NbMET1, and NbHDA6. Our investigation showed that V2 did not interact but competed with NbMET1 for direct binding to NbHDA6. To our knowledge, this is the first report that viral proteins inhibit TGS by interacting with histone deacetylase but not by blocking the methyl cycle. This work provides an additional mechanism for TGS suppression by geminiviruses.
KEYWORDS: Tomato yellow leaf curl virus, V2, histone deacetylase 6 (HDA6), geminivirus, transcriptional gene silencing (TGS), suppressor of RNA silencing, DNA methylation, methyltransferase 1, MET1, suppressor
ABSTRACT
Cytosine DNA methylation is a conserved epigenetic silencing mechanism that defends against biotic stresses such as geminivirus infection. As a countermeasure, geminiviruses encode proteins that inhibit methylation and transcriptional gene silencing (TGS). Previous studies showed that V2 protein of Tomato yellow leaf curl virus (TYLCV) functions as a TGS suppressor. However, how V2 mediates TGS suppression remains unknown. Here we show that V2 interacts directly with a Nicotiana benthamiana histone deacetylase 6 (NbHDA6), a homolog of Arabidopsis HDA6 (AtHDA6), known to be involved in gene silencing in cooperation with methyltransferase 1 (MET1). NbHDA6 genetically complemented a late-flowering phenotype and restored histone deacetylation of an AtHDA6 mutant. Furthermore, our investigation showed that NbHDA6 displayed histone deacetylase enzymatic activity, which was not inhibited by V2. Genetic analysis revealed that silencing of NbHDA6 expression resulted in enhanced susceptibility to TYLCV infection. In addition, methylation-sensitive PCR and bisulfite sequencing analysis showed that silencing of NbHDA6 expression caused reduced DNA methylation of the viral genome in infected plants. HDA6 was previously shown to recruit and physically interact with MET1 to function in gene silencing. Using competitive pulldown and coimmunoprecipitation assays, we demonstrated that V2 did not interact but competed with NbMET1 for direct binding to NbHDA6. These findings suggest that V2 interacts with host HDA6 and interferes with the recruitment of MET1 by HDA6, resulting in decreased methylation of the viral DNA genome by TGS with a concomitant increase in host susceptibility to TYLCV infection.
IMPORTANCE Plants employ repressive viral genome methylation as an epigenetic defense against geminiviruses. In turn, geminiviruses encode proteins that inhibit methylation by TGS. Previous studies showed that TYLCV V2 can efficiently suppress TGS, but the mechanism remains unknown. We showed that V2 interacted with NbHDA6 but did not inhibit its enzymatic activity. As HDA6 is known to be involved in gene silencing in cooperation with MET1, we explored the relationship between V2, NbMET1, and NbHDA6. Our investigation showed that V2 did not interact but competed with NbMET1 for direct binding to NbHDA6. To our knowledge, this is the first report that viral proteins inhibit TGS by interacting with histone deacetylase but not by blocking the methyl cycle. This work provides an additional mechanism for TGS suppression by geminiviruses.
INTRODUCTION
Cytosine DNA methylation is a well-characterized epigenetic silencing mechanism modulating a number of key biological processes, including genomic imprinting, X-chromosome inactivation, suppression of repetitive elements, and carcinogenesis (1, 2). It also acts as an effective defense system against invading nucleic acids, such as those of geminiviruses (3–5).
The Geminiviridae is a plant virus family consisting of members with small, single-stranded DNA (ssDNA) genomes that cause extensive agricultural losses in crops worldwide. Geminiviruses are currently classified into 9 genera (Becurtovirus, Begomovirus, Curtovirus, Eragrovirus, Mastrevirus, Topocuvirus, Turncurtovirus, Capulavirus, and Grablovirus) with the genus Begomovirus containing the largest number of plant viruses in tropical, subtropical, and temperate agroecosystems (6, 7). Viruses belonging to the Geminiviridae specify 4 to 7 proteins, and they replicate in infected-cell nuclei by rolling-circle replication (RCR), complementary-strand replication (CSR), and recombination-dependent replication (RDR) using double-stranded DNA (dsDNA) intermediates (8–12). These dsDNA intermediates associate with cellular histones to form minichromosomes (13, 14), which are potential targets for repression of transcription.
Plants employ transcriptional gene silencing (TGS) as a defense against geminiviruses (15–17). As a counterdefensive measure, geminiviruses produce unique proteins that serve as transcriptional gene silencing suppressors to interfere with this process. Virus-encoded TGS suppressors act through divergent mechanisms. The most extensively studied geminivirus silencing suppressors are the AC2/AL2 protein encoded by members of the genus Begomovirus and the related C2/L2 protein encoded by members of the genus Curtovirus. Both AC2/AL2 and C2/L2 suppress TGS by a mechanism that correlates with methyl cycle interference through inhibition of adenosine kinase (ADK) activity (18–20). In addition, Beet severe curly top virus (BSCTV) C2 interferes with the host epigenetic defense by attenuating the activity of 26S proteasome-mediated degradation of S-adenosyl-methionine decarboxylase 1 (SAMDC1), highlighting the importance of the methyl cycle for defense against geminiviruses (21). More recently, AC2/C2 was shown to interact with and inhibit the H3K9 histone methyltransferase SUVH4/KYP to attenuate TGS (22, 23). Besides AC2/C2, the βC1 protein encoded by Tomato yellow leaf curl China betasatellite (TYLCCNB) interacts with and inhibits the activity of S-adenosyl homocysteine hydrolase (SAHH) to block the methyl cycle (24). Furthermore, replication-associated proteins (Reps, also known as C1, AL1, or AC1) of several geminiviruses suppress TGS by reducing the expression of plant DNA methyltransferases, methyltransferase 1 (MET1) and chromomethylase 3 (CMT3) (25). In addition, the AC5 protein encoded by Mungbean yellow mosaic India virus (MYMIV) was shown to prevent TGS by repressing the expression of domains rearranged methyltransferase 2 (DRM2) (17). Our previous studies have shown that the V2 protein of Tomato yellow leaf curl virus (TYLCV) is a TGS suppressor (26). However, how TYLCV V2 mediates TGS suppression remains unknown.
Histone deacetylase 6 (HDA6) is a well-studied histone deacetylase that has multifaceted roles in regulation of genome maintenance, development, environmental stress responses, gene silencing, and maintenance of cytosine methylation in plants (27–29). HDA6 was first identified to play a role in transgene silencing through an auxin-responsive element mutant screening, where HDA6 mutant alleles axe1-1 to axe1-5 release auxin-responsive reporter gene silencing in the absence of auxin treatment (30). In addition, HDA6 was also required for maintenance of TGS (31). The role of HDA6 in methylation maintenance was further highlighted by the observation that HDA6 was an essential component in the process of RNA-directed DNA methylation (32). Several studies have demonstrated that HDA6 plays an important role in gene silencing in cooperation with MET1. Analysis of locus-directed heterochromatin silencing indicated that HDA6 and MET1 cotarget to heterochromatin sites and maintain heterochromatin silencing (28). Moreover, Liu et al. reported that HDA6 physically interacts with MET1 to function cooperatively in gene silencing (33).
In this study, to determine the mechanism of V2-based TGS suppression, a yeast two-hybrid screening was performed. We show that TYLCV V2 interacts with a Nicotiana benthamiana histone deacetylase 6 (NbHDA6). Using competitive pulldown and coimmunoprecipitation (co-IP) assays, we demonstrate that V2 competes with NbMET1 for direct binding to NbHDA6. The implication of this finding to the geminivirus infection cycle is discussed.
RESULTS
V2 interacts with N. benthamiana HDA6.
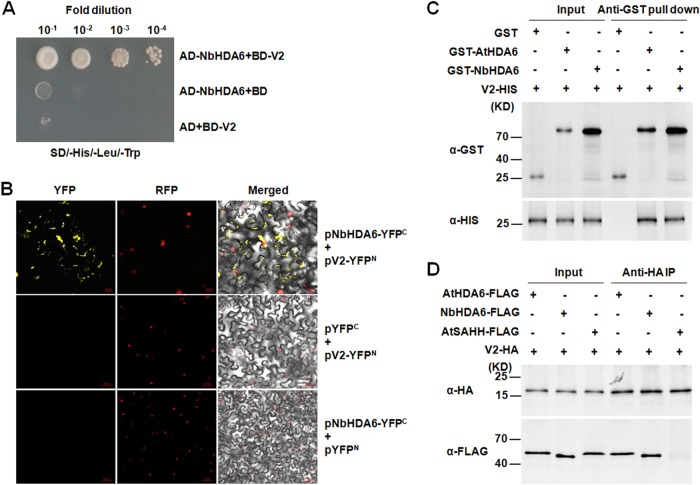
To identify host cell targets of V2, we performed a yeast two-hybrid screen (34) of an N. benthamiana cDNA library using the TYLCV V2 protein as bait. From a total of approximately 7 × 105 transformants assayed for histidine prototrophy and β-galactosidase activity, one cDNA clone, designated NbHDA6 (GenBank accession no. KU170188), was identified as an interacting partner of V2. Amino acid sequence analysis revealed significant homology between NbHDA6 and HDA6s from Nicotiana tomentosiformis, Solanum lycopersicum, Vitis vinifera, Morus notabilis, Theobroma cacao, and Arabidopsis thaliana (85.8%, 93.0%, 82.3%, 84.1%, 83.9%, 78.1%). As is typical for members of the RPD3-like histone deacetylase (HDAC) protein family, both NbHDA6 and AtHDA6 contain a conserved histone deacetylase kinase domain, which is necessary for histone deacetylase activity (35). The full-length coding sequence of NbHDA6 was amplified from N. benthamiana cDNA, and its interaction with V2 was confirmed using the yeast two-hybrid system (Fig. 1A).
FIG 1.
TYLCV V2 physically interacts with NbHDA6. (A) Yeast two-hybrid assay between NbHDA6 and V2. Yeast strain Y2HGold cotransformed with the indicated plasmids was spotted on synthetic depleted (SD) growth medium lacking histidine, leucine, and tryptophan in 10-fold serial dilutions. (B) BiFC visualization of an interaction between V2 and NbHDA6 in N. benthamiana leaves. RFP-histone 2B (RFP-H2B) was used as a marker for the nucleus. Left panels, YFP fluorescence; middle panels, RFP fluorescence; right panels, YFP/RFP/bright-field overlay. Bars, 20 μm. (C) Pulldown assay for detecting an in vitro interaction between NbHDA6/AtHDA6 and V2. Equal molar amounts of GST or GST fusion proteins were used to pull down HIS fusion proteins. Immunoblot assays were performed using anti-HIS or anti-GST antibody to detect the associated proteins. (D) In vivo co-IP assay for the interaction between NbHDA6/AtHDA6 and V2. Approximately 1% of input and one-quarter of eluted protein complex were analyzed by immunoblotting. These experiments were repeated three times with similar results.
A bimolecular fluorescence complementation (BiFC) assay was further performed to test for the interaction between V2 and NbHDA6 in plant cells. V2 and NbHDA6 were fused to the N-terminal fragment of yellow fluorescent protein (YFP) and the C-terminal fragment of YFP, respectively. The corresponding constructs were codelivered into RFP-H2B plant leaves by agroinfiltration, and fluorescence was observed using a confocal microscope at 72 h postinfiltration (hpi). As shown in Fig. 1B, pairwise expression of pV2-YFPN and pNbHDA6-YFPC resulted in a strong YFP fluorescence signal in distinct microbodies throughout the cytoplasm and nucleus periphery, but no fluorescence was observed when pV2-YFPN and pYFPC or pNbHDA6-YFPC and pYFPN were coexpressed, suggesting a tight interaction between the V2 and NbHDA6 proteins in plant cells (Fig. 1B). To further demonstrate the interaction of V2 with NbHDA6 and AtHDA6, in vitro pulldown and co-IP assays were performed. When full-length V2 was fused with the His tag and NbHDA6 and AtHDA6 were fused with glutathione S-transferase (GST) tags, V2-HIS was pulled down by GST-NbHDA6 and GST-AtHDA6, whereas no signal was observed when GST-NbHDA6 or GST-AtHDA6 was replaced by GST (Fig. 1C). Co-IP experiments showed that NbHDA6 or AtHDA6 could interact with V2 in Arabidopsis protoplasts while the negative control, AtSAHH, could not (Fig. 1D). These results indicated a specific and direct association of V2 with NbHDA6 and AtHDA6.
Subcellular localization and expression pattern of NbHDA6.
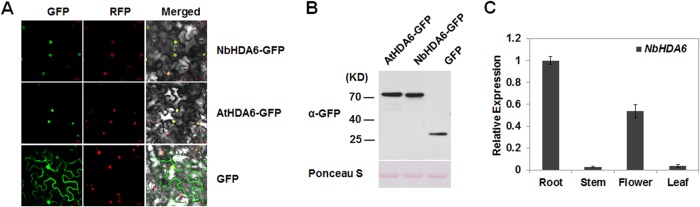
The subcellular localization of NbHDA6 was examined by agroinfiltration of epidermal cells of RFP-H2B plants (Fig. 2A). Consistent with previous studies, green fluorescence was observed in the nuclei of cells expressing green fluorescent protein (GFP) fusion to the C terminus of AtHDA6 (36–38). A similar colocalization of the RFP-H2B marker was observed exclusively in the nuclei of cells expressing NbHDA6-GFP. This indicates that the NbHDA6-GFP fusion protein localizes in the nucleus of N. benthamiana epidermal cells. Western blotting showed that NbHDA6-GFP and AtHDA6-GFP fusion proteins were expressed and remained stable in N. benthamiana cells (Fig. 2B). The expression pattern of NbHDA6 was analyzed by using real-time reverse transcription (RT)-PCR with total RNA isolated from various N. benthamiana tissues as the template. As shown in Fig. 2C, NbHDA6 transcripts were detected in roots, leaves, stems, and flowers of N. benthamiana. The highest accumulation of NbHDA6 mRNA was found in root with an intermediate level in flower, and the lowest levels were in leaf and stem, consistent with previous studies on AtHDA6 mRNA accumulation in Arabidopsis (36).
FIG 2.
NbHDA6 localization and expression. (A) Subcellular localization of HDA6. Micrographs showing cells expressing NbHDA6-GFP, AtHDA6-GFP, or GFP were examined under GFP fluorescence (left panels), RFP fluorescence (middle panels), or an overlay of GFP/RFP/bright illumination (right panels) by confocal microscopy. RFP-H2B was used as a marker for the nucleus. Bars, 20 μm. (B) Western blot analysis to detect NbHDA6-GFP, AtHDA6-GFP, or GFP protein expression using anti-GFP antibody. The gel was stained with Ponceau S to show protein loadings. (C) Real-time RT-PCR analysis of NbHDA6 mRNA levels in N. benthamiana. GAPDH was used as an internal control. Each data set was derived from at least three biological repeats.
NbHDA6 functionally complements an Arabidopsis hda6 mutant.
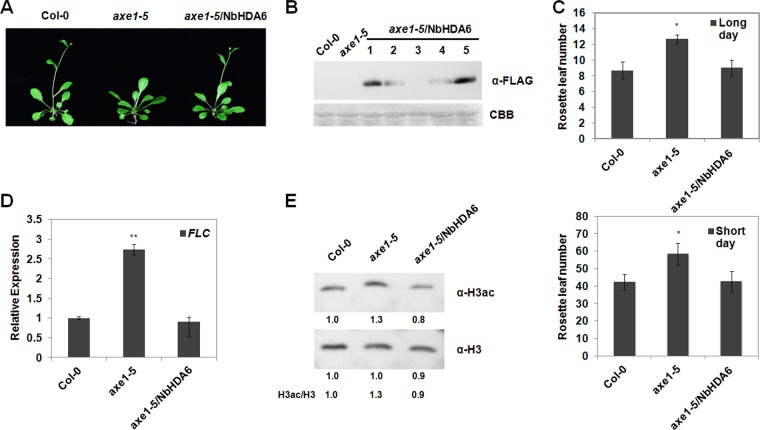
The Arabidopsis hda6 mutant axe1-5 (ecotype Col-0) develops normally under standard growth conditions with the exception of a late-flowering phenotype (31, 36, 39). To directly demonstrate that NbHDA6 in fact encodes a functional HDA6 protein, we examined whether the NbHDA6 gene could complement the late-flowering phenotype of axe1-5. Transgenic plants that constitutively expressed a FLAG-tagged NbHDA6 (axe1-5/NbHDA6) were produced (Fig. 3A). The resultant plants were first examined for the presence and expression of the NbHDA6 transgene. Western blot analysis using an antibody specific for the FLAG tag revealed the presence of the NbHDA6 protein in axe1-5/NbHDA6 transgenic lines, but not in axe1-5 or Col-0 control plants (Fig. 3B). We selected two lines (1# and 5#) for further characterization because of high NbHDA6 expression levels in these plants.
FIG 3.
Genetic complementation of the Arabidopsis axe1-5 mutant by NbHDA6. (A) Comparison of growth phenotypes of the axe1-5 mutant, the complementation line axe1-5/NbHDA6, and Col-0 plants under long-day (LD) conditions. (B) Immunoblot assays were performed using anti-FLAG antibody to detect the NbHDA6 protein expression of axe1-5/NbHDA6 transgenic plants. Five independent axe1-5/NbHDA6 transgenic Arabidopsis lines (lanes 1 to 5) are shown. The gel was stained with Coomassie brilliant blue (CBB) to show protein loadings. Two lines (1 and 5) were selected for further characterization because of high expression levels. (C) Rosette leaf numbers at the initiation of flowering for axe1-5 and axe1-5/NbHDA6 transgenic plants grown under LD or short-day (SD) conditions compared with Col-0 plants. Student's t test was performed using rosette leaf numbers at the initiation of flowering from six individual plants. A single asterisk indicates a significant difference (P < 0.05) between the two-paired samples. (D) Real-time RT-PCR analysis of FLC expression in Col-0, axe1-5, and axe1-5/NbHDA6 plants. Actin was used as an internal control. Student's t test was performed, and double asterisks indicate a significant difference (P < 0.01) between the two-paired samples. (E) Western blot analysis to detect acetylated H3 (H3ac) using H3ac antibody in protein extracts from Col-0, axe1-5, and axe1-5/NbHDA6 transgenic lines. Total H3 protein was used as loading control. Each data set was derived from at least three biological repeats. Numbers below the blots indicate the relative abundances of H3ac or H3 proteins calculated by ImageJ.
Col-0, axe1-5, and axe1-5/NbHDA6 plants were grown under long-day (LD) and short-day (SD) conditions (see Materials and Methods), and the numbers of rosette leaves present at the initiation of flowering were compared. The flowering of axe1-5 plants was greatly delayed in LD as well as in SD, leading to significantly increased numbers of rosette leaves at the time of flowering initiation (Fig. 3A and C). As expected, the delay in flowering time of axe1-5 plants was completely reversed by NbHDA6 expression in axe1-5/NbHDA6 (Fig. 3A and C). The different flowering times of axe1-5 and axe1-5/NbHDA6 plants prompted us to analyze whether the expression of flowering locus C (FLC), a transcription factor that controls the transition from vegetative to reproductive development, was affected. Real-time RT-PCR analysis indicated that the expression of FLC was increased in axe1-5 and returned to normal levels in axe1-5/NbHDA6 compared with Col-0 plants (Fig. 3D). These observations indicate that the NbHDA6 cDNA is able to genetically complement the axe1-5 mutation, suggesting that NbHDA6 represents a functional homolog of AtHDA6.
To further examine whether HDA6 has an HDAC activity, the levels of acetylated histone H3 in Col-0, axe1-5, and axe1-5/NbHDA6 plants were analyzed by Western blot analysis. As shown in Fig. 3E, there were increased levels of acetylated H3 in axe1-5 lines compared with Col-0, which is consistent with previous studies (36). As seen for the late-flowering phenotype, the increased level of acetylated H3 of axe1-5 plants was completely corrected by NbHDA6 expression in axe1-5/NbHDA6. All these observations suggested that NbHDA6 encodes a functional HDA6 protein.
NbHDA6 activity is not affected by V2.
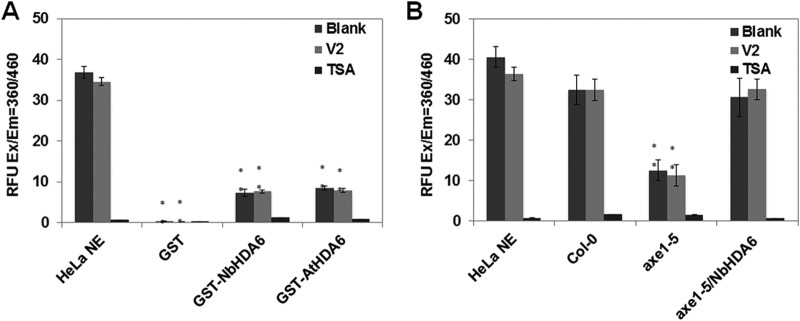
The interaction between V2 and NbHDA6 raises the question of whether this physical interaction affects the activity of the latter. Histone deacetylases catalyze the removal of acetyl groups from acetylated lysine (K) residues, which are active epigenetic marks, in the N termini of nucleosomal histones (41). To investigate whether HDA6 has an HDAC activity, the histone deacetylase activity of GST-tagged NbHDA6 and AtHDA6 was evaluated. HeLa nuclear extract (NE) and purified GST protein served as positive and negative controls, respectively. As shown in Fig. 4A, both NbHDA6 and AtHDA6 displayed weak HDAC enzymatic activity and the HDAC activity was sensitive to the HDAC inhibitor trichostatin A (TSA). To further examine whether V2 affects the activity of NbHDA6, NbHDA6 mixed with large amounts of V2 (32-fold molar excess V2:NbHDA6) were assessed. The results showed that V2 was unable to inhibit the HDAC activity, at least at this molar excess (Fig. 4A). The purified HDA6 proteins having very weak deacetylase activity might be due to the potential cofactors of HDA6 that were lost. Therefore, we repeated the deacetylation assay with nuclear protein extracts from Col-0, axe1-5, and axe1-5/NbHDA6 plants using HeLa NE as the positive control. As expected, the nuclear protein extracts of axe1-5/NbHDA6 plants displayed strong HDAC enzymatic activity, which had no significant difference with the positive control. However, the HDAC enzymatic activity of nuclear protein extracts from axe1-5 plants was significantly lower. Moreover, this strong HDAC enzymatic activity was sensitive to TSA but not V2 (Fig. 4B). Thus, the interaction of V2 with NbHDA6 does not appear to affect the activity of NbHDA6.
FIG 4.
Deacetylase activity of NbHDA6 is not affected by V2. (A) Detection of the histone deacetylase activity of GST-NbHDA6 and GST-AtHDA6 using an HDAC activity fluorometric assay kit. HDA6 mixed with large amounts of V2 (32-fold molar excess V2:HDA6) or HDAC inhibitor TSA were also assessed. HeLa nuclear extract (HeLa NE) served as the positive control, and purified GST protein served as the negative control. (B) Detection of the histone deacetylase activity of nuclear protein extracts from Col-0, axe1-5, and axe1-5/NbHDA6 plants using an HDAC activity fluorometric assay kit. Nuclear protein extracts mixed with large amounts of V2 or HDAC inhibitor TSA were also assessed. HeLa NE served as the positive control. The relative fluorescence units (RFU) generated from the assay for each mixture are compared. Error bars represent the standard errors of the means from independent measurements. Student's t test was performed, and double asterisks indicate a significant difference (P < 0.01) between the two-paired samples. Each data set was derived from at least three biological repeats.
Viral symptoms and DNA accumulation are enhanced in NbHDA6-silenced plants.
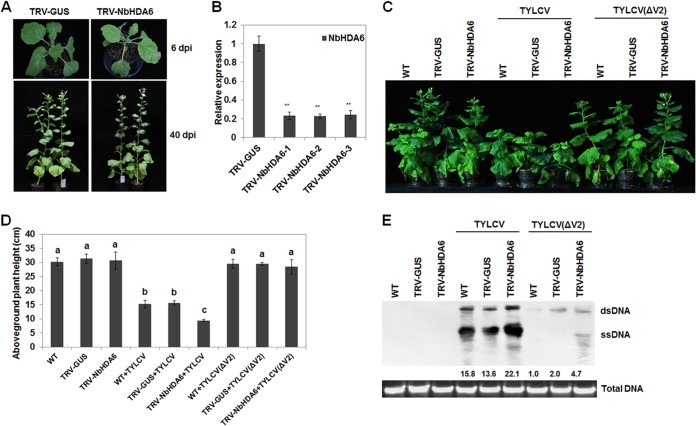
To assess the biological significance of the V2-NbHDA6 interaction in vivo, we silenced the NbHDA6 gene in N. benthamiana using virus-induced gene silencing (VIGS). Real-time RT-PCR results showed that NbHDA6 genes were simultaneously downregulated in the silenced lines compared with the TRV-GUS control (Fig. 5A and B). Wild-type (WT) and NbHDA6-silenced plants were then inoculated with TYLCV, and infection was monitored over time. NbHDA6-silenced plants infected with TYLCV developed more-severe symptoms than did WT plants (Fig. 5C). The aboveground plant height of NbHDA6-silenced plants infected with TYLCV was much lower than that of WT plants (Fig. 5D). Moreover, the viral DNA, as determined by Southern blot analysis, accumulated at greater levels in NbHDA6-silenced plants than in WT plants (Fig. 5E). Furthermore, we constructed an infectious clone [TYLCV(ΔV2)] that contained a nontranslatable V2, and then WT and NbHDA6-silenced N. benthamiana plants were inoculated with TYLCV(ΔV2). Results showed that both WT and NbHDA6-silenced N. benthamiana plants infected with TYLCV(ΔV2) developed very mild symptoms (Fig. 5C). Moreover, as seen with TYLCV, the viral DNA, as determined by Southern blot analysis, accumulated at greater levels in NbHDA6-silenced than in WT plants (Fig. 5E). These results suggest that NbHDA6 may play a critical role in host defense against TYLCV infection.
FIG 5.
N. benthamiana HDA6 silenced lines show enhanced susceptibility to TYLCV infection. (A) Comparison of phenotypes of N. benthamiana plants in which the NbHDA6 gene was silenced (TRV-NbHDA6) and those of plants treated with a control silencing vector (TRV-GUS). (B) Relative NbHDA6 gene expression levels in VIGS and control N. benthamiana determined by real-time RT-PCR. TRV-NbHDA6-1, TRV-NbHDA6-2, and TRV-NbHDA6-3 represented three independent VIGS plants, and TRV-GUS represents the control plants. GAPDH was used as an internal control. Student's t test was performed, and double asterisks indicate a significant difference (P < 0.01) between the two-paired samples. Each data set was derived from at least three biological repeats. (C) Phenotypes of TYLCV or V2-knockout TYLCV [TYLCV(ΔV2)]-infected wild-type, TRV-GUS control, and NbHDA6-silenced plants. Leaves were photographed 30 days after inoculation. (D) Aboveground plant heights of TYLCV- or TYLCV(ΔV2)-infected WT, TRV-GUS, and NbHDA6-silenced plants. Means with the same letter are not significantly different (SPSS, P = 0.05). (E) Southern blot analysis of TYLCV viral DNA accumulation in systemic leaves of N. benthamiana infected with TYLCV or TYLCV(ΔV2). The DNA agarose gel was stained with ethidium bromide for genomic DNA as loading control and then blotted using probes specific for TYLCV. Numbers below the blot indicated relative abundances of TYLCV viral DNA calculated by ImageJ. Similar results were observed in at least three independent experiments.
DNA methylation of the TYLCV genome is reduced in NbHDA6-silenced plants.
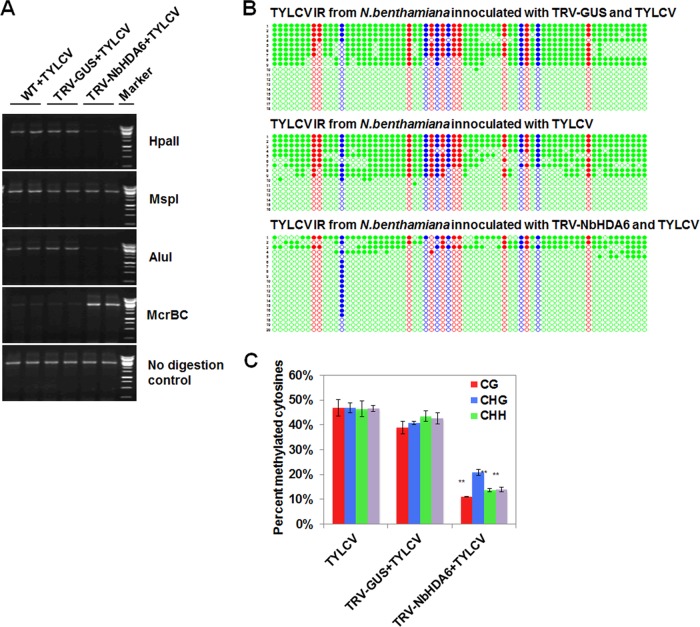
To test whether the enhanced infection efficiency of TYLCV was associated with cytosine methylation, we used methylation-sensitive PCR to examine the methylation status of viral DNA in TYLCV-infected WT and NbHDA6-silenced plants. HpaII, MspI, and AluI are methylation-sensitive endonucleases whose cleavage activities are blocked by methylation of cytosine in their target sites. McrBC is a methylation-dependent endonuclease that preferentially cuts methylated DNAs. There are two restriction sites of HpaII (mCCGG, CmCGG), two restriction sites of MspI (mCCGG), five restriction sites of AluI (AGmCT), and many restriction sites of McrBC [(G/A)mC] within the PCR target regions. Genomic DNA was extracted and digested with these four enzymes, and the digested DNAs were subjected to PCR separately. We observed that the PCR product decreased in HpaII- and AluI-digested DNA extracted from NbHDA6-silenced plants compared to DNA extracted from TYLCV-infected WT plants (Fig. 6A). In addition, McrBC was not able to cleave DNA extracted from NbHDA6-silenced plants, in contrast to DNA extracted from TYLCV-infected WT plants (Fig. 6A). However, the PCR product exhibited no difference using MspI digestion of DNA isolated from NbHDA6-silenced and WT infected plants (Fig. 6A). These results strongly suggest that the reduced cytosine methylation, especially in the CG and CHH (where H is C, A, or T) sites, contributes to the enhanced infection efficiency of TYLCV.
FIG 6.
NbHDA6 positively regulated the cytosine methylation of TYLCV. (A) Analysis of DNA methylation of the TYLCV genome by methylation-sensitive PCR. Genomic DNA was digested with HpaII, MspI, AluI, or McrBC and then loaded into the PCR system. Undigested DNA is shown as a control. (B) Cytosine methylation profiles assessed by bisulfite sequencing. The circles represent cytosine residues and are color coded according to the sequence context (red for CG, blue for CHG, and green for CHH). Solid circles indicate methylated cytosines. Each line represents the sequence of an individual clone. (C) Percentages of methylated cytosines in the TYLCV intergenic regions (IR). Student's t test was performed using the methylation values from individual clones. Double asterisks indicate a significant difference (P < 0.01) between the two-paired samples. Error bars represent the standard errors of the means from independent measurements. Samples were prepared by pooling six leaves from six systemically infected plants at 30 dpi. Similar results were observed in at least three independent experiments.
To investigate the methylation status at a higher resolution, bisulfite sequencing was performed to assess the methylation status of the viral intergenic region (IR). The TYLCV IR (about 398 bp) includes 67 cytosines in different contexts, with 10 CG, 6 CHG, and 51 CHH sites. Following bisulfite treatment to convert unmethylated cytosines to uracil, the viral strand was amplified by PCR, and almost 20 clones were sequenced per treatment. Student's t test was used to compare methylation values from individual clones. As shown in Fig. 6B and C, we found that the RNA silencing of NbHDA6 resulted in a 28% decrease in the total number of cytosine residues methylated, with most reductions occurring at non-CG sites (23% CG and 30% CHH). In comparison, the methylation levels at CHG sites were not significantly different between TYLCV-infected WT and NbHDA6-silenced plants. These results indicate that enhanced disease is accompanied by a substantial reduction in methylation.
V2 competes with NbMET1 for direct binding to NbHDA6.
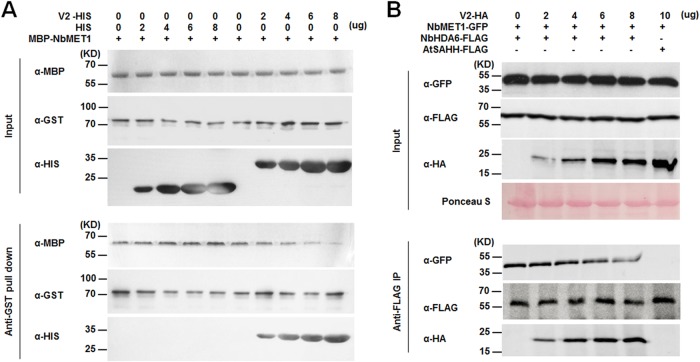
Previous genetic analysis and protein interaction results indicate that MET1 functions together with HDA6 to regulate gene silencing (28, 33). In this study, we found that V2 directly interacted with NbHDA6. These complex interactions prompted us to examine whether V2 could also directly interact with MET1. Unfortunately, despite considerable effort, we were unable to express full-length MET1 protein in N. benthamiana or Escherichia coli cells using standard methods. A truncated MBP-NbMET1 fusion protein was purified from E. coli cells. Although the truncated MET1 (R2FB) contained only the first bromo-adjacent homology (BAH) domain (amino acids 735 to 869) from the MET1 protein, it still can interact with HDA6 (33, 38). Results of in vitro pulldown analysis showed that MBP-NbMET1 specifically interacted with GST-NbHDA6 but not GST-V2.
To test the possibility that V2 may compete with NbMET1 for binding with NbHDA6, we performed competitive pulldown assays. We found that the amounts of MBP-NbMET1 pulled down by GST-NbHDA6 were reduced when increasing amounts of V2 were added to the mixture, suggesting a competition between V2 and NbMET1 in binding to NbHDA6 (Fig. 7A). This was confirmed by in vivo competitive co-IP assays between NbHDA6-FLAG and NbMET1-GFP in the presence of V2-HA. The increasing amount of V2-HA expressed in Arabidopsis protoplasts was accomplished by using an increasing amount of V2-HA recombinant plasmid for transfection. Again, expression of V2 in Arabidopsis protoplasts reduced the association of NbMET1 with NbHDA6 (Fig. 7B). These findings demonstrate that V2 competes with NbMET1 for direct binding to NbHDA6.
FIG 7.
V2 competes with NbMET1 for direct binding to NbHDA6. (A) In vitro competitive pulldown assays. The indicated amounts of V2-HIS or HIS protein were mixed with 2 μg of MBP-NbMET1 and pulled down by 2 μg of GST-NbHDA6. The bound protein was detected by immunoblotting with the indicated antibodies. (B) In vivo competitive co-IP assays. NbHDA6-FLAG and NbMET1-GFP were coexpressed in Arabidopsis protoplasts in the presence or absence of V2-HA. The immune complexes were pulled down by using anti-FLAG agarose beads. These experiments were repeated three times with similar results.
DISCUSSION
Geminiviruses infect a broad variety of plants and induce a wide range of symptoms (6, 7). Members of the Geminiviridae replicate their genome via dsDNA intermediates, which can induce methylation-mediated TGS in infected plants (4, 15). This antiviral response is counteracted by RNA silencing suppressors, which are encoded by several geminiviruses. Until now, a few viral suppressors have been reported to suppress TGS through protein-protein interactions that interfere with the methyl cycle in plants. For example, the AC2 protein encoded by TGMV and CaLCuV, the C2 protein encoded by BCTV, and the βC1 protein encoded by TYLCCNB all interact with host factors to block the methyl cycle (19, 21, 24). TYLCV is a major tomato pathogen worldwide that causes extensive tomato losses (42). We demonstrated previously that V2 of TYLCV can suppress TGS when expressed using a PVX vector or when expressed stably in transgenic Arabidopsis and is presumably responsible for suppression of TGS during TYLCV infection (26). In this study, we screened an N. benthamiana cDNA library using TYLCV V2 as bait in a yeast two-hybrid assay and successfully identified NbHDA6 as a V2-interacting protein. The interaction between V2 and NbHDA6 was confirmed using yeast two-hybrid, BiFC, GST pulldown, and co-IP assays (Fig. 1).
What is worth mentioning is that coexpression of V2 and NbHDA6 in leaf cells resulted in strong yellow fluorescence, indicating an interaction between V2 and NbHDA6 in N. benthamiana (Fig. 1B). Interestingly, the fluorescence accumulated in distinct microbodies throughout the cell cytoplasm and nucleus periphery. Earlier studies on the localization of TYLCV V2 showed that the protein is distributed in the cell cytoplasm, accumulating in inclusion bodies or aggregates that were observed mainly at the cell periphery (43). As we have shown in Fig. 2B, transiently expressed NbHDA6 and AtHDA6 localized in the nucleus of N. benthamiana cells. This is expected both from previous reports showing AtHDA6 localizing to cell nuclei and from the anticipated location of its clients (histones) (36, 37). Based on the changes of subcellular localization, we speculate that the presence of TYLCV V2 in the cell can significantly alter the subcellular localization pattern of NbHDA6. However, although the coexpression of V2 altered the subcellular localization pattern of NbHDA6, it did not completely break the nuclear location of NbHDA6. Further studies are necessary to understand why there is such a change of subcellular localization and how it changes.
Arabidopsis axe1-5 plants contain a splice site mutant of HDA6 that has a base change at an intron splice site resulting in two HDA6 transcripts with altered lengths (30). axe1-5 plants display a late-flowering phenotype, as measured by the number of rosette leaves at the initiation of flowering (31, 33, 36, 39). In addition, increased levels of acetylated H3 were observed in axe1-5 plants compared with Col-0 plants (36, 44). To directly demonstrate that NbHDA6 in fact represents a functional HDA6 protein, we examined whether the NbHDA6 gene can complement the known phenotype of axe1-5 plants. As shown in Fig. 3, the late-flowering phenotype and high levels of H3ac were completely rescued by NbHDA6 expression. These results suggest that NbHDA6 represents a functional homolog of AtHDA6 in plants. It is worth mentioning that our previous studies revealed that flowering in V2 transgenic Arabidopsis was also delayed compared with the Col-0 plants (26). This similar phenotype indicates functional correlations between V2 and NbHDA6.
HDA6 regulates several biological processes through histone deacetylation function (41). By environmental (light, cold, and jasmonic acid) and developmental signals, the chromatin status is activated and HDA6 deacetylates the target histone acetylation of HDA6 target genes. Then, deacetylated chromatins regulate the gene activity of several biological processes in response to the environmental stimuli (45, 46). In addition, HDA6 functions in inactivating and constitutively silencing genome regions with DNA methylation by MET1 and the small interfering RNA (siRNA)-mediated pathway (28, 29, 33). In this study, we demonstrate that NbHDA6 has histone deacetylase activity, but the activity is not influenced by the presence of TYLCV V2 (Fig. 4). Earlier studies revealed that the effect of AtHDA6 on transgene silencing might be independent from its histone deacetylase activity (30). However, HDAC activity of HDA6 has been shown to be required for rRNA gene variant silencing and spurious intergenic spacer (IGS) transcription suppression (47). Together these observations suggest that suppression of TGS by V2 does not occur through inhibition of the deacetylation function of HDA6.
The symptoms on NbHDA6-silenced plants appeared more severe than on WT plants after inoculation with TYLCV (Fig. 5C and D), and viral DNA accumulated to higher levels in NbHDA6-silenced plants (Fig. 5E). Moreover, silencing of NbHDA6 partially restored the pathogenicity of TYLCV(ΔV2), which is nearly nonpathogenic on N. benthamiana plants. These results strongly indicate that NbHDA6 may play a critical role in host defense against TYLCV infection and is specifically targeted by V2 for its virulence function. In addition to viral symptoms, results of methylation-sensitive PCR and bisulfite sequencing showed that DNA methylation levels of the TYLCV genome were reduced in NbHDA6-silenced compared to WT plants (Fig. 6). Several earlier studies have indicated that gene silencing can be reversed or partially reversed by treatment with HDAC inhibitors (48, 49). Earley et al. further showed that decreased cytosine methylation occurs at rRNA gene promoters in HDA6-silenced lines (44). Our results support the above-mentioned studies and indicate that NbHDA6 is one of the critical host factors involved in methylation-mediated TGS. Moreover, analysis with methylation-sensitive restriction enzymes and bisulfite sequencing revealed that methylation at CGs and CHHs was reduced, while levels of CHG methylation remain relatively unchanged in NbHDA6-silenced plants. The analysis suggests that HDA6 is not required for de novo CHG methylation but appears to be required for CG and CHH maintenance methylation during geminivirus infection.
Previous protein-protein interaction analysis indicated that HDA6 physically interacts with MET1 in vitro and in vivo, and the C-terminal region of HDA6 and the first BAH domain of MET1, respectively, were responsible for the interactions (33). These results suggested that HDA6 and MET1 interact directly and act together to function in gene silencing. In this study, we demonstrated that NbHDA6 interacted with V2 directly and V2 did not interact with NbMET1 but competed for direct binding to NbHDA6.
Besides suppressing the plant TGS defense by targeting the host HDA6, which we characterized in this study, several V2-interacting host partners have been identified to manipulate diverse cellular processes. SGS3, a protein that is required to convert ssRNA to dsRNA in the RNA-silencing pathway to produce siRNAs, has been reported to interact with V2. A point mutant of V2 that is unable to bind SlSGS3 also lost its ability to suppress RNA silencing, suggesting that this interaction is necessary for the suppressor activity of V2 (50). V2 also interacts with CYP1, a tomato papain-like cysteine protease that is involved in plant defense against diverse pathogens (51). The V2-CYP1 interaction inhibited the enzymatic activity of CYP1 but did not interfere with the posttranslational maturation of this protein (52). Therefore, during plant-geminivirus interactions, V2 represents a multifunctional viral anti-host-defense factor to help successful infection and subsequent spread by diverse countermeasures.
It is important to note that HDA6 also functions in chromatin histone acetylation through its deacetylation function. Alexiadis et al. directly investigated the influence of histone acetylation on chromatin replication using an in vitro simian virus 40 (SV40) minichromosome replication system (53). Through comparing the replication efficiency of hyperacetylated and normal SV40 minichromosomes, the authors demonstrated that acetylation of the core histones caused a modest but significant stimulation of the elongation of replication. Geminiviruses replicate using double-stranded DNA (dsDNA) intermediates (8–12). These dsDNAs are assembled into host nucleosomes, yielding circular viral minichromosomes (13, 14). A question to be answered in the future is whether HDA6 affects viral replication by directly regulating geminiviral minichromosome acetylation.
Based on our work and results from others, a working model to explain the mechanism of V2 and NbHDA6 in virus-plant interaction is proposed (Fig. 8). HDA6 is a conserved regulator for inactivating and constitutively silencing regions of the genome with DNA methylation. HDA6 recruits and physically interacts with MET1 to function cooperatively in gene silencing. TYLCV V2 is a TGS suppressor that appears to target NbHDA6 to interfere with recruitment of MET1, resulting in decreased methylation of the viral DNA, thereby increasing host susceptibility to TYLCV infection.
FIG 8.
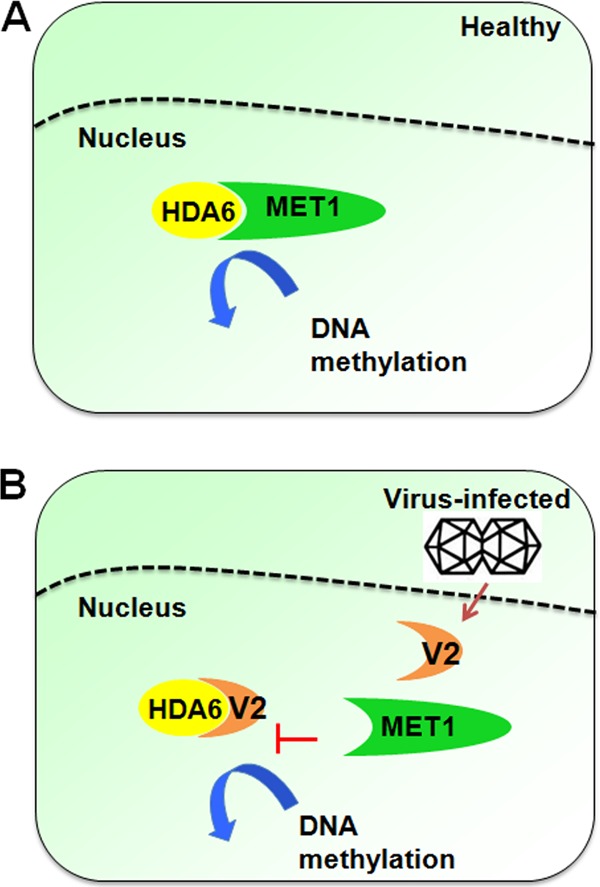
Proposed model for the NbHDA6-V2 interaction in regulation of methylation-mediated TGS during geminivirus infection. (A) HDA6 is a conserved regulator for inactivating and constitutively silencing genomic regions through DNA methylation. HDA6 recruits and physically interacts with MET1 to function cooperatively in gene silencing. (B) The V2 protein encoded by TYLCV is a TGS suppressor. This model proposes that V2 targets NbHDA6, thereby interfering with the recruitment of MET1 by HDA6. This results in a decrease in methylation of the viral DNA, leading to an increase in host susceptibility to TYLCV infection.
In summary, we have demonstrated that the V2 protein encoded by TYLCV interacted directly with N. benthamiana HDA6. Genetic complementation revealed that NbHDA6 complemented the late-flowering phenotype and restored histone deacetylation of an AtHDA6 mutation, indicating that NbHDA6 is in fact a functional homolog of AtHDA6. In addition, NbHDA6 displayed HDAC enzymatic activity, which was not affected by V2 protein. Genetic analysis revealed that silencing of NbHDA6 expression resulted in increased susceptibility to TYLCV infection and reduced DNA methylation of the viral genome in infected plants. Since HDA6 was previously shown to recruit and physically interact with MET1 to function in gene silencing, we further demonstrated that V2 does not interact with NbMET1 but competes for direct binding to NbHDA6. To our knowledge, this is the first report that viral proteins inhibit TGS by interacting with histone deacetylase but not by blocking the methyl cycle. Our studies described here add a further mechanism for TGS suppression by geminiviruses.
MATERIALS AND METHODS
Plant material, virus inoculation, and transformation.
Wild type (WT) and transgenic N. benthamiana plants expressing red fluorescent protein (RFP)-tagged histone 2B (RFP-H2B) (54) were grown at 25°C in growth chambers under long-day (LD; 16-h light/8-h dark) conditions. Arabidopsis was grown at 22°C in growth chambers under LD or short-day (SD; 8-h light/16-h dark) conditions. The hda6 mutant line axe1-5 was originally isolated based on deregulated expression of auxin-responsive transgenes (30) and was outcrossed into Arabidopsis WT Col-0 (ecotype Columbia) three times. N. benthamiana plants at the 4- to 6-leaf stage were agroinoculated with Tobacco rattle virus (TRV) vectors, TYLCV (isolate SH2), and V2 knockout TYLCV [TYLCV(ΔV2)] as previously described (55–57). For genetic complementation assays, agrobacterium-mediated transformation of axe1-5 with Agrobacterium tumefaciens EHA105 carrying pCambia2300-NbHDA6-2×FLAG was performed as previously described (58).
Plasmid construction.
To produce the bait plasmid for yeast two-hybrid screens, the full-length coding sequence for V2 was amplified using an infectious clone of TYLCV in pBinplus (59), and the PCR fragment was then inserted into the EcoRI-BamHI site of the yeast GAL4 DNA binding domain vector pGBKT7 (Clontech). To generate pGADT7-NbHDA6 vector for yeast two-hybrid verification, the full-length coding sequence for NbHDA6 was amplified using N. benthamiana cDNA as the template. The PCR fragment was then inserted into the NdeI-BamHI site of the yeast GAL4 activation domain vector pGADT7 (Clontech). For production of bimolecular fluorescence complementation (BiFC) vectors, the coding sequence of V2 was amplified and cloned into the PacI-AscI site of p2YN (60) as a fusion with the N-terminal fragment of yellow fluorescent protein (YFP), resulting in pV2-YFPN. The coding sequence of NbHDA6 was amplified and cloned into the PacI-AscI site of p2YC (60) as a fusion with the N-terminal fragment of YFP, resulting in pNbHDA6-YFPC. To generate GST-NbHDA6, GST-AtHDA6, GST-V2, HIS-V2, and MBP-NbMET1 expression vectors for GST pulldown assays, the coding sequence of V2 was amplified and cloned into the EcoRI-SalI site of pGEX-6P-1 and pET-32a. The coding sequences of NbHDA6 and AtHDA6 were amplified from N. benthamiana or A. thaliana cDNA and then inserted into the BamHI-SalI or EcoRI-XhoI site of pGEX-6P-1, respectively. The coding sequence of truncated NbMET1 (R2FB, containing the first bromo-adjacent homology [BAH] domain (33) was amplified from N. benthamiana cDNA and cloned into the SalI-XhoI site of pMBP-28. For production of co-IP vectors, the coding sequence of V2 was amplified and cloned into the KpnI-SalI site of pUC19-35S-HA. The coding sequence of full-length NbHDA6, AtHDA6, or AtSAHH was amplified from N. benthamiana or A. thaliana cDNA and cloned into the XhoI-Csp45I site of pUC19-35S-FLAG. The coding sequence of truncated NbMET1 (33) was amplified from N. benthamiana cDNA and cloned into the KpnI-XbaI site of pUC19-35S-GFP. For subcellular localization studies, NbHDA6 and AtHDA6 were tagged at their C terminus with green fluorescent protein (GFP). The PCR-amplified NbHDA6 or AtHDA6 coding sequence was cloned into the KpnI-XbaI site of pCHF3-GFP to produce pCHF3-NbHDA6-GFP or pCHF3-AtHDA6-GFP, respectively. For genetic complementation studies, the coding sequence of NbHDA6 was amplified and cloned into the EcoRI-SalI site of pCambia2300-2×FLAG to produce pCambia2300-NbHDA6-2×FLAG. To generate plasmids for virus-induced gene silencing (VIGS) assays, partial coding sequences of NbHDA6 (C-terminal 330 to 430 amino acids of NbHDA6, which is not conserved in the HDAC family) and GUS were amplified and cloned into the XbaI-BamHI sites of a TRV vector (61). The infectious clone containing a nontranslatable V2 carrying a nonsense mutation, which terminates translation of the V2 protein at amino acid 8 by a point mutation that changes glutamic acid (GAA) to a stop codon (TAA), was generated as previously reported (57). Primers used in plasmid construction are available upon request.
Yeast two-hybrid screen.
The plasmid pGBKT7-V2 and an N. benthamiana cDNA library were cotransformed into Saccharomyces cerevisiae strain Y2HGold (Clontech), and then double transformants were assayed for histidine prototrophy and β-galactosidase activity. One cDNA clone, designated NbHDA6, was selected. The plasmids pGBKT7-V2 and pGADT7-NbHDA6 were then cotransformed into S. cerevisiae strain Y2HGold to verify their interaction. Plasmids pGBKT7-V2 and pGADT7 or pGBKT7 and pGADT7-NbHDA6 were used as negative controls. Transformants were grown at 30°C for 72 h on synthetic medium lacking leucine and tryptophan and then transferred to medium lacking histidine, leucine, and tryptophan to identify binding activity.
BiFC assay.
pV2-YFPN and pNbHDA6-YFPC were introduced individually into A. tumefaciens strain C58C1 by electroporation. BiFC experiments were performed as described previously (60). Leaves of 2- to 4-week-old transgenic N. benthamiana plants expressing RFP-H2B were infiltrated with C58C1 containing the pV2-YFPN and pNbHDA6-YFPC construct pairs. YFP fluorescence was observed and photographed using confocal microscopy (Leica TCS SP5) at 72 h after infiltration (hpi) as described previously (62). RFP-H2B served as a marker for the nucleus (54).
In vitro pulldown assay.
Recombinant proteins were produced in Escherichia coli strain BL21 after induction for 16 h with 1 mM isopropyl β-d-thiogalactoside (IPTG) at 16°C. Bacterial cells were pelleted through centrifugation, resuspended with phosphate-buffered saline (PBS) buffer, and then sonicated for 30 min. The HIS-fused, GST-fused, and MBP-fused proteins were individually purified using a nickel-nitrilotriacetic acid (Ni-NTA) resin binding column (Qiagen), GST binding column (Qiagen), and MBP binding column (Novagen), respectively, according to the manufacturer's instruction. For GST pulldown assays, equal amounts of purified proteins were mixed and incubated on GST binding columns for 1 h at 4°C. After 10 min of centrifugation at 8,000 × g, the mixed proteins were washed 10 times in washing buffer, eluted, and detected by immunoblotting with anti-HIS (ab18184; Abcam), anti-MBP (ab9084; Abcam), or anti-GST (ab19256; Abcam) antibody. Competitive pulldown assays were performed as described previously (63). Briefly, the indicated amounts of V2-HIS or HIS were mixed with 2 μg of GST-NbHDA6 for 1 h before being incubated with 2 μg of MBP-NbMET1 for pulldown assays.
Coimmunoprecipitation assay.
Arabidopsis protoplast preparation and transfection were essentially as previously described (40). Protoplasts were transfected with the indicated constructs and incubated for 12 h, and total protein was isolated with extraction buffer (50 mM HEPES-KOH, pH 7.5, 1 mM EDTA, 150 mM KCl, 1 mM dithiothreitol [DTT], 0.3% Triton X-100, protease inhibitors [Roche]). For immunoprecipitation, total protein was precleared with protein A agarose (Millipore) for 1 h, followed by precipitation with 2 μg anti-hemagglutinin (anti-HA) antibody (AB104; Tiangen) together with protein A agarose for 4 h. Total protein and immunoprecipitates were detected by immunoblotting with anti-FLAG (F7425; Sigma-Aldrich) or anti-HA antibody, respectively. AtSAHH-FLAG was used as a negative control. Approximately 1% of input and one-quarter of eluted protein complex were analyzed by immunoblotting. Competitive co-IP assays were performed as described previously (64). MET1-GFP was coexpressed with NbHDA6-FLAG in the presence of V2-HA constructs in Arabidopsis protoplasts. The experiment was conducted with an increased amount of V2-HA plasmid (0, 2, 4, 6, or 8 μg), while the amounts of MET1-GFP and NbHDA6-FLAG plasmids were 10 μg. Total protein extracts were used in the Flag-IP assay. Purified protein complexes were separated by SDS-PAGE and immunoblotted to detect FLAG, HA, and GFP signals using anti-FLAG, anti-HA, or anti-GFP antibody (AB105-1; Tiangen). AtSAHH-FLAG was used as a negative control.
Subcellular localization of proteins.
Plasmids pCHF3-GFP, pCHF3-NbHDA6-GFP, and pCHF3-AtHDA6-GFP were transformed individually into A. tumefaciens strain C58C1 through electroporation. Leaves of 2- to 4-week-old RFP-H2B plants were infiltrated with A. tumefaciens harboring each construct as described. Approximately 48 hpi, 1-cm2 leaf explants were excised and GFP fluorescence was examined in epidermal cells by confocal microscopy (Leica TCS SP5) as described previously (62). Then, total proteins of infiltrated plants were isolated with extraction buffer (50 mM HEPES-KOH, pH 7.5, 1 mM EDTA, 150 mM KCl, 1 mM DTT, 0.3% Triton X-100, protease inhibitors [Roche]) and detected through Western blot assays using anti-GFP antibody.
RNA extraction and real-time RT-PCR analysis.
RNA extraction and real-time RT-PCR procedures have been described previously (36, 65). For expression levels of NbHDA6 and FLC studies, 4-week-old N. benthamiana plants and 4-week-old Arabidopsis plants were used. For studies of expression levels of NbHDA6 in NbHDA6-silenced lines, upper leaves of 4-week-old N. benthamiana plants inoculated with silencing vectors (TRV-NbHDA6 or TRV-GUS) for 10 days were used. Briefly, 1 ml of TRIzol reagent (Invitrogen) was used to extract RNA from 0.1 g plant tissue. The first-strand cDNA was synthesized in a volume of 10 μl that contained reverse transcriptase (Toyobo) at 37°C for 1 h. Real-time PCR was performed by using SYBR Premix Ex Taq on an Agilent Mx3005P real-time PCR machine according to the manufacturer's instructions. The primers used for real-time RT-PCR amplification are available upon request. Relative transcript levels were calculated using 2−ΔΔCT method with the GAPDH (glyceraldehyde-3-phosphate dehydrogenase gene) or Actin transcript serving as the internal control. Each data set was derived from at least three biological repeats.
Histone extraction and Western blotting.
Total histone proteins were prepared according to the method of Li et al. (47). Briefly, seedlings were ground to powder using liquid nitrogen and then suspended in NIB buffer (250 mM sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 0.8% Triton X-100]. The suspension was centrifuged, and the pellet was resuspended in 0.4 M H2SO4 for 1 h at 4°C and then precipitated by acetone. The histone proteins were detected through Western blot assays using anti-histone H3ac (06-599; Millipore) or anti-histone H3 (06-755; Millipore) antibody.
HDAC activity assay.
The histone deacetylase activity of NbHDA6 was evaluated using an HDAC activity fluorometric assay kit (Biovision) according to the manufacturer's instructions. For in vitro HDAC activity assay, 10 μg of HeLa cell nuclear extract (HeLa NE) was used as a positive control. Purified GST-NbHDA6 or GST-AtHDA6 proteins (100 μg) were used as samples, and 100 μg of purified GST protein was included in the assay as a negative control. For the semi-in vivo HDAC activity assay, the nuclear protein extracts of Col-0, axe1-5, and axe1-5/NbHDA6 plants were obtained by using the plant nucleus isolation/extraction kit (Sigma-Aldrich) according to the manufacturer's instructions. Ten micrograms of HeLa NE was used as a positive control, and 10 μg of the nuclear protein extracts of Col-0, axe1-5, and axe1-5/NbHDA6 plants was used for the samples. Samples were incubated with 5 μl HDAC fluorometric substrate. After 30 min, the reactions were stopped with 10 μl lysine developer. Samples were then read in a fluorescence plate reader with excitation (Ex) and emission (Em) wavelengths of 360 and 460 nm, respectively.
DNA gel blotting.
After either mock inoculation or inoculation with TYLCV or TYLCV(ΔV2) for 30 days, total DNAs were extracted from leaves of N. benthamiana plants as described previously. For each sample, 40 μg DNA was depurinated and denatured within the gel and transferred to Hybond-N+ nylon membranes (GE Healthcare). Gel blotting was performed as described previously (55). Genomic DNA stained with ethidium bromide was used as a loading control.
Methylation-sensitive PCR and bisulfite sequencing.
For methylation-sensitive PCR, genomic DNA was isolated from the symptomatic plants infected by TYLCV (30 days after inoculation [dpi]) using the DNeasy plant minikit (Qiagen). Then, 100 ng of the genomic DNA was digested for 1 h at 37°C with 2 U of HpaII, MspI, AluI, or McrBC in 20-μl reaction mixtures. The enzymes were heat inactivated, and 2 μl of the cleaved DNA was loaded into PCR mixtures containing primers for full-length TYLCV. PCR products from genomic DNA that was not digested served as controls. Bisulfite sequencing was performed using 1 μg genomic DNA isolated from the symptomatic plants infected by TYLCV (30 dpi). Genomic DNA was digested overnight using the restriction enzyme NdeI, followed by overnight treatment with proteinase K (Tiangen). Bisulfite modification was carried out using the EZ DNA methylation gold kit (Zymo Research) according to the manufacturer's instructions. Primers for methylation-sensitive PCR and bisulfite sequencing are available upon request.
ACKNOWLEDGMENTS
We thank Yijun Qi (Tsinghua University) for providing the pCambia2300-2×FLAG vector, David M. Bisaro (The Ohio State University) for providing the p2YN and p2YC vectors, and Michael M. Goodin (University of Kentucky) for providing the transgenic RFP-H2B line.
This research was supported by grants from the National Natural Science Foundation of China (31390422 and 31371914) and the National Key Basic Research and Development Program of China (2012CB114004).
Author contributions: X.Z. and B.W. planned and designed the research. B.W., X.Y., Y.W., and Y.X. performed experiments and analyzed data. B.W. and X.Z. wrote the manuscript.
REFERENCES
- 1.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisaro DM. 2006. Silencing suppression by geminivirus proteins. Virology 344:158–168. doi: 10.1016/j.virol.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 4.Pooggin MM. 2013. How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int J Mol Sci 14:15233–15259. doi: 10.3390/ijms140815233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackel JN, Storer JM, Coursey T, Bisaro DM. 2016. Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on geminivirus chromatin. J Virol 90:7529–7540. doi: 10.1128/JVI.00656-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navas-Castillo J, Fiallo-Olive E, Sanchez-Campos S. 2011. Emerging virus diseases transmitted by whiteflies. Annu Rev Phytopathol 49:219–248. doi: 10.1146/annurev-phyto-072910-095235. [DOI] [PubMed] [Google Scholar]
- 7.Varsani A, Roumagnac P, Fuchs M, Navas-Castillo J, Moriones E, Idris A, Briddon RW, Rivera-Bustamante R, Zerbini FM, Martin DP. 2017. Capulavirus and Grablovirus: two new genera in the family Geminiviridae. Arch Virol 162:1819–1831. doi: 10.1007/s00705-017-3268-6. [DOI] [PubMed] [Google Scholar]
- 8.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- 9.Jeske H. 2009. Geminiviruses. Curr Top Microbiol Immunol 331:185–226. [DOI] [PubMed] [Google Scholar]
- 10.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X. 2013. Advances in understanding begomovirus satellites. Annu Rev Phytopathol 51:357–381. doi: 10.1146/annurev-phyto-082712-102234. [DOI] [PubMed] [Google Scholar]
- 12.Erdmann JB, Shepherd DN, Martin DP, Varsani A, Rybicki EP, Jeske H. 2010. Replicative intermediates of maize streak virus found during leaf development. J Gen Virol 91:1077–1081. doi: 10.1099/vir.0.017574-0. [DOI] [PubMed] [Google Scholar]
- 13.Pilartz M, Jeske H. 1992. Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 189:800–802. doi: 10.1016/0042-6822(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 14.Pilartz M, Jeske H. 2003. Mapping of abutilon mosaic geminivirus minichromosomes. J Virol 77:10808–10818. doi: 10.1128/JVI.77.20.10808-10818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raja P, Sanville BC, Buchmann RC, Bisaro DM. 2008. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol 82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Negrete EA, Carrillo-Tripp J, Rivera-Bustamante RF. 2009. RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J Virol 83:1332–1340. doi: 10.1128/JVI.01474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F, Xu X, Huang C, Gu Z, Cao L, Hu T, Ding M, Li Z, Zhou X. 2015. The AC5 protein encoded by mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol 208:555–569. doi: 10.1111/nph.13473. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Hao LH, Shung CY, Sunter G, Bisaro DM. 2003. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020–3032. doi: 10.1105/tpc.015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. 2009. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol 83:5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackel JN, Buchmann RC, Singhal U, Bisaro DM. 2015. Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J Virol 89:3176–3187. doi: 10.1128/JVI.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, Zhou X, Guo H, Xie Q. 2011. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23:273–288. doi: 10.1105/tpc.110.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castillo-Gonzalez C, Liu XY, Huang CJ, Zhao CJ, Ma ZY, Hu T, Sun F, Zhou YJ, Zhou XP, Wang XJ, Zhang XR. 2015. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife 4:e06671. doi: 10.7554/eLife.06671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun YW, Tee CS, Ma YH, Wang G, Yao XM, Ye J. 2015. Attenuation of histone methyltransferase kryptonite-mediated transcriptional gene silencing by Geminivirus. Sci Rep 5:16476. doi: 10.1038/srep16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X. 2011. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog 7:e1002329. doi: 10.1371/journal.ppat.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Negrete E, Lozano-Duran R, Piedra-Aguilera A, Cruzado L, Bejarano ER, Castillo AG. 2013. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol 199:464–475. doi: 10.1111/nph.12286. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Li F, Huang C, Yang X, Qian Y, Xie Y, Zhou X. 2014. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J Gen Virol 95:225–230. doi: 10.1099/vir.0.055798-0. [DOI] [PubMed] [Google Scholar]
- 27.Aufsatz W, Stoiber T, Rakic B, Naumann K. 2007. Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene 26:5477–5488. doi: 10.1038/sj.onc.1210615. [DOI] [PubMed] [Google Scholar]
- 28.To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, Kimura H, Yokoyama S, Shinozaki K, Seki M. 2011. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JM, To TK, Seki M. 2012. An epigenetic integrator: new insights into genome regulation, environmental stress responses and developmental controls by histone deacetylase 6. Plant Cell Physiol 53:794–800. doi: 10.1093/pcp/pcs004. [DOI] [PubMed] [Google Scholar]
- 30.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. 2001. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13:1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, Vaucheret H, Mittelsten Scheid O. 2004. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. 2002. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. 2012. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol 158:119–129. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fields S, Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 35.Hollender C, Liu Z. 2008. Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 36.Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. 2008. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- 37.Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. 2010. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24:1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang SD, Zhan XQ, Xu XM, Cui P, Zhu JK, Xia YJ, Xiong LM. 2015. Two domain-disrupted hda6 alleles have opposite epigenetic effects on transgenes and some endogenous targets. Sci Rep 5:17832. doi: 10.1038/srep17832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. 2011. Histone deacetylase 6 interacts with flowering locus D and regulates flowering in Arabidopsis. Plant Physiol 156:173–184. doi: 10.1104/pp.111.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Zhang MX, Shen DY, Liu TL, Chen YY, Zhou JM, Dou DL. 2016. A Phytophthora sojae effector PsCRN63 forms homo-/heterodimers to suppress plant immunity via an inverted association manner. Sci Rep 6:26951. doi: 10.1038/srep26951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahbazian MD, Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 42.Moriones E, Navas-Castillo J. 2000. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res 71:123–134. doi: 10.1016/S0168-1702(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 43.Zrachya A, Glick E, Levy Y, Arazi T, Citovsky V, Gafni Y. 2007. Suppressor of RNA silencing encoded by tomato yellow leaf curl virus-Israel. Virology 358:159–165. doi: 10.1016/j.virol.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 44.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. 2006. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RS, van Driel R, Voesenek LACJ, Spillane C, Pikaard CS, Fransz P, Peeters AJM. 2009. Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet 5:e1000638. doi: 10.1371/journal.pgen.1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Z, An F, Feng Y, Li P, Xue LAM, Jiang Z, Kim JM, To TK, Li W, Zhang X, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo H. 2011. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci U S A 108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Qian W, Zhao Y, Wang C, Shen J, Zhu JK, Gong Z. 2012. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci U S A 109:11425–11430. doi: 10.1073/pnas.1208557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen ZJ, Pikaard CS. 1997. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selker EU. 1998. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc Natl Acad Sci U S A 95:9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y. 2008. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A 105:157–161. doi: 10.1073/pnas.0709036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bar-Ziv A, Levy Y, Hak H, Mett A, Belausov E, Citovsky. 2012 The tomato yellow leaf curl virus (TYLCV) V2 protein interacts with the host papain-like cysteine protease CYP1. Plant Signal Behav 7:983–989. doi: 10.4161/psb.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bar-Ziv A, Levy Y, Citovsky V, Gafni Y. 2015. The tomato yellow leaf curl virus (TYLCV) V2 protein inhibits enzymatic activity of the host papain-like cysteine protease CYP1. Biochem Biophys Res Commun 460:525–529. doi: 10.1016/j.bbrc.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexiadis V, Halmer L, Gruss C. 1997. Influence of core histone acetylation on SV40 minichromosome replication in vitro. Chromosoma 105:324–331. doi: 10.1007/BF02529747. [DOI] [PubMed] [Google Scholar]
- 54.Bandyopadhyay A, Kopperud K, Anderson G, Martin K, Goodin M. 2010. An integrated protein localization and interaction map for potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402:61–71. doi: 10.1016/j.virol.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui X, Tao X, Xie Y, Fauquet CM, Zhou X. 2004. A DNAβ associated with tomato yellow leaf curl China virus is required for symptom induction. J Virol 78:13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui X, Li G, Wang D, Hu D, Zhou X. 2005. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J Virol 79:10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hak H, Levy Y, Chandran SA, Belausov E, Loyter A, Lapidot M, Gafni Y. 2015. TYLCV-Is movement in planta does not require V2 protein. Virology 477:56–60. doi: 10.1016/j.virol.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, Gong H, Zhou X. 2009. Molecular characterization and pathogenicity of tomato yellow leaf curl virus in China. Virus Genes 39:249–255. doi: 10.1007/s11262-009-0384-8. [DOI] [PubMed] [Google Scholar]
- 60.Yang X, Baliji S, Buchmann RC, Wang H, Lindbo JA, Sunter G, Bisaro DM. 2007. Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J Virol 81:11972–11981. doi: 10.1128/JVI.00617-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP. 2007. A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145:1161–1170. doi: 10.1104/pp.107.107391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X. 2011. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus beta-satellite. Plant Physiol 157:1394–1406. doi: 10.1104/pp.111.184648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH. 2008. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22:2564–2577. doi: 10.1101/gad.1682208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong L, Qiu X, Kang J, Wang Y, Chen H, Huang J, Qiu M, Zhao J, Kong G, Ma Z, Wang Y, Ye W, Dong S, Ma W, Wang Y. 2017. A phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr Biol 27:981–991. doi: 10.1016/j.cub.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 65.Huang C, Xie Y, Zhou X. 2009. Efficient virus-induced gene silencing in plants using a modified geminivirus DNA1 component. Plant Biotechnol J 7:254–265. doi: 10.1111/j.1467-7652.2008.00395.x. [DOI] [PubMed] [Google Scholar]