MeV and CDV remain important human and animal pathogens. Development of antivirals may significantly support current global vaccination campaigns. Cell entry is orchestrated by two interacting glycoproteins (H and F). The current hypothesis postulates that tetrameric H ectodomains (composed of stalk, connector, and head domains) undergo receptor-induced rearrangements to productively trigger F; these conformational changes may be regulated by the H-stalk C-terminal module (linker) and the following connector domain. Mutagenesis scan analysis of both microdomains revealed that replacing amino acid 146 in the H-linker region with nonhydrophobic residues produced covalent H tetramers which were compromised in triggering membrane fusion activity. However, these mutant proteins retained their ability to traffic to the cell surface and to bind to the virus receptor. These data suggest that the morbillivirus linker module contributes to the folding of functional pre-F-triggering H tetramers. Furthermore, such structures might be critical to convert receptor engagement into F activation.
KEYWORDS: Morbillivirus, host cell invasion, attachment protein, stalk domain, linker module, protein folding, membrane fusion activation
ABSTRACT
Morbillivirus (e.g., measles virus [MeV] and canine distemper virus [CDV]) host cell entry is coordinated by two interacting envelope glycoproteins, namely, an attachment (H) protein and a fusion (F) protein. The ectodomain of H proteins consists of stalk, connector, and head domains that assemble into functional noncovalent dimer-of-dimers. The role of the C-terminal module of the H-stalk domain (termed linker) and the connector, although putatively able to assume flexible structures and allow receptor-induced structural rearrangements, remains largely unexplored. Here, we carried out a nonconservative mutagenesis scan analysis of the MeV and CDV H-linker/connector domains. Our data demonstrated that replacing isoleucine 146 in H-linker (H-I146) with any charged amino acids prevented virus-mediated membrane fusion activity, despite proper trafficking of the mutants to the cell surface and preserved binding efficiency to the SLAM/CD150 receptor. Nondenaturing electrophoresis revealed that these charged amino acid changes led to the formation of irregular covalent H tetramers rather than functional dimer-of-dimers formed when isoleucine or other hydrophobic amino acids were present at residue position 146. Remarkably, we next demonstrated that covalent H tetramerization per se was not the only mechanism preventing F activation. Indeed, the neutral glycine mutant (H-I146G), which exhibited strong covalent tetramerization propensity, maintained limited fusion promotion activity. Conversely, charged H-I146 mutants, which additionally carried alanine substitution of natural cysteines (H-C139A and H-C154A) and thus were unable to form covalently linked tetramers, were fusion activation defective. Our data suggest a dual regulatory role of the hydrophobic residue at position 146 of the morbillivirus head-to-stalk H-linker module: securing the assembly of productive dimer-of-dimers and contributing to receptor-induced F-triggering activity.
IMPORTANCE MeV and CDV remain important human and animal pathogens. Development of antivirals may significantly support current global vaccination campaigns. Cell entry is orchestrated by two interacting glycoproteins (H and F). The current hypothesis postulates that tetrameric H ectodomains (composed of stalk, connector, and head domains) undergo receptor-induced rearrangements to productively trigger F; these conformational changes may be regulated by the H-stalk C-terminal module (linker) and the following connector domain. Mutagenesis scan analysis of both microdomains revealed that replacing amino acid 146 in the H-linker region with nonhydrophobic residues produced covalent H tetramers which were compromised in triggering membrane fusion activity. However, these mutant proteins retained their ability to traffic to the cell surface and to bind to the virus receptor. These data suggest that the morbillivirus linker module contributes to the folding of functional pre-F-triggering H tetramers. Furthermore, such structures might be critical to convert receptor engagement into F activation.
INTRODUCTION
Despite the availability of an effective vaccine, measles virus (MeV), which belongs to the genus Morbillivirus within the family Paramyxoviridae, still kills more than 90,000 people every year (1, 2). Most deaths due to MeV occur in developing countries. However, developed countries still share some vulnerability to the virus because of resistance against vaccination, which has precluded achieving the required herd immunity necessary for eradication. At the same time, the animal pathogen canine distemper virus (CDV), which is closely related to MeV, has been reported to cross species barriers, causing outbreaks in several species, including nonhuman primates (3, 4). Therefore, antiviral drugs, in addition to existing vaccines, may not only offer the opportunity to treat nonvaccinated patients postexposure but also might support global MeV eradication programs by closing the gaps in herd immunity (2).
MeV and CDV use two tightly interacting viral surface glycoproteins to infect a host cell, the attachment (H) protein, which binds to a specific host cell receptor, and the fusion (F) protein, which fuses the lipid bilayer of the viral envelope and the target cell plasma membrane (5, 6). Virus-to-cell membrane fusion is thought to rely on a series of specific structural rearrangements of both surface glycoproteins induced by receptor binding (7, 8). Those conformational changes are thought to be associated with lipid mixing and formation of a fusion pore. H and F proteins are not only mediating virus-to-cell fusion but are also involved in cell-to-cell fusion, which ultimately leads to syncytium formation (9–14).
CDV H consists of 604 or 607 amino acids (depending on the viral strain) and is glycosylated on the extracellular part (15). It is assumed that the physiological oligomeric state of CDV H is a tetramer (16), which, more precisely, results from the noncovalent association of two covalently linked dimers (dimer-of-dimers). Each covalent dimer is composed of disulfide-linked monomers via their Cys139 and Cys154 (17–19). CDV H is a type II transmembrane protein containing an N-terminal cytosolic tail (amino acids [aa] 1 to 35), followed by a single-pass transmembrane domain (aa 36 to 58) and a large extracellular region (ectodomain) (aa 59 to 604 or 607). The ectodomain is further subdivided into three domains: a membrane-proximal stalk region (aa 59 to 154), a connecting region (aa 155 to 187), and a membrane-distal large cubic head domain (aa 188 to 604 or 607). Of note, the MeV H-head domains contain slightly more amino acids, thereby increasing the overall size to 617 amino acids (20).
Although structural data for the CDV H head domain are not yet available, high-resolution structures of the MeV H head domain in the presence or absence of natural ligands were resolved (21–25). While these studies demonstrated that MeV H-heads assumed a six-bladed β-propeller fold, two of them additionally highlighted the precise mode of interaction with the natural morbillivirus receptors SLAM (signaling lymphocyte activation molecule, or CD150) and nectin-4 (or PVRL4) (21–23, 25). It must also be stated that atomic structures of MeV H-head dimers and tetramers were additionally reported; however, the functional relevance and the positioning of the heads with regard to the stalk domains requires further investigations (23).
Thorough mutagenesis analyses led to the definition of three additional modules in the CDV and MeV H-stalk domain (aa 59 to 154): first, an N-terminal lower/central portion, which interacts with and activates prefusion F proteins (aa 85 to 119); second, an intermediate spacer segment (aa 120 to 139), which may allow intracellular H/F glycoproteins assembly; and third, a C-terminal linker region (aa 140 to 154) with basically unknown function (17, 26, 27). Based on the atomic structures of parainfluenza virus type 5 (PIV5) and Newcastle disease virus attachment protein (HN) stalk domains (28–30), we and others suggested that the CDV and MeV H-stalks fold into a similar four-helix bundle (4HB) architecture, at least up to Cys139 (19, 31, 32). Interestingly, while the structure of the entire H-stalk linker module (aa 140 to 154) remains to be determined, the first part of the following MeV H connector domain was successfully crystalized and was shown to contain a short α-helix (H1) contacting the head domain (aa 155 to 166). Interestingly, the length of the following C-terminal unresolved segment (aa 167 to 183) was recently hypothesized to be crucial for the generation of productive H proteins by controlling proper tetrameric oligomerization (33).
The most recent model of membrane fusion triggering indicates that CDV/MeV H-stalk, connector, and head domains fold into native structures that enable intracellular assembly of H/F complexes while temporarily inhibiting the constitutive F-protein activation domain in the central stalk region (referred to as the autorepressed state). Upon receptor engagement, H-heads rearrange relative to each other, as well as relative to the H-stalk, unlocking the inherent F-triggering activity of the stalk (26, 34–38). This so-called safety-catch model suggests some structural freedom within the unresolved H-stalk linker region and/or the following connector module to enable the critical structural rearrangements to occur. It is worth noting that the flexibility of the head-to-stalk region of the PIV5 HN protein was recently inferred from mutagenesis analyses (39), highlighting some similarities in the way paramyxovirus triggers membrane fusion, although slightly different models were proposed (26, 27, 34, 40, 41).
In this study, we report the results obtained by detailed mutagenesis analysis that specifically targeted the linker module of the stalk, as well as the following N-terminal part of the connector region of both CDV and MeV H proteins. Overall, our data provided evidence that the unresolved linker module contributed to the proper assembly of F-triggering-permissive H dimer-of-dimers. Our findings are further suggestive for a potentially critical hydrophobic hinge structure in the linker domain that may undergo conformational change upon H head-to-receptor interaction, in turn leading to F-protein activation.
RESULTS
Two hydrophobic residues within the C-terminal linker region of the CDV H stalk are critical for membrane fusion triggering.
Previous functional screens, carried out by our and other groups, have identified key amino acids governing discrete functions of both MeV and CDV H proteins (7, 17–19, 42–46). Although a better knowledge of the structure and function of H has emerged from these studies, a complete molecular understanding of the morbillivirus entry mechanism is still lacking. Interestingly, the least investigated regions of both H proteins are the linker, located at the C terminus of the stalk domain, and the contiguous connector segment, which bridges the stalk to the head domain (Fig. 1A). Based on our current model of membrane fusion triggering, this region may require structural flexibility to enable receptor binding-induced conformational changes of H, leading to F activation and ensuing fusion pore formation (27, 33).
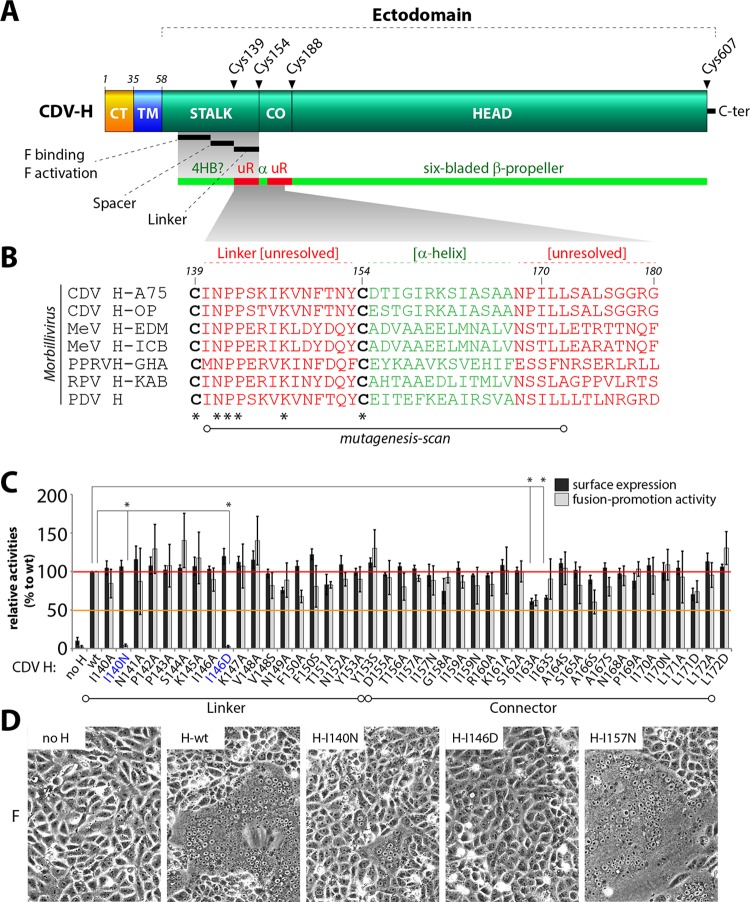
FIG 1.
Mutagenesis screen in CDV H linker and connector region. (A) Schematic representation of the CDV H protein. CT, cytosolic tail; TM, transmembrane domain; CO, connector domain; uR, unresolved region in crystal structures; α, α-helix H1. Structural information derived from MeV H protein (PDB entries 2ZB5 and 4GJT) (24, 25). (B) Sequence comparison of the linker uR, α-helix, and uR regions in various morbilliviruses. Amino acid conservation is indicated by an asterisk. H-A75, CDV H of the A75/17 strain; H-OP, CDV H of the Onderstepoort vaccine strain; H-EDM, MeV H of the Edmonston strain; H-ICB, MeV H of the ICB strain; PPRV H-GHA, peste des petits ruminants virus H of the Ghana strain; RPV H-KAB, Rinderpest virus H of the Kabete strain; H-PDV, phocine distemper virus H protein. (C) Nonconservative mutagenesis scan results for surface expression and fusion promotion activity, both relative to the wild type (for naturally hydrophobic amino acids the alanine mutation also is shown). Surface expression results are derived from nine independent transfections. Fusion promotion capability was determined by quadruplicate measurements of three independent transfections. Mutants showing significant reduction in fusion promotion activity are color coded blue. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.0001). (D) Qualitative fusion assay of selected CDV H mutants (cotransfected with CDV F-wt) in receptor-positive Vero-cSLAM cells.
In this study, we decided to conduct a detailed nonconservative mutagenesis approach targeting the poorly conserved (among morbilliviruses) H fragment encompassing amino acids 140 to 172 to shed light on the functional aspects associated with the linker/connector H regions (Fig. 1B). Note that we used the H protein of the wild-type A75/17 CDV strain (H-wt) as the template for mutagenesis. To this aim, every nonhydrophobic amino acid was replaced with the small hydrophobic alanine (A). Conversely, for positions naturally harboring hydrophobic residues, a polar or charged amino acid (asparagine [N], serine [S], or aspartic acid [D]) was selected for the exchange. The surface expression (investigated by flow cytometry analyses [all H proteins carry a C-terminal FLAG tag]) and the fusion promotion capacity (investigated using a split luciferase-based quantitative fusion assay [qFA]) of these H mutants were determined next. While almost all H mutants displayed phenotypes very similar to that of the H-wt strain, the mutant proteins H-I140N and H-I146D exhibited severe deficits in promoting membrane fusion despite excellent surface expression profiles (Fig. 1C). When those two isoleucine residues were replaced with alanine, no functional impact was recorded (Fig. 1C). We next confirmed the fusion promotion-defective phenotypes of H-I140N and I146D in a regular cell-to-cell fusion assay (Fig. 1D).
Overall, our nonconservative mutagenesis screen identified two hydrophobic residues (aa 140 and aa 146) encompassed within the linker module of the H-stalk that played a regulatory role in fusion promotion. We focused our analyses on residue 146, since H-I146D exhibited the most severe fusion promotion impairments.
Charged amino acids at position 146 prevent morbilliviral H-mediated membrane fusion promotion.
We next were interested in investigating whether defined chemical properties at position 146 of the CDV H-stalk would correlate with bioactivity. To this aim, we replaced the isoleucine at position 146 with amino acids characterized by different chemical properties: charged (D, E, K, and R), polar (T), hydrophobic (A, V, L, and M), and aromatic (F, Y, and W). We also mutated H-I146 into cysteine (C). We then subjected this second generation of H mutants to established functional assays: (i) surface expression efficiency, (ii) qFA, and (iii) SLAM binding ability.
Overall, although slight differences between H mutants were monitored, replacing the isoleucine residue 146 with several other hydrophobic, polar, or aromatic amino acids did not substantially modify either their fusion promotion phenotypes or their SLAM binding and intracellular trafficking abilities (Fig. 2A and B). In sharp contrast, mutating residue I146 of CDV H into any charged amino acid or cysteine led invariably to drastic functional impairments (Fig. 2A and B).
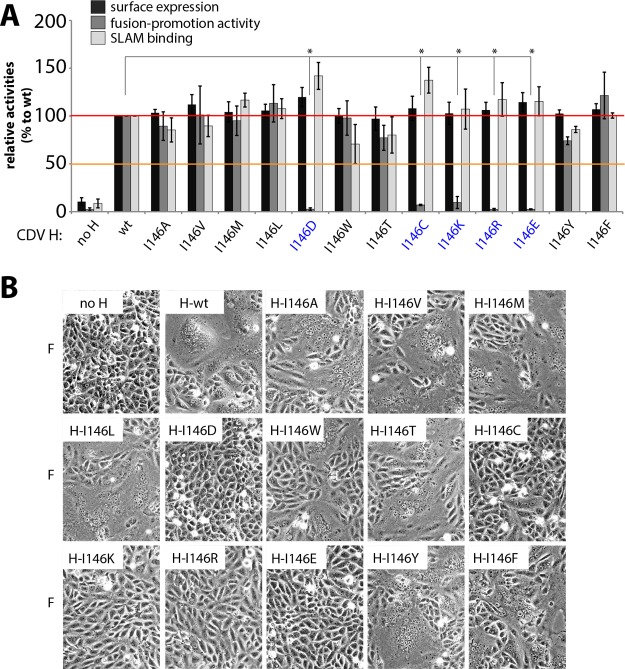
FIG 2.
Detailed mutagenesis scan on CDV H-I146. (A) Surface expression, fusion promotion activity, and SLAM binding of various CDV H-I146 mutations relative to the wild type. Mutants showing significant reduction in fusion promotion activity are color coded blue. Surface expression and SLAM binding results are derived from nine independent transfections. Fusion promotion capability was determined by quadruplicate measurements of three independent transfections. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.0001). (B) Qualitative fusion assay of selected CDV H-I146 mutants (cotransfected with CDV F-wt) in receptor-positive Vero-cSLAM cells.
We next thought to determine whether the regulatory role of residue 146 in fusion promotion was exclusive to CDV H or more generally was shared by the attachment proteins of other morbillivirus members. To tackle this issue, we decided to mutate the isoleucine residue 146 of MeV H into 13 other amino acids (D, E, R, K, T, W, Y, F, V, A, M, L, and C) and subjected the panel of H mutants to functional assays similar to those described above (using MeV F). Note that we used the H protein of the wild-type ICB MeV strain (H-wt) as the template for mutagenesis.
Strikingly, MeV H mutants carrying charged residues at position 146 exhibited similar severe defects in MeV F activation while preserving proper SLAM binding and cell surface expression bioactivities. However, in the case of MeV H, mutating position I146 into threonine (I146T), tryptophan (I146W), and tyrosine (I146Y) also led to fusion promotion deficiency. In sharp contrast, replacing H-I146 with other hydrophobic residues had no significant functional impact (Fig. 3A and B).
FIG 3.
Detailed mutagenesis scan on MeV H-I146. (A) Surface expression, fusion promotion activity, and SLAM binding of various MeV H-I146 mutations relative to the wild type. Mutants showing significant reduction in fusion promotion activity are color coded blue. Surface expression and SLAM binding results are derived from nine independent transfections. Fusion promotion capability was determined by quadruplicate measurements of three independent transfections. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.0001). (B) Qualitative fusion assay of selected MeV H-I146 mutants (cotransfected with MeV F-wt) in receptor-positive Vero-hSLAM cells.
H-I146 mutants do not display drastic impairments in physical interaction with the F protein.
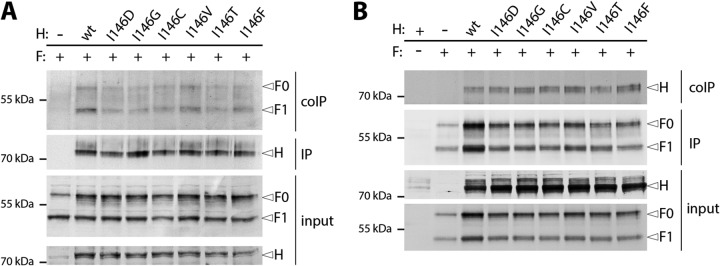
Because the strength of H to F association is a known major factor influencing membrane fusion activity, we performed reciprocal coimmunoprecipitation (coIP) experiments, as previously described (19). Overall, our results indicate that all CDV H mutants preserved F-binding capacity, given that no significant reduction was observed regardless of whether F or H was initially immunoprecipitated (Fig. 4A and B).
FIG 4.
Reciprocal coimmunoprecipitation of the CDV surface glycoproteins H and F. (A) Total lysate of transfected cells is shown (input) as well as immune-precipitated H (IP) and coimmunoprecipitated F (coIP). (B) Total lysate of transfected cells is shown (input) as well as immune-precipitated F (IP) and coimmunoprecipitated H (coIP). The Western blot shows F0 (∼65 kDa), F1 (∼45 kDa), and various H-I146 mutants (∼75 kDa).
Taken together, these data confirmed that CDV H proteins harboring charged residues at position 146 of the H-inker module did not lose their F-triggering function due to a lack of F-binding activity.
Combining charged CDV H-I146 mutants with hyperactive CDV F-proteins results in limited membrane fusion-triggering activity.
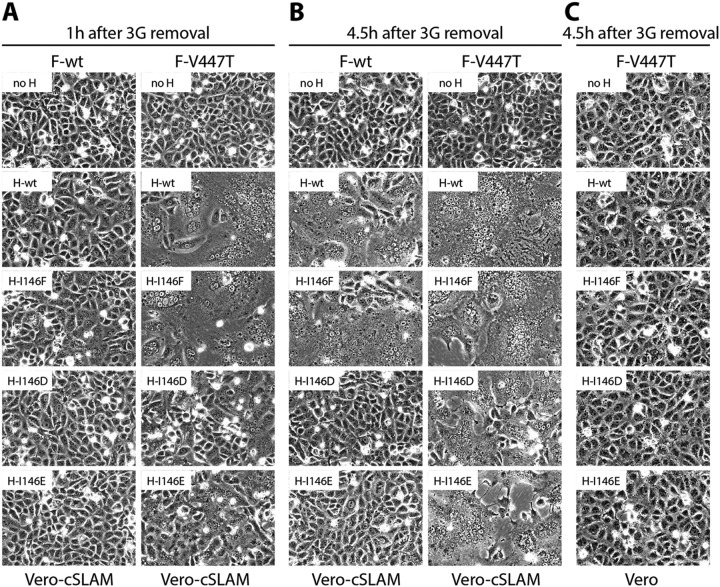
We next investigated whether CDV H mutants carrying charged residues at position 146 of the stalk domain exhibited impairments in CDV F activation as a consequence of drastic conformational differences. Therefore, we made use of a previously reported CDV F mutant (F-V447T) that was characterized by a destabilized prefusion state associated with a hyperfusogenic phenotype (37, 47). Vero-SLAM cells were thus transfected with plasmid DNA encoding H-wt or derivative mutants and combined with F-wt- or F-V447T-expressing plasmids. For better accuracy of membrane fusion assessment, cells were treated overnight with the fusion-inhibitory drug 3G to better synchronize the initiation of fusion activity. 3G is a small chemical compound inhibiting the fusion process by binding to F-trimers at low-micromolar-range concentrations. Although F-V447T exhibits resistance to 3G (47–51), treating the cells with 75 μM the drug still substantially reduced cell-to-cell fusion induction during the first hours of H/F protein expression. Of note, because accurate data acquisition was not possible using the qFA, the fusion promotion profiles of the selected H mutants, when combined with the hyperfusogenic F mutant, were recorded based on the qualitative cell-to-cell fusion assay.
While no significant fusion promotion activity could be visually observed between tested H mutant proteins (I146D and I146E) in combination with F-wt even after 4.5 h after 3G removal (Fig. 5B), cell-to-cell fusion was already visible 1 h after 3G removal using F-V447T (Fig. 5A). Fusion promotion activity induced by those H-I146 mutants nevertheless remained strongly impaired compared to that of H-wt and fully dependent on productive receptor interactions, since coexpressing H proteins and F-V447T in receptor-negative Vero cells did not result in any signs of membrane fusion triggering (Fig. 5C).
FIG 5.
Qualitative fusion assay after cotransfection of CDV H-I146 mutants along with either F-wt or F-V447T (hyperfusogenic F mutant) in receptor-negative Vero cells or receptor-positive Vero-cSLAM cells at various time points after fusion-inhibitory drug (3G) removal. 3G was removed 24 h posttransfection, and pictures were taken at the indicated time points.
These data suggested that since charged H-I146 mutants preserved some fusion promotion activity when combined with a hyperactive F variant, their overall conformation was not critically modified. The latter conclusion was further supported by the (i) proper expression of all mutants at the cell surface and (ii) their productive interaction with SLAM.
The H-I146D substitution impairs headless H-stalk bioactivity but drives the formation of a highly stable tetrameric population.
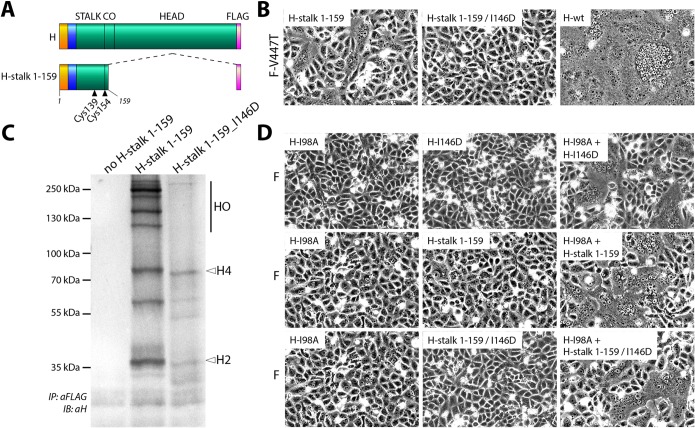
To investigate the impact of the attachment protein head domains on the functional defects recorded with the charged H-I146 mutants, we introduced the I146D substitution in a reportedly bioactive headless CDV H-stalk construct containing the linker region (Fig. 6A, H-stalk 1 to 159). Our previous work demonstrated that, only in the presence of the hyperfusogenic F-V447T mutant, various H-stalk variants could promote (to a limited extent compared to that of H-wt) fusion activity in an H-head- and receptor-independent manner, as also illustrated for headless attachment proteins of other members of the family Paramyxoviridae (34–37, 52).
FIG 6.
Bioactivity investigation of headless H-stalk 1-159 proteins harboring I146 mutation in different assays. (A) Schematic representation of the H-stalk 1-159 construct. CO, connector domain. (B) Qualitative fusion assay after cotransfection of indicated CDV H constructs along with F-V447T in receptor-positive Vero-cSLAM cells. (C) Oligomerization pattern in a nonreducing surface immunoprecipitation experiment using H-stalk 1-159 wild-type or I146D sequences. Three wells of a 6-well plate were transfected per mutant. IB, immunoblot. (D) Qualitative fusion restoration experiment with the cotransfection of various CDV H mutants that are individually fusion-triggering deficient in combination with F-wt in receptor-positive Vero-cSLAM cells. H2, H-stalk 1-159 dimer (∼36 kDa); H4, H-stalk 1-159 tetramer (∼70 kDa); HO, H-stalk 1-159 higher oligomers. Pictures were taken 30 h posttransfection.
In contrast to the CDV H-stalk 1-159 construct, the I146D variant of H-stalk (H-stalk_I146D) exhibited complete deficiency in promoting membrane fusion activity (Fig. 6B). Interestingly, when H-stalk cell surface antigenic materials were immunoprecipitated and subjected to Western blot analyses conducted under nonreducing conditions, the main detected population of the H-stalk 1-159_I146D variant migrated as tetramers (∼75 kDa; Fig. 6C) and, to some extent, dimers (∼36 kDa). Note that H-stalk 1-159_I146D is less expressed than the nonmodified parental H-stalk 1-159. The nonmodified parental H-stalk 1-159 protein exhibited the propensity to form dimers, a few trimers, tetramers, and several prominent populations of high-order oligomers (Fig. 6C). It is worth mentioning that under regular conditions, C139 and C154 are expected to form covalent bonds between two monomers involved in the identical dimeric unit, and therefore a main dimeric population should be detected.
Taken together, since the H-stalk 1-159_I146D variant lost bioactivity compared to the wild-type headless construct, an impact of the mutation on the inherent F-triggering bioactivity of the stalks was inferred. While the H-stalk (wt) version displayed an unexpected propensity to form multiple high-ordered oligomers, the introduction of the I146D mutation did stabilize a tetrameric population.
Although these data did not provide information about which H-stalk (wt) population(s) may have retained bioactivity, the fact that H-stalk 1-159_I146D mainly produced tetramers while exhibiting defective fusion promotion profiles supported the idea that high-order oligomers are F-trigger deficient. The low bioactivity recorded using F-V447T with H-stalk 1-159 compared to that of H-wt may therefore partly result from the amount of noncovalent dimer-of-dimers present at the cell surface (represented by the dimeric population in the gel).
Expressing H-I146D with the reportedly F-trigger-deficient I98A H-mutant and F-wt restores fusion promotion activity.
We next assessed whether the I146D mutation was required in all monomers of H-tetrameric structures to inhibit fusion promotion activity. To test this hypothesis, we adapted a previously described H transcomplementation assay (37). The rationale behind this approach is based on the possible restoration of H protein bioactivity when two discrete inactive mutants are combined. As expected, when H-I98A and H-I146D (two F-trigger-incompetent variants) were expressed alone with F proteins, no membrane fusion activity was recorded (Fig. 6D). In contrast, when both inactive H variants were coexpressed in the presence of F-wt, fusion promotion was readily restored (Fig. 6D). Similar phenotypes were monitored with the H-I98A/H-stalk 1-159 (H-stalk 1-159 does not promote fusion in the presence of wild-type F) and H-I98A/H-stalk 1-159_I146D combinations.
Overall, these data illustrated that all H monomers must harbor the I146D mutation in order to optimally prevent membrane fusion triggering. In turn, this inferred the possibility that I146 of different monomers locates in close proximity to one another within H oligomers. Therefore, replacement of I146 by a charged amino acid might result in repulsive electrostatic interaction and translate into a functional impact. Alternatively, I146 residues may not fold in close proximity, but the charged properties may hamper the linker domain to achieve a productive conformation. In both cases, all four linker domains must be impaired to completely inhibit H bioactivity.
Morbilliviral attachment proteins carrying fusion triggering-defective mutations at position 146 correlate with highly stable H tetramers.
MeV and CDV H tetramers are proposed to result from the assembly of two noncovalently interacting, covalent dimers (dimer-of-dimers). To determine whether fusion promotion deficiency observed with some membrane-anchored CDV and MeV H-I146 mutants correlated with modified oligomerization profiles (as observed for the H-stalk variants), we first expressed a panel of selected H mutants in Vero cells. Cell surface immunoprecipitation of H-antigenic materials then was carried out, and proteins underwent Western blot analyses under nonreducing conditions using Tris-acetate gels. As expected, full-length H-wt proteins mostly migrated as dimers (∼170 kDa). Strikingly, for both CDV and MeV H-I146 nonfunctional mutants, a tetrameric population (∼350 kDa) was readily detected (Fig. 7A and C). Quantification of band intensities clearly demonstrated that all H mutants exhibiting highly stable tetrameric populations displayed functional defects (Fig. 7B and D). Of note, CDV H-I146T and I146Y exhibited only a moderate increase of stabilized tetramers, which correlated with proper fusion promotion activity. In contrast, MeV H-I146 mutants carrying these substitutions folded into unproductive stable tetramers (Fig. 7A to D). However, our data also suggested that CDV H-wt (A75/17 strain) naturally assembles more often into stabilized H-tetramers than MeV H proteins (ICB strain).
FIG 7.
Modified oligomerization pattern of nonfunctional CDV and MeV H-I146 mutants. (A) Oligomerization pattern revealed by immunoblotting (IB) in a nonreducing surface immunoprecipitation experiment (IP) of various CDV H-I146 mutants. (B) Quantification of tetrameric and dimeric band intensities of CDV H-I146 mutants using the Aida Image Analysis software. (C) Oligomerization pattern revealed by IB in a nonreducing surface IP of various MeV H-I146 mutants. (D) Quantification of tetrameric and dimeric band intensities of MeV H-I146 mutants using the Aida Image Analysis software. H2, H dimer (∼170 kDa); H4, H tetramer (∼350 kDa); FPA, fusion promotion activity (red, nonfunctional; green, functional). For quantification shown in panels B and D, at least three independent experiments were analyzed per mutant H. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.005).
Overall, our findings provided a clear, direct correlation between highly stable H-tetramer assembly and fusion-triggering impairments.
Highly stable H tetramers do not reactivate upon dithiothreitol (DTT) treatments and are resistant to prolonged heat shock.
The monitored highly stable CDV H tetramers carrying charged residues at position 146 may have been formed as a consequence of biased disulfide bond formation between different monomers, thus leading to covalent tetramerization. This may occur in two different manners. (i) In regular H oligomers, isoleucine 146 (which may locate in close proximity to each other) could contribute to the folding of the H-linker domains into a specific conformation, leading to the assembly of productive dimer-of-dimers (Fig. 8A). Introducing a charged residue at this position may disrupt this critical linker conformation. The misfolded H-linker may then reorganize the C-terminal region of the stalk, thereby forcing C139 to switch to a monomer other than C154 for disulfide bond formation and consequently trap the tetramers (Fig. 8B). (ii) Alternatively, the linker and/or head domains may switch interaction partners during protein assembly, in turn imposing C154 to covalently link the tetramers (Fig. 8C).
FIG 8.
Investigation of H-I146 mutation-induced oligomeric changes and disulfide bond reduction experiment. (A) Model of wild-type H-linker (blue) contribution to oligomerization into dimer-of-dimers. Disulfide bonds between two natural dimers are indicated in green. Residue I146 is shown in pink. Mutation at position 146 of CDV and MeV H may force unnatural (indicated in red) disulfide bond formation to monomers belonging to another dimer via C139 (B) or C154 (C). Residue I146 mutated in either E or D is shown in orange. (D) Denaturation of potential noncovalent stable tetramers by increasing the boiling incubation time. H2, H dimer (∼170 kDa); H4, H tetramer (∼350 kDa). (E) Disulfide bond reduction experiment using DTT in combination with F-wt in receptor-positive Vero-cSLAM cells to evaluate if covalent oligomerization is the cause of fusion promotion deficiency in H-I146 mutants. H-L105C is the positive control for fusion promotion restoration after DTT treatment.
To exclude any other protein interactions forming the highly stable H tetramers rather than disulfide bonds (induced by charged substitution at position 146), we immunoprecipitated cell surface H-antigenic materials, heated the proteins for prolonged incubation (95°C, 5 to 60 min), and subjected them to Western blot analyses performed under nonreducing conditions. Remarkably, no substantial reduction of H tetramers was observed (Fig. 8D), which indeed argued for the production of covalently linked H tetramers.
We next assessed the impact of covalent H tetramerization on fusion promotion activity. To this aim, we reduced the disulfide bridges of different H-I146 mutants and determined their F-triggering capabilities when coexpressed with F-wt in Vero-SLAM cells. To inhibit premature cell-to-cell fusion induction, cells were treated with the fusion inhibitor 3G (48). One day posttransfection, 3G was removed and cells were treated with a diluted concentration of the reducing agent DTT (25 mM) for 30 min. DTT was then removed and cells were kept in Dulbecco's modified Eagle's medium (DMEM) for 4 h before recording images. Under these experimental conditions, H-wt promoted membrane fusion activity even in the absence of disulfide bonds, whereas H-L105C (a reported covalent H tetramer mutant exhibiting DTT-dependent reversible fusion-triggering inhibition [19]) exclusively displayed F-triggering capacity in treated cells (Fig. 8E). Conversely, CDV H-I146D and I146E remained nonfunctional regardless of the presence or absence of DTT in the medium (Fig. 8E).
Taken together, these data provided strong evidence that charged H-I146 mutants indeed generated covalent H-tetramers. The lack of the restoration of the fusion promotion activity of H-I146D and H-I146E upon DTT treatments, however, suggested that the inability to activate F was not directly caused by the presence of interdimer disulfide bridges. Rather, misfolding (or premature refolding) of key functional microdomains (e.g., H-linker), which indirectly may correlate with incorrect disulfide bond formation during protein assembly, would lead to the generation of F-trigger-deficient H structures.
Covalent H tetramerization is not the unique mechanism leading to functional impairments.
To determine whether charged H-I146 mutants were generating covalent tetramers based on the two cysteines involved in natural dimerization (C139 or C154), we additionally mutated one of these two residues into alanine. These C139A or C154A mutant H proteins are unable to form covalent tetramers, as they can only form one intermonomer disulfide bond. Combining a charged I146 substitution with one of those cysteine-to-alanine mutations resulted in mutant H proteins which should have been able to activate the fusion protein if covalent tetramerization was the cause of triggering deficiency. The panel of newly engineered H mutants was subjected to our various bioassays: cell surface expression, qualitative and quantitative cell-to-cell fusion promotion activity, and oligomeric propensity.
While the single H-mutants C139A and C154A preserved bioactivity, introducing the I146D substitution (generating H-C139A_I146D and H-I146D_C154A) resulted in fusion promotion-deficient H proteins despite proper intracellular trafficking (Fig. 9A and C). As expected, combining them with the hyperfusogenic F mutant only led to slight fusion promotion activity, thereby confirming their severely impaired functional profiles, as well as validating that their overall conformation was not critically changed (Fig. 9B). Interestingly, although I146D- or I146E-containing double H mutants (H-C139A_I146D/E or H-I146D/E_C154A) were unable to generate covalent tetramers, reintroducing a second cysteine at position 146 in the background of C154A only (H-I146C_C154A) readily restored the formation of a covalently linked tetrameric population (Fig. 9D and E). Conversely, absence of covalent tetramers in case of H-C139A_I146C demonstrated that, for this specific mutant, C146 and C154 trapped the identical monomers of the tetramer.
FIG 9.
Validation of phenotypes observed for C139A or C154A harboring mutant H proteins. Qualitative fusion assay after cotransfection of CDV H-I146 mutants additionally having C139 or C154 mutated into alanine along with either F-wt (A) or F-V447T (B) in receptor-positive Vero-cSLAM cells. (C) Surface expression and fusion promotion (in combination with F-wt) of various CDV H mutants relative to the wild type. Surface expression results are derived from nine independent transfections. Fusion promotion capability was determined by quadruplicate measurements of three independent transfections. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.0001). (D) Oligomeric profile of cysteine-to-alanine mutants under nonreducing, denaturing conditions. (E) Quantification of tetrameric and dimeric band intensities of CDV H-I146 mutants using the Aida Image Analysis software. Note that this gel and the one presented in Fig. 10 are identical. For clarification and better interpretation of the data, the gel was divided into two parts. The black line indicates where the image has been cropped for better presentation. For quantification, at least three independent experiments were analyzed per mutant H. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.005).
These findings highlight that the two natural cysteines (C139 and C154) were strictly required to generate covalent H tetramers in the case of inactive CDV H I146X mutants. Although all engineered mutants were unable to form covalent tetramers (apart from H-I146C_C154A), only the mutants H-C139A and H-C154 remained functional when combined with F-wt. Therefore, we conclude that the connection between fusion promotion deficiency of certain H mutants and their covalent tetramerization profile is noncausal.
Despite strong covalent tetramerization propensity, H-I146G (glycine) remains partially competent in F triggering.
Finally, we investigated whether replacing I146 with glycine, which has only a hydrogen as a side chain, resulted in similar phenotypes. Remarkably, H-I146G remained capable of triggering fusion activity to a certain extent (around 30% of the wild-type H protein), despite proper expression at the cell surface, interaction with SLAM, and, most importantly, its strong propensity to generate covalent H tetramers (Fig. 10A to D).
FIG 10.
Investigation of CDV H-I146G phenotypes. (A) Qualitative fusion assay of CDV H-I146G with F-wt in receptor-positive Vero-cSLAM cells. (B) Quantification of SLAM binding, fusion promotion activity, and surface expression of CDV H-I146G mutant proteins relative to the wild type. Surface expression results are derived from nine independent transfections, and fusion promotion capability was determined by quadruplicate measurements of three independent transfections. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.001). (C) Oligomeric patterns of CDV H-I146G under nonreducing but denaturing conditions. (D) Quantification of tetrameric and dimeric band intensities of the CDV H-I146G mutant using the Aida Image Analysis software. This gel and the one presented in Fig. 9 are identical. For clarification and better interpretation of the data, the gel was divided into two parts. For quantification, at least four independent experiments were analyzed. The means and the standard deviations from the means are shown. An unpaired two-tailed t test was performed to assess significant differences compared to the wild type (*, P value of ≤0.005).
Combined with our previous data, these findings suggested that the linker domain represents a key novel component contributing to the regulation of H bioactivity. With regard to the charged H-I146 mutants, inactivity would primarily result from unproductive linkers rather than an achieved completely dysregulated covalent H structure (although the latter may additionally contribute to the functional defects). In contrast, the specific profile of the H-I146G variant, which assumed irregular covalent structures while preserving some F-triggering activity, led us to speculate that, for this specific mutant, the linker domain may have partially preserved its functional role.
DISCUSSION
In this study, we focused our investigations on a domain of the attachment protein H of CDV and MeV, which bridges the stalk to the head domains (H-linker/connector modules) (31, 33). Based on the current model of morbillivirus cell entry, this region may play an important regulatory role (33). Indeed, it is proposed that upon receptor binding by the H-head(s), the latter may undergo structural rearrangements with respect to other heads and/or the stalk domain, which in turn allows stalk rearrangement and subsequent F activation (27, 34). In this regard, the linker/connector fragment may require some structural flexibility for these rearrangements to occur. Interestingly, recent data provided evidence that the head-to-stalk linker region of PIV5 HN proteins may indeed exhibit structural flexibility, thus pointing toward commonalities between the cell entry mechanisms of different paramyxoviruses, although the proposed models of membrane fusion triggering also illustrated some variations (26, 27, 34, 40, 41). Further supporting a potential flexible nature of the head-to-stalk connecting region, only a short domain α-helix (H1) could be resolved in crystal structures of MeV H, possibly due to poor diffraction data collected in this region (24, 25). However, complete flexibility may disturb proper folding of H proteins into F-triggering-permissive architectures, thereby spotlighting the probably critical structural nature of the morbilliviral head-to-stalk bridging region.
To further refine our knowledge on the structure/function of this specific H domain, series of mutants spanning the linker and connector modules were generated. Strikingly, among the 32 targeted residues, only two substituted isoleucines (position 140 and 146) located within the H-linker region did significantly reduce cell-to-cell fusion promotion, with most drastic impairments observed with H variants carrying nonconservative mutations at position 146. In the case of CDV H, although nonhydrophobic residues prevented H protein bioactivity, introducing amino acids with aromatic (W and Y) and/or polar (T and Y) side chains at this position did not result in major functional impact. In contrast, for MeV H, hydrophobicity was strictly required to preserve proper bioactivity, spotlighting slight differences between the two morbilliviral attachment proteins (see below). Additional work is, however, required to precisely understand this discrepancy.
Two lines of evidence supported the notion that mutating residue I146 into charged amino acids led to the formation of covalent H tetramers. First, heating at 95°C the H-I146D/E variants (even up to 1 h) did not substantially influence the dimer-to-tetramer profiles. Second, removing either C139 or C154 from H-I146D (the two natural cysteines producing covalent dimers) prevented the formation of visible tetramers. Additional important mechanistic information was obtained using H transcomplementation analyses. Based on the selected defective mutants and combinations used in our studies, it appeared that only when the fusion-promoting-deficient mutations (e.g., I146D) were present in all four monomers was full inhibition of F triggering recorded. These data supported the possibility that, in standard H tetramers, isoleucine 146 of different monomers may locate in close spatial proximity. Consequently, replacing isoleucine 146 with any charged amino acids may lead to the generation of repulsive electrostatic forces that would cause C139 and C154 to covalently link two distinct monomers, thereby forming irregular (but likely close-to-native) covalent H tetramers. Alternatively, a charged residue at position 146 may disrupt a specific conformation adopted by the H-linker, and all four domains must harbor the destabilizing substitution to display full inactivity. In any case, isoleucine 146 of H proteins appeared to act as a key factor contributing to the folding process of functional dimer-of-dimers.
The data obtained with the H-C139A_I146D and H-I146D_C154A double mutants provided key additional mechanistic information. Indeed, both proteins were properly expressed at the cell surface and did not generate covalent tetramers, but nevertheless they exhibited complete impairments in F-triggering activity. This was unexpected, since the parental single H-C139A and H-C154A mutants were also unable to form covalent tetramers but remained fully functional. Thus, it clearly appeared that covalent tetramerization was not the sole mechanism leading to fusion promotion deficiency. Rather, F-triggering activity might be lost as a consequence of the charged H-I146 mutants folding into close-to-native structures, which would lack the essential conformation of some microdomains. Remarkably, the contrasting phenotypes recorded with the neutral H-I146G variant (strong covalent tetramerization propensity but still capable of F triggering) supported this notion. Based on these observations, we concluded that the linker domain, rather than unnatural covalent H oligomerization per se, is playing a critical regulatory role in F-triggering activity.
The question about how the H-linker module of the morbilliviral attachment protein might contribute mechanistically to membrane fusion triggering can be addressed as follows. First, it appears that the H-linker is composed of a panel of hydrophobic residues potentially interacting with each other. Hence, those hydrophobic amino acids, combined with some structural flexibility, may coordinate the folding of a hydrophobic hinge region linking the 4HB stalk to α-helix H1 of the connector, which contacts the cubic head domain (Fig. 11A). It is interesting that submitting the sequence of the CDV H protein to the Robetta protein structure prediction server (http://robetta.bakerlab.org) returned best hits with the linker module displaying good propensity to assume hydrophobic hinge structures (Fig. 12). Correct folding of the hydrophobic hinge would be crucial in directing the two natural cysteines (C139 and C154) toward the same monomers to ensure correct oligomerization during protein assembly and, most importantly, achieve the autorepressed state. Second, we hypothesize that this hinge domain undergoes a receptor-induced conformational change, ultimately leading to F activation. Such structural rearrangement may allow repositioning of one (or more) head domain(s) with regard to the stalk and thus deactivates the autorepressed state. This model therefore spotlights the H-linker domain as an unrecognized essential component of the complete morbilliviral cell entry machinery (Fig. 11A and B).
FIG 11.
Potential mode of action of morbillivirus linker domain in H-mediated F-triggering activity. For simplicity, only two out of four linkers are shown. The cysteines Cys188 and Cys602 form intramonomer disulfide bonds. (A) Model of H in prereceptor binding state showing the potential hydrophobic hinge region within the linker. The hydrophobic hinge may be essential in preserving the autorepressed state prior to receptor binding. (B) Model of H in postreceptor binding state where one potential hinge region was refolded during the fusion activation signal transduction. Repositioning of H heads with regard to the stalk may in turn enable stalk destabilization and refolding within the central region (not shown). Pm, plasma membrane.
FIG 12.
Robetta tertiary structure prediction for the CDV H amino acids 129 to 607 based on the crystal structure of the measles virus hemagglutinin (PDB entry 2ZB6) by the Ginzu domain prediction method. (A) Amino acids 129 to 607 are shown. Cysteines naturally responsible for covalent dimer formation are shown as yellow sticks. (B) Predicted hydrophobic hinge region involving I146. Hydrophobic amino acids potentially involved in hydrophobic patch formation are shown as sticks.
Introducing charged amino acids at position 146 of the linker domain may thus impact proper folding of the hinge region (an interference that may be even more pronounced if I146 of different monomers were located in close proximity), thereby generating irregular covalent tetrameric H structures featuring a dysregulated autorepressed state. Alternatively, an optimal autorepressed state may have been achieved, but the charged residues may disturb the conformational changes required for F-triggering activity. Evidently, the latter mechanisms would explain our inability to reactivate the fusion promotion-deficient H-I146D/E mutants upon DTT treatments. However, although some fusion promotion activity was recorded when combined with the hyperactive F variant, we cannot yet discriminate between irregular tetramers still exhibiting some productivity or the limited population of dimer-of-dimers that nevertheless may have remained functional.
In line with our hypothesis, the slight differences in fusion promotion activities and oligomeric states induced by several H-I146 mutants (e.g., I146W/Y/T) may result from primary structure differences between CDV and MeV H-linker domains. One of the differences in this area is found at position 150; CDV H contains a phenylalanine at this position, while a tyrosine is found in MeV H. We thus speculate that the putative hydrophobic patch of CDV H would be more stable than the one found in MeV H; therefore, CDV H would better accommodate I146 substitutions with bulky aromatic (W and Y) and/or polar (T and Y) amino acids. In the case of the CDV H-I146G mutant, the linker structure may have been partially preserved. However, although the putative hydrophobic patch might be stabilized by the remaining residues, the interaction strength might be weaker, since glycine does not contribute to the stability with a hydrophobic side chain. Consequently, this would translate into the assembly of H proteins featuring partially impaired F-triggering activity. Additional investigation might also clarify which other amino acids are essential in the formation of this putative patch; one potential candidate is, for example, I140.
Finally, the strong propensity of the H-stalk 1-159 construct to form higher-ordered oligomers suggested that the H-heads also contribute to proper folding of the H-linkers. In the absence of head domains, the linker region may feature unusual flexibility. This would not only prevent folding of the hydrophobic hinge region, and subsequently impair disulfide bond formation of the cysteines toward the same two monomers, but also promote disulfide bridge formation between neighboring H tetrameric and/or dimeric stalk populations. In contrast, introducing the I146D mutation may drive the formation of an inactive covalent tetrameric population. However, the low surface expression level of H-stalk 1-159_I146D may have additionally contributed to the nonfusogenic phenotype.
In summary, we provided data suggestive for a potent dual mechanistic role of the linker module of the morbilliviral attachment protein in membrane fusion activation. On the one hand, by assuming a putative hydrophobic hinge, the linker would regulate the proper folding of productive H-dimer-of-dimers. On the other, it may undergo receptor-induced conformational changes leading to F triggering. Given that growing evidence highlights hydrophobic patches of proteins as attractive templates for drug design (53), targeting the hydrophobic H-linker microdomain may pave the way to the development of potent antimorbillivirus entry inhibitors. If the proposed dual regulatory role of the H-linker is confirmed, then small molecules stabilizing or destabilizing the hydrophobic hinge region may interfere with F activation. Additionally, stabilization of the linker region might facilitate the data acquisition of this region during X-ray diffraction or electron microscopy. If the linker were sufficiently stabilized, this may provide a useful strategy to solve the structure of the entire ectodomain of a morbilliviral attachment protein.
MATERIALS AND METHODS
Cell cultures and transfections.
Depending on the experiments, the cells used included Vero cells (ATCC CCL-81), Vero cells stably expressing canine SLAM (Vero-cSLAM; kindly provided by Yusuke Yanagi, Kyushu University, Japan), or human SLAM (Vero-hSLAM; kindly provided by Yusuke Yanagi, Kyushu University, Japan) or 293T cells (ATCC CRL-11268). Cells of every type were grown in Dulbecco's modified Eagle's medium (Gibco, Invitrogen) containing 10% fetal calf serum (FCS; BioSwissTech) and penicillin-streptomycin at 37°C and 5% CO2. Cells were transfected with TransIT-LT1 (Mirus) by following the manufacturer's instructions.
Plasmids and mutagenesis.
All mutant CDV H protein-expressing plasmids were based on the pCI plasmid containing CDV-H of the A75/17 CDV strain. Most of the CDV mutations were introduced using the QuikChange lightning site-directed mutagenesis kit (Agilent Technologies). To generate the series of MeV H mutants, we applied the same kit to the pCI plasmid expressing the MeV-H protein derived from ICB323 (kindly provided by Jürgen Schneider-Schaulies, Würzburg, Germany). The dual split protein (DSP)-expressing plasmids were kindly provided by Zene Matsuda (Tokyo, Japan) (54).
Surface expression detection.
Vero cells in a 24-well plate were transfected with 1 μg of wt or variant H protein-expressing plasmid DNA per well. Twenty-four hours posttransfection the medium was replaced by Opti-MEM containing anti-FLAG monoclonal antibody (MAb) (1:1,000; F1804; Sigma-Aldrich) and incubated at 4°C for 1 h. The wells were washed with phosphate-buffered saline (PBS) and then incubated with Opti-MEM containing 1:2,500 Alexa Fluor 488 goat anti-mouse IgG (H+L) (ThermoFisher) at 4°C for 1 h. Wells were first washed with PBS, and then PBS containing 0.5 mM EDTA was added to each well prior to maintaining the cells at 4°C until fluorescence-activated cell sorting (FACS) analysis. Cells were finally detached and subjected to flow cytometry analyses. Nine independent transfections per mutant were performed and quantified.
Qualitative fusion assay.
Vero-cSLAM (for CDV H mutants) or Vero-hSLAM (for MeV H mutants) cells in a 6-well plate were transfected with 1 μg of variant H protein-expressing plasmid and 2 μg of homotypic F protein-expressing plasmid. Pictures were taken at 11 to 24 h posttransfection depending on the combination of plasmids (syncytium formation in experiments using CDV F-V447T was much more efficient than when F-wt was used).
Quantitative fusion assay.
Vero cells in a 24-well plate were transfected with 0.5 μg of variant H protein-expressing plasmid, 1 μg of F protein-expressing plasmid, and 1 μg of the second part of the dual-split reporter-expressing plasmid (DPS8-11) per well. At the same time, Vero-cSLAM (or Vero-hSLAM when using MeV H and F) cells in a 24-well plate were transfected with 1 μg of the first half of the dual-split reporter-expressing plasmid (DSP1-7). Twenty-four hours posttransfection the Vero cells expressing the fusion machinery and DPS8-11 were detached with 80 μl trypsin before adding 235 μl DMEM and transferring wells into individual tubes (these cells are the so-called effector cells). Vero-cSLAM cells were detached with 80 μl trypsin before adding 235 μl of DMEM containing 120 μM EnduRen live cell substrate (Promega) and pooling the cells (these cells are the so-called target cells). In the next step, 235 μl of target cells was then aliquoted into the tubes containing effector cells. The mixed cells were aliquoted into a white 96-well plate in quadruplicate for each mutant. The plate was centrifuged for 5 min at 200 × g and then placed in a Cytation 5 cell imaging multimode reader (BioTek). Luminescence intensities were measured every 30 min for 18 h.
Quantification of the luminescence intensity (read-out) was carried out as follows. The highest value of luminescence detected in each well was divided by the time needed to reach that specific read-out. As a result, an indication of kinetics was gained.
Oligomerization state determination.
Vero cells in a 6-well plate were transfected with 3 μg of variant H protein-expressing plasmid DNA per well. At 24 h posttransfection the medium was replaced by Opti-MEM containing 1:2,000 anti-FLAG MAb (Sigma-Aldrich; SAB4200071) and incubated at 4°C for 1 h. Cells were washed with PBS and then incubated with lysis buffer (radioimmunoprecipitation assay [RIPA] buffer and PBS at a ratio of 1:1) containing protease inhibitor (cOmplete mix; Roche) at 4°C. No difference in the oligomeric pattern was observed when 25 mM iodoacetamide was added to the lysis buffer. Cell lysates were transferred into tubes and centrifuged at 4°C at 16,000 × g for 30 min. Supernatants were transferred into new tubes, and 15 μl of Dynabeads protein G for immunoprecipitation (Invitrogen) was added. The tubes were incubated for 4 h at 4°C on a rotating wheel before the beads were washed with PBS containing 0.05% Tween 40 (Merck). The beads were then boiled at 97°C in Laemmli buffer without reducing agent. Gel electrophoresis was performed on a NuPAGE 7% Tris-acetate protein gel (Invitrogen). A nitrocellulose membrane was used for Western blotting.
Coimmunoprecipitation.
Vero cells in a 6-well plate were transfected with 3 μg of variant H protein-expressing plasmid and 3 μg of F protein-expressing plasmid DNA per well. At 24 h posttransfection the cells were washed, and PBS containing 1 mM DTSSP (Sigma-Aldrich) was added for 2 h at 4°C. After removal of the cross-linker, PBS containing 20 mM Tris was added for 20 min at 4°C. The cells were lysed with RIPA buffer containing protease inhibitor (cOmplete mix; Roche) at 4°C. Cell lysates were transferred into tubes and centrifuged at 4°C at 16,000 × g for 30 min. Supernatants were transferred into new tubes, and 15 μl of Dynabeads protein G for immunoprecipitation (Invitrogen) was added, as well as 1:1,000 anti-H MAb 3734 (55) for immunoprecipitation of H. For immunoprecipitations of F, 15 μl of Dynabeads protein G was preincubated with 1:1,000 anti-hemagglutinin (HA) MAb (11867423001; Sigma-Aldrich) after the volume was adjusted to 1 ml using PBS according to the manufacturer's guidelines before incubation with cell lysis supernatant. The tubes were incubated for 4 h at 4°C on a rotating wheel before the beads were washed with PBS containing 0.05% Tween 40 (Merck). The beads were then boiled at 97°C in Laemmli buffer and loaded onto a 10% Tris-glycine SDS-PAGE (Invitrogen). A nitrocellulose membrane was used for Western blotting.
Receptor binding assay.
Soluble SLAM protein (HA tagged) solution was prepared as previously described (37). Vero cells in a 24-well plate were transfected with 1 μg of variant H protein-expressing plasmid DNA per well. Twenty-four h posttransfection 20× concentrated (using a 10-kDa PES centrifuge filter; Sartorius Stedim Biotech) soluble SLAM protein solution was added to each well at 1:20 and incubated for 1 h at 4°C. After washing, the medium was changed to Opti-MEM containing 1:1,000 anti-HA MAb (Lucerna Chem 901 514; Covance) and incubated at 4°C for 1 h. The wells were washed with PBS and then incubated with Opti-MEM containing 1:5,000 Alexa Fluor 488 goat anti-mouse IgG (H+L) (ThermoFisher) at 4°C of 1 h. Wells were washed with PBS before PBS containing 0.5 mM EDTA was added, and cells were stored at 4°C until fluorescence-activated cell sorting (FACS) analysis.
ACKNOWLEDGMENTS
We thank Yusuke Yanagi for kindly donating the receptor-expressing Vero cells. We are also very thankful for receiving plasmids from Jürgen Schneider-Schaulies and Zene Matsuda. We thank Michel Koch and Francesco Origgi for having critically reviewed the manuscript.
This work was supported by the University of Bern, the Swiss National Science Foundation (ref. no. 310030_173185 to P.P.), and the Bangerter-Rhyner Foundation (to P.P.).
REFERENCES
- 1.Simons E, Ferrari M, Fricks J, Wannemuehler K, Anand A, Burton A, Strebel P. 2012. Assessment of the 2010 global measles mortality reduction goal: results from a model of surveillance data. Lancet 379:2173–2178. doi: 10.1016/S0140-6736(12)60522-4. [DOI] [PubMed] [Google Scholar]
- 2.Plemper RK, Hammond AL. 2014. Synergizing vaccinations with therapeutics for measles eradication. Expert Opin Drug Discov 9:201–214. doi: 10.1517/17460441.2014.867324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai K, Nagata N, Ami Y, Seki F, Suzaki Y, Iwata-Yoshikawa N, Suzuki T, Fukushi S, Mizutani T, Yoshikawa T, Otsuki N, Kurane I, Komase K, Yamaguchi R, Hasegawa H, Saijo M, Takeda M, Morikawa S. 2013. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J Virol 87:1105–1114. doi: 10.1128/JVI.02419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roelke-Parker ME, Munson L, Packer C, Kock R, Cleaveland S, Carpenter M, O'Brien SJ, Pospischil A, Hofmann-Lehmann R, Lutz H, Mwamengele GL, Mgasa MN, Machange GA, Summers BA, Appel MJ. 1996. A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature 379:441–445. doi: 10.1038/379441a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol 66:4564–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu XL, Ray R, Compans RW. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol 66:1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. 2009. Probing the spatial organization of measles virus fusion complexes. J Virol 83:10480–10493. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brindley MA, Chaudhury S, Plemper RK. 2015. Measles virus glycoprotein complexes preassemble intracellularly and relax during transport to the cell surface in preparation for fusion. J Virol 89:1230–1241. doi: 10.1128/JVI.02754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496. In Fields B, Knipe DM, Howley PM (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Russell CJ, Jardetzky TS, Lamb RA. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J 20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brindley MA, Plattet P, Plemper RK. 2014. Efficient replication of a paramyxovirus independent of full zippering of the fusion protein six-helix bundle domain. Proc Natl Acad Sci U S A 111:E3795–E3804. doi: 10.1073/pnas.1403609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delpeut S, Rudd PA, Labonte P, von Messling V. 2012. Membrane fusion-mediated autophagy induction enhances morbillivirus cell-to-cell spread. J Virol 86:8527–8535. doi: 10.1128/JVI.00807-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiener D, Plattet P, Cherpillod P, Zipperle L, Doherr MG, Vandevelde M, Zurbriggen A. 2007. Synergistic inhibition in cell-cell fusion mediated by the matrix and nucleocapsid protein of canine distemper virus. Virus Res 129:145–154. doi: 10.1016/j.virusres.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Sawatsky B, von Messling V. 2010. Canine distemper viruses expressing a hemagglutinin without N-glycans lose virulence but retain immunosuppression. J Virol 84:2753–2761. doi: 10.1128/JVI.01813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plemper RK, Hammond AL, Cattaneo R. 2000. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J Virol 74:6485–6493. doi: 10.1128/JVI.74.14.6485-6493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem 283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navaratnarajah CK, Kumar S, Generous A, Apte-Sengupta S, Mateo M, Cattaneo R. 2014. The measles virus hemagglutinin stalk: structures and functions of the central fusion activation and membrane-proximal segments. J Virol 88:6158–6167. doi: 10.1128/JVI.02846-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JP, Orvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2012. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem 287:16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alkhatib G, Briedis D. 1986. The predicted primary structure of the measles virus hemagglutinin. Virology 150:479–490. doi: 10.1016/0042-6822(86)90312-0. [DOI] [PubMed] [Google Scholar]
- 21.Santiago C, Celma ML, Stehle T, Casasnovas JM. 2010. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol 17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 22.Colf LA, Juo ZS, Garcia KC. 2007. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol 14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 23.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol 18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 24.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A 104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. 2013. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol 20:67–72. doi: 10.1038/nsmb.2432. [DOI] [PubMed] [Google Scholar]
- 26.Bose S, Jardetzky TS, Lamb RA. 2015. Timing is everything: fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479-480C:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plattet P, Alves L, Herren M, Aguilar HC. 2016. Measles virus fusion protein: structure, function and inhibition. Viruses 8:112. doi: 10.3390/v8040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol 85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. 2013. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog 9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A 108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navaratnarajah CK, Negi S, Braun W, Cattaneo R. 2012. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J Biol Chem 287:38543–38551. doi: 10.1074/jbc.M112.410563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brindley MA, Takeda M, Plattet P, Plemper RK. 2012. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci U S A 109:E3018–E3027. doi: 10.1073/pnas.1210925109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navaratnarajah CK, Rosemarie Q, Cattaneo R. 2016. A structurally unresolved head segment of defined length favors proper measles virus hemagglutinin tetramerization and efficient membrane fusion triggering. J Virol 90:68–75. doi: 10.1128/JVI.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A 109:E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bose S, Song AS, Jardetzky TS, Lamb RA. 2014. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol 88:3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol 87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ader-Ebert N, Khosravi M, Herren M, Avila M, Alves L, Bringolf F, Orvell C, Langedijk JP, Zurbriggen A, Plemper RK, Plattet P. 2015. Sequential conformational changes in the morbillivirus attachment protein initiate the membrane fusion process. PLoS Pathog 11:e1004880. doi: 10.1371/journal.ppat.1004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. 2011. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol 18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adu-Gyamfi E, Kim LS, Jardetzky TS, Lamb RA. 2016. Flexibility of the head-stalk linker domain of paramyxovirus HN glycoprotein is essential for triggering virus fusion. J Virol 90:9172–9181. doi: 10.1128/JVI.01187-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. 2009. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol 83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plemper RK, Hammond AL, Cattaneo R. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem 276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 42.Apte-Sengupta S, Navaratnarajah CK, Cattaneo R. 2013. Hydrophobic and charged residues in the central segment of the measles virus hemagglutinin stalk mediate transmission of the fusion-triggering signal. J Virol 87:10401–10404. doi: 10.1128/JVI.01547-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu C, Zhang P, Liu X, Qi Y, Zou T, Xu Q. 2004. Characterization of a region involved in binding of measles virus H protein and its receptor SLAM (CD150). Biochem Biophys Res Commun 316:698–704. doi: 10.1016/j.bbrc.2004.02.106. [DOI] [PubMed] [Google Scholar]
- 44.Lin LT, Richardson CD. 2016. The host cell receptors for measles virus and their interaction with the viral hemagglutinin (H) protein. Viruses 8:250. doi: 10.3390/v8090250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navaratnarajah CK, Vongpunsawad S, Oezguen N, Stehle T, Braun W, Hashiguchi T, Maenaka K, Yanagi Y, Cattaneo R. 2008. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150). J Biol Chem 283:11763–11771. doi: 10.1074/jbc.M800896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plemper RK, Brindley MA, Iorio RM. 2011. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog 7:e1002058. doi: 10.1371/journal.ppat.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avila M, Alves L, Khosravi M, Ader-Ebert N, Origgi F, Schneider-Schaulies J, Zurbriggen A, Plemper RK, Plattet P. 2014. Molecular determinants defining the triggering range of prefusion F complexes of canine distemper virus. J Virol 88:2951–2966. doi: 10.1128/JVI.03123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singethan K, Hiltensperger G, Kendl S, Wohlfahrt J, Plattet P, Holzgrabe U, Schneider-Schaulies J. 2010. N-(3-Cyanophenyl)-2-phenylacetamide, an effective inhibitor of morbillivirus-induced membrane fusion with low cytotoxicity. J Gen Virol 91:2762–2772. doi: 10.1099/vir.0.025650-0. [DOI] [PubMed] [Google Scholar]
- 49.Ader N, Brindley M, Avila M, Orvell C, Horvat B, Hiltensperger G, Schneider-Schaulies J, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2013. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J Virol 87:314–326. doi: 10.1128/JVI.01826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plemper RK, Erlandson KJ, Lakdawala AS, Sun A, Prussia A, Boonsombat J, Aki-Sener E, Yalcin I, Yildiz I, Temiz-Arpaci O, Tekiner B, Liotta DC, Snyder JP, Compans RW. 2004. A target site for template-based design of measles virus entry inhibitors. Proc Natl Acad Sci U S A 101:5628–5633. doi: 10.1073/pnas.0308520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun A, Prussia A, Zhan W, Murray EE, Doyle J, Cheng LT, Yoon JJ, Radchenko EV, Palyulin VA, Compans RW, Liotta DC, Plemper RK, Snyder JP. 2006. Nonpeptide inhibitors of measles virus entry. J Med Chem 49:5080–5092. doi: 10.1021/jm0602559. [DOI] [PubMed] [Google Scholar]
- 52.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. 2013. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog 9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fosgerau K, Hoffmann T. 2015. Peptide therapeutics: current status and future directions. Drug Discov Today 20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Ishikawa H, Meng F, Kondo N, Iwamoto A, Matsuda Z. 2012. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng Des Sel 25:813–820. doi: 10.1093/protein/gzs051. [DOI] [PubMed] [Google Scholar]
- 55.Orvell C, Sheshberadaran H, Norrby E. 1985. Preparation and characterization of monoclonal antibodies directed against four structural components of canine distemper virus. J Gen Virol 66(Part 3):443–456. doi: 10.1099/0022-1317-66-3-443. [DOI] [PubMed] [Google Scholar]