Extracellular vesicles (EVs) are released by all types of cells as they constitute major mechanism of intercellular communication and have the capacity to alter the functions of recipient cells despite their limited capacity for cargo. How the EVs released by HSV-infected cells could alter the surrounding microenvironment and influence the infection currently remains unknown. The cargo of EVs reflects the physiological state of the cells in which they were produced, so the content of EVs originating from infected cells is expected to be substantially different from that of healthy cells. Our studies indicate that the EVs released by HSV-1-infected cells carry innate immune components such as STING and other host and viral factors; they can activate innate immune responses in recipient cells and inhibit HSV-1 replication. The implication of these data is that the EVs released by HSV-1-infected cells could control HSV-1 dissemination promoting its persistence in the host.
KEYWORDS: extracellular vesicles, innate immunity, STING, herpes simplex virus, macrophages
ABSTRACT
Herpes simplex virus 1 (HSV-1)-infected cells release extracellular vesicles (EVs) that deliver to uninfected cells viral factors and host components, such as the stimulator of interferon genes (STING), which activates type I interferon upon foreign DNA sensing. The functions of EVs released by HSV-1-infected cells have remained unknown. Here, we describe a procedure to separate the EVs from HSV-1 virions that is based on an iodixanol/sucrose gradient. STING, along with the EV markers CD63 and CD9, was found in light-density fractions, while HSV components accumulated in heavy-density fractions. HSV-1 infection stimulated the release of EVs from the cells. The EVs derived from infected cells, but not from uninfected cells, activated innate immunity in recipient cells and suppressed viral gene expression and virus replication. Moreover, only the EVs derived from infected cells stimulated the expression of a subset of M1-type markers in recipient macrophages. Conversely, EVs derived from STING-knockdown cells failed to stimulate the expression of these M1-type markers, they activated innate immune responses to a lesser extent in recipient cells, and they did not sustain the inhibition of virus replication. These data suggest that STING from the EV donor cells contributes to the antiviral responses in cells receiving EVs from HSV-1-infected cells. Perturbations in the biogenesis of EVs by silencing CD63 or blocking the activity of the neutral spingomyelinase-2 (nSMase-2) increased the HSV-1 yields. Overall, our data suggest that the EVs released from HSV-1-infected cells negatively impact the infection and could control the dissemination of the virus.
IMPORTANCE Extracellular vesicles (EVs) are released by all types of cells as they constitute major mechanism of intercellular communication and have the capacity to alter the functions of recipient cells despite their limited capacity for cargo. How the EVs released by HSV-infected cells could alter the surrounding microenvironment and influence the infection currently remains unknown. The cargo of EVs reflects the physiological state of the cells in which they were produced, so the content of EVs originating from infected cells is expected to be substantially different from that of healthy cells. Our studies indicate that the EVs released by HSV-1-infected cells carry innate immune components such as STING and other host and viral factors; they can activate innate immune responses in recipient cells and inhibit HSV-1 replication. The implication of these data is that the EVs released by HSV-1-infected cells could control HSV-1 dissemination promoting its persistence in the host.
INTRODUCTION
Herpes simplex virus (HSV) infections afflict 80% of the population worldwide (1). Following primary infection and lytic replication in mucosal epithelial cells, HSV-1 enters into sensory neurons that innervate the infected tissues and is transported to the neuronal cell bodies, where a lifelong latent infection is established (1). Periodically, the virus is reactivated in neurons, and anterograde transport of the virus results in its release at or near the portal of entry into the body. The consequences of reactivation range from benign cold sores to fatal encephalitis (1).
The extracellular vesicles (EVs) constitute major mechanism of intercellular communication especially on occasions cellular mobility is limited, such as in neurons where HSV-1 establishes its latent reservoir (2–7). There are three major types of EVs according to their size and origin: the microvesicles (100 to 1,000 nm) that originate from the plasma membrane by outward budding and fission, the exosomes (50 to 150 nm) that originate from endosomes by inward budding and formation of intraluminal vesicles inside multivesicular bodies, and the apoptotic bodies (1.000 to 5.000 nm) that originate from cells undergoing apoptosis (2, 3). The cargo of EVs is composed of nucleic acids and proteins that are mainly cytosolic, and EVs are also enriched in select lipids such as sphingomyelin cholesterol and ceramide (3, 4, 8–10).
A wealth of literature is focused on mechanisms by which HSV combats antiviral responses inside the cells in which it replicates. How the virus influences the messages delivered to uninfected cells through EVs to alter the microenvironment of the infection has become the subject of recent investigation. Previously, we reported that STING is packaged in EVs during HSV-1 infection, and we hypothesized that it could be delivered to uninfected cells to control the dissemination of the virus (6, 11). STING being delivered into the recipient cells is not degraded (6, 11). In addition to STING, its ligand the 2′3′-cGAMP was found inside nonviral particles following exposure of cells to large DNA viruses, such as human cytomegalovirus (12). One hypothesis is that during HSV-1 infection factors exocytosed by infected cells prime uninfected recipient cells to combat the infection. In addition to STING, several HSV-1 miRNAs such as miR-H3, miR-H5, and miR-H6, as well as viral transcripts, were found inside EVs (6, 11). Recently, two more viral miRNAs, miR-H28 and miR-H29, whose transient expression in cell cultures blocks virus replication, were also found inside EVs (13). The role of these viral products delivered within EVs remains currently elusive (6, 11). In another study, analysis of the secretome of HSV-1-infected macrophages revealed that multiple antiviral factors are released with EVs upon infection (14). Finally, the viral glycoprotein B perturbs the major histocompatibility complex class II trafficking in the endocytic pathway and promotes sorting of HLA-DR receptors to exosomes instead of the cell surface (15). Maybe this is part of the strategy of the virus to evade the antigen presentation process.
In addition to HSV-1, other viruses hijack or modulate EV pathways (16, 17). Small RNA viruses such as HIV-1 could be packaged inside EVs and delivered to recipient cells escaping immune surveillance (18–22). Other viruses package their genomes inside EVs and enter nonpermissive cells using noncanonical entry mechanisms (e.g., hepatitis C virus) (23–27). The EVs could also contribute to pathogenesis. For example, EVs released from Epstein-Barr virus-associated nasopharyngeal carcinoma carry the viral oncogene latent membrane protein 1 (LMP1) and manipulate the tumor microenvironment to enhance tumor progression while suppressing immune responses in tumor cells. Similarly, the EVs produced from lymphomas positive for Kaposi's sarcoma-associated herpesvirus appear to exacerbate disease progression and pathogenesis (28–33).
A major obstacle studying the EVs during a productive viral infection is that the sizes of EVs and virions generally coincide; thus, their separation becomes challenging. In this study, we describe a procedure to separate EVs from HSV-1 virions that relies on their density differences. The EVs isolated from infected cells, but not from uninfected cells, activated innate immunity gene transcription in recipient cells and blocked virus replication. In addition, EVs derived from infected cells, but not from uninfected cells, promoted the expression of a subset of M1 markers in recipient macrophages but none of the M2 markers tested. In contrast, EVs derived from STING knockdown cells failed to activate innate immunity gene expression in recipient cells to the same extent and failed to sustain the inhibition of virus replication. Changes in the quality of EVs by depleting the tetraspanin CD63 or blocking the activity of nSMase-2 increased HSV-1 virus yields. Taken together, our data suggest that the EVs released by infected cells can control HSV infection and dissemination.
RESULTS
Separation of extracellular vesicles from HSV-1 virions.
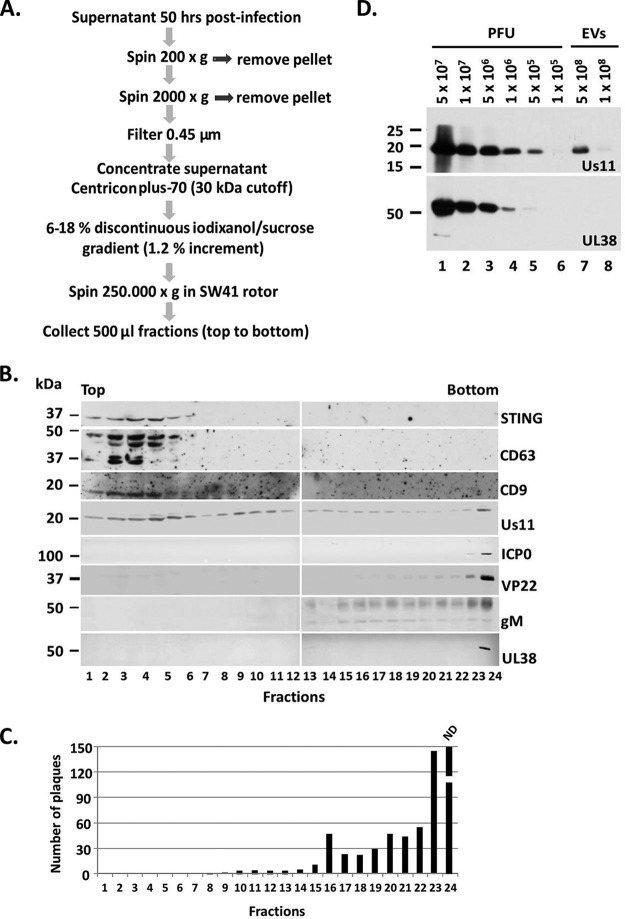
Previously, we demonstrated that during HSV-1 infection the DNA sensor STING is packaged in EVs and is released from the infected cells (6, 11). The study of EVs released by infected cells has been challenging, since EVs and virions copurify in many density gradients, including Dextran-T10 (6, 11). To separate the EVs from HSV virions, we developed a procedure based on their density differences. For this, HEp-2 cells were exposed to HSV-1(F) (0.1 PFU/cell), and the supernatant was collected at 50 h postinfection. The supernatant was subjected to two rounds of low-speed centrifugation to remove cell debris and nuclei and then filtered through 0.45-μm-pore size filters as shown in Fig. 1A. The concentration of the supernatant was performed by centrifugation through 30-kDa cutoff filters. An iodixanol/sucrose discontinuous gradient with iodixanol concentration ranging between 6 and 18% was prepared. The gradient was almost 12 ml in total, with increments of 1.2%. The concentrated supernatant was loaded on the top of the gradient and subjected to high-speed centrifugation, as shown in Fig. 1A. Fractions (500 μl) were collected from the top to the bottom of the gradient and analyzed by immunoblot analysis to identify the fractions containing the EVs and by plaque assays to verify that infectious viral particles were excluded from the fractions with the EVs. The results are summarized as follows. Markers of EVs such as CD63 and CD9 were concentrated in light-density fractions, within the first six fractions of the gradient (Fig. 1B). STING, which was previously found to coprecipitate with the tetraspanin CD9 from semipurified virions, was present in the same fractions as the marker of EVs (Fig. 1B) (11). HSV structural components, such as the capsid protein UL38, the envelope protein gM, and the tegument proteins VP22 and ICP0, accumulated in high-density fractions and were absent from the fractions containing the markers of EVs (Fig. 1B). The tegument protein Us11 was reproducibly found in the fractions containing the markers of EVs, besides the fractions with the virions, suggesting that it could be packaged in the EVs (Fig. 1B). All of the fractions were tested for the presence of infectious viral particles by plaque assays in Vero cells and, consistent with the immunoblot analysis, we found that the infectious particles accumulated in the high-density fractions (Fig. 1C) and started appearing in the gradient between the fractions 8 and 9 (Fig. 1C).
FIG 1.
Separation of EVs from HSV virions. (A) Procedures to separate EVs from HSV-1 virions. (B) The proteins present in the fractions collected from procedures in panel A were separated in denaturing polyacrylamide gels, transferred to nitrocellulose sheets, and immunoblotted with the mouse monoclonal antibodies to STING, Us11, and the rabbit polyclonal antibodies for CD63, CD9, ICP0, VP22, gM, and UL38. (C) The fractions presented in panel B were incubated with Vero cells in a virus plaque assay. The cells exposed to the bottom fraction of the gradient (fraction number 24) were fully infected so plaques could not be determined. ND, not determined. (D) Assessment of the detection limit of the Us11 and UL38 antibodies by immunoblot analysis using different amounts of HSV-1 infectious particles. The viral protein Us11, but not UL38, was found in the EV fractions derived from infected cells.
To ensure that the virus present in the fractions with the CD63-positive EVs was negligible, we performed two additional experiments. In the first, we assessed the infectious virus present in EVs using plaque assays, and we found that 5 × 108 EVs carry as a contaminant approximately 200 infectious particles. We also quantified the viral genome present in 5 × 108 EVs, and we found approximately 2,000 copies. In the second experiment, we assessed the sensitivity of our antibodies against viral proteins that appear to be packaged into EVs. Using the Us11 antibody, we could detect the amount of Us11 protein present in 5 × 105 infectious particles but not in 1 × 105 particles (Fig. 1D). For every infectious particles, we calculated roughly ten times more noninfectious particles. Notably, an adequate amount of Us11 protein was detected in 5 × 108 EVs (Fig. 1D), although a trace amount of virus was present. In a similar way, we analyzed the cutoff for detection of the capsid protein UL38 using a UL38 antibody. We found that the UL38 protein was detectable in 106 infectious particles but undetectable in EVs (Fig. 1D). Other viral proteins tested were absent from the EVs (Fig. 1B). We conclude that separation of EVs from HSV virions is feasible through a 6 to 18% discontinuous iodixanol/sucrose gradient. Our data also indicate that the tegument protein Us11 is packaged in the EVs released by infected cells.
Properties of the extracellular vesicles released from HSV-1-infected cells.
We chose to perform the rest of our studies in human immortalized embryonic fibroblasts (HEL cells) since they are immunocompetent and the STING-dependent pathway is intact (34). To characterize the EVs released during HSV-1 infection, 108 HEL cells were exposed to HSV-1(F) (0.1 PFU/cell) for 50 h prior to the isolation of EVs, as indicated in Fig. 1. The fractions collected after separating the EVs from the virions were analyzed under an electron microscope following negative staining. A representative image of the EVs present in the first six fractions of the gradient is depicted in Fig. 2A (upper panel), while representative viral particles present in the heavier fractions are shown in Fig. 2A (lower panel). The number of EVs isolated from HEL cells at 50 h postinfection and a distribution of their size were determined using nanoparticle tracking analysis (NTA) in samples from three independent isolations of EVs. The infected cells displayed a 2-fold increase in the total number of EVs compared to uninfected cells (Fig. 2B). Particularly, uninfected or infected HEL cells (1 × 108) at 50 h after a medium change had in their supernatants approximately 1 × 1010 and 2 × 1010 EVs, respectively, in total. In addition, the EVs from infected cells displayed a shift toward larger sizes compared to the EVs from uninfected cells (Fig. 2C and D). Thus, approximately 20% fewer EVs from 50 to 150 nm were present in the supernatants of the infected cells compared to uninfected cells, with a concomitant increase in the number of EVs larger than 150 nm (Fig. 2C and D). We also found that, on average, the EVs derived from infected cells were 35 nm larger than those from uninfected cells (Fig. 2E). Notably, filtration through 0.45-μm-pore size filters that our protocol utilizes leads to the loss of larger vesicles and that could confound the results. Another confounding factor is that we focused only on those EVs that are separable from HSV-1 virions through the particular gradient. This does not exclude the possibility of the existence of different types of EVs that are inseparable from the HSV-1 virions and are negative for the EV markers tested. Thus, the measurements shown in Fig. 2 represent only EVs lighter than the HSV-1 virions with a size smaller than 0.45 μm. We conclude that different types of EVs are released by infected and uninfected cells which, based on their sizes, correspond primarily to exosomes and microvesicles. Overall, the HSV-1-infected cells release more and larger EVs than the uninfected cells.
FIG 2.
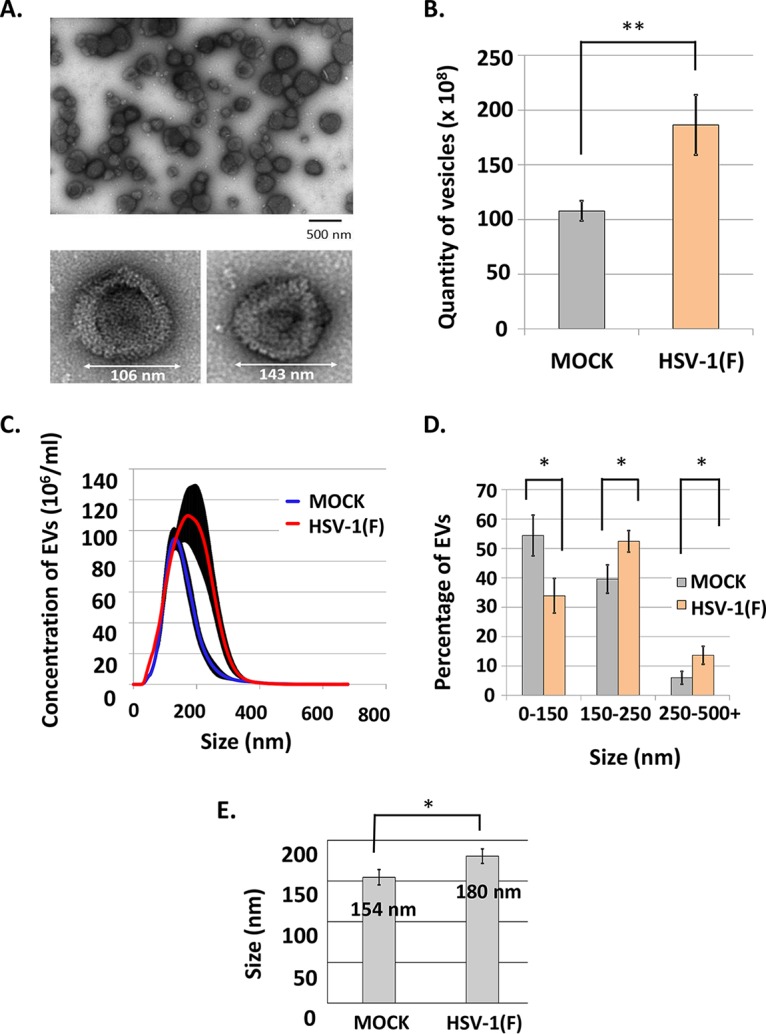
Characterization of the EVs released by HSV-1-infected cells. (A) EVs (upper panel) or virions (lower panel) were derived from HSV-1(F)-infected HEL cells following procedures as in Fig. 1, negatively stained, and analyzed by a JEOL JEM-1400 transmission electron microscope equipped with a Lab6 gun. (B) Quantity of the EVs derived from uninfected and infected HEL cells (108). The P value (P = 0.0089) was calculated using a standard two-tailed unpaired Student t test analysis, as detailed in Materials and Methods. (C) Concentration and size distribution of the EVs from panel B. (D) Size distribution of the EVs from panel B. All data were acquired using the NanoSight LM10 and analyzed with software provided by the supplier. Results represent the means of three independent isolations of EVs. The P values for the differences observed in the size distribution of EVs released from infected versus uninfected cells were calculated as described above: P = 0.018 for EV sizes 0 to 150 nm, P = 0.025 for EV sizes 150 to 250 nm, and P = 0.024 for EV sizes >250 nm. (E) Average size of the EVs released from infected (180 nm) versus uninfected (154 nm) cells following NTA (P = 0.0256).
Perturbation in the biogenesis of EVs enhances HSV-1 virus growth.
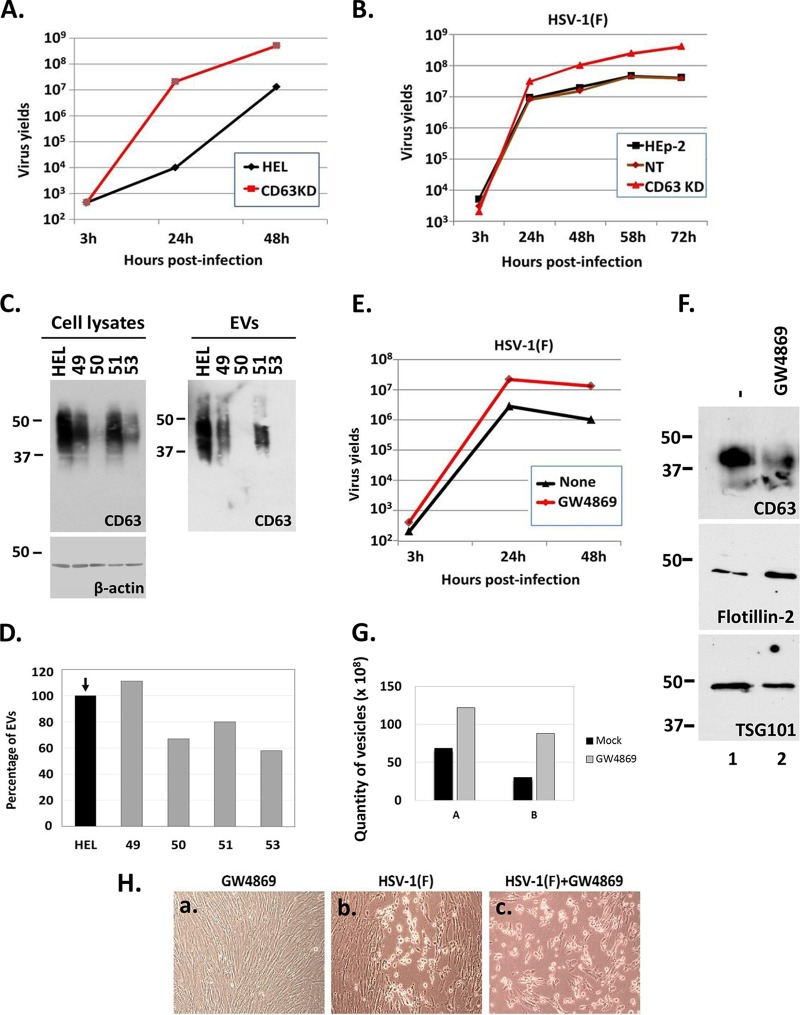
In this series of experiments, we investigated whether disruption of the biogenesis of EVs affects HSV-1 growth. The tetraspanin CD63 and the nSMase-2 are both involved in the biogenesis of EVs, albeit by different mechanisms (35–37). CD63 has a major role in sorting cargo to the EVs, while nSMase-2 hydrolyzes sphingomyelin to produce ceramide, which is an abundant membrane component of the EVs (35–37). First, we developed two CD63 knockdown cell lines with the aid of lentiviral vectors. The first cell line was based on the HEL fibroblasts, and the second cell line was based on human cancer epithelial cells (HEp-2). The efficiency of CD63 knockdown in HEL cells using different shRNAs (i.e., shRNAs 49, 50, 51, and 53) is depicted in Fig. 3C (left panel), and the efficiency of depletion of the CD63-positive population of EVs is depicted in Fig. 3C (right panel). We chose the HEL and HEp-2 cell lines silenced with the CD63 shRNA 50 to perform the next experiments. The HEL and HEp-2 cell lines and their CD63 knockdown derivatives were exposed to HSV-1(F) (0.01 PFU/cell) and harvested 3, 24, 48, 58, and 72 h after infection; titrations were then performed in Vero cells. We found that in CD63 knockdown HEL cells the wild-type virus yields were >20-fold higher at 24 and 48 h postinfection compared to the parental HEL cells (Fig. 3A). Similarly, the yields of wild-type virus in CD63-knockdown HEp-2 cells were at least 10-fold higher than in parental cells or control shRNA-treated cells at 48, 58, and 72 h postinfection (Fig. 3B). The smaller differences observed in the cancer cells HEp-2 versus the immortalized HEL cells could be due to the fact that innate immunity in HEp-2 cells is partially impaired, as discussed previously (38). The CD63 knockdown cell lines with the most efficient knockdown of CD63 displayed a 20 to 40% reduction in the total amount of the released EVs, as shown in Fig. 3D. We conclude that knockdown of CD63 enhances HSV-1 virus yields.
FIG 3.
Perturbations in the biogenesis of EVs released by infected cells enhance HSV virus yields. (A and B) HEL cells, HEp-2 cells, their CD63 knockdown derivatives, and nontargeted shRNA-treated cells (NT) were exposed to HSV-1(F) at 0.01 PFU/cell. The cells were harvested at 3, 24, 48, 58, and 72 h postinfection, and titrations were performed in Vero cells. (C) Development of CD63-knockdown HEL cells with the aid of lentiviral vectors carrying different shRNAs (49, 50, 51, 53) and amounts of CD63 present in equal numbers of EVs derived from these cell lines. (D) Quantification of the EVs released by the different CD63-knockdown clones compared to the parental cells using NTA. Results are presented as a percentage compared to the parental HEL cells. (E) HEL cells were infected with HSV-1(F) at 0.01 PFU/cell in the presence or absence of GW4869 (20 μM). The cells were harvested at 3, 24, and 48 h after infection, and titration of progeny viruses was performed in Vero cells. (F) HEL cells were either treated with GW4869 (20 μM) or remain untreated. The supernatant of the cells was harvested at 24 h posttreatment, and EVs were isolated as described for Fig. 1A. Equal numbers of EVs were analyzed by immunoblot analysis using antibodies against CD63, Flotillin-2, and TSG101. (G) Quantification of EVs from panel E was done by NTA. Results from two independent experiments (panels A and B) are depicted. (H) HEL cells were infected with HSV-1(F) at 0.01 PFU/cell. GW4869 (20 μM) was added to the cells the moment of infection. Images were captured at 24 h after infection using a Nikon Eclipse TE2000-S microscope equipped with a Nikon DS-Fi1 camera.
In the second assay, HEL cells were treated with the nSMase-2 inhibitor GW4869 (20 μM) added to the cultures simultaneously with HSV-1(F) (0.01 PFU/cell). The cells were harvested at 3, 24, and 48 h after infection, and titrations were done in Vero cells. As shown in Fig. 3E, the wild-type virus yields were 10-fold higher in the GW4869-treated cells than in untreated cells at 24 and 48 h postinfection. To verify that the treatment with GW4869 perturbs the biogenesis of EVs, HEL cells were either not treated or treated with GW4869 (20 μM) for 48 h, and the EVs were isolated as described in Fig. 1 and quantified using NTA. The results from two independent experiments (experiments A and B) are presented in Fig. 3G and demonstrate that treatment with GW4869 causes the release of twice the amount of vesicles compared to the untreated cells. To investigate whether the treatment with GW4869 causes quality changes to the EVs, we analyzed known markers of EVs in equal number of vesicles. As shown in Fig. 3F, in the presence of the nSMase-2 inhibitor the amounts of CD63-positive and TSG101-positive EVs decreased, while the amounts of Flotillin-2 -positive EVs increased, and no toxicity was noticed during the time frame of drug treatment of the cells (Fig. 3H, subpanel a). Notably, an accelerated infection was observed in cells treated with the GW4869 drug compared to the untreated cells (Fig. 3H, compare subpanel c to subpanel b). We conclude that perturbations in the biogenesis of EVs by knocking down CD63 or blocking the activity of the nSMase-2 result in increased HSV-1 virus yields.
The EVs released by infected cells activated innate immune responses in recipient cells in a dose-dependent manner.
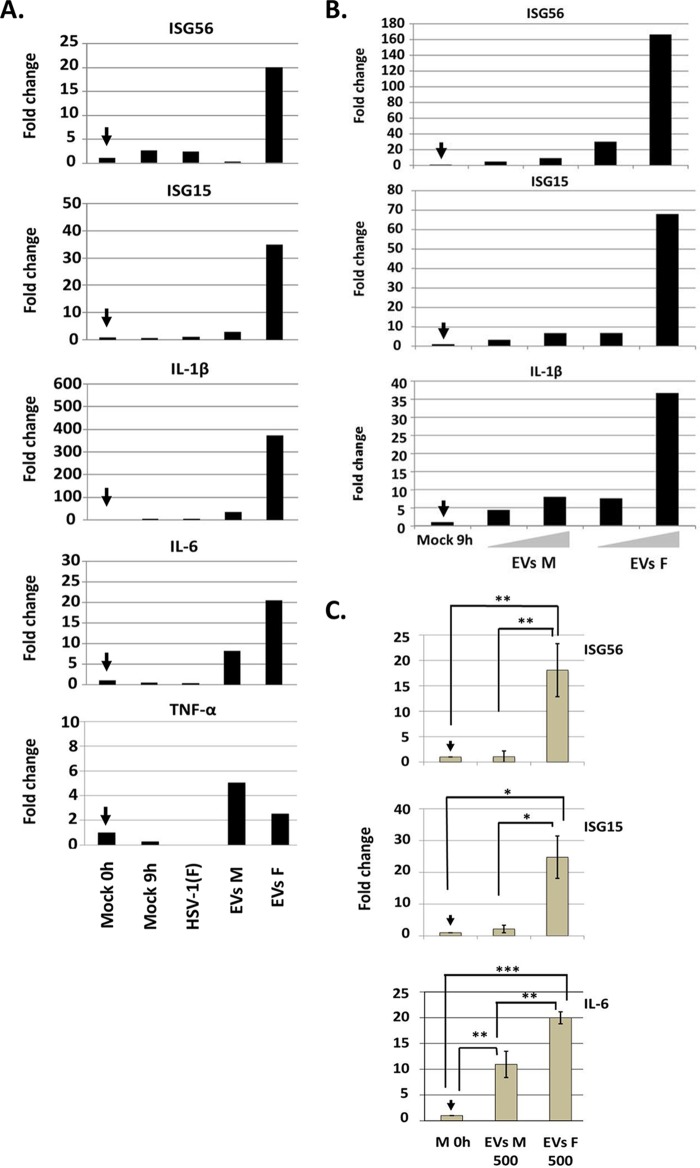
We assessed the ability of the EVs released by infected cells to activate innate immune responses in recipient cells. The EVs were purified from HEL cells that were exposed to HSV-1(F) (0.1 PFU/cell) or remained untreated, as described in Fig. 1A. Replicate cultures of HEL cells seeded in 6-well plates were exposed to EVs corresponding to 10 μg of proteins in cargo and harvested at 8 h postexposure. The results (Fig. 4A) suggest that EVs derived from HSV-1(F)-infected cells activated interferon-stimulated gene transcription, such as ISG56 and ISG15, and inflammatory gene transcription, such interleukin-1β (IL-1β) and IL-6. In contrast, equal amounts of proteins from EVs derived from uninfected cells did not activate innate immunity or inflammation. An exception was the expression of tumor necrosis factor alpha (TNF-α) that was stimulated by EVs derived from uninfected or infected cells at similar but low levels. Infection with the wild-type virus at a low multiplicity of infection (0.1 PFU/cell) did not activate innate immune responses or inflammation (Fig. 4A). We conclude that EVs released by infected cells trigger antiviral responses in recipient cells.
FIG 4.
EVs derived from HSV-1(F)-infected cells activated antiviral responses in recipient cells in a dose-dependent manner. (A) EVs were isolated from HEL cells exposed to HSV-1(F) at 0.1 PFU/cell, following procedures described in Fig. 1A. EVs corresponding to 10 μg of proteins in cargo were added to cells seeded in 6-well plates. All samples were harvested 8 h after the addition of EVs, total RNA was extracted and converted to cDNA, and quantification of the ISG56, ISG15, IL-1β, IL-6, and TNF-α transcripts was done by real-time PCR analysis. 18S rRNA was used for normalization. The fold change is relative to the amount of transcripts present in HEL cells the moment of addition of the EVs (arrow). (B) HEL cells seeded in 6-well plates were exposed to two different doses of EVs derived from uninfected or HSV-1(F)-infected cells that corresponded to 7.5 and 15 μg of proteins, respectively. The cells were harvested at 8 h postexposure to the EVs, and quantification of the ISG56, ISG15, and IL-1β transcripts was done as described above. EVs M, EVs derived from mock-infected cells; EVs F, EVs derived from HSV-1(F)-infected cells; M, mock-infected cells; HSV-1(F), herpes simplex virus type-1 strain F-infected cells. (C) EVs from infected or uninfected HEL cells obtained from three biological replicates were used to expose HEL cells (2 × 105) at a dose of 500 EVs per cell. The cells were harvested at 8 h after exposure to the EVs, and relative quantification of ISG56, ISG15, and IL-6 was done by real-time PCR analysis as described above. P values: for ISG56, P = 0.0018 (Mock versus EVs F) and P = 0.0021 (EVs M versus EVs F); for ISG15, P = 0.0146 (Mock versus EVs F) and P = 0.0177 (EVs M versus EVs F); and for IL-6, P = 0.0001 (Mock versus EVs F), P = 0.0016 (EVs M versus EVs F), and P = 0.0025 (Mock versus EVs M). 18S rRNA was used for normalization.
We then sought to determine whether these effects depend on the quantity of EVs. Thus, HEL cells were exposed to two different amounts of EVs that corresponded to 7.5 and 15 μg of protein in cargo, respectively. The cells were harvested at 8 h postexposure, and the transcription of representative innate immunity and inflammatory genes was quantified by real-time PCR analysis. The results shown in Fig. 4B suggest that the EVs derived from HSV-1(F)-infected cells but not from uninfected cells activated innate immunity and inflammatory gene transcription in a dose-dependent manner.
In the experiments described above, the cells were exposed to equal amounts of proteins, and thus we excluded the possibility that the lack of responses is due to differences in the amount of cargo. In the next set of experiments, we exposed cells to equal numbers of EVs to elucidate whether potential differences in the quantity of the cargo could impact the antiviral responses. For this, we performed three independent isolations of EVs from infected and uninfected cells to obtain biological replicates, quantify the EVs using NTA, and expose HEL cells to equal numbers of EVs per cell (500 EVs/cell). As shown in Fig. 4C and similar to the results depicted in Fig. 4A and B, the EVs from infected cells activated ISG56, ISG15, and IL-6 gene expression but not the EVs from uninfected cells. The differences observed were statistically significant and followed the pattern of the responses described above. We conclude that EVs derived from infected cells trigger innate immune responses in recipient cells.
STING-dependent activation of innate immunity gene expression by EVs derived from HSV-1-infected cells.
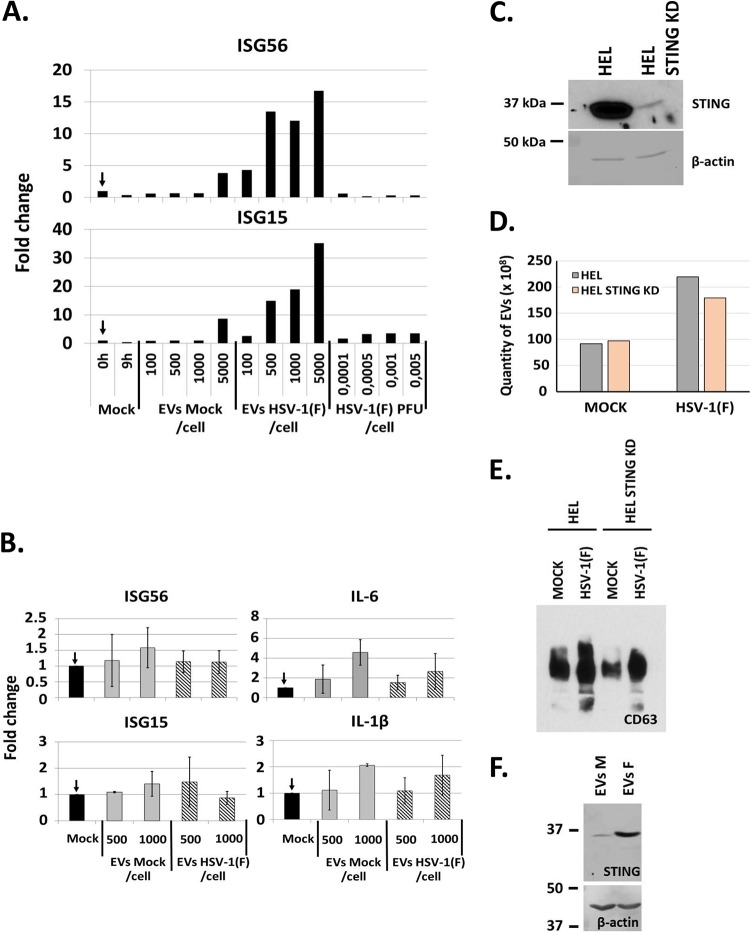
HEL cells were exposed to 100, 500, 1,000, or 5,000 EVs/cell isolated from infected or uninfected HEL cells as in Fig. 1. The cells were harvested at 8 h postexposure, and quantification of ISGs expression was done by real-time PCR analysis. Consistent with Fig. 4, EVs derived from infected cells activated ISG56 and ISG15 transcription in a dose-dependent manner (Fig. 5A). Notably, this isolation of EVs that yielded results similar to those described above was performed independently of the experiments in Fig. 4 and represents another biological replicate. EVs derived from uninfected cells only minimally activated ISG transcription at doses higher than 5,000 vesicles per cell. To exclude the possibility that innate immunity is activated due to the trace amount of virus in the EVs, we exposed HEL cells to the wild-type virus at 0.0001 PFU/cell up to 0.005 PFU/cell. As shown in Fig. 5A, none of the doses of HSV-1(F) activated innate immune responses in HEL cells.
FIG 5.
EVs derived from STING-knockdown (STING-KD) HEL cells failed to activate innate immune responses in recipient HEL cells. (A) EVs were isolated from infected or uninfected cells, as in Fig. 1. HEL cells were exposed to doses of EVs ranging from 100 to 5,000 EVs per cell. The cells were harvested at 8 h postexposure, and quantification of ISG56 and ISG15 transcripts was done by qPCR analysis. In parallel, HEL cells were exposed to HSV-1(F) at 0.0001 up to 0.005 PFU/cell for 8 h, and quantification of ISG56 and ISG15 transcripts was performed as described previously. The fold change is relative to the uninfected HEL cells the moment of exposure to the EVs. (B) EVs were purified from the STING-knockdown cells as in Fig. 1A. HEL cells were exposed to the EVs and processed for quantification of the ISG56, ISG15, IL-1β, and IL-6 transcripts exactly as described for panel A. (C) Efficiency of STING-knockdown in HEL cells. B-actin served as a loading control. STING was detected by ECL reagent, while β-actin by the BCIP/NBT reagent (see Materials and Methods). (D) EVs were isolated from HEL cells and their STING-KD derivatives that were either uninfected or infected with HSV-1 (0.1 PFU/cell) following procedures described in Fig. 1. Quantification of EVs was done by NTA. (E) Equal numbers of EVs from panel D were analyzed for CD63 by immunoblot analysis. (F) STING-KD HEL cells were incubated with equal amount of EVs derived from HSV-1(F)-infected (EVs F) or uninfected (EVs M) cells. The EV recipient cells were harvested 6 h postexposure to the EVs, and equal amounts of proteins were analyzed for STING by immunoblot analysis. B-actin was used as a loading control.
To address the contribution of STING in the activation of innate immunity gene expression in the EV recipient cells, we used in parallel EVs isolated from infected or uninfected STING knockdown (STING-KD) HEL cells. The results, shown in Fig. 5B, demonstrated that EVs derived from the STING-KD infected cells failed to activate innate immunity or inflammatory gene transcription. The efficiency of the knockdown of STING in the STING-KD HEL cells is depicted in Fig. 5C. In addition, we compared the amounts of EVs released by infected and uninfected STING-KD HEL cells to their parental HEL cells and found no significant differences (Fig. 5D). Moreover, we compared the amounts of CD63 present in equal number of EVs released by infected and uninfected STING-KD cells to their parental HEL cells and found no significant differences (Fig. 5E). The uptake of EVs carrying STING by the STING-KD HEL cells is depicted in Fig. 5F and has been described before (11). We conclude that the STING protein from the infected cells contributes to activation of antiviral responses in cells receiving EVs from these infected cells.
The EVs derived from infected fibroblasts activated the expression of a subset of M1-type markers in recipient macrophages in a STING-dependent manner.
Since the EVs derived from infected fibroblasts activated innate immune responses in recipient fibroblasts, we investigated whether they could also trigger antiviral responses in monocytes. Four series of experiments were performed. First, undifferentiated THP-1 monocytes were exposed to different numbers of EVs (100 to 5,000 EVs/cell) derived from infected or uninfected HEL cells. We found that these THP-1 monocytes did not activate innate immune responses after receiving the EVs (data not shown).
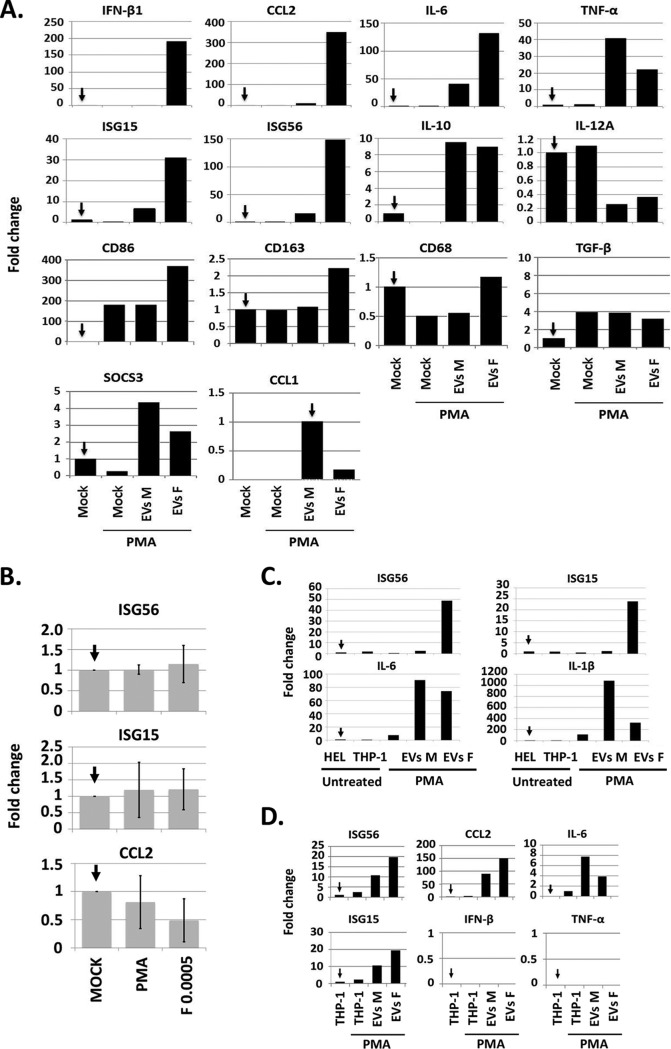
Second, the THP-1 monocytes were pretreated with phorbol 12-myristate 13-acetate (PMA; 5 ng/ml for 24 h) to stimulate their differentiation to M0 macrophage-like cells. The cells were then exposed to 500 EVs per cell derived from HSV-1-infected or uninfected HEL cells, as described above. The cells were harvested at 24 h after the exposure to EVs, and the expression of innate immunity genes was quantified by real-time PCR analysis. As shown in Fig. 6A, the EVs derived from infected cells (EVs F) but not from uninfected cells (EVs M) stimulated the expression of a subset of M1 polarization markers in recipient M0 macrophage-like cells. These markers included the C-C motif chemokine ligand 2 (CCL2), interferon-stimulated genes 56 and 15 (ISG56 and ISG15), beta interferon (IFN-β), and the cluster of differentiation 86 (CD86) (39, 40). Other innate immunity factors that were induced, such as IL-6 and TNF-α, are shared between the different types of macrophages (39, 40). TNF-α was induced after exposure to EVs derived from uninfected fibroblasts, but to a lesser extent in the cells receiving EVs from infected cells. IL-12A, which is a marker of M1-polarized macrophages, was not activated by any of these EVs, whereas IL-10, which is a marker of the M2-polarized macrophages, was induced to the same extent by EVs derived from infected or uninfected fibroblasts. We did not detect induction of the cluster of differentiation 68 (CD68) or the expression of iNOS and IFN-γ, which have been associated with M1 macrophage activation (data not shown). The suppressor of cytokine signaling 3 (SOCS3), a marker expressed predominantly in M1 macrophages, was not significantly stimulated by EVs derived from infected cells. The cluster of differentiation 163 (CD163) is considered generally an M2 marker and displayed a 2-fold increase in expression in cells receiving EVs from infected cells. However, the expression profile of the transforming growth factor β (TGF-β) and the C-C motif chemokine ligand 1 (CCL1) in EVs (F)-treated cells did not support M2 polarization. Infection of the PMA-treated THP-1 cells with HSV-1(F) (0.0005 PFU/cell) did not trigger innate immune responses (Fig. 6B). We conclude that EVs derived from HSV-1-infected fibroblasts stimulate polarization of the M0-like macrophages to express a subset of M1 type markers but none of the M2 markers tested.
FIG 6.
EVs derived from infected cells triggered the expression of a subset of M1-type markers in recipient macrophages in a STING-dependent manner. (A) Replicate cultures of THP-1 cells (2 × 105) were treated with PMA (5 ng/ml) for 24 h to trigger differentiation. The cells were then exposed to 500 EVs/cell derived from infected or uninfected HEL cells, as above, for 24 h. Quantification of IFN-β1, CCL-2, IL-6, IL12A, ISG15, ISG56, TNF-α, IL-10, SOCS3, CD86, CD163, and CD68 was done by qPCR analysis. The fold change is relative to undifferentiated THP-1 cells. (B) THP-1 cells treated with PMA as in panel A were infected with HSV-1(F) at 0.0005 PFU/cell. The cells were harvested at 24 postinfection and ISG56, ISG15, and CCL2 were quantified as in panel A. (C) Conditioned medium from PMA-treated THP-1 cells exposed to EVs derived from infected or uninfected cells from panel A was collected, clarified to remove cells, cell nuclei, and debris, and used to expose HEL cells. The HEL cells were collected at 8 h after exposure to the conditioned medium, and quantification of ISG56, ISG15, IL-6, and IL-1β was done as described above. Treatment with conditioned medium from untreated THP-1 cells, PMA-treated THP-1 cells, or HEL cells served as a control. (D) Procedures were exactly as in panel A except that the EVs were derived from the STING-KD HEL cells.
In the third series of experiments, the supernatant from the PMA-treated THP-1 cells (5 × 105) that had been exposed to EVs derived from infected or uninfected HEL cells for 24 h was collected, clarified to remove cells, nuclei, and cell debris, and used to expose fresh cultures of HEL cells (5 × 105). The recipient HEL cells were harvested at 8 h after exposure to the conditioned medium, and quantification of ISGs or cytokines transcription was performed by real-time PCR analysis. We found that the supernatant of the PMA-treated THP-1 cells that had been exposed to EVs from infected cells activated transcription of ISG56 and ISG15, but not the supernatant of PMA-treated THP-1 cells that had been exposed to EVs from uninfected cells. However, the supernatants from both sources activated cytokines such as IL-6 and IL-1β, albeit at different extents. Control supernatant from PMA-treated THP-1 cells or untreated THP-1 cells did not activate the transcription of ISGs or cytokines (Fig. 6C). The indicated fold change is relative to untreated HEL cells. We conclude that macrophages receiving EVs from infected cells secrete factors that also activate antiviral responses in uninfected recipient cells.
In the last series of experiments, PMA-treated THP-1 cells were exposed to 500 EVs/cell derived from infected or uninfected STING-KD HEL cells, as described in Fig. 6A, followed by quantification of innate immunity gene transcription by real-time PCR analysis (qPCR) analysis. The results shown in Fig. 6D suggest that EVs from infected STING-KD HEL cells failed to activate IFN-β or TNF-α gene transcription in the recipient macrophages but activated ISG56, ISG15, and CCL2, albeit to a lesser extent than the EVs derived from the HEL cells (compare Fig. 6D to A). We conclude that STING and STING-related factors packaged in EVs during HSV-1 infection contribute to the activation of antiviral responses in EV recipient cells.
EVs released by infected cells suppressed viral gene transcription and virus replication.
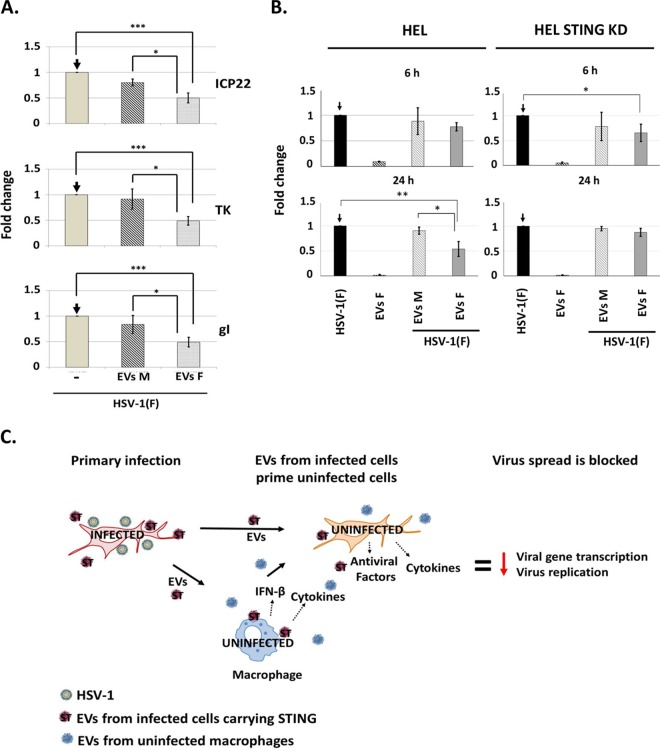
We sought to determine the effect of EVs released by infected cells on HSV-1 infection. The EVs were isolated as in Fig. 1, quantified by NTA, and tested for their ability to activate innate immune responses, as in Fig. 3 and 4 (data not shown). Subsequently, two series of experiments were performed. First, HEL cells were exposed to EVs (500 vesicles/cell) derived from infected or uninfected HEL cells, and HSV-1(F) (0.5 PFU/cell) was added to the cultures simultaneously with the EVs. The cells were harvested at 3 h postinfection, and viral gene transcription was quantified by qPCR analysis. As shown in Fig. 7A, EVs derived from infected cells caused a 50% reduction in the amount of all classes of viral gene transcripts (ICP22; α-gene, TK1; β-gene, gI; and γ-gene), while the EVs from uninfected cells had only a minor effect.
FIG 7.
EVs derived from infected cells blocked HSV-1 infection. (A) HEL cells were exposed to EVs (500 EVs/cell) derived from infected or uninfected cells simultaneously with HSV-1(F) at 0.5 PFU/cell. Viral gene transcription was quantified at 3 h postinfection by qPCR analysis. The fold change is expressed relative to infected but EV-untreated cells. 18S rRNA was used for normalization. For ICP22 levels, P = 0.0009 (Mock versus EVs F) and P = 0.0119 (Mock versus EVs M). For TK levels, P = 0.0004 (Mock versus EVs F) and P = 0.0267 (Mock versus EVs M). For gI levels, P = 0.0007 (Mock versus EVs F) and P = 0.0391 (Mock versus EVs M). (B, left panel) HEL cells were exposed to EVs (500 EVs/cell) derived from infected or uninfected HEL cells simultaneously with HSV-1(F) at 0.01 PFU/cell. The cells were harvested at 6 and 24 h after infection, and quantification of the viral genome was done by qPCR analysis using primer pairs against the HSV-1 TK1 coding region. B-actin DNA was used for normalization. For the differences observed in virus replication, P = 0.0058 for HSV-1(F) versus EVs (F) at 24 postinfection and P = 0.0183 EVs (F) versus EVs (M) at 24 h postinfection. (B, right panel) Procedures were followed in parallel to those described in the left panel except that the HEL cells were exposed to EVs derived from the STING-KD HEL cells. For the differences observed in virus replication, P = 0.026 for HSV-1(F) versus EVs (F) at 6 h postinfection. (C) Proposed model for the functions of EVs during HSV-1 infection. Infected cells release EVs that carry STING and other antiviral factors. The EVs from infected cells activate antiviral responses in uninfected recipient cells that lead to suppression of viral gene expression and virus replication.
In the second series of experiments, we investigated the role of EVs derived from infected cells on virus replication. Thus, HEL cells were exposed to EVs (500 EVs/cell) derived from infected or uninfected cells simultaneously with HSV-1(F) (0.01 PFU/cell). The cells were harvested at 6 and 24 h after infection, and the amount of viral genome was quantified by qPCR analysis. As shown in Fig. 7B, EVs derived from infected HEL cells inhibited the infection, resulting in a decrease in the amount of viral genome by 50% at 24 h postinfection, while EVs derived from uninfected cells did not. Notably, EVs derived from infected HEL cells had negligible amounts of viral genome as a contaminant. In contrast, EVs derived from infected STING-KD HEL cells (500 EVs/cell) displayed no effect on virus replication at 24 h postinfection. A similar inhibition of virus replication was observed using EVs from infected STING-KD cells or their parental cells at 6 h postinfection, but this effect was sustained only in cells treated with EVs from the wild-type HEL cells. These results indicate that STING-dependent and STING-independent factors present in EVs inhibit virus replication. We conclude that EVs derived from infected cells but not from uninfected cells inhibit virus replication and that STING plays an important role in this process.
DISCUSSION
Extracellular vesicles (EVs) have garnered attention over the last decade as it became apparent that they are not a mechanism of cells to dispose of unwanted components, but rather they carry information that influences the surrounding environment and reflects the physiological state of the cells in which they were produced (2, 3, 6, 10, 41–48). Under pathological conditions and depending on the integrity of the host, the EVs could either contribute to the healing process or they exacerbate pathogenesis (2, 3, 6, 10, 41–48). Previously, we reported that EVs released by HSV-1-infected cells carry the DNA sensor STING and viral and host RNAs (6, 11). These components are stable inside the recipient cells (6, 11). We hypothesized that the EVs released by HSV-1-infected cells function to curtail rather than support the dissemination of the virus in the body.
The studies presented here highlight three major findings. First, we discuss the development of a procedure to separate the EVs from HSV-1 virions. The purification process is based on a discontinuous iodixanol/sucrose gradient and takes advantage of the density differences between the HSV-1 virions and the EVs, with the virions being heavier than the EVs. The use of an iodixanol/sucrose gradient in the purification of EVs is labor-intensive, but it provides a high quality of EVs for qualitative assays. Several attempts to separate EVs from HSV virions using immunoaffinity approaches and/or high-speed sedimentation were ineffective, since they led to coaggregation of EVs with virions (6). In addition, the EVs remain intact, which is achieved by avoiding the high-speed pelleting that is known to damage EVs. The purity of the EVs was verified by immunoblot analysis, in which markers of EVs accumulated in fractions preceding the fractions containing viral structural components, and by quantification of the viral genome, in which trace amounts of HSV-1 genome could be detected in the EV fractions. Overall, infectious virus accumulated in heavier density fractions and was largely absent from the fraction containing the EVs. We also found that HSV-1 infection stimulated the release of extracellular vesicles; these vesicles were larger than those from uninfected cells and their size coincided with that of exosomes (50 to 150 nm) and of smaller microvesicles (150 to 500 nm). Interestingly, an abundant tegument protein of the virus, Us11 was consistently found in the fractions containing EVs, but other viral structural components were absent, suggesting that Us11 may be packaged into the EVs. In native gels exosomal Us11 was found in large complexes and not as a monomer (data not shown). Us11 is known as an RNA binding protein that can bind transcripts but also smaller RNAs known as aptamers, and one possibility is that it could deliver RNA cargo to the EVs (49). Notably, UL46, another tegument protein that colocalizes with and interacts with STING, was absent from the fractions with the EVs (data not shown) (34).
Second, we found that EVs derived from infected cells but not from uninfected cells activated innate immune responses in recipient cells. The effect of EVs on recipient cells was dose dependent. The induction of innate immunity in cells receiving EVs from infected cells was observed using independent isolations of EVs. The consistency between biological replicates increases our confidence about the reproducibility of the EV purification protocol and the consistency of the cargo of EVs under these conditions. Analysis that was performed under the same conditions using EVs derived from autologous STING-KD cells demonstrated that innate immune responses were largely but not completely compromised. These data suggest that STING from the EV donor cells triggers antiviral responses in the recipient cells and also imply that STING-related factors may contribute to the innate immune responses in the recipient cells. Activation of STING is known to establish an antimicrobial state in cells by activating the expression of interferon-related genes. Studies are in progress to elucidate whether the lack of activation of innate immunity by the STING-KD EVs is solely due to the absence of STING or because other factors that copackage with STING inside EVs during HSV infection are concurrently depleted. Previous studies demonstrated that the agonist of STING cGAMP is present in nonviral particles upon infection with large DNA viruses (12). The silencing of STING does not affect the generation of cGAMP, but it could affect its packaging into EVs if it copackaged with STING. Thus, cGAMP or foreign DNA potentially present in the EVs derived from STING-KD do not appear to trigger antiviral responses. Therefore, the lack of responses in the case of the STING-depleted EVs indicate that STING from the donor cells either directly or indirectly mediates antiviral responses in EV recipient cells.
Another observation is that the EVs from infected fibroblasts activated a subset of M1-type markers in recipient macrophages but none of the M2-type markers tested. The M1-polarized macrophages are characterized by massive inflammatory cytokine secretion and the production of nitric oxide that lead to pathogen elimination (39, 40). The M2-polarized macrophages are activated by immune complexes, complement, apoptotic cells, or pathogens, they display high phagocytic activity, and their role expands beyond inflammation to wound healing, repair, and morphogenesis (39, 40). The question then arises, “why do the EVs derived from infected cells selectively activate only some pathways involved in M1 polarization?” Perhaps activation of these pathways is sufficient to downmodulate HSV-1 infection, avoiding the consequences of extensive damage to the host due to inflammation. These observations also imply that specific cargo packaged in the EVs triggers these responses. Thus, factors linked to pathways that are inactivated by the virus during the course of the infection are less likely to be present in the EVs. For example, HSV-1 infection leads to downmodulation of the TNFR-1 receptor and the cessation of TNF-α-dependent signaling (50). Therefore, factors linked to the TNF-α pathway are less likely to be present in the EVs. This could explain why EVs from infected cells do not activate TNF-α signaling.
We also observed that EVs derived from infected STING-expressing and STING-knockdown cells both suppressed the onset of virus replication, but only the EVs from STING-expressing cells sustained this suppression. Also, the inhibition in viral gene transcription shortly after exposing cells concurrently to EVs and the virus cannot be attributed to activation of innate immunity, since stimulation of innate immunity genes was found to occur later. One interpretation of these data is that there are two barriers that inhibit virus replication: one STING dependent and one STING independent. EVs carry in addition to STING other factors, including viral miRNAs that could interfere with viral gene expression causing a delay in the replication of the virus, and this is may be the first barrier (11, 13). STING and STING-related factors packaged in the EVs appear to be a second barrier, probably through the activation of innate immunity. If the STING-dependent barrier is missing, the virus replication ensues despite the initial delay. This most likely occurs because our system is “closed,” and we monitor effects after a single exposure of cells to EVs derived from infected cells. Consequently, when viral miRNAs carried in EVs are titrated out by the viral mRNAs, the infection progresses despite the initial delay. In dynamic systems this balance is expected to shift depending on the host responses and the ability of the virus to evade them.
Third, consistent with the findings discussed above, we found that depletion of the CD63-positive population of EVs had a positive impact on HSV yields. To alter the population of EVs, we either knocked down CD63 or we used the neutral sphingomyelinase-2 inhibitor GW4869. CD63 is a tetraspanin that is involved in the biogenesis of EVs (4, 35, 51–54). CD63 has the capacity to interact with other membrane receptors and signaling molecules at the membranes; it is organized in tetraspanin-enriched microdomains and plays roles in EV biogenesis, the sorting of cargo components such as proteins and miRNAs into EVs, and the binding and uptake of EVs by target cells (4, 35, 51–54). The neutral sphingomyelinase-2 is the first molecule reported to be related to miRNA secretion into exosomes (36, 37). Overexpression of nSMase-2 increases the number of exosomal miRNAs, and conversely inhibition of nSMase-2 expression reduced the number of exosomal miRNAs (36). nSMase-2 localizes to the Golgi and also recycles back from the plasma membrane through the endosomal system (36). nSMase-2 is a ceramide-generating enzyme that has been implicated in exosome generation (36, 37). A high-throughput screen for small molecule inhibitors of nSMase-2 identified GW4869 as a selective nSMase2 inhibitor (36, 55). Since its identification, GW4869 has been the most commonly used pharmacological inhibitor to study nSMase-2 function in cellular and animal models (36, 55). Based on these observations, the only known shared function so far between CD63 and nSMase-2 is their role in the biogenesis of EVs. The fact that both approaches that disrupt the CD63-positive population of EVs display the same effect on the virus growth further enhances our hypothesis that the EVs from infected cells have a negative impact on HSV-1 infection. Another observation is that neither the knockdown of CD63 nor the treatment with GW4869 blocked the production of EVs. Usually, shutdown of one EV biogenesis pathway triggers the release of EVs by other pathways. Thus, we found that treatment of cells with GW4869 led to higher number of EVs compared to untreated cells, but these EVs had less CD63 and TSG101 and more Flotillin-2. Also, knockdown of CD63 reduced but did not abolish the production of EVs. Such alterations are expected to cause alterations in the cargo of EVs and by extension to the signals delivered to recipient cells.
Taken together, a proposed function of the EVs released by infected cells could be to control the dissemination of the virus and its persistence in the host. According to our model, during the lytic cycle the EVs released by infected cells, along with other secreted factors and immune cells, could contribute in restricting the dissemination of the virus (Fig. 7C). Some of the viral components that were found in the EVs, such as STING and the viral microRNAs miR-H3, miR-H5, and miR-H6, could have a greater impact during the latent stage of the virus by promoting the establishment and maintenance of latency as they negatively regulate viral gene expression (56, 57). In the sensory ganglia where the virus establishes latent infections, neurons are in association with Schwann cells and satellite cells, and these cells communicate with each other largely through vesicles. The cells surrounding the infected cells or cells that shed the virus could be those controlling the dissemination of the virus and thereby promote its persistence in the host.
MATERIALS AND METHODS
Viruses.
Herpes simplex virus strain F [HSV-1(F)], a limited passage isolate is the prototype strain used in this laboratory (58).
Cells.
The sources and propagation of telomerase transformed human embryonic fibroblasts (HEL cels), HEp-2 cells (American Type Culture Collection [ATCC]), HEK293 cells (ATCC), and Vero cells (ATCC) were reported elsewhere (59, 60). THP-1 cells were kindly provided by Steven Weinman. (University of Kansas Medical Center) and were maintained in RPMI medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific). For differentiation, THP-1 cells were treated with PMA (5 ng/ml; Sigma) for 24 h.
Virus growth curves.
One step growth curves for HSV-1 have been described elsewhere (34). GW4869 (20 μM; Sigma) was added at the moment of infection in medium supplemented with 10% EV-depleted FBS.
EV purification.
HEL or HEp-2 cells were infected at 95% confluence with HSV-1(F) (0.1 PFU/cell). The FBS utilized is EV depleted (Gibco). At 50 h postinfection, the supernatant was collected and centrifuged at 1,200 rpm for 5 min and at 3,500 rpm for 20 min prior to filtration through a 0.45-μm-pore size filter. The supernatant then was concentrated using a Centricon Plus 70 (Millipore) with a 30-kDa cutoff according to the manufacturer's instructions. The supernatant was loaded on top of an ioxidanol-based gradient ranged from 6 to 18%, with a 1.2% increment. The 60% iodixanol was diluted in 10 mM Tris (pH 8) and 0.25 M sucrose. Samples were centrifuged in an SW41 Ti rotor for 2 h at 250,000 × g and 4°C in a Beckman Coulter Optima XPN-80 ultracentrifuge. Fractions (500 μl) were collected from top to bottom and analyzed. Fractions containing the CD63-positive EVs were pulled together, dialyzed against Opti-MEM-I medium (Gibco) using a 3.5-kDa cutoff filter, and concentrated using a 10-kDa cutoff Centricon (Thermo Fisher Scientific) for further analysis.
Nanoparticle tracking analysis.
NTA was performed by applying a monochromatic 404-nm laser to diluted EVs samples and measuring the Brownian movements of each particle with a NanoSight LM10 instrument (NanoSight, Salisbury, United Kingdom). For each sample, nine different acquisitions were obtained of 60 s each. Nanoparticle Tracking Analysis software, version 2.3, was used to analyze 60-s videos of data collection to obtain the mean, median, and mode of vesicle size and the concentration. Results represent the averages of three independent isolations of EVs; the standard deviations (SD) were also determined.
Negative staining.
Extracellular vesicles were fixed for 1 h in 2% glutaraldehyde and incubated 20 min on a discharged carbon-filmed grid. Grids were next washed seven times in water and finally incubated for 10 s in 1% uranyl acetate and allowed to dry for at least 24 h. Grids were then analyzed using a JEOL JEM-1400 transmission electron microscope equipped with a Lab6 gun.
Development of knockdown cell lines with the aid of lentiviral vectors.
The PLKO.1 shRNAs for CD63, the PLKO.1 shRNAs for STING, and the nontargeted control shRNAs were purchased from Sigma. The procedures for the development of the CD63-KD and the STING-KD cells are described elsewhere (38). Briefly, for the development of the respective lentiviral vectors, HEK293 cells seeded in an F 25-cm2 flask at a 60% confluence were cotransfected with 8 μg of the plasmid carrying the shRNA, 8 μg of the Gag-Pol-expressing plasmid, and 1 μg of the VSV-G encoding plasmid with TurboFect (Thermo Fisher Scientific) according to the manufacturer's instructions. At 48 h after transfection, the supernatant from the cultures was collected, filtered through 0.45-μm-pore size filters, and used to infect HEp-2 or HEL cells. Puromycin selection was initiated 24 h after exposing cells to lentiviruses and continued until only resistant clones survived. The clones of HEp-2 and HEL cells with the greater reduction of CD63 or STING expression were used for further experimentation. Puromycin was removed from the cells 24 h before performing any assay.
Immunoblot analysis.
The procedures for immunoblot analysis were described in detail elsewhere (60). Briefly, cells were solubilized in triple-detergent buffer (50 mM Tris-HCl [pH:8], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% sodium deoxycholate, 100 μg of phenylmethylsulfonyl fluoride ml−1) supplemented with phosphatase inhibitors (10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate) and protease inhibitor cocktail (Sigma) and then briefly sonicated. The protein concentration was determined with the aid of Bio-Rad protein assay (Bio-Rad Laboratories). A total of 10 to 40 μg of total protein per sample was subjected to further analysis. Rabbit polyclonal antibodies to ICP0, VP22, UL38, and gM and the mouse monoclonal antibody Us11 were used at a dilution of 1:1,000 and were kindly provided by B. Roizman (University of Chicago). Mouse monoclonal antibodies to β-actin (Sigma), STING (R&D Systems), CD63 (Santa cruz), Flotillin-2 (BD Transduction Laboratories), and TSG101 (Santa Cruz) were used at a 1:1,000 dilution. Rabbit polyclonal antibodies to CD9 (Abcam) and CD63 (Santa Cruz) were used at a 1:500 dilution. Protein bands were visualized with BCIP (5-bromo-4-chloro-3-indolylphosphate)/nitroblue tetrazolium (NBT) (VWR) or with ECL Western blotting detection reagents (Amersham Biosciences) according to the manufacturer's instructions (34).
Real-time PCR analysis.
The procedures for RNA extraction and cDNA synthesis have been described elsewhere (34). Real-time PCR analyses were performed using SYBR green reagent (Invitrogen) or TaqMan (Applied Biosystems) according to the manufacturer's recommendations in a 7500 Fast real-time PCR system (Applied Biosystems). 18S rRNA primers (Universal Primers; Ambion) and the eukaryotic 18S rRNA endogenous control (FAM/MGB probe; Thermo Fisher Scientific) were used for normalization. The primers for ICP22, gI, and TK1 were reported elsewhere (34, 61). Predesigned probes (FAM/MGB) for human ISG15, IFN-β1, IL-1β, CCL2, CCL1, IL-12A, IL-10, SOCS3, CD86, CD163, CD68, and TNF-α transcripts were obtained through Thermo Fisher Scientific. The primers for ISG56, IL-6, and β-actin DNA are listed elsewhere (34, 61). For viral DNA quantification, total DNA was extracted from the cells using a NucleoSpin DNA RapidLyse kit (Macherey-Nagel). To detect the viral DNA, we used primer pairs targeting the area of the viral genome encoding glycoprotein I (gI). The primers used for viral DNA were 5′-CCC ACG GTC AGT CTG GTA TC-3′ (forward) and 5′-TTT GTG TCC CAT GGG GTA GT-3′ (reverse). For normalization, we used the following primers targeting the β-actin DNA: 5′-CAT GTA CGT TGC TAT CCA GGC-3′ (forward) and 5′-CTC CTT AAT GTC ACG CAC GAT-3′ (reverse).
Statistical analysis.
The Prism 7 software (GraphPad) was used for statistical analysis of the NanoSight or qPCR data. P values were calculated using a standard two-tailed unpaired Student t test. P values of ≤0.05 were considered significant, as indicated by asterisks in the figures (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). All statistical analyses were performed using biological replicates.
ACKNOWLEDGMENTS
We thank Bernard Roizman (University of Chicago) for kindly providing the Us11, ICP0, VP22, gM, and UL38 antibodies. The THP-1 cells were kindly provided by Steven Weinman (University of Kansas Medical Center). We thank graduate students Christos Dogrammatzis and Hope Waisner (University of Kansas Medical Center) for providing technical assistance.
M.K. is funded through University of Kansas Medical Center startup funds and through COBRE grant P20GM113117.
REFERENCES
- 1.Knipe DM, Howley PM. 2013. Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Piper RC, Katzmann DJ. 2007. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. 2002. The biogenesis and functions of exosomes. Traffic 3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 4.Raposo G, Stoorvogel W. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budnik V, Ruiz-Canada C, Wendler F. 2016. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci 17:160–172. doi: 10.1038/nrg.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalamvoki M, Deschamps T. 2016. Extracellular vesicles during herpes simplex virus type 1 infection: an inquire. Virol J 13:63. doi: 10.1186/s12985-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajendran L, Bali J, Barr MM, Court FA, Kramer-Albers EM, Picou F, Raposo G, van der Vos KE, van Niel G, Wang J, Breakefield XO. 2014. Emerging roles of extracellular vesicles in the nervous system. J Neurosci 34:15482–15489. doi: 10.1523/JNEUROSCI.3258-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subra C, Laulagnier K, Perret B, Record M. 2007. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Wahlgren J, Statello L, Skogberg G, Telemo E, Valadi H. 2016. Delivery of small interfering RNAs to cells via exosomes. Methods Mol Biol 1364:105–125. doi: 10.1007/978-1-4939-3112-5_10. [DOI] [PubMed] [Google Scholar]
- 11.Kalamvoki M, Du T, Roizman B. 2014. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci U S A 111:E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulen MF, Ablasser A. 2015. Inside job: viruses transfer cGAMP between cells. Cell Host Microbe 18:263–265. doi: 10.1016/j.chom.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B, Zhou G. 2016. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci U S A 113:E894–E901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miettinen JJ, Matikainen S, Nyman TA. 2012. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J Virol 86:12770–12778. doi: 10.1128/JVI.01545-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temme S, Eis-Hubinger AM, McLellan AD, Koch N. 2010. The herpes simplex virus-1 encoded glycoprotein B diverts HLA-DR into the exosome pathway. J Immunol 184:236–243. doi: 10.4049/jimmunol.0902192. [DOI] [PubMed] [Google Scholar]
- 16.Alenquer M, Amorim MJ. 2015. Exosome biogenesis, regulation, and function in viral infection. Viruses 7:5066–5083. doi: 10.3390/v7092862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobrie A, Colombo M, Raposo G, Thery C. 2011. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 18.Aqil M, Naqvi AR, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. 2014. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J Extracell Vesicles 2014:3. doi: 10.3402/jev.v3.23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard MA, Zhao H, Yue SC, Anandaiah A, Koziel H, Tachado SD. 2014. Novel HIV-1 miRNAs stimulate TNFα release in human macrophages via TLR8 signaling pathway. PLoS One 9:e106006. doi: 10.1371/journal.pone.0106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Columba CS, Federico M. 2013. Sequences within RNA coding for HIV-1 Gag p17 are efficiently targeted to exosomes. Cell Microbiol 15:412–429. doi: 10.1111/cmi.12046. [DOI] [PubMed] [Google Scholar]
- 21.Gould SJ, Booth AM, Hildreth JE. 2003. The Trojan exosome hypothesis. Proc Natl Acad Sci U S A 100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madison MN, Okeoma CM. 2015. Exosomes: implications in HIV-1 pathogenesis. Viruses 7:4093–4118. doi: 10.3390/v7072810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. 2014. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog 10:e1004424. doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosset FL, Dreux M. 2014. HCV transmission by hepatic exosomes establishes a productive infection. J Hepatol 60:674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Greenhill C. 2013. Hepatitis: new route of HCV transmission. Nat Rev Gastroenterol Hepatol 10:504. doi: 10.1038/nrgastro.2013.148. [DOI] [PubMed] [Google Scholar]
- 26.Masciopinto F, Giovani C, Campagnoli S, Galli-Stampino L, Colombatto P, Brunetto M, Yen TS, Houghton M, Pileri P, Abrignani S. 2004. Association of hepatitis C virus envelope proteins with exosomes. Eur J Immunol 34:2834–2842. doi: 10.1002/eji.200424887. [DOI] [PubMed] [Google Scholar]
- 27.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin RV, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJ. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ariza ME, Rivailler P, Glaser R, Chen M, Williams MV. 2013. Epstein-Barr virus encoded dUTPase containing exosomes modulate innate and adaptive immune responses in human dendritic cells and peripheral blood mononuclear cells. PLoS One 8:e69827. doi: 10.1371/journal.pone.0069827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceccarelli S, Visco V, Raffa S, Wakisaka N, Pagano JS, Torrisi MR. 2007. Epstein-Barr virus latent membrane protein 1 promotes concentration in multivesicular bodies of fibroblast growth factor 2 and its release through exosomes. Int J Cancer 121:1494–1506. doi: 10.1002/ijc.22844. [DOI] [PubMed] [Google Scholar]
- 30.Keryer-Bibens C, Pioche-Durieu C, Villemant C, Souquere S, Nishi N, Hirashima M, Middeldorp J, Busson P. 2006. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer 6:283. doi: 10.1186/1471-2407-6-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kung CP, Meckes DG Jr, Raab-Traub N. 2011. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCδ. J Virol 85:4399–4408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meckes DG Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meckes DG Jr, Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N. 2013. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 110:E2925–E2933. doi: 10.1073/pnas.1303906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deschamps T, Kalamvoki M. 2017. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J Virol 91:e00535-17. doi: 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pols MS, Klumperman J. 2009. Trafficking and function of the tetraspanin CD63. Exp Cell Res 315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 36.Shamseddine AA, Airola MV, Hannun YA. 2015. Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes. Adv Biol Regul 57:24–41. doi: 10.1016/j.jbior.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 38.Kalamvoki M, Roizman B. 2014. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A 111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez FO, Gordon S. 2014. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray PJ. 2017. Macrophage polarization. Annu Rev Physiol 79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 41.Azmi AS, Bao B, Sarkar FH. 2013. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chahar HS, Bao X, Casola A. 2015. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses 7:3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coleman BM, Hill AF. 2015. Extracellular vesicles: their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol 40:89–96. doi: 10.1016/j.semcdb.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Fleming A, Sampey G, Chung MC, Bailey C, van Hoek ML, Kashanchi F, Hakami RM. 2014. The carrying pigeons of the cell: exosomes and their role in infectious diseases caused by human pathogens. Pathog Dis 71:109–120. doi: 10.1111/2049-632X.12135. [DOI] [PubMed] [Google Scholar]
- 45.Kalani A, Tyagi A, Tyagi N. 2014. Exosomes: mediators of neurodegeneration, neuroprotection, and therapeutics. Mol Neurobiol 49:590–600. doi: 10.1007/s12035-013-8544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kharaziha P, Ceder S, Li Q, Panaretakis T. 2012. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta 1826:103–111. [DOI] [PubMed] [Google Scholar]
- 47.Nazarenko I, Rupp AK, Altevogt P. 2013. Exosomes as a potential tool for a specific delivery of functional molecules. Methods Mol Biol 1049:495–511. doi: 10.1007/978-1-62703-547-7_37. [DOI] [PubMed] [Google Scholar]
- 48.Schwab A, Meyering SS, Lepene B, Iordanskiy S, van Hoek ML, Hakami RM, Kashanchi F. 2015. Extracellular vesicles from infected cells: potential for direct pathogenesis. Front Microbiol 6:1132. doi: 10.3389/fmicb.2015.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryant KF, Cox JC, Wang H, Hogle JM, Ellington AD, Coen DM. 2005. Binding of herpes simplex virus-1 US11 to specific RNA sequences. Nucleic Acids Res 33:6090–6100. doi: 10.1093/nar/gki919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang L, Roizman B. 2006. Herpes simplex virus 1 precludes replenishment of the short-lived receptor of tumor necrosis factor alpha by virion host shutoff-dependent degradation of its mRNA. J Virol 80:7756–7759. doi: 10.1128/JVI.00587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowal J, Tkach M, Thery C. 2014. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Andreu Z, Yanez-Mo M. 2014. Tetraspanins in extracellular vesicle formation and function. Front Immunol 5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rana S, Zoller M. 2011. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem Soc Trans 39:559–562. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 54.Zoller M. 2009. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer 9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 55.Luberto C, Hassler DF, Signorelli P, Okamoto Y, Sawai H, Boros E, Hazen-Martin DJ, Obeid LM, Hannun YA, Smith GK. 2002. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J Biol Chem 277:41128–41139. doi: 10.1074/jbc.M206747200. [DOI] [PubMed] [Google Scholar]
- 56.Du T, Han Z, Zhou G, Roizman B. 2015. Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation. Proc Natl Acad Sci U S A 112:E49–E55. doi: 10.1073/pnas.1422657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piedade D, Azevedo-Pereira JM. 2016. The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses 8:E156. doi: 10.3390/v8060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 59.Kalamvoki M, Roizman B. 2008. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci U S A 105:20488–20493. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalamvoki M, Roizman B. 2010. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A 107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deschamps T, Kalamvoki M. 2017. Impaired STING pathway in the human osteosarcoma U2OS cells contributes to the growth of ICP0-null mutant herpes simplex virus. J Virol 91:e00006-17. doi: 10.1128/JVI.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]