HSV-1 UL51 is conserved in all members of the Herpesviridae family. This viral protein is phosphorylated and functions in viral cell-cell spread and cytoplasmic virion maturation in HSV-1-infected cells. Although the downstream effects of HSV-1 UL51 have been clarified, there is a lack of information on how this viral protein is regulated as well as the significance of the phosphorylation of this protein in HSV-1-infected cells. In this study, we show that the phosphorylation of UL51 at Ser-184 promotes viral replication, cell-cell spread, and nuclear egress in cell cultures and viral pathogenicity in mice. This is the first report to identify the mechanism by which UL51 is regulated as well as the significance of UL51 phosphorylation in HSV-1 infection. Our study may provide insights into the regulatory mechanisms of other herpesviral UL51 homologs.
KEYWORDS: HSV-1, nuclear egress, UL51, cell-cell spread, protein phosphorylation
ABSTRACT
Herpes simplex virus 1 (HSV-1) UL51 is a phosphoprotein that functions in the final envelopment in the cytoplasm and viral cell-cell spread, leading to efficient viral replication in cell cultures. To clarify the mechanism by which UL51 is regulated in HSV-1-infected cells, we focused on the phosphorylation of UL51. Mass spectrometry analysis of purified UL51 identified five phosphorylation sites in UL51. Alanine replacement of one of the identified phosphorylation sites in UL51, serine 184 (Ser-184), but not the other identified phosphorylation sites, significantly reduced viral replication and cell-cell spread in HaCaT cells. This mutation induced membranous invaginations adjacent to the nuclear membrane, the accumulation of primary enveloped virions in the invaginations and perinuclear space, and mislocalized UL34 and UL31 in punctate structures at the nuclear membrane; however, it had no effect on final envelopment in the cytoplasm of HaCaT cells. Of note, the alanine mutation in UL51 Ser-184 significantly reduced the mortality of mice following ocular infection. Phosphomimetic mutation in UL51 Ser-184 partly restored the wild-type phenotype in cell cultures and in mice. Based on these results, we concluded that some UL51 functions are specifically regulated by phosphorylation at Ser-184 and that this regulation is critical for HSV-1 replication in cell cultures and pathogenicity in vivo.
IMPORTANCE HSV-1 UL51 is conserved in all members of the Herpesviridae family. This viral protein is phosphorylated and functions in viral cell-cell spread and cytoplasmic virion maturation in HSV-1-infected cells. Although the downstream effects of HSV-1 UL51 have been clarified, there is a lack of information on how this viral protein is regulated as well as the significance of the phosphorylation of this protein in HSV-1-infected cells. In this study, we show that the phosphorylation of UL51 at Ser-184 promotes viral replication, cell-cell spread, and nuclear egress in cell cultures and viral pathogenicity in mice. This is the first report to identify the mechanism by which UL51 is regulated as well as the significance of UL51 phosphorylation in HSV-1 infection. Our study may provide insights into the regulatory mechanisms of other herpesviral UL51 homologs.
INTRODUCTION
Herpesviruses in the family Herpesviridae are subclassified into three subfamilies: Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (1). Herpes simplex virus 1 (HSV-1) is one of the most commonly studied members of the family Herpesviridae. It causes a variety of human diseases, including mucocutaneous diseases, keratitis, skin diseases, and encephalitis (1). The HSV-1 virion, similar to those of other herpesviruses, contains a linear double-stranded DNA viral genome in an icosahedral capsid, which is enclosed by an envelope, and between the envelope and the nucleocapsid there is a proteinaceous layer consisting of a number of different viral proteins, designated the tegument (1, 2). In HSV-1-infected cells, the packaging of nascent virus genomes into preformed capsids takes place in the nucleus. The nascent progeny nucleocapsids acquire a primary envelope by budding through the inner nuclear membrane (INM) into the perinuclear space between the INM and outer nuclear membrane (ONM) (primary envelopment) (3–5). The enveloped nucleocapsids then fuse with the ONM to release de-enveloped nucleocapsids into the cytoplasm in a process termed de-envelopment (3, 5). Subsequently, the nucleocapsids acquire a final envelope by budding into cytoplasmic vesicles, which are probably membranes derived from the trans-Golgi network and/or endosomes (secondary envelopment). Mature virions then are secreted from the infected cells by exocytosis (1, 3, 5).
HSV-1 UL51, the subject of this study, is conserved in the family Herpesviridae (1) and is incorporated into the tegument of virions, similar to UL51 homologs in other members of the Alpha-, Beta-, and Gammaherpesvirinae subfamilies (6–10). In HSV-1-infected cells, UL51 is posttranslationally modified by phosphorylation and palmitoylation, the latter of which was suggested to have a role in the association of UL51 with cellular membranes (11, 12) and has an important role in viral secondary envelopment (13). The role of UL51 in viral secondary envelopment was also shown in UL51 homologs of pseudorabies virus (PRV), varicella-zoster virus (VZV), and human cytomegalovirus (HCMV) (14–18). In addition, HSV-1 UL51 has a role in cell-cell spread without affecting viral production and release, which is cell type dependent (19). In HSV-1-infected cells, UL51 interacts with other conserved tegument proteins, UL7 (20, 21) and UL14 (13), and envelope glycoprotein E (gE) (19), a well-known HSV-1 regulator that promotes viral cell-cell spread (22, 23). The interaction between UL51 and UL14 is important in HSV-1 secondary envelopment. This is based on observations that mutations in three minimal amino acids in UL51 required for interactions with UL14 induced secondary envelopment defects at levels similar to those of UL51-null, UL14-null, and UL51/UL14 double-null mutations (13). In contrast, similar studies of UL51 interactions with UL7 or gE have not been reported thus far; therefore, the significance of these interactions in HSV-1-infected cells remains to be elucidated. Although the downstream effects of UL51 in HSV-1-infected cells have been clarified, the mechanisms by which UL51 is regulated in HSV-1-infected cells are unclear.
It is well established that protein phosphorylation is a common and effective posttranslational modification that regulates the function of the target protein (24, 25). This is also the case for HSV-1 proteins in infected cells. A previous study reported that the phosphorylation of HSV-1 proteins by viral and cellular protein kinases was important for the regulation of their respective viral proteins in infected cells and that these regulatory effects were critical for viral replication and pathogenesis (26–29). Recently, several large-scale phosphoproteomics analyses of HSV-1-infected cells were reported, and numerous phosphorylation sites in various HSV-1 proteins were identified (30–33). However, few phosphorylation sites whose phosphorylation regulates their respective HSV-1 proteins in infected cells have been reported. Furthermore, many phosphorylation sites have no regulatory effect on viral replication and pathogenesis following their phosphorylation. For example, the phosphorylation of UL51 threonine 190 (Thr-190), which has been detected in all phosphoproteomic analyses of HSV-1-infected cells thus far (30–33), had no effect on viral replication and pathogenesis (34). Therefore, the functional analyses of identified phosphorylation sites are essential to understand the regulatory mechanisms of HSV-1 protein phosphorylation in infected cells.
In this study, we attempted to identify a functional phosphorylation site(s) in UL51 to clarify the mechanism by which UL51 is regulated in HSV-1-infected cells. The high-accuracy mass spectrometry (MS) analysis of UL51 purified from cells ectopically expressing UL51 identified five phosphorylation sites in UL51. We investigated the effects of the phosphorylation of each of the identified phosphorylation sites in cell cultures and in vivo.
RESULTS
Identification of Ser-184, Thr-190, Ser-226, Thr-228, and Ser-235 as phosphorylation sites in UL51.
To identify phosphorylation sites in UL51, we used the tandem affinity purification of transiently expressed UL51 in 293T cells, coupled with MS-based technology (35–39). Five phosphorylation sites were identified in UL51: Ser-184, Thr-190, Ser-226, Thr-228, and Ser-235 (Table 1).
TABLE 1.
UL51 phosphopeptides and phosphorylation sites identified by MS/MS
| Phosphorylation site | Peptide sequencea | Mascot score |
|---|---|---|
| Thr-190 | LGGLGVTEAPSLGHPH(p)TPPPEVTLAPAAR | 63 |
| Ser-184 and Thr-190 | LGGLGVTEAP(p)SLGHPH(p)TPPPEVTLAPAAR | 46 |
| Ser-226 | VSVPRPTA(p)SPTAPRPGPSR | 34 |
| Thr-228 | VSVPRPTASP(p)TAPRPGPSR | 32 |
| Ser-235 | VSVPRPTASPTAPRPGP(p)SR | 31 |
In the peptide sequences, (p) indicates phosphorylated amino acid.
Generation of a self-excisable HSV-1(F) BAC and construction of recombinant UL51 mutant viruses using the newly generated HSV-1(F) BAC.
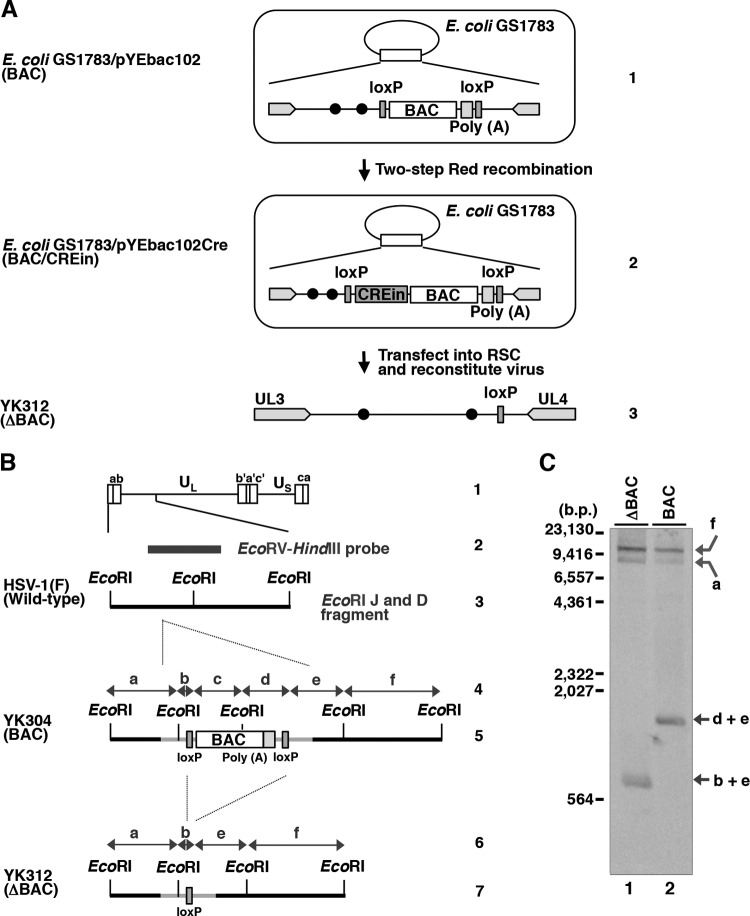
To examine the effects of UL51 and its phosphorylation at the identified phosphorylation sites, we constructed a series of UL51 mutant viruses using a self-excisable HSV-1(F) bacterial artificial chromosome (BAC) clone (pYEbac102Cre) generated in this study. To generate pYEbac102Cre, we modified the original HSV-1(F) BAC clone pYEbac102 (40) by inserting an expression cassette containing a gene encoding Cre recombinase into the intergenic site between the loxP site and the BAC sequence (Fig. 1A). Recombinant viruses reconstituted from pYEbac102Cre and its derivatives were expected to excise the BAC sequences through the functional Cre enzyme in HSV-1-infected cells as described previously (41). When the purified DNAs from YK304 (BAC) and YK312 (ΔBAC), reconstituted from pYEbac102 and pYEbac102Cre, respectively, were digested with EcoRI and analyzed by Southern blotting, the probe (Fig. 1B) hybridized to fragments a (10.6 kbp), f (15.0 kbp), and d+e (1.9 kbp) in YK304 (BAC), whereas fragment d+e was shifted to fragment b+e (1.0 kbp) in YK312 (ΔBAC) as a result of the BAC excision (Fig. 1B and C). Of note, the probe did not detect fragment b+c in YK304 (BAC) (Fig. 1C). These results indicated that the BAC vector sequence can be almost completely removed from the viral genome during the reconstitution of recombinant viruses in this system.
FIG 1.
Strategy to construct a self-excisable HSV-1(F) BAC clone. (A) Lines 1 and 2, schematic diagrams of E. coli plasmid pYEbac102 and pYEbac102Cre, respectively, contained in GS1783; line 3, schematic diagrams of recombinant virus YK312 (ΔBAC). (B) Schematic diagram of the genome structures of wild-type HSV-1(F) and relevant domains of the recombinant viruses. Line 1, sequence arrangement of the HSV-1(F) genome. Line 2, location of the EcoRV-HindIII fragment used as a labeled probe in panel C. Line 3, an enlarged portion of the EcoRI J and D fragments of HSV-1(F). Lines 4 and 6, double-headed arrow represents the fragment region. Lines 5 and 7, summary of DNA fragments from recombinant viruses with or without the BAC sequence and the poly(A) signals. (C) Southern blot analysis of EcoRI-digested DNA isolated from YK312 (ΔBAC)- or YK304 (BAC)-infected cells using the EcoRV-HindIII fragment of pRB442 as a probe. The letters on the right refer to the designations of the DNA fragments generated by restriction endonuclease EcoRI cleavage.
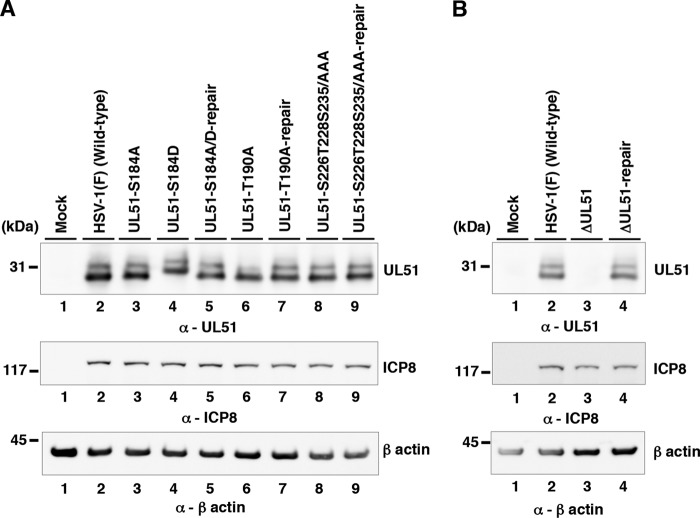
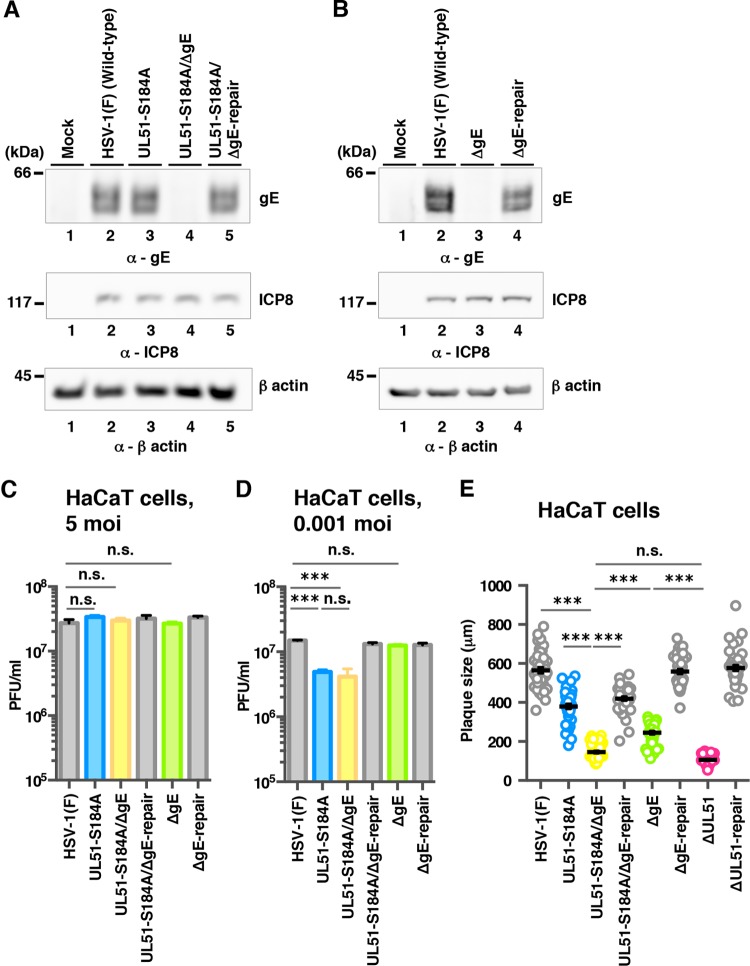
Using pYEbac102Cre, we constructed a series of recombinant viruses: recombinant virus YK631 (UL51-S184A), encoding a mutant UL51 in which Ser-184 was replaced with alanine (S184A), recombinant virus YK632 (UL51-S184D), carrying the phosphomimetic mutation (aspartic acid replacement) at Ser-184 (S184D), recombinant virus YK634 (UL51-T190A), encoding a mutant UL51 in which Thr-190 was replaced with alanine (T190A), recombinant virus YK636 (UL51-S226T228S235/AAA), encoding a mutant UL51 in which Ser-226, Thr-228, and Ser-235 were replaced with alanines (S226T228S235/AAA), and a UL51-null mutant virus YK638 (ΔUL51). Their repaired viruses were also generated: YK633 (UL51-S184A/D-repair), YK635 (UL51-T190A-repair), YK637 (UL51-S226T228S235/AAA-repair), and YK639 (ΔUL51-repair) (Fig. 2). YK633 (UL51-S184A/D-repair) was the repaired virus for both YK631 (UL51-S184A) and YK632 (UL51-S184D). These recombinant viruses were characterized as follows. (i) HaCaT cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK635 (UL51-T190A-repair), YK636 (UL51-S226T228S235/AAA), YK637 (UL51-S226T228S235/AAA-repair), or YK639 (ΔUL51-repair) accumulated UL51 and another HSV-1 protein ICP8 at comparable levels (Fig. 3A). (ii) As expected, HaCaT cells infected with wild-type HSV-1(F) or YK639 (ΔUL51-repair) produced UL51, but those infected with YK638 (ΔUL51) did not. Furthermore, the infected cells produced ICP8 at comparable levels (Fig. 3B). These results indicated that the phosphorylation of UL51 at Ser-184, Thr-190, Ser-226, Thr-228, and Ser-235 was not required for the correct accumulation of UL51 and ICP8 in HSV-1-infected cells. Notably, UL51 from infected cells was detected as doublet bands in denaturing gels as described previously (13), and T190A and S184D mutations in UL51 affected the migration of either or both of the isoforms of UL51 in denaturing gels, whereas other mutations in UL51 had no effect, suggesting that these two mutations altered the overall charge of UL51 and/or molecular mass of the viral protein (Fig. 3A).
FIG 2.
Schematic diagrams of the genome structure of HSV-1(F) and recombinant viruses used in this study. Line 1, wild-type HSV-1(F) genome; line 2, domains of the UL50 to UL52 and Us7 (gI) to Us8A genes; lines 3 to 15, recombinant viruses with mutations in the UL51 and/or Us8 (gE) genes. Stop denotes a stop codon.
FIG 3.
Effects of mutation(s) in UL51 on the steady-state expression levels of UL51. (A) HaCaT cells were mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), YK631 (UL51-S184A) (lane 3), YK632 (UL51-S184D) (lane 4), YK633 (UL51-S184A/D-repair) (lane 5), YK634 (UL51-T190A) (lane 6), YK635 (UL51-T190A-repair) (lane 7), YK636 (UL51-S226T228S235/AAA) (lane 8), or YK637 (UL51-S226T228S235/AAA-repair) (lane 9) at an MOI of 5, harvested at 18 h postinfection, and analyzed by immunoblotting with antibodies to UL51, ICP8, and β-actin. (B) HaCaT cells were mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), YK638 (ΔUL51) (lane 3), or YK639 (ΔUL51-repair) (lane 4) at an MOI of 5 and analyzed as described for panel A. A molecular mass marker is indicated on the left.
Effect of phosphorylation of the identified phosphorylation sites in UL51 on HSV-1 replication in cell cultures.
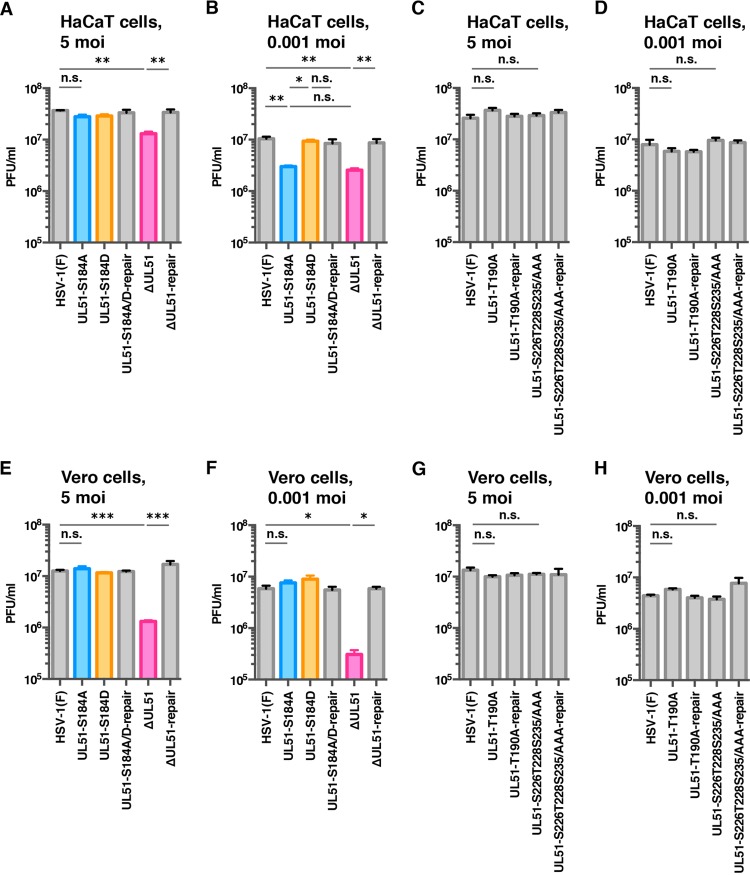
To examine the effect of the phosphorylation of UL51 Ser-184, Thr-190, Ser-226, Thr-228, or Ser-235 on viral replication in cell cultures, we analyzed progeny virus yields in HaCaT (Fig. 4A to D) and Vero (Fig. 4E to H) cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK635 (UL51-T190A-repair), YK636 (UL51-S226T228S235/AAA), YK637 (UL51-S226T228S235/AAA-repair), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at a multiplicity of infection (MOI) of 5 for 24 h (HaCaT cells) and 18 h (Vero cells) or at an MOI of 0.001 for 48 h (HaCaT cells) and 36 h (Vero cells). In agreement with previous reports (13, 19, 21), the progeny virus yields in HaCaT and Vero cells infected with YK638 (ΔUL51) at MOIs of 5 and 0.001 were significantly lower than those in these cells infected with wild-type HSV-1(F) and YK639 (ΔUL51-repair) (Fig. 4A, B, E, and F). In contrast, the progeny virus yields of YK631 (UL51-S184A), YK634 (UL51-T190A), and YK636 (UL51-S226T228S235/AAA) at an MOI of 5 in both HaCaT and Vero cells and at an MOI of 0.001 in Vero cells were similar to those of wild-type HSV-1(F) (Fig. 4A and C to H). Notably, the progeny virus yield of YK631 (UL51-S184A) in HaCaT cells at an MOI of 0.001 was significantly lower than those of wild-type HSV-1(F) and YK633 (UL51-S184A/D-repair) (Fig. 4B), whereas those of YK634 (UL51-T190A) and YK636 (UL51-S226T228S235/AAA) in these cells were similar to that of wild-type HSV-1(F) (Fig. 4D). Furthermore, the wild-type progeny virus yield was restored in HaCaT cells infected with YK632 (UL51-S184D), carrying a phosphomimetic mutation in UL51, at an MOI of 0.001 (Fig. 4B). These results indicated that the phosphorylation of UL51 Ser-184 was required for efficient viral replication in cell cultures but that the effect of phosphorylation on viral replication differed by cell type and MOI.
FIG 4.
Effects of mutation(s) in UL51 on viral replication. (A to H) HaCaT (A to D) or Vero (E to H) cells were infected at an MOI of 5 (A, C, and E) or 0.001 (B, D, and F) with wild-type HSV-1(F) (A to H), YK631 (UL51-S184A) (A, B, E, and F), YK632 (UL51-S184D) (A, B, E, and F), YK633 (UL51-S184A/D-repair) (A, B, E, and F), YK634 (UL51-T190A) (C, D, G, and H), YK635 (UL51-T190A-repair) (C, D, G, and H), YK636 (UL51-S226T228S235/AAA) (C, D, G, and H), YK637 (UL51-S226T228S235/AAA-repair) (C, D, G, and H), YK638 (ΔUL51) (A, B, E, and F), or YK639 (ΔUL51-repair) (A, B, E, and F). Total virus from cell culture supernatants and infected cells was harvested at 24 h (A and C), 48 h (B and D), 18 h (E and G), or 36 h (F and H) postinfection and assayed on Vero cells. Each data point is the mean ± standard error from triplicate or quadruplicate samples. Data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Tukey's test); n.s., not significant.
Effect of phosphorylation of the identified phosphorylation sites in UL51 on HSV-1 cell-cell spread in cell cultures.
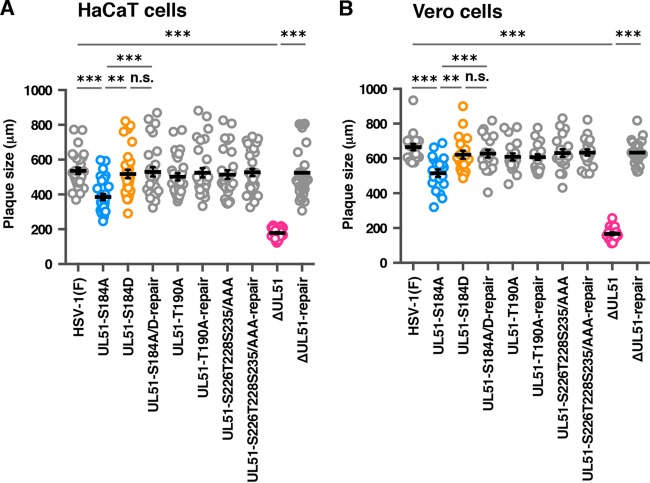
To examine the effect of phosphorylation of UL51 Ser-184, Thr-190, Ser-226, Thr-228, or Ser-235 on HSV-1 cell-cell spread in cell cultures, we analyzed plaque sizes in HaCaT and Vero cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK635 (UL51-T190A-repair), YK636 (UL51-S226T228S235/AAA), YK637 (UL51-S226T228S235/AAA-repair), YK638 (ΔUL51), or YK639 (ΔUL51-repair) using plaque assay conditions. In HaCaT cells, YK631 (UL51-S184A) produced smaller plaques than wild-type HSV-1(F) and YK633 (UL51-S184A/D-repair) and larger plaques than YK638 (ΔUL51) (Fig. 5A). Wild-type plaque sizes were restored in HaCaT cells infected with YK632 (UL51-S184D) (Fig. 5A). YK634 (UL51-T190A) and YK636 (UL51-S226T228S235/AAA) had plaque sizes similar to those of wild-type HSV-1(F) (Fig. 5A). Similar results were obtained with Vero cells infected with each of these viruses (Fig. 5B). Notably, in contrast to the growth properties of YK631 (UL51-S184A) in Vero cells at MOIs of 5 and 0.001, which were similar to those of wild-type HSV-1(F), this mutant virus produced significantly smaller plaques than wild-type HSV-1(F), YK633 (UL51-S184A/D-repair), and YK632 (UL51-S184D) (Fig. 5B). The effects of the null mutation in UL51 on HSV-1 cell-cell spread in these cells agreed with previous reports (19–21). These results suggested that the phosphorylation of UL51 Ser-184 was required for efficient HSV-1 cell-cell spread in both HaCaT and Vero cells.
FIG 5.
Effects of mutation(s) in UL51 on viral cell-to-cell spread. (A and B) HaCaT (A) and Vero (B) cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK635 (UL51-T190A-repair), YK636 (UL51-S226T228S235/AAA), YK637 (UL51-S226T228S235/AAA-repair), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 0.001 (A) or 0.0001 (B) under plaque assay conditions. The diameters of 30 (A) or 20 (B) single plaques for each of the indicated viruses were measured 48 h postinfection. Each data point is the mean ± standard error of the measured plaque sizes. The horizontal bars indicate the mean for each group. **, P < 0.01; ***, P < 0.001 (by Tukey's test); n.s., not significant.
Effects of a null mutation in gE on the replication and cell-cell spread of YK631 (UL51-S184A) in HaCaT cells.
To investigate the relationship between the effects of UL51 phosphorylation at Ser-184 and those of gE on HSV-1 replication and cell-cell spread, we constructed a gE-null mutant virus YK640 (ΔgE), a recombinant virus YK642 (UL51-S184A/ΔgE) carrying both a S184A mutation in UL51 and a null mutation in gE, and their repaired viruses, YK641 (ΔgE-repair) and YK643 (UL51-S184A/ΔgE-repair) (Fig. 2). These recombinant viruses were characterized as follows. (i) As expected, HaCaT cells infected with wild-type HSV-1(F), YK641 (ΔgE-repair), or YK643 (UL51-S184A/ΔgE-repair) expressed gE (Fig. 6A and B), but cells infected with YK640 (ΔgE) or YK642 (UL51-S184A/ΔgE) did not (Fig. 6A and B). (ii) The S184A mutation in UL51 had no effect on the accumulation of gE, and all mutations in gE or UL51 had no effect on the accumulation of ICP8 in HSV-1-infected cells (Fig. 6A and B).
FIG 6.
Effects of null mutation in gE on viral replication and cell-to-cell spread of UL51 mutant virus. (A) HaCaT cells were mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), YK631 (UL51-S184A) (lane 3), YK642 (UL51-S184A/ΔgE) (lane 4), or YK643 (UL51-S184A/ΔgE-repair) (lane 5) at an MOI of 5, harvested at 18 h postinfection, and analyzed by immunoblotting with antibodies to gE, ICP8, and β-actin. (B) HaCaT cells were mock infected (lane 1) or infected with wild-type HSV-1(F) (lane 2), YK640 (ΔgE) (lane 3), or YK641 (ΔgE-repair) (lane 4) at an MOI of 5 and analyzed as described for panel A. (C and D) HaCaT cells were infected at an MOI of 5 (C) or 0.001 (D) with wild-type HSV-1(F), YK631 (UL51-S184A), YK642 (UL51-S184A/ΔgE), YK643 (UL51-S184A/ΔgE-repair), YK640 (ΔgE), or YK641 (ΔgE-repair). Total virus from cell culture supernatants and infected cells was harvested at 24 h (C) or 48 h (D) postinfection and assayed on Vero cells. Each data point is the mean ± standard error from three independent experiments. (E) HaCaT cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK642 (UL51-S184A/ΔgE), YK643 (UL51-S184A/ΔgE-repair), YK640 (ΔgE), YK641 (ΔgE-repair), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 0.001 under plaque assay conditions. The diameters of 40 single plaques for each of the indicated viruses were measured 48 h postinfection. Each data point is the mean ± standard error from the measured plaque sizes. The horizontal bars indicate the mean for each group. ***, P < 0.001 (by Tukey's test); n.s., not significant.
We then analyzed progeny virus yields in HaCaT cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK642 (UL51-S184A/ΔgE), YK643 (UL51-S184A/ΔgE-repair), YK640 (ΔgE), or YK641 (ΔgE-repair). In agreement with previous reports (42–44), the gE-null mutation had no significant effect on HSV-1 replication in HaCaT cells at MOIs of 5 and 0.001 (Fig. 6C and D). Similarly, the progeny virus yields of YK642 (UL51-S184A/ΔgE) in HaCaT cells at MOIs of 5 and 0.001 were similar to those of YK631 (UL51-S184A), although the virus yields of YK631 (UL51-S184A) and YK642 (UL51-S184A/ΔgE) in HaCaT cells at an MOI of 0.001 were significantly lower than those of wild-type HSV-1(F) and YK643 (UL51-S184A/ΔgE-repair) (Fig. 6C and D). We also analyzed plaque sizes in the infected cells. As shown in Fig. 6E, YK631 (UL51-S184A) produced smaller plaques than wild-type HSV-1(F), larger plaques than YK642 (UL51-S184A/ΔgE) and YK640 (ΔgE), and plaques of sizes similar to those of YK643 (UL51-S184A/ΔgE-repair). YK642 (UL51-S184A/ΔgE) produced smaller plaques than YK640 (ΔgE) and plaques of sizes similar to those of YK638 (ΔUL51) (Fig. 6E). Thus, the S184A mutation in UL51 and the gE-null mutation synergistically reduced HSV-1 cell-cell spread in HaCaT cells. Collectively, these results indicated that the phosphorylation of UL51 Ser-184 regulated HSV-1 cell-cell spread in HaCaT cells independent of gE.
Effect of phosphorylation of the identified phosphorylation sites in UL51 on HSV-1 morphogenesis in HaCaT cells.
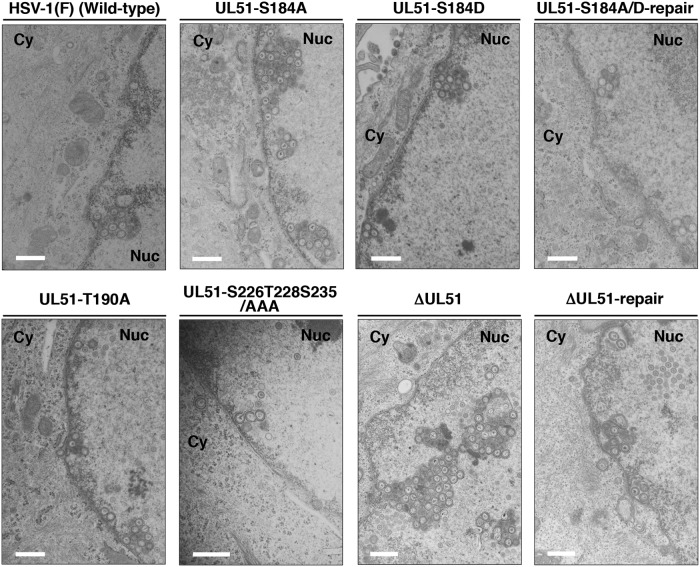
To determine the step(s) at which the phosphorylation of UL51 Ser-184 regulates UL51 during HSV-1 replication in HaCaT cells, we investigated viral morphogenesis by quantitating the number of virus particles by electron microscopy analysis of different morphogenetic stages of HaCaT cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK636 (UL51-S226T228S235/AAA), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 5 for 18 h.
As shown in Fig. 7, in HaCaT cells infected with YK631 (UL51-S184A) or YK638 (ΔUL51), membranous structures containing primary enveloped virions formed by invaginations of the INM into the nucleoplasm were readily observed adjacent to the nuclear membrane. These membranous invagination structures were present at a lower but consistent frequency in HaCaT cells infected with wild-type HSV-1(F), YK633 (UL51-S184A/D-repair), or YK639 (ΔUL51-repair) (Fig. 7), as reported previously (45). The quantification of virus particles at different morphogenetic stages showed that 24.4% and 27.8% of enveloped virions in HaCaT cells infected with YK631 (UL51-S184A) or YK638 (ΔUL51), respectively, were present in the perinuclear space and invagination structures. However, in HaCaT cells infected with wild-type HSV-1(F), YK633 (UL51-S184A/D-repair), or YK639 (ΔUL51-repair), 4.9%, 10.5%, or 10.7% of enveloped virions, respectively, were present in these compartments (Table 2). The wild-type frequency of enveloped virions in the perinuclear space and invagination structures were restored in HaCaT cells infected with YK632 (UL51-S184D) (Fig. 7 and Table 2). The fraction of total virus particles in the nucleus of HaCaT cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK638 (ΔUL51), or YK639 (ΔUL51-repair) was comparable (37.6% to 49.5%) (Table 2).
FIG 7.
Ultrastructural analysis of the effects of mutation(s) in UL51 on viral nuclear egress in HaCaT cells. HaCaT cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK636 (UL51-S226T228S235/AAA), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 5, fixed at 18 h postinfection, embedded, sectioned, stained, and examined by transmission electron microscopy. Nuc, nucleus; Cy, cytoplasm. Scale bars, 500 nm.
TABLE 2.
Effects of the mutations in UL51 on the distribution of viral particles in HSV-1-infected HaCaT cells
| Virus | Avg % of virus particles in morphogenetic stage (no. of particles in stage) |
Total counted (particles/cells) | |||||
|---|---|---|---|---|---|---|---|
| Capsids in nucleus | Enveloped virions in perinuclear space | Unenveloped capsids in cytoplasm | Partially enveloped capsids in cytoplasm | Enveloped virions in cytoplasm | Extracellular enveloped virions | ||
| HSV-1 (F) (wild type) | 39.7 (1,088) | 4.9 (135) | 2.5 (69) | 1.3 (36) | 4.0 (109) | 47.5 (1,301) | 2,738/15 |
| YK631 (UL51-S184A) | 37.6 (1,630) | 24.4 (1,057) | 1.0 (42) | 1.0 (42) | 3.2 (138) | 32.9 (1,428) | 4,337/15 |
| YK632 (UL51-S184D) | 46.4 (1,622) | 7.1 (248) | 2.2 (76) | 1.3 (45) | 5.0 (176) | 38.0 (1,328) | 3,495/15 |
| YK633 (UL51-S184A/D-repair) | 44.4 (1,609) | 10.5 (381) | 1.8 (64) | 1.1 (40) | 5.3 (193) | 36.9 (1,335) | 3,622/15 |
| YK634 (UL51-T190A) | 47.7 (1,731) | 12.5 (455) | 1.8 (64) | 1.2 (42) | 4.1 (149) | 32.8 (1,189) | 3,630/15 |
| YK636 (UL51-S226T228S235/AAA) | 47.9 (1,880) | 9.5 (374) | 1.1 (42) | 1.1 (42) | 4.0 (155) | 36.6 (1,436) | 3,929/15 |
| YK638 (ΔUL51) | 49.5 (1,905) | 27.8 (1,072) | 4.1 (158) | 2.7 (102) | 1.4 (52) | 14.6 (563) | 3,852/15 |
| YK639 (ΔUL51-repair) | 45.8 (1,694) | 10.7 (397) | 1.0 (35) | 1.1 (41) | 4.4 (163) | 37.0 (1,369) | 3,699/15 |
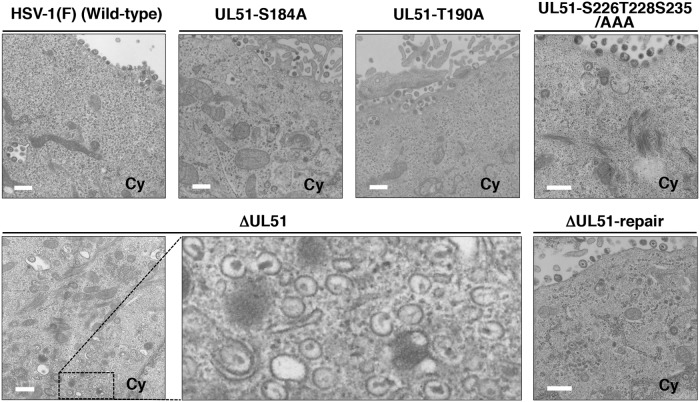
In the cytoplasm of HaCaT cells infected with YK638 (ΔUL51), 2.7% of virus particles were partially enveloped nucleocapsids. In contrast, only 1.3%, 1.0%, or 1.1% of virus particles were partially enveloped nucleocapsids in the cytoplasm of cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), or YK639 (ΔUL51-repair), respectively (Fig. 8 and Table 2). The frequency (6.8%) of unenveloped or partially enveloped nucleocapsids in the cytoplasm of HaCaT cells infected with YK638 (ΔUL51) was higher than the frequency of virions in the cytoplasm of cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), or YK639 (ΔUL51-repair) (3.8%, 2.0%, and 2.1%), respectively (Fig. 8 and Table 2). In contrast, 51.5%, 36.1%, or 41.4% of virus particles in HaCaT cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), or YK639 (ΔUL51-repair), respectively, were enveloped virions in the cytoplasm and extracellular space. However, in cells infected with YK638 (ΔUL51), the percentage of virus particles that were enveloped virions in the cytoplasm and extracellular space decreased to 16.0% (Fig. 8 and Table 2).
FIG 8.
Ultrastructural analysis of the effects of mutation(s) in UL51 on viral final envelopment in the cytoplasm of HaCaT cells. HaCaT cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK636 (UL51-S226T228S235/AAA), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 5, fixed at 18 h postinfection, embedded, sectioned, stained, and examined by transmission electron microscopy. Scale bar, 500 nm.
Results similar to those with wild-type HSV-1(F) were also obtained with YK634 (UL51-T190A)- or YK636 (UL51-S226T228S235/AAA)-infected HaCaT cells (Fig. 7 and 8 and Table 2).
Collectively, these results indicated the following. (i) In HaCaT cells, the UL51-null mutation and preclusion of the phosphorylation of UL51 Ser-184 induced membranous invaginations containing primary enveloped virions adjacent to the nuclear membrane and induced the comparable aberrant accumulation of primary envelope virions in the perinuclear space and the induced invagination structures. (ii) In HaCaT cells, the UL51-null mutation, but not preclusion of the phosphorylation of UL51 Ser-184, induced the comparable aberrant accumulation of unenveloped nucleocapsids and partially enveloped nucleocapsids in the cytoplasm. These phenotypes observed in the cytoplasm of cells with the UL51-null mutation were in agreement with those reported previously (13).
Effects of UL51 and its phosphorylation on the intracellular localization of UL34 and UL31 in HaCaT cells.
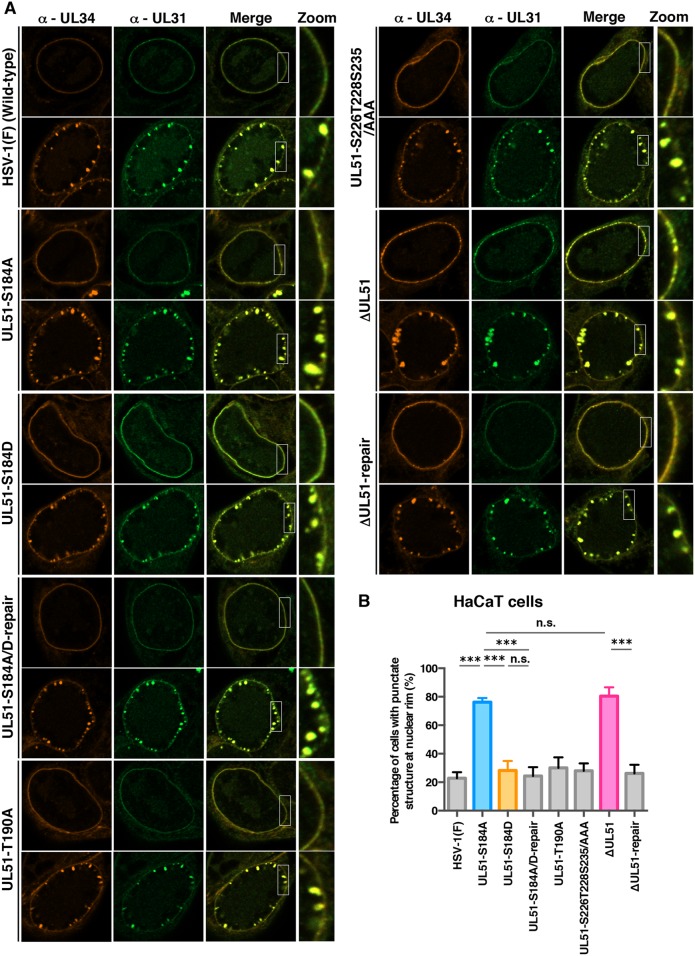
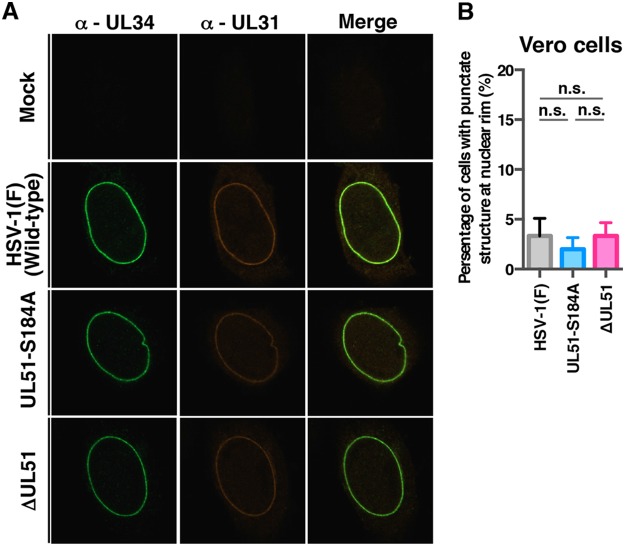
Our data showed that null and S184A mutations in UL51 induced membranous invaginations containing primary enveloped virions adjacent to the nuclear membrane and in the perinuclear space. This led us to investigate the effects of these mutations on the intracellular localization of UL34 and UL31, which are critical for the nuclear egress of HSV-1 nucleocapsids (3–5, 46–49), because the phenotypes observed above were reported to be linked to the aberrant localization of UL34 and UL31 (38, 45, 47, 48, 50). To this end, HaCaT cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK636 (UL51-S226T228S235/AAA), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 5 for 18 h, and the localization of UL34 and UL31 in these infected cells was analyzed by confocal microscopy. Although the UL34 and UL31 proteins were colocalized along the nuclear rim in the majority of HaCaT cells infected with wild-type HSV-1(F), YK633 (UL51-S184A/D-repair), or YK639 (ΔUL51-repair) (69.9% to 77.2%), some UL34 and UL31 proteins were colocalized in punctate structures adjacent to the nuclear rim (22.8% to 30.1%) (Fig. 9), as reported previously (45). In contrast, in HaCaT cells infected with YK631 (UL51-S184A) or YK638 (ΔUL51), UL31 and UL34 were colocalized in punctate structures adjacent to the nuclear rim in the majority of cells (76.2% or 80.5%, respectively) and were colocalized along the nuclear rim in a smaller percentage of cells (Fig. 9). The fraction of YK631 (UL51-S184A)- or YK638 (ΔUL51)-infected HaCaT cells with punctate structures (76.2% or 80.5%, respectively) was significantly larger than that of cells infected with HSV-1(F) or each of the corresponding repair viruses with punctate structures (22.8% to 30.1%) (Fig. 9B). Differences in the frequencies between HaCaT cells with punctate structures infected with YK631 (UL51-S184A) and those with punctate structures infected with YK638 (ΔUL51) were not statistically significant (Fig. 9B). Furthermore, the wild-type frequency of HaCaT cells with punctate structures (28.2%) was restored in HaCaT cells infected with YK632 (UL51-S184D) (Fig. 9B). Results similar to those with wild-type HSV-1(F) were also obtained with YK634 (UL51-T190A)- or YK636 (UL51-S226T228S235/AAA)-infected HaCaT cells (Fig. 9A). In contrast, in Vero cells infected with YK631 (UL51-S184A) or YK638 (ΔUL51), the punctate structures of UL34 and UL31 at the nuclear membrane were barely detectable, similar to Vero cells infected with wild-type HSV-1(F) (Fig. 10A and B). These results indicated that UL51 and its phosphorylation at Ser-184 were required for the correct localization of UL34 and UL31 in HSV-1-infected HaCaT cells but not in Vero cells.
FIG 9.
Effects of mutation(s) in UL51 on the localization of UL31 and UL34 in HaCaT cells. (A) HaCaT cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK636 (UL51-S226T228S235/AAA), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 5, fixed at 18 h postinfection, permeabilized, stained with anti-UL34 and anti-UL31 antibodies, and examined by confocal microscopy. Each image in the far-right column is the magnified image of the boxed area in the image to its left. Because different patterns of UL31 and UL34 localization were observed in cells infected with each of the indicated viruses, images of two of the infected cells are shown here as examples of UL31 and UL34 localization. (B) Quantitation of infected HaCaT cells with aberrant punctate structures of UL31 and UL34 adjacent to the nuclear rim. The experiments were performed as described for panel A, and the percentage of cells with aberrant punctate structures at the nuclear rim was determined. Each value is the mean ± standard error of the results from three independent experiments. ***, P < 0.001 (by Tukey's test); n.s., not significant.
FIG 10.
Effects of mutation(s) in UL51 on the localization of UL31 and UL34 in Vero cells. (A) Vero cells were infected with wild-type HSV-1(F), YK631 (UL51-S184A), or YK638 (ΔUL51) at an MOI of 5, fixed at 12 h postinfection, permeabilized, stained with anti-UL34 and anti-UL31 antibodies, and examined by confocal microscopy. (B) Quantitation of infected Vero cells with aberrant punctate structures of UL31 and UL34 adjacent to the nuclear rim. The experiments were performed as described for panel A, and the percentage of cells with aberrant punctate structures at the nuclear rim was determined. Each value is the mean ± standard error of the results from three independent experiments. n.s., not significant.
Localization of UL51 in HSV-1-infected HaCaT cells.
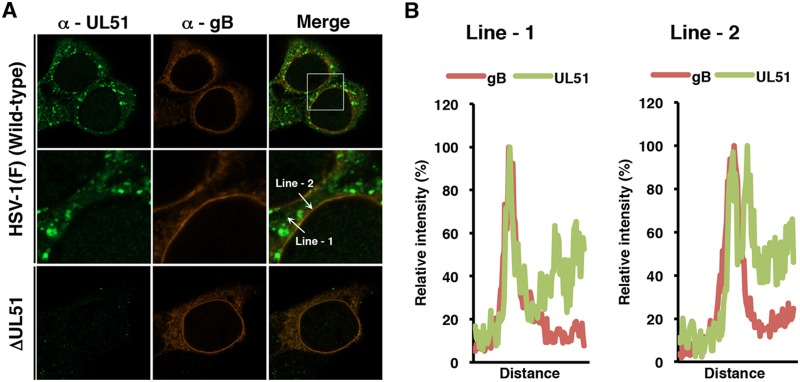
The data described above suggested that UL51 regulated HSV-1 nuclear egress at the nuclear membrane; therefore, we investigated the localization of UL51 in infected HaCaT cells. In agreement with previous reports (11, 13, 19, 21), UL51 accumulated in cytoplasmic punctate structures in HaCaT cells infected with wild-type HSV-1(F) (Fig. 11A). UL51, unlike gB, localized in cytoplasmic perinuclear regions at the nuclear rim and at sites other than the nuclear membrane (Fig. 11A). Of note, UL51 also localized at the nuclear rim and colocalized with gB, which was reported to localize both at the inner and outer nuclear membranes (51, 52) (Fig. 11A). Line plot analyses verified that UL51 was mainly colocalized with gB at the nuclear rim (Fig. 11B), suggesting UL51 localizes at the nuclear membrane in HSV-1-infected HaCaT cells.
FIG 11.
Localization of UL51 and gB in HaCaT cells. (A) HaCaT cells were infected with wild-type HSV-1(F) or YK638 (ΔUL51) at an MOI of 5, fixed at 18 h postinfection, permeabilized, stained with anti-UL51 and anti-gB antibodies, and examined by confocal microscopy. Each image in the middle column is the magnified image of the boxed area in the upper column. (B) Line-scan analysis of colocalization between UL51 and gB. The experiments were performed as described for panel A, and the fluorescence intensity of white arrows in the middle panels was determined.
Effects of UL51 or its phosphorylation at Ser-184 on pathogenesis in mice.
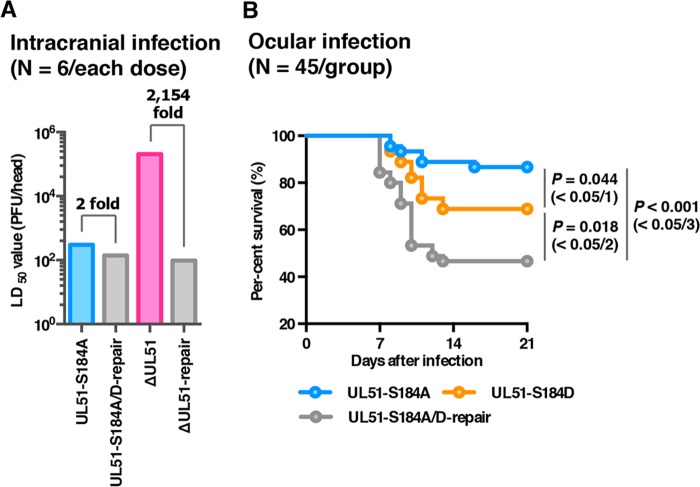
To examine the effects of UL51 and the phosphorylation of UL51 Ser-184 on HSV-1 infection in vivo, 3-week-old female ICR mice were infected intracranially with 10-fold dilutions of YK631 (UL51-S184A), YK633 (UL51-S184A/D-repair), YK638 (ΔUL51), or YK639 (ΔUL51-repair), and mortality was monitored for 14 days postinfection. As shown in Fig. 12A, the 50% lethal dose (LD50) value of YK638 (ΔUL51) was approximately 2 × 103-fold greater than that of YK639 (ΔUL51-repair). In contrast, the LD50 value of YK631 (UL51-S184A) was comparable to that of YK633 (UL51-S184A/D-repair) (Fig. 12A). These results indicated that UL51 was critical for HSV-1 virulence in mice following intracranial infection, whereas the phosphorylation of UL51 Ser-184 was not.
FIG 12.
Effects of mutation(s) in UL51 on the mortality of mice following intracranial or ocular infection. (A) Three-week-old female mice were inoculated intracranially with serial 10-fold dilutions of the indicated viruses in groups of 6 per dilution and were monitored for 14 days. LD50 values were determined by the Behrens-Karber method. (B) Forty-five 3-week-old female ICR mice were infected ocularly with 1 × 106 PFU/eye of each virus and were monitored for 14 days. Differences in the mortality of infected mice were statistically analyzed by the log-rank test, and for three comparison analyses, P values of <0.0167 (0.05/3), <0.025 (0.05/2), or <0.05 (0.05/1) were sequentially considered significant after Holm's sequentially rejective Bonferroni multiple-comparison adjustment.
To further examine the effect of the phosphorylation of UL51 Ser-184 in HSV-1 pathogenesis, 4-week-old female ICR mice were ocularly infected with 1 × 106 PFU/eye of YK631 (UL51-S184A), YK632 (UL51-S184D), or YK633 (UL51-S184A/D-repair), and mortality was monitored for 14 days postinfection. As shown in Fig. 12B, the survival of mice infected with YK631 (UL51-S184A) was significantly lower than that in mice infected with YK633 (UL51-S184A/D-repair) or YK632 (UL51-S184D), although the survival of mice infected with YK632 (UL51-S184D) was significantly greater than that of mice infected with YK633 (UL51-S184A/D-repair). These results suggested that the phosphorylation of UL51 Ser-184 was required for efficient HSV-1 virulence in mice following ocular infection.
DISCUSSION
Although HSV-1 UL51 has long been recognized as a phosphoprotein (12), the regulatory effect of UL51 phosphorylation in HSV-1-infected cells is unclear. In this study, the MS analysis of cells ectopically expressing UL51 identified Ser-184, Thr-190, Ser-226, Thr-228, and Ser-235 as phosphorylation sites in UL51. The phosphorylation of UL51 at Ser-184, Thr-190, and/or Ser-226 in HSV-1-infected cells was previously reported in a large-scale phosphoproteomics analysis of HSV-1-infected cells (30–33); however, the biological significance of these phosphorylation sites in HSV-1 infection is unknown. In contrast, the phosphorylation of UL51 at Thr-228 and Ser-235 has not been reported. Studies with mutant viruses in which the phosphorylation of UL51 Ser-184, Thr-190, Ser-226, Thr-228, and/or Ser-235 was precluded or mimicked by amino acid substitution(s) revealed that among the identified UL51 phosphorylation sites, only UL51 Ser-184 was functional in HSV-1-infected cells, and this was dependent on cell type and MOI. We report that the phosphorylation of UL51 Ser-184 was required for efficient HSV-1 replication in HaCaT cells at an MOI of 0.001 and for efficient viral cell-cell spread in both HaCaT and Vero cells. In contrast, phosphorylation was not required for HSV-1 replication in HaCaT cells at an MOI of 5 and in Vero cells at MOIs of 0.001 and 5. Importantly, we demonstrated that the phosphorylation of UL51 Ser-184 was required for efficient HSV-1 pathogenicity in mice following ocular infection. From these observations, we concluded that UL51 was regulated by its phosphorylation at Ser-184 and that this regulation was critical for HSV-1 replication and pathogenicity. UL51 is conserved in all members of the family Herpesviridae (1). This is the first report clarifying a regulatory mechanism of UL51 in any herpesvirus UL51 orthologs as well as showing the significance of the phosphorylation of UL51 in HSV-1 infection.
Roller et al. previously showed that the UL51 regulation of HSV-1 cell-cell spread might be dependent on gE, based on observations that UL51 interacted with gE and a dominant-negative mutant of UL51 mislocalized gE in HSV-1-infected cells (19). Of note, they also reported that a cell-cell spread defect associated with a UL51-null mutation was more severe than that associated with a gE-null mutation in HSV-1-infected cells, suggesting that UL51 also functions in HSV-1 cell-cell spread independent of gE (19). In this study, we showed that the preclusion of phosphorylation at UL51 Ser-184 and gE-null mutation synergistically reduced HSV-1 cell-cell spread in HaCaT cells. These results clearly support the hypothesis of Roller et al. and indicate that this gE-independent UL51 function for the promotion of HSV-1 cell-cell spread is regulated by phosphorylation at Ser-184. Recently, Albecka et al. reported that both UL51 and UL7 were required to maintain the morphology of HSV-1-infected cells as well as to stabilize focal adhesions, which have an important role in cell attachment (53), and that null mutation(s) in either or both of these HSV-1 proteins equivalently impaired HSV-1 cell-cell spread (21). Based on these observations, they proposed that the UL51-UL7 complex promotes HSV-1 cell-cell spread by stabilizing focal adhesions (21). Therefore, it will be of interest to investigate the effects of the phosphorylation at UL51 Ser-184 on the morphology of HSV-1-infected cells and the stabilization of focal adhesions.
We also identified at which step of HSV-1 replication the phosphorylation of UL51 Ser-184 is regulated in HaCaT cells. Preclusion of phosphorylation at UL51 Ser-184 and the UL51-null mutation in HaCaT cells induced membranous invagination structures adjacent to the nuclear membrane that accumulated primary enveloped virions. Of note, these phenotypes with the UL51-null mutation were not observed in Vero cells in our recent study (13). Aberrant virion accumulation in membranous invagination structures might reflect a decreased rate of virion egress from the perinuclear space, whereas the rate of virion delivery into this space may be unchanged or decreased to a lesser degree. Therefore, factors including HSV-1 Us3, cellular factors, CD98 heavy chain, and p32, whose inactivation is responsible for these phenotypes, might regulate HSV-1 de-envelopment (38, 47, 48, 50). In addition, the inactivation of these known de-envelopment factors resulted in the aberrant localization of UL31 and UL34 in punctate structures adjacent to the nuclear rim (38, 47, 48, 50). Furthermore, in the current study we showed that the preclusion of phosphorylation at UL51 Ser-184 and the UL51-null mutation induced this phenotype in HaCaT cells but not in Vero cells. Importantly, the preclusion of phosphorylation at UL51 Ser-184 induced these phenotypes at levels similar to those of the UL51-null mutation, and the phosphomimetic mutation at UL51 Ser-184 restored the phenotypes to wild-type levels. From these results, we concluded that UL51 promoted HSV-1 de-envelopment during nuclear egress dependent on cell type and that the regulation of UL51 by phosphorylation at Ser-184 was critical for this UL51 function. In agreement with this conclusion, cell type-dependent defects in HSV-1 de-envelopment were reported for simultaneous mutations in HSV-1 gB and gH: the dual mutations induced the aberrant (>60-fold) accumulation of primary enveloped virions in the perinuclear space in HaCaT cells but had little effect (only approximately 2-fold) in Vero cells (51). Interestingly, in addition to defects in HSV-1 de-envelopment, the UL51-null mutation caused defects in viral secondary envelopment in HaCaT cells, as previously observed with this mutation in Vero cells (13). In contrast, the preclusion of phosphorylation at UL51 Ser-184 in HaCaT cells had no effect on virion morphogenesis in the cytoplasm, indicating that UL51 Ser-184 phosphorylation regulated only some UL51 functions.
Early studies testing recombinant PRV and VZV carrying a null mutation in UL51 homologs in mice and SCID-hu dorsal root ganglia xenograft mice, respectively, reported these UL51 homologs were required for efficient viral neurovirulence, neuroinvasiveness, and/or spread in vivo (16, 18, 54). However, a role for HSV-1 UL51 in viral pathogenicity has not been reported thus far. In this study, we showed that the UL51-null mutation remarkably (approximately 2 × 103-fold) impaired survival in mice following intracranial infection. In contrast, the preclusion of phosphorylation at UL51 Ser-184 had no effect on virulence in mice following intracranial infection but significantly reduced viral virulence in mice following ocular infection. In experimental mouse models of HSV-1 infection, virulence following intracranial infection and at peripheral sites, such as the eye and vagina, were important and semi-independent indicators of HSV-1 virulence. Virulence (neurovirulence) in mice following intracranial infection depends on the ability of HSV-1 to replicate in the central nervous system (CNS), whereas virulence (neuroinvasiveness) in mice following ocular infection results from the sum of multiple capacities of HSV-1 to replicate in the eyes, trigeminal ganglia, and brains, thereby gaining access to the trigeminal ganglia from the eye and invading the CNS from the trigeminal ganglia. Our observations in this study indicated that HSV-1 UL51 was a significant neurovirulence factor that has a role in viral neuroinvasiveness. Although the regulation of UL51 by its phosphorylation at Ser-184 did not appear to be required for viral neurovirulence, it was critical for HSV-1 neuroinvasiveness. Thus, the phosphorylation-regulated functions of UL51 reported in this study, including the promotion of de-envelopment, cell-cell spread, and viral replication, might contribute to HSV-1 pathogenicity.
MATERIALS AND METHODS
Cells and viruses.
Simian kidney epithelial Vero cells, human keratinocyte HaCaT cells, human embryonic kidney epithelial HEK293T cells, and rabbit skin cells, as well as HSV-1 wild-type strain HSV-1(F), were described previously (36, 40, 45, 55). Recombinant virus YK304, reconstituted from pYEbac102 containing a complete HSV-1(F) sequence with the BAC sequence inserted into the HSV intergenic region between UL3 and UL4, was described previously (40). YK304 has an phenotype identical to that of wild-type HSV-1(F) in cell cultures and in mouse models (40).
Plasmids.
pcDNA-CREin-KanS, used to generate pYEbac102Cre, was constructed by amplifying the Cre-encoding gene, containing a synthetic intron that prevented the expression of the functional protein in Escherichia coli, by PCR from pBS-CREin-KanS (56) using the primers shown in Table 3. This was then cloned into the EcoRI site of pcDNA3.1(+) (Thermo Fisher Scientific). pcDNA-FEM-KanS was constructed by PCR amplification of the domain containing the FEM tag, I-SceI site, and kanamycin resistance gene from pBS-FEM-KanS and cloning it into the EcoRI and HindIII sites of pcDNA3.1/myc-His(−)-A (Thermo Fisher Scientific). pcDNA-FEM was constructed by digesting pcDNA-FEM-KanS with NheI and self-ligating to remove the I-SceI site and kanamycin resistance gene. pcDNA-FEM-UL51 was constructed by amplifying the entire UL51 open reading frame (ORF) from pYEbac102 by PCR and cloning it into pcDNA-FEM in frame with FEM. pBS1007, used to generate the recombinant virus YK639 (ΔUL51-repair), was described previously (11). pRB442, used to generate the probe for Southern blotting, contains a 6.3-kbp KpnI-HindIII fragment of HSV-1(F), which encompasses the left end of the UL region, including the region between 0.0415 and 0.084 map units, as described previously (57).
TABLE 3.
Oligonucleotide sequences for the construction of plasmids and recombinant viruses
| Plasmid or recombinant virus | Sequence (5′–3′) |
|---|---|
| pcDNA-CREin-KanS | 5′-GCGAATTCATGCCCAAGAAGAAGAGGAAGGTGTCCA-3′ |
| 5′-ACTAGGCGGCCGCGCGAATTCCTAATCGCCATCTTCCAGCA-3′ | |
| pBS-FEM-KanS (1st PCR) | 5′-ATGGACTACAAGGACGACGATGACAAAGATGACCGTGATTATGATATTCCAACTACTGCTAGCGAGAATTTGTATTTTCAGGGTGAAGGATGACGACGATAAGTAGGG-3′ |
| 5′-TCACAAGTCCTCTTCAGAAATGAGCTTTTGCTCCATGGTGGCGGATCCGAGCTCACCCTGAAAATACAAATTCTCGCTAGCAGTAGTTGGAACAACCAATTAACCAATTCTGATTAG-3′ | |
| pBS-FEM-KanS (2nd PCR) | 5′-ATAAGAATGCGGCCGCATGGACTACAAGGACGACG-3′ |
| 5′-GAGAATTCTCACAAGTCCTCTTCAGAAATGAGC-3′ | |
| pYEbac102Cre | 5′-GCCAGTAGTGCTCGCCGCAGTCGAGCGACAGGGCGAAGCCACATTGATTATTGACTAGTTATTAATAG-3′ |
| 5′-ATATAAGTGCTGTTCCCTGGTGCTTCCTCGCTCACTCGAGCCATAGAGCCCACCGCATCC-3′ | |
| YK631 (UL51-S184A) | 5′-GGGAACCCGCTTGGGGGGGCTTGGGGTGACCGAGGCGCCCGCCTTGGGGCACCCCCACACGCCAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CCAGCGTAACCTCCGGGGGCGGCGTGTGGGGGTGCCCCAAGGCGGGCGCCTCGGTCACCCCAACAACCAATTAACCAATTCTGATTAG-3′ | |
| YK632 (UL51-S184D) | 5′-GGGAACCCGCTTGGGGGGGCTTGGGGTGACCGAGGCGCCCGACTTGGGGCACCCCCACACGCCAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CCAGCGTAACCTCCGGGGGCGGCGTGTGGGGGTGCCCCAAGTCGGGCGCCTCGGTCACCCCAACAACCAATTAACCAATTCTGATTAG-3′ | |
| YK633 (UL51-S184A/D-repair) | 5′-GGGAACCCGCTTGGGGGGGCTTGGGGTGACCGAGGCGCCCTCCTTGGGGCACCCCCACACGCCAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CCAGCGTAACCTCCGGGGGCGGCGTGTGGGGGTGCCCCAAGGAGGGCGCCTCGGTCACCCCAACAACCAATTAACCAATTCTGATTAG-3′ | |
| YK634 (UL51-T190A) | 5′-GCTTGGGGGGGCTTGGGGTGACCGAGGCGCCCTCCTTGGGGCACCCCCACGCACCGCCCCCGGAGGTTACAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CGTTTCGGGCGGCAGGCGCCAGCGTAACCTCCGGGGGCGGTGCGTGGGGGTGCCCCAAGGCAACCAATTAACCAATTCTGATTAG-3′ | |
| YK635 (UL51-T190A-repair) | 5′-GCTTGGGGGGGCTTGGGGTGACCGAGGCGCCCTCCTTGGGGCACCCCCACACGCCGCCCCCGGAGGTTACAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CGTTTCGGGCGGCAGGCGCCAGCGTAACCTCCGGGGGCGGCGTGTGGGGGTGCCCCAAGGCAACCAATTAACCAATTCTGATTAG-3′ | |
| YK636 (UL51-S226T228S235/AAA) | 5′-GGAATCGTGTCCCCGCGTGTCCGTTCCCAGACCTACAGCCGCCCCAGCCGCACCTCGCCCTGGCCCAGCTCGAGCAGGATGACGACGATAAGTAGGG-3′ |
| 5′-AAACACGCATTTATTGACCCAAAACACACGGAGCTGCTCGAGCTGGGCCAGGGCGAGGTGCGGCTGGGGCGGCTGCAACCAATTAACCAATTCTGATTAG-3′ | |
| YK637 (UL51-S226T228S235/AAA-repair) | 5′-GGAATCGTGTCCCCGCGTGTCCGTTCCCAGACCTACAGCCTCCCCAACCGCACCTCGCCCTGGCCCATCTCGAGCAGGATGACGACGATAAGTAGGG-3′ |
| 5′-AAACACGCATTTATTGACCCAAAACACACGGAGCTGCTCGAGATGGGCCAGGGCGAGGTGCGGTTGGGGAGGCTGCAACCAATTAACCAATTCTGATTAG-3′ | |
| YK638 (ΔUL51) | 5′-GGAGGCGGAGCCGCGGCTGCAGGAGGCCCTGGTGGTCGTTTAAATGCGTGTTTTCATCCAAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CAAACACAGAACACACGGGTTGGATGAAAACACGCATTTAAACGACCACCAGGGCCTCAACCAATTAACCAATTCTGATTAG-3′ | |
| YK640 (ΔgE) and YK642 (UL51-S184A/ΔgE) | 5′-GGCGGGAACGCCCAAAACGTCCTGGAGACGTAGTTAGTTAGGGTGAGTGTCGGCGAGGACAGGATGACGACGATAAGTAG-3′ |
| 5′-GCAACGAAACGTCCTCGCCGACACTCACCCTAACTAACTACGTCTCCAGGACGTTTTGGGCAACCAATTAACCAATTC-3′ | |
| YK641 (ΔgE-repair) and YK643 (UL51-S184A/ΔgE-repair) | 5′-TATCGTGCTTGGCGGGAACGCCCAAAACGTCCTGGAGACGGGTGAGTGTCGGCGAGGACGAGGATGACGACGATAAGTAGGG-3′ |
| 5′-CGGAGCTGGAAGCAACGAAACGTCCTCGCCGACACTCACCCGTCTCCAGGACGTTTTGGGCAACCAATTAACCAATTCTGATTAG-3′ |
Identification of phosphorylation sites in UL51.
HEK293T cells were transfected with pcDNA-FEM-UL51 using polyethylenimine as described previously (58). These were then harvested at 48 h posttransfection and lysed in 0.5% NP-40 buffer (0.5% NP-40, 120 mM NaCl, 50 mM Tris-HCl [pH 8.0], and 50 mM NaF) containing protease and phosphatase inhibitor cocktails (Nacalai Tesque). After centrifugation, the supernatants were immunoprecipitated with an anti-Myc monoclonal antibody, and the immunoprecipitates were incubated with AcTEV protease (Invitrogen) for 1 h at room temperature. After another centrifugation, the supernatants were immunoprecipitated with an anti-Flag M2 affinity gel (Sigma), and the immunoprecipitates were washed three times with 0.1% NP-40 buffer and two times with wash buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 50 mM NaF). Flag elution buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5 mg Flag peptide/ml) was added and the immunoprecipitates were rotated for 2 h at 4°C. The eluted protein solution was incubated with trypsin and analyzed by nano-liquid chromatography–tandem mass spectrometry (nanoLC-MS/MS) as described previously (35). For this analysis, we used LTQ-Orbitrap Velos (Thermo Fisher Scientific) coupled with Dina-2A (KYA Technologies). The MS/MS signals were then analyzed against the 68,711 protein sequences in the RefSeq human protein database (National Center for Biotechnology Information [NCBI]) and the 74 virus protein sequences based on the complete genome sequence of human herpesvirus 1 strain F (GenBank accession number GU734771) using the Mascot algorithm (version 2.4.1; Matrix Science) with the following parameters: variable modifications, oxidation (Met), protein N-terminal acetylation, pyroglutamination (Gln), phosphorylation (Ser, Thr, and Tyr); maximum missed cleavages, 2; peptide mass tolerance, 3 ppm; MS/MS tolerance, 0.8 Da. For peptide identification, we conducted decoy database searching by Mascot and applied a filter for a false-positive rate of <1%. Determination of phosphorylated sites in the peptides was performed using Proteome Discoverer 1.3 (Thermo Fisher Scientific).
Construction of a self-excisable HSV-1(F) BAC.
To construct an E. coli GS1783 strain harboring a self-excisable HSV-1 BAC plasmid pYEbac102Cre carrying an expression cassette consisting of the CMV promoter, the CREin sequence, BGH poly(A), and transcription termination sequences for enhanced mRNA stability in the intergenic region between loxP and the BAC sequence (pYEbac102Cre) (Fig. 1), a two-step Red-mediated mutagenesis procedure was carried out using the primers listed in Table 3, to generate pcDNA-CREin-Kan and E. coli GS1783 harboring pYEbac102, as described previously (59, 60).
Recombinant viruses YK631 (UL51-S184A), encoding UL51 with alanine substituted for Ser-184, YK634 (UL51-T190A), encoding UL51 with alanine substituted for Thr-190, and YK636 (UL51-S226T228S235/AAA), encoding UL51 with alanine substituted for Ser-226, Thr-228, and Ser-235 (Fig. 2) were generated by the two-step Red-mediated mutagenesis procedure using E. coli GS1783 containing pYEbac102Cre as described previously (59, 60), except the primers listed in Table 3 were used. YK632 (UL51-S184D), encoding UL51 with aspartic acid substituted for Ser-184 (Fig. 2), was generated by the two-step Red-mediated mutagenesis procedure as described previously (59, 60), except for using E. coli GS1783 containing the YK631 (UL51-S184A) genome and the primers listed in Table 3. Recombinant virus YK638 (ΔUL51), in which the UL51 gene was disrupted by deleting UL51 codons 43 to 244, YK640 (ΔgE), in which the Us8 gene was disrupted by the insertion of an 11-bp sequence carrying two stop codons into the Arg-29 locus of the Us8 gene, and YK642 (UL51-S184A/ΔgE), encoding a S184A mutation in UL51 and an 11-bp insertion in gE, were constructed by the two-step Red-mediated mutagenesis procedure using E. coli GS1783 carrying pYEbac102Cre as described previously (59, 60), except for using the primers listed in Table 3. Recombinant viruses YK633 (UL51-S184A/D-repair), YK635 (UL51-T190A-repair), YK637 (UL51-S226T228S235/AAA-repair), YK641 (ΔgE-repair), and YK643 (UL51-S184A/ΔgE-repair), in which the UL51-S184D, UL51-T190A, UL51-S226T228S235/AAA, and gE-null mutations in YK632, YK634, YK636, YK640, and YK642, respectively (Fig. 2), were repaired and generated as described previously (59, 60), except for using the primers listed in Table 3. Recombinant virus YK639 (ΔUL51-repair), in which the mutation in YK638 (ΔUL51) was repaired (Fig. 2), was generated by the cotransfection of rabbit skin cells with the YK638 (ΔUL51) genome and pBS1007 using the calcium phosphate precipitation technique as described previously (40). Plaques of progeny viruses from the transfections and infections were isolated three times on Vero cells, and the restoration of original sequences was confirmed by sequencing (61).
Antibodies.
Commercial antibodies used in this study were mouse monoclonal antibodies to gB (H1817; Virusys), gE (9H3; Virusys), ICP8 (10A3; Millipore), and β-actin (AC15; Sigma). Mouse polyclonal antibodies to HSV-1 UL31 were described previously (62). Rabbit polyclonal antibodies to HSV-1 UL51 or UL34 were described previously (12, 63).
Immunoblotting, immunofluorescence, and determination of plaque size.
Immunoblotting, immunofluorescence, and the determination of plaque size were performed as described previously (45, 61, 64). The fluorescence intensity of immunofluorescence images was quantified using an LSM800 microscope with ZEN2.3 software (Zeiss).
Southern blotting.
Viral DNAs were digested with EcoRI, analyzed by electrophoresis on 0.8% agarose gels, and transferred to Hybond-N+ nylon membranes (GE Healthcare). The bands were hybridized with the appropriate DNA probe labeled with horseradish peroxidase using a direct nucleic acid labeling system (GE Healthcare) by following the manufacturer's instructions.
Electron microscopic analysis.
HaCaT cells infected with wild-type HSV-1(F), YK631 (UL51-S184A), YK632 (UL51-S184D), YK633 (UL51-S184A/D-repair), YK634 (UL51-T190A), YK636 (UL51-S226T228S235/AAA), YK638 (ΔUL51), or YK639 (ΔUL51-repair) at an MOI of 5 for 18 h were examined by ultrathin-section electron microscopy as described previously (45).
Animal studies.
Female ICR mice were purchased from Charles River Laboratories. Three-week-old mice were infected intracranially with 10-fold serial dilutions of the indicated viruses in groups of 6 per dilution as described previously (34, 40). Mice were monitored daily, and mortality occurring from 1 to 14 days postinfection was attributed to the infecting virus. LD50 values were calculated by the Behrens-Karber method. For ocular infection, 4-week-old mice were infected with 1 × 106 PFU/eye of each of the indicated viruses as described previously (56, 65). Mice were monitored daily, and mortality occurring from 1 to 14 days postinfection was attributed to the infecting virus. All animal experiments were carried out in accordance with the guidelines for the proper conduct of animal experiments of the Science Council of Japan. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Medical Science, The University of Tokyo (IACUC protocol approval numbers PA19-26, PA11-81, and PA16-69).
Statistical analysis.
Differences in viral yields, plaque sizes, and percentage of cells with punctate structures containing UL31 and UL34 were statistically analyzed by analysis of variance and Tukey's test. A P value of <0.05 was considered statistically significant. Differences in the mortality of infected mice were statistically analyzed by the log-rank test, and for the three comparison analyses, P values of <0.0167 (0.05/3), <0.025 (0.05/2), or <0.05 (0.05/1) were sequentially considered significant after Holm's sequentially rejective Bonferroni multiple-comparison adjustment.
ACKNOWLEDGMENTS
We thank Tomoko Ando, Yoshie Asakura, and Risa Abe for their excellent technical assistance.
This study was supported by Grants for Scientific Research from the Japan Society for the Promotion of Science (JSPS) to Y.K., A.K., J.A., and N.K., Grants for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports and Technology of Japan (16H06433, 16H06429, and 16K21723 to Y.K. and 18H04968 to A.K.), a contract research fund from the Program of Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from the Japan Agency for Medical Research and Development (AMED) (JP18fm0108006 to Y.K.), the Grant for Joint Research Project of The Institute of Medical Science, The University of Tokyo, and grants from the Takeda Science Foundation to Y.K., A.K., and J.A.
We have no competing financial interests to declare.
A.K. and S.O. conceived, designed, and performed the experiments, analyzed the data, and wrote the manuscript. M.W., M.O., H.K.-H., N.K., Y.M., and J.A. assisted with the experiments and analyzed the data. Y.K. conceived and designed the experiments, analyzed the data, and wrote the manuscript.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott-Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Laine RF, Albecka A, van de Linde S, Rees EJ, Crump CM, Kaminski CF. 2015. Structural analysis of herpes simplex virus by optical super-resolution imaging. Nat Commun 6:5980. doi: 10.1038/ncomms6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 4.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bauerlein FJ, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grunewald K. 2015. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163:1692–1701. doi: 10.1016/j.cell.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mettenleiter TC, Muller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 6.Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J Virol 79:4952–4964. doi: 10.1128/JVI.79.8.4952-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu FX, Chong JM, Wu L, Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J Virol 79:800–811. doi: 10.1128/JVI.79.2.800-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG II, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nozawa N, Daikoku T, Koshizuka T, Yamauchi Y, Yoshikawa T, Nishiyama Y. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J Virol 77:3204–3216. doi: 10.1128/JVI.77.5.3204-3216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol 79(Part 12):3027–3031. doi: 10.1099/0022-1317-79-12-3027. [DOI] [PubMed] [Google Scholar]
- 13.Oda S, Arii J, Koyanagi N, Kato A, Kawaguchi Y. 2016. The interaction between herpes simplex virus 1 tegument proteins UL51 and UL14 and its role in virion morphogenesis. J Virol 90:8754–8767. doi: 10.1128/JVI.01258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–3832. doi: 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meissner CS, Suffner S, Schauflinger M, von Einem J, Bogner E. 2012. A leucine zipper motif of a tegument protein triggers final envelopment of human cytomegalovirus. J Virol 86:3370–3382. doi: 10.1128/JVI.06556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selariu A, Cheng T, Tang Q, Silver B, Yang L, Liu C, Ye X, Markus A, Goldstein RS, Cruz-Cosme RS, Lin Y, Wen L, Qian H, Han J, Dulal K, Huang Y, Li Y, Xia N, Zhu H. 2012. ORF7 of varicella-zoster virus is a neurotropic factor. J Virol 86:8614–8624. doi: 10.1128/JVI.00128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang HF, Wang W, Jiang X, Zeng WB, Shen ZZ, Song YG, Yang H, Liu XJ, Dong X, Zhou J, Sun JY, Yu FL, Guo L, Cheng T, Rayner S, Zhao F, Zhu H, Luo MH. 2017. ORF7 of varicella-zoster virus is required for viral cytoplasmic envelopment in differentiated neuronal cells. J Virol 91:e00127-17. doi: 10.1128/JVI.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp BG, Granzow H, Klopfleisch R, Fuchs W, Kopp M, Lenk M, Mettenleiter TC. 2005. Functional analysis of the pseudorabies virus UL51 protein. J Virol 79:3831–3840. doi: 10.1128/JVI.79.6.3831-3840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roller RJ, Haugo AC, Yang K, Baines JD. 2014. The herpes simplex virus 1 UL51 gene product has cell type-specific functions in cell-to-cell spread. J Virol 88:4058–4068. doi: 10.1128/JVI.03707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roller RJ, Fetters R. 2015. The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J Virol 89:3112–3122. doi: 10.1128/JVI.02799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albecka A, Owen DJ, Ivanova L, Brun J, Liman R, Davies L, Ahmed MF, Colaco S, Hollinshead M, Graham SC, Crump CM. 2017. Dual function of the pUL7-pUL51 tegument protein complex in herpes simplex virus 1 infection. J Virol 91:e02196-16. doi: 10.1128/JVI.02196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol 68:834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol 75(Part 6):1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 24.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 25.Edelman AM, Blumenthal DK, Krebs EG. 1987. Protein serine/threonine kinases. Annu Rev Biochem 56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- 26.Jacob T, Van den Broeke C, Favoreel HW. 2011. Viral serine/threonine protein kinases. J Virol 85:1158–1173. doi: 10.1128/JVI.01369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi Y, Kato K. 2003. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev Med Virol 13:331–340. doi: 10.1002/rmv.402. [DOI] [PubMed] [Google Scholar]
- 28.Deruelle MJ, Favoreel HW. 2011. Keep it in the subfamily: the conserved alphaherpesvirus US3 protein kinase. J Gen Virol 92:18–30. doi: 10.1099/vir.0.025593-0. [DOI] [PubMed] [Google Scholar]
- 29.Kato A, Yamamoto M, Ohno T, Kodaira H, Nishiyama Y, Kawaguchi Y. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J Virol 79:9325–9331. doi: 10.1128/JVI.79.14.9325-9331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell C, Desjardins M, Thibault P, Radtke K. 2013. Proteomics analysis of herpes simplex virus type 1-infected cells reveals dynamic changes of viral protein expression, ubiquitylation, and phosphorylation. J Proteome Res 12:1820–1829. doi: 10.1021/pr301157j. [DOI] [PubMed] [Google Scholar]
- 31.Kato A, Tsuda S, Liu Z, Kozuka-Hata H, Oyama M, Kawaguchi Y. 2014. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral dUTPase and regulates its catalytic activity in infected cells. J Virol 88:655–666. doi: 10.1128/JVI.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi R, Kato A, Oda S, Koyanagi N, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. 2015. Function of the herpes simplex virus 1 small capsid protein VP26 is regulated by phosphorylation at a specific site. J Virol 89:6141–6147. doi: 10.1128/JVI.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulej K, Avgousti DC, Sidoli S, Herrmann C, Della Fera AN, Kim ET, Garcia BA, Weitzman MD. 2017. Time-resolved global and chromatin proteomics during herpes simplex virus type 1 (HSV-1) infection. Mol Cell Proteomics 16:S92–S107. doi: 10.1074/mcp.M116.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato A, Shindo K, Maruzuru Y, Kawaguchi Y. 2014. Phosphorylation of a herpes simplex virus 1 dUTPase by a viral protein kinase, Us3, dictates viral pathogenicity in the central nervous system but not at the periphery. J Virol 88:2775–2785. doi: 10.1128/JVI.03300-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arii J, Goto H, Suenaga T, Oyama M, Kozuka-Hata H, Imai T, Minowa A, Akashi H, Arase H, Kawaoka Y, Kawaguchi Y. 2010. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 467:859–862. doi: 10.1038/nature09420. [DOI] [PubMed] [Google Scholar]
- 36.Maruzuru Y, Fujii H, Oyama M, Kozuka-Hata H, Kato A, Kawaguchi Y. 2013. Roles of p53 in herpes simplex virus 1 replication. J Virol 87:9323–9332. doi: 10.1128/JVI.01581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujii H, Kato A, Mugitani M, Kashima Y, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. 2014. The UL12 protein of herpes simplex virus 1 is regulated by tyrosine phosphorylation. J Virol 88:10624–10634. doi: 10.1128/JVI.01634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Kato A, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. 2015. Role of host cell p32 in herpes simplex virus 1 de-envelopment during viral nuclear egress. J Virol 89:8982–8998. doi: 10.1128/JVI.01220-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato Y, Kato A, Maruzuru Y, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. 2016. Cellular transcriptional coactivator RanBP10 and herpes simplex virus 1 ICP0 interact and synergistically promote viral gene expression and replication. J Virol 90:3173–3186. doi: 10.1128/JVI.03043-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77:1382–1391. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards AL, Sollars PJ, Smith GA. 2016. New tools to convert bacterial artificial chromosomes to a self-excising design and their application to a herpes simplex virus type 1 infectious clone. BMC Biotechnol 16:64. doi: 10.1186/s12896-016-0295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGraw HM, Friedman HM. 2009. Herpes simplex virus type 1 glycoprotein E mediates retrograde spread from epithelial cells to neurites. J Virol 83:4791–4799. doi: 10.1128/JVI.02341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder A, Polcicova K, Johnson DC. 2008. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J Virol 82:10613–10624. doi: 10.1128/JVI.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meignier B, Longnecker R, Mavromara-Nazos P, Sears AE, Roizman B. 1988. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 45.Shindo K, Kato A, Koyanagi N, Sagara H, Arii J, Kawaguchi Y. 2016. Characterization of a herpes simplex virus 1 (HSV-1) chimera in which the Us3 protein kinase gene is replaced with the HSV-2 Us3 gene. J Virol 90:457–473. doi: 10.1128/JVI.02376-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J Virol 74:117–129. doi: 10.1128/JVI.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol 76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirohata Y, Arii J, Liu Z, Shindo K, Oyama M, Kozuka-Hata H, Sagara H, Kato A, Kawaguchi Y. 2015. Herpes simplex virus 1 recruits CD98 heavy chain and beta1 integrin to the nuclear membrane for viral de-envelopment. J Virol 89:7799–7812. doi: 10.1128/JVI.00741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farnsworth A, Wisner TW, Webb M, Roller R, Cohen G, Eisenberg R, Johnson DC. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci U S A 104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imai T, Sagou K, Arii J, Kawaguchi Y. 2010. Effects of phosphorylation of herpes simplex virus 1 envelope glycoprotein B by Us3 kinase in vivo and in vitro. J Virol 84:153–162. doi: 10.1128/JVI.01447-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. 2004. Focal adhesion regulation of cell behavior. Biochim Biophys Acta 1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Klopfleisch R, Klupp BG, Fuchs W, Kopp M, Teifke JP, Mettenleiter TC. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J Virol 80:5571–5576. doi: 10.1128/JVI.02589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 56.Kato A, Ando T, Oda S, Watanabe M, Koyanagi N, Arii J, Kawaguchi Y. 2016. Roles of Us8A and its phosphorylation mediated by Us3 in herpes simplex virus 1 pathogenesis. J Virol 90:5622–5635. doi: 10.1128/JVI.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baines JD, Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol 65:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 60.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 61.Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J Virol 82:5198–5211. doi: 10.1128/JVI.02681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z, Kato A, Shindo K, Noda T, Sagara H, Kawaoka Y, Arii J, Kawaguchi Y. 2014. Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J Virol 88:4657–4667. doi: 10.1128/JVI.00137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiba C, Daikoku T, Goshima F, Takakuwa H, Yamauchi Y, Koiwai O, Nishiyama Y. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J Gen Virol 81:2397–2405. doi: 10.1099/0022-1317-81-10-2397. [DOI] [PubMed] [Google Scholar]
- 64.Kawaguchi Y, Van Sant C, Roizman B. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato A, Liu Z, Minowa A, Imai T, Tanaka M, Sugimoto K, Nishiyama Y, Arii J, Kawaguchi Y. 2011. Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J Virol 85:9599–9613. doi: 10.1128/JVI.00845-11. [DOI] [PMC free article] [PubMed] [Google Scholar]