Human noroviruses (HuNoVs) are the major cause of nonbacterial gastroenteritis worldwide and impose a great burden on patients and health systems every year. So far, no antiviral treatment or vaccine is available. We show that MNV evades the host immune response by reducing the amount of MHC class I proteins displayed on the cell surface. This reduction leads to a decrease in viral antigen presentation and interferes with the CD8+ T cell response. CD8+ T cells respond to foreign antigen by activating cytotoxic pathways and inducing immune memory to the infection. By evading this immune response, MNV is able to replicate efficiently in the host, and the ability of cells to respond to consecutive infections is impaired. These findings have a major impact on our understanding of the ways in which noroviruses interact with the host immune response and manipulate immune memory.
KEYWORDS: mouse norovirus, antigen presentation, MHC I protein, CD8+ T cells, immune evasion
ABSTRACT
Human noroviruses are highly infectious single-stranded RNA (ssRNA) viruses and the major cause of nonbacterial gastroenteritis worldwide. With the discovery of murine norovirus (MNV) and the introduction of an effective model for norovirus infection and replication, knowledge about infection mechanisms and their impact on the host immune response has progressed. A major player in the immune response against viral infections is the group of major histocompatibility complex (MHC) class I proteins, which present viral antigen to immune cells. We have observed that MNV interferes with the antigen presentation pathway in infected cells by reducing the surface expression of MHC class I proteins. We have shown that MNV-infected dendritic cells or macrophages have lower levels of surface expression of MHC class I proteins than uninfected and bystander cells. Transcriptional analysis revealed that this defect is not due to a decreased amount of mRNA but is reflected at the protein level. We have determined that this defect is mediated via the MNV NS3 protein. Significantly, treatment of MNV-infected cells with the endocytic recycling inhibitor dynasore completely restored the surface expression of MHC class I proteins, whereas treatment with the proteasome inhibitor MG132 partly restored such expression. These observations indicate a role for endocytic recycling and proteasome-mediated degradation of these proteins. Importantly, we show that due to the reduced surface expression of MHC class I proteins, antigen presentation is inhibited, resulting in the inability of CD8+ T cells to become activated in the presence of MNV-infected cells.
IMPORTANCE Human noroviruses (HuNoVs) are the major cause of nonbacterial gastroenteritis worldwide and impose a great burden on patients and health systems every year. So far, no antiviral treatment or vaccine is available. We show that MNV evades the host immune response by reducing the amount of MHC class I proteins displayed on the cell surface. This reduction leads to a decrease in viral antigen presentation and interferes with the CD8+ T cell response. CD8+ T cells respond to foreign antigen by activating cytotoxic pathways and inducing immune memory to the infection. By evading this immune response, MNV is able to replicate efficiently in the host, and the ability of cells to respond to consecutive infections is impaired. These findings have a major impact on our understanding of the ways in which noroviruses interact with the host immune response and manipulate immune memory.
INTRODUCTION
Human noroviruses (HuNoVs) cause the majority of nonbacterial gastroenteritis cases worldwide, affecting millions of people every year (1–3). Although HuNoV infections are usually nonlethal, children and the elderly can develop severe infections that lead to hospitalization and death. In developing countries, at least 220,000 deaths per year are associated with HuNoV (4, 5). In addition, HuNoV cases are estimated to lead to a combined yearly burden of 60 billion USD due to health care costs and loss of productivity (6).
NoVs are positive-sense single-stranded RNA (ssRNA) viruses and can be classified into five genogroups, according to the phylogenetic analysis of the viral capsid (VP1) gene. Infections can occur through contact with contaminated food, water, or surfaces, or from person to person (7, 8). So far, no medical treatment against HuNoV infection exists, but vaccines to prevent HuNoV infections are under development (9–11). The current lack of suitable treatment is partly due to the difficulty of cultivating HuNoVs in the laboratory without a robust animal or tissue culture model, although it has recently been discovered that human B cells exposed to enteric bacteria could be infected with HuNoV (12), and enteroid cultures for infection have been developed (13). In 2003, an unknown NoV infecting mice was discovered and was allocated to genogroup V (GV) (14). This murine norovirus (MNV) shares characteristics with HuNoV strains and is cultivable in mice as well as in murine dendritic cells and macrophages (15). Because of its similarity to the human strains, MNV is used as a model to further our knowledge of NoVs (16).
Like HuNoVs, MNV causes an acute and rapid infection, which is dependent on essential pathways of the innate immune response, including the induction of type I and III interferons (IFNs) (15, 17, 18). Another important group of proteins in the surveillance and defense against viral infections is the major histocompatibility complex (MHC) class I proteins. Peptides are processed from proteins found in the cytoplasm and are loaded onto MHC class I molecules, which are subsequently presented on the surfaces of cells. These peptides are usually generated through proteasome-mediated degradation, which affects host proteins as well as viral proteins. Host and viral peptides are transported into the endoplasmic reticulum (ER) via the transporter associated with antigen processing (TAP) (19). In the ER, a complex of MHC class I proteins and peptides is formed with the aid of the peptide-loading complex (PLC; comprising TAP, tapasin, MHC class I proteins, ERp57, and calreticulin), leading to the incorporation of the antigen peptide into the binding groove of MHC class I molecules (20–22). Successfully formed complexes are then transported to the cell membrane via the Golgi network. MHC class I proteins can present viral peptide antigens to CD8+ T cells, initiating the cellular immune response. Additionally, a decrease in MHC class I protein levels on the cell surface alerts NK cells, leading to their activation and the elimination of the affected cell. MHC class I proteins have been the targets of many viruses, because of their importance and impact in triggering an immune response to the infection.
Based on the importance of MHC class I proteins in the immune defense against viruses, we were interested in investigating the interaction of MNV with the MHC class I expression pathway. We reveal that MNV exerts an inhibitory effect on the intracellular as well as the surface expression of MHC class I proteins in infected cells. This phenotype is likely to be caused by the viral protein NS3, since we observed a significant reduction in the level of MHC class I proteins on the surfaces of NS3-transfected cells. Rescue experiments with endocytic recycling and proteasome inhibitors imply a potential degradation of MHC class I proteins during MNV infection, which could be either directly or indirectly initiated by NS3. More significantly, we observed that the reduction in MHC class I surface expression affects the presentation of viral peptides on the surfaces of infected cells and perturbs recognition by antigen-specific CD8+ T cells.
RESULTS
MNV infection reduces the surface expression of MHC class I proteins.
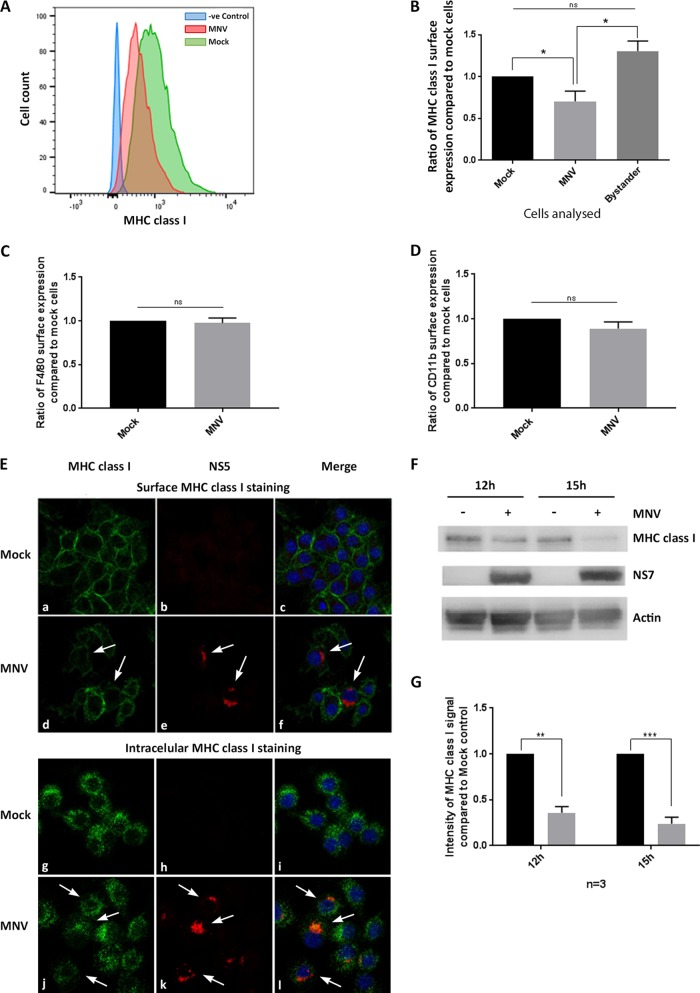
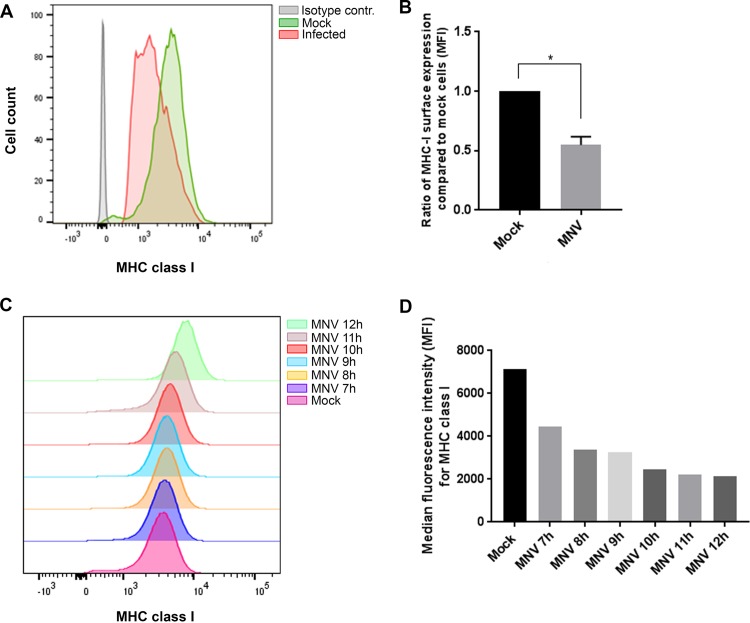
Many viruses, such as human immunodeficiency virus (HIV) and human cytomegalovirus (HCMV) (23, 24), are known to interfere with the transport or synthesis pathways of MHC class I molecules so as to reduce the detection of viral antigens by both the innate and adaptive immune responses. MHC class I proteins play a major role in the presentation and detection of viral antigens and are an essential element for the establishment of an antiviral immune response. Therefore, we aimed to determine if MNV replication alters the synthesis and cell surface expression of MHC class I proteins. To test this hypothesis, we investigated the most important functional expression of MHC class I proteins, their expression on the surfaces of infected cells. For this purpose, RAW264.7 cells were infected with MNV (multiplicity of infection [MOI], 5), and MHC class I expression was analyzed via flow cytometry (Fig. 1A and B) and immunofluorescence (IF) imaging (Fig. 1E). Mock- or MNV-infected cells were harvested at 12 h after infection and were stained with an anti-MHC class I antibody at the cell surface and an anti-MNV NS5 antibody (viral marker) within the cell. Infected cells were selected via positive staining with the anti-NS5 antibody and were analyzed for their MHC class I surface expression.
FIG 1.
MNV infection leads to reduced MHC class I protein levels and reduced surface expression of MHC class I proteins in mouse macrophages. RAW264.7 macrophages were either infected with MNV CW1 (MOI, 5) for 12 h or left untreated and were then stained with FITC-conjugated MHC class I-specific antibodies (H-2Kd/H-2Dd; Bio-Gems) on the cell surface and with antibodies to viral protein NS5 within the cell. MNV-infected cells were gated for the NS5-positive population. (A) Representative histogram of MHC class I surface expression in MNV-infected (red) and uninfected (green) cells. The blue area represents isotype control cells. (B) Ratios of the MFI of the MHC class I signals of MNV-infected and bystander (exposed to virus, but NS5 negative) cells to that of mock-treated cells (n = 4). (C) Ratio of the MFI of the F4/80 signal of MNV-infected RAW264.7 cells to that of mock-treated cells (n = 3). (D) Ratio of the MFI of the CD11b signal of MNV-infected DC2.4 cells to that of mock-treated cells (n = 3). (E) Immunofluorescence analyses of MNV-infected (d, e, f, j, k, and l) and mock-infected (a, b, c, g, h, and i) cells stained with MHC class I antibodies on the surface (a and d) or within the cells (g and j) or with NS5 antibodies within the cell (b, e, h, and k). DAPI served as the nuclear stain for the merged image (c, f, i, and l). Infected cells are indicated by white arrows. (F) Immunoblot analysis of MNV-infected or uninfected macrophages after 12 h and 15 h. Whole-lysate samples were probed with anti-actin, anti-NS7, and anti-MHC class I antibodies (n = 3). (G) Quantitation of MHC class I immunoblot intensity (F) in MNV-infected cell lysates (shaded bars) relative to that in uninfected controls (filled bars) at 12 and 15 hpi (n = 3). Data in all bar graphs are averages ± standard errors of the means. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Our flow cytometry analysis revealed a significant decrease in MHC class I surface expression in cells that were infected with MNV (NS5 positive) from that in mock-infected cells (Fig. 1A). Quantitation of the reduction of MHC class I expression was evaluated by measuring the median fluorescence intensity (MFI), which revealed that only ∼70% of the total amount of MHC class I protein expressed on the surfaces of mock-infected cells was expressed on the surfaces of infected cells (Fig. 1B). The decrease in MHC class I surface expression was further verified by IF analysis (Fig. 1E). Cells staining positive for anti-NS5 again showed a reduction in the MHC class I signal from that in mock-infected cells, in agreement with our flow cytometric analysis. This decrease in MHC class I surface proteins was found to be specific for infected cells, because “bystander” cells, which were exposed to the virus but remained uninfected with MNV (NS5 negative), had significantly larger amounts of MHC class I proteins on their surfaces than MNV-infected cells (Fig. 1B and E). To investigate if the reduction in MHC class I proteins on the cell surface is specific or can be observed for other surface proteins, we analyzed the surface expression of F4/80 and CD11b during MNV infection (Fig. 1C and D, respectively). Neither F4/80 nor CD11b surface expression was reduced in MNV-infected cells from that in uninfected cells, indicating that the reduction in MHC class I surface expression is not due to a global decrease in the levels of cell surface proteins. Additionally, we examined the intracellular signal for MHC class I via IF analysis to assess if MHC class I proteins were inhibited from being transported to the surface and were accumulating within the infected cells (Fig. 1E). In comparison to levels in mock-infected cells, there was no accumulation of intracellular MHC class I proteins in infected cells, indicating that the observed surface reduction of MHC class I proteins is not caused by an inhibition of transport to the cell surface. To further our analysis, we investigated the total amount of MHC class I protein at different time points postinfection by Western blot analysis of cell lysates (Fig. 1F and G). In mock-infected cells, there was consistent production of MHC class I proteins; however, upon infection with MNV, we observed a steady and drastic decline in the amount of MHC class I proteins from the 12-h postinfection (12-hpi) to the 15-hpi time point. These observations indicate that MNV not only reduces the cell surface expression of MHC class I proteins but also appears to reduce MHC class I protein levels in general.
Combined, these results indicate that MNV specifically impedes the cell surface expression of MHC class I proteins in infected macrophages and that this may be mediated via downregulation of the total amount of MHC class I proteins in infected cells.
Transcription of MHC class I mRNA is not affected by MNV infection.
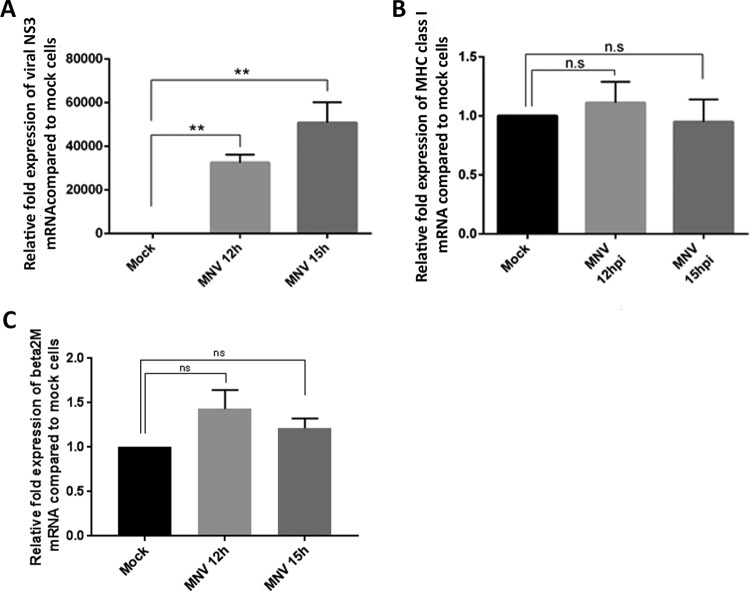
Since we observed that MNV indeed affects MHC class I protein amounts and surface expression (Fig. 1), and we have observed previously that some genes associated with immune recognition and antigen presentation are transcriptionally downregulated in MNV-infected cells (25), we aimed to identify the step in the MHC class I synthesis pathway that MNV was inhibiting. To investigate if the reduction in MHC class I surface expression was associated with a decrease in MHC class I transcription, quantitative reverse transcription-PCR (RT-qPCR) was performed on RNA samples from mock- and MNV-infected cells (Fig. 2A and B). MHC class I mRNA levels were analyzed at two time points (12 and 15 hpi) using MHC class I- and MNV (NS3)-specific primers. We observed a significant increase in viral mRNA (NS3) levels in the infected cells, confirming successful infection of the macrophages (Fig. 2A). However, at neither of the time points investigated did we observe a change (either an increase or a decrease) in MHC class I mRNA levels between infected and uninfected macrophages (Fig. 2B). Furthermore, we examined the mRNA expression levels of β2 microglobulin, which is associated with the MHC class I complex (Fig. 2C). Again, there was no significant change in the mRNA levels of β2 microglobulin in cells infected with MNV for 12 h or 15 h from those in uninfected cells. Therefore, we can conclude that our observed decrease in MHC class I surface protein levels is not based on a reduction in MHC class I or β2 microglobulin transcription and MHC class I mRNA levels.
FIG 2.
Transcription levels of MHC class I mRNA are not reduced during MNV infection. RAW264.7 cells were either infected with MNV CW1 (MOI, 5) or left uninfected (mock). RNA samples were taken 12 h and 15 h after infection and were analyzed via RT-qPCR. (A) Fold expression of viral mRNA (NS3) in MNV-infected cells relative to that in mock-infected cells (n = 4). (B) Fold expression of MHC class I mRNA in MNV-infected cells relative to that in mock-infected cells (n = 3). (C) Fold expression of β2 microglobulin mRNA in MNV-infected cells relative to that in mock-infected cells (n = 4). Data in all panels are averages ± standard errors of the means. ns, P > 0.05; **, P < 0.01.
Treatment with the proteasome inhibitor MG132 and the endocytosis inhibitor dynasore leads to the partial restoration of MHC class I surface expression in MNV-infected cells.
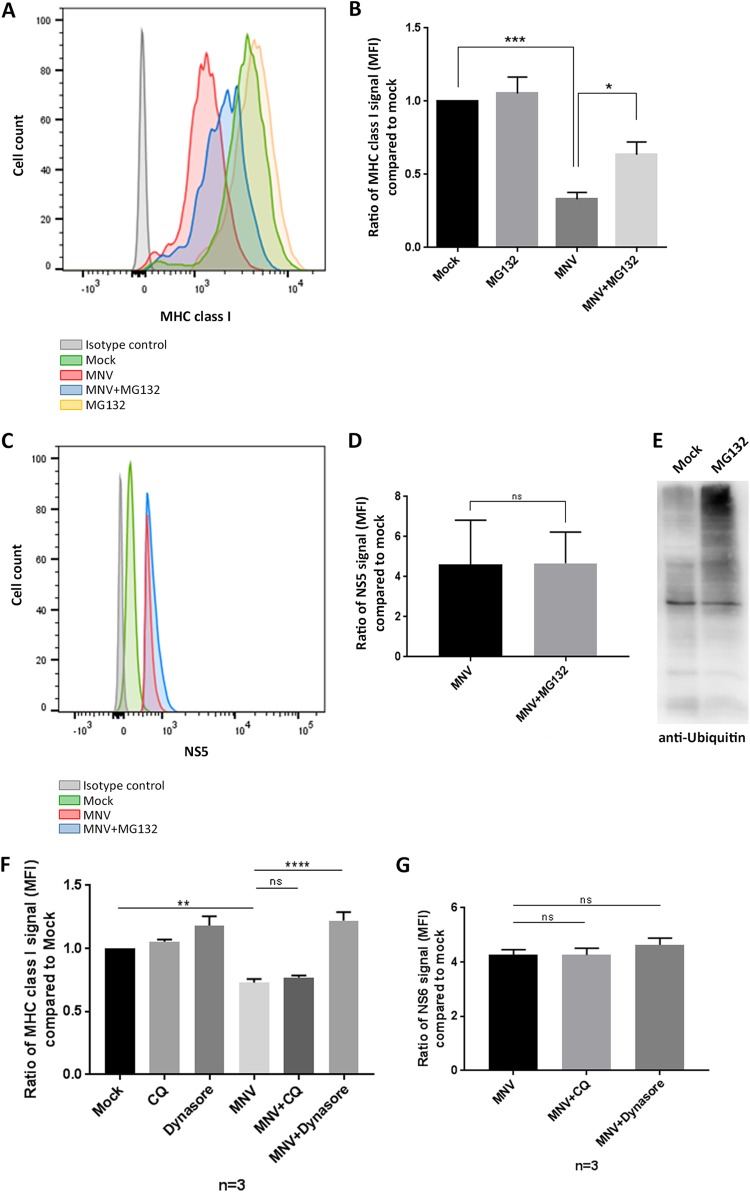
Based on previously documented viral mechanisms interfering with the MHC class I expression pathway, we aimed to test if MNV infection leads to the degradation of MHC class I molecules by either targeting MHC class I proteins for proteasome degradation or actively degrading them (26–31). MG132 is a proteasome inhibitor that specifically acts on the proteolytic activity of the 26S proteasome (32). Therefore, we utilized MG132 treatment to determine if we could rescue the reduced MHC class I surface expression observed during MNV infection. RAW264.7 cells were infected with MNV (MOI, 5) and an hour later were either treated with MG132 (0.5 μM) or left untreated. Cells were harvested at 12 hpi and were analyzed via flow cytometry for MHC class I surface expression (Fig. 3A and B). To exclude a detrimental effect of MG132 on the replication of MNV, we also evaluated the NS5 signal in MNV-infected cells with and without MG132 (Fig. 3C and D). Infected cells were fixed, permeabilized, and stained with an anti-NS5 antibody. MNV-infected cells treated with MG132 showed levels of NS5 similar to those in MNV-infected untreated cells, indicating that MG132 did not adversely affect viral replication. NS5-positive, non-MG132-treated cells showed a reduction in the surface expression of MHC class I proteins similar to that in the uninfected cells we had observed earlier (Fig. 1). When MNV-infected cells were additionally treated with the proteasome inhibitor MG132, we observed a significant increase in MHC class I surface expression over that in infected but untreated cells. The MG132 treatment did not result in a complete restoration of MHC class I surface expression in the infected cells but did appear to restore ∼50% of the amount of MHC class I proteins on the surface (Fig. 3A and B). The activity of MG132 was confirmed by the increased levels of ubiquitinated proteins during MG132 treatment (Fig. 3E).
FIG 3.
Treatment with the proteasome inhibitor MG132 or the endocytosis inhibitor dynasore partly restores MHC class I surface expression. RAW264.7 macrophages were either infected with MNV CW1 (MOI, 5) or left untreated (mock) for 12 h. The proteasome inhibitor MG132 (0.5 μM) was added 1 h postinfection. Cells were stained with an FITC-conjugated MHC class I-specific antibody (H-2Kd/H-2Dd; Bio-Gems) on their surfaces and with antibodies to viral protein NS5 within the cells. MNV-infected cells were gated for the NS5-positive population. (A) Representative histogram of MHC class I surface expression in MNV-infected (red), MNV-infected and MG132-treated (blue), mock-infected (green), mock-infected and MG132-treated (yellow), and isotype control (gray) cells. (B) Ratios of the MFI of the MHC class I signals for different treatments to that for mock treatment (n = 4). (C) Representative histogram of NS5 signals in MNV-infected (red), MNV-infected and MG132-treated (blue), mock-infected (green), and isotype control (gray) cells. (D) Ratios of the MFI of the NS5 signals for MNV-infected cells, either treated with MG132 or left untreated, to that for mock-infected cells (baseline) (n = 4). (E) RAW264.7 cells were either treated with MG132 or left untreated. Cell lysates were analyzed via immunoblotting. Staining with an anti-ubiquitin antibody was used to verify the efficiency of MG132 at inhibiting the proteasome. (F) RAW264.7 cells were infected with MNV (MOI, 5) and either were treated with chloroquine (CQ) or dynasore at 1 hpi or were left untreated. Cells were stained for MHC class I proteins (H-2Kd/H-2Dd; Bio-Gems) on their surfaces at 12 hpi. Cells were fixed, permeabilized, and stained for NS6 within the cells. Infected cells were identified via positive NS6 staining, and ratios of the MFI for MHC class I signals on their surfaces to that for mock-infected cells are depicted (n = 3). (G) Ratios of the MFI for intracellular NS6 signals in MNV-infected cells, either treated with chloroquine or dynasore or left untreated, to that for mock-infected cells (n = 3). Data for all bar graphs are averages ± standard errors of the means. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Additionally, we aimed to uncover if not only degradation of MHC class I proteins via the proteasome, but also degradation via the lysosome, was playing a role during MNV infection. To test this, we treated RAW264.7 cells with the lysosome inhibitor chloroquine at 1 hpi (Fig. 3F). Interestingly, inhibiting degradation via the lysosome did not rescue the surface expression of MHC class I proteins. In contrast to that finding, MNV-infected cells treated with the endocytosis inhibitor dynasore at 1 hpi displayed high levels of MHC class I proteins on their surfaces, which were similar to MHC class I levels on the surfaces of uninfected cells (Fig. 3F). Neither chloroquine nor dynasore treatment interfered with MNV infection, as evidenced by the fact that similar levels of the viral protein NS6 were observed in MNV-infected cells that were treated with either of the inhibitors and those that were left untreated (Fig. 3G).
Overall, these observations indicate that MNV infection induces some degree of proteasome-mediated, lysosome-independent degradation of MHC class I proteins and that the internalization of MHC class I proteins via the endocytic pathway is a major contributor to the reduced surface expression of MHC class I proteins on MNV-infected cells.
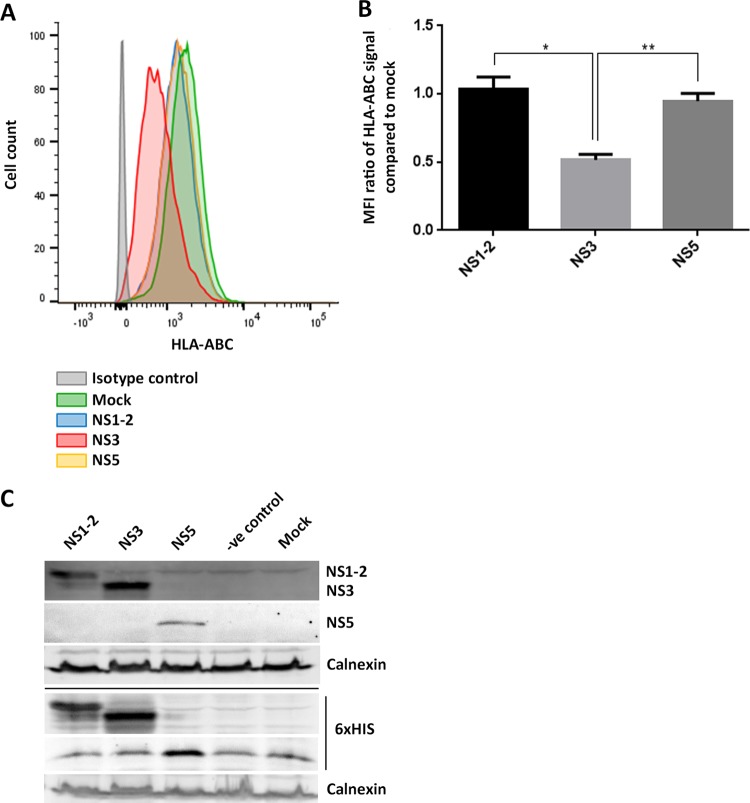
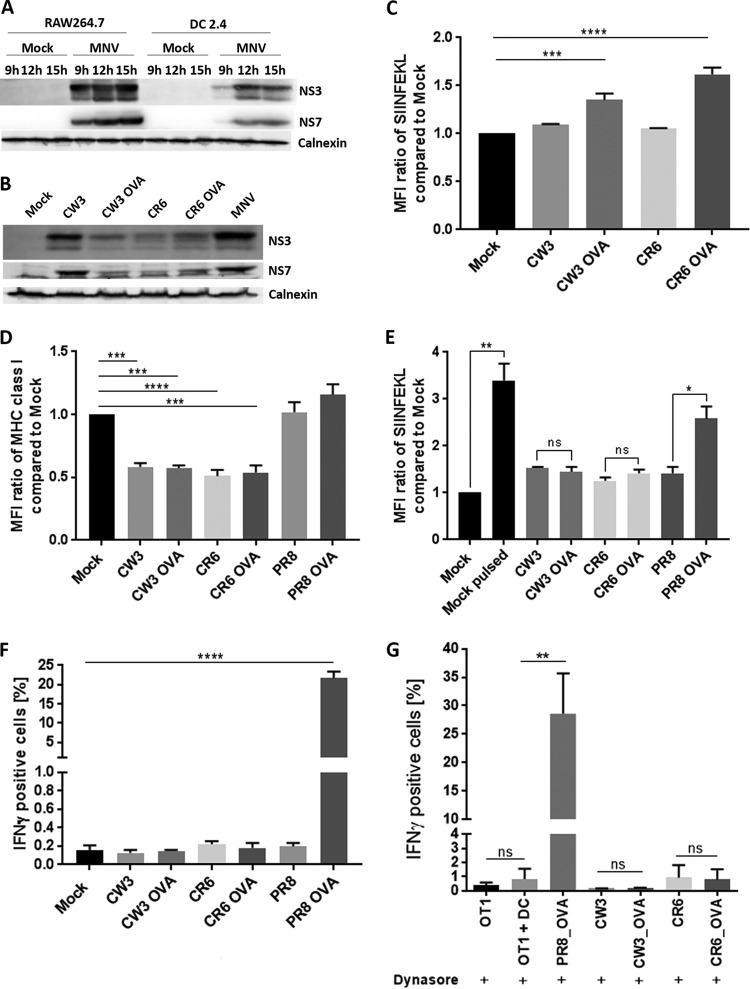
The MNV NS3 protein promotes a reduction in MHC class I surface expression when expressed individually.
Previously published studies on mechanisms of viral interference with MHC class I proteins, and our findings so far, led us to hypothesize that one of the viral nonstructural proteins is responsible for the observed reduction in the surface expression of MHC class I proteins in MNV-infected cells (26–31). To identify the protein potentially interfering with MHC class I expression, we transfected recombinant cDNA plasmids encoding the MNV NS proteins (NS1-2 to NS7 [33]) individually into 293T cells and assessed their abilities to restrict MHC class I surface expression. The transfected cells were harvested 18 h after transfection and were immunostained at their surfaces with an anti-HLA-ABC antibody. Cells were then fixed, permeabilized, and immunostained for the 6×His epitope tag of the expressed MNV NS proteins. Samples were analyzed via flow cytometry, and transfected cells were identified via the positive 6×His stain (Fig. 4). To ensure that the 6×His-positive population would be representative for the expression of NS1-2, NS3, and NS5, we expressed the NS proteins in 293T cells and investigated their expression via immunoblot analysis (Fig. 4C). NS proteins were detected either with an anti-6×His antibody or with a specific anti-NS1-2, anti-NS3, or anti-NS5 antibody and showed similar signal intensities for both antibodies. We can therefore conclude that the 6×His-positive population is representative of the specific NS-positive populations. We observed that the majority of the MNV NS proteins did not perturb MHC class I surface expression, and for simplicity, only the expression of NS1-2, NS3, and NS5 is depicted in Fig. 4. The expression of MNV NS4, NS6, and NS7 was assessed for effects on MHC class I surface expression, but we detected no differences in MHC class I levels from those in mock-transfected cells (data not shown). Interestingly, cells transfected with the plasmid encoding the MNV NS3 protein showed a significant reduction in the surface expression of MHC class I proteins from that in mock-transfected cells, as well as from that in cells transfected with plasmids encoding the other MNV proteins, NS1-2 and NS5 (Fig. 4A and B). Intriguingly, the observed reduction in MHC class I surface expression upon NS3 protein expression was similar to the reduction in cell surface expression of MHC class I proteins that we have observed during MNV infection.
FIG 4.
Expression of MNV NS3 in 293T cells reduces the surface expression of MHC class I proteins. 293T cells either were transfected with MNV CW1 NS1-2, NS3, or NS5 or were left untreated. Cells were stained with an APC-conjugated human MHC class I-specific antibody on their surfaces and the NS protein tag (6×His) within the cells. NS protein-transfected cells were gated for the 6×His-positive population. (A) Representative histogram of MHC class I surface expression in NS protein-transfected 293T cells. Blue, NS1-2; red, NS3; orange, NS5; green, mock-transfected cells; gray, isotype control cells. (B) Ratios of the MFI of the MHC class I signals for the different NS proteins to that for mock-transfected cells (n = 4). Data are averages ± standard errors of the means. *, P < 0.05; **, P < 0.01. (C) Immunoblot analysis of the expression levels of NS1-2, NS3, and NS5, detected via staining with anti-NS1-2, anti-NS3, and anti-NS5 antibodies or with anti-6×His. Calnexin expression levels were detected with an anti-calnexin antibody.
This observation implies that the MNV NS3 protein is potentially responsible for our observed reduction in MHC class I surface expression during MNV infection.
MHC class I surface expression reduction occurs early during MNV infection and can be observed in other cell types.
To investigate if the decrease in MHC class I proteins on the cell surface during MNV infection was specific for macrophages or if it can be observed in other cell types as well, we infected mouse dendritic cells (DC 2.4 cells) with MNV. Cells were stained with an anti-MHC class I antibody on their surfaces at 12 hpi and were analyzed via flow cytometry (Fig. 5A and B). MNV-infected cells were identified by staining for the viral protein NS5 within the cell. Like RAW264.7 cells, DC 2.4 cells that were infected with MNV and were positive for the viral stain displayed smaller amounts of MHC class I proteins on their surfaces than uninfected cells (Fig. 5A). MHC class I levels in infected DC 2.4 cells were ∼50% lower than those in uninfected cells (Fig. 5B), indicating that MNV infection causes a significant decrease in MHC class I levels in dendritic cells as well. Additionally, we analyzed the change in MHC class I levels on the cell surface over time during MNV infection. Cells were infected with MNV and were stained with an anti-MHC class I antibody on their surfaces at 7, 8, 9, 10, 11, and 12 hpi (Fig. 5C and D). In addition, fixed cells were stained with an anti-NS5 antibody within the cell to identify infected cells. From 7 hpi, we observed a continuous decrease in the surface levels of MHC class I proteins in MNV-infected cells. This decreased amount of MHC class I proteins was stable from 8 hpi on, and there was no indication of MHC class I levels reverting to levels similar to those in mock-infected cells at any time point tested.
FIG 5.
MNV causes a decrease in MHC class I levels on the cell surface early during infection and in different cell types. DC 2.4 cells were either infected with MNV CW1 (MOI, 5) for 12 h or left uninfected and were analyzed for MHC class I expression on their surfaces and MNV NS5 within the cell. (A) Representative histogram of MHC class I signals (MHC class I Y3) in MNV-infected (red), uninfected (green), and isotype control (gray) cells. (B) Quantitative analysis of the median fluorescence intensities for MHC class I proteins in MNV-infected and uninfected cells. On average, DC 2.4 cells displayed an infection rate of 30 to 40%. (C) DC 2.4 cells were infected with MNV for 7, 8, 9, 10, 11, or 12 h before being stained with an anti-MHC class I antibody (Y3) on their surfaces. MNV-infected cells were identified via intracellular staining with NS4. Histograms depict MHC class I signals on the surfaces of MNV-infected cells at different times. (D) Median fluorescence intensities for the MHC class I signals in MNV-infected and mock-infected control cells.
Conclusively, we discovered that MNV affects the MHC class I levels not only in macrophages but also in dendritic cells and that this effect is established early during infection.
The reduction of MHC class I surface expression inhibits the presentation of viral peptides and the activation of CD8+ T cells.
Having repeatedly observed the robust phenotype of the reduced surface expression of MHC class I proteins on MNV-infected cells, we were interested in determining whether a decrease in MHC class I expression could potentially have a functional effect on the immune response. For this purpose, we used DC 2.4 cells, which previously showed a similar decrease in MHC class I surface expression after MNV infection (Fig. 5A and B). We additionally show that the infection kinetics in DC 2.4 cells are similar to those in RAW264.7 cells (Fig. 6A). As mentioned above, MHC class I proteins play a major role in presenting antigens to CD8+ T cells. Therefore, we aimed to investigate the effect of reduced MHC class I levels on the presentation of viral peptides and the activation of CD8+ T cells. For this purpose, we used the ovalbumin (OVA) peptide (SIINFEKL)-expressing MNV strains CW3 (acute) and CR6 (persistent) (34), as well as influenza A virus (IAV) PR8 (as a control), to infect DC 2.4 cells. To ensure that all MNV strains (CW3, CW3 expressing the OVA peptide [CW3 OVA], CR6, and CR6 OVA) express the viral protein NS3, we analyzed their expression levels via immunoblot analysis (Fig. 6B). We compared NS3 expression to the expression of the viral polymerase NS7 and observed that all viruses expressed NS3 at levels comparable to that of NS7, indicating that NS3 expression was not impaired. Our earlier results indicated that dynasore stabilized MHC class I expression during MNV infection (Fig. 3F). Therefore, we used dynasore treatment to verify that infection with the CW3 OVA and CR6 OVA strains indeed leads to the expression of SIINFEKL on the surfaces of infected cells when the internalization of MHC class I proteins is inhibited (Fig. 6C).
FIG 6.
The surface presentation of viral peptides and the activation of CD8+ T cells are suppressed in MNV-infected cells. (A) Comparative infection kinetics for MNV CW1-infected RAW264.7 and DC2.4 cells at 9 h, 12 h, and 15 h postinfection. Cell lysates were analyzed via immunoblot analysis with anti-NS3, anti-NS7, and anti-calnexin antibodies. (B) Immunoblot analysis of NS3 and NS7 expression in DC2.4 cells infected with CW3, CW3 OVA, CR6, CR6 OVA, or MNV (CW1). (C) DC2.4 cells either were infected with CW3, CW3 OVA, CR6, or CR6 OVA or were left uninfected. Cells were treated with dynasore (50 μM) at 1 hpi and were stained for SIINFEKL on their surfaces at 12 hpi. The MFI signals for SIINFEKL in infected cells were compared to that for uninfected cells. (D and E) DC 2.4 cells were infected with the acute CW3 strain or the persistent CR6 strain containing the OVA peptide as well as with the corresponding WT strains. As a control, cells either were infected with WT PR8 or PR8 OVA or were left uninfected. DC 2.4 cells were stained with an anti MHC class I (MHC class I M1/42) (D) or anti-SIINFEKL (E) antibody on their surfaces and with an anti-dsRNA antibody within the cells. Infected populations were identified via positive anti-dsRNA staining and were analyzed for their MHC class I and SIINFEKL signals (median fluorescence intensity) via flow cytometry. (F) DC 2.4 cells were exposed to in vitro-activated OT-I-GFP CD8+ T cells and were incubated for 6 h at 37°C in the presence of BFA and monensin. Cells were fixed, stained for IFN-γ within the cells, and analyzed via flow cytometry. The percentage of IFN-γ-positive cells in the OT-I-GFP population (GFP positive) was calculated. n, 3 for all analyses. (G) DC 2.4 cells either were infected with WT CW3, WT CR6, CW3 OVA, CR6 OVA, or PR8 OVA or were left uninfected. Cells were treated with dynasore (50 μM) at 1 hpi and were exposed to OT-I cells for 6 h at 37°C. OT-I activation was analyzed via intracellular IFN-γ staining (n, 4 for all conditions except WT CR6 and CR6 OVA [n = 2]). Data are averages ± standard errors of the means. ns, P > 0.05; *, P < 0.05; **, P < 0.01, ***, P < 0.001; ****, P < 0.0001.
Additionally, we examined the surface expression of MHC class I proteins in cells infected with MNV CW3, MNV CR6, or IAV PR8 and observed that cells infected with any of the MNV strains tested expressed significantly lower levels of MHC class I proteins on their surfaces than uninfected or IAV PR8-infected cells (Fig. 6D). We also investigated the ability of MNV-infected cells to present viral antigens on their surfaces. DC 2.4 cells were either mock infected and left untreated, mock infected and pulsed with SIINFEKL, or infected with OVA-expressing or wild-type (WT) MNV strains, WT PR8, or PR8 OVA, and all cells were stained with an anti-SIINFEKL antibody (Fig. 6E). The SIINFEKL-pulsed uninfected cells showed 4-fold higher expression of SIINFEKL on their surfaces than uninfected and unpulsed cells, confirming that DC 2.4 cells are able to present the peptide. Similarly, PR8 OVA-infected cells displayed larger amounts (∼2-fold) of SIINFEKL on their surfaces than the negative control, WT PR8-infected cells. Interestingly, neither CW3 OVA- nor CR6 OVA-infected cells presented more SIINFEKL peptide than cells infected with WT CW3 or CR6, indicating that SIINFEKL was not exposed on the cell surface (Fig. 6E).
We used in vitro-activated green fluorescent protein (GFP)-tagged OT-I cells (C57BL/6 background, SIINFEKL stimulated) to assess the antigen-specific CD8+ T cell responses to CW3-, CR6-, and PR8-infected cells (Fig. 6F). Successful infection with CW3, CR6, and PR8 was verified via flow cytometry, and activation of OT-I-GFP CD8+ T cells was determined by staining for IFN-γ. OT-I-GFP cells did not react to exposure to uninfected DC 2.4 cells or cells infected with viruses that did not contain the SIINFEKL peptide (i.e., WT CW3, WT CR6, and WT PR8 [Fig. 6F]). In contrast, DC 2.4 cells infected with IAV PR8 OVA activated OT-I-GFP cells (∼25% IFN-γ positive), whereas, strikingly, CW3 OVA- and CR6 OVA-infected cells did not initiate an IFN-γ response in the OT-I-GFP cells, and IFN-γ levels were similar to those in OT-I-GFP cells exposed to uninfected cells or to cells infected with WT CW3 or CR6 (Fig. 6F). These findings indicate that the reduction in MHC class I expression on the cell surface leads to a reduction in viral antigen presentation, rendering CD8+ T cells blind to the infection.
As mentioned above, we observed that treatment with the inhibitor dynasore could prevent the decrease in MHC class I levels in infected cells (Fig. 3F). We therefore examined the effect of dynasore on the ability of OT-I cells to detect CW3 OVA- and CR6 OVA-infected cells. For this purpose, DC 2.4 cells were infected with CW3 OVA or CR6 OVA, as well as with the WT CW3 and CR6 strains, and were treated with dynasore at 1 hpi. To ensure that dynasore treatment does not interfere with SIINFEKL presentation, cells infected with PR8 OVA were used as a positive control. The DC 2.4 cells were added to OT-I cells, and their activation was analyzed via intracellular IFN-γ staining (Fig. 6G). We observed a robust increase in IFN-γ positive OT-I cells exposed to PR8 OVA-infected cells, confirming that dynasore does not interfere with antigen presentation or recognition. Despite the fact that dynasore inhibits the decrease in MHC class I levels in MNV-infected cells, we did not detect a significant increase in the IFN-γ-positive population among OT-I cells that were exposed to CW3 OVA- or CR6 OVA-infected cells treated with dynasore (Fig. 6G). Thus, although dynasore has stabilized MHC class I levels on the surfaces of MNV-infected cells, it appears to have no effect on the intracellular levels of MHC class I proteins, and it cannot restore the MHC class I and MNV-peptide complexes needed for OT-I activation on the cell surface.
In conclusion, we have demonstrated that MNV infection results in a significant surface reduction of MHC class I proteins. This impediment hinders the presentation of viral peptides, preventing recognition of the infected cells by antigen-specific CD8+ T cells and thereby reducing the initiation of the cellular immune response.
DISCUSSION
NoVs cause a quick and acute infection in hosts, seemingly avoiding our rapid immune detection and delaying clearance. MHC class I proteins play a critical role in the immune response against viruses by exposing viral antigens to the innate immune cells and initiating the adaptive T cell response. In this study, we showed that infection of mouse macrophages and dendritic cells with MNV leads to significant reductions in MHC class I expression levels at the surfaces of infected cells, resulting in impairment of CD8+ T cell recognition and activation (Fig. 1, 5 and 6). The ability of MNV to interfere with MHC class I expression may contribute to its ability to quickly establish replication in infected hosts.
This observation was specific for MNV-infected cells, since surrounding “bystander” cells, which had been exposed to the virus but remained uninfected, displayed MHC class I surface expression levels similar to those of mock-infected cells (Fig. 1A to C). The decrease in MHC class I proteins on the cell surface seems to be protein specific and not based on a global reduction in cell surface proteins, because we did not observe changes in the expression of F4/80 or CD11b (Fig. 1C and D). At this point, we cannot exclude the possibility that other surface proteins are targeted during MNV infection, but we can exclude an overall effect on surface protein expression. Most significantly, we have demonstrated that the decreased MHC class I surface expression levels were not due to a reduction in MHC class I transcription (Fig. 2B) but most likely occurred via reductions in intracellular levels of MHC class I proteins (Fig. 1C), potentially mediated via the proteasome and the endocytic pathway (Fig. 3). Importantly, we observed that this reduction in surface expression of MHC class I protein was associated with the inability of CD8+ T cells to recognize surface-expressed viral peptides and, thus, to recognize the infected cell. We also acknowledge that the data presented are from infections of cell lines and that infection of primary cells and of cells from animal tissue would be useful to support our observations.
We hypothesized that MHC class I proteins were degraded in MNV-infected cells, and we therefore attempted to rescue MHC class I protein expression with the proteasome inhibitor MG132 (Fig. 3). Treatment of MNV-infected cells with MG132 led to a partial, but not a complete, rescue of MHC class I surface expression. At this stage, our observations are contradictory to some extent, as there is a close and critical association of the MHC class I pathway with the proteasome. Normally, the proteasome-mediated degradation of proteins provides the source of peptides to be loaded onto the MHC class I molecules in the ER. Thus, inhibiting the proteasome would decrease the amount of peptides generated and imported into the ER, where smaller amounts of MHC class I and peptide complexes can be made and located to the surface. However, we observed that blocking protein degradation via the proteasome had a significant effect on restoring the surface expression of MHC class I proteins in infected cells. Interestingly, MHC class I levels on the cell surface were not restored when lysosome-mediated degradation was inhibited using chloroquine (Fig. 3), indicating that specifically proteasome-mediated degradation is playing a role in MHC class I levels during MNV infection. We additionally show that MHC class I surface expression can be completely restored by blocking the endocytic pathway with dynasore (Fig. 3). At this stage, it is unclear if MNV infection leads to the internalization and subsequent degradation of MHC class I proteins or if inhibiting the endocytic pathway is helping to stabilize existing levels of MHC class I on the cell surface.
Significantly, though, dynasore treatment of MNV-infected cells did not lead to an increase in the number of IFN positive OT-I cells (Fig. 6G), indicating that despite the restored MHC class I surface expression, OT-I cells are unable to recognize MNV-infected cells. This would further support the hypothesis that MNV infection leads to the degradation of MHC class I proteins within the cell, so that MHC class I and MNV-antigen complexes are rarely formed, and the reduced abundance of these complexes is not sufficient to activate CD8+ T cells. Intriguingly, our previous observations have shown that MNV recruits membrane and proteins from the endocytic pathway to the viral replication complex (35), implying a potential impediment to protein and membrane trafficking in this area. Thus, we can only postulate that in the MNV-infected cells, MHC class I molecules are targeted for degradation via the proteasome, possibly via translocation into the cytosol through the endocytic pathway and subsequent ubiquitination, and we aim to further our studies to elucidate this degradation mechanism.
One notable finding was our identification of the MNV NS3 protein as responsible for the reduced surface expression (Fig. 4). NS3 has been characterized as a nucleoside triphosphatase (NTPase), and we and others have recently shown that it is associated with the viral replication complex, the ER, mitochondria, lipids, and microtubules (36–38). NTPases have diverse activities and functions, including an association with microtubule dynamics, intracellular transport, and protein degradation (39–43). Our current studies have not shown if NS3 acts directly on MHC class I proteins or if it affects their expression via manipulation of the MHC class I protein pathway. Intriguingly, our attempts to rescue the MNV NS3 reduction in surface expression of MHC class I protein with MG132 were unsuccessful (data not shown), suggesting that this protein may invoke an alternative intervention to that which we observed during infection. We are actively pursuing this lead. Since we cannot completely rescue MHC class I expression even during infection, there may be additional modalities contributing to these observations.
In contrast, studies on MNV VP2 have identified a crucial role of the protein in MHC class I expression via inhibition of MHC class I transcription (44). Additionally, our recent RNA-seq (transcriptome sequencing) transcriptional profiling of MNV-infected cells revealed that many proteins involved in antigen presentation and immune activation were significantly altered (25). Based on our transcriptional analysis, we cannot exclude the possibility that MNV infection might prevent the upregulation of MHC class I transcription, but so far, we have not been able to detect a specific downregulation of MHC class I transcription during MNV infection (Fig. 2B). If MNV affected only the upregulation of MHC class I transcription, then MNV- and mock-infected cells would display similar levels of MHC class I proteins on their surfaces. Conversely, we have observed a reduction of surface and intracellular MHC class I protein levels in MNV-infected cells (Fig. 1 and 5). Therefore, we speculate that VP2 might prevent the upregulation of MHC class transcription, but MNV NS3 may contribute to the observed reduction in MHC class I surface expression levels.
Our observations presented here are similar to those for other viruses, such as HIV, HCMV, and Epstein-Barr virus (EBV), all of which interfere with the surface expression of MHC class I proteins, albeit via different strategies (23, 24, 28). It is known that viruses utilize several strategies to manipulate MHC class I surface expression, which are employed at different stages throughout the MHC class I protein expression pathway. For example, EBV inhibits proteasome-mediated degradation, preventing the synthesis of viral peptides and thus the formation of viral antigen and MHC class I complexes (26), whereas herpes simplex virus (HSV) and HCMV encode proteins that block the import of peptides, including viral peptides, into the ER (24, 27, 45). Additionally, the Nef protein of HIV causes the downregulation of MHC class I expression and inhibits the trafficking of MHC class I complexes from the Golgi apparatus to the cell surface (23, 46). We can now contribute another mechanism, one whereby MNV induces proteasome-mediated degradation of the MHC class I protein itself, thus resulting in decreased protein levels and surface expression.
The downregulation of MHC class I surface expression has critical effects on the ability of both the innate and the adaptive immune response to detect and clear MNV-infected cells. We demonstrated that the reduction in MHC class I proteins causes no viral peptides, or only small amounts, to be presented on MNV-infected cells (Fig. 5B). Therefore, CD8+ T cells cannot detect the infection and do not become activated (Fig. 5A). Tomov et al. showed previously that the antigen-specific CD8+ T cell response during persistent MNV infection is restrained, reducing clearance of the virus and allowing persistent replication within the host (47, 48). In this study, we have observed that both the acute (CW3) and persistent (CR6) strains of MNV decrease MHC class I surface levels to prevent detection by CD8+ T cells (Fig. 5). Early studies on MNV have demonstrated that the innate, but not the adaptive, immune response is critical for mice to survive the infection (15). These findings could be based on the fact that MNV interferes with MHC class I expression and antigen presentation. Avoiding immune recognition by CD8+ T cells allows the virus to replicate more efficiently and undisturbedly but also affects the immune memory generated in response to an infection. MNV and HuNoV infections have been shown to induce little immune memory to homologous challenges, and the response to vaccine treatment has been very variable so far (49–52). Even though the great strain variety and constant changes in the structural protein VP1 have an impact on immune memory, it needs to be considered that this defect in immune memory could be based on insufficient presentation of MNV antigens due to a reduction in MHC class I surface expression, inhibiting the priming of CD8+ T cells.
Overall, the reduction in MHC class I surface expression levels seems to be highly beneficial for the virus, since smaller amounts of MHC class I peptide complexes are displayed on the cell surface, resulting in reduced presentation of viral antigen and decreased detection by surveillance immune cells, such as CD8+ T cells. Manipulating MHC class I expression on the cell surface helps the virus to hide within the infected cell and dampens the immune response, allowing for efficient and swift viral replication.
MATERIALS AND METHODS
Cell lines.
RAW264.7 cells, a murine macrophage cell line derived from the BALB/c mouse strain, and DC 2.4 cells, a murine dendritic cell line derived from C57BL/6 mice, as well as human 293T cells, were kept in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 2 mM GlutaMAX (all from Gibco). All cell lines are semiadherent and were detached from the culture vessel by using a 18-gauge 1 ½-in syringe (RAW264.7, DC2.4) or by vigorous pipetting (293T). Cells were cultivated at 37°C under 5% CO2.
Plasmids and antibodies.
Plasmids encoding the 6×His-tagged MNV NS proteins (NS1-2 to NS4, NS6, NS7) on a pcDNA3.1 backbone have been generated and published previously (33). pcDNA3.1(+)-MNV-NS5-HIS was generated using the following primers to amplify the MNV NS5 sequence via PCR: forward primer VPG.EcoRV-F (5′-GATATCACCACCATGGGAAAGAAGGGCAAG-3′) and reverse primer VPG.SpeI-R (5′-ACTAGTTCAATGATGATGATGATGATGCTCAAAGTTGATCTT-3′). The PCR product was cut with the restriction enzymes SpeI and EcoRV, whose restriction sites had been incorporated into the forward and reverse primers, respectively. The pcDNA3.1(+) vector was digested with XbaI and EcoRV before ligation of the PCR product and the vector with the T4 DNA ligase (Promega). Antibodies to the following antigens were used: the 6×His tag (Abcam), actin (Sigma), calnexin (Abcam), double-stranded RNA (dsRNA) (Scicons), HLA-ABC, IFN-γ, and SIINFEKL (conjugated with allophycocyanin [APC]; BioLegend), MHC class I (H-2Kd/H-2Dd) (conjugated with fluorescein isothiocyanate [FITC]; Bio-Gems), MHC class I (Y3) (kindly provided by Justine Mintern, Bio21 Institute, Melbourne, Australia), MHC class I (M1/42) (kindly provided by the Villadangos Lab, Doherty Institute, Melbourne, Australia), MNV NS3 and NS6 (kindly provided by Kim Green, NIH, USA), and MNV NS5 and NS7 (manufactured by Invitrogen).
MNV infection and drug treatments.
RAW264.7 macrophages and DC 2.4 dendritic cells were infected at a multiplicity of infection (MOI) of 5 with a tertiary stock of the MNV strain CW1, unless stated otherwise. The culture medium was replaced with serum-free medium with GlutaMAX. Virus was added in a low volume, and culture dishes were rocked regularly for about 60 min to ensure virus binding and to prevent drying of cells. Afterwards, sufficient serum-free medium was added, depending on the culture vessel. Unless otherwise indicated, cells were harvested or fixed 12 h after infection. MG132 is an inhibitor of the proteasome and was used to inhibit protein degradation. MG132 was used at a concentration of 0.5 μM and was added to the culture medium 1 hpi. Chloroquine is an inhibitor of lysosome-mediated degradation and was used at a concentration of 10 μM. Dynasore inhibits endocytosis and was used at a concentration of 50 μM. The SIINFEKL peptide is a short peptide sequence derived from the ovalbumin protein and can be presented by MHC class I proteins and recognized by T cells. SIINFEKL was used at a concentration of 1 μg/ml and was applied for 1 h at 37°C.
Transfection of cell lines.
293T cells were transfected with Lipofectamine 2000, and cells were treated according to the manufacturer's protocol. In brief, plasmid DNA and the transfection reagent were diluted in Opti-MEM, mixed together, and incubated at room temperature. Plasmid DNA and transfection reagent mixtures were added dropwise to the cells, which were kept in DMEM with 10% FBS and 2 mM GlutaMAX (all from Gibco). Cells were incubated at 37°C for 18 h before analysis via flow cytometry.
Immunofluorescence staining and confocal imaging.
Cells intended for confocal imaging analysis were grown on 10-mm round coverslips and treated according to experimental design. Cells were stained with an MHC class I-specific antibody (anti-H-2Kd/H-2Dd, FITC conjugated; Bio-Gems) in 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) on their surfaces prior to fixation on ice for 30 min or were stained for MHC class I proteins intracellularly instead. After the cells were washed in 0.1% BSA (in PBS), the secondary antibody (Alexa Fluor 488-conjugated anti-mouse antibody; Life Technologies) was added. Cells were fixed in 4% (wt/vol) paraformaldehyde (PFA) (Electron Microscopy Sciences) (in PBS) for 10 min and were permeabilized in 0.1% (wt/vol) Triton X-100 (Sigma) and 4% PFA (10 min). After a wash step with PBS, 0.2 M glycine (Sigma) was added to reduce the autofluorescence of formaldehyde groups. Samples used for staining with the GEF-H1 antibody were fixed in ice-cold methanol for 3 min, followed by a 30-s incubation with ice-cold acetone. Fixed cells were stained with relevant primary antibodies in 1% (wt/vol) BSA for 45 to 60 min, washed in 0.1% BSA in PBS, and incubated again with the secondary antibodies for 30 to 45 min. Coverslips were then washed in PBS only, and 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml) was added for 5 min. Final washing steps were performed with Milli-Q water (Millipore), and coverslips were dried and mounted on cover slides with Ultramount 4 (Fronine). Samples were kept cool and dark until imaging with confocal microscopes. Confocal pictures were collected with either the LSM 700 or the LSM 710 confocal microscope using ZEN software (Zeiss).
Immunoblotting.
Samples were lysed in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40) and were centrifuged at 21,000 rcf for 10 min to sediment cell debris. All protein samples were handled at 4°C or on ice and were supplemented with Protease Inhibitor Cocktail III (Astral Scientific). Protein samples were boiled at 95°C in SDS loading buffer (125 mM Tris-HCl, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue) prior to immunoblotting and were kept on ice. Lysates were separated on a polyacrylamide gel (10% or 12%, depending on the size of the protein of interest) via SDS-PAGE (120 V), and afterwards, proteins were transferred to a 0.2-μm polyvinylidene difluoride (PVDF) membrane (Bio-Rad) (100 V, 65 min). Membranes were processed according to the manufacturer's protocol and were incubated in 5% BSA (dissolved in 0.1% Tween in PBS) at 4°C overnight or at room temperature for at least 2 h. Primary antibodies were diluted in 5% BSA and 0.1% Tween in PBS and were incubated with the membrane for 3 to 4 h at room temperature or at 4°C overnight. After wash steps with 0.1% Tween in PBS, the secondary antibodies in 0.1% Tween in PBS were added to the membrane (2 to 3 h). Last, the membrane was washed again; membranes were incubated with ECL Plus Western blotting substrate (Pierce, Thermo Fisher) for 5 min; and signals were visualized with the MF-ChemiBis documentation system (DNR).
Flow cytometry.
Cells were harvested and washed twice in staining buffer (1% FBS in PBS). About 1 × 106 cells were resuspended in a staining buffer containing cell surface antibodies and were incubated at 4°C in the dark for 30 min. To remove residual antibodies, cells were washed in the staining buffer. If unconjugated antibodies were used, the previous steps were repeated with the secondary antibodies. For fixation, cells were thoroughly resuspended in a fixation buffer (2% PFA in PBS) and were kept at room temperature for 15 min. Cells were centrifuged, and the fixation buffer was aspirated, before the permeabilization solution (0.1% Triton X-100 in PBS) was applied. Permeabilized cells were incubated with intracellular staining antibodies for 1 h at room temperature. Cells were washed in staining buffer, resuspended in PBS, and kept in the dark at 4°C until flow cytometry analysis. If unconjugated antibodies were used, the intracellular staining was repeated with the secondary antibodies for 45 min. Flow cytometry data were collected with a BD FACSCanto II or BD LSRFortessa analyzer using BD FACSDiva software (BD Biosciences). Data were analyzed using FlowJo analysis software.
Quantitative RT-PCR.
Cells were harvested and washed in PBS. Cell pellets were lysed in TRIzol reagent (Life Technologies) for 10 min and were stored at −80°C. RNA was extracted by adding chloroform at a ratio of 1:5, mixing thoroughly, and incubating the mixture for 5 min at room temperature to allow separation of the aqueous and organic phases. After centrifugation at 12,000 rcf for 15 min at 4°C, the organic phase was discarded, and 20 ng glycogen was added to the aqueous phase. RNA was precipitated in isopropanol (isopropanol/TRIzol ratio, 1:2) for 10 min and was centrifuged at 12,000 rcf for 10 min at 4°C. The supernatant was removed, and the RNA pellet was washed with 70% ethanol, centrifuged again for 5 min, and, after removal of the ethanol, left at room temperature to dry. RNA pellets were dissolved in autoclaved diethyl pyrocarbonate (DEPC)-treated water at 60°C for 10 min. For consecutive reverse transcription, the RNA concentration was measured using a NanoDrop system (Thermo Scientific). To exclude DNA contamination, 1 μg RNA was treated with RQ1 DNase (Promega) and was incubated for 30 min at 37°C. DNase was heat inactivated at 65°C for 15 min with EDTA. For reverse transcription, SensiFAST reverse transcriptase (Bioline) was used, and the following incubation pattern was applied: 25°C for 10 min, 42°C for 15 min, 85°C for 5 min. iTaq Universal Sybr green supermix (2× concentrated; Bio-Rad) and the following primers were used to set up the qPCR samples in duplicate: GAPDH (F) Ms (5′-CGTCCCGTAGACAAAATGGT-3′), GAPDH (R) Ms (5′-TCAATGAAGGGGTCGTTGAT-3′), MNV-NS3 (F) (5′-TTGTTGGCATCAAGGACACCTG-3′), and MNV-NS3 (R) (5′-TGGATGGAATGAAGGGCTCC-3′). MHC class I primers were purchased from Bio-Rad (PrimePCR for SYBR green assay: H2-D1 mouse and GAPDH mouse), and primers for β2 microglobulin were obtained from Sigma (KSPQ12012G). RNA levels were analyzed with a Stratagene Mx3005P qPCR machine. Fold changes in target genes were calculated using the ΔΔCT method relative to the internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
In vitro activation of CD8+ T cells.
Stimulator cells were obtained from C57BL/6 mouse spleens. For this purpose, a single-cell suspension in RPMI medium (10% fetal calf serum [FCS] with penicillin-streptomycin, l-glutamine, 2-mercaptoethanol) was prepared from spleens and was incubated with the SIINFEKL peptide (0.5 μg/ml) for 1 h at 37°C. Cells were washed in PBS three times before exposure to responder cells. To generate responder cells, a single-cell suspension from the spleens of OT-I-GFP mice (kindly provided by Lauren Holz, University of Melbourne, Melbourne, Australia) was created and added to the stimulator cells. The medium was supplemented with 10 U/ml interleukin 2 (IL-2) and 0.17 μg/ml lipopolysaccharide (LPS), and cells were grown for 4 to 5 days. Additional medium was added to the cells on days 2 and 3 after activation, and cells were washed in PBS multiple times to remove the LPS before they were used in the functional assay. T cell purity was determined via staining with anti-CD8 and anti-Vα antibodies and was analyzed via flow cytometry.
Functional assay for CD8+ T cell activation.
DC 2.4 cells were either infected with the WT CW3, CW3 OVA, WT CR6, or CR6 OVA MNV strain (34) or the WT PR8 or PR8 OVA influenza virus strain (53, 54) (all at an MOI of 5) or left uninfected. Cells were harvested, and the cell concentration was adjusted to 5 × 106 cells/ml. OT-I-GFP responder cells were obtained as described above, adjusted to 10 × 106 cells/ml, and added to target cells at a 2:1 ratio (5 × 105 target cells with 1 × 106 responder cells). Cells were incubated in the presence of brefeldin A (BFA) (5 μg/ml; Sigma-Aldrich) and monensin (5 μg/ml; BD Biosciences) for 6 h at 37°C. To assess the viability and activation of the OT-I-GFP cells, cells were stained on the surface with LIVE/DEAD stain (aqua; excitation at 405 nm; Invitrogen), fixed for 10 min in 1% PFA in PBS, and permeabilized in BD FACS Permeabilizing Solution 2 afterwards. An anti-IFN-γ antibody (BioLegend) was used to stain cells intracellularly for 1 h at room temperature. Cells were analyzed via flow cytometry using the FACSDiva and FlowJo programs. OT-I-GFP cells were identified via positive GFP staining and were analyzed for their IFN-γ expression.
ACKNOWLEDGMENTS
We thank the Biological Optical Microscopy Platform (BOMP) and the ImmunoID Flow Cytometry facility at the University of Melbourne for assistance with this project. A special thank you to Andrew Brooks, Justine Mintern, and José Villadangos for advice and feedback and for the provision of antibodies. We thank Kim Green for generously providing antibodies. We also thank the Heath laboratory and Lauren Holz for providing OT-I-GFP cells and reagents and Tim Nice and Skip Virgin for providing virus constructs expressing WT CW3, WT CR6, CW3 OVA, and CR6 OVA. We also thank the Kedzierska lab (Doherty Institute, Melbourne, Australia) for providing IAV PR8 and PR8 OVA.
This study has been supported by project grant 1083139, awarded to J.M.M. and P.A.W. by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Angulo FJ, Kirk MD, McKay I, Hall GV, Dalton CB, Stafford R, Unicomb L, Gregory J. 2008. Foodborne disease in Australia: the OzFoodNet experience. Clin Infect Dis 47:392–400. doi: 10.1086/589861. [DOI] [PubMed] [Google Scholar]
- 2.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg Infect Dis 5:607. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. 2014. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mead PS, Slutsker L, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States: reply to Dr. Hedberg. Emerg Infect Dis 5:841–842. doi: 10.3201/eid0506.990625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel MM, Hall AJ, Vinje J, Parashar UD. 2009. Noroviruses: a comprehensive review. J Clin Virol 44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. 2016. Global economic burden of norovirus gastroenteritis. PLoS One 11:e0151219. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J Infect Dis 186:1–7. doi: 10.1086/341085. [DOI] [PubMed] [Google Scholar]
- 8.Matthews J, Dickey B, Miller R, Felzer J, Dawson B, Lee A, Rocks J, Kiel J, Montes J, Moe C. 2012. The epidemiology of published norovirus outbreaks: a review of risk factors associated with attack rate and genogroup. Epidemiol Infect 140:1161–1172. doi: 10.1017/S0950268812000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, Estes MK, Graham DY, Opekun AR, Richardson C, Mendelman PM. 2011. Norovirus vaccine against experimental human Norwalk virus illness. N Engl J Med 365:2178–2187. doi: 10.1056/NEJMoa1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein DI, Jackson L, Patel SM, El Sahly HM, Spearman P, Rouphael N, Rudge TL Jr, Hill H, Goll JB. 2014. Immunogenicity and safety of four different dosing regimens of anthrax vaccine adsorbed for post-exposure prophylaxis for anthrax in adults. Vaccine 32:6284–6293. doi: 10.1016/j.vaccine.2014.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein DI, Atmar RL, Lyon GM, Treanor JJ, Chen WH, Jiang X, Vinje J, Gregoricus N, Frenck RW Jr, Moe CL, Al-Ibrahim MS, Barrett J, Ferreira J, Estes MK, Graham DY, Goodwin R, Borkowski A, Clemens R, Mendelman PM. 2015. Norovirus vaccine against experimental human GII.4 virus illness: a challenge study in healthy adults. J Infect Dis 211:870–878. doi: 10.1093/infdis/jiu497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L. 2016. Replication of human noroviruses in stem cell–derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 15.Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wobus CE, Thackray LB, Virgin HW IV. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol 80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 2015. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha-Pereira J, Jacobs S, Noppen S, Verbeken E, Michiels T, Neyts J. 2018. Interferon lambda (IFN-λ) efficiently blocks norovirus transmission in a mouse model. Antiviral Res 149:7–15. doi: 10.1016/j.antiviral.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Vyas JM, Van der Veen AG, Ploegh HL. 2008. The known unknowns of antigen processing and presentation. Nat Rev Immunol 8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wearsch PA, Cresswell P. 2008. The quality control of MHC class I peptide loading. Curr Opin Cell Biol 20:624–631. doi: 10.1016/j.ceb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wearsch PA, Peaper DR, Cresswell P. 2011. Essential glycan-dependent interactions optimize MHC class I peptide loading. Proc Natl Acad Sci U S A 108:4950–4955. doi: 10.1073/pnas.1102524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park B, Lee S, Kim E, Cho K, Riddell SR, Cho S, Ahn K. 2006. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell 127:369–382. doi: 10.1016/j.cell.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz O, Maréchal V, Le Gall S, Lemonnier F, Heard J-M. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med 2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 24.Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJ, Ploegh HL, Peterson PA, Yang Y, Früh K. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613–621. doi: 10.1016/S1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 25.Enosi Tuipulotu D, Netzler NE, Lun JH, Mackenzie JM, White PA. 2017. RNA sequencing of murine norovirus-infected cells reveals transcriptional alteration of genes important to viral recognition and antigen presentation. Front Immunol 8:959. doi: 10.3389/fimmu.2017.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A 94:12616–12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 28.Ahn K, Meyer TH, Uebel S, Sempé P, Djaballah H, Yang Y, Peterson PA, Frueh K, Tampé R. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J 15:3247–3255. [PMC free article] [PubMed] [Google Scholar]
- 29.Hewitt EW, Gupta SS, Lehner PJ. 2001. The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J 20:387–396. doi: 10.1093/emboj/20.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson PG, Efstathiou S, Doherty PC, Lehner PJ. 2000. Inhibition of MHC class I-restricted antigen presentation by γ2-herpesviruses. Proc Natl Acad Sci U S A 97:8455–8460. doi: 10.1073/pnas.150240097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci U S A 97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee DH, Goldberg AL. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8:397–403. doi: 10.1016/S0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 33.Hyde JL, Mackenzie JM. 2010. Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology 406:138–148. doi: 10.1016/j.virol.2010.06.047. [DOI] [PubMed] [Google Scholar]
- 34.Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, Tomov VT, Kobuley D, Tran SV, Bittinger K, Bailey AG, Laughlin AL, Boucher JL, Wherry EJ, Bushman FD, Allen JE, Virgin HW, Artis D. 2014. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 345:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyde JL, Sosnovtsev SV, Green KY, Wobus C, Virgin HW, Mackenzie JM. 2009. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J Virol 83:9709–9719. doi: 10.1128/JVI.00600-0910.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotton B, Hyde JL, Sarvestani ST, Sosnovtsev SV, Green KY, White PA, Mackenzie JM. 2017. The norovirus NS3 protein is a dynamic lipid- and microtubule-associated protein involved in viral RNA replication. J Virol 91:e02138-. doi: 10.1128/JVI.02138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doerflinger SY, Cortese M, Romero-Brey I, Menne Z, Tubiana T, Schenk C, White PA, Bartenschlager R, Bressanelli S, Hansman GS. 2017. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog 13:e1006705. doi: 10.1371/journal.ppat.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen J-B, Wei L-H, Chen L-W, Chen L-Y, Hung C-H, Wang S-S, Chang P-J. 2018. Subcellular localization and functional characterization of GII.4 norovirus-encoded NTPase. J Virol 92:e01824-. doi: 10.1128/JVI.01824-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith DM, Benaroudj N, Goldberg A. 2006. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol 156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Paschal BM, Shpetner HS, Vallee RB. 1987. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol 105:1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshansky V, Futai M. 2008. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol 20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. 1991. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 266:17707–17712. [PubMed] [Google Scholar]
- 43.Rabinovich E, Kerem A, Fröhlich K-U, Diamant N, Bar-Nun S. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu S, Regev D, Watanabe M, Hickman D, Moussatche N, Jesus DM, Kahan SM, Napthine S, Brierley I, Hunter RN III. 2013. Identification of immune and viral correlates of norovirus protective immunity through comparative study of intra-cluster norovirus strains. PLoS Pathog 9:e1003592. doi: 10.1371/journal.ppat.1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehner PJ, Karttunen JT, Wilkinson GW, Cresswell P. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc Natl Acad Sci U S A 94:6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roeth JF, Williams M, Kasper MR, Filzen TM, Collins KL. 2004. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J Cell Biol 167:903–913. doi: 10.1083/jcb.200407031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomov VT, Osborne LC, Dolfi DV, Sonnenberg GF, Monticelli LA, Mansfield K, Virgin HW, Artis D, Wherry EJ. 2013. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol 87:7015–7031. doi: 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomov VT, Palko O, Lau CW, Pattekar A, Sun Y, Tacheva R, Bengsch B, Manne S, Cosma GL, Eisenlohr LC. 2017. Differentiation and protective capacity of virus-specific CD8+ T cells suggest murine norovirus persistence in an immune-privileged enteric niche. Immunity 47:723–738.e5. doi: 10.1016/j.immuni.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinkula J, Ball JM, Löfgren S, Estes MK, Svensson L. 1995. Antibody prevalence and immunoglobulin IgG subclass pattern to Norwalk virus in Sweden. J Med Virol 47:52–57. doi: 10.1002/jmv.1890470111. [DOI] [PubMed] [Google Scholar]
- 50.Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. 1977. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N Engl J Med 297:86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- 51.Graham DY, Jiang X, Tanaka T, Opekun AR, Madore HP, Estes MK. 1994. Norwalk virus infection of volunteers: new insights based on improved assays. J Infect Dis 170:34–43. doi: 10.1093/infdis/170.1.34. [DOI] [PubMed] [Google Scholar]
- 52.El-Kamary SS, Pasetti MF, Mendelman PM, Frey SE, Bernstein DI, Treanor JJ, Ferreira J, Chen WH, Sublett R, Richardson C. 2010. Adjuvanted intranasal Norwalk virus-like particle vaccine elicits antibodies and antibody-secreting cells that express homing receptors for mucosal and peripheral lymphoid tissues. J Infect Dis 202:1649–1658. doi: 10.1086/657087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenkins MR, Webby R, Doherty PC, Turner SJ. 2006. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J Immunol 177:2917–2925. doi: 10.4049/jimmunol.177.5.2917. [DOI] [PubMed] [Google Scholar]
- 54.Stock AT, Jones CM, Heath WR, Carbone FR. 2006. CTL response compensation for the loss of an immunodominant class I-restricted HSV-1 determinant. Immunol Cell Biol 84:543–550. doi: 10.1111/j.1440-1711.2006.01469.x. [DOI] [PubMed] [Google Scholar]