The present study demonstrates that the V protein of SeV, Nipah virus, and human parainfluenza virus type 2 interacts with NLRP3 to inhibit NLRP3 inflammasome activation, potentially suggesting a novel strategy by which viruses evade the host innate immune response. As all members of the Paramyxovirinae subfamily carry similar V genes, this new finding may also lead to identification of novel therapeutic targets for paramyxovirus infection and related diseases.
KEYWORDS: NLRP3, V protein, inflammasome, paramyxovirus
ABSTRACT
Inflammasomes play a key role in host innate immune responses to viral infection by caspase-1 (Casp-1) activation to facilitate interleukin-1β (IL-1β) secretion, which contributes to the host antiviral defense. The NLRP3 inflammasome consists of the cytoplasmic sensor molecule NLRP3, adaptor protein ASC, and effector protein pro-caspase-1 (pro-Casp-1). NLRP3 and ASC promote pro-Casp-1 cleavage, leading to IL-1β maturation and secretion. However, as a countermeasure, viral pathogens have evolved virulence factors to antagonize inflammasome pathways. Here we report that V gene knockout Sendai virus [SeV V(−)] induced markedly greater amounts of IL-1β than wild-type SeV in infected THP1 macrophages. Deficiency of NLRP3 in cells inhibited SeV V(−)-induced IL-1β secretion, indicating an essential role for NLRP3 in SeV V(−)-induced IL-1β activation. Moreover, SeV V protein inhibited the assembly of NLRP3 inflammasomes, including NLRP3-dependent ASC oligomerization, NLRP3-ASC association, NLRP3 self-oligomerization, and intermolecular interactions between NLRP3 molecules. Furthermore, a high correlation between the NLRP3-binding capacity of V protein and the ability to block inflammasome complex assembly was observed. Therefore, SeV V protein likely inhibits NLRP3 self-oligomerization by interacting with NLRP3 and inhibiting subsequent recruitment of ASC to block NLRP3-dependent ASC oligomerization, in turn blocking full activation of the NLRP3 inflammasome and thus blocking IL-1β secretion. Notably, the inhibitory action of SeV V protein on NLRP3 inflammasome activation is shared by other paramyxovirus V proteins, such as Nipah virus and human parainfluenza virus type 2. We thus reveal a mechanism by which paramyxovirus inhibits inflammatory responses by inhibiting NLRP3 inflammasome complex assembly and IL-1β activation.
IMPORTANCE The present study demonstrates that the V protein of SeV, Nipah virus, and human parainfluenza virus type 2 interacts with NLRP3 to inhibit NLRP3 inflammasome activation, potentially suggesting a novel strategy by which viruses evade the host innate immune response. As all members of the Paramyxovirinae subfamily carry similar V genes, this new finding may also lead to identification of novel therapeutic targets for paramyxovirus infection and related diseases.
INTRODUCTION
Recombinant Sendai virus that does not express the V protein [SeV V(−)] is attenuated in vivo but not in vitro (1). Together with previous studies showing that SeV V protein counteracts innate immunity other than that mediated by interferon (2, 3), this finding suggests that SeV V protein may play a role in modulating the immune response. Thus, the host factor responsible for the early clearance of SeV V(−) might be expressed in cells, such as macrophages and dendritic cells, present only in whole organisms, including mice, but not in the standard cell lines used to model infection. The host factor responsible for the early clearance of SeV V(−) remains enigmatic.
We have recently been studying the inhibitory action of SeV against innate immunity (4–9) and found that the interleukin-1β (IL-1β) response to SeV V(−) was greater than that to wild-type SeV (SeV wt) in infected THP1 macrophages. This finding indicates that the V protein may inhibit inflammasome activation and further inflammatory responses. Thus, we decided to study how SeV regulates IL-1β secretion in macrophages during viral infection, as well as the underlying mechanism.
In response to viral infection, macrophages secrete large amounts of cytokines, including IL-1β. Although the production of many proinflammatory cytokines is principally regulated at the transcriptional level, IL-1β requires an additional proteolytic event that is regulated in two steps (10). The first regulatory step involves stimulation via Toll-like receptors (TLRs) or retinoic acid-inducible gene-I like receptors (RLRs), which induces the synthesis of IL-1β as an inactive precursor. The second regulatory step involves posttranslational processing, which is required for the secretion and bioactivity of IL-1β and is catalyzed by inflammasome activation (11–13). The inflammasome operates as a platform for caspase-1 (Casp-1) activation, resulting in Casp-1-dependent proteolytic maturation and secretion of IL-1β. This, in turn, activates the expression of other immune genes and facilitates lymphocyte recruitment to the site of primary infection, thereby controlling invading pathogens. Moreover, inflammasomes counter viral replication and remove infected immune cells through an inflammatory cell death program termed pyroptosis (14). As a countermeasure, viral pathogens have evolved virulence factors to antagonize inflammasome pathways (14–17). Although many RNA viruses are recognized by the NLRP3 and RIG-I inflammasomes (17, 18), SeV has been reported to activate the inflammasome through double-stranded RNA (dsRNA) (19, 20). Therefore, we hypothesized that SeV has evolved the V protein to antagonize the NLRP3 inflammasome pathway.
In the present study, we clarified whether and how SeV V protein inhibits NLRP3 inflammasome activation leading to IL-1β secretion. Specifically, we showed that the V protein of SeV inhibits NLRP3 self-oligomerization by interacting with NLRP3 to block the NLRP3-mediated ASC oligomerization required for full activation of the inflammasome.
RESULTS
V gene knockout Sendai virus activates the NLRP3 inflammasome to induce IL-1β secretion.
The effect of SeV V protein on IL-1β activation was first determined in THP1 macrophages infected with SeV V(−). These differentiated macrophages were generated by stimulation of the human monocytic cell line THP1 with phorbol 12-myristate 13-acetate (PMA) (21, 22).
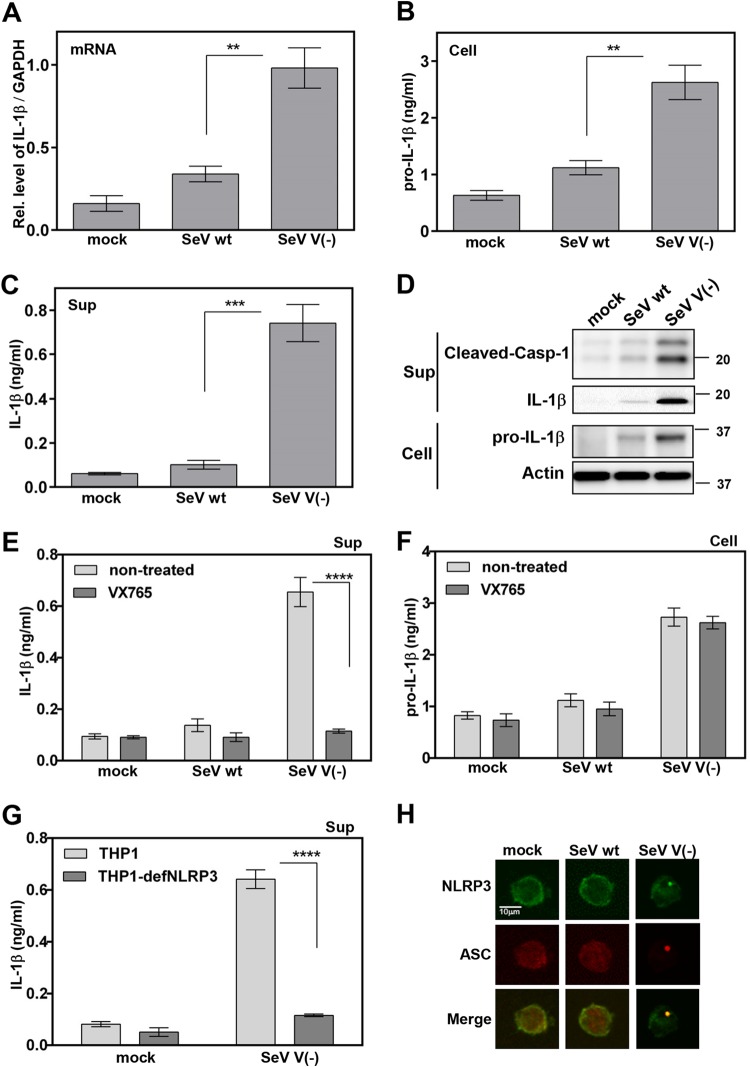
In THP1 macrophages, IL-1β secretion in cell supernatants was induced approximately 7-fold by SeV V(−) (Fig. 1C), whereas pro-IL-1β mRNA and protein production in cell lysates were increased approximately 2.7- and 2.3-fold, respectively, by SeV V(−) (Fig. 1A and B) compared to that by SeV wt. IL-1β maturation and caspase 1 (Casp-1) cleavage in cell supernatants were also activated by SeV V(−) (Fig. 1D). Taken together, these results demonstrate that SeV V(−) activates the production and secretion of IL-1β in infected THP1 macrophages.
FIG 1.
Role of the NLRP3 inflammasome in the regulation of SeV V(−)-induced IL-1β secretion. (A to D and H) PMA-differentiated THP1 (THP1/PMA) cells in a 96-well plate, 24-well plate, or cell chamber were infected with SeV wt or SeV V(−) at a multiplicity of infection (MOI) of 3 for 24 h. (E and F) THP1/PMA cells in a 96-well plate were mock treated or treated with Casp-1 inhibitor VX-765 for 1 h and then infected with SeV wt or SeV V(−) at an MOI of 3 for 24 h. (G) THP1/PMA or THP1-defNLRP3/PMA cells in a 96-well plate were infected with SeV V(−) at an MOI of 3 for 24 h. mRNAs for pro-IL-1β and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were quantified by real-time PCR (A). Pro-IL-1β levels in cell lysates (B and F) or IL-1β levels in cell supernatants (C, E, and G) were determined by ELISA. Mature IL-1β in supernatants or pro-IL-1β in lysates was determined by immunoblot (IB) analysis. (H) Subcellular localizations of NLRP3 and ASC were examined under confocal microscopy. The scale bars represent 10 μm. Data were derived from at least three or four independent experiments and are presented as mean values. SD are shown as error bars. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The activation of IL-1β is regulated by two pathways (10): transcription of mRNA for pro-IL-1β regulated by nuclear factor κB (NF-κB) and processing of IL-1β mediated by Casp-1. In THP1 macrophages, IL-1β secretion was stimulated by SeV V(−), although this activation was repressed by VX-765, a Casp-1 inhibitor (Fig. 1E). However, the level of pro-IL-1β was not affected by VX-765 (Fig. 1F). These results suggest that Casp-1 is involved in SeV V(−)-induced activation of IL-1β.
To assess the role of the NLRP3 inflammasome in SeV V(−)-induced secretion of IL-1β, a THP1 cell line deficient for NLRP3 (THP1-defNLRP3) was used (23, 24). In PMA-differentiated THP1-defNLRP3 macrophages, IL-1β secretion was reduced in the absence of NLRP3 (Fig. 1G), indicating that NLRP3 is required for SeV V(−)-induced secretion of IL-1β.
Punctate localization of NLRP3 is an indication of inflammasome complex formation. We thus investigated the effect of SeV V(−) on NLRP3 inflammasome formation (25). NLRP3 was diffusely distributed in the cytoplasm of uninfected and SeV wt-infected THP1 macrophages but formed distinct small specks in SeV V(−)-infected THP1 macrophages (Fig. 1H), suggesting that SeV V(−) facilitates NLRP3 speck formation to activate the inflammasome. As the oligomerization of ASC is a direct indicator of inflammasome activation (26), we further examined the effect of SeV V(−) ASC pyroptosome formation in THP1 macrophages. ASC was diffusely distributed in the nucleus and cytoplasm in uninfected and SeV wt-infected macrophages but formed distinct small specks in SeV V(−)-infected THP1 macrophages (Fig. 1H), suggesting that SeV V(−) facilitates ASC oligomerization. Taken together, our findings demonstrate that SeV V(−) activates the NLRP3 inflammasome by NLRP3 speck formation and ASC oligomerization to induce IL-1β secretion.
SeV V protein inhibits the activation of the NLRP3 inflammasome to induce IL-1β secretion.
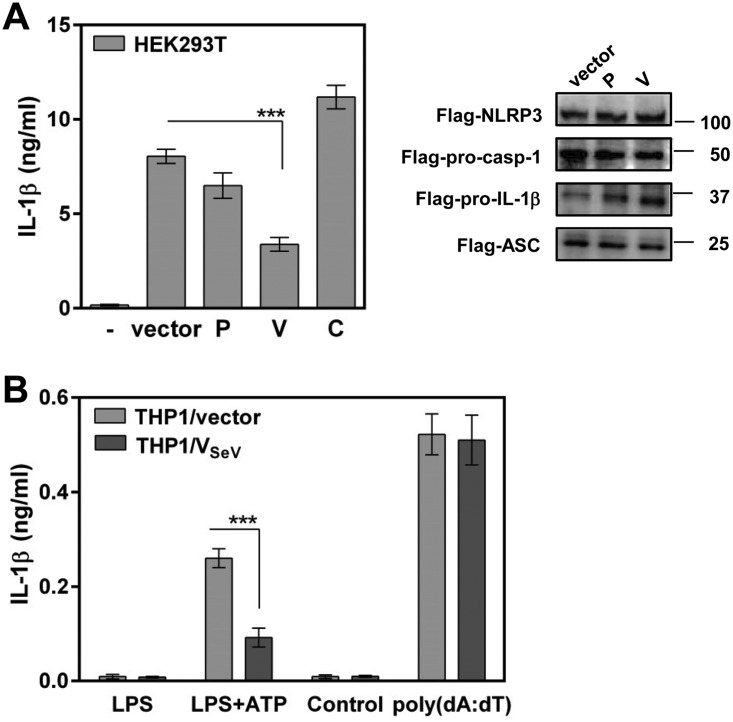
The NLRP3 inflammasome is composed of three major components: the inflammatory signal recognition receptor NLRP3, the adaptor protein ASC, and pro-caspase-1 (pro-Casp-1). On activation, danger signals from damaged cells and pathogens induce the assembly of these components, which, in turn, activates Casp-1, leading to cleavage and maturation of IL-1β for secretion. The ability to inhibit NLRP3 inflammasome-dependent IL-1β secretion without the need for other viral proteins was assessed by using a reconstitution system in which HEK293T cells were transfected with plasmids expressing pro-IL-1β and the NLRP3 inflammasome components (NLRP3, ASC, and pro-Casp-1), together with the P gene products of SeV. HEK293T cells transfected with these plasmids induced a large amount of mature IL-1β secretion (27). This secretion was markedly inhibited (∼60%) in the presence of SeV V protein without affecting the expression of components of the NLRP3 inflammasome, albeit not in the presence of P or C protein (Fig. 2A), suggesting that V protein inhibits NLRP3 inflammasome-dependent IL-1β secretion without the need for other viral proteins.
FIG 2.
Role of SeV V protein in inhibition of the NLRP3 inflammasome. (A) HEK293T cells were transfected with plasmids encoding NLRP3 inflammasome signaling molecules together with the indicated plasmid. At 24 h posttransfection, IL-1β levels in cell supernatants were determined by ELISA, whereas cells were immunoblotted with anti-Flag antibody to measure the expression of NLRP3, pro-Casp-1, pro-IL-1β, and ASC. (B) THP1/vector/PMA and THP1/VSeV/PMA cells were stimulated with LPS (5 ng/ml) plus ATP (0 or 5 mM) or with poly(dA·dT) (0 or 400 ng/ml). IL-1β levels in cell supernatants were determined 24 h after treatment (B). Data were derived from at least three or four independent experiments and are presented as mean values. SD are shown as error bars. ***, P < 0.001.
As THP1 macrophages are responsive to NLRP3 inflammasome ligands, we established THP1 cells stably expressing Flag-tagged SeV V protein (THP1/VSeV) to confirm the inhibitory effect of V protein on NLRP3 inflammasome-dependent signaling. Stimulation with an NLRP3 ligand, lipopolysaccharide (LPS) plus ATP, resulted in IL-1β secretion, whereas stable expression of V protein clearly suppressed the response of THP1/vector macrophages to LPS plus ATP. In contrast, no significant difference in IL-1β secretion was observed between THP1/VSeV and control macrophages when an AIM inflammasome ligand, poly(dA·dT), was used (Fig. 2B), indicating that NLRP3 inflammasome-dependent signaling was prevented in THP1/VSeV macrophages. These results suggest that V protein targets the NLRP3 inflammasome signaling pathway for inhibition of IL-1β secretion.
SeV V protein interacts with NLRP3 to inhibit inflammasome-dependent IL-1β secretion.
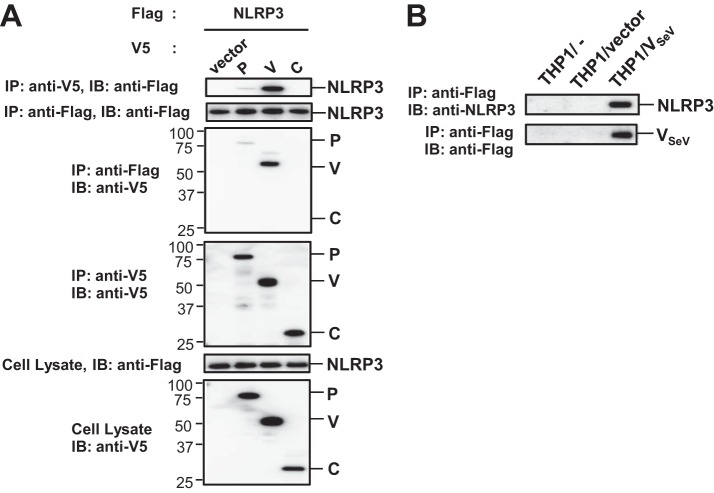
Next, the mechanism by which SeV V protein inhibits the NLRP3 inflammasome was investigated. Coimmunoprecipitation experiments were performed to identify the molecular target of SeV V protein. Flag-tagged NLRP3 (Flag-NLRP3) was transfected into HEK293T cells together with V5-His (VH)-tagged P gene products of SeV, and the resultant cell lysates were subjected to immunoprecipitation (IP). As shown in Fig. 3A, Flag-NLRP3 was coimmunoprecipitated with VH-tagged SeV V (VH-V) and with lesser amounts of VH-tagged SeV P (VH-P), but not with VH-tagged SeV C (VH-C). VH-V was also coimmunoprecipitated with Flag-NLRP3. Similarly, endogenous NLRP3 was coimmunoprecipitated with Flag-V when Flag-V was stably expressed in THP1 macrophages (Fig. 3B). Conversely, when HEK293T cells were transfected with Flag-tagged ASC (Flag-ASC) or Flag-tagged pro-Casp-1 (Flag-pro-Casp-1) together with VH-tagged SeV V protein (VH-V), neither Flag-ASC nor Flag-pro-Casp-1 was coimmunoprecipitated with VH-V in the absence of NLRP3 (data not shown). These results suggest that SeV V protein binds NLRP3.
FIG 3.
Interaction of SeV V protein with NLRP3 protein. (A) HEK293T cells were transfected with various combinations of the indicated plasmids. These cells were then lysed 24 h after transfection and the lysates were subjected to immunoprecipitation (IP) with anti-V5 or Flag antibody, followed by IB analysis with anti-Flag or V5 antibody. The whole-cell lysates prepared for IP were also subjected to IB with anti-Flag or anti-V5 antibody. (B) The THP1/VSeV/PMA cells were lysed, and the lysates were subjected to IP with anti-Flag, followed by IB analysis with anti-NLRP3 or anti-Flag antibody. Data are representative of those from three independent experiments.
The interaction of SeV V protein with NLRP3 is involved in inhibition of the NLRP3 inflammasome.
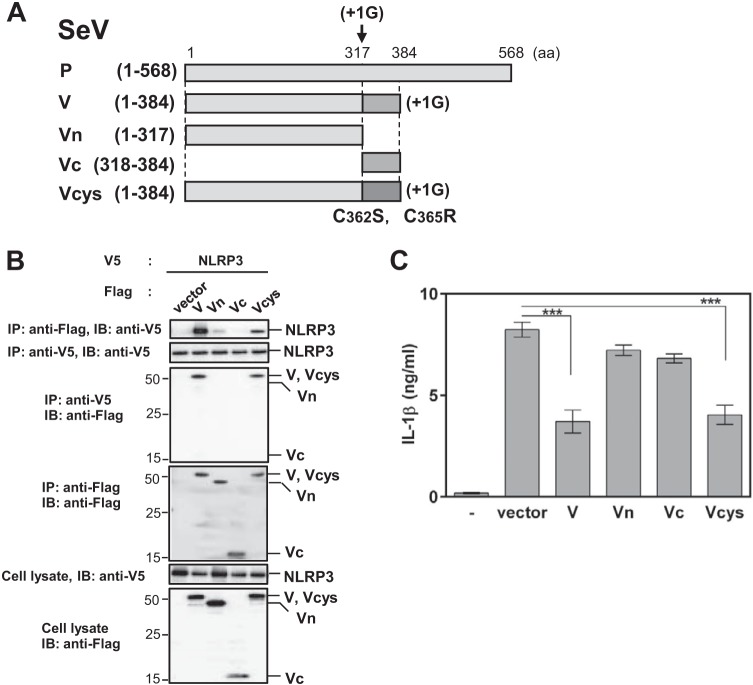
We subsequently examined the role of a functional region of V protein for interaction with NLRP3 by constructing plasmids encoding the Flag-tagged truncated mutants SeV Vn (amino acids [aa] 1 to 317) and SeV Vc (aa 318 to 384), as well as a point mutant, SeV Vcys (C362S and C365R) (Fig. 4A). VH-tagged NLRP3 (VH-NLRP3) was transfected into HEK293T cells together with one of these Flag-tagged mutants, and the resultant cell lysates were subjected to immunoprecipitation. Coimmunoprecipitation experiments showed that Vc essentially lost its NLRP3 binding capacity, whereas Vn exhibited slight binding ability (Fig. 4B). In contrast, there was no significant difference in NLRP3 binding between the Vcys mutant and wild-type V (Fig. 4B), suggesting that both the N and C-terminal regions are required for the interaction of SeV V with NLRP3 but that C362 and C365 in SeV V protein are not critical for the interaction.
FIG 4.
Requirement of the NLRP3-binding ability of V protein for inhibition of the NLRP3 inflammasome. (A) Schematic diagram of the SeV V mutants Vn (aa 1 to 317), Vc (aa 318 to 384), and Vcys (C362S and C365R). (B) HEK293T cells were transfected with plasmids encoding NLRP3 inflammasome signaling molecules together with the indicated plasmid. Immunoprecipitation and immunoblot analysis were performed as described for Fig. 3A. Data are representative of those from three independent experiments. (C) HEK293T cells were transfected with plasmids encoding NLRP3 inflammasome signaling molecules together with the indicated plasmid. IL-1β levels in cell supernatants were determined as described for Fig. 2A. Data were derived from at least three or four independent experiments and are presented as mean values. SD are shown as error bars. ***, P < 0.001.
V interacts with NLRP3 to inhibit inflammasome-dependent IL-1β secretion.
To assess whether the V-NLRP3 interaction is important for the blockade of NLRP3 inflammasome-dependent IL-1β secretion, HEK293T cells were transfected with pro-IL-1β and the NLRP3 inflammasome components, together with either wild-type V or one of the V mutants. Wild-type V and Vcys, but not Vn and Vc, inhibited NLRP3 inflammasome-dependent IL-1β, in agreement with their binding ability (Fig. 4C). These results demonstrate the importance of the V-NLRP3 interaction for the inhibition of NLRP3 inflammasome-dependent IL-1β secretion.
SeV V protein interacts with NLRP3 to inhibit inflammasome assembly.
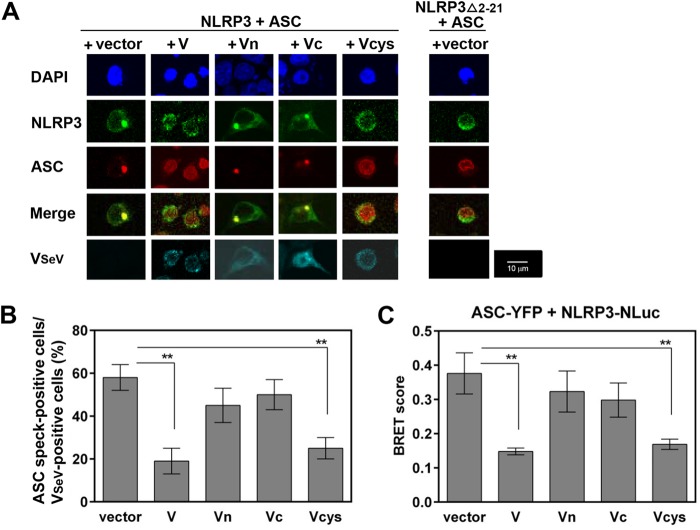
As SeV V protein binds NLRP3, the effects of this interaction on the formation of NLRP3-dependent ASC specks (ASC specks), a key event in NLRP3 inflammasome activation, were explored in HEK293T cells. NLRP3Δ2-21-GFP, a deletion mutant of NLRP3 that lacks 20 amino acids at the N terminus, was used as a positive control to inhibit NLRP3-dependent ASC speck formation (exhibiting loss of ASC specks) (28). When HEK293T cells were transfected with GFP-tagged NLRP3 (GFP-NLRP3) and Flag-ASC, together with the vector, NLRP3 and ASC were colocalized and distributed in the cytosol to form single specks (ASC specks) (Fig. 5A and B, vector), whereas in the presence of V or Vcys, the speck formation was reduced. In contrast, Vn or Vc exhibited little inhibitory effect (Fig. 5A and B). No significant differences in the expression of V were observed at the protein level between V and V mutants in HEK293T cells. V, Vn, and Vcys localized in the cytosol, whereas Vc was observed in both the cytosol and the nucleus (Fig. 5A and B), consistent with previous reports (29). Together, these data suggest that V protein blocks NLRP3 inflammasome complex assembly via V-NLRP3 interaction.
FIG 5.
Effect of SeV V protein on NLRP3 inflammasome assembly. (A and B) HEK293T cells in an 8-well microscope glass slide with a removable 8-well chamber were transfected with plasmids encoding NLRP3-GFP and ASC-Flag, together with the indicated plasmid encoding VSeV-V5His. (A) Subcellular localizations of NLRP3-GFP (green), ASC-Flag (red), VSeV-VH (blue-green), and nuclei (DAPI; blue) were visualized by confocal microscopy. The scale bars represent 10 μm. (B) ASC speck-forming cells and VSeV-positive cells were quantified from five randomly selected fields, and the percentages of ASC speck- and VSeV-positive cells in total VSeV-positive cells are shown. Data were derived from at least three or four independent experiments and are represented as mean values. SD are shown as error bars. **, P < 0.01. (C) Experimental data on BRET signals at 24 h posttransfection were derived from three independent experiments and are represented as mean values. SD are shown as error bars. **, P < 0.01 versus transfection with empty vector.
We then examined whether V protein could affect the association of NLRP3 with ASC, which may be essential for ASC oligomerization. The ability of V protein to block intermolecular interactions between NLRP3 and ASC was assessed by using the bioluminescence resonance energy transfer (BRET) assay (30), in which HEK293T cells were transfected with plasmids encoding yellow fluorescent protein (YFP)-tagged ASC (ASC-YFP) and NLuc-tagged NLRP3 (NLRP3-NLuc), together with V mutants. As shown in Fig. 5C, all combinations provided similar inhibitory patterns, suggesting that V protein disrupts the intermolecular interactions between NLRP3 and ASC. Taken together, these results indicate that SeV V protein interferes with NLRP3-ASC association and subsequent NLRP3-dependent ASC oligomerization by interacting with NLRP3. This finding raises the possibility that SeV V protein may directly interfere with NLRP3-ASC association or inhibit the conversion of inactive NLRP3 to its active form.
V interacts with NLRP3 to inhibit the self-oligomerization of NLRP3.
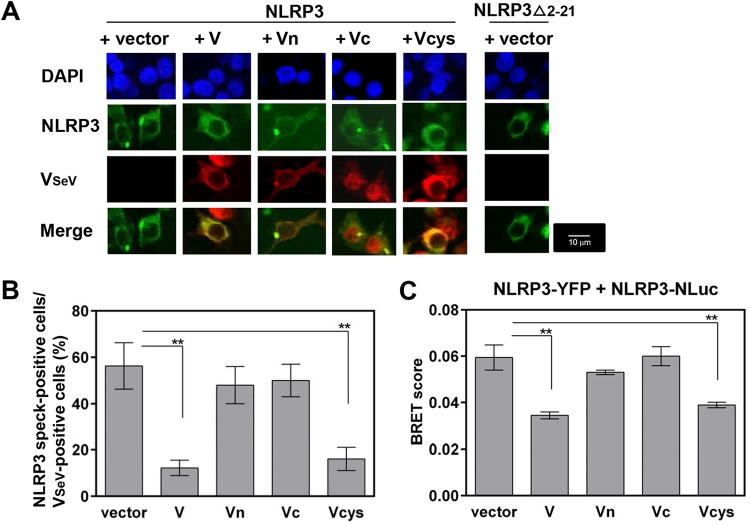
We examined the effect of V protein on the self-oligomerization of NLRP3, which is also required for NLRP3-dependent ASC oligomerization (30). Transfection of NLRP3-GFP led to the formation of small cytosolic aggregates (NLRP3 speck-like structures) (Fig. 6A, vector), consistent with previous reports (28, 31). NLRP3Δ2-21-GFP was also used as a positive control to show loss of NLRP3 speck formation (28). Coexpression of V and Vcys with NLRP3 abolished the NLRP3 speck-like structures. In contrast, little inhibitory effect was observed for Vn and Vc (Fig. 6A and B), suggesting that V protein inhibits the self-oligomerization of NLRP3 to activate inflammasome complex assembly by interacting with NLRP3 (Fig. 6A and B).
FIG 6.
Role of SeV V protein in NLRP3 self-oligomerization. (A and B) HEK293T cells in an 8-well microscope glass slide with a removable 8-well chamber were transfected with plasmids encoding NLRP3-GFP, together with the indicated plasmid encoding VSeV-Flag. (A) Subcellular localizations of NLRP3-GFP (green), VSeV-Flag (red), and nuclei (DAPI; blue) were visualized by confocal microscopy. The scale bars represent 10 μm. (B) NLRP3 speck-forming cells and VSeV-positive cells were quantified from five randomly selected fields, and the percentages of NLRP3 speck- and VSeV-positive cells in the total VSeV-positive cells are shown. Data were derived from at least three or four independent experiments and are presented as mean values. SD are shown as error bars. **, P < 0.01. (C) Experimental data on BRET signals at 24 h posttransfection were derived from three independent experiments and are presented as mean values. SD are shown as error bars. **, P < 0.01 versus transfection with empty vector.
To obtain more direct evidence of the inhibition of NLRP3 self-oligomerization, we assessed the ability of V protein to block intermolecular interactions between NLRP3 molecules by using the BRET assay (31), in which HEK293T cells were transfected with plasmids encoding NLRP3-YFP and NLRP3-Nluc, together with V mutants. As shown in Fig. 6C, all combinations provided a similar inhibitory pattern in the BRET assay, suggesting that V protein disrupts the intermolecular interactions between NLRP3 molecules. Taken together, these results indicate that SeV V protein disrupts NLRP3-NLRP3 intermolecular interactions and subsequent NLRP3 self-oligomerization by interacting with NLRP3.
Inhibition of NLRP3 inflammasome activation is shared by paramyxovirus V proteins.
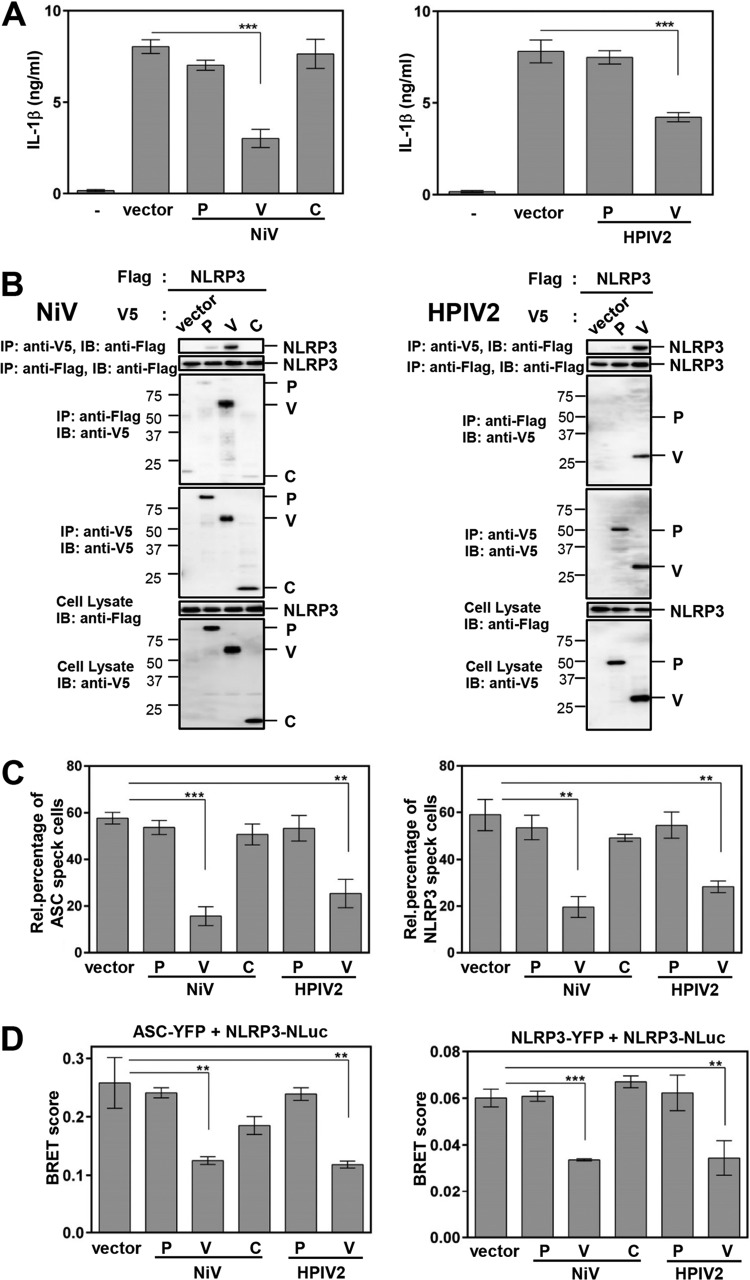
The subfamily Paramyxovirinae consists of seven distinct genera, with all members of the Paramyxovirinae subfamily carrying similar V genes. To determine whether the inhibitory activity described above is shared across genera, we examined the effect of two V proteins, that of Nipah virus (NiV; genus Henipavirus) and that of human parainfluenza virus type 2 (HPIV2; genus Rublavirus), using the same procedure as for SeV V. As shown in Fig. 7A, NiV V and HPIV2 V proteins exhibited similar inhibitory activities against NLRP3-dependent IL-1β secretion. Coimmunoprecipitation experiments and immunoblot (IB) analysis showed that both V proteins interacted with NLRP3 (Fig. 7B). Microscopy analysis showed that both V proteins inhibited NLRP3-dependent ASC oligomerization and NLRP3 self-oligomerization (Fig. 7C). Furthermore, BRET experiments showed that both V proteins inhibited intermolecular interactions between NLRP3 and ASC, as well as NLRP3 and NLRP3 (Fig. 7D). These results revealed that anti-NLRP3 inflammasome activity and its underlying mechanism have been conserved during evolution.
FIG 7.
Inhibition of NLRP3 inflammasome activation is shared by paramyxovirus V proteins. HEK293T cells were transfected with various combinations of the indicated plasmids. (A) The concentration of IL-1β in the culture supernatants was determined as described for Fig. 1C. (B) IP and IB were performed as described for Fig. 3A. (C) The effect of V protein of HPIV2 or NiV on ASC speck formation (left) or NLRP3 speck formation (right) was evaluated as described for Fig. 5B and 6B. (D) The effect of V protein of HPIV2 and NiV on intermolecular interactions between NLRP3 and ASC (right) or NLRP3 and NLRP3 (left) was evaluated as described for Fig. 5C and 6C. SD are shown as error bars. **, P < 0.01, and ***, P < 0.001, versus transfection with empty vector.
DISCUSSION
A fundamental component of the innate immune response of the host to viral infection is the production and secretion of the proinflammatory cytokine IL-1β (14). The activation of IL-1β is regulated by two steps (10): (i) the production of IL-1β as an inactive precursor induced via RLRs or TLRs and (ii) the secretion of IL-1β regulated by the NLRP3 inflammasome complex (18, 25), in which NLRP3 together with ASC promotes the cleavage of pro-Casp-1, which regulates IL-1β maturation. In the present study, a novel mechanism by which SeV V protein suppresses NLRP3 inflammasome activation to facilitate IL-1β maturation was revealed.
SeV V(−) activates the production and secretion of IL-1β in infected THP1 macrophages. This is consistent with previous reports that measles virus (MeV) V(−) activates the production and secretion of IL-1β more than wild-type MeV does (27) and that influenza virus NS1 protein also targets both NF-κB and NLRP3, thus abrogating inflammasome activation (32). As SeV V protein is known as an antagonist of the RIG-I-dependent pathway, similar to MeV V and influenza virus NS1 (33, 34), SeV V(−) thus likely induces production of pro-IL-1β through the RIG-I-NF-κB pathway. However, the modest increase (approximately 2.5-fold) in pro-IL-1β production in SeV V(−)-infected cells alone probably does not explain the marked increase (approximately 7-fold) in IL-1β secretion. Deficiency of NLRP3 in THP1 macrophages leads to downregulation of SeV V(−)-induced IL-1β, demonstrating that NLRP3 is required for SeV V(−)-induced IL-1β secretion. SeV V(−) facilitates NLRP3 speck formation, indicative of inflammasome complex formation (25), as well as ASC oligomerization, a direct indicator of inflammasome activation (26), suggesting that Casp-1 is required for SeV V(−)-induced IL-1β secretion. Therefore, our findings demonstrate that SeV V(−) infection triggers IL-1β secretion by activating the NLRP3 inflammasome.
To reveal the molecular mechanism by which SeV V protein inhibits the inflammasome to facilitate IL-1β maturation, the effects of SeV V protein on NLRP3 inflammasome activation and assembly were analyzed. Overexpression of V protein inhibited NLRP3 inflammasome-dependent IL-1β secretion, demonstrating that V protein targets the NLRP3 inflammasome signaling pathway without requiring other viral proteins. Moreover, SeV V protein inhibited assembly of the NLRP3 inflammasome, including NLRP3-dependent ASC oligomerization, NLRP3-ASC association, NLRP3 self-oligomerization, and intermolecular interactions between NLRP3 molecules. Furthermore, a high correlation between the NLRP3-binding capacity of V protein and the ability to block the inflammasome complex assembly was observed. Therefore, SeV V protein likely inhibited NLRP3 self-oligomerization by interacting with NLRP3 and inhibiting subsequent recruitment of ASC to block NLRP3-dependent ASC oligomerization, in turn blocking full activation of the NLRP3 inflammasome and subsequent IL-1β secretion. Similarly, a previous study indicated that MeV V protein inhibits NLRP3 inflammasome-dependent IL-1β secretion (27). However, that study did not elucidate how V protein inhibits activation of the NLRP3 inflammasome. In this study, we characterized the detailed mechanisms underlying the inhibition of inflammasome activation by paramyxoviruses. Moreover, although the NS1 protein of influenza virus has been previously reported to interact with NLRP3 and to inhibit NLRP3-dependent ASC oligomerization leading to IL-1β secretion (35), we have further demonstrated that SeV V protein disrupts NLRP3-NLRP3 intermolecular interactions to block NLRP3 self-oligomerization and the subsequent recruitment of ASC required for NLRP3-dependent ASC oligomerization.
The V protein is synthesized from an additional mRNA, which is generated from the P gene by insertion of a pseudotemplated G residue at the specific editing site in the middle of the gene. Consequently, P and V proteins share the same 317 residues at the amino terminus (Vn region), whereas the V protein has a unique 67-residue carbonyl terminus (Vc region). We found that V in particular is required for regulation of the NLRP3 inflammasome. SeV V targets RLRs (MDA5 and RIG-I) to inhibit RLRs signaling. Thus, we revealed a novel function of V in regulation of the NLRP3 inflammasome. We showed that V protein inhibits NLRP3 inflammasome activation in a Vc-independent and C362- and C365-independent manner, in agreement with its NLRP3 binding ability.
SeV V protein comprises Vn and Vc domains and has several interacting partners in host cells. Vc interacts with RIG-I, MDA5, and LGP2, whereas the entire V protein interacts with IRF3 and NLRP3. These partner proteins are all involved in antiviral responses, although V protein interacts with these proteins differently, as demonstrated by the V protein mutant C362R C365S. Specifically, based on the crystal structure of parainfluenza virus 5 V protein (36), it was predicted that SeV Vc forms two loops that coordinate two zinc ions via one histidine and seven cysteine residues (37). The first loop likely coordinates zinc via H318, C337, C362, and C365 and the second loop by the central cysteines (C341, C353, C355, and C358). The substitution of C362 and C365 in SeV V protein has been reported to disrupt its interaction with MDA5 (29), suggesting that the first loop is important for the interaction of SeV V protein with MDA5. In contrast, our results showed that mutations at C362 and C365 do not affect the interaction of V with NLRP3, suggesting that the intact second loop of Vc is sufficient for its interaction with NLRP3. Alternatively, a previous study suggested that MeV V protein interacts with NLRP3 through its carboxyl-terminal domain (Vc) and that the intact first loop of Vc is sufficient for its interaction with NLRP3 (27). However, we could not confirm the inhibition of IL-1β secretion by MeV (Edmonton strain) Vc protein and the interaction of MeV (Edmonton strain) Vc protein with NLRP3 (data not shown). The differences in the V protein between the IC-B strain and Edmonton strain remain unclear. Because we could not observe the inhibition of IL-1β secretion by the interaction of MeV (Edmonton strain) Vc protein with NLRP3, it would be of great interest to determine the functional regions driving the V-NLRP3 interactions seen in these paramyxoviruses, including those involving the MeV V protein.
Several viruses have evolved their own strategies to inhibit inflammasome assembly by preventing its oligomerization (16, 17). Additionally, viral proteins can prevent the recruitment of ASC molecules, thus inhibiting the innate immune response and promoting a better environment for viral replication (26). Some viral proteins have been reported to inhibit inflammasome activation by interacting with inflammasome members and thus interfering with inflammasome formation. For example, Kaposi's sarcoma-associated herpesvirus Orf63, a viral homolog of human NLRP1, inhibits inflammasome activation by interacting with NLRP1 (26). In addition, the PYD-containing proteins, Shope fibroma virus PYD-only protein and M13L protein from myxoma virus, a rabbit-specific poxvirus, interact with ASC by PYD-PYD interactions and inhibit inflammasome activation (38, 39). These studies demonstrate that viral proteins contain regions homologous with either NLRP1 or ASC to inhibit inflammasome complex formation. However, the results of a Basic Local Alignment Search Tool for proteins (BLAST) analysis and ClusalW2 alignment did not provide evidence to support the notion that SeV V protein contains regions homologous with NLRP3. Therefore, paramyxovirus likely inhibits inflammasome activation by interacting with NLRP3 independently of homologous conserved sequences.
A critical question raised by these findings is whether the IL-1β antagonism that decreases production and secretion of IL-1β by SeV V contributes to its pathogenicity. The induction of IL-1β by inflammasome activation is required for immune defense of the host against pathogens, including influenza virus. Moreover, the IL-1β response to viroporin SH gene knockout respiratory syncytial virus (RSVΔSH) is greater than that to wild-type RSV, despite a decreased viral load, whereas when IL-1β is blocked in vivo, the viral load returns to wild-type levels (40). Therefore, antagonism of the inflammasome, which decreases IL-1β secretion, is likely involved in viral pathogenicity. This attenuation manifests in vivo but not in vitro, whereas the IL-1β increase manifests even in vitro, similar to that occurring upon SeV V(−) infection. SeV evades the RLR signaling pathway, leading to induction of interferon beta (IFN-β) and probably pro-IL-1β, which is important for host antiviral innate immunity, through interaction of V with RIG-I (3, 41). Indeed, SeV V(−) induced pro-IL-1β mRNA and protein production in the present study. However, SeV V(−) still has functional C proteins to counteract the IFN response; thus, innate immunity other than interferon is thought to be important for early clearance of SeV V(−).
Here we demonstrate that SeV V suppresses IL-1β secretion by preventing NLRP3 inflammasome formation and thus reveal a novel function of V in the regulation of host innate immunity. Consequently, SeV V as an inhibitor of NLRP3 inflammasome-dependent IL-1β secretion might play important roles in SeV infection and associated pathogenicity.
MATERIALS AND METHODS
Plasmid construction.
The cDNA fragments for expressing the P, V, and C proteins of SeV (Z strain), MeV (Edmonton strain), and NiV and HPIV2 (Toshiba strain) with either an N-terminal Flag or V5-His (VH) tag were cloned into the multicloning site of pCA7 (42). The cDNA fragments for mutants of SeV V protein (Vn [aa 1 to 317], Vc [aa 318 to 384], and Vcys [C362S and C365R]), each with either a C-terminal Flag or VH tag, were newly created by PCR using appropriate PCR primers and SeV V cDNA and introduced into the multicloning site of pCA7. The cDNA for human NLRP3 was purchased from the National Institute of Technology and Evaluation, Biological Resource Center, Tokyo, Japan. The plasmids encoding human ASC, pCas-1, and pro-IL-1β were gifts from T. Ichinohe (University of Tokyo, Tokyo, Japan) (27). These cDNA fragments containing an open reading frame with an N- or C-terminal Flag or VH tag were newly introduced into pCA7. The cDNA fragments for NLRP3 or ASC with N- or C-terminal yellow fluorescent protein (YFP), or NanoLuc-luciferase (NLuc) tag, were newly cloned into pCA7. The cDNA fragments for the mutant of NLRP3 [Δ2–21(aa 22–1036)], each with C-terminal green fluorescent protein (GFP), were newly cloned into pCA7. pEBMulti-Puro-Flag-VSeV was created by inserting Flag-tagged VSeV from pCA7-Flag-VSeV (7) into the expression vector pEBMulti-Puro (Wako Pure Chemical, Osaka, Japan). The sequence fidelity of all newly created plasmids was confirmed by sequence analysis.
Cells and viruses.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (FBS). THP1-Null (THP1) and NLRP3-deficient THP1 (THP1-defNLRP3) cells were purchased from InvivoGen (San Diego, CA), and THP1 cells were cultured in RPMI 1640 medium containing 10% FBS according to the manufacturer's instructions. THP1-Null cells stably expressing the SeV V protein (THP1/VSeV) or the empty vector (THP1/vector) were isolated from puromycin-resistant colonies after transfection with pEBMulti-Puro-Flag-VSeV or pEBMulti-Puro, respectively. For macrophage differentiation, THP1-Null and THP1-defNLRP3 cells were treated with phorbol 12-myristate 13-acetate (PMA) (0.5 μg/ml; Sigma, St. Louis, MO) at 37°C for 6 h. SeV wt (cDNA derived from Z strain) and SeV V(−) were propagated in Vero cells in the presence of 3 μg/ml of trypsin (4).
Real-time PCR.
Total RNA was isolated from THP1 macrophages using Nucleospin RNA Plus (TaKaRa, Shiga, Japan), and cDNA was synthesized by reverse transcriptase (Superscript III; Invitrogen, Carlsbad, CA). The level of mRNA expression was determined using TB Green Premix EX Taq, ROX plus (TaKaRa). with a StepOne real-time PCR system (Applied Biosystems, Foster City, CA). Real-time PCR primers used in this study were as previously described (43).
Quantitation of IL-1β and pro-IL-1β.
Cell-free supernatants were collected at 24 h postinfection, at 24 h following transfection with poly(dA·dT) (InvivoGen), or at 24 h after stimulation with LPS plus ATP. IL-1β was quantified using an enzyme-linked immunosorbent assay (ELISA) kit from eBiosciences (number 88-7010; San Diego, CA). To quantify intracellular pIL-1β levels, cells were lysed by repeated cycles of freezing and thawing in phosphate-buffered saline (PBS) containing 2% FBS. The lysates were then analyzed by the same ELISA kit.
Reconstitution of the NLRP3 inflammasome.
Reconstitution was performed according to the procedure of Komune et al. (27). Briefly, HEK293T cells (2 × 105 cells/well) in a 24-well plate were transfected with plasmids encoding NLRP3 inflammasome signaling molecules (NLRP3 [10 ng/well], ASC [2.5 ng/well], pro-Casp-1 [2.5 ng/well], and pro-IL-1β [50 ng/well]), together with the plasmids encoding the viral proteins P, V, C, Vn, Vc, and Vcys (500 ng/well). At 24 h posttransfection, the levels of human IL-1β in the cell-free supernatants were determined by ELISA.
Immunoprecipitation (IP).
HEK293T cells (∼5 × 105 cells/well) in a 6-well plate were transfected with various combinations of plasmids (2 μg/well each) by using polyethyleneimine (PEI) MAX (number 24765; Polysciences Inc., Warrington, PA). After incubation for an appropriate duration, cells were lysed in 400 μl of a lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100) containing protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). Cell lysates were then incubated with anti-Flag antibody or anti-V5 mouse monoclonal antibody (MAb)-coated magnetic beads (MBL, Aichi, Japan) at 4°C for 1 h. Beads were washed five times with the lysis buffer and boiled with Laemmli sample buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10% glycerol, and 5% 2-mercaptoethanol). The eluted proteins were subjected to immunoblot (IB) analysis.
IB analysis.
Samples were resolved by SDS-polyacrylamide (10 to 20%) gel electrophoresis and then electroblotted onto a membrane filter (Immobilon-P; Millipore, Bedford, MA). The membrane was blocked in Blocking One (Nacalai Tesque) for 30 min, followed by incubation at room temperature for another hour with a mouse monoclonal antibody to Flag (MBL), V5 (MBL), His (MBL), NLRP3 (AG-20B-014-C100; AdipoGen, San Diego, CA), ASC (sc-22514-R; Santa Cruz Biotechnology, Dallas, TX), cleaved Casp-1 (number 4199; Cell Signaling Technology, Danvers, MA), or actin (number 4967; Cell Signaling Technology). The membrane was then incubated at room temperature for 30 min with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (GE Healthcare Bio-Science, Little Chalfont, England). Immunoreactive bands were visualized using enhanced chemiluminescence Western Lightning Ultra substrate (PerkinElmer, Waltham, MA) and an LAS4000 imaging system (GE Healthcare).
Immunofluorescence microscopy analysis.
To observe the effects of SeV infection on NLRP3 inflammasome complex formation, THP1 differentiated macrophages were cultured or infected with SeV wt or SeV V(−). At 20 h after infection, cells were fixed and permeabilized with PBS containing 2.5% formaldehyde for 20 min and 0.5% Triton X-100 for 15 min at room temperature. The cells were incubated with 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature and stained with primary antibodies (rabbit monoclonal anti-NLRP3 antibody [AG-20B-014-C100; AdipoGen]) and rabbit monoclonal anti-ASC antibody (sc-22514-R; Santa Cruz), followed by incubation with secondary antibodies [Alexa Fluor 488-conjugated goat polyclonal anti-mouse IgG(H+L) (number 11001; Thermo Fisher, Waltham, MA) and Alexa Fluor 568-conjugated goat polyclonal anti-rabbit IgG(H+L) (number 11011; Thermo Fisher)] for 1 h at room temperature. The cells were washed with 0.01% BSA in PBS between each step. The slide was mounted with SlowFade Diamond antifade mountant (S36967; Thermo Fisher). Images were visualized using a confocal microscope (Olympus Fluoview, Tokyo, Japan).
To observe the effects of wild-type and mutant V protein on the formation of NLRP3-dependent ASC or NLRP3 specks, HEK293T cells in an 8-well microscope glass slide with a removable 8-well chamber (SCS-N28; Matsunami-glass, Osaka, Japan) were transfected with plasmids encoding NLRP3-GFP WT(aa 1–1036) (50 ng/well) plus ASC-Flag (10 ng/well) or NLRP3-GFP (60 ng/well), together with plasmids encoding the viral protein V, Vn, Vc, or Vcys (200 ng/well) using PEI MAX. The mutant NLRP3-GFP Δ2–21(aa 22–1036) (75 ng/well) was used as a positive control to show loss of NLRP3-dependent ASC speck formation and NLRP3 speck formation. At 24 h posttransfection, cells were fixed and permeabilized with PBS containing 4% formaldehyde for 5 min and 0.2% Triton X-100 for 10 min at room temperature. The cells were incubated in 3% BSA in PBS for 1 h at room temperature and stained with anti-Flag tag–Alexa Fluor 594 (M185-A59; MBL), or anti-His tag MAb–Alexa Fluor 647 (D291-A64; MBL) for 1 h at room temperature. The cells were washed with PBS between each step. The cells were counterstained and mounted with 4′,6′-diamidino-2-phenylindole (DAPI) Fluoromount-G (number 0100-20; Southern Biotech, Birmingham, AL). Images were visualized by using a confocal microscope (Olympus Fluoview). Where applicable, the ASC speck-positive cells or NLRP3 speck-positive cells and SeV V (VSeV)-positive cells were counted from five randomly selected fields per condition, and the percentage of ASC speck- and VSeV-positive cells among VSeV-positive cells was calculated.
BRET measurements.
HEK293T cells (1.5 × 105 cells/well) in a 24-well plate were transfected with plasmids encoding the bioluminescence resonance energy transfer (BRET) donor and acceptor NLRP3-NLuc (25 ng/well) and ASC-YFP (75 ng/well) or NLRP3-Nluc (25 ng/well) plus NLRP3-YFP (75 ng/well), together with the plasmids encoding the viral proteins V, Vn, Vc, and Vcys (200 ng/well). Signals were detected by using a fusion universal microplate analyzer (Packard, Salt Lake City, UT) with two filter settings: an NLuc filter (485 ± 20 nm) and a YFP filter (530 ± 25 nm). The BRET ratio was defined as the difference between the emission at 530 nm/485 nm of cotransfected NLuc and YFP fusion proteins and the emission at 530 nm/485 nm of NLuc fusion protein alone, and the ratios were expressed as the BRET score. The amounts of YFP- and NLuc-tagged proteins were determined by reading separate plates at 530 nm after excitation at 485 nm in the presence of coelenterazine-H.
Statistical analysis.
All graphs were generated with GraphPad Prism software (GraphPad Software, Inc., San Diego, CA). Data are presented as means ± standard deviations (SD), and P values were calculated using an unpaired Student t test with two-tailed analysis. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We thank T. Ichinohe (Tokyo) for providing invaluable plasmids, K. Takeuchi (Fukui) for advice about recombinant SeV wt and SeV V(−), and K. Takahashi (Aichi) for her excellent technical assistance.
This work was supported by a KAKENHI Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (C17K08866).
REFERENCES
- 1.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J 16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiyotani K, Sakaguchi T, Kato A, Nagai Y, Yoshida T. 2007. Paramyxovirus Sendai virus V protein counteracts innate virus clearance through IRF-3 activation, but not via interferon, in mice. Virology 359:82–91. doi: 10.1016/j.virol.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 3.Irie T, Kiyotani K, Igarashi T, Yoshida A, Sakaguchi T. 2012. Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein. J Virol 86:7136–7145. doi: 10.1128/JVI.06705-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komatsu T, Takeuchi K, Yokoo J, Gotoh B. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-β production. Virology 325:137–148. doi: 10.1016/j.virol.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu T, Takeuchi K, Gotoh B. 2007. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes Infect 9:954–962. doi: 10.1016/j.micinf.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J Virol 82:10102–10110. doi: 10.1128/JVI.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitagawa Y, Yamaguchi M, Zhou M, Komatsu T, Nishio M, Sugiyama T, Takeuchi K, Itoh M, Gotoh B. 2011. A tryptophan-rich motif in the human parainfluenza virus type 2 V protein is critical for the blockade of Toll-like receptor 7 (TLR7)- and TLR9-dependent signaling. J Virol 85:4606–4611. doi: 10.1128/JVI.02012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitagawa Y, Yamaguchi M, Zhou M, Nishio M, Itoh M, Gotoh B. 2013. Human parainfluenza virus type 2 V protein inhibits TRAF6-mediated ubiquitination of IRF7 to prevent TLR7- and TLR9-dependent interferon induction. J Virol 87:7966–7976. doi: 10.1128/JVI.03525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi M, Kitagawa Y, Zhou M, Itoh M, Gotoh B. 2014. An anti-interferon activity shared by paramyxovirus C proteins: inhibition of Toll-like receptor 7/9-dependent alpha interferon induction. FEBS Lett 588:28–34. doi: 10.1016/j.febslet.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 11.Rietdijk ST, Burwell T, Bertin J, Coyle AJ. 2008. Sensing intracellular pathogens-NOD-like receptors. Curr Opin Pharmacol 8:261–266. doi: 10.1016/j.coph.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Lamkanfi M, Kanneganti TD. 2010. Nlrp3: an immune sensor of cellular stress and infection. Int J Biochem Cell Biol 42:792–795. doi: 10.1016/j.biocel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornung V, Latz E. 2010. Intracellular DNA recognition. Nat Rev Immunol 10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 14.Shrivastava G, León-Juárez M, García-Cordero J, Meza-Sánchez DE, Cedillo-Barrón L. 2016. Inflammasomes and its importance in viral infections. Immunol Res 64:1101–1117. doi: 10.1007/s12026-016-8873-z. [DOI] [PubMed] [Google Scholar]
- 15.Rathinam VAK, Fitzgerald KA. 2010. Inflammasomes and anti-viral immunity. J Clin Immunol 30:632–637. doi: 10.1007/s10875-010-9431-4. [DOI] [PubMed] [Google Scholar]
- 16.Gregory SM, Damania B. 2011. Inhibition of the inflammasome response by a viral protein that interacts with NLRs. Commun Integr Biol 4:416–418. doi: 10.4161/cib.15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen I-Y, Ichinohe T. 2015. Response of host inflammasomes to viral infection. Trends Microbiol 23:55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, Wu Y, Wang J. 2017. Activation and role of NACHT, LRR, and PYD domains-containing protein 3 inflammasome in RNA viral infection. Front Immunol 8:1420. doi: 10.3389/fimmu.2017.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Núñez G. 2006. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem 281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 20.Park S, Juliana C, Hong S, Datta P, Hwang I, Fernandes-Alnemri T, Yu J-W, Alnemri ES. 2013. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J Immunol 191:4358–4366. doi: 10.4049/jimmunol.1301170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. 1982. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res 42:1530–1536. [PubMed] [Google Scholar]
- 22.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. 2007. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res 56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 23.Lerner AG, Upton JP, Praveen PVK, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, Heintz N, Greengard P, Hui S, Tang Q, Trusina A, Oakes SA, Papa FR. 2012. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossol M, Pierer M, Raulien N, Quandt D, Meusch U, Rothe K, Schubert K, Schöneberg T, Schaefer M, Krügel U, Smajilovic S, Bräuner-Osborne H, Baerwald C, Wagner U. 2012. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nat Commun 3:1329. doi: 10.1038/ncomms2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinon F, Mayor A, Tschopp J. 2009. The inflammasomes: guardians of the body. Annu Rev Immunol 27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Mao L, Meng G. 2013. The NLRP3 inflammasome activation in human or mouse cells, sensitivity causes puzzle. Protein Cell 4:565–568. doi: 10.1007/s13238-013-3905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komune N, Ichinohe T, Ito M, Yanagi Y. 2011. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1β secretion. J Virol 85:13019–13026. doi: 10.1128/JVI.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. 2013. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakaguchi T, Irie T, Kuwayama M, Ueno T, Yoshida A, Kawabata R. 2011. Analysis of interaction of Sendai virus V protein and melanoma differentiation-associated gene 5. Microbiol Immunol 55:760–767. doi: 10.1111/j.1348-0421.2011.00379.x. [DOI] [PubMed] [Google Scholar]
- 30.Compan V, Martin-Sanchez F, Baroja-Mazo A, Lopez-Castejon G, Gomez AI, Verkhratsky A, Brough D, Pelegrin P. 2015. Apoptosis-associated speck-like protein containing a CARD forms specks but does not activate caspase-1 in the absence of NLRP3 during macrophage swelling. J Immunol 194:1261–1273. doi: 10.4049/jimmunol.1301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Compan V, Baroja-Mazo A, López-Castejón G, Gomez AI, Martínez CM, Angosto D, Montero MT, Herranz AS, Bazán E, Reimers D, Mulero V, Pelegrín P. 2012. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37:487–500. doi: 10.1016/j.immuni.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Cheong W-C, Kang H-R, Yoon H, Kang S-J, Ting JP-Y, Song MJ. 2015. Influenza A virus NS1 protein inhibits the NLRP3 inflammasome. PLoS One 10:e0126456. doi: 10.1371/journal.pone.0126456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Aparicio MT, Feinman LJ, García-Sastre A, Shaw ML. 2018. Paramyxovirus V proteins interact with the RIG-I/TRIM25 regulatory complex and inhibit RIG-I signaling. J Virol JVI.01960-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, García-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriyama M, Chen I-Y, Kawaguchi A, Koshiba T, Nagata K, Takeyama H, Hasegawa H, Ichinohe T. 2016. The RNA- and TRIM25-binding domains of influenza virus NS1 protein are essential for suppression of NLRP3 inflammasome-mediated IL-1β secretion. J Virol 90:4105–4114. doi: 10.1128/JVI.00120-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 37.Ramachandran A, Horvath CM. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J Virol 84:11152–11163. doi: 10.1128/JVI.01375-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricuttio D, Wang G, McFadden G. 2005. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity 23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Dorfleutner A, Talbott SJ, Bryan NB, Funya KN, Rellick SL, Reed JC, Shi X, Rojanasakul Y, Flynn DC, Stehlik C. 2007. A Shope fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes 35:685–694. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell RF, McDonald JU, Ivanova M, Zhong Z, Bukreyev A, Tregoning JS. 2015. The partial attenuation of small hydrophobic (SH) gene deleted RSV is associated with elevated IL-1β responses. J Virol 89:8974–8981. doi: 10.1128/JVI.01070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez-Aparicio MT, Feinman LJ, García-Sastre A, Shaw ML. 2018. Paramyxovirus V proteins interact with the RIG-I/TRIM25 regulatory complex and inhibit RIG-I signaling. J Virol 92:e01960-17. doi: 10.1128/JVI.01960-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitagawa Y, Zhou M, Yamaguchi M, Komatsu T, Takeuchi K, Itoh M, Gotoh B. 2010. Human metapneumovirus M2-2 protein inhibits viral transcription and replication. Microbes Infect 12:135–145. doi: 10.1016/j.micinf.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Li G, De W, Luo Z, Pan P, Tian M, Wang Y, Xiao F, Li A, Wu K, Liu X, Rao L, Liu F, Liu Y, Wu J. 2018. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1β secretion. Nat Commun 9:106. doi: 10.1038/s41467-017-02645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]