One of the key advantages that distinguish lentiviruses, such as HIV-1, from all other retroviruses is its ability to infect nondividing cells. Interaction of the HIV-1 capsid with Nup153 and CPSF6 is important for nuclear entry and integration; however, the contribution of each of these proteins to nuclear import and integration is not clear. Using genetics, we demonstrated that these proteins contribute to different processes: Nup153 is essential for the HIV-1 nuclear import in nondividing cells, and CPSF6 is important for HIV-1 integration. In addition, nuclear factors such as CPSF6 and the state of the chromatin are known to be important for integration site selection; nevertheless, the preferential determinant influencing integration site selection is not known. This work demonstrates that integration site selection is a multiparameter process that depends upon nuclear factors and the state of the cellular chromatin.
KEYWORDS: HIV-1, Nup153, CPSF6, nondividing cells, capsid binding, nuclear import, integration, HIV integration, HIV nuclear import, NPC
ABSTRACT
Human immunodeficiency virus type 1 (HIV-1) displays the unique ability to infect nondividing cells. The capsid of HIV-1 is the viral determinant for viral nuclear import. To understand the cellular factors involved in the ability of HIV-1 to infect nondividing cells, we sought to find capsid mutations that allow the virus to infect dividing but not nondividing cells. Because the interaction of capsid with the nucleoporin protein 153 (Nup153) is important for nuclear import of HIV-1, we solved new crystal structures of hexameric HIV-1 capsid in complex with a Nup153-derived peptide containing a phenylalanine-glycine repeat (FG repeat), which we used to guide structure-based mutagenesis of the capsid-binding interface. HIV-1 viruses with mutations in these capsid residues were tested for their ability to infect dividing and nondividing cells. HIV-1 viruses with capsid N57 substitutions infected dividing but not nondividing cells. Interestingly, HIV-1 viruses with N57 mutations underwent reverse transcription but not nuclear translocation. The mutant capsids also lost the ability to interact with Nup153 and CPSF6. The use of small molecules PF74 and BI-2 prevented the interaction of FG-containing nucleoporins (Nups), such as Nup153, with the HIV-1 core. Analysis of integration sites in HIV-1 viruses with N57 mutations revealed diminished integration into transcriptionally active genes in a manner resembling that of HIV-1 in CPSF6 knockout cells or that of HIV-1-N74D. The integration pattern of the N57 mutant HIV-1 can be explained by loss of capsid interaction with CPSF6, whereas capsid interaction with Nup153 is required for HIV-1 to infect nondividing cells. Additionally, the observed viral integration profiles suggested that integration site selection is a multiparameter process that depends upon nuclear factors and the state of the cellular chromatin.
IMPORTANCE One of the key advantages that distinguish lentiviruses, such as HIV-1, from all other retroviruses is its ability to infect nondividing cells. Interaction of the HIV-1 capsid with Nup153 and CPSF6 is important for nuclear entry and integration; however, the contribution of each of these proteins to nuclear import and integration is not clear. Using genetics, we demonstrated that these proteins contribute to different processes: Nup153 is essential for the HIV-1 nuclear import in nondividing cells, and CPSF6 is important for HIV-1 integration. In addition, nuclear factors such as CPSF6 and the state of the chromatin are known to be important for integration site selection; nevertheless, the preferential determinant influencing integration site selection is not known. This work demonstrates that integration site selection is a multiparameter process that depends upon nuclear factors and the state of the cellular chromatin.
INTRODUCTION
The critical influence of the physiological state of cells on retroviral replication was initially demonstrated by experiments showing that cell division arrest with X-rays or UV light prevents Rous sarcoma virus replication (1). Subsequent research established the relationship between cell cycle stage and retroviral infection, revealing that retroviruses do not all have the same requirements for productive infection. For example, productive infection by gammaretroviruses, such as murine leukemia virus (MLV), requires host cells to pass through mitosis (2, 3). In contrast, lentiviruses such as human immunodeficiency virus type 1 (HIV-1) show no difference in productive infection of dividing versus nondividing cells (4). This suggests that lentiviruses have developed specific molecular mechanisms for the infection of nondividing cells. The ability of HIV-1 to infect nondividing cells has been attributed to its capacity to transport the preintegration complex (PIC) to the nucleus through nuclear pores (5, 6). Translocation of the HIV-1 PIC into the nucleus is not a simple process, as its size is similar to that of a ribosome, which is at least 56 nm in diameter (5, 7). Because of its large size, it is unlikely that the PIC enters the nucleus by passive diffusion (8). Instead, HIV-1 PIC translocation into the nucleus is likely to be an active process. Several components of the PIC, such as matrix, Vpr, integrase, and the central DNA flap, have been proposed to be involved in PIC nuclear translocation (9–11). Although barely detectable amounts of capsid can be found on the HIV-1 PIC (5, 12–14), recent evidence has shown that the capsid nonetheless plays a role in HIV-1 infection of nondividing cells (15–17). In agreement with this evidence, several studies suggest that the HIV-1 core associates with the nuclear pore (18, 19). While the mechanism used by HIV-1 PIC to enter the nucleus is not completely understood, it is accepted that nuclear import of the complex is an active and energy-dependent process (6).
In addition to the viral determinants involved in HIV-1 PIC nuclear import, several host factors have been implicated in the process: (i) importin 7 (20–22), (ii) importin a3 (23), (iii) importin α/importin β heterodimer (20, 24, 25), (iv) transportin-SR2/TNPO3 (26–33), (v) RanBP2/Nup358 (19, 34), and (vi) Nup98 and Nup153 (19, 32, 35–38).
Recent studies have also proposed an important role for Nup153 in HIV-1 replication (19, 32, 35, 38–40). Nup153 localizes to the nuclear side of the nuclear pore and was initially described as being necessary for HIV-1 replication by three independent genome-wide short interfering RNA (siRNA) screens (28, 32, 33). Although the C-terminal domain of Nup153 has been shown, via use of bacterially purified proteins, to bind the HIV-1 integrase protein (36), the genetic determinant for the Nup153 requirement during HIV-1 infection has been mapped to the capsid protein (38). Interestingly, HIV-1 with capsid mutations showed different infectivities as a function of Nup153 depletion (37, 38, 41). In agreement with these findings, we and others have found that Nup153 interacts directly with the capsid via phenylalanine-glycine (FG) repeats (35, 40), which directly bind to a pocket generated by the so-called N-terminal/C-terminal (NTD-CTD) interface between adjacent subunits in the capsid hexamer (42, 43). Because Nup153 depletion affects HIV-1 nuclear import, these findings suggested that Nup153 interaction with the viral core must be important for infection.
Even though CPSF6 and Nup153 have been shown to compete for the same binding site on the HIV-1 capsid hexamer (42–44), the two proteins appear to mediate distinct processes, with CPSF6 directing HIV-1 integration to transcriptionally active chromatin, whereas Nup153 is important for nuclear translocation (28, 45). It is possible that interaction of Nup153 and CPSF6 with the capsid occurs at different stages of viral replication, governing discrete events at each stage. For example, Nup153 could govern nuclear import, while CPSF6 could govern viral integration. Understanding the precise role of Nup153 and CPSF6 in HIV-1 infection of nondividing cells remains an ongoing challenge in retroviral research.
Although it is generally agreed that Nup153 has a role in HIV-1 nuclear translocation, the pathways and specific proteins on which the virus relies to infect nondividing cells have not been elucidated. In order to address the role of Nup153 in the ability of HIV-1 to infect nondividing cells, we performed structure-based mutagenesis to identify HIV-1 capsid mutations that modulate Nup153-capsid interaction. For this purpose, we solved the crystal structure of a complex between a Nup153-derived peptide containing a capsid-binding FG repeat and the hexameric HIV-1 capsid in order to identify capsid residues that are interacting with Nup153. This allowed us to identify mutant capsid HIV-1 viruses that are unable to infect nondividing cells but can infect dividing cells. The most interesting change was on capsid residue N57, which directly interacts with the FG repeat of Nup153. Infection of nondividing cells by HIV-1 viruses with these capsid mutations was found to be defective for nuclear translocation but unaffected for reverse transcription. HIV-1 capsid with these mutations lost the ability to interact with Nup153, suggesting that this interaction is essential for the HIV-1 infection of nondividing cells. Given the observed role of CPSF6 in determining HIV-1 integration sites and the fact that Nup153 and CPSF6 bind to the capsid at the same site, we also examined integration sites for N57 mutant HIV-1 in dividing cells and found these sites to closely resemble those of wild-type HIV-1 in the absence of CPSF6. The integration pattern of HIV-1 viruses with changes on N57 is explained by the loss of interaction with CPSF6, whereas interaction of the capsid with Nup153 is required for HIV-1 infection of nondividing cells. Taken together, our findings show how different capsid mutations can delineate functional interactions between different host factors that modulate HIV-1 infectivity and help unravel the complex processes that govern nuclear translocation and integration.
RESULTS
Interaction of Nup153 with HIV-1 capsid.
We and others have previously demonstrated the ability of Nup153 to interact with HIV-1 capsid (35, 40, 42). Accordingly, to identify specific residues in the hexameric HIV-1 capsid that interact with Nup153, we attempted to cocrystallize a number of FG-containing peptides derived from Nup153 with the disulfide-stabilized HIV-1 capsid hexamer. Remarkably, only one of the tested peptides was found to bind capsid in the crystals, which we interpret to mean that binding of the FG region is modulated by surrounding residues and that certain FG sequences are preferred over others. Our structures all contained a peptide derived from Nup1531407–1429 (1407TNNSPSGVFTFGANSSTPAASAQ1429), previously described to be a high-affinity motif for the HIV-1 capsid (40). We obtained two different crystal forms distinct from those reported by Price and colleagues (43). Interestingly, even though each hexamer has six potential binding sites, none of our structures showed saturation despite the excess of peptide in the crystallization mixture. Three crystallographically independent hexamers were observed, two of which had three bound peptides while the third contained only two. Our interpretation of these results is that the hexamer architecture is incompatible with saturation binding of FG repeats, perhaps because full occupancy has a destabilizing effect.
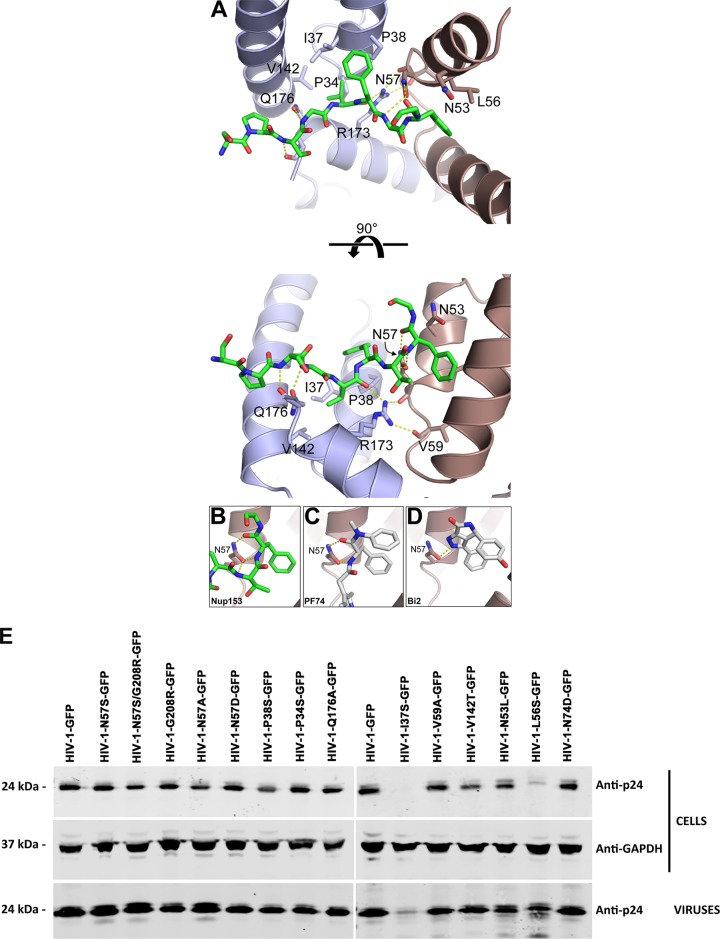
As reported previously (43), the Nup1531407–1429 peptide binds at the NTD-CTD interface, and we found that it interacts with capsid residues P34, I37, P38, N53, L56, N57, V59, V142, and Q176 (Fig. 1A and Tables 1 and 2). Notably, capsid residue N57 is buried within the binding site and makes hydrogen bonds with the FG peptide backbone. The mode of interaction is similar to that observed with HIV-1 inhibitors PF74 and BI-2 (42, 43) (Fig. 1C and D). We also attempted to cocrystallize the hexamer and the Nup1531407–1429 peptide with an N57S mutation; however, repeated screening of multiple crystals failed to reveal any binding. These experiments highlight the importance of the N57 residue for many different interactions involving the capsid. This residue is therefore likely to be an important determinant for translocation of the PIC through nuclear pores.
FIG 1.
Structure of Nup153 FG in complex with hexameric HIV-1 capsid. (A) Structure of NUP153 FG peptide (green) in its binding pocket within the hexameric HIV-1 capsid. The capsid side chains that contact the peptide are shown as sticks and labeled. N57 is binding Nup153 (B), PF74 (C) (42), and BI2 (D) (43). Hydrogen bonds are shown as dashed yellow lines. (E) Production and maturation of HIV-1 capsid mutants viruses. The indicated HIV-1 viruses were produced in human 293T cells. Producer cells and purified viruses were analyzed for HIV-1 capsid expression by Western blotting using anti-p24 antibodies. Producer cells validated for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression were used as loading controls. Experiments were performed three times, and a representative example is shown.
TABLE 1.
Infection of nondividing cells by HIV-1 variantsd
| HIV-1 variant | THP1-SAMHD1-KOa |
HT1080b |
Cf2Thb |
HeLab |
HeLa-Nup153-KOc |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dividing | Nondividing | Dividing | Nondividing | Dividing | Nondividing | Dividing | Nondividing | Dividing | Nondividing | |
| WT | + | + | + | + | + | + | + | + | − | ND |
| N57S | + | − | + | − | + | + | ND | ND | + | ND |
| N57A | + | − | + | − | + | + | ND | ND | ND | ND |
| N57D | + | − | + | − | + | + | ND | ND | ND | ND |
| G208R | + | + | + | + | + | + | ND | ND | ND | ND |
| N57S/G208R | + | − | + | − | + | + | + | − | + | ND |
| P38S | + | + | + | + | + | + | ND | ND | ND | ND |
| Q176A | + | + | + | + | + | + | ND | ND | ND | ND |
| P34S | − | − | − | − | − | − | ND | ND | ND | ND |
| I37S | − | − | − | − | − | − | ND | ND | ND | ND |
| N53L | − | − | − | − | − | − | ND | ND | ND | ND |
| L56S | − | − | − | − | − | − | ND | ND | ND | ND |
| V59A | − | − | − | − | − | − | ND | ND | ND | ND |
| V142T | − | − | − | − | − | − | ND | ND | ND | ND |
| N74D | + | + | + | + | + | + | + | + | − | ND |
PMA-treated (nondividing) or untreated (dividing) THP-1–SAMHD1-KO cells were challenged with increasing amounts of HIV-1-GFP viruses containing the indicated capsid change.
Aphidicolin-treated or untreated HT1080, Cf2Th, or HeLa cells were challenged with increasing amounts of HIV-1-GFP viruses containing the indicated capsid change.
HeLa cells transiently knocked down for expression of Nup153 were challenged with the indicated HIV-1 variants.
Infection was measured 48 hpi by determining the percentage of GFP-positive cells by flow cytometry. A plus or minus indicates infection or the absence of infection, respectively. Experiments were repeated at least three times, and typical results are shown. ND, not determined.
TABLE 2.
Crystallographic statistics
| Parameter | Value(s) fora: |
|
|---|---|---|
| R3 | P1 | |
| Diffraction data | ||
| Beamline | APS 23-ID-D | APS 23-ID-D |
| Wavelength | 1.000 | 1.000 |
| Processing program | HKL2000 | HKL2000 |
| Space group | R3 | P1 |
| Cell dimensions | ||
| a (Å) | 151.924 | 57.333 |
| b (Å) | 151.924 | 91.487 |
| c (Å) | 69.646 | 150.047 |
| Angle (°) | α = β = 90, γ = 120 | α = 89.54, β = 90.94, γ = 96.08 |
| Resolution range (Å) | 50–2.50 (2.54–2.50) | 50–1.90 (1.97–1.90) |
| Rsym/Rmeas/Rpim | 0.09 (0.46)/0.11 (0.64)/0.07 (0.45) | 0.07 (0.75)/0.09 (1.00)/0.07 (0.75) |
| Mean I/σ<I> | 10.6 (1.2) | 9.6 (1.0) |
| Completeness (%) | 75.3 (23.7) | 96.5 (92.0) |
| Avg redundancy | 2.4 (1.5) | 1.7 (1.6) |
| Wilson B factor (Å2) | 31.58 | 19.16 |
| Refinement | ||
| Refinement program | Phenix | Phenix |
| Resolution range | 47.83–2.50 (2.60–2.50) | 45.50–1.90 (1.92–1.90) |
| No. of unique reflections | 12,545 (190) | 205,470 (1,577) |
| Reflections in free set | 1,266 (23) | 10,123 (66) |
| Rwork | 0.20 (0.30) | 0.18 (0.31) |
| Rfree | 0.25 (0.36) | 0.23 (0.29) |
| NCS copies | 2 | |
| No. of nonhydrogen atoms | ||
| Protein | 3,415 | 22,652 |
| Solvent | 0 | 2,179 |
| Avg B factor (Å2) | ||
| Protein | 47.24 | 27.78 |
| Solvent | NA | 29.54 |
| Validation and deposition | ||
| Coordinate deviations | ||
| Bond lengths (Å2) | 0.002 | 0.007 |
| Bond angles (°) | 0.475 | 0.784 |
| Ramachandran plot (%) | ||
| Favored | 96.5 | 98.9 |
| Outliers | 0 | 0 |
| MolProbity clashscore | 1.03 | 1.3 |
| PDB entry | 5TSV | 5TSX |
Values in parentheses are for the highest-resolution shell.
HIV-1 viruses bearing capsid mutations that affect their ability to infect nondividing cells.
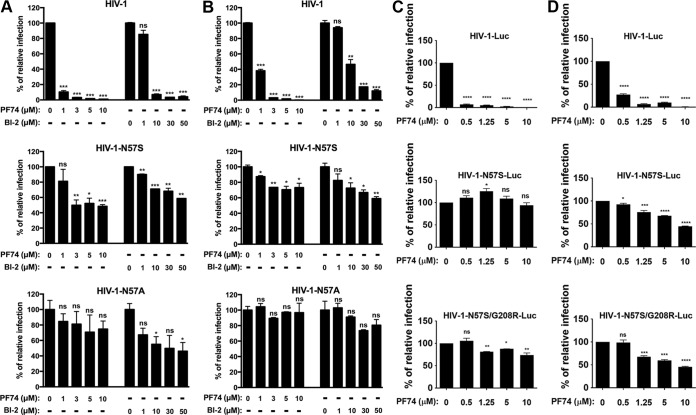
The interaction of capsid with Nups is an important early step in HIV-1 replication. All residues found to interact with Nup153 in our structural studies are highly conserved in primate lentiviruses, such as HIV-1, HIV-2, and simian immunodeficiency virus (SIV) (53). Guided by the crystal structures, we synthesized HIV-1 viruses with the following single-point mutations on each of the residues that directly contact the FG peptide: P34S, I37S, P38S, N53L, L56S, N57S, N57A, N57D, V59A, V142T, and Q176A. As shown in Table 1, we found several HIV-1 viruses containing capsid mutants that infected dividing cells: N57S, N57A, N57D, P38S, and Q176A. In contrast, HIV-1 viruses containing capsid mutants P34S, I37S, N53L, L56S, V59A, and V142T were either not released or were infectivity defective.
We further characterized these capsid mutants by analyzing expression in cells, viral production, viral maturation, and infectivity (Table 1 and Fig. 1E). All HIV-1 capsid variants studied here expressed similar levels of p55 in producer cells. Levels of p24 protein were detected in producer cells for all variants, with the exception of I37S and N53L (Fig. 1E). Analysis of semipurified viral particles revealed that HIV-1 capsid variants showed no defects on maturation, as detected by the levels of p24 in viruses, with the exception of I37S (Fig. 1E). Infectivity assay revealed that HIV-1 capsid variants N57S, N57A, N57D, P38S, and Q176A were infectious; however, HIV-1 capsid variants P34S, I37S, N53L, L56S, V59A, and V142T were not infectious (Table 1).
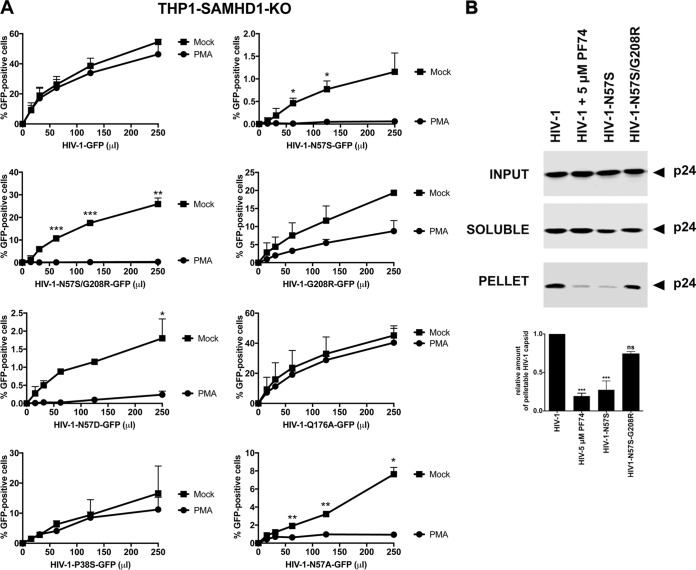
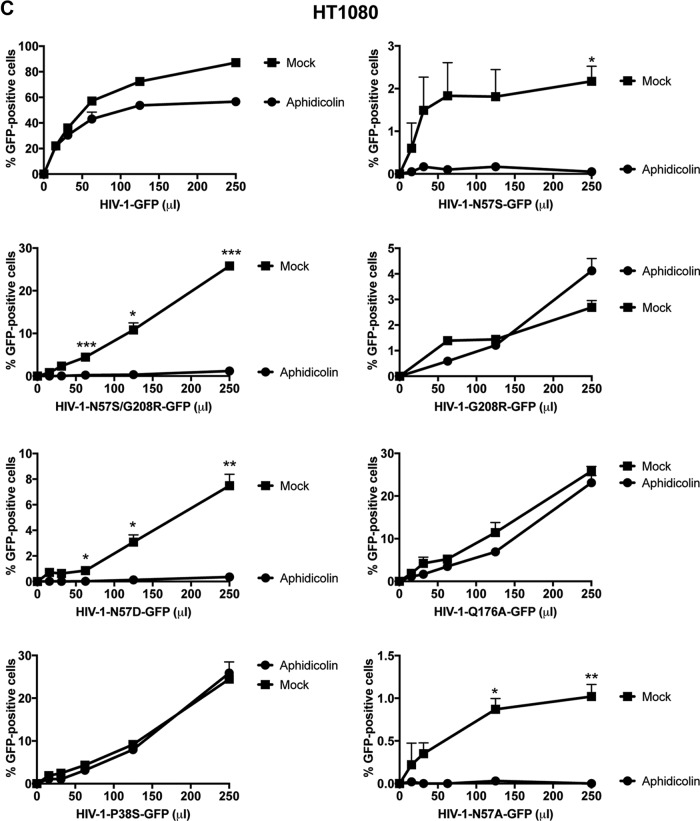
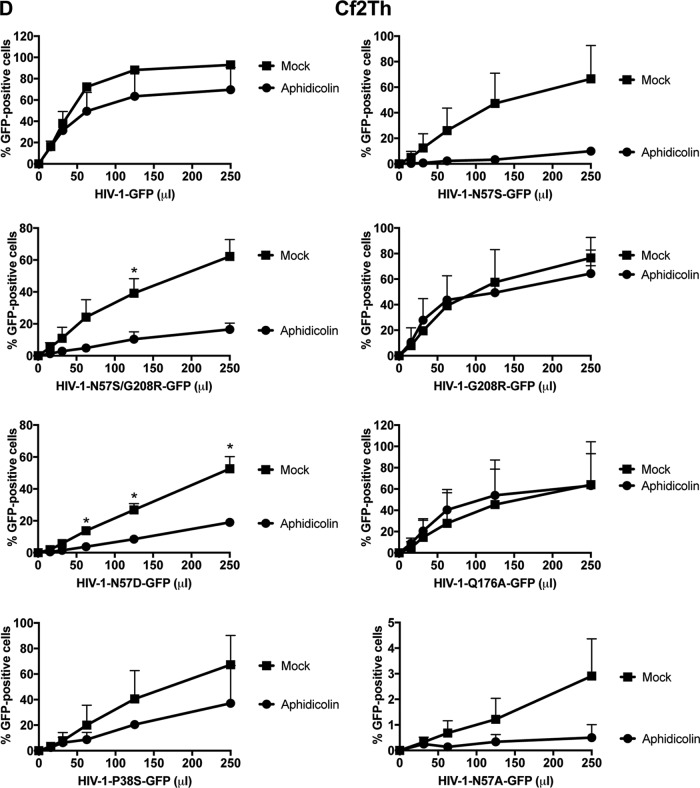
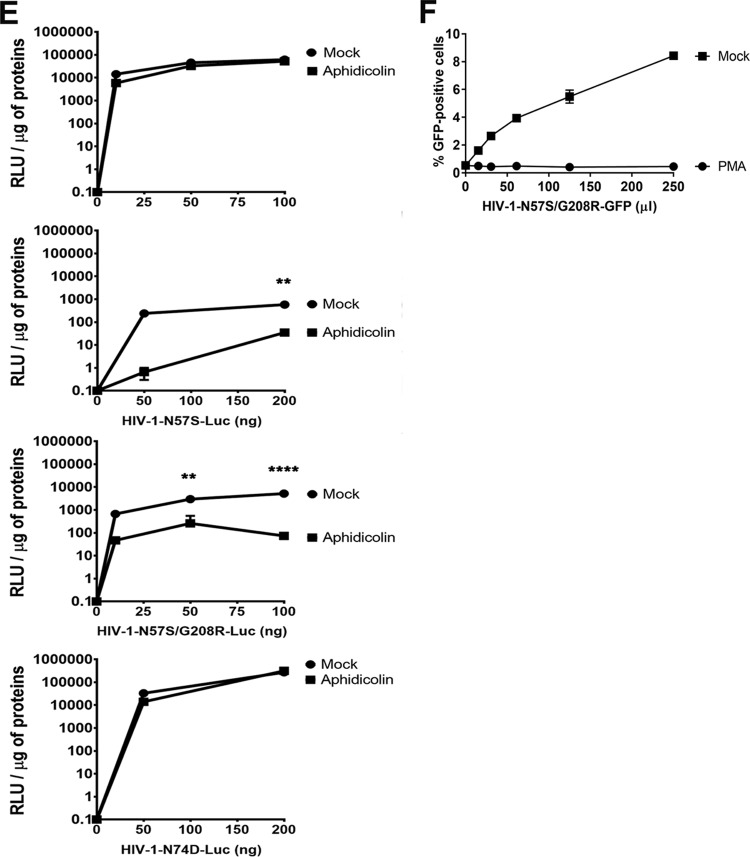
We next wanted to test whether any of these capsid mutants could abrogate the ability of HIV-1 to infect nondividing cells. For this purpose, we tested the ability of p24-normalized HIV-1 viruses with capsid mutations to infect nondividing cells. As a model to study nondividing cells, we treated THP-1 monocytes lacking the restriction factor SAMHD1 (THP-1–SAMHD1-KO) with phorbol 12-myristate 13-acetate (PMA), which induces differentiation of THP-1 cells to a nondividing macrophage-like state (54). The SAMHD1-KO eliminated a restriction factor that would otherwise inhibit viral replication by a mechanism different from the process being studied in this experiment. As shown in Fig. 2A, HIV-1-GFP viruses with capsid mutation N57S, N57A, or N57D poorly infected THP-1–SAMHD1-KO nondividing cells (PMA treated). In contrast, HIV-1 viruses with these capsid changes were able to infect dividing cells (Mock). HIV-1 viruses with mutations on capsid residue N57 resulted in decreased capsid stability, as shown by the “fate of the capsid” assay (Fig. 2B) (55). In an effort to restore this defect in capsid stability, we produced HIV-1 containing the double mutations N57S and G208R, which is known to increase the cytosolic stability of the viral core. Interestingly, HIV-1-N57S/G208R recovered core stability in the cytoplasm of infected cells (Fig. 2B), which allowed us to partially repair the stability defect of HIV-1-N57S. As expected, HIV-1-N57S/G208R viruses behave like N57S in PMA-treated THP-1–SAMHD1 KO cells (nondividing) (Fig. 2A and Table 1). We also tested the ability of p24-normalized HIV-1 with capsid mutations to infect human HT1080 and canine Cf2Th cells in the presence of the cell cycle inhibitor aphidicolin, which blocks the cell cycle at early S phase. As shown in Fig. 2C and D, HIV-1 with capsid mutations N57S, N57S/G208R, N57D, and N57A poorly infected aphidicolin-treated cells. Similar effects were observed for infection of aphidicolin-treated human HeLa cells by HIV-1-N57S and HIV-1-N57S/G208R expressing luciferase as a reporter of infection (Fig. 2E). As a control, we tested the ability of HIV-1-N74D viruses to infect nondividing cells and found that it efficiently infected nondividing cells compared to wild-type virus. The results of these experiments suggested that in contrast to N74, capsid residue N57 confers upon HIV-1 the ability to infect nondividing cells (Fig. 2E). Furthermore, the results raised the possibility that interaction of the HIV-1 capsid with Nup153 is essential for the ability of HIV-1 to infect nondividing cells.
FIG 2.
HIV-1 viruses bearing changes on the capsid residue N57 are not able to infect nondividing cells. (A) PMA-treated (PMA) or untreated (Mock) THP-1–SAMHD1 knockout (KO) cells were challenged with increasing amounts of the indicated HIV-1-GFP viruses. Infection was determined by measuring the percentage of GFP-positive cells at 48 hpi. PMA-treated THP-1–SAMHD1 KO cells are used as a model of noncycling cells. (B) HIV-1-N57S/G208R recovers core stability during infection. Canine Cf2Th cells were challenged with the indicate viruses, and core stability was assayed by performing the fate-of-the-capsid assay. Eight hpi, cells were lysed and a postnuclear fraction (INPUT) was separated into SOLUBLE and PELLET fractions by using a 50% sucrose gradient, as described in Materials and Methods. INPUT, SOLUBLE, and PELLET fractions were analyzed for capsid using anti-p24 antibodies. PELLET represents the fraction of capsid that is forming cores. Experiments were performed at least three times, and a representative example is shown. (C to E) Human HT1080 (C), dog Cf2Th (D), or human HeLa (E) cells pretreated for 12 h with 0.5 μg/ml aphidicolin were challenged with increasing amounts of the indicated HIV-1 viruses. The use of 0.5 μg/ml of aphidicolin for 12 h stops the cell cycle at S phase, as determined by analyzing DNA content per cell using propidium iodide. Infection was determined by measuring the percentage of GFP-positive cells or the luciferase activity at 48 hpi, and standard deviations for triplicates are shown. Results were analyzed using two-tailed Student's t test. Differences were considered statistically significant at a P value of <0.05 (*), <0.01 (**), or <0.001 (***) or were nonsignificant. (F) PMA-treated (PMA) or untreated (Mock) U937 cells were challenged with increasing amounts of HIV-1-N57S/G208R-GFP viruses. Infection was determined by measuring the percentage of GFP-positive cells at 48 hpi. Experiments were performed at least three times, and a representative example is shown.
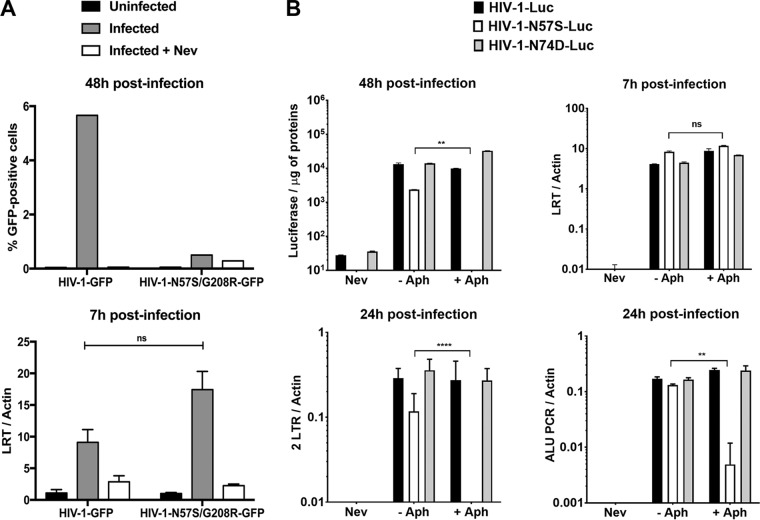
To further expand the number of cell lines used to examine our viruses, we tested the myeloid cell line U937, which does not express SAMHD1. As shown in Fig. 2F, HIV-1-N57S/G208R-GFP viruses poorly infected PMA-treated U937 (nondividing) cells compared to untreated (dividing) cells.
HIV-1 viruses with mutations on capsid residue N57 undergo reverse transcription but not nuclear translocation in nondividing cells.
In the preceding section, we showed that mutations in the capsid residue N57 inhibit infection of nondividing cells by HIV-1. We next tested whether HIV-1-N57S/G208R undergoes reverse transcription in nondividing cells. For this purpose, we challenged PMA-treated THP-1–SAMHD1 KO cells (nondividing cells) and measured the ability of HIV-1-N57S/G208R expressing GFP (HIV-1-N57S/G208R-GFP) as a reporter of infection to undergo reverse transcription at 7 h postinfection (hpi). As shown in Fig. 3A, HIV-1 with the capsid mutations N57S/G208R did not infect nondividing cells (upper) but underwent reverse transcription (lower). As a control, we used nevirapine (Nev), which inhibits HIV-1 reverse transcription. These experiments suggested that HIV-1 with mutations on capsid residue N57 is defective only after reverse transcription in nondividing cells.
FIG 3.
Infection of nondividing cells by HIV-1-N57S is stopped after reverse transcription but prior to nuclear translocation. (A) PMA-treated THP-1–SAMHD1 knockout (KO) cells were challenged with the indicated DNase-pretreated HIV-1-GFP viruses. (Upper) Infection was determined by measuring the percentage of GFP-positive cells by flow cytometry at 48 hpi. (Lower) In parallel, cells from similar infections were lysed at 7 hpi and total DNA extracted. The DNA samples collected at 7 hpi postinfection were used to determine the levels of late reverse transcripts by real-time PCR. As a control, we used 10 μM the reverse transcription inhibitor nevirapine (Nev). Late reverse transcript levels were normalized to actin. (B) Similarly, HeLa cells pretreated with 0.5 μg/ml aphidicolin for 12 h were subsequently infected by the indicated DNase-pretreated HIV-1–Luc viruses. (Upper) Infection was determined by measuring luciferase activity at 48 hpi. In parallel, cells from similar infections were lysed at 7 and 24 hpi, and total DNA was extracted. The DNA samples collected at 7 hpi were used to determine the levels of late reverse transcripts by real-time PCR. (Lower) HIV-1 2-LTR circles, a marker for nuclear import, were quantified by real-time PCR of DNA samples collected at 24 hpi. In addition, integration was measured by Alu-PCR in DNA samples collected at 24 hpi. The levels of late reverse transcripts, 2-LTR circle, and products of Alu-PCR were normalized to actin. Nevirapine was used as a control. Experiments were repeated at least three times, and a representative example is shown. Results were analyzed using two-tailed Student's t test. Differences were considered statistically significant at a P value of <0.05 (*), <0.01 (**), <0.001 (***), or <0.0001 (****) or were nonsignificant (ns).
To further investigate the defect imposed by N57 mutations on HIV-1 replication, we monitored reverse transcription and formation of two long terminal repeat (2-LTR) circles in aphidicolin-treated cells at 7 and 24 hpi, respectively. Aphidicolin-treated human HeLa cells (nondividing) were challenged with luciferase-expressing HIV-1-N57S-Luc, and infection (at 48 hpi), reverse transcription (at 7 hpi), formation of 2-LTR circles (at 24 hpi), and integration by Alu-PCR (at 24 hpi) were measured. In agreement with our previous results, HIV-1-N57S-Luc viruses did not infect aphidicolin-treated cells but underwent normal reverse transcription (Fig. 3B, upper). Interestingly, HIV-1-N57S-Luc viruses were defective for the formation of 2-LTR circles and integration, suggesting a defect in nuclear translocation (Fig. 3B, lower). As controls, we monitored infection, reverse transcription, formation of 2-LTR circles, and integration by HIV-1-N74D-Luc viruses, which are not affected in their ability to infect aphidicolin-treated cells (nondividing) (Fig. 3B, lower). Taken together, these experiments demonstrated that HIV-1 viruses containing mutations on capsid residue N57 caused a defect after reverse transcription but before nuclear translocation in nondividing cells. Interestingly, a less than 2-fold increase in reverse transcription was observed for HIV-1 viruses bearing N57 changes. Although this increase on reverse transcription was highly reproducible, this phenotype remains to be fully investigated.
HIV-1 capsids bearing mutations on residue N57 lose the ability to interact with Nup153.
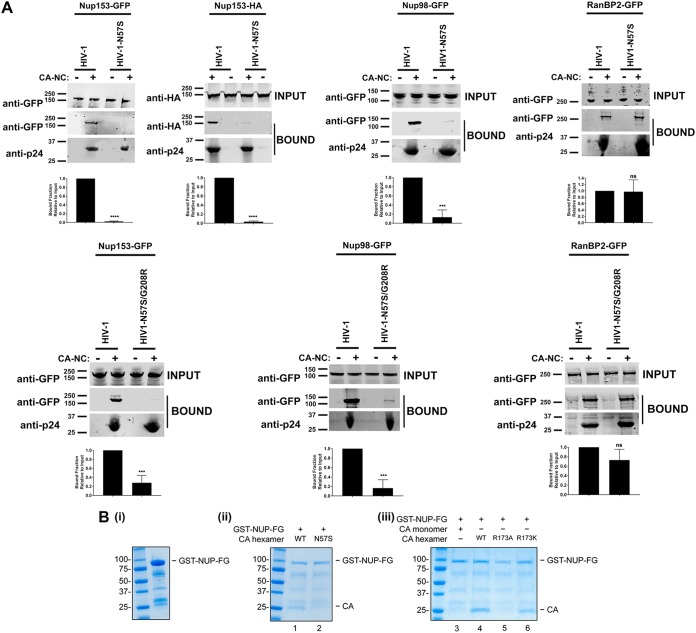
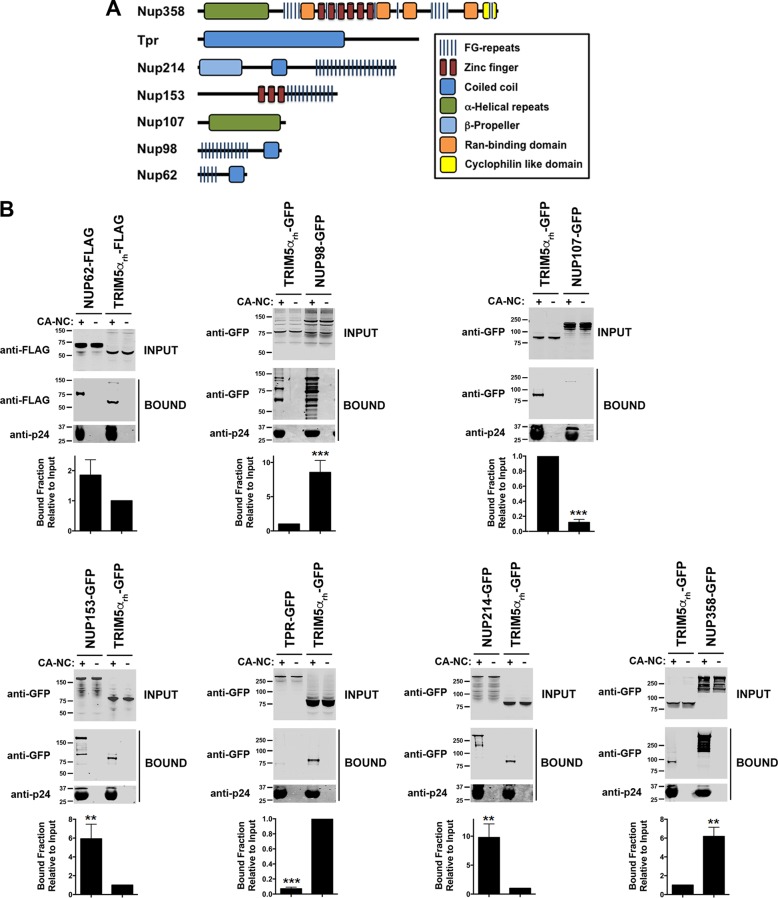
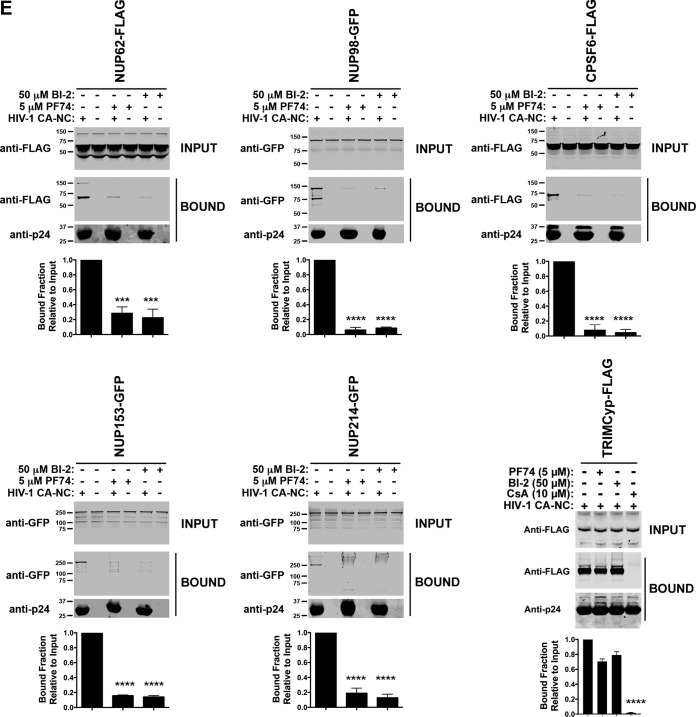
Based on our structure-function studies, we hypothesized that HIV-1 capsid with N57 mutations loses the ability to interact with Nup153, an essential protein for infection (28). To test this hypothesis, we evaluated the ability of Nup153-GFP to bind to HIV-1 capsid-nucleocapsid (CA-NC) complexes assembled in vitro, which recapitulates the surface of the HIV-1 core (35, 46). As shown in Fig. 4A, Nup153-GFP lost its ability to bind in vitro-assembled HIV-1 CA-NC complexes with the N57S mutation. Similarly, Nup153 was unable to interact with complexes bearing the N57S/G208R mutations. In contrast, Nup153-GFP could bind effectively to the wild-type complexes. To rule out the possibility that the GFP moiety of Nup153-GFP is responsible for the binding of Nup153-GFP to capsid, we measured the ability of Nup153-HA, which contains a small hemagglutinin (HA) tag of 8 amino acids, to bind wild-type and mutant in vitro-assembled HIV-1 CA-NC complexes. Similarly, Nup153-HA bound wild-type but not N57S capsids (Fig. 4A). This suggests that the ability of HIV-1 to infect nondividing cells requires the interaction of capsid with Nup153. Interestingly, Nup98-GFP was also defective in its ability to bind complexes with the N57S mutation; however, it should be noted that depletion of Nup98 expression in human cells poorly affects HIV-1 infection compared to Nup153 depletion (35). Nup98, like Nup153, contains FG repeats that are likely to allow capsid interaction, consistent with our own crystallographic analysis indicating that capsid residue N57 interacts with one of the FG repeats of Nup153. We showed in a control experiment that RanBP2/Nup358 interacts with both wild-type and mutant HIV-1 CA-NC complexes (Fig. 4A), which was to be expected, since RanBP2/Nup358 is known to bind to capsid using both FG repeats and a cyclophilin-like domain (19). These experiments suggested that the interaction of capsid with Nup153 is essential for the ability of HIV-1 to infect nondividing cells. By the same token, our results suggested that RanBP2/Nup358 is not important for the ability of HIV-1 with changes on N57 to infect nondividing cells.
FIG 4.
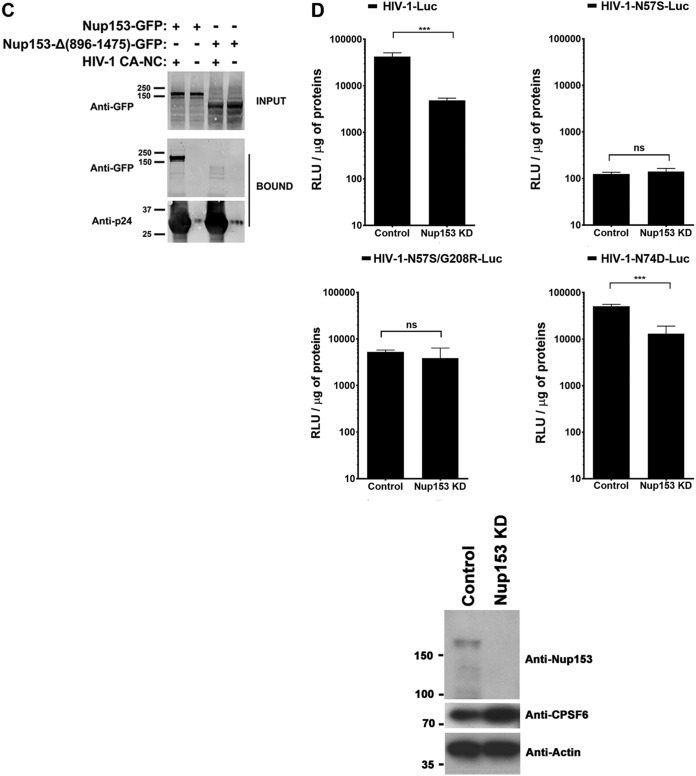
HIV-1 capsids bearing the N57S change do not bind Nup-153. (A) The ability of Nup153-GFP, Nup153-HA, Nup98-GFP, and RanBP2/Nup358-GFP to bind the indicated in vitro-assembled HIV-1 CA-NC complexes was measured as described in Materials and Methods. INPUT and BOUND fractions were analyzed by Western blotting using anti-GFP, anti-HA, or anti-p24 antibodies. Experiments were repeated at least three times, and a representative experiment is shown. (B) GST protein fused to the Nup153 residues 896 to 1475 containing the FG repeat (GST-NUP-FG) binds to HIV-1 hexameric capsid. (i) Purified GST-NUP-FG proteins were incubated with hexameric capsids bearing the indicated changes. (ii and iii) Subsequently, complexes were pulldown using glutathione beads and analyzed by SDS-PAGE. (C) The ability of Nup153 that does not contain FG repeats [Nup153Δ(896-1475)-GFP] to bind the indicated in vitro-assembled HIV-1 CA-NC complexes was measured as described in Materials and Methods. Similarly, INPUT and BOUND fractions were analyzed by Western blotting using anti-GFP or anti-p24 antibodies. Experiments were repeated at least three times, and a representative experiment is shown. (D) HIV-1-N57S virus infection of Nup-153-depleted cells. Nup153-depleted HeLa cells (Nup153 KD) were challenged with the indicated HIV-1-Luc viruses. (Upper) Infection was determined by measuring luciferase activity 48 h postinfection. (Lower) Depletion of Nup153 was achieved transiently using shRNA, and expression knockdown was confirmed by Western blotting using anti-Nup153 antibodies, as described in Materials and Methods. Results were analyzed using two-tailed Student's t test. Differences were considered statistically significant at a P value of <0.05 (*), <0.01 (**), <0.001 (***), or <0.0001 (****) or were nonsignificant (ns). Experiments were repeated at least three times, and a representative example is shown.
To further support these results, we expressed and purified a glutathione S-transferase (GST) fusion protein containing the entire FG repeat region of human Nup153 (residues 896 to 1475) (Fig. 4Bi). As shown in Fig. 4Bii, the GST-NUP-FG fusion protein pulled down wild-type HIV-1 capsid hexamers but not those of the N57S mutant. Importantly, GST-NUP-FG also did not bind monomeric HIV-1 capsid, confirming that the FG repeats bind the NTD-CTD interface that can exist only in fully assembled capsid. These experiments are the first to show direct interaction of the entire FG repeat region of Nup153 with the HIV-1 capsid hexamer using purified proteins.
Similar to our findings on the N57S mutant, hexameric HIV-1 capsids with an R173A mutation did not bind to GST-NUP-FG (Fig. 4Biii). In contrast, the R173K capsid retained the ability to interact with GST-NUP-FG. It is of interest that, even though the capsid mutant R173A did not bind GST-NUP-FG, HIV-1 viruses with the capsid mutation R173A are not infectious (56).
We next measured the capsid-binding ability of Nup153 that does not contain FG repeats [Nup153Δ(896-1475)-GFP]. As shown in Fig. 4C, Nup153Δ(896-1475)-GFP did not bind in vitro-assembled HIV-1 CA-NC complexes, unlike Nup153-GFP, suggesting that FG repeats are necessary for the ability of Nup153 to bind to the HIV-1 core.
We next tested whether Nup153 is required for the cellular infectivity of HIV-1 with the N57S mutation. For this purpose, Nup153-depleted HeLa cells were challenged with HIV-1-N57S-Luc and HIV-1-N57S/G208R-Luc. As shown in Fig. 4D, both viruses were insensitive to Nup153 depletion in dividing cells. These results suggested that HIV-1 with the N57S mutation utilizes a different mode of infection independent of Nup153. As previously shown (35), infection of Nup-153-depleted cells by wild-type and HIV-1-N74D viruses was severely diminished. These experiments raise the possibility that the N57S mutant has gained the ability to infect dividing cells by a mechanism in which Nup153 is not required.
FG repeats mediate the interaction of Nup proteins with HIV-1 capsid.
Our crystallization experiments suggested that the HIV-1 capsid has some preference for certain FG-containing peptides. We therefore tested other Nups for the ability to bind to in vitro-assembled HIV-1 CA-NC complexes. We found that Nups that contain FG repeats (RanBP2/Nup358, Nup214, Nup153, Nup98, and Nup62) bound to the CA-NC complexes (Fig. 5A and B), whereas non-FG Nups (Tpr and Nup107) did not (Fig. 5A and B). As a control, we tested the ability of rhesus monkey TRIM5α (TRIM5αrh) protein to bind to HIV-1 CA-NC complexes; TRIM5αrh is known to bind capsid (57) (Fig. 5B). Binding of the FG-containing Nups was abrogated by the N57S mutation (Fig. 5C), confirming that the same pocket in the capsid protein lattice mediated the interaction.
FIG 5.
HIV-1-N57S viruses do not interact with Nups containing FG repeats. (A) Several Nups that do or do not contain FG repeats are shown. (B) Binding of Nups to HIV-1 capsid. The ability of the indicated Nups to bind in vitro-assembled HIV-1 CA-NC complexes was measured as described in Materials and Methods. INPUT and BOUND fractions were analyzed by Western blotting using anti-GFP or anti-p24 antibodies. As a positive control, we measured the ability of rhesus TRIM5α (TRIM5αrh) to bind in vitro-assembled HIV-1 CA-NC complexes. (C) Binding of Nups to HIV-1 capsids bearing the mutation N57S or N74D. Similarly, the ability of the indicated Nups to bind in vitro-assembled HIV-1 CA-NC bearing the mutation N57S or N74D was measured. INPUT and BOUND fractions were analyzed by Western blotting using anti-GFP or anti-p24 antibodies. As a control, we measured the ability of CPSF6 to bind in vitro-assembled HIV-1 CA-NC complexes bearing the mutation N57S or N74D. Results were analyzed using two-tailed Student's t test. Differences were considered statistically significant at a P value of <0.05 (*), <0.01 (**), <0.001 (***), or <0.0001 (****) or were nonsignificant (ns). (D) HIV-1-N57S virus infection of cells depleted for the expression of different Nups. HeLa cells depleted for the expression of the different Nups were challenged with the indicated HIV-1-Luc viruses. (Upper) Infection was determined by measuring luciferase activity 48 h postinfection. Statistical analysis by one-way analysis of variance was applied (****, P < 0.0001). (Lower) Depletion of the indicated Nup was achieved transiently using shRNA, and expression knockdown was confirmed by Western blotting using the indicated antibodies, as described in Materials and Methods. Experiments were repeated at least three times, and a representative experiment is shown.
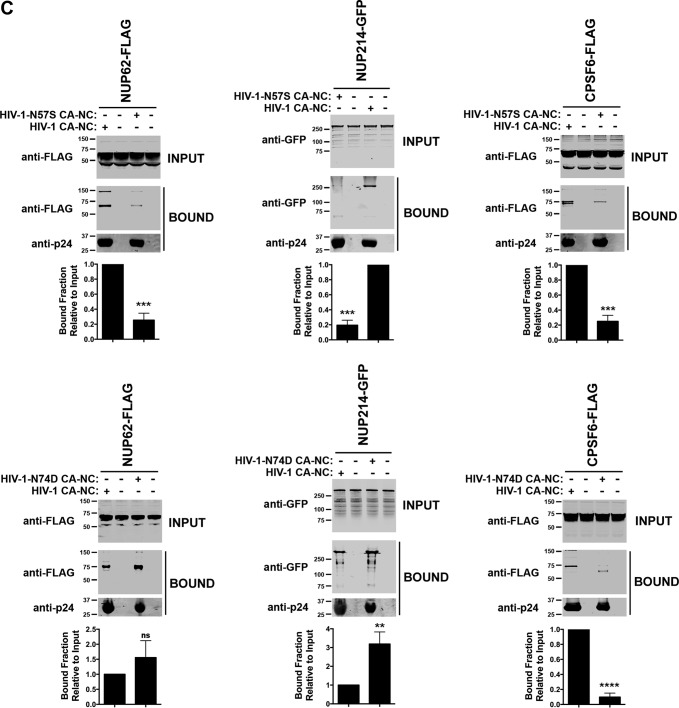
We also tested the ability of CPSF6 to bind to HIV-1 CA-NC complexes with the N57S mutation. As shown in Fig. 5C, CPSF6 similarly lost the ability to bind the N57S mutant capsid, as we have previously shown (58). These results demonstrate that N57S behaves similarly to N74D in terms of binding to CPSF6 (37, 59). As a control, we confirmed that CPSF6 does not bind HIV-1 capsids with the N74D substitution (Fig. 5C) (37). Unlike the N57S mutant, the N74D capsid mutant did not lose the ability to bind Nup153 (35).
We also tested whether the binding of Nup153-HA to the HIV-1 capsid is affected by the use of increasing concentrations of the FG peptide used in our crystallographic studies (Nup1531407–1429) (data not shown). These results showed that increasing concentrations of the peptide containing an FG repeat were not sufficient to outcompete the binding of full-length Nup153 to capsid. One possibility is that the avidity effect present in the binding of full-length Nup153 to capsid, which arises from the presence of multiple capsid-binding segments in Nup153, enhances Nup153 affinity for capsid. This avidity effect is not present in the peptide that only contains one binding site for capsid.
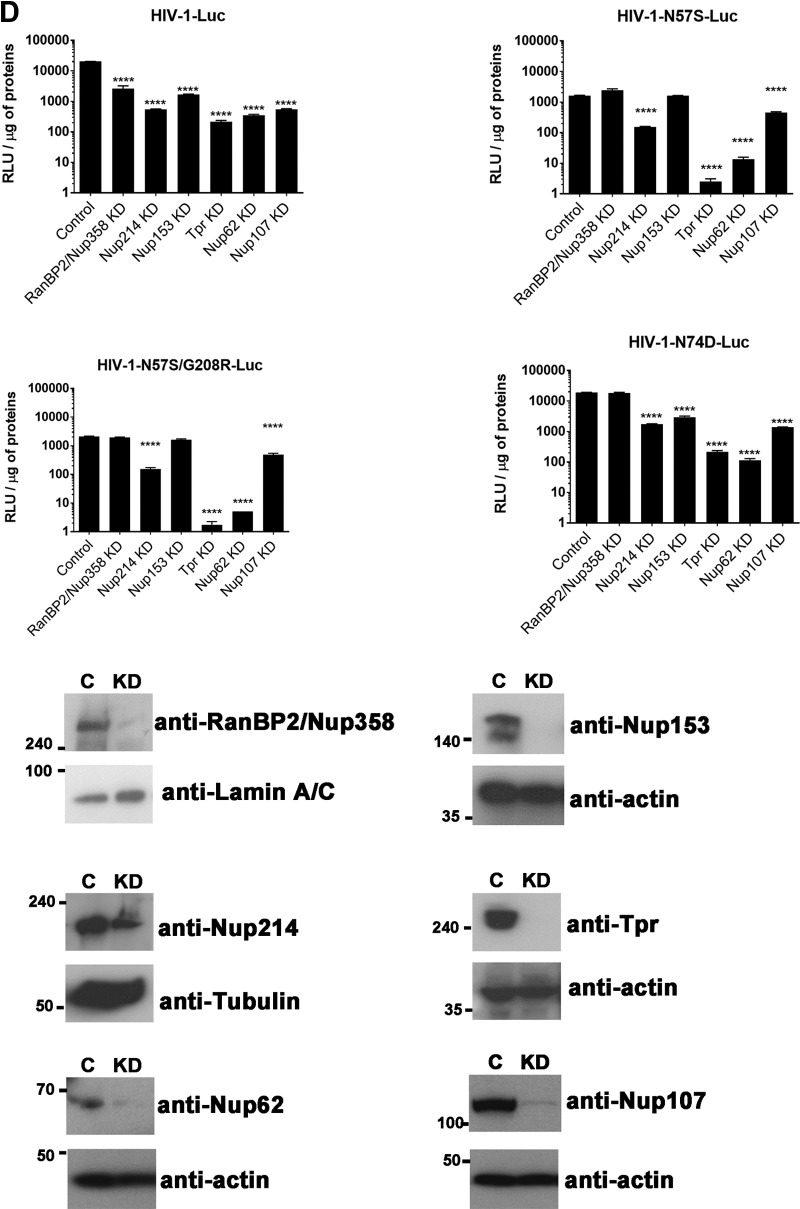
We next tested whether HIV-1 viruses with mutations on capsid residue N57 require Nup153 expression for infectivity. To this end, we transiently knocked down expression of Nup153 in HeLa cells and challenged the cells with HIV-1 viruses containing mutations in capsid residue N57. As shown in Fig. 5D, HIV-1-N57S-Luc and HIV-1-N57S/G208R-Luc viruses were able to infect Nup153-depleted cells, suggesting that these viruses are using a different pathway to infect cells. In contrast, wild-type and HIV-1-N74D-Luc viruses were not able to infect Nup153-depleted cells (28, 35, 40). To understand the effect of depleting other Nups on HIV-1 viruses with mutations on capsid residue N57, we depleted the expression of RanBP2/Nup358, Nup214, and Tpr. In the case of RanBP2/Nup358-depleted cells, we found that, unlike wild-type HIV-1, viruses with mutations on capsid residue N57 were not affected in infectivity (Fig. 5D), as previously shown (37, 41). In contrast, depletion of Nup214, Nup62, and Tpr did affect the infectivity of both wild-type and mutant viruses. In agreement with this result, we and others have previously shown that Nup214, Nup62, and Tpr act after the HIV-1 nuclear translocation step (19, 39, 60, 61). In particular, Nup214 has an indirect role on HIV-1 infection, because it is involved in global mRNA export (19). Nup62 interacts with HIV-1 integrase and is involved in the integration step (60), and Tpr organizes the chromatin underneath the NPC; however, strong depletion of Tpr can interfere with the cell cycle step, explaining the defect in infectivity of all tested viruses (39). These experiments suggested that HIV-1 viruses with a capsid change on N57 are using a mode of infection independent of Nup153 and RanBP2/Nup358. A caveat for these experiments is the possibility that depletion of one Nup is affecting the expression of other Nups, which has not been extensively tested here.
HIV-1 viruses with mutations at capsid residue N57 are not subject to the influence of PF-74 and BI-2.
Previous observations suggested that capsid residue N57 interacts with PF74 and BI-2 (42, 43), raising the possibility that HIV-1 viruses mutated at capsid residue N57 are resistant to the small-molecule inhibitors PF74 and BI-2. To test this hypothesis, we challenged human HT1080 (Fig. 6A), dog Cf2Th (Fig. 6B), human HeLa (Fig. 6C), and human Jurkat cells (Fig. 6D) with HIV-1 viruses mutated at capsid residue N57 in the presence of increasing concentrations of PF74 and BI-2. As shown in Fig. 6, HIV-1-N57S, HIV-1-N57S/G208R, and HIV-1-N57A were unaffected by PF74 and BI-2. We also tested infectivity of HIV-1-N74D in the presence of increasing concentrations of PF74. Infection of HIV-1-N74D viruses was inhibited by PF74, similar to the wild type (data not shown). These experiments suggested that PF74 and BI-2 are preventing the ability of HIV-1 capsid to interact with proteins containing FG repeats, such as Nups and CPSF6. To directly test this hypothesis, we measured the ability of FG repeat-containing proteins Nup62, Nup98, Nup153, Nup214, and CPSF6 to interact with HIV-1 CA-NC complexes in the presence of PF74 and BI-2. As shown in Fig. 6E, PF74 and BI-2 prevented the ability of these proteins to bind to in vitro-assembled HIV-1 CA-NC complexes. To verify that the small molecules PF74 and BI-2 are specifically preventing the binding of Nups that contain FG repeats to capsid, we tested the ability of PF74 and BI-2 to affect the ability of TRIMCyp to bind capsid. As shown in Fig. 6E, the ability of TRIMCyp to bind capsid was affected by cyclosporine (CsA) (62) but not by PF74 or BI-2. These experiments suggested that PF74 and BI-2 affected the ability of capsid residue N57 to interact with various cellular factors, which might more fully explain how these drugs inhibit HIV-1.
FIG 6.
Small-molecule inhibitors PF74 and BI-2 prevent the binding of Nups containing FG repeats to the HIV-1 capsid. (A) HIV-1 viruses bearing changes on residue N57 are resistant to the inhibitory effects of PF74 and BI-2. Human HT1080 cells were challenged with the indicated HIV-1-GFP viruses in the presence of increasing concentrations of PF74 or BI-2. Infection was determined by measuring the percentage of GFP-positive cells 48 h postinfection. The percentage of infection relative to that of untreated samples is shown. Similar experiments were performed using human HeLa (B), dog Cf2Th (C), and human Jurkat (D) as target cells. (E) Binding of Nups to HIV-1 capsid is inhibited by PF74 and BI-2. The ability of the indicated Nups to bind in vitro-assembled HIV-1 CA-NC complexes in the presence of PF74 or BI-2 was measured as described in Materials and Methods. As a specificity control, we tested the ability of TRIMCyp to bind in vitro-assembled HIV-1 CA-NC complexes in the presence of PF74, BI-2, or cyclosporine (CsA). INPUT and BOUND fractions were analyzed by Western blotting using anti-GFP, anti-FLAG, or anti-p24 antibodies. As a positive control, we measured the ability of CPSF6 to bind in vitro-assembled HIV-1 CA-NC complexes in the presence of PF74 or BI-2. Results were analyzed using two-tailed Student's t test. Differences were considered statistically significant at a P value of <0.05 (*), <0.01 (**), <0.001 (***), or <0.0001 (****) or were nonsignificant (ns).
HIV-1 viruses with capsid mutation N57S showed different integration site preferences.
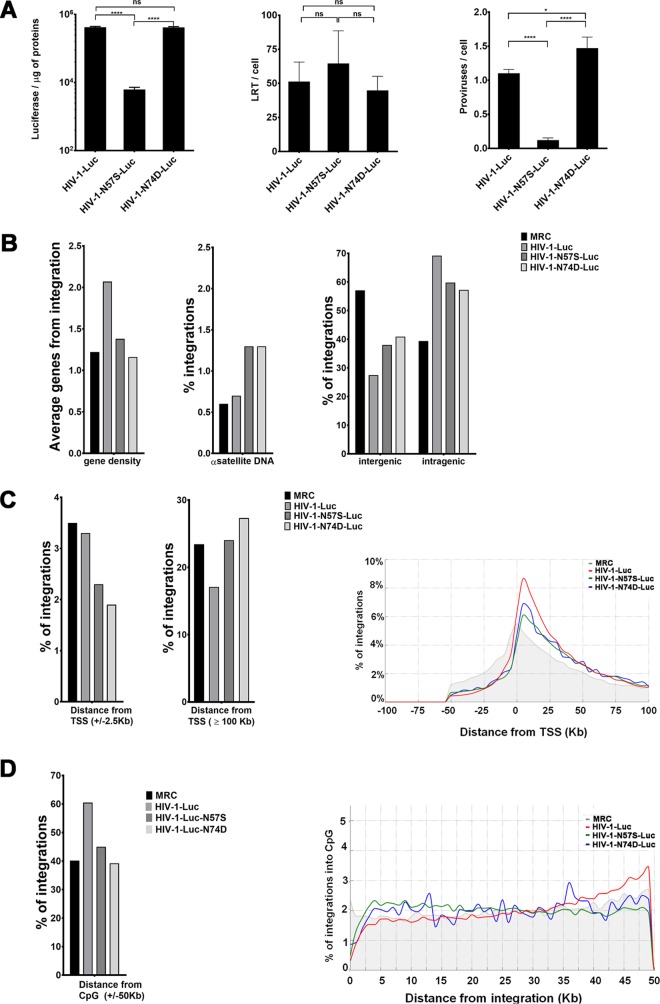
Wild-type HIV-1 integration is favored in chromosomal regions containing high gene density and high transcriptional activity (63). Previously, we showed that depletion of Nup153 or Tpr reduces HIV-1 integration frequency in high-gene-density regions, establishing a link between HIV-1 translocation and integration (35, 39). To investigate the contribution of capsid-Nup153 interactions to HIV-1 integration, we investigated the proviral integration pattern of HIV-1 viruses bearing the change N57S on Jurkat T cells by next-generation sequencing (NGS). As a control, we studied integration of HIV-1 viruses bearing the change N74D. Infection of Jurkat cells by HIV-1-N57S supported levels of reverse transcription similar to those of wild-type HIV-1 (Fig. 7A). We also observed a decrease of luciferase expression exclusively for the virus carrying the mutation N57S, consistent with the number of proviruses integrated as assessed by Alu-PCR (Fig. 7A). Our results revealed that the integration pattern of HIV-1 N57S mutant is very similar to that of HIV-1 N74D (Fig. 7B and Tables 3 and 4). HIV-1-N57S viruses showed a decreased ability to integrate in intragenic regions compared to that of wild-type HIV-1 (Fig. 7B and Table 4). We detected fewer HIV-1-N74D-Luc (57.2%) and HIV-1-N57S-Luc (59.8%) integrations in intragenic regions, mapping inside exons or introns of known genes, than wild-type integrations (69.2%) (Fig. 7B and Table 4). Instead, we detected an increase of integration sites in intergenic regions, upstream or downstream of known genes, for N57S (38%) and N74D (40.9%) mutants compared to those for the wild type (27.5%) (Fig. 7B and Table 4). Consistent with gene-targeting preferences into gene-dense regions, integrations at distances of ≥100 kb from transcription start sites (TSSs) were highly disfavored for wild-type virus, while N57S and N74D mutant viruses were more similar or exceed the matched random control (MRC) (Fig. 7C and Table 4). All of the differences observed among samples were statistically significant (P value of <0.001 by Wilcoxon rank-sum statistical and proportions test). Indeed, the majority of integrations of wild-type HIV-1 occurred ∼2.5 to 50 kb downstream from TSSs (Fig. 7C and Table 4). Integration of capsid mutant viruses occurred less frequently at and near TSSs than for wild-type virus. In contrast, integrations of wild-type and mutant viruses were similarly disfavored ∼50 kb downstream from TSSs (Fig. 7C and Table 4). HIV-1-N57S and -N74D mutant viruses favor integration within transcription units; however, their integration site distributions statistically diverged from that of wild-type virus. Interestingly, we observed that these two mutants, N57S and N74D, integrated in chromatin regions with lower gene density as well as in CpG islands, which are chromatin features associated with promoters (Fig. 7D and Table 4). The differences observed among the three viruses in the percent distribution of integrations in host chromatin regions and chromatin features all were statistically significant. The distribution of all integration sites as a function of gene density for the three viruses was studied. Interestingly, the capsid mutations studied here reduced the ability of HIV-1 to target gene-dense regions in the host chromatin (data not shown).
FIG 7.
HIV-1 infection and integration site distribution. Shown is integration site distribution for wild-type and N57S and N74D HIV capsid mutant viruses: intergenic, intragenic, gene density, and alpha satellite. (A) Jurkat cells were challenged with the indicated HIV-1-Luc viruses. (Left) Infectivity was determined by measuring luciferase reporter activity at 48 hpi. (Middle) Late reverse transcription (LRT) was measured by quantitative PCR at 7 hpi as described in Materials and Methods. (Right) The number of proviruses integrated to the genome per cell (proviruses/cell) was determined using Alu-PCR. Asterisks indicate statistical significance in comparisons of one virus to another virus (determined by paired two-tailed Student's t tests; ns, nonsignificant; *, P < 0.05; ****, P < 0.0001). (B, left) Histograms representing the average gene density. (Middle) Percentage of integrations having an α-satellite region within an interval of 50 kb. (Right) Percentage of integrations classified as inter- or intragenic; intragenic integrations are those localized within 50 kb of a gene. (C, left) Percentage of integrations having a TSS within a 2.5-kb window. (Middle) Histogram recapitulating the percentage of integration sites at a distance of ≥100 kb from the TSS. (Right) Plots of the percentage of integrations grouped by distance from TSS in a 100-kb window; the matched random control (MRC) is represented in gray. (D, left) Percentage of integrations falling within a 50-kb window from any CpG island. (Right) Plot representing the percentage of integrations grouped by their distance from any CpG in a 0- to 50-kb range.
TABLE 3.
Integration of HIV-1 virusesa
| Parameter | WT |
N57S |
N74D |
|||
|---|---|---|---|---|---|---|
| No. | % lost | No. | % lost | No. | % lost | |
| Initial filtered readsb | 1,288,748 | 1,655,553 | 1,517,568 | |||
| Reads matching on human genomec | 1,014,432 | 21.3 | 1,237,632 | 25.2 | 1,222,659 | 19.4 |
| Quality filtered readsd | 771,358 | 24.0 | 912,662 | 26.3 | 931,713 | 23.8 |
| HIV Nef matchese | 323,998 | 58.0 | 71,815 | 92.1 | 395,932 | 57.5 |
| Reads with unique mappingf | 310,619 | 4.1 | 69,787 | 2.8 | 383,341 | 3.2 |
| Genomic integrationsg | 105,329 | 66.1 | 6,137 | 91.2 | 116,138 | 69.7 |
Number or percent lost indicates the number of integrations conserved or lost after the mapping steps.
Raw reads from HIV-1 integration sites library.
Filtered reads of HIV-1 integration sites were mapped against the UCSC hg19 release of the human genome.
Filtered reads of HIV-1 integration sites were mapped against the UCSC hg19 release of the human genome and processed for their quality.
Filtered reads of HIV-1 integration sites were mapped against the UCSC hg19 release of the human genome, processed for their quality, and matched with HIV Nef sequence.
Reads obtained after unique mapping.
The number of reads and percentage of HIV-1 integration sites is indicated for every HIV-1 variant.
TABLE 4.
Effect of capsid mutations on HIV-1 integration sites in Jurkat cells
| Genomic feature | Valuea (%) for: |
|||
|---|---|---|---|---|
| MRCb | WT | N57S | N74D | |
| Intergenic (±50 kb) | 57.1 | 27.5 | 38.0 | 40.9 |
| Intragenic (±50 kb) | 39.4 | 69.2 | 59.8 | 57.2 |
| TSS (±2.5 kb) | 3.5 | 3.3 | 2.3 | 1.9 |
| CpG islands (±50 kb) | 40.2 | 60.5 | 45.0 | 39.2 |
| DNase I HS (±50 kb) | 83.7 | 96.2 | 91.5 | 90.7 |
| α-Satellite (±50 kb) | 0.6 | 0.7 | 1.3 | 1.3 |
| β-Satellite (±50 kb) | 0.2 | 0.2 | 0.2 | 0.2 |
| Avg no. of genes in ±50 kb | 1.22 | 2.07 | 1.38 | 1.16 |
Values are shown relative to the number of unique integration sites per genomic feature, except those for gene-dense regions. Integration of wild-type and mutant HIV-1 viruses in different regions of chromatin from Jurkat T cells is shown as a percentage of total integration: intragenic, intergenic, transcriptional start sites (TSS), CpG, alpha and beta satellites, and DNase I-hypersensitive regions. The predicted integration for an MRC is shown. Pearson's chi-squared test was used (P < 2.2e−16).
MRC, matched random control containing coordinates for 14 million computer-generated integration sites in the vicinity of BfaI and BglII restriction sites in hg19.
The average gene density within a ±50-kb window surrounding integration sites was 2.07 transcription units for wild-type virus, 1.38 transcription units for N57S, and 1.16 transcription units for N74D (Table 4). The two capsid mutants showed an integration curve profile shifted to the left with respect to wild-type virus, consistent with a lower preference for integration into regions containing higher gene density. Wild-type HIV-1 preferentially targeted the midsections of genes roughly encompassing 15 to 60% of the genes' lengths. N74D and N57S capsid mutant viruses showed a less marked preference for integration into the midsection of genes compared to that of the wild type while showing an increase in the targeting of 3′ gene regions (data not shown). The curve of the N74D capsid mutant more closely resembles that of the MRC than that of the N57S mutant (Fig. 7D). Interestingly, N74D and N57S mutant viruses showed reduced integration near CpG islands compared to wild-type HIV-1 (Fig. 7D and Table 4). We also observed a decreased integration of the mutant viruses into DNase I-hypersensitive sites (DHSs), which are characteristic features of active chromatin (Table 4). Finally, we also analyzed proviral integration into chromatin regions that are usually excluded, such as alpha- and beta-satellites, which are enriched in heterochromatin. Both mutants integrated to a greater extent than the wild-type virus into alpha-satellite regions (Fig. 7B and Table 4). In contrast, integration of these mutant viruses was disfavored in beta-satellite regions (Table 4).
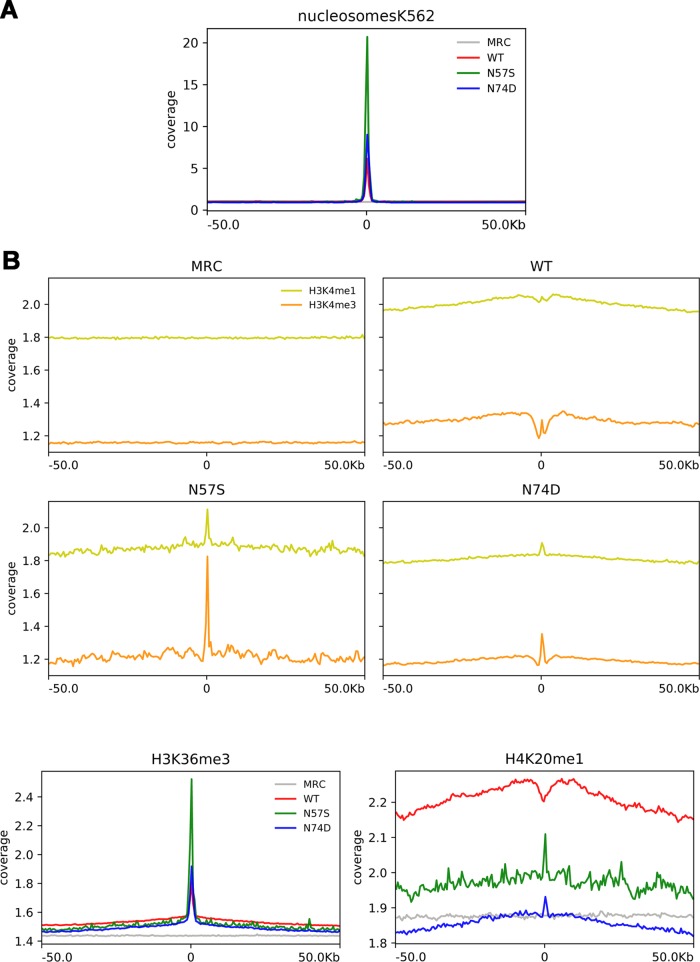
In parallel, we have also performed a nucleosome density analysis around viral integration sites, and we found an increase of the preference for nucleosome-dense regions for all three viruses with respect to MRC (Fig. 8A).
FIG 8.
Analysis of nucleosome density around viral integration sites and the relationship between integration frequency and epigenetic mark density. (A) The plot shows the mean normalized coverage in 100-kb windows around the viral integration sites of each virus (WT, N57S, and N74D), as well as of matched random controls (MRC) for nucleosome density feature (data available for K562 cells in the UCSC database). (B) The plots show the mean normalized coverage for different histone modifications (derived from ChIP-seq data) in 100-kb windows around the viral integration sites of each virus (WT, N57S, and N74D) as well as of matched random controls (MRC). The plots show curve coverage signal for H3K4me1, H3K4me3, H3K36me3, and H4K20me1. Coverage was normalized to the number of tags.
To identify the relationship between viral integration sites and histone modifications, we also analyzed the coverage profile for different histone modifications in a window of ±50 kb around the integration sites for each virus (WT, N57S, N74D, and MRC) (Fig. 8B). We observed that HIV-1 WT provirus accumulation was positively correlated with histone methylation patterns characteristic of active transcription units, such as H3K4me1, H4K20me1, and H3K36me3, as has been previously reported (64). In addition, we found a few histone modifications that show clear differences between wild-type and mutant viruses.
H3K4me3, which is a mark of regulatory regions usually excluded around HIV-1 integration sites, showed a pick enrichment around integration sites for both mutant viruses (N57S and N74D) (39, 64), while we observed a reduction for this histone modification density near the integration sites of the WT virus (± 2.5 kb from integration site). This result suggests a relocation of HIV-1 integrations due the viral capsid mutation.
Interestingly, all three viruses showed a peak of H3K36me3 enrichment around integration sites (Fig. 8B). This phenomenon could be due to the fact that all of these viruses interact with LEDGF/p75, which binds this particular histone modification, helping the virus to integrate in selected sites of the host chromatin (39, 65). The persistent tendency of the tested viruses to integrate into open chromatin probably is due not only to the role of LEDGF/p75 (Table 4) but also to the intrinsic preference of HIV-1 IN for open chromatin structures, as previously reported (66, 67).
DISCUSSION
Nups hold a critical role in HIV-1 nuclear import and integration. Although Nup153 and RanBP2/Nup358 are known to be required for HIV-1 nuclear translocation, the experiments that previously established this requirement were only performed in cycling cells (19, 34, 35, 37, 40). Therefore, the factors required for nuclear translocation in nondividing cells remain unknown. In addition, the study of HIV-1 nuclear translocation typically has been performed in cells that are depleted for Nup153 and RanBP2/Nup358 expression (28, 32, 33), which could disrupt the architectural integrity of nuclear pores or affect expression of other Nups. Moreover, interference with the structural components of nuclear pores could lead to erroneous interpretation of HIV-1 phenotypes. Use of HIV-1 with capsid mutations has the advantage that it does not disrupt the integrity of nuclear pores. Accordingly, we chose HIV-1 capsid mutants to understand the cellular factors involved in lentiviral nuclear translocation in nondividing cells. For this purpose, we solved the structure of the peptide containing the FG repeat of Nup153 (Nup1531407–1429) in complex with the hexameric HIV-1 capsid. Our crystal structure was consistent with previous findings on the interaction of capsid with an FG repeat (43) and highlighted several capsid residues that interact with the FG repeat. We then mutated these residues and screened for mutant viruses that could infect dividing but not nondividing cells. Our results revealed that HIV-1 with capsid mutations N57S, N57A, or N57D can infect dividing cells but are abrogated in their ability to infect nondividing cells. Using the fate-of-the-capsid assay, we demonstrated that mutations on N57 residue decreased the stability of the HIV-1 core during infection. Because core stability during infection might be the reason that N57-mutated HIV-1 viruses cannot infect nondividing cells, we improved core stability by simultaneously introducing the capsid mutations N57S/G208R. We then showed that even though HIV-1-N57S/G208R cores were more stable than HIV-1-N57S cores, these mutants were still unable to infect nondividing cells. Therefore, the inability of HIV-1-N57S to infect nondividing cells cannot be attributed to defects in core stability.
We next examined the viral replication stage at which the infection by HIV-1-N57S and HIV-1-N57S/G208R viruses was inhibited. Interestingly, these viruses underwent reverse transcription but did not generate 2-LTR circles, suggesting a defect in nuclear translocation. This is in agreement with the hypothesis that these mutant viruses are unable to enter the nucleus. One possibility is that the core is no longer interacting with an important Nup needed for HIV-1 infection of nondividing cells. This concept is consistent with our finding that the N57 residue directly interacts with the FG repeats of Nup153. By the same token, this observation raises the possibility that the N57 region of the HIV-1 capsid is important for the ability of the core to generally interact with Nups containing FG repeats.
Depletion of Nup153 or RanBP2/Nup358 expression in human cells prevents HIV-1 nuclear translocation, suggesting a role for these proteins in nuclear entry. We therefore tested the ability of HIV-1 capsid containing N57 mutations to bind to the Nups Nup153 and RanBP2/Nup358, which contain FG repeats and are essential for HIV-1 nuclear translocation in cycling cells. Capsids with N57 mutations lost the ability to bind Nup153 but not RanBP2/Nup358, suggesting that Nup153 is directly involved in HIV-1 nuclear translocation in nondividing cells. Although N57 mutant capsids did not bind any of the tested Nups containing FG repeats, the only functional interactions related to nuclear translocation are capsid-Nup153 and capsid-RanBP2/Nup358, indicating that interaction of capsid with Nup153 plays an important role in HIV-1 nuclear translocation in nondividing cells. One possibility is that the HIV-1 core or preintegration complex interacts with Nup153 in the cytosol, and that it is this interaction that is essential for guiding the complex into the nuclear pore. This is in agreement with evidence suggesting that Nup153 shuttles between the nucleus and cytoplasm (68).
Structural studies revealed that N57 is also used by the HIV-1 capsid to interact with the drugs PF74 and BI-2, suggesting that these inhibitors disrupt the ability of capsid to interact with Nups containing FG repeats. In agreement with this hypothesis, we found that PF74 and BI-2 prevent capsid binding to Nups with FG repeats. One possibility is that PF74 and BI-2 can block HIV-1 infection by preventing the interaction of capsid with FG-containing Nups such as Nup153. This notion is in agreement with independent reports suggesting that PF74 and BI-2 block HIV-1 infection at a step prior to nuclear translocation (69, 70).
Lentiviruses such as HIV-1 have the ability to infect both dividing and nondividing cells. HIV-1 infection of nondividing cells requires the HIV-1 preintegration complexes to cross the nuclear pore. In contrast, infection of dividing cells provides easier access to the cellular genome, where transport across the nucleopore is not required. Gammaretroviruses such as MLV are unable to infect nondividing cells and require mitosis for efficient infection (3). Similarly, mitosis provides gammaretroviruses easier access to the cellular genome. These observations raise the possibility that lentiviruses and gammaretroviruses encounter cellular chromatin in various transcriptional states, resulting in different integration patterns, as previously observed for HIV-1 and MLV (63). Although the chromatin topology is likely to play a role in retroviral integration, several recently described cellular factors have been reported to modulate site selection of retroviral integration: (i) LEDGF/p75 and HRP-2 are important for site selection of HIV-1 integration (71–80), (ii) CPSF6 directs HIV-1 integration into transcriptionally active chromatin (45, 81), and (iii) the BET family of proteins targets murine leukemia virus integration into transcription start sites (82). Taken together, this evidence suggests that integration site selection is a multiparameter mechanism that involves nuclear factors and the state of the chromatin. Unlike HIV-1, which favors integration into transcriptionally active chromatin, MLV favors integration into transcription start sites (63, 81). We have shown that HIV-1 viruses with the N57S capsid mutation integrate differently than wild-type HIV-1. We found that expression of integrated proviruses derived from N57S viruses is decreased by ∼6-fold compared to that of the WT. This observation is in agreement with our results, suggesting that integration of N57S occurs in regions of the chromatin other than those of the wild type. Our results showed that N57S viruses integrate in regions with low gene density. The integration of N57S viruses in different regions of the chromatin explains the decrease in proviral expression.
We observed that the integration pattern of HIV-1-N57S is very similar to that of HIV-1-N74D, which is similar to the HIV-1 integration pattern of HIV-1 in CPSF6 knockout cells (45, 83). This is in agreement with our data showing that N57 mutant capsids do not interact with CPSF6. Thus, the integration pattern observed for N57S is likely due to loss of its ability to interact with CPSF6. Unlike HIV-1-N57S viruses, HIV-1-N74D does not show a defect in ability to infect nondividing cells, which is likely to be due to the fact that the N74D-mutant capsid interacts with Nup153 (35). This combined evidence allowed the following conclusions regarding early steps of HIV-1 infection: (i) the interaction of capsid with CPSF6 is required for achieving normal patterns of HIV-1 integration, since the loss of capsid interaction with CPSF6 by N74D or N57S results in an integration pattern similar to the one produced by HIV-1 viruses in CPSF6 knockout cells (45), and (ii) the interaction of capsid with Nup153 is important solely for the ability of HIV-1 to infect nondividing cells.
MATERIALS AND METHODS
Wild-type and mutant HIV-1 CA-NC expression and purification. HIV-1 CA-NC protein was expressed, purified, and assembled as previously described (46). The pET11a expression vector (Novagen) expressing the CA-NC protein of HIV-1 was used to transform BL-21(DE3) Escherichia coli. CA-NC expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the culture reached an optical density of 0.6 at 600 nm. After 4 h of induction, the cells were harvested and resuspended in 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, 10 mM 2-mercaptoethanol, and protease inhibitors (Roche). Lysis was performed by sonication, and debris was pelleted (35,000 × g, 30 min). Nucleic acids were stripped from the solution by adding 0.11 equivalents of 2 M (NH4)2SO4 and the same volume of 10% polyethylenimine. Nucleic acids were removed by stirring and centrifugation (29,500 × g, 15 min). The protein was recovered by addition of 0.35 equivalents of saturated (NH4)2SO4. The protein was centrifuged at (9,820 × g, 15 min) and resuspended in 100 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, and 10 mM 2-mercaptoethanol. Lastly, the CA-NC protein was dialyzed against 50 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, and 10 mM 2-mercaptoethanol and stored at −80°C.
In vitro assembly of HIV-1 CA-NC complexes.
HIV-1 CA-NC particles were assembled in vitro by diluting the CA-NC protein to a concentration of 0.3 mM in 50 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 2 mg/ml DNA oligo(TG)50. The mixture was incubated at 4°C overnight and centrifuged at 8,600 × g for 5 min. The pellet was resuspended in assembly buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl) at a final protein concentration of 0.15 mM (46, 47) and stored at 4°C until needed.
Binding to HIV-1 capsid complexes.
293T cells were transfected with plasmids expressing the different Nups fused to GFP or the indicated proteins. Forty-eight hours after transfection, cell lysates were prepared by resuspending washed cells in capsid-binding buffer (10 mM Tris, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]). The cell suspension was frozen, thawed, and incubated on ice for 10 min. The lysate next was centrifuged in a refrigerated Eppendorf microcentrifuge (∼14,000 × g, 5 min). The supernatant was supplemented with 1/10 volume of 10× phosphate-buffered saline (PBS) and then used in the binding assay. To test binding, 5 μl of CA-NC particles preassembled in vitro was incubated with 200 μl of cell lysate at room temperature for 1 h. A portion of this mixture, here referred to as INPUT, was stored. The mixture was spun through a 70% sucrose cushion (70% sucrose, 1× PBS, and 0.5 mM DTT) at 100,000 × g in an SW55 rotor (Beckman) for 1 h at 4°C. After centrifugation, the supernatant was carefully removed and the pellet resuspended in 1× SDS-PAGE loading buffer and here is referred to as BOUND. The level of protein was determined by Western blotting using the appropriate antibody. The level of HIV-1 CA-NC protein in the pellet was assessed by Western blotting with an anti-p24 capsid antibody.
Luciferase assays.
Luciferase (Promega) activity was measured at 48 hpi, according to the manufacturer's instructions, using a microplate fluorimeter (Victor, PerkinElmer). Protein quantification by Bio-Rad protein assay was carried out on the same lysates to normalize the luciferase data for protein content.
Lentiviral vector carrying shRNA and HIV-1 infection.
Complementary oligonucleotides coding for a short hairpin RNA (shRNA) cassette against Nup153, Tpr, Nup358/RanBP2, and Nup214 have been previously cloned in the HIV-1-derived vector (LV-shRNA) TRIP-GFP, which is ΔU3. Vector cloning and LV production were performed as recently described (35). Titers of LV-shRNAs carrying GFP were determined in HeLa cells using flow cytometry to assess GFP expression at 3 days postinfection. Capsid N74D, N57S, and N57S/G208R mutations were introduced into pNL4.3 Luc ENV− by site-directed mutagenesis. HeLa and Jurkat cells were transduced with LV-shRNA against Nups at different multiplicities of infection of up to 100 to generate knockdown cells. Single-cycle HIV-1 viruses were produced by transient transfection of 293T cells using calcium phosphate coprecipitation with NL4.3 Luc ENV−, with or without the capsid mutation N74D, N57S, or N57S/G208R (luciferase gene in place of nef) and cotransfection with the vesicular stomatitis virus glycoprotein (VSV-G) envelope expression plasmid pHCMV-G (VSV-G). The viruses harvested from 293T cells 48 h posttransfection were treated with 25 U/ml of DNase I (Roche) and with 100 mM MgCl2 at 37°C for 30 min. Virus normalizations were performed by p24 enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (PerkinElmer).
Quantitative PCR.
Infectivity, DNA synthesis, and integration during acute HIV-1 infection were quantified by real-time PCR. We analyzed for (i) infectivity by luciferase expression (the luciferase gene is a reporter gene inserted in place of nef in the HIV genome), (ii) late reverse transcription (LRT) products representing all near-full-length HIV-1 DNA in the cell, and (iii) integration of proviruses into human genome. Viruses were treated for 30 min at 37°C with 1,000 U of DNase I (Roche), with 10 μM nevirapine used in infected cells as a control. Total cellular DNA was isolated using the QIAamp DNA micro kit (Qiagen) at 7 and 24 hpi. Viral DNA synthesis products at 7 hpi were measured by real-time PCR using Sybr green and luciferase-specific primers (5′GAATCCATCTTGCTCCAACAC and 5′TTCGTCCACAAACACAACTC) located exclusively in the HIV-1-Luc but not in the LV shRNA used to generate Nup knockdown (KD) or control cells. Two long terminal repeat (2-LTR)-containing circles were detected using primers MH535/536 and probe MH603 (48) and for the standard curve the pUC2LTR plasmid, which contains the HIV-1 2-LTR junction. Integration was assessed by Alu-PCR as previously described (35).
Crystallography.
Soluble hexamers were mixed with excess peptides (2- to 5-fold) and then crystallized with tacsimate and polyethylene glycol as precipitants. Binding was determined by molecular replacement phasing with the original hexameric capsid structure followed by examination of electron density maps for the presence of bound peptide. Several FG-containing peptides derived from NUP153 were tested, but only the previously characterized high-affinity peptide (1407TNNSPSGVFTFGANSSTPAASAQ1429) was observed within the crystals. We refined structures containing fluorescein-tagged peptide and also analyzed crystals containing unlabeled peptide. Synchrotron diffraction data were collected at beamline 23-ID-D at the Advanced Light Source and processed with HKL2000. Surprisingly, two crystal forms (R3 and P1) were obtained with the fluorescently labeled peptide, which grew in separate drops but under similar conditions. Crystal packing and peptide occupancies in both crystals were distinct from previously described structures. Molecular replacement phasing, model building, refinement, and model validation were performed using the tools in Phenix. Structure statistics are summarized in Table 2.
GST-NUP-FG purification and pulldowns.
DNA encoding residues 896 to 1475 of human NUP153 was subcloned into the NdeI and BamHI sites of a modified pGEX2T vector, a kind gift of W. I. Sundquist. The fusion protein was expressed in Escherichia coli DH5α cells by isopropyl-β-d-thiogalactopyranoside (IPTG) induction. Cells were lysed in buffer (25 mM Tris, pH 7.5, 200 mM NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA) by lysozyme treatment and sonication. GST-NUP-FG was then bound to glutathione-Sepharose resin and eluted in buffer containing 20 mM reduced glutathione. For pulldown experiments, purified GST-NUP-FG (1.8 mg/ml) was preincubated with 25 μl of beads for 30 min at 4°C. The beads were washed with binding buffer (50 mM Tris, pH 8, 150 mM NaCl, 1 mM EDTA) and then resuspended in binding buffer containing 1% (wt/vol) bovine serum albumin (BSA). Capsid hexamers were then added and incubated for 1 h at 4°C. After extensive washing with binding buffer, bound fractions were eluted and analyzed by SDS-PAGE.
Integration site sequencing and bioinformatics analysis.
Integration site sequencing was performed using 2 × 108 Jurkat cells infected with 10 μg of the p24 antigen of NL4.3-Luc ENV-carrying wild-type or mutant viruses. Two days later, genomic DNA was extracted by using a QIAamp DNA micro kit (Qiagen) and digested with the 4-cutter enzymes BfaI and BglII to prevent amplification of internal 3′-LTR fragments, as previously described (35). 5′-LTR-genome junctions were amplified by ligation-mediated PCR using primers specific for the linker and the HIV 5′ LTR and including overhang adapter sequences compatible with the Illumina sequencing platform. A final indexing PCR (eight cycles) was performed to tag each sample with specific Nextera XT dual barcodes (Illumina, San Diego, CA, USA) before sample pooling and sequencing on a single-read 150-cycle run on the MiSeq platform (Illumina).
Raw reads were filtered and trimmed by a properly developed bioinformatics pipeline to remove HIV LTR proviral and linker sequences from the sequencing data. The mapping procedure was based on that described in Cattoglio et al. (49). Only nonredundant trimmed reads longer than 20 bases were retained. Filtered reads were mapped against human reference genome (UCSC hg19) using gmap software (50). Putative integration sites were filtered for match quality, requiring an identity score of ≥0.9 (identity score calculated as matching nucleotides − [mismatching nucleotides + query gap + tile gap]/query size) and discarding reads with multiple identically scoring matches on the reference genome. We obtained 105,329 unique integration sites for wild-type HIV-1, 116,138 sites for the capsid mutant N74D, and 6,137 sites for the mutant N57S (Table 3). Chromosomal distributions were then compared, and a comparable number of integrations (i.e., 101,879) was selected from an MRC data set containing more than 14 million random integration sites. MRC was generated as in Cattoglio et al. (49). Each integration site was annotated for genes mapping in a ±50-kb interval and for genomic features, such as CpG islands, DNase I-hypersensitive sites, and α- and β-satellite regions, using UCSC Genome Browser tracks. For pairwise statistical comparisons, a two-sample test for equality of proportions with continuity correction was applied, whereas to compare the average number of genes in a ±50-kb interval the Wilcoxon rank-sum test was used. All statistical analyses were performed in MATLAB (Natick, MA, USA). The full filtering, mapping, annotation, and MRC generation pipeline is freely available upon request to the authors.
Analysis of provirus accumulation near histone modification sites.
Chromatin immunoprecipitation sequencing (ChIP-seq) histone modification data were downloaded from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.html as bed files of mapped data, and genome coordinates were converted from hg18 to hg19/GRCh37 genome build by the liftOver utility (51). Low-complexity regions were filtered out using the DAC Blacklist table from the UCSC Mappability track. Bed files were converted to bedGraph using BEDTools (v 2.26.0), and the data value columns were normalized on library size (i.e., total number of mapped reads). The mean signal distributions relative to the viral integration sites of the different viruses as well as of MRC were computed and plotted using deepTools2 (v 3.0.1).
Analysis of nucleosome density around viral integration sites.
MNase-seq nucleosome positioning data publicly available for K562 cells from the ENCODE Project were downloaded from the UCSC database as a bigwig file (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeSydhNsome/wgEncodeSydhNsomeK562Sig.bigWig) (52). Data were normalized, processed, and plotted as mentioned above for ChIP-seq data.
Accession number(s).
Coordinates and structure factors were deposited in the PDB database under accession codes 5TSV and 5TSX, respectively. Raw sequence data of integration experiments were uploaded to the NCBI Short Reads Archive under accession number SRP096351. The full data set, including information about the samples, can be retrieved under BioProject accession number PRJNA358663.
ACKNOWLEDGMENTS
We are thankful to the NIH/AIDS Repository Program for providing valuable reagents, such as antibodies and drugs. We thank Jonathan Wagner for assistance with crystallography and for performing early crystallization screens. We also gratefully acknowledge technical support by Demetrio Turati and Philippe Souque.
The work was funded by NIH grants R01-GM123540 and R01-AI087390 to F.D.-G., R01-AI129678 to O.P. and B.K.G.-P., and R01-AI120956 to F.D.-G. and O.P. The work was also supported by grants from the Agence Nationale des Recherches Scientifiques (ANRS ECTZ4469), Sidaction/FRM and the Pasteur Institute, and the Italian Ministry of Health (GR-2011-02352026).
REFERENCES
- 1.Rubin H, Temin HM. 1959. A radiological study of cell-virus interaction in the Rous sarcoma. Virology 7:75–91. doi: 10.1016/0042-6822(59)90178-3. [DOI] [PubMed] [Google Scholar]
- 2.Lewis PF, Emerman M. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol 68:510–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roe T, Reynolds TC, Yu G, Brown PO. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12:2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis P, Hensel M, Emerman M. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J 11:3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki Y, Craigie R. 2007. The road to chromatin–nuclear entry of retroviruses. Nat Rev Microbiol 5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 7.Bowerman B, Brown PO, Bishop JM, Varmus HE. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev 3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 8.Mattaj IW, Englmeier L. 1998. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 9.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 10.De Rijck J, Vandekerckhove L, Christ F, Debyser Z. 2007. Lentiviral nuclear import: a complex interplay between virus and host. Bioessays 29:441–451. doi: 10.1002/bies.20561. [DOI] [PubMed] [Google Scholar]
- 11.Fassati A. 2006. HIV infection of non-dividing cells: a divisive problem. Retrovirology 3:74. doi: 10.1186/1742-4690-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. 2006. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology 3:4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci U S A 90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita M, Emerman M. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita M, Emerman M. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol 78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita M, Perez O, Hope TJ, Emerman M. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog 3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. 2007. HIV-1 DNA flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J 26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Nunzio F, Danckaert A, Fricke T, Perez P, Fernandez J, Perret E, Roux P, Shorte S, Charneau P, Diaz-Griffero F, Arhel NJ. 2012. Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration. PLoS One 7:e46037. doi: 10.1371/journal.pone.0046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fassati A, Gorlich D, Harrison I, Zaytseva L, Mingot JM. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J 22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaitseva L, Cherepanov P, Leyens L, Wilson SJ, Rasaiyaah J, Fassati A. 2009. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology 6:11. doi: 10.1186/1742-4690-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. 2007. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J Biol Chem 282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- 23.Ao Z, Danappa Jayappa K, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X. 2010. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol 84:8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallay P, Hope T, Chin D, Trono D. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci U S A 94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hearps AC, Jans DA. 2006. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem J 398:475–484. doi: 10.1042/BJ20060466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valle-Casuso JC, Di Nunzio F, Yang Y, Reszka N, Lienlaf M, Arhel N, Perez P, Brass AL, Diaz-Griffero F. 2012. TNPO3 is required for HIV-1 replication after nuclear import but prior to integration and binds the HIV-1 core. J Virol 86:5931–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, Debyser Z. 2008. Transportin-SR2 imports HIV into the nucleus. Curr Biol 18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 28.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. 2010. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol 84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thys W, De Houwer S, Demeulemeester J, Taltynov O, Vancraenenbroeck R, Gerard M, De Rijck J, Gijsbers R, Christ F, Debyser Z. 2011. Interplay between HIV entry and transportin-SR2 dependency. Retrovirology 8:7. doi: 10.1186/1742-4690-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin A, Hayouka Z, Friedler A, Loyter A. 2010. Transportin 3 and importin alpha are required for effective nuclear import of HIV-1 integrase in virus-infected cells. Nucleus 1:422–431. doi: 10.4161/nucl.1.5.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Ocwieja KE, Brady TL, Ronen K, Huegel A, Roth SL, Schaller T, James LC, Towers GJ, Young JA, Chanda SK, Konig R, Malani N, Berry CC, Bushman FD. 2011. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog 7:e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Nunzio F, Fricke T, Miccio A, Valle-Casuso JC, Perez P, Souque P, Rizzi E, Severgnini M, Mavilio F, Charneau P, Diaz-Griffero F. 2013. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 440:8–18. doi: 10.1016/j.virol.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodward CL, Prakobwanakit S, Mosessian S, Chow SA. 2009. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J Virol 83:6522–6533. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol 85:7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz-Griffero F, Zimmer C, Charneau P, Di Nunzio F. 2015. Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matreyek KA, Yucel SS, Li X, Engelman A. 2013. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M. 2014. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A 111:18625–18630. doi: 10.1073/pnas.1419945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price AJ, Jacques DA, McEwan WA, Fletcher AJ, Essig S, Chin JW, Halambage UD, Aiken C, James LC. 2014. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog 10:e1004459. doi: 10.1371/journal.ppat.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price AJ, Fletcher AJ, Schaller T, Elliott T, Lee K, KewalRamani VN, Chin JW, Towers GJ, James LC. 2012. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog 8:e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sowd GA, Serrao E, Wang H, Wang W, Fadel HJ, Poeschla EM, Engelman AN. 2016. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci U S A 113:E1054–E1063. doi: 10.1073/pnas.1524213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 47.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol 78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat Med 7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 49.Cattoglio C, Pellin D, Rizzi E, Maruggi G, Corti G, Miselli F, Sartori D, Guffanti A, Di Serio C, Ambrosi A, De Bellis G, Mavilio F. 2010. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- 50.Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 51.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 52.ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J 17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. 2007. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res 56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Luban J, Diaz-Griffero F. 2014. The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol Biol 1087:29–36. doi: 10.1007/978-1-62703-670-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kortagere S, Madani N, Mankowski MK, Schon A, Zentner I, Swaminathan G, Princiotto A, Anthony K, Oza A, Sierra LJ, Passic SR, Wang X, Jones DM, Stavale E, Krebs FC, Martin-Garcia J, Freire E, Ptak RG, Sodroski J, Cocklin S, Smith AB III. 2012. Inhibiting early-stage events in HIV-1 replication by small-molecule targeting of the HIV-1 capsid. J Virol 86:8472–8481. doi: 10.1128/JVI.05006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J. 2009. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol 83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fricke T, Valle-Casuso JC, White TE, Brandariz-Nunez A, Bosche WJ, Reszka N, Gorelick R, Diaz-Griffero F. 2013. The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology 10:46. doi: 10.1186/1742-4690-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ao Z, Jayappa KD, Wang B, Zheng Y, Wang X, Peng J, Yao X. 2012. Contribution of host nucleoporin 62 in HIV-1 integrase chromatin association and viral DNA integration. J Biol Chem 287:10544–10555. doi: 10.1074/jbc.M111.317057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F, Giacca M, Lusic M. 2015. Nuclear architecture dictates HIV-1 integration site selection. Nature 521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 62.Diaz-Griffero F, Vandegraaff N, Li Y, McGee-Estrada K, Stremlau M, Welikala S, Si Z, Engelman A, Sodroski J. 2006. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology 351:404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G, Tebas P, June CH, Bushman FD. 2009. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther 17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 66.Benleulmi MS, Matysiak J, Henriquez DR, Vaillant C, Lesbats P, Calmels C, Naughtin M, Leon O, Skalka AM, Ruff M, Lavigne M, Andreola ML, Parissi V. 2015. Intasome architecture and chromatin density modulate retroviral integration into nucleosome. Retrovirology 12:13. doi: 10.1186/s12977-015-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, Arneodo A, Andreola ML, Lavigne M, Parissi V. 2011. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog 7:e1001280. doi: 10.1371/journal.ppat.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakielny S, Shaikh S, Burke B, Dreyfuss G. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fricke T, Buffone C, Opp S, Valle-Casuso J, Diaz-Griffero F. 2014. BI-2 destabilizes HIV-1 cores during infection and prevents binding of CPSF6 to the HIV-1 capsid. Retrovirology 11:120. doi: 10.1186/s12977-014-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamorte L, Titolo S, Lemke CT, Goudreau N, Mercier JF, Wardrop E, Shah VB, von Schwedler UK, Langelier C, Banik SS, Aiken C, Sundquist WI, Mason SW. 2013. Discovery of novel small-molecule HIV-1 replication inhibitors that stabilize capsid complexes. Antimicrob Agents Chemother 57:4622–4631. doi: 10.1128/AAC.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh PK, Plumb MR, Ferris AL, Iben JR, Wu X, Fadel HJ, Luke BT, Esnault C, Poeschla EM, Hughes SH, Kvaratskhelia M, Levin HL. 2015. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev 29:2287–2297. doi: 10.1101/gad.267609.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. 2013. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, Jurado KA, Wu X, Shun MC, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P, Kvaratskhelia M, Hughes SH, Engelman A. 2012. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res 40:11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schrijvers R, Vets S, De Rijck J, Malani N, Bushman FD, Debyser Z, Gijsbers R. 2012. HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology 9:84. doi: 10.1186/1742-4690-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levin A, Rosenbluh J, Hayouka Z, Friedler A, Loyter A. 2010. Integration of HIV-1 DNA is regulated by interplay between viral rev and cellular LEDGF/p75 proteins. Mol Med 16:34–44. doi: 10.2119/molmed.2009.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Levin A, Hayouka Z, Friedler A, Loyter A. 2010. Peptides derived from the HIV-1 integrase promote HIV-1 infection and multi-integration of viral cDNA in LEDGF/p75-knockdown cells. Virol J 7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, Agapkina J, Ziganshin R, Gottikh M, Nazabal A, Emiliani S, Benarous R, Moras D, Schultz P, Ruff M. 2009. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J 28:980–991. doi: 10.1038/emboj.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Botbol Y, Raghavendra NK, Rahman S, Engelman A, Lavigne M. 2008. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res 36:1237–1246. doi: 10.1093/nar/gkm1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raghavendra NK, Engelman A. 2007. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology 360:1–5. doi: 10.1016/j.virol.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, Green PL, Wu L, Roth MJ, Bushman FD, Kvaratskhelia M. 2013. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A 110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K, Gijsbers R, Debyser Z. 2013. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep 5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koh Y, Wu X, Ferris AL, Matreyek KA, Smith SJ, Lee K, Kewalramani VN, Hughes SH, Engelman A. 2013. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol 87:648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]